Abstract
Chemotherapy serves as the first choice in clinic to treat advanced gastric cancer. However, emerging evidence indicated the induction of drug resistance and cancer stem cells occasionally by chemotherapy, which seriously limit the therapeutic effects, but the regulatory mechanism remains unclear. Here we treated two human gastric cancer cell lines SGC7901 and BGC823 with 5-Fluorouracil (5-Fu) or Cisplatin (DDP) in vitro. The survived cells showed significant increase of drug resistance, cell stemness and cytokine GM-CSF expression and secretion. As such, GM-CSF was applied to stimulate gastric cancer cells, followed by the subpopulation of CD133 + CSC analysis, sphere formation assay and stemness genes expression analysis. As a result, CSCs showed induction by GM-CSF treatment. A gastric cancer animal model further indicated that the gastric cancer cells significantly promoted tumor growth after GM-CSF treatment in vivo. High-throughput miRNA and mRNA sequencing analyses identified a subset of miRNAs and mRNAs under regulation of both 5-Fu and GM-CSF in gastric cancer cells, including upregulation of miR-877-3p and downregulation of SOCS2. Targeted overexpression or knockdown of miR-877-3p in gastric cancer cells revealed the oncogenic function of miR-877-3p in regulating gastric cancer by suppressing target gene SOCS2. Jak2/Stat3 signaling pathway, as a downstream target of SOCS2, showed activation in vitro and in vivo after treatment with miR-877-3p or GM-CSF. Our findings not only revealed a novel mechanism through which chemotherapy induced CSCs in gastric cancer via GM-CSF-miRNA-Jak2/Stat3 signaling, but also provided an experimental evidence for appropriate dose reduction of adjuvant chemotherapy in treatment of cancer patients.
Keywords: gastric cancer, cancer stem cells, GM-CSF, miR-877–3p, chemotherapy
Introduction
Gastric cancer (GC), as the fourth leading cause of cancer death all over the world (Sung et al., 2021), shows good clinical outcomes to chemotherapy including 5-Fluoride (5-FU) and Cisplatin (DDP) (Seo et al., 2019). However, chemo-resistance is commonly observed in patients with GC after chemotherapy (Choi et al., 2002). Chemotherapy-induced resistance was reported to be related to the acquisition of stem cell-like properties in cancer cells (Li and Clevers, 2010; Xu et al., 2015). This type of cell is called cancer stem cells (CSCs), which are characterized by self-renewal, differentiation, strong tumor-regenerative ability and resistance to therapy. CSCs are believed to play important roles in tumor invasion, cancer metastasis and cancer recurrence (Alison et al., 2012; Schulenburg et al., 2015). Since the first identification of CSCs in myeloid leukemia in 1997, numerous studies have identified CSCs in multiple types of solid cancer tumors including breast cancer, brain cancer, prostate cancer and GC (Bonnet and Dick, 1997; Al-Hajj et al., 2003; Ricci-Vitiani et al., 2007; Takaishi et al., 2009). Although the regulation of chemo-resistance and cancer relapse by CSCs has been well demonstrated, the molecular mechanisms remain unclear.
Tissue damage and inflammatory response caused by chemotherapy are believed as one of the main causes of chemo-resistance. In addition to kill cancer cells, chemotherapy causes the abundance changes of a variety of inflammatory factors in the microenvironment, affecting the chemotherapeutic outcomes (Edwardson et al., 2019). For example, granulocyte-macrophage colony-stimulating factor (GM-CSF), as a monomeric cytokine involving in the immune modulation and hematopoiesis, can be induced by chemotherapy (Hong, 2016; O'Shaughnessy et al., 1994). GM-CSF is mostly secreted by activated monocytes, macrophages, T cells, B cells, fibroblasts, mast cells, vascular endothelial cells, and a variety of cancer cells (Shi et al., 2006), regulating proliferation and maturation of immune cells including dendritic cells, granulocytes and macrophages (McLeish et al., 1998; Pei et al., 1999; Ju et al., 2016). Emerging evidence indicates GM-CSF acting as a tumor-driver in some cases by promoting tumor growth and progression in multiple cancer types, such as meningiomas, gliomas, skin cancer, head and neck cancer, lung cancer, and so on (Pei et al., 1999; Obermueller et al., 2004; Gutschalk et al., 2006; Uemura et al., 2006; Hong, 2016; Sielska et al., 2020).
MicroRNAs (miRNAs) are a class of highly conserved small non-coding RNA with 18–24 nucleotides in length. Typically, miRNAs bind to the 3′-untranslated region (3′-UTR) of target mRNAs, directing the formation of miRNA-mRNA silencing complexes and leading to degradation or translational inhibition of the targeted mRNAs (Bartel, 2009; Su et al., 2015). MiRNAs play an important role in regulating cancer cell stemness, tumor regeneration, metastasis and chemo-resistance during the development and progression of cancer (Sun et al., 2014; Rupaimoole and Slack, 2017) via targeting various signaling pathways including Wnt, Akt, Jak/Stat, et al. (Gomes et al., 2016; Matsui, 2016; Mihanfar et al., 2019). For example, miR-106a-3p induced apatinib resistance in gastric cancer cells by targeting the Cytokine signaling (SOCS) system and activating Jak2/Stat3 signaling (Guo et al., 2019). Activation of the Jak2/Stat3 signaling promotes cell proliferation and cell stemness in cancer (Yu et al., 2014; Park et al., 2019). SOCS proteins function as negative regulators of cytokine-triggered cell signaling. In gastric cancer, Jak/Stat signal pathway is frequently deregulated by the SOCS family and miRNAs (Zhou et al., 2015; Guo et al., 2019).
In the current study, we demonstrated the increased level of GM-CSF both inside and outside of the survived gastric cancer cells after treatment with 5-FU or DDP, which was associated with promoted drug resistance and cell stemness. In order to determine the relationship between the increased GM-CSF level and promoted cell stemness after chemotherapy in GC, exogenous GM-CSF was applied to the culture medium of GC cells, followed by the analysis of CD133 + CSC subpopulation, indicating positive regulation of cancer cell stemness by GM-CSF stimulation in vitro. A GC animal model further demonstrated increased growth of tumors derived from the GM-CSF-treated GC cells in vivo. To further reveal the regulatory mechanism, high-throughput miRNA and mRNA sequencing analyses were applied to the GC cells before and after chemotherapy or GM-CSF treatment. As a result, a subset of miRNAs was identified with deregulation upon treatment with 5-FU or GM-CSF, including upregulation of miR-877-3p and downregulation of SOCS2. Functional assays demonstrated that miR-877-3p is capable to promote GC cell proliferation and cell stemness. SOCS2 was identified as a key direct target gene of miR-877-3p in GC, where miR-877-3p suppressed the expression of SOCS2 and promoted cancer cell stemness and chemoresistance subsequently by activating Jak2/Stat3 signaling. The current study is the first to demonstrate a mechanism through which GM-CSF-miRNA-Jak/Stat signaling mediates chemotherapy-induced cell stemness and drug resistance in gastric cancer.
