Abstract

Ag3PO4 nanostructures (APNs) containing silver (Ag metal; of the noble metal families) have the potential to exhibit enzyme-mimetic activity. A nanostructure shape, including its surface facets, can improve the bioactivity of enzyme mimicry, yet the molecular mechanisms remain unclear. Herein, we report facet-dependent peroxidase and oxidase-like activity of APNs with both antibacterial and biofilm degrading properties through the generation of reactive oxygen species. Cubic APNs had superior antibacterial effects than rhombic dodecahedral shapes when inhibiting Gram-positive and Gram-negative bacterial pathogen proliferation and biofilm degradation. A similar performance was observed for rhombic dodecahedral shapes, being greater than tetrahedral-shaped APNs. The extent of enzyme-mimetic activity is attributed to the facets {100} present in cubic APNs that led the peroxide radicals to inhibit the proliferation of bacteria and degrade biofilm. These facets were compared to rhombic dodecahedral APNs {110} and tetrahedral APNs {111}, respectively, to reveal a facet-dependent enhanced antibacterial activity, providing a plausible mechanism for shape-dependent APNs material enzyme-mimetic effects on bacteria. Thus, our research findings can provide a direction to optimize bactericidal materials using APNs in clinically relevant applications.
1. Introduction
An array of antibiotics have been developed since the discovery of penicillin and are necessary to medically manage diseases caused by diverse bacterial infections. These modern antibiotics have transformed the world of medicine and saved millions of lives and played a critical role in advancing medicine and surgery recovery.1,2 Emergence of antibiotic resistance is a natural process and should occur only after a significant duration of time. However, over-prescribing and inappropriate use of antibiotics, including in agriculture as a means of weight stimulation and veterinary treatment, accelerated a new problem of bacterial resistance and multidrug resistance (MDR) in the environment and clinics.3 This global issue is threatening generations of medical progress, and alternative solutions for bactericidal options are necessary for various applications to avoid MDR and provide alternatives for policy making.
A significant driver of bacterial resistance is the formation of a heterogeneous and dynamic complex system of biofilms—densely packed communities of microbial cells protected from environmental degradation and oxidation by secretions of extracellular polymeric substances (EPSs).4 Moreover, EPSs act as a physical barrier to effectively prevent the penetration of any external physical or chemical agent, including antibiotics. Biofilms are usually coupled with a devastating number of microbial infections, such as chronic lung infections in cystic fibrosis and other pathophysiological conditions, such as periodontitis and endocarditis. Prosthetic implants and medical devices are very prone to colonies forming biofilms. Biofilms can form on living or inert surfaces, making them a potential hazard in surgical spaces and hospitals, where the routine use of bactericidal agents encourages resistant biofilm formation.
Medical implant failure due to the formation of biofilms on implanted devices causes device breakdown or else chronic-recurrent or lethal infections.5 Their treatment requires a high dose of antibiotics and/or replacement of implanted devices with high economic costs and elevated risk to the patients. Bacterial biofilms can be very easily colonized on different medical devices, such as ventricular assistance devices, dental implants, and orthopedic devices, and are very difficult to treat.6 Biofilms contribute to the emergence of antibiotic resistance by slowing down the distribution and penetrance of antibiotics, contributing to the exacerbation of several diseases, such as tuberculosis, listeriosis, and salmonellosis.7 Such bacteria colonize different kinds of niches in the body with variable EPSs, having several inherent properties that protect them from the host immune system.8 Resistance from MDR and/or biofilm adaptations has been reported for two-thirds of the currently available antibiotics.9
Chronic infections related to biofilms and intracellular bacterial pathogens are difficult to eradicate due to attenuated host immune response and resistance to different antibiotics. If the problem of antibiotic resistance persists, then treatment of common bacterial infections could be lost in a practical sense within a generation.10 Hence, there is an urgent need to develop novel alternate approaches to conventional antibiotics to cope with the crisis. Nanomaterials have recently received much attention due to their extensive antibacterial properties. Various nanostructured materials such as Ag, Au, Zn, Cu, Fe, ZnO, and TiO2 have demonstrated biocidal activity.11 Among these materials, silver nanostructures have received considerable attention from centuries-old attributions to antibacterial effects, relatively low cost, and stability.12 However, the mechanisms by which nanostructures eliminate pathogens have not been thoroughly investigated. It has been proposed that the released Ag+ ions from nanostructures can bind and destabilize the bacterial membrane surface, causing proton leakage.13 Comparatively, such low concentrations of silver nanostructures are nontoxic to human and animal cells. In this regard, silver/silver phosphate nanostructures are considered less risky and less toxic to the environment.14 In this context, recent developments in the field of nanotechnology have created immense opportunities to design new biomaterials and surfaces with bactericidal and antibiofilm properties. Nanostructures entering the bacterial cytoplasm can interfere with different metabolic substrates and hinder their normal pathway for survival. Ag3PO4 nanostructures (APNs) with different facets must be engineered to improve their properties and make them suitable for therapeutic use. Currently, silver orthophosphate (Ag3PO4) nanostructures are being developed for their enhanced enzyme-mimetic catalytical properties.15 In the present study, we have synthesized APNs with different exposed facets and effectively established their antibacterial activity against Gram-positive and Gram-negative bacteria with enhanced capacity for biofilm erosion.
2. Materials and Methods
2.1. Materials
Silver nitrate (AgNO3; 99.9%, Spectrochem), ammonium nitrate (NH4NO3, 99%, Ranbaxy), potassium hydrogen phosphate (K2HPO4, 99.0%, Himedia), sodium hydroxide (NaOH, 98.2%, Merck), and ethanol (C2H5OH, 99.9%, Merck) were procured. All chemicals were utilized as received without further purification, unless otherwise specified. Furthermore, ultrapure distilled and deionized water (18.2 MΩ) was used to prepare all solutions. All culture growth media were obtained from Himedia Laboratories Pvt., Mumbai, India.
2.2. Synthesis of APNs with Tunable Shapes
The synthesis of APNs and tuning of cubic (CU), rhombic dodecahedron (RD), and tetrahedral (TH) APNs was reported by Huang et al.(16) In brief, CU, RD, and TH APNs were prepared using 8.93, 8.42, and 1.887 mL of deionized water, respectively, added in vials, followed by the addition of 100, 600, and 218.8 μL of NH4NO3 (0.4 M) and 180, 180, and 393.8 μL of NaOH (0.2 M) solutions and further introduction of 400, 400, and 500 μL AgNO3 (0.05 M) in the respective vials under vigorous stirring, which resulted in the formation of the [Ag(NH3)2]+ complex in ∼10 min. Finally, after the addition in the respective vials of 400 and 400 μL of K2HPO4 (0.1 M) and 7000 μL of K2HPO4 (0.7 M) to the complex solution under continuous stirring for 2 min, the solution color changed from colorless to light yellow, indicating the formation of CU-, RD-, and TH-shaped APNs, respectively. Unreacted chemicals were then removed through centrifugation at 7500 rpm for 10 min and washed twice with deionized water and 95% ethanol having a 1:1 (v/v) ratio; furthermore, the collected APNs were then dispersed into ethanol and stored at 4 °C until further use.
