Abstract
Optimization of metabolism to maximize production of bio-based chemicals must consistently balance cellular resources for biocatalyst growth and desired compound synthesis. This mini-review discusses synthetic biology strategies for dynamically controlling expression of genes to enable dual-phase fermentations in which growth and production are separated into dedicated phases. Emphasis is placed on practical examples which can be reliably scaled to commercial production with the current state of technology. Recent case studies are presented, and recommendations are provided for environmental signals and genetic control circuits.
Keywords: Synthetic biology, Genetic circuits, Dynamic control
Introduction
Commercial bioprocesses require reliable and practical control strategies to realize their tremendous potential. A recent report from the McKinsey Institute looking at applications of biological innovations stated that biology could be used to produce “60 percent of the physical inputs to the global economy” and that the applications studied could have a $4 trillion direct economic impact within the next 20 years (Chui et al., 2020). Many of the cell factory-dependent bioprocesses needed to produce those physical inputs will rely on tight control of host metabolism to be successful. While production of high-cost compounds such as those for the therapeutics market might permit less cost-efficient processes, production of commodity chemicals will not. A significant portion of the forecasted impact of biological innovation on the global economy will depend upon a cost advantage from biological production of commodity chemicals.
Optimization of metabolism to maximize production efficiency must consistently balance cellular resources for biocatalyst and desired compound production. Toxicity of products and intermediates may need to be managed, often dynamically. Metabolic control strategies must be precisely and reliably matched to the process input and infrastructure constraints. Unlike infrastructure and process control design (Crater & Lievense, 2018; Hill et al., 2020), genetic control of production host metabolism is scientifically feasible but not currently precise or predictably engineered at industrial scale. How can the progress and promise of synthetic and systems biology for genetic control strategies be translated into practical and scalable engineering solutions for commercially successful bioprocesses? What tools are available and how ready are they for industrial application? What are the key considerations in choosing components of a metabolic control strategy for different types of processes? This review addresses these and other related questions, providing overall guidance towards developing a reliable, dynamically controlled biocatalyst that can be successfully taken to scale in industrial bioreactors. For the sake of brevity, the examples presented are limited to Escherichia coli; however, the same overall design principles can be adapted to other host organisms.
Considerations for Scale-Up
The high capital and operating costs of commercial scale fermentation and downstream processing unit operations demand that the development of bioprocesses “begin with the end in mind” (Covey, 1989; Crater & Lievense, 2018; Grotkjaer, 2015; Hill et al., 2020). The most desirable end, in this case, is a process for producing a commodity chemical at any standard facility, with minimal custom engineering, protocols, and logistics. The facility should be able to be operated safely and efficiently by those without a background in the biological sciences. Operational failures and performance variability create significant risk to current and future investment in bio-based chemicals.
Synthetic biology has an important role in enabling the deployment of biomanufacturing processes, but homogeneous small scale conditions used to characterize synthetic control elements are often not representative of industrial scale operations conditions. Some characteristics of a system which could be desirable for synthetic biology proof of concept projects may need additional consideration when designing for use at commercial scale. For example, undesirable gradients of pH, temperature, dissolved gases, or nutrient concentrations are common challenges encountered in large fermenters where cells are grown to high densities and kept under carbon and/or oxygen limitation. These environmental heterogeneities can lead not only to nutrient limitations that trigger cellular stress responses, but also could alter the induction response of genetic control systems due to uneven distribution of inducer molecules or inducing signals, and increased stochasticity of gene expression, resulting in inefficient production (Delvigne & Goffin, 2014; Lara et al., 2006). Designing robust control elements that behave predictably and require minimal operator interaction, and applying established methods to diagnose scalability will help mitigate the risks.
For fermentations where shifts from growth into production phase are achieved by genetic switches, slower or longer transitions may be more compatible with plant operation, as corrections to avoid process upsets may be easier to manage. Some lag in transition from growth to production phase may be beneficial when switching is applied upon transfer from seed tank to production. Allowing some cellular resources to be allocated to growth and protein production in the production tank can provide more biocatalyst for greater productivity. It also decreases the cell mass required to transfer from the seed to achieve desired volumetric productivity.
