Abstract
Comparative sequence analysis of a 16S rRNA gene clone library from the chemocline of the meromictic Lake Cadagno (Switzerland) revealed the presence of a diverse number of phototrophic sulfur bacteria. Sequences resembled those of rRNA of type strains Chromatium okenii DSM169 and Amoebobacter purpureus DSM4197, as well as those of four bacteria forming a tight cluster with A. purpureus DSM4197 and Lamprocystis roseopersicina DSM229. In situ hybridization with fluorescent (Cy3 labeled) oligonucleotide probes indicated that all large-celled phototrophic sulfur bacteria in the chemocline of Lake Cadagno were represented by C. okenii DSM169, while small-celled phototrophic sulfur bacteria consisted of four major populations with different distribution profiles in the chemocline indicating different ecophysiological adaptations.
Lake Cadagno is an alpine lake situated 1,923 m above sea level in the Piora valley in the south of Switzerland (46°33′N, 8°43′E). The lake has a surface area of 26 by 105 m2 and a maximum depth of 21 m. Due to the infiltration of water through dolomite rich in gypsum, Lake Cadagno is a meromictic lake characterized by a high salinity of the monimolimnion and a permanent chemocline at a depth of between 9 and 14 m separating the aerobic epilimnion from the anaerobic, sulfidogenic hypolimnion (22, 33). A turbidity maximum in this chemocline suggests that a bacterial community making use of energy gradients is present. It is assumed that such a community may mainly consist of sulfate-reducing bacteria (10) and lithotrophic sulfur bacteria (14, 17), as well as phototrophic sulfur bacteria if light reaches the anoxic part of the lake (11, 18, 21).
Since many of these organisms require complex gradients of, e.g., light, sulfide, oxygen, and pH for optimal growth (24), studies of their ecology are often impeded by the fact that they are difficult to isolate or even resist cultivation, which is an essential prelude to characterization by traditional laboratory methods. Therefore, studies often focus on bacteria with distinctive morphological features which can be analyzed by light or epifluorescence microscopy after staining with fluorochromes such as acridine orange (13) or 4′-6-diamidino-2-phenylindole (DAPI) (12, 23). This approach, however, is restricted to a few organisms with highly distinctive morphologies. Based on a combination of cell size and autofluorescence as distinctive parameters for different populations of phototrophic sulfur bacteria, the major populations in the chemocline of Lake Cadagno, for example, are identified as Chromatium okenii and Amoebobacter purpureus (7, 22).
The aim of our study was to analyze populations of phototrophic sulfur bacteria in the chemocline of Lake Cadagno by molecular methods. These studies were based on comparative sequence analysis of a 16S rRNA gene clone library (19, 29) from the chemocline and 16S ribosomal DNA (rDNA) of phototrophic sulfur bacteria, including C. okenii DSM169, Lamprocystis roseopersicina DSM229, A. purpureus DSM4197, and Amoebobacter roseus DSM235. Specific oligonucleotide probes were subsequently designed and used to enumerate specific populations of phototrophic sulfur bacteria in the chemocline.
Sequence analysis.
Nucleic acids from C. okenii DSM169, L. roseopersicina DSM229, A. purpureus DSM4197, and A. roseus DSM235, as well as from bacterioplankton in water samples from the chemocline, were isolated by the method described by Ausubel et al. (4). 16S rRNA genes were amplified by PCR on a Perkin-Elmer (Rotkreuz, Switzerland) GeneAmp 2400 thermal cycler with primers UNI16SRNA-3F (5′ ATT CTA GAG TTT GAT CAT GGC TCA) and UNI16SRNA-1419R (5′ ATG GTA CCG TGT GAC GGG CGG TGT GTA), respectively. Thirty-five rounds of temperature cycling (94°C for 30 s, 52°C for 30 s, and 72°C for 1 min) were followed by a final 7-min incubation at 72°C. Amplified fragments were cloned into Phagemidvector Bluescript and transformed into Escherichia coli TOP-10 (Stratagene, Heidelberg, Germany). Plasmid DNA was extracted and purified with the QIAprep-spin plasmid purification procedure kit (Qiagen, Inc., Chatsworth, Calif.). Three hundred fifty clones of the rDNA clone library were screened by restriction analysis with the endonucleases HaeIII and MboI (Promega, Wallisellen, Switzerland), respectively, to determine phylotype distribution (16). rDNA of representative clones of the phylotypes as well as of C. okenii DSM169, L. roseopersicina DSM229, A. purpureus DSM 4197, and A. roseus DSM235 was reamplified, and the amplification products were purified by using Centri-Sep columns (Princeton Separations, Inc., Adelphia, USA) and sequenced with an ABI PRISM Ready Reaction dye deoxy terminator cycle sequencing kit and an ABI Prism 310 automated sequencer (Perkin-Elmer).