2.3. Characterization of the APNs
The resultant APNs phase and morphology were determined by powdered X-ray diffraction (XRD, Malvern PANalytical BV, Almelo, The Netherlands) at a scan rate of 0.018 in the range of 15–90° using Cu Kα radiation. The nanomaterial shape, size, and morphology were analyzed using a JSM 7610F Schottky field emission scanning electron microscope (JEOL USA, Inc., MA, USA. The functional groups were identified by Fourier transformation infrared (FTIR, Bruker, Alpha, PN: 1010951/07, SN: 200470) spectroscopy. Shape-dependent catalytic peroxidase activity was determined using the 3,3,5,5-tetramethylbenzidine (TMB) and ascorbic acid (AA) assay using a UV–visible spectrophotometer (Shimadzu, model UV-3600). The release of silver ions from the APNs was quantified by inductively coupled plasma optical emission spectrometry (ICP–OES) (icap-6500 DUO-Thermo Fisher Scientific).
2.4. Shape-Dependent Oxidase-like Catalytical Activity
The enzyme-mimetic oxidase activity of the APNs and their ability to produce reactive oxygen species (ROS) in the dispersed aqueous phase were assessed through AA oxidation.17 The produced ROS from shaped APNs oxidize the AA, which results in the production of dehydroascorbic acid with quantifiable absorption band maxima at 266 nm. The dispersed nanostructures were serially diluted with a phosphate-buffered saline (PBS) (pH 7.4) solution over a range of concentrations: 10, 20, and 40 μg/mL, followed by the addition of AA [PBS-dissolved AA] to a final concentration and volume of 60 μM and 1 mL, respectively. Furthermore, the resultant dispersions were incubated at 37 °C for 1 h and then centrifuged at 7000 rpm. After centrifugation, the supernatant solutions were collected and subjected to UV–visible absorption spectroscopy to determine the APNs’ shape-dependent catalytical performance. The reported results are from two independent trials.
2.5. Shape-Dependent Peroxidase-like Catalytical Activity
The shape-dependent peroxidase-like mimetic activity of the APNs was assessed through the TMB assay, as described by Shen et al. with minor modification.18 The TMB solution was made with a final concentration of 2 mg/mL using an acetic acid (NaAc/HAc) buffer at a pH around 5.2. To this, 1 μL of 10 mM H2O2 was added, followed by the addition of 5–10 μg of shaped APNs. All assays were performed in triplicate.
2.6. Shape-Dependent Hydroxyl Radical (•OH) Generation Catalytical Activity
The formation of hydroxyl radicals was monitored by fluorescence using terephthalate (TA), which reacts with hydroxyl radicals to form fluorescent 2-hydroxy terephthalate.19 TA (5 mM), H2O2 (10 mM) + TA, and shaped APNs + TA in the presence and absence of H2O2 samples were monitored to determine the formation of hydroxyl radicals. The resultant samples were subjected to incubation at room temperature for 3 h. After completion of incubation, the mixtures were removed and the fluorescence was quantified at an excitation wavelength of 285 nm and an emission wavelength of 420 nm.
2.7. Biological Evaluation
2.7.1. Bacterial Strains
Two Gram-positive and two Gram-negative bacterial strains were evaluated: Staphylococcus aureus (MTCC 96), K. planticola (MTCC 530), E. coli (MTCC 739), and M. luteus (MTCC 2470) obtained from the Microbial Type Culture Collection and Gene Bank (MTCC), CSIR-Institute of Microbial Technology, Chandigarh, India.
2.7.2. Bacterial Culture Growth
All four strains were individually plated on Mueller–Hinton agar (MHA) plates and cultured in Mueller–Hinton broth (MHB) media for 24 h at 37 °C. After picking up a single colony of the respective bacteria from an MHA plate, the bacteria were seeded/incubated in 10 mL of MHB culture media at 37 °C for 12 h with 150 rpm agitation. Furthermore, the bacterial concentration was empirically determined by density absorbance at 600 nm at an optical density of 0.4–0.6 (OD600).
(a). Antibacterial Activity Assay
As reported by Wang et al.,20 the antibacterial properties of the APNs were evaluated by the microdilution method. Rifampicin (positive control) and a saline solution (negative control) were used in the microdilution method in MHB media for all four bacterial strains. All of the experiments were carried out using a 96-well plate. Following this, the individual grouping of APNs in the first well of each group with the highest concentration (500 μg/mL) was carried out; each well of each group was supplemented with 100 μL of the MHB medium. The solutions were mixed, and 100 μL of the sample solution was transferred from the first well of each group to the second, resulting in a twofold dilution. Except for the last two wells, which served as negative controls, similar dilutions were used. Bacterial strains with 106 CFU/mL in MHB media and their respective wells were inoculated with 10 μL of pathogen suspensions and incubated at 37 °C for 18 h. The treatments were carried out in triplicate, and the absorbance was calculated using an Infinite M200 Proplate reader (Tecan Group Ltd. Mannedorf, Switzerland).
(b). Colony Counting Antibacterial Activity
The bacterial suspensions (200 μL, 1 × 106 CFL/mL) were mixed with APNs [MIC to inhibit 50% of growth (MIC50)] in 1.5 mL centrifuge tubes. As a control, sterile saline was used instead of APNs. Then, 200 μL of the bacterial suspensions was streaked on Luria–Bertani agar plates and subjected to incubation at 37 °C for 24 h.20 Furthermore, the formation of colony units was counted to compare shape-dependent APNs antibacterial activity.
(c). Biofilm Inhibition Activity
The formation of mature biofilms was achieved by inoculating 96-well plates with 20 μL of the bacterial suspension (1 × 107 CFU/mL) and 180 μL of the MHB medium at 37 °C for 48 h. The culture medium was aspirated from wells and washed three times with 1× PBS after 48 h of incubation.21 The mature biofilm that formed was visible on the well’s rim. The biofilms were then incubated for 24 h at 37 °C in 200 μL of the MHB medium containing APNs at a twofold dilution in all wells of the 96-well plate except the last two. The wall-adhering biofilms were washed twice with sterile 1× PBS after the unanchored cells and media were removed. Crystal violet (1%, 200 μL per well) was used to stain the adherent biofilms for 40 min at room temperature. Both pathogenic strain crystal violet-stained biofilms were washed with 1× PBS to eliminate the excess stain and allowed to dry overnight before being solubilized in 120 μL of 90% ethanol. The absorbance was measured at OD600 for quantification of the biofilm density. All studies were carried out in triplicate.
(d). ROS Assay
The nitroblue tetrazolium (NBT) assay was used to estimate intracellular ROS accumulation in bacterial strains due to the impact of APNs. The bacteria were cultured and diluted to a McFarland standard of 0.5 before being suspended in a 96-well microtiter plate. The bacterial cells were stained with NBT (0.1% in PBS) at 37 °C for 40 min after being treated with various formed nanostructures. As a control, the cells that had not been treated were used. A spectrophotometer set to 570 nm was used to calculate the intracellular ROS levels. All of the tests were done in triplicate, and the standard deviation was calculated using the mean value.
(e). Bacterial Morphology by FESEM
The bacteria were cultured in MHB for 48 h at 37 °C, then treated with APNs, and incubated for 3 h, with untreated cells serving as a control. After the treatment, the corresponding cells were collected and centrifuged for 10 min at 6000 rpm. The cells were washed in 1× PBS before being fixed with 2.5% glutaraldehyde and incubated at 4 °C overnight. The cells were dehydrated using a graded ethanol solution after three PBS washes (10, 30, 70, and 100%).19
(f). Determination of the Biofilm Dry Weight
Lee et al. calculated the dry weight of bacterial biofilm.22 For the development of mature biofilms, all bacterial strains were cultured in six-well plates and treated with APNs at 37 °C for 48 h without shaking. As a positive control, an untreated bacterial culture was used. The suspension of cells was discarded after 48 h, and they were rinsed in 1× PBS. Biofilm-forming cells were removed by pipetting vigorously two times. The biofilm cells were collected using a 0.22 μm-pore-size preweighed filter syringe. These filters were then dried for 8 h at 60 °C, and the dry weight of the cells was determined.
