Graphical abstract
Abbreviations: DMA, dimethylarsinic acid; DEGs, Differentially Expressed Genes; ER, endoplasmic reticulum; EXT, extinction (a measure of optical density); GO, gene ontology; HgCl2, mercury(ii) chloride; iAs, inorganic arsenic; L1, first larval stage C. elegans; LD50, the median lethal dose per kilogram of body weight; LOEL, lowest observed effect level; meHgCl, methylmercury chloride; NaAsO2, sodium (meta)arsenite; NOEL, no observed effect level; OxStrR, Oxidative Stress Response; TOF, time of flight (a measure of size); UPR, Unfolded Protein Response
Keywords: Alternative Toxicity Model, Arsenic, Inorganic, Mercury, Organic, Predictive Toxicology
Highlights
-
•
Similar developmental delays and oxidative stress with 20x DMA relative to NaAsO2.
-
•
Dissimilar gene expression and locomotion with organic vs. inorganic mercury.
-
•
Dissimilar unfolded protein responses for organic vs. inorganic arsenic and mercury.
-
•
Across phyla, methylation has opposite effects on arsenic vs. mercury toxicity.
Abstract
Exposures to mercury and arsenic are known to pose significant threats to human health. Effects specific to organic vs. inorganic forms of these toxic elements are less understood however, especially for organic dimethylarsinic acid (DMA), which has recently been detected in pups of rodent dams orally exposed to inorganic sodium (meta)arsenite (NaAsO2). Caenorhabditis elegans is a small animal alternative toxicity model. To fill data gaps on the effects of DMA relative to NaAsO2, C. elegans were exposed to these two compounds alongside more thoroughly researched inorganic mercury chloride (HgCl2) and organic methylmercury chloride (meHgCl). For timing of developmental milestone acquisition in C. elegans, meHgCl was 2 to 4-fold more toxic than HgCl2, and NaAsO2 was 20-fold more toxic than DMA, ranking the four compounds meHgCl > HgCl2 > NaAsO2 ≫ DMA for developmental toxicity. Methylmercury induced significant decreases in population locomotor activity levels in developing C. elegans. DMA was also associated with developmental hypoactivity, but at >100-fold higher concentrations than meHgCl. Transcriptional alterations in native genes were observed in wild type C. elegans adults exposed to concentrations equitoxic for developmental delay in juveniles. Both forms of arsenic induced genes involved in immune defense and oxidative stress response, while the two mercury species induced proportionally more genes involved in transcriptional regulation. A transgenic bioreporter for activation of conserved proteosome specific unfolded protein response was strongly activated by NaAsO2, but not DMA at tested concentrations. HgCl2 and meHgCl had opposite effects on a bioreporter for unfolded protein response in the endoplasmic reticulum. Presented experiments indicating low toxicity for DMA in C. elegans are consistent with human epidemiologic data correlating higher arsenic methylation capacity with resistance to arsenic toxicity. This work contributes to the understanding of the accuracy and fit-for-use categories for C. elegans toxicity screening and its usefulness to prioritize compounds of concern for further testing.
1. Introduction
The Agency for Toxic Substances and Disease Registry (ATSDR) prioritizes chemical compounds of concern based on their toxicity and potential for human exposure. Arsenic and mercury are at or near the top of this list of hazardous substances and are known to be especially harmful during early development (ATSDR., 2019, Congress, 2021, FDA, 2020). The bioavailability and effects of arsenic and mercury are known to vary based on chemical form to the extent that safety and mode of action assessments for one form do not apply to other forms, highlighting the need to better understand their relative toxicities (Davidson et al., 2004, Luvonga et al., 2020). The Food and Drug Administration’s (FDA’s) “Closer to Zero” initiative identifies actions the agency will take towards reducing exposure to toxic elements from foods consumed by babies and young children and supports continuing research on the impacts of toxic elements during development (FDA, 2021). For mercury, lower limits on intake levels are recommended for methylmercury relative to mercury chloride due to the neurodevelopmental toxicity of organic mercury (Atsdr, 1999, Efsa, 2012). For inorganic arsenic (iAs), the provisional tolerable weekly intake (PTWI) established by the World Health Organization in 1986 of 15 µg/kg body weight was withdrawn in 2009 due to carcinogenic effects at lower exposures, and a new PTWI has not been set (Naess et al., 2020). Recently, dimethylarsinic acid (DMA) was found in the pups of rodent dams fed iAs (Twaddle et al., 2018), raising concerns about the developmental effects of DMA. iAs is generally considered more toxic than organic forms of arsenic, however mammalian oral toxicity data for organic arsenic species is limited, particularly for DMA (EFSA., 2009, Faoun, who. , 2011, FDA, 2016). DMA has been detected in rice, and since rice flour is an ingredient in many baby foods (FDA, 2016), more information on the developmental effects of DMA is needed.
The FDA’s Predictive Toxicology Roadmap supports the identification, development, and assessment of emerging methods and new technologies for use in risk assessments for regulatory purposes (FDA, 2017). The qualification and progress towards utilization of these leading-edge predictive toxicology tools are priorities for FDA’s Center for Food Safety and Applied Nutrition (CFSAN, 2015). Caenorhabditis elegans is a small, non-pathogenic nematode that can be maintained at low cost and handled using standard in vitro equipment and techniques, and its 3-day development from egg to egg-laying adult allows for rapid developmental toxicity testing. C. elegans assays provide data from a whole animal with intact and metabolically active digestive, reproductive, sensory, and neuromuscular systems (Hunt et al., 2020). There is significant conservation between C. elegans and humans for biological pathways involved in organismal development and neurotransmission (Leung et al., 2008, Ruszkiewicz et al., 2018), and the toxicodynamics for many types of chemicals is similar in C. elegans and mammals (Hartman et al., 2021). Conserved alimentary features such as acidic and non-acidic portions of the digestive tract, digestive enzymes, and brush boarder function make C. elegans a potential model for predictive oral toxicity assessment (Hunt, 2017). A single C. elegans technician can assess a dozen or more compounds or concentrations in a week for endpoints such as viability, locomotor activity, developmental timing, or pathway of toxicity specific transgene expression. While this type of low-to-medium-throughput C. elegans screening cannot replace a descriptive toxicology study in lab mammals, it is very rapid and inexpensive by comparison, and can provide useful information on conserved modes of toxic action and apical endpoint responses (Avila et al., 2020, Hartman et al., 2021, Masjosthusmann et al., 2018, Parish et al., 2020). Several studies have demonstrated that toxicity ranking screens in C. elegans can predict developmental toxicity or LD50 ranking in mammals (Boyd et al., 2010, Boyd et al., 2016, Hunt et al., 2012, Li et al., 2013), indicating that C. elegans has the potential to provide a bridge between in vitro human cell based assays and mammalian in vivo oral toxicity testing (Lagido et al., 2015).
Several lines of evidence indicate that C. elegans is a good model to study conserved aspects of toxic responses to arsenic. From worms to mammalian cell cultures, exposure to iAs activates the p38 mitogen-activated protein kinase cascade (Inoue et al., 2005, Kang and Lee, 2008, Wang et al., 2013), induces Nrf/SKN-1 regulated phase II detoxification enzymes and protein folding‐sensitive molecular chaperones (Del Razo et al., 2001, Oliveira et al., 2009, Tam and Wang, 2020, Yu et al., 2016), alters innate immune function (Dangleben et al., 2013, Escudero-Lourdes, 2016, Ma et al., 2021), and has similar effects on mitochondrial function (Luz et al., 2016, Luz et al., 2017, Tam and Wang, 2020). The C. elegans model reflects some of the differing mammalian effects of organic vs. inorganic mercury. For rodents and C. elegans, meHgCl is more toxic than HgCl2 during early development, and has greater effects on locomotor activity (McElwee and Freedman, 2011, Wyatt et al., 2016). For mammals and C. elegans, methylmercury is a developmental neurotoxin, and the very young are more sensitive than adults to its effects (Helmcke et al., 2009, Ruszkiewicz et al., 2018). Effects of organic vs. iAs may also be conserved. Therefore, we assessed the effects of DMA, sodium (meta)arsenite (NaAsO2), methylmercury chloride (meHgCl), and mercury chloride (HgCl2) on C. elegans development and stage specific locomotor activity, adult native gene expression, and pathway of toxicity specific transgene expression.
2. Material and methods
2.1. Chemicals
Test chemicals (Table 1) were purchased from MilliporeSigma (Burlington, MA). Fresh dosing solutions were prepared for each experiment in Milli-Q purified water within two hours prior to dosing. Test chemicals readily dissolved in water with exception of meHgCl, which was first dissolved in water at 200 µg/ml for 30 min with stirring prior to dilution and dosing.
Table 1.
Chemicals Selected for Testing.
| Compound * | Abbreviation | CAS RN | LD50 | Oral RfD | TWI |
|---|---|---|---|---|---|
| sodium (meta)arsenite | NaAsO2 | 7784–46-5 | 41 mg/kg (rat, oral) | 0.3 µg/kg-day (IRIS, 1991) | withdrawn # |
| dimethylarsinic acid (DMAV) | DMA | 75–60-5 | 700 mg/kg (rat, oral) 1.2 g/kg (mouse, oral) |
not established | not established |
| mercury chloride | HgCl2 | 7487–94-7 | 1 mg/kg (rat, oral) 6 mg/kg (mouse, oral) |
0.3 µg/kg-day (IRIS, 1995) | 4 µg Hg/kg-week (EFSA, 2012) |
| methylmercury chloride | meHgCl | 115–09-3 | 30 mg/kg (rat, oral) 58 mg/kg (mouse, oral) |
0.1 µg/kg-day (IRIS, 2001) | 1.3 µg Hg/kg-week (EFSA, 2012) |
CAS RN: The Chemical Abstracts Service (CAS) Registration Number (RN) is a unique numerical identifier designating a specific substance.
LD50: the median lethal dose per kilogram of body weight.
RfD: Reference Dose for chronic oral exposure, an estimate of a daily exposure that is likely to be without.
an appreciable risk of deleterious effects during a lifetime.
TWI: tolerable weekly intake, the amount per kilogram of body weight that can be ingested per week without risk of adverse health effects.
# Provisional TWI of 15 µg As/kg was withdrawn in 2009 as too high for safety (Naess et al., 2020).
* Compounds and CAS RNs listed are those purchased and used in this study. Reference values (LD50, RfD, and TWI) may be for slightly different compounds, for example the available oral RfD is for methylmercury, CAS RN 22967–92-6.
2.2. Worm maintenance
C. elegans wild type N2, LD1171 (ldIs3 [gcs-1p::GFP + rol-6(su1006)]), CL2166 (dvIs19 [(pAF15)gst-4p::GFP::NLS] III), SJ4005 (zcIs4 [hsp-4p::GFP] V), and SJ4003 (zcIs3 [aip-1p::GFP] I) strains were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Strains were grown under continuously well-fed conditions on agar plates with OP50 E. coli as a feeder organism for three generations prior to hypochlorite treatment to isolate eggs and transfer to C. elegans Habitation Medium (CeHM) as previously described (Nass and Hamza, 2007). Growth in CeHM containing 20% non-fat cows’ milk allows C. elegans larvae to develop at the same rate as with E. coli (Clegg et al., 2002, Sprando et al., 2009). Aliquots were frozen at −80 °C soon after transfer to CeHM, and fresh aliquots were thawed every 6 months to avoid genetic drift in test cohorts. Cultures were fed fresh CeHM regularly in a biological cabinet using sterile technique and maintained at 20 °C on shakers in hot/cold incubators in vented, polystyrene flasks. Thawed aliquots were allowed a minimum of three weeks growth in CeHM prior to toxicity testing to minimize effects of freeze-thawing on stress resistance and gene expression. For test cohorts, well-fed gravid C. elegans were subjected to hypochlorite treatment to isolate eggs. Eggs were allowed to hatch overnight in non-nutrient M9 buffer for 19 +/- 1 hr to obtain age-synchronized first larval stage (L1) worms. Isolates containing > 1 dauer per approximately 5,000 eggs were discarded along with the culture flasks they were obtained from to ensure cohorts originated from well-fed cultures and were exposed to minimal amounts of dauer pheromone. Synchronized L1s were centrifuged, resuspended to approximately 900 worms per mL CeHM, and then either dosed immediately for developmental toxicity assessment, or maintained with regular media changes for dosing at later stages.
2.3. Developmental toxicity testing
3-day exposures of synchronized L1s were conducted in sterile CELLSTAR 12-well polystyrene plates with lids and carried out in two wMicroTrackers (InVivo Biosystems, Eugene, OR) side-by-side inside a 20 °C incubator. 100 µL of water or 10x dosing solution in water were added to 900 µL of L1s in CeHM in a diagonal pattern, with one water control and three dosed conditions per plate, and three replicate wells per condition. Each worm Development and Activity Test (wDAT) trial was performed as four independent experiments using freshly prepared L1 cohorts and dosing solutions. Over the four experiments, each well placement was used once and only once for each condition to account for small differences in wMicroTracker (wMT) infrared beam placement. wMT output was read in 30 min increments as mean beam-interruptions per condition over the three replicate wells. Peaks were smoothed by taking the mean of seven half-hour timepoints, and the highest value at the third larval stage (L3) was recorded as peak height for population locomotor activity, and the corresponding time as time to reach L3. Values were normalized to in-plate controls and presented as relative time to peak (delay) and relative peak height (population locomotor activity). Students’ T-test p-values of < 0.05 were considered statistically significant. Based on differences between controls (listed as 0 L-R in graphs) from simultaneously run plates over the study, changes of > 4% for developmental delay and > 5% for locomotor activity were considered biologically significant.
2.4. Adult gene expression – Dosing
Worms were age-synchronized at L1 as described (section 2.2) and maintained until the fourth larval stage (L4). Wildtype N2 C. elegans reach mid-L4 at about 50 h post-L1 hatching at 20 °C, but transgenic strains can differ from the wildtype in the time to reach L4 due to their genetic alterations. Therefore instead of dosing at a standard timepoint, developmental morphology was closely monitored as previously described (Seydoux et al., 1993) in order to dose at mid-L4. All exposures were carried out in sterile Falcon 24-well polystyrene plates with lids and maintained on shakers in a 20 °C incubator. After the exposure period, worms were washed twice with M9 nematode buffer, and then either resuspended in TRIzol Reagent (ThermoFisher Scientific, Waltham, MA) and frozen for subsequent microarray assessments, or directly run through a Complex Object Parametric Analyzer and Sorter (COPASTM, Union Biometrica, Holliston, MA) for transgene expression analysis. For microarray assessment, N2 C. elegans were exposed from mid-L4 to 8 µM 5-Fluoro-2′-deoxyuridine (FUdR, a contraceptive used to avoid confounding embryonic and larval gene expression), and 24 h later 900 µL of adults in CeHM plus FUdR were dosed with 100 µL of 10x dosing solutions and exposed for 24 h. For COPAS assessment, C. elegans were exposed without FUdR from mid-L4 for 24 h. For SJ4005 hsp-4p::GFP assessment, both heat shock and tunicamycin (30 µg/ml in 0.1% DMSO) were tested for use as positive controls (Hou et al., 2014, Tsialikas and Argon, 2017). Heat stress treatments of C. elegans in CeHM at 30 °C for 1, 3 and 5 h did not induce a change in GFP levels, while 30 µg/ml tunicamycin resulted in an average 1.7x increase in expression, so tunicamycin was utilized as the positive control (Supplemental Fig. 1A).
2.5. RNA extraction and gene expression analysis using microarray technology.
Total RNA was extracted from C. elegans samples using miRNeasy Mini kits (Qiagen, Valencia, CA). Whole genome expression in the C. elegans treated with water (control group), 10 µg/ml NaAsO2, 200 µg/ml DMA, 2 µg/ml HgCl2, or 0.5 µg/ml meHgCl for 24 h was determined by using Cellegans_UnrestrictedGE_G2519F_020186 Microarray (Agilent Technologies, Santa Clara, CA). The C. elegans genome codes for approximately 20,000 genes (Hillier et al., 2005), and according to the manufacturer, this chip represents > 18,000 unique genes, providing > 90% coverage. Sample labeling and microarray processing were performed as detailed in the manufacturer’s protocol. The hybridized slides were scanned with an Agilent SureScan DNA Microarray Scanner (Agilent Technologies, Santa Clara, CA) at 3 µm resolution. The resulting images were analyzed by determining the Cy3 fluorescence intensity of all gene spots (features) on each array using the Agilent Feature Extraction Software (Version 11.5). The raw data was uploaded into the ArrayTrack database (Fang et al., 2017). The median fluorescence intensity of all the pixels within one feature was taken as the intensity value for that feature. The raw intensity values were then normalized using 75 percentile channel scaling normalization within ArrayTrack. To identify genes that were differentially expressed between the control group and each experimental group, Benjamini-Hochberg adjusted p-values were calculated (Benjamini and Hochberg, 1995). Genes that passed the 1.5-fold change criterion and had Benjamini-Hochberg adjusted p-value < 0.1 were considered statistically significant. Microarray results are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE196891.
2.6. Adult native gene expression – Microarray data mining
Standard software tools for differential expression analysis DAVID (Huang da et al., 2009) and Ingenuity Pathway Analysis (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA) yielded little information with the C. elegans differentially expressed genes (DEGs). Therefore, two different methods for expression analysis were also utilized, 1. analysis of WormBase generated Expression Clusters and 2. curation (section 2.7). With the first method, sequence identifiers for all DEGs were entered into WormBase’s SimpleMine software (WormBase, 2021) for gene ontology (GO) associations, description, expression cluster summary, human homologs, and associated human diseases. DEGs without any information within these parameters were excluded from further analysis. WormBase expression cluster summaries were used to identify DEGs also affected by other chemicals and drugs. To account for variability in the number of DEGs induced by the different conditions, data are presented as a percent or fraction of all DEGs for each test article. Some evaluation was required for inclusion into GO term categories. For the Innate Immune response, search terms ‘innate,’ ‘defense’ and ‘bacteri’ were utilized to screen DEGs for inclusion in this category. The Locomotion category included DEGs with ‘locomot’ or ‘behavior’ in the WormBase gene description or GO terms. For the transcriptional regulation (Transcriptional Reg.) category, DEGs with GO terms including ‘transcription’ and ‘histone’ were considered for inclusion. The terms ‘unfolded,’ ‘folding,’ ‘chaperone,’ ‘heat,’ and ‘hsp’ were used to search for DEGs to include in the Unfolded Protein Response (UPR) category.
2.7. Adult native gene expression – Curated DEGs
In the second method for differential expression analysis, genes with the greatest fold change (the 100 most downregulated for all four conditions and the 100 most upregulated, or all upregulated genes if <100) were further assessed for function. A literature search was conducted for each of these genes to determine if they fit into any patterns or pathways. Unlike the GO term analysis of all DEGs above, in this curated analysis, genes were included in the Immune Resp. category only if they were reported to be regulated by 1.5-fold or more in the same direction with exposure to at least two pathogens or in at least two separate studies, or if RNAi indicated that they were required for microbial defense (Block et al., 2015, Bolz et al., 2010, Engelmann et al., 2011, Evans et al., 2008, Irazoqui et al., 2010, Muir and Tan, 2008, O'Rourke et al., 2006, Pujol et al., 2008, Pukkila-Worley et al., 2011, Ren et al., 2009, Troemel et al., 2006, Wong et al., 2007, Yang et al., 2016). For the Nrf/SKN-1 and DAF-16 response pathway response categories, DEGs were included only if they were shown to be regulated in the same direction by SKN-1 (Hasegawa et al., 2010, van der Hoeven et al., 2011, Jones et al., 2013, Miller et al., 2011, Oliveira et al., 2009, Park et al., 2009, Przybysz et al., 2009, Steinbaugh et al., 2015, Yanase et al., 2020) or DAF-16 (Kaplan et al., 2019, Kim and Sun, 2007, McElwee et al., 2003, Minniti et al., 2009, Murphy et al., 2003, Nag et al., 2017, Oh et al., 2006, Tepper et al., 2013).