High similarity values were obtained for sequences of clones compared to sequences of representative cultures of phototrophic sulfur bacteria (Table 1) when the phylogenetic positions of C. okenii DSM169, L. roseopersicina DSM229, A. purpureus DSM4197, A. roseus DSM235 and selected clones after EMBL/GenBank database searches with FASTA (20) were preceded by alignment of sequences of selected bacteria by using the CLUSTAL W program (version 1.6) (30). Clone 359 showed a nearly identical sequence to C. okenii DSM169, differing in just one base, whereas clone 345 showed a sequence identical to that of A. purpureus DSM4197. A high similarity value of 99.2% was also obtained for rRNA sequences of A. purpureus DSM4197 and L. roseopersicina DSM229. Sequences of representative clones of four phylotypes, clones 136, 261, 335, and 371, respectively, displayed similarity values of between 98.3 and 99.9% to each other and to sequences obtained from both A. purpureus DSM4197 and L. roseopersicina DSM229. This indicated the occurrence of a phylogenetically very tight cluster of different phototrophic sulfur bacteria in the chemocline of Lake Cadagno. Similarity values of all clones to other phototrophic sulfur bacteria such as, e.g., A. roseus, Chromatium vinosum, Thiorhodococcus minus, or Thiorhodococcus gelatinosa were generally below 95% (Table 1).
TABLE 1.
Percent similarity between the 16S rRNAs of clones from the chemocline of Lake Cadagno and those of closely related organismsa
| Organism (accession no.) | % Similarity to 16S rRNA of organism:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| 1 Clone 359 (AJ006221) | 100 | |||||||||||||||||||
| 2 Clone 136 (AJ006057) | 93.5 | 100 | ||||||||||||||||||
| 3 Clone 261 (AJ006058) | 93.4 | 98.5 | 100 | |||||||||||||||||
| 4 Clone 335 (AJ006059) | 94.1 | 96.4 | 95.0 | 100 | ||||||||||||||||
| 5 Clone 345 (AJ006060) | 93.5 | 98.3 | 99.3 | 95.3 | 100 | |||||||||||||||
| 6 Clone 371 (AJ006061) | 94.2 | 96.5 | 95.1 | 99.9 | 95.3 | 100 | ||||||||||||||
| 7 Amoebobacter purpureus (AJ223235) | 93.6 | 98.4 | 99.4 | 95.3 | 100 | 95.3 | 100 | |||||||||||||
| 8 Amoebobacter roseus (AJ006062) | 93.4 | 94.2 | 94.7 | 94.7 | 94.5 | 94.7 | 94.5 | 100 | ||||||||||||
| 9 Lamprocystis roseopersicina (AJ006063) | 93.4 | 98.8 | 99.3 | 95.3 | 99.1 | 95.4 | 99.2 | 94.4 | 100 | |||||||||||
| 10 Chromatium okenii (AJ223234) | 100 | 93.5 | 93.4 | 94.1 | 93.5 | 94.2 | 93.6 | 93.4 | 93.4 | 100 | ||||||||||
| 11 Chromatium purpuratum (Af001581) | 91.4 | 92.9 | 92.7 | 92.3 | 92.3 | 92.5 | 92.3 | 91.7 | 92.4 | 91.4 | 100 | |||||||||
| 12 Chromatium tepidum (M59150) | 91.5 | 90.2 | 90.5 | 90.9 | 90.0 | 91.0 | 90.1 | 92.1 | 90.2 | 91.5 | 88.7 | 100 | ||||||||
| 13 Chromatium vinosum (M26629) | 94.1 | 92.9 | 92.8 | 93.5 | 92.7 | 93.5 | 92.8 | 93.4 | 92.8 | 94.1 | 92.2 | 93.4 | 100 | |||||||
| 14 Thiorhodococcus minus (Y11316) | 93.2 | 94.3 | 93.9 | 93.7 | 94.2 | 93.8 | 94.3 | 93.7 | 94.2 | 93.2 | 93.6 | 90.9 | 94.8 | 100 | ||||||
| 15 Thiocystis gelatinosa (Y11317) | 95.3 | 93.9 | 94.2 | 94.2 | 93.7 | 94.2 | 93.7 | 94.3 | 93.7 | 95.2 | 91.8 | 91.7 | 94.8 | 93.0 | 100 | |||||
