Abstract
Background
There is a paucity of data regarding blood culture utilization and antimicrobial-resistant (AMR) infections in low and middle-income countries (LMICs). In addition, there has been a concern for increasing AMR infections among COVID-19 cases in LMICs. Here, we investigated epidemiology of AMR bloodstream infections (BSI) before and during the COVID-19 pandemic in the Indonesian national referral hospital.
Methods
We evaluated blood culture utilization rate, and proportion and incidence rate of AMR-BSI caused by WHO-defined priority bacteria using routine hospital databases from 2019 to 2020. A patient was classified as a COVID-19 case if their SARS-CoV-2 RT-PCR result was positive. The proportion of resistance was defined as the ratio of the number of patients having a positive blood culture for a WHO global priority resistant pathogen per the total number of patients having a positive blood culture for the given pathogen. Poisson regression models were used to assess changes in rate over time.
Results
Of 60,228 in-hospital patients, 8,175 had at least one blood culture taken (total 17,819 blood cultures), giving a blood culture utilization rate of 30.6 per 1,000 patient-days. A total of 1,311 patients were COVID-19 cases. Blood culture utilization rate had been increasing before and during the COVID-19 pandemic (both p < 0.001), and was higher among COVID-19 cases than non-COVID-19 cases (43.5 vs. 30.2 per 1,000 patient-days, p < 0.001). The most common pathogens identified were K. pneumoniae (23.3%), Acinetobacter spp. (13.9%) and E. coli (13.1%). The proportion of resistance for each bacterial pathogen was similar between COVID-19 and non-COVID-19 cases (all p > 0.10). Incidence rate of hospital-origin AMR-BSI increased from 130.1 cases per 100,000 patient-days in 2019 to 165.5 in 2020 (incidence rate ratio 1.016 per month, 95%CI:1.016–1.017, p < 0.001), and was not associated with COVID-19 (p = 0.96).
Conclusions
In our setting, AMR-BSI incidence and etiology were similar between COVID-19 and non-COVID-19 cases. Incidence rates of hospital-origin AMR-BSI increased in 2020, which was likely due to increased blood culture utilization. We recommend increasing blood culture utilization and generating AMR surveillance reports in LMICs to inform local health care providers and policy makers.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-022-01114-x.
Keywords: Antimicrobial resistance, Blood culture, Blood culture utilization, Bloodstream infection, COVID-19, Indonesia
Background
Antimicrobial-resistant (AMR) bacterial infections pose an emerging health problem globally, with a disproportionate impact in low and middle-income countries (LMICs) [1, 2]. The COVID-19 pandemic has potentially escalated this problem due to increased use of antibiotics in patients hospitalized with COVID-19 [3, 4].
Microbiology laboratories with blood culture facility hold a critical function of diagnosing the bacterial cause of infection and monitoring the AMR situation. The Surviving Sepsis Campaign International Guidelines recommend performing blood culture before starting antimicrobial therapy in patients presenting with sepsis [5]. Blood culture can be used to identify pathogenic organisms causing either community or hospital-acquired bloodstream infections (BSI); hence, blood culture results can guide definitive antimicrobial choices for each individual patient. In addition, cumulative antibiogram reports can be used to monitor the epidemiology of AMR infections and guide empirical antimicrobial choices to population [6].
There is a paucity of systematic surveillance networks evaluating blood culture utilization and burden of AMR infections in LMICs, including Indonesia. Indonesia is a lower-middle-income country in Southeast Asia with the world’s fourth largest population. A range of complex factors, e.g. limited laboratory infrastructure and limited specialized health care practitioners, lack of regulations on antimicrobial use and high burden of infectious diseases have hampered the implementation of the Indonesian National Action Plan for AMR [7–9]. Indonesia first reported AMR surveillance key indicators to the World Health Organization (WHO) Global Antimicrobial Resistance Surveillance System (GLASS) in 2021 [10]. The blood culture utilization in Indonesia is low (9% patients sampled for blood cultures out of all inpatients in Makassar versus 21% in Thailand in 2015) [11], which could lead to an underestimation of incidence rates and an overestimation of the proportion of AMR infection [12]. Thus, it is crucial to evaluate blood culture utilization rate together with the trend of AMR infections, particularly in LMICs [12].
Indonesia has been highly impacted by the COVID-19 pandemic. Following the first two confirmed cases of SARS-CoV-2 infection in Indonesia on 2 March 2020, there was a rapid increase with three pandemic waves of COVID-19 patients reaching 5.8 million confirmed cases and 1506,000 deaths countrywide at March 2022 [13]. Here, we evaluate blood culture utilization and epidemiology of AMR bloodstream infections in the Indonesian national referral hospital before and during the COVID-19 pandemic.
Methods
Study design, setting and population
We conducted a retrospective hospital-wide surveillance study by using routine data of all patients hospitalized at Cipto Mangunkusumo Hospital, the Indonesian national referral hospital, Jakarta, Indonesia, from 1 January 2019 to 31 December 2020. In response to increase number of COVID-19 cases in Indonesia, the hospital has expanded its capacity from 1,000 beds in 2019 to 1,125 beds in 2020, allocating 238 beds for COVID-19 cases and 887 beds for non-COVID-19 cases.
Data collection
At the hospital, blood culture collection was determined by attending physicians based on the national standard practice [14]. Blood cultures were routinely performed at the microbiology laboratory of the Department of Clinical Pathology (International Organization for Standardization [ISO] 15,189, ISO 17205 and Joint Committee International accredited). A BacT/ALERT 3D automated microbial detection system machine expanded with additional incubator module (bioMerieux, Inc. Durham, USA) which can incubate up to 360 bottles was used. Isolated bacteria were identified using conventional bacterial identification methods and Vitek®2 (bioMerieux, Inc. Durham, USA). Antimicrobial susceptibility testing (AST) was performed using the Kirby-Bauer disc diffusion method according to Clinical and Laboratory Standards Institute guidelines [15].
Blood culture data were obtained through the Hospital Information System Management including the medical record number (MRN), admission date, specimen type, specimen date, culture and AST result. Hospital admission data were collected from the routine in-patient electronic records, and included MRN, admission date, discharge date and healthcare reimbursement program.
Definitions
The blood culture utilization rate was defined as the ratio of the number of blood cultures per 1,000 patient-days [12]. Blood culture contamination was defined as the isolation of one or more common commensal organisms listed on National Healthcare Safety Network the Centers for Disease Control and Prevention list 2022 in only one set of blood culture or one of a series of two or more blood culture [16]. The blood culture contamination rate is defined as the ratio of the number of blood culture contamination per number of total blood cultures [17].
We used the definitions of infection origin as proposed by WHO GLASS. Community-origin (or hospital-origin) BSI was defined for patients in the hospital less (or more) than the first two calendar days of admission when the first blood specimen culture positive for a pathogen were taken, with calendar day one equal to the day of admission. For deduplication purposes, only the first isolate per patient, per pathogen, per year period was included in the analyses [18].
