Abstract
Background
Recombinant human growth hormone (rhGH) has shown a great growth-promoting potential in children with idiopathic short stature (ISS). However, the response to rhGH differs across individuals, largely due to genetic and epigenetic heterogeneity. Since epigenetic marks on the methylome can be dynamically influenced by GH, we performed a comprehensive pharmacoepigenomics analysis of DNA methylation changes associated with long-term rhGH administration in children with ISS.
Results
We measured DNA methylation profiles before and after GH treatment (with a duration of ~ 18 months in average) on 47 healthy children using customized methylC-seq capture sequencing. Their changes were compared and associated with changes in plasma IGF1 by adjusting sex, age, treatment duration and estimated blood proportions. We observed a considerable inter-individual heterogeneity of DNA methylation changes responding to GH treatment. We identified 267 response-associated differentially methylated cytosines (DMCs) that were enriched in promoter regions, CpG islands and blood cell-type-specific regulatory elements. Furthermore, the genes associated with these DMCs were enriched in the biology process of “cell development,” “neuron differentiation” and “developmental growth,” and in the TGF-beta signaling pathway, PPAR Alpha pathway, endoderm differentiation pathway, adipocytokine signaling pathway as well as PI3K-Akt signaling pathway, and cAMP signaling pathway.
Conclusion
Our study provides a first insight in DNA methylation changes associated with rhGH administration, which may help understand mechanisms of epigenetic regulation on GH-responsive genes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-022-01281-z.
Keywords: Pharmacoepigenomics, Growth hormone, DNA methylation, Intervention epigenetics, Idiopathic short stature
Introduction
Growth hormone (GH) acts directly on intracellular pathways downstream of the GH receptor or via the stimulation and action of IGF1 [1]. Physiological effects of GH are mainly on growth, body composition and metabolism. In the epiphyseal growth plate, GH effects are largely mediated by promoting IGF1 production to stimulate skeletal growth. In addition, GH is known to increase muscle mass. GH also contributes to the acute metabolic response to stressful situations, such as fasting [2, 3], exercise [4], injuries, critical illnesses [5] and infectious diseases [6] by acting on the liver, pancreatic beta cells, adipose tissue and muscle. Overall, most of these GH effects are mediated by activating the transcription of numerous genes.
Non-physiological exposure to GH occurs in the human clinic when children with short stature (SS) receive supra-physiological doses of recombinant human GH (rhGH) to stimulate their skeletal growth. These cases differ widely in the level and duration of exposure to rhGH which may leave lasting marks in different tissues that could conceivably be based on epigenetic mechanisms.
Changes in CpG methylation of the genome are one of the most studied epigenetic mechanisms. They can facilitate or reduce the transcription of certain genes when they occur in the regulatory zones of these genes. Once installed, some of them cannot be erased. However, they can show a certain plasticity and vary with age or environmental factors [7–11]. Most of them are variable depending on the tissue [12–14]. Modifications of CpG methylation have been associated with cancer, metabolic and cardiovascular diseases [15–17]. Following binding to its receptor, GH activates GHR-associated Src family kinases, acting via other intracellular pathways [18], such as ERK and Jun, which are known to affect CpG methylation [19].
It was therefore interesting to investigate whether GH was capable of producing its own epigenetic marks on the methylome. The only opportunity to detect them in the human clinic is to study the methylome before and after exposure to the hormone. This is made possible in one particular circumstance, the application of rhGH treatment to normal small children. Given the considerable individual variability of methylation marks, it would have been difficult to compare rhGH treated with untreated children. Therefore, we searched for acquired CpG modifications by studying the same children before and during rhGH exposure. As for most DNA methylation studies in humans, it was only possible to study blood cells in these children, with the expectation that changes in blood cells may reflect those occurring in the physiological target tissues of GH action. It is probably important to clarify that our study does not address at all the epigenetic variations that could contribute to the mechanisms of short stature, which have been the subject of a number of previous studies. Such methylation marks are not expected to vary upon GH administration and are therefore outside the scope of our approach devoted to the epigenetic consequences of exposure to GH action.
Methods
Cohort—sample collection
DNA samples were obtained from 47 children with non-pathological idiopathic short stature (ISS) recruited among 317 children who served as controls in a study of type 1 diabetes (T1D) epigenetics [20]. The 47 studied children with short stature all had ISS, as defined [21]. Briefly, this diagnosis requires that child’s height is < − 2SD (standard deviation) from the population average, with a birth length > − 2SD for gestational age (none of them had intrauterine growth retardation) and appropriate for parents’ height, then linear and normal growth rate, and no detectable etiology for the short stature, such as chromosomal, endocrine, or skeletal diseases. Classically, testing of such short children includes clinical examination, bone X-rays, IGF1measurement and/or GH stimulation tests (to exclude GH deficiency). The 47 studied children all went through these screening analyses, and none of them showed any abnormality in the cited parameters. This study was supported by the Programme Hospitalier de Recherche Clinique of the French Ministry of Health according to the French bioethics law with the objective of studying gene–environment factors in young patients with T1D and age-matched controls. Families were carefully informed about the investigational nature of the study and signed an informed consent agreed by CPP (number DC-2008-693; NI 2620, Comité de Protection des Personnes). The studied children had received rhGH treatment for a duration of 6–38.4 months.
None had deficiencies of GH or other hormones, chromosomal disorders or syndromes, skeletal dysplasia or metabolic disease. All children received rhGH as daily sc injections 6 days/wk, starting with a 40 µg/kg day rhGH dose and following a target-to-treat rhGH dosing protocol based on the growth response to treatment, so that individual average dose ranged 40–113 µg/kg day across the studied children. Children gave two blood samples, one before onset of rhGH treatment and the other after 6–38.4 months of rhGH treatment. All were seen as outpatients every 6 months for clinical examination and measurements of serum IGF-I. Children were healthy at time of study, with no sign of viral or other intercurrent infection. Blood samples were collected in the course of routine medical evaluation of patients. DNA methylation was measured in peripheral blood mononuclear cells (PBMC) samples from these patients before and during treatment.
Isolation of genomic DNA
Peripheral blood mononuclear cells (PBMC) were isolated from fresh blood using a density gradient. Five milliliters of fresh blood were mixed with 5 ml of NaCl 154 mM, and 5 ml of Lymphoprep solution (Eurobio, Paris, France) was added to the diluted blood and centrifuged for 20 min at room temperature at 800 g. After centrifugation, the interphase containing PBMC was carefully aspirated and the cells were mixed with NaCl. The cell suspension was centrifuged at 300 g and the cell pellet washed with PBS. PBMC were frozen at − 80 °C. Nucleic acids were extracted from PBMC using Gentra Puregene blood kit (Qiagen, Hilden, Germany).
Measurement of IGF1 concentrations
IGF1 concentrations were measured in serum samples around 07.00 am to 08.00 pm before breakfast using a chemiluminescent immunometric assay after pre-treatment with acid using Immulite®2000 (Siemens Healthcare Diagnostics Products Llanberis, UK). IGF1 values under treatment were averaged for analysis.
