Abstract
Background & Aims
Hyperbaric oxygen therapy (HBOT) is a promising treatment for moderate-to-severe ulcerative colitis. However, our current understanding of the host and microbial response to HBOT remains unclear. This study examined the molecular mechanisms underpinning HBOT using a multi-omic strategy.
Methods
Pre- and post-intervention mucosal biopsies, tissue, and fecal samples were collected from HBOT phase 2 clinical trials. Biopsies and fecal samples were subjected to shotgun metaproteomics, metabolomics, 16s rRNA sequencing, and metagenomics. Tissue was subjected to bulk RNA sequencing and digital spatial profiling (DSP) for single-cell RNA and protein analysis, and immunohistochemistry was performed. Fecal samples were also used for colonization experiments in IL10-/- germ-free UC mouse models.
Results
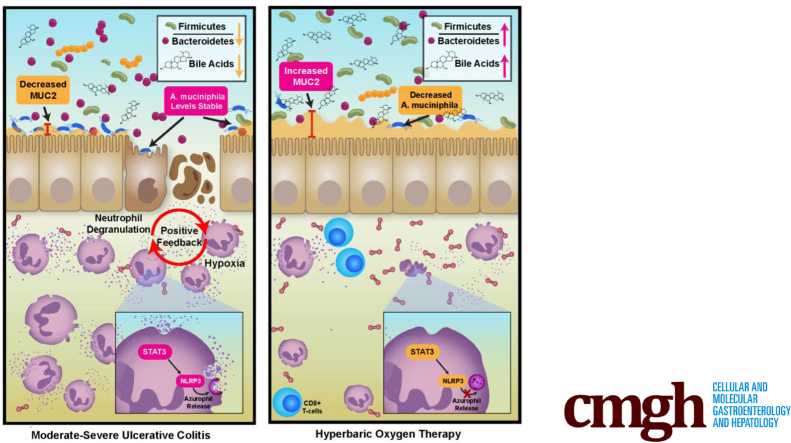
Proteomics identified negative associations between HBOT response and neutrophil azurophilic granule abundance. DSP identified an HBOT-specific reduction of neutrophil STAT3, which was confirmed by immunohistochemistry. HBOT decreased microbial diversity with a proportional increase in Firmicutes and a secondary bile acid lithocholic acid. A major source of the reduction in diversity was the loss of mucus-adherent taxa, resulting in increased MUC2 levels post-HBOT. Targeted database searching revealed strain-level associations between Akkermansia muciniphila and HBOT response status. Colonization of IL10-/- with stool obtained from HBOT responders resulted in lower colitis activity compared with non-responders, with no differences in STAT3 expression, suggesting complementary but independent host and microbial responses.
Conclusions
HBOT reduces host neutrophil STAT3 and azurophilic granule activity in UC patients and changes in microbial composition and metabolism in ways that improve colitis activity. Intestinal microbiota, especially strain level variations in A muciniphila, may contribute to HBOT non-response.
Abbreviations used in this paper: BCLXL, B-cell lymphoma-extra large; BIM, bcl-2 interacting protein; DSP, digital spatial profiling; FDR, false discovery rate; HBOT, hyperbaric oxygen therapy; HIF, hypoxia inducible factor; IL, interleukin; LCA, lithocholic acid; MAPK, mitogen-activated protein kinase; MS, mass spectrometry; NLRP3, NOD-, LRR- and pyrin domain-containing protein 3; ROI, regions of interest; ROS, reactive oxygen species; STAT3, signal transducer and activator of transcription 3; TMT, tandem mass tag; UC, ulcerative colitis
Graphical abstract
Summary.
Hyperbaric oxygen therapy improves disease activity in ulcerative colitis through a reduction in STAT3 mediated neutrophil degranulation and shifts in microbial composition and metabolism. Treatment resistance is mediated by microbial adaptations to oxygen exposure, particularly for Akkermansia muciniphila.
Maintenance of oxygen homeostasis is essential to survival, and the human body has developed adaptative programs of transcriptional response to hypoxia that allow for maintenance. Dysregulation of these responses is a core feature of ulcerative colitis (UC), where tissue hypoxia results in epithelial barrier disruption, microbial dysbiosis, recruitment and activation of neutrophils and pathogenic T cells, and overproduction of inflammatory cytokines.1 These changes further augment microenvironmental hypoxia and ultimately lead to worsening colonic inflammation.1 Therapeutics are in development targeting aspects of these dysregulated hypoxia response pathways.2 However, it remains to be defined what the key mediators of this complex host-microbe response are.
In a phase 2A multicenter, double-blind, sham-controlled, randomized trial for moderate-severe hospitalized UC patients, we demonstrated that the intermittent delivery of high amounts of oxygen to the colon through hyperbaric oxygen therapy (HBOT) resulted in significantly greater rates of clinical response and remission and significantly lower rates of progression to infliximab or colectomy during the hospitalization.3 In a subsequent phase 2B multicenter, dose-finding, randomized trial, we further confirmed treatment efficacy of HBOT in hospitalized UC patients and observed 5 HBOT sessions to be superior to 3 HBOT sessions for improving disease outcomes.4 In the current study we aimed to define the host-microbe response to HBOT in UC to better understand mechanisms through which targeting hypoxia may result in disease remission. Insights gained from our work will be of importance as we target hypoxia for the treatment of UC and other inflammatory disorders. The multi-omics approach and digital spatial profiling (DSP) performed will be of value to researchers looking to apply these technologies to other diseases, particularly when attempting to study the role of neutrophils in intestinal inflammation because of technical challenges and complexities with isolating these immune cells from mucosal tissue.
Results
HBOT Stabilizes Mucosal Hypoxia Response Pathways Independently of Clinical Response Status
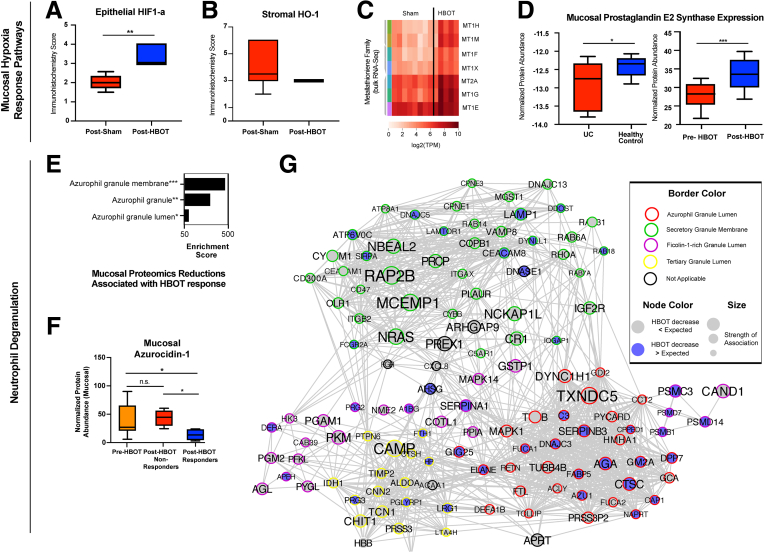
HBOT resulted in (1) increase in epithelial hypoxia inducible factor (HIF)-1α activity relative to sham (Figure 1A), (2) a more uniform distribution in activity for stromal heme oxygenase-1 relative to sham (Figure 1B), and (3) an increase in gene expression for metallothionein (MT1 and 2) family members (Figure 1C). Proteomics of paired biopsies also revealed an increase in mucosal prostaglandin E2 synthase (Figure 1D). However, the effect of HBOT on these mucosal hypoxia response pathways was independent of response status and was observed in both HBOT responders and non-responders.
Figure 1.
Translational sequencing studies identify effect for hyperbaric oxygen on hypoxia response pathways and neutrophil degranulation. (A and B) Boxplots of immunohistochemistry score for epithelial HIF-1α and HO-1 activity for sham (n = 8) and HBOT (n = 10) treated UC patients. Upper and lower limits represent sample score range (A, P < .001, unpaired t test; B, not significant). (C) Post-treatment bulk RNA sequencing results of mucosal biopsies for sham (n = 8) and HBOT (n = 5) treated UC patients enrolled in the phase 2A clinical trial. Metallothioneins are a family of highly conserved stress-induced proteins that, by control of cellular zinc homeostasis, protect against oxidative stress. (D) Boxplot of prostaglandin E2 synthase proteome abundance in mucosal biopsies is demonstrated to be significantly lower in UC patients compared with healthy controls using a publicly available proteomics dataset (left bar graph, n = 20; P = .03, unpaired t test), and prostaglandin E2 synthase expression in mucosal biopsies is demonstrated to be significantly increased with HBOT exposure in the phase 2 trials (right bar graph, n = 20; P < .0001, paired t test). Ranges shown represent minimum and maximum. Sample measurement range. (E) TMT-multiplexed proteomics-derived enrichments in proteins reduced by HBOT treatment were strongly associated in proteins associated with azurophilic granule membranes, granules, and granule lumen. Enrichment strength (plotted) is derived from the odds ratio and adjusted P value, output from Enrichr algorithm. (F) Day 10 pre- and post-HBOT mucosal proteome demonstrated a significant decrease in azurocidin-1 protein abundance in HBOT responders (n = 5) compared with pre-HBOT (analysis of variance–adjusted group-wise comparison, P = .04) and non-responders (P = .01), whereas non-responders were not significantly different from pre-HBOT condition (n = 5, P > .99). (G) Fecal proteome dataset-based neutrophil-associated protein network generated by protein-protein interaction engine STRING (reformatted in CytoScape), segregated by subnetworks of granule type-specific proteins. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, analysis of variance adjusted. Range represents minimum and maximum sample measurements.
HBOT Response Status Associated With Reductions in Azurophilic Granule Related Proteins
Proteomics of mucosal tissue and fecal samples both identified an enrichment of effect for HBOT on reducing neutrophil degranulation, with specificity of effect for reducing azurophilic granule related proteins (P < .001 for both mucosal and fecal proteome datasets). This effect was more pronounced among HBOT responders compared with non-responders (Figure 1E–G). In mucosal samples, a significant negative correlation was observed between response to HBOT and azurocidin-1 (r = –0.65, P = .04), and HBOT responders had significantly lower normalized protein abundances for azurocidin-1 at day 10 follow-up compared with HBOT non-responders (Figure 1F). In fecal samples, reductions in azurophilic granule proteins observed with HBOT exposure were greater than would be expected with changes in disease activity alone, and this was observed in both HBOT clinical trial datasets independently (Figure 1G). Separate analyses linked proteins related to NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) with a positive response to HBOT (data not shown).
