Abstract
The high lipid content of the brain, coupled with its heavy oxygen dependence and relatively weak antioxidant system, makes it highly susceptible to oxidative DNA damage that contributes to neurodegeneration. This study is aimed at identifying specific ROS-responsive miRNAs that modulate the expression and activity of the DNA repair proteins in human astrocytes, which could serve as potential biomarkers and lead to the development of targeted therapeutic strategies for neurological diseases. Oxidative DNA damage was established after treatment of human astrocytes with 10μM sodium dichromate for 16 h. Comet assay analysis indicated a significant increase in oxidized guanine lesions. RT-qPCR and ELISA assays confirmed that sodium dichromate reduced the mRNA and protein expression levels of the human base-excision repair enzyme, 8-deoxyguanosine DNA glycosylase 1 (hOGG1). Small RNAseq data were generated on an Ion Torrent™ system and the differentially expressed miRNAs were identified using Partek Flow® software. The biologically significant miRNAs were selected using miRNet 2.0. Oxidative-stress-induced DNA damage was associated with a significant decrease in miRNA expression: 231 downregulated miRNAs and 2 upregulated miRNAs (p < 0.05; >2-fold). In addition to identifying multiple miRNA-mRNA pairs involved in DNA repair processes, this study uncovered a novel miRNA-mRNA pair interaction: miR-1248:OGG1. Inhibition of miR-1248 via the transfection of its inhibitor restored the expression levels of hOGG1. Therefore, targeting the identified microRNA candidates could ameliorate the nuclear DNA damage caused by the brain’s exposure to mutagens, reduce the incidence and improve the treatment of cancer and neurodegenerative disorders.
Keywords: astrocytes, oxidative stress, DNA repair, microRNA
Introduction
Impairment and mutations in the DNA repair pathways in glial cells are associated with neurodegenerative disorders and premature aging (Borgesius et al., 2011). The brain is a prime target of reactive oxygen species (ROS), because of its heavy oxygen dependence, high energy requirements (high glucose metabolism), extensive lipid composition, and more importantly, its relatively weak antioxidant system in comparison to other tissues (Yamamoto et al., 2007). Despite the body’s antioxidant defense, an imbalance between the production of ROS and the repair of oxidative damage can potentially result in DNA mutations. Oxidative DNA damage and mutations are implicated in cancer development and progression, as well as aging and age-related neurological disorders (Cooke et al., 2003). Glial cells are continuously attacked by ROS, and effective repair of ROS-induced 8-OHdG accumulation is critical to maintaining brain function and reducing the rate of C: G to A: T transversion mutation. Astrocytes play a critical role in modulating synaptic transmissions, regulating energy metabolism, water, and ion homeostasis, protecting neurons from oxidative stress, and regeneration of the CNS (Bélanger et al., 2011; Pfrieger, 1997). Therefore, human astrocytes are an ideal biological model to investigate the relationship between oxidative stress and DNA damage and repair mechanisms. Understanding the epigenetic regulation of astrocyte DNA repair mechanisms could lead to the identification of novel targets for the treatment of CNS disorders. Considering the important role in maintaining brain activity, astrocytes can be regarded as a potential target for therapies aimed at the prevention and cure of age-related neurodegenerative diseases (Kim & M. Wilson III, 2012).
Nucleic acids are susceptible to oxidation by environmental and endogenous factors. ROS can induce oxidative modification of the DNA bases, producing lesions including the ring-opened formamide-pyrimidine derivatives of guanine and adenine, 8-hydroxy-2′-deoxyguanosine (8-OHdG), formyluracil, dihydroxyuracil, and thymine glycol (Perlow-Poehnelt et al., 2004). Among the five nucleobases, guanine is the most susceptible to oxidation because of its high electron density. Thus, the most common biomarker of ROS-induced DNA damage is the mutagenic base 8-OHdG, an oxidized derivative of deoxyguanosine, which causes DNA mutations by incorporation of adenine instead of cytosine during DNA replication resulting inC: G to A: T substitutions in the DNA (Valavanidis et al., 2009). The concentration of 8-OHdG within a cell is a well-established marker of nucleic acid oxidative damage and an indicator of oxidative stress (Kasai, 1997). Elevated levels of 8-OHdG have been reported in leukemia (Sentürker et al., 1997), breast cancer (Malins & Haimanot, 1991), colorectal cancer (Oliva et al., 1997), lung cancer (Vulimiri et al., 2000), Parkinson disease (Zhang et al., 1999), and Alzheimer’s disease (Lezza et al., 1999).
To preserve genomic integrity, eukaryotic cells need a complex but synergistic DNA damage response (DDR) network of proteins. The DDR loop is a kinase-based signal transduction network that initiates phosphorylation-driven cascades (Tessitore et al., 2014) that acts by transient coordination of DNA repair, replication, cell cycle progression, telomere homeostasis, and the subsequent induction of permanent cell cycle arrest, or apoptosis if the damage cannot be repaired (Rinaldi et al., 2021). Base excision repair (BER) is the mechanism for the excision and replacement of oxidative DNA damage. Single nucleotides modified by methylation, alkylation, deamination, or oxidation are removed by BER (Natarajan & Palitti, 2008). DNA glycosylases recognize and remove oxidative DNA base damage in a substrate-specific manner. One type of DNA glycosylase is the human 8-deoxyguanosine DNA glycosylase 1 (hOGG1), a bi-functional 8-OHdG-specific glycosylase that plays an important role in reducing the rate of mutation. By providing the first line of defense against oxidative DNA damage, OGG1 plays an important role in decreasing the detrimental effects of oxidative DNA damage (Liu et al., 2011). In addition, OGG1 is involved in oxidative stress-induced DNA demethylation by interacting and recruiting TET1, a DNA-binding protein that modulates DNA methylation, to the 8-oxoG lesion (Zhou et al., 2016). A single nucleotide polymorphism changing serine to a cysteine residue at position 326 [Cys-326] has been linked to decreased OGG1 activity, and thus, has been used as a marker of individual susceptibility to sodium dichromate induced DNA damage and reduced antioxidant capacity (Lee et al., 2005). However, the focus of these published studies was on the genetic changes that influence OGG1 activity in cells and did not consider non-coding RNA regulation.
MicroRNAs (miRNAs) are small non-coding RNAs (19–23nts) that inhibit gene expression at the post-transcriptional level (Kim & Nam, 2006), and play a pivotal role in the DNA Damage Response (DDR) that leads to changes in miRNA expression (Liu & Lu, 2012). MiR-182 expression is inversely correlated with breast cancer type 1 susceptibility (BRCA1) protein levels that impair homologous recombination-mediated DNA repair pathway (Moskwa et al., 2011). MiR-203 inhibits DNA damage repair via the PI3K/AKT and JAK/STAT3 pathways and contributes to the modulation of radiation sensitivity in human malignant glioma cells (Chang et al., 2016). Overexpression of miR-92 is associated with an increased accumulation of oxidative DNA damage and immortalization in hepatocellular carcinoma (Romilda et al., 2012). Astrocyte-derived miR-181, miR-29, and miR-146a enhance neuron survival after cerebral ischemia (Ouyang et al., 2014). Although much progress has been made in elucidating the function of various miRNAs, there is a paucity of scientific data regarding the specific miRNAs that regulate the proteins involved in the base excision repair pathway. Changes in microRNA expression profile and ROS signaling have been associated with tumor development, progression, metastasis, and therapeutic response prompting investigations into possible crosstalk between ROS and microRNAs. Emerging evidence suggests a reciprocal connection between ROS signaling and the miRNA pathway whereby ROS induces epigenetic alterations of miRNA genes (Lu et al., 2020). It has been proposed that ROS are involved in various steps of miRNA biogenesis via several mechanisms which include: inhibition and enhancement of the expression of certain miRNA genes through the epigenetic regulatory enzymes, DNMT1 and HDACs, the regulatory activation of stress-related transcription factors such as p53, nuclear factor (NF)-κB to induce miRNA expression (Dansen & Burgering, 2008; Reuter et al., 2010), and the recruitment of the miRNA processing machinery, Drosha, and Dicer, which can be directly or indirectly regulated by ROS (He & Jiang, 2016). DNA damage also regulates the biogenesis of miRNA expression at the transcriptional level via the p53 pathway (Hu & Gatti, 2011).
Currently, there are no reports on the high-throughput analysis of miRNA expression following exposure of human astrocytes to oxidative DNA damage. With the cross-talk between ROS and miRNA-processing enzymes (He & Jiang, 2016), there is a very high possibility that closely related miRNAs and their target genes would be involved in mediating the oxidative stress-induced cellular response. Therefore, it is hypothesized that ROS-induced DNA damage will alter the epigenomic (miRNA expression) patterns of affected cells and modulate functionally related DDR-associated genes. This scientific hypothesis was tested by performing high-throughput small RNA sequencing and functional analysis to identify novel ROS-induced differentially expressed miRNAs that modulate the expression of genes that are associated with the DNA repair mechanism in human astrocytes (Figure 1). The discovery of ROS-responsive miRNAs provides a potential novel strategy to specifically overcome ROS-mediated pathological conditions. An increased understanding of how miRNAs regulate DNA repair in astrocytes will expand the potential for miRNA-based therapeutics that will substantially improve the outlook for patients with cancer and age-related neurodegenerative disorders.
