Abstract
Goose astrovirus (GoAstV) is a new Avastrovirus of the genus astrovirus causing gout, hemorrhage, and swellings of kidneys that have affected goslings around the major goose-producing regions in China. The GoAstV is divided into goose astrovirus type 1 (GoAstV-1) and goose astrovirus type 2 (GoAstV-2). Although GoAstV-2 is known to be the causative agent of goose gout, little published information about the relationship between GoAstV-1 and goose gout is unknown. In this study, we investigated the presence of GoAstV-1 in 293 visceral tissue/dead embryos samples with gout on different farms in Jiangxi province, China. A survey result indicated that the mono-infection of GoAstV-1 (32.08%) and co-infection of GoAstV-1 (12.28%) with GoAstV-2 in gout goslings in Jiangxi, China. JXGZ, a GoAstV-1 strain, was effectively isolated from the visceral tissue of gosling gout and serially propagated for more than 25 passages in a goose embryo. The JXGZ strain's whole genome was sequenced and investigated. Phylogenetic analysis of complete genome and capsid protein sequences of JXGZ strain show that it was more closely related to GoAstV-1 strain than GoAstV-2 strain and was grouped within the GoAstV-1 cluster. These findings will aid in the development of efficient diagnostic reagents and possible vaccinations by providing insight into the prevalence and genetic evolution of GoAstV-1 in China.
Key words: isolation, phylogenetic analysis, goose astrovirus type 1, goose gout
Introduction
Astroviruses are nonenveloped, single-stranded, positive-sense RNA viruses belonging to the Astroviridae family. Based on the host range, astroviruses have been divided into 2 genera, Mamastroviruses (MAstVs) and Avastroviruses (AAstVs) (Wohlgemuth et al., 2019). Viruses from the genus MAstVs are identified in various mammal species, including humans, pigs, cattle, dogs, cats, sheep, and bats, and have been closely related to viral diarrhea and encephalitis in animals (Boujon et al., 2017; Johnson et al., 2017). Genus AAstVs also have a wide range of hosts and can cause enteritis and death in turkeys, diarrhea, and nephritis in infected chickens, growth inhibition in young chickens, enteritis in pearl chickens, and viral hepatitis in ducks, which caused huge economic losses in the poultry industry (Smyth, 2017; Yin et al., 2021).
Since 2016, an infectious disease with visceral urate deposition as the main symptom and joint urate deposition have been occurring in goslings in Shandong, Anhui, Guangxi, Guangdong, and other provinces in China (Jin et al., 2018; Liu et al., 2018; Niu et al., 2018; Wan et al., 2018; Zhang et al., 2018). The disease mainly occurs in the goose within 3 weeks mortality rate of 10 to 50% (Wan et al., 2018; Zhang et al., 2018). Through pathogen isolation and animal regression experiments, the pathogen causing the disease was determined to be goose astrovirus type 2 (GoAstV-2) (Wang et al., 2020, Yang et al., 2018; Yin et al., 2021). Recently, Clinical reports indicated that goose astrovirus type 1 (GoAstV-1) could cause kidney swelling and visceral gout in goslings, symptomatically indistinguishable from those caused by GoAstV-2 (Wang et al., 2021). However, little information is available because GoAstV-1 is a new viral disease. There are currently no effective GoAstV-1 therapies or vaccinations available.
GoAstV-1 is characterized by an approximately 30 nm diameter. GoAstV-1’s full-length genome is approximately 7.3 kb in size and is organized in the following order: 5′ untranslated region (UTR), open reading frames 1a and 1b (ORF1a/b), open reading frame 2 (ORF2), 3′UTR and a poly (A) tail. ORF1a and ORF1b encode nonstructural protein for viral RNA replication and transcription, and ORF2 encodes capsid proteins associated with the virion formation (Wang et al., 2021; Zhang et al., 2017). The astrovirus polymerase has no proofreading function during the replication cycle, so the virus is prone to error, increasing the mutation rate (Wohlgemuth et al., 2019). Astroviruses' high genetic diversity and recombination potential imply that cross-species transmission may be frequent (Chen et al., 2021; Chen et al., 2020; Li et al., 2021). Although various papers have documented the identification of the GoAstV-1 genome in goslings in mainland China, only a handful have reported on GoAstV-1 isolation or the features of those isolates.
In March 2020, some goose farms in Jiangxi province experienced disease outbreaks in goslings aged 1 to 2 weeks. The anatomical lesions were characterized by severe visceral gout and articular gout, with mortality rates of affected flocks up to 40%. In this study, a GoAstV-1 strain, JXGZ, was identified and serially reproduced in healthy goose embryos. The JXGZ strain's whole genome was sequenced and investigated.
