Abstract
In the glucose-rich milieu of red blood cells, asexually replicating malarial parasites mainly rely on glycolysis for ATP production, with limited carbon flux through the mitochondrial tricarboxylic acid (TCA) cycle. By contrast, gametocytes and mosquito-stage parasites exhibit an increased dependence on the TCA cycle and oxidative phosphorylation for more economical energy generation. Prior genetic studies supported these stage-specific metabolic preferences by revealing that six of eight TCA cycle enzymes are completely dispensable during the asexual blood stages of Plasmodium falciparum, with only fumarate hydratase (FH) and malate–quinone oxidoreductase (MQO) being refractory to deletion. Several hypotheses have been put forth to explain the possible essentiality of FH and MQO, including their participation in a malate shuttle between the mitochondrial matrix and the cytosol. However, using newer genetic techniques like CRISPR and dimerizable Cre, we were able to generate deletion strains of FH and MQO in P. falciparum. We employed metabolomic analyses to characterize a double knockout mutant of FH and MQO (ΔFM) and identified changes in purine salvage and urea cycle metabolism that may help to limit fumarate accumulation. Correspondingly, we found that the ΔFM mutant was more sensitive to exogenous fumarate, which is known to cause toxicity by modifying and inactivating proteins and metabolites. Overall, our data indicate that P. falciparum is able to adequately compensate for the loss of FH and MQO, rendering them unsuitable targets for drug development.
Keywords: malaria, Plasmodium falciparum, mitochondrion, tricarboxylic acid cycle, aspartate shuttle, DiCre, metabolomics, oxaloacetate
Abbreviations: AAT, aspartate aminotransferase; ADSS, adenylosuccinate synthase; ARSL, argininosuccinate lyase; aTC, anhydrotetracycline; CMA, Complete Medium with Albumax; DHODH, dihydroorotate dehydrogenase; diCre, dimerizable Cre; FC, fold change; FH, fumarate hydratase; ΔFM, double knockout mutant of FH and MQO; gRNA, guide RNA; HA, homology arm; IDC, intraerythrocytic developmental cycle; iRBC, infected red blood cell; mETC, mitochondrial electron transport chain; MQO, malate–quinone oxidoreductase; PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase; RBC, red blood cell; RT, room temperature; SFG, superfolder GFP; S7P, sedoheptulose 7-phosphate; TCA, tricarboxylic acid; uRBC, uninfected red blood cell
The mitochondrion of the malarial parasite, Plasmodium falciparum, is a validated drug target, with atovaquone and several experimental quinolone analogs serving as potent inhibitors of the parasite’s mitochondrial electron transport chain (mETC) (1, 2, 3, 4, 5, 6). During cellular respiration, the mETC receives electrons from many metabolic processes including the mitochondrial tricarboxylic acid (TCA) cycle, which catalyzes the oxidative breakdown of glycolytic pyruvate. The electrons are funneled to oxygen through a series of membrane-bound mETC protein complexes, creating a proton gradient across the organelle’s inner membrane, which in turn supports bulk energy generation by ATP synthase. In the process, the mtETC regenerates the electron carrier ubiquinone, which participates in redox reactions catalyzed by various mitochondrial dehydrogenases including DHODH (dihydroorotate dehydrogenase), an essential enzyme in de novo pyrimidine synthesis (7, 8).
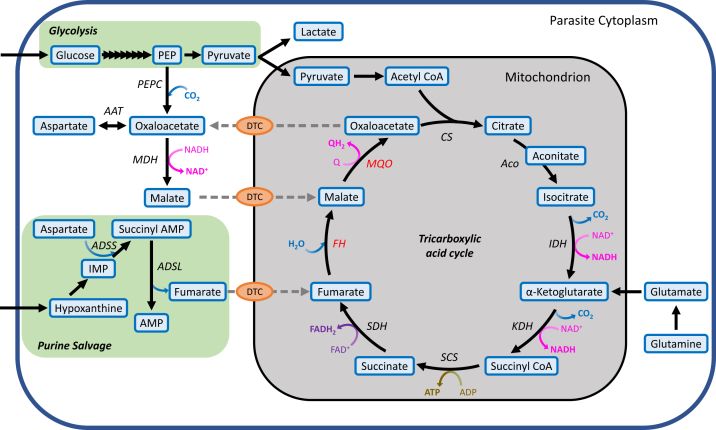
Interestingly, asexual intraerythrocytic P. falciparum parasites do not rely on mitochondrial oxidative phosphorylation for ATP production, instead directing more than 70% of glycolytic pyruvate into anaerobic lactate fermentation (9, 10). This heavy dependence on glycolysis for energy generation is supported by several additional lines of evidence: (1) mitochondria in asexual parasites are largely devoid of cristae, which contain the majority of mETC complexes and ATP synthase (11, 12); (2) they consume minimal levels of oxygen and actually prefer a microaerophilic environment in vitro (13, 14); (3) mETC inhibitors are ineffective in the presence of a yeast DHODH copy that does not require ubiquinone, which indicates that the primary function of the mETC in these stages is to support pyrimidine synthesis (15); and (4) most enzymes of the TCA cycle are dispensable in asexual blood stages (Fig. 1) (16). Only two TCA cycle enzymes have been refractory to disruption in P. falciparum—fumarate hydratase (FH) and malate–quinine oxidoreductase (MQO) (16, 17). Since asexual parasites can survive without an operational TCA cycle, it has been assumed that these two proteins have alternative essential roles, leading to considerable interest in their potential as drug targets (18, 19, 20).
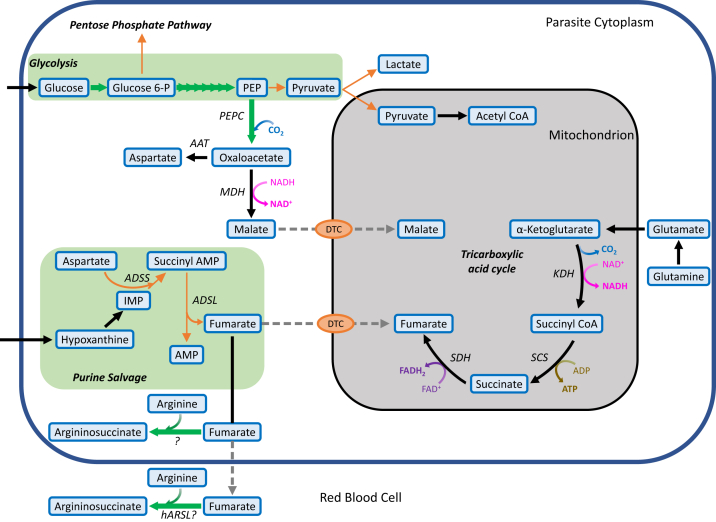
Figure 1.
Schematic of the mitochondrial TCA cycle and its various carbon sources in Plasmodium falciparum. Glucose is imported into the parasite via a hexose transporter (67) and is converted to pyruvate by glycolysis. While majority of the pyruvate is fermented anaerobically to lactate, a small fraction is transported to the mitochondrion where it is converted into the two-carbon acetyl CoA (68), which then enters the TCA cycle. The main source of carbon for the TCA cycle is glutamine, which feeds the cycle as α-ketoglutarate (69). Fumarate, generated as a byproduct of purine salvage, and cytosolic malate, are predicted to serve as anaplerotic inputs and likely enter the mitochondrion through a di/tricarboxylic acid carrier (DTC), which may also function as a malate/oxaloacetate antiporter. Gray dotted lines indicate the predicted transport of metabolites. AAT, aspartate aminotransferase; Aco, aconitase; ADSL, adenylosuccinate lyase; ADSS, adenylosuccinate synthetase; CS, citrate synthase; FAD+/FADH, flavin adenine dinucleotide/reduced; FH, fumarate hydratase; IDH, isocitrate dehydrogenase; KDH, α-ketoglutarate dehydrogenase; MDH, malate dehydrogenase; MQO, malate–quinone oxidoreductase; PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase; Q/QH2, ubiquinone/ubiquinol; SCS, succinate synthase; SDH, succinate dehydrogenase; TCA, tricarboxylic acid.
FH converts fumarate to malate, which is then used as a substrate by MQO to yield oxaloacetate in an irreversible ubiquinone-dependent step (17) (Fig. 1). Apart from the TCA cycle, fumarate is also generated as a byproduct of purine salvage in a reaction catalyzed by the cytosolic enzyme adenylosuccinate lyase (21). Metabolic labeling studies have confirmed that fumarate added to erythrocyte-free parasites enters the mitochondrial TCA cycle (21). In many organisms including humans, an absence or decrease in FH activity can lead to the accumulation of millimolar levels of fumarate, which causes cytotoxicity by irreversibly modifying cysteine residues of proteins and metabolites in a process called succination (22, 23, 24). The FH enzyme in P. falciparum may similarly play a vital role in preventing fumarate buildup and associated toxicity.
FH and MQO have also been postulated to participate in a shuttle across the parasite’s mitochondrial membranes to transport reducing equivalents (25). In this proposed model, oxaloacetate generated by MQO is exported from the mitochondrion to the cytosol by a di/tricarboxylic acid carrier (26). A cytosolic enzyme, malate dehydrogenase, converts oxaloacetate to malate while consuming NADH (27). Malate is then imported into the mitochondrion by di/tricarboxylic acid carrier and reutilized by MQO with the concomitant reduction of ubiquinone to ubiquinol (Fig. 1). Similar shuttles in other organisms are important for maintaining redox balance across subcellular compartments (28). In addition, FH and MQO may play an important role in contributing to the cytosolic oxaloacetate pool. In the absence of the cytosolic enzyme phosphoenolpyruvate carboxylase (PEPC), which converts glycolytic phosphoenolpyruvate (PEP) to oxaloacetate, supplementation with fumarate or malate is required for optimal parasite growth (25). Oxaloacetate is used by aspartate aminotransferase (AAT) to generate aspartate, which is incorporated into proteins and nucleic acids (21, 29). Although aspartate can be obtained from the digestion of host hemoglobin, it may be insufficient to meet the needs of rapidly replicating blood-stage parasites, thus requiring an additional route of production through the combined actions of PEPC, FH/MQO, and AAT (29).
