Summary
In addition to cooperatively driving transcriptional programs, emerging evidence supports transcription factors interacting with one another to modulate the outcome of binding events. As such, transcription factor interactions fine-tune the unique gene expression profiles required for developmental progression. Using human-induced pluripotent stem cells as a model of human endoderm lineage commitment, we reveal that GATA6 transiently co-localizes with EOMES at regions associated with non-endodermal lineages and is required for the repression of chromatin opening at these loci. Our results indicate that GATA6-dependent repression of chromatin remodeling, which is potentially mediated via the recruitment of NCOR1 to the EOMES interactome, contributes to definitive endoderm commitment. We anticipate that similar mechanisms are common during human development, furthering our understanding of the complex mechanisms that define cell fate decisions.
Subject areas: Molecular biology, Molecular mechanism of gene regulation, Developmental biology
Graphical abstract
Highlights
-
•
GATA6 transiently occupies a subset of loci during early endoderm formation
-
•
Chromatin accessibility increased at the identified regions in GATA6−/− cells
-
•
EOMES binds independently of GATA6 at the identified sites
-
•
Recruitment of NCOR1 to the EOMES interactome is reduced in GATA6−/− cells
Molecular biology; Molecular mechanism of gene regulation; Developmental biology
Introduction
Investigation into the cell fate decisions that underpin mammalian development continues to identify evermore complex mechanisms of transcriptional control (Zaret, 2020). It is widely accepted that transcription factors are induced at precisely controlled stages of development to establish lineage-specific transcriptional profiles. Emerging evidence has highlighted that the capacity of a transcription factor to occupy its target binding sites is dependent on external parameters, such as chromatin state and the presence of additional transcription factors (Cernilogar et al., 2019; Donaghey et al., 2018; Fernandez Garcia et al., 2019; Heslop et al., 2021; Lee et al., 2019). Although the availability of additional regulators is known to influence the binding profile of a transcription factor, the molecular basis of these interactions remains incompletely characterized.
Induced pluripotent stem cells (iPSCs) offer an ideal model to interrogate the molecular basis of human development (Heslop and Duncan, 2019). iPSCs can be expanded indefinitely and genetically manipulated with ease using CRISPR/Cas9 (Liu et al., 2020; Ran et al., 2013). Moreover, developmental processes can be reproducibly modeled with iPSCs by induction of known developmental signaling cascades through the supplementation of growth factors and small molecules. Differentiations are relatively synchronous, efficient, and reproducible, which permits the high-resolution investigation of an individual factor’s contribution to cell fate decisions at selected time points during development.
Previous work using iPSCs as a model of early foregut development revealed that GATA6 is essential for definitive endoderm formation (Chia et al., 2019; Fisher et al., 2017; Shi et al., 2017; Tiyaboonchai et al., 2017). Moreover, we subsequently reported that GATA6 is required to establish the accessible chromatin profile within the definitive endoderm, thereby permitting developmental progression (Heslop et al., 2021).
In the present study, we examine the complex relationship between GATA6 and another transcription factor—Eomesodermin (EOMES)—during cell differentiation. We identified sites of transient GATA6 binding during the establishment of definitive endoderm and hypothesized that transient GATA6 occupancy could also influence chromatin accessibility. In support of this model, we identified subsets of sites transiently occupied by GATA6 in wild-type cells that increased in chromatin accessibility in the absence of GATA6. Importantly, these loci were commonly bound by EOMES independent of GATA6 and neighbored hallmark neuronal and mesoderm-enriched genes. Further investigation revealed that GATA6 directly interacts with the EOMES protein and that the loss of GATA6 resulted in the significant loss of NCOR1, a corepressor protein, from the EOMES protein interactome. We hypothesize a model whereby GATA6 is required to restrict chromatin remodeling events that favor non-endodermal lineages to facilitate endodermal fate commitment.
Results
GATA6 binding is dynamic during definitive endoderm formation
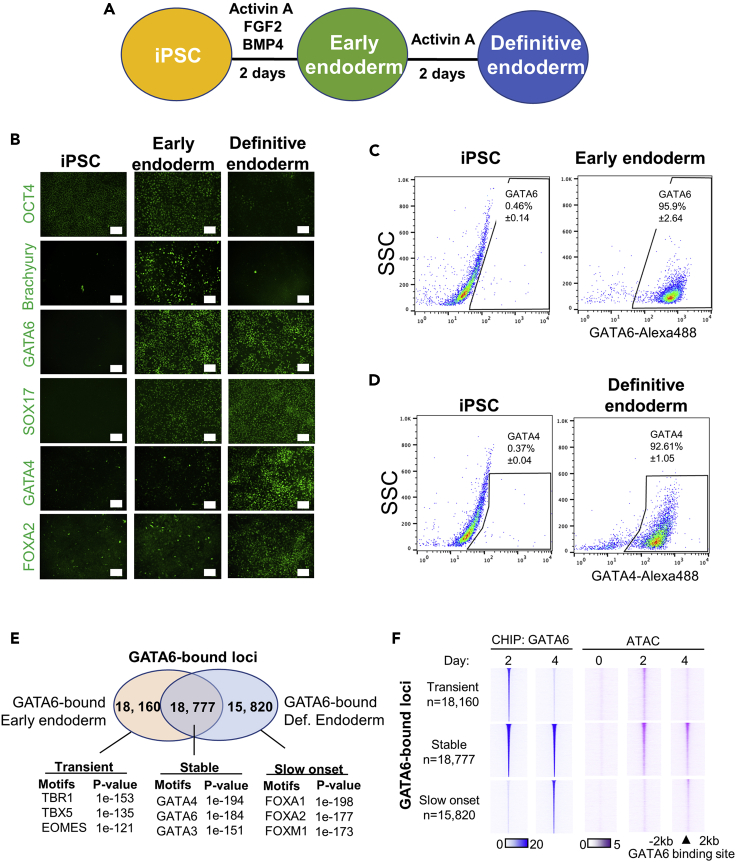
To further our understanding of the molecular basis of definitive endoderm formation, we utilized human iPSCs as a model of early development. Using the early stages of a differentiation protocol that robustly generates hepatocyte-like cells (Si-Tayeb et al., 2010a), we confirmed that the iPSC-derived populations expressed stage-specific markers as they formed definitive endoderm (Figures 1A and 1B). Using this approach, definitive endoderm populations were generated at >90% efficiency and recapitulated normal developmental transitions (Figures 1B–1D, S1A, and S1B).
Figure 1.
Dynamic GATA6 occupancy during definitive endoderm formation
(A–F) The definitive endoderm differentiation protocol; (B) Immunofluorescence analysis of stage-enriched markers during definitive endoderm formation. Sale bar: 100μM; (C) Flow cytometry analysis for GATA6 expression in iPSCs and early endoderm (day 2) cell populations, n = 2, mean ± SD; (D) Flow cytometry analysis for GATA4 expression in iPSCs and definitive endoderm (day 4) cell populations, n = 3, mean ± SD; (E) Venn diagram depicting the number of peaks detected by GATA6 ChIP-seq analysis at day 2 (early endoderm) and day 4 (definitive endoderm) of differentiation (upper panel). Hypergeometric motif enrichment analysis of DNA regions uniquely enriched within each subset (lower panel); (F) Heatmap depicting GATA6 ChIP-seq and ATAC-seq signal intensity at different subsets of GATA6 binding during definitive endoderm formation.
Previous work from our laboratory found that GATA6 is essential to establish the definitive endoderm and the associated chromatin accessibility profile (Fisher et al., 2017; Heslop et al., 2021). Subsequent analysis of the GATA6 ChIP-seq datasets revealed an underappreciated dynamism to the binding profile of GATA6 during definitive endoderm formation (Figure 1E). We found that GATA6 occupies similar numbers of loci in transient (Day 2 only), stable (Day 2 and 4), or slow onset (Day 4 only) profiles as the iPSCs transitioned to a definitive endoderm fate (Figures 1E, S1C, and Table S1).