2.8. Adult transgene expression
The parameters of fluorescence, time of flight (TOF, an indicator of size) and extinction (EXT, an indicator of optical density) were assessed with a COPAS™. Green fluorescent protein (GFP) fluorescence was used to measure oxidative stress response (OxStrR, strains LD1171 gcs-1p::GFP and CL2166 gst-4p::GFP) and unfolded protein response (UPR, strains SJ4003 aip-1p::GFP and SJ4005 hsp-4p::GFP), while TOF and EXT, which can be adversely affected by toxic substances, were used to define population health for accurate analysis and phenotypic comparison to controls. Because progeny can be excluded from the analysis by gating, FUdR was not used. Compared to the OxStrR biosensor strains used in this study, both UPR transgenic strains were difficult to grow. SJ4003 and SJ4005 took longer to reach L4, and populations were not as abundant as with the wildtype N2 and OxStrR transgenic strains Cl2166 and LD1171, necessitating extra care in baseline phenotype assessment in order to consistently dose at mid-L4. Concentrations tested were determined through range finding experiments. Dosed populations with TOF or EXT morphology values that differed from matched controls by > 20% were excluded from analyses. These changes in morphology parameters were detected at lower concentrations in the UPR strains than in the wild type and OxStrR strains. Because adverse conditions can influence autofluorescence levels, which are not distinguishable from GFP by COPAS, wildtype N2 worms were also evaluated for every condition. Tested concentrations did not affect autofluorescence levels in N2 (data not shown). Data shown are the mean green fluorescence values relative to control and include no effect levels where available up to the highest concentration obtainable that maintained TOF and EXT within 80% of control. Significance was determined from a minimum of three independent experiments per exposure group for Student’s T-test p-values *<0.05, ** <0.005.
3. Results
3.1. Developmental toxicity and stage-specific activity changes
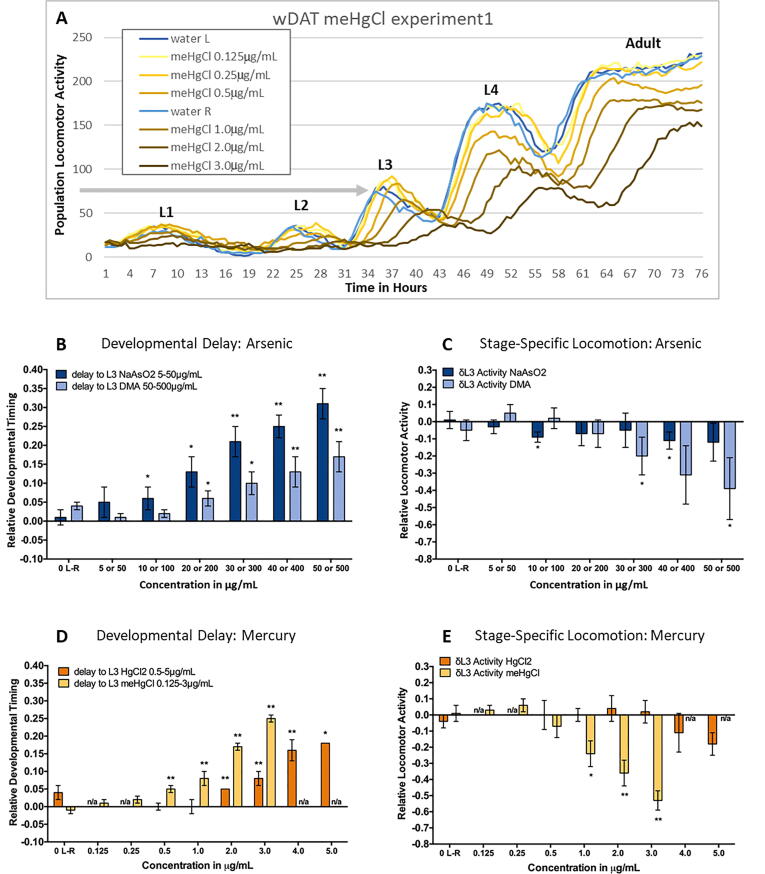
C. elegans develop through four distinct larval stages (L1 – L4) prior to adulthood, with a period of inactivity during cuticle molting between each stage. wMicroTrackers (wMTs) use infrared beam interruptions to assess movement within wells of multi-well plates (Simonetta, 2010). Using wMTs, the worm Development and Activity Test (wDAT) tracks these periods of activity and inactivity to simultaneously monitor both the timing of developmental stage acquisition and population locomotor activity levels (Hunt et al., 2018). A wDAT right shift in peak timing indicates developmental delay, and a change in stage-specific peak height indicates hyper- or hypoactivity (Fig. 1A). Over the course of the study, the maximum differences between controls in plates run side by side in the time to reach the 3rd larval stage (L3) peak and the height of L3 peaks was 4% and 5% respectively, providing an indication of assay variability. This experimental variability among negative controls for milestone acquisition timing and stage-specific locomotion is indicated by 0 L-R (Fig. 1B-1E) and was used as a cut-off for wDAT biological significance.
Fig. 1.
Developmental toxicity of inorganic vs. organic arsenic and mercury. (A) A representative example of the readout from a single worm Development and Activity Test (wDAT) for methylmercury chloride is shown. The y-axis indicates population locomotor activity levels as measured by mean infrared beam interruption values per well from a single experiment, graphed over half hour time increments on the x-axis. L1 through L4 indicates the four larval stages of C. elegans development. There are two separate sets of water controls (L and R), represented by blue and light blue lines for the left and right plates run simultaneously. The horizontal gray arrow from the y-axis indicates the control peak height of the 3rd larval stage (L3). The corresponding times for the two controls to reach the peak height for L3 are 35.5 and 36 h in this experiment. The variability for L3 peak height and timing between controls in separate plates run side-by-side was used as a measure of experimental variability and is shown in 1B-E as ‘0 L-R’. (B-E) Bars and error bars represent the means and standard deviations from four independent wDAT experiments, with the exception of those for 5.0 µg/mL HgCl2 for which there were data from only two of four experiments. (B&D) The delay in the time to reach L3 relative to controls is plotted. (C&E) L3 locomotor activity peak level relative to controls. Not assessed (n/a). T-test p-values *<0.05, ** <0.005. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The lowest observed effect level (LOEL) of 6% for delay in the time to reach L3 was observed at 10 µg/ml (76 µM) sodium arsenite (NaAsO2) or 200 µg/ml (1.4 mM) dimethylarsinic acid (DMA). The delay was 13% with 20 µg/ml (154 µM) NaAsO2 or 400 µg/ml (2.8 mM) DMA (Fig. 1B), indicating that about 20 times more DMA was required to achieve the similar levels of developmental delay as with NaAsO2. DMA concentrations that induced 10% to 17% delays in time to reach L3 (300–500 µg/ml) were also associated with reduced population locomotor activity in a dose–response curve from −20% to −39%. There was some hypoactivity with NaAsO2, but the changes were smaller, in the range of −5% to −12%, and not in a dose response pattern (Fig. 1C).
A delay in developmental timing of 5% was observed at 0.5 µg/ml (2 µM) methylmercury chloride (meHgCl) or 2.0 µg/ml (7.5 µM) mercury chloride (HgCl2), and an approximately 16% delay at 2 µg/ml (8 µM) meHgCl or 4 µg/ml (15 µM) HgCl2 (Fig. 1D), indicating that for developmental delay, organic meHgCl is two to four times more toxic than inorganic HgCl2. At 1.0 to 3.0 µg/ml meHgCl, concentrations above the LOEL for delay, stage specific population locomotor activity levels decreased with dose, from −24% to −53%. There was some hypoactivity with HgCl2, but the effects were smaller (-11% to −18%), and too variable to be considered significant (Fig. 1E). These results indicate that at concentrations relevant to developmental delay, organic forms of arsenic and mercury induce additional modes of toxic action beyond those that affect developmental timing, though there is a two orders of magnitude difference in the range of concentrations at which hypoactivity was detected for DMA vs. meHgCl.
The distinctive patterns seen with the wDAT (Fig. 1A) are dependent on the population in each well progressing synchronously through the four larval stages of development. Asynchronous development results in a wDAT readout without distinguishable peaks and valleys, and C. elegans larvae observed by microscopy at a variety of developmental stages at the end of the three-day test (Hunt et al., 2018). This loss of synchronous development occurred in two of four HgCl2 experiments at 5 µg/ml, so data presented are from only two experiments (Fig. 1D-E).
3.2.1. Adult native gene expression – Microarray analysis
Gene expression patterns in C. elegans larvae change rapidly as they progress through their developmental stages. Assessment in C. elegans larvae of toxic effects on gene expression at test chemical concentrations that also delay development is complicated by larval stage specific gene expression (our unpublished observations). For test chemical specific mode of action evaluation, we therefore exposed adult C. elegans for 24 h to concentrations that were equitoxic LOELs for developmental delay in juveniles, 10 µg/ml (76 µM) NaAsO2, 200 µg/ml (1.4 mM) DMA, 2.0 µg/ml (7.5 µM) HgCl2, or 0.5 µg/ml (2 µM) meHgCl.
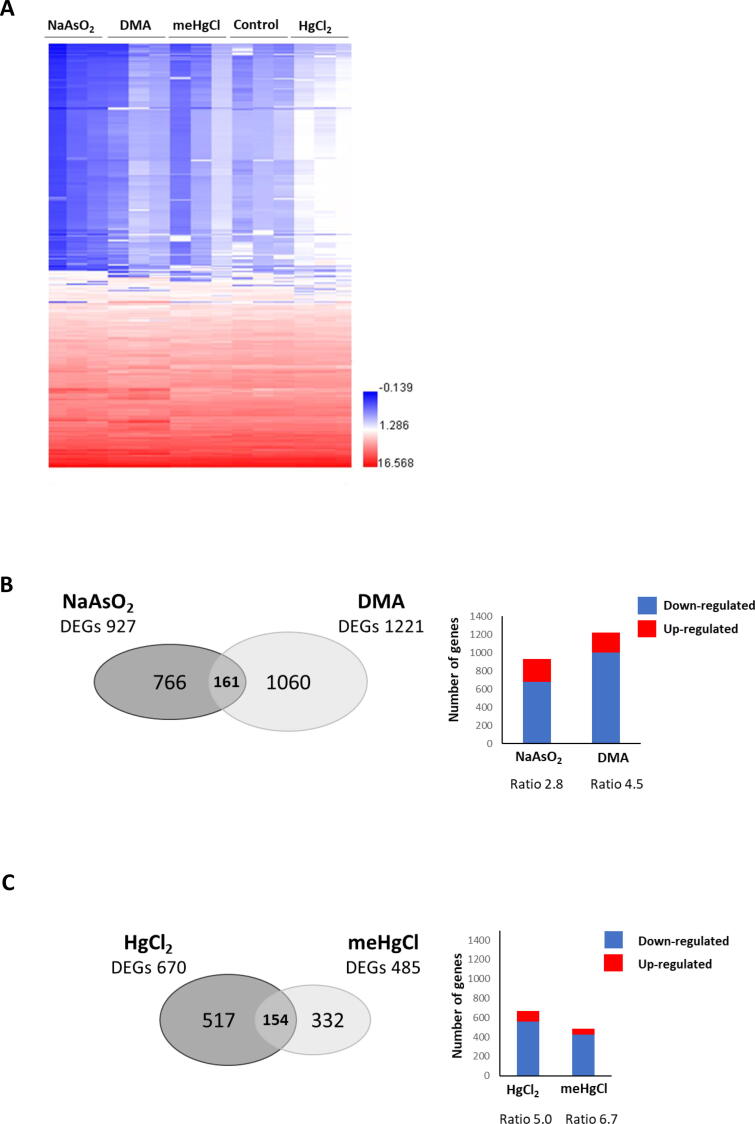
The results of high-throughput whole genome microarray analyses showed changes in the transcriptome of adult C. elegans exposed to NaAsO2, DMA, HgCl2, or meHgCl relative to the water control group. An unsupervised hierarchical clustering of the gene expression data shows that the different treatments could be distinguished by gene expression profile (Fig. 2A). To identify genes that were differentially expressed between the control group and each experimental group, Benjamini-Hochberg adjusted p-values were calculated. Genes that passed 1.5-fold change criterion and had Benjamini-Hochberg adjusted p-value < 0.1 were considered statistically significant.
Fig. 2.
Microarray analysis of gene expression with inorganic vs. organic arsenic and mercury. Whole-genome microarray transcriptomic analysis of C. elegans exposed to 10 µg/ml NaAsO2, 200 µg/ml DMA, 0.5 µg/ml meHgCl, or 2.0 µg/ml HgCl2. A) Heat maps illustrating differences in global gene expression profiles between control and exposed adult C. elegans. B) Venn diagram illustrating the number of differentially expressed genes and their analysis in C. elegans exposed to NaAsO2 or DMA. C) Venn diagram illustrating the number of differentially express genes and their analysis in C. elegans exposed to meHgCl or HgCl2.
A total of 927 and 1221 differentially expressed genes (DEGs) were found in C. elegans treated with 10 µg/ml NaAsO2 or 200 µg/ml DMA, respectively (Fig. 2B). Interestingly, only 161 DEGs were common for these two chemicals. The treatment with NaAsO2 and DMA induced down-regulation of a greater number of genes in comparison with up-regulated genes, however the organic form of arsenic induced more down-regulated genes than the inorganic arsenic. This was shown by the ratio of down-regulated to up-regulated DEGs of 2.8 for NaAsO2 and 4.5 for DMA.
Adult C. elegans exposed to 2 µg/ml HgCl2 or 0.5 µg/ml meHgCl induced 670 and 485 DEGs, respectively. Out of these genes, 154 were commonly expressed by the two treatments. Both chemicals induced a greater number of down-regulated genes in comparison with up-regulated genes, but the organic form of mercury induced a greater number of down-regulated genes than its inorganic form. The ratio of down-regulated/up-regulated DEGs was 5 for HgCl2 and 6.7 for meHgCl (Fig. 2C).
3.2.2. Adult native gene expression – Analysis of all differentially expressed genes
DEGs for each condition were entered into WormBase’s SimpleMine software (WormBase, 2021), and all genes for which there was any expression or functional information, 1073 for 200 µg/ml DMA, 721 for 10 µg/ml NaAsO2, 582 for 2.0 µg/ml HgCl2, and 410 for 0.5 µg/ml meHgCl, were included in the following analyses. At tested concentrations, few genes with apoptosis or programmed cell death (Apoptosis/PCD) related GO terms were differentially regulated by the two forms of arsenic (Fig. 3A). In contrast, while the numbers were small, HgCl2 affected the highest proportion of genes in this category. Apoptosis/PCD genes ced-1 (CEll Death abnormality 1), drp-1 (Dynamin-Related Protein 1), dyn-1 (Dynamin 1), mcd-1 (Modifier of Cell Death phenotype), and apoptotic cell clearance regulator lst-4 (Almendinger et al., 2011, WormBase., 2021) were all upregulated by HgCl2 (Fig. 3B & Table 2).
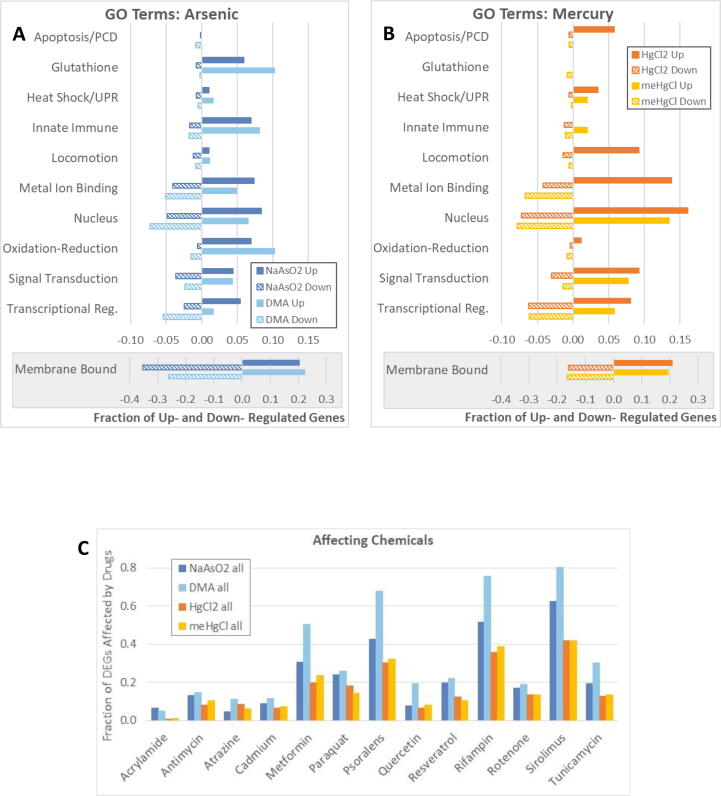
Fig. 3.
Gene Ontology (GO) terms, and genes also affected by other chemicals. WormBase’s GO terms and descriptions relating to specific functions were used to assign into categories all differentially expressed genes (DEGs) for which there was expression, function, and/or homology information in WormBase. (A-B) The number of DEGs varied among conditions, therefore bars represent the fraction of all DEGs that were in each category. Many DEGs fell into multiple categories and were therefore counted more than once. Note that the fraction of DEGs with the GO term “integral component of membrane” (Membrane Bound) are shown with a different scale. (C) The fraction of DEGs (y-axis) also altered by exposure to listed drugs (x-axis) is graphed. WormBase’s expression cluster summary only indicates that a gene’s expression is affected by a chemical, but not in which direction, therefore the fraction of DEGs also affected by various chemicals is presented for up- and downregulated DEGs together. Abbreviations: Upregulated (Up), Downregulated (Down), Programmed Cell Death (PCD), Unfolded Protein Response (UPR), Transcriptional Regulation plus Histone Modification (Transcriptional Reg.).
Table 2.