| 16 Thiocystis violacea (Y11315) | 94.7 | 93.9 | 93.9 | 94.4 | 93.6 | 94.6 | 93.7 | 93.5 | 93.9 | 94.7 | 91.3 | 92.8 | 94.6 | 93.7 | 95.4 | 100 | ||||
| 17 Rhabdochromatium marinum (X11316) | 92.3 | 93.3 | 93.9 | 92.9 | 93.6 | 92.8 | 93.7 | 93.3 | 93.4 | 92.3 | 92.0 | 90.8 | 92.4 | 93.7 | 92.6 | 93.1 | 100 | |||
| 18 Symbiont in I. leukodermatus (U24110) | 90.3 | 91.9 | 92.4 | 91.4 | 92.1 | 91.4 | 92.1 | 91.4 | 92.1 | 90.3 | 92.0 | 89.1 | 91.6 | 92.7 | 90.8 | 91.9 | 92.7 | 100 | ||
| 19 Methylocaldum szegediense (U89300) | 89.2 | 89.8 | 90.1 | 89.8 | 90.1 | 89.6 | 90.2 | 89.3 | 89.9 | 89.2 | 88.4 | 89.1 | 89.4 | 89.5 | 89.0 | 89.9 | 89.6 | 89.3 | 100 | |
| 20 Pseudomonas aeruginosa (M34133) | 87.3 | 87.2 | 87.6 | 87.1 | 87.6 | 87.1 | 87.6 | 87.4 | 87.6 | 87.3 | 86.9 | 85.7 | 88.2 | 87.7 | 86.8 | 87.7 | 87.4 | 88.3 | 88.0 | 100 |
Similarity was calculated from data obtained from the EMBL nucleotide sequence database by using the GAP program with a creation penalty of 5.0 and an extension penalty of 0.3.
The formation of a tight cluster of clones 136, 261, and 345 with sequences from A. purpureus and L. roseopersicina, with clones 371 and 335 still closely related to this cluster, was further shown in a phylogenetic tree which was calculated by the neighbor-joining method (26) in CLUSTAL W (Fig. 1). These results suggested that small-celled phototrophic sulfur bacteria in the chemocline of Lake Cadagno consisted of at least four major populations, while large-celled phototrophic sulfur bacteria were represented only by C. okenii.
FIG. 1.
Neighbor-joining tree based on the aligned sequences of selected clones from the 16S rRNA gene library of the chemocline of Lake Cadagno and of selected bacteria searched from the EMBL/GenBank databases by FASTA through the GCG package. The distance scale indicates the expected number of changes per sequence position. Bars and probe designations indicate target groups of phototrophic sulfur bacteria for specific oligonucleotide probes.
Probe design.
In order to investigate the significance of the sequence information obtained from the environment, specific oligonucleotide probes were designed that allowed us to detect all sequences within the cluster representing the small-celled phototrophic sulfur bacteria (Fig. 1). All probes were targeting 16S rRNA at the position 453 to 478 according to the E. coli numbering (5). Sequence comparison in this highly variable region revealed at least five differences on a small stretch of 26 bases between sequences of the phototrophic sulfur bacteria investigated which allowed us to design highly specific probes (Table 2). Probe Apur453 targeted A. purpureus, but it also targeted the rRNA of bacteria harboring the sequence of clone 345. Probe Laro453 targeted L. roseopersicina, as well as the rRNA of bacteria harboring the sequence of clone 136, while probe S453D targeted the 16S rRNA of bacteria harboring the sequence of clone 261, and probe S453F targeted the rRNA of bacteria harboring the sequences of clones 335 and 371.