To further probe neutrophil response, DSP on formalin-fixed paraffin-embedded mucosal tissue was then performed with a specific focus on neutrophils because of observed effects of HBOT on neutrophil degranulation. When comparing differences between groups (HBOT and controls) for within-group (post- vs pre-) changes in neutrophil gene expression, a significant reduction in CD68 (logFC –0.53, adjusted P = .012) and integrin alpha X (logFC –0.65, adjusted P = .002) was observed with HBOT relative to controls. Integrin alpha X is part of the neutrophil degranulation pathway, and CD68 is a membrane bound protein that is found in primary (azurophilic) neutrophil granules.5
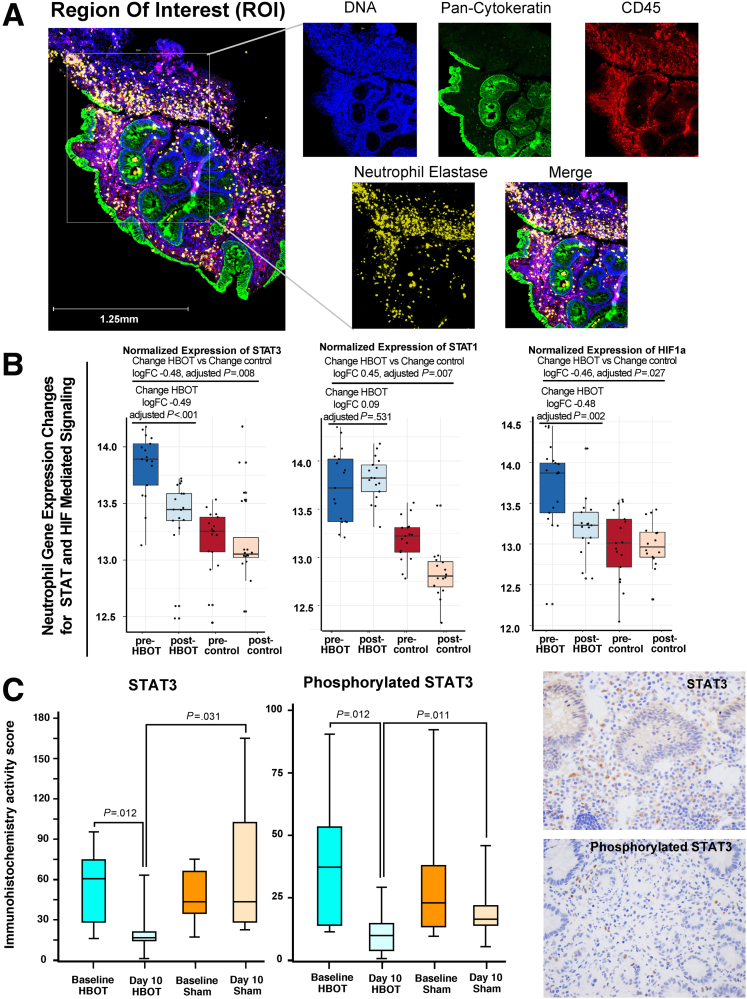
HBOT Reduces STAT3 Activity in Neutrophils
DSP of mRNA transcripts revealed an enrichment of signal transducer and activator of transcription 3 (STAT3) mediated interleukin (IL) signaling related to HBOT (P < .001). HBOT-treated patients had significantly reduced STAT3 and HIF-1α compared with controls treated with biologics (Figure 2A and B). Each patient sample had between 5 and 7 neutrophil regions analyzed with DSP, allowing for an assessment of heterogeneity in effect and changes. HBOT resulted in a significant decrease in STAT3 across all neutrophil regions at follow-up for all treated patients; however, this consistency in reduction was not observed for HIF-1α. Other notable neutrophil gene expression changes related to the STAT3 pathway with HBOT versus controls (change HBOT versus change control) included IFNGR1 (logFC –0.89, adjusted P < .001), FAS (logFC –0.60, adjusted P = .003), IFNG (logFC –0.88, adjusted P = .008), IL12B (logFC –0.38, adjusted P = .024), CSFR1 (logFC –0.66, adjusted P = .007), B2M (logFC 1.27, adjusted P = .006), and IFNAR1 (logFC –0.38, adjusted P = .024). We observed no significant effect for HBOT on STAT3 or HIF-1α in the non-neutrophil immune cell regions (CD45+elastase-) (data not shown), demonstrating this effect of HBOT on STAT3 to be neutrophil specific.
Figure 2.
HBOT reduces STAT3. (A) Representative image for defining ROI from mucosal tissue in patients with UC receiving HBOT, used for DSP (blue = DNA, green = Pan-Cytokeratin, red = CD45, yellow = Neutrophil Elastase). (B) Single-cell gene expression changes of STAT3 (left), STAT1 (center), and HIF-1α (right) in neutrophil-specific ROI (CD45+elastase+) in HBOT-treated vs matched control patients with UC not treated with HBOT. Each dot represents a unique ROI within a single sample, and on average 6 neutrophil-specific ROIs were available per sample per patient. Ranges represent maximum and minimum calculated values. (C) Boxplot of immunohistochemistry scores and staining for STAT3 and phosphorylated STAT3 done for mucosal biopsies. Measurements were taken before and after exposure to sham (n = 8) or HBOT (n = 22) in the phase 2 trials. Ranges represent absolute minimum and maximum. Sample values. Statistical significance derived using analysis of variance–adjusted P value calculations.
This effect of HBOT on STAT3 was confirmed by using immunohistochemistry and observed that HBOT-treated patients had a significant reduction in both STAT3 and phosphorylated STAT3 at study day 10 compared with baseline, with no significant changes in these markers in the sham-treated patients, and day 10 values for STAT3 and phosphorylated STAT3 were significantly lower in HBOT versus sham-treated patients from the randomized phase 2 clinical trials (Figure 2C).
HBOT Reduces Mitogen-Activated Protein Kinase and Markers of Reactive Oxygen Species and Increases the Pro-apoptotic BIM and the NLRP3 Inhibitor BCLXL
In the neutrophil transcriptome datasets, a significant decrease was observed in CXCR6 with HBOT (logFC –0.61, adjusted P = .002), known to be involved in reactive oxygen species (ROS) mediated injury via mitogen-activated protein kinase (MAPK).6 In the neutrophil proteome datasets, HBOT-treated patients had a significant reduction in protein abundance for phosphorylated p38 MAPK (logFC –1.11, adjusted P = .007), phosphorylated ERK (logFC –0.95, adjusted P = .016), and phosphorylated JNK (logFC –0.70, adjusted P = .032) relative to controls. In the proteome datasets, a significant effect for HBOT was observed for neutrophil apoptosis (P < .001) and programmed cell death (P < .01). Alongside the above noted neutrophil changes in MAPK and ROS mediated pathways, we observed a significant effect for HBOT on increasing neutrophil protein expression for B-cell lymphoma-extra large (BCLXL) (logFC 0.69, adjusted P = .008) and bcl-2 interacting protein (BIM) (logFC 0.44, adjusted P = .033). No significant effect was seen for HBOT on neutrophil protein expression for bcl-2–associated death promoter (adjusted P = .62), and in the neutrophil transcriptome dataset no significant effect was seen for HBOT on neutrophil gene expression for bcl-2 (adjusted P = .18).
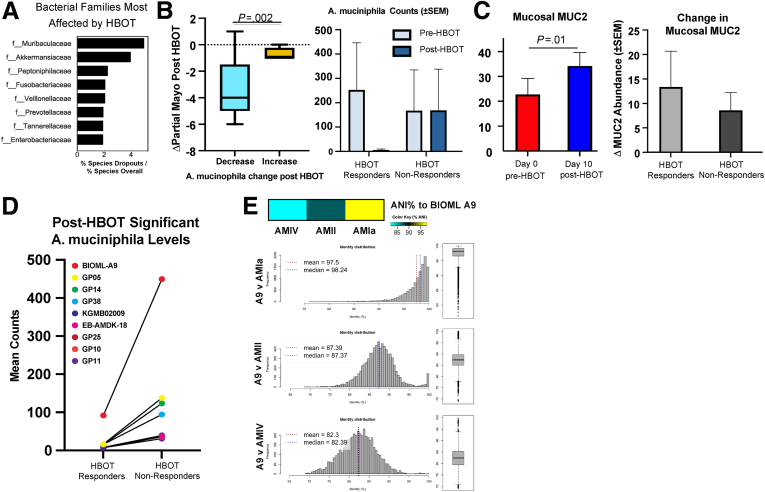
HBOT Reduces Akkermansia and Increases MUC2 in UC Patients
HBOT resulted in a relative increase in the Firmicutes:Bacteroidetes ratio within the metagenome (P = .076 for linear trend over time) and a reduction in host proteins relative to Firmicutes in the metaproteome (P = .013 for linear trend over time). Fecal metabolomics observed an association between HBOT response status and lithocholic acid (LCA) levels (r = 0.9 change in partial Mayo and LCA, P = .0002). Within the 16S dataset, we observed this proportional increase in Firmicutes to be a result of a significant reduction in Shannon diversity with HBOT responders relative to sham-treated patients. The degree of reduction in Shannon diversity was linearly related to degree of HBOT response (r = –0.72, P = .04) and number of HBOT treatment sessions used (r = –0.56, P = .03).
The 2 most reduced families by HBOT were Muribaculaceae (formerly Bacteroidales S24-7) and Akkermansiaceae (Figure 3A), both known to be mucinophiles. Reductions in A muciniphila specifically were significantly associated with reductions in partial Mayo scores with HBOT (Figure 3B). We also observed a significant accompanying increase in MUC2 with HBOT in the mucosal proteomics dataset (Figure 3C, left), and the relative increase in MUC2 was greater in HBOT responders compared with HBOT non-responders (Figure 3C, right). Fecal proteomics confirmed an increase in MUC2 with HBOT, which was incremental, that was based on duration of exposure (data not shown). In the fecal proteomic and metagenomic datasets we observed an association between reductions in A mucinophilia and higher MUC protein values (data not shown). HBOT response may therefore be mediated in part by the dropout of mucus digesting microbes, allowing for enhanced mucus layer strength, strengthening of the epithelial barrier, and repopulation of Firmicutes and Firmicute-related bile acid metabolism. Together, these effects may promote improvement in colitis.
Figure 3.
Multi-omic characterization of fecal samples identifies HBOT response status associated increases in Firmicutes and bile acid production. (A) Top 8 taxonomic families most affected by HBOT treatment calculated ratiometrically by comparing the percentage of families with species negatively altered by HBOT treatment with their overall species-level percentage representation in metagenomic features. (B) Group-wise comparison of changes in partial Mayo score as a function of post-HBOT loss in the genus Akkermansia (Welch t test, P = .014). Patients showing a decrease (<50 counts compared with their pre-HBOT condition) or no measurable levels were in decreased group, whereas no change or increase was included in the increased group. Range represents maximum calculated values. (C) Left: Group-wise comparison (paired t test, error bars plotted using ± standard error of the mean) of mucosal proteomics abundance changes in MUC2 comparing pre-HBOT (day 0, n = 10) and post-HBOT (day 10, n = 10). Right: Group-wise comparison of post-HBOT mucosal biopsy levels of MUC2 by response group. Results were not significant (unpaired t test, P = .57). (D) A muciniphila strains significantly (P < .05, uncorrected) higher post-exposure in HBOT non-responders compared with HBOT responders. (E) Heatmap generated using mean percentage scores of Average Nucleotide Identity (ANI) between BIOML A9 compared with several canonical phylogroups, with accompanying histograms of ANI scores comparing BIOML A9 and several phylogroups (left) and their boxplot distributions (bottom).
Strain-Level Variation of A muciniphila Comparing HBOT Non-Responders With Responders
To better characterize the lack of A muciniphila reduction in HBOT non-responders, we assessed A muciniphila strain variation in inflammatory bowel disease patients. Shotgun metagenomic data were compared with 279 unique reference A muciniphila strains, including strains of A muciniphila previously identified to be oxygen tolerant and/or resistant (AMIa and AMII phylogroups).7,8 We identified 10 strains significantly greater in post-HBOT non-responders compared with the post-HBOT responders. Six of these strains (GP05, GP14, GP38, GP25, GP10, and GP11) are already known to belong to the oxygen-resistant AMIa or AMII phylogroups. One of the strains not previously classified (BIOML-A9) was 5-fold higher in HBOT non-responders versus HBOT responders (Figure 3D). A genomic comparison of BIOML-A9 with different phylogroups demonstrated it strongly aligned with the AMIa and AMII phylogroups (Figure 3E). Together, these data suggest that A muciniphila adaptation to oxygen exposure with ongoing mucus digestion may play an important role in treatment resistant to HBOT.
