Abstract
A new microscopic method for simultaneously determining in situ the identities, activities, and specific substrate uptake profiles of individual bacterial cells within complex microbial communities was developed by combining fluorescent in situ hybridization (FISH) performed with rRNA-targeted oligonucleotide probes and microautoradiography. This method was evaluated by using defined artificial mixtures of Escherichia coli and Herpetosiphon aurantiacus under aerobic incubation conditions with added [3H]glucose. Subsequently, we were able to demonstrate the potential of this method by visualizing the uptake of organic and inorganic radiolabeled substrates ([14C]acetate, [14C]butyrate, [14C]bicarbonate, and 33Pi) in probe-defined populations from complex activated sludge microbial communities by using aerobic incubation conditions and anaerobic incubation conditions (with and without nitrate). For both defined cell mixtures and activated sludge, the method proved to be useful for simultaneous identification and analysis of the uptake of labeled substrates under the different experimental conditions used. Optimal results were obtained when fluorescently labeled oligonucleotides were applied prior to the microautoradiographic developing procedure. For single-cell resolution of FISH and microautoradiographic signals within activated sludge flocs, cryosectioned sample material was examined with a confocal laser scanning microscope. The combination of in situ rRNA hybridization techniques, cryosectioning, microautoradiography, and confocal laser scanning microscopy provides a unique opportunity for obtaining cultivation-independent insights into the structure and function of bacterial communities.
In most natural and engineered systems offering a sufficient nutrient supply, microorganisms grow as spatially organized, matrix-enclosed, multispecies communities in biofilms, aggregates, or flocs rather than as single planktonic cells (11). When studying these systems, microbial ecologists attempt to determine population structure and dynamics, as well as the spatial distribution of species and the function of species within communities which are characterized both by various cell-cell interactions (13) and by pronounced architectural and chemical heterogeneity (7, 10, 33). Since a rigorous biomass disaggregation step is a common feature of all traditional cultivation-based microbiological approaches used for analysis of complex microbial communities, spatial information is lost. In addition, biases introduced by conventional cultivation methods result in pronounced population shifts in the community structure and lead to a failure to detect dominant bacterial community members (50, 51, 56). Determining the physiological and biochemical properties of a single species in a laboratory pure culture may also bias the phenotype since gene expression is strongly influenced by environmental constraints and the growth mode of the cells (12). Since isolated strains are known to adapt genetically to the environmental conditions which they encounter, they may also not be genetically representative of their environmental counterparts (8).
With the advent of rRNA in situ hybridization techniques in microbial ecology, cultivation-independent quantitative examination of the structure and dynamics of complex microbial communities became possible (for a review see reference 2). Confocal laser scanning microscopic detection of signals conferred by a variety of fluorescent strains or fluorescently labeled RNA-targeted nucleic acid probes also allows analysis of the architecture of bacterial aggregates (for a recent review see reference 28) and the spatial arrangement of bacterial species within their habitats (4, 16, 18, 35, 41, 50–54).
Keeping in mind the physiological versatility of many prokaryotes, it is apparent that identification of a bacterial species in situ does not provide much information about its function in its habitat, except for members of a few highly physiologically restricted bacterial lineages like the ammonia oxidizers belonging to the beta subclass of the Proteobacteria. While in situ identification of uncultured microorganisms may allow assumptions concerning possible physiological characteristics if known physiological properties of the most closely phylogenetically related species which can be cultivated are taken into account (26), a more encompassing in situ analytical approach that yields information about the identity and function of a cell in its environmental niche would be desirable. Such information not only would allow a more detailed understanding of the ecology of complex microbial communities but also might provide information which could be used for the development of new, more appropriate enrichment and isolation methods for molecularly identified but uncultured prokaryotes.
Recently, an elegant combination of fluorescent oligonucleotide probing and microelectrode measurements has been used for structure-function analysis in microbial ecology. The relationship between chemical gradients and the stratification of sulfate-reducing (38) and nitrifying bacteria (41, 42) in trickling filters and a fluidized bed reactor has been examined by this method. Microelectrodes, however, measure chemical profiles only at a single point and consequently do not describe two-dimensional or three-dimensional chemical profiles in heterogeneous communities. The spatial resolution provided by microsensors is also above the single-cell level. Other attempts have been made to develop methods for functionally based microbial community analysis on a single-cell level. One approach has focused on the detection of specific gene expression in microbial cells by exploiting fusions of a gene and the coding sequence of a reporter protein (19, 27, 30, 45, 57). While techniques involving reporter proteins provide valuable information, they are limited in their applicability to microbial ecology as they require (i) the availability of genetic tools for the microorganism being studied and (ii) the introduction of genetically engineered microorganisms into the environment. Other approaches for demonstrating cell-specific gene expression involve the use of electron microscopic immunocytochemistry for detection of specific enzymes (44) or the use of nucleic acid-based techniques for visualization of specific mRNA (17, 20, 21, 23, 48, 55). These methods require either cultivation of the microorganism being studied for protein purification or detailed information concerning the target mRNA sequences. Use of these methods has been restricted to pure or simple mixed cultures because of the requirement for cell permeabilization procedures tailored to the bacteria being studied.
The use of radiolabeled substrates in combination with microautoradiography does allow analysis of the metabolic activity of prokaryotes under conditions that approach in situ conditions by direct visualization of microorganisms with active substrate uptake systems within a complex community (6). Microautoradiography has been successfully used in many ecological studies to measure activities of members of different autotrophic and heterotrophic prokaryotic groups (9, 37). Microautoradiography has been combined with microelectrodes (37) or simple staining techniques (3, 34, 36, 47) for characterization of microbial communities. A major limitation of microautoradiography has been an inability to correlate the activity detected with identification and detection of a responsible organism. One attempt to solve this problem has involved combining microautoradiography with fluorescently labeled antibody (FA) techniques (14, 16). However, considering the low percentage of bacteria successfully cultured from most environments and the enormous structural complexity of many microbial communities, the immunofluorescence approach is limited in its applicability for the following reasons: first, the FA approach is not completely cultivation independent due to its requirement for a pure culture of the microorganism being studied for antibody production; second, the FA approach can be disturbed by the presence of detritus particles, fungal spores, and extracellular polymeric substances that have been reported to cause unspecific binding and to hinder antibody penetration (46); and third, due to the great serological diversity of complex microbial communities, antibody specificity is for the most part restricted to the species level or below, which implies that hundreds if not thousands of antibodies must be used for an encompassing structural analysis of a complex microbial community.