Spatial population heterogeneity can be minimized when the characteristic time of switching response is slower than that of mixing; that is, there are no gradients of cells in different metabolic states within the fermenter. Differences in mixing frequency and mass transfer between laboratory and commercial scales constitute the greatest challenge to developing a reliable process that performs as intended by design (Crater & Lievense, 2018; Rugbjerg & Olsson, 2020). In addition to matching induction response time to production tank mixing time, removal or rewiring of native regulatory feedback loops can minimize heterogeneous induction. Particularly, inducer-responsive expression of high affinity transporter genes can be replaced with inducer-independent expression. Native transporters of inducers that are positively regulated by the inducer create positive feedback loops and bimodal populations (Wang & Dunlop, 2019).
Motivation
Constitutive expression of all pathways required for optimal growth as well as production may seem like the best choice to maximize productivity, but such an approach is not always optimal. In early approaches to metabolic engineering, much focus was spent on growth-coupled production so that these competing objectives were aligned rather than opposed (Burgard & Maranas, 2003; Yim et al., 2011). Metabolic pathways were deleted in such a way that forced chemical production in order for the cell to grow, generally resulting in high yield. Although this approach led to some significant commercial successes, it became clear that this strategy was unsuitable for many products. It often would require deletion of major pathways, leading to poor growth; thus productivity would be poor even when yield is high. In extreme cases, the desired deletions were essential for growth so they could not be implemented. This incompatibility between robust growth and production led to the development of alternate strategies with separate, dedicated phases for growth and production with dynamic switching between them (Burg et al., 2016; Hartline et al., 2021). The burden of pathway expression can be silenced during the growth phase so all resources are dedicated to robust biomass production. Subsequently, growth essential pathways that may be detrimental to product synthesis can be turned down or off once growth is complete, so all intracellular resources can be directed to production.
Three basic steps must be taken to develop a dynamic control system. First, pathways are selected to be “metabolic valves” for dynamic control (Dinh & Prather, 2019; Venayak et al., 2018). This includes pathway genes that must be turned on as well as native pathways to be silenced once growth is complete. Computational modeling approaches can be used to select the latter (Venayak et al., 2018). Second, an environmental signal must be chosen that can enable switching at the right time. Finally, genetic circuits are developed to serve as an actuator, turning pathways on or off in response to the signal. This control can occur at the transcriptional, translational, or post-translational levels. Technologies that can be applied to achieve control at each of these levels are described below, followed by specific examples of how these can be combined to provide effective dynamic control.
Gene Expression Control
Decoupling growth from production has become a standard practice in fermentations to decrease the burden of engineered activities during growth. Transitioning from the growth phase to the production phase involves redirection of carbon feedstocks and nutrients towards synthesis of the target chemicals. This may include not only induction of biosynthetic pathways but also attenuation of reactions competing for substrates and resources. Switching from off to on production states as well as the dynamic regulation of activities along the fermentation process to minimize cellular metabolic burden and prevent strain genetic instability requires the ability to control gene expression and enzyme activities deliberately. A vast array of tools exists that allows regulatory control at any level of expression, including transcription, translation, and protein degradation (Hartline et al., 2021; Kent & Dixon, 2020). Engineered regulatory systems take advantage of key control points during gene expression. For instance, control of transcription initiation can be exerted by engineering the affinity of transcriptional factors (positive or negative regulators) to a ligand effector (inducer) (Meyer et al., 2018; Scholz et al., 2004; Taylor et al., 2016) to control expression of promoters, which might also be engineered to alter deoxyribonucleic acid (DNA) operator binding affinity (Daber & Lewis, 2009). Trans- (Cho et al., 2018; Kim et al., 2016; Wu et al., 2015) and cis-active regulatory elements (Felleti et al., 2016; Scull et al., 2021), as well as protein degradation tags (Cameron & Collins, 2014; Li, S et al., 2020) enable expression control at transcriptional, translational, and post-translational levels, and have been used to fine-tune central metabolism and efficiently channel carbon towards small molecule production. Simple off/on systems or genetic regulatory circuits to carry out controlled and dynamic repression and derepression of genes and protein activities can be designed and optimized for product synthesis while minimizing negative impacts on cellular homeostasis. However, complex regulatory networks, when not properly regulated, can represent a load to the producing cells and impose selective pressure, resulting in genotypic drift and selection of non-producers (Rugbjerg et al., 2018; Wehrs et al., 2019). An ideal gene expression control system will be such that it shows tight regulation (low expression under off state), wide range of tunable expression, strong and rapid response to the desired induction stimuli, and orthogonality (limited or no response to other engineered or native expression systems). In addition, synthetic biology tools used in production strains should be developed considering their applicability and robustness at commercial-scale operations.
