Abstract
Kinesin-1 is an ATP-driven, two-headed motor protein that transports intracellular cargoes (loads) along microtubules. The movement of kinesin-1 has generally been modeled according to its correlation with ATP cleavage (forward movement), synthesis (backward movement), or unproductive cleavage (futile consumption). Based on recent experimental observations, we formulate a mechanochemical model for this movement in which the forward/backward/futile cycle can be realized through multiple biochemical pathways. Our results show that the backward motion of kinesin-1 occurs mainly through backward sliding along the microtubule and is usually also coupled with ATP hydrolysis. We also found that with a low external load, about 80% of ATP is wasted (futile consumption) by kinesin-1. Furthermore, at high ATP concentrations or under high external loads, both heads of kinesin-1 are always in the ATP- or ADP Pi-binding state and tightly bound to the microtubule, while at low ATP concentrations and low loads, kinesin-1 is mainly in the one-head-bound state. Unless the external load is near the stall force, the motion of kinesin-1 is almost deterministic.
Keywords: Biochemical pathways, Mean run time, Mean run length, Effective diffusion constant, Mean velocity
Abbreviations: MT, microtubule; NL, neck linker
Kinesin-1 plays a central role in the intracellular transport of various vesicles and organelles (1, 2, 3, 4). Each kinesin-1 dimer consists of two motor domains (heads), which are connected to a coiled-coil stalk through a 14-amino-acid-long sequence known as the neck linker (NL) (5, 6, 7, 8, 9). Kinesin proceeds unidirectionally toward the plus ends of a microtubule (MT) by an asymmetric hand-over-hand fashion, hydrolyzing an ATP molecule for each 8-nm step (10, 11, 12, 13, 14, 15, 16). Motion of kinesin-1 is significantly processive (17, 18), taking about 100 steps before detaching from the MT (1, 19, 20). At low loads, kinesin-1 always steps forward to the plus ends of the MT, while at high loads, it can also result in backward steps (21, 22, 23).
In previous studies, kinds of theoretical models have been presented to understand the mechanism of kinesin-1 motion (refer to the studies by Parrondo and Cisneros (24), Kolomeisky and Fisher (25), Chowdhury (26), and Mugnai and Hyeon (27) for details). Generally, the motion of kinesin-1 can be regarded as a Markov process with multiple biophysical and biochemical states, and in a steady state, meaningful quantities can be obtained by using the Fokker–Planck equation, master equation, Langevin dynamics, or any other complicated hybrid models (28, 29). Among most of these models, a backward step of kinesin-1 is usually thought to occur by directional reversal of a forward step and therefore happens with ATP synthesis.
Generally, both forward and backward steps of kinesin-1 may be accomplished through different biochemical pathways, and backward motion of kinesin-1 may be resulted simply from pure biophysical slips along the MT (30). According to a mechanochemical model (see Fig. 1), this study shows that there are two biochemical pathways for forward step. At low ATP concentration and low external load, kinesin-1 will spend more time waiting for ATP, and its MT-bound head might be in the nucleotide-free state. However, at high ATP concentration or high external load, all MT-bound heads are usually in ATP or ADP Pi-binding state, and ATP binding to the front head of kinesin-1 is earlier than the release of Pi and the detachment of the trailing head from the MT. There are altogether six possible pathways for backward motion, unless at some extreme conditions the backward motion of kinesin-1 is through backward slips along the MT when it is in a semidetach state and usually coupled with ATP hydrolysis. Directional reversal of forwarding pathways is only non-negligible when the ATP molecule is scarce, while the external load is very high. One surprising finding is that about 80% of ATP is wasted by kinesin-1, and this proportion of waste is almost independent of ATP concentration.
Figure 1.
The mechanochemical model of kinesin-1.A, the depiction of detailed configurations of the two heads of kinesin-1. There are six possible configurations at each binding site of the MT. B, mechanochemical model used in this study to describe the periodic motion of kinesin-1, where denotes the state that kinesin-1 is at position n and in configuration j, with . MT, microtubule.
Results
Biophysics of kinesin-1 motion along the MT
Using parameter values listed in Table 1, our model can reproduce the related experimental results well (see Fig. 2). For detailed methods of theoretical predictions, see the supplemental information. In the following, we will always use parameter values listed in Table 1 to discuss the mechanochemical properties of kinesin-1.
Table 1.
