Abstract
Objective
New York City (NYC) experienced a large first wave of coronavirus disease 2019 (COVID-19) in the spring of 2020, but the Health Department lacked tools to easily visualize and analyze incoming surveillance data to inform response activities. To streamline ongoing surveillance, a group of infectious disease epidemiologists built an interactive dashboard using open-source software to monitor demographic, spatial, and temporal trends in COVID-19 epidemiology in NYC in near real-time for internal use by other surveillance and epidemiology experts.
Materials and methods
Existing surveillance databases and systems were leveraged to create daily analytic datasets of COVID-19 case and testing information, aggregated by week and key demographics. The dashboard was developed iteratively using R, and includes interactive graphs, tables, and maps summarizing recent COVID-19 epidemiologic trends. Additional data and interactive features were incorporated to provide further information on the spread of COVID-19 in NYC.
Results
The dashboard allows key staff to quickly review situational data, identify concerning trends, and easily maintain granular situational awareness of COVID-19 epidemiology in NYC.
Discussion
The dashboard is used to inform weekly surveillance summaries and alleviated the burden of manual report production on infectious disease epidemiologists. The system was built by and for epidemiologists, which is critical to its utility and functionality. Interactivity allows users to understand broad and granular data, and flexibility in dashboard development means new metrics and visualizations can be developed as needed.
Conclusions
Additional investment and development of public health informatics tools, along with standardized frameworks for local health jurisdictions to analyze and visualize data in emergencies, are warranted.
Keywords: COVID-19, surveillance, public health informatics, data visualization
Lay Summary
New York City (NYC) experienced a large first wave of COVID-19 in the spring of 2020, but the Health Department lacked tools to easily visualize and analyze incoming surveillance data to inform response activities. To streamline ongoing surveillance, a group of infectious disease epidemiologists built an interactive dashboard using open-source software to monitor demographic, spatial, and temporal trends in COVID-19 epidemiology in NYC in near real-time for internal use by other surveillance and epidemiology experts. The dashboard allows key staff to quickly identify concerning trends and easily maintain granular situational awareness of COVID-19 epidemiology in NYC, and has alleviated the burden of manual report production on infectious disease epidemiologists. The system was built by and for epidemiologists, which is critical to its utility and functionality. Interactivity allows users to understand broad and granular data, and flexibility in dashboard development means new metrics and visualizations can be developed as needed. Additional investment and development of public health informatics tools, along with standardized frameworks for local health jurisdictions to analyze and visualize data in emergencies, are warranted.
INTRODUCTION
New York City (NYC) was an early epicenter of the coronavirus disease 2019 (COVID-19) outbreak in the United States beginning in March of 2020.1 The initial surge in cases quickly threatened to overwhelm the health care system in NYC and posed significant challenges for public health officials to rapidly import, clean, and report on surveillance data to inform situational awareness and guide response activities for an outbreak of a novel pathogen.
Local public health agencies are critical for managing outbreak response activities as they are typically the first level of collation of large volumes of surveillance data for an entire jurisdiction. Specifically, local public health informaticians and epidemiologists are uniquely placed to rapidly integrate and interpret many streams of data to identify trends and significant aberrations in the epidemiological elements of person, place, and time. As the first wave of COVID-19 in NYC began to decline, the NYC Department of Health and Mental Hygiene (DOHMH) recognized the need to improve systems for monitoring surveillance data to track the progress and spread of the outbreak and to plan for a second wave. DOHMH has well-developed electronic surveillance data systems and infrastructure; however, at the onset of the COVID-19 outbreak, these did not include tools to easily visualize or analyze incoming surveillance data nor to rapidly and frequently report key metrics. Early public-facing dashboards comparing basic numbers across states and countries did not provide adequate detail to inform response activities.2–4 DOHMH published data and visualizations daily onto an external-facing website designed for public consumption4,5; however, this was of limited value for internal decision-making because of the aggregated nature of the data to protect patient privacy and the lack of flexibility for rapidly updating data elements on the public website. NYC DOHMH prioritized the development of an internal surveillance dashboard designed by and for epidemiologists that was accessible, automated, and routinely updated to enable a rapid and thorough understanding of the local COVID-19 epidemiology by key surveillance team members. This would, in turn, help direct response resources, plan for a second wave, and minimize burden of this unprecedented pandemic on staff.6
OBJECTIVE
Using existing surveillance data infrastructure, we built an interactive dashboard to monitor demographic, spatial, and temporal trends in COVID-19 epidemiology in NYC in near real-time. The dashboard was targeted toward surveillance and epidemiology data team staff and leadership, with the goal of improving the understanding of the full epidemiologic situation of COVID-19 in NYC by surveillance experts familiar with the local setting. The overarching question driving the design of the dashboard was “who is getting sick and where?”. The dashboard was built iteratively using RStudio,7 and designed to minimize additional information technology infrastructure needs and expertise.