In this work, we used recently established genetic methods to explore the essentiality of FH and MQO in P. falciparum. We found that both proteins are in fact dispensable for parasite survival in the blood stage. However, the FH gene was challenging to disrupt without the provision of a conditional second copy, which suggests that parasites had to adapt to the loss of the enzyme. Using metabolomic analyses, we identified a subset of differently regulated pathways in parasites lacking FH and MQO, and we discuss the implications of these changes. Overall, our results indicate that metabolic plasticity enables parasites to grow robustly even in the absence of FH and MQO, which argues against further exploration of these two proteins as antimalarial drug targets.
Results
Knockdown of FH does not affect parasite fitness
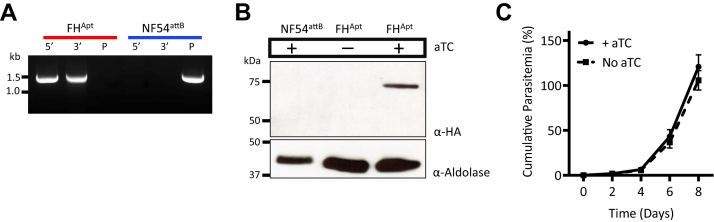
Since previous studies reported that P. falciparum FH could not be disrupted (16, 17), we attempted to conditionally knock down FH expression using the TetR-DOZI system (30, 31). A linear plasmid carrying a 3× hemagglutinin tag and a 10× aptamer array was inserted at the 3′ end of the FH gene in NF54attB parasites using CRISPR/Cas9 (Fig. S1). The genotype of the resulting parasite line (referred to as FHApt) was confirmed by PCR (Fig. 2A). FHApt parasites were continually maintained in medium containing the TetR ligand anhydrotetracycline (aTC), and they robustly expressed hemagglutinin-tagged protein (Fig. 2B). To knock down FH expression, aTC was removed from the culture medium. In the absence of aTC, TetR-DOZI can bind the aptamers in the 3′-UTR of FH mRNA molecules and block their translation. As expected, FH protein could no longer be detected in parasites after the removal of aTC (Fig. 2B). Despite successful knockdown of FH, the parasites continued to grow at a rate comparable with cultures maintained in aTC over an 8-day period (Fig. 2C). This suggested that FH is either dispensable for blood-stage parasites, or alternatively, trace levels of FH are sufficient to support parasite growth.
Figure 2.
TetR-DOZI mediated knockdown of fumarate hydratase (FH).A, integration of the pKDFH plasmid into the 3′-UTR of the FH gene was confirmed by PCR amplification of the 5′ and 3′ loci at the insertion site. PCR for the unmodified locus failed to detect any residual parental (P) parasites in the FHApt population (red); the NF54attB parent line served as a control (blue). B, Western blotting using antihemagglutinin antibody to probe for FH revealed a lack of tagged protein in NF54attB or in FHApt parasites cultured without aTC for 8 days. Anti–aldolase antibody was used as a loading control. C, growth of synchronized FHApt parasites in the presence or the absence of aTC was monitored by flow cytometry for 8 days. The parasites were diluted 1:10 on day 4. Removal of aTC did not cause significant growth inhibition of FHApt parasites. Three biological experiments were conducted in quadruplicate. Growth curves were plotted using GraphPad Prism 9 (GraphPad Software, Inc). Error bars represent the SEM. aTC, anhydrotetracycline.
FH knockout parasites are readily obtained in the presence of a second copy
The absence of an obvious phenotype upon depleting FH in P. falciparum parasites led us to revisit gene knockout strategies. Prior attempts at disrupting FH may have failed because of the use of traditional homologous recombination methods, which rely on random and infrequent dsDNA breaks at the target site (16). We therefore employed CRISPR/Cas9 for targeted deletion of the FH locus (Fig. S2). Despite eight transfection attempts with two different guide RNAs (gRNAs), knockout parasites were not recovered (Table 1).
Table 1.
Summary of transfection outcomes to delete the native FH gene by CRISPR/Cas9
| Parasite line | gRNA (a or b) | Transfection attempts | Successful transfections | |
|---|---|---|---|---|
 |
Nf54attB | a | 4 | 0 |
| Nf54attB | b | 4 | 0 | |
| FH∗-SFG | a | 3 | 3 | |
 |
Nf54attB | a | 4 | 1 |
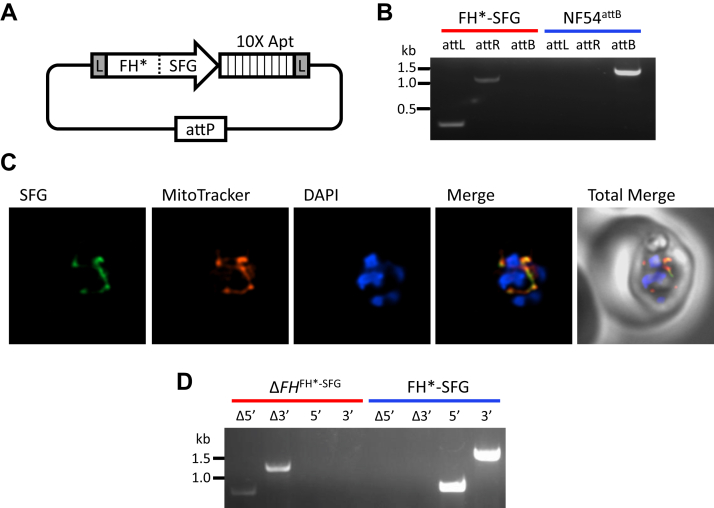
The successful insertion of the TetR-DOZI plasmid into the FH 3′-UTR (as shown in Fig. 2) indicated that the locus is amenable to genetic manipulation. We therefore asked if the FH gene could be deleted in the presence of a second copy. A recodonized version of FH (denoted as FH∗) with a C-terminal superfolder GFP (SFG) tag, a 10× aptamer array, and flanking LoxP sites were inserted into the expression cassette of a plasmid called pCre. The plasmid also carries components of the dimerizable Cre (diCre) conditional knockout system (32, 33) and TetR-DOZI (Figs. 3A, S3 and File S1). The pCre plasmid was introduced into the cg6 locus of NF54attB parasites (34) via attP/attB recombination (35, 36) (Figs. 3B and S4). The parasites were cultured in media containing aTC to maintain elevated levels of FH∗-SFG protein. Using live microscopy, we confirmed that FH∗-SFG localized to the mitochondrion (Fig. 3C). We then used the same CRISPR knockout construct illustrated in Fig. S2 along with one of the gRNAs to delete native FH. All attempted transfections successfully yielded knockout parasites (Fig. 3D) in the FH∗-SFG background (knockout line referred to as ΔFHFH∗-SFG) (Table 1).
Figure 3.
Deletion of native FH locus in the presence of FH∗-SFG.A, schematic of the pCre-FH∗-SFG plasmid displaying a recodonized copy of FH fused to SFG. A 10× aptamer array is located immediately downstream of FH∗-SFG, and the entire locus is flanked by loxP (L) sites. The attP sequence facilitates plasmid integration into NF54attB parasites. B, integration of the pCre-FH∗-SFG plasmid into the attB site of NF54attB parasites was confirmed by PCR amplification of the attL and attR sites. PCR for the unmodified locus failed to detect intact attB sequence in the FH∗-SFG population (red); parental NF54attB parasites served as a control (blue). C, representative immunofluorescence microscopy images of FH∗-SFG parasites show that FH∗-SFG (green) colocalizes with Mitotracker (red) (Mander’s coefficient M1 [green in red] = 0.96, SD = 0.038, n = 28). Images represent fields that are 10 μm long by 10 μm wide. D, disruption of the FH gene in FH∗-SFG parasites was confirmed by PCR amplification (Δ5′ and Δ3′). PCR for the unmodified locus (5′ and 3′) failed to detect native FH in the ΔFHFH∗-SFG population (red); parental FH∗-SFG parasites served as a control (blue). FH, fumarate hydratase; SFG, superfolder GFP.
P. falciparum can survive in the absence of FH
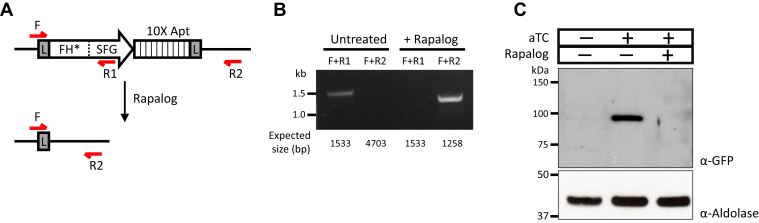
To determine if the ΔFHFH∗-SFG line relies on the second copy of FH for survival, we treated the parasites with rapalog (A/C heterodimerizer) to remove the floxed FH∗-SFG gene (Fig. 4A). Complete excision of the locus was confirmed by PCR (Fig. 4B), and FH∗-SFG protein could no longer be detected in rapalog-treated parasites (Fig. 4C). Surprisingly, the parasites continued to grow despite loss of the FH∗-SFG copy. This indicated that FH is in fact dispensable in P. falciparum, but the genetic route taken to disrupt the locus appears to matter. We performed more transfections in the parental NF54attB line without a second copy of FH and eventually obtained ΔFH parasites from one of a total of 12 attempts (Fig. S5A) (Table 1).
Figure 4.
Excision of FH∗-SFG from ΔFH parasites.A, schematic illustrating the excision of the loxP-flanked FH∗-SFG locus upon treatment with rapalog. The red arrows represent binding sites for primers that were utilized in PCRs shown in (B) to confirm genome rearrangement after rapalog treatment. Anticipated sizes of PCR products for each primer combination in untreated and treated parasites are indicated. The 4.7 kb product expected from F and R2 primers could not be amplified from untreated parasites given the large amplicon size. Rapalog treatment abolishes the R1 primer-binding site from the parasite genome; therefore, the F + R1 primer combination no longer yields a PCR product in treated parasites. The following are the full names of the primers: F, Cre-F; R1, FH∗.SFG-R; and R2, Ter-R. C, Western blotting using anti-GFP antibody to probe for FH∗-SFG could detect protein only in the presence of aTC and without rapalog treatment. Anti–aldolase antibody was used as a loading control. aTC, anhydrotetracycline; FH, fumarate hydratase; SFG, superfolder GFP.