DNA methylation capture sequencing
Methylation capture sequencing (MCC-Seq) was performed as previously described [22–24]. In brief, the MCC-Seq protocol was carried out using the SeqCap Epi Enrichment System protocol (Roche NimbleGen) [22–24]. Specifically, a whole-genome sequencing library was prepared and bisulfite converted, amplified and then a capture enriching for targeted bisulfite-converted DNA fragments was carried out. Equal amounts of multiplexed libraries (12 samples per capture) were combined and were further amplified. Lastly, the MCC-Seq libraries were sequenced on the Illumina HiSeq4000 or NovaSeq 6000 system using 100 bp paired-end sequencing. More specifically, whole-genome sequencing libraries were generated from 700 to 1000 ng of genomic DNA spiked with 0.1% (w/w) unmethylated λ DNA (Promega) previously fragmented to 300–400 bp peak sizes using the Covaris focused-ultrasonicator E210. Fragment size was controlled on a Bioanalyzer DNA 1000 Chip (Agilent), and the KAPA High Throughput Library Preparation Kit (KAPA Biosystems) was applied. End repair of the generated dsDNA with 3′- or 5′-overhangs, adenylation of 3′-ends, adaptor ligation and clean-up steps were carried out as per KAPA Biosystems' recommendations. The cleaned-up ligation product was then analyzed on a Bioanalyzer High Sensitivity DNA Chip (Agilent) and quantified by PicoGreen (Life Technologies). Samples were then bisulfite converted using the Epitect Fast DNA Bisulfite Kit (Qiagen), according to the manufacturer's protocol. Bisulfite-converted DNA was quantified using OliGreen (Life Technologies) and, based on quantity, amplified by 9–12 cycles of PCR using the Kapa Hifi Uracil + DNA polymerase (KAPA Biosystems), according to the manufacturer's protocol. The amplified libraries were purified using Ampure Beads and validated on Bioanalyzer High Sensitivity DNA Chips, and quantified by PicoGreen.
The hybridization procedure of the amplified bisulfite-converted library was performed as described by the manufacturer, using 1 μg of total input of library, which was evenly divided by the libraries to be multiplexed, and incubated at 47 °C for 72 h. Washing and recovering of the captured library, as well as PCR amplification and final purification, were carried out as recommended by the manufacturer. Quality, concentration and size distribution of the captured library were determined by Bioanalyzer High Sensitivity DNA Chips.
This blood MCC-Seq panel covers (1) the majority of human gene promoters, blood-cell-lineage-specific enhancer regions and methylation footprint regions [25] observed in blood, (2) CpGs from Illumina Human Methylation 450 Bead Chips and (3) published autoimmune-related SNPs as well as SNPs in their LD regions with r2 > 0.8. Overall, it covers 4,861,805 CpGs which have been applied to multiple blood-based epigenome-wide association studies [24, 26].
MCC-Seq data process
Targeted MCC-Seq HiSeq and NovaSeq reads were aligned using the Epigenome Pipeline available from the DRAGEN Bio-IT platform (Edico Genomics/Illumina). Specifically, the MCC-Seq paired-end raw reads were first demultiplexed into FASTQ files using Illumina’s bcl2Fastq2-2.19.1 software. Reads were then trimmed for quality (phred33 ≥ 20) and Illumina adapters using trimgalore v.0.4.2 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), a wrapper tool around Cutadapt [27] and FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, the trimmed reads were aligned, per sequencing lane, to the bisulfite-converted GRCh37 reference genome using DRAGEN EP v2.6.3 or later in paired-end mode using the directional/Lister methylation protocol presets; alignments were calculated for both strands, and the unique alignment with highest quality was retained. Lane bam files were merged and then de-duplicated using Picard (version 2.9). A genome-wide cytosine methylation report was generated by DRAGEN to record counts of methylated and unmethylated cytosines at each cytosine position in the genome. Methylation counts are provided for the CpG, CHG and CHH cytosine contexts. DNA methylation level of each CpG was calculated by the number of methylated reads over the total number of sequenced reads. CpGs that were located within sex chromosomes, overlapping with SNPs (dbSNP 137), the DAC Blacklisted Regions or Duke Excluded Regions (generated by the ENCODE project) were removed. CpG sites with less than 20X read coverage were also discarded.
Variants/SNPs (including homozygous alternate and heterozygous genotypes) were inferred using Bis-SNP (version 0.82.2) [28] on the de-duplicated bam files. The homozygous reference genotypes of individuals on these SNPs were extracted from the aligned bam files by requiring > = 10X read coverage aligned to the reference allele. Hierarchical clustering was performed based on the genotype profiles of SNPs on chromosome 1 where genotypes were inferred from all samples.
Statistical analysis
A linear regression model (LM) was built to investigate the association between the changes of DNA methylation level before and after treatment (delta beta) and the changes of IGF1 level (delta IGF1) by correcting age onset, the treatment duration and the estimated blood cell proportions. We used the R function lm() to fit the model and calculated p values for variables of interest. Due to limited sample size, the nominal p value < 1e−4 was used as a threshold of statistical significance to determine differentially methylated CpGs (DMCs). To reduce the impact of zero-inflated methylation differences on inferring the regression, we limited our analysis on CpGs that have > = 30% of pairs with nonzero methylation differences. Blood deconvolution was done using constrained linear projection [29] via the projectMix function of the RefFreeEWAS package, using a custom panel of 30,455 cell-type-specific hypo-methylated and hyper-methylated CpGs. The blood reference epigenome profiles include neutrophil, monocyte, B cell and T cell.
Genome features and function enrichment analysis
We downloaded the genome feature annotation tables, including transcription start sites (TSSs), 3’UTRs, 5’UTRs, first exons, exons, introns and transcription end sites (TESs), from the UCSC genome browser with the hg19 build version (https://genome.ucsc.edu/). We considered both TSS200 (200 bp from TSSs) and TSS1500 (1500 bp from TSSs) for the promoter regions. We also downloaded the CpG islands (CGI) annotation table from the UCSC genome browser. Furthermore, CGI north and south shores were defined as the 2-kb flanking sequences on upstream and downstream of CGIs, respectively, and north and south shelves were defined as the 2-kb flanking sequences beyond the shores. Genome feature enrichment analyses of GH response DMCs were performed using Fisher’s exact test for significance where the background set was the all testable CpGs. The gene ontology enrichment analyses were performed using homer [30] (version 4.11) with gene sets detected from the immune panel as the background set.
Results
Descriptions of the GH cohort
Main characteristics of the 47 subjects (pairs) included in this study are shown in Table 1. All had ISS as defined by [31]. Briefly, ISS is a condition in which the height of the individual is more than 2 standard deviations (SD) below the corresponding mean height for a given age, sex and population, in whom no identifiable disorder is present.
Table 1.
Main characteristics of the studied children
| Mean ± SD | |
|---|---|
| N | 47 |
| Sex (M/F) | 23/24 |
| Age at rhGH onset (year) | 9.8 ± 1.8 |
| Tanner stage (stage 0/stage 1) | 36/11 |
| Mean rhGH dose (µg/kg day) | 70.8 ± 19 |
| Treatment duration (months) | 18.1 ± 7.3 |
| Growth rate before treatment (cm/year) | 4.6 ± 1.0 |
| Growth rate during treatment (cm/year) | 9.1 ± 1.5 |
| Plasma IGF1 at baseline (ng/ml) | 167 ± 80 |
| Mean IGF1during treatment (ng/ml) | 402 ± 123 |
| Delta IGF1 under treatment (ng/ml) | 235 ± 117 |
The sex-matched participants (24 females vs. 23 males) were aged 4.9 to 13.1 years at time of starting rhGH treatment. They received a mean supra-physiological dose of rhGH of 70.8 μg/kg per day for a duration range from 6 to 38.4 months. During rhGH therapy, the mean increase in plasma IGF1 across children ranged from 4 to 658 ng/ml.