Our target pathogens were 12 bacteria species in the WHO global priority pathogens list; including carbapenem-resistant Acinetobacter spp. (CRACI), carbapenem-resistant Pseudomonas aeruginosa (CRPA), carbapenem-resistant or 3rd generation cephalosporin-resistant Klebsiella pneumoniae (CRKP or 3GCRKP), carbapenem-resistant or 3rd generation cephalosporin-resistant Escherichia coli (CREC or 3GCREC), vancomycin-resistant Enterococcus faecium, methicillin-resistant Staphylococcus aureus (MRSA), Helicobacter pylori, clarithromycin, fluoroquinolone-resistant Campylobacter, fluoroquinolone-resistant Salmonella spp, 3rd generation cephalosporin-resistant or fluoroquinolone-resistant Neisseria gonorrhoeae, penicillin-non-susceptible Streptococcus pneumoniae, ampicillin-resistant Haemophilus influenzae, fluoroquinolone-resistant Shigella spp. [19].
The proportion of resistance was defined as the ratio of the number of patients having a positive blood culture for a WHO global priority resistant pathogen per the total number of patients having a positive blood culture for the given pathogen [18]. The incidence rate of community-origin AMR BSI is defined as the ratio of the number of patients with community-origin AMR BSI per 1,000 admissions. The incidence rate of hospital-origin AMR BSI is defined as the ratio of the number of patients with hospital-origin AMR BSI for each pathogen and antibiotic per 100,000 bed-days at risk of hospital-acquired infection. Moreover, as proposed by the WHO GLASS [18], we also estimated the incidence rate of AMR BSI per 100,000 tested patients as described previously [12].
A patient was classified as a COVID-19 case if their SARS-CoV-2 RT-PCR result was positive at any point during the admission period. We identified PCR-positive COVID-19 cases using the data of the healthcare imbursement program (Indonesian Case Based Groups [INA-CBG]) code of B34.2. which will cover confirmed COVID-19 patient expanses until cure). The year 2019 and 2020 was regarded as before and during the COVID-19 pandemic, respectively.
Ethics
The study was approved by the Faculty of Medicine Universitas Indonesia Ethics Committee (KET-115/UN2.F1/ETIK/PPM.00.02/2021) and Oxford Tropical Research Ethics Committee (Reference: 503-22). The requirement for patient consent was waived as this was a secondary analysis of anonymised routine surveillance data. Permission was obtained from the hospital’s Innovation and Intellectual Property Directorate to use the routine hospital database for this study.
Data analysis
Pearson’s chi-squared test and Fisher’s Exact test were used to compare categorical variables between groups. Kruskal Wallis test was used to compare continuous variables between groups.
We compared the blood culture utilization rate, contamination rate, isolated pathogens, and proportions and incidence rates of AMR BSI between COVID-19 and non-COVID-19 cases and between patients admitted in 2019 and 2020. Poisson regression models were used to assess changes in rate over time. All data analyses were performed using the STATA version 15.1 (StataCorp, College Station, TX, USA). We visualized figures with GraphPad Prism version 8.3.0 (La Jolla, California, USA). We also generated an overall AMR surveillance report using “AutoMated tool for Antimicrobial resistance Surveillance System (AMASS)” [20].
Results
Baseline characteristics
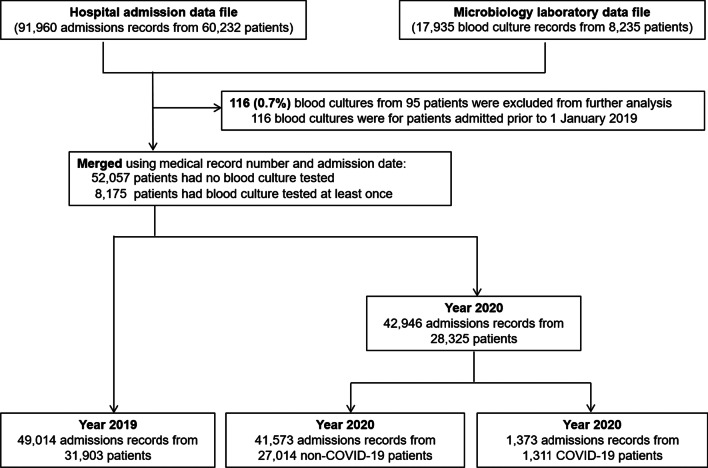
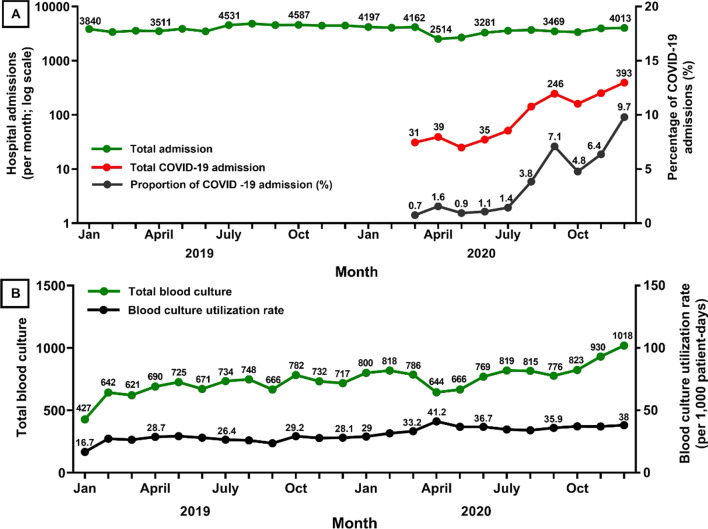
Of 91,960 admissions (from 60,228 patients) admitted during the study period, 1,373 (from 1,311 patients) were COVID-19 cases (Fig. 1 and Table 1). In 2019, prior to the COVID-19 pandemic, the number of hospital admissions per month was relatively stable with a mean of 4,085 (range 3,374–4,818; Fig. 2A). At the start of the COVID-19 pandemic, the number of hospital admissions per month decreased sharply from 4,162 in March 2020 to 2,514 in April 2020 (39% decrease). The proportion of COVID-19 admissions per total admissions increased from 0.7% (31 admissions) in March 2020 and reached the highest to 9.7% (393 admissions) in December 2020. Overall, the total numbers of admissions per year was higher in 2019 at 49,014 (of 31,903 patients) than in 2020 at 42,946 (of 28,325 patients) (12% difference; Table 1).
Fig. 1.
Title: Flow diagram
Table 1.