Figure 1.
Schematics of experimental workflow that includes induction of oxidative DNA damage using 10µM sodium dichromate treatment followed by small RNA sequencing, computational analysis of differentially expressed targets, and miR inhibition functional analysis.
Materials & Methods
Cell Culture and Sodium Dichromate Treatment
Human primary astrocytes (Sciencell, Carlsbad, CA) were cultured in T-25 flasks containing 5 mL of Astrocyte Medium (Sciencell, #1801), supplemented with 1% penicillin/streptomycin solution (Sciencell, #0503), 2% Fetal Bovine Serum (Sciencell), and 1% astrocyte growth supplement (Sciencell #1852). The cells were grown at 37°C, 95% humidity, and 5.0% CO2/air until they reached 80% confluence. A uniform seeding density of 2.1 × 106 cells per flask was maintained during subculture using Countess II FL Automated Cell Counter (ThermoFisher Scientific, Waltham, MA). All experiments were performed using cells with a maximum passage of three. The cells were treated with 10 µM sodium dichromate and incubated for 16 h before being harvested for analysis. Non-treated cells were maintained in parallel and used as control. Before cell harvest, microscopic images of control (non-treated) and treated cells were collected to evaluate the changes in cell morphology following treatments. Both treated and untreated cells were detached using trypsin, collected, and counted using Countess automated cell counter (ThermoFisher Scientific). Each experimental group included three biological replicates.
Comet Assay and ROS Assessment
Human astrocytes were grown in T-25 flasks to 80% confluency and treated with 10 µM sodium dichromate for 16 h. Trypsin was added to both the control (non-treated) and treated cells for 5 min at 37°C to detach the cells. 15,000 cells were then collected and centrifuged for 5 min at 4°C. The cell pellets were suspended in 200 µL of 6% low-melting-point agarose (pre-warmed to 37°C). The agarose cell suspension (60 µL) was pipetted onto an agarose-coated coverslip and placed over a slide. The coverslip was removed after 10 min and the samples were placed in lysis buffer (100 mM EDTA, 2.5 M NaCl, 10 mM Tris-HCl, 1% Triton-X, pH 10) at 4°C overnight. The slides were placed in chilled ddH2O for 5 min, rinsed one more time with chilled ddH2O for 10 min, and washed with enzyme reaction buffer (40 mM HEPES, 0.1 M KCL, 0.6 mM EDTA, 0.2 mg mL−1 BSA, pH 8). Formamidopyrimidine [fapy]-DNA glycosylase (FPG, New England Biolabs) was diluted in the enzyme reaction buffer to a final concentration of 8 U mL−1. Both treated and control samples were incubated with either enzyme reaction buffer or FPG for 30 min at 37°C. The slides were placed in the electrophoresis buffer for 20 min to allow equilibration and gel electrophoresis was performed for 20 min at 30 V and 300 mA. The slides were placed in a neutralization buffer (0.4M Tris-HCL, pH 7.5) for 20 min, rinsed with ddH2O for 10 min, and allowed to dry overnight at 37°C. The samples were rehydrated in water the next day and stained with 2.5mg mL−1 propidium iodide (ThermoFisher Scientific, cat. #P1304MP). After 20 min of incubation with the dye, the slides were rinsed with ddH2O for 10 min and dried overnight at 37°C. Comets were visualized using a Nikon Eclipse Ti microscope at 150× magnification. The OpenComet plugin for Image J was used to quantify the tail moment from the tail length and staining intensity of the head and tail (Gyori et al., 2014; Xiao et al., 2019). At least 70 cells were analyzed for each condition.
The ROS Assay Kit (OZ Bioscience) measures ROS activity within the cell using the cell-permeable fluorogenic probe. The cells were seeded in a 96-well plate and the two experimental groups included 10 replicates each. Briefly, the cells were incubated with 12mM fluorogenic probe 2′, 7′- Dichlorodihydrofluorescein diacetate (DCF-DA) for 30 min at 37°C followed by a PBS wash. The fluorescence was measured using a Tecan Infinite F200 Fluorescence Microplate Reader (ex.485nm/ em.535nm). The fluorescent intensity was normalized to the total number of cells in each well.
Small RNA Sequencing and Bioinformatics Analysis
The human astrocytes (untreated and 10 µM Sodium Dichromate-treated) were lysed and the small RNA fraction was enriched using mirPremier® MicroRNA Isolation Kit (Sigma-Aldrich, Cat. # SNC10). Six samples were processed for Small RNA sequencing analysis that includes 3 biological replicates in the treatment and control. The concentration of small RNA fraction was determined using a Qubit RNA HS Assay Kit (ThermoFisher Scientific, Cat. # Q32852) using Qubit 4 Fluorometer (ThermoFisher Scientific, cat. #Q33238). Next-generation sequencing (NGS) small RNA libraries were size-selected to enrich for constructs containing mature miRNAs. The sequencing was performed on Ion Torrent™ next-generation sequencing systems (ThermoFisher Scientific, Waltham, MA). Computational analysis of the FASTQ data files that include the raw data was performed using the microRNA-Seq algorithms of the Partek ® Flow® software, v10.0 (Partek Inc., 2020). The reads were aligned using the Bowtie algorithm and processed via Partek’s default MicroRNA-Seq pipeline that mapped the reads to a reference genome (Homo sapiens, hg38) indexed to miRBase Mature MicroRNA (version 21). The differential gene expression analysis was performed using Partek’s Gene Specific Analysis (GSA) algorithm to identify a list of significantly differentially expressed miRNAs (≥ 2-fold, FDR< 0.01, p-value < 0.05). Hierarchical clustering and visualization plots were generated on the Partek ® platform to aid the visualization and interpretation of the data. The biological processes and genes targeted by the differentially expressed miRNAs were identified using miRNet 2.0 (Chang et al., 2020). Bioinformatics analysis for differentially expressed (DE) miRNA species that selectively hybridize to the 3′ UTR of OGG1 mRNA was carried out, and a hit in at least two of the used miRNA-mRNA interaction databases was set as the satisfying criterion.
Reverse Transcription Quantitative PCR (RT-qPCR)
RT-qPCR was performed to validate the changes in the expression levels of the identified miRNAs and their mRNA targets. The MicroRNAs, hsa-miR-335-5p (Sigma Aldrich, cat. #MIRAP00323) and hsa-miR-1248 (Sigma Aldrich, cat. #MIRAP00761), were validated using the MystiCq® MicroRNA Quantitation System (Sigma-Aldrich, St. Louis, MO). Briefly, cDNA was synthesized from the treated and untreated small RNA samples using MystiCq™ microRNA cDNA Synthesis Mix Kit according to the manufacturer’s instructions. The cDNA concentration was measured using a Qubit 4 Fluorometer (Thermo Fisher Scientific, cat. #Q32854). Equal amounts of each cDNA (2ng), mixed with 10 µM of each MystiCq microRNA qPCR Assay Primer and 10 µM of MystiCq Universal PCR Primer plus MystiCq microRNA SYBR Green qPCR Ready-mix, were amplified as recommended by the manufacturer on a LightScanner 32 real-time PCR instrument. Relative quantification of miRNA expression was determined by using the Livak-Schmidt (2−ΔΔCT) method and normalized against SNORD 44 provided with the MicroRNA Quantitation System (Masè et al., 2017; Morata-Tarifa et al., 2017). The results are represented as mean ± standard error calculated from three biological replicates.
The mRNA targets, OGG1 and PARP1, were reverse-transcribed separately using the TaqMan™ RNA-to-CT™ 1-Step Kit (ThermoFisher Scientific, cat. # 4392653) and LUNA One-step RT-qPCR kit (New England BioLabs Inc., cat. #E3005S) respectively, on a QuantStudioTM 3 Real-time PCR system (Applied Biosystems, Cat. # A28567). Total RNA was harvested individually from treated and control cells with an RNA Miniprep Kit (Zymo Research, cat. #R1054), while the quantity and quality of the extracted RNA were determined by the Nanodrop 2000 system (Thermo Fisher Scientific, Waltham, MA). Equal amounts of RNA (6 ng) were mixed with 10 µM forward and reverse primers (PARP 1 and GAPDH) to 0.5 µM final concentration, and reverse-transcribed for 10 min at 55°C, followed by inactivation of reverse transcriptase and cDNA denaturation at 95°C for 1 min. The subsequent amplification followed the vendor-recommended 40-cycle program of 10 s at 95°C, 30 s at 60°C, and 10 s at 95°C per cycle at a ramp heating/cooling rate of 20°C sec−1. The GAPDH and PARP1 gene primer sequences (Integrated DNA Technologies, Coralville, ID) are shown in Table 1. OGG1 mRNA expression level was analyzed using OGG1 and GAPDH TaqMan™ Gene Expression Assay (FAM) (ThermoFisher Scientific cat. # 4453320, 4448892), with an initial reverse transcription of 6 ng RNA at 48°C for 15 min, followed by AmpliTaq Gold® DNA polymerase activation at 95°C for 10 min, and a final 40-cyle amplification of 95°C for 15 s and 60°C for 1 min. Data were normalized to the CT values of the internal control gene GAPDH, and analyzed using the 2−ΔΔCT method (Livak & Schmittgen, 2001).
