Abstract
Antimicrobial peptides (AMPs) are widely studied as therapeutic agents due to their broad-spectrum efficacy against infections. However, their clinical use is hampered by the low in vivo bioavailability and systemic toxicity. Such limitations might be overcome by using appropriate drug delivery systems. Here, the preparation of a drug delivery system (DDS) by physical conjugation of an arginine-rich peptide and hydrothermal carbon nanoparticles (CNPs) has been explored, and its antimicrobial efficacy against Eschericia coli (E. coli) and Staphylococcus aureus investigated in comparison with the unloaded carrier and the free peptide. The mechanism of interaction between CNPs and the bacteria was investigated by scanning electron microscopy and a combined dielectrophoresis–Raman spectroscopy method for real-time analysis. In view of a possible systemic administration, the effect of proteins on the stability of the DDS was investigated by using albumin as a model protein. The peptide was bounded electrostatically to the CNPs surface, establishing an equilibrium modulated by pH and albumin. The DDS exhibited antimicrobial activity toward the two bacterial strains, albeit lower as compared to the free peptide. The decrease in effectiveness toward E. coli was likely due to the rapid formation of a particle-induced extracellular matrix. The present results are relevant for the future development of hydrothermal CNPs as drug delivery agents of AMPs.
1. Introduction
Infectious diseases remain an important cause of morbidity and mortality worldwide, especially in low-income countries.1 Despite the positive impact of antibiotics and the advances of research over the years, the control and eradication of infectious diseases are still challenging. In particular, antibiotic overuse and misuse have promoted the onset of antibiotic resistance with potentially devastating consequences for human health.2 Thus, novel antibiotic treatments are urgently needed.
Recent studies bring attention to the development of peptide-based drugs possessing the ability to kill bacteria resistant to common antibiotics.3 Cationic antimicrobial peptides (AMPs) have been discovered more than four decades ago. They are constituted by a low number of amino acids and are characterized by a net positive charge, as well as a high proportion of hydrophobic residues, which together provide them with amphipathic properties.4 Thanks to their rapid and high efficacy as compared with other antimicrobial agents,5 and their low susceptibility to induce drug resistance due to their multiple sites of action, AMPs are less likely to promote resistance and have been proposed as a valid alternative to conventional antibiotics. It was shown that AMPs display a broad-spectrum action toward bacteria, fungi, and viruses and demonstrated a capacity to eradicate biofilms.6
In spite of their promising properties, AMPs suffer from reduced in vivo antimicrobial action due to their low plasma half-life and degradation by proteolytic enzymes or low-pH environments, e.g., in the stomach or infected tissues. Several approaches have been proposed to avoid proteolysis, such as the use of d-amino acids or synthetic analogues of peptides. Another approach is the use of nanoparticles-based drug delivery systems (DDSs), which can enhance the antimicrobial efficacy and selectivity of the peptide, decrease the systemic toxicity, and extend their biostability.7−11 A wide number of DDSs have been proposed, such as lipid-based nanoparticles, polymers, or metals.9
Carbon-based nanomaterials (CNMs) are a class of materials attracting interest as multifunctional drug delivery systems due to their biocompatibility and rich chemistry that allow functionalization with targeting agents and drugs.12,13 CNMs have been extensively proposed both as antimicrobial agents on their own or as drug delivery systems. However, few examples of conjugation with AMPs exist.11 Carbon nanoparticles (CNPs) obtained by hydrothermal carbonization possess several advantages over other CNMs as they are easy to prepare with a defined size by a one-pot synthesis.14 CNPs are composed of elemental carbon, mainly amorphous, and are decorated by acidic carboxylic groups at the surface.14 Exhibiting a high negative surface charge, CNPs form highly stable suspensions at all pH values.14 These functionalities also make them suitable as carriers of cationic peptides that might be bonded to the nanoparticle surface by electrostatic interactions. Other important advantages of CNPs are their biocompatibility. In fact, they exhibit no cytotoxicity against the cells of the immune system14 and are hemocompatible.15 Finally, previous data suggest that CNPs are not biopersistent due to degradation by neutrophils.16
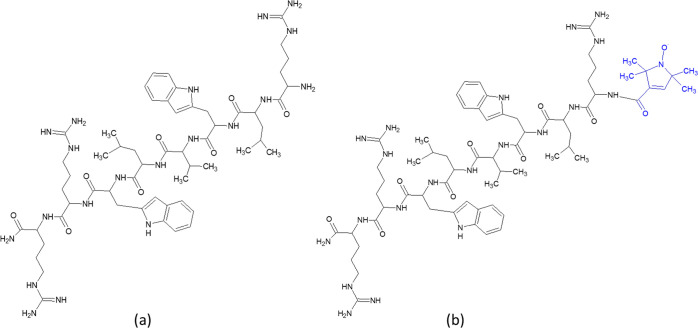
The aim of this study was the preparation of a nanoformulation based on the physical conjugation of CNPs with the antimicrobial peptides Bac8c2,5 Leu, and the evaluation of the antibacterial activity of CNPs and peptide-loaded CNPs against Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli). Both strains are already well characterized in literature and usually used as models for Gram-positive (Gram+) and Gram-negative (Gram−) organisms, respectively. Furthermore, they are both involved in frequent diseases and infections such as those associated with the urinary tract,17 skin lesions, or endocarditis18 which are all commonly treated with antibiotics contributing to the insurgency of resistance. The cationic peptide Bac8c2,5Leu (Scheme 1) has been chosen. This AMP has been synthesized for the first time in 2014,19 starting from the sequence of Bac2A,20 a modified variant of the natural peptide Bactenecin, a decapeptide discovered in bovine neutrophils.21 It exhibits a low minimal inhibitory concentration against S. aureus (8 μg/mL, 6.75 μM) and other medically relevant bacteria, and it is effective in eradicating S. aureus biofilm infection in vitro.3 The presence of three positively charged arginine residues connected by five nonpolar amino acids makes this peptide a good candidate for the physical conjugation with CNPs.
Scheme 1. Structures of (a) Bac8c2,5 Leu and (b) SL-Bac8c2,5 Leu.
The effect of albumin on the release of the peptide has also been studied to get insight into the stability of the DDS in biological fluids.
2. Results
2.1. Synthesis of CNPs
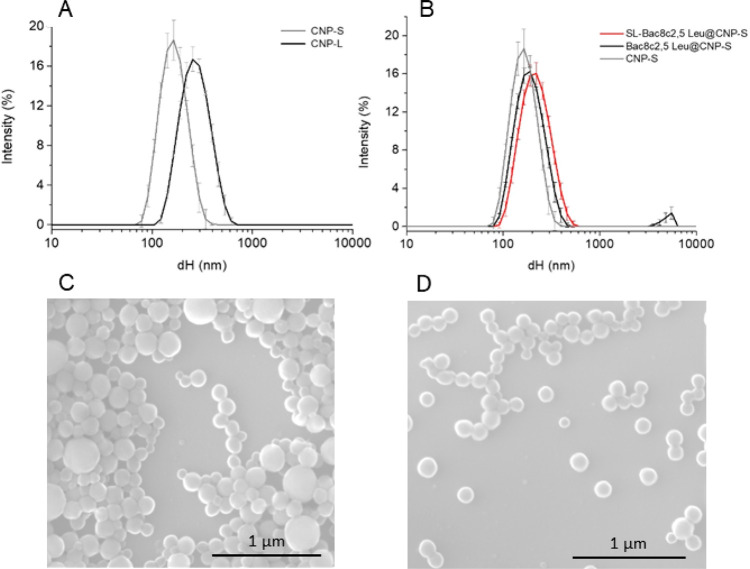
Hydrothermal CNPs were synthesized in two different sizes, hereafter referred to as small (CNP-S) and large (CNP-L), by modifying the synthetic parameters as described in the Experimental Section. The hydrodynamic diameters (dH) distribution in water of the two CNPs batches are compared in Figure 1A. CNP-S appeared monodisperse (polydispersion index, PDI < 0.1), while CNP-L exhibited a slightly wider size range. The mean dH were 132 and 243 nm, respectively. The colloidal suspensions in water were very stable, because of the highly negative ζ-potential of the particles (−52,7 mV).