Enrichment analysis identifying binding motifs overrepresented within each GATA6-binding subset demonstrated that stably occupied regions were enriched for GATA motifs. Conversely, transiently occupied sites were more significantly enriched for T-BOX motifs than the other subsets and slow onset binding loci had greater enrichment of FOXA binding motifs (Figure 1E, Table S2). Therefore, the dynamic aspects of the GATA6 binding profile are likely mediated via interactions with co-localized transcriptional regulators enriched during the different stages of endoderm formation.
We have previously demonstrated that stable GATA6 binding sites are associated with GATA6-dependent increases in chromatin accessibility related to the commitment and specification of definitive endoderm (Heslop et al., 2021); however, the role of transiently bound GATA6 during early endoderm formation is uncharacterized. Therefore, we investigated whether transient GATA6 binding contributes to endoderm fate commitment.
First, we established a temporal chromatin accessibility profile of the GATA6 bound subsets using ATAC-seq analysis during definitive endoderm formation. We found that the accessibility of the bound regions mirrored the dynamic pattern of GATA6 occupancy (Figures 1F and S1D). Because we have previously shown that stable GATA6 binding is consequential to the correct patterning of chromatin state during endoderm formation, we predicted that transient GATA6 occupancy may also modulate chromatin accessibility.
GATA6-dependent restriction of chromatin opening occurs at sites of transient occupancy
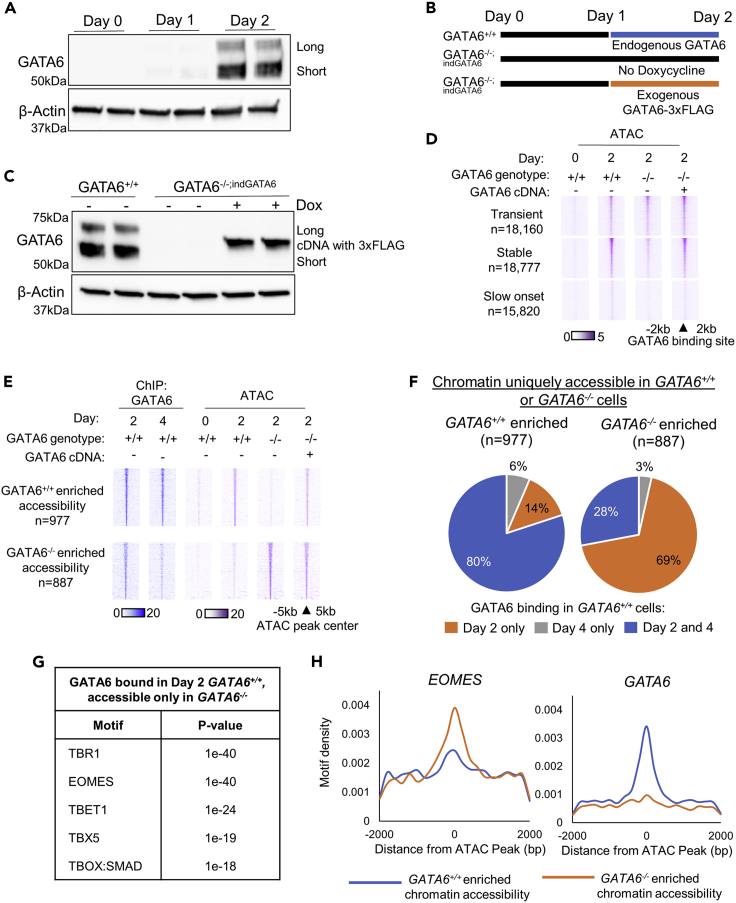
To mechanistically evaluate the function of transient GATA6 binding, we utilized a previously reported GATA6Ex4Δ1/Δ2;ind GATA6 iPSC line (Heslop et al., 2021), referred to as GATA6−/− hereafter. The GATA6−/−iPSC line contains a frameshift deletion within exon four of the GATA6 gene and a doxycycline-inducible GATA6-3xFLAG cDNA vector (Figures S1E and S1F). Therefore, GATA6 expression is strictly dependent on the supplementation of doxycycline to the cell culture medium. GATA6−/− cells fail to form a definitive endoderm (Heslop et al., 2021; Shi et al., 2017; Tiyaboonchai et al., 2017). Importantly, the GATA6−/− phenotype can be rescued through the induction of ectopic GATA6-3xFLAG cDNA expression (Figures S1G and S1H) (Fisher et al., 2017; Heslop et al., 2021). Using this model, we replicated the GATA6 expression profile of wild-type cells (Figures 2A and S2A) through the addition of doxycycline at day 1 of differentiation, with GATA6-3xFLAG cDNA induced for 24 h at endogenous levels (Figures 2B and 2C).
Figure 2.
Aberrantly accessible chromatin in day 2 GATA6−/− populations is enriched at sites of transient GATA6 binding
(A–H); Western blot analysis for GATA6 and β-actin during early endoderm formation, quantification Figure S2A; (B) Schematic representation of doxycycline supplementation experimental plan; (C) Western blot analysis for GATA6 and β-actin at day 2 of differentiation in GATA6+/+ and GATA6−/− cells ± doxycycline; (D) Heatmaps depicting ATAC-seq signal intensity at subsets of GATA6 binding in GATA6+/+ and GATA6−/− cells ± doxycycline during early endoderm formation; (E) Heatmaps depicting GATA6 ChIP- and ATAC-seq signal intensity during early endoderm formation in GATA6+/+ and GATA6−/− cells at high confidence GATA6 occupied regions of differentially accessible chromatin identified between GATA6+/+ and GATA6−/− cells ± doxycycline at day 2 of differentiation; (F) Pie chart showing the percentage of sites with increased chromatin accessibility in GATA6+/+ or GATA6−/− cells at day 2 of differentiation that are bound by GATA6 at 1) day 2 only, 2) day 4 only, or 3) both day 2 and day 4 in wild-type cells; (G) Hypergeometric motif enrichment analysis of DNA regions enriched in chromatin that is uniquely accessible GATA6−/− cells; (H) Histograms showing density of GATA6 and EOMES motifs and their proximity to the regions of chromatin with differential accessibility between GATA6+/+ and GATA6−/− cells at day 2 of differentiation.
Analysis of ATAC-seq datasets in GATA6+/++/+ and GATA6−/−-/- with and without doxycycline revealed that the chromatin accessibility of regions of stable GATA6 occupancy was diminished in GATA6−/−-/- (-dox) compared to GATA6+/++/+ and GATA6−/−-/-(+dox) cell populations (Figure 2D). Therefore, at regions of stable GATA6 binding, chromatin accessibility has a GATA6-dependent profile. Conversely, the overall accessibility profile of regions transiently bound by GATA6 was not reduced in the absence of GATA6 (Figure 2D), indicating that transient GATA6 binding is not required for the overall increases in chromatin accessibility observed at these loci. It is becoming increasingly accepted that transcription factors occupy many poorly accessible regions of chromatin but only remodel a subset of these loci (Donaghey et al., 2018). We postulated that transient GATA6 binding may have a more targeted role in modulating chromatin accessibility. Therefore, we compared sites with differential accessibility between GATA6+/+ or GATA6−/− cells at day 2 of differentiation (p < 0.05, Fold-change >1.5; Table S3). Our results demonstrated that the majority of chromatin domains that are accessible in GATA6+/+, but not GATA6−/−, are bound by GATA6 at both day 2 and 4 in wild-type cells (Figures 2E and 2F). Conversely, the majority of chromatin regions accessible in GATA6−/−, but not GATA6+/+ cells, are bound by GATA6 at day 2 of differentiation but not day 4 in wild-type cells (Figures 2E and 2F). Therefore, at a subset of loci, transient GATA6 binding appears related to the repression of chromatin opening. To understand what set the identified group of GATA6 bound loci apart, we performed motif enrichment analysis. Interestingly, chromatin domains bound by GATA6 in wild-type cells but aberrantly accessible in day 2 GATA6−/− cells were significantly enriched for T-BOX motifs, including EOMES; however, GATA binding motifs were poorly enriched (Figures 2G and 2H).