Differentially methylated CpGs in response to GH treatment
The average sequence genome coverage in targeted regions for these 94 samples was 25-fold (Additional file 1: Table S1). Close to 5.3 million CpGs (including those out of the targeted panels) were captured at autosomes with more than 20-fold read coverage and in at least one sample. When restricting attention to CpGs with good sample coverage in at least 10 before and after treatment pairs, 3,342,494 CpGs at autosomes remained for downstream analysis. In addition, the paired samples before and after GH treatment for 47 children were clustered well according to their genotype profiles which were inferred from the methylation sequencing data (Additional file 2: Figure S1).
We first compared the DNA methylation difference of each CpG per individual pair. For these ~ 3.3 M CpGs across 47 individual pairs, 68.1% of all comparisons were measured, see the sample (pair) coverage distribution of CpGs in Additional file 3: Figure S2A. A majority of the CpGs showed low mean absolute methylation difference (e.g., > 95% of these CpGs are showing mean methylation differences ≤ 5%) and low standard deviation (Fig. 1A, B). Specifically, it showed majority of them have DNA methylation difference of zero as indicated in the middle peak of Fig. 1A. Meanwhile, it also showed two lower sub-peaks which indicated the potential epigenetic responses to the GH treatment and represent the average methylation changes (i.e., ~ ± 5%). When considering each of the measured CpG pairs across 47 children (n = 107,019,585), the mean absolute value of the methylation changes between the baseline and rhGH stimulated samples is 4.8 ± 6.6% (SD). There is no difference between male and female groups (Additional file 3: Figure S2B). By requiring ≥ 10% methylation difference between the baseline and rhGH stimulated samples, 304,528 CpGs showed either ≥ 10 hypo-methylated samples or ≥ 10 hyper-methylated samples. Among those CpGs, 37,128 CpGs showed discordant responses to rhGH treatment (i.e., ≥ 10 hypo-methylated samples and ≥ 10 hyper-methylated samples). Furthermore, the corresponded numbers of CpGs drop to 11,398 and 0 if requiring ≥ 20% methylation changes. Overall, we observed quite heterogeneous DNA methylation responses to rhGH treatment. Additional file 3: Figure S2C demonstrates the methylation changes pattern for the top 5% most variable CpGs whose methylation profiles were measured for all individuals.
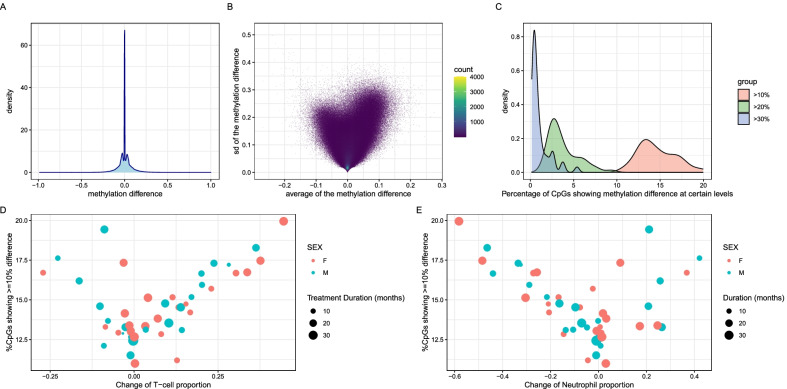
Fig. 1.
Characterization of DNA methylation changes before and after GH stimulation treatment. A The distribution of the DNA methylation changes. B The scatter plot between the DNA methylation changes and standard deviation of DNA methylation changes over all CpGs. The density of CpGs was also illustrated using different colors as indicated in the legend. C The distribution of the percentage of CpGs showing different level of differential methylation changes (> 10%, > 20% and > 30%) across samples. D, E The correlation between the percentage of DMCs (at > 10% methylation level difference) and the changes of T cell proportion (D) and neutrophil proportion (E). The sex and treatment duration were indicated with different colors and different sizes of the dots
We observed that roughly 14.64%, 3.76% and 1.07% of the CpGs showed methylation difference > 10%, 20% and 30% comparing the baseline and after treatment samples, respectively (Fig. 1C). We then correlated the ratio of CpGs showing methylation difference > 10% of individuals with other phenotypes such as onsite age, difference of blood proportions and changes in plasma IGF1 concentration under treatment. We observed that the proportion changes of T cell and neutrophil are significantly corrected with the ratio of CpGs showing > = 10% differences (r = 0.39 and − 0.32, with p = 0.006 and 0.03, respectively) (Fig. 1D, E), but not for others parameters. This correlation trends are further increased when checking the ratio of CpGs showing > = 20% or 30% methylation level changes (Additional file 1: Table S2). Furthermore, we also observed that the proportion changes of B cell are showing significant correlation with the ratio CpGs showing 30% methylation level changes. Additional file 1: Table S2 presents the detailed correlations between CpG ratios at different levels of methylation changes and various phenotypic features. This implied that the methylation level changes might be confounded by the blood proportions changes.
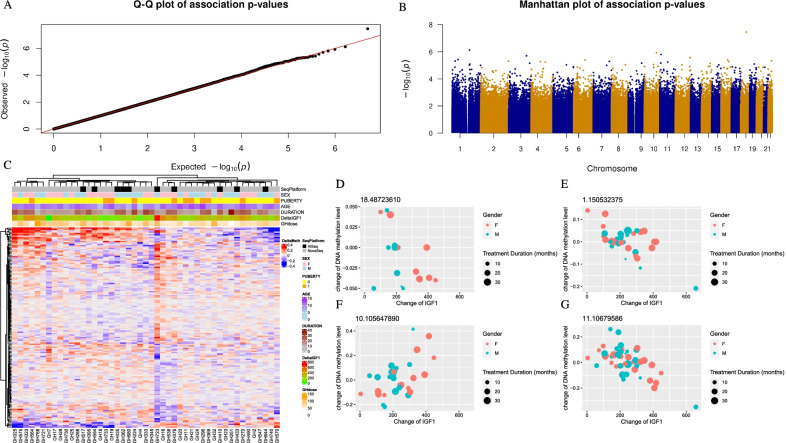
To identify the differentially methylated CpGs upon long-term exposure to rhGH, we fit a linear regression model on the difference between the DNA methylation values of all baseline and post-treatment PBMC samples and the difference in IGF1 values (Delta-IGF1) by adjusting the age at onset of treatment, treatment duration and differences in blood cell proportions before and after treatment. Figure 2A, B illustrates the QQ-plot and the Manhattan plot for this analysis. We did not observe any significant associations between Delta-IGF1 and the methylation level changes of CpGs (response DMCs) between baseline and post-treatment samples after multiple test corrections at a FDR of 0.05. However, we identified 2599 CpGs showing response DMCs (p < 1e−3) among baseline and post-treatment samples where 1317 response DMCs are negatively correlated and 1282 DMCs are positively correlated. With a more stringent p value threshold, we identified 267 DMCs at p value < 1e−4 with 123 negatively correlated and 144 positively correlated response DMCs. The DNA methylation change’s pattern of the DMCs at p < 1e−3, which were also measured by all the individuals, is illustrated in Fig. 2C. Table 2 lists the top 20 significant response DMCs with the top examples demonstrated in Fig. 2D–G, and Additional file 1: Table S3 lists the full list of these 267 response DMCs.