Colonization of Mice With HBOT-Shaped Microbial Communities From Responders Reduces Colitis
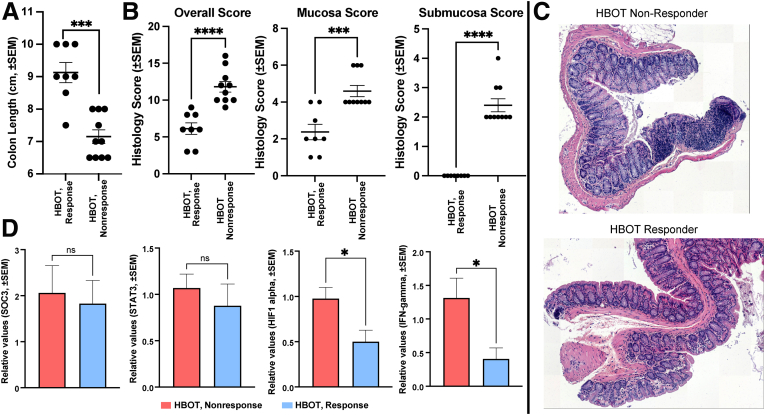
IL10-/- mice demonstrate a colitis phenotype driven by NLRP3 inflammasome activity9 and thus resemble phenotypes observed in UC patients. Germ-free IL10-/- mice were colonized with post-treatment stool from 4 HBOT-treated patients, 2 clinical responders (n = 5 mice per patient sample) and 2 clinical non-responders (n = 5 mice per patient sample) to characterize the role HBOT-resulting alterations in gut microbe composition had on host inflammation. All 4 patients had persistent endoscopic inflammation at the time of stool collection. Despite all 4 patient stool donors having active endoscopic inflammation, mice colonized with stool from the 2 HBOT clinical responders had longer colon length and less overall inflammation (Figure 4A–C). Polymerase chain reaction confirmed differences in colitis activity were not related to induction of the STAT3 pathway (Figure 4D). No significant differences were observed by flow cytometry analysis of T cells (data not shown). Altogether, these data demonstrate a central role played by the intestinal microbiota in driving HBOT-induced phenotypical response.
Figure 4.
Fecal colonization of IL10-/- mice with stool from responders and non-responders to HBOT demonstrates differential modulation of colitis. Group-wise comparison (Welch unpaired t test, ± standard error of the mean) of (A) colon length (P = .0004) and (B) histology scores between IL10-/- germ-free mice colonized with post-HBOT stool from 2 non-responder donors (n = 10 mice) and 2 responder donors (n = 8 mice) measuring overall (eg, summed) histologic inflammation (P < .0001), mucosa (P = .0004), and submucosa (P < .0001). (C) Representative colon histology images from mice colonized with stool from HBOT non-responders or HBOT responders. (D) Bar plot of expression differences between mouse colonized with responder or non-responder stool (n = 8 replicates, non-responders = 10 replicates) of SOCS3 (not significant), STAT3 (not significant), HIF-1 (Welch unpaired t test, P = .017), and interferon-γ (Welch t test, P = .043) as measured by quantitative polymerase chain reaction on colon tissue.
Discussion
HIF-1α is important in UC pathogenesis, and targeting this may be of therapeutic benefit. Using targeted and untargeted sequencing approaches, which benefited from novel proteomics and transcriptomics technology, we identified a specific effect for HBOT on HIF-1α activity. In the epithelium HBOT resulted in significantly higher HIF-1α expression, and in neutrophil HBOT resulted in significantly lower HIF-1α expression; however, these changes were not consistent across all regions of neutrophilic infiltrate, and they did not directly correlate to HBOT response status. Furthermore, we were not able to identify consistency in associations or directionality in changes between HBOT and HIF-1α across other approaches (bulk RNA sequencing or mucosal tandem mass tag [TMT]-multiplexed proteomics). Taken together, HBOT impacts HIF-1α, but changes in HIF-1α alone do not explain mechanisms underlying improvements in UC disease activity with HBOT.
Hypoxic microenvironments enhance neutrophil degranulation, particularly for azurophilic granules, and this has been thought to be partially HIF-dependent.10 HIF-1α is the predominant HIF subtype found in neutrophils; however, hypoxia-augmented degranulation could not be re-demonstrated with pharmacologic HIF-1α stabilization and activation alone.10 HIF-independent pathways involved in neutrophil degranulation under hypoxic conditions include ROS, MAPK, and STAT3 signalling.10,11 Using TMT-multiplexed proteomics, we identified a consistent and highly significant association between HBOT response status and neutrophil degranulation, with specificity of effect for the STAT3-NLRP3 inflammasome-related azurophilic granules in both the fecal and mucosal proteomics datasets. NLRP3 is directly linked to azurophilic granules, with mutations of NLRP3 being responsible for hypersensitive inflammasome function disorders through an azurophilic granule-selective process.12 STAT3 plays a central role in NLRP3 inflammasome-mediated inflammation, and prior studies have demonstrated that STAT3 inhibition abrogates NLRP3 protein expression and IL1ß release.13,14 In the DSP dataset, we observed a significant reduction in neutrophil STAT3 mRNA abundance, phosphorylated MAPK protein expression, and markers of ROS-mediated injury. STAT3 and NLRP3 are known targets of JNK, and inhibition of MAPK signalling in neutrophils abrogates the release of azurophilic granule contents.11,13 At the single-cell level in the proteome dataset, we observed HBOT to significantly increase the pro-apoptotic protein BIM alongside BCLXL. Hypoxia is known to prolong neutrophil survival and inhibit apoptosis,15 and HBOT has been shown to increase neutrophil apoptosis and enhance their clearance.16 BIM, a member of the Bcl-2 family, is known to promote neutrophil apoptosis and has been demonstrated to limit cytokine-mediated prolonged survival of neutrophils.17 BCLXL-mediated regulation of neutrophil apoptosis is ROS-dependent,18 and BCLXL has been demonstrated to inhibit NLRP3 inflammasome formation and IL1ß activation.19 Prior in vitro studies have demonstrated an increase in BCLXL and reduction in ROS, MAPK, STAT3, and NLRP3 with HBOT,20, 21, 22, 23, 24 further supporting our observations. Together these data, in combination with prior in vitro studies from other groups supporting our observations, suggest that HBOT improves UC disease activity in part through the neutrophil STAT3 pathway.
Recognizing that a microbiome-neutrophil crosstalk exists,25 the microbiome has been observed to regulate STAT3 expression specifically,26 and HBOT significantly alters colonic microbial composition and function in animal models,27 we sought to further assess the effects of HBOT on the microbiome and whether microbial changes may have contributed to observed host effects of HBOT on STAT3. We observed an acute dropout of bacterial subpopulations after HBOT, with an accompanying proportional increase in Firmicutes and a key secondary bile acid, LCA. UC patients have reduced LCA, this deficiency is due to a reduction in Firmicutes, and LCA supplementation is protective against colitis in animal models.28 Several of the Firmicutes increased with HBOT in our trial are known producers of secondary bile acids,29 and in healthy volunteers hyperbaric conditions have been observed to increase bile acid producing Firmicutes.30 The specific microbes most affected by HBOT, which allowed for this proportional increase in Firmicutes, included known mucinophiles with an association between A muciniphila and HBOT response status. Akkermansia is associated with risk of inflammatory bowel disease development,31 but data are conflicting on whether A muciniphila is pathogenic or protective against colitis.32, 33, 34 Strain-specific anti- or pro-inflammatory properties exist for A muciniphila.35,36 Although A muciniphila is considered to be a strict anaerobe, it is capable of adapting to oxygen exposure through increased expression of respiration genes, allowing it to take advantage of epithelial oxygen gradients.7 We observed persistence of A muciniphila strains known to be oxygen tolerant and/or resistant (AMIa and AMII phylogroups) in the HBOT non-responders, suggesting adapation to oxygen may explain HBOT non-response. Furthermore, we observed an association between HBOT and MUC2 protein levels. MUC2 is the core structural protein for the inner mucus layer, and weakening of this barrier is an early event in UC pathogenesis in humans and animal models (IL10-/-).37 Mutations in MUC2 determine early dysbiosis in colitis models,38 and MUC2-/- models are characterized by increased JAK/STAT activity with specificity toward IL6/STAT3 activity.39,40 MUC2 deficiency–related dysbiosis is characterized by reduction in Firmicutes and increases in the Muribaculaceae and Akkermansiaceae families.38 Therefore, our observed changes with HBOT for a reduction in Muribaculaceae and Akkermansiaceae families, increase in Firmicutes, and increase in MUC2 are in line with expected pathologic mechanisms. Using animal models, we were able to confirm that global changes in microbial composition and metabolite alterations were associated with differential colitis induction; however, there were no differences in induction of STAT3. This suggests that HBOT response mechanisms involve direct effects on host neutrophil STAT3 pathways and separate effects on the microbiome, which may further augment improvements in disease activity.
Our study has multiple strengths, which include the use of baseline and follow-up mucosal and fecal samples from 2 prospective clinical trials, separate healthy and disease severity matched control samples, use of novel and emerging techniques for proteomics and mRNA transcriptomics, our ability to study tissue neutrophils, and the consistency of observation and strength of associations for our proposed mechanisms of action across sequencing datasets and prior literature. Most notably, we provide the first application of DSP in UC to study neutrophils, and it was this approach that linked observations toward a unifying central hypothesis related to neutrophil degranulation and STAT3. Furthermore, our proposed host response mechanism is confirmed by multiple prior in vitro studies by other groups, and the contribution of changes in microbial composition and metabolism to improving disease activity was assessed through dedicated in vivo colonization experiments. Our study is not without limitations; the most notable is the lack of in vivo confirmation of STAT3 pathway related mechanisms for HBOT. Prior neutrophil STAT3 knockout models that developed spontaneous chronic enterocolitis also resulted in knockout of STAT3 in macrophages,41,42 and it was the macrophage STAT3 deficiency that contributed to development of colitis.43 A more neutrophilic-specific STAT3 knockout model could be made through crossing STAT3flox mice with MRP8-Cre mice; however, this will still result in knockout of STAT3 in up to 20% of macrophages, depending on the tissue of interest.44 Prior work has suggested that neutrophil STAT1 activity may be a determinant of UC activity,45 and conditional deletion of STAT3 may result in increased activity of STAT1 through heterodimer receptor occupancy, which may further compound phenotypic presentations of animal models.46 Further confirmation of our observations is best done within a larger phase 3 clinical trial program where comparisons of neutrophil expression profiles can be made between randomized HBOT and sham-treated UC patients.
Conclusion
Our multi-omics analysis has observed 2 complementary mechanisms through which HBOT may improve UC activity. First, we observed a reduction in neutrophil degranulation through the STAT3-NLRP3-azurophilic granule pathways. Second, we observed a decrease in mucus digesting bacteria, with an accompanying increase in MUC2, epithelial HIF-1α, Firmicutes, and bile acid production. Further investigation of A muciniphila strain level adaptations are needed to understand mechanisms of HBOT non-response.
Methods
Clinical Trial Overview
We used clinical data and human biospecimens collected from phase 2 clinical trials3,47 for HBOT in UC patients hospitalized for a medically refractory moderate to severe UC flare (Full Mayo score ≥6, all subscores 2–3). All patients received intravenous steroids along with either HBOT (2.4 atmospheres absolute (sea level), 100% oxygen, 90-minute sessions, one session per day) or sham air (1.34 atmospheres absolute, 21% oxygen, 90-minute sessions, one session per day) over 3–5 days. Disease activity was measured by using the Mayo score. Endoscopy subscores were available at study days 0 and 10, with rectal bleeding and stool frequency subscores available at days 0, 3, 5, and 10. The institutional review board at each center approved the protocols, and all patients gave written informed consent for participation in the trial and the translational research outlined using human biospecimens collected. Trial details can be found in the original publications.3,4 Trial registration can be found at clinicaltrials.gov (#NCT02144350). Patient demographics from the trial and additional translational samples used can be found in Tables 1 and 2.
Table 1.
Demographics of UC Patients Included
| Phase 2A trial: HBOT- or sham-treated UC (n = 13) | Phase 2B trial: HBOT-treated UC (n = 20) | Disease severity matched UC without HBOT (n = 16) | |
|---|---|---|---|
| Age, mean, y (SD) | 44 (19) | 37 (15) | 43 (16) |
| Male gender, n (%) | 6 (46) | 10 (50) | 13 (81) |
| Prior anti-TNF, n (%) | 7 (53) | 15 (75) | 10 (63) |
| Prior vedolizumab, n (%) | — | 7 (35) | 6 (38) |
| Prior tofacitinib, n (%) | — | 5 (25) | 1 (6) |
| Mayo endoscopic subscore of 3, n (%) | 8 (62) | 17 (85) | 7 (43) |
| CRP, median (IQR) | 93 (15–123) | 14.4 (3–51) | — |
| Albumin, median (IQR) | 3.3 (3.1–3.6) | 3.5 (3.1–3.6) | — |
CRP, C-reactive protein; IQR, interquartile range; TNF, tumor necrosis factor.
Table 2.