Table 1.
Forward and Reverse Primer sequences.
| Primer | Oligonucleotide sequences |
|---|---|
| GAPDH | F 5’– ACA TCG CTC AGA CAC CAT G – 3’ |
| R 5’– TGT AGT TGA GGT CAA TGA AGG G – 3’ | |
| PARP-1 | F 5’-CGC ATA CTC CAT CCT CAG TG- 3’ |
| R 5’–GGA TCA GGG TGT AAA AGC GAT – 3’ |
miR-1248 Knockdown: PCR Analysis of OGG1 mRNA Expression Level
Human astrocytes were seeded at a uniform density of 3 × 105 cells per well in 6-well plates and incubated for 72 h. At 80% confluency, the cells were transfected separately in triplicates with mirVana® hsa-miR-1248 inhibitor (ThermoFisher Scientific, Cat. # 4464084) and with mirVana® negative control #1 (ThermoFisher Scientific, Cat. # 4464076), according to manufacturer's protocol. Briefly, the miR-1248 inhibitor and the negative control #1 were reconstituted to 10 µM stock concentrations with nuclease-free water, and then 3 µL of each was diluted in 150 µL Opti-MEMTM I Reduced Serum Medium (ThermoFisher Scientific, Cat. # 31985062) to obtain 30 pmol per well. 9 µL LipofectamineTM RNAiMAX Transfection Reagent (ThermoFisher Scientific, Cat. # 13778100) was diluted in 150 µL Opti-MEMTM I Reduced Serum Medium. Both diluted reagents were mixed at a 1:1 ratio and the resulting miRNA-lipid complex was incubated for 5 min at room temperature. The transfection reagent mix was added to cells at 250 µL per well, so that the final amount of miRNA inhibitor or negative control used per well is 25 pmol, while the final amount of LipofectamineTM RNAiMAX Transfection Reagent per well was 7.5 µL. The transfected cells were incubated at 37°C for 48 h. Control experiments of non-transfected cells were also maintained in triplicate. The efficacy of the transfection protocol was confirmed by assessing the expression levels of miR-1248 post-inhibition, using an hsa-miR-1248 primer (Sigma Aldrich, cat. #MIRAP00761) and the MystiCq® MicroRNA Quantitation System (Sigma-Aldrich, St. Louis, MO), as earlier described, on a QuantStudioTM 3 Real-time PCR system (Applied Biosystems, Cat. # A28567).
To assess the expression levels of the OGG1 mRNA after inhibitions of miR-1248, total RNA was extracted from the cells using the Quick-RNATM Miniprep Kit (Zymo Research, Cat. # R1054), and quantified with the QuibitTM RNA HS Assay Kit (ThermoFisher Scientific, Cat. # Q32852), used with the QuibitTM 4 Fluorometer (ThermoFisher Scientific, Cat. # Q33238). RT-qPCR was performed with a TaqMan™ RNA-to-CT™ 1-Step Kit (ThermoFisher Scientific, cat.# 4392653) to analyze the gene expression level of OGG1 using GAPDH as an internal control (ThermoFisher Scientific cat.# 4453320, 4448892).
miR-1248 Knockdown: Proteomics Analysis of OGG1 Expression
Protein samples from three experimental groups (untreated, 10μM sodium dichromate-treated, and miR-1248 transfected) were concentrated at 5mg mL−1 using Protein-Concentrate Kit Micro (Millipore, cat.#2100). The OGG1 protein expression in the samples was measured using the Human OGG1 Sandwich ELISA Kit (LS-BIO, cat. # LS-F6751), following the vendor’s protocol. Briefly, 100 µL of each sample, standard reagent, or blank was added to separate wells, covered with a plate sealer. After incubation for 1 h at 37°C, the liquid was aspirated and the wells were incubated with 100 µL of Detection Reagent A for 1 h at 37°C. After a wash step, 100 µL Detection Reagent B was added for 30 min at 37°C followed by a 90 µL TMB substrate solution. The wells were incubated at 37°C for 15 min, in the dark and a 50 µL Stop solution was added to each well. The optical density of each well was measured at 450 nm using a Chromate 4300 microplate reader. The OGG1 protein quantification for each sample was extrapolated from a standard curve of concentration vs OD450.
Statistical Analysis
Statistical analyses were performed using GraphPad 6.0.1 software (GraphPad Software Inc., San Diego, CA, USA). A p-value less than 0.05 was considered significant and the results are expressed as mean ± SEM.
Results
Sodium Dichromate Induces Oxidative DNA Damage in Human Astrocytes
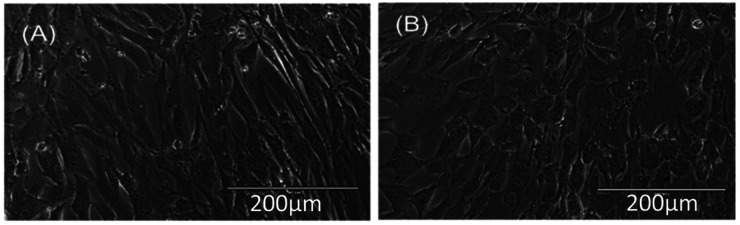
Sodium dichromate treatment was associated with reduced growth and distinct morphological changes in human astrocytes. Phase-contrast microscope images of the untreated spindle-like astrocytes displayed a typical cellular structure (Figure 2A) while 16 h of treatment with 10 µM sodium dichromate (Figure 2B) results in cytomorphological changes, cytoplasmic granulation, and reduced density. The average number of attached cells in the untreated and treated groups were 5.15 × 105 cells and 2.34 × 105 cells indicating a 45% reduction in adhesion.
Figure 2.
Bright-field image of human astrocytes, 10× magnification (A) control, and (B) treated with 10µM sodium dichromate.
A formamidopyrimidine-DNA glycosylase (FPG) modified comet assay was performed to assess the sodium dichromate-induced oxidative DNA damage. The comet assay detects DNA strand breaks by measuring the migration of DNA from individual nuclei in an alkaline environment, and can also detect oxidative DNA damage when the nuclei are treated with a DNA glycosylase enzyme (Lee et al., 2004). The average tail moment was used as a measurement of the magnitude of strand breakage (-FPG) or oxidative DNA damage (+FPG). The sodium dichromate-treated cells displayed longer tail lengths and tail moments in comparison to the control samples (Figure 3). Elongated tails and larger tail moments are consistent with the fragmentation of nuclei due to strand breakage or recognition and removal of damaged bases by FPG. FPG recognizes 7, 8-dihydro-8-oxoguanine (8-oxoguanine), 8-oxoadenine, fapy-guanine, methy-fapy-guanine, fapy-adenine, 5-hydroxy-cytosine, and 5-hydroxy-uracil (Tchou et al., 1994). The sodium dichromate treatment was associated with an increased accumulation of cellular ROS (Figure 3C).
Figure 3.
Sodium dichromate increases oxidative DNA base damage. The alkaline comet assay with FPG treatment was used to detect oxidative base damage following 10 μM Na2Cr2O7 treatment for 16 h (A) and the tail moment was measured using OpenComet (B). Analysis was performed on one experiment with at least 70 cells in each experimental group. Error bars represent SD and **** represents P < 0.0001 using a Student's t-test. (C) ROS levels in untreated and treated cells, n = 10.
Small RNA Sequencing Identifies a Large Number of Differentially Downregulated MicroRNAs
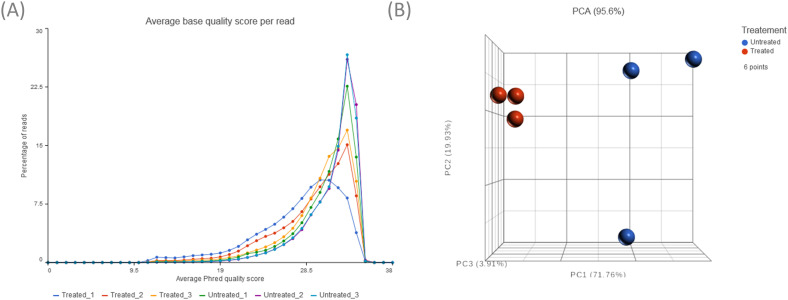
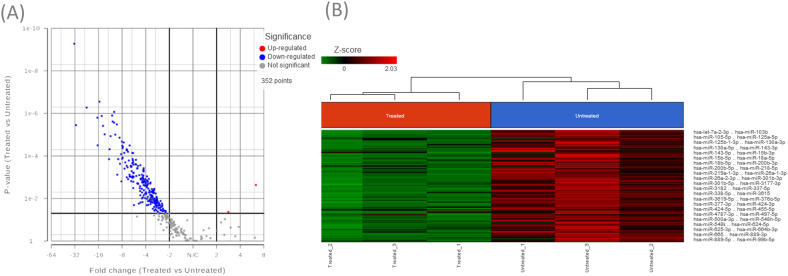
This study identified a subset of differentially expressed miRNAs after 10 µM sodium dichromate treatment. Quality control analysis was performed on Partek® Flow® software, v10.0 to obtain the average base quality score per reading. The average Phred quality score ranged from 28% to 31% (Figure 4A). Phred score of 20% signifies that the specific base call is 99% accurate, while 30% means it is 99.9% accurate, confirming that the accuracy of the raw reads is within acceptable limits. A principal component analysis (PCA) plot was generated to identify outliers among the two treatment groups (Figure 4B). A Volcano plot was generated to show the distribution of upregulated and downregulated miRNA genes (Figure 5A). Hierarchical sample clustering via heatmap diagram was generated to visualize the sample clustering of the differentially expressed miRNAs (Figure 5B). A coverage breakdown for each sample confirmed that on average 20% of the reads mapped fully or partly within a microRNA (see supplemental figure 1). This is not unexpected, especially for the human genome that includes large segments of intronic regions interspersing the gene-coding sequences (Palazzo & Gregory, 2014). After filtering based on ≥ 2-fold and p < 0.05 statistical significance, 231 miRNAs were found to be downregulated, while only 2 were upregulated (supplemental Table 1).