Baseline characteristics by year and by COVID-19 status
| Parameters | Year 2019 | Year 2020 | P values | Non-COVID-19 cases | COVID-19 cases* | P values |
|---|---|---|---|---|---|---|
| Total number of admissions | 49,014 | 42,946 | – | 90,587 | 1,373 | – |
| Total number of inpatients (de-duplicated) | 31,903 | 28,325 | – | 58,917 | 1,311 | – |
| Number of patient-days | 308,926 | 274,322 | – | 571,707 | 11,541 | – |
| Number of blood culture specimens received | 8,155 | 9,664 | – | 17,286 | 533 | – |
| Number of blood culture positive for any organism | 1,589 | 2,025 | – | 3,513 | 101 | – |
| Blood culture positivity rate | 19.5% | 20.9% | 0.02 | 20.3% | 18.9% | 0.44 |
| Number of blood culture positive for commensal bacteria** | 438 | 570 | – | 977 | 32 | – |
| Blood culture contamination rate** | 5.4% | 5.9% | 0.13 | 5.7% | 6.0% | 0.73 |
| Number of patients sampled for blood cultures (de-duplicated) | 4,026 | 4,501 | – | 7,973 | 348 | – |
| Prevalence of patients sampled for blood cultures among all inpatients | 12.6% | 15.9% | < 0.001 | 13.5% | 26.5% | < 0.001 |
| Average number of blood culture specimens sampled per admission | 1.6 | 1.7 | – | 1.7 | 1.5 | – |
| Total number of admissions that had at least two blood culture specimens sampled (%) | 2,472 (5.0%) | 2,889 (6.7%) | < 0.001 | 5,174 (5.7%) | 187 (13.6%) | < 0.001 |
| Median duration between the first and second blood culture specimen (days, IQR)*** | 5 (3–9) | 5 (3–8) | 0.52 | 5 (3–8) | 4 (2–7) | 0.57 |
| Blood culture utilization rate (per 1,000 patient-days) | 26.4 | 35.1 | < 0.001 | 30.2 | 43.5 | < 0.001 |
| Blood culture utilization for community-origin BSI | ||||||
| Prevalence of blood culture specimens being collected within the first 2 calendar days of hospital admission | 34.3% (2,801/8,155) | 31.9% (3,087/9,664) | 0.01 | 32.7% (5,650/17,286) | 45.0% (240/533) | < 0.001 |
| Number of patients tested for community-origin BSI (de-duplicated) **** | 1,747 | 2,481 | – | 4,458 | 176 | – |
| Blood culture utilization for hospital-origin BSI | ||||||
| Prevalence of blood culture specimens being collected after the first 2 calendar days of hospital admission | 65.7% (5,354/8,155) | 68.1% (6,577/9,664) | 0.01 | 67.3% (11,636/17,286) | 55.0% (293/533) | < 0.001 |
| Number of patients tested for hospital-origin BSI (de-duplicated) **** | 2,556 | 2,385 | – | 4,175` | 176 | – |
BSI Bloodstream infections
*All COVID-19 cases were in 2020
**Commensal bacteria included coagulase-negative Staphylococci, viridans group Streptococci, Propionibacterium acnes, Corynebacterium spp., and Bacillus spp.
***Among admissions that had at least two blood culture specimens sampled
****Patients tested for community-origin BSI were defined as patients with the first blood culture performed within the first two calendar days of admissions during the reporting period. Patients tested for hospital-origin BSI were defined as patients with the first blood culture performed after the first two calendar days of admissions during the reporting period
Fig. 2.
Numbers of patient admissions (A) and blood cultures (B) among inpatients from 2019 to 2020
Blood culture utilization
Of 60,228 patients, 8,175 had at least one blood culture taken (total 17,819 blood cultures). Total patient-days during the study period were 583,248, giving a blood culture utilization rate of 30.6 per 1,000 patient-days. 2,735 patients had at least two blood cultures sampled within a single admission, and the median duration between the first and second blood culture was 5 calendar days (IQR 3–8 calendar days).
The blood culture utilization rate showed an increasing trend over 2019, before the COVID-19 pandemic, from 16.7 per 1,000 patient-days in January 2019 to 28.1 in December 2019 (utilization rate ratio [URR] 1.02 per month; 95%CI 1.01–1.02, p < 0.001, Fig. 2B). This increasing trend continued throughout 2020 (URR 1.02 per month; 95%CI 1.01–1.02, p < 0.001).
The blood culture utilization rate was higher among COVID-19 cases compared to non-COVID-19 cases (43.5 vs. 30.2 per 1,000 patient-days; p < 0.001; Table 1). In a multivariable Poisson regression model, the blood culture utilization rate was independently associated with time (adjusted URR [aURR]: 1.02 per month, 95% CI 1.01–1.02, p < 0.001) and COVID-19 cases (aURR 1.19, 95% CI 1.09–1.30, p < 0.001).
Isolated organisms
Of 17,819 blood cultures, 1,008 were positive for commensal bacteria, giving a blood culture contamination rate of 5.6% during the study period. Of 8,175 patients who had at least one blood culture taken, 1,895 (23.1%) had at least one blood culture positive for one or more pathogenic organisms.
Among patients with BSI, 1,342 (70.8%) were Gram-negative bacteria, 296 (15.6%) were Gram-positive bacteria, 205 (10.8%) were fungi and 52 (2.8%) were polymicrobial infections (Table 2 and Additional file 1: Table S1). The most common pathogens identified were K. pneumoniae (23.3%; n = 442), Acinetobacter spp. (13.9%; n = 263), E. coli (13.1%; n = 249), S. aureus (11.6%; n = 219) and P. aeruginosa (8.6%; n = 163).
Table 2.