Categorized Differentially Regulated Genes.
| sequence | gene name | human homolog(s) | NaAsO2 | DMA | HgCl2 | meHgCl | category | references |
|---|---|---|---|---|---|---|---|---|
| PAR2.3 | aak-1 | PRKAA1, PRKAA2 | – | – | – | 1.5 | AMPK, hhDG | (Kuo et al., 2020, Narbonne and Roy, 2009, WormBase., 2021) |
| ZK909.2 | kin-1 | PRKACA | −1.8 | −1.8 | −1.7 | – | AMPK | (WormBase, 2021) |
| K09E9.4 | – | PKIA, PKIB, PKIG | – | – | −1.7 | −2.0 | AMPK | (WormBase, 2021) |
| ZK1320.4 | cyp-13A10 | CYP3A4, CYP3A43 | – | −2.1 | −1.8 | −2.0 | CYP, hhDG, mib | (WormBase, 2021) |
| T10B9.5 | cyp-13A3 | CYP3A4, CYP3A43 | −1.8 | – | – | – | CYP, hhDG, mib | (WormBase, 2021) |
| K09A11.3 | cyp-14A2 | CYP2U1 | – | −2.1 | – | – | CYP, hhDG, mib | (WormBase, 2021) |
| K09A11.4 | cyp-14A3 | CYP2U1 | – | −1.8 | – | – | CYP, hhDG, mib | (WormBase, 2021) |
| R04D3.1 | cyp-14A4 | CYP2U1 | – | 4.6 | – | – | CYP, iIR, hhDG, mib | (Engelmann et al., 2011, WormBase., 2021) |
| T10H4.10 | cyp-34A1 | CYP2U1 | – | −2.5 | – | – | CYP, DrugR hhDG, mib | (WormBase, 2021) |
| K09D9.2 | cyp-35A3 | CYP2U1 | – | −2.7 | – | – | CYP, DrugR, hhDG, mib | (WormBase, 2021) |
| C49G7.8 | cyp-35A4 | CYP2U1 | 1.6 | – | – | CYP, DrugR, hhDG, mib | (WormBase, 2021) | |
| K07C6.2 | cyp-35B3 | CYP2U1 | – | 1.9 | – | – | CYP, DrugR, hhDG, mib | (WormBase, 2021) |
| F14H3.10 | cyp-35D1 | CYP2U1 | – | 5.6 | – | – | CYP, iIR, DrugR, hhDG, mib | (Troemel et al., 2006, Wong et al., 2007, WormBase., 2021) |
| F01D5.9 | cyp-37A1 | – | – | – | – | −1.5 | CYP | (WormBase, 2021) |
| F55A8.2d | egl-4 | PRKG1 | – | – | −1.8 | – | DAF-2/IIS, Loco, hhDG, mib | (WormBase, 2021) |
| ZK1251.2 | ins-7 | – | −1.9 | −1.9 | – | – | DAF-2/IIS, rSKN-1 | (Murphy et al., 2003, Oliveira et al., 2009) |
| H42K12.1b | pdk-1 | PDPK1 | – | 1.5 | 1.5 | – | DAF-2/IIS, hhDG, OxStrR | (WormBase, 2021) |
| B0454.7 | clec-2 | – | 7.1 | 1.8 | – | – | rDAF-16 | (Nag et al., 2017, WormBase., 2021) |
| F55G11.5 | dod-22 | – | – | −4.2 | – | – | rDAF-16 | (Tepper et al., 2013, Watanabe et al., 2020) |
| F37F2.3 | gst-25 | GSTP1 | 6.9 | 10.4 | – | – | rDAF-16, hhDG | (Minniti et al., 2009) |
| R02D3.6 | grl-19 | – | – | 2.3 | – | 1.6 | rDAF-16, OxStrR | (Kim and Sun, 2007) |
| ZK75.2 | ins-2 | – | −1.6 | −2.2 | −1.8 | – | rDAF-16 | (Murphy et al., 2003) |
| R02D3.1 | aass-1 | AASS | – | – | 1.6 | – | hhDG | (WormBase, 2021) |
| Y74C10AR.3 | abtm-1 | ABCB7 | – | – | 1.6 | – | hhDG, OxStrR | (Gonzalez-Cabo et al., 2011, WormBase., 2021) |
| T11B7.4 | alp-1 | LDB3, PDLIM5 | – | – | – | −2.0 | hhDG | (WormBase, 2021) |
| Y71G12B.1 | chaf-2 | CHAF1B | – | – | – | 1.6 | hhDG | (WormBase, 2021) |
| E04F6.11 | clh-3 | CLCN2 | – | – | 1.7 | – | hhDG | (WormBase, 2021) |
| ZC116.3 | cubn-1 | CUBN | −2.7 | −1.7 | – | −2.5 | hhDG | (WormBase, 2021) |
| F11H8.4 | cyk-1 | DIAPH1, DIAPH2 | – | – | – | −2.3 | hhDG | (WormBase, 2021) |
| ZC477.9 | deb-1 | VCL | – | – | 1.6 | – | hhDG | (WormBase, 2021) |
| T12E12.4 | drp-1 | DNM1L | – | – | 1.6 | – | hhDG | (WormBase, 2021) |
| F43G9.6 | fer-1 | DYSF | 2.5 | −1.5 | – | – | hhDG | (WormBase, 2021) |
| ZK180.1 | gbb-2 | GABBR2 | – | −2.1 | – | – | hhDG, DrugR | (WormBase, 2021) |
| F45H7.2 | gei-1 | DLC1, STARD13 |
– | – | −2.1 | −2.1 | hhDG | (WormBase, 2021) |
| F54D7.3 | gnrr-1 | GNRHR | – | – | 1.8 | – | hhDG | (WormBase, 2021) |
| R03E9.4 | irk-1 | KCNJ14, KCNJ4 | – | 2.1 | – | – | hhDG | (WormBase, 2021) |
| R13A1.2 | kcc-1 | SLC12A4, SLC12A6 | – | – | −2.1 | – | hhDG | (WormBase, 2021) |
| M01B2.1 | kin-30 | KIT, PDGFRA | – | – | – | −3.3 | hhDG | (WormBase, 2021) |
| B0457.1 | lat-1 | ADGRL3 | 1.6 | – | 1.7 | – | hhDG | (WormBase, 2021) |
| R05C11.3 | mca-2 | ATP2B2, ATP2B3 | – | – | 1.5 | – | hhDG | (WormBase, 2021) |
| F32D1.10 | mcm-7 | MCM7 | – | – | – | 1.5 | hhDG, DNArep | (WormBase, 2021) |
| F20B6.3 | mrp-6 | ABCC4 | – | – | – | 2.7 | hhDG | (WormBase, 2021) |
| Y43F8C.12 | mrp-7 | ABCC2 | 1.9 | – | 1.8 | 1.8 | hhDG | (WormBase, 2021) |
| T22E5.5.1 | mup-2 | TNNT1, TNNT3 | – | – | 1.6 | – | hhDG | (WormBase, 2021) |
| Y37A1C.1 | nkcc-1 | SLC12A2 SLC12A3 |
– | – | 1.5 | – | hhDG | (WormBase, 2021) |
| F31B9.1 | npr-33 | AGTR1, GPR15 | – | – | −2.3 | – | hhDG | (WormBase, 2021) |
| C01F6.6 | nrfl-1 | SLC9A3R1 | – | – | 1.6 | – | hhDG | (WormBase, 2021) |
| H12C20.2 | pms-2 | PMS2 | – | – | – | −2.5 | hhDG, DNArep | (WormBase, 2021) |
| K07A3.2 | ptr-12 | PTCHD3 | – | – | – | 2.0 | hhDG | (WormBase, 2021) |
| Y18D10A.7 | ptr-17 | PTCHD3 | – | – | – | 2.1 | hhDG | (WormBase, 2021) |
| F25H5.3 | pyk-1 | PKLR, PKM | – | – | 1.9 | – | hhDG, mib | (WormBase, 2021) |
| T08B2.5 | rbm-5 | RBM10, RBM5 | – | – | 1.7 | – | hhDG, mib | (WormBase, 2021) |
| F25B3.3 | rgef-1 | RASGRP3 | – | – | −2.8 | −3.1 | hhDG | (WormBase, 2021) |
| C06E1.10 | rha-2 | DHX37 | – | – | 1.8 | – | hhDG | (WormBase, 2021) |
| T01H8.1 | rskn-1 | RPS6KA1, RPS6KA2 | – | – | 1.5 | – | hhDG | (WormBase, 2021) |
| C06E7.1 | sams-3 | MAT1A, MAT2A | – | – | 1.7 | – | hhDG, mib | (WormBase, 2021) |
| K11G12.3 | smf-2 | SLC11A1, SLC11A2 |
– | – | – | −2.1 | hhDG | (WormBase, 2021) |
| H21P03.3 | sms-1 | SGMS2 | – | – | 1.5 | – | hhDG | (WormBase, 2021) |
| F55H12.1 | snf-2 | SLC6A11, SLC6A6 | – | – | – | 2.1 | hhDG | (WormBase, 2021) |
| Y46G5A.30 | snf-5 | SLC6A8, SLC6A12 | 2.1 | – | – | – | hhDG, mib | (WormBase, 2021) |
| Y48E1B.14 | snx-14 | SNX14 | – | – | 1.9 | – | hhDG | (WormBase, 2021) |
| T19H12.11 | ugt-10 | UGT1A9, UGT2B28 | 4.5 | – | – | – | hhDG | (WormBase, 2021) |
| F29F11.2 | ugt-34 | UGT1A8, UGT2B15 | 94.5 | – | – | – | hhDG | (WormBase, 2021) |
| ZK1151.1 | vab-10 | DST | 1.7 | 2.0 | – | 2.9 | hhDG | (WormBase, 2021) |
| T08G11.1 | – | VPS13A | – | – | 1.7 | – | hhDG | (WormBase, 2021) |
| Y37A1A.4 | – | RIPOR2, RIPOR3 | −1.5 | – | – | −1.8 | hhDG | (WormBase, 2021) |
| Y48G1C.10 | – | MTMR10, MTMR12 |
– | – | – | 1.6 | hhDG | (WormBase, 2021) |
| Y71H10B.1 | – | NT5C2 | – | – | 1.6 | – | hhDG, OxStrR | (Wang et al., 2010, WormBase., 2021) |
| ZK1073.1 | – | NDRG1, NDRG2 | – | – | 1.5 | – | hhDG | (WormBase, 2021) |
| R05F9.12 | aagr-2 | MGAM | – | – | 1.6 | – | iIR, hhDG | (Engelmann et al., 2011, WormBase., 2021) |
| K02G10.7 | aqp-8 | AQP9 | – | 2.2 | – | – | iIR | (Irazoqui et al., 2010, O'Rourke et al., 2006, WormBase., 2021) |
| ZK896.7 | clec-186 | – | – | – | – | −1.9 | iIR, rMAPK | (Block et al., 2015, WormBase., 2021) |
| C41H7.7 | clec-3 | – | 9.9 | 5.8 | – | – | iIR | (Engelmann et al., 2011, Evans et al., 2008, Muir and Tan, 2008, Troemel et al., 2006) |
| Y46C8AL.3 | clec-70 | – | – | −8.6 | −5.6 | iIR | (WormBase, 2021) | |
| F58E6.7 | hrg-3 | – | −1.9 | −2.3 | −1.9 | −2.0 | iIR | (Engelmann et al., 2011, WormBase., 2021) |
| F26A1.10 | nspd-9 | 1.5 | 2.1 | – | – | iIR | (Engelmann et al., 2011) | |
| C05A9.1 | pgp-5 | ABCB1 | – | −2.3 | – | – | iIR, DrugR | (WormBase, 2021) |
| C05C10.4 | pho-11 | – | 1.6 | 2.2 | – | – | iIR, rDAF-16 | (Bolz et al., 2010, McElwee et al., 2003, Troemel et al., 2006, Wong et al., 2007) |
| C43H6.6 | – | – | – | – | – | 1.7 | iIR | (Engelmann et al., 2011) |
| F01D5.2 | – | – | 4.9 | 3.1 | – | – | iIR | (Engelmann et al., 2011, Ren et al., 2009, Troemel et al., 2006) |
| F26D2.3 | – | GCNT1, GCNT3 | – | – | – | 2.0 | iIR, hhDG | (Engelmann et al., 2011, WormBase., 2021) |
| F47B3.7 | – | – | – | 2.6 | – | – | iIR | (Engelmann et al., 2011, Wong et al., 2007) |
| F48E3.9 | – | – | 2.2 | – | – | – | iIR | (Engelmann et al., 2011) |
| H12I13.5 | – | – | 1.8 | – | – | 1.7 | iIR | (Engelmann et al., 2011) |
| K02D3.1 | – | – | −3.3 | – | – | – | iIR | (Irazoqui et al., 2010, Troemel et al., 2006) |
| R04B5.11 | – | – | – | 2.6 | – | – | iIR | (Engelmann et al., 2011) |
| R09H10.5.2 | – | – | – | – | 2.1 | – | iIR | (Engelmann et al., 2011) |
| T01B6.1 | – | SAPCD2 | 2.1 | – | – | – | iIR | (Engelmann et al., 2011) |
| T28A11.2 | – | – | – | 3.6 | – | – | iIR | (Engelmann et al., 2011) |
| B0207.12 | avr-14 | GLRA3, GLRA4 | 2.0 | 2.6 | 1.6 | – | Loco, DrugR | (WormBase, 2021) |
| K04F10.4 | bli-4 | PCSK5, PCSK6 | −1.5 | – | – | – | Loco, hhDG | (WormBase, 2021) |
| F46C8.5 | ceh-14 | LHX-3, LHX4 | – | – | −1.7 | – | Loco, TxR, hhDG, mib | (WormBase, 2021) |
| C55B7.12 | che-1 | ZNF500 | – | −2.6 | – | – | Loco, TxR, mib | (WormBase, 2021) |
| Y41C4A.4 | crh-1 | ATF1, CREB1, CREM | – | – | −2.5 | – | Loco, TxR, hhDG | (WormBase, 2021) |
| F15A8.5 | dop-1 | DRD1, DRD5 | – | 1.6 | – | – | Loco, hhDG | (WormBase, 2021) |
| C16C2.2 | eat-16 | RGS9, RGS11 | – | – | −1.6 | – | Loco, hhDG | (WormBase, 2021) |
| C06A12.4 | gcy-27 | GUCY2D | – | – | −2.2 | – | Loco, hhDG | (WormBase, 2021) |
| T15H9.3 | hlh-6 | ASCL3 | – | −1.8 | – | – | Loco, TxR | (WormBase, 2021) |
| F58A3.1 | ldb-1 | LDB1, LDB2 | – | – | 1.6 | – | Loco, TxR, rDAF-16 | (Oh et al., 2006, WormBase., 2021) |
| T07H8.4 | mec-1 | – | −1.5 | −1.7 | 1.5 | – | Loco | (WormBase, 2021) |
| E01F3.1 | pde-3 | PDE3A | −1.6 | – | – | – | Loco, TxR, hhDG | (WormBase, 2021) |
| Y11D7A.4 | rab-28 | RAB28 | – | −2.4 | – | – | Loco, hhDG | (WormBase, 2021) |
| M01G5.5 | snf-6 | SLC6A5, SLC6A14 | – | – | 1.6 | – | Loco, hhDG | (WormBase, 2021) |
| C18A3.5 | tiar-1 | TIA1, TIAL1 | – | – | 1.9 | – | Loco, OxStrR | (WormBase, 2021) |
| F56A12.1 | unc-39 | SIX4, SIX6 | – | −1.9 | – | – | Loco, TxR, hhDG | (WormBase, 2021) |
| ZC101.2 | unc-52 | HSPG2 | −1.6 | – | – | – | Loco, hhDG | (WormBase, 2021) |
| C11D2.6 | unc-77 | NALCN | – | – | – | −2.0 | Loco | (WormBase, 2021) |
| C09D1.1 | unc-89 | SPEG | – | – | −1.6 | – | Loco, hhDG | (WormBase, 2021) |
| F35C8.3 | jkk-1 | MAP2K7 | – | – | 1.9 | – | MAPK, Loco, mib | (WormBase, 2021) |
| K02H8.1 | mbl-1 | MBNL1, MBNL2 | −2.0 | 1.9 | – | – | MAPK, TlR, hhDG, mib | (Matilainen et al., 2021, WormBase., 2021) |
| ZC504.4 | mig-15 | MINK1, TNIK | – | – | −1.9 | −2.1 | MAPK, hhDG | (WormBase, 2021) |
| Y95B8A.10 | pde-6 | PDE8A, PDE8B | – | – | – | −2.2 | MAPK, hhDG, mib | (WormBase, 2021) |
| K11E8.1 | unc-43 | CAMK2D | – | – | −2.0 | – | MAPK, TxR, hhDG, mib | (WormBase, 2021) |
| W02G9.2.3 | kel-8 | KLHL8 | – | – | – | 1.6 | rMAPK | (Cui et al., 2007) |
| F21F8.4 | asp-12 | CTSE, PGA4, PGC | – | −1.9 | – | −1.5 | PCD | (WormBase, 2021) |
| Y39B6A.23 | asp-16 | CTSE, PGA4, PGC | – | −2.3 | – | – | PCD | (WormBase, 2021) |
| Y47H9C.4 | ced-1 | MEGF10, MEGF11 | – | – | 1.5 | – | PCD, UPR, iIR, hhDG | (Haskins et al., 2008, WormBase., 2021) |
| F08F1.5 | ced-8 | XKR4, XKR6, XKR7 | – | −1.5 | – | – | PCD | (WormBase, 2021) |
| C09G12.8 | ced-10 | RAC1, RAC2 | – | – | −1.6 | – | PCD, hhDG | (WormBase, 2021) |
| F43G9.11 | ces-1 | SCRT1, SCRT2 | – | −3.4 | – | – | PCD, TxR | (WormBase, 2021) |
| ZK909.4 | ces-2 | DBP,TEF | – | −3.4 | – | – | PCD, TxR, hhDG | (WormBase, 2021) |
| T21H3.3 | cmd-1 | CALM1 | – | – | −1.5 | – | PCD, hhDG, mib | (WormBase, 2021) |
| T12E12.4 | drp-1 | DNM1L | – | – | 1.6 | – | PCD, hhDG | (WormBase, 2021) |
| C02C6.1 | dyn-1 | DNM1, DNM2 | – | – | 1.5 | – | PCD, Loco, hhDG | (WormBase, 2021) |
| C49A1.4 | eya-1 | EYA1, EYA2, EYA4 | – | −1.7 | – | – | PCD, TxR, OxStrR, Loco, UPR, DNArep, hhDG, mib | (WormBase, 2021) |
| M05B5.5 | hlh-2 | TCF3, TCF4, TCF12 | −1.7 | – | – | – | PCD, TxR, hhDG | (WormBase, 2021) |
| Y37A1B.2 | lst-4 | SNX18, SNX33 | – | – | 1.9 | – | PCD | (WormBase, 2021) |
| Y51H1A.6 | mcd-1 | – | – | – | 1.6 | – | PCD, mib | (WormBase, 2021) |
| C33A11.1 | nfki-1 | NFKBIZ | – | – | 1.5 | – | PCD, TxR | (WormBase, 2021) |
| ZK524.1 | spe-4 | – | – | −1.7 | – | – | PCD, Notch | (WormBase, 2021) |
| F11F1.7 | ttr-52 | – | – | −2.4 | −2.0 | −1.8 | PCD | (WormBase, 2021) |
| T19E7.2 | skn-1 | NFE2L1, NFE2L2, NFE2L3 |
– | – | – | 1.5 | SKN-1, TxR, OxStrR, UPR, iIR, rMAPK, hhDG | (WormBase, 2021) |
| K10C2.3 | asp-14 | CTSE, PGA3 | 2.3 | 2.3 | – | – | rSKN-1, iIR, rAMPK, rMAPK | (Bolz et al., 2010, Oliveira et al., 2009, Shin et al., 2011, Troemel et al., 2006, WormBase., 2021) |
| C12C8.2 | cbl-1 | – | 2.1 | – | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009) |
| F35E8.11 | cdr-1 | FAXC | – | 5.0 | – | – | rSKN-1, iIR, rDAF-16, | (Engelmann et al., 2011, Evans et al., 2008, McElwee et al., 2003, Oliveira et al., 2009, Troemel et al., 2006, WormBase., 2021) |
| ZK666.6 | clec-60 | – | −2.2 | −2.2 | – | – | rSKN-1, iIR | (Oliveira et al., 2009, WormBase., 2021) |
| ZK673.9 | clec-143 | – | 2.8 | – | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009, Troemel et al., 2006) |
| K10B2.2 | ctsa-4.1 | CTSA | 1.9 | – | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009, Troemel et al., 2006, Wong et al., 2007) |
| F02C12.5 | cyp-13B1 | CYP3A4, CYP3A5 | – | −3.0 | – | – | rSKN-1, CYP, hhDG, mib | (WormBase, 2021) |
| K10H10.3 | dhs-8 | WWOX | 1.8 | 4.3 | – | – | rSKN-1, hhDG | (Oliveira et al., 2009, WormBase., 2021) |
| K10D11.1 | dod-17 | – | 6.4 | 2.6 | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009, Troemel et al., 2006, WormBase., 2021) |
| C32H11.12 | dod-24 | – | 5.3 | 1.7 | – | – | rSKN-1, iIR | (Block et al., 2015, Bolz et al., 2010, Oliveira et al., 2009, Troemel et al., 2006, WormBase., 2021) |
| Y4C6B.6 | gba-4 | GBA | – | – | – | −4.7 | rSKN-1, iIR, hhDG | (Paek et al., 2012, WormBase., 2021, Zárate-Potes et al., 2020) |
| F37B12.2 | gcs-1 | GCLC | 1.8 | 1.6 | – | – | rSKN-1, OxStrR | (Hasegawa et al., 2008, Oliveira et al., 2009, WormBase., 2021) |
| R107.7 | gst-1 | GSTP1 | 1.6 | 1.7 | – | – | rSKN-1, UPR, hhDG | (Oliveira et al., 2009, Settivari et al., 2013) |
| R03D7.6 | gst-5 | HPGDS | – | 3.0 | – | – | rSKN-1, iIR | (Evans et al., 2008, Oliveira et al., 2009, Troemel et al., 2006, Wong et al., 2007) |
| F11G11.3 | gst-6 | HPGDS | – | 1.8 | – | – | rSKN-1 | (Oliveira et al., 2009, WormBase., 2021) |
| F11G11.2 | gst-7 | HPGDS | – | 1.5 | – | – | rSKN-1 | (Oliveira et al., 2009, WormBase., 2021) |
| F11G11.1 | gst-8 | HPGDS | – | 1.6 | – | – | rSKN-1 | (Oliveira et al., 2009, WormBase., 2021) |
| Y45G12C.2 | gst-10 | GSTP1 | 2.1 | 2.1 | – | – | rSKN-1, OxStrR, hhDG | (Oliveira et al., 2009, Park et al., 2009, Tullet et al., 2008, WormBase., 2021) |
