Abstract
Background
Lymphatic metastasis has been associated with poor prognosis in bladder cancer patients with limited therapeutic options. Emerging evidence shows that heat shock factor 1 (HSF1) drives diversified transcriptome to promote tumor growth and serves as a promising therapeutic target. However, the roles of HSF1 in lymphatic metastasis remain largely unknown. Herein, we aimed to illustrate the clinical roles and mechanisms of HSF1 in the lymphatic metastasis of bladder cancer and explore its therapeutic potential.
Methods
We screened the most relevant gene to lymphatic metastasis among overexpressed heat shock factors (HSFs) and heat shock proteins (HSPs), and analyzed its clinical relevance in three cohorts. Functional in vitro and in vivo assays were performed in HSF1‐silenced and ‐regained models. We also used Co‐immunoprecipitation to identify the binding proteins of HSF1 and chromatin immunoprecipitation and dual‐luciferase reporter assays to investigate the transcriptional program directed by HSF1. The pharmacological inhibitor of HSF1, KRIBB11, was evaluated in popliteal lymph node metastasis models and patient‐derived xenograft models of bladder cancer.
Results
HSF1 expression was positively associated with lymphatic metastasis status, tumor stage, advanced grade, and poor prognosis of bladder cancer. Importantly, HSF1 enhanced the epithelial‐mesenchymal transition (EMT) of cancer cells in primary tumor to initiate metastasis, proliferation of cancer cells in lymph nodes, and macrophages infiltration to facilitate multistep lymphatic metastasis. Mechanistically, HSF1 interacted with protein arginine methyltransferase 5 (PRMT5) and jointly induced the monomethylation of histone H3 at arginine 2 (H3R2me1) and symmetric dimethylation of histone H3 at arginine 2 (H3R2me2s). This recruited the WD repeat domain 5 (WDR5)/mixed‐lineage leukemia (MLL) complex to increase the trimethylation of histone H3 at lysine 4 (H3K4me3); resulting in upregulation of lymphoid enhancer‐binding factor 1 (LEF1), matrix metallopeptidase 9 (MMP9), C‐C motif chemokine ligand 20 (CCL20), and E2F transcription factor 2 (E2F2). Application of KRIBB11 significantly inhibited the lymphatic metastasis of bladder cancer with no significant toxicity.
Conclusion
Our findings reveal a novel transcriptional program directed by the HSF1‐PRMT5‐WDR5 axis during the multistep process of lymphatic metastasis in bladder cancer. Targeting HSF1 could be a multipotent and promising therapeutic strategy for bladder cancer patients with lymphatic metastasis.
Keywords: HSF1, PRMT5, KRIBB11, transcriptional program, bladder cancer, lymphatic metastasis, prognostic factor, targeted therapy
Abbreviations
- LN
lymph node
- BCa
bladder cancer
- TCGA
The Cancer Genome Atlas
- HSF1
heat shock factor 1
- HSF
heat shock factor
- HSP
heat shock protein
- NAT
normal adjacent tissues
- LN+ CA
lymphatic metastasis‐positive BCa tissues
- LN‐ CA
lymphatic metastasis‐negative BCa tissues
- LN+
metastasic lymph node tissues
- NMIBC
non‐muscle‐invasive bladder cancer
- MIBC
muscle‐invasive bladder cancer
- siRNAs
small interfering RNAs
- PRMT5
protein arginine methyltransferase 5
- WDR5
WD repeat domain 5
- MLL
mixed‐lineage leukemia
- LEF1
lymphoid enhancer‐binding factor 1
- MMP9
matrix metallopeptidase 9
- CCL20
C‐C motif chemokine ligand 20
- E2F2
E2F transcription factor 2
- H3R2me1
monomethylation of histone H3 at arginine 2
- H3R2me2s
symmetric dimethylation of histone H3 at arginine 2
- H3R8me2s
symmetric dimethylation of histone H3 at arginine 8
- H4R3me2s
symmetric dimethylation of histone H4 at arginine 3
- H3K4me3
trimethylation of histone H3 at lysine 4
- EMT
epithelial‐mesenchymal transition
- IVIS
in vivo imaging system
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- EdU
ethynyl deoxyuridine
- DAPI
4,6‐diamidino‐2‐phenylindole
- HSP90
heat shock protein 90
- HSP70
heat shock protein 70
- BCG
bacillus Calmette‐Guerin
- PIM2
proviral integration site of Moloney virus‐2
- PD‐L1
programmed death‐ligand 1
- Co‐IP
Co‐immunoprecipitation,
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative real‐time polymerase chain reaction
- PDX
patient‐derived xenograft
- CCR6
C‐C motif chemokine receptor 6
- CD163
CD163 molecule
- CD204
CD204 molecule
- CD206
CD206 molecule
- IL‐6
interleukin 6
- IL‐10
interleukin 10
- TGF‐β
transforming growth factor‐beta 1
- TME
tumor microenvironment
- TAM
tumor‐associated macrophage
- VEGF‐C
vascular endothelial growth factor C
- p‐TEFb
positive transcription elongation factor b
- Treg
regulatory T cells
- Th17
T helper type 17 cells
- MORC2
MORC family CW‐type zinc finger 2
- PRC2
polycomb repressive complex 2
- EZH2
enhancer of zeste homolog 2
- ArgBP2
Arg kinase‐binding protein 2
- H3K27me3
trimethylation of histone H3 at lysine 27
1. BACKGROUND
Metastasis is the leading cause of poor outcomes in patients with cancer [1]. For most epithelial malignancies, lymphatic metastasis is the primary approach used by cancers to metastasize away from their primary location [2]. Among urinary carcinomas, lymphatic metastasis is most prone to bladder cancer (BCa). Once lymphatic metastasis occurs, the 5‐year disease‐specific survival rate of BCa patients decreases significantly from 81.4% in patients with N0 to 29.3% in patients with N1, 18.2% with N2, and 0% with N3. However, neither chemotherapy nor radical cystectomy has demonstrated significant improvement [3, 4]. Therefore, there is an urgent need to clarify the mechanisms involved in lymphatic metastasis of BCa and explore novel therapeutic strategies targeting this condition.
Heat shock transcription factors (HSFs) and heat shock proteins (HSPs) were initially identified as proteins sustaining healthy proteome under environmental challenges [5]. Intriguingly, distinct from the phenomenon that HSFs and HSPs are impaired in most neurodegenerative diseases, these genes are overexpressed in many malignancies [6, 7]. In BCa, HSP90 was found to be highly expressed, while dual targeting of HSP90 and HSP70 enhanced the anticancer effect of chemotherapeutic agents [8]. HSP90 was also reported as a useful predictor for the failure of bacillus Calmette‐Guerin (BCG) therapy in BCa [9]. However, no study has described the roles of HSFs or HSPs in the lymphatic metastasis of BCa.
HSF1 is the master regulator among HSFs [10]. Increasing evidence shows that HSF1 is important in cancer growth and progression via directing transcriptional programs regulating the expression of oncogenes instead of HSPs [11, 12]. Yang et al. [13] demonstrated that the proviral integration site of Moloney virus‐2 (PIM2) phosphorylated HSF1 on Thr120 and bonded to the programmed death‐ligand 1 (PD‐L1) promoter to promote breast tumor growth. A recent study also found that HSF1 regulated the expression of a subset of E2F transcription factor family gene targets, which enhanced brain metastasis in lung adenocarcinoma [14]. Meanwhile, inhibitors targeting HSF1 were developed and showed encouraging prospects in cancer therapy, especially in myeloma [15, 16]. However, whether and how HSF1 is involved in BCa, especially in lymphatic metastasis, remains largely unknown. The effect of HSF1 inhibitors in blocking lymphatic metastasis of BCa still needs further clarifications.
In this study, we hypothesized that one or some HSFs or HSPs might play key roles in the lymphatic metastasis of BCa. To address this, we performed a stepwise selection of the upregulated proteins among all HSFs and HSPs in an online database and BCa tissues from our hospital. We identified that HSF1 had the highest clinical relevance and exerted oncogenic roles in lymphatic metastasis of BCa. Further, we investigated the transcriptional mechanism directed by HSF1 in BCa cells and the efficiency of HSF1 inhibitors in blocking lymphatic metastasis.
2. MATERIALS AND METHODS
2.1. Human tissue samples
All cancer tissue samples included in this study were pathologically diagnosed with bladder cancer between January 2004 and August 2019. Samples without clear pathological, clinical and survival information were excluded. A total of 104 formalin‐fixed, paraffin‐embedded BCa tissues, 15 metastatic lymph node tissues (LN+), and 11 normal adjacent tissues (NAT), termed Cohort 1, were obtained from patients undergoing surgery at the Sun Yat‐sen Memorial Hospital (Guangzhou, Guangdong, China). Meanwhile, 96 BCa tissues and 13 NAT, named Cohort 2, were acquired from the Sun Yat‐sen University Cancer Center (Guangzhou, Guangdong, China). Two pathologists confirmed each sample pathologically and classed them into lymphatic metastasis‐positive BCa tissues (LN+ CA) and lymphatic metastasis‐negative BCa tissues (LN‐ CA). Tissue microarrays containing 60 BCa tissues and 11 NAT, termed Cohort 3, were purchased from Avila Biotechnology (BL601b, Xi'an, Shaanxi, China).
2.2. Immunohistochemistry (IHC) analyses
IHC was conducted as described previously [17]. Briefly, the specimens were dewaxed, rehydrated, and incubated with protease K at 37°C for 15 minutes for antigen retrieval. Then, the specimens were incubated with a solution of 3% H2O2 for 10 minutes at 25°C to block endogenous peroxidase activity. After that, the specimens were incubated with the primary antibodies overnight at 4°C. The antibodies used to evaluate corresponding proteins expression in tissue samples are listed in Supplementary Table S1. After washing for 3 times with PBS, the specimens were incubated with biotinylated secondary antibodies for 1 hour at 25°C, followed by color development through DAB solutions (ZSGB‐BIO, Beijing, China). After washing, the tissues were counterstained with hematoxylin.
IHC analyses were conducted as previously described [18]. Two pathologists blindly quantified the expression of HSF1 in specimens according to a staining scoring system. Briefly, the proportion of positively stained cancer cells was assessed as a percentage. The intensity of immunostaining in each sample was graded as negative = 0, weak = 1, moderate = 2, or strong = 3. The staining score, termed H‐score, was then calculated as the numbers representing intensity multiples by the percentage of cells stained (H‐score = Intensity × percentage of positive cells). The samples were classed with low (score < 150) or high (score ≥ 150) HSF1 expression. The same method was used to assess the expression of LEF1, MMP9, CCL20, E2F2, Ki67, E‐cadherin, N‐cadherin and vimentin. Images were visualized using a Nikon ECLIPSE Ti microscope system (Tokyo, Japan) and processed with Nikon software.
2.3. TCGA, Oncomine and R2 genomics platform data mining
Patients’ clinical profiles in The Cancer Genome Atlas (TCGA) Bladder Cancer (BLCA) cohort are available at https://cancergenome.nih.gov/ [19]. Kaplan‐Meier survival analysis of HSF1, and the correlation between HSF1 and PRMT5 in the TCGA BLCA cohort was obtained from GEPIA (http://gepia.cancer‐pku.cn/index.html) [20]. The clinical profiles of patients in the Lee bladder cohort were available in the Oncomine database (https://www.oncomine.org). Patients' information in the Hoglund cohort are available at the R2 genomics platform (https://r2platform.com).
2.4. Cell cultures
The human uroepithelial cell line SV‐HUC‐1, embryonic kidney cell line HEK‐293T, and bladder cancer cell lines UM‐UC‐3, T24, 5637 and TCCSUP were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). All cells were cultured as previously described [21]. They were tested negative for mycoplasma contamination and had no misidentification or contamination with other cells after short tandem repeat (STR) authentication (IGE biotechnology, Guangzhou, Guangdong, China).
2.5. RNA interference
The small interfering RNA (siRNA) oligonucleotides targeting HSF1, LEF1, PRMT5, WDR5 and negative control siRNA were purchased from GenePharma (Shanghai, China) and are listed in Supplementary Table S2. The siRNA transfections were performed according to the manufacturer's instructions and as previously described [22]. Briefly, 5 μL dissolved siRNA was incubated with 3 μL Lipofectamine RNAiMAX (Invitrogen, Carlsbad, California, USA) in 200 μL OPTI‐MEM (Gibco, Carlsbad, California, USA) at 25°C for 20 minutes. Then, the mixture was added to cells and incubated for 48 hours.