Materials and Methods
Animals. Six-week-old immune-deficient female nude mice were purchased from the SiPeiFu Animal Company (Beijing, China) for in vivo assays. 2×106 SGC7901 cells with or without GM-CSF stimulation were transplanted per mouse by subcutaneous injection to establish the animal model with gastric cancer. All animal studies were performed following the relevant guidelines, regulations and protocols approved by our Institutional Animal Care and Use Committee.
Cells. Human gastric cancer cell lines SGC7901 and BGC823 were purchased from the cell bank of the Chinese Academy of Sciences at Shanghai, China, maintained in our lab, and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 1% penicillin-streptomycin and 10% fetal bovine serum (Gibco, United States). All of these cells were cultured at 37°C with 5% CO2 in a humidified environment.
RNA Extraction, miRNA and mRNA sequencing, Bioinformatics analysis. Total RNA was extracted using Trizol reagent (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. The quantity of the total RNA was accessed by NanoDrop One spectrophotometer (Thermo Fisher Scientific), and the integrity of the RNA was assessed by Bioanalyzer 2,100 (Agilent, CA, United States) with RIN number >7.0, and confirmed by electrophoresis with denaturing agarose gel. After quality check, approximately 1 μg of total RNA was used to prepare small RNA library according to protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, United States), and approximately 1 μg of total RNA was used for mRNA library. In two libraries, we performed the single-end sequencing (1 × 50 bp) on an Illumina Hiseq2500 and paired-end sequencing (2 × 150 bp) on an illumine Novaseq™ 6000 LC-Bio Technology Company, Ltd., (Hangzhou, China) following the vendor’s recommended protocol. Differentially expressed miRNAs based on normalized deep-sequencing counts were analyzed using Student’s t-test. The screening criteria were a fold change >−2 and p < 0.01. The differentially expressed mRNAs were selected with log2 (fold change) > 1 or log2 (fold change) <−1 and with statistical significance (p-value < 0.05) by the edgeR package. After quality control, bioinformatics analyses (Heatmaps and Venn diagram) were performed with the online OmicStudio tools at http://www.omicstudio.cn/tool.
Plasmids, oligos, and transfection. miR-877-3p mimics, anti-miR-877-3p inhibitors, and corresponding negative controls were synthesized by RiboBio Co., Ltd. (Guangzhou, China). Firefly luciferase reporter plasmids carrying either wild type or mutated SOCS2 3′UTR were constructed by Genomeditech company (Shanghai, China). Oligo transfection was performed using lipofectamine 2000 (Invitrogen, United States) following the manufacturer’s instructions. A final concentration of 30 nM of miRNA mimic or negative control was used in all in vitro assays.
First strand cDNA preparation and Real-Time PCR. Total RNAs were extracted by using Trizol reagent (Invitrogen, Thermo Fisher Scientific). The method of adding a poly A tail to small RNAs was used for reverse transcription of miRNAs. Prime script™ RT Reagent kit with gDNA Eraser (Takara, Japan) was used for reverse transcription of mRNAs. Power Up SYBR Green Master Mix (Applied Biosystem, Thermo Fisher Scientific) and Applied Biosystems QuantStudio 6 (Applied Biosystem, Thermo Fisher Scientific) were used for real-time PCR assays. GAPDH and 5s rRNA were used for mRNA and miRNA normalization. GAPDH forward: 5’-GGAGCGAGATCCCTCCAAAAT-3’; reverse: 5’-GGCTGTTGTCATACTTCTCATGG-3’; 5s forward: 5’-AGTACTTGGATGGGAGACCG-3’; miR-877-3p forward: 5’-UCCUCUUCUCCCUCCUCCCAG-3’.
Quantitative analysis of GM-CSF. Secreted GM-CSF in the supernatant of SGC7901 or BGC823 cells before or after treatment with 5-FU or DDP was quantified using sandwich ELISA following the manufacturer’s instructions (Multi Sciences, Hangzhou, China).
Western Blot. Cells were lysed in RIPA buffer (Beyotime, China), and protein concentration was measured using a BCA Assay Kit (Beyotime, China). 50μg protein lysates were prepared and resolved by 8–12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE) and transferred onto an Immuno-Blot Polyvinylidene difluoride (PVDF) membrane (Millipore, United States). The membranes were then blocked with 5% non-fat milk in TBST for 1 h at room temperature and subsequently incubated with the primary antibodies in 1:1,000 dilution overnight at 4°C. After washing with TBST three times, then the membrane was incubated with the secondary antibody for 1 h at room temperature. Protein bands were visualized using the Minichemi chemiluminescence Imaging System (Beijing Sage Creation Science Co., Ltd., China). The following antibodies were used for Western blot: anti-SOCS2 (2779T, Cell Signaling Technology), anti-JAK2 (3230T, Cell Signaling Technology), anti-p-JAK2 (4406T, Cell Signaling Technology), anti-STAT3 (9139T, Cell Signaling Technology), anti-p-STAT3 (9145T, Cell Signaling Technology), anti-OCT4 (2750S, Cell Signaling Technology), anti-NANOG (4903S, Cell Signaling Technology), anti-GAPDH (sc-47724, Santa Cruz), anti-KLF4 (sc-393462, Santa Cruz), anti-h-TERT (sc-377511, Santa Cruz), anti-GM-CSF (sc-32753, Santa Cruz) and anti-β-tubulin (ab18207, Abcam). Secondary antibodies (1:10,000) were HRP-linked anti-rabbit IgG (7074S, Cell Signaling Technology) and HRP-linked anti-mouse IgG (7076S, Cell Signaling Technology).
Cell proliferation assay. For proliferation assay, 3,000 cells per well were seeded into 96-well culture plates in triplicate. and incubated for 2 days at 37°C in a humidified incubator with 5% CO2. Every 24 h interval, each well was added with 10 μL CCK-8 solution (SB-CCK8, Sharebio, Shanghai, China), then cultured for 3 h at cell culturing condition followed by measurement of OD value at 450 nm wavelength (SpectraMax M5, MolecularDevices, United States).
Colony formation assay. Cancer cells were plated into a 6-well plate at a density of 2,000 cells/well, and after 7-14 days culture until visible colonies were formed. Then, colonies were washed with PBS and fixed with 4% paraformaldehyde. Finally, the visible colonies were stained with 0.5% crystal violet for 20 min. All experiments have three repetitions.