Figure 1.
Size distribution of CNPs monitored by dynamic light scattering (DLS) and scanning electron microscopy (SEM). (A) Hydrodynamic diameter distribution of CNP-S and CNP-L in water. (B) Hydrodynamic diameter distribution changes following functionalization of CNP-S with Bac8c2,5Leu or SL-Bac8c2,5Leu. Hydrodynamic diameters (dH) distribution (% intensity) is expressed as the mean value of 3 measurements ± SD. Representative SEM images of (C) CNP-L and (D) CNP-S.
SEM analysis (Figure 1C,D) showed spherical particles, with geometrical diameters of 129 ± 22 and 239 ± 85 nm, respectively, compatible with the hydrodynamic diameter values (Figure 1A). CNP-L appeared to be less homogeneous than CNP-S, being composed of different populations of particles of different sizes.
2.2. Effects of CNP-S and CNP-L on the S. aureus and E. coli Cell viability
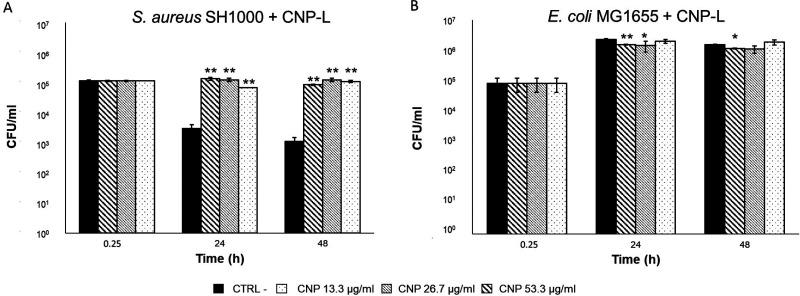
The vitality of bacteria was analyzed after 0.25, 24, and 48 h of incubation with three concentrations of CNPs in PBS. This test is a standard methodology22 used to evaluate possible bactericidal effects induced by antibiotics in a simplified and controlled system. This medium was chosen since standard culture media contain proteins, carbohydrates, and salts, which could adsorb onto the CNPs surface, thus modifying their action on bacterial cells. Figure 2 shows the average of the vital colonies counted at each time point and expressed as CFUs/mL of S. aureus and E. coli, respectively, alone or in the presence of CNP-L.
Figure 2.
Effect of CNP-L on the vitality of S. aureus SH1000 and E. coli MG1655. Viable counts (CFUs) of (A) S. aureus SH1000 and (B) E. coli MG1655 after 0, 24, and 48 h. Bacteria were grown in PBS without CNPs as CTRL—or in the presence of CNPs at concentrations of 53.3, 26.7, and 13.3 μg/mL. Each experiment was replicated three independent times. *, p value < 0.05; **, p value < 0.01.
S. aureus showed a decrease in the number of vital bacterial cells over time in the absence of particles, likely due to the lack of nutrients. This was in fact not observed in standard bacterial culture medium (MH broth) (Supporting Information, Figure S1). CNP-L do not demonstrate bactericidal effects, but instead, they seem to maintain a bacterial growth similar to that of the inoculum (Figure 2A). These results suggest that CNPs may have been used by Gram+ bacteria as a carbon and energy source. Even though no data are present in literature on the possible degradation by S. aureus of hydrothermal carbon nanoparticles, few studies report the capability of some bacteria communities to degrade other carbon nanomaterials.23−25
E. coli appeared more resistant to the lack of nutrients (Figure 2B) compared to S. aureus. A slight but statistically significant decrease of the vitality in comparison with the negative control was observed at the highest CNPs concentrations, albeit the effect decreased over time. The antimicrobial activity of CNP-L was also evaluated in MH broth (Supporting Information, Figure S1). In this case, CNP-L did not elicit any activity on both strains.
CNP-S elicited similar effects on both the bacterial strains (Supporting Information, Figure S2).
2.3. Mechanism of Interaction of CNP-L with S. aureus and E. coli
To investigate whether the observed effect on the bacteria vitality was due to a direct interaction of the CNPs with the cells, Raman spectroscopy and SEM were used. Unfortunately, both bacteria and CNPs concentrations were too low in our experimental conditions to be detected with conventional Raman spectroscopy. The dielectrophoresis (DEP) technique overcame this problem by increasing the local bacteria concentration in specific volumes of a specially conceived cell, which can be then analyzed with Raman spectroscopy.26
CNPs are mainly composed of amorphous elemental carbon, and therefore their detection in cells is not straightforward. However, vibrational bands generated by few crystalline domains present in the CNPs bulk structure allow their identification.14 Raman spectroscopy also allows rapid detection and characterization of bacterial chemical fingerprints directly in suspension without the need of any chemical labels or complex sample preparation. The Raman fingerprints of the two bacteria, of PBS and of a suspension of CNP-L in water are shown in the Supporting Information (Figure S2).
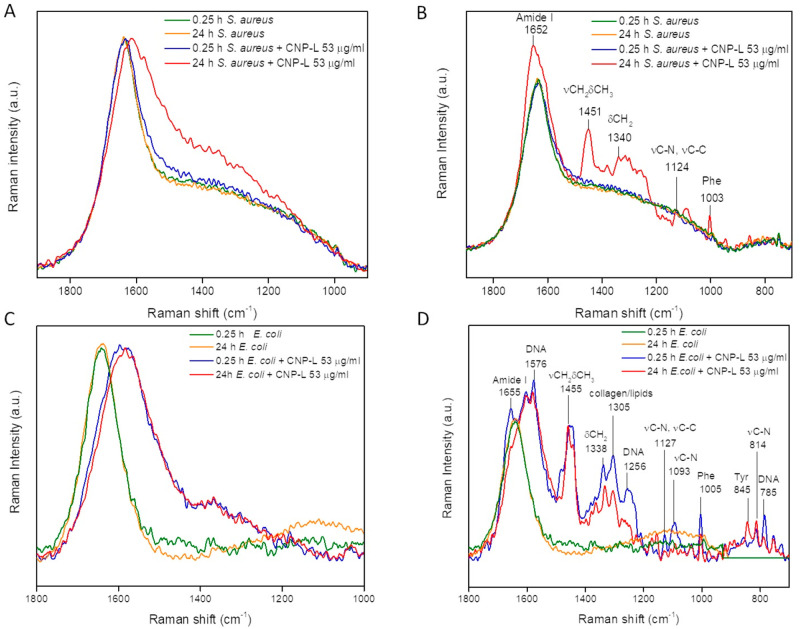
Representative DEP–Raman measurements on S. aureus and E. coli exposed to the CNP-L for 0.25 and 24 h are shown in Figure 3.
Figure 3.
DEP–Raman analysis of the interaction of CNP-L with S. aureus and E. coli. DEP–Raman spectra of (A, B) S. aureus SH1000 and (C, D) E. coli MG1655 in the absence or presence of CNP-L (53 μg/mL) after 0.25 and 24 h of incubation. For each bacterial strain two different signals were recorded, focalizing the Raman microscope in regions of the same sample in which the ECM was (B, D) or was not (A, C) present. Each line represents the normalized average of three Raman spectra recorded during three independent experiments.
The Raman spectra of the samples not exposed to the CNPs showed no differences at the two time points for both strains (Figure 3, yellow and green lines), while two different Raman signals ascribable to CNPs (Figure 3A,C) or to extracellular matrix (ECM) components (Figure 3B,D) were detected in the Raman spectra of both bacteria.27 For S. aureus both CNPs and ECM signals were visible only after 24 h of exposition to the CNPs (Figure 3A,B, red lines), while in the case of E. coli (Figure 3C,D) these signals were already visible after a few minutes of incubation with CNPs (Figure 3C,D, blue lines), indicating a faster interaction between cells and CNPs for E. coli than for S. aureus. The observed CNPs Raman signal is evidenced by an increase of the band intensity in the 1600–1200 cm–1 region. This increase is ascribable to the summation of the G and D bands distinctive of the CNPs (1585 and 1360 cm–1)14 to the background due to the medium and indicates the association between bacteria and CNPs. In fact, the dielectrophoresis (DEP) forces act selectively on the bacterial cells and not on free CNPs. Moreover, since the samples were washed to remove unbound CNPs from the suspension, any Raman signal ascribable to CNPs corresponds to particles either bound to the bacterial surface or internalized. This interaction between bacteria and CNPs was further confirmed by the presence of ECM Raman signals, which were observed in different regions of each of the two bacterial samples.