(g). Confocal Laser Scanning Microscopy
The bacterial strain’s cell membrane integrity and viability were evaluated by staining them with acridine orange (AO) and propidium iodide (PI) dye using confocal laser scanning microscopy (CLSM).23 First, the bacteria were cultured in MHB in CLSM dishes (SPL Scientific, Gyeonggi-do, South Korea) for 48 h at 37 °C; thereafter, the bacterial strains were treated with APNs for 12 h, and the untreated group was used as the control. 2 μL of each of the AO and PI mixture was added and incubated for 20 min at room temperature, and then they were visualized using an Eclipse Ti confocal laser scanning microscope (Nikon Corporation, Tokyo, Japan) interfaced with an argon-ion laser at the green and red channels. The images were analyzed using Ti Control software ver. 4.4.4, and the scale bar on the image represents a distance of 5 μm.
(h). Theoretical Modeling
First-principles calculations were carried out to evaluate the distinct catalytic activities of the APNs, and all calculations were carried out using the Vienna ab initio simulation package.24 The Perdew–Burke–Ernzerhof25,26 exchange–correlation functional was employed under the framework for the generalized-gradient approximation. The projector-augmented wave potential was employed for the ion-electron terms, with the valence configurations of Ag (s1d10), P (s2p3), and O (s2p4), according to the implementation of Kresse and Joubert.27 All surface slabs, reactants, and complexes were optimized using an energy cutoff of 520 eV. The Ag3PO4(100), Ag3PO4(110), and Ag3PO4(111) surfaces were modeled; for geometry optimization, all layers of the (100), (110), and (111) surfaces were fully relaxed without any constraints; the conjugated gradient algorithm was employed. Structural optimization was performed until the twin convergence criteria of energy and force reached 1 × 10–5 eV and 0.02 eV/Å, respectively. Brillouin-zone integrations were performed using k-point grids using a 4 × 4 × 1 mesh for relaxation of the slabs. A period slab with a vacuum space of 15 Å in the direction of the surface was normal to avoid the interactions.
(i). Statistical Analysis
All of these studies were done in triplicate. The mean values were used to represent the results. In addition to respective controls, a one-way analysis of variance (ANOVA) and a two-way ANOVA were used to assess the statistical significance.
3. Results and Discussion
The synthesis of CU, RD, and TH APNs was carried out using aqueous solutions of NH4NO3, NaOH, AgNO3, and K2HPO4 by incubating them at room temperature under dark conditions as described by Huang et al.(15) Adjusting the reagent amounts allows tuning of the nanostructure shape.15Figure 1A–C depicts the FESEM morphological images of the synthesized APNs shapes after tuning to CU, RD, and TH APNs. All these samples exhibited facets of uniform shape, size, and morphology for each APN type. In this study, the TH facet had a curved contour due to the presence of shorter tips in the facets in comparison with regular TH.15 Moreover, CU APNs were exclusively enclosed by the {100} surface facet.14 The RD and TH APNs were bound by enclosed surface facets {110} and {111}, respectively.15 Furthermore, the CU, RD, and TH APNs had similar sizes from ∼675 to 800 nm.
Figure 1.
FESEM images of CU- (A), RD- (B), and TH- (C) shaped APNs and powder XRD patterns of APNs, where the black line represents the CU, the red line represents the TH, and the blue color line represents the RD shape (D), and the respective FTIR spectra are depicted in (E).
Figure 1D illustrates the powdered XRD pattern of the APNs and is consistent with a standard pattern of Ag3PO4 (JCPDS no. 06-0505).14 However, notable changes were observed in the orientations and their respective dimensions, resulting in peak intensity elevation and a conspicuous reflected surface facet. More specifically, in the case of CU APNs, a reflection peak diffraction (200) was remarkably enhanced by the intensity in comparison with RD- and TH-shaped APNs, confirming that the CU APNs are primarily composed of the {100} crystalline plane.22 Similarly, RD- and TH-shaped APNs exhibited enhanced reflection diffractions of (110) and (222), respectively, suggesting that their surface is dominated by the {110} and {111} crystalline planes.14,15 Furthermore, we assessed the PO43– functional groups of these samples and confirmed (Figure 1E) the depicted peaks in the range of 890–1100 cm–1 by FTIR spectroscopy, consistent with those that have been previously reported.28 Functional group analysis of the APNs was carried out using the FTIR spectroscopy technique. The bands appeared from 541 to 559 and from 988 to 1074 cm–1 due to the presence of asymmetric O=P–O and P–O–P bending and stretching vibrations, respectively. The bands at 1663–1671 and 3010–3370 cm–1 are due to the presence of bending and stretching vibrations from water. Furthermore, the weak intensified peaks at 2346–2398 cm–1 are due to the presence of hydrogen bonds between water and phosphate functional groups.29,30 Moreover, various parameters such as particle size, shape, and morphology can influence the nanostructured materials’ catalytic activity. Therefore, the shape tuning of various APN materials may result in superior catalytic performance due to the presence of surface facets.
Oxidase-like activity was screened for the various APNs shapes by using a natural, physiologically relevant, antioxidant substrate, that is, AA (vitamin C), which exhibited an absorption maximum at 266 nm, which dissipates upon oxidation.17,19 The efficient antioxidant agent of this AA biomolecule played an important role in the cyclical redox reactions occurring in the cellular system.17 After incubation of these nanostructures with the AA probe in PBS, the shaped nanostructures’ catalytic activity was observed by reducing the AA absorbance maxima at 266 nm and it varied depending on the extent of the active facets of the APNs, and similar results were observed by Fang et al. (Figure 2A–C).19 These results indicate that the production of ROS varied with the shape (i.e., active facets present in the nanocrystal) of the APNs that make contact with the substrate AA. Typically, the CU APNs exhibited higher catalytical activity than the RD and TH APNs to oxidize AA. Interestingly, the catalytic oxidation process generates hydrogen peroxide.17 Notably, an increased level of H2O2 was detected in the presence of APNs in comparison with AA (control). A significantly higher amount of H2O2 was generated in the case of CU APNs, and its level was higher than in the case of RD and TH APNs, and a similar higher performance was observed between RD- and TH-shaped APNs.
Figure 2.
UV absorption spectra of time dependence-catalyzed AA-measured oxidase-like properties in the presence of (A) CU, (B) RD, and (C) TH shape-dependent APNs, where (a) 0, (b) 5, (c) 15, (d) 30, (e) 35, and (f) 40 min. Peroxidase property of the APNs-monitored TMB probe formation of intermediate of TMB+ in the presence of H2O2 and (D) CU, (E) RD, and (F) TH shapes of APNs, whereas (a) 0, (b) 15, and (c) 20 min respective color vials are presented in the inset.
We then investigated the intrinsic peroxidase-like properties of the shape-dependent APNs, utilizing the peroxidase substrate TMB-catalyzed oxidation with H2O2. During the catalytic redox reaction, the substrate oxidized into TMB+ (one-electron oxidation intermediate) and exhibited a characteristic band around 652 nm.19Figure 2D–F illustrates this compared with the control, and time-dependent augmentation of the absorption spectral band at 652 nm implied that the APNs are capable of catalyzing the oxidation of the TMB substrate via H2O2, indicating the generation of an intermediate product (characteristic light- and dark-blue color and inset: colored change solution). These results indicate the peroxidase property of the APNs. Notably, the amount of substrate intermediate produced by the CU APNs was significantly greater than that produced by RD and TH APNs. Moreover, the RD APNs had the similar potential to form TMB intermediates but less than CU APNs, yet more than TH APNs. The peroxidase property is greater for the {100}-faceted CU APNs with significantly higher efficacy to generate the intermediate products than {110} and {111} facets of the RD and TH APNs. As expected, a similar analysis corroborates the RD facet, which has higher and lower efficacy than TH and CU APNs, respectively, for peroxidase-like activity.
