Abstract
The existence of strains of Vibrio vulnificus serovar E that are avirulent for eels is reported in this work. These isolates were recovered from water and oysters and differed from eel virulent strains in (i) fermentation and utilization of mannitol, (ii) ribotyping after HindIII digestion, and (iii) susceptibility to eel serum. Lipopolysaccharide of these strains lacked the highest molecular weight immunoreactive bands, which are probably involved in serum resistance.
Vibrio vulnificus is a bacterial species that includes strains virulent for humans and aquatic animals (1, 18, 19, 20). Classically, the strains had been grouped into two biotypes (20). Biotype 1 was believed to include environmental (mainly from marine water and oysters) and clinical isolates (18), and biotype 2 was thought to include strains of obligately pathogenic character for eels (20). This classification was supported by physiological, biochemical (mainly indole test), and serological differences (2, 10, 20) and differences in host range (20).
Recent investigations have revealed that (i) several clinical strains belong to biotype 2 (1, 6, 8), (ii) both biotypes can infect cultured shrimps (6, 8), and (iii) no phenotypic trait can with certainty be associated with any biotype (6, 8). Based on these data, we have proposed a subspecific classification in serovars that ascribes the biotype 2 strains to serovar E (8). The specific antigen of this serogroup is the lipopolysaccharide (LPS) (10), which acts as a virulence determinant for eels (3).
All known serovar E strains have been isolated from infected animal (mainly eel) tissues. However, it is reasonable to think that they can be found in marine and brackish waters because they can survive in water for years (16) and use it as an infective route (5). In 1993, several V. vulnificus strains negative for indole production were isolated from water and oysters in Taiwan (13). Since indole-negative strains classically belonged to serovar E, these strains would represent the first strains isolated from water. In consequence, the objective of the present study was to serotype them and to compare them with well-characterized serovar E strains.
The strains used in this study were identified by PCR by using the cytolysin-hemolysin gene and confirmed by biochemical tests (13) (Table 1). Strains were routinely cultured on tryptone soya agar and in tryptone soya broth (Oxoid), both supplemented with 0.5% (wt/vol) NaCl (TSA-1 and TSB-1, respectively), at 25°C for 24 h.
TABLE 1.
Strains, sources, and results of the serological characterization
| Strain | Source | Sampling site, yr | Serovar | Microagglutination titera | ELISA titers |
|---|---|---|---|---|---|
| ATCC 27562T | Human blood | United States | Non-E | <1 | 100b/10c |
| E86 | Diseased eel | Spain, 1990 | E | 256 | 64,000/10,000 |
| E105 | Diseased eel | Spain, 1990 | E | 256 | 64,000/10,000 |
| CG100 | Oyster | Taiwan, 1993 | E | 128 | 64,000/10,000 |
| CG106 | Oyster | Taiwan, 1993 | E | 64 | 30,000/5,000 |
| CG110 | Seawater | Taiwan, 1993 | E | 64 | 24,000/5,000 |
| CG111 | Seawater | Taiwan, 1993 | E | 64 | 20,000/1,800 |
| CG118 | Seawater | Taiwan, 1993 | E | 64 | 15,000/NTd |
Reciprocal of the highest dilution of serum giving total agglutination. Serum was obtained by using whole cells of strain E86 as an antigen.
Reciprocal of the highest dilution of serum which produced a reaction at an optical density at 405 nm of >0.2 after subtraction of the control values. Serum was obtained by using whole cells of strain E86 as an antigen.
Reciprocal of the highest dilution of serum which produced a reaction at an optical density at 405 nm of >0.2 after subtraction of the control values. Serum was obtained by using LPS of strain E86 as an antigen.
NT, not tested.