2.6. RNA isolation, qPCR, and Western blotting
RNA isolation, quantitative real‐time polymerase chain reaction (qPCR) and Western blotting were performed as previously described [17]. Briefly, total RNA was extracted using TRIzol Reagent (Takara, Kusatsu, Shiga, Japan). A total of 1 μg RNA was transcribed to complementary DNA (cDNA) using PrimerScript RT‐PCR kit (Takara). The qPCR was conducted using an SYBR Green reaction mix (Vazyme, Nanjing, Jiangsu, China) with a LightCycler 96 System (Roche, Basel, Switzerland). Relative expression was calculated using the 2−ΔΔCt method (Ct, cycle threshold). All specific primers are listed in Supplementary Table S3.
As for Western blotting, the cells were lysed in RIPA lysis buffer (CWBIO, Beijing, China) with protease and phosphatase inhibitors (CWBIO). After identifying quantities of proteins using Pierce BCA Protein Assay Kit (Invitrogen), the proteins were electrophoresed using SDS‐PAGE and transferred to PVDF membranes (Merck, Burlington, Massachusetts, USA), blocked and incubated with primary antibodies at 4°C overnight. Primary antibodies specific to HSF1, LEF1, MMP9, E2F2, E‐cadherin, N‐cadherin, vimentin, PRMT5, FLAG, GAPDH are listed in Supplementary Table S1. After incubation, the membranes were incubated with HRP‐conjugated secondary antibodies at 25°C for 1 hour. Protein bands were visualized using enhanced chemiluminescence.
2.7. Wound healing, migration and invasion assays
BCa cells used for wound healing assays were cultured in six‐well plates until the cell density reached 100%. We then scratched the plates with a sterile p200 pipette tip, and the cells were maintained in a serum‐free medium. Afterward, image collection and migration distance measurements were conducted at 0 and 36 hours for UM‐UC‐3 cells and at 24 hours for T24 cells.
Migration assays were performed according to our previous study [23]. Briefly, 6 × 104 BCa cells were added to the upper chambers with 8 μm pores, whereas lower chambers were filled with conditioned medium. After incubating UM‐UC‐3 cells for 21 hours and T24 cells for 8 hours, the cells on the lower surface were fixed (4% paraformaldehyde), stained (0.1% crystal violet) and counted. For invasion assays, the chambers were coated with 25 μg Matrigel (Corning, Bedford, Massachusetts, USA) before adding the cells, and UM‐UC‐3 cells were incubated for 23 hours and T24 cells for 10 hours. The assay was then performed following similar procedures described above.
2.8. Plasmids and transfection
Two short hairpin RNA (shRNA) sequences specifically targeting HSF1 were cloned into the pLKO.1‐Puro vector. The open reading frame (ORF) of HSF1 (full, synonymous mutant according to the shRNA sequences), LEF1 (full) were cloned into pCDH‐CMV‐MCS‐EF1‐Puro, while HSF1 (truncated as shown in Figure 5E), PRMT5 (full) were cloned into pCDNA3.1 vector. The LEF1, MMP9, CCL20 and E2F2 promoter regions were subcloned in pGL3‐control vector. All vectors were purchased from IGE (Guangzhou, Guangdong, China). Bidirectional sequencing was performed to verify the correct sequences. The sequences of all shRNAs are listed in Supplementary Table S2. The transient transfection, lentivirus production and infection were performed as previously described [24]. For transient transfection, the plasmids were incubated with X‐tremeGENE (Invitrogen) at 25°C for 20 minutes. After incubation, the mixture was added to cells and treated for 24‐48 hours. To package lentivirus, HEK‐293T cells were transfected with psPAX2 (IGE), PMD2.G (IGE) and stably silenced or overexpressed vectors using X‐tremeGENE. After incubation for 48 hours, lentiviruses were harvested, filtered and concentrated. The cells were then infected with viruses using polybrene (IGE) and selected through puromycin.
FIGURE 5.
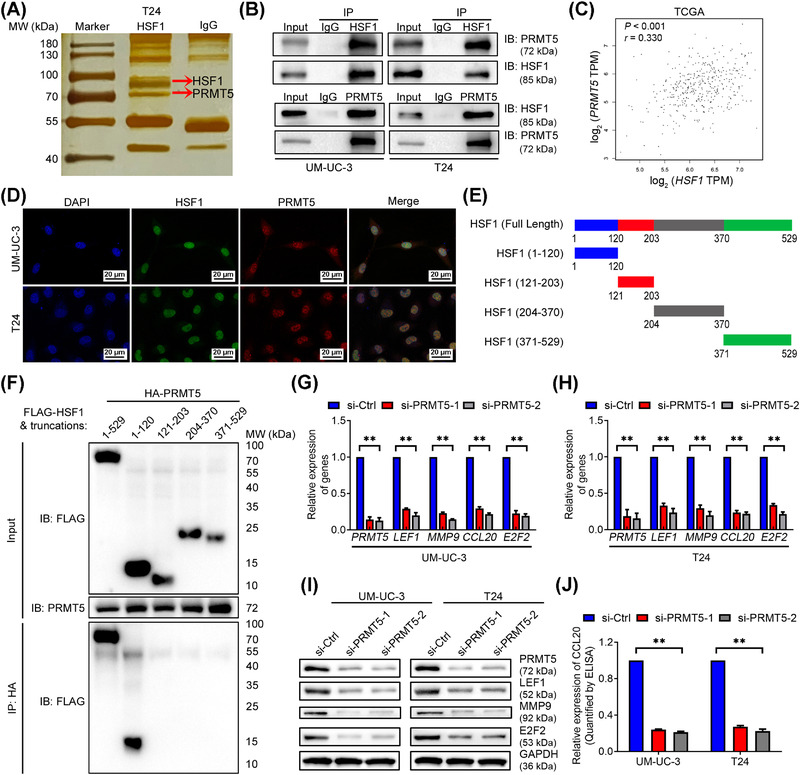
HSF1 regulated target genes expression by interacting with PRMT5. (A) Co‐immunoprecipitation (Co‐IP) was performed in T24 cells using anti‐HSF1 antibody or negative control IgG followed by silver staining. The red arrows show the positions of HSF1 (above) and PRMT5 (below). (B) Co‐IP and Western blotting analysis showing the interaction between endogenous HSF1 and PRMT5. (C) Pearson correlations between the expression of HSF1 and PRMT5 in the TCGA BLCA cohort. (D) Representative immunofluorescence (IF) images of HSF1 and PRMT5 colocalization in the nuclei of bladder cancer cells. Blue, nuclei; green, HSF1; red, PRMT5. Scale bars: black, 20 μm. (E) HSF1‐truncated mutants used in this study. (F) HEK‐293T cells were transfected with HA‐PRMT5 and Flag‐HSF1 or Flag‐HSF1‐truncations as indicated. Cell lysates were immunoprecipitated with anti‐HA antibody, and protein levels were analyzed by Western blotting with the indicated antibodies. (G, H) Quantification of LEF1, MMP9, CCL20, and E2F2 mRNA expression by qPCR after PRMT5 knockdown in (G) UM‐UC‐3 and (H) T24 cells. (I) Western blotting assay of the protein levels of LEF1, MMP9, and E2F2, and (J) ELISA measuring CCL20 secretion in PRMT5‐silenced UM‐UC‐3 and T24 cells. The error bars mean the standard deviations of three experiments independently. * P < 0.05 and ** P < 0.01. Abbreviations: Co‐IP, Co‐immunoprecipitation; TCGA, The Cancer Genome Atlas; BLCA, bladder cancer; qPCR, quantitative real‐time polymerase chain reaction
2.9. Cell proliferation assays
MTT (3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide) assays and colony formation assays were performed to detect cell viability. Meanwhile, as EdU (ethynyl deoxyuridine) could bind to the DNA instead of thymidine during DNA synthesis in the S phase of cell cycle, we used EdU assays to detect cell populations at the S phase. The experiments were performed as described in our previous study [25]. Briefly, for MTT assays, 2 × 103 cells of different groups were seeded in each well of a 96‐well plate. Then, the cells were incubated with 20 μL MTT solution (Sigma‐Aldrich, St Louis, MO, USA) for 3 hours. After that, the MTT solution was discarded, and 150 μL dimethyl sulfoxide was added to each well. The optical density (OD) value was assessed at 490 nm.
Colony formation assays were performed by adding a total of 1000 BCa cells from scrabmle, HSF1‐sh1, HSF1‐sh2 and HSF1‐sh+HSF1 groups or transfected using siRNAs in 6‐well plates. After culture for 10 days, the colonies were fixed, stained and counted.
EdU assays were performed using an EdU Kit (C10310‐1, RIOBIO, Guangzhou, Guangdong, China). Briefly, BCa cells after different treatments mentioned above were plated in 96‐well plates (3000 cells/well). After incubation with EdU for 4 hours, the cells were fixed, stained by Apollo®567, and analyzed using fluorescence microscopy.
2.10. Flow cytometry analysis
The cell cycle and apoptosis analysis were performed using the Cell Cycle Detection Kit (KGA512, KeyGEN, Nanjing, Jiangsu, China) and Annexin V‐FITC/PI Apoptosis Detection Kit (KGA107, KeyGEN) according to the corresponding protocol. Briefly, 1 × 105 cells were collected, fixed and labeled DNA with propidium iodide (PI). After that, the cell cycle was evaluated using flow cytometry (BD Biosciences, San Jose, California, USA). For apoptosis assay, the cells were collected and subsequently stained with FITC‐annexin V and PI. The stained cells were analyzed using flow cytometry (BD Biosciences).
2.11. In vivo popliteal LN metastasis and tumorigenesis assays, and KRIBB11 treatment
In vivo popliteal LN metastasis and tumorigenesis assays were established using male BALB/c nude mice (4‐5 weeks old) purchased from the Experimental Animal Center of Sun Yat‐sen University (Guangzhou, Guangdong, China). All mice were housed in Specific Pathogen Free (SPF) barrier facilities. The popliteal LN metastasis and tumorigenesis assays were performed as previously described [26]. Briefly, we used six mice in each group of control, HSF1 knockdown and HSF1 restoration cells, respectively. The corresponding UM‐UC‐3 cells (3 × 106 cells) with stably expressed firefly luciferase were injected into the footpads of mice to establish the popliteal LN metastasis models. The status of the popliteal lymph node was monitored and imaged using IVIS (in vivo imaging system) Spectrum Imaging System (PerkinElmer, Waltham, Massachusetts, USA) after the injection for 6 weeks. For further study, the primary footpad tumors and popliteal lymph nodes were enucleated and embedded in paraffin.
For the in vivo tumorigenesis assays, a total of 1 × 106 corresponding UM‐UC‐3 cells were subcutaneously injected into the right side of the dorsum. The mice were euthanized by cervical dislocation following the Declaration of Helsinki after 4 weeks, and tumors were surgically dissected. The tumor specimens were fixed and embedded in paraffin for further study.
After the injections of BCa cells for about 1 week, the footpad tumors or subcutaneous tumors were visible. KRIBB11 (Selleck Chemicals, Shanghai, China; 70 mg/kg in 10% dimethylacetamide, 50% PEG300 and 40% distilled water) or vehicle were injected intraperitoneally into different groups of mice. The subsequent procedures were performed as described above.
2.12. Establishment of bladder cancer patient‐derived xenografts (PDX)
The male, 4‐5 weeks old, NOD‐SCID mice (Beijing Vital River Laboratory Animal Technology, Beijing, China) and BALB/c nude mice (Experimental Animal Center of Sun Yat‐sen University) used in establishing the PDX models were housed in SPF barrier facilities. Two tumor samples obtained from patients diagnosed with high‐grade, lymphatic metastasis‐positive BCa between January 2019 to August 2019 were acquired from the Sun Yat‐sen Memorial Hospital. Once the surgical resection of BCa from patients was finished, the tumor sample was stored in sterile DMEM on ice. Within 6 hours after resection, the tumor sample was cut into approximately 3‐5 mm diameter and implanted subcutaneously into the right side of the dorsum of NOD‐SCID mice to generate F1 tumors. After 4 weeks, when the F1 tumors had reached approximately 400 mm3 in size, the mice were euthanized by cervical dislocation, and xenografts were surgically dissected and subsequently implanted into BALB/c nude mice through the same procedure. When the tumors were visible and reached a diameter of about 2‐3 mm, the mice were randomized into two groups of 6 mice each. The mice in each group had similar average xenograft tumor volumes and were assigned to receive treatment with KRIBB11 or vehicle, as described above.