Pathogenic organisms isolated from 1,895 patients with bloodstream infections between 2019 and 2020*
| Pathogens | Year 2019 (N = 828) | Year 2020 (N = 1067) | P values | Non COVID-19 cases (N = 1838) | COVID-19 cases** (N = 57) | P values |
|---|---|---|---|---|---|---|
| Gram negative bacteria | ||||||
| Escherichia coli | 115 (13.9%) | 115 (10.7%) | 0.04 | 221 (12%) | 9 (15.8%) | 0.39 |
| Klebsiella pneumonia | 201 (24.3%) | 207 (19.4%) | 0.01 | 398 (21.7%) | 10(17.5%) | 0.45 |
| Klebsiella spp | 16 (1.9%) | 18 (1.7%) | 0.69 | 32 (1.7%) | 2 (3.5%) | 0.32 |
| Proteus spp | 7 (0.9%) | 12 (1.1%) | 0.54 | 19 (1%) | 0 (0%) | 0.44 |
| Salmonella spp | 21 (2.5%) | 12 (1.1%) | 0.02 | 33 (1.8%) | 0 (0%) | 0.62 |
| Salmonella enterica | 3 (0.4%) | 1 (0.1%) | 0.32 | 4 (0.2%) | 0 (0%) | > 0.99 |
| S. enterica serovar typhi | 2 (0.2%) | 4 (0.4%) | 0.70 | 5 (0.3%) | 1 (1.8%) | 0.05 |
| Shigella spp | 1 (0.1%) | 0 (0%) | 0.43 | 1 (0.1%) | 0 (0%) | > 0.99 |
| Pseudomonas aeruginosa | 59 (7.1%) | 98 (9.2%) | 0.10 | 152 (8.3%) | 5 (8.8%) | 0.89 |
| Pseudomonas spp | 1 (0.1%) | 3 (0.3%) | 0.63 | 4 (0.2%) | 0 (0%) | > 0.99 |
| Acinetobacter spp | 102 (12.3%) | 151 (14.1%) | 0.24 | 245 (13.3%) | 8 (14%) | 0.87 |
| Aeromonas spp | 5 (0.6%) | 5 (0.5%) | 0.75 | 10 (0.5%) | 0 (0%) | > 0.99 |
| Burkholderia cepacia | 6 (0.8%) | 7 (0.7%) | 0.85 | 13 (0.7%) | 0 (0%) | > 0.99 |
| Citrobacter spp | 4 (0.5%) | 3 (0.3%) | 0.70 | 7 (0.4%) | 0 (0%) | > 0.99 |
| Serratia spp | 11 (1.3%) | 6 (0.6%) | 0.07 | 17 (0.9%) | 0 (0%) | > 0.99 |
| Other Gram-negative bacteria | 63 (7.6%) | 83 (7.7%) | 0.89 | 143 (7.8%) | 3 (5.3%) | 0.48 |
| Gram positive bacteria | ||||||
| Staphylococcus aureus | 88 (10.6%) | 128 (12%) | 0.35 | 213 (11.6%) | 3 (5.3%) | 0.13 |
| Streptococcus pneumoniae | 2 (0.2%) | 1 (0.1%) | 0.58 | 2 (0.1%) | 1 (1.8%) | 0.08 |
| Streptococcus pyogenes | 2 (0.2%) | (0.2%) | > 0.99 | 4 (0.2%) | 0 (0%) | > 0.99 |
| Enterococcus faecium | 6 (0.8%) | 5 (0.5%) | 0.54 | 11 (0.6%) | 0 (0%) | > 0.99 |
| Enterococcus faecalis | 22 (2.7%) | 39 (3.6%) | 0.22 | 55 (3.0%) | 6 (10.4%) | 0.01 |
| Lactococcus garvieae | 0 (0%) | 1 (0.1%) | > 0.99 | 1 (0.1%) | 0 (0%) | > 0.99 |
| Fungi | ||||||
| Candida albicans | 7 (0.9%) | 30 (2.8%) | 0.01 | 34 (1.8%) | 3 (5.3%) | 0.67 |
| Non-albicans Candida spp | 53 (6.4%) | 110 (10.3%) | 0.01 | 157 (8.5%) | 6 (10.5%) | 0.59 |
| Cryptococcus spp | 2 (0.2%) | 0 (0%) | 0.19 | 2 (0.1%) | 0 (0%) | > 0.99 |
| Other fungi | 0 (0%) | 3 (0.3%) | 0.26 | 3 (0.3%) | 0 (0%) | > 0.99 |
| Polymicrobial infections*** | 29 (3.5%) | 23 (2.2%) | 0.07 | 52 (2.8%) | 0 (0%) | 0.40 |
BSI Bloodstream infections
*Only the first pathogenic isolate per patient during the study period was included
**All COVID-19 cases were in 2020
***Three most common polymicrobial infections were Escherichia coli + Klebsiella pneumoniae (10 patients), Klebsiella pneumoniae + Other Gram-negative bacteria (10 patients), Acinetobacter spp. + Klebsiella pneumoniae (7 patients). Polymicrobial infections are described in Addition file 1: Table S2
The proportion of isolated pathogens among BSI patients was moderately different between 2019 and 2020 (Table 2). The isolated pathogens were not different between COVID-19 and non-COVID-19 cases (all p > 0.05), except that the proportion of Enterococcus faecalis was lower in non-COVID-19 than COVID-19 cases (3.0% vs. 10.4% p = 0.01).
The most common pathogens identified as the cause of community-origin BSI was E. coli (20%; n = 103/515), followed by S. aureus (16.9%; n = 87/515) and K. pneumoniae (11.1%; n = 57/515), while the most common pathogens identified as the cause of hospital-origin BSI was K. pneumoniae (25.4%; n = 351/1,380), followed by Acinetobacter spp. (14%; n = 193/1,380) and non-albicans Candida (10.7%; n = 16/1,380) (Additional file 1: Table S1 and S2).
Proportion of AMR BSI
Of 442 patients with BSI caused by K. pneumoniae, 371 (83.9%) and 160 (36.2%) were caused by 3GCRKP and CRKP, respectively (Table 3). Of 249 patients with BSI caused by E. coli, 187 (76.1%) and 34 (13.6%) were caused by 3GCREC and CREC, respectively. All CREC and CRKP were also resistant to 3GC. The proportion of CRACI was 46.8% (123/263).
Table 3.
Proportion of WHO global priority AMR pathogens causing bloodstream infections
| Priority AMR pathogens* | Year 2019 | Year 2020 | P values | Non COVID-19 cases | COVID-19 cases** | P values |
|---|---|---|---|---|---|---|
| Carbapenem resistant Acinetobacter spp. | 46% (48/105) |
48.7% (77/158) |
0.56 |
48.2% (123/255) |
25% (2/8) |
0.29 |
|
Carbapenem resistant P. aeruginosa |
27% (17/64) |
24.2% (24/99) |
0.74 |
26% (41/158) |
0% (0/5) |
0.33 |
| Carbapenem resistant *** K. pneumoniae |
34% (75/218) |
38% (85/224) |
0.44 |
35.9% (155/432) |
50% (5/10) |
0.51 |
|
3rd Cephalosporin resistant *** K. pneumoniae |
85.3% (186/218) |
82.5% (185/224) |
0.43 |
83.8% (362/432) |
90% (9/10) |
> 0.99 |
|
Carbapenem resistant *** E. coli |
16.3% (21/129) |
10.8% (13/120) |
0.21 |
13% (31/240) |
34% (3/9) |
0.11 |
|
3rd Cephalosporin resistant *** E. coli |
76% (98/129) |
74.2% (89/120) |
0.74 |
75% (180/240) |
77.8% (7/9) |
> 0.99 |
|
Vancomycin resistant E. faecium |
0% (0/7) |
20% (1/5) |
0.42 |
8.3% (1/12) |
0% (0/0) |
|
|
Methicillin resistant S. aureus |
3.4% (3/88) |
9.2% (12/131) |
0.11 |
6.9% (15/216) |
0% (0/3) |
> 0.99 |
|
Fluoroquinolone resistant Salmonella spp. |
17.9% (5/28) |
5.9% (1/17) |
0.38 |
13.3% (6/45) |
0% (0/0) |
> 0.99 |
|
Fluoroquinolone resistant Shigella spp. |
100% (1/1) |
0% (0/0) |
100% (1/1) |
0% (0/0) |
||
|
Penicillin resistant S. pneumoniae |
50% (1/2) |
0% (0/1) |
> 0.99 |
50% (1/2) |
0% (0/1) |
> 0.99 |
| Overall**** |
55.9% (359/642) |
51.5% (389/755) |
0.10 |
53.6% (730/1361) |
50% (18/36) |
0.67 |
CO Community-origin, HO Hospital-origin
CO and HO were defined as proposed by WHO GLASS [18])
*Only the first pathogenic isolate per patient during the study period was included
**All COVID-19 cases were in 2020
***All carbapenem-resistant E. coli and K. pneumoniae were also resistant to 3rd cephalosporin cephalosporin
****Among patients with blood culture positive for Acinetobacter spp., P. aeruginosa, K. pneumoniae, E. coli, E. faecium, S. aureus, Salmonella spp, Shigella spp or S. pneumoniae
The proportion of AMR for each priority pathogen was not different between 2019 and 2020 (all p > 0.10), and between COVID-19 and non-COVID-19 cases (all p > 0.10; Table 3). However, the proportion of AMR for each priority pathogen were different between community-origin BSI and hospital-origin BSI (Additional file 1: Table S3). For example, the proportion of 3GCRKP (61.5% vs. 87.8%, p < 0.001), 3GCREC (67.8% vs. 77.6%, p = 0.02) and CRACI (16.4% vs. 56.9%, p < 0.001) were lower among community-origin BSI compared to those of hospital-origin BSI. Additional file 1: Table S4 provides additional details on proportion of AMR stratified by infection origin (community-origin vs. hospital-origin) and COVID-19 status. The overall AMR surveillance report is provided in Additional file 2.