| F37B1.2 | gst-12 | HPGDS | 3.8 | 5.9 | – | – | rSKN-1, OxStrR | (Oliveira et al., 2009, Park et al., 2009, VanDuyn et al., 2010) |
| T26C5.1 | gst-13 | HPGDS | 1.6 | 2.0 | – | – | rSKN-1, iIR | (Oliveira et al., 2009, Park et al., 2009, Troemel et al., 2006, WormBase., 2021) |
| F37B1.3 | gst-14 | HPGDS | 2.8 | 4.0 | – | – | rSKN-1, iIR, OxStrR | (Engelmann et al., 2011, Oliveira et al., 2009, Park et al., 2009) |
| F37B1.5 | gst-16 | HPGDS | 5.8 | 6.8 | – | – | rSKN-1 | (Przybysz et al., 2009) |
| F37B1.8 | gst-19 | HPGDS | −2.2 | −2.2 | – | −3.2 | rSKN-1 | (Oliveira et al., 2009) |
| ZK546.11 | gst-30 | HPGDS | 5.3 | 7.5 | – | – | rSKN-1 | (Przybysz et al., 2009) |
| Y1H11.2 | gst-35 | HPGDS | 2.1 | 6.4 | – | – | rSKN-1 | (Oliveira et al., 2009) |
| C02D5.3 | gsto-2 | GSTO1, GSTO2 | 4.1 | 5.3 | – | – | rSKN-1, OxStrR | (Oliveira et al., 2009, WormBase., 2021) |
| ZK84.3 | ins-5 | – | 1.8 | 2.3 | −1.8 | 2.3 | rSKN-1, rMAPK, rDAF-16 | (Kaplan et al., 2019, Yanase et al., 2020) |
| F35E12.5 | irg-5 | – | 1.9 | 2.7 | – | – | rSKN-1, iIR, rMAPK | (Bolz et al., 2010, Foster et al., 2020, O'Rourke et al., 2006, Oliveira et al., 2009, Pukkila-Worley et al., 2011, Wan et al., 2021) |
| T22G5.6 | lbp-8 | FABP5, PMP2 | – | 7.2 | – | – | rSKN-1, hhDG | (Steinbaugh et al., 2015) |
| K08H10.2 | lea-1 | PLIN4 | – | – | 1.5 | – | rSKN-1, rUPR | (Oliveira et al., 2009, WormBase., 2021) |
| ZK1058.6 | nit-1 | – | 3.6 | 2.2 | – | – | rSKN-1, rMAPK | (Miller et al., 2011, Oliveira et al., 2009) |
| H12D21.1 | nspa-1 | – | 10.7 | 8.7 | – | – | rDAF-16 | (McElwee et al., 2003) |
| ZC412.6 | nspa-5 | – | 9.4 | 7.5 | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009) |
| W06A7.5 | nspa-8 | – | 34.0 | 21.8 | – | – | rSKN-1 | (Oliveira et al., 2009) |
| F57C9.1 | pdxk-1 | PDXK | 2.0 | 2.0 | – | – | rSKN-1, mib | (Oliveira et al., 2009, WormBase., 2021) |
| C55A6.5 | sdz-8 | CBR3 | – | 2.2 | – | – | rSKN-1 | (Oliveira et al., 2009) |
| C15F1.7 | sod-1 | SOD1 | – | 1.7 | – | – | rSKN-1, OxStrR, mib | (Park et al., 2009, WormBase., 2021) |
| H23N18.1 | ugt-13 | UGT1A1, UGT2B28, UGT2B7 | 2.3 | – | – | – | rSKN-1, iIR, hhDG | (Engelmann et al., 2011, Hasegawa et al., 2010, Jones et al., 2013, Troemel et al., 2006) |
| B0348.2 | – | – | −1.5 | −1.9 | – | – | rSKN-1, mib | (Oliveira et al., 2009, WormBase., 2021) |
| C17H12.6 | – | – | 2.2 | – | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009, Troemel et al., 2006) |
| C32H11.3 | – | – | 10.1 | 6.6 | – | – | rSKN-1, iIR | (Engelmann et al., 2011, Oliveira et al., 2009, Troemel et al., 2006) |
| C32H11.4 | – | – | 11.7 | 6.7 | – | – | rSKN-1, iIR | (Muir and Tan, 2008, Oliveira et al., 2009, Yang et al., 2016) |
| F01D5.3 | – | – | 3.4 | 3.3 | – | – | rSKN-1, iIR | (Oliveira et al., 2009, Ren et al., 2009) |
| F01D5.5 | – | – | 2.9 | – | – | – | rSKN-1, iIR | (O'Rourke et al., 2006, Oliveira et al., 2009, Ren et al., 2009, Troemel et al., 2006) |
| F39B2.3 | – | CRYZ | 1.9 | 2.0 | – | – | rSKN-1, TlR | (Oliveira et al., 2009, WormBase., 2021) |
| F55G11.2 | – | – | 10.4 | 2.9 | – | – | rSKN-1, iIR | (Block et al., 2015, Engelmann et al., 2011, Evans et al., 2008, Oliveira et al., 2009, Troemel et al., 2006) |
| F56D5.3 | – | – | 3.1 | 1.8 | – | – | rSKN-1, OxStrR | (Oliveira et al., 2009, Park et al., 2009) |
| H25K10.1 | – | ACP7 | 1.9 | – | – | – | rSKN-1, iIR, mib | (Engelmann et al., 2011, Oliveira et al., 2009, WormBase., 2021) |
| W06H8.2 | – | – | 4.9 | 4.4 | – | – | rSKN-1 | (Przybysz et al., 2009) |
| Y73B6BL.14 | – | – | 2.1 | 2.0 | – | – | rSKN-1, DNArep | (Oliveira et al., 2009, WormBase., 2021) |
| ZK742.4 | – | – | 2.3 | 2.8 | – | – | rSKN-1, rDAF-16 | (McElwee et al., 2003, Oliveira et al., 2009) |
| R186.8 | – | – | −1.8 | – | – | – | TlR | (WormBase, 2021) |
| Y80D3A.2 | emb-4 | AQR | 1.9 | – | – | – | TlR | (WormBase, 2021) |
| Y48G10A.4 | ints-8 | INTS8 | – | – | 1.5 | 1.5 | TlR | (WormBase, 2021) |
| D1007.12 | rpl-24.1 | RPL24 | 1.9 | – | – | – | TlR | (WormBase, 2021) |
| Y92C3B.2 | uaf-1 | U2AF2 | – | – | – | 1.6 | TlR | (WormBase, 2021) |
| Y53C12B.3 | nos-3 | – | – | −2.1 | 2.1 | 2.3 | TlR, iIR | (O'Rourke et al., 2006, Pujol et al., 2008, WormBase., 2021) |
| F29C12.3 | rict-1 | RICTOR | – | – | 1.8 | – | TOR | (Blackwell et al., 2019, WormBase., 2021) |
| F56A3.5 | bro-1 | CBFB | – | −2.0 | – | −2.8 | TxR | (WormBase, 2021) |
| H25P06.2 | cdk-9 | CDK9 | – | – | 1.7 | – | TxR, hhDG | (WormBase, 2021) |
| C16C2.1 | ceh-5 | NOTO, VAX1 | – | −2.8 | – | −1.8 | TxR, hhDG | (WormBase, 2021) |
| ZK265.4 | ceh-8 | RAX | – | −3.1 | – | – | TxR, hhDG | (WormBase, 2021) |
| C13G5.1 | ceh-16 | EN1, EN2 | – | – | −1.8 | – | TxR, hhDG | (WormBase, 2021) |
| C37E2.4 | ceh-36 | OTX1, OTX2 | – | −2.1 | – | – | TxR, hhDG | (WormBase, 2021) |
| ZK993.1 | ceh-45 | GSC | – | – | −2.2 | – | TxR | (WormBase, 2021) |
| F34D6.2 | ceh-87 | ZHX1 | −1.9 | – | – | – | TxR | (WormBase, 2021) |
| T23D8.8 | cfi-1 | ARID3A, ARID3C | 1.7 | 2.1 | – | – | TxR | (WormBase, 2021) |
| F44B9.3 | cit-1.2 | CCNT1 | – | – | 1.8 | – | TxR | (WormBase, 2021) |
| C15C8.2 | cky-1 | NPAS4 | – | −2.1 | – | – | TxR | (WormBase, 2021) |
| C34E10.7 | cnd-1 | NEUROD1 | – | −2.4 | – | – | TxR, hhDG | (WormBase, 2021) |
| F25E2.5 | daf-3 | SMAD4 | – | −1.6 | −2.4 | −2.3 | TxR, TGFβ, hhDG, mib | (WormBase, 2021) |
| C27C12.6 | dmd-4 | DMRT3 | – | −1.8 | – | – | TxR | (WormBase, 2021) |
| F54F7.7 | dot-1.2 | DOT1L | 1.6 | – | – | – | TxR, DNArep | (WormBase, 2021) |
| F49E12.6 | efl-3 | E2F7 | – | −1.9 | – | – | TxR | (WormBase, 2021) |
| C08C3.1 | egl-5 | HOXB8 | – | – | −1.7 | – | TxR | (WormBase, 2021) |
| F40H3.4 | fkh-8 | FOXR1 | – | −2.7 | – | −1.6 | TxR | (WormBase, 2021) |
| C14B9.6 | gei-8 | NCOR1 | 1.7 | – | – | – | TxR | (WormBase, 2021) |
| ZK131.3 | his-9 | – | – | – | – | −1.7 | TxR | (WormBase, 2021) |
| ZK131.5 | his-11 | H2BC1 | – | 1.7 | – | −1.6 | TxR | (WormBase, 2021) |
| F38C2.2 | hlh-17 | BHLHE23 | −1.7 | −1.7 | – | – | TxR | (WormBase, 2021) |
| T01D3.2 | hlh-34 | NPAS1 | – | −3.7 | – | −2.1 | TxR, hhDG | (WormBase, 2021) |
| F43G6.6 | jmjd-1.1 | KDM7A | – | – | −2.1 | – | TxR, hhDG, mib | (WormBase, 2021) |
| F23D12.5 | jmjd-3.2 | KDM6A, UTY | – | −1.7 | – | – | TxR, hhDG | (WormBase, 2021) |
| F20H11.2 | let-765 | SBNO1 | – | – | – | 1.5 | TxR | (WormBase, 2021) |
| T14F9.5 | lin-32 | ATOH1 | – | −1.8 | – | – | TxR, hhDG | (WormBase, 2021) |
| C01H6.5 | nhr-23 | RORC | 1.8 | – | – | – | TxR, hhDG | (WormBase, 2021) |
| C25E10.1 | nhr-30 | – | – | – | −2.5 | – | TxR, mib | (WormBase, 2021) |
| F44C4.2 | nhr-37 | PPARA | – | – | – | −2.2 | TxR, hhDG, mib | (WormBase, 2021) |
| T09A12.4 | nhr-66 | – | – | – | 1.5 | – | TxR, mib | (WormBase, 2021) |
| H12C20.3 | nhr-68 | – | – | – | – | −1.7 | TxR, mib | (WormBase, 2021) |
| C47F8.8 | nhr-81 | – | 1.9 | – | – | – | TxR, mib | (WormBase, 2021) |
| T13F3.3 | nhr-127 | PPARA | – | −4.0 | – | −2.0 | TxR, mib | (WormBase, 2021) |
| C03G6.8 | nhr-147 | HNF4A, HNF4G | – | −3.1 | −2.0 | −2.1 | TxR, hhDG, mib | (WormBase, 2021) |
| C06B8.1 | nhr-150 | PPARA | – | −2.4 | −1.8 | −1.6 | TxR, hhDG, mib | (WormBase, 2021) |
| C54F6.8 | nhr-171 | – | – | −2.4 | −1.9 | −1.8 | TxR, mib | (WormBase, 2021) |
| F47C10.4 | nhr-187 | – | – | −2.3 | −1.7 | – | TxR, mib | (WormBase, 2021) |
| F47C10.7 | nhr-188 | – | – | −1.7 | −1.7 | TxR, mib | (WormBase, 2021) | |
| R07B7.14 | nhr-207 | HNF4A, HNF4G | – | – | – | −1.6 | TxR, hhDG, mib | (WormBase, 2021) |
| T07C5.3 | nhr-214 | – | – | −4.4 | – | – | TxR | (WormBase, 2021) |
| T13F3.2 | nhr-218 | PPARA | – | −3.8 | – | – | TxR, mib | (WormBase, 2021) |
| Y116A8C.18 | nhr-229 | – | – | – | – | −2.6 | TxR, mib | (WormBase, 2021) |
| F21D12.5 | npax-1 | – | – | −1.7 | −1.8 | −1.8 | TxR | (WormBase, 2021) |
| M6.3 | pha-2 | HHEX | −1.6 | −1.9 | – | −1.8 | TxR | (WormBase, 2021) |
| F38A6.1 | pha-4 | FOXA1, FOXA2 | −1.5 | – | – | – | TxR | (WormBase, 2021) |
| C47C12.3 | ref-2 | ZIC1 | – | −2.0 | −1.7 | – | TxR, hhDG | (WormBase, 2021) |
| F26A3.8 | rrf-1 | – | – | – | 1.8 | – | TxR | (WormBase, 2021) |
| T05A10.1 | sma-9 | HIVEP1 | – | – | −1.6 | – | TxR, TGFβ, hhDG | (WormBase, 2021) |
| Y43F4B.3 | set-25 | – | – | – | 1.8 | – | TxR, mib | (Klosin et al., 2017, WormBase., 2021) |
| W02H5.7 | sknr-1 | NFE2L1 | – | – | −1.5 | – | TxR, hhDG | (WormBase, 2021) |
| T08A11.2 | sftb-1 | SF3B1 | – | – | 1.7 | – | TxR, hhDG | (WormBase, 2021) |
| F31E8.3 | tab-1 | BSX | −1.6 | −2.6 | −2.0 | −1.7 | TxR | (WormBase, 2021) |
| T07C4.2 | tbx-8 | EOMES, TBR1 |
– | – | −2.3 | – | TxR, hhDG | (WormBase, 2021) |
| D2030.9 | wdr-23 | DCAF11 | 1.5 | 1.5 | – | – | TxR | (WormBase, 2021) |
| ZC123.3 | zfh-2 | ZFHX3 | – | – | 1.5 | – | Txr, hhDG | (WormBase, 2021) |
| Y48G8AL.10 | znf-236 | ZNF729, ZNF99 | – | – | – | 1.9 | TxR, hhDG | (WormBase, 2021) |
| D1044.6 | – | ZNF318 | 2.1 | – | – | – | TxR | (WormBase, 2021) |
| F26A10.2 | – | ZNF148 | – | – | −2.3 | – | TxR | (WormBase, 2021) |
| M03D4.4 | – | ZNF653 | – | −3.3 | – | – | TxR | (WormBase, 2021) |
| R10E4.11 | – | – | – | – | −1.6 | – | TxR | (WormBase, 2021) |
| Y111B2A.10 | – | ZNF689 | – | −1.7 | −2.4 | −2.6 | TxR | (WormBase, 2021) |
| Y53C10A.15 | – | – | – | – | – | −2.2 | TxR | (WormBase, 2021) |
| Y56A3A.28 | – | – | – | – | −2.4 | – | TxR | (WormBase, 2021) |
| Y57G11C.9 | – | PPHLN1 | – | – | −2.0 | – | TxR | (WormBase, 2021) |
| C03A7.14 | abu-8 | – | – | −2.5 | – | – | UPR | (WormBase, 2021) |
| T10B5.5 | cct-7 | CCT7 | – | – | 1.9 | – | UPR, Loco | (WormBase, 2021) |
| ZK970.2 | clpp-1 | CLPP | −1.8 | – | – | – | UPR, hhDG | (WormBase, 2021) |
| F38A5.13 | dnj-11 | DNAJC2 | – | – | −2.0 | – | UPR | (WormBase, 2021) |
| F39B2.10 | dnj-12 | DNAJA1, DNAJA4 | – | – | 1.6 | – | UPR, mib | (WormBase, 2021) |
| ZK1193.5 | dve-1 | SATB2 | – | – | −1.7 | −1.6 | UPR, TxR, hhdG | (WormBase, 2021) |
| T12D8.8 | hip-1 | ST13 | – | −1.6 | – | – | UPR | (WormBase, 2021) |
| T22A3.2 | hsp-12.1 | HSPB2 | – | −2.0 | – | – | UPR | (WormBase, 2021) |
| Y46H3A.2 | hsp-16.41 | CRYAB, HSPB6 | – | – | −1.6 | – | UPR, iIR, hhDG | (O'Rourke et al., 2006, Wong et al., 2007, WormBase., 2021) |
| T27E4.3 | hsp-16.48 | – | −1.5 | – | – | – | UPR | (WormBase, 2021) |
| C15H9.6.3 | hsp-3 | HSPA5 | – | – | 1.7 | – | UPR | (WormBase, 2021) |
| ZK256.1 | pmr-1 | ATP2C1 | – | – | 1.5 | – | UPR, iIR, OxStrR, hhDG | (Kourtis et al., 2012, Schifano et al., 2019, WormBase., 2021) |
| T02C5.5 | unc-2 | CACNA1A | −1.5 | – | – | – | UPR, TGFβ, hhDG, Loco, mib | (WormBase, 2021) |
| W01B6.1 | cwn-2 | WNT5A | – | −1.7 | −1.6 | – | Wnt, hhDG | (WormBase, 2021) |
| F58G4.4 | sdz-23 | – | −3.4 | −2.6 | −2.3 | −2.7 | rWnt | (Lezzerini and Budovskaya, 2014) |
AMPK, effector of AMP-activated Protein Kinase Signaling; rAMPK, regulated by effectors of AMPK signaling; DAF-2/IIS, effector of the Insulin/Insulin-like Growth Factor Signaling Pathway; rDAF-16, regulated by DAF-16 in the same direction; DNArep, DNA replication/repair; DrugR, response to drug, xenobiotic metabolic process; hhDG, close homology to human disease gene(s); iIR, innate Immune Response (≥1.5x change in the same direction with exposure to ≥ two different pathogens or in ≥ two studies, or role in iIR described); Loco, involved in locomotion or behavior; MAPK, effector of Mitogen-Activated Protein Kinase Signaling (ERK, p38, JUK/SAPK); rMAPK, regulated by effectors of MAPK signaling; mib, metal ion binding; OxStrR, oxidative stress response; PCD, effector of programmed cell death and/or apoptotic process; rSKN-1, regulated by Nrf/SKN-1 in the same direction; TGFβ, effector of transforming growth factor beta receptor signaling pathway; TlR, Translational regulation and machinery; TOR, effector of TOR signaling; TxR, transcriptional regulation and/or histone modification; UPR, effector of unfolded protein/heat shock response; rUPR, regulated by UPR effectors; Wnt, effector of Wnt signaling; rWnt, regulated by Wnt signaling.
From worms to humans, oxidative stress is a major effector of arsenite toxicity due to arsenic’s ability to bind with free thiols in proteins and glutathione (Cartwright, 2016, Oliveira et al., 2009, Sahu et al., 2013). Consistent with this, 6–10% of all DEGs upregulated by NaAsO2 or DMA were involved in glutathione related processes (Fig. 3A). Upregulated DEGs in this category for both forms of arsenic included glutathione-s-transferases gst-10, gst-12, gst-14, gst-16, gst-25, gst-30, gst-35, and gsto-2. Additionally, gcs-1, the C. elegans ortholog of human GCLC, a subunit of the enzyme that catalyzes the first step in glutathione synthesis (An and Blackwell, 2003) was also upregulated by both forms of arsenic. With the exception of gst-25, all of these gst genes, as well as gcs-1, are upregulated by SKN-1 activity, the C. elegans ortholog of mammalian Nrf transcriptional regulators that act in multiple stress resistance pathways, including oxidative stress (Jones et al., 2013, Oliveira et al., 2009, Park et al., 2009, Przybysz et al., 2009). DMA upregulated seven additional gst genes unaffected by NaAsO2, as well as oxidative stress response gene sod-1 (SuperOxide Dismutase 1). At the tested concentrations, no gst genes were upregulated by either form of mercury (Fig. 3B). Of note, gst-19, which is suppressed by SKN-1 (Miller et al., 2011), was downregulated with both forms of arsenic and by meHgCl (Table 2). Consistent with arsenic inducing oxidative stress, the GO term “oxidation-reduction process” was associated with 7% and 10% of DEGs upregulated by NaAsO2 and DMA, respectively. Genes in this category upregulated by both forms of arsenic included dhs-8 and K10H10.6, both homologs of human WWOX oxidoreductase, and F30B5.4, a homolog of human OSGIN1 (oxidative stress induced growth inhibitor) (Table 2). In contrast to arsenic, at concentrations tested, the two mercury compounds affected the expression of few, if any, genes with associated glutathione or oxidation–reduction related GO terms.