Materials and Methods
Ethics statement
The Experimental Animal Ethics Committee in Jiangxi Academy of Agricultural Sciences (JXAAS 2020-0025) approved this study. All procedures involving animals were conducted according to the guidelines for the care and use of experimental animals established by the Ministry of Agriculture of China.
Sample collection and Clinical diagnostics
A deadly infectious illness with gout epidemic occurred at a goose farm, a small-scale independent commercial farm with around 12,000 geese, in Jiangxi, China, in March 2020. Clinical signs of the disease included profuse wasted, sluggish movement, swollen joints, instability, and severe mortality (>60%) in goslings, similar to GoAstV-2 disorders. Visceral tissue/dead embryos samples (n = 293) from goslings with gout from different farms in Jiangxi province, China, between February 2021 and January 2022, were submitted to the Jiangxi Academy of Agricultural Sciences' Institute of Animal Husbandry and Veterinary Medicine for identification of the etiologic agent (s). RT-PCR tests were used to first look at GoAstV-2. Only 115 of 293 (39.25 percent) of the samples examined were positive for GoAstV-2. We postulated that GoAstV-1 would contribute to the goose gout epidemic in goslings based on the characteristics of the gout-like condition and sought to identify and characterize the putative GoAstV-1. (Figure 1, Figure 2)
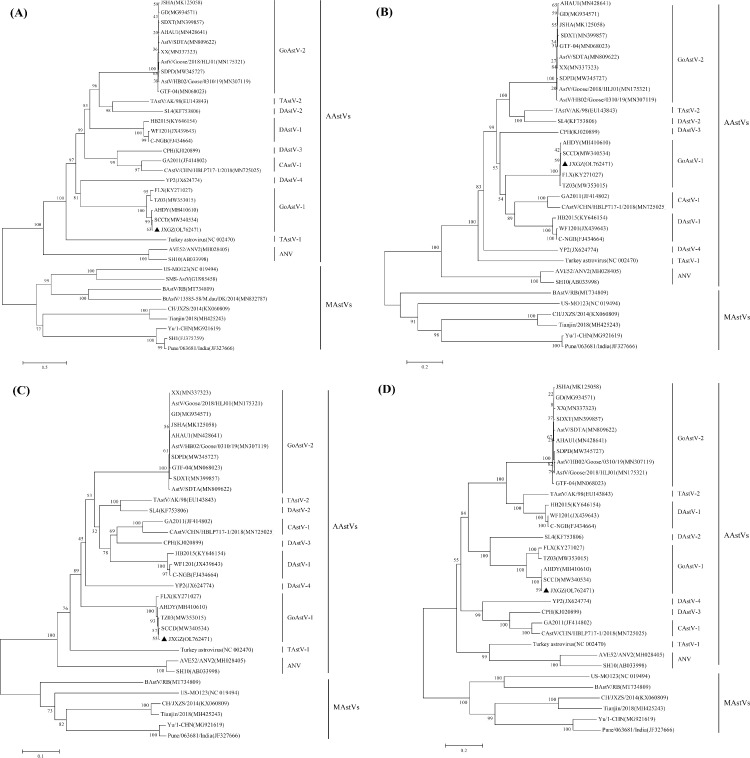
Figure 1.
Phylogenetic tree of the nucleotide sequences of AstVs isolates based on the complete genome (A), ORF1a (B), ORF1b (C), and ORF2 (D). All the reference sequences used in this study were obtained from the GenBank database. The Tree was constructed by the neighbor-joining method with 1,000 bootstrap replications using MEGA 7.0.14 software. The JXGZ strains (GenBank accession no. OL762471) were marked with a black triangle.
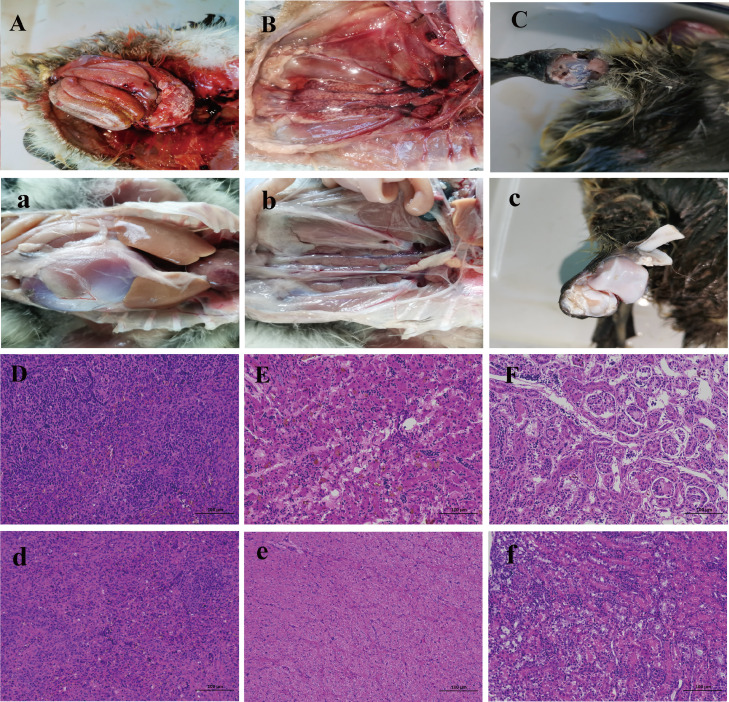
Figure 2.