The objective of this study was to develop and evaluate a new method that involves a direct combination of fluorescent in situ hybridization (FISH) performed with rRNA-targeted oligonucleotide probes and microautoradiography for simultaneous in situ identification and determination of substrate uptake patterns of individual microbial cells within complex microbial consortia. Liquid scintillation counting of samples and pasteurized control samples was performed in all experiments in order to confirm that biologically induced uptake of radiolabeled substrates occurred.
MATERIALS AND METHODS
Bacterial strains.
Mixtures of Escherichia coli ATCC 11775T and Herpetosiphon aurantiacus ATCC 23779T were used in a defined system. E. coli was cultivated at 37°C on Luria-Bertani agar (Difco), and H. aurantiacus was cultivated at 30°C on R2A agar (Difco). Prior to the start of the experiment, both strains were separately cultivated on R2A broth at 30°C and harvested after incubation for 12 h (E. coli) and 48 h (H. aurantiacus). The cells were washed twice with glucose-free R2A broth (40) and were diluted to a concentration of approximately 1 mg (dry weight) ml−1 in sterile, modified glucose-free R2A broth.
Activated sludge samples.
Activated sludge samples were collected from two wastewater treatments plants. Activated sludge containing high levels of phosphorus-accumulating and nitrifying bacteria was obtained from the Malmö Water and Sewage Works (Sweden), a municipal nutrient-removing wastewater pilot plant (50 population equivalents) with high biological phosphorus- and nitrogen-removing activities. In addition, grab samples of activated sludge were collected in October 1997 and March 1998 from the intermittently aerated nitrification-denitrification basin of an industrial wastewater treatment plant receiving sewage from an animal carcass-processing facility (Tierkörperbeseitigungsanstalt Kraftisried, Kraftisried, Germany; 6,000 population equivalents). In March 1998, incomplete nitrification, most probably caused by nonoptimal pH values (pH >8) and low temperatures, was observed at the Kraftisried plant. All activated sludge samples were stored at 4°C before they were used. Samples were incubated with radioactive substrates within 3 to 24 h after collection. Prior to incubation, sludge samples were diluted so that they contained 2 and 1 g of dry matter (suspended solids) per liter with nitrate-free water obtained from filtration (Whatman GF/C glass microfiber filter; pore size, 1 μm) of activated sludge in order to study heterotrophic activity and autotrophic activity (with [14C]bicarbonate), respectively. A dry matter content between 1 and 4 g per liter is typically found in activated sludge from wastewater treatment plants.
Incubation with radioactive and nonradioactive compounds.
The following substrates, labeled with three different isotopes, were used: (i) d-[6-3H]glucose (specific activity, 15.6 mCi/mmol), (ii) sodium [14C]bicarbonate (specific activity, 5.0 mCi/mmol), (iii) [2-14C]acetate (sodium salt; specific activity, 58.0 mCi/mmol), (iv) [n-14C]butyrate (sodium salt; specific activity, 10 mCi/mmol), and (v) 33Pi (specific activity, 3 mCi/mmol). Radioactive chemicals were obtained from Amersham (Buckinghamshire, United Kingdom), NEN Life Science Products (Boston, Mass.), and Sigma (Deisenhofen, Germany).
Ten different experimental systems with different types of radioactive and nonradioactive substrates and different incubation conditions were studied (Table 1). For each experimental system, 1.9-ml portions of sample material were transferred to 9-ml serum bottles. The serum bottles were sealed with gas-tight rubber stoppers. Activated sludge samples that were going to be incubated anaerobically were flushed with oxygen-free nitrogen gas to remove the oxygen and then were preincubated for 1 h to ensure that oxygen was completely removed. Subsequently, samples were supplemented with sterile radioactive and nonradioactive substrates (Sigma); the final volume of each sample was 2 ml. For anaerobic incubation oxygen-free substrates were injected anaerobically with syringes through the rubber stoppers into the serum bottles. To study microbial phosphorus uptake, activated sludge was subjected to anaerobic and aerobic conditions by opening the rubber stopper after 2 h of anaerobic incubation and shaking the serum bottle thoroughly for 1 min before sealing it again. During the 2-h aerobic incubation, the serum bottle was vigorously shaken. All experiments were performed in duplicate.
TABLE 1.
Incubation conditions used for experimentsa
| Expt | Sample | Concn of suspend-ed solids (g/liter) | Nonradioactive substrate(s) | Radioactive tracer
|
Incubation conditions | Incubation time (h) | |
|---|---|---|---|---|---|---|---|
| Compound | Concn (μCi/mg of suspended solids) | ||||||
| 1 | Pure cultures of E. coli and H. aurantiacus | 1 | Glucose (1 mM)b | d-[6-3H]glucose | 20 | Aerobic | 3–24 |
| 2 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) | Sodium [14]bicarbonate | 20 | Aerobic | 8 |
| 3 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) | Sodium [14C]bicarbonate | 20 | Aerobic | 8 |
| 4 | Activated sludge (Malmö)c | 2 | Monobasic phosphate (0.3 mM) + sodium acetate (1 mM) | 33Pi | 10 | Anaerobic | 2 |
| Aerobic | 2 | ||||||
| 5 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) + sodium acetate (1 mM) | Sodium [2-14C]acetate | 10 | Aerobic | 2 |
| 6 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) + sodium acetate (1 mM) + potassium nitrate (2 mM) | Sodium [2-14C]acetate | 10 | Anaerobic with nitrate | 2 |
| 7 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) + sodium acetate (1 mM) | Sodium [2-14C]acetate | 10 | Anaerobic | 2 |
| 8 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) + sodium butyrate (1 mM) | Sodium [2-14C]butyrate | 10 | Aerobic | 2 |
| 9 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) + sodium butyrate (1 mM) + potassium nitrate (2 mM) | Sodium [2-14C]butyrate | 10 | Anaerobic with nitrate | 2 |
| 10 | Activated sludge (Kraftisried) | 1 | Ammonia (3 mM) + sodium butyrate (1 mM) | Sodium [2-14C]butyrate | 10 | Anaerobic | 2 |
All experiments were performed at room temperature at pH 7 to 7.5 when substrates were added.