GATA6 and EOMES co-localize during early definitive endoderm formation
EOMES has recently been shown to work in concert with Brachyury during mesoderm formation (Tosic et al., 2019). EOMES has also been shown to cooperate with GATA6 and SMAD2/3 to set up the transcriptional profile of definitive endoderm (Chia et al., 2019). Cumulatively, these studies indicate that EOMES actively contributes to both mesoderm and endoderm commitment, and its function is dictated by the cofactors enriched within each lineage.
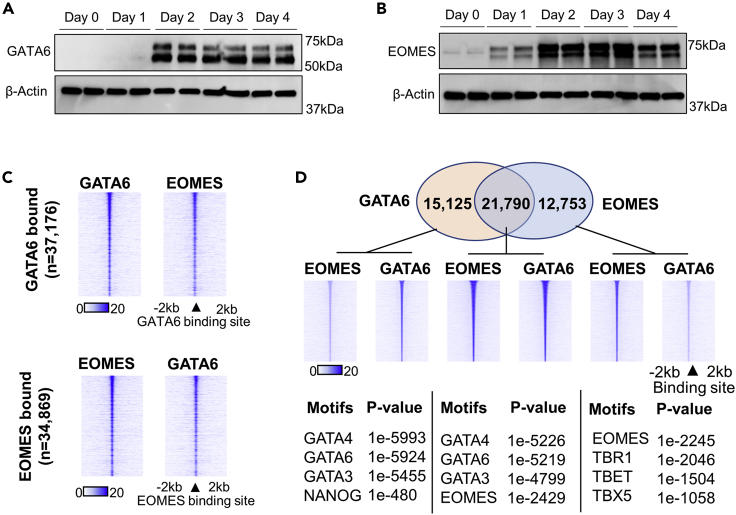
We hypothesized that EOMES function is altered in GATA6−/− cells, resulting in the aberrant opening of chromatin domains relating to alternate lineages. Therefore, we investigated the relationship between GATA6 and EOMES in our iPSC model of early definitive endoderm formation. mRNA and protein expression analysis revealed that EOMES expression preceded that of GATA6, with robust expression of both factors first observable at day 2 of differentiation (Figures 3A, 3B, and S2A–S2C).
Figure 3.
GATA6 and EOMES co-occupancy during early endoderm formation
(A–D) Western blot analysis for GATA6 and β-actin during endoderm formation, quantification Figure S2A; (B) Western blot analysis for EOMES and β-actin during early endoderm formation, quantification Figure S2B; (C) Heatmap intensity of GATA6 and EOMES ChIP-seq signal depicting overlap of GATA6 and EOMES binding patterns at all GATA6 bound loci (left panel) and all EOMES bound loci (right panel); (D) Venn diagram of the overlap between GATA6 and EOMES peaks as determined by ChIP-seq (upper panel), heatmaps depicting GATA6 and EOMES ChIP-seq intensity within each subset (middle panel). Hypergeometric motif enrichment analysis of DNA regions uniquely enriched within each subset (lower panel).
ChIP-seq analysis at day 2 of differentiation identified remarkable overlap in the DNA-binding profiles of GATA6 and EOMES in wild-type cells (Figure 3C). Using peak overlap analyses, we established the subsets of sites bound by GATA6 and EOMES (Figure 3D). Motif enrichment analysis of the peak subsets revealed that loci bound by EOMES alone were enriched for T-BOX motifs; however, GATA6-only sites were enriched for GATA motifs (Figure 3D). At GATA6-EOMES co-bound sites, the ChIP-seq signal intensity for both factors was greater than at sites bound by a single factor. At GATA6-bound sites where EOMES occupancy was below the threshold for peak detection, subthreshold EOMES ChIP-seq signals could still be identified (Figure 3D). The same was true for GATA6 signals at EOMES bound sites lacking identifiable GATA6 peaks.
Therefore, GATA6 and EOMES are both robustly expressed at day 2 of differentiation, occupy very similar binding profiles and demonstrate enhanced occupancy at co-localized sites during early endoderm commitment.
Regions of increased chromatin accessibility in GATA6−/− cells are occupied by EOMES
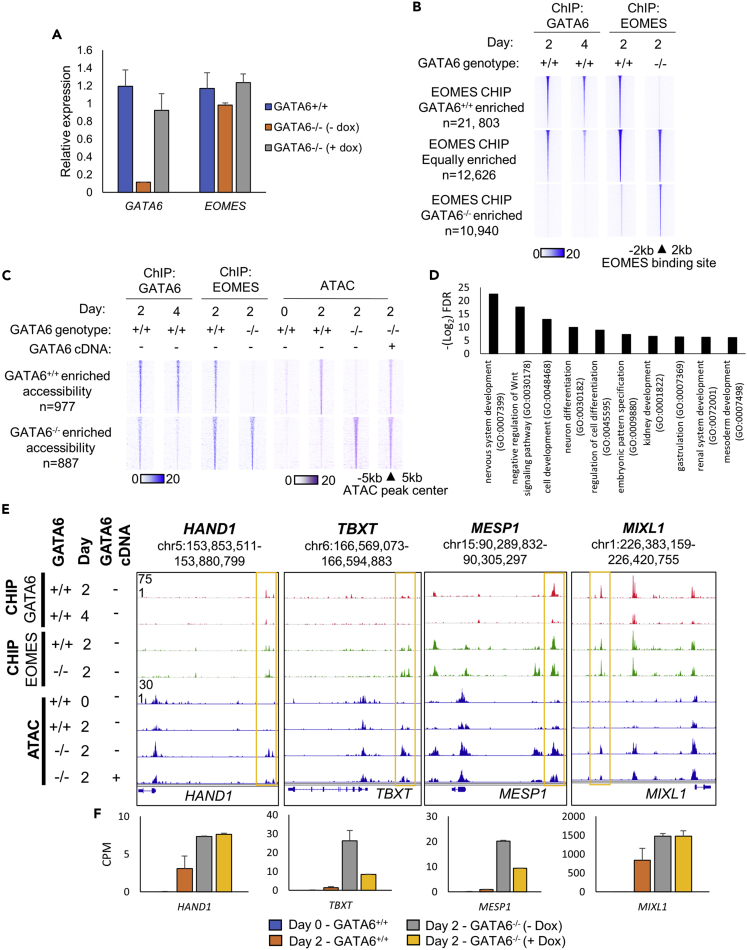
We next investigated how the EOMES expression and binding profile were affected by the absence of GATA6. Comparisons of GATA6+/+ and GATA6−/− cells with and without doxycycline revealed that GATA6 genotype did not result in a marked reduction in EOMES expression at day 2 of differentiation (Figures 4A and S2D).
Figure 4.
EOMES occupies regions of increased chromatin accessibility in GATA6−/− cells
(A–F) RT-qPCR analysis of GATA6 and EOMES mRNA levels at day 2 of differentiation in GATA6+/+ and GATA6−/− cells ± doxycycline. Values normalized to the housekeeping mRNA RPL13a and shown relative to GATA6+/+ cells, n = 3 mean ± SD; (B) Heatmap intensity of GATA6 and EOMES ChIP-seq signal at subsets of EOMES binding that are GATA6-dependent and GATA6-independent; (C) Heatmaps depicting EOMES and GATA6 ChIP- and ATAC-seq signal intensity in GATA6+/+ and GATA6−/− cells at GATA6 occupied regions of differentially accessible chromatin identified between GATA6+/+ and GATA6−/− cells ± doxycycline during early endoderm formation; (D) GO biological process analysis. Selected significantly-enriched developmental pathways related to the genes that neighbor regions of chromatin uniquely accessible in GATA6−/− cells; (E) Genome viewer representation of the HAND1, TBXT, MESP1, and MIXL1 genomic loci aligned with GATA6 and EOMES ChIP-seq and ATAC-seq signal during early endoderm formation in GATA6+/+ and GATA6−/− cells; (F) mRNA expression of the genes that neighbor the selected examples of differentially accessible regions of chromatin, RNA-seq data displayed as counts per million (CPM), n = 2 mean ± SD.