Fig. 2.
The distribution of response-dependent differentially methylated CpGs. A The QQ-plot of p values from the analysis of the CpGs respond to the GH treatment. B Manhattan plot of p values from the response analysis. C The heatmap of response DMCs at p value < 1e−3 whose methylation profiles were measured for all individuals. Different phenotype features (including different sequencing platforms, sex, puberty, age onset, treatment duration, changes of IGF1 concentration and GH dose) are illustrated in the top plots. D–G Scatter plot for the examples of top response DMCs. The sex and treatment duration were indicated with different colors and different sizes of the dots
Table 2.
The top 20 response DMCs list with p value < 1e−4. The response DMCs were sorted by the p value. CpG chromosome and position, regression p value, beta value (coefficient) and the annotated closest gene information (including genomic Annotation, Distance to TSS, Gene Name, Gene Type, and Gene Description of the closest gene) were provided
| chr.position | p value | Beta | Annotation | Distance to TSS | Gene Name | Gene type | Gene description |
|---|---|---|---|---|---|---|---|
| chr18.48723610 | 3.49E−08 | − 0.22733 | exon (NM_016626, exon 1 of 2) | 440 | MEX3C | Protein-coding | mex-3 RNA binding family member C |
| chr1.150532375 | 7.49E−07 | − 0.39527 | TTS (NR_104133) | 7971 | MIR4257 | ncRNA | microRNA 4257 |
| chr10.105647890 | 1.17E−06 | 0.893509 | intron (NM_024928, intron 9 of 9) | 30,051 | STN1 | Protein-coding | STN1 subunit of CST complex |
| chr11.10679586 | 1.56E−06 | − 0.58556 | intron (NM_001206880, intron 1 of 19) | − 5739 | MRVI1 | Protein-coding | Murine retrovirus integration site 1 homolog |
| chr3.156848563 | 1.97E−06 | − 0.14999 | Intergenic | − 7773 | LINC00880 | ncRNA | Long intergenic non-protein-coding RNA 880 |
| chr12.129252225 | 2.68E−06 | − 1.11703 | Intergenic | 56,277 | SLC15A4 | Protein-coding | Solute carrier family 15 member 4 |
| chr1.50834156 | 3.76E−06 | 0.153745 | Intergenic | 54,958 | DMRTA2 | Protein-coding | DMRT-like family A2 |
| chr16.56553641 | 3.90E−06 | − 0.13719 | intron (NM_031885, intron 1 of 16) | 294 | BBS2 | Protein-coding | Bardet-Biedl syndrome 2 |
| chr6.99283220 | 3.95E−06 | − 0.12451 | exon (NM_005604, exon 1 of 1) | 771 | POU3F2 | Protein-coding | POU class 3 homeobox 2 |
| chr22.46519089 | 4.61E−06 | − 1.1146 | Intergenic | 9524 | MIRLET7B | ncRNA | microRNA let-7b |
| chr1.25228801 | 4.78E−06 | − 0.30407 | exon (NM_004350, exon 5 of 5) | 17,105 | MIR6731 | ncRNA | microRNA 6731 |
| chr2.241976241 | 4.88E−06 | − 0.48453 | exon (NM_001080437, exon 5 of 32) | 38,207 | SNED1 | Protein-coding | Sushi, nidogen and EGF-like domains 1 |
| chr7.139187227 | 5.05E−06 | − 0.39751 | Intergenic | − 18,809 | KLRG2 | Protein-coding | Killer cell lectin-like receptor G2 |
| chr4.79545349 | 5.32E−06 | 0.366908 | Intergenic | − 21,798 | LINC01094 | ncRNA | Long intergenic non-protein-coding RNA 1094 |
| chr16.66554996 | 5.35E−06 | 0.684785 | intron (NR_073520, intron 5 of 8) | 29,028 | TK2 | Protein-coding | Thymidine kinase 2 |
| chr11.68608981 | 5.63E−06 | − 0.13725 | intron (NM_001876, intron 1 of 18) | 402 | CPT1A | Protein-coding | Carnitine palmitoyltransferase 1A |
| chr4.122871430 | 5.71E−06 | 0.274039 | intron (NM_001366479, intron 1 of 10) | 1784 | TRPC3 | Protein-coding | Transient receptor potential cation channel subfamily C member 3 |
| chr11.8364991 | 6.74E−06 | 0.562738 | Intergenic | − 74,658 | LMO1 | Protein-coding | LIM domain only 1 |
| chr9.117026652 | 6.75E−06 | 1.475667 | exon (NM_032888, exon 29 of 61) | 54,939 | MIR455 | ncRNA | microRNA 455 |
| chr3.183873096 | 6.93E−06 | 0.345301 | promoter-TSS (NM_004423) | − 68 | DVL3 | Protein-coding | Disheveled segment polarity protein 3 |
Genome feature and function enrichment analysis of response DMCs
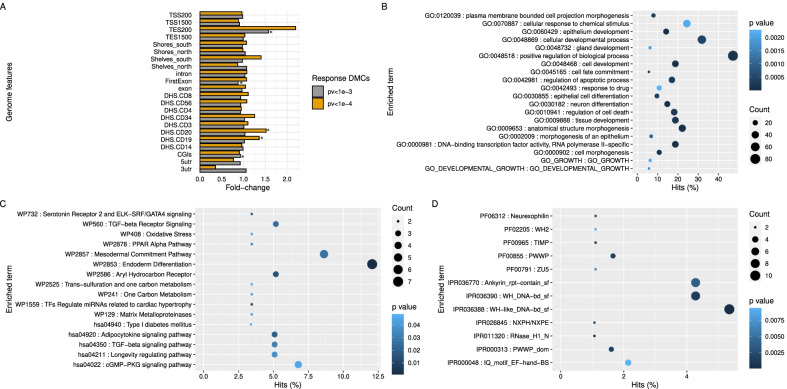
We first performed a genomic feature enrichment analysis of response DMCs with the p value < 1e−4 and p value < 1e−3. We observed that the response DMCs with p value < 1e−3 were slightly enriched in the first exon, TES200 and CGIs (Fig. 3A) but no significant regions for the response DMCs with p value < 1e−4 (Fig. 3A). Furthermore, we also observed slightly enrichments on the blood-specific regulatory elements—DNase I hypersensitive sites (DHSs) regions of CD19 and CD20 for the response DMCs with p value < 1e−4 but not for the response DMCs with p value < 1e−3 (Fig. 3A).
Fig. 3.