Model parameter values obtained by fitting to experimental data of kinesin-1 purified from wildtype Drosophila measured in the studies by Schnitzer et al (44) Visscher et al (33), see Figures 1 and 2
| Parameter | Value | Parameter | Value |
|---|---|---|---|
| , | 9.827 μM−1 s−1 | k21, k43 | 5074.875 s−1 |
| 1627.099 s−1 | δ23 | 0.055 | |
| 0.006 s−1 | δ32 | 0.416 | |
| 137.582 s−1 | δ31 | 0 | |
| δ42 | 0.006 | 1.666 s−1 | |
| δ13 | 0.529 | δ24 | 0.523 |
| 5146.371 s−1 | 77.252 s−1 | ||
| 4.344 s−1 | 1455.909 s−1 | ||
| 345.215 s−1 | 2923.252 s−1 | ||
| 0.001 s−1 | 5.286 s−1 | ||
| 0.803 nm |
Figure 2.
Theoretical predictions (solid lines) and experimental data (markers) of various biophysical properties of kinesin-1 purified from wildtype Drosophila. A–E, the data in (A, B, D, and E) are from the study by Schnitzer et al (44), and in (C and F) are from the study by Visscher et al (33). In (D), the left axis is for 5 μM ATP, while the right axis is for 2 mM ATP. v is the mean velocity of kinesin-1 along the MT, is the mean run length of kinesin-1 before its detachment from MT, r is the randomness, and D is the diffusion constant which is estimated from with nm the step size of kinesin-1. For methods of theoretical predictions, see the supplemental information. MT, microtubule.
From Table 1, , which implies that there may be two substeps within each 8-nm step of kinesin-1, with size 3.76 nm and 4.23 nm, respectively, as previously observed in the studies by Coppin et al (31) and Nishiyama et al (32). According to Figure 1A, these two substeps are caused by binding of the nucleotide-free head to the front site of the MT and detachment of the ADP-bound rear head from the MT, respectively. But, for high ATP concentration, rates , and are very large and the substep coupled with transition is difficult to be observed in experiments.
The load distribution factor means the energy barrier between state 3 and 1 is very close to state 3. Transition rate is independent of external load F, while increases rapidly with F. The process is similar since is also very small.
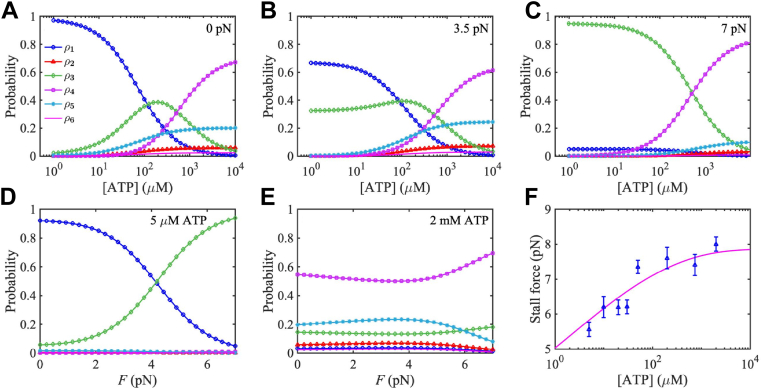
Figure 3, A and D show that, at low load F and low ATP concentration [ATP], kinesin-1 mainly stays in biochemical state 1, i.e., with only one nucleotide-free head bound to the MT while the other ADP-bound head detached. At high ATP concentration, kinesin-1 stays mainly in state 4, with both heads in ATP or ADP Pi-bound state and bound to the MT tightly. Meanwhile, if the load F is not high, the probability that kinesin-1 in the two ADP-bound state 5 is also relatively high (about 1/5) (see Fig. 3, A, B and E). At low [ATP] while high load F, kinesin-1 stays mainly in state 3 with both two heads bound to the MT but one in nucleotide-free state (see Fig. 3, C and D). Provided the load F is high, kinesin-1 stays mainly in two head-bound states and stays mainly in state 4 when [ATP] is high and in state 3 otherwise (see Fig. 3C).
Figure 3.
Probability ρi of kinesin-1 in the six biochemical states.A–C, probability of kinesin-1 in the six biochemical states as depicted in Figure 1, versus ATP concentration [ATP] at different load. D–E, probability of kinesin-1 versus load F at different ATP concentrations. F, stall force of kinesin-1 as a function of [ATP], where the solid line shows the theoretical predictions using parameters listed in Table 1, and triangles denote experimental data measured in the study by Visscher et al (33).