ChIP-seq analysis comparing the EOMES occupancy profile in GATA6+/+ and GATA6−/− day 2 cell populations revealed a pronounced shift in EOMES binding, with EOMES occupancy in wild-type cells established as being either GATA6-dependent or GATA6-independent (Figure 4B). EOMES co-binding with GATA6 was most commonly GATA6-dependent and located at sites of stable GATA6 binding, but a large subset of EOMES binding sites were GATA6-independent and related to transient GATA6 occupancy (Figure S2E). Further analysis of the GATA6-independent EOMES occupied sites identified subsets of sites within low accessibility chromatin. Motif enrichment comparisons of the identified EOMES-bound subsets revealed that GATA6-independent, low accessibility EOMES occupancy had the greatest enrichment for the canonical EOMES binding motif (Figure S2F). These results indicate that EOMES occupancy is more dependent on the presence of the canonical EOMES motif at regions of poor chromatin accessibility and the absence of cofactor binding. As such, the different binding characteristics of EOMES appear, at least in part, to be motif-encoded. Interestingly, EOMES also occupied a number of loci in GATA6−/− cells that were not bound in GATA6+/+ cells, indicating an overall shift in the binding profile of EOMES (Figure 4B). Motif enrichment analysis revealed that these GATA6−/− specific loci were uniquely enriched for TEAD and AP-1-related motifs when compared to EOMES binding sites in wild-type cells (Table S2).
Importantly, day 2 and day 4 GATA6 ChIP-seq read depth analysis across the subsets of EOMES bound loci revealed a reduction in GATA6 ChIP-seq read depth at sites of GATA6-independent EOMES occupancy between day 2 and day 4 of differentiation (Figure 4B). Therefore, at sites of GATA6-independent EOMES occupancy, GATA6 binding is commonly transient, binding more robustly during the early stages of endoderm commitment.
We next assessed whether EOMES occupied regions of differentially accessible chromatin between GATA6+/+ and GATA6−/− cells. Strikingly, EOMES does bind independently of GATA6 at loci with greater accessibility in GATA6−/− than GATA6+/+ in day 2 cell populations (Figure 4C). Therefore, regions of increased chromatin accessibility in GATA6−/− cells are bound by EOMES irrespective of GATA6 expression. Interestingly, the chromatin accessibility profile and the mRNA expression of the neighboring gene sets displayed similar patterns, indicating that the observed changes in accessibility correlate with corresponding changes in gene expression (Figures S3A and S3B).
GO biological processes analyses of genes that neighbored the GATA6−/− accessible chromatin revealed pathways relating to developmental processes to be significantly enriched (Figures 4D and S3C). Importantly, these pathways did not include endoderm-related processes but did include pathways relating to WNT repression and neuronal and mesoderm development. Furthermore, when the overall mRNA expression profile of the GO endoderm and mesoderm pathways was compared, mesoderm-related gene expression increased in the absence of GATA6, whereas endoderm-related gene expression decreased (Figure S3D). By targeting specific genes with established importance during mesoderm formation, we found that the sites bound by EOMES independent of GATA6 with increased chromatin accessibility in GATA6−/− cells neighbored TBXT, HAND1, MESP1, and MIXL1 (Figure 4E). mRNA expression of the selected mesoderm-enriched genes also increased in the absence of GATA6 (Figure 4F).
Taken together, these datasets reveal that GATA6 transiently co-localizes with EOMES at a subset of sites associated with non-endoderm lineage commitment. The absence of GATA6 does not prevent EOMES binding and at a subset of loci culminates in chromatin opening and increased mRNA expression of the neighboring genes. These results imply that GATA6 encourages endoderm lineage commitment in part by restricting accessibility at enhancers relating to alternative lineages.
GATA6 directly interacts with EOMES and is required for the recruitment of corepressor complexes to the EOMES interactome
We previously defined the GATA6 interacting proteins in definitive endoderm by rapid immunoprecipitation of endogenous proteins (RIME analysis) (Heslop et al., 2021). Importantly, EOMES was identified as a high-confidence GATA6 interacting protein as part of this analysis. Therefore, we hypothesized that GATA6 directly interacts with EOMES and influences the binding outcome through modulation of the EOMES protein interactome.
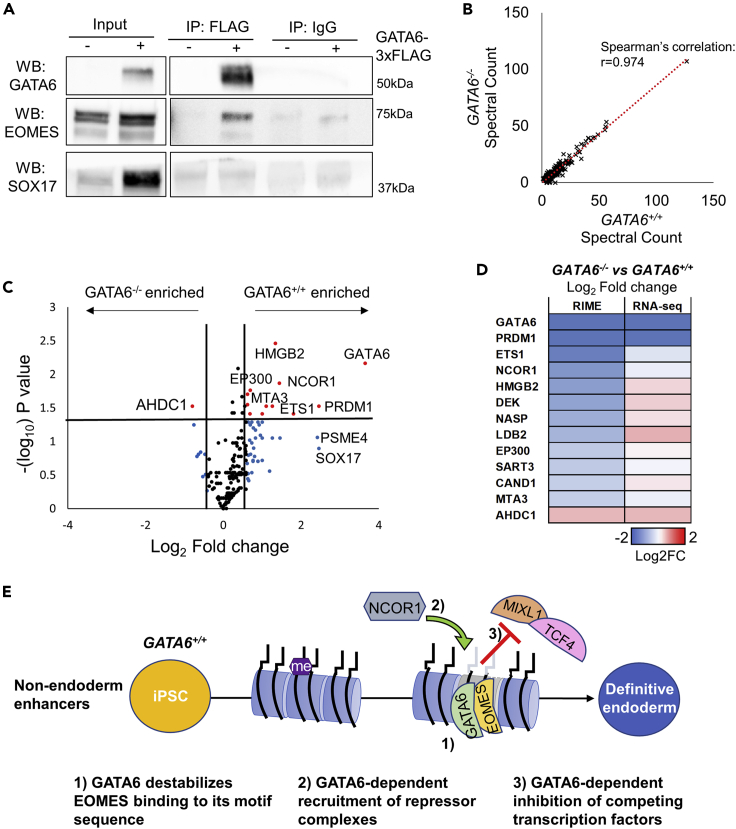
To confirm whether GATA6 and EOMES physically interact, we used the FLAG epitope at the N terminus of the GATA6-3xFLAG cDNA expression construct inserted into the GATA6−/− iPSC line. We performed FLAG immunoprecipitation on samples derived from GATA6−/− cells with and without doxycycline at day 2 of differentiation. Western blot analysis revealed that EOMES was enriched in the FLAG immunoprecipitate compared to control conditions (Figures 5A and S4A). Of note, we could not detect enrichment for another endoderm-enriched transcription factor, SOX17, at the same time point using this model (Figure 5A).
Figure 5.
EOMES interactome is altered in the absence of GATA6
(A–E) Western blot analysis of GATA6, EOMES, and SOX17 following immunoprecipitation of GATA6-3xFLAG protein in GATA6−/− day 2 cultures differentiated ± doxycycline, quantification Figure S4A; (B) Correlation analysis of the spectral counts of proteins enriched within the EOMES interactome in GATA6+/+ and GATA6−/− cells at day 2 of differentiation, n = 2 immunoprecipitations; (C) Proteins differentially enriched within the EOMES interactome between GATA6+/+ and GATA6−/− cells, n = 2 immunoprecipitations; (D) Heatmaps representing the relative log2 fold change in protein enrichment and mRNA expression of proteins with p < 0.05 and −0.6 > log2 fold change >0.6 differential enrichment in GATA6−/− compared to GATA6+/+ cells at day 2 of differentiation., mean of n = 2 immunoprecipitations, or n = 2 RNA-seq samples; (E) Potential mechanisms of GATA6-dependent repression of chromatin opening non-endoderm-associated enhancers during early endoderm formation.