Figure 4.
A) Pre-alignment QA/QC showing average base quality score per reading. The Phred quality scores of the analyzed samples ranged from 28% to 31%. B) Principal Component Analysis (PCA) plot showing clustering of the treated and control samples.
Figure 5.
A) Volcano plot depicting the distribution of upregulated and downregulated miRNA genes in treated samples relative to controls. (B) Unsupervised hierarchical clustering using the differentially expressed miRNAs between treated and control samples represented as a heat map. The heat map colors correspond to microRNA expression as indicated in the color key: Red (up-regulated) and Green (down-regulated).
MiRNet Functional Enrichment Analysis Identifies Physiological and Pathological Processes That are Regulated by Differentially Expressed miRNAs
Gene Ontology analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database identified 49 miRNAs and 13,585 target genes involved in molecular pathways associated with signaling, cell cycle control, and DNA damage and repair (supplemental tables 1 and 2). The most statistically significant pathways include cancerogenesis and cell cycle control (Table 2). These major pathways were obtained from three combined databases (miRTarBase v8.0, TarBase v8.0, and miRecords) hosted on the miRNet 2.0. Further analysis of the miRNA-disease associations identified 21 mRNAs that were experimentally validated to be linked to 143 pathologies (supplemental Table 3). MiR-107 was linked to more brain-linked pathologies than all of the other miRNAs, followed by miR-9-5p, miR-128-3p, miR-125b-5p, miR-181b-5p, and miR-139-5p. Empirically proven protein-protein interactions (PPIs) were also investigated on miRNet-hosted STRING Interactome, v11 (Szklarczyk et al., 2019), set at a confidence score cutoff of 500. A list of 18,553 proteins interacting with one another was generated (supplemental Table 4).
Table 2.
Top pathways enriched in putative miRNA-targeted genes.
| Biological Process/Pathway | Adj. p-Value | Genes |
|---|---|---|
| Pathways in cancer | 2.11E-16 | 285 |
| Cell cycle | 2.79E-11 | 120 |
| Neurotrophin signaling pathway | 3.24E-07 | 114 |
| Wnt signaling pathway | 3.19E-06 | 129 |
| ErbB signaling pathway | 8.73E-06 | 81 |
| RNA transport | 1.15E-05 | 113 |
| Glioma | 1.29E-05 | 62 |
| Axon guidance | 1.71E-05 | 106 |
| p53 signaling pathway | 3.11E-05 | 64 |
| Jak-STAT signaling pathway | 0.000241 | 88 |
| MAPK signaling pathway | 0.000362 | 218 |
| mTOR signaling pathway | 0.001303 | 42 |
| Pyrimidine metabolism | 0.001896 | 87 |
| RNA degradation | 0.001896 | 54 |
| Ribosome biogenesis in eukaryotes | 0.004891 | 49 |
| Purine metabolism | 0.004891 | 134 |
| Apoptosis | 0.006644 | 71 |
| Alzheimer's disease | 0.015065 | 43 |
| Prion diseases | 0.016875 | 20 |
| mRNA surveillance pathway | 0.027838 | 68 |
| Huntington's disease | 0.03759 | 25 |
| VEGF signaling pathway | 0.05427 | 62 |
MiRNAs and DNA Repair Genes: Computational Analysis and Target Validation
Among the differentially expressed microRNAs (supplemental table 1), 10 downregulated miRNAs were identified using miRNet 2.0 to target DNA repair proteins. These include miR-15a-5p, miR-16-5p, miR-17-5p, miR-93-5p, miR-125a-5p, miR-125b-5p, miR-128-3p, miR-155-5p, miR-335-5p, let-7b-5p (see Figure 6, Table 3). The annotations for two upregulated miRNAs, miR-1248 and miR-4284, were discovered on miRDB (Chen & Wang, 2020). In silico prediction of OGG1-targeting microRNA candidates via a search on miRDB (supplemental Table 5), filtered by a target prediction score of ≥75 and excluding miRNAs with more than 2000 predicted targets in the human genome, returned 32 positive results which included the differentially upregulated miR-1248 in our RNA-seq data set (ranked 10th with a score of 88). miR-1248 got a second positive call on miRWalk (supplemental Table 6).
Figure 6.
A coherent group of miRNAs target DNA repair proteins. Network constructed on miRNet 2.0.
Table 3.
List of Differentially Expressed BER-Associated MicroRNAs (p < 0.05).
| Regulation Post-Treatment | miRNA ID | Fold Change | Experimentally validated DNA repair gene targets |
|---|---|---|---|
| Downregulated | hsa-miR-16-5p | 10.1 | UNG/UDG, MBD4, NTHL1 |
| hsa-miR-128-3p | 8.05 | APEX1 | |
| hsa-miR-17-5p | 7.97 | APEX1 | |
| hsa-miR-93-5p | 5.89 | NTHL1, MPG | |
| hsa-miR-125b-5p | 5.78 | UNG/UDG, TDG | |
| hsa-miR-125a-5p | 4.29 | TDG | |
| hsa-miR-15a-5p | 4.28 | MBD4 | |
| hsa-miR-155-5p | 3.19 | MUT | |
| hsa-miR-335-5p | 2.81 | PARP1 | |
| hsa-let-7b-5p | 2.48 | MPG, FEN1 | |
| Upregulated | hsa-miR-4284i | 6.33 | No Hit |
| hsa-miR-1248i | 2.82 | OGG1 i |
No hits were found on the miRNet-hosted databases as the miRNAs have not been annotated. Both miRNAs had several hits on miRDB. Only miR-1248 had a BER target protein, OGG1.
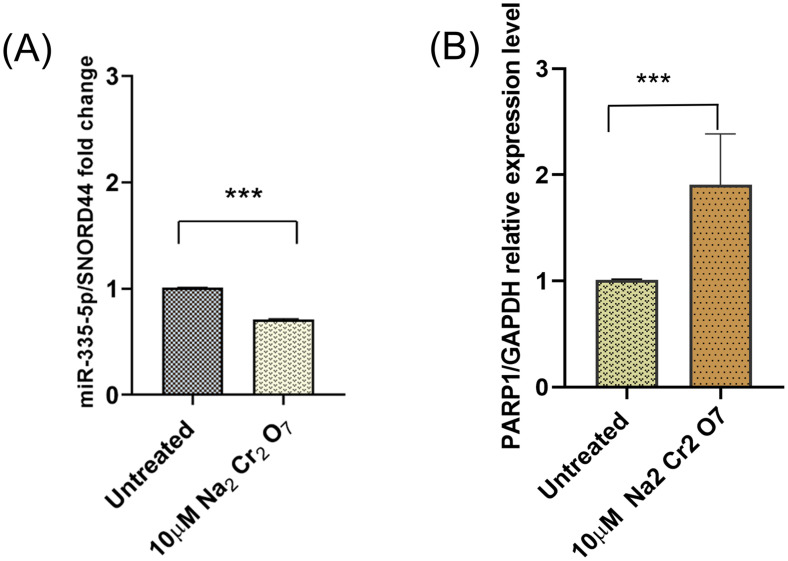
Selected miRNA-mRNA target pairs were further analyzed to validate the predicted regulatory network. The expression levels of OGG1, a canonical BER enzyme that is directly involved in base-excision, and Poly-(ADP-Ribose) polymerase 1 (PARP-1), an auxiliary DNA damage repair protein (Lai et al., 2019), were analyzed. Interestingly, OGG1 binding to PARP-1 plays a functional role in the repair of oxidative DNA damage (Hooten et al., 2011). The relationship between miR-335 and PARP1 was previously experimentally validated (Luo et al., 2017). The computational analysis predicts miR-1248 binding with the 3′ UTR region of OGG1 and the miRNA-mRNA relationship was validated via miRNA inhibitor transfection experiments. RT-qPCR of the expression levels of the selected miRNAs and their target genes validate the opposite expression pattern that is consistent with the negative regulatory effect of miRNAs on their mRNA targets. PARP-1 was significantly upregulated and miR-335 downregulated in treated astrocytes compared to the non-treated samples. PARP-1 is a target of miR-335 (Figure 6, Table 3) and the PCR results (Figure 7) validated the trend obtained from the small RNA-seq analysis.