Sphere formation assay. After GC cells were transfected with miRNA-877-3p (mimic, negative control and inhibitor) for 24h, 2,000 GC cells per well were seeded into a 6-well ultra-low attachment cell culture plate (Corning, United States), and cultured with 20 ng/ml of bFGF (R&D Systems, United States), 20 ng/mL EGF (Sigma, United States), and 1×B27 supplement (Invitrogen, United States) in stem cell medium DMEM/F12. The number and sizes of tumorsphere in each well were determined after incubation for 10 days.
Luciferase reporter assay. pGL-3 luciferase reporter plasmids carrying either wild type or mutated SOCS2 3’UTR and Renilla luciferase plasmid (pRL-TK) were co-transfected into 293T cells with miR-877-3p mimic or negative control in a 24-well plate. After 18-h transfection. Luciferase activities were determined with the Dual-Luciferase Reporter Assay kit (Promega, USA).
Statistical analysis. Quantitative data are expressed as mean ±SEM unless otherwise stated. Statistical significance was determined using Student’s t-test followed by least-significant difference (LSD). The data were considered to be significant when the P < 0.05.
Results
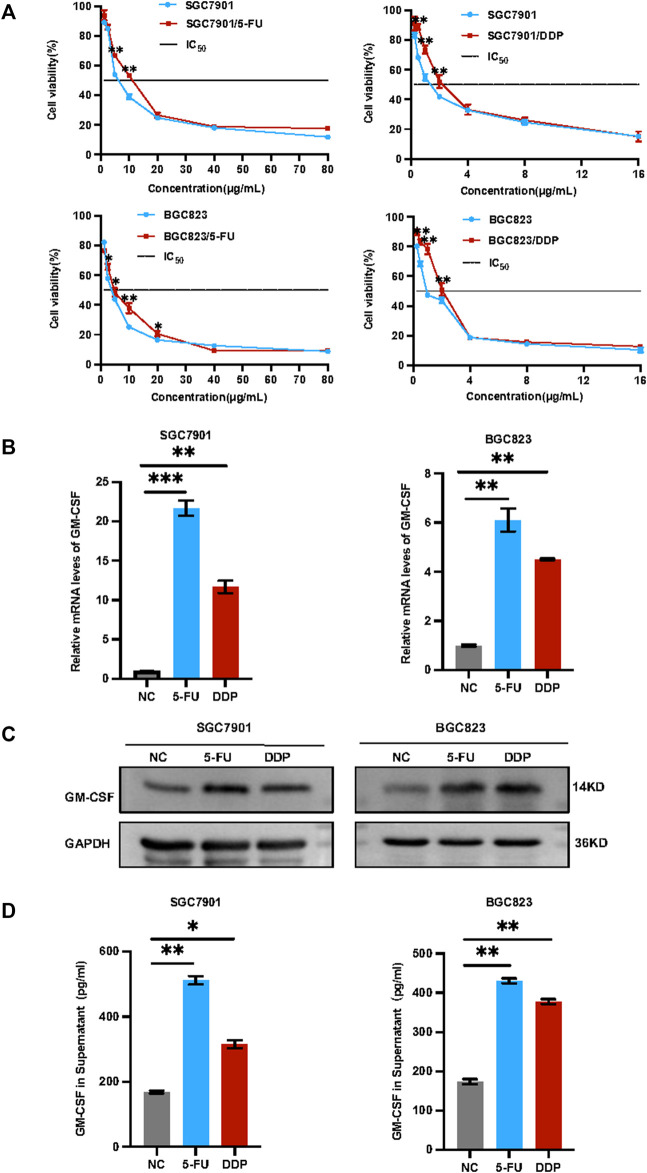
Induction of drug resistance and GM-CSF expression/secretion by chemotherapy in gastric cancer. In view of observation GM-CSF is overexpressed in tumor cells after radiotherapy and induced tumor migration (Vilalta et al., 2014; Vilalta et al., 2018). GM-CSF combined with chemoradiation could trigger abscopal effect (Benna et al., 2020). Highly expressed granulocyte colony-stimulating factor (G-CSF) and granulocyte colony-stimulating factor receptor (G-CSFR) leads to poor survival in gastric cancer (Fan et al., 2018). Tumor-derived GM-CSF promotes gastrointestinal tumorigenesis (Wang et al., 2014), we herein applied in vitro and in vivo assays to validate the phenotypes and determine the regulatory mechanism. Human gastric cancer cells SGC7901 and BGC823 were treated with a low concentration of 5-FU or DDP for 72 h in vitro. Survived cells were collected for further analysis including IC50, cell stemness, as well as GM-CSF levels. As shown in Figure 1A, both survived SGC7901 and BGC823 cells showed increased IC50 and drug resistance, associated with increased GM-CSF levels at both mRNA and protein levels in cells (Figures 1B,C) and in secretion in the supernatant (Figure 1D).
FIGURE 1.
Induction of drug resistance and GM-CSF expression/secretion in the survived gastric cancer cells after chemotherapy. (A): IC50 analysis of the survived SGC7901 and BGC823 cells after treatment with 5-FU or DDP for 72 h. (B): QRT-PCR analysis of the GM-CSF mRNA levels in the survived SGC7901 and BGC823 cells. (C): Western blot analysis of the GM-CSF protein levels in the survived SGC7901 and BGC823 cells. (D): ELISA analysis of the GM-CSF levels in supernatants of the survived SGC7901 and BGC823 cells. Data are presented as the mean ± SEM (N = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
GM-CSF treatment promoted cancer cell stemness in vitro and tumorigenesis in vivo. SGC7901 and BGC823 cells were stimulated with exogenous GM-CSF by adding into the cell culture medium, followed by the CD133 + CSC subpopulation analysis. As a result, The CD133 + CSC subpopulation increased from 2.69% to 9.09% in SGC7901 cells (Figures 2A,B), and from 2.72% to 12.52.% in BGC823 cells (Figures 2C,D) after stimulation, respectively. In addition, a group of well-defined stemness genes including h-Tert, Klf4, Nanog and Oct4 was examined by quantitative RT-PCR and western blot analyses in the 2 GC cell lines before or after treatment with GM-CSF. In consistent with the results in Figures 2A–D, these stemness genes showed induction in expression at both mRNA and protein levels by GM-CSF stimulation (Figures 2E–G).
FIGURE 2.
GM-CSF treatment promoted cancer cell stemness in vitro and tumorigenesis in vivo. (A,C): Flow cytometry analysis of CD133 + CSCs in SGC7901 (A) and BGC823 (C) cells before and after stimulation with exogenous GM-CSF in the cell culture medium. B and D: Quantitative analysis of A (B) and C (D). (E,F): QRT-PCR analysis of the stemness genes expression (h-Tert, Klf4, Nanog and Oct4) in SGC7901 (E) and BGC823 (F) cells with or without treatment with GM-CSF. (G): Western blot analysis of the stemness genes expression in SGC7901 and BGC823 cells with or without treatment with GM-CSF. (H): A gastric cancer xenograft model by transplantation of SGC7901 cells with or without stimulation with GM-CSF into nude mice (n = 10 in each group). (I): Tumor growth curves in (H). (J): Tumor images in (H). (K): Tumor weight in (H). Data are presented as the mean ± SEM (N = 3 for in vitro assays, and N = 10 for in vivo assay). *p < 0.05, **p < 0.01, ***p < 0.001.