The APNs were then shown to be capable of converting H2O2 species into hydroxyl ion (•OH) free radicals, which might occur due to the exhibited peroxidase-like property. To corroborate this mechanism, TA was used as a substrate to monitor the formation of (•OH), which would react to form 2-hydroxy TA (TAOH) and exhibit bright fluorescence.19 Fluorescence analysis identified the •OH and corresponding samples’ fluorescence spectra, as depicted in Figure S1. The intensified fluorescence was observed in the presence of APNs, TA, and H2O2, whereas fluorescence was not observed in the absence of either H2O2 or APNs. This in turn is an indication of •OH species generation from H2O2 catalyzed by the APNs (data not shown). Moreover, this fluorescence intensity corroborates the fact that the facet {100} of CU APNs exhibited more efficiency to generate •OH than the RD {110} and TH {111} facets present in those APNs. Similarly, the RD facet {110} present in the APNs exhibited higher efficiency for the production of •OH than the TH facet {111} APNs.
3.1. Antibacterial Activity of Different Facet Nanostructures of the APNs
We have established that the APNs of different facets show oxidase- and peroxidase-like properties by successively generating •OH radicals, which indicates potential for biocidal activity. To confirm this, we evaluated the antibacterial properties using two representative strains of Gram-negative (E. coli and K. planticola) and Gram-positive (S. aureus and M. luteus) bacteria and performed a microdilution method to determine their MIC and minimum bactericidal concentration (MBC) value. We observed that different APNs efficiently inhibited the growth and proliferation of all representative Gram-negative and Gram-positive bacterial strains at different concentrations (Figure S2). Interestingly, a facet-dependent antibacterial activity was observed where the CU APNs showed better growth inhibition as compared to RD and TH APNs in all representative bacterial strains (Table 1). We observed that CU APNs inhibited the growth of E. coli and S. aureus at 3.9 μg/mL for both as compared to the rifampicin positive control, which was required at concentrations of 31.25 and 15.62 μg/mL for E. coli and S. aureus, respectively (Figure 3). This shows that the CU APNs have superior biocidal activity against both Gram-negative and Gram-positive bacteria as compared to a therapeutically proven antibiotic (rifampicin) and might be considered similarly for use as a broad-spectrum antibiotic. The results of the MBC were consistent with those of the MIC, thereby indicating that the APNs had facet-dependent antibacterial properties (Table 2). To avoid any misinterpretation in the evaluation of antibacterial properties due to turbidity in the broth medium, we also checked the antibacterial activity of different APNs using an agar medium. In that case, different agar plates of E. coli and S. aureus having a concentration of 103 CFU mL–1 were evaluated for visualized colonies on a cultured agar plate. We observed dense colony formation in the control plates (bacterial agar plates without any treatment with APNs after 12 h). However, APNs-treated plates showed facet-dependent antibacterial activity and negligible colony formation. In the case of E. coli, no colonies were observed in plates treated with the CU APNs and RD APNs, while a single colony was observed in the TH APNs-treated plate. Similar results were observed in the S. aureus-cultured plates, depicting their facet-dependent antibacterial activity (Figure S3). Furthermore, the excellent superior catalytic activity is due to the release of silver ions from CU, RD, and TH APNs that were incubated in pathogen broth media subjected to ICP–OES. In the case of E. coli broth media, a higher release of silver ions was observed for CU APNs and their concentration was found to be 12.08 mg/L, whereas the leached silver ion concentration decreased to 4.789 mg/L for the RD APNs, and even lower to 3.403 mg/L for the TH APNs. A similar concentration trend for the CU, RD, and TH APNs was exhibited due to the release of silver ions, and the concentrations were found to be 15.59, 12.32, and 2.241 mg/L, respectively, for S. aureus. These results suggested that the release of silver ions from nanostructures played a crucial role against antibacterial response.31,32
Table 1. Antimicrobial Activity of Shape-Dependent APNsa.
| MIC (μg/mL) |
||||
|---|---|---|---|---|
| Gram-negative |
Gram-positive |
|||
| APN shape | E.cb | K.pc | S.ad | M.le |
| CU | 3.90 | 7.81 | 3.90 | 3.90 |
| RD | 7.81 | 15.62 | 7.81 | 7.81 |
| TH | 15.62 | 31.25 | 31.25 | 15.62 |
| rifampicin | 31.25 | 15.62 | 15.62 | 7.81 |
MIC (μg/mL) of the shape-dependent APNs that inhibit the visible growth of bacterial strains.
E. coli.
K. planticola.
S. aureus.
M. luteus and rifampicin/control.
Figure 3.
MIC to inhibit 50% of growth (MIC50) by shape-dependent APNs compounds (CU, RD, and TH APNs), which inhibit the visible growth of Gram-negative (A) E. coli and (B) K. planticola and Gram-positive (C) M. luteus and (D) S. aureus. Data are expressed as statistically significant (p* ≤ 0.05, p** ≤ 0.005, and p*** ≤ 0.0001).
Table 2. MBC (μg/mL) of Shape-Dependent APNs-Limited Growth of Bacterial Strains.
| MBC (μg/mL) |
||||
|---|---|---|---|---|
| Gram-negative |
Gram-positive |
|||
| APN shape | E.ca | K.pb | S.ac | M.ld |
| CU | 7.81 | 15.62 | 7.81 | 7.81 |
| RD | 15.62 | 31.25 | 15.62 | 15.62 |
| TH | 31.25 | 62.50 | 62.50 | 31.25 |
| rifampicin | 62.50 | 31.62 | 31.62 | 15.62 |
E. coli.
K. planticola.
S. aureus.
M. luteus and rifampicin/control.
To determine if the APNs are capable of eliminating the formation of bacterial biofilm (EPS matrix). We observed that a detectable biofilm was generated in the absence of APNs (Figure 4) and that no noticeable changes in the biofilm were found when the APN concentrations were less than 3.9 μg/mL in most samples. However, biofilm was eliminated when the concentration of the APNs reached 31.25 μg/mL. At this concentration, the biofilms no longer showed any intense crystal violet staining, which indicates the successful removal of the biofilm (Figure 4). Moreover, we found that the different APNs were not equivalent in biofilm eradication at particular concentrations (Figure 5). This also reflected the facet-dependent antibacterial activity against biofilm elimination.
Figure 4.
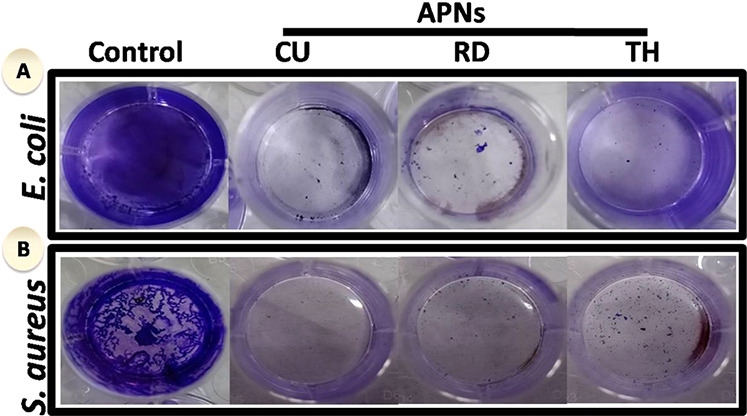
Influence of shape-dependent APNs on the formation of biofilms: (A) crystal violet-stained biofilm of E. coli alone (control), biofilm treated with CU-, RD-, and TH-shaped APNs; (B) color of crystal violet-stained biofilm of S. aureus alone (control) and those treated with CU-, RD-, TH-shaped APNs.