For serotyping we used a microagglutination test and an enzyme-linked immunosorbent assay (ELISA) with O antigens and polyclonal antibodies against whole cells and crude LPS from strain E86 (9, 10). Titers were defined as the reciprocals of the highest dilutions that agglutinated cells or gave an optical density at 405 nm of ≥0.2 after subtraction of control values (values for wells without antigen). The non-serovar E strain ATCC 27562T was used as a negative control. The strains were biochemically characterized by using the API 20E system (BioMérieux) and conventional tests including drug resistance assays (Oxoid or BBL) for penicillin (10 μg/ml), amoxicillin (50 μg/ml), ampicillin (50 μg/ml), kanamycin (50 μg/ml), gentamicin (10 μg/ml), amikacin (25 μg/ml), erythromycin (15 μg/ml), chloramphenicol (25 μg/ml), tetracycline (20 μg/ml), oxolinic acid (10 μg/ml), nitrofurantoin (10 μg/ml), rifampin (30 μg/ml), sulfamethoxazole/trimethoprim (25 μg/ml), polymyxin B (300 U), sulfanilamide (300 μg/ml), streptomycin (25 μg/ml), and nalidixic acid (50 μg/ml) according to previously described procedures (10). The LPS was extracted from whole cells according to the procedure of Hitchcock and Brown as modified by Biosca et al. (10). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed according to the method of Laemmli (15). LPS bands were visualized by immunostaining after being transferred to nitrocellulose sheets as described before (10). The ribotypes were analyzed by a previously described method (13) by using HindIII. DNA fragments were hybridized with Escherichia coli ribosomal RNA (Boehringer GmbH, Mannheim, Germany) end-labeled with [γ-32P]ATP. The enzymatic and toxic activities were evaluated after the extraction of the extracellular products (ECPs) by the cellophane plate technique (2). The assays were made on agarose (Oxoid) plates (0.8% [wt/vol] in phosphate-buffered saline solution [pH 7]) supplemented with skimmed milk (2% [wt/vol]), Tween 80 (1% [wt/vol]), egg yolk emulsion (2% [wt/vol]), fibrinogen (0.28% [wt/vol]), elastin (0.1% [wt/vol]), and eel erythrocytes (1% [vol/vol]) as previously described (10). The highest dilutions of ECPs that gave a positive response were recorded. The sensitivity to eel serum was evaluated with bacteria grown to stationary phase in microtiter plates. In each well, a volume of 20 μl of serum was mixed with 20 μl of a suspension of bacteria (105 to 106 CFU/ml) in saline solution (1.5% NaCl [pH 7]). Assays were made in triplicate, by taking samples at 0, 1, 2, and 4 h of incubation at room temperature. Viable counts were determined by drop plating on TSA-1. Assays for pathogenicity were made on elvers (average weight, 10 g) with and without iron pretreatment with hemin (0.28 μg of Fe as hemin component/g of fish) or deferrioxamine B mesylate (Desferal) (250 μg/g of fish) by intraperitoneal inoculation (4, 11). The toxicity of ECPs was evaluated by intraperitoneal injection of 0.1 ml of crude and diluted (1/10) ECP samples per fish. Experiments were made in triplicate and included groups of noninfected fish (inoculated with saline solution) as negative controls.
The subdivision of the species into serovars is based on a serotyping scheme that uses O antigens and antisera against whole cells (10). The O antigens from all Taiwanese isolates gave a clearly positive agglutination with antisera raised against whole cells and LPS from strain E86 (Table 1). No agglutination was detected with the strain used as a negative control (ATCC 27562). Titers measured by microagglutination and ELISA indicated that all Taiwanese strains belonged to serovar E (Table 1).
All marine isolates gave the same profiles in the API system and in conventional tests (Table 2). Like the reference strain E105, these isolates were positive for ornithine decarboxylation, a phenotypic trait that is displayed by more than 20% of characterized strains of serovar E (8, 10). All Taiwanese strains were indole negative both in the API system and in Luria broth. In the other tests, the Taiwanese strains gave the same results as the reference serovar E strains except for fermentation and utilization of d-mannitol (Table 2). The biochemical characterization also included an analysis of resistance to drugs described as serogroup specific (10). The isolates presented a resistance pattern identical to that of reference strains (data not shown); they were resistant to penicillin, ampicillin, polymyxin B, and streptomycin and sensitive to the rest of the drugs tested.
TABLE 2.