In order to further determine the effects of GM-CSF on tumorigenesis in vivo, a gastric cancer xenograft model was established by transplantation of SGC7901 cells with or without stimulation with GM-CSF into immunodeficient female nude mice through via subcutaneous injection, followed by continuous tracking of the tumor growth (Figure 2H). The tumor growth curves (Figure 2I), tumor images (Figure 2J) and tumor weight (Figure 2K) indicated significant promotion of tumor growth by GM-CSF stimulation.
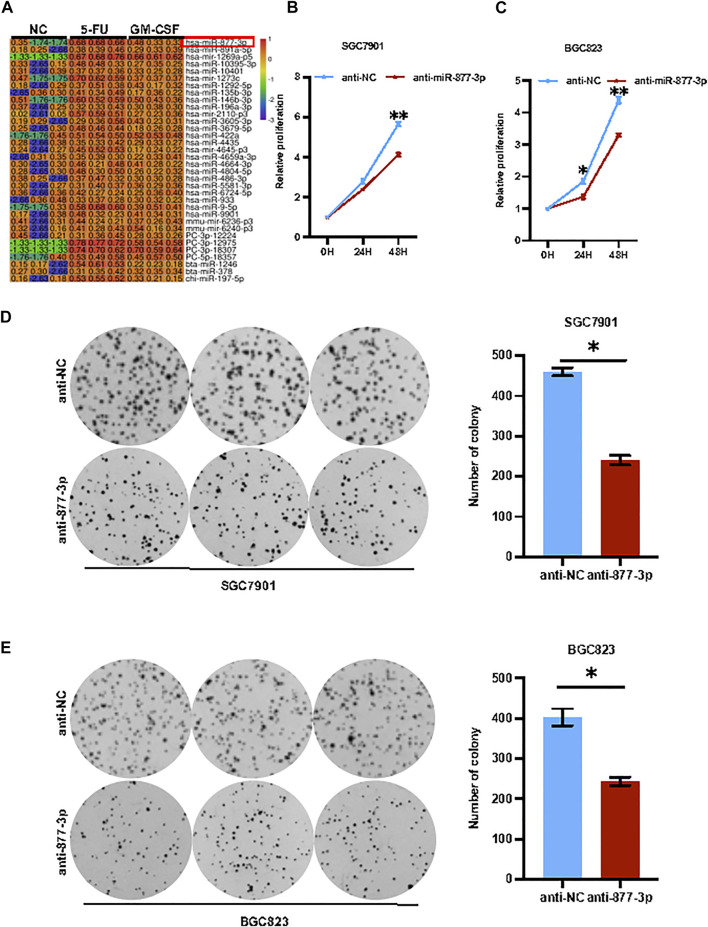
miR-877-3p mediated chemotherapy and GM-CSF induced tumor progression in gastric cancer. To identify the key genes regulating chemotherapy-induced or GM-CSF-induced tumor progression in gastric cancer, SGC7901 cells with or without stimulation with 5-FU or GM-CSF were applied for a high-throughput miRNA sequencing analysis. As a result, a subset of miRNAs was identified with a differential expression upon treatment with 5-FU and GM-CSF, respectively (Figure 3A). Some miRNAs, such as miR-9-5p, miR-196a and miR-422a, have been well documented to regulate tumorigenesis and cancer stem cells in gastric cancer (Pan et al., 2017; He et al., 2018; Liu et al., 2020), while the function of miR-877-3p remains unclear in GC. As shown in Supplementary Figure S1, quantitative real-time PCR analysis validated of miR-877-3p overexpression in both SGC7901 and BGC823 cells were treated with 5-FU and DDP respectively. Therefore, we focused on miR-877-3p to determine the relationship between upregulation of miR-877-3p and chemotherapy-induced drug resistance and cell stemness. Overexpression or knockdown of miR-877-3p was applied to gastric cancer cells (Supplementary Figures S2, S3), followed by CCK8 cell proliferation and colony formation assay. As shown in Figures 3B–E, knockdown of miR-877-3p suppressed cell proliferation and colony formation in both SGC7901 and BGC823 cells, respectively. Whereas overexpression of miR-877-3p dramatically increased cell proliferation and colony formation in both SGC7901 and BGC823 cells (Supplementary Figure S6).
FIGURE 3.
Knockdown of miR-877-3p suppressed gastric cancer cell proliferation. (A): High-throughput miRNA sequencing analyses on SGC7901 cells with or without stimulation with 5-FU or GM-CSF identified a group of deregulated miRNAs, including miR-877-3p. (B,C): Knockdown of miR-877-3p in SGC7901 (B) and BGC823 (C) cells suppressed cell proliferation assayed by CCK8. (D,E): Knockdown of miR-877-3p in SGC7901 (D) and BGC823 (E) cells suppressed the cellular colony formation. Data are presented as the mean ± SEM (N = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
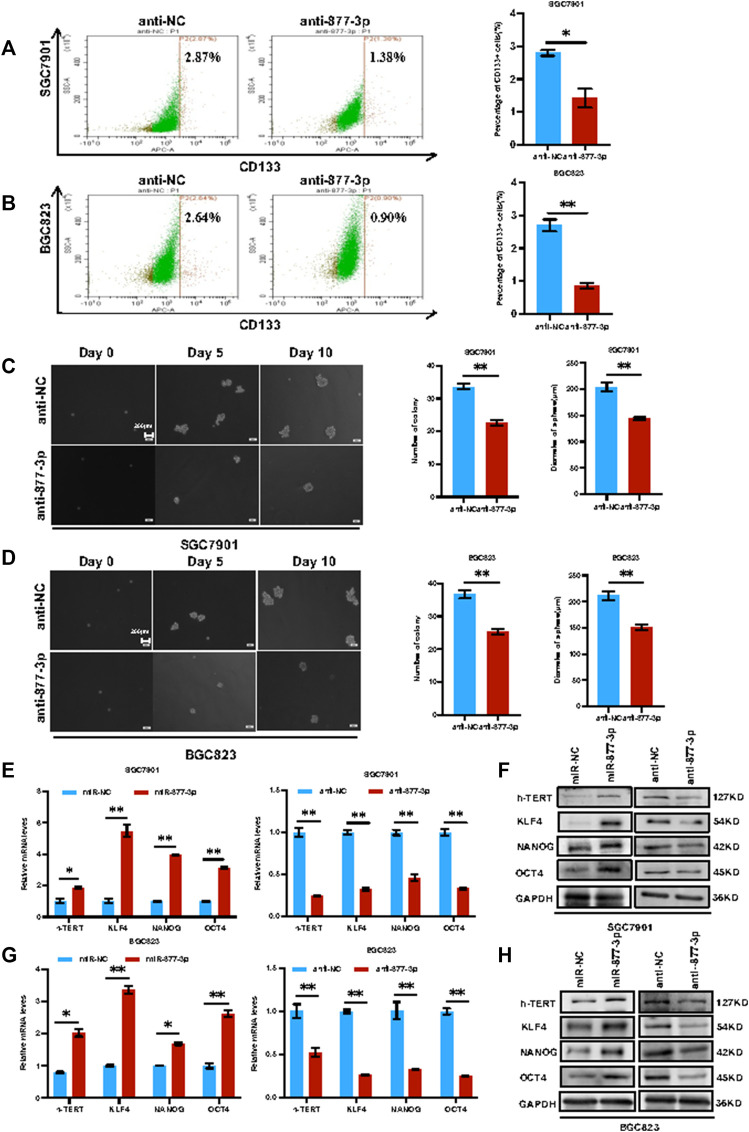
Overexpression of miR-877-3p promoted the cell stemness in both SGC7901 and BGC823 cells. After overexpression or knockdown of miR-877-3p in both SGC7901 and BGC823 cells, the changes of the CD133+ CSC percentage were determined by flow cytometry analysis. As shown in Figure 4A, knockdown of miR-877-3p in SGC7901 cells decreased CD133+ CSC subpopulation. Similar results were obtained from BGC823cells (Figure 4B). In addition, As shown in Figures 4C,D, sphere formation assays were performed to further determine the stemness changes after knockdown of miR-877-3p in both SGC7901 and BGC823 cells. Quantitative analysis indicated that knockdown of miR-877-3p decreased both sphere number and sphere size. Whereas