Figure 5.
Antibiofilm activity of CU-, RD-, and TH-shaped APNs at different concentrations. Biofilm inhibition of compounds against Gram-negative (E. coli and K. planticola) and Gram-positive (M. luteus and S. aureus). p > 0.05 indicates nonsignificant variance (ns), p < 0.05 indicates significant variance (*), p < 0.01 indicates significant variance (**), and p < 0.001 indicates significant variance (***) between the variant concentrations of APNs with the untreated group (control) by two-way ANOVA.
The result of biofilm elimination by the different APNs was further reinforced by the evaluation of the dry weight of different biofilms formed by representative Gram-negative and Gram-positive bacteria. We found that the dry weight of different biofilms decreased variably with different APNs (Figure 6). The extent of decrease was significantly higher in the case of the CU APNs, consistent with the previous findings. This result suggests that the CU APNs could be used as a potent antibacterial agent for the elimination of different biofilms formed by various strains of bacteria.
Figure 6.
ROS quantification in Gram-negative (E. coli and K. planticola) and Gram-positive (M. luteus and S. aureus) bacteria treated with CU-, RD-, and TH-shaped APNs. Rifampicin was used as a positive control, and untreated strains were used as a negative control. p indicates a significant difference between the APNs ROS with the untreated group (negative control); p > 0.5 nonsignificant variance (ns), p* ≤ 0.05, p** ≤ 0.005, and p*** ≤ 0.0001 by one-way ANOVA.
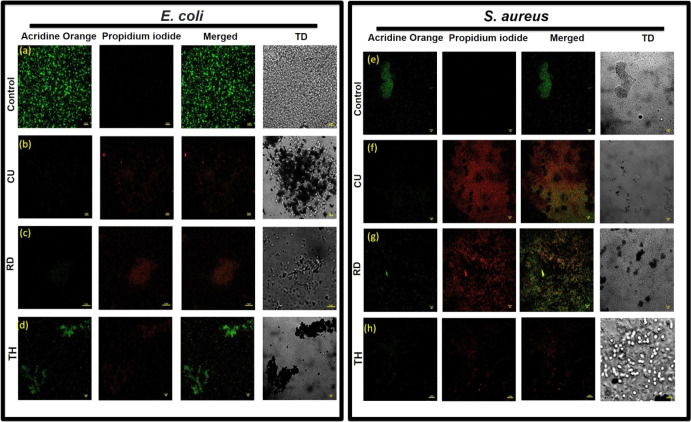
To further examine the biocidal properties of APNs, the bacteria were stained with fluorescence-based dyes such as AO and PI to measure bacterial viability. The outer surface of dead bacteria becomes permeable to PI, turning red. We observed that bacteria without treatment with any APNs grow successfully and appear intense green due to the fluorescence of AO. However, different APNs-treated bacteria appear red due to biocidal activity (Figure 7). The intensity of red fluorescence was highest with the treatment of CU APNs compared to that with the RD and TH APNs in the biofilm of E. coli and S. aureus bacteria (Figure 8). These results further support the facet-dependent antibacterial activity of the APNs.
Figure 7.
Dry weight percentage of biofilm for Gram-negative (E. coli and K. planticola) and Gram-positive (M. luteus and S. aureus) bacteria treated with CU-, RD-, and TH-shaped APNs. Rifampicin was used as a positive control, and untreated strains were used as a negative control. p > 0.05 indicates nonsignificant variation (ns), and p < 0.001 indicates significant variation (***) in comparison to the untreated group (control) by two-way ANOVA.
Figure 8.
Confocal images of the biofilm formed by Gram-negative (a–d) E. coli and Gram-positive (e–h) S. aureus after 24 h of growth. (a) Live cells of E. coli without treatment showed an intact cell membrane; (b–d) E. coli coincubated with CU-, RD-, and TH-shaped APNs induce lysis of the bacterial cell membrane. (e) Live cells of S. aureus without treatment showed an intact cell membrane; (f–h) S. aureus coincubated with CU-, RD-, and TH-shaped APNs induce lysis of the bacterial cell membrane.
These results indicate that the MIC and MBC for different facets of CU, RD, and TH APNs vary significantly in the Gram-negative and Gram-positive strains of bacteria, but all three APNs exert biocidal activity in terms of planktonic and biofilm elimination. This effect differential can be explained based on the morphological differences in the cell wall of each strain.33 Usually, there is an outer membrane in the Gram-negative bacteria that enables resistance at a morphological level that provides better protection against different antibiotics and chemicals as compared to Gram-positive bacteria.34 On the other hand, the cell wall of Gram-positive bacteria is thicker as compared to that of Gram-negative bacteria, offering greater resistance to membrane stability versus permeability. Therefore, our results emphasize the morphological and physiological differences of the cell membrane of Gram-positive and Gram-negative bacteria.
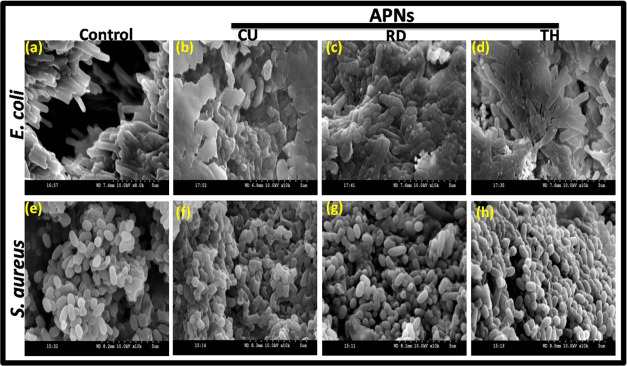
To determine membrane damage and cell membrane integrity, scanning electron microscopy was used. The sub-MIC concentrations of different APNs were used to treat the biofilm formed by E. coli and S. aureus bacteria; each strain displayed characteristic rod-like and spherical morphologies, respectively. However, both bacterial cells were damaged and disrupted after the treatment with APNs (Figure 8). The extent of damage to the morphology was significantly higher in the CU APNs-treated bacterial biofilm as compared to biofilms treated with other APNs. These morphological changes potentiate the antibacterial effect of CU APNs.