The embrace of technologies for bio-based production of chemicals depends on their cost-competitiveness against established non-renewable, petrochemical operations. It is well known that the costs of the raw materials such as carbon feedstock can represent up to half the cost of goods in bio-fermentations (Claassen et al., 1999; Grotkjaer, 2015). Genetic control relies on molecular and/or environmental induction signals or effectors. Inducers may be compounds added to the fermentation medium at the appropriate time or could be endogenously produced by shifts in the process conditions. Added inducers, such as isopropyl β-D-1-thiogalactopyranoside, could also account for a significant fraction of production costs, making them uneconomical for industrial applications, and highlighting the need to develop alternative scalable control systems. Non-metabolized inducers may also require costly removal and safe disposal. Therefore, for synthetic biology-enabled technologies to be applied at industrial scale, they need to be cost-effective, support high yield product and productivity by cutting fermentation time, and avoid inducers that interfere with or complicate downstream purification processes.
Guidance Example
A metabolic control strategy matching the best synthetic biology tools with the driving economic and engineering considerations for a successful bioprocess could include the following components. Depletion of a nutrient or other growth medium component during the initial (growth) phase can provide the primary switch. Such a component should be low cost and readily, broadly available. It should already be part of the desired process, or easily incorporated. Examples include the depletion of, or switching between inexpensive fed sugars (Fig. 1; Bothfeld et al., 2017; Meyer et al., 2018), phosphate limitation (Nitta et al., 2021; Trung et al., 2019), or oxygen supply (Lange et al., 2017). Alternatively, pH (Li, C et al., 2020) or temperature shift (Wang et al., 2021), if consistent with the ideal process and infrastructure, could be employed.
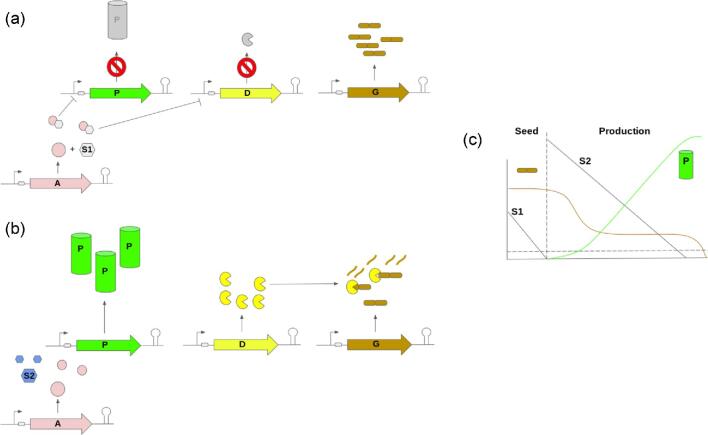
Fig. 1.
Example of a practical genetic control strategy for an industrial bioprocess. Switch is based on depletion of sugar 1 (S1) in this example. a. In the seed tank, sugar 1 (S1) binds anti-repressor (A, results in negative control of gene expression upon substrate binding) and negatively regulates product (P) pathway associated gene expression, promoting growth. Targeted degradation of growth-associated proteins (from G) is also negatively controlled by (via repression of protease gene D). b. Prior to inoculation of the production tank, S1 is depleted to below switching threshold concentration. Without S1 bound to A, repression of both P and D is removed. Sugar 2 (S2, not a ligand for A) is used as a carbon source, P and D are expressed, and targeted proteolysis of key growth associated proteins maximizes product formation. c. Schematic illustrating trends of B and G concentrations throughout the fermentation.