Generally, probability decreases while increases with ATP concentration. At high load F, decreases with [ATP] monotonically; otherwise, may increase first and then decrease with [ATP]. At low ATP concentration, kinesin-1 mainly stays in the ATP waiting state 1 or 3. But with the increase of load F, kinesin-1 switches from the one head-bound state 1 to the two-head-bound state 3 (see Fig. 3, A–D). When ATP concentration is high, all probabilities change only slightly with the load F (see Fig. 3E).
Figure 3F shows the stall force , under which the mean velocity of kinesin-1 vanishes, increases slightly with ATP concentration and eventually approaches to a constant between 7 and 8 pN, which agrees well with experimental measurements (33, 34, 35).
Figure 4, A and B show the mean run time of kinesin-1 decreases with ATP concentration monotonically, while it first decreases slightly and then increases rapidly with load F. These results seem surprising. One possible explanation is that, in general, kinesin-1 does not detach from the MT unless it has transported its cargo to the corresponding destination (18, 36). So, with low ATP concentration, kinesin-1 will stay on the MT for more time, since the period of single biochemical cycle becomes long due to the lack of ATP molecule. Actually, Figures 4B and 2B show both mean run time and mean run length tend to limit constants at high ATP concentration. Possibly, in cells, such run time/length is enough to transport any cargoes to their corresponding destinations. At low ATP concentration and high external load, increases with F, which implies kinesin-1 will try to run more time along the MT to complete its task when its motion is slowed down by resistance (see Fig. 4, A and B).
Figure 4.
Mean and variance of run timet and run length l of kinesin-1 along MT.A and B, mean run time with the change of load F and [ATP]. C, variance of run length with the change of F. D, variance of run time with the change of [ATP]. E–F, probability flux of kinesin-1 detachment from the MT, , with the change of load F and [ATP]. In (A and E), the left axis is for [ATP] μM and right axis is for [ATP] mM. For methods of theoretical predictions of , , and , see the supplemental information. MT, microtubule.
Generally, both mean value and variance decrease with load F. If F is not too large, both and increase with [ATP] (see Figs. 2, B and E, 4C, and S1D). The variance of run time decreases with ATP concentration, while it decreases first and then increases with the load F (see Figs. 4D and S1A). Further calculations show both coefficient of variation and almost equal to 1 (see Fig. S1, B, C, E and F). So both the run time of kinesin-1 along the MT and its run length are approximately exponentially distributed.
The mean run time is actually the time spent by kinesin-1 in average to detach from the MT. So, is related to probability flux of detachment . Intuitively, should change with F and [ATP] in a similar manner as . In contrast to Figure 4, A, B, E and F show detachment flux increases monotonically with [ATP], while it increases first and then decreases with load F.
Pathways of forward/backward/futile biochemical cycle of kinesin-1
Given the model depicted in Figure 1, there is no evident information about the coupling between the biochemical cycle of ATP hydrolysis and mechanical step of kinesin-1. As pointed out before, there are generally three categories of the biochemical cycle, forward/backward/futile cycle. Mean velocity v is actually the sum of probability fluxes of different biochemical cycles, weighted by mechanical step size. It is generally difficult to sort out these three categories of biochemical cycle and then calculate their corresponding fluxes since net flux between any two adjacent biochemical states i and j, , usually changes with external load F and ATP concentration [ATP]. Fortunately, thorough numerical calculations show that, with parameter values listed in Table 1 and biophysically meaningful load F and [ATP], there are only five different cases for probability flux and in each case, the direction of net flux between any two adjacent states does not change (see Figs. S2–S4). For each case, forward/backward/futile cycle can be phenomenologically sorted out by analyzing all possible biochemical pathways. The results are summarized in Table 2.
Table 2.