Due to the loss of membrane integrity, the APNs may cross through the damaged membrane and gain entry into the bacterial cytoplasm, whereupon they interact with the internal cellular environment and produce oxidative stress by generating ROS.18,35 We determined the intracellular level of ROS produced by the different APNs in the Gram-negative and Gram-positive bacterial strains. ROS generation is primarily responsible for oxidative stress and causes oxidative damage to the different cellular organelles of bacteria. ROS is a broad term for all types of chemical molecules that have very high positive redox potential, and we demonstrate that the primarily antibacterial activity of the APNs is from oxidative stress by catalyzed ROS formation.34 The amount of ROS generated in E. coli, K. planticola, M. luteus, and S. aureus after the treatment with different APNs was determined by the NBT assay. We observed that all types of APNs generate ROS as compared to the control group (Figure 9). However, the extent of ROS generation is found to be a facet-dependent phenomenon.36 The CU APNs showed more absorbance than the RD APNs. The absorbance of TH APNs lay in between those of the CU and RD APNs. Moreover, the same pattern of absorbance was observed for all three different kinds of facets of the APNs in four different types of bacteria used in our study. Hence, we have used the same concentration of NBT throughout the assay. Therefore, these absorbance values reflect that more NBT is being redox-reduced in the case of CU APNs, hence producing more ROS in the treated bacteria than RD and TH APNs. There was a significantly lower amount of ROS generated in bacteria without any APNs treatment, which was considered a negative control in this study. The basal level of absorbance by the negative control group was assumed to reflect the endogenous ROS. In contrast, the ROS generated in the different groups of bacteria treated with rifampicin was considered the positive control. The CU APNs generate around 6 times more ROS in both Gram-positive and Gram-negative bacteria as compared to the negative control. However, the extent of ROS generation by the RD APNs is different in both Gram-positive and Gram-negative bacteria. Similar results as RD APNs were also observed in the case of TH APNs, which generated the least amount of ROS in treated bacteria as compared to the negative control. On the basis of the above observations, we infer that the CU APNs might be used as a potential antibacterial agent against Gram-positive and Gram-negative bacteria.
Figure 9.
SEM images of biofilms formed by (a–d) E. coli and (e–h) S. aureus untreated and those treated with CU-, RD-, and TH-shaped APNs. (a) Untreated E. coli as the control with the biofilm matrix and undamaged cell morphology, (b) E. coli treated with CU APNs, (c) E. coli treated with RD APNs, (d) E. coli treated with TH APNs, showing the damaged cell morphology and disrupted biofilm with CU > RD > TH effects. (e) Untreated S. aureus as the control with an intact biofilm and undamaged cell morphology, (f) S. aureus treated with CU APNs, (g) S. aureus treated with RD APNs, and (h) S. aureus treated with TH APNs showing a damaged cell morphology and disrupted biofilm with CU > RD > TH effects.
In general, the APNs crossed the cell membrane to become intracellular and generated the ROS inside the bacterial cell to create oxidative stress and cause damage to lipids, DNA, RNA, and proteins.37 We also observed that the APNs of different facets were able to generate a significant amount of ROS in different Gram-negative and Gram-positive bacteria to a variable extent. The produced ROS in the bacteria causes oxidative stress that has been assumed as a major contributor in changing the permeability of the cell membrane, which can damage the integrity of the bacterial cell membrane.38 Under normal conditions, bacterial cells encounter the ROS with the help of different intrinsic enzymes and try to balance the internal environment of the cell for their successful growth and reproduction.34
Our results are in line with a different study, in which APNs of varying particle sizes effectively produced antibacterial effects against Gram-negative and Gram-positive bacteria.39−43 With oxidative stress, several biomolecules, such as proteins, DNA, and so forth, which are the building blocks of any living organism, can undergo various degrees of oxidation. Moreover, ROS is also responsible for inducing the gene expression of different oxidative proteins, which helps to facilitate the death processes of the bacterial cell.44 Several enzymes are present in the periplasmic space of the bacterial cell, which are essential for maintaining their normal morphology and physiology and are also adversely affected by the high level of ROS activity.45 Thus, it seems that a high amount of ROS generated in the bacterial cell due to the treatment of APNs of different facets could cause bacterial cell death (Figure 10) by altering their membrane integrity and DNA damage.
Figure 10.
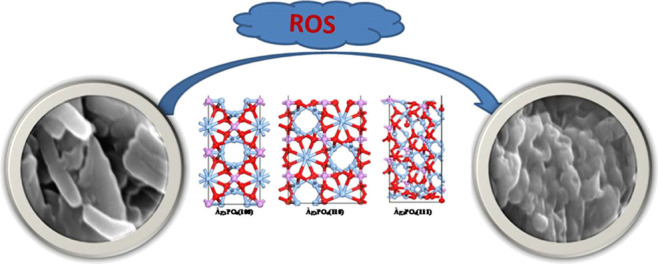
Schematic representation of facet-dependent ROS-like activities of APNs conferring outstanding antibacterial capabilities by producing ROS.
3.2. DFT Simulations on the Catalytic Activities of APNs
To provide detailed mechanistic insights of O2 adsorption and dissociation on the Ag3PO4 surfaces, we performed first-principles calculations on distinct catalytic activities of Ag3PO4. We have computed O2 dissociation on three different Ag3PO4 surfaces, namely, Ag3PO4(100), Ag3PO4(110), and Ag3PO4(111).
Mainly, DFT studies estimate the energies involved during the oxidase- and peroxidase-like processes. Earlier, Li and Fang et al.(19,35) have reported the O2 oxidase and peroxidase mechanisms on distinct Pd surface slabs; the same strategy was applied here to estimate the dissociative adsorption of the O2 species on the respective Ag3PO4 surfaces. Initially, we computed the O2 dissociation energy barriers for the O2 molecule on the three distinct Ag3PO4 surfaces. The dissociation mechanism involves three consecutive steps:19 in the first step, O2 molecule absorption occurs on the Ag3PO4(100) surface, followed by O–O bond dissociation, and finally O–O bond cleavage to yield single O atoms (Figure 11). We have computed O2 dissociation reaction pathways on Ag3PO4(100), Ag3PO4(110), and Ag3PO4(111) on three surfaces; among them, the lower dissociation reaction energy pathway is shown in Figure 11 and those for Ag3PO4(110) and Ag3PO4(111) are provided in the Supporting Information (Figure S4). Purified TA (PTA) in PBS, similarly treated but without nanoparticles, served as the control. The adsorption energies of O2 on all three surfaces are −1.05, −0.9, and −1.75 eV, respectively. O2 binds tightly to the Ag3PO4(111) surface, with the corresponding O2 dissociation energies of 0.06, 0.47, and 3.03 eV. PTA in PBS, similarly treated but without nanoparticles, served as the control. We can see from these activation energies that dissociative adsorption of O2 is more favorable on the Ag3PO4(100) and Ag3PO4(110) surfaces than on Ag3PO4(111) due to the high dissociative activation energy.
Figure 11.
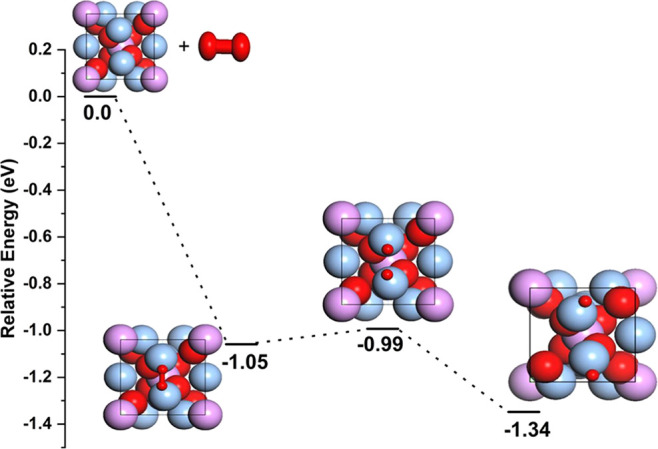
Energy profiles of O2 dissociative adsorption on Ag3PO4(100) surfaces and relative energies (eV) are shown.