The concentrations in parentheses are the absolute amounts added.
Two parallel incubations were performed in order to differentiate between chemically and biologically induced P removal by using different washing procedures (see Materials and Methods).
As a control for possible adsorption phenomena, sample material pasteurized at 70°C for 10 min was incubated with the radioactive and nonradioactive substrates in parallel in all experimental systems. Paraformaldehyde-fixed sample material was not used as a control in this study, since we observed, compared to the pasteurized samples, slightly enhanced levels of adsorption (probably caused by fixative-induced chemical modification of bacterial surfaces) in pilot experiments (data not shown). In addition, control samples without added radioactivity were always prepared as controls to check for chemography.
Sample fixation, washing procedures, and DAPI staining of control samples.
After incubation with radioactive and nonradioactive substrates, samples were fixed with 6 ml of 4% paraformaldehyde for 3 h at 4°C. Subsequently, the samples were washed three times with 5 ml of phosphate-buffered saline (PBS) (10 mM sodium phosphate buffer, 130 mM sodium chloride; pH 7.2) in order to remove the excess soluble radioactive substrate and fixative. Alternatively, one of the activated sludge samples incubated with 33Pi was washed twice with 0.1 M sodium citrate-HCl buffer (pH 2) prior to the washing procedure described above for removal of chemically bound phosphorus (29). All washing steps consisted of centrifugation of the sample at 37,100 × g for 5 min, removal of the supernatant, addition of further washing buffer, and mixing. After the washing procedure, the samples used as controls for adsorption and chemography were stained with 4′,6-diamidino-2-phenylindole (DAPI) by resuspension of the pellet in 1 ml of a DAPI solution (50 μg/ml in PBS) and incubation for 5 min. Excess DAPI was removed by a subsequent PBS washing step.
Homogenization of activated sludge flocs.
Fixed, PBS-washed, radioactive, pure cultures and activated sludge pellets were resuspended in 5 ml of PBS, homogenized with a tissue grinder (Thomas Scientific, Swedesbro, N.J.) for 2 min, and then used for liquid scintillation analysis. For in situ hybridization, aliquots of the homogenized sample material were spotted onto poly-l-lysine (Sigma)-coated slides or coverslips, dried at 46°C, and dehydrated in an ethanol series (50, 80, and 96% ethanol, 3 min each).
Liquid scintillation counting.
Liquid scintillation counting of homogenized sample materials and pooled wash solutions was performed in all experiments. A 2-ml portion of a wash solution or dispersed sample material was added to 2 ml of scintillation liquid (Ultima Gold XR; Packard Instrument Co., Meriden, Conn.). After thorough mixing, the sample was allowed to react for at least 3 h at room temperature prior to counting with a Packard model 1600 TR liquid scintillation analyzer as recommended by the manufacturer.
Cryosectioning.
For cryosectioning, nonhomogenized, fixed, and PBS-washed radioactive activated sludge pellets were resuspended in 500 μl of Tissue-Tek (Microm, Walldorf, Germany) and frozen at −20°C for 1 h. Sections that were 5 to 10 μm thick were cut with a cryomicrotome (model MIKROM HM500; Microm), transferred onto poly-l-lysine-coated coverslips, and dehydrated in an ethanol series (50, 80, and 96% ethanol, 3 min each).
Oligonucleotide probes.
The following rRNA-targeting oligonucleotide probes were used for FISH: (i) EUB 338 (5′-GCTGCCTCCCGTAGGAGT-3′), which is complementary to a conserved region of most bacterial 16S rRNA molecules (1); (ii) NEU (5′-CCCCTCTGCTGCACTCTA-3′), which is complementary to a signature region of the 16S rRNA of most halophilic and halotolerant ammonia oxidizers belonging to the beta subclass of the Proteobacteria (54); (iii) CTE (5′-TTCCATCCCCCTCTGCCG-3′), which was used as an unlabeled competitor oligonucleotide probe to ensure specific hybridization of probe NEU (54); and (iv) BET42a (5′-GCCTTCCCACTTCGTTT-3′) and GAM42a (5′-GCCTTCCCACATCGTTT-3′), which target 23S rRNA regions conserved in members of the beta and gamma subclasses of the Proteobacteria, respectively (32). Oligonucleotides labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS) or with the monofunctional, hydrophilic, sulfoindocyanine dyes Cy3 and Cy5 were purchased from Interactiva, Ulm, Germany.
DAPI staining and in situ hybridization.
In situ hybridization of the samples was performed as described previously (32). Briefly, 9 μl of hybridization solution containing 50 ng of probe (FLUOS-labeled probes) or 30 ng of probe (Cy3- and Cy5-labeled probes) was applied to a sample immobilized on a slide or coverslip and incubated for 90 min at 46°C in an isotonically equilibrated humid chamber. The probes were removed from the slides or coverslips with 2 ml of washing solution and were immediately immersed in 50 ml of prewarmed (48°C) washing solution and incubated for 15 min. The slides and coverslips were rinsed briefly with ice-cold distilled water and dried with pressurized air. After in situ hybridization, the cells were stained with DAPI (0.33 μg/ml in H2O) for 5 min. The excess DAPI was removed by rinsing the slides or coverslips with ice-cold double-distilled H2O.
Autoradiographic procedure and combination with in situ hybridization.
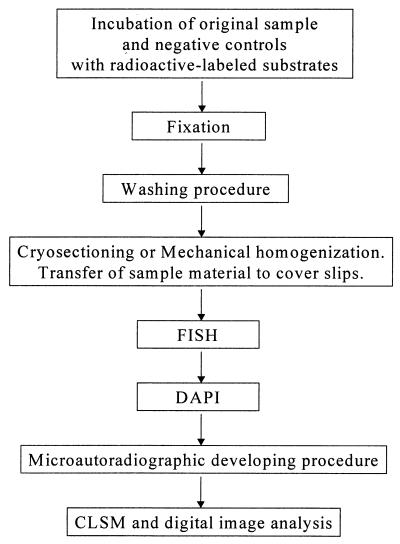
An autoradiographic procedure in which autoradiographic emulsion LM-1 (Amersham International) was used was performed as described by Andreasen and Nielsen (3). The optimum exposure time for the samples was between 3 and 7 days. Kodak D19 developer was used as the developer (2 min), and after each preparation was washed in demineralized water for 1 min, it was fixed in 30% (wt/vol) thiosulfate for 4 min. The slides and/or coverslips were washed for 10 min in gently running tap water and then were washed twice (2 min each) in demineralized water. The slides and coverslips were allowed to air dry. Application of the in situ hybridization protocol before and after the autoradiographic procedure was tested. A summary of the method used is presented as flow scheme in Fig. 1.