Pathway details of forward/backward/futile biochemical cycle of kinesin-1
| Conditions | Cycles | Pathway | ATP | Steps | Total flux |
|---|---|---|---|---|---|
| + + ++ (Case 1) | Forward | 1 | 1 | flux23 | |
| 1 | 1 | ||||
| Backward | 1 | −1 | flux61 | ||
| Futile | 1 | 0 | flux51 | ||
| − + ++ (Case 2) | Forward | 1 | 1 | flux23 | |
| Backward | 1 | −1 | flux61 | ||
| 1 | −1 | ||||
| Futile | 1 | 0 | flux51 | ||
| 1 | 0 | ||||
| − − ++ (Case 3) | Forward | — | — | — | 0 |
| Backward | 1 | −1 | flux61 + flux51 | ||
| 1 | −1 | ||||
| 0 | −1 | ||||
| 0 | −2 | ||||
| Futile | 1 | 0 | flux51 | ||
| 1 | 0 | ||||
| − − − + (Case 4) | Forward | — | — | — | 0 |
| Backward | 1 | −1 | flux61 + flux24 +flux51 | ||
| 0 | −2 | ||||
| 0 | −1 | ||||
| −1 | −1 | ||||
| Futile | 1251 | 1 | 0 | flux51 | |
| − − − − (Case 5) | Forward | — | — | — | 0 |
| Backward | −1 | −1 | flux32 + flux61 | ||
| 0 | −2 | ||||
| −1 | −1 | ||||
| 0 | −1 | ||||
| Futile | — | — | — | 0 |
Column ‘Conditions’ lists the signs of , and , respectively. Column ‘ATP’ lists the number of ATP molecule consumed in the corresponding pathway, where ‘’ means one ATP is synthesized. Column ‘Steps’ gives the number of mechanical step coupled with the pathway, with one step 8 nm. Column ‘Total flux’ lists the total flux of the forward/backward/futile cycle. denotes the biochemical pathway . It is evident from Figure 1 that . Flux , , and are always nonnegative for pN and 1 μM [ATP] 10,000 μM, see Fig. S2.
At low load F, the flux of backward cycle is negligible, indicating the impossibility of backward motion (18, 36) (see Figs. 5, A, B, D and E, S5). Both forward flux and futile flux increase with [ATP]. At low load F, proportions of and are almost independent of [ATP], which implies that the utilization ratio of ATP is independent of [ATP] (see Fig. 5, D and E). At super stall load, kinesin-1 motion is dominated by backward cycle when [ATP] is small, while it is dominated by futile cycle when [ATP] is large. Particularly, for pN, the proportion of backward flux is almost 1 when [ATP] μM, indicating that kinesin-1 moves backward almost deterministically. For large enough [ATP], the lower limit value of the proportion of is almost the same as the upper limit value of the proportion of , so the motion of kinesin-1 is completed stalled (see Fig. 5, C and F). With increasing [ATP], the proportion of rises rapidly, until 263.47 μM, at which the sign of changes from negative to positive, switching from Case 3 to Case 2, producing the flux of forward cycle (see Figs. S3C, 5, C and F). On the whole, at high load and low ATP concentration, kinesin-1 can only move backward. As [ATP] increases, the futile flux begins to rise rapidly, and then forward flux is generated. For large enough [ATP], backward flux is balanced by forward flux, and the motion of kinesin-1 is stalled completely.
Figure 5.
Probability flux of forward/backward/futile cycle of kinesin-1, as well as their proportions, versus [ATP], with loadpN, 4 pN and 8 pN, respectively.
Further calculations show that both forward flux and its proportion decrease, while the proportion of increases, with load F, which implies the utilization ratio of ATP decreases with load F (see Fig. S5). At [ATP] = 5 μM and [ATP] = 2 mM, the intersection points of and are found at pN and pN, respectively, which are exactly the stall forces of kinesin-1 at [ATP] = 5 μM and [ATP] = 2 mM (see Fig. 3F). One surprising result is that, except the extreme cases with high load F and very low [ATP], the biochemical process of kinesin-1 is mainly dominated by futile cycle, and utilization ratio of ATP is only about 20% (see Fig. 5). At low load F, the backward flux is almost zero. While for load F larger than stall force, the forward flux decreases rapidly to zero. This indicates that, unless at external load which is near the stall force, the motion of kinesin-1 is almost deterministic. At low load, kinesin-1 hardly steps backward (18). This is different with the conclusion drawn from Brownian ratchet models (37, 38).