Gross lesions and pathologic changes of goslings infected with GoAstV-1 strain JXGZ. Macroscopic lesions of visceral organs (A, a), kidneys(B, b), articular cavity(C, c) from the infected gosling (A, B, C), and uninfected (a, b, c) at 5 dpi. The tissues from the goslings incubated with JXGZ or PBS were collected at 5 DPI and were stained with hematoxylin and eosin. Severe cell necrosis with inflammatory cell infiltration in the GoAstV-1 infected goslings' spleen, liver, and kidney, is presented.
Reverse transcription-polymerase chain reaction
A reverse transcription-polymerase chain reaction (RT-PCR) was performed to detect GoAstV-1 from visceral tissue/dead embryos samples of goslings. A pair of primers targeting the ORF1b gene of GoAstV-1 (5′-TTCGCTGAGACTCCTATGTCA-3′ and 5′-GCCCAACTTCTGGTAGCTTGAT-3′) were designed according to the sequence of FLX strain (GenBank accession no. KY271027), and the anticipated sizes of amplicons from the primers are 119 bp for the ORF1b gene of GoAstV-1. Viral RNAs from samples were isolated using the RNA-easy Isolation Reagent (Vazyme, Nanjing, China) and reversely transcribed with a random primer according to the HiScript II first Strand cDNA-Synthesis Kit's instructions (Vazyme, Nanjing, China). The PCR was carried out in a 25 μL volume that contained 2.5 μL of cDNA, 12.5 μL of 2 × Rapid Taq Master Mix, 1 μL of each primer (10 μm/L), and 8 μL ddH2O. The reactions were treated to 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension step of 5 min at 72°C. The PCR products were purified, cloned, and sequenced.
Viral isolation, propagation, and titration
GoAstV-1 isolation was tried on samples that tested positive for GoAstV-1 alone. Kidney, spleen, and liver tissue samples were homogenized with sterile phosphate-buffered saline (PBS, pH 7.4) to a 20% suspension (w/v) and centrifuged at 8,000 g for 10 min 4°C to get the inoculum for virus isolation. The supernatants were separated and filtered through a 0.22 μm filter, and the filtrate was inoculated into six 10-day-old goose embryos (0.2 mL/egg) via the chorioallantoic membrane route. The goose embryos are placed in a fully automatic incubator at 37.8°C, 55% humidity, and the interval for egg turning is set at 2 h.
Embryos that died beyond 24 h and those that survived until 5 days after inoculation were chilled to 4 °C overnight. As previously disclosed, allantoic fluids were collected and injected into new cells. Nucleic acid was isolated from the allantoic fluids after 5 passages to detect the presence of GoAstV-1, and subsequent passages were performed for the GoAstV-1 positive allantoic fluids. These samples served as seed stocks for further passage and detection of GoAstV-1, and other goose viruses (e.g., GoAstV-2, goose parvovirus [GPV], avian influenza virus [AIV], goose circovirus [GocV], goose reovirus [GRV], goose pegivirus [GoPgV], Tembusu virus [TMUV], goose hemorrhagic polyomavirus [GHPV], and fowl adenovirus [FAdV]) and successive passages were performed for the only GoAstV-1 positive allantoic fluids (other goose viruses primer were listed in Table S1) (Liu et al., 2020; Wei et al., 2020; Zhen et al., 2021). The virus suspension was 10-fold serially diluted with PBS and inoculated into 10-day-old goose embryos via the chorioallantoic membrane route to determine the infectious titers of the 5, 15, and 25th passage. As previously described, the embryos were incubated for 7 d at a fully automatic incubator. The mean embryo lethal dose (ELD50) of infectious virus was calculated using the Reed–Muench method.