Genome feature and functional enrichment analysis of the response DMCs. A Genomic features and blood regulatory element enrichment analysis of the response DMCs with p value < 1e−3 and p value < 1e−4. Fisher test: *: p value < 0.01. B–D Functional enrichment analysis of the response DMCs. Enrichment of functional grouping of genes through the biological process, groups of the genes in the same pathway through KEGG, pathway interaction database as well as the WikiPathways, and the similar domain and features of the gene’s product proteins through PFAM and Interpro domain database were illustrated in (B), (C) and (D), respectively. The number of genes in each item and p value of the enrichment analysis was shown in the legend
We then performed a gene function enrichment analysis on 265 genes which are associated with 267 p value < 1e−4 response DMCs using Homer (annotatepeaks function) [30]. It revealed that these genes were enriched in the Biological process GO term of “neuron differentiation” (p value < 1e−05), “cell development” and “cell morphogenesis” (p value < 1e−4), as well as other developmental process-related terms, including the “Development growth” and “Growth” terms (p value < 3e−3) (Fig. 3B). Strikingly, these genes were observed to be enriched in the “Endoderm differentiation pathway” (p value < 1e−3), “Adipocytokine signaling pathway” (p value = 0.019), “TGF-beta signaling pathway” (p value = 0.02) and “PPAR Alpha pathway” (p value < 0.03) as well as other pathways related to growth factor receptor or stimulating hormone signaling pathway (Fig. 3C). Furthermore, the common domain families of “Rnase H1,” “WH/WH-like DNA” (p value < 1e−4), “TIMP,” “PWWP,” etc. (p value < 1e−3), were observed to be over-represented in the proteins of these response DMCs-associated genes (Fig. 3D). In addition, when exploring the enrichment for genes associated with response DMCs at p < 1e−3 (n = 2247 genes), we observed that these genes were enriched in “PI3K-Akt signaling pathway,” “Sphingolipid signaling pathway” and “cAMP signaling pathway” (Additional file 4: Figure S3).
We compared our identified DMCs with previously reported GH-related DMCs (n = 239, [32]). We did not find that any reported significant DMCs were replicated at our study, but when expanded to 1 kb distance-based neighboring CpGs, we observed a slight enrichment (fold-change of 1.73) of these reported DMCs at our response DMCs (p value < 0.001) compared with our non-significant CpGs (p value > 0.001) although this enrichment was not significant (p value = 0.15) due to the small number of overlapped CpGs being counted. Interestingly, one response DMC (with p value = 1e−5) at chr11:68608981, which is located within the CTP1A gene, was within 200 bp to the reported GH DMC at chr11:68609166 (p value = 1e−5, [32]).
It was reported that the effect of GH treatment on growth is potentially influenced by individual’s SNPs [33]. We first linked our response DMCs to 37 potential SNPs which were reported to be GH-response-associated SNPs. We observed that only 14 SNPs have their surrounding CpGs in our tested CpG sets within 5 kb distance. Interestingly, three of these 14 potential SNPs were showing significant response DMCs (p value < 0.01) within a 5 kb distance to these SNPs. Particularly, one SNP (rs6600230, chr16:738477) is overlapping with the gene WDR24 where multiple CpGs were showing p value < 0.05 with the significant one (chr16:739598) located at the first exon region (p value = 0.01).
Discussion
The emerging field of pharmacoepigenomics will provide promising insights into the role drugs play in modulating the host epigenome and in addressing inter-individual variability in drug response and adverse effects. Although there is growing evidence that pharmacoepigenetics has the potential to become an important element of personalized medicine, we know of no study that has evaluated the changes in the individual methylome of the same group of patients undergoing a treatment, as performed in the current study. An additional advantage of our pharmacoepigenetic study is that clinical (growth rate, height) and biological outcomes (IGF1) can be quantified in response to precise rhGH dosing carefully injected by parents. More specifically, our data provide the first comprehensive pharmacoepigenomics analysis on rhGH treatment in children with ISS by comparing DNA methylation marks before and after several months of rhGH treatment. We identified 267 response DMCs which are associated with 265 genes and these genes were enriched in the biological process of cell differentiation, system development and different growth-related pathways such as endoderm differentiation, adipocytokine signaling, PPAR alpha and TGF-beta signaling pathways. This pilot study thus supports the existence of dynamic epigenetic changes in response to rhGH treatment. Again, it should be recalled that these are methylation changes induced by prolonged GH administration, and not epigenetic marks associated with short stature, an example of which can be found in one of our previous studies [34].
Our customized methylation sequencing panel captured more than 5 million CpGs, which is much larger than the previously used 450 K array data (i.e., > 10 folder larger), representing an unprecedented level of resolution. After quality control, more than 3.3 million CpGs remained for the response association analysis, providing the potential to discover novel signals. Indeed, of identified 267 response DMCs (with p value < 1e−4), only 114 (43%) of them are located within 100 bp distance to the known 450 K loci and only 25 (9.4%) of them are exactly located at the 450 K loci. Among the top 20 response DMCs listed in Table 2, half of them does not have neighboring CpGs (with 100 bp distance) in 450 K loci.
Previous studies had shown that the TGF-beta signaling pathway plays an important role in regulating osteoblast differentiation and could inhabit IGF-1/Akt signaling pathway [35]. The adipocytokine pathway and cAMP-signaling pathway are downstream signaling pathways upon activation of IGF1 receptor and contribute to the signal transduction of insulin-like growth factors on growth [36]. Another study reported that the GH modulates EGFR expression and signaling and further activates PI3K-Akt signaling, which was enriched in our response DMCs (p value < 1e−3) [37]. Moreover, our response DMC-associated genes were enriched in DNA-binding transcription factors as well as proteins with the common domain families of “WH/WH-like DNA” and “TIMP.” Particularly, TIMP3 is known to modulate GHR abundance and GH sensitivity [38], and NFKB1 is a known gene associated with short stature [39] and the growth-promoting effects of the transcription factor family of NFKB seems to be facilitated by GH and IGF-1[40], while FOXA1, FOXN1 are regulators for GH activation [41]. Here, our identified genes with rhGH-associated methylation changes were enriched in these pathways, supporting the biological relevance of our findings. The genes involved in these pathways include CDKN2B, LEFTY2, PPP2R1B, CPT1A, RXRA, NFKB1, KCNMA1, BORCS8-MEF2B, MRVI1, PPIF and GATA4 (See the full response DMC list at p value < 1e−3, Additional file 1: Table S4).
The current study identified marked intra-individual responses of DNA-methylation to long-term rhGH treatment. A study by Kolarova et al. investigated 24 patients at baseline and after only 4 days of rhGH administration [32]. The studied patients had various forms of GH deficiency (N = 13) or other pathological conditions, which could influence the epigenetic responses to rhGH and complicate the interpretation. In comparison, only healthy children were selected for the current study and were either prepubertal or with minimal manifestations of puberty in order to avoid epigenetic changes that are associated in blood cells with advancing puberty [42].
Array-based DNA-methylation profiling of paired peripheral blood mononuclear cell samples in the Kolarova et al.’s study revealed clustering according to individuals rather than treatment [32]. Supervised analysis identified 239 CpGs as significantly differentially methylated between baseline and acutely GH-stimulated samples, which nevertheless did not retain significance after adjustment for multivariate analysis. In a companion study, Kolarova et al. investigated the long-term effects of prolonged rhGH treatment on the DNA-methylome and analyzed peripheral blood cells from an independent cohort of 36 rhGH-treated children born small for gestational age (SGA) compared to 18 untreated controls. These were not paired samples which had to face major unwanted inter-individual variance of children methylome. No differentially methylated targets reached the level of significance in this long-term rhGH-treated cohort [32]. Our study did not replicate any of these 239 DMCs but observed a slight enrichment if considering significant response DMCs in 1 kb distance to them. The lack of high replicates might due to different etiologies of short stature (intrauterine growth retardation may influence epigenetic marks in the Kolarova et al.’s study) design, different treatment durations and dosing, different ages and more importantly the considerable inter-individual heterogeneity, while our study investigated paired intra-individual changes in methylation.