MQO is dispensable individually and together with FH
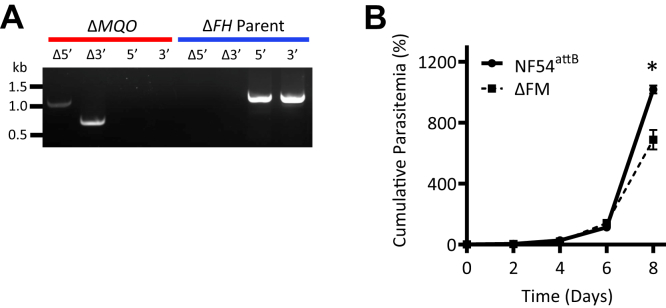
We next targeted the MQO gene for disruption in the parental NF54attB line using CRISPR/Cas9 (Fig. S1). ΔMQO parasites were easily obtained (Fig. S5B), indicating that previous difficulties in disrupting this gene were likely a consequence of the genetic technique employed (16). Since MQO has been postulated to participate in a malate shuttle together with FH, we examined if both gene products could be dispensed with simultaneously or if such an attempt would result in synthetic lethality. We employed the same MQO CRISPR construct to disrupt the gene in the ΔFH background and were successful in obtaining a double knockout mutant of FH and MQO (ΔFM) (Fig. 5A). Growth comparisons of the ΔFM mutant against the parental NF54attB line over an 8-day period revealed a minor fitness defect (Fig. 5B). Despite this, ΔFM parasites continued to grow indefinitely in culture. Together, these results indicate that blood-stage P. falciparum parasites do not require FH and/or MQO for survival.
Figure 5.
Dispensability of malate–quinone oxidoreductase (MQO).A, disruption of the MQO locus in ΔFH parasites was confirmed by PCR amplification (Δ5′ and Δ3′). An intact MQO locus was not detected in the ΔMQO population (red); ΔFH parent parasites served as a control (blue). B, growth of synchronized ΔFM and NF54attB parasites was monitored by flow cytometry for 8 days. Cultures were seeded at 1% parasitemia on day 0 and diluted 1:5 every other day. Three biological experiments were conducted in quadruplicate (two-way ANOVA computed using GraphPad Prism 9 [GraphPad Software, Inc], followed by Bonferroni’s correction; ∗, p < 0.05). Error bars represent the SEM. FH, fumarate hydratase; ΔFM, double KO mutant of FH and MQO.
Metabolomic analysis of ΔFM parasites reveals perturbations in fumarate-generating pathways
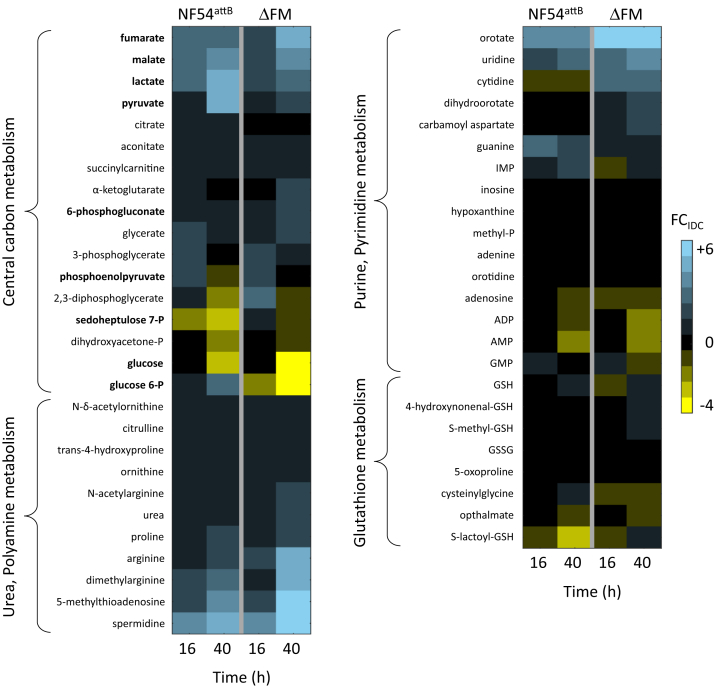
To determine if global changes occurred to accommodate the loss of FH and MQO in ΔFM parasites, we performed metabolomic analysis of synchronized parasite samples collected at 16 and 40 h postinvasion of red blood cells (RBCs) (denoted as infected RBC [iRBC]), which corresponded to early and late stages of the 48 h intraerythrocytic developmental cycle (IDC), respectively. Samples from parallel cultures of uninfected RBCs (uRBC) were also collected at the same time points for metabolomic analysis. Of 552 identified metabolites in iRBC, 413 were common to our previously published control (NF54attB) dataset in which samples were collected under identical conditions (File S2) (37). The fold change (FCIDC) between metabolite levels in iRBC and uRBC samples was computed to determine parasite-specific contributions to the metabolome (Fig. 6 and File S3). The metabolic profile of ΔFM parasites was largely unchanged when compared against the control dataset. Surprisingly, we observed only slight increases in the relative abundance of fumarate and malate in late-stage ΔFM parasites.
Figure 6.
Heat map of select metabolite classes in earlystage and late-stage Plasmodium falciparum parasites. Fold changes (FCIDC) of metabolite abundances in parasite-infected RBCs (iRBC) against uninfected cultures (uRBC) are shown for parental NF54attB and ΔFM parasites. Data are presented as the means of quadruplicate experiments. Metabolites in bold font are discussed in the text. See File S3 for the calculated FCIDC values of all metabolites. ΔFM, double knockout mutant of FH and MQO; IDC, intraerythrocytic developmental cycle; iRBC, infected red blood cell; P, phosphate; uRBC, uninfected red blood cell.
There was a notable difference in FCIDC between ΔFM and control parasites among certain glycolytic pathway metabolites, with a decrease noted in the abundance of glucose, glucose 6-phosphate, and pyruvate in the 40 h ΔFM samples. Lower glucose levels imply greater consumption; we looked for some evidence of this in the two parasite pathways that use glucose, namely, glycolysis and the pentose phosphate pathway. The levels of sedoheptulose 7-phosphate (S7P), a pentose phosphate pathway metabolite, were higher at 16 h in ΔFM parasites (38). However, in a recent analysis of variability in the uRBC metabolome across five independent studies, S7P emerged as one of few RBC metabolites that were highly variable in abundance across time and different studies. We therefore are not confident that the observed change in S7P abundance is meaningful. We detected a substantial increase in the glyoxylase pathway intermediate, S-lactoylglutathione at 40 h, which typically arises from high rates of glycolysis. The glyoxylase system uses GSH to detoxify methylglyoxal, which is formed by the spontaneous elimination of phosphate from the glycolytic triose phosphates, glyceraldehyde 3-phosphate and dihydroxyacetone phosphate. Despite the predicted increase in glycolysis, the end products—pyruvate and lactate—were lower in ΔFM parasites. Since levels of PEP were mostly unchanged in the mutant, we speculate that more PEP is redirected into oxaloacetate production (via PEPC) rather than pyruvate.
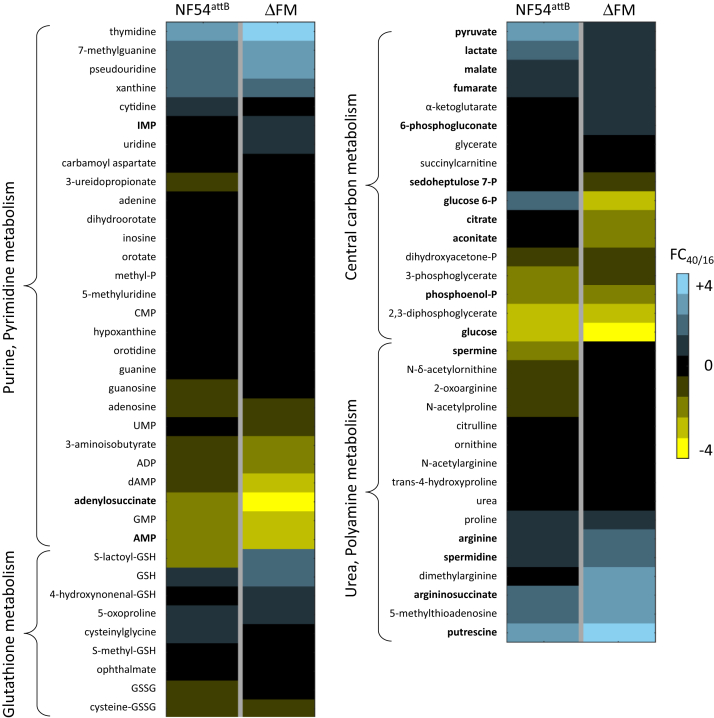
Since several metabolites present in iRBC were not detected in uRBC, we calculated the FC of iRBC metabolite abundances at 40 h relative to 16 h (FC40/16) and compared these values against the control data. Of the 413 common metabolites, 58 had substantially altered FC40/16 values (≥2-fold difference) in ΔFM iRBC, implying that the associated pathways progressed differently in ΔFM parasites during the IDC (File S4). We also computed Z-scores to identify significantly different FC40/16 values between the Nf54attB and ΔFM datasets (File S4). We observed a decrease in the FC40/16 ratios for citrate and aconitate in ΔFM parasites, which are metabolites generated by TCA cycle enzymes immediately downstream of FH and MQO (Fig. 7 and File S4). Similar to our observations from iRBC/uRBC comparisons, we did not observe a significant difference in the levels of fumarate and malate in ΔFM samples. In the absence of FH, the parasites should have experienced an accumulation of fumarate generated from purine salvage (21). This pathway involves the conversion of inosine monophosphate from salvaged hypoxanthine to succinyl-AMP by the enzyme adenylosuccinate synthase (ADSS). Succinyl-AMP in turn is cleaved into AMP and fumarate by adenylosuccinate lyase. Interestingly, late-stage ΔFM parasites displayed a twofold increase in inosine monophosphate and a fourfold decrease in succinyl-AMP ratios, indicative of a decrease in ADSS activity (Fig. 7 and File S4). This suggests that the amount of fumarate generated through the purine salvage pathway is lower in the ΔFM mutant. Both FCIDC and FC40/16 analyses revealed increases in pyrimidine metabolites in ΔFM samples, which could be in response to reduced purine salvage in these parasites.