An allosteric transcription factor such as the lac repressor, engineered to respond to the desired process-driven switch (effector) and to function as an anti-repressor (effector-binding increases the anti-repressor's affinity for DNA, e.g. lactose-bound anti-LacI repression, or the S1-bound “A” repression in Fig. 1). Such an engineered anti-repressor can serve as a simple actuator and is based on well-studied, proven systems (Groseclose et al., 2020). This primary switch and actuator pairing can be used to open key metabolic valves (Venayak et al., 2018). It can also be used to initiate targeted proteolysis (Cameron & Collins, 2014; Li, S et al., 2020) in order to close other valves.
From the perspective of synthetic biology, this kind of circuitry is extremely simple. Additional circuit components, using similarly robust parts, could be added as needed. However, applying the scale up engineering considerations, the genetic control system (circuit) complexity should be limited to what is sufficient to provide stability (Bothfeld et al., 2017; Greco et al., 2021) and desired process dynamics. Its net function should only strengthen the overall process and it should require no user input outside of the established and standardized process inputs.
Example 1: Dynamic Control Using Quorum Sensing
The E. coli quorum sensing system can be leveraged to modulate expression of genes in a growth phase-dependent manner. Dinh and Prather (2019) demonstrate use of two independently tunable quorum sensing control modes to implement dynamic regulation for two model products, salicylic acid and naringenin. The system relies on two constitutively expressed regulator proteins, LuxR and EsaR, which respond to the common signaling molecule 3-oxohexanoyl homoserine lactone (AHL) to activate or repress their cognate promoters, respectively. The synthesis of AHL is constitutive, so as the cells grow and reach a threshold density the promoters can be either activated or repressed. The luxR and esaR expression levels were tuned by modulating promoter or ribosome binding site (RBS) strength, to obtain a range of switching dynamics.
The first two steps for naringenin biosynthesis from tyrosine were expressed from the PesaR promoter, so that initially the pathway was not active and all resources could be focused on cell growth. In addition, clustered regularly interspaced short palindromic repeats-interference (CRISPRi) coupled to the PluxR promoter was used to repress fatty acid synthesis genes. This results in less malonyl-CoA directed away from the naringenin pathway as the cells enter the production phase. By tuning the promoter driving eraR and the RBS for luxR, over 100x increase in titer was achieved relative to the static expression approach. A similar approach was used for salicylic acid, where CRISPRi targeted the pheA and tyrA genes to prevent loss of the precursor chorismate to aromatic amino acid synthesis during the growth phase. Neither of these examples was scaled beyond bench-scale, so it is conceivable that quorum sensing behavior could change at larger scales. However, the ability to modulate expression of AHL synthase and the transcriptional regulators enable flexibility to accommodate scale-up challenges. The model systems developed in this work would provide an excellent opportunity to better understand the volume-dependent behavior of quorum sensing and its impact on luxR/esaR promoters.
Example 2: Switching by Phosphate Starvation
One of the most extensive developments in dynamic control for chemical production is exhibited in works by DMC Biotechnologies and Duke University. These groups have applied control at the transcriptional and post-translational levels, using phosphate limitation as the master signal, to production of metabolites derived from pyruvate (Li, S et al., 2020; Lynch & Ye, 2019, US patent app. 2019/0390232) and malonyl-coenzyme A (malonyl-CoA) (Lynch et al., 2020, US patent app. 2020/0347388). Their strategy involves both induction of downstream pathway genes using promoters responsive to phosphate starvation, and controlled proteolysis combined with gene silencing of central metabolic pathways to create synthetic valves (Li, 2020). To determine the most suitable phosphate starvation promoters, 16 different promoters of the pho regulon (Wanner, 1993) were screened for activity in well plates as well as bioreactors (Moreb et al., 2020). Only four promoters had suitable activity and dynamic range, and behavior that also scaled to bioreactors: ugpB, yibD, phoA, and phoB promoters. Controlled gene silencing of central metabolism genes was obtained using the native E. coli type I-E Cascade/CRISPR system. Guide RNAs targeting the relevant gene were expressed by one of the above promoters, so that gene expression would be reduced as the cells enter stationary phase due to phosphate depletion. Finally, proteins encoded by these genes were appended with a degradation tag, which is recognized by the native protease SspB. The gene encoding SspB was then expressed under control of a phosphate promoter, so that these “valve” enzymes would be degraded at the same time the gene is silenced, creating a more rapid “closing” of the valve (Li, S et al., 2020).