Of interest, MEX3C, the top gene in our response association analysis, was reported to be a translational regulator of IGF expression in mice [43]. IGF1 protein expression in bone cells was decreased upon MEX3C deficiency in Mex3c homozygous mutant mice. Given that MEX3C is highly conserved among mammalian species, the observation in mice might be relevant to the human IGF1 regulation and warrants further investigation. Among the top 20 signals, the response DMC at CPT1A (chr11:68608981) was close to the reported locus (cg20228509, chr11:68609166) within 1 kb in the Kolarova et al.’s short-term rhGH treatment study [32]. CPT1A was observed to be a genetic regulation of fatty acid metabolism, and missense mutation reduces height [44]. Although this evidence was not revealed in epigenetic studies, the potential pathway CPT1A involved (such as the adipocytokine signaling pathway, an important pathway related to IGF signaling) might indicate its indirect association with GH. In the same study, more than 3 CpGs in SLC15A4 were identified as differentially methylated loci and two of them were further validated with bisulfite pyrosequencing [32]. Our data showed the top signal was at the downstream of SLC15A4, and a few CpGs with nominal significance at p value < 0.01 were located in the intron of SLC15A4 gene. In addition, we also observed a couple of non-coding RNAs at our top signals list. Their functions related to GH are currently unknown and need further exploration.
As in almost all epigenetic studies in humans, we were only able to characterize DNA methylation in blood cells, which may not recapitulate all GH-induced changes that may occur in other target cells. However, PBMC are sensitive to the GH/IGF1 axis [45] and may thus reveal epigenetic changes triggered by GH or IGF1. B-lymphocytes and monocytes as well as T-lymphocytes and natural killer cells express GH receptors on their cell surfaces [46, 47]. These cells also express IGF1 receptors [48], which activate the mTOR pathway and can subsequently induce epigenomic changes [49]. GH [50–52] and IGF1 signaling [53, 54] have been studied in PBMC and lymphocytes. Since the top variable CpGs in our study were highly associated with the proportion changes of T cell and neutrophils, we applied a well-established computational approach to deconvolute the PBMC blood compositions and included them as covariates in our analysis model, which would effectively remove the confounders due to dynamic blood cell proportion changes. Finally, the current study supports the utility of PBMC to detect DNA methylation changes responding to rhGH treatment.
In summary, we have identified multiple response DMCs that are associated with rhGH treatment although none of them show the FDR significance after multiple testing correction. This is most likely due to the limited sample size given the large inter-individual variation in DNA methylation changes, which restricted our power to detect significant associations at FDR q-value threshold of 0.05. The downstream functional analysis revealed that the response DMCs were enriched in many pathways biologically relevant to GH. Larger sample sizes will be needed to more definitively identify epigenetic changes arising from rhGH administration. Further functional genomics investigations are also encouraged for validation of our discoveries, particularly for the top signals.
Supplementary Information
Additional file 1: Table S1. Sequence statistics for the GH samples. Table S2. Correlations between DMC ratios and other meta data features. The ratio of CpGs showing > = 10%, > = 20%, > = 30% methylation level changes against other features. Table S3. Full list of response DMCs at p value < 1e−4. The chromosome and position of CpGs, regression p value, beta value (coefficient), and the annotated closest gene information (including genomic Annotation, Distance to TSS, Gene Name, Gene Type, and Gene Description of the closest gene) were provided. Table S4. Full list of response DMCs at p value < 1e−3. The chromosome and position of CpGs, regression p value, beta value (coefficient), and the annotated closest gene information (including genomic Annotation, Distance to TSS, Gene Name, Gene Type, and Gene Description of the closest gene) were provided.
Additional file 2: Figure S1. Hierarchical clustering of 94 samples based on their genotype profiles. Genotype profiles of SNPs measured for all the 94 samples were used to perform the hierarchical clustering. Manhattan distance was used to calculate the distance between two samples. T0: baseline; T1: second time point (treatment).
Additional file 3: Figure S2. Characterization of DNA methylation changes. (A) Individual coverage of CpGs which have both before and after treatment measures in this cohort. (B) The distribution of the DNA methylation changes per sex. Male and female were indicated in the legend. (C) The heatmap of top 5% variable CpGs whose methylation profiles were measured for all individuals. Different phenotype features (including different sequencing platforms, sex, puberty, age onset, treatment duration, changes of IGF1 concentration and GH dose) are illustrated in the top plots.
Additional file 4: Figure S3. Functional enrichment analysis of the response DMCs at p value < 1e−3. (A-C) Functional enrichment analysis of the response DMCs at p value < 1e−3. Enrichment of functional grouping of genes through the biological process, groups of the genes in the same pathway through KEGG, pathway interaction database as well as the WikiPathways, and the similar domain and features of the gene’s product proteins through PFAM and Interpro domain database were illustrated in (A), (B) and (C), respectively. The number of genes in each item and p value of the enrichment analysis were shown in the legend.
Acknowledgements
We thank the team at the McGill University Genome Centre for performing the MCC-Seq library preparations and sequencing. We also thank for the computational resources provided in part by Calcul Québec and Compute Canada.
Abbreviations
- cAMP
Cyclic adenosine 3′, 5′-monophosphate
- CGIs
CpG islands
- DHS
DNase I hypersensitive sites
- DMCs
Differentially methylated CpG sites
- ENCODE
Encyclopedia of DNA elements
- FDR
False discovery rate
- GH
Growth hormone
- GO
Gene ontology
- IGF1
Insulin-like growth factor 1
- ISS
Idiopathic short stature
- LM
Linear regression
- MCC-Seq
MethylC-capture sequencing
- PBMC
Peripheral blood mononuclear cells
- PDGF
Platelet-derived growth factor
- PKC
Protein kinase C
- rhGH
Recombinant human growth hormone
- SD
Standard deviation
- SGA
Small for gestational age
- T1D
Type 1 diabetes
- TGF-beta
Transforming growth factor beta
- TIMP
Tissue inhibitors of matrix metalloproteinase
- TSS
Transcription start site
Author contributions
PB and TP designed the study. XS and PB wrote the paper. XS wrote codes, performed the data analyses and created tables/figures. WC and TK performed the data processing and contributed to the sample sequencing and data analysis. CLeS and PB provided samples from the patients. CLeS and ML edited the paper and provided insight on Methods section. All the authors have reviewed results and drafts of the paper and agreed to the published version of the manuscript.
Funding
Funds were provided by the RESET-AID Trilateral Consortium involving Canadian Institutes of Health Research (CIHR) and Agence Nationale de la Recherche (ANR). Xiaojian Shao is supported by the National Research Council Canada through the Artificial Intelligence for Design program.
Availability of data and materials
All the raw read files were submitted to European Genome-phenome Archive under the accession number EGAS00001006115.
Declarations
Ethics approval and consent to participate
Families were carefully informed about the investigational nature of the study and signed an informed consent agreed by CPP (number DC-2008-693; NI 2620, Comité de Protection des Personnes).
Consent for publications
Not applicable.
Competing interests
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaojian Shao, Email: Xiaojian.shao@nrc-cnrc.gc.ca.
Tomi Pastinen, Email: tpastinen@cmh.edu.
Pierre Bougnères, Email: pierre.bougneres@inserm.fr.