To determine how interactions between GATA6 and EOMES may modulate the protein complexes that co-localize and interact with EOMES, we next performed RIME analysis for EOMES in GATA6+/+ and GATA6−/− day 2 cell populations. Our results identified successful immunoprecipitation of the EOMES protein within both cell populations (Figure S4B, Table S4). Importantly, the majority of the proteins that were co-immunoprecipitated with EOMES were similarly enriched in GATA6+/+ and GATA6−/− cell populations (Figure 5B), indicating that the EOMES interactome does not undergo substantial rewiring in the absence of GATA6.
Despite the majority of interactions being sustained, a subset of proteins did demonstrate differential enrichment between the experimental groups (Figure 5C, Table S4; p < 0.05, −0.6 > log2 fold change >0.6). As expected, GATA6 was absent from the EOMES interactome in GATA6−/− cells. In addition, proteins including PRDM1, NCOR1, EP300, and HMGB2 had reduced enrichment in GATA6−/− cells compared to the GATA6+/+ control (Figures 5C and 5D). AHDC1 had greater enrichment within GATA6−/− cells (Figures 5C and 5D). Using less stringent statistical cut offs (−0.3 > log2 fold change >0.3), we identified additional proteins of interest, including reduced enrichment of SOX17, SUMO2, PATZ1, and RCOR2 in GATA6−/− cells (Figure S4C). Conversely, TCF4 and MIXL1 had greater enrichment in GATA6−/− cells compared to wild-type controls (Figure S4C).
The differentially enriched proteins were further triaged based on RNA-seq analysis, to establish whether the change in enrichment could be attributed to differential recruitment or altered levels of expression (Figures 5D and S4C). SOX17, PRDM1, PSME4, TCF4, and MIXL1 demonstrated differential mRNA expression in GATA6−/− cells in line with the change in enrichment observed within the EOMES interactome (Figures 5D and S4C). Therefore, the differential co-immunoprecipitation of this protein subset is likely the result of altered availability rather than perturbed GATA6-dependent recruitment.
Importantly, we were able to identify differentially enriched proteins that did not correspond to an equivalent change in mRNA expression in GATA6−/− cells. This group included the corepressors NCOR1, RCOR2, and PATZ1, in addition to the chromatin remodeling proteins HMGB2, EP300, MTA3, and BRD4 (Figures 5D and S4C). These results indicate that GATA6 is required for the interactions between EOMES and important regulators of transcription factor function. In particular, the significantly perturbed recruitment of NCOR1 to the EOMES interactome provides one potential mechanism through which GATA6 could mediate repressive chromatin remodeling to facilitate endoderm fate commitment (Figure 5E).
Discussion
Significant progress has been made in understanding how transcription factors establish the accessible chromatin domains that underpin foregut development. Importantly, chromatin opening events occur at only a subset of bound loci, with the majority of binding events not resulting in detectable chromatin remodeling. Despite continued efforts, the role of these ‘silent’ binding events remains incompletely characterized.
In the present study, we identify a role for a selection of sites ‘silently’ occupied by GATA6 during the early stages of endoderm commitment. GATA6 transiently engaged with sites occupied by EOMES that neighbored hallmark mesodermal-enriched genes, despite there being little enrichment for GATA binding motifs. In wild-type cells, the chromatin at these sites remained poorly accessible; however, in the absence of GATA6, the chromatin became accessible. The EOMES binding motif was enriched with high statistical confidence at the regions of interest and the EOMES protein robustly occupied these sites independent of GATA6. Growing evidence points toward chromatin remodeling by transcription factors being highly influenced by motif-encoded binding stability and stabilizing transcription factors (Donaghey et al., 2018; Geusz et al., 2021; Meers et al., 2019). As such, the presented evidence for aberrant chromatin opening in GATA6−/− cells being EOMES-dependent is provocative. We predict that EOMES occupancy will be a requirement for the identified chromatin remodeling events; however, further empirical assessment using combinational EOMES-GATA6 knockout models and investigation of known co-regulators, such as SMAD2/3 (Teo et al., 2011), will be required to prove this definitively.
It will also be important to establish the molecular basis of GATA6-dependent repression of chromatin opening. Through protein interaction-based analysis, we have identified several mechanisms through which GATA6 may repress chromatin accessibility (Figure 5E). The direct interaction identified between GATA6 and EOMES alone may be sufficient to destabilize EOMES occupancy and prevent the recruitment of chromatin remodeling machinery at sites lacking a canonical GATA binding site. Using RIME protein interaction analysis, we additionally established proteins that are differentially available within the EOMES interactome between GATA6+/+ and GATA6−/− cells in early endoderm. NCOR1 was one such protein. NCOR1 has previously been associated with transcriptional repression following recruitment to loci by a range of hormone receptors (Mottis et al., 2013) and transcription factors (Jepsen et al., 2008; Li et al., 2019). Previous work by this laboratory looking at the GATA6 protein interactome in definitive endoderm (day 4) populations by RIME analysis identified the NCOR1 protein as 2.5-fold enriched over IgG controls, but this fell below the 5-fold enrichment required to be classed a high-confidence interacting protein (Heslop et al., 2021). Therefore, GATA6-dependent recruitment of NCOR1 to the EOMES interactome would likely be transient or indirect. Finally, we identified increases in both the GATA6−/− EOMES interactome enrichment and mRNA expression of the TCF4 and MIXL1 transcription factors. Therefore, the GATA6-dependent repression of chromatin opening may also be mediated through the transcriptional repression of transcription factors that go on to stabilize EOMES occupancy at the regions of interest. Establishing how GATA6-dependent repression of chromatin remodeling primarily occurs will be of significant interest for our understanding of lineage restriction during human development.
It is important to acknowledge that although exogenous expression of GATA6-3xFLAG cDNA efficiently rescues GATA6-dependent chromatin opening, we found it was less effective at reducing the aberrantly accessible chromatin in GATA6−/− cells. Our experimental model was designed to recapitulate GATA6 expression during wild-type endoderm formation. Therefore, GATA6-3xFLAG cDNA was induced between day 1 and 2 of differentiation, mirroring the wild-type GATA6 expression profile. We predict that during the 24-h period preceding robust GATA6 induction, lower levels of GATA6 perform a restrictive role at the sites identified in this study. In the absence of GATA6, open chromatin domains are established that cannot be rescued by the subsequent expression of exogenous GATA6. Cumulatively, our results suggest that transient GATA6 recruitment restricts the initiation of chromatin opening events rather than actively reducing chromatin accessibility at regions of established euchromatin.
In summary, we report GATA6-dependent restriction of chromatin remodeling and propose that this mechanism contributes to definitive endoderm commitment. We predict that the repressive function of GATA6 will be common to many transcription factors but remains underappreciated because of the transient nature of the binding events and the requirement for complex loss-of-function studies to be conducted in developmentally relevant models. As such, we anticipate that the findings will be broadly applicable to studies of cell fate and lineage restriction during human development and in disease processes.