Forward cycle consists of two possible pathways, and , both of which include hydrolysis of one ATP and a forward mechanical step of 8 nm (see Table 2). Futile cycle consists of two possible pathways, and , both of which include hydrolysis of one ATP but without a mechanical step. Backward cycle consists of six possible pathways, which are , , , , , and . These six pathways can be further classified into three categories, (i) and , which are mainly induced by backward sliding 6 1 through the semidetach state, (ii) , , and , which can be roughly regarded as the reversal of corresponding forward pathways, and (iii) , which includes both the reversal of a forward pathway and the backward sliding 6 1, and therefore coupled with a backward motion of total 16 nm. Pathway and include the synthesis of ATP, while includes the hydrolysis of ATP. Note that pathway can be denoted equivalently as , , etc. For convenience, we denote the probability flux through pathway by .
With small [ATP] and load , the forward cycle is mainly realized through pathway (see Figs. S6, A and D, S8, A and D, S9, A and D). Otherwise, the forward cycle is mainly realized through pathway (Figs. S6–S10, A and D). During pathway , binding of ATP to the front head of kinesin-1 is earlier than the release of phosphate Pi from the rear head, while during pathway , ATP binds to the front head only after the detachment of rear head from the MT. So, our results imply that at low external load and low ATP concentration, the MT-bound head might be in the nucleotide-free state, while at high ATP concentration or high load, the head bound to the MT is always in ATP or ADP Pi-binding state. This is consistent with results shown in Figure 3.
Except some extreme cases, futile cycle is mainly realized through pathway . Moreover, for small load F or small [ATP], the probability that the futile cycle is realized through is almost negligible (see Figs. S6–S10, B and E).
If load F is small, backward cycle is mainly realized through pathway and other pathways are all negligible (see Figs. S6–S9, C and F). If both F and [ATP] are large, the backward cycle is mainly realized through pathway and pathway and also with the most prominent one (see Figs. S7, C and F, S10, C and F). If F is large but [ATP] is small, pathways , , , and are all important, while both pathway and pathway are negligible (see Figs. S6, C and F, S10, E and F). For large F and small [ATP], prominent pathways of backward cycle are roughly related to the reversal of corresponding pathways of forward cycle and consequently might lead to the synthesis of ATP (see Table 2). However, this is difficult to observe experimentally since kinesin-1 may have already detached from the MT before achieving such harsh conditions.
During pathways and , backward motion of kinesin-1 is induced only by the backward sliding of 8 nm through transition 6 1. But during pathway , the backward motion is accomplished by both one backward sliding of 8 nm and one backward step of 8 nm. Figure 6 shows that the probability flux through pathway is very rare and the backward step of 16 nm is observable only under the very harsh condition that external load F is high but ATP is scarce. If ATP concentration reach saturating, both probability flux of 8-nm backstep and 16-nm backslip decrease to zero, with only backslip 8 nm existing (see Fig. 6, B and C).
Figure 6.
Probability flux of 8-nm backstep, 8-nm backslip, and 16-nm backslip, as well as their corresponding proportions at various [ATP] and load F. (A–C) show the probability flux at [ATP] = 5 μM, [ATP] = 2 mM, and pN, respectively, and (D–F) show their corresponding proportions. The flux of 8-nm backstep is obtained by , the flux of 8-nm backslip is obtained by , and the flux of 16-nm backslip is .
Discussion
A mechanochemical model of kinesin-1 is constructed, in which the motor can step forward/backward through multiple biochemical pathways. Biophysical meaningful quantities of kinesin-1, including its mean velocity, diffusion constant, and mean run time/length, are obtained theoretically.
Our study shows that mean run time of kinesin-1 along the MT decreases with ATP concentration monotonically and finally tends to an external load-dependent limit value but decreases first and then increases rapidly with external load, which are different with properties of the mean run length observed in experiments. However, both distributions of run time and run length are approximately exponential.
Forward motion of kinesin-1 can be realized through two possible biochemical pathways. At high ATP concentration or under high external load, the kinesin head bound to the MT will always be in ATP or ADP Pi-binding state. But at low ATP concentration and low load, new ATP molecule will not bind to the nucleotide-free head before the release of phosphate Pi and the detachment of the trailing head from the MT. Kinesin-1 may stay at the one-head-bound state for a long time to wait for ATP arrival.
Backward motion of kinesin-1 is mainly caused by backward sliding along the MT through a semidetach state and hardly through directional reversal of forward pathways as usually believed in previous studies. Large backward step and directional reversal of forward pathways can only happen in very harsh environments with scarce ATP molecules but under high external load. Usually, kinesin-1 has already been pulled down from the MT before achieving such conditions.