Biochemical and physiological profiles of the Taiwanese strains and the reference strains determined by API 20E and conventional tests
| Characteristic | E86 | E105 | ATCC 27562 | Taiwanese strains |
|---|---|---|---|---|
| API code | 5206005 | 5306005 | 5146105 | 5306105 |
| Sensitivity to O129 (150 μg) | + | + | + | + |
| Acid production from glycerol | − | − | + | − |
| Growth at 42°C | − | − | + | − |
| Growth in 6% NaCl | + | + | − | + |
| Utilization of: | ||||
| d-Mannitol | − | − | + | + |
| Lactose | − | − | − | − |
| Hydrolysis of: | ||||
| DNA | + | + | − | + |
| Casein | + | + | + | + |
| Elastin | + | − | + | + (80%) |
| Tween 80 | + | + | − | + |
| Phospholipids | + | + | + | + |
| Fibrinogen | + | + | + | + |
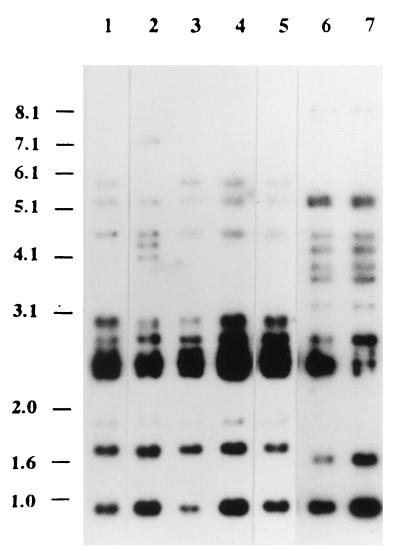
To date, the eel serovar E strains have presented a highly homogeneous ribopattern after digestion with HindIII (8). The Taiwanese strains all displayed a group of four bands typical of the species (8, 13) and showed two profiles which differed from those of reference strains (Fig. 1) but were similar to those of nonserovar E marine isolates (13). These results suggest that the Taiwanese strains have clonal origins different from those of the eel serovar E strains and that ribotyping is more related to the source of the strain than to the serovar. Similar results have recently been obtained with other serovar E strains, which displayed different ribotypes depending on their source: eel or clinical isolate (6).
FIG. 1.
Ribotype profiles of the Taiwanese strains and strains E86 and E105 digested with HindIII. From lane 1 to lane 7, strains CG100, CG106, CG110, CG111, CG118, E86, and E105 are shown. Molecular sizes are indicated in kilobases.
All the well-characterized serovar E strains, irrespective of their sources, are pathogenic for eels and produce lethal exotoxins (2, 7, 8, 10, 20). The virulence degree is dependent on iron availability in host fluids and achieves the highest values in eels treated with hemin, hemoglobin, or Desferal (11). Surprisingly, the marine isolates were avirulent for eels (50% lethal dose [LD50], >108 CFU/fish), even for pretreated eels (LD50, >108 CFU/fish). However, the ECPs of the isolates with the highest proteolytic and hemolytic activities were lethal (Table 3). Therefore, the Taiwanese strains’ lack of virulence cannot be explained by the absence of toxic activity.
TABLE 3.
Enzymatic and lethal activities displayed by ECPs from the Taiwanese strains and the control strains
| Strain | [Protein] (mg/ml)a | Production ofb:
|
Hemolytic titerc | Lethality for eelsd | ||||
|---|---|---|---|---|---|---|---|---|
| Cas | Elas | Fib | Phos | Lip | ||||
| E86 | 1.1 | + (16) | + (8) | + (10) | + (1) | + (2) | 64 | + (1.2) |
| CG100 | 1.04 | + (32) | + (2) | + (8) | − | − | 64 | + (16.4) |
| CG106 | 0.06 | + (64) | + (8) | + (128) | + (1) | + (1) | 128 | + (0.3) |
| CG110 | 1.3 | + (10) | + (2) | + (10) | − | − | 128 | + (20) |
| CG111 | 0.61 | + (8) | + (2) | + (10) | + (1) | + (1) | 256 | + (9.4) |
| CG118 | 0.85 | − | − | + (2) | + (2) | + (5) | 32 | − (>20) |
Protein content of undiluted ECP samples.
Data in parentheses indicate the reciprocal of the highest dilution of ECP with positive activity. Cas, caseinase; Elas, elastase; Fib, fibrinogenolysin; Phos, phospholipase; Lip, lipase.
Titer of hemolysis of eel erythrocytes was expressed as the reciprocal of the highest dilution of ECP with hemolytic activity.
Lethal effect of undiluted ECP. Data in parentheses are the LD50s expressed in micrograms of ECP protein per gram of fish.