Figure 7.
RT-qPCR confirmed (A) downregulation of miR-335, and (B) upregulation of its target mRNA, PARP-1 (p < <0.001), n = 3.
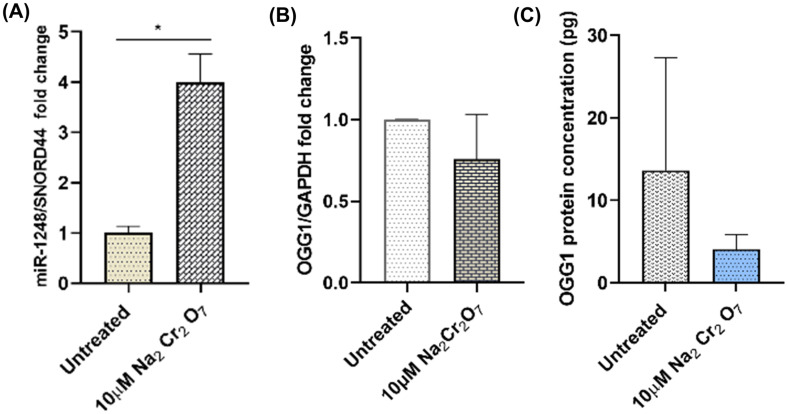
PCR analysis indicates that miR-1248 is upregulated while its computationally predicted target mRNA, OGG1 is decreased by 25% (Figure 8A, B). The OGG1 protein level also decreased (Figure 8C). These findings are in agreement with the established inverse expression relationship between miRNAs and their target mRNAs.
Figure 8.
Effect of sodium dichromate treatment (10µM, 16 h) on (A) miR-1248, P < 0.05, (B) OGG1 mRNA, and (C) OGG1 protein expression levels (n = 3).
Functional Analysis of miR-1248: OGG1
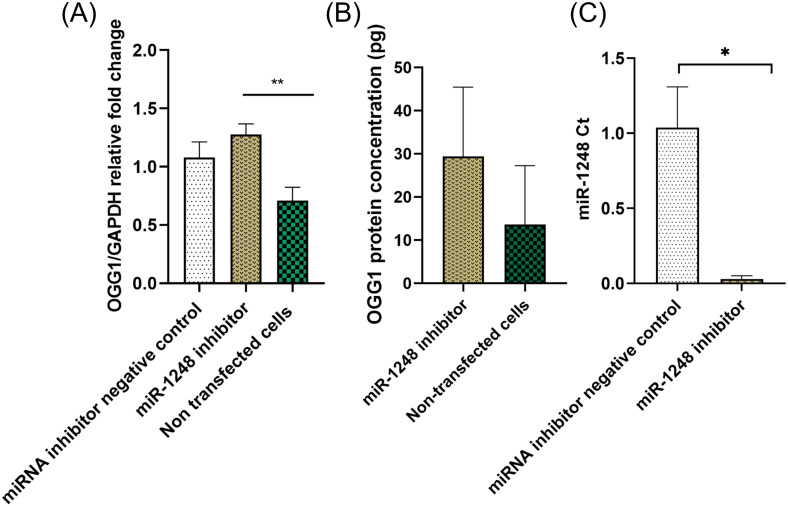
The knockdown experiment showed that the inhibition of miR-1248 via transfection of human astrocytes is associated with an upregulation of OGG1 mRNA and protein expressions relative to the non-treated controls (Figure 9A, B). Experiments were performed to assess the expression level of miR-1248 after the transfection to validate the successful inhibition of the targeted miRNA. As shown in Figure 9C, the expression of miR-1248 was downregulated after transfection with its inhibitor.
Figure 9.
OGG1 and miR-1248 expression analysis after inhibition experiments. (A) OGG1 mRNA upregulation (p < 0.01), (B) increased OGG1 protein expression, and (C) miR-1248 downregulation (p < 0.05), after inhibition of miR-1248 in human astrocytes (n = 3).
Discussion
The brain is intrinsically vulnerable to ROS-induced oxidative stress. Cell shrinkage, cytoplasmic granulation, nuclear fragmentations, and cell lysis are the major visible hallmarks of apoptosis (Saraste & Pulkki, 2000). This study indicated that sodium dichromate-induced oxidative DNA damage resulted in varying degrees of cell shrinkage and changes in cell morphology in the human astrocytes. The incorporation of mismatched base pairs related to oxidative stress-induced ROS activates the proapoptotic cellular machinery (Matés & Sánchez-Jiménez, 2000). Previous work confirmed the relationship between the TNF-related apoptosis-inducing ligand (TRAIL) expression in human astroglial cells and immune cell effector functions (Kwon & Choi, 2006). Other members of the death ligand/receptor pairs like the Fas/Faslg pair are also recruited following oxidative stress, and this is supported by our PPI data (supplemental Table 4). The apoptotic mechanisms are synergistic and act either through sensitivity priming (Denning et al., 2002) or through bystander cytotoxicity (Kwon et al., 2001).
The PCA plot compresses the expression data from thousands of genes to represent the overall variation in expression. Samples with similar noncoding RNA expressions that correlate with the treatment conditions clustered together on the PCA plot and heat map, as shown in Figures 4B and 5B, respectively. The small RNA sequencing results indicate that a significant number of miRNAs are downregulated (231) following sodium dichromate treatment, suggesting an underlying upregulation in the transcription of multiple genes. On the other hand, miR-1248 and miR-4284 expressions were upregulated (> 2.0-fold) due to cellular stress. logically, at the center of this phenomenon could be the involvement of ROS in the pathways of microRNA biogenesis, especially at the transcription level via p53′s regulatory effect on the Drosha enzyme, as has been reported (He & Jiang, 2016; Hu & Gatti, 2011). Since the p53 signaling pathway is among the top pathways highlighted by our Gene Ontology analysis (Table 2), p53 could be responsible for the global downregulation of miRNAs through a negative feedback loop mechanism. In a previous study, exogenous H2O2 exposure has been shown to cause a significant decrease in Dicer expression resulting in the downregulation of 89% of microRNAs that are normally expressed in endothelial cells (Ungvari et al., 2013).
Functional enrichment analysis identified physiological and pathological processes that are regulated by differentially expressed miRNAs. The biological processes that are highly regulated by the miRNA network include genes regulating cell cycle control, protein processing in the endoplasmic reticulum, axon guidance, and neurotrophin signaling which promotes survival, growth, and differentiation of neurons and neurites (Table 2). In addition to supporting neuronal growth, neurotrophins play an important role in the pathophysiology of many neurodegenerative and psychiatric disorders (Mitre et al., 2017). This suggests that genes involved in the cell cycle and CNS response may be regulated by stress-induced miRNAs. Pathways in cancer accounted for the highest hits (285 genes), supporting the correlation between oxidative stress and cancer that has been established by multiple groups (Reuter et al., 2010). Noncoding RNA-mediated expression of the genes associated with apoptosis includes the Caspase protein family, TP53, BAX, BCL2, FADD, ATM, FAS, FASLG, which corresponded with the decrease in cell density and cytoskeleton architectural changes in treated cells (Figure 1). Genes involved in RNA metabolism and transport were also deregulated. These included UPF1, the gene that codes for RNA helicase and translocase enzyme that is essential for nonsense-mediated decay, mediating both mRNA nuclear export and mRNA surveillance through ribonucleoprotein (RNP) remodeling (Fiorini et al., 2015). Post-transcriptional regulation of gene expression in response to cellular stress occurs on RNP granules (Kucherenko & Shcherbata, 2018). This could be used for the development of stress-adaptive miRNA-based therapeutics by recreating the concentration- and phase-dependent assembly of these membranous compartments in cells of interest.
This study identified numerous, and often, overlapping targets for the dysregulated miRNAs (see Table 2). It is a well-established fact that miRNAs are pleiotropic and redundant regulators, and therefore one miRNA can target multiple genes, as well as multiple miRNAs can target a single gene (Baulina et al., 2016). MicroRNAs that target genes within the DNA damage repair (DDR) loop were mostly downregulated, indicating an activation of the DDR transcriptome. Functional enrichment analysis showed that the majority of the targeted genes are associated with signaling, cell cycle control, and maintenance of DNA integrity. This supports our hypothesis that oxidative stress alters the miRNA expression signature to reflect bloc activation of coherent, functionally related pathways. The DDR loop is a kinase-based functional network that initiates phosphorylation-driven cascades (Tessitore et al., 2014), and this gene set was over-represented in the analyzed data. Key players within the DDR loop that were captured in our data set include PI3K, ATM, ATR, which are essential in maintaining genome stability and reducing pathological processes (Blackford & Jackson, 2017; Gobbini et al., 2016; Rinaldi et al., 2021). This presents a veritable lead for the investigation and development of molecular mechanistic models for DDR loop activation. DNA-damage response RNAs (DDRNAs) have been proposed as a requirement to fully activate the DDR response (d’Adda di Fagagna, 2014; Francia et al., 2016). These DDRNAs, like miRNAs, are generated by Drosha and Dicer processing, and they have the same sequence as their cognate damaged DNA. Thus, this sequence specificity may help them to act as guides for the localization and activation of DDR proteins.