The JXGZ strain complete genome sequencing and phylogenetical analysis
In the 5th passage, the GoAstV-1 JXGZ allantoic fluids sample was centrifuged at 8,000 g for 5 min. The RNA-easy Isolation Reagent was used to extract viral RNA from a volume of 200 L of cleared supernatants according to the manufacturer's instructions (Vazyme, Nanjing, China). The RNA samples were kept at −80°C and utilized as the template for the RT-PCR. A set of overlapping primers was synthesized according to the GoAstV-1 strain, FLX, to amplify the nearly complete GoAstV-1 genome (Table 1) and characterize the GoAstV-1 JXGZ genome. The approach mentioned above was used to extract viral RNA. Fragments encompassing the whole genome of GoAstV-1 were amplified using denaturation conditions of 95°C for 5 min, 35 cycles (95°C 30 s, 53°C 30 s, and 72°C 2 min), and a final extension of 72°C for 5 min. The 5′- and 3′-RACE for terminal sequence determination were carried out using the 5′/3′ SMARTer RACE kit (Clontech, Beijing, China) according to the manufacturer′s instructions. According to the manufacturer's protocol, the PCR product was cloned into the pEASY-T1 cloning vector (TransGen Biotech, Beijing, China). Three to5 positive clones of each amplicon were sent for Sanger sequencing in both directions (Sangon Biotech, Shanghai, China). For assembly and annotation, the raw genomic sequence fragments were imported to SeqMan in DNAStar Lasergene V 7.10 (DNAStar, Inc., Madison, WI). Nucleotide sequences of the virus genome and the deduced amino acids of the ORFs were compared with known AstV ORF sequences retrieved from the GenBank database. The phylogenetic tree of the complete genome nucleotide sequences, ORF1b, and ORF2 amino acid sequences was constructed using the neighbor-joining method with 1,000 bootstrap replications by the MEGA 7.0.14 software.
Table 1.
Oligonucleotide primers for amplification of the complete genome of GoAstV-1 strain JXGZ by RT‐PCR.
| Primer | Sequence(5′-3′) | Nucleotide positiona | Product size(bp) |
|---|---|---|---|
| GoAstV-1 1F | CCGAAAGCGTTGGTGAGAG | 1-19 | 1552 |
| GoAstV-1 1R | GGTCCCACAGCTCATTGAAAT | 1532-1552 | |
| GoAstV-1 2F | CAGTACCACTTCAGATCTTACT | 1497-1518 | 1632 |
| GoAstV-1 2R | GCGGTTTTGGGTTCATGCAC | 3109-3128 | |
| GoAstV-1 3F | CCTTGGAGCAGAGGAAGA | 3081-3098 | 1481 |
| GoAstV-1 3R | ATGACAATCCTTCAGGTACCA | 4541-4561 | |
| GoAstV-1 4F | GAAGAGAATGTTAAGGTGCAG | 4519-4539 | 1517 |
| GoAstV-1 4R | CTGCTTAACTTGTTGGAAGATCTTT | 6011-6035 | |
| GoAstV-1 5F | GAAGATCTTCGGTGCTGCAGG | 5930-5950 | 1337 |
| GoAstV-1 5R | TTGGTTCAAAAACAGAACCG | 7247-7266 | |
| GoAstV-1 5′RACE | CCAGCATCATCACTCCAGGTT | 283-303 | |
| GoAstV-1 3′RACE | CCAGGCCTGGTCCGCTTTGA | 6948-6967 |
Nucleotide position is numbered based on the FLX strain (KY271027).
GoAstV-1, goose astrovirus type 1.
Pathogenicity of GoAstV-1 Strain JXGZ in goslings
Twenty-four one-day-old healthy goslings were randomly divided into 2 groups (A and B, n = 12/group) to validate the pathogenicity of the isolated GoAstV-1 strain. The experimental goslings were inoculated subcutaneous injection with 0.2 mL of allantoic fluids containing 104.32 ELD50/0.2 mL GoAstV-1 JXGZ from passage 5, the control goslings received 0.2 mL of sterile PBS (pH 7.2) subcutaneous injection; experimental, and control goslings were housed in separate rooms. All the goslings were monitored daily for clinical signs of infection, and tissues were collected for histological examination using hematoxylin and eosin (HE) staining.
Results
RT-PCR
An RT-PCR for GoAstV-1 detection was developed and used to evaluate 24 gout samples from goslings obtained by our laboratory. The detection limit of RT-PCR assay was 1 × 103 copies of GoAstV-1, and the assay could specifically amplify GoAstV-1 and had no cross-amplification with GoAstV-2, goose parvovirus, goose circovirus, avian influenza virus, tembusu virus, goose reovirus, goose pegivirus, and Fowl Adenovirus. Nineteen of these samples yielded the required RT-PCR results and matched sequences via cloning and sequencing. The BLAST search results of these fragments amplified by RT-PCR revealed that the sequences acquired had 99.16% nucleotide (nt) similarity with GoAstV-1 strain SCCD, suggesting that the virus discovered was GoAstV-1.