FIG. 1.
Flow scheme showing the method which allowed direct combination of FISH and microautoradiography. CLSM, confocal laser scanning microscopy.
Microscopy.
A model LSM 510 scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) equipped with a UV laser (351 and 364 nm), an Ar ion laser (450 to 514 nm), and two HeNe lasers (543 and 633 nm) was used to record optical sections. The formation of silver grains in the autoradiographic film covering a sample was observed by using the transmission mode of the instrument. Image processing was performed with the standard software package delivered with the instrument (version 1.6). Reconstructed and processed images were printed by using the software package Microsoft Power Point (version 7.0; Microsoft, Redmond, Wash.) and a Kodak model 8650 PS printer.
RESULTS AND DISCUSSION
Combination of microautoradiography and FISH: method development and evaluation of pure cultures.
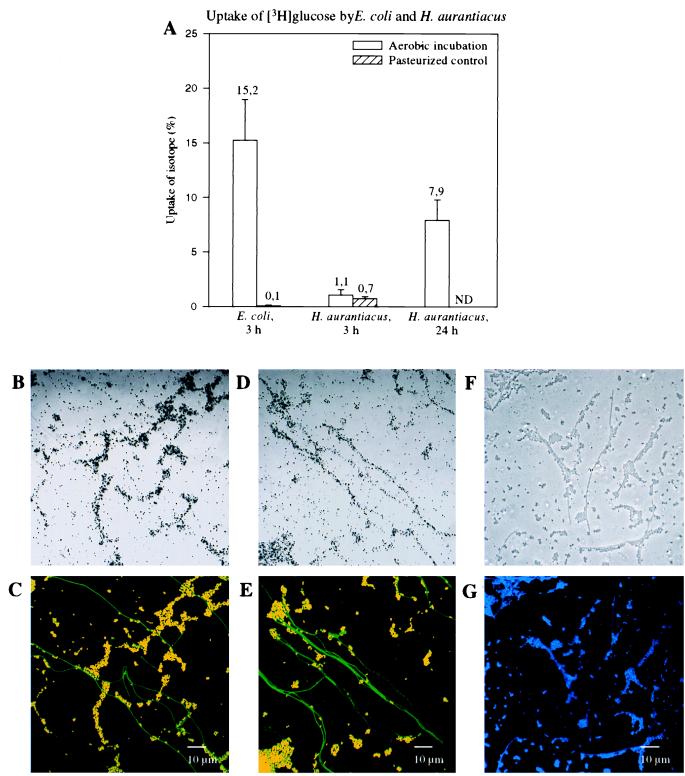
Separate cultures of E. coli and H. aurantiacus, a nonphotosynthetic member of the green nonsulfur bacteria, were incubated aerobically with a mixture of unlabeled glucose and [3H]glucose (Table 1). Uptake of radioactive glucose was initially monitored after homogenization by liquid scintillation counting. After 3 h of incubation the E. coli and H. aurantiacus cells contained 15.2 and 1.1% of the added radioactively labeled glucose, respectively (Fig. 2A). After 24 h of incubation, a significant increase in glucose uptake by H. aurantiacus (7.9%) (Fig. 2A) was observed. Differences in growth rate, glucose uptake kinetics, and glucose degradation kinetics between E. coli and H. aurantiacus might have contributed to the differences detected in the amounts of radioactive glucose per gram of biomass of the two species. It should be noted that liquid scintillation counting underestimates the actual amount of glucose taken up since a fraction of the radioactive label is excreted as 3H2O due to metabolic glucose transformations. In control experiments performed with pasteurized E. coli and H. aurantiacus cells, only traces of radioactively labeled glucose were detected (E. coli, 0.1%; H. aurantiacus, 0.7%) (Fig. 2A), which demonstrated that there was no adsorption.
FIG. 2.
Radiometry and microautoradiography combined with FISH performed with pure cultures of E. coli and H. aurantiacus. (A) Liquid scintillation counts for viable and pasteurized E. coli and H. aurantiacus cells after incubation with [3H]glucose for 3 h. For H. aurantiacus [3H]glucose uptake was also determined after 24 h. ND, not determined. (B through E) Confocal laser scanning microscopic images of artificial mixtures of E. coli and H. aurantiacus incubated with [3H]glucose and analyzed by a combination of microautoradiography and FISH by using Cy3-labeled probe GAM42a (red) and Cy5-labeled probe EUB338 (colored green by image analysis). (B) Microautoradiographic image of E. coli and H. aurantiacus after 3 h of incubation with [3H]glucose. (C) Whole-cell hybridization of the microscopic field in panel B. E. coli cells appear yellow because of the overlapping labels. (D) Microautoradiographic image of E. coli and H. aurantiacus after 3 h (E. coli) and 24 h (H. aurantiacus) of incubation with [3H]glucose. (E) Whole-cell hybridization of the microscopic field in panel D. (F and G) Confocal laser scanning microscopic images of an artificial mixture of pasteurized E. coli and H. aurantiacus cells incubated with [3H]glucose for 3 h and analyzed by a combination of microautoradiography (F) and DAPI staining (G). Since no silver grain formation was observed in the pasteurized control sample, cell morphologies were determined by using the transmission mode of the confocal laser scanning microscope.