Incidence rates of AMR BSI
The incidence rate of community-origin AMR BSI per 1,000 admissions was not significantly different between year 2019 and 2020 (1.6 to 1.6 per 1,000 admissions, p = 0.97; Table 4), while the incidence rate of hospital-origin AMR BSI per 100,000 patient-days in 2020 (165.5 per 100,000 patient-days at risk) was higher than 2019 (130.1 per 100,000 patient-days at risk) (p = 0.003). No specific outbreaks of AMR BSI were observed during the study period.
Table 4.
Incidence rate of WHO global priority AMR pathogens causing bloodstream infections
| Year 2019 | Year 2020 | P values | Non COVID-19 cases | COVID-19 cases** | P values | |
|---|---|---|---|---|---|---|
| Incidence rate of community-origin BSI caused by WHO global priority AMR pathogens | ||||||
| per 1,000 admissions |
1.6 (76/49,014) |
1.6 (67/42,946) |
0.97 |
1.6 (141/90,587) |
1.5 (2/1,373) |
> 0.99 |
| per 100,000 patients tested for community-origin BSI* | 4,350.3 (76/1,747) | 2,700.5 (67/2,481) | 0.004 |
3,162.8 (141/4,458) |
1,136.3 (2/176) | 0.11 |
| Incidence rate of hospital-origin BSI caused by WHO global priority AMR pathogens | ||||||
| per 1,000 admissions at risk of hospital-origin BSI* | 8.5 (283/33,226) | 10.9 (322/29,510) | 0.003 |
9.5 (589/61,758) |
14.6 (16/1,089) |
0.10 |
| per 100,000 patient-days at risk of hospital-origin BSI* |
130.1 (283/217,398) |
165.5 (322/194,486) |
0.003 | 146.9 (589/400,750) | 143.7 (16/11,134) | 0.96 |
| per 100,000 patients tested for hospital-origin BSI* |
11,071.9 (283/2,556) |
13,501.1 (322/2,385) |
0.01 |
14,107.7 (589/4,175) |
9,090.9 (16/176) |
0.07 |
BSI Bloodstream infections
*Patients tested for community-origin BSI were defined as patients with the first blood culture performed within the first two calendar days of admissions during the reporting period. Patients were considered at risk of hospital-origin BSI after they stayed in the hospital for more than 2 days. Patients tested for hospital-origin BSI were defined as patients with the first blood culture performed after the first two calendar days of admissions during the reporting period
**All COVID-19 cases were in 2020
The incidence rate of community-origin AMR BSI per 1,000 admissions was not different between COVID-19 and non-COVID-19 cases during 2019 and 2020 (1.5 vs. 1.6, p > 0.99). The incidence rate of hospital-origin AMR BSI per 100,000 patient-days at risk was also not different between COVID-19 and non-COVID-19 cases (143.7 vs. 146.9, p = 0.96).
We observed that the incidence rate of community-origin AMR BSI per 100,000 tested patients was higher in 2019 compared with 2020 (4350.3 vs. 2,700.5 per 100,000 tested patients, p = 0.004), while incidence rate of hospital-origin AMR BSI per 100,000 tested patients was lower in 2019 (11,071.9 vs. 13,501.1 per 100,000 tested patients, p = 0.01; Table 4). We found that incidence rate of community-origin AMR BSI and of hospital-origin AMR BSI per 100,000 tested patients was not significantly different between non-COVID-19 and COVID-19 cases (p = 0.11 and p = 0.07, respectively).
Discussion
This study illustrates that the use of readily available electronic hospital databases could provide robust and useful information on blood culture utilization and burden of AMR infections before and during the COVID-19 pandemic in LMICs. Although several reports have recently described an increase in AMR infections among COVID-19 cases [21, 22], we did not observe a difference of AMR BSI between COVID-19 and non-COVID-19 cases during the same time period in our setting. Strikingly, our study showed that the blood culture utilization rate had been increasing at the hospital before the COVID-19 pandemic (in 2019) and during the pandemic (in 2020), and, furthermore, that it was higher in COVID-19 cases than non-COVID-19 cases.
We did not observe a clear difference in the proportion and incidence rate of AMR infections between COVID-19 and non-COVID-19 cases, which is consistent with a study from Singapore [23]. Improved infection prevention control in hospitals and communities, and reduced mobilization in community could hypothetically explain this finding [24]. Nonetheless, studies from China [25], India [22], Italy [26], Taiwan [21], reported an increase in the proportion or incidence rates of AMR infections in COVID-19 patients. Multiple possible reasons for an increase include the high antibiotic use, predominance of severe COVID-19 patients in intensive care unit (ICU) with multiple predispositions towards AMR infections and protracted hospital stay, overcrowding of patients, and limited guideline adherence [27–31]. Therefore, appropriate antimicrobial prescribing, accurate diagnosis and appropriate infection prevention control are crucial for both COVID-19 and non-COVID-19 patients.
The observed increase in the incidence rate of hospital-origin AMR BSI during the COVID-19 pandemic is most likely due to the increase in blood culture utilization rate. A simulation study showed that observed incidence rate of AMR BSI (per 100,000 patient-days) could considerably increase if a hospital improves their blood culture utilization rate even if there are no changes in true susceptibility profiles of pathogenic organisms and in true infection rates in that environment over time [12]. We did not observe changes in proportion of AMR BSI and specific outbreaks of AMR infections during the study period, as was noticed by Hospital Infection Prevention Control Committee.