MATERIALS AND METHODS
Existing public health infrastructure, including electronic reporting and database systems, was a critical foundation. Results from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid amplification testing (NAAT) and antigen testing are reported to DOHMH electronically through the New York State Electronic Clinical Laboratory Reporting System (ECLRS).8 All positive test results are sent to the DOHMH Bureau of Communicable Disease surveillance and case management system, Maven®, which matches incoming reports to existing person records using a combination of patient name, date of birth, social security number, residential address, phone number, and medical record number. Maven either merges reports with an existing event or creates a new case of COVID-19.9,10 Person-level deduplication is performed automatically within Maven, with a small subset of cases requiring manual human review. Additional information for cases that are investigated is entered directly into case records in Maven. Cases of COVID-19 are matched by patient identifiers with regional health information organizations and hospital systems weekly, and the syndromic surveillance system daily, for hospitalization status. Cases are matched with the DOHMH electronic vital registry system daily for occurrence of death.1,11 Data on hospitalization and vital status, as well as information from these sources on underlying medical conditions or race/ethnicity, are imported directly into Maven. Testing data typically have a 1–3-day lag between specimen collection and receipt in ECLRS, and hospitalization and death data have longer lags because of time needed to issue and process hospital records and death certificates.
The high volume of negative SARS-CoV-2 test results makes importation of all negative results into Maven computationally prohibitive; negative test information is imported for a subset of cases but is otherwise only accessible in ECLRS and not processed by Maven’s matching and deduplication algorithms. Three analytic line-level datasets for cases, NAAT, and antigen testing are created daily using SAS Enterprise Guide (v7.1), with person-level identifiers, demographics, and key fields for analysis. Patient, provider, and facility addresses are geocoded using R and the NYC Department of City Planning Geosupport.12
Using the case and testing analytic datasets, a dataset of metrics aggregated by a rolling 7-day period is generated daily, including patient age group, sex, race/ethnicity, borough, and modified ZIP code tabulation area (modZCTA), a geographic approximation of ZIP codes, with areas with small population combined to allow for more stable estimates. The dashboard analytic dataset includes counts of confirmed and probable COVID-19 cases, hospitalizations, deaths, NAAT and antigen tests, and NAAT and antigen-positive results (Table 1). This dataset excludes residents of long-term care and correctional facilities to focus on community transmission of COVID-19. Testing data are deduplicated by person and week, such that one person is counted once per 7-day period, preferentially selecting a positive result in the event of multiple tests. Tests among persons who have previously tested positive (ie, been a case of COVID-19) are excluded. Aggregation of data into 7-day periods was necessary to reduce processing time and avoid unstable daily estimates. Data include a 3-day lag to allow for testing turnaround time and reporting; we found this balanced recent data with reliably complete information.
Crude rates of COVID-19 cases, hospitalizations, deaths, and persons tested were calculated per 100 000 population using population estimates for 2019 for age, sex, borough (county) of residence, and race/ethnicity produced by DOHMH based on the US Census Bureau Population Estimate Program files.13 Age-adjusted rates were calculated using direct standardization for age and weighting by the US 2000 standard population.14 Percent positivity was calculated as the number of persons testing positive divided by the number of persons tested, deduplicated for a given time period and geography, excluding persons after their first positive test. For recent data, persons who meet the case definition of a confirmed or probable COVID-19 case ≥90 days after a previous COVID-19 diagnosis or probable COVID-19 onset date are counted as new cases.