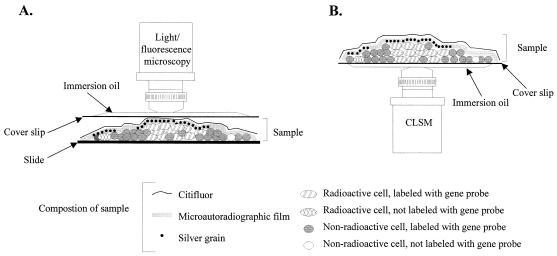
In order to develop a protocol for combining FISH performed with rRNA-targeted oligonucleotide probes and microautoradiography, artificial mixtures of E. coli and H. aurantiacus were prepared on poly-l-lysine-coated slides by applying homogenized subsamples of the cell pellets used for scintillation counting (see above). Two types of FISH-microautoradiography combinations were tested; cells were hybridized with fluorescently labeled oligonucleotide probes either prior to or after the microautoradiographic developing step. Both combinations allowed simultaneous in situ detection of DAPI signals, probe-conferred fluorescence, and silver grain formation (mediated by the presence of radioactive compounds within the fixed cells) at a single-cell level. However, detachment of biomass and autoradiographic film was frequently observed if FISH and DAPI staining were performed after the developing procedure. In addition, the cellular signal intensities obtained by FISH were significantly higher if the FISH procedure was performed prior to the developing protocol (data not shown). The low signal intensities obtained by FISH after the developing step might be explained by hampered probe penetration through the microautoradiographic film. Even though improved results were obtained if FISH and DAPI staining were performed prior to the developing procedure, the DAPI and probe-conferred signals of radioactively labeled cells were still masked by the overlying silver grains within the autoradiographic film. To solve this problem, the microscopic observation technique described by Fuhrmann and Azam (15) was modified by spreading radioactively labeled cells onto poly-l-lysine-coated coverslips (instead of slides). After FISH (and DAPI staining) and the autoradiographic development procedure were performed, fluorescently labeled microorganisms and silver grains could be simultaneously observed at a high resolution by inverse confocal laser scanning microscopy. In contrast to conventional microscopic observation (Fig. 3A), inverse microscopic analysis of the processed sample materials on coverslips (Fig. 3B) also avoided detachment or disruption of the autoradiographic film by movement of the objective, thereby improving the assignment of silver grains to individual microbial cells. In the artificial mixture containing E. coli and H. aurantiacus, most cells labeled by [3H]glucose after 3 h of incubation were identified as E. coli cells by simultaneous hybridization with probes EUB338-Cy5 and GAM42a-Cy3 (Fig. 2B and C). Consistent with the results obtained by scintillation counting, [3H]glucose uptake could also be visualized with the majority of filamentous Herpetosiphon cells after 24 h of incubation (Fig. 2D and E), demonstrating the importance of a timed series of measurements to determine substrate uptake kinetics. No silver grain-coated microorganisms were observed in the sample without added [3H]glucose and in the pasteurized control sample (Fig. 2F and G), which showed that chemographic and nonspecific adsorption did not occur.
FIG. 3.
Schematic comparison of conventional microscopic analysis (A) and the inverse confocal laser scanning microscopic observation technique (B) for viewing samples analyzed by microautoradiography and FISH. During conventional analysis a processed sample is viewed through a coverslip placed on top of the microautoradiographic film, while our new technique allows analysis of a sample through a coverslip located below the processed sample. CLSM, confocal laser scanning microscope.
Combination of microautoradiography and FISH: use with complex microbial communities.
After we developed the protocol for combining FISH and microautoradiography, this technique was used to document in situ substrate uptake in several probe-defined microbial populations within different complex activated sludge microbial consortia by using different types of radioactively labeled substrates.
[14C]bicarbonate.
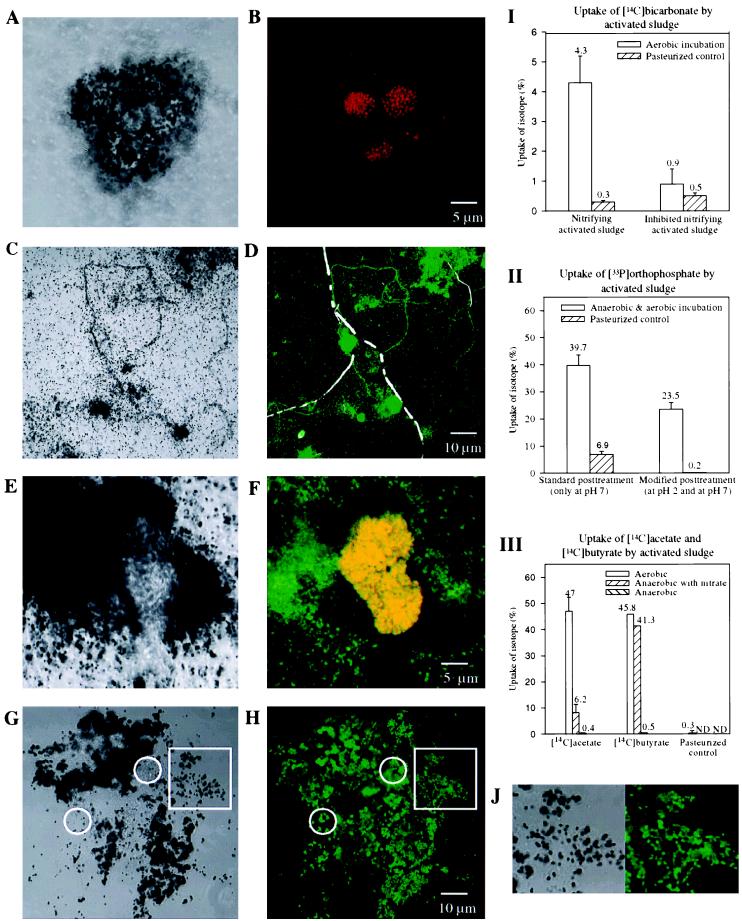
Comparative experiments were performed by using [14C]bicarbonate and probe NEU targeting halophilic and halotolerant members of the beta subclass ammonia oxidizers (54) with activated sludge with high nitrifying activity (Kraftisried, October 1997; more than 80% N removal) and low nitrifying activity (Kraftisried, March 1998; approximately 10% N removal). Experimental details are given in Table 1 (experiments 2 and 3). After FISH and autoradiographic development, most silver grains accumulated close to clusters of ammonia oxidizers detectable with probe NEU in the homogenized sample obtained in October 1997 (Fig. 4A and B), reflecting the autotrophic activity of the ammonia oxidizers. While all of the NEU-positive cell clusters were labeled with radioactive bicarbonate, DAPI-positive cell clusters that were labeled with radioactive bicarbonate but were not stained with probe NEU were also detected (data not shown). These clusters most likely represented additional autotrophic nitrifying bacteria (e.g., ammonia oxidizers which were not detectable with probe NEU, as well as nitrite-oxidizing bacteria belonging to the genus Nitrospira [25]). In the sample obtained in March 1998, which displayed low nitrifying activity, very few NEU-positive and/or radioactively labeled cells were visualized. In pasteurized controls for both samples no uptake of radioactively labeled bicarbonate was observed. These results were confirmed by liquid scintillation counting of subsamples in both experiments (Fig. 4I).