Limitations of the study
Within this study, we use an iPSC model of early endoderm formation to demonstrate that GATA6 is required for the repression of chromatin opening at a range of loci that neighbor genes related to non-endodermal cell fates. We identified the putative mechanisms through which GATA6 may function; however, further mechanistic studies are required to validate the roles of EOMES and the chromatin modulating complexes in this process.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GATA6 | Cell Signaling | 5851 RRID:AB_10705521 |
| GATA4 | Santa Cruz | SC-1237 RRID:AB_2108747 |
| SOX17 | RnD | AF1924 RRID:AB_355060 |
| FOXA2 | RnD | AF2400 RRID:AB_2294104 |
| EOMES | Cell Signaling | 66325 RRID: Unknown |
| EOMES (RIME analysis) | Abcam | Ab23345 RRID: AB_778267 |
| FLAG | Sigma-Aldrich | F1804 RRID:AB_262044 |
| Normal Mouse IgG | Sigma-Aldrich | 12-371 RRID:AB_145840 |
| β-Actin | Sigma-Aldrich | A1978 RRID:AB_476692 |
| Protein G Dynabeads | ThermoFisher Scientific | 10004D |
| Bacterial and virus strains | ||
| pzbFGF BL21 Star E. coli | (Ludwig et al., 2006) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| mTesR | (Ludwig et al., 2006) | N/A |
| E-cadherin | (Nagaoka et al., 2010) | N/A |
| RPMI 1640 | ThermoFisher Scientific | 22400089 |
| Geltrex | ThermoFisher Scientific | A1413302 |
| B-27 Supplement (50x), minus insulin | ThermoFisher Scientific | A1895601 |
| Activin A Recombinant Human Protein | ThermoFisher Scientific | PHC9563 |
| FGF-Basic (AA 10-155) Recombinant Human Protein | ThermoFisher Scientific | PHG0023 |
| Purified recombinant zebrafish FGF-Basic | (Ludwig et al., 2006) | N/A |
| BMP4 Recombinant Human Protein | ThermoFisher Scientific | PHC9533 |
| Formaldehyde 16%(w/v) | ThermoFisher Scientific | 28906 |
| Puromycin | Sigma-Aldrich | P9620 |
| Doxycycline hyclate | Sigma-Aldrich | D9891 |
| ROCK inhibitor Y27632 | StemRD | 146986-50-7 |
| DAPI | Sigma-Aldrich | D1388 |
| Critical commercial assays | ||
| RNeasy mini Kit | QIAGEN | 74106 |
| TURBO DNA-freeTM Kit | ThermoFisher/Ambion | AM1907 |
| M-MLV Reverse Transcriptase | ThermoFisher/Invitrogen | 28025-013 |
| TaqMan Gene Expression | ThermoFisher/Applied Biosystems | 4369016 |
| Pierce BCA protein assay kit | ThermoFisher Scientific | 23227 |
| Pierce co-immunoprecipitation kit | ThermoFisher Scientific | 87787 |
| SimpleChIP Plus Enzymatic ChIP Kit | Cell Signaling | 9005 |
| Transcription Factor Staining Buffer Set | ThermoFisher Scientific/eBioscience | 00-5523-00 |
| Experimental models: cell lines | ||
| IPSC-K3 | (Si-Tayeb et al., 2010b) | N/A |
| IPSC-K3 GATA6Ex4Δ1/Δ2;indGATA6 | (Heslop et al., 2021) | N/A |
| Oligonucleotides | ||
| GATA6 | Fwd: TTCGTTTCCTGGTTTGAATTCC Probe: TCATAGCAAGTGGTCTGGGCACC Rev: TGCAATGCTTGTGGACTCTAC |
Integrated DNA Technologies |
| EOMES | Fwd: GTGGCAAAGCCGACAATAAC Probe: CCGAATGAAATCTCCTGTCTCA Rev: ACCCAGAGTCTCCTAATACTGGTTCCC |
Integrated DNA Technologies |
| RPL13a | Fwd: GCCTTCACAGCGTACGA Probe: AGCAGTACCTGTTTAGCCACGATGG Rev: CGAAGATGGCGGAGGTG |
Integrated DNA Technologies |
| Software and algorithms | ||
| Graphpad with PRISM | https://www.graphpad.com/ | Version 8.4.2 |
| FlowJo | https://www.flowjo.com/ | Version 10.5.0 |
| ImageJ | https://imagej.nih.gov | Version 1.53q |
| EaSEQ | (Lerdrup et al., 2016); https://easeq.net/ | Version 1.111 |
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | Version 0.11.7 |
| Partek Flow | https://www.partek.com/partek-flow/ | Version 9.0.20.0526 |
| STAR | Partek Flow | Version 2.6.1d |
| BOWTIE2 | Partek Flow | Version 2.2.5 |
| MACS2 | Partek Flow | Version 2.1.1 |
| DiffBind | (Ross-Innes et al., 2012); https://bioconductor.org/packages/DiffBind/ | Version 3.13 |
| PANTHER Classification system | (Mi et al., 2013) http://pantherdb.org | Version 17.0 |
| HOMER | (Heinz et al., 2010); http://homer.ucsd.edu/homer/ | Version 4.10 |
| Other | ||
| ATAC-seq data | (Heslop et al., 2021) | GSE156021 |
| RNA-seq data | (Heslop et al., 2021) | GSE156021 |
| CHIP-seq: GATA6 | (Heslop et al., 2021) | GSE156021 |
| CHIP-seq: EOMES | (Heslop et al., 2021) | GSE156021 |
Resource availability
Lead contact
Information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stephen A. Duncan (duncanst@musc.edu).
Materials availability
All reagents generated in this study will be made available upon request to the lead contact.
Experimental model and subject details
Cell lines
Human K3 pluripotent stem cells were previously reprogramed from human foreskin fibroblasts (Si-Tayeb et al., 2010b). GATA6Ex4Δ1/Δ2;indGATA6 cells were generated by CRISPR/Cas9 based gene-editing (Heslop et al., 2021). Pluripotent cells were cultured in low oxygen (37°C, 4% O2, 5% CO2) on E-cadherin-IgG Fc fusion protein matrix (Nagaoka et al., 2010), in mTeSR culture medium containing with zebrafish FGF (40 ng/ml) (Ludwig et al., 2006). Culture medium was changed daily and screening for mycoplasma completed at regular intervals.
Method details
Differentiation of pluripotent stem cells
iPSCs were differentiated according to a previously published protocol (Mallanna and Duncan, 2013). Cells were plated at 80% confluency on Geltrex-coated tissue culture plates. After 24 h, the media was replaced with RPMI medium containing 1x B27 minus insulin (ThermoFisher Scientific, NY, #A1895601), Activin A (100 ng/ml; ThermoFisher Scientific, NY, #PHC9563), fibroblast growth factor 2 (FGF2 20 ng/ml; ThermoFisher Scientific, NY, #PHG0023), and bone morphogenetic protein 4 (BMP4, 10 ng/ml; ThermoFisher Scientific, NY, #PHC9533) for 48 h with daily media changes. To commit the populations to a definitive endoderm fate, cells were then cultured in RPMI medium containing 1x B27 minus insulin and Activin A (100 ng/ml) for 48 h with daily media changes. For hepatic endoderm differentiation, cells were maintained in RPMI medium containing 1x B27 minus insulin and Activin A (100 ng/ml) for a further 24 h, before switching to RPMI medium containing 1x B27 (ThermoFisher Scientific, NY, #17504044), supplemented with BMP4 (20 ng/ml) and FGF2 (10 ng/ml) in low oxygen (37°C, 4% O2, 5% CO2) for 72 h, with daily media changes.
Immunofluorescence
Cells were differentiated to the stage of interest and then fixed with 4% paraformaldehyde (w/v; Santa Cruz, TX, #sc-281692) for 30 min. Samples were washed three times with PBS and then permeabilized using 0.5% Triton X-100 for 15 min. 3% BSA (w/v) in PBS was then added for 1 h to block the samples. Blocking solution was removed and primary antibodies added to the samples and incubated overnight at 4°C in 1% BSA (w/v). The next day, the cells were washed three times with 1% BSA (w/v) in PBS and stained with DAPI (1 μg/mL; Sigma Aldrich, MO, #D1388) and Alexafluor488 conjugated secondary antibodies (1:1000; ThermoFisher Scientific, NY) for 2 h at room temperature. After three PBS washes, imaging of the samples was completed using the ZOE fluorescent cell imager (BioRad, CA). All images were processed identically.