TABLE 2.
Probes for the analysis of phototrophic sulfur bacteria in Lake Cadagno, Switzerland
| Probe | Target | Sequencea |
|---|---|---|
| Cmok453 | C. okenii DSM169 and clone 359b | 5′AGCCGATGGGTATTAACCACGAGGTT |
| Cmvi453 | C. vinosum DSM180 | 5′TACACGGAGGTATTAGCCCCGTGCTT |
| Apur453 | A. purpureus DSM4197 and clone 345b | 5′TCGCCCAGGGTATTATCCCAAACGAC |
| Laro453 | L. roseopersicina DSM229 and clone 136b | 5′CATTCCAGGGTATTAACCCAAAATGC |
| S453Dc | Clone 261b | 5′CAGCCCAGGGTATTAACCCAAGCCGC |
| S453Fc | Clones 335 and 371b | 5′CCCTCATGGGTATTARCCACAAGGCG |
Positions 453 to 478 on the 16S rRNA according to the E. coli numbering (5).
Clones of a 16S rRNA gene clone library obtained from DNA extracted from bacteria of the chemocline in Lake Cadagno, Switzerland.
Design of probes based on clones of a 16S rRNA gene clone library obtained from DNA extracted from bacteria of the chemocline in Lake Cadagno, Switzerland.
Large-celled phototrophic sulfur bacteria were analyzed with probe Cmvi453 targeting C. vinosum and with probe Cmok453 targeting C. okenii and the rRNA of bacteria harboring the sequence in clone 359, although it had a mismatch to probe Cmok453 at position 474 (Table 2). Probe specificity with reference to the available 16S rRNA sequences was checked with the ARB program (28) and in the EMBL/GenBank databases by using FASTA (20) through the GCG package (Genetics Computer Group, University of Wisconsin, Madison). Pure cultures of phototrophic sulfur bacteria, such as C. okenii DSM169, C. vinosum DSM180, L. roseopersicina DSM229, A. purpureus DSM4197, and A. roseus DSM235, as well as those of bacteria from other phyla, like Desulfotomaculum orientis DSM765, Desulfovibrio desulfuricans DSM642, Burkholderia cepacia DSM50181, Brevundimonas diminuta DSM1635, and Campylobacter jejuni DSM4688 were used to test probe specificity and to establish appropriate in situ hybridization conditions for the specific detection. The specificity of the hybridization was then adjusted by the addition of 40% formamide to the hybridization buffer (except for probe Cmok453, for which the formamide concentration was 35%) and by a reduction of NaCl in the washing buffer to 80 or 56 mM, depending on the formamide concentration during hybridization (35 and 40%, respectively) (35).
Population analysis of phototrophic sulfur bacteria in the chemocline.
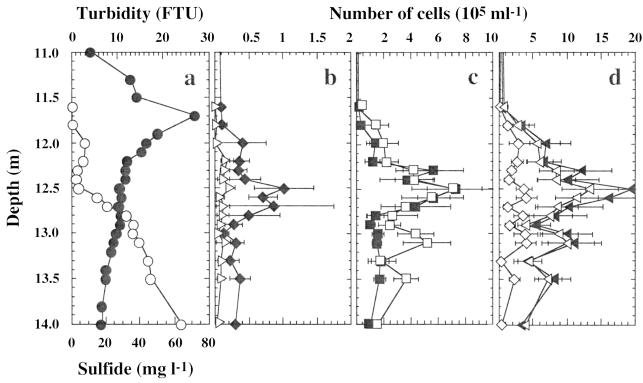
Bacteria were analyzed in water samples from the chemocline obtained with a thin-layer pneumatic multisyringe sampler (University of Zürich, Institute of Microbiology, Zurich, Switzerland) (31). During sampling, the physicochemical parameters (temperature, conductivity, pH, dissolved oxygen, and turbidity) were measured with a Hydropolytester HPT-C profiler (Züllig AG, Rheineck, Switzerland) and showed the characteristic stratification profile of Lake Cadagno (22, 33). The rapid decrease of oxygen (data not shown) and the increase in sulfide concentrations (Fig. 2a) determined colorimetrically in water samples (31) indicated the formation of a condensed chemocline at a depth of between 11.5 and 14 m. The maximum turbidity was found at a depth of 11.7 m (Fig. 2a), suggesting the presence of bacterioplankton taking advantage of the downward O2 or light fluxes and the upward fluxes of reduced compounds (11).