overexpression of miR-877-3p dramatically increased CD133+ CSC subpopulation and sphere formation in both SGC7901 and BGC823 cells (Supplementary Figure S7). Moreover, a group of well-defined stemness genes including h-Tert, Klf4, Nanog and Oct4 was examined in both SGC7901 and BGC823 cells by quantitative RT-PCR and western blot analyses. The results showed that overexpression or knockdown of miR-877-3p remarkably increased or decreased the expression of h-Tert, Klf4, Nanog and Oct4 at both mRNA (Figures 4E,G) and protein levels (Figures 4F, 4H).
FIGURE 4.
Knockdown of miR-877-3p suppressed gastric cancer cell stemness. (A,B): Knockdown of miR-877-3p in SGC7901 and BGC823 cells suppressed the subpopulation of CD133 + CSCs. (C,D): Knockdown of miR-877-3p in SGC7901 and BGC823 cells suppressed the sphere formation ability in the serum-free culture condition. (E,F): QRT-PCR (E) and western blot (F) analyses of the stemness genes including h-Tert, Klf4, Nanog and Oct4 in SGC7901 cells with or without overexpression or knockdown of miR-877-3p. (G,H): QRT-PCR (G) and western blot (H) analyses of the stemness genes in BGC823 cells with or without overexpression or knockdown of miR-877-3p. Data are presented as the mean ± SEM (N = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
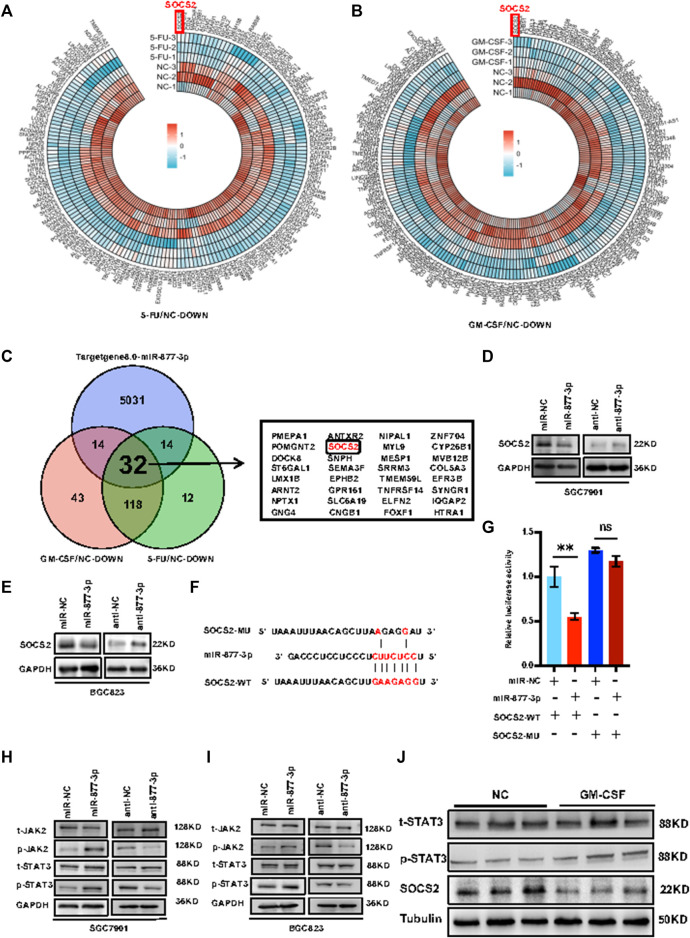
miR-877-3p activated Jak2/Stat3 signaling through targeting SOCS2. In order to determine the molecular mechanism(s) by which miR-877-3p promotes gastric cancer development and progression, RNA-seq was applied to the SGC7901 cells with or without stimulation with 5-FU or GM-CSF, deriving 176 downregulated genes by 5-FU treatment and 207 downregulated genes by GM-CSF treatment (Figures 5A,B). Bioinformatic analysis was using TargetScan Human8.0 (http://www.targetscan.org/vert_80/) predicted 5,091 potential target genes of miR-877-3p. From these three groups of genes, 32 genes were overlapped including SOCS2 (Figure 5C). Quantitative real-time PCR analysis validated downregulation of SOCS2 at the mRNA levels by overexpression of miR-877-3p in both SGC7901 and BGC823 cells (Supplementary Figure S4). Upregulation of SOCS2 was shown after knockdown of miR-877–3p in both SGC7901 and BGC823 cells (Supplementary Figure S5). Western blot analysis further demonstrated downregulation or upregulation of SOCS2 at the protein levels by overexpression or knockdown of miR-877-3p in both SGC7901 and BGC823 cells (Figures 5D,E). In order to demonstrate the direct interaction between SOCS2 and miR-877-3p, luciferase (Luc) reporter constructs carrying either wide type (WT) or miR-877-3p-binding sites-mutated (MU) 3′UTR of SOCS2 were co-transfected with miR-877-3p mimics into 293T cells (Figure 5G). As a result, WT-SOCS2-Luc was inhibited by miR-877-3p, but MU-SOCS2-Luc was not, supporting the target interaction between SOCS2 3′-UTR and miR-877-3p via sequence complementarity (Figures 5F,G). In view of the well-defined tumor-suppressing function of SOCS2 by inhibiting Jak2/Stat3 signaling (Uen et al., 2018; Ren et al., 2019), we detected the effects of miR-877-3p on Jak2/Stat3 signaling in GC. As shown in Figures 5H,I, p-Stat3 and p-Jak2 were induced by overexpression of miR-877-3p, and suppressed by knockdown of miR-877-3p in both SGC7901 and BGC823 cells, which was further validated by western blot analysis on the tumor samples derived from the mouse model (Figure 5J).