The dashboard was developed in an iterative process in R, using the RMarkdown15 and flexdashboard16 packages to produce a standalone, interactive HTML document which could be accessed by users with permissions to view a local network folder. All interactivity was client-side, taking place in the web browser (via JavaScript and HTML), independently of R.
RESULTS
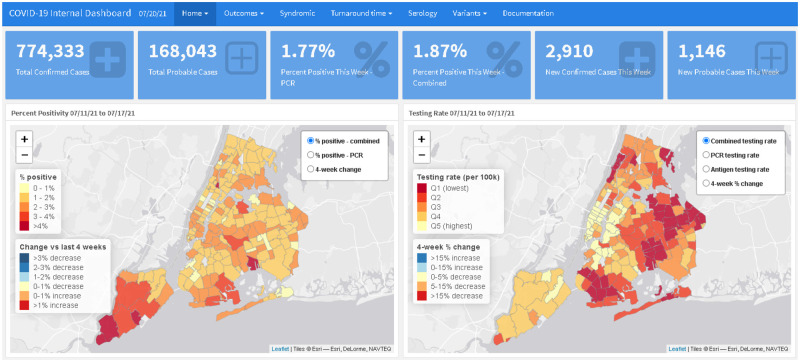
The initial internal surveillance dashboard was launched in August 2020 and displayed data for all reported cases of COVID-19 in NYC residents not living in long-term care or correctional facilities. The initial version included (1) maps showing percent positivity and testing rate by modZCTA, overall and by age group, with a corresponding table with values and recent trends (Figure 1), (2) line graphs showing trends in citywide percent positivity, case rate, and testing rate by age group, and (3) a citywide daily epidemiologic curve of cases and line graph of daily and 7-day percent positivity. Because of geographic disparities in testing patterns and resulting case counts being biased toward areas with higher access to testing, the primary metrics used to assess transmission of COVID-19 and areas of concern were testing rate and percent positivity. These metrics were prioritized in the initial phase of the dashboard and remained as the dashboard home page throughout subsequent updates.
Figure 1.
Home page of the internal surveillance dashboard, showing overall key metrics, maps of COVID-19 percent positivity and testing rate by modZCTA, and table with most recent metrics and trends by modZCTA.
Creation and maintenance of the dashboard by epidemiologists allowed for an iterative and exploratory process, adaptable to a rapidly evolving situation, which would not have been possible with a public-facing or externally managed tool. As users became familiar with the dashboard, additional suggestions and ongoing questions about the epidemiology of COVID-19 in NYC prompted the addition of more metrics, analyses, data sources, and features most relevant to the epidemiologic situation at a given time. Analyses added using the primary analytic dataset included: a map of confirmed, probable, and total case rate by modZCTA, with an accompanying table; trends of crude and age-adjusted rates of hospitalization and death, citywide and by modZCTA (maps and table); and crude and age-adjusted trends in the rate of cases, hospitalization, and death by race/ethnicity group. Additional features included more granular interactivity, allowing users to filter by multiple demographic stratifications simultaneously (eg, by geography and age group); information on seropositivity and serology testing over time, by modZCTA and demographic group; and PCR only and combined PCR and antigen testing and percent positivity as antigen testing became available.
Multiple ad hoc datasets were also incorporated to display additional epidemiologic and operational metrics. These included laboratory test volume and median turnaround time overall and by facility type (eg, hospitals vs urgent care) and laboratory; rate of emergency department visits, admissions, and percent positivity among persons with COVID-like illness from the syndromic surveillance program; the number of estimated but not yet reported daily cases, hospitalizations, and deaths, using nowcasting methods to account for lags in reporting17; counts of hospital admissions and deaths by hospital; and resurgence monitoring metrics to enable early detection of the second wave (Table 1).
Table 1.