Western blotting
Cell lysates were collected using 1x RIPA buffer (Millipore, MA, #20-188) containing HALT protease inhibitor cocktail (ThermoFisher Scientific, NY, #78443) and protein content was determined by BCA assay (ThermoFisher/Pierce, IL, #23227). 10 μg total protein was separated by SDS–PAGE in 4%–15% Mini-protean TGX stain-free precast gels (BioRad, CA, #4568184). Separated proteins were transferred to PVDF membranes using the Trans-Blot Turbo Transfer System (BioRad, CA, #1704155). 5% BSA (w/v) in TBS with 0.1% tween (TBS-T) was added to block the membrane. Primary antibodies were incubated overnight in 1% BSA in TBS-T at 4°C. The next day, membranes were washed with 1% BSA (w/v) in TBS-T. The membrane was then probed with HRP-conjugated secondary antibodies for 2 h at room temperature, followed by washing three times with TBS-T. Clarity chemiluminescent reagent (BioRad, CA, #1705061) was incubated with the membrane for 5 min. Blot images were taken using the Chemidoc Touch gel imager (BioRad, CA). Blots quantified using ImageJ software.
Co-immunoprecipitation
GATA6-/- cells were differentiated to day 2 of the Duncan lab differentiation protocol. After 24 h of differentiation, the cells were cultured in the absence or presence of doxycycline. Samples were collected at day 2. Cells were lysed with IP lysis buffer (ThermoFisher/Pierce, IL, #87787) containing 1x HALT proteinase inhibitor cocktail (ThermoFisher/Pierce, IL, #78443) and Universal Nuclease (ThermoFisher/Pierce, IL, #88701). Lysates were pre-cleared with Protein G Dynabeads (ThermoFisher Scientific, NY, #10004D) for 2 h at 4°C with shaking. The precleared samples were split equally and incubated with either 1 μg of FLAG M2 (Sigma-Aldrich, MO, #F1804) or 1 μg whole mouse IgG (Sigma-Aldrich, MO, #12-371) antibodies overnight at 4°C. Protein G Dynabeads were then added and the mixture was incubated for 2 h at 4°C. IP lysis buffer was used to wash the beads five times, before elution in 1x Lamelli buffer (BioRad, CA, #1610747) at 95°C for 10 min before analysis by western blot.
Flow cytometry
Wild type K3 iPSCs were differentiated day 2 and day 4 of the Duncan lab differentiation protocol. Cell populations were detached using 0.25% trypsin (ThermoFisher Scientific, NY, #25200056). Cell pellets were washed twice with PBS and then incubated for 30 min at room temperature in fixation buffer from the Transcription Factor Staining Buffer Set (ThermoFisher/eBioscience, #00-5523-00). The cell pellet was washed once and then incubated in permeabilization buffer (ThermoFisher/eBioscience, #00-5523-00) with primary antibody for 1 h at room temperature. Next, samples were washed once before a 30-min incubation in permeabilization buffer containing Alexafluor 488 secondary antibody (ThermoFisher Scientific, NY). Percentage of cells positive for markers at each stage were determined by analysis using the Guava EasyCyte Mini (Millipore, MA) and FlowJo software (Version 10.5.0; FlowJo, OR).
Quantitative real-time PCR analysis
RNA was isolated using the RNeasy Mini Kit following the manufacturer’s instructions (QIAGEN, #74106). RNA was then treated with TURBO DNase (ThermoFisher/Ambion, NY, #AM1907). M-MLV Reverse Transcriptase (ThermoFisher/Invitrogen, NY, #28025-013) was used to generate cDNA. Quantitative real-time PCR (qRT-PCR) analysis was performed on a CFX384 real-time PCR machine (BioRad, CA) using TaqMan® Gene Expression assays (ThermoFisher/Applied Biosystems, NY, #4369016). Primer sets were purchased from Integrated DNA Technologies (Integrated DNA Technologies, IA).
RIME analysis
At day 2 of differentiation, GATA6+/+ and GATA6-/- were cross-linked with 1% formaldehyde at room temperature. After incubating for 10 min, the reaction was quenched through the addition 125 mM glycine. After 5 min, the quenching solution was aspirated and crosslinked cells were then collected by scraping into PBS and centrifuged for 10 min at 800 x g. The cell pellet was washed once with PBS containing 0.5% IGEPAL-CA630 (Sigma Aldrich, MO, #I302) and then PBS-IGEPAL with 1 mM PMSF (Sigma Aldrich, MO, #P7626) before being snap frozen on dry ice. RIME analysis was performed by Active Motif, CA., according to published protocols (Mohammed et al., 2016) using EOMES and IgG antibodies. EOMES interacting proteins were identified as proteins with 5-fold enrichment over IgG controls, > 1 spectral count and > 1 unique peptide fragments.
Next generation sequencing sample generation
RNA-seq, ATAC-seq and ChIP-seq analysis sample preparation were completed as part of a previously published study (Heslop et al., 2021). RNA-seq samples were derived from two independent differentiation experiments before collection and processing as described for quantitative real-time PCR analysis. Library preparation and sequencing was completed by Beijing Genomics Institute (BGI). CHIP-seq samples were pooled from n = 4 independent differentiations. CHIP-seq samples were processed using the SimpleChIP Enzymatic Magnetic CHIP kit (Cell Signaling, MA, #9005) before library preparation and sequencing by BGI. ATAC-seq samples were pooled from two independent differentiation experiments and analysis was performed QuickBiology, CA.
Quantification and statistical analysis
Alignment, annotation, and quantification
FastQC software (Version: 0.11.7) was used to assess FASTQ files. The FASTQ files were then aligned to the hg19 genome. RNA-seq FASTQ files were aligned using STAR (Version 2.6.1d) with default settings. Duplicate and mismatched alignments were removed before annotation and counts per millions normalization by Partek Flow software (Partek, MO). ChIP-seq FASTQ files were aligned with Bowtie2 (Version: 2.2.5) and peak calling performed using MACS2 (Version: 2.1.1) with default settings. Peaks were annotated and quantified using Partek Flow software. ATAC-seq FASTQ files were aligned to the hg19 reference genome using Bowtie2 (Version: 2.2.5). Mitochondria and duplicate reads were removed. Uniquely mapped reads were used for peak calling by MACS2 using BAMPE mode (Version: 2.1.1).
Differential analysis of samples
RNA-seq: Differential analysis of normalized RNA-seq data was completed using gene-specific analysis (GSA) in the Partek Flow software suite. GSA analysis calculates the sample distribution of each gene and accordingly applies the appropriate statistical test with multiple correction. Differential analysis of ATAC-seq quantified peaks were identified using Diffbind software (Version 3.13). Peaks with p < 0.05 and > 1.5-fold change were considered differentially accessible. The mergePeaks HOMER script (Version 4.10) with ‘-d given’ settings aligned ChIP-seq peaks. Differential binding between ChIP-seq datasets was determined using the mergePeaks function. The same settings were used to identify CHIP-seq and ATAC-seq overlapping peaks. The ‘-venn’ script established the number of overlapping sites in each category.
Graphs and statistical analysis
HOMER software was used for combined analysis of ATAC- and ChIP-seq datasets. The findMotifsGenome.pl script identified transcription factor sequences enriched within differentially accessible regions of chromatin using a hypergeometric enrichment test. When comparing between subsets of binding sites, the comparative dataset was used as the control group with the ‘-bg’ function to remove motifs equally present in all experimental groups. For all other hypogeometric analysis, HOMER default control settings were used.