Prevalence of GoAstV-1 in clinical diarrhoeal samples
Pathogen identification and isolation were performed on clinical samples taken from gosling gout from various farms in Jiangxi, China, between February 2021 and January 2022. Two hundred ninty-three geese visceral tissue/dead embryos samples were examined, 173 (59.04%) were GoAstV-1 positive, 115 (39.25%) were GoAstV-2 positive, and 79 of 173 (45.66%) GoAstV-1 positive samples were also shown to be positive for GoAstV-2. As for sample type, GoAstV-1 was discovered in 54.98% (127/231) of visceral tissue samples and 74.19% (46/62) in dead embryos samples, respectively (Table S2).
Viral isolation and propagation in goose
The 3 GoAstV-1‐positive samples were inoculated on goose embryos, respectively. However, only the GoAstV-1 strain JXGZ‐inoculated into 10-day-old healthy goose embryos showed severe edema and diffuse hemorrhages of dead embryos. The allantoic fluids were collected, and PCR/RT-PCR was performed using specific primers listed in Table S1 to detect potential viral pathogens. Only GoAstV-1 was found in the JXGZ inoculation cell cultures, which were negative for other prevalent viruses (GoAstV-2, GPV, AIV, APMV-1, GcoV, GRV, GoPgV, TMUV, GHPV, and FAdV). Eventually, GoAstV-1 strains causing goslings' gout were successfully isolated and named JXGZ. The isolate has been serially propagated in goose embryos for >25 passages.
Virus titration performed with the ELD50 protocol
The GoAstV-1 infectious titers were determined using an ELD50 assay on healthy goose embryos during serial passages. 3 isolate passages (P5, P15, and P25) were found. The viral titers for these 3 passages were found to be 104.32 ELD50/0.2 mL, 105.17 ELD50/0.2 mL, and 105.50 ELD50/0.2 mL, respectively.
GoAstV-1 strain JXGZ genome sequence alignment and phylogenetic analysis
The 4 typical GoAstV-1 strains JXGZ whole-genome was sequenced with primers in Table 1. The results revealed that the whole genome of the GoAstV-1 JXGZ strain (GenBank accession no. OL762471) was 7,290 nt long and had the identical order of a 22-nt 5′UTR, 3,282-nt ORF1a, 1542-nt ORF1b, 2,115-nt ORF2 gene, and 307-nt 3′UTR. A 3-nt insertion in the ORF2 (1781ATC1782) and 12-nt deletion (1792AAC1794), (1944AAAA1947), and (1964CT11965), (2115GTT2117), when compared with the other GoAstV-1 strains (FLX, TZ03). JXGZ has the most nt identity (98.4%) with AHDY strain at the whole-genome level, while it was most closely linked to SCCD at the aa level of the Cap protein (99.9%). JXGZ's ORF1a, ORF1b, and Cap proteins shared 98.7% 99.8%, 98.7% 99.6%, and 84.1% 99.9% aa identities with reference GoAstV-1 strains, respectively (Table 2). As is the case with other known avian AstVs, the nonstructural protein contained a trypsin-like peptidase domain, and there was an overlapping region between ORF1a and ORF1b, which contains the highly conserved ribosome frameshift sequence. Two stem-loop-II-like (s2m) motif consisting of 43 nt was revealed adjacent to 10 nt and 105 nt of ORF2 in the 3′-UTR by Rfam analysis, respectively.
Table 2.
Sequence identities between goose astrovirus strain (JXGZ) and members of Avastrovirus genus.
| Strain | GoAstV-1 JXGZ |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Complete sequence | ORF1a |
ORF1b |
ORF2 |
|||||||
| nt | nt | aa | nt | aa | nt | aa | ||||
| GoAstV-1 FLX | 90.4 | 97.8 | 99.8 | 93.4 | 98.7 | 77.8 | 84.3 | |||
| GoAstV-1 TZ03 | 91.4 | 98.7 | 98.7 | 98.9 | 99.6 | 77.6 | 84.1 | |||
| GoAstV-1 AHDY | 98.4 | 98.6 | 99.8 | 98.6 | 99.6 | 98.5 | 99.5 | |||
| GoAstV-1 SCCD | 98.1 | 98.2 | 99.8 | 99.0 | 99.5 | 99.3 | 99.9 | |||
| GoAstV-2 AHAU1 | 55.9 | 59.7 | 53.9 | 65.1 | 63.0 | 58.9 | 49.8 | |||
| GoAstV-2 GD | 55.8 | 59.8 | 54.1 | 65.1 | 63.0 | 58.7 | 49.8 | |||
| GoAstV-2 GTF-04 | 55.8 | 59.7 | 54.1 | 65.1 | 63.0 | 58.9 | 49.9 | |||
| GoAstV-2 SDPD | 55.6 | 59.7 | 53.8 | 65.3 | 63.0 | 59.0 | 50.2 | |||
| DAstV-2 SL4 | 57.4 | 56.6 | 54.6 | 67.6 | 68.0 | 59.0 | 60.1 | |||
| TAstV-2 AK-98 | 55.5 | 56.7 | 54.1 | 65.3 | 65.7 | 54.8 | 47.0 | |||
| DAstV-1 C-NGB | 53.5 | 45.4 | 54.7 | 64.5 | 64.7 | 55.4 | 47.1 | |||
| DAstV-3 CPH | 54.1 | 55.0 | 52.8 | 67.2 | 65.6 | 53.7 | 45.4 | |||
| DAstV-4 YP2 | 58.2 | 58.4 | 49.4 | 63.8 | 62.2 | 54.3 | 47.7 | |||
| CAstV-1 GA2011 | 53.9 | 56.5 | 54.6 | 68.0 | 68.3 | 53.6 | 47.3 | |||
| TAstV-1 | 51.3 | 58.6 | 48.4 | 62.0 | 59.6 | 51.5 | 50.6 | |||
| SH10 | 46.2 | 49.9 | 36.8 | 60.0 | 56.1 | 48.3 | 37.7 | |||
Abbreviations: aa, amino acid sequence; nt, nucleotide sequence.