FIGURE 5.
miR-877-3p activated Jak2/Stat3 signaling through targeting SOCS2 in gastric cancer. (A,B): RNA-seq analysis of the SGC7901 cells with or without stimulation with 5-FU (A) or GM-CSF (B) identified a list of differentially expressed downregulated genes. (C): 32 genes were overlapped from the 176 downregulated genes by 5-FU treatment, 207 downregulated genes by GM-CSF treatment, and 5,091 potential target genes of miR-877-3p predicted by TargetScan Human8.0, including SOCS2. (D,E): Western blot demonstrated inhibition of SOCS2 by miR-877-3p overexpression and promotion of SOCS2 by miR-877-3p knockdown in both SGC7901 and BGC823 cells. (F): Sequence alignment of wide type (WT) or miR-877-3p-binding sites-mutated (MU) 3′UTR of SOCS2. (G): luciferase reporter assay demonstrated inhibition of WT-SOCS2-3′UTR by miR-877-3p, but not MU-SOCS2-3′UTR. (H,I): Western blot demonstrated positive or negative regulation of p-Jak2 and p-Stat3 by overexpression or knockdown of miR-877-3p in both SGC7901 and BGC823 cells. (J): Western blot demonstrated downregulation of SOCS2 and activation of Jak2/Stat3 signaling by GM-CSF treatment in the tumors from the mice model. Data are presented as the mean ± SEM (N = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
FIGURE 6.
Working Model. Schematic representation of the mechanism through which GM-CSF increased the expression of miR-877-3p in gastric cancer cells, which suppressed the expression of SOCS2 as a target gene. SOCS2, as a suppressor gene of Jak2/Stat3 signaling, mediated the chemotherapy-induced cancer cell stemness and drug resistance.
Discussion
Since GM-CSF is able to induce pluripotent stem cells to differentiate into mature granulocytes, macrophages and T cells in bone marrow, it has been used in clinic to protect cancer patients against chemotherapy- or radiotherapy-induced neutropenia. However, emerging evidence found that application of GM-CSF therapy occasionally promoted tumor progression (Uemura et al., 2004; Metcalf, 2010), indicating complexity of the GM-CSF-based cancer therapy. Herein, we experimentally demonstrated a mechanism through which chemotherapy or GM-CSF-based therapy of gastric cancer may induce cancer cell stemness and drug resistance.
Activation of Jak/Stat3 signaling pathway plays a critical role in promoting tumorigenesis, epithelial and mesenchymal transition (EMT), chemo-resistance, and cancer cell stemness (Jin, 2020). In gastric cancer, overexpression of p-Stat3 increased sphere formation from CD44 + CSCs (Hajimoradi et al., 2016). In the current study, we are the first to identify miR-877-3p with upregulation in the chemo survived gastric cancer cells, which was mediated by GM-CSF induction but in turn suppressed SOCS2 and activated Jak/Stat3 signaling. This is in consistence with the literature about the oncogenic function of miR-877-3p in Pancreatic Cancer by interacting with STARD13 (Xu and Zheng, 2020). In addition to Jak/Stat3, PI3k/Akt and Erk signaling pathways have been reported to have interacted with GM-CSF in regulating tumor cell proliferation and migration (Kawaguchi et al., 2004; Carlson et al., 2011). Although we did not analyze in the current study whether these two pathways are involved in regulating the GM-CSF-induced cell stemness and drug resistance, our high-throughput RNA sequencing data analyses suggested activation of PI3k/Akt signaling after GM-CSF treatment in gastric cancer.
In conclusion, CSCs are believed to be the main source of cancer initiation, relapse, and drug resistance. Therapeutic strategies targeting CSCs hold great promise in the fight against cancer. The current study demonstrated a novel mechanism regulating chemotherapy-induced CSCs and drug resistance in gastric cancer.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA811393.
Ethics Statement
The animal study was reviewed and approved by All animal studies were performed following the relevant guidelines, regulations and protocols approved by our Institutional Animal Care and Use Committee.
Author Contributions
HC and YY designed the research and wrote the paper. XX, H-ZM, Y-qC, D-zZ, S-xM and H-jW performed the in vitro and in vivo experiments. XX and D-mL did data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Central to guide local scientific and Technological Development (ZYYDDFFZZJ-1), Gansu Provincial Youth Science and Technology Fund Program (21JR7RA642), Gansu Provincial Hospital project (2019–206), Lanzhou talent innovation and Entrepreneurship Project (2016-RC-56), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (21GSSYC-2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.855351/full#supplementary-material
References
- Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003). Prospective Identification of Tumorigenic Breast Cancer Cells. Proc. Natl. Acad. Sci. USA. 100 (7), 3983–3988. 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alison M. R., Lin W. R., Lim S. M., Nicholson L. J. (2012). Cancer Stem Cells: in the Line of Fire. Cancer Treat. Rev. 38 (6), 589–598. 10.1016/j.ctrv.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell 136 (2), 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benna M., Guy J. B., Bosacki C., Jmour O., Ben Mrad M., Ogorodniitchouk O., et al. (2020). Chemoradiation and Granulocyte-Colony or Granulocyte Macrophage-Colony Stimulating Factors (G-CSF or GM-CSF): Time to Think Out of the Box? Br. J. Radiol. 93 (1109), 20190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J. E. (1997). Human Acute Myeloid Leukemia Is Organized as a Hierarchy that Originates from a Primitive Hematopoietic Cell. Nat. Med. 3 (7), 730–737. 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Carlson S. M., Chouinard C. R., Labadorf A., Lam C. J., Schmelzle K., Fraenkel E., et al. (2011). Large-scale Discovery of ERK2 Substrates Identifies ERK-Mediated Transcriptional Regulation by ETV3. Sci. Signal. 4 (196), rs11. 10.1126/scisignal.2002010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. H., Lim H. Y., Joo H. J., Kim H. S., Yi J. W., Kim H. C., et al. (2002). Expression of Multidrug Resistance-Associated protein1,P-Glycoprotein, and Thymidylate Synthase in Gastric Cancer Patients Treated with 5-fluorouracil and Doxorubicin-Based Adjuvant Chemotherapy after Curative Resection. Br. J. Cancer 86 (10), 1578–1585. 10.1038/sj.bjc.6600305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwardson D. W., Parissenti A. M., Kovala A. T. (2019). Chemotherapy and Inflammatory Cytokine Signalling in Cancer Cells and the Tumour Microenvironment. Adv. Exp. Med. Biol. 1152, 173–215. 10.1007/978-3-030-20301-6_9 [DOI] [PubMed] [Google Scholar]