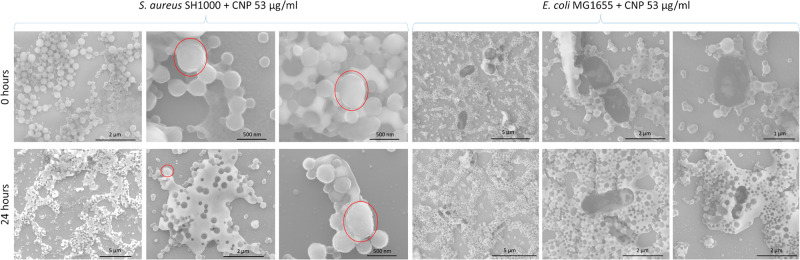
To confirm the physical interaction of CNP-L with bacterial cells, SEM analysis was performed immediately after the inoculum and after 24 h of incubation in the presence of the highest concentration of CNP-L (53.3 μg/mL). The SEM images are shown in Figure 4. S. aureus cells exhibit a spherical morphology similar to CNP-L but can be distinguished because they are three times bigger in size than CNP-L (red circles).
Figure 4.
SEM analysis of the interaction of CNP-L withS. aureus and E. coli. Representative SEM images of S. aureus SH1000 (left) and E. coli MG1655 (right) after 0.25 h (top) and 24 h (bottom) of incubation with 53.3 μg/mL of CNP-L. The red circles indicate the bacterial cells of S. aureus SH1000.
For both bacteria the formation of a large amount of extracellular matrix that englobes both CNPs and bacteria is visible and more evident for E. coli especially after 24 h of incubation. These results are in line with the DEP–Raman findings and confirm the fast interaction between bacteria and CNPs leading to a chemical signaling that induces bacteria to produce ECM, which is enhanced over time.
2.4. Preparation of Bac8c2,5Leu @CNP-S and Evaluation of the Desorption Index
Bac8c2,5Leu (H-RLWVLWRR-NH2) was synthesized by microwave assisted solid-phase synthesis and characterized by analytical RP-HPLC and mass spectrometry (ESI+-MS) (Supporting Information, Figure S4A,B). Mass spectrometric analysis revealed a molecular weight of Bac8c2,5Leu equal to 1183.4, a peak at 395.5 corresponding to the [Bac8c2,5Leu + 3H+]3+ ion, and a peak at 592.6 of the [Bac8c2,5Leu + 2H+]2+ ion. The absence of other peaks in the MS spectra furtherly indicates the successful assembly and purification of the peptide.
CNP-S were selected as nanocarriers for the peptide because they exhibit a larger loading surface area with respect to CNP-L. Loading was performed by physical adsorption (simple incubation of the peptide with CNP-S), exploiting the electrostatic interaction between the cationic peptide and the negatively charged surface of CNPs. Different concentrations of AMP were tested in order to achieve a drug delivery system (DDS) stable in aqueous media, and characterized by the highest possible loading capacity. At the highest concentrations, loading-induced visible aggregation of the CNPs due to the shift of the nanoparticles ζ-potential toward less negative values, reducing the electrostatic repulsion among particles and, in turn, the colloidal stability (data not shown). By decreasing the concentration, a stable colloidal suspension was obtained (PDI = 0.11). A mean hydrodynamic diameter slightly higher than the unloaded sample (203.8 nm) was obtained. The ζ-potential was less negative (−25.9 mV) than the pristine CNPs confirming the actual presence of the peptide at the surface.
The size distribution was further evaluated by nanoparticle tracking analysis (Supporting Information, Figure S5) that revealed a major population in the 50–200 nm range and a mean hydrodynamic diameter of 119.8 nm.
The quantification of the amount of peptide loaded onto CNPs was performed by using an indirect method. After incubation of CNPs in the peptide solution, the nanoparticles were separated by centrifugation and the residual peptide in the supernatant was quantified by means of fluorescence (Supporting Information, Figure S5). The peptide was almost totally absorbed onto the nanoparticles, thus resulting in a DDS with a final concentration of 0.3 mg o Bac8c2,5Leu/(mg of CNPs), equal to 5.5 molecules/(nm2 of CNPs surface area).
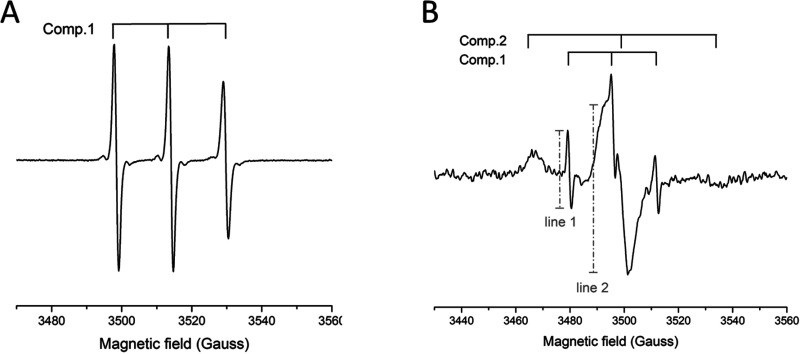
Evidence of the affinity of the peptide for the surface of the CNPs was obtained by electronic paramagnetic resonance (EPR) spectroscopy, by using the spin-labeled peptide (SL-Bac8c2,5Leu) (Scheme 1b). Synthesis and characterization of the labeled peptide are reported in the Supporting Information (Figure S3C). CNP-S loaded with the labeled peptide resulted in a colloidal suspension with a dH distribution completely overlapped with those of Bac8c2,5Leu- @CNP-S (Figure 1B). Figure 5A reports the EPR spectra of the free peptide SL-Bac8c2,5Leu in PBS 10 mM at pH 7.4, which is typical of a labeled peptide freely tumbling in solution, characterized by narrow line widths and short correlation times (τC ∼ 0.2 ns). Figure 5B shows the EPR pattern of the SL-Bac8c2,5Leu @CNPs suspended in PBS 10 mM at pH 7.4. This derives from the overlap of two different components characterized by distinct line widths and rotational correlation times, τC. The first one is analogous to the freely tumbling peptide in solution (component 1), whereas the second is characterized by large line width and high correlation times τC ∼ 3.0 ns (component 2): these last features are typical of a strongly immobilized peptide. This demonstrates the presence of an adsorption equilibrium between the free peptide and the peptide immobilized onto the CNPs. A rough estimate of the relative proportion of the two components is expressed by the intensity ratio of the narrow left line of the first component vs the large central line of the second component (line 1/line 2 in panel B); this ratio has the meaning of a desorption index (DI).
Figure 5.
EPR analysis of SL-Bac8c2,5Leu and SL-Bac8c2,5Leu@CNP-S. EPR spectra of (A) the free peptide SL-Bac8c2,5Leu in PBS 10 mM pH 7.4 and (B) the suspension of SL-Bac8c2,5Leu@CNP-S in PBS 10 mM pH 7.4. Component 1 corresponds to SL-Bac8c2,5Leu desorbed from the CNPs surface; component 2 corresponds to the adduct SL-Bac8c2,5Leu @CNPs (peptide adsorbed onto the CNPs surface). A desorption index (DI) has been calculated as the intensity ratio of line 1 over line 2.
The SL-Bac8c2,5Leu @CNP-S system was monitored by EPR spectroscopy up to 160 h, to get information about the stability of the SL-Bac8c2,5Leu@CNPs adduct at pH 7.4. The plot of DI vs time shows that a slow peptide release occurred in the observed time range (Supporting Information, Figure S6); nevertheless, EPR data prove that a significant peptide fraction kept bound to CNPs along the whole time range. Interestingly, the comparison with a sample kept at pH 4 shows a marked peptide desorption at acidic pH value (Supporting Information, Figure S8). Because the surface of CNPs is rich in protonable residues, this evidence suggests the electrostatic nature of the interaction between Bac8c2,5Leu and CNP-S. The SL-Bac8c2,5Leu@CNP-S system is stabilized by ionic pairs involving the negatively charged residues on the CNPs surface and the positively charged side chains of the amino acids found in Bac8c2,5Leu. As pH is lowered, the rate of unprotonated groups on the CNPs surface decreases and the interaction between Bac8c2,5Leu and CNP-S is destabilized.