FIG. 4.
Combination of microautoradiography and FISH for studying complex microbial communities in activated sludge. Since for all substrates only single time point incorporation measurements were obtained, nonradioactively labeled cells have to be viewed with some caution. (A) Microautoradiographic image of mechanically homogenized nitrifying activated sludge from the Kraftisried plant incubated with [14C]bicarbonate under aerobic conditions. (B) In situ hybridization of the microscopic field in panel A with Cy3-labeled probe NEU (red). (I) Liquid scintillation counts for native and pasteurized activated sludge samples from the Kraftisried plant with high nitrifying activity (left) and low nitrifying activity (right). The nitrifying activities of the sludge samples corresponded to the amounts of detectable radioactivity after incubation with [14C]bicarbonate. (C) Microautoradiographic image of mechanically homogenized activated sludge from the Malmö plant incubated with 33Pi under anaerobic-aerobic conditions. (D) In situ hybridization of the microscopic field panel in C with Cy3-labeled probe GAM42a (colored white by image analysis) and Cy5-labeled probe BET42a (colored green by image analysis). In contrast to the filamentous bacterium belonging to the gamma subclass of the Proteobacteria, various floc-forming and filamentous bacteria belonging to the beta subclass of the Proteobacteria were able to take up Pi. (II) Liquid scintillation counts for native and pasteurized activated sludge samples from the Malmö plant. The effectiveness of the additional pH 2 wash step is reflected in the lower amount of 33Pi detected in the pasteurized control sample. (E) Microautoradiographic image of mechanically homogenized nitrifying activated sludge from the Kraftisried plant incubated with [14C]acetate under aerobic conditions. (F) In situ hybridization of the microscopic field in panel F with Cy3-labeled probe NEU (red) and Cy5-labeled probe BET42a (colored green by image analysis). Ammonia oxidizers belonging to the beta subclass of the Proteobacteria which appear yellow due to the overlap of labels did not take up radioactively labeled acetate. (G) Microautoradiographic image of cryosectioned nitrifying activated sludge from the Kraftisried plant incubated with [14C]acetate under aerobic conditions. (H) In situ hybridization of the microscopic field in panel G with FLUOS-labeled probe BET42a (green). The circles indicate the localization of members of the beta subclass of the Proteobacteria which did not take up radioactively labeled acetate. To demonstrate that the spatial resolution of the silver grains was significantly enhanced compared to the spatial resolution of the mechanically homogenized sample (E), two magnified sections (indicated by squares in panels G and H) are shown in panel J. (III) Liquid scintillation counts for native and pasteurized activated sludge samples from the Kraftisried plant incubated under aerobic conditions and anaerobic conditions (with and without nitrate) with [14C]acetate and [14C]butyrate. ND, not determined.
33Pi.
Phosphate removal from wastewater is a key feature of modern sewage treatment plants. Traditionally, this removal was achieved by chemical phosphate precipitation with metal salts. Over the last few decades chemical phosphate precipitation has been replaced or supplemented by the so-called enhanced biological phosphorus removal (EBPR) process in many sewage treatment plants (24). Despite the importance of the EBPR process, the microorganism(s) responsible for it has still not been identified (49, 51). Here we investigated the possibility of identifying phosphorus-accumulating microorganisms in situ by a combination of FISH and microautoradiography. Activated sludge obtained from the nutrient removal plant at Malmö was analyzed by anaerobic-aerobic incubation with 33Pi (Table 1, experiment 4) and subsequent in situ hybridization with probes BET42a and GAM42a which are specific for the members of the beta and gamma subclasses of the Proteobacteria, respectively. When these procedures were used, sludge flocs labeled with radioactive phosphate were also detected in the pasteurized negative control, indicating that chemical phosphate precipitation occurred (data not shown). Consequently, an additional wash step performed at pH 2 was included to remove chemically bound phosphorus prior to FISH and the autoradiographic development procedure. Following application of the modified protocol, radioactive phosphate was no longer microscopically detected in the pasteurized control sludge. The influence of the pH of the wash buffers on the amount and nature of the radioactive phosphate detected in the activated sludge was also demonstrated by the results of the liquid scintillation counting procedure. While significant relative amounts of radioactive phosphate (6.9%) were detected in the pasteurized control after the standard pH 7 washing procedure, negligible relative amounts of radioactive phosphate (0.2%) remained after the pH 2 washing step (Fig. 4II). These results demonstrate that proper negative control reactions are important to differentiate between chemical adsorption and biological uptake of the labeled substrate. Once chemical adsorption has been observed for a specific substrate, adequate additional washing procedures must be used in the protocol.
In the homogenized unpasteurized sludge, several morphotypes belonging to the beta subclass of the Proteobacteria labeled with radioactive phosphate were detected, while no representatives of the gamma subclass of the Proteobacteria were found to contain radioactive phosphate (Fig. 4C and D). These preliminary findings confirmed the results of previous molecular studies which indicated that members of the beta subclass of the Proteobacteria (as well as gram-positive bacteria with a high DNA G+C content) are more important than Acinetobacter spp. and other members of the gamma subclass of the Proteobacteria in the EPBR process (5, 51). We are currently intensifying the search for the phosphorus-accumulating organism(s) by (i) establishing phylogenetic 16S rRNA-based inventories of microbial communities involved in EBPR, (ii) designing a probe set for the bacteria, and (iii) combining FISH performed with these probes and microautoradiography, using 33Pi as the substrate.
14C-labeled fatty acids.