Demographics for DSP UC Patients
| Case | Baseline endoscopy | Description | Follow-up endoscopy and histology | Clinical outcome |
|---|---|---|---|---|
| UC patients treated with hyperbaric oxygen during hospitalization for acute severe flare | ||||
| 1 | MES 3 with ulcers | Male, biologic and immunomodulator naive before hospitalization, clinical response to HBOT by days 3 and 5 | MES 1, no friability; histology with persistence of neutrophils | Relapse requiring infliximab in 3 months |
| 2 | MES 3 with ulcers | Female, 1 prior anti-TNF agent (adalimumab) which patient was failing at time of hospitalization, clinical response to HBOT by days 3 and 5 | MES 0, normal vascular pattern; histology with persistence of neutrophils | Able to maintain response for 3 months with continued adalimumab, later relapsed requiring vedolizumab |
| 3 | MES 3 with ulcers | Male, prior failure of 2 anti-TNF agents (infliximab, adalimumab), vedolizumab, and failing 10 mg PO BID tofacitinib at time of hospitalization, clinical response to HBOT by days 3 and 5 | MES 1, no friability; histology with persistence of neutrophils | Able to remain colectomy free on 5 mg PO BID tofacitinib |
| UC patients treated with standard of care in outpatient setting for acute severe flare | ||||
| 4 | MES 3 with ulcers | Female, biologic and immunomodulator naive, started on vedolizumab after baseline endoscopy | MES 1, no friability; histology with persistence of neutrophils | Good clinical response with later requirement for Q4 week vedolizumab |
| 5 | MES 3 with ulcers | Female, prior failure of 2 anti-TNF agents (infliximab, adalimumab), and failing vedolizumab at time of baseline endoscopy, started on golimumab after baseline endoscopy | MES 1, no friability; histology with persistence of neutrophils | Good clinical response with maintenance on golimumab |
| 6 | MES 3 with ulcers | Female, 1 prior anti-TNF agent (infliximab) which patient was failing at time of baseline endoscopy, started vedolizumab after baseline endoscopy | MES 0, normal vascular pattern; histology with persistence of neutrophils | Good clinical response with maintenance on vedolizumab |
Anti-TNF, anti–tumor necrosis factor antagonist; BID, twice daily; MES, Mayo endoscopic subscore; PO, oral; Q4, every 4.
Mucosal Tissue Bulk RNA Sequencing
Bulk RNA sequencing was done for available pre- and post-intervention mucosal biopsies (n = 5 HBOT and n = 8 sham-treated patients) from the sham-controlled phase 2A trial based on standard pipelines at the University of California San Diego Institute for Genomic Medicine facility. Biopsy samples were homogenized manually on ice in TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA) and processed according to the manufacturer’s instructions. cDNA libraries were prepared using TruSeq non-Stranded Total RNA Sample Prep Kit (Illumina, San Diego, CA) according to manufacturer’s instructions. cDNA libraries were sequenced with a HiSeq2500 (Illumina) at the University of California San Diego Institute for Genomic Medicine facility. RNAseq reads were processed by first removing adapter sequences with cutadapt (1.14.0) and mapped to a database of repetitive elements (RepBase 18.05) using STAR (2.4.0i). Reads that went unmapped to repeat elements were then mapped to the human genome (hg19) using default parameters. Aligned reads were annotated using featureCounts (v1.5.3) with Gencode V19 annotations. Differential expression was performed with DESeq2 (1.22.1). Gene Ontology enrichment was performed against significant (adjusted P ≤ .05) dysregulated genes using a background of all expressed genes (average DESeq2 normalized count ≥ 1). Ontology data were downloaded from the Ensembl (GRCh37) Biomart API.
Mucosal Tissue Digital Spatial Transcriptomics and Proteomics
DSP was done for pre- and post-intervention mucosal biopsies from 3 hospitalized UC patients treated with HBOT in the phase 2 trials and 3 disease-severity and response-status matched outpatient UC controls not treated with HBOT. All 3 of the HBOT-treated patients included in the DSP cohort had severe endoscopic activity (Mayo endoscopic subscore 3) at day 0 with endoscopic improvement (Mayo endoscopic subscore 0–1) at day 10. The 3 outpatient UC controls were matched to the 3 HBOT UC cases for baseline disease severity, biologic exposure status, and response status, with all 3 having severe endoscopic activity (Mayo endoscopic subscore 3) at baseline and endoscopic improvement (Mayo endoscopic subscore 0–1) at follow-up. The GeoMx DSP developed by NanoString Technologies (Seattle, WA) enables spatially resolved, high-plex (10s–10,000s) digital quantitation of proteins and mRNA in tissue. The assay uses unique reagents (antibodies or RNA probes) coupled to UV photocleavable oligonucleotide barcodes. After incubation and hybridization of the GeoMx reagents along with visualization reagents to slide-mounted formalin-fixed paraffin-embedded tissue, the DSP scans the slides and presents a high-quality image for region of interest (ROI) selection. The oligonucleotide tags are released from user-selected regions of the tissue with focused UV light based on the user region and segmentation selections. The digital micromirror device of the DSP instrument tunes the UV light with 1-μm resolution, allowing great flexibility for region selection including irregularly shaped and noncontinuous segments. Released tags are quantitated with nCounter optical barcodes or an Illumina next-generation sequencer. Counts are mapped back to the tissue location by the software, resulting in a spatially resolved digital profile of protein or mRNA abundance. For the current study, protocols were followed according to the manufacturer’s instructions and in accordance with prior published work using this platform.5,6 Briefly, a 5–10 μm thick formalin-fixed paraffin-embedded tissue section was stained with oligo-tagged antibodies to detect a predetermined panel of markers (Table 3).
Table 3.
RNA and Protein Sequencing Panels for DSP
| RNA sequencing panel | ||||||||
| BATF3 | CMKLR1 | SDHA | BCL2 | CD86 | ITGB2 | VSIR | CSF1R | CXCL10 |
| TIGIT | UBB | FAS | CD276 | VEGFA | PDCD1LG2 | PTEN | CXCL9 | IL15 |
| ICOSLG | ITGAM | CD47 | MS4A1 | TBX21 | STAT3 | EPCAM | CD274 | CD3E |
| IFNAR1 | CCL5 | TNFRSF9 | HAVCR2 | HLA-E | STAT1 | PSMB10 | POLR2A | pan-melanocyte |
| CCND1 | NKG7 | PECAM1 | ICAM1 | HLA-DQ | STAT2 | CD74 | CXCR6 | CTNNB1 |
| PDCD1 | ITGB8 | ITGAX | RAB7A | IFNG | IL6 | TNF | KRT | IFNGR1 |
| HIF1A | OAZ1 | MKI67 | CD8A | IL12B | B2M | DKK2 | CD40LG | |
| Protein sequencing panel | ||||||||
| CD4 | Histone H3 | SMA | HLA-DR | Ms IgG2a | CD20 | S6 | Rb IgG | CTLA4 |
| Ki-67 | Fibronectin | MsIgG1 | CD3 | CD68 | GZMB | CD8 | panCk | CD11c |
| CD56 | GAPHD | PD-1 | CD45 | PD-L1 | Beta-2 microglobulin | PD-L2 | CD127 | ICOS |
| CD80 | CD25 | CD40 | CD27 | CD44 | CD34 | FAP-alpha | CD66b | CD45RO |
| FOXP3 | CD14 | CD14 | CD163 | BIM | PARP | GZMA | Cleaved caspase 9 | BCL6 |
| BAD | NF1 | BCLXL | P53 | CD95 | Phos-Tuberin | Phos-GSK3a | PLCG1 | Phos-GSK3B |
| Phos-AKT | INPP4B | MET | Phos-AKT1 | Phos-PRAS40 | Pan-AKT | Phos-ERK | Phos-JNK | Phos-p38 MAPK |
| Phos-RSK | Pan-RAS | MAPK ERK | BRAF | EGFR | Phos-MEK1 | Phos-c-RAF | 4-1BB | Tim-3 |
| B7-H3 | STING | LAG3 | IDO1` | ARG1 | GITR | OX40L | VISTA | |
Oligo tags are attached to the antibody and ISH probes via a UV-photocleavable linker. ROI were selected on the visualized tissue on the basis of cell positivity for elastase and CD45. After ROI selection, oligos from the selected regions were released upon focused exposure to UV light using the Nanostring DSP instrument, and photocleaved oligos were aspirated via a microcapillary tube and stored in microplate wells. Last, the photocleaved oligos from the spatially resolved ROI in the microplate were quantified using next-generation sequencing Illumina workflows.
Data were pre-processed in Nanostring DSP Software v 2.1.0.106 normalizing to ERCC counts and housekeeping genes to adjust for systemic variations to enable expression counts to be compared across genes and samples. Quality control metrics were assessed, and 2 low-quality regions were removed because of low nuclei counts <20. The distribution of the pre-processed count data reflected a negative binomial distribution as expected, with a need for further normalization with TMM and voom. After TMM and voom normalization, the data met the assumptions necessary for downstream statistical analyses including differential expression analysis using limma-voom accounting for within case repeated measures (ROIs) and spatial deconvolution analysis.
The R BioConductor packages edgeR and limma were used to implement the limma-voom method for differential expression analysis. We normalized and analyzed Neutrophil and Immune Cell ROIs separately and subset RNA only and Protein only for downstream analysis. Trimmed mean of M-values (TMM) normalization was applied. The experimental design was modeled on treatment and time accounting for repeated measures using consensus correlation with the design (∼0 + treatment_group_time). The voom method was used to model the mean-variance relationship, after which lmFit was used to fit per-gene linear models, and empirical Bayes moderation was applied with the eBayes function. Significance was defined by using an adjusted P value cutoff <.05 after multiple testing correction using a moderated t statistic in limma. Cell-type deconvolution was performed using the log-normal regression algorithm of Danaher et al48 and its implementation in Bioconductor. Inputs were the normalized expression values (cpm), the background matrix of background values set to a value of 6, and the training matrix containing log2 expression values in the training set, which is part of the R package and named “Human_Cell_Landscape”.
Mucosal Tissue Proteomics
Each biopsy specimen was initially isolated from storage buffer, and 500 μL of lysis buffer was added (7% sodium dodecyl sulfate, 50 mmol/L Tris, 6 mol/L urea, pH 8.1). Samples were then sonicated for 10 seconds on 10-second off cycles for approximately 6 cycles or until specimen was completely lysed (QSonica, Newtown, CT). Samples were then reduced with 10 μL of 50 mmol/L dithiothreitol (Sigma-Aldrich) for 30 minutes at 56°C and subsequently alkylated with 30 μL of 50 mmol/L iodoacetamide (Sigma-Aldrich) for 1 hour at room temperature in the dark. To each sample, S-trap “binding buffer” (a 7:1 solution of 90% methanol, 10% 1M TEAB pH adjusted to 7.1 with phosphoric acid) was added along with 50 μL 12% phosphoric acid. Samples were washed 5× with S-trap binding buffer and digested with 5 μg trypsin (for 3 hours at 47°C, as described in the S-trap protocol [https://cdn.shopify.com/s/files/1/0271/1964/8832/files/S-Trap_micro_protocol_long.4.6.pdf]). Peptides were then eluted with 50 μL 50 mmol/L TEAB, followed by 50 μL of 0.2% formic acid, and finally 100 μL of 50% acetonitrile, 0.2% formic acid. Eluate was captured, and the volume was evaporated using a Thermo Scientific Speedvac. Dried samples were then resuspended 500 μL 0.2% formic acid and desalted using Seppak tC18 cartridges and subsequently dried down (Waters WAT036820). Each sample was then resuspended in 1000 μL 50% acetonitrile, and peptide concentration was determined using Pierce Quantitative Colorimetric Peptide Assay (catalog #23275). Fifty micrograms of peptide was dried down and labeled using TMT10plex Isobaric Labeling reagents (catalog #90406) as described in Campeau et al.49 Labeled peptides were subsequently dried down, desalted, and fractionated using high pH reverse phase fractionation (Thermo Ultimate 3000 HPLC off-line fraction collector and c18 column, 37 s per well, 96 wells total). After fractionation samples were concatenated into 24 samples. 12 fractions were used in the analysis of each sample.