For heat shock or unfolded protein response (Heat Shock/UPR), though the numbers were small overall, HgCl2 upregulated the highest proportion of DEGs in this category (Fig. 3B), including dnj-12 and hsp-3, homologs of human Hsp40 and Hsp70, respectively. Heat shock genes hsp-12.1, hsp-16.41, and hsp-16.48 were downregulated by DMA, HgCl2, and NaAsO2, respectively. HgCl2 and meHgCl both downregulated dve-1, a transcription factor involved in chaperone binding and mitochondrial UPR (Tian et al., 2016).
Exposure to arsenic or mercury alters mitogen-activated protein kinase (MAPK) pathway activity in human cells, rodents, and C. elegans (Aguado et al., 2013, Hao et al., 2009, Inoue et al., 2005, Wyatt et al., 2017, Yang and Frenkel, 2002). Consistent with a previous report of C. elegans MAPK pathway genes sek-1 and pmk-1 being downregulated with meHgCl and HgCl2 exposure (Wyatt et al., 2017), in this study mig-15, pde-6, and unc-4 were downregulated by one or both forms of mercury. In contrast, human MAP2K7 homolog jkk-1 was upregulated, but only by HgCl2 (Table 2). No MAPK pathway genes were altered by either form of arsenic. However, mbl-1, the sole C. elegans ortholog of mammalian Muscleblind-like proteins and MAPK regulator (Matilainen et al., 2021), was upregulated by DMA but downregulated by NaAsO2.
Arsenic is a known immune modulator in mammals (Dangleben et al., 2013). Consistent with this, GO terms related to immune function including ‘innate immune response,’ ‘defense response to bacterium,’ and ‘defense response to fungus,’ were associated with 7–8% of genes upregulated by NaAsO2 and DMA, while few genes affected by the two mercury species fell in this category. DEGs with locomotion related GO terms or WormBase descriptions were associated with 9% of DEGs upregulated by HgCl2 including ldb-1, involved in mechanosensory behavior, and snf-6, used to study Duchenne muscular dystrophy (WormBase, 2021). DEGs with the GO term “metal ion binding” were induced and inhibited to a similar extent by the two forms of arsenic (Fig. 3A). In contrast, 14% of genes upregulated by HgCl2 were in this category while no DEGs upregulated by meHgCl had “metal ion binding” as a GO term (Fig. 3B).
The GO term “signal transduction” was associated with about 4% of the DEGs upregulated by the two forms of arsenic versus 8–9% of DEGs upregulated by the two forms of mercury. Genes in this category included avr-14, an extracellular glutamate-gated chloride channel subunit upregulated by NaAsO2, DMA and HgCl2, and daf-38, a neuronal G protein-coupled receptor upregulated by NaAsO2, DMA and meHgCl (WormBase, 2021). Both forms of mercury downregulated mig-15 and rgef-1, involved in MAPK and Ras signaling, respectively. A regulator of Rho protein signal transduction, gei-1, was also downregulated by both HgCl2 and meHgCl. Additionally, several insulin genes were in the signal transduction category, including ins-2, downregulated by NaAsO2, DMA, and HgCl2, and ins-5 which was affected by all four conditions (Table 2).
For genes with GO terms or WormBase descriptions relating to transcriptional regulation and/or histone modification (Transcriptional Reg.), the two forms of mercury induced proportionally more DEGs than did the two forms of arsenic, and this was also true for DEGs with known or predicted nuclear localization of their gene product (Fig. 3). Predicted transcription factor tab-1 (Touch Abnormal), homologous to human BSX, was downregulated by all four conditions (Table 2). Known or predicted transcription factors daf-3, nhr-147, nhr-150, nhr-171, npax-1, and Y111B2A.10 were all downregulated by DMA, HgCl2 and meHgCl. Transcriptional regulator pha-2 was downregulated by NaAsO2, DMA, and meHgCl. Regulators of transcription by RNA polymerase II ceh-5, fkh-8, hlh-34, and nhr-127 were downregulated by both DMA and meHgCl. Controllers of DNA and transcription factor binding bro-1, ceh-16, ref-2 and nhr-187 were downregulated by both DMA and HgCl2. Both DMA and NaAsO2 upregulated suppressors of RNA polymerase II activity cfi-1 and wdr-23, and downregulated transcriptional activator hlh-17. HgCl2 and meHgCl downregulated transcriptional regulator nhr-188. Several of the genes in this category affect histone methylation, including jmjd-3.2 and jmjd-1.1 (Agger et al., 2007, Lee et al., 2015), histone demethylases downregulated by DMA and HgCl2, respectively. NaAsO2 upregulated dot-1.2 and met-1, genes predicted to encode histone methyltransferases (Andersen and Horvitz, 2007, Cecere et al., 2013), and gei-8, a histone deacetylase and transcription repressor (Mikoláš et al., 2013). Overall, across all four tested conditions, there was a trend towards downregulation of transcriptional activators, and upregulation of negative regulators, consistent with a higher proportion of genes being downregulated for each chemical.
The GO terms “integral component of membrane” or “integral component of plasma membrane” (Membrane Bound) were associated with about 20% of upregulated DEGs for all four contidions. About 16% of DEGs downregulated by either form of mercury were in this category. In contrast, 35% of genes downregulated by NaAsO2 and 26% of genes downregulated by DMA were known or predicted to encode membrane proteins.
One way to assess the specificity of response to a condition is to assess the proportion of DEGs that were also affected by other chemicals and the pathways those chemicals affect. 50–80% of DEGs affected by DMA were also affected by metformin, psoralens, rifampin, and sirolimus, drugs that influence energy production, mutagenesis, bacterial RNA synthesis, and inflammation, respectively (Fig. 3C). Sharing gene expression patterns with drugs that alter such a wide variety of processes is a likely indication of a generalized toxic response. In contrast to DMA, 31–62% of DEGs induced by NaAsO2 were also influenced by these same drugs, and this was the case for only 20–42% of DEGs for the two mercury compounds. By this measure, the specificity of toxic response would be ranked meHgCl = HgCl2 ≥ NaAsO2 > DMA.
3.2.3. Adult native gene expression – Curated genes
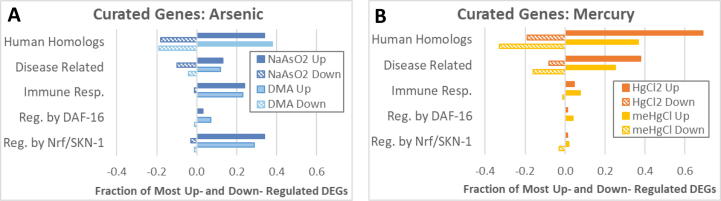
Genes with the greatest fold-change up or down in each set were further curated for references in the literature to clarify function. Only genes for which some functional or expression information could be identified were included in this count. These curated genes included 100 up and down DEGs from each condition, with the exception of HgCl2 and meHgCl which only had 87 and 52 informative upregulated DEGs respectively. Therefore, all upregulated genes for the two mercury compounds were included in the curated analysis. According to WormBase, 34 to 38 of the 100 DEGs most highly upregulated by NaAsO2 and DMA had human homologs, and about a third of these were human disease related. Given that C. elegans homologs have been identified for 60–80% of human genes (Kaletta and Hengartner, 2006), it was surprising that only about 20% of the 100 genes most highly downregulated by the two forms of arsenic had human homologs (Fig. 4A). HgCl2 upregulated DEGs had the highest proportion of human homologs at 69%, and over half of these human homologs, or 38% of all genes upregulated by HgCl2, were human disease related (Fig. 4B). For meHgCl upregulated genes, 36% had human homologs and two thirds of these were human disease related. Among highly downregulated genes with human homologs, meHgCl had the most at 33 of 100, and half of these were human disease related.
Fig. 4.
Curation of DEGs with the greatest fold-change. (A-B) A subset of the most highly differentially regulated genes were further researched for functional information. Abbreviations: regulated (Reg.) in the same direction by DAF-16 or Nrf/SKN-1, genes that respond in the same direction to pathogen exposure (Immune Resp.).
These curated genes were further evaluated in the literature for functional information. In Section 3.2.2 and in Fig. 3, all DEGs with an immune related GO term were included in the Innate Immune category. Here, for this curated set of high-fold-change genes, DEGs were instead put in an immune responder category (Immune Resp.) only if they were reported to be regulated in the same direction by at least 1.5-fold in response to two or more pathogens, or to a single pathogen in at least two studies, or to be required for immune defense (see Methods, Section 2.7). By this measure, 24 and 23 of the 100 highest fold change DEGs upregulated by NaAsO2 and DMA, respectively, are innate immune responders (Fig. 4A). In contrast, only 5–8% of DEGs upregulated by the two forms of mercury were immune responders (Fig. 4B).
Alternatively spliced isoforms of skn-1 are orthologous to the mammalian transcription factors Nrf1 and Nrf2 (NF-E2-related factors), and Nrf/SKN-1 regulated pathways include oxidative stress response and immune function (Blackwell et al., 2019, Oliveira et al., 2009). Curated DEGs were categorized as Nrf/SKN-1 pathway responders if they had been reported to be regulated in the same direction by SKN-1. DEGs controlled by Nrf/SKN-1 made up 34% and 29% of the 100 genes most highly upregulated by NaAsO2 and DMA, respectively (Fig. 4A). Nrf/SKN-1 regulated DEGs highly induced by both NaAsO2 and DMA in the immune pathway responder category included asp-14, dod-17, dod-24, C32H11.3, C32H11.4, F01D5.3, and F55G11.2. Genes highly upregulated by both forms of arsenic that are in the oxidative stress responder category and known to be induced by Nrf/SKN-1 included gcs-1, gst-10, gst-12, gst-13, gst-14, gst-16, gst-25, gst-30, gst-35, and gsto-2 (Table 2). Few Nrf/SKN-1 regulated genes were affected by exposure to the two mercury compounds at assessed concentrations (Fig. 4B). However, consistent with previous reports in C. elegans and mammals (Martinez-Finley et al., 2013a, Ruszkiewicz et al., 2018), skn-1 itself was upregulated 1.5x by meHgCl (Table 2).
DAF-16 and SKN-1 are transcription factors for separate but interacting pathways that regulate stress resistance and immune defense, among other functions. Few curated DEGs for any condition were regulated by DAF-16 or its insulin/IGF-1 signaling (IIS) regulator DAF-2, indicating that IIS does not play a large role in the transcriptional response to arsenic or mercury at tested concentrations (Fig. 4). Similarly, few curated genes were found to be either effectors of, or downstream responders to, other pathways of toxicity such as epigenetic regulation, DNA damage response, the MAPK cascade, osmotic stress, lipid metabolism, or unfolded protein response (Supplemental Table 1).
3.3.1. Transgene expression – Oxidative stress response
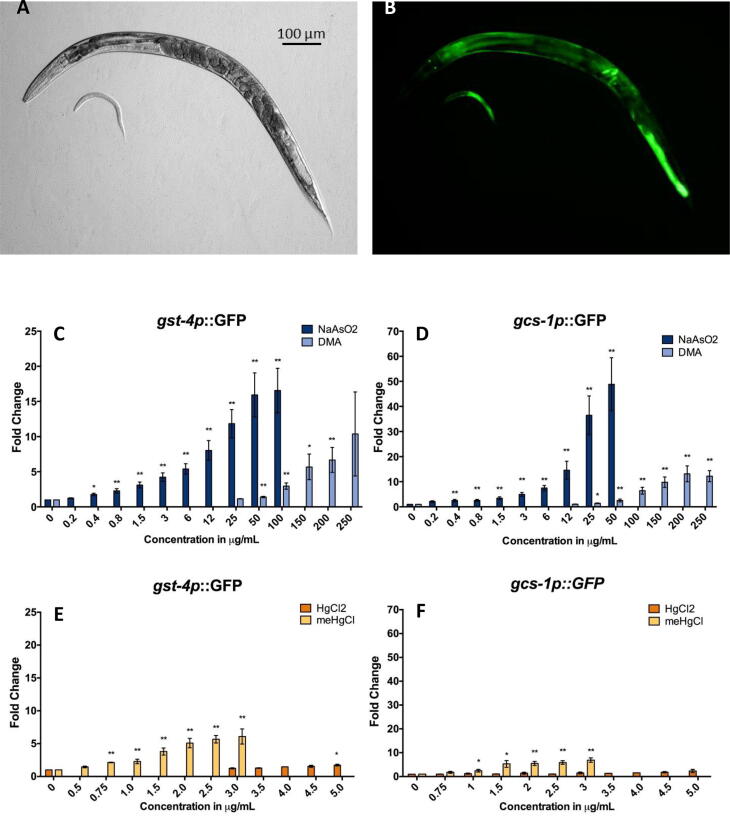
Strains containing integrated GFP transgenes controlled by the promoters for glutathione-S-transferase 4 (Leiers et al., 2003) (gst-4p::GFP, Fig. 5A-B) and gamma glutamylcysteine synthetase (gcs-1p::GFP), which catalyzes the first rate-limiting step of glutathione synthesis (An and Blackwell, 2003), were used to assess OxStrR in adult C. elegans. At concentrations relevant to developmental delay, NaAsO2 strongly induced both OxStrR reporters, while concentrations in the range of 20-fold higher DMA were required to induce similar levels of transgene induction (Fig. 5C-D). Concentrations of meHgCl above the developmental delay LOEL of 0.5 µg/ml induced both OxStrR biomarkers, while HgCl2 only induced gst-4p::GFP, and only at 5.0 µg/ml (Fig. 5E-F). At concentrations from 1.5 to 3.0 µg/ml, meHgCl induced slightly higher GFP fold changes than did NaAsO2 at the same concentrations, indicating a high OxStrR with meHgCl. At higher concentrations of HgCl2 there was a continued trend towards increased OxStrR with gst-4::GFP (data not shown), however changes in time-of-flight and extinction indicated that mercury had adversely affected C. elegans length and optical density, whole body morphology markers that reflect general health, making results less reliable.
Fig. 5.
Assessment of Oxidative Stress Response (OxStrR) Biomarkers. (A-B) Examples of bright field and fluorescence images of C. elegans carrying an integrated transgene with the promoter for phase II enzyme gst-4 fused to GFP to monitor OxStrR. (C-F) Biosensor fold-change relative to water control, from at least three independent experiments per exposure group. Top concentrations shown are maximum concentration not altering adult size and optical density below 80% of control. Error bars: standard error of the mean (SEM), T-test p-values *<0.05, ** <0.005.
3.3.2. Transgene expression – Unfolded protein response
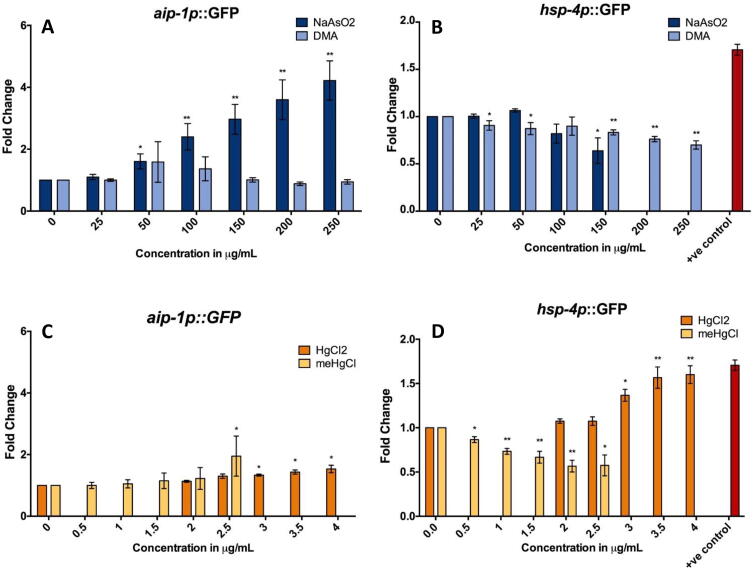
Unfolded Protein Response (UPR) pathways are a key protective response for resisting the effects of some toxic agents. AIRAP/aip-1 encodes an arsenite-inducible proteasome subunit required for resistance to arsenic toxicity in mammals and C. elegans (Sok et al., 2001, Stanhill et al., 2006). NaAsO2 induced aip-1p::GFP in a clear dose–response curve from 50 to 250 µg/ml (0.4–2 mM). In contrast to arsenite, DMA had no consistent effect on this proteasome specific UPR (UPRPS) biomarker at any tested concentration (Fig. 6A).
Fig. 6.
Assessment of Unfolded Protein Response (UPR) Biomarkers. (A-D) UPR response biosensor fold change relative to water control, from at least three independent experiments per exposure group. (A&C) Proteasome specific UPR reporter aip-1p::GFP. (B&D) Endoplasmic reticulum specific UPR reporter hsp-4p::GFP (+ve control: 30 µg/ml Tunicamycin). Top concentrations shown are the maximum concentration not altering adult size or optical density below 80% of control. Error bars: standard error of the mean (SEM), T-test p-values *<0.05, ** <0.005.
C. elegans hsp-4 is a close homolog of the mammalian ER-localized Hsp70 chaperone BiP, and activation of the transgene hsp-4p::GFP is used as a biomarker of endoplasmic reticulum specific UPR (UPRER). UPRER is upregulated in response to ER stress and to tunicamycin, an inhibitor of N-linked glycosylation of nascent proteins (Bull and Thiede, 2012). Both forms of arsenic decreased expression of hsp-4p::GFP. NaAsO2 significantly decreased expression of this UPRER marker only at the highest assessed concentration of 150 µg/ml (1.15 mM), while expression was decreased in a dose response fashion with DMA from 25 to 250 µg/ml (180 µM to 1.8 mM) (Fig. 6B).
HgCl2 slightly but significantly increased UPRPS at 3–4 µg/ml (11–15 µM) while meHgCl increased it only at the highest assessed concentration of 2.5 µg/ml (10 µM) (Fig. 6C). The UPRER marker induced opposite effects for HgCl2 and meHgCl, with HgCl2 increasing expression at 3–4 µg/ml (11–15 µM) and meHgCl decreasing expression between 0.5 and 3.0 µg/ml (2–12 µM) (Fig. 6D).
The two UPR biomarker strains used in this study took longer to grow from egg to egg-laying adult than wild type N2 C. elegans and had much lower reproductive outputs. Additionally, they were more sensitive to toxic stress from compounds tested here, as indicated by reduced TOF and EXT values (Supplemental Fig. 1B). Therefore, more care was required to accurately evaluate life stage for consistent dosing, and fewer concentrations could be assessed, limiting their value for higher-throughput toxicity analyses.
4. Discussion
Various forms of arsenic occur naturally in food and drinking water sources, but risk assessment and regulation are complicated by the fact that the toxicity of arsenic varies widely depending on its chemical composition (Benramdane et al., 1999, Feldmann and Krupp, 2011, Luvonga et al., 2020, Thomas et al., 2001). Many adverse effects of sodium arsenite (NaAsO2) are well understood, but there is little experimental data on the developmental toxicity of dimethylarsinic acid (DMA) (FDA, 2016). DMA has been found in some foods marketed to babies and children, and DMA was detected rodent pups whose dams had been exposed to arsenite in drinking water (FDA, 2016, Twaddle et al., 2018), indicating the importance of filling data gaps on the effects of oral exposure to DMA. In a large study of about 900 chemicals, C. elegans was nearly as good at predicting developmental toxicity in rats or rabbits as those two species were at predicting each other (Boyd et al., 2016). Additionally, the toxicodynamics and apical effects of many types of chemicals are similar in C. elegans and mammals, indicating that the C. elegans model has the potential to contribute useful information to human predictive integrated testing strategies (Hartman et al., 2021, Masjosthusmann et al., 2018, Parish et al., 2020).
With the worm Development and Activity Test (wDAT), the time for developing C. elegans to reach the third larval stage was used as a benchmark developmental milestone to assess the effects of organic and inorganic forms of arsenic and mercury. Across a range of concentrations, similar levels of developmental delay were associated with 20-fold higher concentrations of DMA relative to NaAsO2, with lowest observed effect levels (LOELs) at 10 µg/ml (76 µM) NaAsO2 or 200 µg/ml (1.4 mM) DMA (Fig. 1B). While the endpoint is different, this toxicity ratio is consistent with relative reported rat LD50s (Table 1).