The observed increase in blood culture utilization rate before and during the COVID-19 pandemic could be due to several reasons. Before the COVID-19 pandemic, a new national clinical practice guideline on sepsis was launched including the recommendation to take blood cultures prior to start of empirical antibiotic therapy [14]. Adoption of this guideline in the hospital is likely to have contributed to the increase observed. Nonetheless, the blood culture utilization rate in non-COVID-19 cases was still lower than those reported in Thailand and many other high-income countries (e.g. 307.7, 86.5, 65.4 per 1,000 patient-days in United States, France and United Kingdom, respectively) [11, 32, 33]. Direct comparison of the rate with other LMICs could not be performed due to limited existing publication [34]. The low culture rate could be explained by lack of physician awareness of the sepsis guidelines, misperceptions that blood culture will add health care cost, among other factors [35]. Previous studies have reported contrasting findings on blood culture utilization in COVID-19 patients [22, 36, 37]. The increase in blood culture utilization among COVID-19 cases in our hospital is probably because the national referral hospital manages mostly COVID-19 patients with comorbidities and severe conditions. Given these are very sick people empirical antibiotic treatment is commonly recommended [38]. However, a recent meta-analysis has concluded that antibiotics are heavily overused in COVID-19 cases [39].
Local reporting on the hospital AMR epidemiology allows us to understand the local situation and support local actions. The top three pathogens causing BSIs are similar with findings in other countries in the region [10, 40]. However, we did not observe higher rate of S. aureus co-infection in COVID-19 patients, contrary to several reports from past viral and COVID-19 pandemic worldwide [41]. In addition, our analysis shows BSI cases with Salmonella enterica serovar Typhi, Shigella spp., and Burkholderia pseudomallei. Those pathogens are the cause of typhoid, shigellosis and melioidosis, respectively, and are notifiable pathogens in many countries [42, 43]. This information can support the decision making of the Ministry of Health in Indonesia, where a system of notifiable pathogens is not officially established. Our study also found that 8.6% of hospital-acquired BSI was caused by non-albicans Candida spp. There is limited information of fungal infections, particularly of non-albicans Candida infections, as the cause of BSI in LMICs [44–47] The relatively high proportion of fungemia in our data, compared with 1.1% in Thailand [46], could be due the complex, immunocompromised patient populations, with common invasive procedures and high antibiotic use, all of which are risk factors of invasive candidiasis [31, 48]. Available data worldwide suggest increasing incidence of fungemia caused by non-albicans Candida species [44, 49, 50] together with increasing resistance. We reported our findings to the hospital Infection and Antimicrobial Resistance Control Committee, and these are used to support local guidelines for the prevention and treatment of hospital acquired invasive fungal infections [50, 51].
Our study has some limitations. First, we could not determine whether a blood culture was taken before or after failure of empirical treatment as there was a low adherence to take blood culture prior to antibiotic treatment. Some patients may also be treated with parenteral antibiotics without blood culture taken. Implementation of case-based instead of laboratory-based surveillances could improve data representativeness in the future. Second, our AMR surveillance reports should not be used to guide empirical antibiotics without careful consideration. Hospitals in LMICs with a low blood culture utilization rate should use AMR surveillance reports stratified by exposure to an empirical antibiotic at the study hospital to guide choice of first-line empiric antimicrobial therapy rather than the total antibiogram [12]. Third, although large, our study may lack of power to observe a difference in AMR BSI between COVID-19 and non-COVID-19 cases. Fourth, we use calendar days of admission as a surrogate for defining origin of infection and data of patient transfers are not available. Therefore, a proportion of community-origin BSIs reported could be hospital-origin BSIs transferring from other hospitals. Lastly, the findings may not be generalizable to all other hospitals or the country at large.
Conclusions
In our setting, AMR BSIs were not different between COVID-19 and non-COVID-19 cases. Increased incidence rates of hospital-origin AMR infections observed in 2020 could be due to increasing blood culture utilization rate. Systematic, representative AMR data are required to better estimate the extent of the problem, and adequately inform antibiotic guidelines and stewardship programs. We recommend hospitals in LMICs to increase blood culture utilization and generate annual AMR surveillance reports together with parameters representing blood culture utilization.
Supplementary Information
Additional file1: Supplementary Table 1: Polymicrobial Pathogenic organisms isolated from 52 patients with bloodstream infections at Cipto Mangunkusumo National Hospital, Indonesia, between 2019 and 2020. Supplementary Table 2: Pathogenic organisms isolated from 1,895 patients with bloodstream infections at Cipto Mangunkusumo National Hospital, Indonesia, between 2019 and 2020. Supplementary Table 3: Prevalence of WHO global priority AMR pathogens causing bloodstream infections stratified by infection origin. Supplementary Table 4. Proportion of WHO global priority AMR pathogens causing bloodstream infections stratified by infection origin and COVID-19 status
Additional file2: Antimicrobial Resistance (AMR) Surveillance Report. Antimicrobial Resistance (AMR) Surveillance Report of the study site generated by AMASS
Acknowledgements
We thank the Hospital Information System Management Unit, Cipto Mangunkusumo National Referral Hospital for the support for this project. We thank Samuel Susanto for technical assistance.
Abbreviations
- AMR
Antimicrobial-resistant
- AST
Antimicrobial susceptibility testing
- AMASS
AutoMated tool for Antimicrobial resistance Surveillance System
- BSI
Bloodstream infections
- CRACI
Carbapenem-resistant Acinetobacter spp.
- CREC
Carbapenem-resistant Escherichia coli
- CRKP
Carbapenem-resistant Klebsiella pneumoniae
- CRPA
Carbapenem-resistant Pseudomonas aeruginosa
- GLASS
Global antimicrobial resistance surveillance system
- INA-CBG
Indonesian case based groups
- ICU
Intensive care unit
- LMIC
Low and middle-income country
- MRN
Medical record number
- MRSA
Methicillin-resistant Staphylococcus aureus
- 3GCREC
3rd generation cephalosporin-resistant Escherichia coli
- 3GCRKP
3rd generation cephalosporin-resistant Klebsiella pneumoniae
- WHO
World Health Organization
Author contributions
RS, DL, KCL, SSe conceptualized, designed and developed study. RS and DL wrote the original draft. RS, DL, KCL, SSe, SSuw, EJN, DHD, MRK, AP, SSum, CEM, RLH, NPJD provided valuable input, participated in drafting, editing, revising and approved the final version of the manuscript.
Funding
This study is funded by Research Fund of Division of Tropical and Infectious Diseases, Department of Internal Medicine, Cipto Mangunkusumo National Hospital. RS is funded by Indonesian Education Scholarship (Beasiswa Pendidikan Indonesia [BPI]) [202101182688] from the Ministry of Education, Culture, Research, and Technology Republic of Indonesia: Directorate General of Higher Education, Research, and Technology (Direktorat Jenderal Pendidikan Tinggi, Riset dan Teknologi Kementerian Pendidikan, Kebudayaan, Riset dan Teknologi Republik Indonesia) and Indonesian Endowment Fund for Education (Lembaga Pengelola Dana Pendidikan [LPDP]). DL is supported by the Wellcome Trust [220211]. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. The funders of the investigators and study had no role in the study design, data collection, data analysis, data interpretation, or writing of the manuscript. The corresponding author had the final responsibility for the decision to submit for publication.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by The Faculty of Medicine Universitas Indonesia Ethics Committee (KET-115/UN2.F1/ETIK/PPM.00.02/2021) and Oxford Tropical Research Ethics Committee (Reference: 503-22). The requirement for patient consent was waived as this was a secondary analysis of anonymised routine surveillance data.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Robert Sinto, Email: robert.sinto01@ui.ac.id, Email: robert.sinto@ndm.ox.ac.uk.