Labeled peptides were resuspended in 8 μL 5% formic acid, 5% acetonitrile wherein 3 μL was loaded onto an in-house laser-pulled 100 μm ID nanospray column packed to ∼30 cm with 1.8 μm C18 beads (Sepax Technologies, Newark, DE). Peptides were separated by reversed-phase chromatography on a Thermo Easy Nano-LC for 3 hours per fraction. Buffer A of the mobile phase contained 0.1% formic acid in high-performance liquid chromatography–grade water, whereas buffer B contained 0.1% formic acid in acetonitrile. The high-performance liquid chromatography flow rate was 0.300 μL/minute. Samples were run on a Thermo Fusion mass spectrometer that collected mass spectrometry (MS) data in positive ion mode within the 600–1200 m/z range (+2 charge) or 500–1200 m/z (+3/4 charge). A top-10 MS3 method was used on the Fusion with an initial Orbitrap scan resolution of 60,000. This was followed by high-energy collision-induced dissociation in the ion trap and subsequent reporter ion analysis in the orbitrap. Dynamic exclusion was enabled (repeat count of 1, exclusion duration of 90 s). The automatic gain control for FT full MS was set to 4e5 and for ITMSn was set to 1e4. ITCID was used at MS2 method, and the MS3 AGC was set to 2e5.
Spectra were searched using Proteomics Discoverer 2.1 and the SEQUEST algorithm using a target decoy strategy as previously described.8 Static modifications were set to carbamidomethylation (Δ57.021) and TMT tags (Δ229.163), whereas dynamic modifications were set to methionine oxidation (Δ15.995) and phosphorylation (Δ79.966) of serine, threonine, or tyrosine. Precursor tolerance was set to 50.00 ppm, and fragment tolerance was set to 0.6 Da. Resulting peptide spectral matches were filtered at a 0.01 false discovery rate (FDR) by the Percolator module against the decoy database. Peptide intensities were summed to the protein level.
Statistics were calculated and visualized using R and the following packages: ggplot2 2.2.1, Hmisc 4.0- 3, psych 1.7.8, ggpubr 0.1.5, and RColorBrewer 1.1-2. Univariate statistical analysis (P < .05) for HBOT (repeated-measures t test) samples and the public dataset (independent t test) were done by GraphPad Prism 9.0.0 (San Diego, CA).
Protein abundance was normalized to a pooled TMT “bridge” channel. In addition, each channel was further median scaled. Proteins with more than 50% of abundance values initially present in pre- and post-HBOT samples were subjected to imputation using the missForest R package (v1.4). Enrichment analysis was done by first recording statistically significant (P < .05, paired significance test) proteins that were negatively correlated with disease severity score post-HBOT and submitting this list of proteins to Enrichr, where the calculated “Combined Enrichment Score” was visualized.9
Immunohistochemistry
Immunohistochemistry was performed for HIF-1α and heme oxygenase 1 with an a priori hypothesis that HBOT response status would correlate to changes in these hypoxia response pathways. Post hoc immunohistochemistry was done for STAT3 and phosphorylated STAT3 to confirm observations made through DSP and proteomics. Formalin-fixed paraffin-embedded tissue sections of 4-μm thickness were cut and placed on glass slides coated with poly-L-lysine, followed by deparaffinization and hydration. Heat-induced epitope retrieval was performed using citrate buffer (pH 6.0) in a pressure cooker. Tissue sections were incubated with 0.3% hydrogen peroxidase for 15 minutes to block endogenous peroxidase activity, followed by incubation with primary antibodies overnight in a humidified chamber at 4°C. Antibodies used for immunostaining were anti-HIF-1α [EP1215Y] [1:100, rabbit monoclonal antibody] and anti- anti-heme oxygenase 1 antibody [EP1391Y] [1:50, rabbit monoclonal antibody]. Immunostaining was visualized with a labeled streptavidin-biotin using 3,3′-diaminobenzidine as a chromogen and counterstained with hematoxylin. Samples were quantitatively analyzed and scored on the basis of presence (positive) or absence (negative) of staining by a blinded pathologist with expertise in gastrointestinal diseases. Data are displayed as frequency of staining score, and a χ2 test was used to determine significance. Immunohistochemical slides were cut at 4 μm from formalin-fixed paraffin-embedded tissue and air dried at room temperature before baking at 60°C for 30 minutes. Automated protocol was performed on the Leica Bond Rx and included paraffin dewax, antigen retrieval, and staining. Heat-induced epitope retrieval using Bond Epitope Retrieval 1, pH6 (Leica Biosystems AR9640; Wetzlar, Germany) or Bond Epitope Retrieval 1, pH9 (Leica Biosystems AR9961) was incubated at 100°C for 20 minutes. Primary antibody staining was applied and incubated at room temperature for 30 minutes for STAT 3 (1:800). Primary antibody binding is detected and visualized using the Leica Bond Polymer Refine Detection Kit (DS9800) with DAB chromogen and hematoxylin counterstain. The automated Stainer/Detection system used was from Leica Biosystems Inc, and the antibodies used included Phospho-Stat3 (Tyr705, D3A7, XP Rabbit mAb #9145, 1:400 dilution) and Stat3 (D3Z2G, Rabbit mAb #12640, 1:800 dilution).
Fecal Multi-Omics Experiments
Fecal samples from 43 participants (n = 8 sham and n = 25 HBOT from phase 2 trials, n = 16 outpatient UC severity matched controls [IRB#150675], n = 4 healthy controls recruited through the American Gut Project50) underwent 16S rRNA gene amplicon sequencing, shotgun metagenomics, metabolomics, and metaproteomics. Samples in the phase 2A sham-controlled trial were available at days 0 and 10, and samples from the phase 2B dose-finding trial were available from days 0, 3, 5, and 10. Analyses were adjusted for shifts in disease severity over time and matched against healthy controls and the outpatient UC cohort not treated with HBOT or sham air. In addition to our standard Qiime2-based shotgun metagenomic search pipeline, samples were further searched with a targeted database containing 279 strains of A muciniphila in addition to the previous species-level entries found in our metagenomic database.
Fecal Proteomics
Fecal samples were measured out to ∼0.5 g and suspended in 5 mL of ice-cold, sterile tris-buffered saline. Samples were vortexed until completely suspended. Two 20 μmol/L vacuum, steriflip (Millipore, Burlington, MA) filters were used per sample to remove particulate. Cells were pelleted through centrifugation at 4000 rpm for 10 minutes at 4°C. Next, cells were lysed in 2 mL of buffer containing 75 mmol/L NaCl (Sigma-Aldrich), 3% sodium dodecyl sulfate (Fisher), 1 mmol/L NaF (Sigma-Aldrich), 1 mmol/L beta-glycerophosphate (Sigma-Aldrich), 1 mmol/L sodium orthovanadate (Sigma-Aldrich), 10 mmol/L sodium pyrophosphate (Sigma-Aldrich), 1 mmol/L phenylmethylsulfonyl fluoride (Sigma-Aldrich), and 1X Complete Mini EDTA-free protease inhibitors (Roche) in 50 mmol/L HEPES (Sigma-Aldrich), pH 8.5. An equal volume of 8 mol/L urea in 50 mmol/L HEPES, pH 8.5 was added to each sample. Cell lysis was achieved through two 15-second intervals of probe sonication at 25% amplitude. Proteins were then reduced with dithiothreitol (Sigma-Aldrich), alkylated through iodoacetamide (Sigma-Aldrich), and quenched. Proteins were next precipitated via chloroform-methanol precipitation, and protein pellets were dried. Protein pellets were resuspended in 1 mol/L urea in 50 mmol/L HEPES, pH 8.5 and digested overnight at room temperature with LysC (Wako, Huntleigh, IL). A second, 6-hour digestion using trypsin at 37°C was performed, and the reaction was stopped through addition of 10% trifluoroacetic acid (Pierce).
Samples were then desalted through C18 Sep-Paks (Waters, Milford, MA) and eluted with 40% and 80% acetonitrile solution containing 0.5% acetic acid. Concentration of desalted peptides was determined, and 50 μg aliquots of each sample were dried in a speed-vac. In addition, bridge channels consisting of 25 μg from each sample were created, and 50 μg aliquots of this solution were used in the 126 channels for each TMT (Thermo Fisher Scientific) 10 plex MS experiment. These bridge channels were used to control for labeling efficiency, inter-run variation, mixing errors, and the heterogeneity present in each sample. Mass defects for each TMT set were accounted for in the database searches according to manufacturer’s report per lot number. The lot numbers for TMT reagents were TA262347 for the first cohort of samples and VA296083 for the second cohort of samples. Each sample or bridge channel was resuspended in 30% dry acetonitrile in 200 mmol/L HEPES, pH 8.5 for TMT labeling with 8 μL of the appropriate TMT reagent. Reagents in the 126 channels were used to bridge between mass spec runs, and remaining reagents were used to label samples in random order. Labeling was carried out for 1 hour at room temperature and quenched by adding 9 μL of 5% hydroxylamine (Sigma-Aldrich). Labeled samples were acidified by adding 50 μL of 1% trifluoroacetic acid. After TMT labeling each 10-plex experiment was combined and desalted through C18 Sep-Paks and dried in a speed-vac.
Basic pH reverse-phase liquid chromatography (LC) followed by data acquisition through LC-MS2/MS3 was performed. Briefly, 60-minute linear gradients of acetonitrile were performed on C18 columns using an Ultimate 3000 HPLC (Thermo Fisher Scientific). Subsequently, 96 fractions were combined, and further separation of fractions was performed with an in-line Easy-nLC 1000 (Thermo Fisher Scientific) and a chilled autosampler. LC-MS2/MS3 data were collected on an Orbitrap Fusion (Thermo Fisher Scientific) mass spectrometer with acquisition and separation settings.
Data were processed using Proteome Discoverer 2.1 (Thermo Fisher Scientific). MS2 data were searched against both a public repository of microbial gut genes and the human proteome (www.uniprot.org). The Sequest searching algorithm was used to align spectra to database peptides. A precursor mass tolerance of 50 parts per million (ppm) was specified and 0.6 Da tolerance for MS2 fragments. Included in the search parameters was static modification of TMT 10-plex tags on lysine and peptide n-termini (+229.162932 Da), carbamidomethylation of cysteines (+57.02146 Da), and variable oxidation of methionine (+15.99492 Da). Raw data were searched at a peptide and protein FDR of 1% using a reverse database search strategy. A second search was performed taking proteins assigned to either the forward or reverse database into a subset database to increase the spectral match rate within complex data types like the fecal proteome.
TMT reporter ion intensities were extracted from MS3 spectra for quantitative analysis, and signal-to- noise values were used for quantitation. Additional stringent filtering was used to remove any moderate confidence peptide spectral matches or ambiguous peptide spectral match assignments. In addition, any peptides with a spectral interference above 25% were removed, as well as any peptides with an average signal-to-noise ratio less than 10. All signals from peptide spectral matches assigned to the same protein group were summed to represent protein abundance.
Protein relative abundances were normalized first to the pooled standards for each protein and then to the median signal across the pooled standard. An average of these normalizations was used for the next step. To account for slight differences in amounts of protein labeled, these values were then normalized to the median of the entire dataset and reported as final normalized summed signal-to-noise ratios per protein per sample. Hits to microbial proteins were removed to focus analyses toward host proteins. Proteomic datasets generated from inflammatory bowel disease patient samples resulted in final data tables containing 1803 proteins for cohort 1 fecal samples and 2928 proteins for cohort 2 fecal samples.