2.5. Stability of the SL-Bac8c2,5Leu@CNP-S Adduct in the Presence of Proteins
Bovine serum albumin (BSA) was used as a model protein to investigate protein competition with Bac8c2,5Leu peptide for the CNPs surface. In fact, albumin is the most abundant protein in plasma, and it has been shown abundant in the hard corona of CNPs.15 The competition of BSA toward SL-Bac8c2,5Leu for the adsorption sites on the surface of CNPs was investigated through EPR spectroscopy, DLS, and electrophoretic light scattering (ELS).
The SL-Bac8c2,5Leu@CNP-S system was incubated either with an equimolar amount of BSA or a 6 M excess of BSA, respectively, at the physiological pH 7.4 and at pH 4.0, which is typical of inflamed tissues. The measured DI are reported in Table S1.
At pH 4.0 the system undergoes peptide desorption to a much higher extent in the absence than in the presence of BSA; in addition, the DI values with equimolar and excess BSA are comparable (Supporting Information, Table S1). These data suggest that, at pH 4, BSA inhibits peptide desorption to some extent, possibly through the formation of a BSA layer on top of the peptide layer; this would explain the absence of any significant competition between BSA and SL-Bac8c2,5Leu for the CNPs surface. Interestingly, the EPR spectral pattern of SL-Bac8c2,5Leu is not affected by BSA adsorption: both spectral line width and correlation time are almost unchanged. Hence, BSA adsorption does not foster significant conformational changes in the adsorbed peptide (data not shown).
A rather distinct behavior is observed at pH 7.4. The DI value of the sample incubated with a molar excess of BSA is significantly higher as compared to the sample with an equimolar amount of BSA. In addition, peptide desorption in the presence of BSA is markedly higher at this pH value as compared to pH 4.0. These data suggest a competitive effect of BSA toward the peptide, not observed at pH 4.0. As BSA becomes predominantly negatively charged above its isoelectric point (IP) value (4.7), it may compete with CNPs for the peptide.
In order to confirm the molecular mechanism of interaction of BSA with CNPs, DLS and ELS analyses were performed (Supporting Information, Figure S8). The presence of BSA does not significantly modify the hydrodynamic diameter of the system, indicating the absence of agglomeration processes. On the other hand, a marked shift of the ζ-potential toward less negative values was observed, induced by the progressive covering of the nanoparticles surface by BSA. In conclusion, at neutral pH, BSA increased the rate of peptide release, whereas its release was inhibited. These results are relevant in terms of bioavailability of the peptide in different human body compartments.
2.6. In Vitro Antibacterial Efficacy of Bac8c2,5Leu@CNP-S
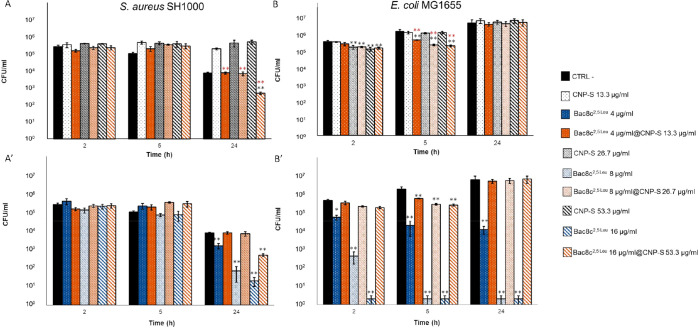
The antibacterial activities of the free peptide and of the Bac8c2,5Leu@CNP-S system toward E. coli and S. aureus were evaluated and compared with that of CNP-S alone. In Figure 6 the kinetics of the interactions (2, 5, and 24 h) at different concentrations are reported. The data expressed as logarithmic differences of bacterial viability against CTRL- and CNP-S are in Tables S2 and S3, Supporting Information. Three different concentrations of CNP-S and Bac8c2,5Leu@CNP-S (13.3, 26.7, and 53.3 μg/mL), were used, while the free Bac8c2,5Leu was tested at concentrations identical to that loaded onto the surface of the CNP-S (4, 8, and 16 μg/mL).
Figure 6.
Bactericidal effect of Bac8c2,5Leu and Bac8c2,5Leu@CNP-S against bacteria. (A, A′) S. aureus SH1000 and (B, B′) E. coli MG1655. Upper panels, Bac8c2,5Leu@CNP-S vs unloaded CNP-S; lower panels, Bac8c2,5Leu@CNP-S vs free peptide. Bacteria were grown in the presence of CNP-S (white bars) at concentrations of 53.3, 26.7, and 13.3 μg/mL or free Bac8c2,5Leu (blue bars) at concentrations equal to those loaded onto CNPs (16, 8, and 4 μg/mL, respectively), or in the presence of Bac8c2,5Leu@CNP-S system (orange bars) at the three combined concentrations of AMP and CNPs. *, p value < 0.05; **, p value < 0.01 Red stars, significance against CNP-S; black stars, significance against CTRL-.
As previously observed for the CNP-L, a statistically significant maintenance of the bacterial vitality over time compared to the negative controls was observed for S. aureus already after 5 h (0.6 log) following treatment with CNP-S, while E. coli exhibited a significant decrease of vitality after shorter time (0.35 log after 2 h) at the highest CNP-S concentration which is lost over time. This confirms a rapid interaction of the latter bacteria with CNPs (Figure 6A,B).
The free peptide was more active toward E. coli than S. aureus (Figure 6A′,B′), inducing a significant (p < 0.05) reduction of cellular viability (1 log) in comparison with the negative control already after 2 h from the inoculum already at the lowest concentration (4 μg/mL). Different reasons might account for these differences. Being positively charged, peptide Bac8c2,Leu is expected to target the negatively charged outer membrane of bacterial cells, with a lower efficiency toward Gram-negative due to their complex surface organization.21 There are, however, examples of antimicrobial peptides having high affinity for the negatively charged LPS present in large quantities in the outer membrane of Gram-negative bacteria.28 Wu and Hancock28 proposed that this electrostatic interaction is fundamental to enhance the initial approach with the bacterial cells, which promotes the permeabilization of the bacterial outer membrane leading to an uptake of the AMP. This might explain the observed higher bactericidal effects detected at shorter times on E. coli with respect to S. aureus. Moreover, the high ratio of hydrophobic to charged units in Bac8c2,Leu should benefit the anti-Gram-negative activity.29 Previous studies reported that E. coli exposed to sublethal concentrations of Bac8c peptides resulted in deleterious downstream events on the cell membrane almost immediately after the inoculum. However, the bacteria defense systems were sufficient for full recovery with time.30
Bac8c2,5Leu@CNP-S system induced a significant reduction of vitality of S. aureus (p value < 0.01) after 24 h from the inoculum at the highest concentration (Figure 6A). This effect was significantly higher than that obtained with CNP-S alone at all concentrations, but lower than the free peptide (Figure 6A′). Oppositely to the free peptide (Figure 6B′), it displayed on E. coli a very small but significant reduction of bacterial growth after a short time of exposure to the system, an effect that disappeared after 24 h of incubation (Figure 6B).