2.13. RNA sequencing analysis
The cells were transfected with HSF1 siRNA (si‐HSF1‐1 and ‐2) or control siRNA (si‐Ctrl) for 48 hours. Then, total RNA was extracted from the cells using Trizol (Invitrogen). Library construction and sequencing were performed by Annoroad Gene Technology (Beijing, China). The libraries were sequenced on an Illumina NovaSeq 6000 platform, and 100 bp paired‐end reads were generated. All primary data in RNA sequencing (RNA‐seq) analysis had been uploaded to the Gene Expression Omnibus (GEO) at GSE185986 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE185986).
2.14. ELISA‐based quantification of secreted CCL20
The cell culture supernatant was collected, and the secreted CCL20 was quantified using Human MIP3a ELISA Kit (ab178015, Abcam, Cambridge, UK) according to the manufacturer's instructions. Briefly, the supernate from BCa cells was collected, diluted at 1:2, and added into the wells coated with CCL20 antibody. After incubation in a 37°C incubator for 30 minutes, the absorbance of each well at 450 nm was measured. We then calculated each well's content of CCL20 according to the standard curve.
2.15. Immunofluorescence (IF) staining
IF staining was carried out according to the description of our previous study [27]. Briefly, BCa cells were planted in confocal dishes, fixed with 4% paraformaldehyde and pre‐hybridized with 0.5% Triton X‐100. After that, the cells were blocked and incubated with primary antibodies at 4°C overnight. The primary antibodies used in this study for anti‐HSF1, anti‐PRMT5 and anti‐N‐cadherin are listed in Supplementary Table S1. After incubation, the dishes were washed with PBS for 3 times and incubated with secondary antibodies at 25°C for 1 hour. Then, the cells were incubated with DAPI (Solarbio, Beijing, China) for 5 minutes at 25°C for nuclear counterstaining. The images were captured using the confocal microscope (Zeiss, Munich, Germany).
2.16. Isolation and recruitment assay of monocytes
Human fresh monocytes were isolated from buffy coats prepared from healthy volunteer donors as previously described [28]. In brief, human fresh peripheral blood was collected in an anticoagulant tube and diluted with PBS at 1:1. Then, the sample was added to the Ficoll separation medium (Merck) and centrifugated at 750 g for 25 minutes. After centrifugation, we collected the cells from the interface layer. Monocytes were further enriched through CD11b microbeads (Miltenyi, Bergisch Gladbach, Germany).
After isolation, monocytes (5 × 105) were added to the upper transwell chambers consisting of 8 μm membrane filter inserts, while lower chambers were filled with fresh DMEM with 0 / 5 / 10 ng/mL of recombinant human CCL20 (rhCCL20), or conditioned medium collected from the BCa cells in scramble, HSF1‐sh1, HSF1‐sh2, HSF1‐sh+HSF1, and HSF1‐sh+HSF1+ CCL20 antibody (Ab) groups. After 12 hours of incubation, the cells in the upper chamber were carefully removed, and the cells that had migrated through the membrane to the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The migrated cells were counted as the migration assay previously described.
2.17. Co‐immunoprecipitation (Co‐IP) and mass spectrometry (MS) analysis
Co‐IP was performed according to our previous work [29]. The interaction between endogenous HSF1 and PRMT5 was investigated in wild‐type UM‐UC‐3 and T24 cells. Briefly, cellular nuclear extracts were incubated with anti‐HSF1, anti‐PRMT5, anti‐HA or control IgG (Supplementary Table S1) at 4°C overnight and then treated with A/G magnet beads for 2 hours at 25°C. Immunoreactive proteins in the lysates were detected by Western blotting. MS analysis was performed by the Bioinformatics and Omics Center of Sun Yat‐Sen Memorial Hospital.
2.18. Chromatin immunoprecipitation (ChIP)
ChIP was conducted according to previously described methods [22]. Briefly, transfected cells were treated with 1% formaldehyde for 10 minutes and lysed with SDS lysis buffer. Following ultrasonication, equal aliquots of chromatin supernatants were immunoprecipitated with anti‐HSF1, anti‐PRMT5, anti‐WDR5, anti‐monomethylation of histone H3 at arginine 2 (H3R2me1), anti‐symmetric dimethylation of histone H3 at arginine 2 (H3R2me2s), anti‐symmetric dimethylation of histone H3 at arginine 8 (H3R8me2s), anti‐symmetric dimethylation of histone H4 at arginine 3 (H4R3me2s), anti‐trimethylation of histone H3 at lysine 4 (H3K4me3), or anti‐RNA polymerase II antibodies or with negative control, IgG (Supplementary Table S1) at 4°C overnight. Then, Protein A/G bead‐antibody‐chromatin complexes were washed with low‐salt, high‐salt, LiCl buffer and elution buffer to harvest the chromatin fragments. The DNA‐protein complexes were reversely cross‐linked, and the DNA was purified through spin columns. The enrichment of purified DNA was examined using qPCR. The primers used in ChIP‐qPCR are listed in Supplementary Table S3.
2.19. Statistical analysis
Data was presented as the mean ± standard deviation (SD) of the values obtained in at least three independent experiments. Two‐tailed Student's t‐tests and one‐way analysis of variance (ANOVA), followed by Dunnett's tests for multiple comparisons, were used to evaluate the data. Clinical variables were assessed using Pearson's chi‐square test, and the correlations between two variables were calculated using Spearman's correlation analysis. Overall survival time was calculated from the date of surgical resection to the date of death or follow‐up, while the disease‐free survival time was calculated from the date of surgical resection to the date of death, recurrence, metastasis or follow‐up. Cumulative survival time was determined using the Kaplan‐Meier method and analyzed by the log‐rank test. Multivariate Cox proportional hazards model was used to estimate the adjusted hazard ratios and 95% confidence intervals (CIs) and to identify independent prognostic factors. All statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS) software (version 20.0; IBM, Armonk, New York, USA). Differences were considered statistically significant at P < 0.05.
3. RESULTS
3.1. HSF1 was positively associated with lymphatic metastasis and poor prognosis in BCa
To identify the key HSFs and HSPs that were dysregulated in lymphatic metastasis of BCa, we first analyzed the expression of all HSFs and HSPs in the TCGA‐BLCA cohort and found that HSF1 and 15 other HSPs were overexpressed in BCa (Supplementary Figure S1A). Next, we examined the expression of HSF1 and these 15 upregulated HSPs in 9 paired tissues by qPCR, including NAT, lymphatic metastasis‐positive BCa tissues (LN+ CA), and metastatic lymph nodes tissues (LN+). Notably, HSF1 was the most significant gene upregulated in LN+ CA and LN+, compared with NAT (Figure 1A‐B). Subsequently, we investigated the protein levels of HSF1 in two independent cohorts from Sun Yat‐sen Memorial Hospital and Sun Yat‐sen University Cancer Center by immunohistochemistry (IHC). We found that the expression of HSF1 was lowest in NAT and gradually increased from non‐metastatic tumors, LN‐metastatic tumors, and metastatic lymph nodes in cohort 1 (Figure 1C‐D and Supplementary Figure S1B‐C). Consistent results were also observed in cohort 2 and tissue microarray (termed cohort 3) (Figure 1E and Supplementary Figure S1D). Analysis of the RNA sequencing statistic in Lee BCa cohort obtained from the Oncomine database showed that HSF1 expression was higher in LN‐positive BCa tissues than in LN‐negative tissues (Figure 1F). Meanwhile, we also found that HSF1 was upregulated in muscle‐invasive BCa (MIBC) compared with non‐muscle‐invasive BCa (NMIBC) in cohort 1‐3 (Figure 1G‐H and Supplementary Figure S1E), as well as in high‐grade (HG) BCa compared with lower‐grade (LG) BCa (Supplementary Figure S1F‐G). Kaplan‐Meier survival analysis of cohort 1, 2 and TCGA BLCA cohorts revealed that patients with high HSF1 expression had shorter overall survival (OS) and disease‐free survival (DFS) (Figure 1I‐N). Moreover, univariate and multivariate Cox regression analyses confirmed that high HSF1 expression in BCa tissues was an independent prognostic factor for shorter OS and DFS (Supplementary Tables S4‐7). Taken together, these data indicated that HSF1 expression was not only associated with lymphatic metastasis status, tumor stage and advanced grade but also an independent indicator of poor prognosis in BCa.
FIGURE 1.
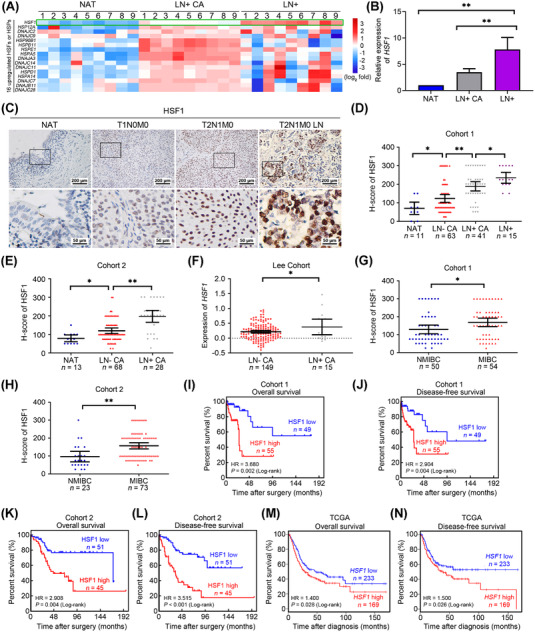
HSF1 was associated positively with lymphatic metastasis and poor prognosis in bladder cancer (BCa). (A) An unsupervised hierarchical clustering of the overexpressed heat shock transcription factors (HSFs) and heat shock proteins (HSPs) expressed in normal adjacent tissues (NAT), lymphatic metastasis‐positive BCa tissues (LN+ CA), and metastatic lymph node tissues (LN+) detected by qPCR. The fold change was calculated by regarding the median of the relative expression of each gene in LN+ CA was 1. The pseudo‐color represents the intensity scale generated by a log2 transformation. (B) Histogram analysis of the HSF1 mRNA in the evaluated tissues detected by qPCR. The error bars represent standard deviations of the relative expression values of HSF1 in each group. (C) Representative immunohistochemistry (IHC) images of HSF1 expression in paraffin‐embedded NAT and BCa tissues. (D, E) Protein levels of HSF1 in (D) NAT, lymphatic metastasis‐negative BCa (LN‐ CA), LN+ CA, and LN+ of cohort 1 and (E) NAT, LN‐ CA, and LN+ CA of cohort 2 assessed by IHC. Statistical significance was assessed through one‐way analysis of variance (ANOVA). (F) HSF1 mRNA expression in LN‐ CA and LN+ CA of the Lee cohort from the Oncomine database. (G, H) Comparison of HSF1 expression in non‐muscle‐invasive BCa (NMIBC) and muscle‐invasive BCa (MIBC) tissues in (G) cohort 1 and (H) cohort 2. Statistical significance was evaluated using two‐tailed t tests in (F‐H). The error bars mean standard deviations of the expression values of HSF1 in each group. (I‐N) Kaplan‐Meier curves for (I) overall survival (OS) and (J) disease‐free survival (DFS) of BCa patients with high (H‐score ≥ 150) vs. low expression of HSF1 (H‐score < 150) in cohort 1 (OS, HR = 3.680, 95% CI = 1.730 ‐ 7.827; DFS, HR = 2.904, 95% CI = 1.442 ‐ 5.846); (K, L) cohort 2 (OS, HR = 2.908, 95% CI = 1.496 ‐ 5.654; DFS, HR = 3.515, 95% CI = 1.916 ‐ 6.447). The (M) OS and (N) DFS from the TCGA BLCA cohort were obtained from GEPIA. *P < 0.05 and **P < 0.01. Abbreviations: BCa, bladder cancer; HSF, heat shock transcription factor; HSP, heat shock protein; NAT, normal adjacent tissue; LN+ CA, lymphatic metastasis‐positive BCa; LN– CA, lymphatic metastasis‐negative BCa; LN+, metastatic lymph node (LN+); NMIBC, non‐muscle‐invasive BCa; MIBC, muscle‐invasive BCa; IHC, immunohistochemistry; TCGA, The Cancer Genome Atlas; BLCA, bladder cancer; qPCR, quantitative real‐time polymerase chain reaction
3.2. HSF1 promoted BCa cell migration and invasion in vitro and lymphatic metastasis in vivo
To investigate the roles of HSF1 in lymphatic metastasis of BCa, we first detected the expression of HSF1 in different BCa cells. To silence or regain the expression of HSF1 with higher efficiencies, as well as providing reliable conclusion through validating the function of HSF1 in two cell lines, we chose the two cell lines with moderate HSF1 expression, UM‐UC‐3 and T24, to perform further functional assays (Supplementary Figure S2A‐B) and transfected two independent siRNAs to knockdown HSF1 in BCa cells (Supplementary Figure S2C‐D). We found the migration speed and number of migrated cells were significantly inhibited in HSF1‐silenced BCa cells by wound healing and transwell assays (Supplementary Figure S2E‐I). Meanwhile, as shown in Supplementary Figure S2J‐K, BCa cells' invasion was markedly decreased after silencing HSF1. To further confirm the role of HSF1 in BCa cells, we performed functional assays in these four groups of BCa cells: scramble, two HSF1‐knockdown cells using shRNAs, and HSF1‐restoration cells using an shRNA‐resistant synonymous mutant of HSF1 after shRNA transfection (Figure 2A). Consistently, the in vitro migration experiments revealed that HSF1 knockdown markedly inhibited the migration speed and number of migrated BCa cells, while HSF1 restoration greatly reversed these effects (Figure 2B‐F). Furthermore, the invasive capability of BCa was significantly suppressed by HSF1 knockdown but mostly rescued by HSF1 restoration (Figure 2G‐H).