References
- 1.Rotwein P. Mapping the growth hormone–Stat5b–IGF-I transcriptional circuit. Trends Endocrinol Metab. 2012;23:186–193. doi: 10.1016/j.tem.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Møller N, Jørgensen JOL. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev. 2009;30:152–177. doi: 10.1210/er.2008-0027. [DOI] [PubMed] [Google Scholar]
- 3.Ho KY, Veldhuis JD, Johnson ML, Furlanetto R, Evans WS, Alberti KG, et al. Fasting enhances growth hormone secretion and amplifies the complex rhythms of growth hormone secretion in man. J Clin Investig. 1988;81:968–975. doi: 10.1172/JCI113450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins PJ. Growth hormone and exercise. Clin Endocrinol. 1999;50:683–689. doi: 10.1046/j.1365-2265.1999.00784.x. [DOI] [PubMed] [Google Scholar]
- 5.Van den Berghe G, de Zegher F, Bouillon R. Acute and prolonged critical illness as different neuroendocrine paradigms. J Clin Endocrinol Metab. 1998;83:1827–1834. doi: 10.1210/jcem.83.6.4763. [DOI] [PubMed] [Google Scholar]
- 6.Lang Ch, Frost RA. Role of growth hormone, insulin-like growth factor-I, and insulin-like growth factor binding proteins in the catabolic response to injury and infection. Curr Opin Clin Nutr Metab Care. 2002;5:271–279. doi: 10.1097/00075197-200205000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Flores KB, Wolschin F, Amdam GV. The role of methylation of DNA in environmental adaptation. Integr Comp Biol. 2013;53:359–372. doi: 10.1093/icb/ict019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin EM, Fry RC. Environmental Influences on the epigenome: exposure-associated DNA methylation in human populations. Annu Rev. 2018;39:309–333. doi: 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- 9.Shao X, Zhang C, Sun MAM-A, Lu X, Xie H. Deciphering the heterogeneity in DNA methylation patterns during stem cell differentiation and reprogramming. BMC Genom. 2014;15:978. [DOI] [PMC free article] [PubMed]
- 10.Xiao FH, Wang HT, Kong QP. Dynamic DNA methylation during aging: a “prophet” of age-related outcomes. Front Genet. 2019;107. www.frontiersin.org. Cited 28 Apr 2021. [DOI] [PMC free article] [PubMed]
- 11.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015 doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LTY, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. [DOI] [PMC free article] [PubMed]
- 13.Busche S, Shao X, Caron M, Kwan T, Allum F, Cheung WA, et al. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol. 2015;16:290. [DOI] [PMC free article] [PubMed]
- 14.Yang X, Shao X, Gao L, Zhang S. Systematic DNA methylation analysis of multiple cell lines reveals common and specific patterns within and across tissues of origin. Hum Mol Genet. 2015;24:4374–4384. doi: 10.1093/hmg/ddv172. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. New Engl J Med. 2018;378:1323–1334. doi: 10.1056/NEJMra1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Wilson R, Heiss J, Breitling LP, Saum K-U, Schöttker B, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. doi: 10.1038/ncomms14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg AP, Fallin MD. Epigenetics at the crossroads of genes and the environment. JAMA. 2015;314:1129–1130. doi: 10.1001/jama.2015.10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks AJ, Waters MJ. The growth hormone receptor: mechanism of activation and clinical implications. Nat Rev Endocrinol. 2010;6:515–525. doi: 10.1038/nrendo.2010.123. [DOI] [PubMed] [Google Scholar]
- 19.MacLeod AR, Szyf M. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis (∗) J Biol Chem. 1995;270:8037–8043. doi: 10.1074/jbc.270.14.8037. [DOI] [PubMed] [Google Scholar]
- 20.Fradin D, Le Fur S, Mille C, Naoui N, Groves C, Zelenika D, et al. Association of the CpG methylation pattern of the proximal insulin gene promoter with type 1 diabetes. PLoS ONE. 2012;7:e36278. doi: 10.1371/journal.pone.0036278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen P, Rogol AD, Deal CL, Saenger P, Reiter EO, Ross JL, et al. Consensus statement on the diagnosis and treatment of children with idiopathic short stature: a summary of the Growth Hormone Research Society, the Lawson Wilkins Pediatric Endocrine Society, and the European Society for Paediatric Endocrinology Workshop. J Clin Endocrinol Metab. 2008;93:4210–4217. doi: 10.1210/jc.2008-0509. [DOI] [PubMed] [Google Scholar]
- 22.Allum F, Shao X, Guénard F, Simon M-M, Busche S, Caron M, et al. Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat Commun. 2015;6:7211. doi: 10.1038/ncomms8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung WA, Shao X, Morin A, Siroux V, Kwan T, Ge B, et al. Functional variation in allelic methylomes underscores a strong genetic contribution and reveals novel epigenetic alterations in the human epigenome. Genome Biol. 2017;18:50. doi: 10.1186/s13059-017-1173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao X, Hudson M, Colmegna I, Greenwood CMT, Fritzler MJ, Awadalla P, et al. Rheumatoid arthritis-relevant DNA methylation changes identified in ACPA-positive asymptomatic individuals using methylome capture sequencing. Clin Epigenet. 2019;11:110. doi: 10.1186/s13148-019-0699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burger L, Gaidatzis D, Schübeler D, Stadler MB. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 2013;41:e155–e155. doi: 10.1093/nar/gkt599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allum F, Hedman ÅK, Shao X, Cheung WA, Vijay J, Guénard F, et al. Dissecting features of epigenetic variants underlying cardiometabolic risk using full-resolution epigenome profiling in regulatory elements. Nat Commun. 2019;10:1209. [DOI] [PMC free article] [PubMed]
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 28.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012;13:1–14. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13(1):2012. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wit JM, Clayton PE, Rogol AD, Savage MO, Saenger PH, Cohen P. Idiopathic short stature: definition, epidemiology, and diagnostic evaluation. Growth Horm IGF Res. 2008;18:89–110. doi: 10.1016/j.ghir.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Kolarova J, Ammerpohl O, Gutwein J, Welzel M, Baus I, Riepe FG, et al. In vivo investigations of the effect of short- and long-term recombinant growth hormone treatment on DNA-methylation in humans. PLoS ONE. 2015;10:e0120463. doi: 10.1371/journal.pone.0120463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dauber A, Meng Y, Audi L, Vedantam S, Weaver B, Carrascosa A, et al. A genome-wide pharmacogenetic study of growth hormone responsiveness. J Clin Endocrinol Metab. 2020;105:3203–3214. doi: 10.1210/clinem/dgaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouni M, Gunes Y, Belot MP, Castell AL, Fradin D, Bougnères P. The igf 1 p2 promoter is an epigenetic qtl for circulating igf 1 and human growth. Clin Epigenet. 2015;7:2–12. doi: 10.1186/s13148-015-0062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochiai H, Okada S, Saito A, Hoshi K, Yamashita H, Takato T, et al. Inhibition of insulin-like growth factor-1 (IGF-1) expression by prolonged transforming growth factor-β1 (TGF-β1) administration suppresses osteoblast differentiation. J Biol Chem. 2012;287:22654. doi: 10.1074/jbc.M111.279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hakuno F, Takahashi S-I. 40 years of IGF1: IGF1 receptor signaling pathways. J Mol Endocrinol. 2018;61:T69–86. doi: 10.1530/JME-17-0311. [DOI] [PubMed] [Google Scholar]