One surprising finding is that the utilization ratio of ATP is only about 20%, and about 80% of ATP is consumed in the futile cycle, which consists of two possible biochemical pathways. Unless ATP concentration is extremely low but external load is high, ATP is required in almost all backward steps of kinesin-1, though these backward steps are mostly accomplished through the sliding process in its semidetach state.
Experimental procedures
Based on experimental observations in the study by Toleikis et al (30), this study uses the mechanochemical model as depicted in Figure 1. The biochemical cycle of kinesin-1 is assumed to begin with the one-head-bound, ATP-waiting state, with the nucleotide-free front head tightly bound to the MT while the ADP-bound rear head unbound (39). ATP binding induces NL docking, and ATP hydrolysis completes NL docking (5, 7, 8) (see Fig. 1A 1 2). In either ATP or ADP Pi-bound states, the head binds tightly to the MT, so these two states are assumed to be the same and denoted as state 2 in Figure 1. Due to NL docking, the unbound head of kinesin-1 swings forward and becomes the front head. ADP release facilitates front head binding to the MT, leading kinesin-1 changes from state 2 to state 3, generating a two-head-bound state. At low ATP concentration, the release of phosphate ion Pi from the rear head usually occurs before an ATP molecule reaches the newly bound front head. Then dissociation of the ADP-bound rear head from the MT leads kinesin-1 to return to the biochemical state 1 again. However, if ATP concentration reaches saturation, after ADP releases from the front head, another ATP molecule will bind to the nucleotide-free head immediately, leading kinesin-1 to enter the biochemical state 4. Next, the release of Pi from the rear head leads kinesin-1 to return to state 2. In either case, 1 2 3 1 or 2 3 4 2, one biochemical cycle of kinesin-1 is completed once the ADP-bound rear head dissociates from the MT. Meanwhile, kinesin-1 makes an 8-nm forward step along the MT and is ready to start a new mechanochemical cycle again (12, 13, 15).
Recent experiments discovered that backward step originates from a different and later state in the biochemical cycle than the one that generates forward steps, and backward step and detachment might emanate from the same state (30). So, our model assumes that forward step can only occur before Pi release from the ADP Pi-bound head (see state 2 in Fig. 1), whereas backward step/slipping and detachment can only occur after Pi release. The release of Pi leads to a weakly bound ADP state (state 5). With the increase of external load, forward step of motor becomes difficult to complete before Pi release, while backward step may occur from state 5. If the rear head releases its ADP before it is semidetached from the MT, kinesin-1 in state 5 will return to state 1. Evidently, one ATP is hydrolyzed during cycle 1 2 5 1 but without output of mechanical work, which we called futile cycle. As concluded in the study by Clancy et al (40), the inclusion of futile cycle is experimentally meaningful and theoretically essential to make the model fit the experimental data well.
Semidetach state 6 is the weakest binding state, and any backward slipping necessarily requires passage through this state. From state 6, kinesin-1 may dissociate completely from the MT and enter into the detachment state, denoted by ‘Off’ in Figure 1. As stated in the study by Toleikis et al (30), kinesin-1 in state 6 may also slip backward along the MT. Our model assumes that unless it is completely detached from the MT, kinesin-1 always slips backward nm in each slipping process, then releases the ADP and returns to state 1. This assumption can be relaxed to allow kinesin-1 to slip backward any distance of integer multiple of 8 nm. But results of fitting to experimental data show that the load free rate is very small, so the possibility of backward sliding is small, and this simplification is reasonable (see Table 1). Meanwhile, kinesin-1 in semidetach state 6 may also slide forward along the MT. But since the probability of kinesin-1 in state 6 is very small (see Fig. 3), compared with the total forward probability flux, the flux produced by forwarding sliding is negligible.
The model used in this study is similar to the one as presented in the study by Toleikis et al (30). One of the key features distinguishing it from earlier ones is the inclusion of semidetach state 6, from which kinesin-1 can either slip backward to state 1 or dissociate from the MT completely. Generally, biochemical cycles of kinesin-1 in this model can be classified into three categories: (i) forward cycle coupled with one forward mechanical step, (ii) backward cycle coupled with one backward mechanical step, and (iii) futile cycle with one ATP hydrolyzed but without change of position.