FIG. 2.
Vertical distribution of physicochemical parameters and bacteria in the chemocline of Lake Cadagno at a depth of between 11 and 14 m. (a) Sulfide (○) and turbidity (●). (b) Cells detectable after in situ hybridization with probes Cmok453 (▹) and Laro453 (⧫). (c) Cells detectable after in situ hybridization with probes S453D (■) and Apur453 (□). (d) Cells detectable after in situ hybridization with probe S453F (◊); the sum of cells detectable after in situ hybridization with probes Apur453, Laro453, S453D, and S453F (◂); and the number of small-celled phototrophic sulfur bacteria determined by using autofluorescence and cell size as distinctive criteria (◃). The data, determined from 40 microscopic fields of three samples, are expressed as means ± standard errors.
In situ hybridization with fluorescent (Cy3-labeled) oligonucleotide probes was performed with aliquots (3 μl) of paraformaldehyde-fixed water samples (n = 3) spotted onto gelatin-coated slides [0.1% gelatin, 0.01% KCr(SO4)2] (9) and hybridized and concomitantly stained with DAPI according to the method of Zarda et al. (35). The analysis was performed in a top-to-bottom approach, initially detecting members of the domain Bacteria (probe EUB338) (3); subsequently detecting bacteria of the α (ALF1b) (15), β (BET42a) (15), γ (GAM42a) (15), and δ (SRB385) (2) subdivisions of Proteobacteria; and finally detecting different populations of phototrophic sulfur bacteria (Table 2). The slides were examined by epifluorescence microscopy with filter sets F31 (AHF Analysentechnik, Tübingen, Germany [D360/40, 400DCLP, D460/50]) and F41 (AHF Analysentechnik [HQ535/50, Q565LP, HQ610/75]). Microorganisms were counted at ×1,000 magnification in 40 fields covering an area of 0.01 mm2 each (8). Numbers were expressed as means ± standard errors. The sum of all bacteria detected with probes targeting phototrophic sulfur bacteria was finally compared to the numbers of autofluorescent bacteria determined by epifluorescence microscopy with filter set F41. Autofluorescence was taken as a distinctive parameter characteristic of all phototrophic sulfur bacteria.
In situ hybridization with probe EUB338 allowed us to detect between 38 and 90% of the DAPI-stained cells in the chemocline (31), which was comparable to other aquatic environments, such as, e.g., anoxic water of a stratified fjord (20 to 50%) (25), lake snow (55 to 100%) (34), the winter cover and pelagic zone of a high mountain lake (40 to 81%) (1), and water from two artificial ponds (35 to 67%) (12). Averaged over the whole chemocline, cells hybridizing with probes ALF1b, BET42a, GAM42a, and SRB385 accounted for 23, 17, 45, and 15% of the DAPI-stained bacteria, respectively (31). On average, 33% of the DAPI-stained bacteria were represented by autofluorescent cells which hybridized to probe GAM42a (31) and morphologically resembled the large-celled phototrophic sulfur bacterium C. okenii, with a cell size of 4.5 to 6 by 8 to 15 μm (32), and the smaller-celled species A. purpureus, with a cell size of 3.3 to 3.8 by 3.5 to 4.5 μm (6).