4. Conclusions
In recent times, many bacteria have developed resistance to multiple types of antibiotics. This emergence has created a crisis in medicine with more difficult-to-eliminate bacterial infections, risking very high morbidity and mortality. Our present study offers one approach to facilitate overcoming this emerging crisis by using non-conventional methods to effectively eliminate different bacterial infections. Our findings suggested the feasibility of using APNs for the biocidal effect that is facet-dependent in terms of activity. Our results revealed that the CU APNs had higher antibacterial activity against selected models of Gram-negative and Gram-positive bacteria than RD or TH APNs. The antibacterial activity of these nanostructures is attributed to the generation of ROS upon the interaction of the bacterial surface, including erosion of the biofilm, resulting in damage to the integrity of their cell membrane, followed by penetrating ROS generation within the cellular microenvironment, leading to bacterial death. This work may open a novel avenue toward the use of APNs to eliminate MDR bacteria and better secure sterile devices, instruments, or operating environments.
Acknowledgments
We gratefully acknowledge support from the CSIR–Indian Institute of Chemical Technology, Hyderabad, India, and N.P. acknowledges the DST-Inspire faculty scheme for financial support (GAP-0631). N.P. thanks the Gujarat State Biotechnology Mission, Govt. of Gujarat, India, grant number (GSBTM/JD(R&D)/618/21-22/00003673). K.S. and R.G. acknowledge the CSIR-UGC (SRF) and DBT (SRF), respectively. B.S. gratefully acknowledges the financial support of the Fundamental Research Funds for the Provincial Universities of Zhejiang GK209907299001-022 and Prof. Niall J English for his computational specialty. The authors express appreciation to Dr. Keith R. Brunt, Dalhousie University, Canada, for the constructive inputs and edits. This work is financially supported by the CSIR-IICT communication no. IICT/Pubs./2022/020.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00864.
TA probe used for the analysis of fluorescence spectra of hydroxyl radicals in the presence of H2O2 and shaped APNs; evolution of the MIC in terms (%) of bacterial survivability of Gram-negative and Gram-positive bacterial strains treated with the APNs—CU, RD, and TH; agar plate images with the antibacterial effect of shape-dependent APNs—CU, RD, and TH—of bacterial strains; and energy profiles of O2 dissociative adsorption on Ag3PO4(110) and Ag3PO4(111) surfaces (PDF)
Author Contributions
K.S. and R.G. performed all chemical and biological experiments. S.B. performed the molecular modelling studies. K.S.B., S.M., and N.P. designed the study and supervised the necessary work. All authors contributed inputs and have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Golkar Z.; Bagasra O.; Pace D. G. Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J. Infect. Dev. Countries 2014, 8, 129–136. 10.3855/jidc.3573. [DOI] [PubMed] [Google Scholar]
- Gould I. M.; Bal A. M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. 10.4161/viru.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyi-Loh C.; Mamphweli S.; Meyer E.; Okoh A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y.; Xia G.; Shi C.; Wan J.; Liu L.; Chen Y.; Wu Y.; Zhang W.; Zhou M.; He H.; Liu R. Therapeutic strategies against bacterial biofilms. Fundam. Res. 2021, 1, 193–212. 10.1016/j.fmre.2021.02.003. [DOI] [Google Scholar]
- Sohns J. M.; Bavendiek U.; Ross T. L.; Bengel F. M. Targeting Cardiovascular Implant Infection: Multimodality and Molecular Imaging. Circ. Cardiovasc. Imaging 2017, 10, e005376 10.1161/CIRCIMAGING.117.005376. [DOI] [PubMed] [Google Scholar]
- Khatoon Z.; McTiernan C. D.; Suuronen E. J.; Mah T.-F.; Alarcon E. I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067 10.1016/j.heliyon.2018.e01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joller N.; Weber S. S.; Müller A. J.; Sporri R.; Spörri P.; Sander P.; Hilbi H.; Oxenius A. Antibodies protect against intracellular bacteria by Fc receptor-mediated lysosomal targeting. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 20441–20446. 10.1073/pnas.1013827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F.; Coque T. M.; Galán J. C.; Martinez J. L. The Origin of Niches and Species in the Bacterial World. Front. Microbiol. 2021, 12, 657986. 10.3389/fmicb.2021.657986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. S.; Cooper M. A. Antibiotics in the clinical pipeline in 2011. J. Antibiot. 2011, 64, 413–425. 10.1038/ja.2011.44. [DOI] [PubMed] [Google Scholar]
- Cox E.; Nambiar S.; Baden L. Needed: Antimicrobial Development. N. Engl. J. Med. 2019, 380, 783–785. 10.1056/nejme1901525. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Majhi R. K.; Singh A.; Mishra M.; Tiwari A.; Chawla S.; Guha P.; Satpati B.; Mohapatra H.; Goswami L.; Goswami C. Carbohydrate-Coated Gold-Silver Nanoparticles for Efficient Elimination of Multidrug Resistant Bacteria and in Vivo Wound Healing. ACS Appl. Mater. Interfaces 2019, 11, 42998–43017. 10.1021/acsami.9b17086. [DOI] [PubMed] [Google Scholar]
- Burduşel A.-C.; Gherasim O.; Grumezescu A. M.; Mogoantă L.; Ficai A.; Andronescu E. Biomedical Applications of Silver Nanoparticles: An Up-to-Date Overview. Nanomaterials 2018, 8, 681. 10.3390/nano8090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavin Y. N.; Asnis J.; Häfeli U. O.; Bach H. Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. 10.1186/s12951-017-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y.; Ouyang S.; Umezawa N.; Cao J.; Ye J. Facet Effect of Single-Crystalline Ag3PO4 Sub-microcrystals on Photocatalytic Properties. J. Am. Chem. Soc. 2011, 133, 6490–6492. 10.1021/ja2002132. [DOI] [PubMed] [Google Scholar]
- Hsieh M.-S.; Su H.-J.; Hsieh P.-L.; Chiang Y.-W.; Huang M. H. Synthesis of Ag3PO4 Crystals with Tunable Shapes for Facet-Dependent Optical Property, Photocatalytic Activity, and Electrical Conductivity Examinations. ACS Appl. Mater. Interfaces 2017, 9, 39086–39093. 10.1021/acsami.7b13941. [DOI] [PubMed] [Google Scholar]
- Huang W.-C.; Lyu L.-M.; Yang Y.-C.; Huang M. H. Synthesis of Cu2O Nanocrystals from Cubic to Rhombic Dodecahedral Structures and Their Comparative Photocatalytic Activity. J. Am. Chem. Soc. 2012, 134, 1261–1267. 10.1021/ja209662v. [DOI] [PubMed] [Google Scholar]
- Gao F.; Shao T.; Yu Y.; Xiong Y.; Yang L. Surface-bound reactive oxygen species generating nanozymes for selective antibacterial action. Nat. Commun. 2021, 12, 745. 10.1038/s41467-021-20965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.; Liu W.; Gao X.; Lu Z.; Wu X.; Gao X. Mechanisms of Oxidase and Superoxide Dismutation-like Activities of Gold, Silver, Platinum, and Palladium, and Their Alloys: A General Way to the Activation of Molecular Oxygen. J. Am. Chem. Soc. 2015, 137, 15882–15891. 10.1021/jacs.5b10346. [DOI] [PubMed] [Google Scholar]