Data elements and metrics in the New York City Department of Health and Mental Hygiene internal surveillance dashboard for COVID-19
| Category | Element | Time | Stratifications | Data source |
|---|---|---|---|---|
| Testing | Percent positivity: PCR, antigen, combined | Week | modZCTA, borough, age group, sex | ELR |
| Test rate: PCR, antigen, combined | Week | modZCTA, borough, age group, sex | ELR | |
| Cases | Case count, crude and age-adjusted rate | Week | modZCTA, borough, age group, sex, race/ethnicity | ELR |
| Outcomes | Hospitalization count, crude and age-adjusted rate | Week | modZCTA, borough, age group, sex, race/ethnicity | Hospitalization match: RHIO, hospital systems, syndromic surveillance; vital registry |
| Death count, crude and age-adjusted rate | Week | modZCTA, borough, age group, sex, race/ethnicity | Vital registry | |
| Hospitalization and death counts by facility | Month | Hospital | Hospitalization match, vital registry | |
| Syndromic | ED visits and admits: count | Day | Syndromic surveillance | |
| ED visits and admits: count CLI | Day | Syndromic surveillance | ||
| CLI visits and admits: proportion COVID+ | Day | Syndromic surveillance | ||
| CLI visit and admit rate | Day | Borough, age group | Syndromic surveillance | |
| Nowcasting | Nowcasted cases, hospitalizations, deaths | Day | Nowcasting17 | |
| Turnaround time | Testing volume | Week | Facility type, laboratory | ELR |
| Median turnaround time, interquartile range | Week | Test type, facility type, laboratory | ELR | |
| Serology | Testing rate | Week | modZCTA, borough, age group, sex | ELR |
| Percent positivity | Week | modZCTA, borough, age group, sex | ELR | |
| Variants | Number and % of cases sequenced | Week | modZCTA | ELR, laboratories |
| Count and proportion by VOI/VOC | Week | modZCTA | ELR, laboratories |
CLI: COVID-19-like illness; ED: emergency department; ELR: electronic laboratory reporting; modZCTA: modified ZIP code tabulation area; RHIO: regional health information organization; VOI/VOC: variant of interest/variant of concern.
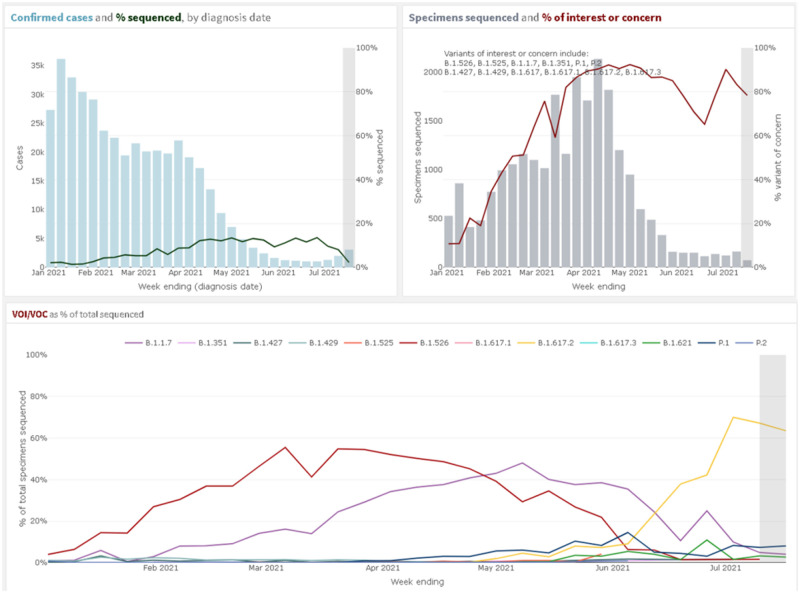
The emergence of multiple variants of interest and variants of concern (VOI/VOC) beginning in late 2020 and the identification of the novel B.1.526 lineage in NYC prompted additional descriptive figures on variant prevalence and distribution (Figure 2).18 Using the line-level case analytic dataset, a new dashboard section was created to show: the weekly count and proportion of confirmed cases that had been sequenced for the purpose of routine surveillance, the proportion attributable to each variant, maps of the proportion of confirmed cases sequenced and proportion of each VOI/VOC by modZCTA, and proportion of cases that were hospitalized or died by VOI/VOC.
Figure 2.
Variants of interest and concern on the internal surveillance dashboard. Upper Left: showing the count of confirmed cases in blue and the percent of all confirmed cases that were sequenced by week of specimen collection. Upper Right: showing the count of specimens sequenced in grey and the percent of sequenced specimens that were variants of interest (VOI) or variants of concern (VOC) in red by week of specimen collection. Bottom: percent of all sequenced specimens that were individual VOI/VOC in different colors by week of specimen collection.