HOMER annotatePeaks.pl script was used to analyze the fragment depth and motif density of ChIP or ATAC-seq samples at subsets of transcription factor binding sites. Heatmaps were created with default parameters using EaSEQ software (Version 1.111). BedGraph files were generated using Bowtie2 and uploaded to the EaSEQ software with genomic regions of interest. Heatmaps for comparisons were generated using identical settings.
Statistical comparisons of proteins identified by RIME analysis completed using GraphPad Prism software (Version 8.4.2) using repeated t-tests with correction for multiple testing. Spearman correlation analysis was also completed using GraphPad software. Gene ontology analysis was completed using PANTHER software (Version 17). Differentially accessible regions of chromatin were annotated by HOMER software and the list of neighboring genes were uploaded to the PANTHER software. The significantly enriched biological processes were identified using Fisher exact T test, FDR < 0.05.
Acknowledgments
Work was supported by funds from the National Institutes of Health (DK102716, DK119728, DK123704, GM130457, and CA138313) and American Liver Foundation, (2018 Charles Trey MD Memorial Postdoctoral Research to J.H.). The project was supported by the Cell Models and Imaging Cores of the MUSC COBRE in Digestive and Liver Disease and the MUSC Digestive Disease Research Core Center.
Author contributions
Conceptualization, J.A.H. and S.A.D.; Methodology, J.A.H. and S.A.D.; Formal Analysis, J.A.H. and B.P.; Investigation, J.A.H. and B.P.; Resources, J.A.H. and S.A.D.; Writing – Original Draft, J.A.H.; Visualization, J.A.H. and B.P.; Writing – Review & Editing, S.A.D.; Supervision, S.A.D.; Project Administration, S.A.D.; Funding Acquisition, S.A.D.
Declaration of interests
S.A.D. is the founder of Grùthan Biosciences, LLC. None of the authors declare conflicting interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104300.
Supplemental information
Peaks identified by MACS2 and overlaps established by the HOMER mergePeaks function.
Also contains complete Gene Ontology analysis results.
p < 0.05, Fold-change >1.5.
Data and code availability
-
•
ATAC-seq, RNA-seq, and ChIP-seq datasets used as part of this study are deposited in the GEO database under the GEO ID code: GEO: GSE156021.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- Cernilogar F.M., Hasenöder S., Wang Z., Scheibner K., Burtscher I., Sterr M., Smialowski P., Groh S., Evenroed I.M., Gilfillan G.D., et al. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 2019;47:9069–9086. doi: 10.1093/nar/gkz627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia C.Y., Madrigal P., Denil S.L.I.J., Martinez I., Garcia-Bernardo J., El-Khairi R., Chhatriwala M., Shepherd M.H., Hattersley A.T., Dunn N.R., Vallier L. GATA6 cooperates with EOMES/SMAD2/3 to deploy the gene regulatory network governing human definitive endoderm and pancreas formation. Stem Cell Rep. 2019;12:57–70. doi: 10.1016/j.stemcr.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghey J., Thakurela S., Charlton J., Chen J.S., Smith Z.D., Gu H., Pop R., Clement K., Stamenova E.K., Karnik R., et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 2018;50:250–258. doi: 10.1038/s41588-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Garcia M., Moore C.D., Schulz K.N., Alberto O., Donague G., Harrison M.M., Zhu H., Zaret K.S. Structural features of transcription factors associating with nucleosome binding. Mol. Cell. 2019;75:921–932.e6. doi: 10.1016/j.molcel.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J.B., Pulakanti K., Rao S., Duncan S.A. GATA6 is essential for endoderm formation from human pluripotent stem cells. Biol. Open. 2017;6:1084–1095. doi: 10.1242/bio.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geusz R.J., Wang A., Lam D.K., Vinckier N.K., Alysandratos K.D., Roberts D.A., Wang J., Kefalopoulou S., Ramirez A., Qiu Y., et al. Sequence logic at enhancers governs a dual mechanism of endodermal organ fate induction by FOXA pioneer factors. Nat. Commun. 2021;12:6636–6719. doi: 10.1038/s41467-021-26950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop J.A., Duncan S.A. The use of human pluripotent stem cells for modeling liver development and disease. Hepatology. 2019;69:1306–1316. doi: 10.1002/hep.30288. [DOI] [PubMed] [Google Scholar]
- Heslop J.A., Pournasr B., Liu J.-T., Duncan S.A. GATA6 defines endoderm fate by controlling chromatin accessibility during differentiation of human-induced pluripotent stem cells. Cell Rep. 2021;35:109145. doi: 10.1016/j.celrep.2021.109145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K., Gleiberman A.S., Shi C., Simon D.I., Rosenfeld M.G. Cooperative regulation in development by SMRT and FOXP1. Genes Dev. 2008;22:740–745. doi: 10.1101/gad.1637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Cho H., Rickert R.W., Li Q.V., Pulecio J., Leslie C.S., Huangfu D. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. 2019;28:382–393.e7. doi: 10.1016/j.celrep.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerdrup M., Johansen J.V., Agrawal-Singh S., Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 2016;23:349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- Li C., Sun X.-N., Chen B.-Y., Zeng M.-R., Du L.-J., Liu T., Gu H.-H., Liu Y., Li Y.-L., Zhou L.-J., et al. Nuclear receptor corepressor 1 represses cardiac hypertrophy. EMBO Mol. Med. 2019;11:e9127. doi: 10.15252/emmm.201809127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.T., Corbett J.L., Heslop J.A., Duncan S.A. Enhanced genome editing in human iPSCs with CRISPR-CAS9 by co-targeting ATP1a1. PeerJ. 2020;8:e9060. doi: 10.7717/peerj.9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., Thomson J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Mallanna S.K., Duncan S.A. Differentiation of hepatocytes from pluripotent stem cells. Stem Cell Biol. 2013;26 doi: 10.1002/9780470151808.sc01g04s26. Unit 1G.4. 24510789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers M.P., Janssens D.H., Henikoff S. Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol. Cell. 2019;75:562–575.e5. doi: 10.1016/j.molcel.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D’Santos C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- Mottis A., Mouchiroud L., Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka M., Si-Tayeb K., Akaike T., Duncan S.A. Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev. Biol. 2010;10:60. doi: 10.1186/1471-213x-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.-D., Lee K., Yang D., Amin S., Verma N., Li Q.V., Zhu Z., Soh C.-L., Kumar R., Evans T., et al. Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell Stem Cell. 2017;20:675–688.e6. doi: 10.1016/j.stem.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Sepac A., Sedlic F., Bosnjak Z.J., Lough J.W., Duncan S.A. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev. Biol. 2010;10:81. doi: 10.1186/1471-213X-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K.K., Arnold S.J., Trotter M.W.B., Brown S., Ang L.T., Chng Z., Robertson E.J., Dunn N.R., Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyaboonchai A., Cardenas-Diaz F.L., Ying L., Maguire J.A., Sim X., Jobaliya C., Gagne A.L., Kishore S., Stanescu D.E., Hughes N., et al. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Rep. 2017;8:589–604. doi: 10.1016/j.stemcr.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosic J., Kim G.J., Pavlovic M., Schröder C.M., Mersiowsky S.L., Barg M., Hofherr A., Probst S., Köttgen M., Hein L., Arnold S.J. Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 2019;21:1518–1531. doi: 10.1038/s41556-019-0423-1. [DOI] [PubMed] [Google Scholar]
- Zaret K.S. Pioneer transcription factors initiating gene network changes. Annu. Rev. Genet. 2020;54:367–385. doi: 10.1146/annurev-genet-030220-015007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peaks identified by MACS2 and overlaps established by the HOMER mergePeaks function.
Also contains complete Gene Ontology analysis results.
p < 0.05, Fold-change >1.5.
Data Availability Statement
-
•
ATAC-seq, RNA-seq, and ChIP-seq datasets used as part of this study are deposited in the GEO database under the GEO ID code: GEO: GSE156021.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request