FIGURE 2.

HSF1 promoted bladder cancer (BCa) cells migration and invasion in vitro and lymphatic metastasis in vivo. (A) Western blotting analysis of HSF1 expression levels in scramble, HSF1‐sh1, HSF1‐sh2 and HSF1‐sh+HSF1 BCa cells. The HSF1‐sh+HSF1 BCa cells were generated by transfecting an shRNA‐resistant synonymous mutant of HSF1 in HSF1‐sh1 BCa cells. (B‐D) Representative images of wound healing assays in (B) UM‐UC‐3 and (C) T24 cells showing cell migration after knockdown or re‐expression of HSF1; (D) a histogram showing cell migration distance. (E) Representative images of migration assays of UM‐UC‐3 and T24 cells showing cell migratory capacity after knockdown or re‐expression of HSF1, and (F) histogram analysis of the number of migratory cells. (G) Representative images of UM‐UC‐3 and T24 cells showing cell invasion after silencing or restoration of HSF1, and (H) histogram analysis of the numbers of invasive cells. (I) Representative bioluminescence images and, (J) histogram analysis of popliteal metastatic lymph nodes (LNs) in the popliteal LN metastasis model generated by different groups of cells (n = 6 per group). (K) Representative image of the popliteal LN metastasis model. (L) Representative image of dissected popliteal LNs and (M) histogram analysis of LNs volumes (n = 6 per group). (N) Representative images obtained by hematoxylin‐eosin (H.E.) staining and immunohistochemistry (IHC) confirming the LN status of the experimental animals (n = 6 per group). (O) LN status percentages in all groups (n = 6 per group). (P) Kaplan‐Meier survival analysis of mice that were inoculated with control, HSF1‐silenced and HSF1‐restored UM‐UC‐3 cells. The error bars stand for the standard deviations of three independent experiments. * P < 0.05 and ** P < 0.01. Abbreviations: BCa, bladder cancer; LN, lymph node; H.E., hematoxylin‐eosin; IHC, immunohistochemistry
Next, we established popliteal LN metastasis models with stable HSF1‐knockdown and HSF1‐restored UM‐UC‐3 cells expressing firefly luciferase. Silencing of HSF1 significantly decreased the luminescence of footpads and popliteal LNs, while HSF1 restoration ameliorated this effect (Figure 2I‐J). In addition, the volumes of the popliteal LNs were significantly smaller in the HSF1‐silenced group and significantly enlarged in HSF1‐restored mice compared to the corresponding control mice (Figure 2K‐M). We also used hematoxylin‐eosin (H&E) staining and IHC for luciferase to confirm the presence of metastasis in LNs. As shown in Figure 2N and O, there was a significant decrease in the rate of lymphatic metastasis from 100.0% in the control to 33.3% in the HSF1 knockdown group, and returned to 100% after re‐expression of HSF1. Moreover, the survival of mice harboring HSF1‐knockdown tumors was much longer than those in the control or HSF1 restoration group (Figure 2P). Collectively, these findings indicated that HSF1 facilitated the migration and invasion in vitro, as well as the lymphatic metastasis in vivo of BCa cells.
3.3. HSF1 enhanced BCa cell proliferation in vitro and tumor growth in vivo
Previous studies showed that metastatic tumor cells in LNs served as a major source of lymphangiogenic factors to accelerate lymphangiogenesis [2]. Therefore, the growth of the metastatic tumor cells in LN was another key factor in promoting lymphatic metastasis [30]. Interestingly, the metastatic UM‐UC‐3 cells in the LNs enucleated from the popliteal LN metastasis models derived by the HSF1‐knockdown cells exhibited lower expression of the proliferation marker Ki67 than either control or HSF1‐restored group (Supplementary Figure S3), suggesting that HSF1 may participate in BCa cells proliferation. Therefore, we conducted MTT and colony formation assays in stable HSF1‐knockdown and HSF1‐restored BCa cells. As shown in Figure 3A‐C, silencing of HSF1 significantly reduced the proliferation of BCa cells, whereas restoration of HSF1 alleviated this effect. Notably, no difference in cells viability was observed after seeding for 48 hours, indicating that the differences in migratory and invasive abilities observed in migration and invasion assays of BCa cells were caused by the intrinsic metastatic behaviors of cancer cells. Furthermore, flow cytometry assays showed that the cells were arrested in G0/G1 phase after silencing HSF1 and were reversed in HSF1‐restored cells (Figure 3D‐E and Supplementary Figure S4A‐B). The results of EdU assays confirmed that HSF1 directly regulated the proportion of cells in the S phase (Figure 3F‐G and Supplementary Figure S4C‐D). However, no difference in the apoptosis rate between HSF1‐knockdown cells and the control group was observed (Supplementary Figure S4E‐F). Therefore, HSF1 might promote BCa cells proliferation by regulating the expression of genes involved in cell cycle transition instead of cell apoptosis.
FIGURE 3.
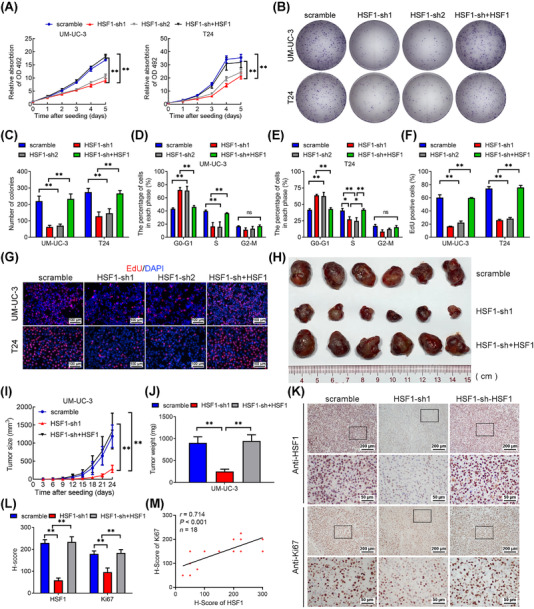
HSF1 enhanced bladder cancer (BCa) cells proliferation in vitro and tumor growth in vivo. (A) Cell viability was evaluated in scramble, HSF1‐sh1, HSF1‐sh2 and HSF1‐sh+HSF1 UM‐UC‐3 and T24 cells by MTT assays. (B) Colony formation assays were performed in HSF1 knockdown and HSF1 re‐expressing UM‐UC‐3 and T24 cells, and (C) a histogram analysis of the numbers of colonies. (D, E) Flow cytometry analysis of (D) UM‐UC‐3 and (E) T24 cells in which HSF1 had been stably silenced or restored compared with control cells. The percentages (%) of the cell population at different stages of the cell cycle are illustrated in the panels. (F, G) Measurement of the cell population in S phase by ethynyl deoxyuridine (EdU) assays and a histogram analysis of the numbers of EdU‐positive cells. Blue, nuclei; red, S‐phase cells. Scale bars: black, 100 μm. The error bars mean the standard deviations of three independent experiments. (H) Representative image of the subcutaneous tumors in the control, HSF1 knockdown, and HSF1 restoration groups. (I) The growth of the tumors in the control, HSF1 knockdown, and HSF1 restoration groups were measured every 3 days, and tumor growth curves were calculated. The mean ± standard deviation (SD) of the tumor volumes measured in 6 mice is shown. (J) Histogram showing the tumor weights in the control, HSF1 knockdown, and HSF1 restoration groups after surgical dissection. (K) Immunohistochemical staining showing HSF1 and Ki67 expression in tumors. (L) Histogram showing the H‐scores in the control, HSF1 knockdown and HSF1 re‐expression groups. The error bars represent the standard deviations of values in each group (n = 6). (M) Pearson correlation analysis between HSF1 and Ki67 expression. * P < 0.05 and ** P < 0.01. Abbreviations: BCa, bladder cancer; MTT, 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide; EdU, ethynyl deoxyuridine
Next, we implanted stable HSF1‐silenced and HSF1‐restored UM‐UC‐3 cells subcutaneously into nude mice. Knockdown of HSF1 significantly slowed tumor growth compared with the control, while HSF1 restoration markedly accelerated tumor growth. The sizes and weights of the tumors in the HSF1‐silenced group were smaller than those in the control group and the HSF1‐restored group (Figure 3H‐J). Further, the tumors that originated from the HSF1 knockdown cells exhibited the lowest expression of Ki67, but HSF1 re‐expression restored Ki67 expression (Figure 3K‐M). Collectively, these results revealed that HSF1 fostered BCa cells proliferation in vitro and tumor growth in vivo.
3.4. Identification of HSF1 target genes in BCa cells
To further identify the underlying mechanisms of HSF1 in lymphatic metastasis, we performed genome‐wide RNA expression profile screening in HSF1‐knockdown and control UM‐UC‐3 and T24 cells (Figure 4A). Considering that HSF1 was associated with specific transcriptome during malignancy progression [11], we mainly focused on the downregulated protein‐coding genes with fold change smaller than 0.6 after silencing HSF1 in both BCa cell lines. We found that the expression of 60 genes were significantly decreased (Figure 4B), and we further verified the 14 cancer‐related genes in control and HSF1‐silenced UM‐UC‐3 and T24 cells via qPCR. As shown in Supplementary Figure S5A‐B, lymphoid enhancer‐binding factor 1 (LEF1), matrix metallopeptidase 9 (MMP9), C‐C motif chemokine ligand 20 (CCL20) and E2F transcription factor 2 (E2F2) were four of the most significantly altered genes. Additionally, previous studies reported that LEF1, MMP9 and CCL20 were involved in cancer metastasis [38, 55, 56], while E2F2 was necessary for cell cycle progression [31]. We also found that patients with high LEF1, MMP9, CCL20 and E2F2 expression showed poor prognosis in the Hoglund BCa cohort (Supplementary Figure S5C). Hence, these four genes were chosen for further study. We confirmed the secreting level of CCL20 through ELISA and the protein levels of LEF1, MMP9 and E2F2 by Western blotting. These genes were downregulated in HSF1‐silenced BCa cells and their expression were regained after HSF1 restoration (Figure 4C‐D). We then analyzed the expression of HSF1, LEF1, MMP9, CCL20 and E2F2 in tissue microarrays by IHC and found a positive correlation between HSF1 expression and these four target genes (Figure 4E‐I). Similarly, the protein levels of LEF1, MMP9, CCL20 and E2F2 were markedly decreased in the HSF1‐silenced xenograft tumors and re‐increased in the HSF1‐restored group (Supplementary Figure S5D‐I).