Data analysis was performed in python, and records of the code are available in corresponding Jupyter Notebooks for this project (https://github.com/rhmills/HBOT_Multiomics). Individual protein abundances from day 1 samples were compared with the abundances of day 10 samples using independent t tests of unequal variance in scipy (www.scipy.org). Enriched or depleted proteins were determined by π score, which accounts for both fold change and P value. A statistical cutoff for highly ranked associations was set to |π| > 1, as previously performed. Datasets generated for the 2 cohorts of patients were analyzed independently. Human protein gene functional enrichment analysis was performed by using Database for Annotation, Visualization and Integrated Discovery, with all human proteins identified as a background list. The python package, Seaborn (version 0.9.0), was used to generate bar plots comparing the –log10 transformed FDR P values provided by Database for Annotation, Visualization and Integrated Discovery for each functional grouping. To account for protein changes related to severity, data were analyzed from a prior project of our group with data available online at www.massive.ucsd.edu (study ID MSV000082094). For this, an equal number of UC patient samples were analyzed by identical methods using a subset of samples with the same average disease activity (determined by partial Mayo scores) as the day 1 and day 10 samples from the hyperbaric oxygen–treated patients of this study. A final enrichment score was compiled for each term by comparing the –log10 transformed FDR P values related HBOT treatment and subtracting the –log10 transformed FDR P values related to the mock severity-matched samples.
Protein-protein interaction networks were created through STRING-db. The association of each protein to hyperbaric oxygen was determined by subtracting the π of proteins in the mock severity-matched samples to the π related to hyperbaric oxygen treatment for proteins identified in both groups. These composite scores were determined as increased or decreased more than expected by using a cutoff of |1|. All significant associations were analyzed for protein-protein interactions using networks created through STRING-db. Associations between proteins were determined through default settings, accounting for textmining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. After identifying a strong association to neutrophil degranulation, proteins specific to this subnetwork were further analyzed. Connections were restricted to interactions between proteins within the query list only. Networks were subsequently visualized through Cytoscape (version 3.5.1). Edges within protein networks were based on the combined evidence scores, with the transparency of the edge indicating higher confidence in the association. Granule type associations were determined by comparing the percentage of significant or not significant proteins that each granule type occupied in relation to the percentage of proteins that each granule type occupied among the total list of human proteins identified (% Expected). Ratios were log2 transformed and plotted through a Seaborn bar plot.
16S Gene Amplicon Sequencing
Frozen samples were thawed and transferred into 96-well plates containing garnet beads and extracted using Qiagen MagAttract (Hilden, Germany) DNA kit adapted for magnetic bead purification. DNA was eluted in 100 μL Qiagen elution buffer. 16S rRNA gene amplicon sequencing was performed according to the Earth Microbiome Project. Briefly, the V4 region of the 16S rRNA gene (515f/806r) was amplified from 1 μL DNA per sample in triplicate.
Amplicons were quantified with Quant-iT PicoGreen dsDNA Assay Kit, and 240 ng, or maximum 15 μL, of each sample was pooled into a final library and cleaned using the QIAquick PCR Purification Kit. Paired-end sequencing was performed on the Illumina MiSeq using MiSeq Reagent Kit v3 (300-cycle). 16S fastq were split, demultiplexed, trimmed to 150 base pairs, and processed through deblur to generate amplicon sequencing varients using QIITA (Study ID 11149).
Metabolomics
Frozen human stool samples were lyophilized using a CentriVap Benchtop Centrifugal Vacuum Concentrator (Labconco, Kansas City, MO) attached to a Savant Ultra Low Pressure Refrigerated Vapor Trap RVT5105 (Thermo Fisher Scientific). Dried stool samples were weighed out to 30 mg (± 0.5 mg) dried weight, and 1 mL extraction solvent (1:1 methanol:water spiked with 0.625 μmol/L sulfamethazine) was added to each sample. Fecal samples were homogenized for 5 minutes at 25 Hz using a TissueLyser II (Qiagen), followed by a 15-minute centrifugation at 14,000 rpm. Nine hundred μL of supernatant was transferred to new microcentrifuge tubes, which were snap frozen at –80°C before another lyophilization. Fecal metabolite extracts were resuspended with 150 μL of resuspension solvent (1:19 acetonitrile:water spiked with 1 μmol/L sulfadimethoxine), transferred to sample injection vials, and stored at 4°C before LC-MS/MS analysis.
Untargeted metabolomics analysis was performed using an ultra high performance liquid chromatography system (Vanquish; Thermo Fisher Scientific) coupled to a quadrupole-Orbitrap mass spectrometer (Q Exactive; Thermo Fisher Scientific). A Phenomenex Kinetex column (C18: 1.7 μm, 2.1 mm × 50 mm) and a mobile flow rate of 0.500 mL were used for all analysis. A 5 μL injection volume was used for all samples. The mobile phase composition was the following: (A) 100% LC-MS grade water spiked with 0.1% formic acid (v/v) and (B) 100% LC-MS grade acetonitrile spiked 0.1% formic acid (v/v). The chromatographic gradient was 0.0–1.0 min, 5% B; 1.0–9.0 min, 5%–100% B; 9.0–11.0 min, 100% B; 11.0–11.5 min, 100%–5% B; 11.5–12.5 min, 5% B. The following heated electrospray ionization parameters were used: auxiliary gas flow rate, 14.0 (arb. units); auxiliary gas heater temperature, 435.0°C; capillary temperature, 268.0°C; sheath gas flow rate, 52.0 (arb. units); spray voltage, 3.5 kV; and S-lens RF, 50 (arb. units). Positive mode MS data were acquired using a data dependent acquisition method where the 5 most abundant ions are identified and subsequently scanned for MS/MS fragmentation via collision-induced dissociation. MS1 data were collected at a resolution of 35,000 and spanned an m/z range of 100–1500. MS2 data were collected at a resolution of 17,500.
Thermo proprietary ms files(.raw) were converted to a GNPS compatible format (.mzXML) using the ProteoWizard program MSConvert. The open-source software MZmine version 2.37 was used for feature detection. MZmine modules were used with the following settings. Mass detection (1E5 MS1 noise level, 1E2 MS2 noise level, MS1 min time span [min] = 0.05, MS1 min height = 3E5, MS1 m/z absolute tolerance = 0.005, MS1 ppm tolerance = 10 ppm), deconvolution (min peak height = 5E3, peak duration = 0–10 min, m/z range for MS2 scan pairing = 0.005 Da, RT range for MS2 scan pairing = 0.2 min), isotope grouper (m/z absolute tolerance = 0.05 Da or 10 ppm, RT tolerance = 0.1 min, maximum charge = 4, representative isotope = most intense), join aligner (m/z tolerance = 0.005 Da or 10 ppm), rows filter (minimum peaks in a row = 2), gap filling (intensity tolerance = 0.2, m/z tolerance = 0.005 Da or 10 ppm, RT tolerance = 0.1 min).
Metagenomics
Metagenomic samples were initially extracted as dictated by the Earth Microbiome Project protocol using a Qiagen MagAttract PowerSoil DNA kit as previously described. Briefly, swabbed fecal material was plated into 96-well PowerBead DNA plates containing garnet beads. DNA was extracted in accordance with manufacturer’s suggested protocol, with an additional incubation at 65°C for 10 minutes after the addition of lysis solution and immediately before shaking (Qiagen TissueLyser II, product 85300). The KingFisher Flex automated system (Thermo Fisher Scientific) was used for magnet-based DNA purification of all samples. From the purified samples, whole-genome shotgun libraries were generated using Kapa Hyperplus DNA library preparation kits (Roche, Indianapolis, IN) and a 1:10 miniaturized-reaction volume. Libraries were subsequently sequenced using Illumina NovaSeq paired-end sequencing and processed using the Qiita processing and analysis platform.
Akkermansia spp. Targeted Database
A set of Akkermansia spp. genomes were selected from NCBI RefSeq and GenBank databases on October 8, 2021. To assess the completeness and contamination of the Akkermansia genomes, we used the lineage-specific workflow from CheckM v1.1.02 with the default database. We then performed a quality filtering of genomes (contamination >5% and completeness <90%). Closely related genomes may present a problem for our database because they increase the ambiguity during read classification. Therefore, we identified and removed such genomes. For this, we first estimated the distance between high-quality genomes with Mash v1.13, testing multiple k-mer sizes. We then performed a hierarchical clustering over the mash distance matrix using the average linkage method, implemented in SciPy v1.7.1. Considering the strains of interest for this study, we chose an optimal distance cutoff value to determine the number of clusters. We then selected a representative genome for each cluster as the genome with the lowest distance to other genomes of the same cluster. Finally, we merged the high-quality and non-redundant Akkermansia spp. genomes with the current Web of Life database.
Sequence Alignment and Classification of Akkermansia Strains
FASTQ sequences of studies 11149 and 12675 were downloaded from the Qiita web server. We aligned all sequences against the updated Web of Life database, containing the high-quality and non-redundant Akkermansia spp. genomes with Bowtie 2 v2.4.4. Woltka v0.1.3 was used to classify aligned sequences at the species and strain level, filtering features (species and strains) whose abundance in particular samples was below 0.01% because very low abundance hits are false-positive assignments.
Fecal Colonization Experiment
To confirm independence of effect for HBOT on host and microbe responses and effects of microbial changes on host immune responses, we performed fecal colonization in germ-free IL10 knockout (IL10-/-) mice using post-intervention stool from patients treated with HBOT stratified by response status. Two groups of mice (n = 8–10 mice per group) were colonized; group 1 was colonized with stool collected from 2 UC patients who responded clinically to HBOT (at least a 2-point reduction in partial Mayo with at least 1-point reduction in both rectal bleeding and stool frequency subscores), and group 2 was colonized with stool collected from 2 UC patients who did not respond clinically to HBOT (0- or 1-point reduction in partial Mayo with no change in rectal bleeding subscore). To ensure differences observed were not simply a function of mucosal remission status, the 2 HBOT responders chosen were patients who had begun to respond clinically to HBOT but who had persistent endoscopic inflammation (Mayo endoscopic subscore 2) at the time of stool collection. This was to allow for an assessment of whether the evolving changes in microbial composition in these clinical responders were less colitogenic compared with the non-responders.
Germ-free C57BL/6 IL10-/- male mice (C57BL/6NTac-Il10em8Tac; Taconic reference GF-16006) were maintained in isolated ventilated cages (Isocages; Techniplast, West Chester, PA). At 6–7 weeks of age, mice were orally administered with 200 μL of fecal suspension from 2 patients who responded clinically to HBOT and 2 patients who did not respond to HBOT. Transplanted mice were group-housed (n = 5) in isolated ventilated cages and fed autoclaved Purina Rodent Chow #5021 at Cochin Institute (INSERM U1016 Paris, France) under institutionally approved protocols (APAFIS#24788-2019102806256593 v8). Mice were weighed, and fresh feces were collected at weeks 4 and 8. Eight weeks after microbiota transplantation, mice were weighed and euthanized, and tissue and feces were collected for further analysis.
H&E Staining of Colonic Tissue and Histopathologic Analysis
Mouse colons were fixed in 4% PFA solution and embedded in paraffin. Tissues were sectioned at 5-μm thickness and stained with H&E using standard protocols. Images were acquired using a Lamina (PerkinElmer, Waltham, MA) at the Hist’IM platform (INSERM U1016, Paris, France). Histologic scoring was determined on each colon as previously described. Briefly, each colon was assigned 4 scores based on the degree of epithelial damage and inflammatory infiltrate in the mucosa, submucosa, and muscularis/serosa. Each of the 4 scores was multiplied by a coefficient 1 if the change was focal, 2 if it was patchy, and 3 if it was diffuse, and the 4 individual scores per colon were added.
Quantification of Fecal Lipocalin-2 by Enzyme-Linked Immunosorbent Assay
For quantification of fecal lipocalin-2 by enzyme-linked immunosorbent assay, frozen fecal samples were reconstituted in phosphate-buffered saline containing to a final concentration of 100 mg/mL and vortexed for 20 minutes to get a homogenous fecal suspension. These samples were then centrifuged for 10 minutes at 14,000g and 4°C. Clear supernatants were collected and stored at –20°C until analysis. Lipocalin-2 levels were estimated in the supernatants using Duoset murine Lcn-2 enzyme-linked immunosorbent assay kit (R&D Systems) using the colorimetric peroxidase substrate tetramethylbenzidine, and optical density was read at 450 nm (SpectraMax ABS Plus microplate reader; Molecular Devices, San Jose, CA).