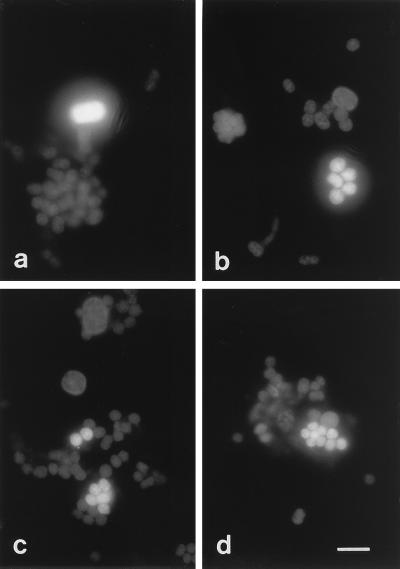
All large-celled phototrophic bacteria enumerated by using a combination of cell size and autofluorescence as a distinctive parameter hybridized to probe Cmok453 targeting the C. okenii type strain, DSM169 (Fig. 3a). None of these cells showed hybridization signals with probe Cmvi453 targeting C. vinosum (data not shown). These results demonstrated that type strain DSM169, which was isolated from a pond, really represented the large-celled species C. okenii in Lake Cadagno. C. okenii occurred in numbers of between (3 ± 4) × 103 and (24 ± 7) × 103 cells ml−1 in the chemocline, with a maximum occurrence at a depth of 12.5 m (Fig. 2b). Maximum numbers were comparable to those of earlier studies which were (48 ± 13) × 103 cells ml−1 (22) or to those in other lakes, such as Lake Belovod, with approximately 50 × 103 cells ml−1 (27). The occurrence of C. okenii correlated with environmental parameters, including anoxic conditions and high sulfate and low sulfide concentrations (Fig. 2a).
FIG. 3.
In situ detection of phototrophic sulfur bacteria with Cy3-labeled probes Cmok453, targeting C. okenii (a); Apur453, targeting A. purpureus (b); S453D, targeting clone 261 (c); and S453F, targeting clones 335 and 371 (d). Bar, 10 μm.
Compared to the vertical distribution of C. okenii, A. purpureus-like cells showed a broader distribution, with high cell densities in the lower part of the chemocline (Fig. 2b to d) which might be due to their high sulfide tolerance (6). In situ hybridization with probes Apur453, Laro453, S453D, and S453F targeting a phylogenetically and morphologically very tight cluster of different small-celled phototrophic sulfur bacteria resulted in the detection of distinct populations of phototrophic sulfur bacteria in the chemocline of Lake Cadagno (Fig. 3b to d). Probe Apur453 detected up to (7 ± 2) × 105 cells ml−1 at a depth of 12.5 m, whereas probes Laro453, S453D, and S453F detected (1 ± 0) × 105, (7 ± 3) × 105, and (4 ± 1) × 105 cells ml−1 at their maximum occurrence at a depth of 12.5 m, respectively (Fig. 2b to d).
These results showed that the type strain of A. purpureus, DSM4197, did not represent the major population of Amoebobacter-like cells in the chemocline of Lake Cadagno and that different populations were present. This result was not surprising, because earlier studies with an isolate obtained from Lake Cadagno (LcCAD1) had already indicated some size differences from the type strain that was isolated from Lake Schleinsee in Germany (6). Populations of small-celled phototrophic sulfur bacteria not only differed with respect to total population sizes but also differed with respect to population profiles in depth. While populations detected with probe S453D only revealed a distinct maximum occurrence at a depth of 12.5 m, populations detected with probe Apur453 showed a second maximum occurrence at a depth of 13.1 m, and those detected with probe S453F showed an evenly high distribution at a depth of between 12 and 13.1 m (Fig. 2b and c).
In the overall depth profile, the sum of the individual numbers detected after hybridization with probes Apur453, Laro453, S453D, and S453F was comparable to the cell numbers determined by using a combination of cell size and autofluorescence as a distinctive parameter for small-celled phototrophic sulfur bacteria (Fig. 2d). Between 96.4 and 100% of autofluorescent cells were further detected throughout the whole chemocline when a combination of all probes was used. These results suggested that sequence information for the major populations of small-celled phototrophic sulfur bacteria was obtained and retrieved in the 16S rRNA gene clone library from bacterioplankton in the chemocline of Lake Cadagno. Small-celled phototrophic sulfur bacteria in the chemocline of Lake Cadagno comprised four major populations with different distribution profiles indicating different ecophysiological adaptations and requirements (32). Future studies of populations of small-celled phototrophic sulfur bacteria in the chemocline of Lake Cadagno will therefore include an analysis of temporal and spatial distributions of specific populations in relation to environmental factors such as light, electron donors, and oxygen and carbon sources (21, 32).
Nucleotide sequence accession number.
The 16S rDNA sequences determined in this study have been deposited in the EMBL/GenBank databases under accession no. AJ223234, AJ223235, AJ006221, and AJ006057 to AJ006063, respectively.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (NF31-46855.96), the Swiss Federal Office of Environment, Forests and Landscape (BUWAL) and the canton of Ticino (Switzerland).