FIGURE 4.
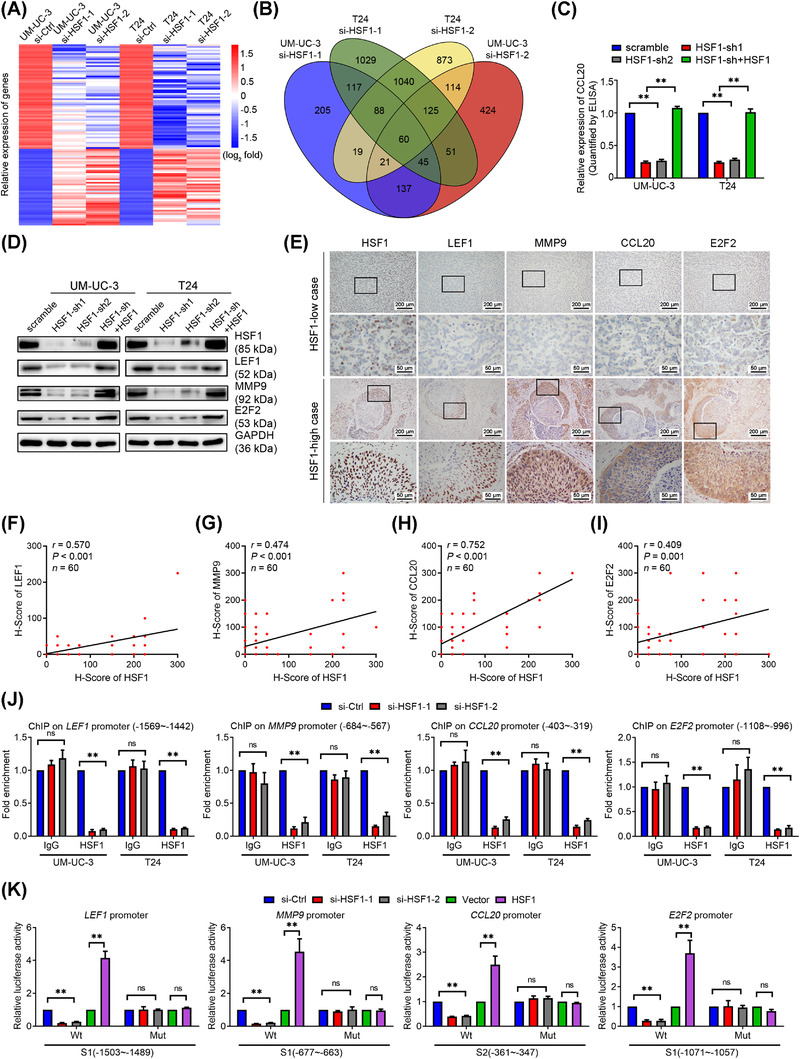
Identification of the target genes of HSF1 in bladder cancer (BCa) cells. (A) Heatmap showing mRNA levels in UM‐UC‐3 and T24 cells transfected for 48 hours with control or small interfering RNAs (siRNAs) targeting HSF1. Red and blue represent the log2 fold change of upregulation and downregulation of the genes expression, respectively. (B) Venn diagram showed the overlapping protein‐coding genes which were downregulated in UM‐UC‐3 and T24 cells after silencing HSF1. (C) The CCL20 secretion level was determined by ELISA, and (D) the protein levels of LEF1, MMP9, and E2F2 were detected by Western blotting in scramble, HSF1‐sh1, HSF1‐sh2 and HSF1‐sh+HSF1 BCa cells. (E) Representative immunohistochemistry (IHC) images of HSF1, LEF1, MMP9, CCL20 and E2F2 in BCa tissues, and Pearson correlations between the expression levels of HSF1 and (F) LEF1, (G) MMP9, (H) CCL20 and (I) E2F2 in 60 BCa tissues in microarrays. (J) Chromatin immunoprecipitation (ChIP) ‐qPCR analysis of negative control IgG and HSF1 enrichment at the promoters of HSF1 target genes in UM‐UC‐3 and T24 cells. (K) Relative luciferase activities associated with the wild‐type (Wt) and site‐mutation (Mut) of the HSF1 binding sequences on target genes promoters in HSF1 knockdown and overexpressing HEK‐293T cells. The error bars represent the standard deviations of three experiments independently. * P < 0.05, ** P < 0.01. Abbreviations: BCa, bladder cancer; siRNAs, small interfering RNAs; ChIP, chromatin immunoprecipitation; qPCR, quantitative real‐time polymerase chain reaction; Wt, wild‐type; Mut, site‐mutation; ns, not statistically significant
To investigate how HSF1 regulates the expression of these genes, we analyzed the promoter regions of the four target genes using the JASPAR program (http://jaspar.genereg.net) to predict potential HSF1 binding sites (Supplementary Figure S6A‐D). Further, ChIP‐qPCR assays indicated that silencing HSF1 decreased the enrichment of HSF1 on one of the target promotors, while negative control IgG showed no differences (Figure 4J and Supplementary Figure S6E). Dual‐luciferase reporter assays revealed that the luciferase activities driven by the binding sites of target genes were significantly decreased in HSF1‐knockdown HEK‐293T cells but increased after HSF1 overexpression. Point mutations of the binding sequences abrogated these effects (Figure 4K). Thus, these results showed that HSF1 directly targeted the promoters to regulate the transcription of LEF1, MMP9, CCL20 and E2F2.
3.5. HSF1 interacted with PRMT5 to promote metastasis and proliferation of BCa cells
Given that HSF1 was a transcription factor that bonded to specific proteins to regulate transcription [32], we performed Co‐IP to identify the binding proteins of HSF1 in BCa cells. Intriguingly, we found an overtly differential band between 70 and 100 kDa after silver staining, and it was identified as PRMT5 by MS (Figure 5A and Supplementary Figure S7A‐B). Further, Co‐IP and Western blotting experiments confirmed the interaction between endogenous HSF1 and PRMT5 in wild‐type UM‐UC‐3 and T24 cells (Figure 5B). Meanwhile, there was a positive correlation between HSF1 and PRMT5 expression in the TCGA BLCA cohort (Figure 5C). Immunofluorescence (IF) assays also showed that HSF1 and PRMT5 colocalized in the nuclei of BCa cells (Figure 5D). To identify the precise domain(s) of HSF1 responsible for its interaction with PRMT5, we generated four HSF1 truncations (Figure 5E) and separately transfected them with PRMT5. The Co‐IP assay showed that PRMT5 bounded specifically to the DNA‐binding domain (DBD) of HSF1 (Figure 5F). These results jointly confirmed that HSF1 interacted with PRMT5 in BCa cells.
Previous studies have demonstrated that PRMT5 promoted the lymphatic metastasis and proliferation of laryngeal carcinoma [33]. Similarly, the migration, invasion and proliferation of BCa cells were markedly attenuated by knockdown of PRMT5 in vitro (Figure S7C‐J). Meanwhile, silencing PRMT5 could also inhibit the expression of LEF1, MMP9, CCL20 and E2F2 (Figure 5G‐J). In conclusion, HSF1 interacted with PRMT5 to enhance the migration, invasion and proliferation of BCa cells in vitro.
3.6. HSF1 regulated LEF1, MMP9, CCL20 and E2F2 expression via PRMT5‐WDR5‐mediated histone modifications
PRMT5 was a key factor in transcriptional activation or repression depending on diverse histone methylation modifications [34]. To determine how PRMT5 regulated LEF1, MMP9, CCL20 and E2F2 expression, we screened several substrates that have been reported to be methylated by PRMT5. ChIP‐qPCR showed increased enrichment of H3R2me1 and H3R2me2s, but not H3R8me2s or H4R3me2s, on the promoters of LEF1, MMP9, CCL20 and E2F2 (Figure 6A‐B and Supplementary Figure S8A‐B). Additionally, a previous study revealed that transcriptional activation mediated by H3R2me1 and H3R2me2s was recognized by WDR5, resulting in the recruitment of the SET1/MLL complex and H3K4me3 [35]. Interestingly, as shown in Figure 6C and D, high enrichment of WDR5 and H3K4me3 on the promoters of these genes were observed. Knockdown of WDR5 also decreased the expression of these four target genes (Figure 6E‐G). Moreover, our ChIP‐qPCR results showed that silencing of HSF1 or PRMT5 significantly decreased the occupancy of the promoters of LEF1, MMP9, CCL20 and E2F2 by H3R2me1, H3R2me2s, WDR5, H3K4me3 and Pol‐II, but no differences by IgG, compared with control cells. Intriguingly, we found that on the promoters of target genes, the knockdown of HSF1 abolished the binding of both HSF1 and PRMT5 (Figure 6H‐K, and Supplementary Figure S8C‐G), while silencing PRMT5 only abrogated the occupancy of PRMT5 but not HSF1 (Figure 6L‐O, and Supplementary Figure S8H‐L). These results indicated that HSF1 recruited PRMT5 to the promoter of target genes to enhance their expression. Taken together, HSF1 upregulated the expression of LEF1, MMP9, CCL20 and E2F2 via PRMT5‐WDR5‐mediated histone modifications.
FIGURE 6.
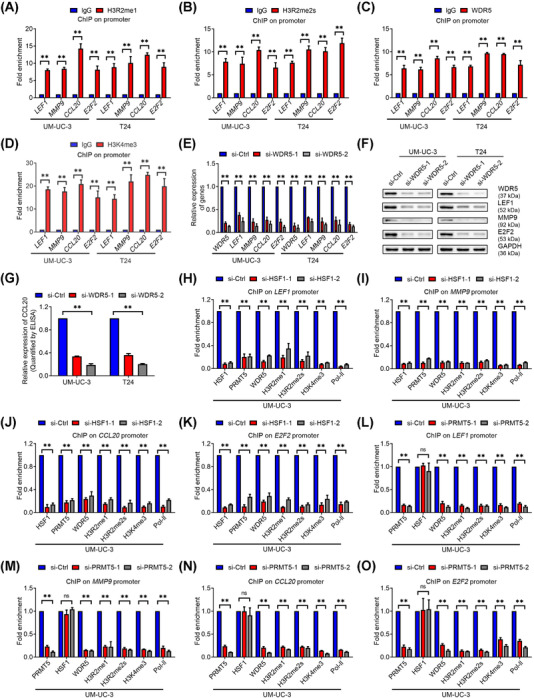
HSF1 regulated target genes expression via PRMT5‐WDR5 axis‐mediated histone methylation. (A‐D) Chromatin immunoprecipitation (ChIP) ‐qPCR analysis of (A) H3R2me1, (B) H3R2me2s, (C) WDR5, and (D) H3K4me3 enrichment on the promoters of LEF1, MMP9, CCL20 and E2F2 in UM‐UC‐3 and T24 cells. (E) Quantification of LEF1, MMP9, CCL20 and E2F2 mRNA expression by qPCR after WDR5 knockdown in UM‐UC‐3 and T24 cells. (F) Western blotting assay of the protein levels of LEF1, MMP9, and E2F2, and (G) ELISA measuring CCL20 levels in WDR5‐silenced UM‐UC‐3 and T24 cells. (H‐K) ChIP‐qPCR analysis of HSF1, PRMT5, WDR5, H3R2me1, H3R2me2s, H3K4me3 and RNA polymerase‐II (Pol‐II) status at the (H) LEF1, (I) MMP9, (J) CCL20 and (K) E2F2 promoters in HSF1‐silenced UM‐UC‐3 cells. (L‐O) ChIP‐qPCR analysis of PRMT5, HSF1, WDR5, H3R2me1, H3R2me2s, H3K4me3 and Pol‐II enrichment at candidate HSF1 target genes promoters in PRMT5‐silenced UM‐UC‐3 cells. The error bars represent the standard deviations of three independent experiments. * P < 0.05 and ** P < 0.01. Abbreviations: ChIP, chromatin immunoprecipitation; qPCR, quantitative real‐time polymerase chain reaction; Pol‐II, RNA polymerase‐II; ns, not statistically significant
3.7. HSF1 facilitated EMT of BCa cells in a LEF1‐dependent manner
LEF1 was shown to be involved in EMT to accelerate cancer metastasis [36]. Silencing of LEF1 significantly inhibited the migratory and invasive ability of BCa cells (Supplementary Figure S9). Then, we overexpressed LEF1 in HSF1‐silenced BCa cells (Supplementary Figure S10A). Intriguingly, the migratory and invasive behaviors inhibited by HSF1 repression were mainly rescued by LEF1 overexpression (Supplementary Figure S10B‐E). Further, overexpressing LEF1 increased both LN volumes and metastatic rate in HSF1‐knockdown cells in vivo (Figure 7A‐C and Supplementary Figure S10F‐I).