To further explore the characteristics of the newly identified GoAstV-1 strains, amino acid polymorphism of the viral proteins was compared with other GAstV-1 strains. The results revealed that most of the amino acid mutations in the newly identified GAstV-1 JXGZ strains located in the Cap C-terminus (Figure S1), such as G595S, V610I, L642F, and S651V. Remarkably, the mutations of ORF2 located in the capsid p2 domain of virion spike indicate that these mutations might alter the viral protease capacity and antigenicity.
Pathogenicity of GoAstV-1 Strain JXGZ in goslings
The pathogenicity of goose embryos cultured GoAstV-1 strain JXGZ from passage 5 was demonstrated in the goslings. The geese of the infected group began to show symptoms such as malaise and loss of appetite on the fifth day after being exposed to the poison, and the infected group experienced a significant peak of death during 7 to10 d. After that, the mortality rate of the flock gradually decreases (Figure S2). The gout symptoms were similar to those seen in clinical cases, with urate deposits in the liver, ureter, kidney, arthrosis, and other visceral organs, severe bleeding, and swellings of kidneys and spleens observed in dead geese. Histopathologically, the spleen showed diffuse hemorrhage and necrosis. The kidneys showed interstitial hemorrhage and necrosis in kidney tubular epithelium and interstitial lung congestion and hemorrhage with a small infiltration of inflammatory cells. JXGZ was also recovered from infected/dead geese and validated by RT-PCR and sequencing.
Discussion
Gooses in China, a major goose-raising country in the world, have been suffering from severe gout in recent years, but only about 30% to 70% of the goslings gout samples were confirmed as GoAstV-2 infection, indicating that other infectious/noninfectious factors may be involved in the outbreaks of gout in gooses (Liu et al., 2019; Wu et al., 2020; Yuan et al., 2019). In 2019, a disease outbreak occurred in a commercial geese farm located in Jiangsu Province. The pathogen was GoAstV-1 by Next-Generation Sequencing (NGS) and replication experiments (Wang et al., 2021). However, there has been minimal coverage on GoAstV-1 in China. It is critical to develop assays to identify GoAstV-1, a newly discovered gout-related virus in China that might shed light on gout epidemics in goslings.
This study established an RT-PCR for detecting the newly emerged GoAstV-1, and its pathogenicity was evaluated in replication experiments. Our investigation demonstrated a significant incidence of GoAstV-1 in Jiangxi gout goslings, consistent with previous independent studies from China. The overall positive percentage of GoAstV-1 infection in Jiangxi geese was 59.04%, according to RT-PCR detection of 293 gout goslings samples collected between February 2021 and January 2022 and was common in Jiangxi gout goslings, and 79 of 173 (45.66%) GoAstV-1 positive samples were proved to be positive for GoAstV-2. The GoAstV-1 existence in the goose herds needs to be verified by evaluating more samples to limit the collected sample sizes. In addition, we only collected the samples from goslings and dead embryos with gout; however, samples from geese without such symptoms were not gathered to identify GoAstV-1 infection. We will also collect samples from geese that do not have gout to thoroughly assess the prevalence of GoAstV-1 in Jiangxi goose herds. We postulate that GoAstV-1 has been a neglected pathogen linked with gout goslings in China for years and that GoAstV-1/GoAstV-2 co-infection may result in significant mortality in gout goslings.