3. Discussion
Among the different approaches proposed to improve the in vivo bioavailability of antimicrobial peptides (AMPs) and to reduce their systemic toxicity, delivery systems appear to have the highest potential.7−11,31 AMPs delivery systems may be produced by encapsulation, covalent conjugation, or surface attachment of the peptide to the nanocarrier.31 In the present study, the production of delivery systems by physical adsorption of the cationic peptides with biocompatible17,19 hydrothermal carbon nanoparticles (CNPs) has been explored. Physical adsorption has the advantage with respect to covalent grafting or encapsulation because of the simpler methodology, higher yield, and, possibly, higher bioavailability of the peptide in the resulting product. CNPs are composed of elemental carbon but exhibit at the surface acidic groups that are dissociated at pH range 2–10.14 The nanoparticles are, therefore, negatively charged. The surface density of the acidic groups has been estimated to be 3 groups/nm2, leading to a highly negative ζ-potential.14
The Bac8c2,5Leu peptide was chosen not just because of its high antimicrobial activity but also for its structure. The presence of three positively charged arginine residues allows the formation of strong ionic bonds with the surface, whereas the nonpolar amino acids may interact with hydrophobic patches on the CNPs’ surface. EPR spectroscopy showed that the adsorption of the Bac8c2,5Leu peptide onto the surface of CNPs is partially reversible: this implies its slow release in solution. The presence of an adsorption/desorption equilibrium supports the hypothesis that electrostatic forces are mainly responsible for the formation of the Bac8c2,5Leu@CNP-S system; this is further confirmed by the pH dependence of the peptide release rate from the surface of CNPs. However, we cannot exclude the contribution of hydrophobic interactions, as a relevant amount of peptide keeps bound to CNPs over time. Overall, these data indicate that the stability of the Bac8c2,5Leu@CNP-S system is influenced by acid–base equilibria involving protonable moieties on the surface of CNPs. Interestingly, the system is pH responsive and exhibits a higher desorption rate of the peptide at acidic pH, which is typical of inflamed tissues.
The antimicrobial activity of the nanocarrier alone or loaded with the peptide has been investigated against Staphylococcus aureus and Escherichia coli.
CNPs synthesized in two different sizes exhibit no or transient bactericidal effect on both strains. These results were expected since the CNPs used in the present study are spherical and exhibit a smoothed, negatively charged and highly hydrophilic surface.14 In fact, 1D/2D carbon structures such as CNTs,32 graphene, or graphene oxide33 have been shown to elicit bactericidal activity by extracting phospholipids from the bacterial membranes inducing cell death, a property that is strictly related to their sheet-or needle-like shape.34
CNPs get rapidly in contact with both Gram+ and Gram– bacteria, albeit with a different rate of association, and induce the production of ECM that englobes nanoparticles. Even if SEM analysis does not allow identifying possible uptake of particles by the bacteria, it demonstrates strong interaction between CNPs and bacteria that might favor the transport of the loaded AMP close to the bacterial cells.
The Bac8c2,5Leu@CNP-S system induced a significant reduction of viability of S. aureus after 24 h from the inoculum at the highest concentration, which was cytotoxic on E. coli only at a short time of exposure. As compared to the free peptide, a decrease of activity was found with both bacteria. This might be due to the lower availability of the bonded peptide with respect to the free peptide. However, albeit the free peptide demonstrated a higher bactericidal activity against E. coli, the Bac8c2,5Leu@CNP-S system resulted to be more active against S. aureus. This result suggests a different mechanism of action involving the whole DDS. One possibility is that the high amount of ECM matrix produced by E. coli, acting as protective coating, might counterbalance Bac8c2,5Leu@CNP toxicity, thus accounting for the reversal of selectivity toward the two strains. In fact, a previous study showed that Bac8c2,5Leu peptide (albeit in the D form) was significantly less active against bacteria in biofilms rather than in their planktonic form.3 In perspective, modulation of the nanocarriers properties (such as size or surface chemistry) aimed at reducing bacteria adhesion might allow an increase of efficacy.
Nanoparticles in biological fluids are rapidly covered by a layer of proteins generally referred to as “protein-corona”.35 Depending on the surface properties of the materials and the kind of proteins employed, irreversible or reversible adsorption may occur.36 Proteins with high affinity for surfaces can mask molecules appositely bound to the surface of nanoformulations,37 or compete with adsorbed molecules for the surface adsorption sites.
Proteins are abundant in mammalian cells or extracellular matrices, bacteria, and biofilms. Therefore, the formation of a bio corona is expected, possibly affecting the stability or bioactivity of the drug delivery systems.37 In the present case, bovine serum albumin (BSA), used as model protein, affected the peptide release rate in a pH-dependent mode. In fact, the increase of peptide desorption rate at neutral pH might be explained by the competition between negatively charged BSA and CNPs for the positively charged peptide. Conversely, as BSA displays a net positive charge at acidic pH values, it may adsorb onto the Bac8c2,5Leu@CNPs system inhibiting the peptide release. These results indicate that the bioavailability of the peptide in different human body compartments is likely to be strongly affected by proteins. This suggests that strategies to avoid protein adsorption could improve the efficacy of the DDS.
4. Conclusion
The results reported herein pave the way for the development of hydrothermal CNPs-based drug delivery systems of antimicrobial cationic peptides. The results obtained suggest that the modulation of properties of CNPs to suppress bacterial and proteins adhesion might result in enhanced effectiveness of this DDS.
5. Experimental Section
5.1. Synthesis of CNP-S and CNP-L and Formulation of Bac8c2,5Leu@CNP-S
Carbon nanoparticles were produced starting from glucose using a one-step hydrothermal process as previously described by Kokalari et al.14 Briefly, 2 g of glucose was dissolved in 50 mL of ultrapure water followed by the addition of 15 mg of sodium polyacrylate. The solution was introduced in a pressure reactor system (Büchi AG) and heated at 190 °C for 3 (CNP-S) or 8 h (CNP-L). The CNPs were then purified with ultrapure water either by centrifugation for large carbon nanoparticles (CNP-L) or by tangential flow ultrafiltration (Vivaflow 50R; MW, 30 kDa) for the small carbon nanoparticles (CNP-S).
The antimicrobial peptide Bac8c2,5Leu was synthesized following the procedure described in the Experimental Section and characterized by mass spectrometry (Figure S4). The DDS was prepared by incubating 60 μg of Bac8c2,5Leu with 200 μg of CNP-S in 2 mL of 10 mM PBS, pH 7.4, under shaking at 400 rpm, for 1 h. Then, the suspension was subjected to centrifugation, 18.000 rpm for 30 min, to prove the successful loading of Bac8c2,5Leu onto the nanoparticles surface. For this purpose, the concentration of the free peptide in the supernatant was quantified by fluorescence spectroscopy. A solution of Bac8c2,5Leu in PBS, at the same concentration, was subjected to the identical treatment (incubation and centrifugation) and used as control.
5.2. Hydrodynamic Diameter Distribution, ζ-Potential and SEM
The hydrodynamic diameter of CNPs and peptide-loaded CNPs was evaluated by using dynamic light scattering technique (DLS, ZetaSizer Nano, Malvern, U.K.), while the ζ-potential was determined using electrophoretic light scattering (ELS, ZetaSizer Nano). The results were expressed as mean hydrodynamic diameter (dH), dH distribution, Polydispersion Index (PDI) or mean ζ-potential. The PDI is a dimensionless measure of the degree of polydispersion, and it is calculated by the cumulative analysis of the autocorrelation function. It ranges from 0 to 1. Nanoparticle tracking analysis (NTA) was performed by using a Nanosight NS300 (Malvern, U.K.) instrument equipped with a blue laser (488 nm).
The size distribution of CNPs was evaluated by scanning electron microscopy. A 10 μL aliquot of the CNPs suspension was spotted on virgin silicon wafers, which were previously cleaned in hydrofluoric acid 15% and left to air-dry. The images were acquired using a SEM FEI Inspect F in UHV with an acceleration potential of 10 kV, with a spot of 3.5 and a magnification of 10000×. The mean size was measured as the mean of at least 50 particles.
5.3. Synthesis and Characterization of Bac8c2,5Leu and SL-Bac8c2,5Leu
The antimicrobial peptide sequence (H-RLWVLWRR-NH2) was synthesized at a 0.1 mmol scale, following the protocol described in [Forde 2014]. The l-form of the peptide was chosen because of its lower production cost with respect to the d-form.