Khie Chen Lie, Email: chen_tropik@hotmail.com.
Siti Setiati, Email: s_setiati@yahoo.com.
Suhendro Suwarto, Email: suhendro.dr@gmail.com.
Erni J. Nelwan, Email: ejnelwan@yahoo.com
Dean Handimulya Djumaryo, Email: deanhandimulya@gmail.com.
Mulya Rahma Karyanti, Email: karyanti@ikafkui.net.
Ari Prayitno, Email: ariprayitno@yahoo.com.
Sumariyono Sumariyono, Email: sumariyono0704@gmail.com.
Catrin E. Moore, Email: camoore@sgul.ac.uk
Raph L. Hamers, Email: raph.hamers@ndm.ox.ac.uk
Nicholas P. J. Day, Email: nickd@tropmedres.ac
Direk Limmathurotsakul, Email: direk@tropmedres.ac.
References
- 1.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Neill J. Review on antimicrobial resistance 2014. Available from: http://amr-review.org/. Accessed 1 March 2022.
- 3.Lobie TA, Roba AA, Booth JA, Kristiansen KI, Aseffa A, Skarstad K, et al. Antimicrobial resistance: a challenge awaiting the post-COVID-19 era. Int J Infect Dis. 2021;111:322–325. doi: 10.1016/j.ijid.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kariyawasam RM, Julien DA, Jelinski DC, Larose SL, Rennert-May E, Conly JM, et al. Antimicrobial resistance (AMR) in COVID-19 patients: a systematic review and meta-analysis (November 2019-June 2021) Antimicrob Resist Infect Control. 2022;11(1):45. doi: 10.1186/s13756-022-01085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit Care Med. 2021;49(11):e1063–e1143. doi: 10.1097/CCM.0000000000005337. [DOI] [PubMed] [Google Scholar]
- 6.WHO . Global action plan on antimicrobial resistance. Geneva: World Health Organization; 2015. [Google Scholar]
- 7.Limato R, Nelwan EJ, Mudia M, de Brabander J, Guterres H, Enty E, et al. A multicentre point prevalence survey of patterns and quality of antibiotic prescribing in Indonesian hospitals. JAC Antimicrob Resist. 2021;3(2):dlab047. doi: 10.1093/jacamr/dlab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parathon H, Kuntaman K, Widiastoety TH, Muliawan BT, Karuniawati A, Qibtiyah M, et al. Progress towards antimicrobial resistance containment and control in Indonesia. BMJ. 2017;358:j3808. doi: 10.1136/bmj.j3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Bank. Indonesia-World Bank Data 2022. Available from: https://data.worldbank.org/country/ID. Accessed 1 March 2022.
- 10.WHO . Global antimicrobial resistance and use surveillance system (GLASS) report 2021. Geneva: World Health Organization; 2021. [Google Scholar]
- 11.Teerawattanasook N, Tauran PM, Teparrukkul P, Wuthiekanun V, Dance DAB, Arif M, et al. Capacity and utilization of blood culture in two referral hospitals in Indonesia and Thailand. Am J Trop Med Hyg. 2017;97(4):1257–1261. doi: 10.4269/ajtmh.17-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim C, Hantrakun V, Teerawattanasook N, Srisamang P, Teparrukkul P, Sumpradit N, et al. Impact of low blood culture usage on rates of antimicrobial resistance. J Infect. 2021;82(3):355–362. doi: 10.1016/j.jinf.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO. Indonesia: WHO coronavirus disease (COVID-19) Dashboard 2022. Available from: https://covid19.who.int/region/searo/country/id. Accessed 18 March 2022.
- 14.Indonesia KKR. Pedoman Nasional Pelayanan Kedokteran Tata Laksana Sepsis. Jakarta: Kementerian Kesehatan Republik Indonesia; 2017. [Google Scholar]
- 15.CLSI. Document M39-A4: Analysis and presentation of cumulative antimicrobial susceptibility test data. Wayne, PA: CLSI; 2014.
- 16.CDC. Bloodstream Infection Event (Central Line-Associated Bloodstream Infection and Non-central Line Associated Bloodstream Infection) 2022. Available from: https://www.cdc.gov/nhsn/pdfs/pscmanual/4psc_clabscurrent.pdf. Accessed 1 March 2022.
- 17.Doern GV, Carroll KC, Diekema DJ, Garey KW, Rupp ME, Weinstein MP, et al. Practical guidance for clinical microbiology laboratories: a comprehensive update on the problem of blood culture contamination and a discussion of methods for addressing the problem. Clin Microbiol Rev. 2019. 10.1128/CMR.00009-19.
- 18.WHO . Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2020. Geneva: World Health Organization; 2020. [Google Scholar]
- 19.WHO . Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Geneva: World Health Organization; 2017. [Google Scholar]
- 20.Lim C, Miliya T, Chansamouth V, Aung MT, Karkey A, Teparrukkul P, et al. Automating the generation of antimicrobial resistance surveillance reports: proof-of-concept study involving seven hospitals in seven countries. J Med Internet Res. 2020;22(10):e19762.
- 21.Lai CC, Chen SY, Ko WC, Hsueh PR. Increased antimicrobial resistance during the COVID-19 pandemic. Int J Antimicrob Agents. 2021;57(4):106324.
- 22.Saini V, Jain C, Singh NP, Alsulimani A, Gupta C, Dar SA, et al. Paradigm shift in antimicrobial resistance pattern of bacterial isolates during the COVID-19 pandemic. Antibiotics (Basel). 2021;10(8):954.
- 23.Ong CCH, Farhanah S, Linn KZ, Tang YW, Poon CY, Lim AY, et al. Nosocomial infections among COVID-19 patients: an analysis of intensive care unit surveillance data. Antimicrob Resist Infect Control. 2021;10(1):119.
- 24.Collignon P, Beggs JJ. CON: COVID-19 will not result in increased antimicrobial resistance prevalence. JAC Antimicrob Resist. 2020;2(3):dlaa051. doi: 10.1093/jacamr/dlaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wang J, Yang Y, Cai P, Cao J, Cai X, et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9(1):153. doi: 10.1186/s13756-020-00819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiri B, Sensi E, Marsiliani V, Cantarini M, Priante G, Vernelli C, et al. Antimicrobial stewardship program, COVID-19, and infection control: spread of carbapenem-resistant Klebsiella Pneumoniae colonization in ICU COVID-19 patients. What did not work? J Clin Med. 2020;9(9):2744.