- Fang G.; Li W.; Shen X.; Perez-Aguilar J. M.; Chong Y.; Gao X.; Chai Z.; Chen C.; Ge C.; Zhou R. Differential Pd-nanocrystal facets demonstrate distinct antibacterial activity against Gram-positive and Gram-negative bacteria. Nat. Commun. 2018, 9, 129. 10.1038/s41467-017-02502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Song Z.; Gu J.; Li S.; Wu Y.; Han H. Nitrogen-Doped Carbon Quantum Dots for Preventing Biofilm Formation and Eradicating Drug-Resistant Bacteria Infection. ACS Biomater. Sci. Eng. 2019, 5, 4739–4749. 10.1021/acsbiomaterials.9b00583. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Bao C.; Li F.; Hou E.; Qin S.; Zhang Q.; Liu J. Discovery, Synthesis, and Biological Evaluation of Dunnianol-Based Mannich Bases against Methicillin-Resistant Staphylococcus aureus (MRSA). ACS Infect. Dis. 2020, 6, 2478–2489. 10.1021/acsinfecdis.0c00377. [DOI] [PubMed] [Google Scholar]
- Lee J.-H.; Kim Y.-G.; Khadke S. K.; Yamano A.; Watanabe A.; Lee J. Correction to Inhibition of Biofilm Formation by Candida albicans and Polymicrobial Microorganisms by Nepodin via Hyphal-Growth Suppression. ACS Infect. Dis. 2020, 6, 1283. 10.1021/acsinfecdis.0c00146. [DOI] [PubMed] [Google Scholar]
- Khan M. F.; Husain F. M.; Zia Q.; Ahmad E.; Jamal A.; Alaidarous M.; Banawas S.; Alam M. M.; Alshehri B. A.; Jameel M.; Alam P.; Ahamed M. I.; Ansari A. H.; Ahmad I. Anti-quorum Sensing and Anti-biofilm Activity of Zinc Oxide Nanospikes. ACS Omega 2020, 5, 32203–32215. 10.1021/acsomega.0c03634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse G.; Furthmüller J. Efficient iterative schemes forab initiototal-energy calculations using a plane-wave basis set. Phys. Rev. B: Condens. Matter Mater. Phys. 1996, 54, 11169–11186. 10.1103/physrevb.54.11169. [DOI] [PubMed] [Google Scholar]
- Kresse G.; Furthmüller J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comput. Mater. Sci. 1996, 6, 15–50. 10.1016/0927-0256(96)00008-0. [DOI] [PubMed] [Google Scholar]
- Perdew J. P.; Burke K.; Ernzerhof M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. 10.1103/physrevlett.77.3865. [DOI] [PubMed] [Google Scholar]
- Kresse G.; Joubert D. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys. Rev. B: Condens. Matter Mater. Phys. 1999, 59, 1758–1775. 10.1103/physrevb.59.1758. [DOI] [Google Scholar]
- Puvvada N.; Panigrahi P. K.; Pathak A. Room temperature synthesis of highly hemocompatible hydroxyapatite, study of their physical properties and spectroscopic correlation of particle size. Nanoscale 2010, 2, 2631–2638. 10.1039/c0nr00611d. [DOI] [PubMed] [Google Scholar]
- Botelho G.; Andres J.; Gracia L.; Matos L. S.; Longo E. Photoluminescence and Photocatalytic Properties of Ag3PO4Microcrystals: An Experimental and Theoretical Investigation. ChemPlusChem 2016, 81, 202–212. 10.1002/cplu.201500485. [DOI] [PubMed] [Google Scholar]
- Kalita H.; Pal P.; Dhara S.; Pathak A. Fabrication and characterization of polyvinyl alcohol/metal (Ca, Mg, Ti) doped zirconium phosphate nanocomposite films for scaffold-guided tissue engineering application. Mater. Sci. Eng. C 2017, 71, 363–371. 10.1016/j.msec.2016.09.063. [DOI] [PubMed] [Google Scholar]
- Sinclair T. R.; van den Hengel S. K.; Raza B. G.; Rutjes S. A.; de Roda Husman A. M.; Peijnenburg W. J. G. M.; Roesink H. D. W.; de Vos W. M. Surface chemistry-dependent antiviral activity of silver nanoparticles. Nanotechnology 2021, 32, 365101. 10.1088/1361-6528/ac03d6. [DOI] [PubMed] [Google Scholar]
- Shin J. U.; Gwon J.; Lee S.-Y.; Yoo H. S. Silver-Incorporated Nanocellulose Fibers for Antibacterial Hydrogels. ACS Omega 2018, 3, 16150–16157. 10.1021/acsomega.8b02180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston S. D.; Willis L. M. Racing to build a wall: glycoconjugate assembly in Gram-positive and Gram-negative bacteria. Curr. Opin. Struct. Biol. 2021, 68, 55–65. 10.1016/j.sbi.2020.11.013. [DOI] [PubMed] [Google Scholar]
- Wang L.; Hu C.; Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. 10.2147/ijn.s121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhang W.; Niu J.; Chen Y. Mechanism of photogenerated reactive oxygen species and correlation with the antibacterial properties of engineered metal-oxide nanoparticles. ACS Nano 2012, 6, 5164–5173. 10.1021/nn300934k. [DOI] [PubMed] [Google Scholar]
- Janani S.; Stevenson P.; Veerappan A. Activity of catalytic silver nanoparticles modulated by capping agent hydrophobicity. Colloids Surf. B Biointerfaces 2014, 117, 528–533. 10.1016/j.colsurfb.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Schieber M.; Chandel N. S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloni G.; Marti E.; Slaveykova V. I. Interactive effects of copper oxide nanoparticles and light to green alga Chlamydomonas reinhardtii. Aquat. Toxicol. 2016, 170, 120–128. 10.1016/j.aquatox.2015.11.018. [DOI] [PubMed] [Google Scholar]
- Dong Y.; Zhu H.; Shen Y.; Zhang W.; Zhang L. Antibacterial activity of silver nanoparticles of different particle size against Vibrio Natriegens. PLoS One 2019, 14, e0222322 10.1371/journal.pone.0222322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anees Ahmad S.; Sachi Das S.; Khatoon A.; Tahir Ansari M.; Afzal M.; Saquib Hasnain M.; Kumar Nayak A. Bactericidal activity of silver nanoparticles: A mechanistic review. Mater. Sci. Energy Technol. 2020, 3, 756–769. 10.1016/j.mset.2020.09.002. [DOI] [Google Scholar]
- Lee N.-Y.; Ko W.-C.; Hsueh P.-R. Nanoparticles in the Treatment of Infections Caused by Multidrug-Resistant Organisms. Front. Pharmacol. 2019, 10, 1153. 10.3389/fphar.2019.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. M.; Polash S. A.; Takikawa M.; Shubhra R. D.; Saha T.; Islam Z.; Hossain S.; Hasan M. A.; Takeoka S.; Sarker S. R. Investigation of the Antibacterial Activity and in vivo Cytotoxicity of Biogenic Silver Nanoparticles as Potent Therapeutics. Front. Bioeng. Biotechnol. 2019, 7, 239. 10.3389/fbioe.2019.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallo da Silva B.; Caetano B. L.; Chiari-Andréo B. G.; Pietro R. C. L. R.; Chiavacci L. A. Increased antibacterial activity of ZnO nanoparticles: Influence of size and surface modification. Colloids Surf. B Biointerfaces 2019, 177, 440–447. 10.1016/j.colsurfb.2019.02.013. [DOI] [PubMed] [Google Scholar]
- Wu B.; Zhuang W.-Q.; Sahu M.; Biswas P.; Tang Y. J. Cu-doped TiO2 nanoparticles enhance survival of Shewanella oneidensis MR-1 under Ultraviolet Light (UV) exposure. Sci. Total Environ. 2011, 409, 4635–4639. 10.1016/j.scitotenv.2011.07.037. [DOI] [PubMed] [Google Scholar]
- Padmavathy N.; Vijayaraghavan R. Interaction of ZnO Nanoparticles with Microbes-A Physio and Biochemical Assay. J. Biomed. Nanotechnol. 2011, 7, 813–822. 10.1166/jbn.2011.1343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.