Figure 7.
Heat map of select metabolite classes in Plasmodium falciparum–infected RBC cultures. Fold changes (FCs) of metabolite abundances at 40 h relative to 16 h (FC40/16) for parental NF54attB and ΔFM parasites are shown. Data are presented as the means of quadruplicate experiments. Metabolites in bold font are discussed in the text. See File S3 for the calculated FC40/16 values of all metabolites. ΔFM, double knockout mutant of FH and MQO; P, phosphate; RBC, red blood cell.
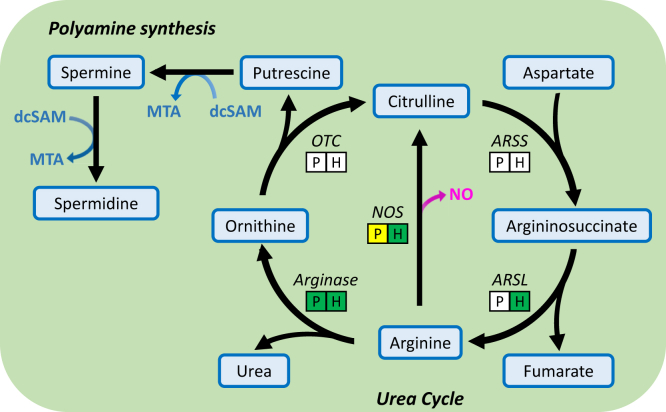
We also observed a higher FC40/16 ratio for argininosuccinate, a urea cycle metabolite, in ΔFM parasites (Fig. 7). The robust detection of argininosuccinate was surprising since neither P. falciparum nor human RBCs contain a complete urea cycle (39) (Fig. 8). However, moderate levels of the urea cycle enzyme argininosuccinate lyase (ARSL) are found in RBCs (40), which converts argininosuccinate to arginine and fumarate. In human cells with deleterious FH mutations, an intracellular buildup of fumarate leads to argininosuccinate production from the reversed activity of ARSL (41). The presence of argininosuccinate in iRBC and its observed increase in late-stage ΔFM iRBC could be indicative of a similar phenomenon whereby excess fumarate in the parasite is secreted and utilized as a substrate by the ARSL enzyme of RBCs. Fumarate generated from purine salvage has been detected at low quantities in culture media, indicating that it can be secreted from parasites into the RBC compartment (25). The level of fumarate secretion is presumably higher when there is no FH to incorporate fumarate into parasite metabolism. The FC40/16 ratio for arginine was observed to be higher in ΔFM iRBC, which could signify increased import of the amino acid from the culture medium to offset fumarate accumulation (39, 42). FH-deficient human cell lines become auxotrophic for arginine, implying that the reverse reaction catalyzed by ARSL plays an important role in fumarate detoxification and is a key compensatory feature in these cells (41). The higher abundance of arginine in ΔFM samples is reflected in the downstream polyamine synthesis pathway, which incorporates carbons from arginine into putrescine, spermine, and spermidine while generating 5′-methylthioadenosine (43) (Fig. 8 and File S4). Together, these results suggest that fumarate-associated pathways can modulate the levels of intracellular fumarate in ΔFM-infected RBCs.
Figure 8.
Schematic of the urea cycle and polyamine synthesis. Arginine acquired from the culture medium or from protein degradation can be converted to ornithine by parasite or human arginase. Ornithine feeds the polyamine synthesis pathway. Relative abundances of putrescine, spermine, spermidine, and MTA are higher in ΔFM parasites. ARSL catalyzes the conversion of argininosuccinate to arginine, generating fumarate as a byproduct. In human cells, ARSL operates in the reverse direction in the presence of excess fumarate. Rectangles under the urea cycle enzymes indicate if a homolog is present (green), predicted (yellow), or absent (white) in Plasmodium falciparum (P) and in the human RBC (H). ARSL, argininosuccinate lyase; ARSS, argininosuccinate synthetase; dcSAM, decarboxylated S-adenosylmethionine; ΔFM, double knockout mutant of FH and MQO; MTA, 5′-methylthioadenosine; NO, nitric oxide; NOS, nitric oxide synthase; OTC, ornithine transcarbamylase; RBC, red blood cell.
ΔFM parasites are sensitive to excess fumarate
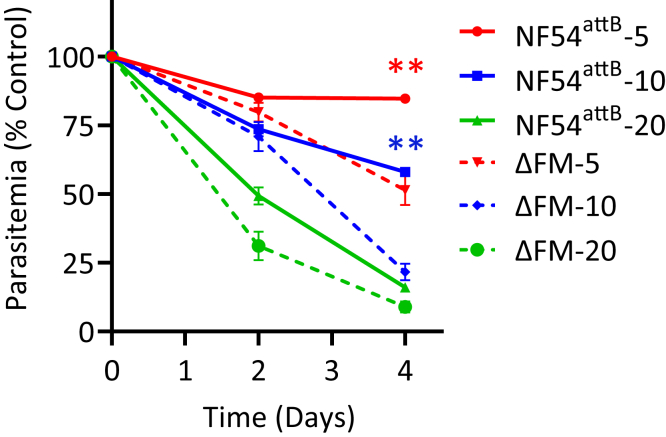
The observed metabolic adjustments to control fumarate levels in ΔFM parasites led us to examine if high concentrations of fumarate were toxic to P. falciparum in the absence of FH enzyme. Parental NF54attB and ΔFM parasites were exposed to various concentrations of fumarate, and their growth was monitored over a 4-day period. Since fumarate has low membrane permeability (44), the metabolite was added to cultures at millimolar concentrations in these experiments. We found that the presence of 5 mM or 10 mM fumarate repressed the growth of ΔFM parasites to a greater degree than that of the parental population (Fig. 9). The highest tested concentration of 20 mM fumarate inhibited both parasite lines to a similar extent. These results indicate that the loss of FH reduces the tolerance of ΔFM parasites to added fumarate.
Figure 9.
Response of ΔFM parasites to fumarate. The growth of synchronized ΔFM and NF54attB parental parasites in the presence of 5, 10, or 20 mM fumarate was monitored by flow cytometry over a 4-day period. Two biological experiments were conducted in quadruplicate (two-way ANOVA computed using GraphPad Prism 9 [GraphPad Software, Inc], followed by Bonferroni’s correction; ∗∗p < 0.01). Error bars represent the SEM. ΔFM, double knockout mutant of FH and MQO.
Discussion
It is well established that a complete turning of the TCA cycle is not required for the survival of blood-stage P. falciparum parasites (16). A previous report found that six of eight TCA cycle enzymes are dispensable with little effect on parasite replication in erythrocytes (16). Metabolomic studies using stable-isotope labeled glucose and glutamine showed that these genetic disruptions resulted in largely predictable losses of carbon flux through the TCA cycle, arguing against the existence of unidentified proteins with redundant functions. It was therefore surprising that two participating enzymes, FH and MQO, were refractory to deletion. There is some evidence that FH and MQO play roles in other aspects of cellular metabolism (21, 25). Our study, however, shows that their functions are nonessential for blood-stage survival. Using recently developed CRISPR-based methods (45), we were able to disrupt the coding regions for FH and MQO with limited fitness costs to the parasite.
Although FH was ultimately found to be dispensable, deletion of the enzyme was not straightforward. After multiple failed attempts to delete FH in the NF54attB (parent) strain, we tried to knock out the gene encoding FH in a parasite line that expressed a second recodonized copy of FH fused to SFG. FH was readily deleted in the presence of the second copy, but surprisingly, these parasites continued to survive even after the diCre-mediated excision of FH∗-SFG. It is unclear why this circuitous approach was required to obtain an FH-null line. It is possible that addition of the C-terminal SFG tag and/or expression from a non-native promoter resulted in suboptimal activity of the second FH enzyme. Upon deletion of the native FH, the parasites may have undergone metabolic rewiring to adjust to the underperforming FH∗-SFG enzyme. These metabolically altered parasites could then have been better positioned to survive the subsequent loss of FH∗-SFG. We eventually obtained ΔFH parasites from one of many transfections in the parent line without a second copy, which suggested that the starting population had cell-to-cell metabolic variability with a very small subset capable of tolerating FH deletion. Studies of the FH homolog in the rodent malarial parasite Plasmodium berghei have yielded similarly intriguing results in which the enzyme was shown to be dispensable for blood-stage infection in C57BL/6 mice but not in the BALB/c strain (17, 46). P. berghei preferentially grows within reticulocytes, which have a more complex repertoire of metabolites than mature RBCs (29). It is likely that host strain differences in metabolism permitted the loss of FH in one genetic background but not the other.
Given the difficulty in disrupting FH without a second copy, we were surprised to witness no measurable impact on parasite growth when FH expression was knocked down by TetR-DOZI. Immunoblotting demonstrated successful knockdown of FH when parasites were depleted of the aTC ligand; however, some protein below the detection limit could be keeping these parasites alive. Alternatively, the FHApt line may have undergone compensatory changes to account for the unnatural C terminus and 3′-UTR of the FH locus. Modifying the 3′-UTR of certain genes in Plasmodium has been shown to adversely affect expression (47, 48). The C-terminal hemagglutinin tag may have also interfered with FH enzymatic activity. Either scenario could have resulted in an FHApt line that had to adapt to insufficient levels of protein or an inefficient enzyme. If such compensation did occur, it could explain why we do not see a difference in parasite growth when aTC is withdrawn.