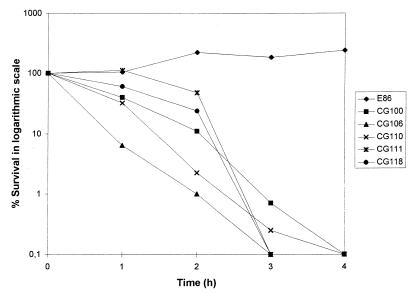
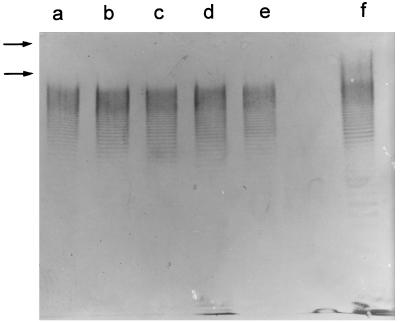
We have previously demonstrated that serum plays an important role in the natural defense of eels against bacterial infections; serovar E strains resist, whereas strains of other serovars activate, the alternative pathway of complement (1). Survival curves of the Taiwanese strains in nonimmune serum showed a dramatic decrease in viable numbers (Fig. 2). Therefore, the susceptibility of the Taiwanese strains to serum could explain their avirulence for eels (remember that vibriosis is a septicemia). Since the O side chain of the serovar E LPS determines, at least in part, the resistance to serum complement (1), we analyzed the LPS of Taiwanese strains. The stained molecule showed the majority of the O side chain bands but did not show those of the highest molecular weight (Fig. 3). Thus, it seems probable that the lack of these bands is related to the susceptibility to serum and, consequently, explains their avirulence for eels. In fact, in bacterial pathogens whose LPS molecules confer resistance to serum, the high-molecular-weight portion is usually the one responsible (14, 17).
FIG. 2.
Survival curves in fresh eel serum. A volume of 20 μl of bacteria at the stationary phase of growth in saline solution (105 cells per ml) was mixed with 20 μl of serum. Viable counts were determined by drop plating of serial dilutions on TSA-1. Each point is the average value for three independent experiments.
FIG. 3.
LPS immunostaining patterns of the Taiwanese strains and strain E86. LPS was immunostained with antiserum against whole cells of strain E86. From lane a to lane f, strains CG110, CG111, CG1118, CG106, CG100, and E86 are shown. The portion of the LPS with the highest molecular weight is indicated by arrows.
In conclusion, the results of this work suggest that the serovar E of V. vulnificus is heterogeneous. It seems to include a pathogenic group, which comprises the strains previously classified as biotype 2, and a nonpathogenic group, which comprises the strains studied in this work and probably other environmental isolates that are as yet uncharacterized (i.e., putative biotype 2 strains isolated from samples collected off the coast of Denmark that are still serologically uncharacterized [12]). The avirulence for eels of these strains seems to be due to their sensitivity to the bactericidal action of serum.
Acknowledgments
This work was partially supported by grant AGF95-1085-CO2-O1 from the Comisión Interministerial de Ciencia y Tecnología (CICYT) and NSC84-2321-B-006-001-B06 from the National Science Council, Taiwan, Republic of China. E. Marco-Noales thanks the Generalitat Valenciana (Plan Valenciano de Ciencia y Tecnología) for a predoctoral fellowship.
We thank Barraclough-Donellan for help with the English text.