Peptide synthesis was based on the classical 9-fluorenylmethoxycarbonyl (Fmoc) method, by performing a high-efficiency solid-phase peptide synthesis process (HE-SPPS) on a Liberty Blue automated microwave peptide synthesizer (CEM Corp., Buckingham, U.K.). A rink amide MBHA (4-methylbenzhydrylamine) resin (Novabiochem, Germany) and l-amino acids (Fmoc-l-Arg(Pbf)–OH, Fmoc-l-Leu-OH, Fmoc-l-Val-OH, Fmoc-l-Trp(Boc)–OH, from CEM Corp.) were used. The couplings of amino acids were performed in DMF using DIC/OxymaPure activation. Fmoc removal was done using a solution of 20% piperidine in DMF. The cleavage used to deprotect and remove the peptide for the synthesis resin was performed manually at room temperature for 4 h, using a cleavage cocktail composed by 80% trifluoroacetic acid, 5% thioanisole, 5% H2O, 5% ethanedithiol, and 5% triisopropylsilane.
Following cleavage, the crude peptide was precipitated and washed twice with diethyl ether, dried, dissolved in H2O, and freeze-dried. The lyophilized powder was stored at −20 °C. The peptide was then purified by reverse-phase high-performance liquid chromatography (RP-HPLC) on a Shimazu CBM-20A, equipped with a photodiode array detector SPD-M20A.
The purified peptide was characterized by analytical HPLC and by mass spectroscopy (ESI+-MS, Advion CMS).
HPLC buffers used were mobile phase A (0.1% trifluoroacetic acid [TFA] in water) and mobile phase B (0.1% TFA in acetonitrile) with a gradient of 5–65% buffer B in 18 column volumes (analytical) or 5 column volumes (semipreparative) with a flow rate of 1 mL/min (analytical) or 5 mL/min (semipreparative) and main wavelength detection at 214 nm.
To determine an easy and low-cost method for the quantification of Bac8c2,5Leu in aqueous solution, the peptide was tested for its possible fluorescence, due to the presence of tryptophan in its structure. The calibration curve in aqueous media was built on the basis of the fluorescence spectra registered at different concentrations using a Varian Cary Eclipse fluorescence spectrophotometer. Fluorescence was tested in ultrapure water and PBS, and in both cases, it well correlates with the concentration.
5.4. Labeling Protocol
The following solutions were prepared: (i) 2.0 mg of peptide Bac8c2,5Leu dissolved in 780 μL of a 1:1 (v/v) mixture of acetonitrile/50 mM borate buffer, pH 10; (ii) 3.5 mg spin–label (SL) dissolved in 610 μL of acetonitrile. Solutions i and ii were mixed and 610 μL of 50 mM borate buffer, pH 10, were further added to the mixture. The labeling mixture was kept under stirring in the dark, overnight, at RT and subsequently lyophilized; the powder was resuspended in a 1:1 (v/v) mixture of acetonitrile/10 mM PBS, pH 7.4, up to a final concentration of 1.21 mg/mL (raw batch of SL-Bac8c2,5 Leu). Acetonitrile was employed because spin–labeling turned out to lower the water solubility of the peptide. Conjugation of SL to Bac8c2,5Leu was verified by ESI+-MS (ESI+-MS, Orbitrap Fusion, Thermo Fisher).
5.5. Investigation of the CNPs/AMP Interaction by SDSL-EPR Spectroscopy
In order to enable EPR investigations on the interaction between peptide Bac8c2,5Leu and CNPs, the spin–label (SL) 1-oxyl-2,2,5,5-tetramethylpyrroline-3-carboxylate N-hydroxysuccinimide ester (Acros Organics) was conjugated to the N-terminus of the peptide according to the protocol described in the Supporting Information to obtain the labeled peptide SL-Bac8c2,5Leu.
5.5.1. Formulation of SL-Bac8c2,5Leu@CNP-S
The adduct SL-Bac8c2,5Leu@CNP-S was prepared according to the following protocol: 50 μL of raw batch SL-Bac8c2,5Leu was incubated with 100 μg of carbon nanoparticles in 2 mL of 10 mM PBS pH 7.4 for 1 h at 37 °C under stirring. To get rid of the unconjugated spin–label, which would interfere with EPR measurements, the mixture was subsequently dialyzed against 10 mM PBS, pH 7.4 (tube cutoff, 14 kDa; Sigma-Aldrich) for 20 h, with three buffer changes. Samples were finally concentrated by centrifugation on Vivaspin 500 (cutoff, 10 KDa) and resuspended in 180 μL of PBS buffer, before undergoing EPR analysis.
5.5.2. EPR Spectra of SL-Bac8c2,5Leu@CNP-S
EPR measurements were performed in a flat cell at RT on a ESP300E Bruker X-band machine equipped with a 4103 cylindrical cavity. The following instrumental setting was employed: microwave frequency, 9.3 GHz; modulation amplitude, 1 G; modulation frequency, 100 kHz; microwave power, 5 mW; time constant 163 ms; 30 scans.
5.5.3. Desorption Kinetics of SL-Bac8c2,5Leu@CNP-S
Samples of SL-Bac8c2,5Leu@CNP-S obtained by incubation for 1 h at 37 °C in 10 mM PBS at pH 7.4 (as previously described) underwent dialysis against PBS buffer at three different pH values: 4.0, 7.4, and 9.0 for 20 h. Samples were centrifuged and resuspended against the same buffer employed for dialysis, according to the above-described protocol. The samples were subsequently monitored by EPR spectroscopy along 160 h.
5.6. In Vitro Characterization of the Interaction of CNPs with Bacteria
5.6.1. Bacterial Strains and Cultivation
Frozen stock cultures of Escherichia coli MG1655 and of Staphylococcus aureus SH1000 were revitalized on Muller Hilton agar (MHA) and a single-well separated colony was selected from each culture, suspended in 5 mL of MH broth, and allowed to grow overnight at (37 ± 1) °C under agitation at 150 rpm. The optical density of each bacterial suspension was measured using UV–vis spectrophotometer (Lange DR500) in the single wavelength mode at 600 nm (OD600). MH culture medium without bacterial inoculation was employed as blank, and the OD600 was adjusted to 0.05 in MH broth. Then bacteria were allowed to grow at 37 ± 1 °C under agitation (150 rpm) at least for 1.5 h until they both reached an OD600 of about 0.1, corresponding to 1 × 108 CFUs/ml. Finally, they were inoculated in all of the samples to be tested in a final concentration of 1 × 105 CFU/mL.
5.6.2. Bacterial Viability Assay
A 1 mL aliquot of the synthesis batch of CNP-L was sonicated at 180 W for 20 min at 37000 Hz and was diluted in six Falcon conical tubes containing 5 mL of MH broth to obtain final CNPs concentrations of 53.3, 26.7, and 13.3 μg/mL in duplicate, one for each bacterial strain. Three Falcon conical tubes containing each one of the three different CNPs concentrations were inoculated with S. aureus SH1000, prepared as described in Section 5.6.1, to obtain a final bacterial concentration of 1 × 106 CFU/ml. The remaining three samples were inoculated with E. coli MG1655 at the same final concentration of S. aureus. All of the samples were incubated at 37 ± 1 °C under agitation (150 rpm) for 24 h. Then, 1 mL of each sample was collected and serially diluted in PBS, plated on MHA, and incubated overnight at 37 ± 1 °C. Bacterial colonies were counted, and CFUs/mL were plotted against CNPs concentration.
5.6.3. Bacteria–CNPs Kinetic Interaction Assay
Measurements of the association between bacteria and CNPs were conducted dynamically by a combination of alternating voltage dielectrophoresis (DEP) and Raman microspectroscopy. A cell to conduct DEP–Raman experiments, previously described by Barzan et al.,26 was employed to manipulate bacteria in liquid by DEP to maximize their Raman signal by locally concentrating suspended bacteria in the microscope focal volume. Samples of the three selected concentrations of CNP-L (13.3, 26.7, 53.3 μg/mL) and a negative control with PBS only were inoculated with 1 × 105 CFUs/mL of the two bacterial strains and incubated at 37 ± 1 °C under agitation (150 rpm). Samples with CNPs only, without bacteria, were also prepared in the same way and analyzed as controls. An aliquot of each sample was collected after 0 h, 24 and 48 h to be analyzed with the DEP-Raman method to investigate CNP–bacteria interaction. Before every DEP–Raman measurement, bacteria were precipitated at 3000 rpm for 5 min and 5-fold concentrated to reach the limit of detection of the technique. Bacteria were then washed twice at 14000 rpm for 1.30 min with PBS 0.5×, resuspended in 200 μL of this buffer and 100 μL of each sample were injected in the DEP cell. The agglomeration conditions of the DEP cell for E. coli were 5 V peak-to-peak sinusoidal voltage between the electrodes with a frequency of 800 kHz, while for S. aureus the same waveform with an amplitude of 4 V peak-to-peak and a frequency of 1 MHz was employed; for both strains, the accumulation time before Raman measurements was 6 min. The Raman microspectrometer was a Thermo Fisher Scientific DXR dispersive Raman microscope; the acquisition conditions were a 532 nm Nd:YAG excitation laser radiating 10 mW power at the sample, an Olympus 60× water immersion microscope objective with 1.1 NA (model LUMFLN60XW). The integration times for each Raman spectrum were 60 scans of 2.5 s each (2.5 min total integration time per spectrum).