Complementary sequence matching predicted that miR-1248 binds to the 3′-UTR of OGG1, a DNA glycosylase that is involved directly in the BER pathway (Table 3). The fact that all the miRNAs participating in the BER network (Figure 5) are downregulated suggests that their target DNA repair enzymes are upregulated. However, miR-1248 is upregulated and the mRNA expression level of OGG1 is decreased (Figure 8). The relationship between OGG1 and miR-1248 is not been previously published in the literature, and that prompted us to follow that lead to successfully establish a novel miR-1248:OGG1 interaction via miR-1248 inhibitor transfection experiments (Figure 9). The inverse relationship between miR-1248 and OGG1 (Figure 8) and the upregulation of the mRNA and protein target (OGG1) after miR-1248 inhibition (Figure 9) validates the computationally predicted regulatory relationship between the miRNA:mRNA pair. The TaqMan RT-qPCR set of probes targets a region that spans exon 2-3 and detects the expression level of two splice variants of the gene. NM_016819 codes for OGG1 transcript variant 1b is localized in the mitochondria, while NM_002542 for OGG1 variant 1a is localized in the nucleus ( Furihata, 2015 ). Therefore, miR1248 may target predominantly one of the isoforms of OGG1 due to the different mRNA 3′UTR regions.
MiR-1248 was previously reported to be involved in post-transcriptional regulation of IL-5 inflammatory response (Panganiban et al., 2012), activation of IFN production with modulation of calcium signaling (Jang et al., 2019), and TRIM24-mediated proliferation, and invasion of non-small cell lung cancer cells (Yang et al., 2020). Based on our experimental results, miR-1248 was upregulated after sodium dichromate-induced oxidative stress, and that causes reduced expression of OGG1 and increased accumulation of 8-OHdG in the DNA. Normally, the glycosylase would excise the oxidized adduct to reduce cellular damage. Interestingly, OGG1 deficiency has also been associated with a protective role against inflammatory lesions and mutagenic effects associated with Helicobacter pylori infection in a mouse model (Touati et al., 2006). Since age-associated cellular inflammation contributes to neurodegeneration, understanding the epigenetic mechanisms by which oxidative stress triggers chronic inflammation can lead to the development of therapeutics for dementia and associated diseases. The scores of protein-protein interactions (supplemental Table 4) involved the amyloid precursor protein (APP), a synaptic protein driving sporadic and familial Alzheimer’s disease (Hébert et al., 2009; Patel et al., 2008), lends credence to that possibility.
Most degenerative diseases have underlying genetic etiologies linked to oxidative stress and resultant chronic inflammation. Both are closely related pathophysiological processes, one of which can be easily induced by the other (Bu et al., 2017; Hardbower et al., 2013; Mateescu et al., 2011; Subrata Kumar Biswas, 2016). Oxidative stress is associated with mitochondrial dysfunction (Hutson et al., 2021), progressive neurodegeneration, and neuronal death (Konovalova et al., 2019). The induced expression of a relatively large number of pro-inflammatory proteins in our PPI data set (supplemental Table 4) may reflect a requirement for the activation of astrocytes by cytokines produced from other immune cells during oxidative stress, possibly brain microglia. Emerging evidence continue to support a strong relationship between microRNAs and oxidative stress – and the associated DNA damage response pathways (Sharma & Misteli, 2013; Tessitore et al., 2014; Tinaburri et al., 2018; Wang & Taniguchi, 2013). MiR-1248 is pro-inflammatory, regulating the expression of mRNAs involved in chronic inflammatory reactions in an age-dependent fashion (Hooten et al., 2013).
Our results (Figure 7) support a previous finding (Luo et al., 2017) that oxidative stress caused the downregulation of miR-335, leading to increased expression of PARP-1. PARP-1 maintains open chromatin architecture and positively regulates gene expression (Krishnakumar & Kraus, 2010). Interestingly, OGG1 binding to PARP-1 has been shown to play a functional role in the recognition and repair of oxidative DNA damage by stimulating its poly(ADP-ribosyl)ation activity (Hooten et al., 2011). Conversely, and very importantly, activated PARP-1 has been reported to exert inhibitory effects on the activity of OGG1 (Lebedeva et al., 2021). This negative feedback loop could well contribute to the increase in 8-OHdG accumulation in the cells undergoing oxidative stress, as observed in the Comet assay. The PARP-1-DNA trapping ability of certain PARP-1 inhibitors has been employed successfully in cancer therapy. There is an ongoing debate on their appropriateness and safety in neurodegenerative disorders due to the cytotoxic effects on astrocytes (Sinha et al., 2021). In addition to blocking enzymatic activity, some of these inhibitors alter the way PARP-1 interacts mechanistically with DNA – and this could be deleterious if this interaction involves OGG1. Therefore, the selective use of PARP inhibitors to treat neurodegenerative disorders should be investigated further, keeping in mind the polypharmacological properties of PARP-1 inhibitors and the proposed reciprocal interaction with OGG1.
Conclusion
Small RNA sequencing was used to identify miRNAs that are differentially expressed in human astrocytes during DNA oxidative damage which is a major hallmark of cancer and other neurodegenerative disorders. These findings indicate that micromolar concentrations of sodium dichromate induce 8-OHdG and this is associated with significant downregulation of multiple miRNAs (with only a small subset upregulated). MicroRNA functional analysis identified a subset of non-coding RNAs that modulate DNA repair pathways and a novel miRNA-mRNA pair interaction between miR-1248 and OGG1. The altered miRNA expression profile strongly points to a non-coding RNA-mediated DDR loop activation. This supports our hypothesis that oxidative stress alters the miRNA expression profile in such a way that coherent, functionally related pathways are turned on/off either singly or as an assembly complex. The miRNA candidates identified in this study could serve as potential miRNA therapeutic targets for patients with cancer and neurodegenerative disorders. Collectively, the results from this work provide evidence that oxidative damages affect DNA repair pathways in astroglial cells through non-coding RNA-controlled mechanisms.
Supplemental Material
Supplemental material, sj-png-1-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-2-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-3-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-4-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-5-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xls-6-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xls-7-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P2O GM103424-19.
ORCID iD: Gergana G. Nestorova https://orcid.org/0000-0002-9960-9435
Supplemental material: Supplemental material for this article is available online.