- Fan Z., Li Y., Zhao Q., Fan L., Tan B, Zuo J., et al. (2018). Highly Expressed Granulocyte Colony-Stimulating Factor (G-CSF) and Granulocyte Colony-Stimulating Factor Receptor (G-CSFR) in Human Gastric Cancer Leads to Poor Survival. Med. Sci. Monit. 24, 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes B. C., Rueff J., Rodrigues A. S. (2016). MicroRNAs and Cancer Drug Resistance. Methods Mol. Biol. 1395, 137–162. 10.1007/978-1-4939-3347-1_9 [DOI] [PubMed] [Google Scholar]
- Guo W., Li W., Yuan L., Mei X., Hu W. (2019). MicroRNA-106a-3p Induces Apatinib Resistance and Activates Janus-Activated Kinase 2 (JAK2)/Signal Transducer and Activator of Transcription 3 (STAT3) by Targeting the SOCS System in Gastric Cancer. Med. Sci. Monit. 25, 10122–10128. 10.12659/MSM.919610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschalk C. M., Herold-Mende C. C., Fusenig N. E., Mueller M. M. (2006). Granulocyte colony-stimulating Factor and Granulocyte-Macrophage colony-stimulating Factor Promote Malignant Growth of Cells from Head and Neck Squamous Cell Carcinomas In Vivo . Cancer Res. 66 (16), 8026–8036. 10.1158/0008-5472.CAN-06-0158 [DOI] [PubMed] [Google Scholar]
- Hajimoradi M., Mohammad Hassan Z., Ebrahimi M., Soleimani M., Bakhshi M., Firouzi J., et al. (2016). STAT3 Is Overactivated in Gastric Cancer Stem-like Cells. Cell J 17 (4), 617–628. 10.22074/cellj.2016.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Li Z., Zhang X., Yin K., Wang W., Xu Z. (2018). MiR-422a Regulates Cellular Metabolism and Malignancy by Targeting Pyruvate Dehydrogenase Kinase 2 in Gastric Cancer. Cell Death Dis 9 (5), 505. 10.1038/s41419-018-0564-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong I. S. (2016). Stimulatory versus Suppressive Effects of GM-CSF on Tumor Progression in Multiple Cancer Types. Exp. Mol. Med. 48 (7), e242. 10.1038/emm.2016.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W. (2020). Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial-Mesenchymal Transition. Cells 9 (1), 217. 10.3390/cells9010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H., Xing W., Yang J., Zheng Y., Jia X., Zhang B., et al. (2016). An Effective Cytokine Adjuvant Vaccine Induces Autologous T-Cell Response against colon Cancer in an Animal Model. BMC Immunol. 17 (1), 31. 10.1186/s12865-016-0172-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M., Kokubu F., Odaka M., Watanabe S., Suzuki S., Ieki K., et al. (2004). Induction of Granulocyte-Macrophage colony-stimulating Factor by a New Cytokine, ML-1 (IL-17F), via Raf I-MEK-ERK Pathway. J. Allergy Clin. Immunol. 114 (2), 444–450. 10.1016/j.jaci.2004.03.047 [DOI] [PubMed] [Google Scholar]
- Li L., Clevers H. (2010). Coexistence of Quiescent and Active Adult Stem Cells in Mammals. Science 327 (5965), 542–545. 10.1126/science.1180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Y., Wei C., Yang Z., Chang W., Zhang X. (2020). LncRNA HULC Promotes the Progression of Gastric Cancer by Regulating miR-9-5p/MYH9 axis. Biomed. Pharmacother. 121, 109607. 10.1016/j.biopha.2019.109607 [DOI] [PubMed] [Google Scholar]
- Lu J., Wang Y. H., Yoon C., Huang X. Y., Xu Y., Xie J. W., et al. (2020). Circular RNA Circ-RanGAP1 Regulates VEGFA Expression by Targeting miR-877-3p to Facilitate Gastric Cancer Invasion and Metastasis. Cancer Lett. 471, 38–48. 10.1016/j.canlet.2019.11.038 [DOI] [PubMed] [Google Scholar]
- Matsui W. H. (2016). Cancer Stem Cell Signaling Pathways. Medicine (Baltimore) 95 (1 Suppl. 1), S8–S19. 10.1097/MD.0000000000004765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeish K. R., Knall C., Ward R. A., Gerwins P., Coxon P. Y., Klein J. B., et al. (1998). Activation of Mitogen-Activated Protein Kinase Cascades during Priming of Human Neutrophils by TNF-Alpha and GM-CSF. J. Leukoc. Biol. 64 (4), 537–545. 10.1002/jlb.64.4.537 [DOI] [PubMed] [Google Scholar]
- Metcalf D. (2010). The colony-stimulating Factors and Cancer. Nat. Rev. Cancer 10 (6), 425–434. 10.1038/nrc2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihanfar A., Fattahi A., Nejabati H. R. (2019). MicroRNA-mediated Drug Resistance in Ovarian Cancer. J. Cel Physiol 234 (4), 3180–3191. 10.1002/jcp.26060 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy J. A., Denicoff A. M., Venzon D. J., Danforth D., Pierce L. J., Frame J. N., et al. (1994). A Dose Intensity Study of FLAC (5-fluorouracil, Leucovorin, Doxorubicin, Cyclophosphamide) Chemotherapy and Escherichia Coli-Derived Granulocyte-Macrophage colony-stimulating Factor (GM-CSF) in Advanced Breast Cancer Patients. Ann. Oncol. 5 (8), 709–716. 10.1093/oxfordjournals.annonc.a058975 [DOI] [PubMed] [Google Scholar]
- Obermueller E., Vosseler S., Fusenig N. E., Mueller M. M. (2004). Cooperative Autocrine and Paracrine Functions of Granulocyte colony-stimulating Factor and Granulocyte-Macrophage colony-stimulating Factor in the Progression of Skin Carcinoma Cells. Cancer Res. 64 (21), 7801–7812. 10.1158/0008-5472.CAN-03-3301 [DOI] [PubMed] [Google Scholar]