As illustrated in Figure 1A, biochemical transitions 2 3, 3 1, and 4 2 are coupled with head attachment/detachment to/from the MT-binding site, which may result in the change of mass center of kinesin-1. So rates of these transitions, i.e., , , , , , , are assumed to be external load F dependent. Similar as in the studies by Fisher and Kolomeisky (35) and Zhang (41), this study assumes that
| (1) |
where are load-free transition rates, F is the external load (positive if it points to the minus end of the MT), is the Boltzmann constant, T is the absolute temperature, and nm is the step size of kinesin-1. is called the load distribution factor that reflects how external load affects the rate of transition from state i to state j, which satisfies
Since transitions 4 2 and 3 1 describe the same biochemical process, during which phosphate Pi is released and then the trailing head is detached from the MT, we let and . However, the position of energy barrier between states 4 and 2 might be different from that between states 3 and 1, so load distribution factors are generally different from (1, 35).
The detachment rate of kinesin-1 from the MT is assumed to be load F dependent and formulated as , with a characteristic distance. This expression is actually equivalent to the one used previously in the studies by Müller et al (42) and Zhang and Fisher (43), where the detachment rate is assumed the be the form with parameter called detachment force. Meanwhile, it is biophysically reasonable to assume the sliding process from states 6 to 1 is load-dependent, and the rate is given as .
Both transitions 1 2 and 3 4 describe ATP binding to the nucleotide-free head of kinesin-1. So, rates and depend on ATP concentration. In our model, we assume . The inverse transitions 2 1 and 4 3 describe the disassociation of ATP. So, rates and are independent of load F and [ATP]. Moreover, we let .
Rates describe the release/recruiting of phosphate Pi or ADP. For simplicity, we assume the change of concentrations of ADP and Pi in environment is negligible, and all these rates are assumed to be constants, though more sophisticated methods can be used to handle their dependence on [ATP] (35).
Data availability
The data for kinesin-1 purified from wildtype Drosophila that support the findings of this study are available within the published articles (33, 44), where the data of mean velocity and mean run length are from the study by Schnitzer et al (44), and the data of randomness and stall force are from the study by Visscher et al (33).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
B. S. and Y. Z. conceptualization; B. S. and Y. Z. methodology; B. S. writing–original draft; Y. Z. writing–review and editing.
Edited by Enrique De La Cruz
Supporting information
References
- 1.Howard J. Sinauer Associates; Sunderland, MA: 2001. Mechanics of Motor Proteins and the Cytoskeleton. [Google Scholar]
- 2.Vale R.D. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Schliwa M. Wiley-Vch; Weinheim: 2003. Molecular Motors. [Google Scholar]
- 4.Kolomeisky A.B. CRC Press, Taylor & Francis Group; Boca Raton, FL: 2015. Motor Proteins and Molecular Motors. [Google Scholar]
- 5.Case R.B., Rice S., Hart C.L., Ly B., Vale R.D. Role of the kinesin neck linker and catalytic core in microtubulebased motility. Curr. Biol. 2000;380:157. doi: 10.1016/s0960-9822(00)00316-x. [DOI] [PubMed] [Google Scholar]
- 6.Al-Bassam J., Cui Y., Klopfenstein D., Carragher B.O., Vale R.D., Milligan R.A. Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J. Cell Biol. 2003;163:743. doi: 10.1083/jcb.200308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomishige M., Stuurman N., Vale R.D. Single-molecule observations of neck linker conformational changes in the kinesin motor protein. Nat. Struct. Mol. Biol. 2006;13:887. doi: 10.1038/nsmb1151. [DOI] [PubMed] [Google Scholar]
- 8.Kalchishkova N., Böhm K.J. The role of kinesin neck linker and neck in velocity regulation. J. Mol. Biol. 2008;382:127. doi: 10.1016/j.jmb.2008.06.092. [DOI] [PubMed] [Google Scholar]
- 9.Hariharan V., Hancock W.O. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol. Bioeng. 2009;2:177. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asbury C.L., Fehr A.N., Block S.M. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yildiz A., Tomishige M., Vale R.D., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 12.Schnitzer M.J., Block S.M. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 13.Coy D.L., Wagenbach M., Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J. Biol. Chem. 1999;274:3667. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 14.Svoboda K., Schmidt C.F., Schnapp B.J., Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi H., Bronner C.E., Park H.W., Endow S.A. Rapid double 8-nm steps by a kinesin mutant. EMBO J. 2004;23:2993. doi: 10.1038/sj.emboj.7600306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guydosh N.R., Block S.M. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter N.J., Cross R.A. Mechanics of the kinesin step. Nature. 2005;435:308. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 18.Toprak E., Yildiz A., Hoffman M.T., Rosenfeld S.S., Selvin P.R. Why kinesin is so processive. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12717. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clancy B.E., Behnke-Parks W.M., Andreasson J.O., Rosenfeld S.S., Block S.M. A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 2011;18:1020. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ariga T., Tomishige M., Mizuno D. Nonequilibrium energetics of molecular motor kinesin. Phys. Rev. Lett. 2018;121:218101. doi: 10.1103/PhysRevLett.121.218101. [DOI] [PubMed] [Google Scholar]
- 21.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 22.Visscher K., Schnitzer M.J., Block S.M. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 23.Block S.M., Asbury C.L., Shaevitz J.W., Lang M.J. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2351. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrondo J.M.R., Cisneros B.J.D. Energetics of brownian motors: a review. Appl. Phys. A. 2002;75:179. [Google Scholar]
- 25.Kolomeisky A.B., Fisher M.E. Molecular motors: A theorist’s perspective. Ann. Rev. Phys. Chem. 2007;58:675. doi: 10.1146/annurev.physchem.58.032806.104532. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury D. Stochastic mechano-chemical kinetics of molecular motors: A multidisciplinary enterprise from a physicist perspective. Phys. Rep. 2013;529:1. [Google Scholar]
- 27.Mugnai M.L., Hyeon C., Hinczewski M., Thirumalai D. Theoretical perspectives on biological machines. Rev. Mod. Phys. 2020;92 [Google Scholar]
- 28.Kampen N.G.V. Elsevier Science & Technology Books; North-Holland, Netherlands: 2007. Stochastic Processes in Physics and Chemistry. [Google Scholar]
- 29.Gardiner C.W. Springer-Verlag; Berlin Heidelberg, Germany: 2010. Handbook of Stochastic Methods. [Google Scholar]
- 30.Toleikis A., Carter N.J., Cross R.A. Backstepping mechanism of kinesin-1. Biophys. J. 2020;119:1984. doi: 10.1016/j.bpj.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coppin C.M., Finer J.T., Spudich J.A., Vale R.D. Detection of sub-8-nm movements of kinesin by high-resolution optical-trap microscopy. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1913. doi: 10.1073/pnas.93.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama M., Muto E., Inoue Y., Yanagida T., Higuchi H. Substeps within the 8-nm step of the ATPase cycle of single kinesin molecules. Nat. Cell Biol. 2001;3:425. doi: 10.1038/35070116. [DOI] [PubMed] [Google Scholar]
- 33.Visscher K., Schnitzer M.J., Block S.M. Single kinesin molecules studied with a molecular force clam. Nature. 1999;400:184. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 34.Coppin C.M., Pierce D.W., Hsu L., Vale R.D. The load dependence of kinesin’s mechanical cycle. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8539. doi: 10.1073/pnas.94.16.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher M.E., Kolomeisky A.B. Simple mechanochemistry describes the dynamics of kinesin molecules. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7748. doi: 10.1073/pnas.141080498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toba S., Watanabe T.M., Yamaguchi-Okimoto L., Toyoshima Y.Y., Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5741. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Astumian R.D. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y. Three phase model of the processive motor protein kinesin. Biophys. Chem. 2008;136:19. doi: 10.1016/j.bpc.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Block S.M. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 2007;92:2986. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clancy B.E., Behnke-Parks W.M., Andreasson J.O., Rosenfeld S.S., Block S.M. A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 2011;18:1020. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y. Phenomenological analysis of ATP dependence of motor proteins. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller M.J.I., Klumpp S., Lipowsky R. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4609. doi: 10.1073/pnas.0706825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Fisher M.E. Dynamics of the tugof- war model for cellular transport. Phys. Rev. E. 2010;82 doi: 10.1103/PhysRevE.82.011923. [DOI] [PubMed] [Google Scholar]
- 44.Schnitzer M.J., Visscher K., Block S.M. Force production by single kinesin motors. Nat. Cell Biol. 2000;2:718. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for kinesin-1 purified from wildtype Drosophila that support the findings of this study are available within the published articles (33, 44), where the data of mean velocity and mean run length are from the study by Schnitzer et al (44), and the data of randomness and stall force are from the study by Visscher et al (33).