References
- Baulina N. M., Kulakova O. G., Favorova O. O. (2016). MicroRNAs: the role in autoimmune inflammation. Acta Naturae, 8(1(28)), 21–33. 10.32607/20758251-2016-8-1-21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger M., Allaman I., Magistretti P. J. (2011). Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metabolism, 14(6), 724–738. 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Biswas Subrata Kumar. (2016). Does the interdependence between oxidative stress and inflammation explain the antioxidant paradox? Oxidative Medicine and Celluar Longevity, 5698931, 1–9. 10.1155/2016/5698931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford A. N., Jackson S. P. (2017). ATM, ATR, and DNA-PK: the TRinity at the heart of the DNA damage response. Molecular Cell, 66(6), 801–817. 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Borgesius N. Z., de Waard M. C., van der Pluijm I., Omrani A., Zondag G. C. M., van der Horst G. T. J., Melton D. W., Hoeijmakers J. H. J., Jaarsma D., Elgersma Y. (2011). Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. Journal of Neuroscience, 31(35), 12543–12553. 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu H., Wedel S., Cavinato M., Jansen-Dürr P. (2017). MicroRNA regulation of oxidative stress-induced cellular senescence. Oxidative Medicine and Cellular Longevity, 2017, 1–12. 10.1155/2017/2398696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. H., Hwang Y. H., Lee D. J., Kim D. H., Park J. M., Wu H.-G., Kim I. A. (2016). MicroRNA-203 modulates the radiation sensitivity of human malignant glioma cells. International Journal of Radiation Oncology*Biology*Physics, 94(2), 412–420. 10.1016/j.ijrobp.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Chang L., Zhou G., Soufan O., Xia J. (2020). miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Research, 48(W1), W244–W251. 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang X. (2020). miRDB : an online database for prediction of functional microRNA targets. 48(August 2019), 127–131. 10.1093/nar/gkz757. [DOI]
- Cooke M. S., Evans M. D., Dizdaroglu M., Lunec J. (2003). Oxidative DNA damage: Mechanisms, mutation, and disease. The FASEB Journal, 17(10), 1195–1214. 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. (2014). A direct role for small non-coding RNAs in DNA damage response. Trends in Cell Biology, 24(3), 171–178. 10.1016/j.tcb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Dansen T. B., Burgering B. M. T. (2008). Unravelling the tumor-suppressive functions of FOXO proteins. Trends in Cell Biology, 18(9), 421–429. 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Denning T. L., Takaishi H., Crowe S. E., Boldogh I., Jevnikar A., Ernst P. B. (2002). Oxidative stress induces the expression of Fas and Fas ligand and apoptosis in murine intestinal epithelial cells. Free Radical Biology and Medicine, 33(12), 1641–1650. 10.1016/S0891-5849(02)01141-3. [DOI] [PubMed] [Google Scholar]
- Fiorini F., Bagchi D., Le Hir H., Croquette V. (2015). Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nature Communications, 6(7581), 1–10. 10.1038/ncomms8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S., Cabrini M., Matti V., Oldani A., d’Adda di Fagagna F. (2016). DICER, DROSHA and DNA damage-response RNAs are necessary for the secondary recruitment of DNA damage response factors. Journal of Cell Science, 129(7), 1468‒1476. 10.1242/jcs.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata C. (2015). An active alternative splicing isoform of human mitochondrial 8-oxoguanine DNA glycosylase (OGG1). Genes and Environment, 37(1), 21. 10.1186/s41021-015-0021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbini E., Cassani C., Villa M., Bonetti D., Longhese M. (2016). Functions and regulation of the MRX complex at DNA double-strand breaks. Microbial Cell, 3(8), 329–337. 10.15698/mic2016.08.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyori B. M., Venkatachalam G., Thiagarajan P. S., Hsu D., Clement M.-V. (2014). OpenComet: An automated tool for comet assay image analysis. Redox Biology, 2, 457–465. 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardbower D. M., de Sablet T., Chaturvedi R., Wilson K. T. (2013). Chronic inflammation and oxidative stress: The smoking gun for helicobacter pylori-induced gastric cancer? Gut Microbes, 4(6), 475–481. 10.4161/gmic.25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Jiang B.-H. (2016). Interplay between reactive oxygen Species and MicroRNAs in cancer. Current Pharmacology Reports, 2(2), 82–90. 10.1007/s40495-016-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert S. S., Horré K., Nicolaï L., Bergmans B., Papadopoulou A. S., Delacourte A., De Strooper B. (2009). MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiology of Disease, 33(3), 422–428. 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Hooten N. N., Fitzpatrick M., Wood W. H., De S., Ejiogu N., Zhang Y., Mattison J. A., Becker K. G., Zonderman A. B., Michele K. (2013). Age-related changes in microRNA levels in serum research. Aging, 5(10), 725–740. 10.18632/aging.100603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooten N. N., Kompaniez K., Barnes J., Lohani A., Evns M. K. (2011). Poly(ADP-ribose) polymerase 1(PARP_1) binds to 8-oxoguanine-DNA glycosolase (OGG1). Journal of Biological Chemistry, 286(52), 44679–44690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Gatti R. A. (2011). MicroRNAs: New players in the DNA damage response. Journal of Molecular Cell Biology, 3(3), 151–158. 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson K. H., Willis K., Nwokwu C. D., Maynard M., Nestorova G. G. (2021). Photon versus proton neurotoxicity: Impact on mitochondrial function and 8-OHdG base-excision repair mechanism in human astrocytes. NeuroToxicology, 82, 158–166. 10.1016/j.neuro.2020.12.011. [DOI] [PubMed] [Google Scholar]
- Inc., P. (2020). Partek® Flow® (Version 10.0) [Computer software]. https://www.partek.com/partek-flow/.
- Jang S. I., Tandon M., Teos L., Zheng C., Warner B. M., Alevizos I. (2019). Dual function of miR-1248 links interferon induction and calcium signaling defects in Sjögren’s syndrome. EBioMedicine, 48, 526–538. 10.1016/j.ebiom.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H. (1997). Analysis of a form of oxidative DNA damage, 8-hydroxy-2′-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutation Research/Reviews in Mutation Research, 387(3), 147–163. 10.1016/S1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- Kim V. N., Nam J.-W. (2006). Genomics of microRNA. Trends in Genetics, 22(3), 165–173. 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., M. Wilson III D. (2012). Overview of base excision repair biochemistry. Current Molecular Pharmacology, 5(1), 3–13. 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova J., Gerasymchuk D., Parkkinen I., Chmielarz P. (2019). Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases . [DOI] [PMC free article] [PubMed]
- Krishnakumar R., Kraus W. L. (2010). PARP-1 Regulates chromatin structure and transcription through a KDM5B-dependent pathway. Molecular Cell, 39(5), 736–749. 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherenko M. M., Shcherbata H. R. (2018). miRNA targeting and alternative splicing in the stress response - events hosted by membrane-less compartments. Journal of Cell Science, 131(4). 10.1242/jcs.202002. [DOI] [PubMed] [Google Scholar]
- Kwon D., Choi C., Lee J., Kim K. O., Kim J. D., Kim S. J., Choi I. H. (2001). Hydrogen peroxide triggers the expression of Fas/FasL in astrocytoma cell lines and augments apoptosis. Journal of Neuroimmunology, 113(1), 1–9. 10.1016/S0165-5728(00)00321-0. [DOI] [PubMed] [Google Scholar]
- Kwon D., Choi I. H. (2006). Hydrogen peroxide upregulates TNF-related apoptosis-inducing ligand (TRAIL) expression in human astroglial cells, and augments apoptosis of T cells. Yonsei Medical Journal, 47(4), 551–557. 10.3349/ymj.2006.47.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Yang H., Zhu Y., Ruan M., Huang Y., Zhang Q. (2019). MiR-7-5p-mediated downregulation of PARP1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer, 19(1), 1–9. 10.1186/s12885-019-5798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva N. A., Rechkunova N. I., Endutkin A. V., Lavrik O. I. (2021). Apurinic/apyrimidinic endonuclease 1 and tyrosyl-DNA phosphodiesterase 1 prevent suicidal covalent DNA-protein crosslink at apurinic/apyrimidinic site. Frontiers in Cell and Developmental Biology, 8,1629. 10.3389/fcell.2020.617301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. J., Hodges N. J., Chipman J. K. (2004). Modified comet assay as a biomarker of sodium dichromate-induced oxidative DNA damage: Optimization and reproducibility. Biomarkers, 9(2), 103–115. 10.1080/13547500410001704891. [DOI] [PubMed] [Google Scholar]
- Lee A. J., Hodges N. J., Chipman J. K. (2005). Interindividual variability in response to sodium dichromate-induced oxidative DNA damage: Role of the Ser326Cys polymorphism in the DNA-repair protein of 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA glycosylase 1. Cancer Epidemiology Biomarkers and Prevention, 14(2), 497–505. 10.1158/1055-9965.EPI-04-0295. [DOI] [PubMed] [Google Scholar]
- Lezza A. M. S., Mecocci P., Cormio A., Beal M. F., Cherubini A., Cantatore P., Senin U., Gadaleta M. N. (1999). Area-Specific differences in OH8dG and mtDNA4977 levels in Alzheimer disease patients and aged controls. Journal of Anti-Aging Medicine, 2(3), 209–216. 10.1089/rej.1.1999.2.209. [DOI] [Google Scholar]
- Liu D., Croteau D. L., Souza-Pinto N., Pitta M., Tian J., Wu C., Jiang H., Mustafa K., Keijzers G., Bohr V. A., Mattson M. P. (2011). Evidence that OGG1 glycosylase protects neurons against oxidative DNA damage and cell death under ischemic conditions. Journal of Cerebral Blood Flow & Metabolism, 31(2), 680–692. 10.1038/jcbfm.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lu X. (2012). Non-coding RNAs in DNA damage response. American Journal of Cancer Research, 2(6), 658–675. [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods (San Diego, Calif.), 25(4), 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu C., Zhou D., Wang Q., Liu W., Yu F., Wu F., Chen C. (2020). Crosstalk of MicroRNAs and oxidative stress in the pathogenesis of cancer. Oxidative Medicine and Cellular Longevity, 2020, 1–13. 10.1155/2020/2415324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Tong L., Meng H., Zhu W., Guo L., Wei T., Zhang J. (2017). MiR-335 regulates the chemo-radioresistance of small cell lung cancer cells by targeting PARP-1. Gene, 600, 9–15. 10.1016/j.gene.2016.11.031. [DOI] [PubMed] [Google Scholar]