Colonic RNAs Extraction and Quantitative Real-Time Polymerase Chain Reaction Analysis
Distal colon was collected during euthanasia and placed in RNAlater. Total RNAs were isolated from colonic tissues using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and as previously described. Quantitative real-time polymerase chain reaction was performed using the QIAGEN kit QuantiFast SYBR Green RT-PCR in a CFX96 apparatus (Bio-Rad, Hercules, CA) with specific mouse oligonucleotides (Table 4). Gene expressions are presented as relative values using the DDCt approach with 36B4 housekeeping gene as reference (Table 4).
Table 4.
Table of qPCR Primers Used for Expression Quantification in Mouse Model.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Shp | AGGAACCTGCCGTCCTTCTG | CTCAGCCACCTCGAAGGTCA |
| Fxr | CCTGAGAACCCACAGCATTT | GTGTCCATCACTGCACATCC |
| Socs3 | GGAACTTGTTTGCGCTTTGATT | TCACACACCCTTTTCTCTTCCAT |
| Stat3 | CTTGTCTACCTCTACCCCGACAT | GATCCATGTCAAACGTGAGCG |
| Hif1 | ATCAAGTCAGCAACGTGGAA | AATGGGTTCACAAATCAGCAC |
| TNF | AGGCTGCCCCGACTACGT | GACTTTCTCCTGGTATGAGATAGCAAA |
| IL-6 | ACAAGTCGGAGGCTTAATTACACAT | TTGCCATTGCACAACTCTTTTC |
| IL-17 | TGAGCTTCCCAGATCACAGA | TCCAGAAGGCCCTCAGACTA |
| IFN | AGCTGCAGGCCTTCAAAAAG | TGGGAGTGAATGTGGCTCAG |
Cells Isolation and Flow Cytometry
Spleen and mesenteric lymph nodes cells were collected after euthanasia in Hanks’ balanced salt solution. Tissues were then grinded successively through a 100-μm filter and a 40-μm filter to generate single-cell suspensions, and splenocytes were treated with red blood cells lysis buffer. Cells were stained 30 minutes with zombie fixable viability kit (BioLegend, San Diego, CA) to differentiate live cells from dead cells. Cells were further preincubated with Fc-Block for 15 minutes at 4°C and stained for 1 hour at 4°C with antibodies to surface markers. For intracellular staining, cells were fixed and permeabilized with a commercially available fixation/permeabilization buffer. Cells were separated and stained using 2 panels. Extracellular staining was performed in the first panel with PerCP-conjugated CD45 (clone 30-F11), APC-H7-conjugated CD4 (clone GK1-5), BV711-conjugated CD3 (clone 17A2), FITC-conjugated CD69 (cloneH1.2F3), BV421-conjugated CD62L (clone MEL-14), BV786-conjugated CD44 (clone IM7), V500-conjugated CD8 (clone 53-6.7), and BV650-conjugated CD25 (clone PC61) and in the second panel with PerCP-conjugated CD45 (clone 30-F11), APC-H7-conjugated CD4 (clone GK1-5), BV711-conjugated CD3 (clone 17A2), BV786-conjugated CD44 (clone IM7), V500-conjugated CD8 (clone 53-6.7), and BV650-conjugated CD25 (clone PC61). Intracellular staining was performed in the first panel with APC-conjugated Foxp3 (clone FJK-16s), AF700-conjugated Gata3 (clone TWAJ), PE-conjugated RorɣT (clone Q31-378), and PE-texas red-conjugated Tbet (clone O4-46) and in the second panel with APC-conjugated Foxp3 (clone FJK-16s), AF700-conjugated Gata3 (clone TWAJ), PE-conjugated RorɣT (clone Q31-378), PE-texas red-conjugated Tbet (clone O4-46), FITC-conjugated IFNɣ (clone XMG1.2), and BV421-conjugated IL17A (clone TCII-18H10). Samples were analyzed on a BD LSRFortessa Cell Analyzer (BD Biosciences, Franklin Lake, NJ), and data were analyzed by using FlowJo v10.8 software.
Acknowledgments
CRediT Authorship Contributions
Carlos G. Gonzalez, PhD (Data curation: Lead; Formal analysis: Equal; Investigation: Lead; Visualization: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Robert H. Mills, PhD (Data curation: Supporting; Formal analysis: Supporting; Visualization: Supporting)
Melissa C. Kordahi, PhD (Formal analysis: Supporting; Investigation: Supporting)
Marvic Carrillo-Terrazas (Formal analysis: Supporting; Investigation: Supporting)
Henry Secaira-Morocho (Formal analysis: Supporting)
Christella E. Widjaja, PhD (Investigation: Supporting)
Matthew S. Tsai, PhD (Investigation: Supporting)
Yash Mittal, MD (Investigation: Supporting)
Brian A. Yee, MS (Formal analysis: Supporting; Investigation: Supporting)
Fernando Vargas (Investigation: Supporting)
Kelly Weldon (Investigation: Supporting)
Julia M. Gauglitz, PhD (Investigation: Supporting)
Clara Delaroque, PhD (Formal analysis: Supporting; Investigation: Supporting)
Consuelo Sauceda (Formal analysis: Supporting; Investigation: Supporting)
Leigh-Ana Rossitto (Investigation: Supporting)
Gail Ackermann, PhD (Data curation: Equal; Project administration: Supporting)
Gregory Humphrey (Investigation: Supporting; Methodology: Supporting)
Austin D. Swafford, PhD (Investigation: Supporting)
Corey A. Siegel, MD (Conceptualization: Supporting; Investigation: Supporting)
Jay C. Buckey, MD (Investigation: Supporting)
Laura E. Raffals, MD (Investigation: Supporting)
Charlotte Sadler, MD (Investigation: Supporting)
Peter Lindholm, MD, PhD (Investigation: Supporting)
Kathleen M. Fisch, PhD (Investigation: Supporting)
Mark Valaseck, MD, PhD (Investigation: Supporting)
Arief Suriawinata, MD (Investigation: Supporting)
Gene W. Yeo, PhD, MBA (Investigation: Supporting)
Pradipta Ghosh, MD (Investigation: Supporting)
John T. Chang, PhD (Investigation: Supporting)
Hiutung Chu, PhD (Investigation: Supporting)
Pieter Dorrestein, PhD (Data curation: Supporting; Investigation: Supporting)
Qiyun Zhu, PhD (Data curation: Supporting; Investigation: Supporting; Software: Supporting)
Benoit Chassaing, PhD (Investigation: Supporting; Methodology: Supporting; Resources: Supporting)
Rob Knight, PhD (Data curation: Supporting; Investigation: Supporting; Resources: Supporting)
David J. Gonzalez, PhD (Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Resources: Supporting)
Parambir S. Dulai, MD (Conceptualization: Lead; Formal analysis: Supporting; Funding acquisition: Lead; Investigation: Equal; Resources: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was directly supported by an American Gastroenterology Association Research Scholar Award and NIDDK U34 Planning Grant (DK126626) to Parambir S. Dulai. This work was also supported by the Crohn's and Colitis Foundation through a Litwin IBD Pioneers Program Grant, the UCSD Collaborative Center for Multiplexed Proteomics, the San Diego Digestive Disease Center (P30 DK120515), UCSD Gastroenterology T32 training grant (DK 0070202; P.S.D., M.T., R.H.M, C.S., and Y.M.), and UCSD Graduate Training Program in Cellular and Molecular Pharmacology (T32 GM007752; L.R.). Y.M. was also supported by an NIH CTSA-funded career-development award (1TL1TR001443). P.G. is supported by the NIH (UG3TR003355, AI155696, and AI141630). J.T.C. is supported by the NIH (AI123202, AI129973, AI132122, and DK119724). C.G.G. is funded by Institutional Research and Academic Career Development Awards (K12GM068524). C.S. is supported by NIH Training grant (T32, DK007202). K.F. is supported by the NIH (2UL1TR001442-06). B.C. is supported by a Starting Grant from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. ERC-2018-StG- 804135), a Chaire d’Excellence from IdEx Université de Paris - ANR-18-IDEX-0001, an Innovator Award from the Kenneth Rainin Foundation, an ANR grant EMULBIONT ANR-21-CE15-0042-01, and the national program “Microbiote” from INSERM. P.C.D. is supported by the Crohn's and Colitis Foundation (grant #675191).
References
- 1.Glover L.E., Colgan S.P. Hypoxia and metabolic factors that influence inflammatory bowel disease pathogenesis. Gastroenterology. 2011;140:1748–1755. doi: 10.1053/j.gastro.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colgan S.P. Targeting hypoxia in inflammatory bowel disease. J Investig Med. 2016;64:364–368. doi: 10.1097/JIM.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dulai P.S., Buckey J.C., Jr., Raffals L.E., Swoger J.M., Claus P.L., O’Toole K., Ptak J.A., Gleeson M.W., Widjaja C.E., Chang J.T., Adler J.M., Patel N., Skinner L.A., Haren S.P., Goldby-Reffner K., Thompson K.D., Siegel C.A. Hyperbaric oxygen therapy is well tolerated and effective for ulcerative colitis patients hospitalized for moderate-severe flares: a phase 2A pilot multi-center, randomized, double-blind, sham-controlled trial. Am J Gastroenterol. 2018;113:1516–1523. doi: 10.1038/s41395-018-0005-z. [DOI] [PubMed] [Google Scholar]
- 4.Dulai P.S., Raffals L.E., Hudesman D., Chiorean M., Cross R., Ahmed T., Winter M., Chang S., Fudman D., Sadler C., Chiu E.L., Ross F.L., Toups G., Murad M.H., Sethuraman K., Holm J.R., Guilliod R., Levine B., Buckey J.C., Jr., Siegel C.A. A phase 2B randomised trial of hyperbaric oxygen therapy for ulcerative colitis patients hospitalised for moderate to severe flares. Aliment Pharmacol Ther. 2020 doi: 10.1111/apt.15984. [DOI] [PubMed] [Google Scholar]
- 5.Sheshachalam A., Srivastava N., Mitchell T., Lacy P., Eitzen G. Granule protein processing and regulated secretion in neutrophils. Front Immunol. 2014;5:448. doi: 10.3389/fimmu.2014.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu G.W., Ju M.J., Zheng Y.J., Hao G.W., Ma G.G., Hou J.Y., Zhang X.P., Luo Z., Lu L.M. CXCL16/CXCR6 is involved in LPS-induced acute lung injury via P38 signalling. J Cell Mol Med. 2019;23:5380–5389. doi: 10.1111/jcmm.14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouwerkerk J.P., van der Ark K.C.H., Davids M., Claassens N.J., Finestra T.R., de Vos W.M., Belzer C. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl Environ Microbiol. 2016;82:6983–6993. doi: 10.1128/AEM.01641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becken B., Davey L., Middleton D.R., Mueller K.D., Sharma A., Holmes Z.C., Dallow E., Remick B., Barton G.M., David L.A., McCann J.R., Armstrong S.C., Malkus P., Valdivia R.H. Genotypic and phenotypic diversity among human isolates of Akkermansia muciniphila. mBio. 2021;12 doi: 10.1128/mBio.00478-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L., Dong Y., Ye M., Jin S., Yang J., Joosse M.E., Sun Y., Zhang J., Lazarev M., Brant S.R., Safar B., Marohn M., Mezey E., Li X. The pathogenic role of NLRP3 inflammasome activation in inflammatory bowel diseases of both mice and humans. J Crohns Colitis. 2017;11:737–750. doi: 10.1093/ecco-jcc/jjw219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodge K.M., Cowburn A.S., Li W., Condliffe A.M. The impact of hypoxia on neutrophil degranulation and consequences for the host. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21041183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mócsai A., Jakus Z., Vántus T., Berton G., Lowell C.A., Ligeti E. Kinase pathways in chemoattractant-induced degranulation of neutrophils: the role of p38 mitogen-activated protein kinase activated by Src family kinases. J Immunol. 2000;164:4321–4331. doi: 10.4049/jimmunol.164.8.4321. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J.L., Ramadass M., Haimovich A., McGeough M.D., Zhang J., Hoffman H.M., Catz S.D. Increased neutrophil secretion induced by NLRP3 mutation links the inflammasome to azurophilic granule exocytosis. Front Cell Infect Microbiol. 2017;7:507. doi: 10.3389/fcimb.2017.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuya M.Y., Asano T., Sumichika Y., Sato S., Kobayashi H., Watanabe H., Suzuki E., Kozuru H., Yatsuhashi H., Koga T., Ohira H., Sekine H., Kawakami A., Migita K. Tofacitinib inhibits granulocyte-macrophage colony-stimulating factor-induced NLRP3 inflammasome activation in human neutrophils. Arthritis Res Ther. 2018;20:196. doi: 10.1186/s13075-018-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwan J., Chae J.J., Goldbach-Mansky R., Colbert R.A. Evidence that STAT3 controls NLRP3 inflammasome-dependent release of IL-1β and pyronecrosis through regulation of mitochondrial activity. Arthritis Rheum. 2013;65 [Google Scholar]