5.6.4. SEM Characterization
The interaction between bacteria and CNPs was analyzed also by scanning electron microscopy to better appreciate bacterial behavior in the presence of the nanoparticles. Aliquots of the same samples prepared for DEP–Raman analysis were collected after 0 and 24h from the inoculum. Bacteria and the interacting CNPs were precipitated by centrifugation at 3000 rpm for 5 min. Then, the supernatant was discarded, and bacteria were washed resuspending them in 500 μL of ultrapure water and by a second centrifugation at 14000 rpm for 1.30 min. The pellets were resuspended in 40 μL of ultrapure water and 10 μL were spotted on virgin silicon wafers, which were previously cleaned in hydrofluoric acid 15% and left to air-dry. The images were acquired using a SEM FEI Inspect F in UHV with an acceleration potential of 10 kV, with a spot of 3.5 and a magnification of 10000x.
5.7. In Vitro Testing of the Antimicrobial Activity of Bac8c2,5Leu@CNPs
To assess the bactericidal potential of the novel DDS Bac8c2,5Leu@CNPs over time, a time-kill kinetic assay has been performed in line with the standard procedure developed by the Clinical and Laboratory Standards Institute (CLSI), USA.23 The three CNPs concentrations used for the bacteria interaction (53.3, 26.7, and 13.3 μg/mL) corresponded to 16, 8, and 4 μg/mL of the Bac8c2,5 Leu, respectively. The same concentrations of the antimicrobial peptide (AMP) alone and of the CNPs unloaded were tested. All the CNPs suspensions were prepared in PBS and incubated at 37 ± 1 °C for 1 h under agitation at 150 rpm. The bactericidal activity of the nanosystem was tested toward the two selected bacteria prepared as described before.
For each bacterial strain, an inoculum of 1.0 × 105 CFU/mK was added to all the suspensions (the DDS, the AMP only and the CNPs unloaded) including a sample of PBS as negative control. For all samples 2 mL aliquots were placed in triplicate in 12 well plates and were incubated at 37 ± 1 °C, under orbital shaking at 150 rpm. Aliquots of 100 μL of each sample were taken at time intervals of 2, 5, and 24 h, serially diluted in PBS, spread aseptically on Muller Hinton agar plates (Sigma-Aldrich), and finally incubated overnight at 37 ± 1 °C. Then, the colony forming units were counted. The CFU/mL were plotted against their collection time point.
5.8. Statistical Analysis
In all of the microbiological vitality assays the statistical significance of the differences between the mean of the CFU counted in all of the experimental replicates between the samples tested and the negative controls were determined performing a t test comparing the means of the negative control and one sample at time obtaining the relative p-values.
5.9. Interaction of SL-Bac8c2,5Leu@CNP-S with Proteins
A 50 μL aliquot of SL-Bac8c2,5Leu was incubated with 100 μg of carbon nanoparticles in 1 mL of 10 mM PBS pH 7.4, under stirring at 400 rpm, for 1 h at 37 ± 1 °C. Afterward, each sample was further incubated with BSA either at equimolar concentration with the peptide (60 μg/mL) or in large excess (50 mg/mL) for 1 h at 37 ± 1 °C, under stirring. A control sample without BSA was also prepared according to the same protocol. To get rid of the unbound BSA or peptide, the samples were dialyzed in float-A-lyzer G2 tubes (cutoff, 1000 kDa) against 10 mM PBS, pH 7.4, for 42 h, with three buffer changes. The samples were finally concentrated by centrifugation on Vivaspin 500 (cutoff, 10 kDa) and resuspended in 180 μL of PBS buffer, before undergoing EPR, DLS, and ELS analysis.
Acknowledgments
This work was funded by the Science Foundation Ireland (SFI) under Equipment Grant No. 06/RFP/CHO024/602 EC07 for the peptide synthesis equipment. I.K. was recipient of a fellowship from the Compagnia di San Paolo (Bando per il finanziamento Ex-Post di progetti di ricerca di Ateneo—Anno 2018), Italy. We thank Ms. Siobhan O’Flaherty for her assistance in the synthesis and characterization of the peptides.
Glossary
Abbreviations
- AMPs
antimicrobial peptides
- BSA
bovine serum albumin
- CNMs
carbon-based nanomaterials
- CNPs
carbon nanoparticles
- dH
hydrodynamic diameters
- DLS
dynamic light scattering technique
- ECM
extracellular matrix
- ELS
electrophoretic light scattering
- EPR
electroparamagnetic resonance
- IP
isoelectric point
- nDEP
negative dielectrophoresis
- NTA
nanoparticle tracking analysis
- OD
optical density
- PBS
phosphate buffer saline
- PDI
polydispersion index
- SEM
scanning electron microscopy
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00305.
(Table S1) Desorption indexes; (Tables S2 and S3) analysis of the logarithmic differences of bacterial viability; (Figure S1) viability assays; (Figure S2) effects of CNP-S on vitality; (Figure S3) Raman spectra; (Figure S4) chromatograms and mass spectra; (Figure S5) size distributions; (Figure S6) peptide loading of CNP-S; (Figure S7) desorption kinetics; (Figures S8 and S9) effects of pH on desorption and BSA on hydrodynamic diameter and z-potential (PDF)
Author Present Address
# Department of Chemical Engineering, Delft University of Technology, Van der Maasweg, 92629 HZ Delft, The Netherlands
Author Contributions
⊥ G.B. and I.K. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- World Health Organization . Top 10 causes of death, December 9, 2020; https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed 2021-09-27).