The first version of the dashboard was developed over 2 months by a team of 4 DOHMH staff members. One epidemiologist with knowledge of R built the initial dashboard, with design and guidance from the 3 others. After the first version was implemented, one supporting analyst with extensive knowledge of R joined the team to help troubleshoot and build the ad hoc analyses and visualizations. Another of the initial team members with basic familiarity with R programming began reviewing the dashboard code, making modifications and additions, and eventually took over primary coding responsibilities. This learning and handover process took approximately 6 months, but likely could have been completed in 2–3 months had shifting primary responsibility been a higher priority.
The dashboard, in its early phases and after iterative developments, allowed for key surveillance team leadership to easily and routinely maintain granular situational awareness of COVID-19 epidemiology in NYC. The dashboard provided a single location where surveillance team leadership and staff could review basic situational data, identify concerning trends, and determine which geographic areas and demographic subgroups were most concerning for COVID-19 transmission. A small team of infectious disease epidemiologists began meeting weekly to identify concerning trends and outline main points to summarize and raise with agency and city leadership, relying primarily on the dashboard to assess the current epidemiologic situation in NYC. The “top takeaways” each week were shared in an easy-to-understand manner with the Surveillance and Epidemiology emergency response group, the DOHMH COVID-19 Incident Commander and Commissioner of Health, and with key leadership at NYC City Hall. These weekly “top takeaway” reports have been used to identify neighborhoods in need of additional resources, to direct neighborhood-based testing and vaccination initiatives, and have focused leadership attention on the most important trends and developments amidst an overwhelming data environment related to COVID-19.
DISCUSSION
The dashboard was a key tool in DOHMH’s epidemiologic response to COVID-19 in NYC after the first wave in the spring of 2020. Interactivity allowed users to drill down to geographic areas or subpopulations, explore trends over time, and formulate and respond to questions about where and when there might be increased transmission in NYC. Rapid, near real-time access to data is critical in any emergency outbreak response, and the dashboard allowed a large number of epidemiologists to easily access and interact with the data, understand the epidemiologic situation, and make recommendations for targeting of resources to leadership. While NYC is larger than many other local or state health departments, the basic question of “who is getting sick and where?” and the need for timely information on local COVID-19 transmission applies for all agencies involved in surveillance and response activities for COVID-19.
Key to the success of this dashboard was that it was built by epidemiologists, who were also the primary end users. Having epidemiologic expertise and an intimate understanding of the inherent limitations of surveillance data allowed the team to address the most pressing internal questions, quickly experiment and develop new metrics and visualizations, and limited the need for extensive documentation or explanation of complex surveillance data systems or caveats given the target audience was other experts in surveillance and epidemiology. The flexibility and independence in designing and modifying the dashboard ensured that the dashboard met the needs of DOHMH’s surveillance and epidemiology team more effectively than other routine reports and public-facing tools available for tracking COVID-19. The high-pressure, fast paced, and often political nature of the COVID-19 response in NYC led to many competing data demands and priorities in the context of a finite number of epidemiologists with expertise in infectious disease outbreak response. Reducing the burden on epidemiologists to respond to urgent, manual labor-intensive, custom requests for situational awareness freed up staff capacity for activities such as more detailed analyses of specific topics and improvements in data systems and quality, and possibly reduced staff burnout.
For monitoring trends in COVID-19 transmission over space and time, aggregate data were adequate to answer the relevant questions and provide situational awareness. Rolling up data by demographic groups and by week reduced computationally intensive processes, simplified modifications to the dashboard, and provided a template that can be expanded to future outbreaks and routine surveillance of other communicable diseases. Creating plans and templates for rapid, interactive data analyses and visualizations is prudent in emergency response plans and in the early stages of outbreak response. While DOHMH has a sophisticated surveillance data management system, the dashboard relied on free, open-source software and was built without the need for additional infrastructure or technical support from personnel with expertise in information technology or informatics. One DOHMH staff member with existing training in R developed the first version of the dashboard in less than 2 months, and another was able to learn R to supplement and eventually take over dashboard development and modifications. Even without a large pool of staff with strong technical skills, other jurisdictions should be able to prioritize R training for a small number of staff and build a simple tool for monitoring surveillance data, which can be enhanced based on emerging needs and as epidemiologists gain experience using R.