In the case of pyruvate production, the synthetic valves chosen were the genes encoding citrate synthase (gltA) and glucose-6-phosphate dehydrogenase (zwf), thus preventing flux into the TCA cycle and pentose phosphate pathway, respectively, when the phosphate limitation response was triggered. Such a strain produced over 25 g/L pyruvate in fermentation, compared to negligible production in the wild-type strain, with approximately 30% reduction in total biomass (Li, S et al., 2020). Reduced a-ketoglutarate production due to the gltA valve also resulted in increased glucose uptake. Furthermore, by also expressing the citramalate synthase cimA3.7, which produces citramalate from one mole each of pyruvate and acetyl-CoA, under control of a phosphate starvation promoter, over 100 g/L citramalate was produced in fed-batch fermentation. Separately, the researchers created an L-alanine production strain using the same synthetic valves, and expressing a Bacillus subtilis alanine dehydrogenase gene (alaDH) from a phosphate starvation promoter (Lynch et al., 2020, US patent app. 2020/0347388). DMC recently demonstrated scale-up of alanine fermentation (“DMC demonstrates full commercial scale production of L-alanine at record speed, 2021”), and announced plans to commercialize this process in a partnership with Conagen (“DMC and Conagen partner on commercial L-alanine manufacturing, 2021”). Although the identity of the production strain has not been disclosed, it likely uses synthetic valves controlled by phosphate limitation as described above.
Concluding Remarks/Future Directions
Examples given above use E. coli as production host and specific genetic control components discussed are from bacterial systems, but the concepts are more broadly applicable. Amyris has used a maltose switch-based system with multilevel control using yeast in commercial scale fermentation processes (Chua et al., 2019). Additionally, non-genetic metabolic control strategies providing separation of growth and production phases and strain stability have been successfully applied to improve industrial fermentation processes (Pooth et al., 2020; Sandoval et al., 2014).
While the primary focus of this review is on highlighting proven genetic control strategies available as scalable engineering solutions for commercial bioprocesses, many other tools are in development and may soon find their way into industrial processes. The phosphate limitation switch-based multilevel control strategy employed by DMC in commercial application provides the first published example of its kind, but a commercial process using a well-developed system such as quorum sensing could reasonably be anticipated in the near future. Other technologies, such as optogenetics (Pouzet et al., 2020), may be further from commercial application but offer great potential for processes with tight, dynamic control far beyond what is currently possible.
The use of contract genetic control system development and scale-down testing services to access synthetic biology expertise in development and validation of robust genetic control for a planned commercial process may enable more deployment of the technology in the near future. Large companies seeking competitive advantages for their bioprocesses but lacking the requisite expertise in these areas may benefit from these services, along with startups with ideas and funding looking to minimize research and development overhead and time to market.
Contributor Information
Jonathan C Moore, No current affiliation, Encinitas, CA 92024, USA.
Itzel Ramos, BP Biosciences Center, San Diego, CA 92121, USA.
Stephen Van Dien, Persephone Biosciences, San Diego, CA 92121, USA.
Conflict of Interest and Funding
The authors have no conflict of interest or funding sources to report.