REFERENCES
- 1.Amaro C, Biosca E G. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl Environ Microbiol. 1996;62:1454–1457. doi: 10.1128/aem.62.4.1454-1457.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaro C, Biosca E G, Esteve C, Fouz B, Toranzo A E. Comparative study of phenotypic and virulence properties in Vibrio vulnificus biotypes 1 and 2 obtained from a European eel farm experiencing mortalities. Dis Aquat Org. 1992;13:29–35. [Google Scholar]
- 3.Amaro C, Fouz B, Biosca E G, Marco-Noales E, Collado R. The lipopolysaccharide O side chain of Vibrio vulnificus serogroup E is a virulence determinant for eels. Infect Immun. 1997;65:2475–2479. doi: 10.1128/iai.65.6.2475-2479.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaro C, Biosca E G, Fouz B, Toranzo A E, Garay E. Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice. Infect Immun. 1994;62:759–763. doi: 10.1128/iai.62.2.759-763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaro C, Biosca E G, Fouz B, Alcaide E, Esteve C. Evidence that water transmits Vibrio vulnificus biotype 2 infections to eels. Appl Environ Microbiol. 1995;61:1133–1137. doi: 10.1128/aem.61.3.1133-1137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arias C R, Verdonck L, Swings J, Garay E, Aznar R. Intraspecific differentiation of Vibrio vulnificus biotypes by amplified fragment length polymorphism and ribotyping. Appl Environ Microbiol. 1997;63:2600–2606. doi: 10.1128/aem.63.7.2600-2606.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biosca E G, Amaro C. Toxic and enzymatic activities of Vibrio vulnificus biotype 2 with respect to host specificity. Appl Environ Microbiol. 1996;62:2331–2337. doi: 10.1128/aem.62.7.2331-2337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biosca E G, Amaro C, Larsen J L, Pedersen K. Phenotypic and genotypic characterization of Vibrio vulnificus: proposal for the substitution of the subspecific taxon biotype for serovar. Appl Environ Microbiol. 1997;63:1460–1466. doi: 10.1128/aem.63.4.1460-1466.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biosca E G, Marco-Noales E, Amaro C, Alcaide E. An enzyme-linked immunosorbent assay for detection of Vibrio vulnificus biotype 2: development and field studies. Appl Environ Microbiol. 1997;63:537–542. doi: 10.1128/aem.63.2.537-542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biosca E G, Oliver J D, Amaro C. Phenotypic characterization of Vibrio vulnificus biotype 2, a lipopolysaccharide-based homogeneous O serogroup within Vibrio vulnificus species. Appl Environ Microbiol. 1996;62:918–927. doi: 10.1128/aem.62.3.918-927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouz B, Mazoy R, Lemos M L, del Olmo M J, Amaro C. Utilization of hemin and hemoglobin by Vibrio vulnificus biotype 2. Appl Environ Microbiol. 1996;62:2806–2810. doi: 10.1128/aem.62.8.2806-2810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Høi L, Larsen J L, Dalsgaard I, Dalsgaard A. Occurrence of Vibrio vulnificus biotypes in Danish marine environments. Appl Environ Microbiol. 1998;64:7–13. doi: 10.1128/aem.64.1.7-13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hor L I, Goo C T, Wan L. Isolation and characterization of Vibrio vulnificus inhabiting the marine environments of the Southwestern area of Taiwan. J Biomed Sci. 1995;2:384–389. doi: 10.1007/BF02255226. [DOI] [PubMed] [Google Scholar]
- 14.Joiner K A, Grossman N, Schmetz M, Levine L. C3 binds preferentially to long-chain lipopolysaccharide during alternative pathway activation by Salmonella montevideo. J Immunol. 1986;136:710–715. [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Marco-Noales E, Biosca E G, Amaro C. Abstracts of the Seventh International Conference on Diseases of Fish and Shellfish. Palma de Mallorca, Spain: European Association of Fish Pathologists; 1995. Long-term survival of Vibrio vulnificus biotype 2 at different salinities; p. 75. [Google Scholar]
- 17.Merino S, Albertí S, Tomás J M. Aeromonas salmonicida resistance to complement-mediated killing. Infect Immun. 1994;62:5483–5490. doi: 10.1128/iai.62.12.5483-5490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver J D. Vibrio vulnificus. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 570–600. [Google Scholar]
- 19.Song Y-L, Cheng W, Shen C H, Ou Y C, Song H B. Occurrence of Vibrio vulnificus infections in cultured shrimp and eel in Taiwan. NSC Symp Ser. 1990;16:172–179. [Google Scholar]
- 20.Tison D L, Nishibuchi M, Greenwood J D, Seidler R J. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl Environ Microbiol. 1982;44:640–646. doi: 10.1128/aem.44.3.640-646.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veenstra J, Rietra J P G M, Coster J M, Stoutenbeek C P, Ter Laak E A, Haenen O L M, De Hier H H W, Dirsks-Go S. Human Vibrio vulnificus infections and environmental isolates in The Netherlands. Aquacult Fish Manag. 1993;24:119–122. [Google Scholar]