On the other hand, ΔMQO mutants of P. falciparum were easily obtained, which suggests that the usage of traditional gene manipulation methods was responsible for past failures to delete MQO. Knockout mutants of MQO have also been established in P. berghei (46). Our finding validates the inferences that were drawn from prior experiments with atovaquone. This drug targets complex III of the mETC and blocks the regeneration of the electron carrier ubiquinone, effectively inactivating five mitochondrial enzymes that depend on ubiquinone (8). These include MQO and another TCA cycle enzyme succinate dehydrogenase, DHODH, type II NADH dehydrogenase, and glycerol 3-phosphate dehydrogenase. The expression of a yeast DHODH, which uses fumarate as an electron acceptor instead of ubiquinone, can bypass atovaquone inhibition of P. falciparum in RBCs, implying that DHODH-mediated pyrimidine synthesis is the only vital process that requires ubiquinone (15). This was supported by gene deletion studies that showed that succinate dehydrogenase and type II NADH dehydrogenase are dispensable in asexual intraerythrocytic parasites (16, 49). To reconcile the atovaquone data with the previously predicted essentiality of MQO, the protein was postulated to have an important structural role instead (16). This idea was reinforced by the observation that MQO homologs have been retained in the related apicomplexans Cryptosporidium parvum and Cryptosporidium hominis, which lack all other TCA cycle enzymes (50, 51). Based on these assumptions, MQO was considered a promising drug target. Several drug screens against recombinant MQO were conducted, with some hit compounds displaying antimalarial activity at low to submicromolar concentrations (18, 19, 20). In light of the results from this study, we suspect that the compounds had off-target effects.
Our analyses of ΔFM (double knockout) parasites revealed metabolic changes around enzymatic steps that involve fumarate, specifically those catalyzed by ADSS and ARSL, which participate in purine salvage and the urea cycle, respectively. Both enzymes are members of a superfamily that generate fumarate as a byproduct during transelimination reactions (52). Our data suggest that ΔFM iRBC has adopted different ways to reduce fumarate levels through these two reactions: (1) decreased activity of ADSS and (2) reversed activity of ARSL, which leads to fumarate consumption as opposed to generation. The kinetic parameters and catalytic mechanism of the P. falciparum ADSS enzyme have been well characterized (53, 54, 55, 56). However, the parasite genome does not appear to encode an ARSL (57). The substrate for its forward reaction, argininosuccinate, could not be robustly detected in uRBC in our current or previous studies, but it has been consistently found in iRBC (37, 58, 59, 60, 61). It is possible that an unidentified parasite enzyme catalyzes this step of the urea cycle. Alternatively, excess fumarate secreted from the parasite may be utilized by the ARSL enzyme of RBCs (40, 41). Fumarate excretion itself could be the primary strategy for parasites to get rid of excess metabolite, with argininosuccinate merely being an indicator of the levels of excretion. The combined metabolic adjustments likely allow P. falciparum to overcome FH deficiency, which can be lethal or mutagenic in some organisms because of fumarate accumulation and protein/metabolite succination (22, 23, 24).
In the absence of FH and MQO, the mitochondrion cannot generate oxaloacetate. We see some evidence of compensation for this loss in our metabolomic data. ΔFM parasites exhibited an increase in glycolysis but a substantial decrease in the pathway product pyruvate. Since levels of PEP (the precursor to pyruvate) were essentially unchanged in the mutant parasites, we speculate that PEP was routed into oxaloacetate production via PEPC. We obtained more support for this hypothesis by computationally removing FH and MQO enzymatic activity in a whole-genome metabolic network model that we have employed in previous studies (37, 59). We found that it was not possible to knock out both genes unless a cytosolic fumarate sink was introduced. We therefore modeled reverse flux through host ARSL to use fumarate generated from purine salvage (Fig. 10). The computational model arrived at the same conclusion, that is, ΔFM parasites increase metabolic flux through PEPC to produce oxaloacetate. Combined with the earlier finding that the ΔPEPC mutant grows better in the presence of malate or fumarate, our data strongly suggest that cytosolic and mitochondrial oxaloacetate production are linked by a malate shuttle (25). Oxaloacetate is subsequently converted in the cytosol to aspartate by AAT, and this step has been shown to be important for parasite survival (29, 62, 63).
Figure 10.
Model of metabolic changes in Plasmodium falciparum–infected RBCs to accommodate the loss of FH and MQO. The disruption of FH and MQO abolishes oxaloacetate production in the mitochondrion and downstream steps in the TCA cycle. Since parasite-generated fumarate can no longer be consumed in the mitochondrion, ΔFM-infected RBCs reduce cellular fumarate levels by lowering flux through the purine salvage pathway and by reverse flux through hARSL or a functionally similar parasite enzyme. Gray dotted lines indicate the predicted transport of metabolites; wide green arrows and narrow orange arrows represent an increase and decrease in pathway flux, respectively. AAT, aspartate aminotransferase; ADSL, adenylosuccinate lyase; ADSS, adenylosuccinate synthetase; DTC, di/tricarboxylic acid transporter; FAD+/FADH, flavin adenine dinucleotide/reduced; FH, fumarate hydratase; ΔFM, double knockout mutant of FH and MQO; hARSL, human argininosuccinate lyase; KDH, α-ketoglutarate dehydrogenase; MDH, malate dehydrogenase; MQO, malate–quinone oxidoreductase; PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase; RBC, red blood cell; SCS, succinate synthase; SDH, succinate dehydrogenase; TCA, tricarboxylic acid.
In summary, this study shows that FH and MQO, like other enzymes in the TCA cycle, are dispensable in blood-stage P. falciparum parasites. Loss of FH and MQO elicited compensatory changes in fumarate-associated pathways and central carbon metabolism. The latter alterations imply the existence of a functional malate shuttle in P. falciparum that contributes to the cellular oxaloacetate pool. While inhibitors targeting FH and MQO are unlikely to be effective against asexual blood stages of the parasites, it will be of interest to examine the impact of further disruption to oxaloacetate production.
Experimental procedures
Parasite culture
P. falciparum NF54attB parasites were cultured in RBCs at 2% hematocrit in CMA (Complete Medium with Albumax) (CMA) containing RPMI1640 medium with l-glutamine (USBiological Life Sciences), supplemented with 20 mM Hepes, 0.2% sodium bicarbonate, 12.5 μg/ml hypoxanthine, 5 g/l Albumax II (Life Technologies), and 25 μg/ml gentamicin. Cultures were maintained at 37 °C in a gas mixture containing 94% N2, 3% O2, and 3% CO2.
Construction of gene knockout and knockdown plasmids
All molecular cloning steps were performed using ligation-independent cloning methods with InFusion (Clontech), unless specified as ligation reactions.
Gene deletion constructs for FH and MQO were generated using the pRSng (60) and pRSng-Neo plasmids, respectively. To generate pRSng-Neo, the hDHFR cassette in pRSng was removed by digestion with BamHI and HindIII. The neomycin resistance cassette was amplified from pINT (35) using the primers pRSng-NeoF and pRSng-NeoR (Table S1) and cloned into the digested pRSng vector. Homology arms 1 (HA1) and 2 (HA2) for FH and MQO were amplified using primers listed in Table S1. HA1 was inserted into the NotI site and HA2 into the NgoMIV site of pRSng or pRSng-Neo.
To create the FH knockdown construct, the pKD plasmid (31) was digested with AscI and AatII. HA1 and HA2 amplicons were amplified using the primer pairs FH.kd.HA1-F + FH.kd.HA1-R and FH.kd.HA2-F + FH.kd.HA2-R, respectively (Table S1). The HAs were fused in a final PCR using the primer pair FH.kd.HA2-F + FH.kd.HA1-R to generate a HA2–HA1 fragment separated by two EcoRV sites. This fragment was inserted into the digested pKD plasmid to generate pKDFH. Prior to transfection, the pKDFH plasmid was linearized with EcoRV.
To generate plasmids with gRNA sequences that target FH and MQO for knockout and/or knockdown, pCasG-LacZ (31) was digested with BsaI. Gene-specific gRNAs were synthesized as oligos (Table S1), annealed, and inserted into the digested plasmid.
Construction of pCre-FH∗-SFG plasmid
Plasmid pCre-FH∗-SFG was created to express a second copy of FH that could be controlled by TetR-DOZI knockdown (30) or by a diCre excision method (32, 33). Plasmid p15-EcDPCK-mCh (64) was modified for this purpose. This plasmid contains an attP site for integration into the genome of any parasite line that contains an attB site. A floxed sequence containing recodonized FH fused to SFG and a 10× aptamer array for TetR-DOZI knockdown was inserted on one side of the plasmid’s bidirectional Cam/HOP promoter. First, the 5′ LoxP site was inserted with Quick Ligase (NEB) into the AvrII/BspEI sites of p15-EcDPCK-mCh using dsDNA made from annealing phosphorylated primers LoxP.Avr.Bsp-F and LoxP.Avr.Bsp-R. The SFG gene was then amplified from a synthetic sequence described previously (65) with primers SFG.Sal-F and SFG.Psp-R and ligated into the SalI/PspOMI sites. The 875 bp aptamer array found in p15-EcDPCK-mCh was replaced with the 631 bp array designed to reduce recombination between array elements (31) using the PspOMI/XmaI sites. A 3′ LoxP site was ligated into the XmaI/AflII sites using dsDNA made from annealing phosphorylated primers LoxP.Xma.Afl-F and LoxP.Xma.Afl-R. Finally, the AvrII and SalI sites were used to insert FH with a recodonized gRNA sequence (FH∗). To recode the sequence, the entire FH coding region was amplified using primers FH.pRSng-F and FH.pRSng-R and inserted into the NotI site of pRSng. The plasmid was digested with SphI and PvuII, which targeted sites proximal to the gRNA sequence. The excised region was replaced with an amplicon generated using primers FH.SphI-F and FH.PvuII-R, with the reverse primer changing the original sequence 5′-CCA ATA GGA ATC GGA GTA AGT-3′ to the recoded sequence 5′-CCT ATC GGT ATA GGT GTT TCA-3′. The recodonized FH gene was amplified using primers FH.pCre-F and FH.pCre-R and inserted into the digested pCre plasmid.