- 15.Hannah S., Mecklenburgh K., Rahman I., Bellingan G.J., Greening A., Haslett C., Chilvers E.R. Hypoxia prolongs neutrophil survival in vitro. FEBS Lett. 1995;372:233–237. doi: 10.1016/0014-5793(95)00986-j. [DOI] [PubMed] [Google Scholar]
- 16.Almzaiel A.J., Billington R., Smerdon G., Moody A.J. Hyperbaric oxygen enhances neutrophil apoptosis and their clearance by monocyte-derived macrophages. Biochem Cell Biol. 2015;93:405–416. doi: 10.1139/bcb-2014-0157. [DOI] [PubMed] [Google Scholar]
- 17.Andina N., Conus S., Schneider E.M., Fey M.F., Simon H.U. Induction of Bim limits cytokine-mediated prolonged survival of neutrophils. Cell Death Differ. 2009;16:1248–1255. doi: 10.1038/cdd.2009.50. [DOI] [PubMed] [Google Scholar]
- 18.Perskvist N., Long M., Stendahl O., Zheng L. Mycobacterium tuberculosis promotes apoptosis in human neutrophils by activating caspase-3 and altering expression of Bax/Bcl-xL via an oxygen-dependent pathway. J Immunol. 2002;168:6358–6365. doi: 10.4049/jimmunol.168.12.6358. [DOI] [PubMed] [Google Scholar]
- 19.Vince J.E., De Nardo D., Gao W., Vince A.J., Hall C., McArthur K., Simpson D., Vijayaraj S., Lindqvist L.M., Bouillet P., Rizzacasa M.A., Man S.M., Silke J., Masters S.L., Lessene G., Huang D.C.S., Gray D.H.D., Kile B.T., Shao F., Lawlor K.E. The mitochondrial apoptotic effectors BAX/BAK activate caspase-3 and -7 to trigger NLRP3 inflammasome and caspase-8 driven IL-1β activation. Cell Rep. 2018;25:2339–2353.e4. doi: 10.1016/j.celrep.2018.10.103. [DOI] [PubMed] [Google Scholar]
- 20.Vlodavsky E., Palzur E., Feinsod M., Soustiel J.F. Evaluation of the apoptosis-related proteins of the BCL-2 family in the traumatic penumbra area of the rat model of cerebral contusion, treated by hyperbaric oxygen therapy: a quantitative immunohistochemical study. Acta Neuropathol. 2005;110:120–126. doi: 10.1007/s00401-004-0946-8. [DOI] [PubMed] [Google Scholar]
- 21.Qi Y., Guo Z., Meng X., Lv Y., Pan S., Guo D. Effects of hyperbaric oxygen on NLRP3 inflammasome activation in the brain after carbon monoxide poisoning. Undersea Hyperb Med. 2020;47:607–619. [PubMed] [Google Scholar]
- 22.Qian H., Li Q., Shi W. Hyperbaric oxygen alleviates the activation of NLRP-3-inflammasomes in traumatic brain injury. Mol Med Rep. 2017;16:3922–3928. doi: 10.3892/mmr.2017.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selvendiran K., Kuppusamy M.L., Ahmed S., Bratasz A., Meenakshisundaram G., Rivera B.K., Khan M., Kuppusamy P. Oxygenation inhibits ovarian tumor growth by downregulating STAT3 and cyclin-D1 expressions. Cancer Biol Ther. 2010;10:386–390. doi: 10.4161/cbt.10.4.12448. [DOI] [PubMed] [Google Scholar]
- 24.Grimberg-Peters D., Büren C., Windolf J., Wahlers T., Paunel-Görgülü A. Hyperbaric oxygen reduces production of reactive oxygen species in neutrophils from polytraumatized patients yielding in the inhibition of p38 MAP kinase and downstream pathways. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D., Frenette P.S. Cross talk between neutrophils and the microbiota. Blood. 2019;133:2168–2177. doi: 10.1182/blood-2018-11-844555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Kundu P., Seow S.W., de Matos C.T., Aronsson L., Chin K.C., Kärre K., Pettersson S., Greicius G. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012;33:1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 27.Albenberg L., Esipova T.V., Judge C.P., Bittinger K., Chen J., Laughlin A., Grunberg S., Baldassano R.N., Lewis J.D., Li H., Thom S.R., Bushman F.D., Vinogradov S.A., Wu G.D. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha S.R., Haileselassie Y., Nguyen L.P., Tropini C., Wang M., Becker L.S., Sim D., Jarr K., Spear E.T., Singh G., Namkoong H., Bittinger K., Fischbach M.A., Sonnenburg J.L., Habtezion A. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–670.e5. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo P., Zhang K., Ma X., He P. Clostridium species as probiotics: potentials and challenges. J Anim Sci Biotechnol. 2020;11:24. doi: 10.1186/s40104-019-0402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oya M., Tadano Y., Takihata Y., Murakami W., Fujii S., Tamai K., Morimoto Y., Ikomi F., Tokunaga T. Effects of hyperbaric conditions on fecal microbiota. Biosci Microbiota Food Health. 2019;38:35–39. doi: 10.12938/bmfh.18-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z.J., Qu H.L., Zhao N., Wang J., Wang X.Y., Hai R., Li B. Assessment of causal direction between gut microbiota and inflammatory bowel disease: a mendelian randomization analysis. Front Genet. 2021;12:631061. doi: 10.3389/fgene.2021.631061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Earley H., Lennon G., Balfe Á., Coffey J.C., Winter D.C., O’Connell P.R. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Scientific Reports. 2019;9:15683. doi: 10.1038/s41598-019-51878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bian X., Wu W., Yang L., Lv L., Wang Q., Li Y., Ye J., Fang D., Wu J., Jiang X., Shi D., Li L. Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Frontiers in Microbiology. 2019;10 doi: 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seregin S.S., Golovchenko N., Schaf B., Chen J., Pudlo N.A., Mitchell J., Baxter N.T., Zhao L., Schloss P.D., Martens E.C., Eaton K.A., Chen G.Y. NLRP6 protects Il10(-/-) mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep. 2017;19:2174. doi: 10.1016/j.celrep.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 35.Zhai R., Xue X., Zhang L., Yang X., Zhao L., Zhang C. Strain-specific anti-inflammatory properties of two Akkermansia muciniphila strains on chronic colitis in mice. Frontiers in Cellular and Infection Microbiology. 2019;9 doi: 10.3389/fcimb.2019.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q., Lu W., Tian F., Zhao J., Zhang H., Hong K., Yu L. Akkermansia muciniphila exerts strain-specific effects on DSS-induced ulcerative colitis in mice. Frontiers in Cellular and Infection Microbiology. 2021;11 doi: 10.3389/fcimb.2021.698914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Post S., Jabbar K.S., Birchenough G., Arike L., Akhtar N., Sjovall H., Johansson M.E.V., Hansson G.C. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. 2019;68:2142–2151. doi: 10.1136/gutjnl-2018-317571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liso M., De Santis S., Verna G., Dicarlo M., Calasso M., Santino A., Gigante I., Eri R., Raveenthiraraj S., Sobolewski A., Palmitessa V., Lippolis A., Mastronardi M., Armentano R., Serino G., De Angelis M., Chieppa M. A specific mutation in Muc2 determines early dysbiosis in colitis-prone winnie mice. Inflamm Bowel Dis. 2020;26:546–556. doi: 10.1093/ibd/izz279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu P., Burger-van Paassen N., van der Sluis M., Witte-Bouma J., Kerckaert J.P., van Goudoever J.B., Van Seuningen I., Renes I.B. Colonic gene expression patterns of mucin Muc2 knockout mice reveal various phases in colitis development. Inflamm Bowel Dis. 2011;17:2047–2057. doi: 10.1002/ibd.21592. [DOI] [PubMed] [Google Scholar]
- 40.Shan Y.S., Hsu H.P., Lai M.D., Yen M.C., Fang J.H., Weng T.Y., Chen Y.L. Suppression of mucin 2 promotes interleukin-6 secretion and tumor growth in an orthotopic immune-competent colon cancer animal model. Oncol Rep. 2014;32:2335–2342. doi: 10.3892/or.2014.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda K., Clausen B.E., Kaisho T., Tsujimura T., Terada N., Förster I., Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 42.Kasembeli M.M., Bharadwaj U., Robinson P., Tweardy D.J. Contribution of STAT3 to inflammatory and fibrotic diseases and prospects for its targeting for treatment. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reindl W., Weiss S., Lehr H.A., Förster I. Essential crosstalk between myeloid and lymphoid cells for development of chronic colitis in myeloid-specific signal transducer and activator of transcription 3-deficient mice. Immunology. 2007;120:19–27. doi: 10.1111/j.1365-2567.2006.02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stackowicz J., Jönsson F., Reber L.L. Mouse models and tools for the in vivo study of neutrophils. Front Immunol. 2019;10:3130. doi: 10.3389/fimmu.2019.03130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber S., Rosenstiel P., Hampe J., Nikolaus S., Groessner B., Schottelius A., Kühbacher T., Hämling J., Fölsch U.R., Seegert D. Activation of signal transducer and activator of transcription (STAT) 1 in human chronic inflammatory bowel disease. Gut. 2002;51:379–385. doi: 10.1136/gut.51.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qing Y., Stark G.R. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- 47.Dulai P., Raffals L.E., Hudesman D., Chiorean M., Cross R., Ahmed T., Winter M., Chang S., Fudman D., Sadler C., Chiu E.L., Ross F.L., Toups G., Hassan Murad M., Sethuraman K., Holm J.R., Guilliod R., Levine B., Buckey J.C., Jr., Siegel C.A. A phase 2B randomized trial of hyperbaric oxygen therapy for ulcerative colitis patients hospitalized for moderate to severe flares. Aliment Pharmacol Ther. 2020;52:955–963. doi: 10.1111/apt.15984. [DOI] [PubMed] [Google Scholar]
- 48.Danaher P, Kim Youngmi, Nelson B, et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nature Communications. 2022 doi: 10.1038/s41467-022-28020-5. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campeau A, Mills R, Stevens Toer, et al. Multi-omics of human plasma reveals molecular features of dysregulated inflammation and accelerated aging in schizophrenia. Molecular Psychicatry. 2021 doi: 10.1038/s41380-021-01339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald D., Hyde E., Debelius J.W., Morton J.T., Gonzalez A., Ackermann G., Aksenov A.A., Behsaz B., Brennan C., Chen Y., DeRight Goldasich L., Dorrestein P.C., Dunn R.R., Fahimipour A.K., Gaffney J., Gilbert J.A., Gogul G., Green J.L., Hugenholtz P., Humphrey G., Huttenhower C., Jackson M.A., Janssen S., Jeste D.V., Jiang L., Kelley S.T., Knights D., Kosciolek T., Ladau J., Leach J., Marotz C., Meleshko D., Melnik A.V., Metcalf J.L., Mohimani H., Montassier E., Navas-Molina J., Nguyen T.T., Peddada S., Pevzner P., Pollard K.S., Rahnavard G., Robbins-Pianka A., Sangwan N., Shorenstein J., Smarr L., Song S.J., Spector T., Swafford A.D., Thackray V.G., Thompson L.R., Tripathi A., Vazquez-Baeza Y., Vrbanac A., Wischmeyer P., Wolfe E., Zhu Q., American Gut C., Knight R. American Gut: an open platform for citizen science microbiome research. mSystems. 2018;3 doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]