Both the U.S. EPA’s reference doses for oral exposure and the European Food Safety Authority’s tolerable weekly intakes (Table 1) set limits for methylmercury exposure at about one third that of inorganic mercury based on the documented detrimental effects of methylmercury at low concentration on the developing nervous system (Atsdr, 1999, Davidson et al., 2004, Efsa, 2012, Tucker and Nowak, 2018). Methylmercury chloride (meHgCl) is also more toxic than mercury chloride (HgCl2) to developing C. elegans (Helmcke et al., 2009, McElwee and Freedman, 2011, Wyatt et al., 2016), though to our knowledge the relative toxicity of these two forms of mercury across a range of concentrations has not been established. Consistent with the difference in recommended safe exposures for humans, we found similar levels of developmental delay in C. elegans with 2 to 4-fold lower concentrations of meHgCl relative to HgCl2. The LOELs for developmental delay as assessed by the wDAT were 0.5 µg/ml (2 µM) for meHgCl and 2.0 µg/ml (7.5 µM) for HgCl2 (Fig. 1D).
While safe levels for oral exposure to inorganic arsenic (iAs) are a current matter of debate and experimental data on DMA in mammals is sparse, there is ample human epidemiologic evidence that robust metabolism of iAs to DMA is protective against arsenic toxicity (FDA, 2016, Schlebusch et al., 2015). In terms of human health it has been noted that, in general, methylation of inorganic species increases the toxicity of mercury but decreases the toxicity of arsenic (Francesconi, 2007), and this was also true in C. elegans, with a developmental toxicity ranking of meHgCl > HgCl2 > NaAsO2 ≫ DMA.
In developing mammals, the nervous system is one of the primary targets for both arsenic and mercury toxicity (Chandravanshi et al., 2018, Davidson et al., 2004), and altered motor activity is a frequently used measure of neurotoxicity (Aschner et al., 2017, Crofton et al., 1991, Mullenix, 1989). With inorganic HgCl2 and NaAsO2, there were low levels of hypoactivity at some concentrations in developing C. elegans, but consistent changes in population locomotor activity levels were not observed. For organic meHgCl and DMA however, there was a non-significant trend towards hyperactivity at below LOEL concentrations for developmental delay, and significant hypoactivity that increased with dose at concentrations above developmental delay LOELs (Fig. 1). Exposure to methylmercury during development has long been known to be associated with juvenile hypoactivity in mammals (Fredriksson et al., 1993, Schalock et al., 1981) and this effect was conserved in C. elegans. It is noteworthy that a. organic arsenicals are not known to be mammalian developmental neurotoxins while methylmercury is a known mammalian developmental neurotoxin, and b. in C. elegans, for meHgCl and DMA, the lowest concentrations at which significant developmental hypoactivity was observed differed by 300-fold. Therefore, it is possible that the exposure concentration at which changes in locomotor activity are observed during C. elegans development could be used to infer developmental neurotoxicity, however current data are insufficient to determine a range for this type of correlation.
For the purpose of identifying modes of toxic action that are specific to the test articles in question, it is important to avoid high concentrations that induce nonspecific toxicity and general cellular stress. Additionally, global gene expression assessment in C. elegans juveniles is complicated by the fact that expression levels of many genes fluctuate dramatically through the course of development, and therefore changes may be due to the developmental delay rather than directly due to the toxic agent. Therefore, LOEL concentrations for developmental delay in juveniles were utilized to assess gene expression changes in adult C. elegans using gene expression microarrays. NaAsO2 at 10 µg/ml and DMA at 200 µg/ml induced 927 and 1,221 differentially expressed genes (DEGs), respectively (Fig. 2B). For DMA, that is an approximately 30% increase in the number of affected genes relative to NaAsO2 at a 20-fold higher DMA concentration. This is consistent with both relatively low developmental toxicity of DMA in C. elegans, and with human epidemiologic data correlating toxicity with consumption of iAs, but not with organic arsenic (Atsdr, 2007, Filippini et al., 2018, Hackethal et al., 2021). Only 161 of the DEGs induced by NaAsO2 or DMA were shared, however the patterns for GO terms were similar (Fig. 3A). Many of the DEGs that were induced by both arsenic compounds are involved in immune and oxidative stress responses, indicating that for these pathways their effects overlap, albeit at very different exposure levels.
2.0 µg/ml HgCl2 and 0.5 µg/ml meHgCl induced 670 and 485 DEGs, respectively. In contrast to arsenic, the GO term category patterns for the two mercury compounds varied considerably, consistent with differing effects. These results with synchronized adult cohorts are consistent with a previous report of disparate gene expression patterns with HgCl2 vs. meHgCl in human cell lines and in mixed population C. elegans (McElwee et al., 2013). While any chemical that adversely affects the development of an organism is considered a developmental toxin, the dose makes the poison. The relative number of DEGs induced in adults by concentrations that were equitoxic in juveniles can be an indicator of the specificity of a chemical’s effects on developmental processes. The four compounds were tested in adults at concentrations that induced similar developmental delays in juveniles, yet the number of induced DEGs differed by 2.5-fold, with DMA inducing the most DEGs and meHgCl inducing the fewest. This likely reflects the specificity of methylmercury’s developmental toxicity. Additionally, relative to the other compounds tested, a far higher proportion of genes influenced by DMA were also reported to be differentially regulated by a variety of chemicals and drugs with disparate cellular effects (Fig. 3C), consistent with DMA at high concentration inducing a more general form of toxicity relative to the other assessed conditions. Together, these results are consistent with greater mercury toxicity in juvenile mammals relative to adults and mercury’s designation as a developmental toxin (WHO, 1991), and also suggest that the developmental delays and hypoactivity associated with high concentrations of DMA were likely related to more general modes of toxic action that are not specific to developmental processes.
Arsenic exposure affects mammalian immune function, with immune regulators and oxidative stress contributing to this mode of toxic action (Dangleben et al., 2013). Consistent with this, relative to the two mercury species, NaAsO2 and DMA induced changes in far more genes associated with glutathione, innate immune, and oxidation–reduction reaction related GO terms than did HgCl2 or meHgCl (Fig. 3A). In contrast, the proportion of both up- and downregulated genes with GO terms associated with nuclear localization and transcriptional regulation was higher for the two mercury species (Fig. 3B). While 21 DEGs encoding known or predicted transcription factors were regulated in the same direction by two or more tested conditions, the only histone modification related DEG shared was his-11, which was up with DMA but down with meHgCl, consistent with chemical specific modulation of genes related to histone modification. For all four compounds, there was a trend towards downregulation of genes involved with histone demethylation, and upregulation of histone methyltransferases. Overall, the direction of expression of genes involved in transcriptional regulation was towards condensation of chromatin and repression of transcription, potentially affecting biological processes including reproduction and transgenerational epigenetic regulation. Histone modification and reproductive toxicity assessment on exposed populations and their progeny will be conducted in a follow-up study to further evaluate these potential epigenetic effects.
Genes with the highest fold changes were further curated to identify trends or patterns. The proportion of the most highly upregulated genes with human homologs was greatest for HgCl2, and over half of these are associated with human diseases (Fig. 4B). Many of the genes most highly regulated by HgCl2 exposure are associated with muscle and gait related diseases in humans including clh-3 (CLCN2, myotonia congenita), dyn-1 (DNM-1, Charcot-Marie-Tooth disease), mup-2 (TNNT1, nemaline myopathy), snf-6 (SLC6A14, Duchenne muscular dystrophy), and unc-89 (SPEG, centronuclear myopathy). These changes in C. elegans are consistent with mammalian data linking mercury exposure to cardiomyopathies and neurological problems including tremors, paresthesia, weakness, slowed response, and unsteady gait (Ayensu and Tchounwou, 2006, Marsh et al., 1995, Myers et al., 1995, Prince and Rand, 2018). Relative to HgCl2, meHgCl affected fewer DEGs with homology to human disease genes, likely due to the four-fold lower tested concentration of methylmercury.
Conserved transcription factors Nrf1 and Nrf2, encoded in C. elegans as alternatively spliced isoforms of skn-1, are involved in regulating several pathways including those for metabolic and immune functions as well as oxidative stress resistance (Blackwell et al., 2019). Arsenite and methylmercury activate Nrf/SKN-1 in mammalian cell cultures and in C. elegans (Aono et al., 2003, Branco et al., 2014, Lau et al., 2013, Martinez-Finley et al., 2013b, Oliveira et al., 2009, VanDuyn et al., 2010). Genes known to be regulated by Nrf/SKN-1 were well represented among the curated DEGs most highly upregulated by NaAsO2 and DMA (Fig. 4A). C. elegans WDR-23 acts as a repressor for SKN-1, and wdr-23 is activated by SKN-1 in a feed-back loop (Blackwell et al., 2015). Though it was not among the 100 most upregulated genes, wdr-23 was upregulated 1.5-fold by both NaAsO2 and DMA (Table 2). Several genes known to be downregulated by SKN-1, including clec-60, gst-19, and ins-7 (Miller et al., 2011, Oliveira et al., 2009), were also downregulated with both forms of arsenic. Together with innate immune and oxidative stress pathway responder genes being upregulated by both NaAsO2 and DMA at LOELs for developmental delay, these data suggest that Nrf/SKN-1 mediated activation of oxidative stress and innate immune response pathways are modes of toxic action for arsenic induced developmental delay.
At assessed concentrations, few curated DEGs affected by HgCl2 or meHgCl were found to be regulated by Nrf/SKN-1 (Fig. 4B). It is interesting to note however that in this study, only meHgCl upregulated expression of skn-1 itself, which is consistent with previous findings that meHgCl upregulates skn-1 and that Nrf/SKN-1 activity is essential for resistance to methylmercury toxicity in C. elegans and in mammalian cells (Branco et al., 2014, Martinez-Finley et al., 2013a, Martinez-Finley et al., 2013b, Toyama et al., 2007, VanDuyn et al., 2010).
C. elegans glutathione synthetase subunit gcs-1 and glutathione-s-transferase gst-4 are both direct targets of SKN-1, and transgenic strains with GFP expression controlled by the promoters for these two genes are used as biomarkers of Oxidative Stress Response (OxStrR) (Detienne et al., 2016). OxStrR is associated with mammalian exposures to organic and inorganic forms of mercury both in vitro and in vivo (Antunes Dos Santos (Antunes dos Santos et al., 2018, Teixeira et al., 2018). In this study, microarray analysis of C. elegans gene expression at single, relatively low concentrations of the two mercury compounds did not induce genes associated with glutathione or oxidation–reduction processes (Fig. 3). Methylmercury concentrations above the 0.5 µg/ml tested with microarrays did activate both OxStrR reporters, however. In contrast, HgCl2 induced a significant increase in expression for only one of the two OxStrR reporters tested, and only at 5 µg/ml, the highest concentration that did not induce significant changes in adult length and optical density (Fig. 5). For arsenic, both OxStrR reporters were upregulated to a similar extent over a range of exposures by approximately 20-fold higher concentrations of DMA relative to NaAsO2 (Fig. 5). Interestingly, meHgCl at 1 to 3 µg/ml induced slightly higher expression levels with the two OxStrR reporters than did NaAsO2 at the same concentrations, consistent with methylmercury being an inducer of oxidative stress at low concentration. Together with developmental delay and microarray data, these results indicate that a. OxStrR biosensors can accurately reflect altered native gene expression, b. exposure to at least an order of magnitude more DMA is required relative to NaAsO2 for similar effects on multiple endpoints, c. for the two forms of arsenic, significant OxStrR was seen at LOEL concentrations for developmental delay, and d. as with mammals, biomarkers of OxStrR in C. elegans are associated with poor early developmental outcomes (Chandravanshi et al., 2018, Rains et al., 2021, Torres-Cuevas et al., 2017).
Unfolded Protein Response (UPR) pathways are activated with exposure to some chemicals and pathogen-produced toxins. Conserved arsenite-inducible proteasomal 19S regulatory particle-associated protein AIRAP/AIP-1 is involved in proteasome specific UPR (UPRPR) and protects worms and mammalian cells from arsenite toxicity (Sok et al., 2001, Stanhill et al., 2006). As expected, NaAsO2 upregulated a UPRPR reporter for AIRAP/AIP-1 expression in a strong dose response curve, but with a LOEL for UPRPR induction in adults five times the NaAsO2 LOEL concentration for developmental delay. This is consistent with a previous report of DEGs associated with protein folding being induced in C. elegans exposed to NaAsO2 at 300 µg/ml but not at 30 µg/mL (Sahu et al., 2013). In contrast, DMA did not significantly upregulate this UPRPR bioreporter at any concentration tested (Fig. 6A). HgCl2 and meHgCl increased expression of the transgenic UPRPR reporter, but only slightly and also with LOELs for induction higher than LOELs for developmental delay.
Binding immunoglobulin protein (BiP) is a highly conserved molecular chaperone that acts in the endoplasmic reticulum (ER), and expression of the C. elegans BiP homolog hsp-4 is used as a biomarker of ER specific UPR (UPRER) (Kapulkin et al., 2005). Under normal conditions, BiP/HSP-4 is closely regulated to align the folding capacity of the endoplasmic reticulum to the level of unfolded proteins present (Calfon et al., 2002, Taylor and Dillin, 2013). In contrast, UPRER is downregulated during aging, in response to certain chemicals, and in some disease states (Katayama et al., 1999, Naidoo et al., 2018, Tan et al., 2015, Yamagishi et al., 2012). Additionally, knockdown of BiP and decreased UPRER inhibits some viral infections, indicating that downregulation of UPR can be an effective immune response (Goodwin et al., 2011, Limjindaporn et al., 2009). The UPRER biomarker under the control of the hsp-4 promoter was slightly but significantly downregulated by both meHgCl and DMA at LOEL concentrations for developmental delay. NaAsO2 also downregulated UPRER, but only at 150 µg/ml, a concentration far above those needed to induce developmental delay. In contrast to UPRER downregulation by meHgCl, HgCl2 upregulated the UPRER biomarker at concentrations relevant to developmental toxicity. These results further highlight the distinct stress responses to different chemical species of the same toxic elements.
Current data do not support the use of C. elegans toxicity responses alone to predict safe exposure levels in other species, however this work does suggest a preliminary ranking scheme. If toxicity in C. elegans were categorized as very toxic for responses at<5 µg/ml, toxic for 5 to 20 µg/ml, moderate toxicity for 20 to 100 µg/ml, and low toxicity for 100 µg/ml to 1 mg/ml, then for developmental toxicity both meHgCl and HgCl2 would be categorized as very toxic, NaAsO2 as toxic, and DMA as low toxicity. For oxidative stress, the categories would be slightly different, with NaAsO2 and meHgCl in the very toxic category, HgCl2 in the toxic category, and DMA in the moderate toxicity category.
In vertebrates, arsenite-3-methyltransferase (AS3MT) is considered by many to be the primary enzyme that methylates iAs to form organic metabolites, though there is growing evidence of other genes playing a significant role as well (Zdraljevic et al., 2019). While the C. elegans genome encodes homologs to the majority of human genes, it lacks a close AS3MT homolog (Cartwright, 2016). N-6-adenine-specific DNA methyltransferase 1 (N6AMT1) has been found to methylate arsenite in vitro, and polymorphisms of N6AMT1 are associated with changes in arsenic metabolism in humans (Chen et al., 2017, Harari et al., 2013). The C. elegans gene mtq-2 (MethylTransferase modifying glutamine (Q)) is a BLASTP match for N6AMT1 for 85.5% of length. Additionally, a search of Wormbase.org (Version: WS280) for methyltransferase genes in C. elegans yields nearly two hundred entries, so it is possible that one or more of these gene products can methylate iAs. If C. elegans do not methylate NaAsO2, then they model the toxicity of iAs without methylated metabolites. If C. elegans do methylate iAs, then they are a closer model to mammalian arsenic metabolism. This question will be answered with a follow-up study where we will assess arsenic uptake and speciation in C. elegans, as well as reproductive and germline toxicities.
5. Conclusions
Developmental toxicity was ranked meHgCl > HgCl2 > NaAsO2 ≫ DMA in C. elegans. In contrast, oxidative stress response was ranked NaAsO2 = meHgCl > HgCl2 > DMA in C. elegans adults. Relative to NaAsO2, exposures to approximately 20-fold higher concentrations of DMA induced similar levels of both developmental delay and oxidative stress. A bioreporter for conserved proteasome specific unfolded protein response (UPRPR) was strongly induced by NaAsO2 but was not significantly altered by DMA, consistent with disparate effects on UPRPR for these two forms of arsenic. Transgene expression of a conserved marker for ER specific unfolded protein response (UPRER) was increased by HgCl2 but decreased by meHgCl, consistent with disparate effects on UPRER for these two forms of mercury. As in juvenile rodents, C. elegans developmental exposure to meHgCl induced hypoactivity, indicating conservation of toxic effect. For both developmental toxicity and oxidative stress response in C. elegans, monomethylation increased mercury toxicity while dimethylation decreased arsenic toxicity. Transgene expression correlated well with native gene expression, indicating that fluorescence assessment using C. elegans GFP transgenic strains can accurately reflect pathway activation, however optimization of some strains would be required prior to use in higher-throughput analyses. Our findings in C. elegans reflect findings in mammals for which there is data, stress the disparity of effects from exposure to different chemical forms of arsenic and mercury, and indicate low oral toxicity for DMA relative to inorganic arsenic in this model organism.
Funding
All authors are FDA employees. This work was funded and conducted within the context of regular operating budgets and employment duties.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling Editor Thomas B Knudsen
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2022.100071.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agger K., Cloos P.A.C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Aguado A., Galan M., Zhenyukh O., Wiggers G.A., Roque F.R., Redondo S., Pecanha F., Martin A., Fortuno A., Cachofeiro V., Tejerina T., Salaices M., Briones A.M. Mercury induces proliferation and reduces cell size in vascular smooth muscle cells through MAPK, oxidative stress and cyclooxygenase-2 pathways. Toxicol. Appl. Pharmacol. 2013;268:188–200. doi: 10.1016/j.taap.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Almendinger, J., K. Doukoumetzidis, J.M. Kinchen, A. Kaech, K.S. Ravichandran, and M.O. Hengartner, 2011. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS One. 6:e18325. https://doi.org/10.1371/journal.pone.0018325. [DOI] [PMC free article] [PubMed]
- An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E.C., Horvitz H.R. Two C. elegans histone methyltransferases repress lin-3EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- Antunes dos Santos A., Ferrer B., Marques Gonçalves F., Tsatsakis A., Renieri E., Skalny A., Farina M., Rocha J., Aschner M. Oxidative Stress in Methylmercury-Induced Cell Toxicity. Toxics. 2018;6(3):47. doi: 10.3390/toxics6030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono J., Yanagawa T., Itoh K., Li B., Yoshida H., Kumagai Y., Yamamoto M., Ishii T. Activation of Nrf2 and accumulation of ubiquitinated A170 by arsenic in osteoblasts. Biochem. Biophys. Res. Commun. 2003;305:271–277. doi: 10.1016/s0006-291x(03)00728-9. [DOI] [PubMed] [Google Scholar]
- Aschner M., Ceccatelli S., Daneshian M., Fritsche E., Hasiwa N., Hartung T., Hogberg H.T., Leist M., Li A., Mundi W.R., Padilla S., Piersma A.H., Bal-Price A., Seiler A., Westerink R.H., Zimmer B., Lein P.J. Reference compounds for alternative test methods to indicate developmental neurotoxicity (DNT) potential of chemicals: example lists and criteria for their selection and use. ALTEX. 2017;34:49–74. doi: 10.14573/altex.1604201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsdr . U.S; Agency for Toxic Substances and Disease Registry: 1999. Toxicological Profile for Mercury. https://permanent.access.gpo.gov/gpo31588/tp46.pdf. [Google Scholar]
- Atsdr . U.S; Agency for Toxic Substances and Disease Registry: 2007. Toxicological Profile for Arsenic. https://www.atsdr.cdc.gov/toxprofiles/tp2.pdf. [PubMed] [Google Scholar]
- ATSDR. 2019. Substance Priority List. https://www.atsdr.cdc.gov/spl/index.html#2019spl.