Activated sludge obtained from the Kraftisried plant in October 1997 was incubated with 14C-labeled acetate and 14C-labeled butyrate. These two fatty acids were selected because they comprise a main fraction of the organic load of the influent wastewater of the Kraftisried plant. Uptake of both fatty acids under aerobic incubation conditions and anaerobic incubation conditions (with and without nitrate) was studied in order to determine the ability of the combined technique to monitor differences in the uptake of substrates by the microbial consortium under different metabolic conditions. Experimental details are given in Table 1 (experiments 5 to 10). Subsequent in situ hybridization was performed with probes NEU and BET42a, which are specific for halophilic and halotolerant beta subclass ammonia oxidizers and members of the beta subclass of the Proteobacteria, respectively. Ammonia oxidizers stained during in situ hybridization with probe NEU failed to take up acetate or butyrate under the different conditions (Fig. 4E and F), suggesting that they were not able to grow mixotrophically. After aerobic incubation with radioactively labeled acetate or butyrate, most sludge flocs, which were composed mainly of members of the beta subclass of the Proteobacteria, were heavily coated with silver grains (Fig. 4E and F), while under anaerobic conditions only insignificant uptake of these fatty acids, comparable to the uptake observed in pasteurized control samples, was detected. Interestingly, under denitrifying conditions (anaerobic conditions with nitrate) the microbial activated sludge population took up significantly more butyrate than acetate (data not shown). The substrate uptake patterns of the activated sludge under different conditions observed microscopically were confirmed by parallel liquid scintillation counting of subsamples (Fig. 4III).
As observed in the [14C]bicarbonate and 33Pi experiments (Fig. 4A and C), insufficient dispersion of cell aggregates and activated sludge flocs by the homogenization procedure hampered single-cell resolution by microautoradiography (Fig. 4E). To improve the spatial resolution, we replaced the homogenization step in the protocol by using cryosections (thickness, 5 to 10 μm) of the radioactively labeled activated sludge flocs for in situ hybridization and subsequent autoradiographic development. This modification allowed us to correlate silver grain formation with individual probe-stained microbial cells, even within voluminous sludge flocs (Fig. 4G, H, and J). However, it should be noted that the native architecture of the sludge flocs might still have been disrupted to some extent by sample handling (e.g., fixation, centrifugation, and resuspension) prior to the cryosectioning step.
Conclusions.
Over the last decade molecular methods have revealed new and fascinating insights into the microbial diversity present in environmental samples (22, 31, 43, 56). Probe-based techniques have also allowed detailed quantitative studies of environmental microbial community structures and dynamics to be performed (1, 38, 39, 53). Probe-based techniques have also allowed detailed quantitative studies of environmental microbial community structures and dynamics to be performed (1, 38, 39, 50, 51). While they have provided important information, such studies have failed to reveal the functions of the microorganisms within their ecological niches. The combination of FISH and microautoradiography described in this paper allowed us to directly analyze the in vivo uptake of substrates by probe-identified microorganisms under different environmental conditions. While microautoradiography does not provide quantitative data, the information obtained from experiments which combined the two techniques should help identify key functional microbial groups that are active in the environment. The possible applications range from fundamental studies in microbial ecology to identifying bacteria involved in biotechnological processes, such as sewage treatment and bioremediation. In addition, information concerning the in vivo physiology of novel bacteria taxa identified by molecular techniques should facilitate the design of appropriate isolation techniques for many of these hitherto uncultured microorganisms.
ACKNOWLEDGMENTS
This work was supported by postdoctoral grant 97-256, from the Swedish Foundation for International Cooperation in Research and Higher Education to N.L., by the Körber Award, by the Danish Technical Research Council (Activity and Diversity in Complex Microbial Systems framework program), and by Sonderforschungsbereich SFB411 from the Deutsche Forschungsgemeinschaft (Research Center for Fundamental Studies of Aerobic Biological Wastewater Treatment). K.H.A. was supported by an industrial Ph.D. fellowship.
We thank Thomas Fritsche for critically reading the manuscript. We gratefully acknowledge the contribution of David Stahl in formulating concepts used in this study.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreasen K, Nielsen P H. Application of microautoradiography for the study of substrate uptake by filamentous microorganisms in activated sludge. Appl Environ Microbiol. 1997;63:3662–3668. doi: 10.1128/aem.63.9.3662-3668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence J R, Hartmann A. In situ localization of Azospirillum brasiliense in the rhizosphere of wheat with fluorescently labeled rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brock T D, Brock M L. Autoradiography as a tool in microbial ecology. Nature. 1968;209:734–736. doi: 10.1038/209734a0. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell D E, Korber D R, Lawrence J R. Confocal laser microscopy and digital image analysis in microbial ecology. Adv Microb Ecol. 1992;12:1–67. [Google Scholar]
- 8.Caldwell D E, Wolfaardt G M, Korber D R. Do bacterial communities transcend Darwinism? Adv Microb Ecol. 1997;15:105–191. [Google Scholar]
- 9.Carman K R. Radioactive labeling of a natural assemblage of marine sedimentary bacteria and microalgae for the trophic studies: an autoradiographic study. Microb Ecol. 1990;19:279–290. doi: 10.1007/BF02017172. [DOI] [PubMed] [Google Scholar]
- 10.Costerton J W, Lewandowski Z, de Beer D, Caldwell D E, Korber D R, James G A. Biofilms: the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 12.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dworkin M. Introduction. In: Dworkin M, editor. Microbial cell-cell interactions. Washington, D.C: American Society for Microbiology; 1991. pp. 1–6. [Google Scholar]