On the other side of the bidirectional Cam/HOP promoter, the two components of the Cre recombinase required for the diCre conditional excision method (Cre59 and Cre60) were added. We used the NgoMIV and HindIII sites flanking the sequence encoding BSD to replace this sequence with a 979 bp synthetic construct made by LifeSct. The sequence encodes BSD followed by a 2A skip peptide, the nuclear localization sequence from SV40, FKBP, a 12 amino acid linker, and the amino-terminal Cre59 region (residues 19–59) of the Cre recombinase. The sequence from the 2A skip peptide to Cre59 was identical to that found in Addgene plasmid #85797 pSkipFlox (66) with the exception of two nucleotide changes that were introduced to remove unwanted BamHI and NsiI sites. We next amplified the Cre60 (residues 60–343 of the Cre recombinase) sequence from pSkipFlox using primers Cre60.AatII-F and Cre60.BspHI-R for ligation into the AatII/BspHI sites upstream of the sequence encoding TetR-DOZI. Finally, the sequence encoding FRB was amplified from pSkipFlox using primers FRB.Bam-F and FRB.Aat-R for insertion into the BamHI/AatII sites upstream of Cre60. This region of plasmid pCre-FH∗-SFG now encodes four proteins separated by three 2A skip peptides: FRB-Cre60, TetR-DOZI, BSD, and SV40-FKBP-Cre59. All primer sequences are listed in Table S1. The sequence of the entire plasmid is available in File S1.
Parasite transfections
For the generation of knockout and knockdown P. falciparum parasites, 350 μl volumes of RBCs were electroporated with 65 μg each of the appropriate pCasG-gRNA plasmid and the repair plasmid. The electroporated RBCs were mixed with 1.5 ml of parasite culture at 3 to 5% parasitemia and maintained in 10 ml culture volumes. Drugs and ligands were acquired from the following vendors: DSM1 (Calbiochem), WR99210 (Jacobus Pharmaceutical Company, Inc), Blasticidin (Corning), G418 (Corning), and aTC (Cayman Chemical).
Cultures transfected with knockout constructs were maintained in CMA for 2 days, after which they were cultured in selective media containing 1.5 μM DSM1 and either 2.5 nM WR99210 (for FH knockout) or 0.5 mg/ml G418 (for MQO knockout) for 7 days. Cultures were then switched to CMA until parasites reappeared, after which they were transferred to CMA containing WR99210 (ΔFH) or G418 (ΔMQO).
Cultures transfected with FH knockdown plasmids were maintained in CMA with 0.5 μM aTC for 2 days, after which they were cultured in media containing 2.5 μg/ml Blasticidin and 1.5 μM DSM1 along with aTC for 7 days. Cultures were then switched to CMA with aTC until the reemergence of parasites, upon which they were transferred to CMA containing Blasticidin and aTC.
To create the FH∗-SFG parasite line, RBCs were electroporated with 65 μg each of the pINT plasmid (encoding the mycobacteriophage Bxb1 integrase) (35) and the pCre-FH∗-SFG construct. The electroporated RBCs were mixed with 1.5 ml of a mixed-stage NF54attB culture at 3% parasitemia. Transfected cultures were maintained in CMA for 2 days and then transferred to selective media containing 2.5 nM WR99210 for 7 days. Following this, cultures were maintained in CMA until parasite reappearance, at which time WR99210 was added back to the cultures.
Genotyping PCR
A 3 μl volume of parasite culture was used in 25 μl PCRs with Phusion High-Fidelity DNA polymerase (NEB). Primer combinations and expected sizes of PCR products are indicated in Figs. S1, S2, 4 and S4. Primer sequences are included in Table S1.
Growth assays
Late-stage parasites were magnetically sorted from mixed-stage cultures using MACS LS columns (Miltenyi Biotech). They were used to seed 250 μl culture volumes/well in a 96-well plate (Corning) at a 0.25% starting parasitemia (unless otherwise indicated) and 2% hematocrit. The plates were incubated in chambers gassed with 94% N2, 3% O2, and 3% CO2 at 37 °C. A 10 μl volume of each parasite sample was collected every other day, diluted 1:10 in PBS, and stored in a 96-well plate at 4 °C. The culture medium was exchanged every other day.
On day 4 and/or day 8, samples were prepared for flow cytometry as described previously (64). The samples were analyzed with an Attune Nxt Flow Cytometer (Thermo Fisher Scientific) at a running speed of 25 μl/min to acquire 10,000 events.
Western blotting
Parasite samples were centrifuged at 500g for 5 min at room temperature (RT). The pellets were resuspended in 0.15% saponin for 5 min at RT to isolate parasites from the RBCs. Following three washes with 1× PBS, parasite pellets were resuspended in NuPAGE LDS sample buffer (Thermo Fisher Scientific) containing 2% β-mercaptoethanol and boiled for 5 min. Proteins were resolved by SDS-PAGE on 4 to 12% gradient gels and transferred to nitrocellulose membranes. Membranes were blocked in 5% milk and probed overnight at 4 °C with 1:2500 rat antihemagglutinin monoclonal antibody 3F10 (Roche) or 1:2500 rabbit anti-GFP pAb (65). They were then incubated for an hour at RT with 1:5000 antirat or anti-rabbit horseradish peroxidase–conjugated antibodies, respectively (GE Healthcare). Proteins were detected on X-ray film using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific), according to the manufacturer’s protocol. For loading controls, membranes were stripped of antibody with 200 mM glycine (pH 2.0) for 5 min and reprobed overnight at 4 °C with 1:25,000 mouse anti–aldolase monoclonal antibody 2E11. After incubation with 1:10,000 antimouse horseradish peroxidase–conjugated antibody (GE Healthcare), proteins were detected as described previously.
Fluorescence microscopy
Samples were prepared for live fluorescence microscopy by incubating 100 μl of parasite culture with 1 μg/ml 4′,6-diamidino-2-phenylindole for 30 min at 37 °C. Following three washes with CMA, each sample was resuspended in 10 μl CMA and transferred to a glass slide. A cover slip was affixed to the slide and sealed with wax. A series of images spanning 5 μm was acquired with 0.2 μm spacing using a Zeiss AxioImager M2 microscope. Following deconvolution with VOLOCITY software (PerkinElmer), a single image in the z-plane was reported.
DiCre activation
To excise FH∗-SFG from the ΔFHFH∗-SFG line, diCre was activated by the addition of A/C heterodimerizer (Clontech) to culture medium at a concentration of 250 nM. After 2 days of treatment, cultures were washed and resuspended in fresh CMA. Excision of the gene encoding FH∗-SFG was confirmed by PCR using primer combinations illustrated in Figure 4A. Primer sequences are listed in Table S1.
Sample collection for metabolomic analysis
ΔFM cultures were passed through an XS magnetic column (Miltenyi Biotech), and late-stage parasites were eluted upon column removal from the magnet. The process was repeated once every cycle three more times to obtain a highly synchronized culture. After the final elution of schizonts from the column, four flasks with 25 ml culture volumes were seeded at 0.8% parasitemia and 2% hematocrit (iRBC). Four flasks with 25 ml volumes of uRBCs at 2% hematocrit were set up in parallel. The blood for these cultures was treated with a NEO High Efficiency Leukocyte Reduction Filter (Haemonetics) to remove contaminating leukocytes. At 0 to 2 h postmerozoite invasion of RBCs, the culture media were replaced with fresh media. At 16 and 40 h, 10 ml of each iRBC and uRBC culture was collected and spun down. The pellets were transferred to fresh tubes, flash-frozen in an ethanol/dry-ice bath, and stored at −80 °C. Samples were sent to Metabolon for metabolite analysis.
Analysis of metabolomic data
To compute FCIDC values of metabolites at 16 and 40 h, we used the bootstrap procedure described by Tewari et al. (37). We used the built-in MATLAB function anova1 to identify statistically significant FCIDC values of metabolites detected in the NF54attB and ΔFM iRBC. We tested the hypothesis that the abundances of each metabolite at 16 and 40 h are drawn from a population with the same mean. File S3 lists the p values associated with the FCIDC values of metabolites at the 16 and 40 h time points.
The FC40/16 values of metabolites having raw counts >1000 in quadruplicate samples of at least one time point were determined. To compute the FC40/16 values, the raw count of each metabolite detected in each sample was first normalized using its Bradford protein concentration (File S2), and any missing values were then imputed using the built-in MATLAB function knnimpute. Next, we used the built-in MATLAB function bootstrp to generate 1000 bootstrap samples from the abundances detected in the quadruplicate samples at each time point for each metabolite and computed an FC in average abundance of each metabolite for all the bootstrap samples, that is, . Here, N denotes the total number of bootstrap samples; and , respectively, represent average abundance of a metabolite in the ith bootstrap sample at the 16 and 40 h time point of the experiment. Finally, FC40/16 of a metabolite was computed by:
| (1) |
To identify statistically different FC40/16 values between the NF54attB and ΔFM samples, we used a two-sample Z-test that compared means and standard deviations of the estimated FC40/16 values for each robustly detected metabolite between the two cultures. Mathematically, we used the following equation to compute the Z-score:
| (2) |
Here, and denote the estimated value of a metabolite x in the NF54attB and ΔFM cultures, and represents the standard error of the estimated means and is equal to:
| (3) |
where and are the estimated standard deviation of the FC40/16 value of the metabolite x in the NF54attB and ΔFM cultures, and denotes the number of bootstrap samples, defined in Equation 1. We identified the critical Z-score such that the probability of is less than 0.05.
Data availability
All data generated or analyzed during this study are included in this article and its supporting information files.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank David Fidock (Columbia University) for providing the NF54attB strain of P. falciparum parasites and David Sullivan (Johns Hopkins Bloomberg School of Public Health) for mouse anti–aldolase monoclonal antibody 2E11. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the US Army, the US Department of Defense, or The Henry M. Jackson Foundation for Advancement of Military Medicine, Inc.
Author contributions
K. R. and S. T. P. conceptualization; K. R. methodology; K. R. validation; S. G. T. formal analysis; K. R. investigation; S. G. T. data curation; K. R. and S. G. T. writing–original draft; A. W. and S. T. P. writing–review & editing; K. R. and S. G. T. visualization; A. W. and S. T. P. supervision; A. W. and S. T. P. project administration; S. T. P. funding acquisition.