- Avila A.M., Bebenek I., Bonzo J.A., Bourcier T., Davis Bruno K.L., Carlson D.B., Dubinion J., Elayan I., Harrouk W., Lee S.L., Mendrick D.L., Merrill J.C., Peretz J., Place E., Saulnier M., Wange R.L., Yao J., Zhao D., Brown P.C. An FDA/CDER perspective on nonclinical testing strategies: Classical toxicology approaches and new approach methodologies (NAMs) Regul. Toxicol. Pharm. 2020;114 doi: 10.1016/j.yrtph.2020.104662. [DOI] [PubMed] [Google Scholar]
- Ayensu W.K., Tchounwou P.B. Microarray analysis of mercury-induced changes in gene expression in human liver carcinoma (HepG2) cells: importance in immune responses. Int. J. Environ. Res. Public Health. 2006;3:141–173. doi: 10.3390/ijerph2006030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc.: Ser. B (Methodol.) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Benramdane L., Bressolle F., Vallon J.J. Arsenic speciation in humans and food products: a review. J. Chromatogr. Sci. 1999;37:330–344. doi: 10.1093/chromsci/37.9.330. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K., Sewell A.K., Wu Z., Han M. TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging. Genetics. 2019;213:329–360. doi: 10.1534/genetics.119.302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T.K., Steinbaugh M.J., Hourihan J.M., Ewald C.Y., Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med. 2015;88:290–301. doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block D.H.S., Twumasi-Boateng K., Kang H.S., Carlisle J.A., Hanganu A., Lai T.-J., Shapira M., Tan M.-W. The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult C. elegans. PLoS Genet. 2015;11(5):e1005265. doi: 10.1371/journal.pgen.1005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz D.D., Tenor J.L., Aballay A. A conserved PMK-1/p38 MAPK is required in caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J. Biol. Chem. 2010;285:10832–10840. doi: 10.1074/jbc.M109.091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd W.A., McBride S.J., Rice J.R., Snyder D.W., Freedman J.H. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol. Appl. Pharmacol. 2010;245:153–159. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd W.A., Smith M.V., Co C.A., Pirone J.R., Rice J.R., Shockley K.R., Freedman J.H. Developmental Effects of the ToxCast Phase I and Phase II Chemicals in Caenorhabditis elegans and Corresponding Responses in Zebrafish, Rats, and Rabbits. Environ. Health Perspect. 2016;124:586–593. doi: 10.1289/ehp.1409645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco V., Godinho-Santos A., Goncalves J., Lu J., Holmgren A., Carvalho C. Mitochondrial thioredoxin reductase inhibition, selenium status, and Nrf-2 activation are determinant factors modulating the toxicity of mercury compounds. Free Radic Biol Med. 2014;73:95–105. doi: 10.1016/j.freeradbiomed.2014.04.030. [DOI] [PubMed] [Google Scholar]
- Bull V.H., Thiede B. Proteome analysis of tunicamycin-induced ER stress. Electrophoresis. 2012;33:1814–1823. doi: 10.1002/elps.201100565. [DOI] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cartwright, I.L. 2016. Invertebrate Models in Arsenic Research: Past, Present, and Future. In Arsenic: Exposure Sources, Health Risks, and Mechanisms of Toxicity. J.C. States, editor. John Wiley & Sons, Inc. 469-493.
- Cecere G., Hoersch S., Jensen M.B., Dixit S., Grishok A. The ZFP-1(AF10)/DOT-1 Complex Opposes H2B Ubiquitination to Reduce Pol II Transcription. Mol. Cell. 2013;50:894–907. doi: 10.1016/j.molcel.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cfsan . U.S; Food and Drug Administration: 2015. 2015–2018 Science and Research Strategic Plan. https://www.fda.gov/Food/FoodScienceResearch/ResearchStrategicPlan/default.htm. [Google Scholar]
- Chandravanshi L.P., Gupta R., Shukla R.K. Developmental Neurotoxicity of Arsenic: Involvement of Oxidative Stress and Mitochondrial Functions. Biol. Trace Elem. Res. 2018;186:185–198. doi: 10.1007/s12011-018-1286-1. [DOI] [PubMed] [Google Scholar]
- Chen X., Guo X., He P., Nie J., Yan X., Zhu J., Zhang L., Mao G., Wu H., Liu Z., Aga D., Xu P., Smith M., Ren X. Interactive Influence of N6AMT1 and As3MT Genetic Variations on Arsenic Metabolism in the Population of Inner Mongolia. China. Toxicol Sci. 2017;155:124–134. doi: 10.1093/toxsci/kfw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg, E.D., H.F. Lapenotiere, D.Y. French, and M. Szilagyi. 2002. Use of CeHR axenic medium for exposure and gene expression studies. In 2002 East Coast Worm Meeting, Reproductive Hazards Laboratory, US Army Center for Environmental Health Research, Fort Detrick, MD.
- Congress U.S. C.o.O.a.R. Subcommittee on Economic and Consumer Policy U.S; House of Representatives: 2021. Baby Foods Are Tainted with Dangerous Levels of Arsenic, Lead, Cadmium, and Mercury. https://www.eenews.net/assets/2021/02/04/document_gw_03.pdf. [Google Scholar]
- Crofton K.M., Howard J.L., Moser V.C., Gill M.W., Reiter L.W., Tilson H.A., MacPhail R.C. Interlaboratory comparison of motor activity experiments: implications for neurotoxicological assessments. Neurotoxicol. Teratol. 1991;13:599–609. doi: 10.1016/0892-0362(91)90043-v. [DOI] [PubMed] [Google Scholar]
- Cui Y., McBride S.J., Boyd W.A., Alper S., Freedman J.H. Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol. 2007;8:R122. doi: 10.1186/gb-2007-8-6-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangleben N.L., Skibola C.F., Smith M.T. Arsenic immunotoxicity: a review. Environ Health. 2013;12:73. doi: 10.1186/1476-069X-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.W., Myers G.J., Weiss B. Mercury exposure and child development outcomes. Pediatrics. 2004;113:1023–1029. [PubMed] [Google Scholar]
- Del Razo L.M., Quintanilla-Vega B., Brambila-Colombres E., Calderon-Aranda E.S., Manno M., Albores A. Stress proteins induced by arsenic. Toxicol. Appl. Pharmacol. 2001;177:132–148. doi: 10.1006/taap.2001.9291. [DOI] [PubMed] [Google Scholar]
- Detienne G., Van de Walle P., De Haes W., Schoofs L., Temmerman L. SKN-1-independent transcriptional activation of glutathione S-transferase 4 (GST-4) by EGF signaling. Worm. 2016;5 doi: 10.1080/21624054.2016.1230585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA. 2009. EFSA Panel on Contaminants in the Food Chain (CONTAM): Scientific Opinion on Arsenic in Food. European Food Safety Authority (EFSA). [DOI] [PMC free article] [PubMed]
- Efsa Scientific Opinion on the risk for public health related to the presence of mercury and methylmercury in food. European Food Safety Authority. 2012 https://efsa.onlinelibrary.wiley.com/doi/abs/10.2903/j.efsa.2012.2985 [Google Scholar]
- Engelmann I., Griffon A., Tichit L., Montañana-Sanchis F., Wang G., Reinke V., Waterston R.H., Hillier L.W., Ewbank J.J., Lehner B. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE. 2011;6(5):e19055. doi: 10.1371/journal.pone.0019055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero-Lourdes C. Toxicity mechanisms of arsenic that are shared with neurodegenerative diseases and cognitive impairment: Role of oxidative stress and inflammatory responses. Neurotoxicology. 2016;53:223–235. doi: 10.1016/j.neuro.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Evans, E.A., T. Kawli, and M.W. Tan, 2008. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4:e1000175. https://doi.org/10.1371/journal.ppat.1000175. [DOI] [PMC free article] [PubMed]
- Fang, H., S.C. Harris, Z. Su, M. Chen, F. Qian, L. Shi, R. Perkins, and W. Tong. 2017. ArrayTrack: An FDA and Public Genomic Tool. In Biological Networks and Pathway Analysis. Vol. Methods in Molecular Biology. T. T. and N. Y., editors. Humana Press, New York, NY.
- Faoun, who. Evaluation of Certain Contaminants in Food: Seventy-second report of the Joint FAO/WHO Expert Committee on Food Additives. World Health. Organization. 2011 https://apps.who.int/iris/handle/10665/44520 [Google Scholar]
- FDA, U.S. 2016. Arsenic in Rice and Rice Products Risk Assessment Report. Center for Food Safety and Applied Nutrition https://www.fda.gov/files/food/published/Arsenic-in-Rice-and-Rice-Products-Risk-Assessment-Report-PDF.pdf.
- FDA, U.S. 2017. FDA's Predictive Toxicology Roadmap. https://www.fda.gov/ScienceResearch/AboutScienceResearchatFDA/ucm601090.htm.
- FDA, U.S. 2020. Metals and Your Food. https://www.fda.gov/food/chemicals-metals-pesticides-food/metals-and-your-food.
- FDA, u.s. U.S; Food and Drug Administration: 2021. Closer to Zero: Action Plan for Baby Foods. https://www.fda.gov/food/metals-and-your-food/closer-zero-action-plan-baby-foods. [Google Scholar]
- Feldmann J., Krupp E.M. Critical review or scientific opinion paper: arsenosugars–a class of benign arsenic species or justification for developing partly speciated arsenic fractionation in foodstuffs? Anal. Bioanal. Chem. 2011;399:1735–1741. doi: 10.1007/s00216-010-4303-6. [DOI] [PubMed] [Google Scholar]
- Filippini T., Malavolti M., Cilloni S., Wise L.A., Violi F., Malagoli C., Vescovi L., Vinceti M. Intake of arsenic and mercury from fish and seafood in a Northern Italy community. Food Chem. Toxicol. 2018;116:20–26. doi: 10.1016/j.fct.2018.04.010. [DOI] [PubMed] [Google Scholar]
- Foster K.J., Cheesman H.K., Liu P., Peterson N.D., Anderson S.M., Pukkila-Worley R. Innate Immunity in the C. elegans Intestine Is Programmed by a Neuronal Regulator of AWC Olfactory Neuron Development. Cell Rep. 2020;31(107478) doi: 10.1016/j.celrep.2020.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi K.A. Toxic metal species and food regulations–making a healthy choice. Analyst. 2007;132:17–20. doi: 10.1039/b610544k. [DOI] [PubMed] [Google Scholar]
- Fredriksson A., Gardlund A.T., Bergman K., Oskarsson A., Ohlin B., Danielsson B., Archer T. Effects of maternal dietary supplementation with selenite on the postnatal development of rat offspring exposed to methyl mercury in utero. Pharmacol. Toxicol. 1993;72:377–382. doi: 10.1111/j.1600-0773.1993.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cabo P., Bolinches-Amoros A., Cabello J., Ros S., Moreno S., Baylis H.A., Palau F., Vazquez-Manrique R.P. Disruption of the ATP-binding cassette B7 (ABTM-1/ABCB7) induces oxidative stress and premature cell death in Caenorhabditis elegans. J. Biol. Chem. 2011;286:21304–21314. doi: 10.1074/jbc.M110.211201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, E.C., A. Lipovsky, T. Inoue, T.G. Magaldi, A.P. Edwards, K.E. Van Goor, A.W. Paton, J.C. Paton, W.J. Atwood, B. Tsai, and D. DiMaio, 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio. 2:e00101-00111. https://doi.org/10.1128/mBio.00101-11 [DOI] [PMC free article] [PubMed]
- Hackethal C., Kopp J.F., Sarvan I., Schwerdtle T., Lindtner O. Total arsenic and water-soluble arsenic species in foods of the first German total diet study (BfR MEAL Study) Food Chem. 2021;346 doi: 10.1016/j.foodchem.2020.128913. [DOI] [PubMed] [Google Scholar]
- Hao C., Hao W., Wei X., Xing L., Jiang J., Shang L. The role of MAPK in the biphasic dose-response phenomenon induced by cadmium and mercury in HEK293 cells. Toxicol. In Vitro. 2009;23:660–666. doi: 10.1016/j.tiv.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Harari F., Engstrom K., Concha G., Colque G., Vahter M., Broberg K. N-6-adenine-specific DNA methyltransferase 1 (N6AMT1) polymorphisms and arsenic methylation in Andean women. Environ. Health Perspect. 2013;121:797–803. doi: 10.1289/ehp.1206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J.H., Widmayer S.J., Bergemann C.M., King D.E., Morton K.S., Romersi R.F., Jameson L.E., Leung M.C.K., Andersen E.C., Taubert S., Meyer J.N. Xenobiotic metabolism and transport in Caenorhabditis elegans. J. Toxicol. Environ Health B Crit. Rev. 2021;24:51–94. doi: 10.1080/10937404.2021.1884921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Isomura K., Tsutsumiuchi K., Taniguchi H., Miwa J. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol. Sci. 2008;101:215–225. doi: 10.1093/toxsci/kfm276. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Tsutsumiuchi K., Miwa J., Dalmasso G. Allyl isothiocyanate that induces GST and UGT expression confers oxidative stress resistance on C. elegans, as demonstrated by nematode biosensor. PLoS ONE. 2010;5(2):e9267. doi: 10.1371/journal.pone.0009267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins K.A., Russell J.F., Gaddis N., Dressman H.K., Aballay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev. Cell. 2008;15:87–97. doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmcke K.J., Syversen T., Miller D.M., 3rd, Aschner M. Characterization of the effects of methylmercury on Caenorhabditis elegans. Toxicol. Appl. Pharmacol. 2009;240:265–272. doi: 10.1016/j.taap.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L.W., Coulson A., Murray J.I., Bao Z., Sulston J.E., Waterston R.H. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 2005;15:1651–1660. doi: 10.1101/gr.3729105. [DOI] [PubMed] [Google Scholar]
- van der Hoeven R., McCallum K.C., Cruz M.R., Garsin D.A., Ausubel F.M. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 2011;7(12):e1002453. doi: 10.1371/journal.ppat.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou N.S., Gutschmidt A., Choi D.Y., Pather K., Shi X., Watts J.L., Hoppe T., Taubert S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci U S A. 2014;111:E2271–2280. doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P.R. The C. elegans model in toxicity testing. J. Appl. Toxicol. 2017;37:50–59. doi: 10.1002/jat.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P.R., Camacho J.A., Sprando R.L. Caenorhabditis elegans for predictive toxicology. Current Opinion in Toxicology. 2020;23–24:23–28. doi: 10.1016/j.cotox.2020.02.004. [DOI] [Google Scholar]
- Hunt P.R., Olejnik N., Bailey K.D., Vaught C.A., Sprando R.L. C. elegans Development and Activity Test detects mammalian developmental neurotoxins. Food Chem. Toxicol. 2018;121:583–592. doi: 10.1016/j.fct.2018.09.061. [DOI] [PubMed] [Google Scholar]
- Hunt P.R., Olejnik N., Sprando R.L. Toxicity ranking of heavy metals with screening method using adult Caenorhabditis elegans and propidium iodide replicates toxicity ranking in rat. Food Chem. Toxicol. 2012;50:3280–3290. doi: 10.1016/j.fct.2012.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Hisamoto N., An J.H., Oliveira R.P., Nishida E., Blackwell T.K., Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J.E., Troemel E.R., Feinbaum R.L., Luhachack L.G., Cezairliyan B.O., Ausubel F.M., Guttman D.S. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6(7):e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRIS. 1991. Chemical Risk Assessment Summary: Inorganic arsenic; CASRN 7440-38-2. U.S. Environmental Protection Agency, Integrated Risk Information System (IRIS) https://iris.epa.gov/ChemicalLanding/&substance_nmbr=278
- IRIS. 1995. Chemical Assessment Summary: Mercuric chloride (HgCl2) CASRN 7487-94-7. U.S. Environmental Protection Agency, Integrated Risk Information System (IRIS) https://iris.epa.gov/ChemicalLanding/&substance_nmbr=692
- IRIS. 2001. Chemical Assessment Summary: Methylmercury (MeHg); CASRN 22967-92-6. U.S. Environmental Protection Agency, Integrated Risk Information System (IRIS) https://iris.epa.gov/ChemicalLanding/&substance_nmbr=73
- Jones, L.M., S.J. Rayson, A.J. Flemming, and P.E. Urwin, 2013. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS One. 8:e69956. https://doi.org/10.1371/journal.pone.0069956. [DOI] [PMC free article] [PubMed]
- Kaletta T., Hengartner M.O. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- Kang Y.H., Lee S.J. The role of p38 MAPK and JNK in Arsenic trioxide-induced mitochondrial cell death in human cervical cancer cells. J. Cell. Physiol. 2008;217:23–33. doi: 10.1002/jcp.21470. [DOI] [PubMed] [Google Scholar]
- Kaplan R.E.W., Maxwell C.S., Codd N.K., Baugh L.R. Pervasive Positive and Negative Feedback Regulation of Insulin-Like Signaling in Caenorhabditis elegans. Genetics. 2019;211:349–361. doi: 10.1534/genetics.118.301702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulkin W.J., Hiester B.G., Link C.D. Compensatory regulation among ER chaperones in C. elegans. FEBS Lett. 2005;579:3063–3068. doi: 10.1016/j.febslet.2005.04.062. [DOI] [PubMed] [Google Scholar]
- Katayama T., Imaizumi K., Sato N., Miyoshi K., Kudo T., Hitomi J., Morihara T., Yoneda T., Gomi F., Mori Y., Nakano Y., Takeda J., Tsuda T., Itoyama Y., Murayama O., Takashima A., St George-Hyslop P., Takeda M., Tohyama M. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat. Cell Biol. 1999;1:479–485. doi: 10.1038/70265. [DOI] [PubMed] [Google Scholar]
- Kim Y., Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell. 2007;6:489–503. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- Kourtis N., Nikoletopoulou V., Tavernarakis N. Small heat-shock proteins protect from heat-stroke-associated neurodegeneration. Nature. 2012;490:213–218. doi: 10.1038/nature11417. [DOI] [PubMed] [Google Scholar]
- Kuo C.T., You G.T., Jian Y.J., Chen T.S., Siao Y.C., Hsu A.L., Ching T.T. AMPK-mediated formation of stress granules is required for dietary restriction-induced longevity in Caenorhabditis elegans. Aging Cell. 2020;19 doi: 10.1111/acel.13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagido, C., D. McLaggan, and L.A. Glover, 2015. A Screenable In Vivo Assay for Mitochondrial Modulators Using Transgenic Bioluminescent Caenorhabditis elegans. J Vis Exp:e53083. https://doi.org/10.3791/53083. [DOI] [PMC free article] [PubMed]
- Lau A., Whitman S.A., Jaramillo M.C., Zhang D.D. Arsenic-mediated activation of the Nrf2-Keap1 antioxidant pathway. J. Biochem. Mol. Toxicol. 2013;27:99–105. doi: 10.1002/jbt.21463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Hong S., Lee M.H., Koo H.-S., Huen M.-Y. A PHF8 Homolog in C. elegans Promotes DNA Repair via Homologous Recombination. PLoS ONE. 2015;10(4):e0123865. doi: 10.1371/journal.pone.0123865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiers B., Kampkotter A., Grevelding C.G., Link C.D., Johnson T.E., Henkle-Duhrsen K. A stress-responsive glutathione S-transferase confers resistance to oxidative stress in Caenorhabditis elegans. Free Radic Biol Med. 2003;34:1405–1415. doi: 10.1016/s0891-5849(03)00102-3. [DOI] [PubMed] [Google Scholar]
- Leung M.C., Williams P.L., Benedetto A., Au C., Helmcke K.J., Aschner M., Meyer J.N. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezzerini M., Budovskaya Y. A dual role of the Wnt signaling pathway during aging in Caenorhabditis elegans. Aging Cell. 2014;13:8–18. doi: 10.1111/acel.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gao S., Jing H., Qi L., Ning J., Tan Z., Yang K., Zhao C., Ma L., Li G. Correlation of chemical acute toxicity between the nematode and the rodent. Toxicol. Res. 2013;2:403–412. doi: 10.1039/c3tx50039j. [DOI] [Google Scholar]
- Limjindaporn T., Wongwiwat W., Noisakran S., Srisawat C., Netsawang J., Puttikhunt C., Kasinrerk W., Avirutnan P., Thiemmeca S., Sriburi R., Sittisombut N., Malasit P., Yenchitsomanus P.T. Interaction of dengue virus envelope protein with endoplasmic reticulum-resident chaperones facilitates dengue virus production. Biochem. Biophys. Res. Commun. 2009;379:196–200. doi: 10.1016/j.bbrc.2008.12.070. [DOI] [PubMed] [Google Scholar]