- 14.Fliermans C B, Schmidt E L. Autoradiography and immunofluorescence combined for autoecological study of single cell activity with Nitrobacter as a model system. Appl Microbiol. 1975;30:676–684. doi: 10.1128/am.30.4.676-684.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrmann J A, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- 16.Ghiorse W C, Miller D N, Sandoli R L, Siering P L. Applications of laser scanning microscopy for analysis of aquatic microhabitats. Microsc Res Tech. 1996;33:73–86. doi: 10.1002/(SICI)1097-0029(199601)33:1<73::AID-JEMT7>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Hahn D, Amann R I, Zeyer J. Detection of mRNA in Streptomyces cells by whole-cell hybridization with digoxigenin-labeled probes. Appl Environ Microbiol. 1993;59:2753–2757. doi: 10.1128/aem.59.8.2753-2757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen H J M, Akkermans A D L, Stams A J M, De Vos W M. Population dynamics of propionate-oxidizing bacteria under methanogenic and sulfidogenic conditions in anaerobic granular sludge. Appl Environ Microbiol. 1996;62:2163–2168. doi: 10.1128/aem.62.6.2163-2168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harry E J, Pogliano K, Losick R. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodson R E, Dustman W A, Garg R P, Moran M A. In situ PCR for visualization of microscale distribution of specific genes and gene products in prokaryotic communities. Appl Environ Microbiol. 1995;61:4074–4082. doi: 10.1128/aem.61.11.4074-4082.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hönerlage W, Hahn D, Zeyer J. Detection of mRNA of nprM in Bacillus megaterium ATCC 14581 grown in soil by whole-cell hybridization. Arch Microbiol. 1995;163:235–241. [Google Scholar]
- 22.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the beta subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins D, Tandoi V. The applied microbiology of enhanced biological phosphate removal—accomplishments and needs. Wat Res. 1991;25:1471–1478. [Google Scholar]
- 25.Juretschko S, Timmerman G, Schmid M, Schleifer K-H, Pommerening-Röser A, Koops H-P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane M D, Poulsen L K, Stahl D A. Monitoring of the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer L, Baulard A, Estaquier J, Poulain-Godefroy O, Locht C. Green fluorescent protein as a new expression marker in mycobacteria. Mol Microbiol. 1995;17:913–922. doi: 10.1111/j.1365-2958.1995.mmi_17050913.x. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence J R, Wolfaardt G M, Neu T R. The study of biofilms using confocal laser scanning microscopy. In: Wilkinson M H F, Schut F, editors. Digital image analysis of microbes—imaging, morphometry, fluorometry and motility techniques and applications. Chichester, United Kingdom: John Wiley & Sons; 1998. pp. 431–465. [Google Scholar]
- 29.Lee, N., M. Wagner, and P. H. Nielsen. Unpublished results.
- 30.Lewis P J, Nwoguh C E, Barer M R, Harwood C R, Errington J. Use of digitized video microscopy with a fluorogenic enzyme substrate to demonstrate cell- and compartment-specific gene expression in Salmonella enteritidis and Bacillus subtilis. Mol Microbiol. 1994;13:655–662. doi: 10.1111/j.1365-2958.1994.tb00459.x. [DOI] [PubMed] [Google Scholar]
- 31.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 33.Massol-Deyá A A, Whallon J, Hickey R F, Tiedje J M. Channel structures in aerobic biofilms of fixed-film reactors treating contaminated groundwater. Appl Environ Microbiol. 1995;61:769–777. doi: 10.1128/aem.61.2.769-777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer-Reil L A. Autoradiography and epifluorescence microscopy combined for the determination of number and spectrum of actively metabolizing bacteria in natural waters. Appl Environ Microbiol. 1978;36:506–512. doi: 10.1128/aem.36.3.506-512.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Møller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen P H, Andreasen K, Wagner M, Blackall L L, Lemmer H, Seviour R J. Variability of type 021N in activated sludge as determined by in situ substrate uptake pattern and in situ hybridization with fluorescent rRNA targeted probes. Wat Sci Technol. 1998;37:423–430. [Google Scholar]
- 37.Paerl H W. Alteration of microbial metabolic activities in association with detritus. Bull Mar Sci. 1984;35:393–408. [Google Scholar]
- 38.Ramsing N B, Kühl M, Jörgensen B B. Distribution of sulfate-reducing bacteria, O2 and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3820–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reasoner D J, Geldreich E E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schramm A, DeBeer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spieck E, Ehrich S, Ammand J, Bock E. Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch Microbiol. 1998;169:225–230. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 45.Stewart G S, Williams P. lux genes and the applications of bacterial bioluminescence. J Gen Microbiol. 1992;138:1289–1300. doi: 10.1099/00221287-138-7-1289. [DOI] [PubMed] [Google Scholar]
- 46.Szwerinski H, Gaiser S, Bardtke D. Immunofluorescence for the quantitative determination of nitrifying bacteria: interference of the test in biofilm reactors. Appl Microbiol Biotechnol. 1985;21:125–128. [Google Scholar]
- 47.Tabor P S, Neihof R A. Direct determination of activities for microorganisms of Chesapake Bay populations. Appl Environ Microbiol. 1984;48:1012–1019. doi: 10.1128/aem.48.5.1012-1019.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolker-Nielsen T, Holmström K, Molin S. Visualization of specific gene expression in individual Salmonella typhimurium cells by in situ PCR. Appl Environ Microbiol. 1997;63:4196–4203. doi: 10.1128/aem.63.11.4196-4203.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Loosdrecht M C M, Smolders G, Kuba T, Heijnen J J. Metabolism of micro-organisms responsible for enhanced biological phosphorus removal from wastewater. Use of dynamic enrichment cultures. Antonie Leeuwenhoek. 1997;71:109–116. doi: 10.1023/a:1000150523030. [DOI] [PubMed] [Google Scholar]
- 50.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner M, Amann R, Kämpfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K-H. Identification and in situ detection of Gram-negative filamentous bacteria in activated sludge. Syst Appl Microbiol. 1994;17:405–417. [Google Scholar]
- 53.Wagner M, Assmus B, Hartmann A, Hutzler P, Amann R. In situ analysis of microbial consortia in activated sludge using fluorescently labelled, rRNA-targeted oligonucleotide probes and confocal laser scanning microscopy. J Microsc. 1994;176:181–187. doi: 10.1111/j.1365-2818.1994.tb03513.x. [DOI] [PubMed] [Google Scholar]
- 54.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;17:251–264. [Google Scholar]
- 55.Wagner M, Schmid M, Juretschko S, Trebesius K H, Bubert A, Goebel W, Schleifer K-H. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol Lett. 1998;160:159–168. doi: 10.1111/j.1574-6968.1998.tb12906.x. [DOI] [PubMed] [Google Scholar]
- 56.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 57.Webb C D, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]