Funding and additional information
This work was supported by the National Institutes of Health grant R01 AI125534 (to S. T. P.), the Johns Hopkins Malaria Research Institute, the Bloomberg Philanthropies, and the Network Science Initiative of the US Army Medical Research and Development Command, Ft. Detrick, Maryland (Award no.: W81XWH-15-C-0061; to S. T. P.). K. R. was supported by the National Institutes of Health training grant T32AI007417. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This article has been approved for public release with unlimited distribution. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Edited by Enrique De La Cruz
Supporting information
References
- 1.Fry M., Pudney M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4'-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80) Biochem. Pharmacol. 1992;43:1545–1553. doi: 10.1016/0006-2952(92)90213-3. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava I.K., Rottenberg H., Vaidya A.B. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J. Biol. Chem. 1997;272:3961–3966. doi: 10.1074/jbc.272.7.3961. [DOI] [PubMed] [Google Scholar]
- 3.Nixon G.L., Pidathala C., Shone A.E., Antoine T., Fisher N., O'Neill P.M., Ward S.A., Biagini G.A. Targeting the mitochondrial electron transport chain of Plasmodium falciparum: New strategies towards the development of improved antimalarials for the elimination era. Future Med. Chem. 2013;5:1573–1591. doi: 10.4155/fmc.13.121. [DOI] [PubMed] [Google Scholar]
- 4.Nilsen A., Miley G.P., Forquer I.P., Mather M.W., Katneni K., Li Y., Pou S., Pershing A.M., Stickles A.M., Ryan E., Kelly J.X., Doggett J.S., White K.L., Hinrichs D.J., Winter R.W., et al. Discovery, synthesis, and optimization of antimalarial 4(1H)-quinolone-3-diarylethers. J. Med. Chem. 2014;57:3818–3834. doi: 10.1021/jm500147k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stocks P.A., Barton V., Antoine T., Biagini G.A., Ward S.A., O'Neill P.M. Novel inhibitors of the Plasmodium falciparum electron transport chain. Parasitology. 2014;141:50–65. doi: 10.1017/S0031182013001571. [DOI] [PubMed] [Google Scholar]
- 6.Stickles A.M., Smilkstein M.J., Morrisey J.M., Li Y., Forquer I.P., Kelly J.X., Pou S., Winter R.W., Nilsen A., Vaidya A.B., Riscoe M.K. Atovaquone and ELQ-300 combination therapy as a novel dual-site cytochrome bc1 inhibition strategy for malaria. Antimicrob. Agents Chemother. 2016;60:4853–4859. doi: 10.1128/AAC.00791-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dooren G.G., Stimmler L.M., McFadden G.I. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 2006;30:596–630. doi: 10.1111/j.1574-6976.2006.00027.x. [DOI] [PubMed] [Google Scholar]
- 8.Painter H.J., Morrisey J.M., Mather M.W., Vaidya A.B. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 9.Jensen M.D., Conley M., Helstowski L.D. Culture of Plasmodium falciparum: The role of pH, glucose, and lactate. J. Parasitol. 1983;69:1060–1067. [PubMed] [Google Scholar]
- 10.Sherman I.W. Malaria: Parasite Biology, Pathogenesis and Protection. ASM Press; WA DC: 1998. Carbohydrate metabolism of asexual stages; pp. 135–144. [Google Scholar]
- 11.Rudzinska M.A. The fine structure of malaria parasites. Int. Rev. Cytol. 1969;25:161–199. doi: 10.1016/s0074-7696(08)60203-x. [DOI] [PubMed] [Google Scholar]
- 12.Evers F., Cabrera-Orefice A., Elurbe D.M., Kea-Te Lindert M., Boltryk S.D., Voss T.S., Huynen M.A., Brandt U., Kooij T.W.A. Composition and stage dynamics of mitochondrial complexes in Plasmodium falciparum. Nat. Commun. 2021;12:3820. doi: 10.1038/s41467-021-23919-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krungkrai J., Burat D., Kudan S., Krungkrai S., Prapunwattana P. Mitochondrial oxygen consumption in asexual and sexual blood stages of the human malarial parasite, Plasmodium falciparum. Southeast. Asian J. Trop. Med. Public Health. 1999;30:636–642. [PubMed] [Google Scholar]
- 14.Scheibel L.W., Ashton S.H., Trager W. Plasmodium falciparum: Microaerophilic requirements in human red blood cells. Exp. Parasitol. 1979;47:410–418. doi: 10.1016/0014-4894(79)90094-8. [DOI] [PubMed] [Google Scholar]
- 15.Ganesan S.M., Morrisey J.M., Ke H., Painter H.J., Laroiya K., Phillips M.A., Rathod P.K., Mather M.W., Vaidya A.B. Yeast dihydroorotate dehydrogenase as a new selectable marker for Plasmodium falciparum transfection. Mol. Biochem. Parasitol. 2011;177:29–34. doi: 10.1016/j.molbiopara.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke H., Lewis I.A., Morrisey J.M., McLean K.J., Ganesan S.M., Painter H.J., Mather M.W., Jacobs-Lorena M., Llinas M., Vaidya A.B. Genetic investigation of tricarboxylic acid metabolism during the Plasmodium falciparum life cycle. Cell Rep. 2015;11:164–174. doi: 10.1016/j.celrep.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaraman V., Suryavanshi A., Kalale P., Kunala J., Balaram H. Biochemical characterization and essentiality of Plasmodium fumarate hydratase. J. Biol. Chem. 2018;293:5878–5894. doi: 10.1074/jbc.M117.816298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartuti E.D., Inaoka D.K., Komatsuya K., Miyazaki Y., Miller R.J., Xinying W., Sadikin M., Prabandari E.E., Waluyo D., Kuroda M., Amalia E., Matsuo Y., Nugroho N.B., Saimoto H., Pramisandi A., et al. Biochemical studies of membrane bound Plasmodium falciparum mitochondrial L-malate:quinone oxidoreductase, a potential drug target. Biochim. Biophys. Acta Bioenerg. 2018;1859:191–200. doi: 10.1016/j.bbabio.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Miyazaki Y., Inaoka D.K., Hartuti E.D., Watanabe Y.I., Shiba T., Harada S., Saimoto H., Burrows J.N., Benito F.J.G., Nozaki T., Kita K. Identification of Plasmodium falciparum mitochondrial malate: Quinone oxidoreductase inhibitors from the pathogen box. Genes (Basel) 2019;10:471. doi: 10.3390/genes10060471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endang Ariyani S., Alim I., Tjut Sugandawaty D., Raden Wisnu N. Antimalarial activity of microalgae extracts based on inhibition of PfMQO, a mitochondrial Plasmodium falciparum enzyme. Pharmacognosy J. 2019;11:1477–1482. [Google Scholar]
- 21.Bulusu V., Jayaraman V., Balaram H. Metabolic fate of fumarate, a side product of the purine salvage pathway in the intraerythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 2011;286:9236–9245. doi: 10.1074/jbc.M110.173328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruecker N., Jansen R., Trujillo C., Puckett S., Jayachandran P., Piroli G.G., Frizzell N., Molina H., Rhee K.Y., Ehrt S. Fumarase deficiency causes protein and metabolite succination and intoxicates Mycobacterium tuberculosis. Cell Chem. Biol. 2017;24:306–315. doi: 10.1016/j.chembiol.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlinson I.P., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., Roylance R.R., Olpin S., Bevan S., Barker K., Hearle N., et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 24.Bardella C., El-Bahrawy M., Frizzell N., Adam J., Ternette N., Hatipoglu E., Howarth K., O'Flaherty L., Roberts I., Turner G., Taylor J., Giaslakiotis K., Macaulay V.M., Harris A.L., Chandra A., et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 2011;225:4–11. doi: 10.1002/path.2932. [DOI] [PubMed] [Google Scholar]
- 25.Storm J., Sethia S., Blackburn G.J., Chokkathukalam A., Watson D.G., Breitling R., Coombs G.H., Müller S. Phosphoenolpyruvate carboxylase identified as a key enzyme in erythrocytic Plasmodium falciparum carbon metabolism. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nozawa A., Fujimoto R., Matsuoka H., Tsuboi T., Tozawa Y. Cell-free synthesis, reconstitution, and characterization of a mitochondrial dicarboxylate-tricarboxylate carrier of Plasmodium falciparum. Biochem. Biophys. Res. Commun. 2011;414:612–617. doi: 10.1016/j.bbrc.2011.09.130. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi A.K., Desai P.V., Pradhan A., Khan S.I., Avery M.A., Walker L.A., Tekwani B.L. An alpha-proteobacterial type malate dehydrogenase may complement LDH function in Plasmodium falciparum. Cloning and biochemical characterization of the enzyme. Eur. J. Biochem. 2004;271:3488–3502. doi: 10.1111/j.1432-1033.2004.04281.x. [DOI] [PubMed] [Google Scholar]
- 28.Borst P. The malate-aspartate shuttle (Borst cycle): How it started and developed into a major metabolic pathway. IUBMB Life. 2020;72:2241–2259. doi: 10.1002/iub.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava A., Creek D.J., Evans K.J., De Souza D., Schofield L., Müller S., Barrett M.P., McConville M.J., Waters A.P. Host reticulocytes provide metabolic reservoirs that can be exploited by malaria parasites. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesan S.M., Falla A., Goldfless S.J., Nasamu A.S., Niles J.C. Synthetic RNA-protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat. Commun. 2016;7:10727. doi: 10.1038/ncomms10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajaram K., Liu H.B., Prigge S.T. Redesigned TetR-aptamer system to control gene expression in Plasmodium falciparum. mSphere. 2020;5 doi: 10.1128/mSphere.00457-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knuepfer E., Napiorkowska M., van Ooij C., Holder A.A. Generating conditional gene knockouts in Plasmodium - a toolkit to produce stable DiCre recombinase-expressing parasite lines using CRISPR/Cas9. Sci. Rep. 2017;7:3881. doi: 10.1038/s41598-017-03984-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins C.R., Das S., Wong E.H., Andenmatten N., Stallmach R., Hackett F., Herman J.-P., Müller S., Meissner M., Blackman M.J. Robust inducible Cre recombinase activity in the human malaria parasite Plasmodium falciparum enables efficient gene deletion within a single asexual erythrocytic growth cycle. Mol. Microbiol. 