Challenges in hosting a dashboard on a server (eg, security considerations, informatics infrastructure, lack of flexibility) and limited staff expertise precluded the use of the popular Shiny19 package for complex interactivity, where user-defined inputs trigger live R code to run on a server. Building the dashboard using Shiny would have allowed live processing of data, and more complex interactivity, such as linked visualizations and additional levels of filters. Saving the dashboard as a standalone HTML file also precluded tracking of usage metrics, such as the number of users and most viewed pages, limiting any quantitative monitoring of dashboard usage. While a server-based setup may have provided additional interactivity, we found it was possible to build a fully functional, interactive tool for monitoring epidemiologic data without additional technical knowledge. Because of the ad hoc and iterative nature of the development process, no formal user testing was conducted during dashboard development. Although formal user testing may have resulted in improved accessibility and more intuitive design, the dashboard was designed by a subset of the end users, and the priority was rapid development and implementation in an emergency context. Feedback from all users was solicited and incorporated into the dashboard on an ongoing basis.
Challenges related to data quality, timeliness, completeness, staff technical skills, and flexibility in developing new informatics systems underscores the need for ongoing focus and investment in informatics-related training, staff, and infrastructure at local health departments. While the dashboard relied on sophisticated data management and matching processes, DOHMH’s existing surveillance systems include only limited integrated features, such as interactivity and functionality for monitoring and visualizing trends. Future informatics developments should include considerations for how the data will be used, ideally available in an interactive format allowing users to drill down to areas or subpopulations of interest. The speed of dashboard development and complexity of features was limited by the small number of DOHMH surveillance team staff with strong skills in R; future investments should prioritize staff training in public health informatics.
CONCLUSION
The internal surveillance dashboard, designed by epidemiologists for epidemiologists, was and continues to be a critical tool in the unprecedented COVID-19 epidemic. The challenges of comparing data across jurisdictions have been mentioned frequently, as data and metrics were not standardized at the start of the outbreak.20 State health departments and the Centers for Disease Control and Prevention could consider developing standard frameworks and toolsets for local health jurisdictions to use in monitoring surveillance data and responding to outbreaks. In the absence of this, the requirements for this internal surveillance dashboard could be modified and adopted by other jurisdictions, to provide near real-time situational awareness of their own data in preparation for future waves of COVID-19 or other outbreaks. The basic structure underlying the internal surveillance dashboard approach to data visualization could be standardized and adapted to other existing communicable diseases as well as for future emerging outbreaks. Ongoing development, investment, and training in health informatics infrastructure and expertise is critical to ensure health departments’ ability to rapidly identify, characterize, and respond to emerging outbreaks in an efficient manner.
FUNDING
This work received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
AUTHOR CONTRIBUTIONS
The dashboard project was conceived by CNT, EM, and SN. KJ, JB, MH, RM, GC, and GVW wrangled and provided the data. The dashboard was developed by JS, SB, SN, MI, and GC, with input on design and visualizations by MM and ME. CNT, ADF, and EM provided critical leadership and guidance throughout the process. All authors contributed to revisions and have approved the final version of this manuscript.
ACKNOWLEDGMENTS
The authors would like to thank Mr. Angel Aponte for his help administering the NYC DOHMH R Server, Dr. Sharon K. Greene for the original conceptualization of the manuscript, and Dr. Marci Layton for her leadership. We would also like to thank the EpiData team in the Surveillance and Epidemiology Emergency Response Group at NYC DOHMH for their commitment and resiliency.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article cannot be shared publicly because of patient confidentiality. The data will be shared on reasonable request to the NYC DOHMH.
REFERENCES
- 1.Thompson CN, Baumgartner J, Pichardo C, et al. COVID-19 outbreak—New York City, February 29–June 1, 2020. MMWR Morb Mortal Wkly Rep 2020; 69 (46): 1725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L.. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20 (5): 533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wissel BD, Van Camp P, Kouril M, et al. An interactive online dashboard for tracking COVID-19 in U.S. counties, cities, and states in real time. J Am Med Inform Assoc 2020; 27 (7): 1121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.New York City Department of Health and Mental Hygiene. COVID-19: Data. 2021. https://www1.nyc.gov/site/doh/covid/covid-19-data.page. Accessed May 5, 2022.