References
- Bothfeld W., Kapov G., Tyo K. (2017). A glucose-sensing toggle switch for autonomous, high productivity genetic control. ACS Synthetic Biology, 6, 1296–1304. 10.1021/acssynbio.6b00257. [DOI] [PubMed] [Google Scholar]
- Burg J. M., Cooper C. B., Ye Z., Reed B. R., Moreb E. A., Lynch M. D. (2016). Large-scale bioprocess competitiveness: the potential of dynamic metabolic control in two-stage fermentations. Current Opinion in Chemical Engineering, 14, 121–136. 10.1016/j.coche.2016.09.008. [DOI] [Google Scholar]
- Burgard A. P., Maranas C. D. (2003). Optimization-based framework for inferring and testing hypothesized metabolic objective functions. Biotechnology and Bioengineering, 82, 670–677. 10.1002/bit.10617. [DOI] [PubMed] [Google Scholar]
- Cameron D. E., Collins J. J. (2014). Tunable protein degradation in bacteria. Nature Biotechnology, 32, 1276–1281. 10.1038/nbt.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Shin J., Cho B. K. (2018). Applications of CRISPR/Cas system to bacterial metabolic engineering. International Journal of Molecular Sciences, 19, 1089. 10.3390/ijms19041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Jiang H, Meadows AM (2019). A genetic switch for stable, long-term fermentative production of anabolic products in yeast. In Presentation to Biochem Molec Eng XXI, Mont Tremblant [Google Scholar]
- Chui M., Evers M., Zheng A. (2020). How the bio revolution could transform the competitive landscape. https://www.mckinsey.com/business-functions/mckinsey-digital/our-insights/how-the-bio-revolution-could-transform-the-competitive-landscape. [Google Scholar]
- Claassen P. A. M., Van Lier J. B., Lopez-Contreras A. M., Van Niel E. W. J., Sijtsma L., Stams A. J. M., De Vries S., Weusthuis R. A. (1999). Utilisation of biomass for the supply of energy carriers. Applied Microbiology and Biotechnology, 52, 741–755. 10.1007/s002530051586. [DOI] [Google Scholar]
- Covey S. (1989). The 7 Habits of Highly Effective People: Powerful Lessons in Personal Change. Simon and Schuster. [Google Scholar]
- Crater J. S., Lievense J. C. (2018). Scale-up of industrial microbial processes. FEMS Microbiology Letters, 365, 1–5. 10.1093/femsle/fny138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daber R., Lewis M. (2009). Towards evolving a better repressor. Protein Engineering, Design & Selection: PEDS, 22, 673–683. 10.1093/protein/gzp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvigne F., Goffin P. (2014). Microbial heterogeneity affects bioprocess robustness: dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnology Journal, 9, 61–72. 10.1002/biot.201300119. [DOI] [PubMed] [Google Scholar]
- Dinh C. V., Prather K. (2019). Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proceedings of the National Academy of Sciences, 116, 25562–25568. 10.1073/pnas.1911144116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DMC and Conagen partner on commercial L-alanine manufacturing . (2021). Bioenergy International. https://bioenergyinternational.com/biochemicals-materials/dmc-conagen-partner-commercial-l-alanine-manufacturing. [Google Scholar]
- DMC demonstrates full commercial scale production of L-alanine at record speed . (2021). Bioenergy International. https://bioenergyinternational.com/biochemicals-materials/dmc-demonstrates-full-commercial-scale-production-l-alanine-record-speed. [Google Scholar]
- Felletti M., Stifel J., Wurmthaler L. A., Geiger S., Hartig J. S. (2016). Twister ribozymes as highly versatile expression platforms for artificial riboswitches. Nature Communications, 7, 12834. 10.1038/ncomms12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco F. V., Pandi A., Erb T. J., Grierson C. S., Gorochowski T. E. (2021). Harnessing the central dogma for stringent multi-level control of gene expression. Nature Communications, 12, 1–11. 10.1038/s41467-021-21995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseclose T. M., Rondon R. E., Herde Z. D., Aldrete C. A., Wilson C. J. (2020). Engineered systems of inducible anti-repressors for the next generation of biological programming. Nature Communications, 11, 1–15. 10.1038/s41467-020-18302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkjaer T. (2015). Commercial development of fermentation processes. In Villadsen J. (Ed.), Fundamental Bioengineering (1st ed., pp. 499–545). Wiley-VCH Verlag GmbH & Co. KGaA. 10.1002/9783527697441.ch17. [DOI] [Google Scholar]