2013;88:687–701. doi: 10.1111/mmi.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adjalley S.H., Johnston G.L., Li T., Eastman R.T., Ekland E.H., Eappen A.G., Richman A., Sim B.K., Lee M.C., Hoffman S.L., Fidock D.A. Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E1214–E1223. doi: 10.1073/pnas.1112037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nkrumah L.J., Muhle R.A., Moura P.A., Ghosh P., Hatfull G.F., Jacobs W.R., Jr., Fidock D.A. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat. Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spalding M.D., Allary M., Gallagher J.R., Prigge S.T. Validation of a modified method for Bxb1 mycobacteriophage integrase-mediated recombination in Plasmodium falciparum by localization of the H-protein of the glycine cleavage complex to the mitochondrion. Mol. Biochem. Parasitol. 2010;172:156–160. doi: 10.1016/j.molbiopara.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tewari S.G., Swift R.P., Reifman J., Prigge S.T., Wallqvist A. Metabolic alterations in the erythrocyte during blood-stage development of the malaria parasite. Malar. J. 2020;19:94. doi: 10.1186/s12936-020-03174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tewari S.G., Rajaram K., Swift R.P., Kwan B., Reifman J., Prigge S.T., Wallqvist A. Inter-study and time-dependent variability of metabolite abundance in cultured red blood cells. Malar. J. 2021;20:299. doi: 10.1186/s12936-021-03780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cobbold S.A., Llinas M., Kirk K. Sequestration and metabolism of host cell arginine by the intraerythrocytic malaria parasite Plasmodium falciparum. Cell Microbiol. 2016;18:820–830. doi: 10.1111/cmi.12552. [DOI] [PubMed] [Google Scholar]
- 40.Tomlinson S., Westall R.G. Argininosuccinic aciduria. Argininosuccinase and arginase in human blood cells. Clin. Sci. 1964;26:261–269. [PubMed] [Google Scholar]
- 41.Zheng L., MacKenzie E.D., Karim S.A., Hedley A., Blyth K., Kalna G., Watson D.G., Szlosarek P., Frezza C., Gottlieb E. Reversed argininosuccinate lyase activity in fumarate hydratase-deficient cancer cells. Cancer Metab. 2013;1:12. doi: 10.1186/2049-3002-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olszewski K.L., Morrisey J.M., Wilinski D., Burns J.M., Vaidya A.B., Rabinowitz J.D., Llinas M. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meireles P., Mendes A.M., Aroeira R.I., Mounce B.C., Vignuzzi M., Staines H.M., Prudencio M. Uptake and metabolism of arginine impact Plasmodium development in the liver. Sci. Rep. 2017;7:4072. doi: 10.1038/s41598-017-04424-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werdenberg D., Joshi R., Wolffram S., Merkle H.P., Langguth P. Presystemic metabolism and intestinal absorption of antipsoriatic fumaric acid esters. Biopharm. Drug Dispos. 2003;24:259–273. doi: 10.1002/bdd.364. [DOI] [PubMed] [Google Scholar]
- 45.Ghorbal M., Gorman M., Macpherson C.R., Martins R.M., Scherf A., Lopez-Rubio J.-J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 46.Niikura M., Komatsuya K., Inoue S.I., Matsuda R., Asahi H., Inaoka D.K., Kita K., Kobayashi F. Suppression of experimental cerebral malaria by disruption of malate:quinone oxidoreductase. Malar. J. 2017;16:247. doi: 10.1186/s12936-017-1898-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waller K.L., Muhle R.A., Ursos L.M., Horrocks P., Verdier-Pinard D., Sidhu A.B., Fujioka H., Roepe P.D., Fidock D.A. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J. Biol. Chem. 2003;278:33593–33601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- 48.Yeoh S., O'Donnell R.A., Koussis K., Dluzewski A.R., Ansell K.H., Osborne S.A., Hackett F., Withers-Martinez C., Mitchell G.H., Bannister L.H., Bryans J.S., Kettleborough C.A., Blackman M.J. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007;131:1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 49.Ke H., Ganesan S.M., Dass S., Morrisey J.M., Pou S., Nilsen A., Riscoe M.K., Mather M.W., Vaidya A.B. Mitochondrial type II NADH dehydrogenase of Plasmodium falciparum (PfNDH2) is dispensable in the asexual blood stages. PLoS One. 2019;14 doi: 10.1371/journal.pone.0214023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mogi T., Kita K. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol. Int. 2010;59:305–312. doi: 10.1016/j.parint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Abrahamsen M.S., Templeton T.J., Enomoto S., Abrahante J.E., Zhu G., Lancto C.A., Deng M., Liu C., Widmer G., Tzipori S., Buck G.A., Xu P., Bankier A.T., Dear P.H., Konfortov B.A., et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 52.Puthan Veetil V., Fibriansah G., Raj H., Thunnissen A.M., Poelarends G.J. Aspartase/fumarase superfamily: A common catalytic strategy involving general base-catalyzed formation of a highly stabilized aci-carboxylate intermediate. Biochemistry. 2012;51:4237–4243. doi: 10.1021/bi300430j. [DOI] [PubMed] [Google Scholar]
- 53.Raman J., Mehrotra S., Anand R.P., Balaram H. Unique kinetic mechanism of Plasmodium falciparum adenylosuccinate synthetase. Mol. Biochem. Parasitol. 2004;138:1–8. doi: 10.1016/j.molbiopara.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Mehrotra S., M B.N., Raman J., Anand R.P., Balaram H. Mutational analysis of cysteine 328 and cysteine 368 at the interface of Plasmodium falciparum adenylosuccinate synthetase. Biochim. Biophys. Acta. 2012;1824:589–597. doi: 10.1016/j.bbapap.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Jayalakshmi R., Sumathy K., Balaram H. Purification and characterization of recombinant Plasmodium falciparum adenylosuccinate synthetase expressed in Escherichia coli. Protein Expr. Purif. 2002;25:65–72. doi: 10.1006/prep.2001.1610. [DOI] [PubMed] [Google Scholar]
- 56.Eaazhisai K., Jayalakshmi R., Gayathri P., Anand R.P., Sumathy K., Balaram H., Murthy M.R. Crystal structure of fully ligated adenylosuccinate synthetase from Plasmodium falciparum. J. Mol. Biol. 2004;335:1251–1264. doi: 10.1016/j.jmb.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 57.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Paulsen I.T., James K., Eisen J.A., Rutherford K., Salzberg S.L., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tewari S.G., Rajaram K., Swift R.P., Reifman J., Prigge S.T., Wallqvist A. Metabolic survival adaptations of Plasmodium falciparum exposed to sub-lethal doses of fosmidomycin. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.02392-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tewari S.G., Rajaram K., Schyman P., Swift R., Reifman J., Prigge S.T., Wallqvist A. Short-term metabolic adjustments in Plasmodium falciparum counter hypoxanthine deprivation at the expense of long-term viability. Malar. J. 2019;18:86. doi: 10.1186/s12936-019-2720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swift R.P., Rajaram K., Liu H.B., Dziedzic A., Jedlicka A.E., Roberts A.D., Matthews K.A., Jhun H., Bumpus N.N., Tewari S.G., Wallqvist A., Prigge S.T. A mevalonate bypass system facilitates elucidation of plastid biology in malaria parasites. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Counihan N.A., Chisholm S.A., Bullen H.E., Srivastava A., Sanders P.R., Jonsdottir T.K., Weiss G.E., Ghosh S., Crabb B.S., Creek D.J., Gilson P.R., de Koning-Ward T.F. Plasmodium falciparum parasites deploy RhopH2 into the host erythrocyte to obtain nutrients, grow and replicate. Elife. 2017;6 doi: 10.7554/eLife.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wrenger C., Muller I.B., Schifferdecker A.J., Jain R., Jordanova R., Groves M.R. Specific inhibition of the aspartate aminotransferase of Plasmodium falciparum. J. Mol. Biol. 2011;405:956–971. doi: 10.1016/j.jmb.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 63.Lunev S., Butzloff S., Romero A.R., Linzke M., Batista F.A., Meissner K.A., Muller I.B., Adawy A., Wrenger C., Groves M.R. Oligomeric interfaces as a tool in drug discovery: Specific interference with activity of malate dehydrogenase of Plasmodium falciparum in vitro. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swift R.P., Rajaram K., Liu H.B., Prigge S.T. Dephospho-CoA kinase, a nuclear-encoded apicoplast protein, remains active and essential after Plasmodium falciparum apicoplast disruption. EMBO J. 2021;40 doi: 10.15252/embj.2020107247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts A.D., Nair S.C., Guerra A.J., Prigge S.T. Development of a conditional localization approach to control apicoplast protein trafficking in malaria parasites. Traffic. 2019;20:571–582. doi: 10.1111/tra.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Birnbaum J., Flemming S., Reichard N., Soares A.B., Mesén-Ramírez P., Jonscher E., Bergmann B., Spielmann T. A genetic system to study Plasmodium falciparum protein function. Nat. Methods. 2017;14:450–456. doi: 10.1038/nmeth.4223. [DOI] [PubMed] [Google Scholar]
- 67.Joët T., Eckstein-Ludwig U., Morin C., Krishna S. Validation of the hexose transporter of Plasmodium falciparum as a novel drug target. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7476. doi: 10.1073/pnas.1330865100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cobbold S.A., Vaughan A.M., Lewis I.A., Painter H.J., Camargo N., Perlman D.H., Fishbaugher M., Healer J., Cowman A.F., Kappe S.H., Llinas M. Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J. Biol. Chem. 2013;288:36338–36350. doi: 10.1074/jbc.M113.503557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.MacRae J.I., Dixon M.W., Dearnley M.K., Chua H.H., Chambers J.M., Kenny S., Bottova I., Tilley L., McConville M.J. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol. 2013;11:67. doi: 10.1186/1741-7007-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supporting information files.