FIGURE 7.
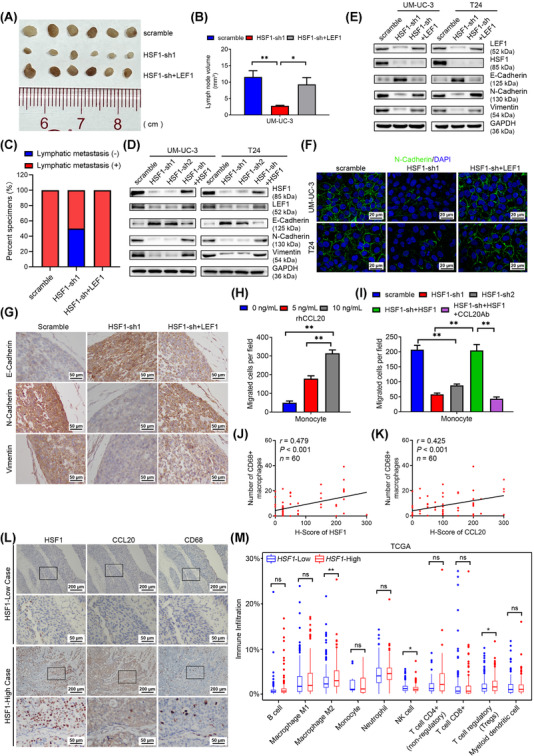
HSF1 facilitated epithelial‐mesenchymal transition (EMT) in bladder cancer (BCa) cells depending on LEF1 and increased macrophages infiltration via CCL20. (A) Representative image and (B) histogram analysis of the dissected popliteal lymph nodes (LNs) volumes from scramble, HSF1‐sh1 and HSF1‐sh+LEF1 groups (n = 6 per group). Statistical significance was calculated by one‐way analysis of variance (ANOVA). The error bars mean standard deviations of values in each group. (C) The percentage of LN status in all groups (n = 6 per group). (D) Western blotting analysis of the protein expression of LEF1 and EMT markers in scramble, HSF1‐sh1, HSF1‐sh2 and HSF1‐sh+HSF1 BCa cells. (E) Western blotting analysis of the protein expression of LEF1, HSF1 and EMT markers in scramble, HSF1‐sh1 and HSF1‐sh+LEF1 BCa cells. (F) Representative immunofluorescence (IF) images showing N‐cadherin expression in scramble, HSF1‐sh1 and HSF1‐sh+LEF1 BCa cells. Blue, nuclei; green, N‐cadherin. Scale bars: black, 20 μm. (G) Representative immunohistochemistry (IHC) images showing E‐cadherin, N‐cadherin and vimentin expression in footpad tumors of the indicated groups. Scale bars: black, 50 μm. (H) Histogram showing the numbers of migratory monocytes treated with various concentrations of recombinant human CCL20 (rhCCL20), and (I) histogram showing the numbers of migratory monocytes after treatment with conditioned medium (CM) from UM‐UC‐3 cells with indicated treatment. The error bars stand for the standard deviations of three independent experiments. (J, K) Pearson correlation analysis of the relationship between HSF1, CCL20 expression and macrophage infiltration determined by anti‐CD68 from 60 BCa tissues in microarrays. (L) Representative IHC images of HSF1, CCL20 and CD68 staining in BCa tissues in microarrays. (M) Demonstration of different immune cells of BCa tissues with high vs. low HSF1 expression by QUANTISEQ in the TCGA BLCA cohort (Wilcox text). * P < 0.05, ** P < 0.01. Abbreviations: EMT, epithelial‐mesenchymal transition; BCa, bladder cancer; LN, lymph node; rhCCL20, recombinant human CCL20; CM, conditioned medium; TCGA, The Cancer Genome Atlas; BLCA, bladder cancer; ns, not statistically significant
To further clarify whether LEF1 participates in EMT, we measured the expression of E‐cadherin, N‐cadherin and vimentin in LEF1‐silenced BCa cells. Interestingly, the expression of E‐cadherin was elevated, while N‐cadherin and vimentin were decreased after silencing of LEF1, suggesting inhibition of EMT (Supplementary Figure S11A). Additionally, as shown in Figure 7D and Supplementary Figure S11B, EMT was aborted after downregulation of HSF1 but was regained in HSF1‐overexpressing cells. We also performed IHC assays of footpad tumor tissues and found a negative correlation between HSF1 and E‐cadherin levels, as well as a positive correlation between HSF1 and N‐cadherin, vimentin levels, respectively (Supplementary Figure S11C‐F). Meanwhile, inhibition of the EMT by HSF1 silencing was greatly recovered after overexpression of LEF1 (Figure 7E, F, and Supplementary Figure S11G, H). We also found that the increase in E‐cadherin, together with the attenuation of N‐cadherin and vimentin levels, in the footpad tumors from the HSF1‐silenced group was reversed in tumors from the LEF1‐overexpressing group (Figure 7G). These data indicated that HSF1 facilitated migration and invasion of BCa cells via LEF1‐dependent EMT.
3.8. HSF1 increased macrophage infiltration via CCL20
A previous study showed that CCL20 promoted ovarian cancer cell migration [37]. However, after knockdown of CCL20 in BCa cells, we found no difference in migration between CCL20‐silenced and control cells in vitro (Supplementary Figure S12A‐D), indicating that CCL20 might influence lymphatic metastasis independent of autocrine signaling. Considering that CCL20 was reported to promote metastasis by recruiting tumor‐associated macrophages (TAMs) [38], we used various concentrations of recombinant human CCL20 (rhCCL20) to recruit fresh human peripheral monocytes. Interestingly, rhCCL20 treatment increased the number of migrated monocytes in a dose‐dependent manner (Figure 7H and Supplementary Figure S12E). Next, we collected conditioned medium (CM) from HSF1‐silenced and HSF1‐restored BCa cells and used it to recruit monocytes. The results showed that the migratory ability of monocytes exposed to HSF1‐silenced CM was weaker than that of monocytes exposed to control CM, while HSF1‐restored CM significantly reversed this effect. The effect of HSF1 re‐expressing CM on monocyte recruitment was largely attenuated by adding a neutralizing anti‐CCL20 antibody to about 20%. These results indicated that CCL20 contributed to HSF1‐induced monocyte recruitment (Figure 7I and Supplementary Figure S12F). Combined with the previous conclusion that CCL20 increases macrophages infiltration in vivo [39], we hypothesized that tumoral secretion of CCL20 mediated by HSF1 might recruit TAMs in BCa. Consequently, as shown in Figure 7J‐L, the number of TAMs in the BCa tissues was positively correlated with HSF1 and CCL20 expression. Furthermore, we analyzed the immune cell infiltration data of the TCGA BLCA cohort and found that the M2 macrophages infiltration rate was significantly higher in groups with HSF1 high‐expressing BCa (Figure 7M). The expression of C‐C motif chemokine receptor 6 (CCR6, specific receptor of CCL20) was positively correlated with the expression of the M2 macrophages markers CD163 molecule (CD163), CD204 molecule (CD204), CD206 molecule (CD206), and the secreted factors interleukin‐6 (IL‐6), interleukin‐10 (IL‐10) and transforming growth factor beta 1 (TGF‐β) in the TCGA BLCA cohort (Supplementary Figure S12G‐L), indicating that the TAMs recruited by BCa cell‐derived CCL20 might be M2 macrophages. Taken together, our results suggested that HSF1 accelerated the infiltration of TAMs, especially M2 macrophages, in a CCL20‐dependent manner.
3.9. Pharmacological inhibition of HSF1 blocked lymphatic metastasis of BCa cells with no significant toxicity
To determine the value of HSF1 in translational applications, we investigated the role of KRIBB11, a pharmacological inhibitor of HSF1, in lymphatic metastasis. As shown in Figure 8A, BCa cell viability was suppressed in a dose‐dependent manner by KRIBB11 treatment. The half‐maximal inhibitory concentration (IC50) was approximately 12 μmol/L, which was similar to that in myeloma [16]. The expression of HSF1, LEF1, MMP9, CCL20 and E2F2, and EMT were significantly inhibited after treatment with KRIBB11 (Figure 8B‐C). Moreover, Co‐IP assays revealed that the binding between HSF1 and PRMT5 was abolished by KRIBB11 (Supplementary Figure S13A). We also performed ChIP‐qPCR and found that KRIBB11 reduced the recruitment of HSF1 on the promoter of target genes, in line with PRMT5, H3R2me1, H3R2me2, WDR5 and H3K4me3 in BCa cells (Supplementary Figure S13B‐I).
FIGURE 8.
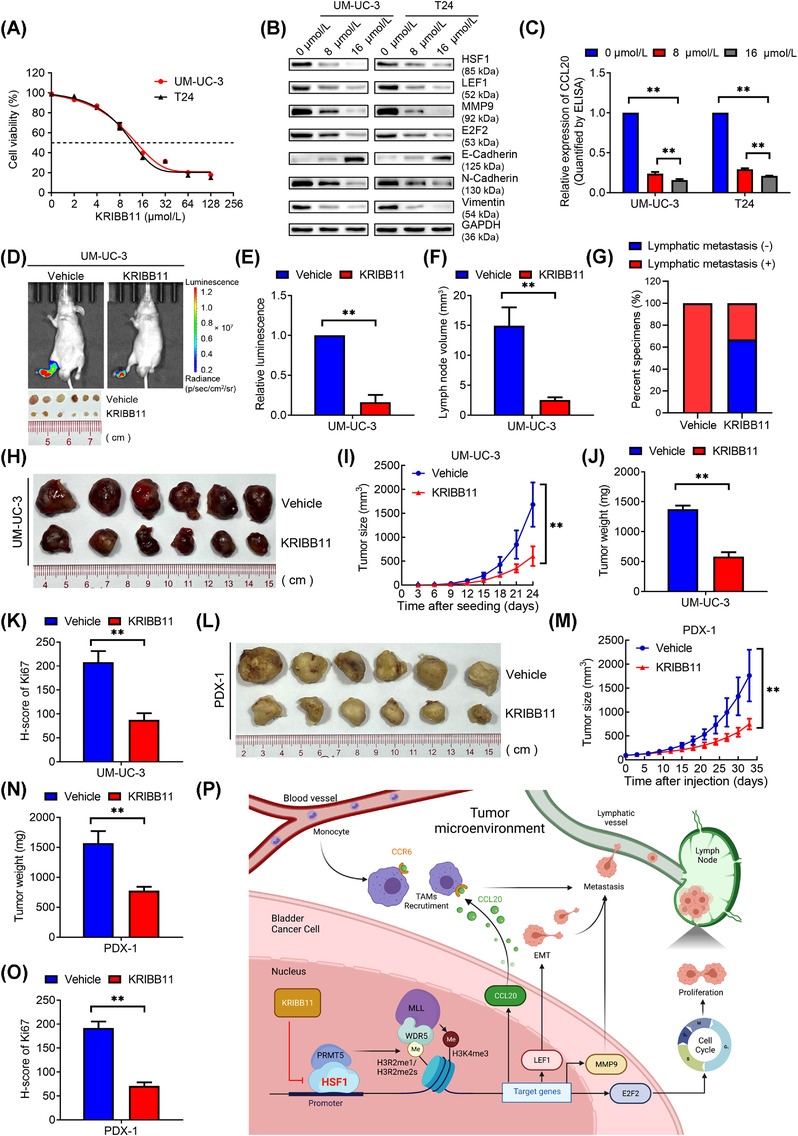
Pharmacological inhibition of HSF1 blocked lymphatic metastasis and tumor growth in bladder cancer (BCa). (A) Viability of UM‐UC‐3 and T24 cells after KRIBB11 treatment. The cells were treated with the indicated concentrations of KRIBB11 for 48 hours, and viability was measured using the MTT assay. (B) Western blotting analysis of the protein levels of HSF1, LEF1, MMP9, E2F2, and EMT markers in BCa cells treated with the indicated concentrations of KRIBB11 for 48 hours. (C) ELISA of CCL20 expression in BCa cells treated with the indicated concentrations of KRIBB11 for 48 hours. The error bars mean the standard deviations of three independent experiments. (D) Representative images of bioluminescence and dissected popliteal lymph nodes (LNs) and (E, F) histogram analysis of the bioluminescence and LN volumes (n = 6 per group). (G) Percentages of LN status in the indicated groups (n = 6 per group). (H) Representative image of the subcutaneous tumors of vehicle and KRIBB11 groups. (I) Tumor growth curves and (J, K) histogram analyses of tumor weights and Ki67 H‐scores in the two indicated groups (n = 6 per group). (L) Representative image of the subcutaneous tumors in BCa patient‐derived xenograft (PDX) models after treatment with vehicle or KRIBB11. (M) Tumor growth curves and (N, O) histogram analysis of tumor weights and Ki67 H‐scores in PDX models after the indicated treatments. The error bars mean the standard deviations of values in each group (n = 6). (P) Illustrative model showing the underlying mechanism by which HSF1 promoted lymphatic metastasis and proliferation in BCa via a PRMT5‐WDR5‐dependent transcriptional program. The image was created using BioRender.com. * P < 0.05, ** P < 0.01. Abbreviations: BCa, bladder cancer; LN, lymph node; H.E., hematoxylin‐eosin; MTT, 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide; PDX, patient‐derived xenograft
Next, we performed metastasis assays and found that KRIBB11 blocked the wound healing, migration and invasion of BCa cells in vitro (Supplementary Figure S14A‐G). We further generated popliteal LN metastasis models and intraperitoneally injected KRIBB11 every 2 days (Supplementary Figure S14H). Consistent with the in vitro data, the relative luminescence measured by IVIS and the volume of popliteal LNs were significantly smaller in the KRIBB11‐treated group than in the control group (Figure 8D‐F). The lymphatic metastatic rate decreased from 100.0% in the vehicle group to 33.3% in the KIRBB11 group (Figure 8G and Supplementary Figure S14I). Meanwhile, the expression of Ki67 in LNs from the KRIBB11 group was significantly lower than that in the vehicle group (Supplementary Figure S14J‐K). Notably, there was no significant difference between these two groups in body weight, liver or kidney functions (Supplementary Figure S15A‐F). Histological analysis using H&E staining revealed no histological alterations in the liver, kidney, lung, heart or spleen after treatment of KRIBB11 (Supplementary Figure S15G). Based on these results, we concluded that KRIBB11 prevented lymphatic metastasis of BCa cells by harmlessly blocking the transcription of metastatic genes induced by HSF1.