- Hartline C. J., Schmitz A. C., Han Y., Zhang F. (2021). Dynamic control in metabolic engineering: theories, tools, and applications. Metabolic Engineering, 63, 126–140. 10.1016/j.ymben.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P., Benjamin K., Bhattacharjee B., Garcia F., Leng J., Liu C.-L., Murarka A., Pitera D., Rodriguez Porcel E. M., da Silva I., Kraft C. (2020). Clean manufacturing powered by biology: how Amyris has deployed technology and aims to do it better. Journal of Industrial Microbiology & Biotechnology, 47, 965–975. 10.1007/s10295-020-02314-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent R., Dixon N. (2020). Contemporary tools for regulating gene expression in bacteria. Trends in Biotechnology, 38, 316–333. 10.1016/j.tibtech.2019.09.007. [DOI] [PubMed] [Google Scholar]
- Kim S. K., Han G. H., Seong W., Kim H., Kim S. W., Lee D. H., Lee S. G. (2016). CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metabolic Engineering, 38, 228–240. 10.1016/j.ymben.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Lange J., Takors R., Blombach B. (2017). Zero-growth bioprocesses: a challenge for microbial production strains and bioprocess engineering. Engineering in Life Sciences, 17, 27–35. 10.1002/elsc.201600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara A. R., Galindo E., Ramírez O. T., Palomares L. A. (2006). Living with heterogeneities in bioreactors: understanding the effects of environmental gradients on cells. Molecular Biotechnology, 34, 355–382. 10.1385/MB:34:3:355. [DOI] [PubMed] [Google Scholar]
- Li C., Gao X., Peng X., Li J., Bai W., Zhong J., He M., Xu K., Wang Y., Li C. (2020). Intelligent microbial cell factory with genetic pH shooting (GPS) for cell self-responsive base/acid regulation. Microbial Cell Factories, 19, 1–13. 10.1186/s12934-020-01457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ye Z., Lebeau J., Moreb E. A., Lynch M. D. (2020). Dynamic control over feedback regulation identifies pyruvate-ferredoxin oxidoreductase as a central metabolic enzyme in stationary phase E. coli. bioRxiv. 10.1101/2020.07.26.219949. [DOI] [Google Scholar]
- Lynch M. D., Trahan A., Rodriguez D., Ye Z., Cooper C., Bozdag A. (2020). Compositions and methods for rapid and dynamic flux control using synthetic metabolic valves [2020/0347388]. United States Patent Application. [Google Scholar]
- Lynch M. D., Ye Z. (2019). Compositions and methods for the production of pyruvic acid and related products using dynamic metabolic control [2019/0390232]. United States Patent Application. [Google Scholar]
- Meyer A. J., Segall-Shapiro T. H., Glassey E., Zhang J., Voigt C. A. (2018). Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature Chemical Biology, 15, 196–204. 10.1038/s41589-018-0168-3. [DOI] [PubMed] [Google Scholar]
- Moreb E. A., Ye Z., Efromson J. P., Hennigan J. N., Menacho-Melgar R., Lynch M. D. (2020). Media robustness and scalability of phosphate regulated promoters useful for two-stage autoinduction in E. coli. ACS Synthetic Biology, 9, 1483–1486. 10.1021/acssynbio.0c00182. [DOI] [PubMed] [Google Scholar]
- Nitta N., Tajima Y., Yamamoto Y., Moriya M., Matsudaira A., Hoshino Y., Nishio Y., Usuda Y. (2021). Fermentative production of enantiopure (S)-linalool using a metabolically engineered Pantoea ananatis. Microbial Cell Factories, 20, 1–14. 10.1186/s12934-021-01543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouzet S., Banderas A., Le Bec M., Lautier T., Truan G., Hersen P. (2020). The promise of optogenetics for bioproduction: dynamic control strategies and scale-up instruments. Bioengineering, 7, 1–17. 10.3390/bioengineering7040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooth V., van Gaalen K., Trenkamp S., Wiechert W., Oldiges M. (2020). Comprehensive analysis of metabolic sensitivity of 1,4-butanediol producing Escherichia coli toward substrate and oxygen availability. Biotechnology Progress, 36, e2917. 10.1002/btpr.2917. [DOI] [PubMed] [Google Scholar]