- Luvonga C., Rimmer C.A., Yu L.L., Lee S.B. Analytical Methodologies for the Determination of Organoarsenicals in Edible Marine Species: A Review. J. Agric. Food Chem. 2020;68:1910–1934. doi: 10.1021/acs.jafc.9b04525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz A.L., Godebo T.R., Bhatt D.P., Ilkayeva O.R., Maurer L.L., Hirschey M.D., Meyer J.N. From the Cover: Arsenite Uncouples Mitochondrial Respiration and Induces a Warburg-like Effect in Caenorhabditis elegans. Toxicol. Sci. 2016;152:349–362. doi: 10.1093/toxsci/kfw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz A.L., Godebo T.R., Smith L.L., Leuthner T.C., Maurer L.L., Meyer J.N. Deficiencies in mitochondrial dynamics sensitize Caenorhabditis elegans to arsenite and other mitochondrial toxicants by reducing mitochondrial adaptability. Toxicology. 2017;387:81–94. doi: 10.1016/j.tox.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Yue J., Liang S., Gao M., Wang X., Cui N., Li H., Zhi D. Realgar increases defenses against infection by Enterococcus faecalis in Caenorhabditis elegans. J. Ethnopharmacol. 2021;268 doi: 10.1016/j.jep.2020.113559. [DOI] [PubMed] [Google Scholar]
- Marsh D.O., Clarkson T.W., Myers G.J., Davidson P.W., Cox C., Cernichiari E., Tanner M.A., Lednar W., Shamlaye C., Choisy O., et al. The Seychelles study of fetal methylmercury exposure and child development: introduction. Neurotoxicology. 1995;16:583–596. [PubMed] [Google Scholar]
- Martinez-Finley E.J., Caito S., Slaughter J.C., Aschner M. The Role of skn-1 in methylmercury-induced latent dopaminergic neurodegeneration. Neurochem. Res. 2013;38:2650–2660. doi: 10.1007/s11064-013-1183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Finley E.J., Chakraborty S., Slaughter J.C., Aschner M. Early-life exposure to methylmercury in wildtype and pdr-1/parkin knockout C. elegans. Neurochem. Res. 2013;38:1543–1552. doi: 10.1007/s11064-013-1054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masjosthusmann S., Barenys M., El‐Gamal M., Geerts L., Gerosa L., Gorreja A., Kühne B., Marchetti N., Tigges J., Viviani B., Witters H., Fritsche E. Literature review and appraisal on alternative neurotoxicity testing methods. EFS3. 2018;15(4) [Google Scholar]
- Matilainen O., Ribeiro A.R.S., Verbeeren J., Cetinbas M., Sadreyev R.I., Garcia S.M.D.A. BioRxiv Cold Spring Harbor Laboratory; 2021. Loss of Muscleblind Splicing Factor Shortens C. elegans Lifespan by Reducing the Activity of p38 MAPK/PMK-1 and Transcription Factors ATF-7 and Nrf/SKN-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J., Bubb K., Thomas J.H. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2(2):111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- McElwee M.K., Freedman J.H. Comparative toxicology of mercurials in Caenorhabditis elegans. Environ. Toxicol. Chem. 2011;30:2135–2141. doi: 10.1002/etc.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee M.K., Ho L.A., Chou J.W., Smith M.V., Freedman J.H. Comparative toxicogenomic responses of mercuric and methyl-mercury. BMC Genomics. 2013;14:698. doi: 10.1186/1471-2164-14-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikoláš, P., J. Kollárová, K. Šebková, V. Saudek, P. Yilma, M. Kostrouchová, M.W. Krause, Z. Kostrouch, and M. Kostrouchová, 2013. GEI-8, a Homologue of Vertebrate Nuclear Receptor Corepressor NCoR/SMRT, Regulates Gonad Development and Neuronal Functions in Caenorhabditis elegans. PLOS ONE. 8:e58462. https://doi.org/10.1371/journal.pone.0058462. [DOI] [PMC free article] [PubMed]
- Miller, D.L., M.W. Budde, and M.B. Roth, 2011. HIF-1 and SKN-1 coordinate the transcriptional response to hydrogen sulfide in Caenorhabditis elegans. PLoS One. 6:e25476. https://doi.org/10.1371/journal.pone.0025476. [DOI] [PMC free article] [PubMed]
- Minniti A.N., Cataldo R., Trigo C., Vasquez L., Mujica P., Leighton F., Inestrosa N.C., Aldunate R. Methionine sulfoxide reductase A expression is regulated by the DAF-16/FOXO pathway in Caenorhabditis elegans. Aging Cell. 2009;8:690–705. doi: 10.1111/j.1474-9726.2009.00521.x. [DOI] [PubMed] [Google Scholar]
- Muir R.E., Tan M.W. Virulence of Leucobacter chromiireducens subsp. solipictus to Caenorhabditis elegans: characterization of a novel host-pathogen interaction. Appl. Environ. Microbiol. 2008;74:4185–4198. doi: 10.1128/AEM.00381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenix P.J. Evolution of motor activity tests into a screening reality. Toxicol. Ind. Health. 1989;5:203–219. doi: 10.1177/074823378900500206. [DOI] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Myers G.J., Davidson P.W., Cox C., Shamlaye C.F., Tanner M.A., Choisy O., Sloane-Reeves J., Marsh D., Cernichiari E., Choi A., et al. Neurodevelopmental outcomes of Seychellois children sixty-six months after in utero exposure to methylmercury from a maternal fish diet: pilot study. Neurotoxicology. 1995;16:639–652. [PubMed] [Google Scholar]
- Naess S., Aakre I., Lundebye A.K., Ornsrud R., Kjellevold M., Markhus M.W., Dahl L. Mercury, lead, arsenic, and cadmium in Norwegian seafood products and consumer exposure. Food Addit Contam Part B Surveill. 2020;13:99–106. doi: 10.1080/19393210.2020.1735533. [DOI] [PubMed] [Google Scholar]
- Nag P., Aggarwal P.R., Ghosh S., Narula K., Tayal R., Maheshwari N., Chakraborty N., Chakraborty S. Interplay of neuronal and non-neuronal genes regulates intestinal DAF-16-mediated immune response during Fusarium infection of Caenorhabditis elegans. Cell Death Discov. 2017;3:17073. doi: 10.1038/cddiscovery.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N., Zhu J., Galante R.J., Lian J., Strus E., Lee A., Keenan B.T., Pack A.I. Reduction of the molecular chaperone binding immunoglobulin protein (BiP) accentuates the effect of aging on sleep-wake behavior. Neurobiol. Aging. 2018;69:10–25. doi: 10.1016/j.neurobiolaging.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- Nass R., Hamza I. The nematode C. elegans as an animal model to explore toxicology in vivo: solid and axenic growth culture conditions and compound exposure parameters. Curr Protoc Toxicol. Chapter. 2007;1(Unit1):9. doi: 10.1002/0471140856.tx0109s31. [DOI] [PubMed] [Google Scholar]
- O'Rourke D., Baban D., Demidova M., Mott R., Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.W., Mukhopadhyay A., Dixit B.L., Raha T., Green M.R., Tissenbaum H.A. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Oliveira R.P., Porter Abate J., Dilks K., Landis J., Ashraf J., Murphy C.T., Blackwell T.K. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek J., Lo J.Y., Narasimhan S.D., Nguyen T.N., Glover-Cutter K., Robida-Stubbs S., Suzuki T., Yamamoto M., Blackwell T.K., Curran S.P. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 2012;16:526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish S.T., Aschner M., Casey W., Corvaro M., Embry M.R., Fitzpatrick S., Kidd D., Kleinstreuer N.C., Lima B.S., Settivari R.S., Wolf D.C., Yamazaki D., Boobis A. An evaluation framework for new approach methodologies (NAMs) for human health safety assessment. Regul. Toxicol. Pharm. 2020;112 doi: 10.1016/j.yrtph.2020.104592. [DOI] [PubMed] [Google Scholar]
- Park S.K., Tedesco P.M., Johnson T.E. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince L.M., Rand M.D. Methylmercury exposure causes a persistent inhibition of myogenin expression and C2C12 myoblast differentiation. Toxicology. 2018;393:113–122. doi: 10.1016/j.tox.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybysz A.J., Choe K.P., Roberts L.J., Strange K. Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mech. Ageing Dev. 2009;130:357–369. doi: 10.1016/j.mad.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N., Zugasti O., Wong D., Couillault C., Kurz C.L., Schulenburg H., Ewbank J.J., Ausubel F.M. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008;4(7):e1000105. doi: 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley, R., F.M. Ausubel, and E. Mylonakis, 2011. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 7:e1002074. https://doi.org/10.1371/journal.ppat.1002074. [DOI] [PMC free article] [PubMed]
- Rains M.E., Muncie C.B., Pang Y.i., Fan L.-W., Tien L.-T., Ojeda N.B. Oxidative Stress and Neurodevelopmental Outcomes in Rat Offspring with Intrauterine Growth Restriction Induced by Reduced Uterine Perfusion. Brain Sci. 2021;11(1):78. doi: 10.3390/brainsci11010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Feng H., Fu Y., Land M., Rubin C.S. Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity. 2009;30:521–532. doi: 10.1016/j.immuni.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkiewicz J.A., Pinkas A., Miah M.R., Weitz R.L., Lawes M.J.A., Akinyemi A.J., Ijomone O.M., Aschner M. C. elegans as a model in developmental neurotoxicology. Toxicol. Appl. Pharmacol. 2018;354:126–135. doi: 10.1016/j.taap.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu, S.N., J. Lewis, I. Patel, S. Bozdag, J.H. Lee, R. Sprando, and H.N. Cinar, 2013. Genomic analysis of stress response against arsenic in Caenorhabditis elegans. PLoS One. 8:e66431. https://doi.org/10.1371/journal.pone.0066431. [DOI] [PMC free article] [PubMed]
- Schalock R.L., Brown W.J., Kark R.A., Menon N.K. Perinatal methylmercury intoxication: behavioral effects in rats. Dev. Psychobiol. 1981;14:213–219. doi: 10.1002/dev.420140310. [DOI] [PubMed] [Google Scholar]
- Schifano E., Ficociello G., Vespa S., Ghosh S., Cipollo J.F., Talora C., Lotti L.V., Mancini P., Uccelletti D. Pmr-1 gene affects susceptibility of Caenorhabditis elegans to Staphylococcus aureus infection through glycosylation and stress response pathways' alterations. Virulence. 2019;10:1013–1025. doi: 10.1080/21505594.2019.1697118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch C.M., Gattepaille L.M., Engstrom K., Vahter M., Jakobsson M., Broberg K. Human adaptation to arsenic-rich environments. Mol. Biol. Evol. 2015;32:1544–1555. doi: 10.1093/molbev/msv046. [DOI] [PubMed] [Google Scholar]
- Settivari R., VanDuyn N., LeVora J., Nass R. The Nrf2/SKN-1-dependent glutathione S-transferase pi homologue GST-1 inhibits dopamine neuron degeneration in a Caenorhabditis elegans model of manganism. Neurotoxicology. 2013;38:51–60. doi: 10.1016/j.neuro.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Savage C., Greenwald I. Isolation and characterization of mutations causing abnormal eversion of the vulva in Caenorhabditis elegans. Dev. Biol. 1993;157:423–436. doi: 10.1006/dbio.1993.1146. [DOI] [PubMed] [Google Scholar]
- Shin H., Lee H., Fejes A.P., Baillie D.L., Koo H.S., Jones S.J. Gene expression profiling of oxidative stress response of C. elegans aging defective AMPK mutants using massively parallel transcriptome sequencing. BMC Res Notes. 2011;4(34) doi: 10.1186/1756-0500-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta S.H. Towards an automatic method for toxicity and pharmacological testing in C. elegans. The Worm Breeder's. Gazette. 2010;18(10) [Google Scholar]
- Sok J., Calfon M., Lu J., Lichtlen P., Clark S.G., Ron D. Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones. 2001;6:6–15. doi: 10.1379/1466-1268(2001)006<0006:airapa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprando R.L., Olejnik N., Cinar H.N., Ferguson M. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a Complex Object Parametric Analyzer and Sorter, and axenic liquid media. Food Chem. Toxicol. 2009;47:722–728. doi: 10.1016/j.fct.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Stanhill A., Haynes C.M., Zhang Y., Min G., Steele M.C., Kalinina J., Martinez E., Pickart C.M., Kong X.P., Ron D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell. 2006;23:875–885. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Steinbaugh M.J., Narasimhan S.D., Robida-Stubbs S., Moronetti Mazzeo L.E., Dreyfuss J.M., Hourihan J.M., Raghavan P., Operana T.N., Esmaillie R., Blackwell T.K. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife. 2015;4 doi: 10.7554/eLife.07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L.M., Wang Y. Arsenic Exposure and Compromised Protein Quality Control. Chem. Res. Toxicol. 2020;33:1594–1604. doi: 10.1021/acs.chemrestox.0c00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Wang S., Wang Y., He M., Liu D. Bisphenol A exposure accelerated the aging process in the nematode Caenorhabditis elegans. Toxicol. Lett. 2015;235:75–83. doi: 10.1016/j.toxlet.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Taylor R.C., Dillin A. XBP-1 Is a Cell-Nonautonomous Regulator of Stress Resistance and Longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.B., de Oliveira A.C.A., Leao L.K.R., Fagundes N.C.F., Fernandes R.M., Fernandes L.M.P., da Silva M.C.F., Amado L.L., Sagica F.E.S., de Oliveira E.H.C., Crespo-Lopez M.E., Maia C.S.F., Lima R.R. Exposure to Inorganic Mercury Causes Oxidative Stress, Cell Death, and Functional Deficits in the Motor Cortex. Front. Mol. Neurosci. 2018;11:125. doi: 10.3389/fnmol.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R.G., Ashraf J., Kaletsky R., Kleemann G., Murphy C.T., Bussemaker H.J. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.J., Styblo M., Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicol. Appl. Pharmacol. 2001;176:127–144. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- Tian Y., Garcia G., Bian Q., Steffen K.K., Joe L., Wolff S., Meyer B.J., Dillin A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt) Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cuevas I., Parra-Llorca A., Sanchez-Illana A., Nunez-Ramiro A., Kuligowski J., Chafer-Pericas C., Cernada M., Escobar J., Vento M. Oxygen and oxidative stress in the perinatal period. Redox Biol. 2017;12:674–681. doi: 10.1016/j.redox.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama T., Sumi D., Shinkai Y., Yasutake A., Taguchi K., Tong K.I., Yamamoto M., Kumagai Y. Cytoprotective role of Nrf2/Keap1 system in methylmercury toxicity. Biochem. Biophys. Res. Commun. 2007;363:645–650. doi: 10.1016/j.bbrc.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Troemel E.R., Chu S.W., Reinke V., Lee S.S., Ausubel F.M., Kim D.H. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2(11):e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas, J., and Y. Argon, 2017. xbp-1 mRNA splicing is attenuated under prolonged exposure to ER stress. MicroPubl Biol. 2017. https://doi.org/10.17912/W2707X. [DOI] [PMC free article] [PubMed]
- Tucker E.K., Nowak R.A. Methylmercury alters proliferation, migration, and antioxidant capacity in human HTR8/SV-neo trophoblast cells. Reprod. Toxicol. 2018;78:60–68. doi: 10.1016/j.reprotox.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet J.M., Hertweck M., An J.H., Baker J., Hwang J.Y., Liu S., Oliveira R.P., Baumeister R., Blackwell T.K. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twaddle N.C., Vanlandingham M., Beland F.A., Doerge D.R. Metabolism and disposition of arsenic species after repeated oral dosing with sodium arsenite in drinking water. II. Measurements in pregnant and fetal CD-1 mice. Food Chem. Toxicol. 2018;115:178–184. doi: 10.1016/j.fct.2018.03.010. [DOI] [PubMed] [Google Scholar]
- VanDuyn N., Settivari R., Wong G., Nass R. SKN-1/Nrf2 inhibits dopamine neuron degeneration in a Caenorhabditis elegans model of methylmercury toxicity. Toxicol. Sci. 2010;118:613–624. doi: 10.1093/toxsci/kfq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Yuan L., Jing H., Zheng Q., Xiao H. Defective apoptotic cell clearance activates innate immune response to protect Caenorhabditis elegans against pathogenic bacteria. Virulence. 2021;12:75–83. doi: 10.1080/21505594.2020.1857982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Liu S., Xi S., Yan L., Wang H., Song Y., Sun G. Arsenic induces the expressions of angiogenesis-related factors through PI3K and MAPK pathways in SV-HUC-1 human uroepithelial cells. Toxicol. Lett. 2013;222:303–311. doi: 10.1016/j.toxlet.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Wang J., Robida-Stubbs S., Tullet J.M.A., Rual J.-F., Vidal M., Blackwell T.K., Kim S.K. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6(8):e1001048. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Morimatsu M., Fujita A., Teranishi M., Sudevan S., Watanabe M., Iwasa H., Hata Y., Kagi H., Nishiyama M., Naruse K., Higashitani A. Increased hydrostatic pressure induces nuclear translocation of DAF-16/FOXO in C. elegans. Biochem. Biophys. Res. Commun. 2020;523:853–858. doi: 10.1016/j.bbrc.2020.01.047. [DOI] [PubMed] [Google Scholar]
- WHO. 1991. Inorganic mercury. United Nations Environment Programme http://www.inchem.org/documents/ehc/ehc/ehc118.htm
- Wong D., Bazopoulou D., Pujol N., Tavernarakis N., Ewbank J.J. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WormBase. 2021. SimpleMine. https://wormbase.org//tools/mine/simplemine.cgi.
- Wyatt L.H., Diringer S.E., Rogers L.A., Hsu-Kim H., Pan W.K., Meyer J.N. Antagonistic Growth Effects of Mercury and Selenium in Caenorhabditis elegans Are Chemical-Species-Dependent and Do Not Depend on Internal Hg/Se Ratios. Environ. Sci. Technol. 2016;50:3256–3264. doi: 10.1021/acs.est.5b06044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt L.H., Luz A.L., Cao X., Maurer L.L., Blawas A.M., Aballay A., Pan W.K., Meyer J.N. Effects of methyl and inorganic mercury exposure on genome homeostasis and mitochondrial function in Caenorhabditis elegans. DNA Repair (Amst). 2017;52:31–48. doi: 10.1016/j.dnarep.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N., Ueda T., Mori A., Saito Y., Hatayama T. Decreased expression of endoplasmic reticulum chaperone GRP78 in liver of diabetic mice. Biochem. Biophys. Res. Commun. 2012;417:364–370. doi: 10.1016/j.bbrc.2011.11.118. [DOI] [PubMed] [Google Scholar]
- Yanase S., Yasuda K., Ishii N. Interaction between the ins/IGF-1 and p38 MAPK signaling pathways in molecular compensation of sod genes and modulation related to intracellular ROS levels in C. elegans. Biochem. Biophys. Rep. 2020;23(100796) doi: 10.1016/j.bbrep.2020.100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Frenkel K. Arsenic-mediated cellular signal transduction, transcription factor activation, and aberrant gene expression: implications in carcinogenesis. J. Environ. Pathol. Toxicol. Oncol. 2002;21:331–342. [PubMed] [Google Scholar]
- Yang W., Dierking K., Rosenstiel P.C., Schulenburg H. GATA transcription factor as a likely key regulator of the Caenorhabditis elegans innate immune response against gut pathogens. Zoology (Jena). 2016;119:244–253. doi: 10.1016/j.zool.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Yu C.W., How C.M., Liao V.H. Arsenite exposure accelerates aging process regulated by the transcription factor DAF-16/FOXO in Caenorhabditis elegans. Chemosphere. 2016;150:632–638. doi: 10.1016/j.chemosphere.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Zárate-Potes A., Yang W., Pees B., Schalkowski R., Segler P., Andresen B., Haase D., Nakad R., Rosenstiel P., Tetreau G., Colletier J.-P., Schulenburg H., Dierking K., Collins J.J. The C. elegans GATA transcription factor elt-2 mediates distinct transcriptional responses and opposite infection outcomes towards different Bacillus thuringiensis strains. PLoS Pathog. 2020;16(9):e1008826. doi: 10.1371/journal.ppat.1008826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Fox B.W., Strand C., Panda O., Tenjo F.J., Brady S.C., Crombie T.A., Doench J.G., Schroeder F.C., Andersen E.C. Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism. Elife. 2019;8 doi: 10.7554/eLife.40260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.