3.10. KRIBB11 inhibited tumor growth of BCa
Considering that HSF1 promoted proliferation, we performed MTT and colony formation assays in BCa cells treated with KRIBB11. Similar to our previous results of silencing HSF1, KRIBB11 significantly suppressed the proliferation of UM‐UC‐3 and T24 cells (Supplementary Figure S16A‐C). Flow cytometry and EdU assays revealed that KRIBB11 restricted the transition from G0/G1 to S phase (Supplementary Figure S16D‐I). Next, we intraperitoneally injected KRIBB11 into subcutaneous xenografts carrying UM‐UC‐3 cells and found that KRIBB11 significantly inhibited tumor growth (Figure 8H‐K, and Supplementary Figure S16J).
We further determined the efficiency of KRIBB11 in BCa PDX models. PDX models were relevant preclinical models that could represent the intrinsic molecular subtypes of cancers and were widely used to evaluate therapeutic approaches for malignancies [40]. We performed intraperitoneal injection of KRIBB11 in PDX models derived from two patients with lymphatic metastasis. As expected, injection of KRIBB11 significantly repressed tumor growth and expression of Ki67 compared to the vehicle group (Figure 8L‐O and Supplementary Figure S17). Thus, KRIBB11 showed promising inhibition of BCa growth.
4. DISCUSSION
HSF1 has been studied for decades and shown to be essential for carcinogenesis and tumor growth. However, only two studies have reported that HSF1 was clinically associated with lymphatic metastasis[41, 42], and the underlying mechanisms remain unknown. Herein, we revealed that HSF1 was overexpressed in BCa with lymphatic metastasis and was an independent predictor of poor prognosis in BCa patients. Mechanistically, HSF1 upregulated the expression of a series of oncogenes in BCa cells via histone methylation mediated by the PRMT5‐WDR5 axis, which facilitated BCa cells EMT and metastatic BCa cells proliferation in LNs. Meanwhile, HSF1 enhanced the secretion of CCL20 from BCa cells to promote macrophages infiltration. Blocking HSF1 by KRIBB11 significantly prevented BCa cells lymphatic metastasis harmlessly (Figure 8P). Taken together, these findings could contribute to better understanding lymphatic metastasis and be used to develop new strategies for its early diagnosis and precision treatment in BCa.
Lymphatic metastasis of cancer cells begins with detachment from the primary tumor, travel through lymphatic vessels, and settlement and growth in LNs [30]. Therefore, targeting multi‐functional molecules could have more advantages in preventing the metastasis of cancer cells. Herein, we found that HSF1 played key roles in many steps of lymphatic metastasis. It not only induced the EMT via LEF1 and dissolved the cell‐matrix via MMP9 [43] but also accelerated the proliferation of metastatic cells via E2F2, a proto‐oncogene that facilitates tumor cell proliferation by regulating the G1‐to‐S phase transition [44].
In terms of the tumor microenvironment (TME), we found that HSF1‐mediated CCL20 secretion in BCa cells was responsible for TAMs recruitment. TAMs, a special group of macrophages that infiltrate tumor tissues and populate the microenvironment of solid tumors, have been extensively studied and serve as prominent metastasis promoters in the TME [45]. In this study, we identified a positive correlation between HSF1 expression and M2 macrophages infiltration intensity and a positive correlation between CCR6 expression and M2 markers expression in the TCGA BLCA cohort. Combined with our previous work showing that M2 TAMs facilitated lymphangiogenesis to enhance lymphatic metastasis by secreting vascular endothelial growth factor C (VEGF‐C) [28] and a previous report which showed that TAMs mediated the metastasis of BCa cells through C‐X‐C motif chemokine ligand 8 (CXCL8) [46], we speculated that overexpression of HSF1 in BCa cells increased M2 TAMs recruitment in a CCL20‐dependent manner, resulting in the promotion of lymphatic metastasis. Previous studies also proved that cancer cells‐derived CCL20 recruited TAMs to accelerate tumor progression in pancreatic and colon cancer [38, 39, 47]. However, recruitment of regulatory T (Treg) cells and T helper type 17 (Th17) cells, both of which are involved in tumor immune evasion, are also the main characteristics of CCL20 [47]. Additionally, in vivo experiments to confirm the roles of HSF1 in TAMs, Tregs and Th17 cells recruitment and identification of key molecules that polarize the TAMs recruited by HSF1‐mediated CCL20 secretion remain a major aim of our future research.
Previous studies showed that the key factors influencing the HSF1‐mediated transcriptome were molecules that bind cooperatively with HSF1 on target genes promoters. Upon stress condition, transcriptional activation of HSPs by HSF1 depends on cooperation with chromatin modulators and transcription elongation factors [48]. However, the molecules that cooperate with HSF1 in tumor progression might be cancer‐specific. They could be epigenetic modulators or elongation factors [5]. For example, the HSF1‐ MORC family CW‐type zinc finger 2 (MORC2) complex increases the recruitment of the polycomb repressive complex 2 (PRC2), especially enhancer of zeste homolog 2 (EZH2), to the Arg kinase‐binding protein 2 (ArgBP2) promoter, thereby increasing the trimethylation of histone H3 at lysine 27 (H3K27me3) levels and transcriptionally repressing the expression of ArgBP2 [49]. In this work, we identified a specific interaction between HSF1 and PRMT5 in BCa cells. PRMT5, a member of the methyltransferase family, transfers methyl groups to arginine residues in target proteins. Emerging studies report that PRMT5 participates in transcriptional programs by epigenetically modifying four arginine residues in histones, namely H4R3, H2AR3, H3R8 and H3R2. Of those, H4R3me2s and H3R8me2s are mostly associated with transcriptional repression, whereas methylation of H3R2 generally causes transcriptional activation. Moreover, methylation of H3R2 recruits the WDR5/MLL co‐activator complex to yield H3K4me3 [50]. Herein, we reported abundant enrichment of H3R2me1, H3R2me2s, together with WDR5 and H3K4me3 on the promoters of HSF1 target genes in BCa cells. Taken together, our results revealed the critical mechanism by which HSF1 enhanced target genes expression through PRMT5‐WDR5‐mediated histone methylation.
Previously, we identified several key factors that regulate BCa cells' metastatic ability [23, 26] and lymphangiogenesis [51]. However, subsequent transformation of these findings into clinical use was unsatisfactory. Although RNA interference (RNAi)‐based cancer treatment showed promising clinical value, safe and efficacious delivery of RNA payloads and targetability of these agents remains a major challenge [52]. In this respect, small molecular inhibitors are more promising, and an increasing number of molecular inhibitors are evaluated in clinical trials for use against tumors [53]. In the present study, we demonstrated that KRIBB11 could inhibit the expression of HSF1 and targeting HSF1 through KRIBB11 significantly prevented lymphatic metastasis of BCa cells in vivo with no significant side effects. Further application of KRIBB11 in PDX models also showed a significant decrease in tumor growth, providing reliable evidence for further clinical use of HSF1 inhibitors. Consistent with our results, KRIBB11 also shows significant inhibition of myeloma growth in vivo [16]. Notably, KRIBB11 was reported to inhibit the expression of HSP70 under heat shock by preventing the recruitment of positive transcription elongation factor b (p‐TEFb) to the HSP70 promoter [54]. Theoretically, targeting HSF1 by KRIBB11 not only inhibited the expression of LEF1, MMP9, CCL20 and E2F2 by downregulating HSF1 but could also block the recruitment of other co‐activator to the promoters of HSPs and decrease their expressions, such as HSP70 and HSP90. Considering that HSP70 and HSP90 were overexpressed and played key roles in BCa, targeting HSF1 by KRIBB11 was a multipotent and promising therapeutic strategy for BCa patients and may shed light on the novel treatment of lymphatic metastasis of BCa.
5. CONCLUSIONS
Our work demonstrated the potential clinical value of HSF1 in lymphatic metastasis of BCa, and described a novel transcriptional mechanism mediated by the HSF1‐PRMT5‐WDR5 axis. Treatment with KRIBB11 was associated with significant and potentially harmless multistep lymphatic metastasis inhibition of BCa. Understanding the precise role of HSF1 in BCa may accelerate the development of diagnostic approaches and therapeutic strategies for BCa patients with lymphatic metastasis.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The ethical consent of this study was approved by Sun Yat‐sen University Committees for Ethical Review of Research involving Human Subjects. All human tissue samples were obtained from patients with written informed consent. All animal studies were conducted with the approval of the Institutional Animal Care and Use Committee of Sun Yat‐sen University (Approval Number SYSU‐IACUC‐2021–000261) and were performed following established guidelines.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE185986. TCGA, Oncomine and R2 Genomics Platform data mining descriptions are included in the MATERIALS AND METHODS.
COMPETING INTERESTS
The authors declare no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
XC, WD and TXL designed this study. MH, RHX and JLW conducted the main experiments and performed data analysis. QS, WGL, WWL performed PDX models construction. KY, YLC performed the analysis of clinical character. QHZ, QZ performed in vitro and in vivo functional experiments. LC and STC performed RNA‐sequencing bioinformatic analysis. SMP performed statistical analyses. XC, JH and TXL wrote and reviewed the manuscript. All authors read and approved the final manuscript. Authorship order among the co‐first authors was determined according to their relative contributions.