We are indebted to N. Ruggeri and A. Caminada for technical support.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F-O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and Wiley Interscience; 1989. [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Eichler B, Pfennig N. A new purple sulfur bacterium from stratified freshwater lakes, Amoebobacter purpureus sp. nov. Arch Microbiol. 1988;149:395–400. [Google Scholar]
- 7.Fischer C, Wiggli M, Schanz F, Hanselmann K W, Bachofen R. Light environment and synthesis of bacteriochlorophyll by populations of Chromatium okenii under natural environmental conditions. FEMS Microbiol Ecol. 1996;21:1–9. [Google Scholar]
- 8.Fischer K, Hahn D, Amann R I, Daniel O, Zeyer J. In situ analysis of the bacterial community in the gut of the earthworm Lumbricus terrestris L. by whole-cell hybridization. Can J Microbiol. 1995;41:666–673. [Google Scholar]
- 9.Glöckner F O, Amann R I, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An optimized in situ hybridization protocol for planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 10.Guerrero R, Abella C, Miracle M R. Spatial and temporal distribution of bacteria in a meromictic karsic lake basin; relationships with physicochemical parameters and zooplankton. Verh Int Verein Limnol. 1978;29:2264–2271. [Google Scholar]
- 11.Guerrero R, Montesinos E, Pedros-Alio C, Esteve I, Mas J, van Gemerden H, Hofman P A G, Bakker J F. Phototrophic sulfur bacteria in two Spanish lakes: vertical distribution and limiting factors. Limnol Oceanogr. 1985;30:919–931. [Google Scholar]
- 12.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbie J E, Daley R J, Jasper S. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jørgensen B B, Kuenen J G, Cohen Y. Microbial transformations of sulfur compounds in a stratified lake (Solar Lake, Sinai) Limnol Oceanogr. 1979;24:799–822. [Google Scholar]
- 15.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 16.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overmann J, Beatty J T, Krouse H R, Hall K J. The sulfur cycle in the chemocline of a meromictic salt lake. Limnol Oceanogr. 1996;41:147–156. [Google Scholar]
- 18.Overmann J, Beatty T, Hall K J, Pfennig N, Northcote T G. Characterization of a dense, purple sulfur bacterial layer in a meromictic lake. Limnol Oceanogr. 1991;36:846–859. [Google Scholar]
- 19.Øvreås L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedrós-Alió C, Montesinos E, Guerrero R. Factors determining annual changes in bacterial photosynthetic pigments in holomictic Lake Cisó, Spain. Appl Environ Microbiol. 1983;46:999–1006. doi: 10.1128/aem.46.5.999-1006.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peduzzi R, Demarta A, Tonolla M. Dynamics of the autochthonous and contaminant bacterial colonization of lakes (Lake of Cadagno and Lake of Lugano as model systems) Stud Environ Sci. 1993;55:323–335. [Google Scholar]
- 23.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 24.Pringault O, de Wit R, Caumette P. A benthic gradient chamber for culturing phototrophic sulfur bacteria on reconstituted sediments. FEMS Microbiol Ecol. 1996;20:237–250. [Google Scholar]
- 25.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Sorokin Y I. Interrelations between sulfur and carbon turnover in meromictic lakes. Arch Hydrobiol. 1979;66:391–446. [Google Scholar]
- 28.Strunk O, Ludwig W. ARB. Munich, Germany: Computer program distributed by the Technical University Munich; 1996. [Google Scholar]
- 29.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonolla M, Demarta A, Hahn D, Peduzzi R. Microscopic and molecular in situ characterization of bacterial populations in the meromictic Lake Cadagno. Doc Ist Ital Idrobiol. 1998;63:31–44. [Google Scholar]
- 32.van Gemerden H, Mas J. Ecology of phototrophic sulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Press; 1995. pp. 49–85. [Google Scholar]
- 33.Wagener S, Schulz S, Hanselmann K. Abundance and distribution of anaerobic protozoa and their contribution to methane production in Lake Cadagno (Switzerland) FEMS Microbiol Ecol. 1990;74:39–48. [Google Scholar]
- 34.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]