- Pan Y., Shu X., Sun L., Yu L., Sun L., Yang Z., et al. (2017). miR-196a-5p M-odulates G-astric C-ancer S-tem C-ell C-haracteristics by T-argeting Smad4. Int. J. Oncol. 50 (6), 1965–1976. 10.3892/ijo.2017.3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y., Lee C. J., Choi J. H., Kim J. H., Kim J. W., Kim J. Y., et al. (2019). The JAK2/STAT3/CCND2 Axis Promotes Colorectal Cancer Stem Cell Persistence and Radioresistance. J. Exp. Clin. Cancer Res. 38 (1), 399. 10.1186/s13046-019-1405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X. H., Nakanishi Y., Takayama K., Bai F., Hara N. (1999). Granulocyte, Granulocyte-Macrophage, and Macrophage colony-stimulating Factors Can Stimulate the Invasive Capacity of Human Lung Cancer Cells. Br. J. Cancer 79 (1), 40–46. 10.1038/sj.bjc.6690009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Wu S., Wu Y., Liu T., Zhao X., Li Y. (2019). MicroRNA-196a/-196b Regulate the Progression of Hepatocellular Carcinoma through Modulating the JAK/STAT Pathway via Targeting SOCS2. Cel Death Dis 10 (5), 333. 10.1038/s41419-019-1530-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., et al. (2007). Identification and Expansion of Human colon-cancer-initiating Cells. Nature 445 (7123), 111–115. 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- Rupaimoole R., Slack F. J. (2017). MicroRNA Therapeutics: towards a new era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 16 (3), 203–222. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- Schulenburg A., Blatt K., Cerny-Reiterer S., Sadovnik I., Herrmann H., Marian B., et al. (2015). Cancer Stem Cells in Basic Science and in Translational Oncology: Can We Translate into Clinical Application? J. Hematol. Oncol. 8, 16. 10.1186/s13045-015-0113-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Ryu M. H., Park Y. S., Ahn J. Y., Park Y., Park S. R., et al. (2019). Loss of HER2 Positivity after Anti-HER2 Chemotherapy in HER2-Positive Gastric Cancer Patients: Results of the GASTric Cancer HER2 Reassessment Study 3 (GASTHER3). Gastric Cancer 22 (3), 527–535. 10.1007/s10120-018-0891-1 [DOI] [PubMed] [Google Scholar]
- Shi Y., Liu C. H., Roberts A. I., Das J., Xu G., Ren G., et al. (2006). Granulocyte-macrophage colony-stimulating Factor (GM-CSF) and T-Cell Responses: what We Do and Don't Know. Cell Res 16 (2), 126–133. 10.1038/sj.cr.7310017 [DOI] [PubMed] [Google Scholar]
- Sielska M., Przanowski P., Pasierbińska M., Wojnicki K., Poleszak K., Wojtas B., et al. (2020). Tumour-derived CSF2/granulocyte Macrophage colony Stimulating Factor Controls Myeloid Cell Accumulation and Progression of Gliomas. Br. J. Cancer 123 (3), 438–448. 10.1038/s41416-020-0862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Yang Z., Xu Y., Chen Y., Yu Q. (2015). MicroRNAs in Apoptosis, Autophagy and Necroptosis. Oncotarget 6 (11), 8474–8490. 10.18632/oncotarget.3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jiao X., Pestell T. G., Fan C., Qin S., Mirabelli E., et al. (2014). MicroRNAs and Cancer Stem Cells: the Sword and the Shield. Oncogene 33 (42), 4967–4977. 10.1038/onc.2013.492 [DOI] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71 (3), 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Takaishi S., Okumura T., Tu S., Wang S. S., Shibata W., Vigneshwaran R., et al. (2009). Identification of Gastric Cancer Stem Cells Using the Cell Surface Marker CD44. Stem Cells 27 (5), 1006–1020. 10.1002/stem.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura Y., Kobayashi M., Nakata H., Harada R., Kubota T., Taguchi H. (2004). Effect of Serum Deprivation on Constitutive Production of Granulocyte-colony Stimulating Factor and Granulocyte Macrophage-colony Stimulating Factor in Lung Cancer Cells. Int. J. Cancer 109 (6), 826–832. 10.1002/ijc.20023 [DOI] [PubMed] [Google Scholar]
- Uemura Y., Kobayashi M., Nakata H., Kubota T., Bandobashi K., Saito T., et al. (2006). Effects of GM-CSF and M-CSF on Tumor Progression of Lung Cancer: Roles of MEK1/ERK and AKT/PKB Pathways. Int. J. Mol. Med. 18 (2), 365–373. 10.3892/ijmm.18.2.365 [DOI] [PubMed] [Google Scholar]
- Uen Y. H., Fang C. L., Lin C. C., Hseu Y. C., Hung S. T., Sun D. P., et al. (2018). Ceramide Synthase 6 Predicts the Prognosis of Human Gastric Cancer: It Functions as an Oncoprotein by Dysregulating the SOCS2/JAK2/STAT3 Pathway. Mol. Carcinog 57 (12), 1675–1689. 10.1002/mc.22888 [DOI] [PubMed] [Google Scholar]
- Vilalta M., Rafat M., Giaccia A. J., Graves E. E. (2014). Recruitment of Circulating Breast Cancer Cells is Stimulated by Radiotherapy. Cell Rep. 8 (2), 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilalta M., Brune J., Rafat M., Soto L., Graves E. E. (2018). The Role of Granulocyte Macrophage Colony Stimulating Factor (GM-CSF) in Radiation-Induced Tumor Cell Migration. Clin. Exp. Metastasis 35 (4), 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Han G., Wang K., Liu G., Wang R., Xiao H., et al. (2014). Tumor-derived GM-CSF Promotes Inflammatory colon Carcinogenesis via Stimulating Epithelial Release of VEGF. Cancer Res. 74 (3), 716–726. 10.1158/0008-5472.CAN-13-1459 [DOI] [PubMed] [Google Scholar]
- Xu X., Zheng S. (2020). MiR-887-3p Negatively Regulates STARD13 and Promotes Pancreatic Cancer Progression. Cancer Manag. Res. 12, 6137–6147. 10.2147/CMAR.S260542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. Y., Tang J. N., Xie H. X., Du Y. A., Huang L., Yu P. F., et al. (2015). 5-Fluorouracil Chemotherapy of Gastric Cancer Generates Residual Cells with Properties of Cancer Stem Cells. Int. J. Biol. Sci. 11 (3), 284–294. 10.7150/ijbs.10248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Lee H., Herrmann A., Buettner R., Jove R. (2014). Revisiting STAT3 Signalling in Cancer: New and Unexpected Biological Functions. Nat. Rev. Cancer 14 (11), 736–746. 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- Zhou X., Xia Y., Li L., Zhang G. (2015). MiR-101 Inhibits Cell Growth and Tumorigenesis of Helicobacter pylori Related Gastric Cancer by Repression of SOCS2. Cancer Biol. Ther. 16 (1), 160–169. 10.4161/15384047.2014.987523 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA811393.