- Rugbjerg P., Myling-Petersen N., Porse A., Sarup-Lytzen K., Sommer M. (2018). Diverse genetic error modes constrain large-scale bio-based production. Nature Communications, 9, 787. 10.1038/s41467-018-03232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugbjerg P., Olsson L. (2020). The future of self-selecting and stable fermentations. Journal of Industrial Microbiology & Biotechnology, 47, 993–1004. 10.1007/s10295-020-02325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval C. M., Ayson M., Moss N., Lieu B., Jackson P., Gaucher S. P., Horning T., Dahl R. H., Denery J. R., Abbott D. A., Meadows A. L. (2014). Use of pantothenate as a metabolic switch increases the genetic stability of farnesene producing Saccharomyces cerevisiae. Metabolic Engineering, 25, 215–226. 10.1016/j.ymben.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Scholz O., Henssler E. M., Bail J., Schubert P., Bogdanska-Urbaniak J., Sopp S., Reich M., Wisshak S., Kostner M., Bertram R., Hillen W. (2004). Activity reversal of Tet repressor caused by single amino acid exchanges. Molecular Microbiology, 53, 777–789. [DOI] [PubMed] [Google Scholar]
- Scull C. E., Dandpat S. S., Romero R. A., Walter N. G. (2021). Transcriptional riboswitches integrate timescales for bacterial gene expression control. Frontiers in Molecular Biosciences, 7, 607158. 10.3389/fmolb.2020.607158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. D., Garruss A. S., Moretti R., Chan S., Arbing M. A., Cascio D., Rogers J. K., Isaacs F. J., Kosuri S., Baker D., Fields S., Church G. M., Raman S. (2016). Engineering an allosteric transcription factor to respond to new ligands. Nature Methods, 13, 177–183. 10.1038/nmeth.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trung N. T., Hung N. M., Thuan N. H., Canh N. X., Schweder T., Jürgen B. (2019). An auto-inducible phosphate-controlled expression system of Bacillus licheniformis. BMC Biotechnology, 19, 1–8. 10.1186/s12896-018-0490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venayak N., von Kamp A., Klamt S., Mahadevan R. (2018). MoVE identifies metabolic valves to switch between phenotypic states. Nature Communications, 9, 1–9. 10.1038/s41467-018-07719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Dunlop M. J. (2019). Controlling and exploiting cell-to-cell variation in metabolic engineering. Current Opinion in Biotechnology, 57, 10–16. 10.1016/j.copbio.2018.08.013. [DOI] [PubMed] [Google Scholar]
- Wang X., Han J.-N., Zhang X., Ma Y.-Y., Lin Y., Wang H., Li D.-J., Zheng T.-R., Wu F.-Q., Ye J.-W., Chen G.-Q. (2021). Reversible thermal regulation for bifunctional dynamic control of gene expression in Escherichia coli. Nature Communications, 12, 1–13. 10.1038/s41467-021-21654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. (1993). Gene regulation by phosphate in enteric bacteria. Journal of Cellular Biochemistry, 51, 47–54. 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- Wehrs M., Tanjore D., Eng T., Lievense J., Pray T. R., Mukhopadhyay A. (2019). Engineering robust production microbes for large-scale cultivation. Trends in Microbiology, 27, 524–537. 10.1016/j.tim.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Wu J., Du G., Chen J., Zhou J. (2015). Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Scientific Reports, 5, 13477. 10.1038/srep13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J. D., Osterhout R. E., Stephen R., Estadilla J., Teisan S., Schreyer H. B., Andrae S., Yang T. H., Lee S. Y., Burk M. J., Van Dien S. (2011). Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nature Chemical Biology, 7, 445–452. 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]