Although the prevalence of GoAstV-1 in China is confirmed, the characteristics of the GoAstV-1 have not been determined. This study used 5 positive samples to isolate GoAstV-1 in LMH cells and healthy goose embryos to isolate culture-adapted GoAstV-1 strains. However, the GoAstV-1 JXGZ strain was successfully isolated from visceral tissue samples in the goose embryos. These results indicate that GoAstV-1 cannot grow in LMH cells and has a different cell receptor from GoAstV-2. In previous studies, most of the GoAstV strains have been isolated from visceral tissue samples, and this was also reported in their attempts to isolate GoAstV-1 from different samples(Wang et al., 2021; Yin et al., 2021; Zhang et al., 2018). Based on these results, we speculate that visceral tissue is ideal for GoAstV-1 and GoAstV-2. The availability of GoAstV-1 isolates is a valuable resource for GoAstV-1 pathogenesis research, virological and serological test development, and vaccine development. In the JXGZ strain pathogenicity testing, infected goslings had clinical signs identical to GoAstV-2. These findings are consistent with earlier research (Wang et al., 2021; Yin et al., 2021; Zhang et al., 2018).
The isolate's whole genome was sequenced to understand the genetic properties of the JXGZ isolate, and phylogenetic analysis was done using known sequences from the GenBank database. Based on whole-genome sequencing, the JXGZ strain was shown to be the most closely related to SCCD and AHDY strains and was distinct from GoAstV-2 and other Astroviruses and clustered within the genus GoAstV-1. Based on the ORF2 gene, JXGZ has the greatest aa identity (99.9%) with the SCCD strain, whereas only 50.2% with the GAstV-2 strain SDPD. Both strains also feature 4 aa mutations and one aa deletion. Previous studies have shown that the determinants of astroviruses tropism are located at the C-terminal region of the Cap protein. The Cap protein of the AstVs might be strongly associated with the pathogenicity and virulence of the virus (Arias & Dubois, 2017; Krishna, 2005). Five TM domains are present in the ORF1a protein as reported in FLX, whereas only 4 TM domains are present in the ORF1a protein of GAstV-2 strains (Zhang et al., 2017).
Moreover, 2 s2m motifs exist in the 3ʹ-UTR, whereas only one s2m motif exists in other goose astroviruses (De Benedictis et al., 2011). Further analysis showed that JXGZ had 2 nucleotide insertions in the 3ʹ-UTR. Whether the stem-loop-II-like motif differences affect the replication of the virus requires further exploration.
In conclusion, this is the first report of a gout epidemic in goslings in Jiangxi, China, and the disease was effectively isolated and serially passaged in goose embryos. JXGZ's whole genome was sequenced and described. These findings will be useful for future studies into the genetic evolution of GoAstV-1. The immunogenicity and pathogenicity of the JXGZ strain warrant more investigation to create efficient diagnostic reagents, tests, and possible vaccines against newly discovered GoAstV-1 strains.
Acknowledgments
Acknowledgments
This research was supported by Jiangxi Special Fund for Agro-scientific Research in the Collaborative Innovation Project (JXXTCXQN202101), the earmarked fund for Jiangxi Agriculture Research System (JXARS-09), and the earmarked fund for the National Waterfowl-industry Technology Research System of China (CARS-42-43).
Data Availability Statement: All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosures
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2022.101800.
Appendix. Supplementary materials
References
- Arias C.F., DuBois R.M. The astrovirus capsid: a review. Viruses. 2017;9:15. doi: 10.3390/v9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boujon C.L., Koch M.C., Seuberlich T. The expanding field of mammalian astroviruses: opportunities and challenges in clinical virology. Adv. Virus Res. 2017;99:109–137. doi: 10.1016/bs.aivir.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang B., Yan M., Diao Y., Tang Y. First report of a novel goose astrovirus outbreak in Cherry Valley ducklings in China. Transbound. Emerg. Dis. 2020;67:1019–1024. doi: 10.1111/tbed.13418. [DOI] [PubMed] [Google Scholar]
- Chen Q., Yu Z., Xu X., Ji J., Yao L., Kan Y., Bi Y., Xie Q. First report of a novel goose astrovirus outbreak in Muscovy ducklings in China. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis P., Schultz-Cherry S., Burnham A., Cattoli G. Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Wang X., Ning K., Liu N., Zhang D. Genetic characterization of a new astrovirus in goslings suffering from gout. Arch. Virol. 2018;163:2865–2869. doi: 10.1007/s00705-018-3932-5. [DOI] [PubMed] [Google Scholar]