- Malins D. C., Haimanot R. (1991). Major alterations in the nucleotide structure of DNA in cancer of the female breast. Cancer Research, 51(19), 5430–5432. http://www.ncbi.nlm.nih.gov/pubmed/1655250 [PubMed] [Google Scholar]
- Masè M., Grasso M., Avogaro L., D’Amato E., Tessarolo F., Graffigna A., Denti M. A., Ravelli F. (2017). Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Scientific Reports, 7(1), 41127. 10.1038/srep41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B., Batista L., Cardon M., Gruosso T., De Feraudy Y., Mariani O., Nicolas A., Meyniel J. P., Cottu P., Sastre-Garau X., Mechta-Grigoriou F. (2011). MiR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nature Medicine, 17(12), 1627–1635. 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- Matés J. M., Sánchez-Jiménez F. M. (2000). Role of reactive oxygen species in apoptosis: Implications for cancer therapy. International Journal of Biochemistry and Cell Biology, 32(2), 157–170. 10.1016/S1357-2725(99)00088-6. [DOI] [PubMed] [Google Scholar]
- Mitre M., Mariga A., Chao M. V. (2017). HHS Public Access. 131(1), 13–23. 10.1042/CS20160044.Neurotrophin. [DOI]
- Morata-Tarifa C., Picon-Ruiz M., Griñan-Lison C., Boulaiz H., Perán M., Garcia M. A., Marchal J. A. (2017). Validation of suitable normalizers for miR expression patterns analysis covering tumour heterogeneity. Scientific Reports, 7(1), 39782. 10.1038/srep39782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa P., Buffa F. M., Pan Y., Panchakshari R., Gottipati P., Muschel R. J., Beech J., Kulshrestha R., Abdelmohsen K., Weinstock D. M., Gorospe M., Harris A. L., Helleday T., Chowdhury D. (2011). MiR-182-Mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Molecular Cell, 41(2), 210–220. 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A. T., Palitti F. (2008). DNA repair and chromosomal alterations. Mutation Research/Genetic Toxicology and Environmental Mutagenesis, 657(1), 3–7. 10.1016/j.mrgentox.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Oliva M. R., Ripoll F., Muñiz P., Iradi A., Trullenque R., Valls V., Drehmer E., Sáez G. T. (1997). Genetic alterations and oxidative metabolism in sporadic colorectal tumors from a Spanish community. Molecular Carcinogenesis, 18(4), 232–243. [DOI] [PubMed] [Google Scholar]
- Ouyang Y. B., Xu L., Yue S., Liu S., Giffard R. G. (2014). Neuroprotection by astrocytes in brain ischemia: importance of microRNAs. Neuroscience Letters, 565, 53–58. 10.1016/j.neulet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A. F., Gregory T. R. (2014). The Case for Junk DNA. 10(5). 10.1371/journal.pgen.1004351. [DOI]
- Panganiban R. P. L., Pinkerton M. H., Maru S. Y., Jefferson S. J., Roff A. N., Ishmael F. T. (2012). Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. American Journal of Clinical and Experimental Immunology, 1(2), 154–165. PMCID:PMC3714196. [PMC free article] [PubMed] [Google Scholar]
- Patel N., Hoang D., Miller N., Ansaloni S., Huang Q., Rogers J. T., Lee J. C., Saunders A. J. (2008). MicroRNAs can regulate human APP levels. Molecular Neurodegeneration, 3(1), 1–6. 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlow-Poehnelt R. A., Zharkov D. O., Grollman A. P., Broyde S. (2004). Substrate discrimination by formamidopyrimidine-DNA glycosylase: Distinguishing interactions within the active site †. Biochemistry, 43(51), 16092–16105. 10.1021/bi048747f. [DOI] [PubMed] [Google Scholar]
- Pfrieger F. W. (1997). Synaptic efficacy enhanced by glial cells in vitro. Science (New York, N.Y.), 277(5332), 1684–1687. 10.1126/science.277.5332.1684. [DOI] [PubMed] [Google Scholar]
- Reuter S., Gupta S. C., Chaturvedi M. M., Aggarwal B. B. (2010). Oxidative stress, inflammation, and cancer: How are they linked? Free Radical Biology and Medicine, 49(11), 1603–1616. 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi C., Pizzul P., Longhese M. P., Bonetti D. (2021). Sensing R-loop-associated DNA damage to safeguard genome stability. Frontiers in Cell and Developmental Biology, 8, 1657. 10.3389/fcell.2020.618157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romilda C., Marika P., Alessandro S., Enrico L., Marina B., Andromachi K., Umberto C., Giacomo Z., Claudia M., Massimo R., Fabio F. (2012). Oxidative DNA damage correlates with cell immortalization and mir-92 expression in hepatocellular carcinoma. BMC Cancer, 12(1), 177. 10.1186/1471-2407-12-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste A., Pulkki K. (2000). Morphologic and biochemical hallmarks of apoptosis. Cardiovascular Research, 45(3), 528–537. https://pubmed.ncbi.nlm.nih.gov/10728374/ 10.1016/S0008-6363(99)00384-3 [DOI] [PubMed] [Google Scholar]
- Sentürker S., Karahalil B., Inal M., Yilmaz H., Müslümanoglu H., Gedikoglu G., Dizdaroglu M. (1997). Oxidative DNA base damage and antioxidant enzyme levels in childhood acute lymphoblastic leukemia. FEBS Letters, 416(3), 286–290. 10.1016/S0014-5793(97)01226-X. [DOI] [PubMed] [Google Scholar]
- Sharma V., Misteli T. (2013). Non-coding RNAs in DNA damage and repair. FEBS Letters, 587(13), 1832–1839. 10.1016/j.febslet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A., Katyal S., Kauppinen T. M. (2021). PARP-DNA trapping ability of PARP inhibitors jeopardizes astrocyte viability: Implications for CNS disease therapeutics. Neuropharmacology, 187, 108502. 10.1016/j.neuropharm.2021.108502. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., Mering C. v. (2019). STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47(D1), D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J., Bodepudi V., Shibutani S., Antoshechkin I., Miller J., Grollman A. P., Johnson F. (1994). Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. Journal of Biological Chemistry, 269(21), 15318–15324. 10.1016/s0021-9258(17)36608-5. [DOI] [PubMed] [Google Scholar]
- Tessitore A., Cicciarelli G., Del Vecchio F., Gaggiano A., Verzella D., Fischietti M., Vecchiotti D., Capece D., Zazzeroni F., Alesse E. (2014). MicroRNAs in the DNA damage/repair network and cancer. International Journal of Genomics, 2014. 10.1155/2014/820248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinaburri L., D’Errico M., Sileno S., Maurelli R., Degan P., Magenta A., Dellambra E. (2018). miR-200a modulates the expression of the DNA repair protein OGG1 playing a role in aging of primary human keratinocytes. Oxidative Medicine and Cellular Longevity, 2018, 1–17. 10.1155/2018/9147326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati E., Michel V., Thiberge J.-M., Ave P., Huerre M., Bourgade F., Klungland A., Labigne A. (2006). Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter, 11(5), 494–505. 10.1111/j.1523-5378.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- Ungvari Z., Tucsek Z., Sosnowska D., Toth P., Gautam T., Podlutsky A., Csiszar A., Losonczy G., Valcarcel-Ares M. N., Sonntag W. E., Csiszar A. (2013). Aging-Induced dysregulation of Dicer1-dependent MicroRNA expression impairs angiogenic capacity of rat cerebromicrovascular endothelial cells. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 68(8), 877–891. 10.1093/gerona/gls242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valavanidis A., Vlachogianni T., Fiotakis C. (2009). 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. Journal of Environmental Science and Health, Part C, 27(2), 120–139. 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- Vulimiri S. V., Wu X., Baer-Dubowska W., Andrade M. d., Detry M., Spitz M. R., DiGiovanni J. (2000). Analysis of aromatic DNA adducts and 7,8-dihydro-8-oxo- 2′-deoxyguanosine in lymphocyte DNA from a case–control study of lung cancer involving minority populations. Molecular Carcinogenesis, 27(1), 34. [DOI] [PubMed] [Google Scholar]
- Wang Y., Taniguchi T. (2013). MicroRNAs and DNA damage response. Cell Cycle, 12(1), 32–42. 10.4161/cc.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A. Y., Maynard M. R., Piett C. G., Nagel Z. D., Alexander J. S., Kevil C. G., Berridge M. V., Pattillo C. B., Rosen L. R., Miriyala S., Harrison L. (2019). Sodium sulfide selectively induces oxidative stress, DNA damage, and mitochondrial dysfunction and radiosensitizes glioblastoma (GBM) cells. Redox Biology, 26, 101220. 10.1016/j.redox.2019.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Nakamura Y., Kobayashi N., Iwamoto T., Yoshioka A., Kuniyasu H., Kishimoto T., Mori T. (2007). Neurons and astrocytes exhibit lower activities of global genome nucleotide excision repair than do fibroblasts. DNA Repair, 6(5), 649–657. 10.1016/j.dnarep.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Yang T., Li M., Li H., Shi P., Liu J., Chen M. (2020). Downregulation of circEPSTI1 represses the proliferation and invasion of non-small cell lung cancer by inhibiting TRIM24 via miR-1248 upregulation. Biochemical and Biophysical Research Communications, 530(1), 348–354. 10.1016/j.bbrc.2020.06.106. [DOI] [PubMed] [Google Scholar]
- Zhang J., Perry G., Smith M. A., Robertson D., Olson S. J., Graham D. G., Montine T. J. (1999). Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in Substantia Nigra neurons. The American Journal of Pathology, 154(5), 1423–1429. 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhuang Z., Wang W., He L., Wu H., Cao Y., Pan F., Zhao J., Hu Z., Sekhar C., Guo Z. (2016). OGG1 is essential in oxidative stress induced DNA demethylation. Cellular Signalling, 28(9), 1163–1171. 10.1016/j.cellsig.2016.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-png-1-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-2-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-3-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-4-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xlsx-5-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xls-6-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro
Supplemental material, sj-xls-7-asn-10.1177_17590914221101704 for Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes by Chukwumaobim Daniel Nwokwu, Adam Y. Xiao, Lynn Harrison and Gergana G. Nestorova in ASN Neuro