- 5.Montesano MPM, Johnson K, Tang A, et al. Successful, easy to access, online publication of COVID-19 data during the pandemic, New York City, 2020. Am J Public Health 2021; 111 (S3): S193–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peddireddy AS, Dawen X, Patil P, et al. From 5Vs to 6Cs: operationalizing epidemic data management with COVID-19 surveillance. medRxiv 2020. doi: 10.1101/2020.10.27.20220830. [DOI] [Google Scholar]
- 7.RStudio PBC. RStudio: Integrated Development Environment for R. 2021. https://www.rstudio.com/products/rstudio/. Accessed September 21, 2021.
- 8.Health Advisory: Reporting Requirements for ALL Laboratory Results for SARS-CoV-2, Including all Molecular, Antigen, and Serological Tests (including “Rapid” Tests) and Ensuring Complete Reporting of Patient Demographics. 2020. https://coronavirus.health.ny.gov/system/files/documents/2020/04/doh_covid19_reportingtestresults_rev_043020.pdf. Accessed September 21, 2021.
- 9.Troppy S, Haney G, Cocoros N, Cranston K, DeMaria A.. Infectious disease surveillance in the 21st century: an integrated web-based surveillance and case management system. Public Health Rep 2014; 129 (2): 132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Council of State and Territorial Epidemiologists. Coronavirus Disease 2019 (COVID-19) Case Definition. 2021. https://ndc.services.cdc.gov/conditions/coronavirus-disease-2019-covid-19/. Accessed September 21, 2021.
- 11.New York City Department of Health and Mental Hygiene (DOHMH) COVID-19 Response Team. Preliminary estimate of excess mortality during the COVID-19 outbreak—New York City, March 11–May 2, 2020. MMWR Morb Mortal Wkly Rep 2020; 69 (19): 603–5. [DOI] [PubMed] [Google Scholar]
- 12.New York City Department of Health and Mental Hygiene. rBES: a package of functions built by Bureau of Epidemiology Services staff, version 0.0.0.9000. 2021. https://github.com/gmculp/rBES. Accessed May 5, 2022.
- 13.NYC Department of Health and Mental Hygiene. NYC DOHMH population estimates, modified from US Census Bureau interpolated intercensal population estimates, 2000–2018 (updated August 2019). 2020.
- 14.Klein RJ, Schoenborn CA.. Age adjustment using the 2000 projected U.S. population. Healthy People 2000 Stat Notes 2001. (20): 1–9. [PubMed] [Google Scholar]
- 15.RStudio PBC. Rmarkdown: Dynamic Documents for R; v2.10.6. 2021. https://rmarkdown.rstudio.com/docs/. Accessed September 21, 2021.
- 16.The Comprehensive R Archive Network (CRAN). flexdashboard: R Markdown Format for Flexible Dashboards; v0.5.2.9000. 2021. https://pkgs.rstudio.com/flexdashboard/. Accessed September 21, 2021.
- 17.Greene SK, McGough SF, Culp GM, et al. Nowcasting for real-time COVID-19 tracking in New York City: an evaluation using reportable disease data from early in the pandemic. JMIR Public Health Surveill 2021; 7 (1): e25538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson CN, Hughes S, Ngai S, et al. Rapid emergence and epidemiologic characteristics of the SARS-CoV-2 B.1.526 variant—New York City, New York, January 1–April 5, 2021. MMWR Morb Mortal Wkly Rep 2021; 70 (19): 712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Comprehensive R Archive Network (CRAN). Shiny from R Studio: Web Application Framework for R. 2021. https://shiny.rstudio.com/reference/shiny/1.4.0/shiny-package.html. Accessed September 21, 2021.
- 20.Schechtman K, Simon S. America’s Entire Understanding of the Pandemic Was Shaped by Messy Data. The Atlantic 2021. https://www.theatlantic.com/science/archive/2021/05/pandemic-data-america-messy/618987/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article cannot be shared publicly because of patient confidentiality. The data will be shared on reasonable request to the NYC DOHMH.