- Roberts S. C.; Zembower T. R. Global increases in antibiotic consumption: a concerning trend for WHO targets. Lancet Infect. Dis. 2021, 21, 10–11. 10.1016/S1473-3099(20)30456-4. [DOI] [PubMed] [Google Scholar]
- Zapotoczna M.; Forde É.; Hogan S.; Humphreys H.; O’gara J. P.; Fitzgerald-Hughes D.; Devocelle M.; O’Neill E. Eradication of staphylococcus aureus biofilm infections using synthetic antimicrobial peptides. J. Infect. Dis. 2017, 215, 975–983. 10.1093/infdis/jix062. [DOI] [PubMed] [Google Scholar]
- Lombardi L.; Maisetta G.; Batoni G.; Tavanti A. Insights into the antimicrobial properties of hepcidins: Advantages and drawbacks as potential therapeutic agents. Molecules 2015, 20, 6319–6341. 10.3390/molecules20046319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P. Y.; Khanum R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. 10.1016/j.jmii.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Sahl H. G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Devocelle M. Targeted antimicrobial peptides. Front. Immunol. 2012, 3, 309. 10.3389/fimmu.2012.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney E. F.; Straus S. K.; Hancock R. E. W. Reassessing the Host Defense Peptide Landscape. Front. Chem. 2019, 7, 43. 10.3389/fchem.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayton M.; Kizhakkedathu J. N.; Straus S. K. Towards Robust Delivery of Antimicrobial Peptides to Combat Bacterial Resistance. Molecules 2020, 25, 3048. 10.3390/molecules25133048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayton M.; Deisinger J. P.; Ludwig K. C.; Raheem N.; Müller A.; Schneider T.; Straus S. K. Host Defense Peptides: Dual Antimicrobial and Immunomodulatory Action. J. Mol. Sci. 2021, 22, 11172. 10.3390/ijms222011172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.; Ma Q.; Chen X.; Chen T.; Ying Y.; Xi X.; Wang L.; Ma C.; Shaw C.; Zhou M. Recent Advances and Challenges in Nanodelivery Systems for Antimicrobial Peptides (AMPs). Antibiotics 2021, 10, 990. 10.3390/antibiotics10080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.; Shen H.; Mao J.; Zhang L.; Jiang Z.; Sun T.; Lan Q.; Zhang Z. Transferrin modified graphene oxide for glioma-targeted drug delivery: In vitro and in vivo evaluations. ACS Appl. Mater. Interfaces 2013, 5, 6909–6914. 10.1021/am402128s. [DOI] [PubMed] [Google Scholar]
- Xu G.; Liu S.; Niu H.; Lv W.; Wu R. Functionalized mesoporous carbon nanoparticles for targeted chemo-photothermal therapy of cancer cells under near-infrared irradiation. RSC Adv. 2014, 4, 33986–33997. 10.1039/C4RA03993A. [DOI] [Google Scholar]
- Kokalari I.; Gassino R.; Giovannozzi A. M.; Croin L.; Gazzano E.; Bergamaschi E.; Rossi A. M.; Perrone G.; Riganti C.; Ponti J.; Fenoglio I. Pro- and anti-oxidant properties of near-infrared (NIR) light responsive carbon nanoparticles. Free Radic. Biol. Med. 2019, 134, 165–176. 10.1016/j.freeradbiomed.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Soddu L.; Trinh D. N.; Dunne E.; Kenny D.; Bernardini G.; Kokalari I.; Marucco A.; Monopoli M. P.; Fenoglio I. Identification of physicochemical properties that modulate nanoparticle aggregation in blood. Beilstein J. Nanotechnol. 2020, 11, 550–567. 10.3762/bjnano.11.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokalari I.; Keshavan S.; Rahman M.; Gazzano E.; Barzan G.; Mandrile L.; Giovannozzi A. M.; Ponti J.; Antonello G.; Monopoli M.; Perrone G.; Bergamaschi E.; Riganti C.; Fadeel B.; Fenoglio I. Efficacy, biocompatibility and degradability of carbon nanoparticles for photothermal therapy of lung cancer. Nanomedicine 2021, 16, 689–707. 10.2217/nnm-2021-0009. [DOI] [PubMed] [Google Scholar]
- Komp Lindgren P.; Karlsson Å.; Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob. Agents Chemother. 2003, 47, 3222–3232. 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M.; Mennitti C.; Laneri S.; Franco A.; De Biasi M. G.; Cesaro A.; Fimiani F.; Moscarella E.; Gragnano F.; Mazzaccara C.; Limongelli G.; Frisso G.; Lombardo B.; Pagliuca C.; Colicchio R.; Salvatore P.; Calabrò P.; Pero R.; Scudiero O. Methicillin-resistant staphylococcus aureus: Risk for general infection and endocarditis among athletes. Antibiotics 2020, 9, 332. 10.3390/antibiotics9060332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde É.; Humphreys H.; Greene C. M.; Fitzgerald-Hughes D.; Devocelle M. Potential of host defense peptide prodrugs as neutrophil elastase-dependent anti-infective agents for cystic fibrosis. Antimicrob. Agents Chemother. 2014, 58, 978–985. 10.1128/AAC.01167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilpert K.; Volkmer-Engert R.; Walter T.; Hancock R. E. W. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005, 23, 1008–1012. 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- Romeo D; Skerlavaj B; Bolognese M; Gennaro R Structure and bactericidal activity of an antibiotic dodecapeptide purified from bovine neutrophils. J. Biol. Chem. 1988, 263, 9573–9575. 10.1016/S0021-9258(19)81553-3. [DOI] [PubMed] [Google Scholar]
- Noviello S.; Ianniello F.; Leone S.; Esposito S. Comparative in vitro bacteriostatic and bactericidal activity of levofloxacin and ciprofloxacin against urinary tract pathogens determined by MIC, MBC, Time-kill curves and bactericidal index analysis. Infez. Med. 2002, 2, 100–106. [PubMed] [Google Scholar]
- Zhang L.; Petersen E. J.; Habteselassie M. Y.; Mao L.; Huang Q. Degradation of multiwall carbon nanotubes by bacteria. Environ. Pollut. 2013, 181, 335–339. 10.1016/j.envpol.2013.05.058. [DOI] [PubMed] [Google Scholar]
- You Y.; Das K. K.; Guo H.; Chang C. W.; Navas-Moreno M.; Chan J. W.; Verburg P.; Poulson S. R.; Wang X.; Xing B.; Yang Y. Microbial Transformation of Multiwalled Carbon Nanotubes by Mycobacterium vanbaalenii PYR-1. Environ. Sci. Technol. 2017, 51, 2068–2076. 10.1021/acs.est.6b04523. [DOI] [PubMed] [Google Scholar]
- Chen M.; Qin X.; Zeng G. Biodegradation of Carbon Nanotubes, Graphene, and Their Derivatives. Trends Biotechnol. 2017, 35, 836–846. 10.1016/j.tibtech.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Barzan G.; Sacco A.; Mandrile L.; Giovannozzi A. M.; Brown J.; Portesi C.; Alexander M. R.; Williams P.; Hardie K. R.; Rossi A. M. New frontiers against antibiotic resistance: A Raman-based approach for rapid detection of bacterial susceptibility and biocide-induced antibiotic cross-tolerance. Sens. Actuators, B 2020, 309, 127774. 10.1016/j.snb.2020.127774. [DOI] [Google Scholar]
- McEwen G. D.; Wu Y.; Zhou A. Probing nanostructures of bacterial extracellular polymeric substances versus culture time by Raman microspectroscopy and atomic force microscopy. Biopolymers 2010, 93, 171–177. 10.1002/bip.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.; Hancock R. E. W. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 1999, 274, 29–35. 10.1074/jbc.274.1.29. [DOI] [PubMed] [Google Scholar]
- Haug B. E.; Stensen W.; Stiberg T.; Svendsen J. S. Bulky nonproteinogenic aminoacids permit the design of very small and effective cationic antibacterial peptides. J. Med. Chem. 2004, 47, 4159–4162. 10.1021/jm049582b. [DOI] [PubMed] [Google Scholar]
- Spindler E. C.; Hale J. D. F.; Giddings T. H.; Hancock R. E. W.; Gill R. T. Deciphering the mode of action of the synthetic antimicrobial peptide bac8c. Antimicrob. Agents Chemother. 2011, 55, 1706–1716. 10.1128/AAC.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Hong T.; Cui P.; Wang J.; Xia J. Antimicrobial peptides towards clinical application: Delivery and formulation. Adv. Drug Delivery Rev. 2021, 175, 113818. 10.1016/j.addr.2021.05.028. [DOI] [PubMed] [Google Scholar]
- Kang S.; Pinault M.; Pfefferle L. D.; Elimelech M. Single-walled carbon nanotubes exhibit strong antimicrobial activity. Langmuir 2007, 23, 8670–8673. 10.1021/la701067r. [DOI] [PubMed] [Google Scholar]
- Zhao C.; Deng B.; Chen G.; Lei B.; Hua H.; Peng H.; Yan Z. Large-area chemical vapor deposition-grown monolayer graphene-wrapped silver nanowires for broad-spectrum and robust antimicrobial coating. Nano Res. 2016, 9, 963–973. 10.1007/s12274-016-0984-2. [DOI] [Google Scholar]
- Tu Y.; Lv M.; Xiu P.; Huynh T.; Zhang M.; Castelli M.; Liu Z.; Huang Q.; Fan C.; Fang H.; Zhou R. Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets. Nat. Nanotechnol. 2013, 8, 594–601. 10.1038/nnano.2013.125. [DOI] [PubMed] [Google Scholar]
- Lundqvist M.; Stigler J.; Elia G.; Lynch I.; Cedervall T.; Dawson K. A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 14265–14270. 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio I.; Fubini B.; Ghibaudi E. M.; Turci F. Multiple aspects of the interaction of biomacromolecules with inorganic surfaces. Adv. Drug Delivery Rev. 2011, 63, 1186–209. 10.1016/j.addr.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Åberg C.; Mahon E.; Dawson K. A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.