- Johnson C., Hargest V., Cortez V., Meliopoulos V.A., Schultz-Cherry S. Astrovirus pathogenesis. Viruses. 2017;9:22. doi: 10.3390/v9010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna N.K. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 2005;18:17–26. doi: 10.1089/vim.2005.18.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Hu W.Q., Liu T.N., Zhang H.H., Opriessnig T., Xiao C.T. Isolation and evolutionary analyses of gout-associated goose astrovirus causing disease in experimentally infected chickens. Poult. Sci. 2021;100:543–552. doi: 10.1016/j.psj.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hu D., Zhu Y., Xiong H., Lv X., Wei C., Liu M., Yin D., He C., Qi K., Wang G. Coinfection of parvovirus and astrovirus in gout-affected goslings. Transbound. Emerg. Dis. 2020;67:2830–2838. doi: 10.1111/tbed.13652. [DOI] [PubMed] [Google Scholar]
- Liu M., Zhao Y., Hu D., Huang X., Xiong H., Qi K., Liu H. Clinical and histologic characterization of co-infection with astrovirus and goose parvovirus in goslings. Avian Dis. 2019;63:731–736. doi: 10.1637/aviandiseases-D-19-00110. [DOI] [PubMed] [Google Scholar]
- Liu N., Jiang M., Dong Y., Wang X., Zhang D. Genetic characterization of a novel group of avastroviruses in geese. Transbound. Emerg. Dis. 2018;65:927–932. doi: 10.1111/tbed.12873. [DOI] [PubMed] [Google Scholar]
- Niu X., Tian J., Yang J., Jiang X., Wang H., Chen H., Yi T., Diao Y. Novel goose astrovirus associated gout in Gosling, China. Vet. Microbiol. 2018;220:53–56. doi: 10.1016/j.vetmic.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Smyth V.J. A review of the strain diversity and pathogenesis of chicken astrovirus. Viruses. 2017;9:29. doi: 10.3390/v9020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C.H., Chen C.T., Cheng L.F., Liu R.C., Shi S.H., Fu G.H., Fu Q.L., Chen H.M., Huang Y. A novel group of avian Avastrovirus in domestic geese, China. J. Vet. Med. Sci. 2018;80:798–801. doi: 10.1292/jvms.18-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bai C., Zhang D., Yang K., Yu Z., Jiang S., Ge K., Li Y. Genomic and phylogenetic characteristics of a novel goose astrovirus in Anhui Province, Central-Eastern China. Gene. 2020;756:44898. doi: 10.1016/j.gene.2020.144898. [DOI] [PubMed] [Google Scholar]
- Wang A.P., Zhang S., Xie J., Gu L.L., Wu S., Wu Z., Liu L., Feng Q., Dong H.Y., Zhu S.Y. Isolation and characterization of a goose astrovirus 1 strain causing fatal gout in Goslings, China. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F., Yang J., Wang Y., Chen H., Diao Y., Tang Y. Isolation and characterization of a duck-origin goose astrovirus in China. Emerg. Microbes Infect. 2020;9:1046–1054. doi: 10.1080/22221751.2020.1765704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth N., Honce R., Schultz-Cherry S. Astrovirus evolution and emergence. Infect. Genet. Evol. 2019;69:30–37. doi: 10.1016/j.meegid.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Xu R., Lv Y., Bao E. Goose astrovirus infection affects uric acid production and excretion in goslings. Poult. Sci. 2020;99:1967–1974. doi: 10.1016/j.psj.2019.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tian J., Tang Y., Diao Y. Isolation and genomic characterization of gosling gout caused by a novel goose astrovirus. Transbound Emerg Dis. 2018;65:1689–1696. doi: 10.1111/tbed.12928. [DOI] [PubMed] [Google Scholar]
- Yin D., Tian J., Yang J., Tang Y., Diao Y. Pathogenicity of novel goose-origin astrovirus causing gout in goslings. BMC Vet. Res. 2021;17:40. doi: 10.1186/s12917-020-02739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Zhou Q., Mai K., Huang J., Yan Z., Wei X., Shen H., Li Q., Chen L., Zhou Q. Isolation and characterization of a novel chicken astrovirus in China. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Meng K., Zhang Y., Yu Z., Ai W., Wang Y. Genome analysis of newly emerging goose-origin nephrotic astrovirus in China reveals it belongs to a novel genetically distinct astrovirus. Infect. Genet. Evol. 2019;67:1–6. doi: 10.1016/j.meegid.2018.10.014. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Cao Y., Wang J., Fu G., Sun M., Zhang L., Meng L., Cui G., Huang Y., Hu X., Su J. Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg. Microbes Infect. 2018;7:71. doi: 10.1038/s41426-018-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren D., Li T., Zhou H., Liu X., Wang X., Lu H., Gao W., Wang Y., Zou X., Sun H., Ye J. An emerging novel goose astrovirus associated with gosling gout disease. China. Emerg. Microbes Infect. 2018;7:152. doi: 10.1038/s41426-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang F., Liu N., Yang L., Zhang D. Complete genome sequence of a novel avastrovirus in goose. Arch. Virol. 2017;162:2135–2139. doi: 10.1007/s00705-017-3297-1. [DOI] [PubMed] [Google Scholar]
- Zhen W., Wu Y., Zhang W., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Liu Y., Zhang L., Yu Y., Pan L., Chen S., Cheng A. Emergence of a novel pegivirus species in southwest China showing a high rate of coinfection with parvovirus and circovirus in geese. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.