- 37.Díaz ME, González L, Miquet JG, Martínez CS, Sotelo AI, Bartke A, et al. Growth hormone modulation of EGF-induced PI3K-Akt pathway in mice liver. Cell Signal. 2012;24:514–523. doi: 10.1016/j.cellsig.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Wang X, Loesch K, May LA, Davis GE, Jiang J, et al. TIMP3 modulates GHR abundance and GH sensitivity. Mol Endocrinol. 2016;30:587–599. doi: 10.1210/me.2015-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clayton P, Bonnemaire M, Dutailly P, Maisonobe P, Naudin L, Pham E, et al. Characterizing short stature by insulin-like growth factor axis status and genetic associations: results from the prospective, cross-sectional, epidemiogenetic EPIGROW study. J Clin Endocrinol Metab. 2013;98:E1122–E1130. doi: 10.1210/jc.2012-4283. [DOI] [PubMed] [Google Scholar]
- 40.De Luca F. Regulatory role of NF-κB in growth plate chondrogenesis and its functional interaction with growth hormone. Mol Cell Endocrinol. 2020;514:1116. doi: 10.1016/j.mce.2020.110916. [DOI] [PubMed] [Google Scholar]
- 41.Chia DJ. Minireview: mechanisms of growth hormone-mediated gene regulation, 2014. https://academic.oup.com/mend/article/28/7/1012/2623318. Cited 27 Mar 2022. [DOI] [PMC free article] [PubMed]
- 42.Almstrup K, Lindhardt Johansen M, Busch AS, Hagen CP, Nielsen JE, Petersen JH, et al. Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Sci Rep. 2016;6:1. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiao Y, Bishop CE, Lu B. Mex3c regulates insulin-like growth factor 1 (IGF1) expression and promotes postnatal growth. Mol Biol Cell. 2012;23:1404–1413. doi: 10.1091/mbc.e11-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skotte L, Koch A, Yakimov V, Zhou S, Søborg B, Andersson M, et al. CPT1A missense mutation associated with fatty acid metabolism and reduced height in greenlanders. Circ Cardiovasc Genet. 2017 doi: 10.1161/CIRCGENETICS.116.001618. [DOI] [PubMed] [Google Scholar]
- 45.Weigent DA. Lymphocyte GH-axis hormones in immunity. Cell Immunol. 2013;285:118–132. doi: 10.1016/j.cellimm.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Rapaport R, Sills IN, Green L, Barrett P, Labus J, Skuza KA, et al. Detection of human growth hormone receptors on IM-9 cells and peripheral blood mononuclear cell subsets by flow cytometry: correlation with growth hormone-binding protein levels. J Clin Endocrinol Metab. 1995;80:2612–2619. doi: 10.1210/jcem.80.9.7673403. [DOI] [PubMed] [Google Scholar]
- 47.Badolato R, Bond HM, Valerio G, Petrella A, Morrone G, Waters MJ, et al. Differential expression of surface membrane growth hormone receptor on human peripheral blood lymphocytes detected by dual fluorochrome flow cytometry. J Clin Endocrinol Metab. 1994;79:984–990. doi: 10.1210/jcem.79.4.7962309. [DOI] [PubMed] [Google Scholar]
- 48.Stentz FB, Kitabchi AE. Transcriptome and proteome expression in activated human CD4 and CD8 T-lymphocytes. Biochem Biophys Res Commun. 2004;324:692–696. doi: 10.1016/j.bbrc.2004.09.113. [DOI] [PubMed] [Google Scholar]
- 49.Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden CDCC, Li Y, et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell Cell Press. 2018;172:135–146.e9. doi: 10.1016/j.cell.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 50.Whatmore AJ, Patel L, Clayton PE. A pilot study to evaluate gene expression profiles in peripheral blood mononuclear cells (PBMCs) from children with GH deficiency and Turner syndrome in response to GH treatment. Clin Endocrinol. 2009;70:429–434. doi: 10.1111/j.1365-2265.2008.03477.x. [DOI] [PubMed] [Google Scholar]
- 51.Fernández-Pérez L, Nóvoa J, Ståhlberg N, Santana-Farré R, Boronat M, Marrero D, et al. The effect of in vivo growth hormone treatment on blood gene expression in adults with growth hormone deficiency reveals potential biomarkers to monitor growth hormone therapy. Clin Endocrinol. 2010;72:800–806. doi: 10.1111/j.1365-2265.2009.03732.x. [DOI] [PubMed] [Google Scholar]
- 52.Welzel M, Appari M, Bramswig N, Riepe FG, Holterhus PM. Transcriptional response of peripheral blood mononuclear cells to recombinant human growth hormone in a routine four-days IGF-I generation test. Growth Horm IGF Res. 2011;21:336–342. doi: 10.1016/j.ghir.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 54.Lotton C, Rodrigue D, Elie C, Rothenbühler A, Lahlou N, Le Stunff C, et al. Akt phosphorylation in lymphocytes provides an index of in vitro insulin-like growth factor i sensitivity associated with growth hormone-induced growth. J Clin Endocrinol Metab. 2008;93:1458–1463. doi: 10.1210/jc.2007-2575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequence statistics for the GH samples. Table S2. Correlations between DMC ratios and other meta data features. The ratio of CpGs showing > = 10%, > = 20%, > = 30% methylation level changes against other features. Table S3. Full list of response DMCs at p value < 1e−4. The chromosome and position of CpGs, regression p value, beta value (coefficient), and the annotated closest gene information (including genomic Annotation, Distance to TSS, Gene Name, Gene Type, and Gene Description of the closest gene) were provided. Table S4. Full list of response DMCs at p value < 1e−3. The chromosome and position of CpGs, regression p value, beta value (coefficient), and the annotated closest gene information (including genomic Annotation, Distance to TSS, Gene Name, Gene Type, and Gene Description of the closest gene) were provided.
Additional file 2: Figure S1. Hierarchical clustering of 94 samples based on their genotype profiles. Genotype profiles of SNPs measured for all the 94 samples were used to perform the hierarchical clustering. Manhattan distance was used to calculate the distance between two samples. T0: baseline; T1: second time point (treatment).
Additional file 3: Figure S2. Characterization of DNA methylation changes. (A) Individual coverage of CpGs which have both before and after treatment measures in this cohort. (B) The distribution of the DNA methylation changes per sex. Male and female were indicated in the legend. (C) The heatmap of top 5% variable CpGs whose methylation profiles were measured for all individuals. Different phenotype features (including different sequencing platforms, sex, puberty, age onset, treatment duration, changes of IGF1 concentration and GH dose) are illustrated in the top plots.
Additional file 4: Figure S3. Functional enrichment analysis of the response DMCs at p value < 1e−3. (A-C) Functional enrichment analysis of the response DMCs at p value < 1e−3. Enrichment of functional grouping of genes through the biological process, groups of the genes in the same pathway through KEGG, pathway interaction database as well as the WikiPathways, and the similar domain and features of the gene’s product proteins through PFAM and Interpro domain database were illustrated in (A), (B) and (C), respectively. The number of genes in each item and p value of the enrichment analysis were shown in the legend.
Data Availability Statement
All the raw read files were submitted to European Genome-phenome Archive under the accession number EGAS00001006115.