- 27.Adebisi YA, Alaran AJ, Okereke M, Oke GI, Amos OA, Olaoye OC, et al. COVID-19 and antimicrobial resistance: a review. Infect Dis (Auckl). 2021;14:11786337211033870.
- 28.Knight GM, Glover RE, McQuaid CF, Olaru ID, Gallandat K, Leclerc QJ, et al. Antimicrobial resistance and COVID-19: intersections and implications. Elife. 2021. 10.7554/eLife.64139.
- 29.Rodriguez-Bano J, Rossolini GM, Schultsz C, Tacconelli E, Murthy S, Ohmagari N, et al. Key considerations on the potential impacts of the COVID-19 pandemic on antimicrobial resistance research and surveillance. Trans R Soc Trop Med Hyg. 2021;115(10):1122–9.
- 30.Clancy CJ, Buehrle DJ, Nguyen MH. PRO: The COVID-19 pandemic will result in increased antimicrobial resistance rates. JAC Antimicrob Resist. 2020;2(3):dlaa049. doi: 10.1093/jacamr/dlaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoma R, Seneghini M, Seiffert SN, Vuichard Gysin D, Scanferla G, Haller S, et al. The challenge of preventing and containing outbreaks of multidrug-resistant organisms and Candida auris during the coronavirus disease 2019 pandemic: report of a carbapenem-resistant Acinetobacter baumannii outbreak and a systematic review of the literature. Antimicrob Resist Infect Control. 2022;11(1):12. doi: 10.1186/s13756-022-01052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen AI, Bilker WB, Hamilton KW, O’Donnell JA, Nachamkin I. Blood culture utilization at an academic hospital: addressing a gap in benchmarking. Infect Control Hosp Epidemiol. 2018;39:1353–9.
- 33.Cassini A, Hogberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66.
- 34.Hemlock C, Luby SP, Saha S, Qamar F, Andrews JR, Saha SK, et al. Utilization of blood culture in South Asia for the diagnosis and treatment of febrile illness. Clin Infect Dis. 2020;71(Suppl 3):S266–75.
- 35.Ombelet S, Barbe B, Affolabi D, Ronat JB, Lompo P, Lunguya O, et al. Best practices of blood cultures in low- and middle income countries. Front Med (Lausanne). 2019;6:131.
- 36.Mormeneo Bayo S, Palacian Ruiz MP, Moreno Hijazo M, Villuendas Uson MC. Bacteremia during COVID-19 pandemic in a tertiary hospital in Spain. Enferm Infecc Microbiol Clin (Engl Ed). 2021;40:183–6.
- 37.Sepulveda J, Westblade LF, Whittier S, Satlin MJ, Greendyke WG, Aaron JG, et al. Bacteremia and blood culture utilization during COVID-19 surge in New York City. J Clin Microbiol. 2020. 10.1128/JCM.00875-20.
- 38.Alhazzani W, Evans L, Alshamsi F, Moller MH, Ostermann M, Prescott HC, et al. Surviving sepsis campaign guidelines on the management of adults with Coronavirus disease 2019 (COVID-19) in the ICU: first update. Crit Care Med. 2021;49(3):e219–e234. doi: 10.1097/CCM.0000000000004899. [DOI] [PubMed] [Google Scholar]
- 39.Langford BJ, So M, Raybardhan S, Leung V, Soucy JR, Westwood D, et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect. 2021;27(4):520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim C, Takahashi E, Hongsuwan M, Wuthiekanun V, Thamlikitkul V, Hinjoy S, et al. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. Elife. 2016. 10.7554/eLife.18082.
- 41.Adalbert JR, Varshney K, Tobin R, Pajaro R. Clinical outcomes in patients co-infected with COVID-19 and Staphylococcus aureus: a scoping review. BMC Infect Dis. 2021;21(1):985.
- 42.Hantrakun V, Kongyu S, Klaytong P, Rongsumlee S, Day NPJ, Peacock SJ, et al. Clinical epidemiology of 7126 melioidosis patients in Thailand and the implications for a national notifiable diseases surveillance system. Open Forum Infect Dis. 2019;6(12):ofz498.
- 43.CDC. National Notifiable Diseases Surveillance System (NNDSS): Notifiable Infectious Disease Data Tables 2022. Available from: https://www.cdc.gov/nndss/data-statistics/infectious-tables/index.html. Accessed 18 March 2022.
- 44.Cortes JA, Reyes P, Gomez C, Buitrago G, Leal AL, Group G. Fungal bloodstream infections in tertiary care hospitals in Colombia. Rev Iberoam Micol. 2011;28(2):74–8.
- 45.Dat VQ, Vu HN, Nguyen The H, Nguyen HT, Hoang LB, Vu Tien Viet D, et al. Bacterial bloodstream infections in a tertiary infectious diseases hospital in Northern Vietnam: aetiology, drug resistance, and treatment outcome. BMC Infect Dis. 2017;17(1):493.
- 46.Kanoksil M, Jatapai A, Peacock SJ, Limmathurotsakul D. Epidemiology, microbiology and mortality associated with community-acquired bacteremia in northeast Thailand: a multicenter surveillance study. PLoS One. 2013;8(1):e54714. doi: 10.1371/journal.pone.0054714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denning DW, Perlin DS, Muldoon EG, Colombo AL, Chakrabarti A, Richardson MD, et al. Delivering on antimicrobial resistance agenda not possible without improving fungal diagnostic capabilities. Emerg Infect Dis. 2017;23(2):177–83.
- 48.Logan C, Martin-Loeches I, Bicanic T. Invasive candidiasis in critical care: challenges and future directions. Intensive Care Med. 2020;46(11):2001–14.
- 49.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, et al. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol. 2012;50(11):3435–42.
- 50.Deshpande A, Gaur S, Bal AM. Candidaemia in the non-neutropenic patient: a critique of the guidelines. Int J Antimicrob Agents. 2013;42(4):294–300. doi: 10.1016/j.ijantimicag.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Ture Z, Alp E. Infection control measures to prevent hospital transmission of Candida. Hosp Pract. 2018;46(5):253–7.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file1: Supplementary Table 1: Polymicrobial Pathogenic organisms isolated from 52 patients with bloodstream infections at Cipto Mangunkusumo National Hospital, Indonesia, between 2019 and 2020. Supplementary Table 2: Pathogenic organisms isolated from 1,895 patients with bloodstream infections at Cipto Mangunkusumo National Hospital, Indonesia, between 2019 and 2020. Supplementary Table 3: Prevalence of WHO global priority AMR pathogens causing bloodstream infections stratified by infection origin. Supplementary Table 4. Proportion of WHO global priority AMR pathogens causing bloodstream infections stratified by infection origin and COVID-19 status
Additional file2: Antimicrobial Resistance (AMR) Surveillance Report. Antimicrobial Resistance (AMR) Surveillance Report of the study site generated by AMASS
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.