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
The authors thank Dr. Bo Wang, Dr. Wenlong Zhong, Dr. Weibin Hou, Dr. Meihua Yang (Sun Yat‐sen Memorial Hospital) for generous help in technical support in isolation and recruitment assay of monocytes. We sincerely thank Prof. Xiaojuan Wang and Tian Qiao from the Bioinformatics and Omics Center in Sun Yat‐sen Memorial Hospital for their assistance in mass spectrometry analysis. This study was supported by the National Key Research and Development Program of China (Grant No. 2018YFA0902803), the National Natural Science Foundation of China (Grant No. 81825016, 82072827, 81961128027, 81702523, 81972383, 82102957), Guangdong Basic and Applied Basic Research Foundation (Grant No. 2021B1515020009, 2020A1515010888, 2019A1515010188), Science and Technology Program of Guangzhou (Grant No. 202102010002), Guangdong Special Support Program (2017TX04R246), Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (for Tianxin Lin), Guangdong Provincial Clinical Research Center for Urological Diseases (2020B1111170006), Guangdong Science and Technology Department (2020B1212060018, 2018B030317001,2017B030314026).
Huang M, Dong W, Xie R, Wu J, Su Q, Li W, et al. HSF1 facilitates the multistep process of lymphatic metastasis in bladder cancer via a novel PRMT5‐WDR5‐dependent transcriptional program. Cancer Commun. 2022;42:447–470. 10.1002/cac2.12284
Contributor Information
Wen Dong, Email: dongwen@mail.sysu.edu.cn.
Xu Chen, Email: chenx457@mail.sysu.edu.cn.
Tianxin Lin, Email: lintx@mail.sysu.edu.cn.
REFERENCES
- 1. Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352(6282):169‐75. [DOI] [PubMed] [Google Scholar]
- 2. Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124(3):922‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hautmann RE, de Petriconi RC, Pfeiffer C, Volkmer BG. Radical cystectomy for urothelial carcinoma of the bladder without neoadjuvant or adjuvant therapy: long‐term results in 1100 patients. Eur Urol. 2012;61(5):1039‐47. [DOI] [PubMed] [Google Scholar]
- 4. Chen X, Zhang J, Ruan W, Huang M, Wang C, Wang H, et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J Clin Invest. 2020;130 (12):6278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in Stress, Development, and Organismal Health. Trends Cell Biol. 2017;27(12):895‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomez‐Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol. 2018;19(1):4‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S. Heat Shock Proteins and Cancer. Trends Pharmacol Sci. 2017;38(3):226‐56. [DOI] [PubMed] [Google Scholar]
- 8. Ma L, Sato F, Sato R, Matsubara T, Hirai K, Yamasaki M, et al. Dual targeting of heat shock proteins 90 and 70 promotes cell death and enhances the anticancer effect of chemotherapeutic agents in bladder cancer. Oncol Rep. 2014;31(6):2482‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lebret T, Watson RW, Molinié V, Poulain JE, O'Neill A, Fitzpatrick JM, et al. HSP90 expression: a new predictive factor for BCG response in stage Ta‐T1 grade 3 bladder tumours. Eur Urol. 2007;51(1):161‐6; discussion 6‐7. [DOI] [PubMed] [Google Scholar]
- 10. Dai C, Sampson SB. HSF1: Guardian of Proteostasis in Cancer. Trends Cell Biol. 2016;26(1):17‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang T, Ren C, Lu C, Qiao P, Han X, Wang L, et al. Phosphorylation of HSF1 by PIM2 Induces PD‐L1 Expression and Promotes Tumor Growth in Breast Cancer. Cancer Res. 2019;79(20):5233‐44. [DOI] [PubMed] [Google Scholar]
- 14. Hoj JP, Mayro B, Pendergast AM. The ABL2 kinase regulates an HSF1‐dependent transcriptional program required for lung adenocarcinoma brain metastasis. Proc Natl Acad Sci U S A. 2020;117(52):33486‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dong B, Jaeger AM, Thiele DJ. Inhibiting Heat Shock Factor 1 in Cancer: A Unique Therapeutic Opportunity. Trends Pharmacol Sci. 2019;40(12):986‐1005. [DOI] [PubMed] [Google Scholar]
- 16. Fok JHL, Hedayat S, Zhang L, Aronson LI, Mirabella F, Pawlyn C, et al. HSF1 Is Essential for Myeloma Cell Survival and A Promising Therapeutic Target. Clin Cancer Res. 2018;24(10):2395‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Q, Chen X, He H, Peng S, Zhang Y, Zhang J, et al. WD repeat domain 5 promotes chemoresistance and Programmed Death‐Ligand 1 expression in prostate cancer. Theranostics. 2021;11(10):4809‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X, Gu P, Xie R, Han J, Liu H, Wang B, et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med. 2017;21(7):1266‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robertson AG, Kim J, Al‐Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle‐Invasive Bladder Cancer. Cell. 2017;171(3):540–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98‐W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Z, Chen X, Xie R, Huang M, Dong W, Han J, et al. DANCR Promotes Metastasis and Proliferation in Bladder Cancer Cells by Enhancing IL‐11‐STAT3 Signaling and CCND1 Expression. Mol Ther. 2019;27(2):326‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen X, Xie R, Gu P, Huang M, Han J, Dong W, et al. Long Noncoding RNA LBCS Inhibits Self‐Renewal and Chemoresistance of Bladder Cancer Stem Cells through Epigenetic Silencing of SOX2. Clin Cancer Res. 2019;25(4):1389‐403. [DOI] [PubMed] [Google Scholar]
- 23. Xie R, Chen X, Cheng L, Huang M, Zhou Q, Zhang J, et al. NONO Inhibits Lymphatic Metastasis of Bladder Cancer via Alternative Splicing of SETMAR. Mol Ther. 2021;29(1):291‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu P, Chen X, Xie R, Han J, Xie W, Wang B, et al. IncRNA HOXD‐AS1 Regulates Proliferation and Chemo‐Resistance of Castration‐Resistant Prostate Cancer via Recruiting WDR5. Mol Ther. 2017;25(8):1959‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J, Zhou Q, Xie K, Cheng L, Peng S, Xie R, et al. Targeting WD repeat domain 5 enhances chemosensitivity and inhibits proliferation and programmed death‐ligand 1 expression in bladder cancer. J Exp Clin Cancer Res. 2021;40(1):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie R, Chen X, Chen Z, Huang M, Dong W, Gu P, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31‐44. [DOI] [PubMed] [Google Scholar]
- 27. He H, Gao Y, Fu J, Zhou Q, Wang X, Bai B, et al. VISTA and PD‐L1 synergistically predict poor prognosis in patients with extranodal natural killer/T‐cell lymphoma. Oncoimmunology. 2021;10(1):1907059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen C, He W, Huang J, Wang B, Li H, Cai Q, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9(1):3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Wang Q, Peng S, Yao K, Chen J, Tao Y, et al. The metastatic promoter DEPDC1B induces epithelial‐mesenchymal transition and promotes prostate cancer cell proliferation via Rac1‐PAK1 signaling. Clin Transl Med. 2020;10(6):e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5(5):402‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clijsters L, Hoencamp C, Calis JJA, Marzio A, Handgraaf SM, Cuitino MC, et al. Cyclin F Controls Cell‐Cycle Transcriptional Outputs by Directing the Degradation of the Three Activator E2Fs. Mol Cell. 2019;74(6):1264‐77.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barna J, Csermely P & Vellai T. Roles of heat shock factor 1 beyond the heat shock response. Cell Mol Life Sci. 2018;75 (16):2897–916. [DOI] [PMC free article] [PubMed]
- 33. Wang N, Yan H, Wu D, Zhao Z, Chen X, Long Q, et al. PRMT5/Wnt4 axis promotes lymph‐node metastasis and proliferation of laryngeal carcinoma. Cell Death Dis. 2020;11(10):864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chiang K, Zielinska AE, Shaaban AM, Sanchez‐Bailon MP, Jarrold J, Clarke TL, et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep. 2017;21(12):3498‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao L, Wu G, Zhu J, Tan Z, Shi D, Wu X, et al. Genotoxic stress‐triggered β‐catenin/JDP2/PRMT5 complex facilitates reestablishing glutathione homeostasis. Nat Commun. 2019;10(1):3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zirkel A, Lederer M, Stöhr N, Pazaitis N, Hüttelmaier S. IGF2BP1 promotes mesenchymal cell properties and migration of tumor‐derived cells by enhancing the expression of LEF1 and SNAI2 (SLUG). Nucleic Acids Res. 2013;41: (13):6618. –36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu W, Wang W, Wang X, Xu C, Zhang N, Di W. Cisplatin‐stimulated macrophages promote ovarian cancer migration via the CCL20‐CCR6 axis. Cancer Lett. 2020;472:59‐69. [DOI] [PubMed] [Google Scholar]
- 38. Jiang S, Zhu L, Zhang M, Li R, Yang Q, Yan J, et al. GABRP regulates chemokine signalling, macrophage recruitment and tumour progression in pancreatic cancer through tuning KCNN4‐mediated Ca2+ signalling in a GABA‐independent manner. Gut. 2019;68(11):1994‐2006. [DOI] [PubMed] [Google Scholar]
- 39. Nandi B, Shapiro M, Samur MK, Pai C, Frank NY, Yoon C, et al. Stromal CCR6 drives tumor growth in a murine transplantable colon cancer through recruitment of tumor‐promoting macrophages. Oncoimmunology. 2016;5(8):e1189052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, et al. Patient‐Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014;4(9):998‐1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin Q, Xiao G, Wang G, He Q, Xu L, Qiu P, et al. Heat Shock Factor 1 in Relation to Tumor Angiogenesis and Disease Progression in Patients With Pancreatic Cancer. Pancreas. 2020;49(10):1327‐34. [DOI] [PubMed] [Google Scholar]
- 42. Dai W, Ye J, Zhang Z, Yang L, Ren H, Wu H, et al. Increased expression of heat shock factor 1 (HSF1) is associated with poor survival in gastric cancer patients. Diagn Pathol. 2018;13(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szarvas T, vom Dorp F, Ergün S, Rübben H, . Matrix metalloproteinases and their clinical relevance in urinary bladder cancer. Nat Rev Urol. 2011;8(5):241‐54. [DOI] [PubMed] [Google Scholar]
- 44. Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23(23):4615‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Y, Xu J, Lan H. Tumor‐associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu H, Zhang X, Han D, Cao J, Tian J. Tumour‐associated macrophages mediate the invasion and metastasis of bladder cancer cells through CXCL8. PeerJ. 2020;8:e8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Korbecki J, Grochans S, Gutowska I, Barczak K, Baranowska‐Bosiacka I. CC Chemokines in a Tumor: A Review of Pro‐Cancer and Anti‐Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int J Mol Sci. 2020;21(20).7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duarte FM, Fuda NJ, Mahat DB, Core LJ, Guertin MJ, Lis JT. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress‐induced gene activation. Genes Dev. 2016;30(15):1731‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tong Y, Li Y, Gu H, Wang C, Liu F, Shao Y, et al. HSF1, in association with MORC2, downregulates ArgBP2 via the PRC2 family in gastric cancer cells. Biochim Biophys Acta Mol Basis Dis. 2018;1864(4 Pt A):1104‐14. [DOI] [PubMed] [Google Scholar]
- 50. Kim H, Ronai ZA. PRMT5 function and targeting in cancer. Cell Stress. 2020;4(8):199‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang C, Liu Q, Huang M, Zhou Q, Zhang X, Zhang J, et al. Loss of GATA6 expression promotes lymphatic metastasis in bladder cancer. FASEB J. 2020.34 (4):5754–66. [DOI] [PubMed] [Google Scholar]
- 52. Revia RA, Stephen ZR, Zhang M. Theranostic Nanoparticles for RNA‐Based Cancer Treatment. Acc Chem Res. 2019;52(6):1496‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ, et al. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem. 2011;286(3):1737‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Santiago L, Daniels G, Wang D, Deng F, Lee P. Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am J Cancer Res. 2017;7 (6):1389–406. [PMC free article] [PubMed] [Google Scholar]
- 56. Lin Y, Lee L, Lee W, Chu C, Tan P, Yang Y, et al. Melatonin inhibits MMP‐9 transactivation and renal cell carcinoma metastasis by suppressing Akt‐MAPKs pathway and NF‐κB DNA‐binding activity. J Pineal Res. 2016;60 (3):277–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Data Availability Statement
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE185986. TCGA, Oncomine and R2 Genomics Platform data mining descriptions are included in the MATERIALS AND METHODS.


