Abstract
Induced pluripotent stem cell (iPSC) and gene editing technologies have revolutionized the field of in vitro disease modeling, granting us access to disease-pertinent human cells of the central nervous system. These technologies are particularly well-suited for studying diseases with strong monogenic etiologies. Epilepsy is one of the most common neurological disorders in children, with approximately half of all genetic cases caused by mutations in ion-channel genes. These channelopathy-associated epilepsies are clinically diverse, mechanistically complex, and hard to treat. Here, we review the genetic links to epilepsy, the opportunities, and challenges of iPSC-based approaches for developing in vitro models of channelopathy-associated disorders, the available tools for effective phenotyping of iPSC-derived neurons and discuss the potential therapeutic approaches for these devastating diseases.
Keywords: Ion channel genes, induced pluripotent stem cells (iPSCs), developmental and epileptic encephalopathies (DEEs), SCN1A, SCN2A, KCNQ2
Epilepsy: clinical presentation and genetic causes
Epilepsy is primarily characterized by unprovoked recurrent seizures caused by abnormal synchronized electrical discharges of cerebral neurons reflected in abnormal electroencephalographic (EEG) recordings [1,2]. Mutations in ion channel genes can cause a diverse class of diseases known as channelopathy-associated epilepsies that range from mild self-limiting seizures that resolve in adulthood, to severe developmental and epileptic encephalopathies (DEEs) [3,4]. DEEs are associated with serious comorbidities including intellectual disability, movement disorders and other multi-organ dysfunctions (for in-depth reviews on clinical seizure classifications, see [2,5,6]), that are particularly resistant to available pharmacological treatments. The advent of iPSC technology has enabled the generation of patient-specific cellular of the CNS, providing a platform for the study of the complex pathophysiological mechanisms that occur in DEEs and other types of epilepsy.
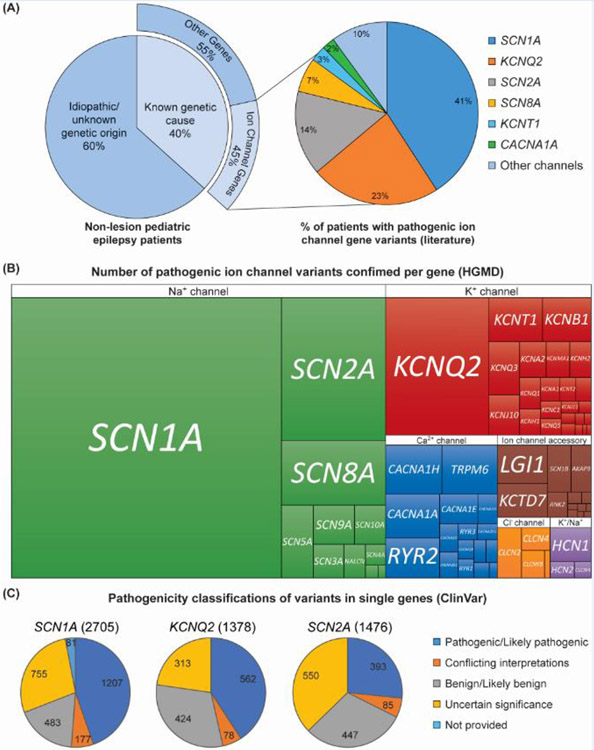
Next-generation sequencing (NGS) technology has led to the identification of pathogenic variants in multiple genes and enabled the development of clinical diagnostic panels targeting specific epilepsy-associated genes. We selected and reviewed 19 recent genetic studies of epilepsy with cohort sizes ranging from 70 to 8565 individuals, screened by whole exome or whole genome sequencing (WES and WGS, see Glossary), or by epilepsy gene panels (see references within Table S1 [7-25]). Importantly, these studies excluded patients with epilepsy secondary to metabolic disorders, brain injury/hypoxia or brain malformation. This integrated review as illustrated in Figure 1, revealed the identification of causative genetic variants in approximately 40% of study participants (Figure 1A; Table S1; [7-25]). However, it is important to consider that in most cases a variant is defined as “causative” based on in silico algorithms that predict a pathogenic effect on protein structure and function [26], as well as allele frequency in the population [27]. Thus, it is likely that the genetic etiology of 40% is an underestimate. Of persons with confirmed pathogenic variants, approximately 45% were in genes encoding ion channels or accessory proteins (excluding ligand-gated ion channels) [7-25]. Epilepsy patients with pathogenic variants in the sodium channel gene SCN1A, associated with Dravet syndrome, were the most common, followed by KCNQ2, SCN2A, SCN8A, KCNT1, CACNA1A and SCN1B [7-25].
Figure 1. Genetic etiology of epilepsy.
(A) 19 studies were reviewed in which patients with epilepsy were screened for genetic etiology, with cohort sizes ranging from 70 to 8565 individuals. See Table S1 for study details. Pathogenic variants were identified in 40% of patients. 45% of patients with pathogenic variants were identified in genes encoding ion channels and their accessory proteins (ligand gated ion channels were not included in this tally). Individuals with pathogenic variants in SCN1A were the most common followed by KCNQ2 and SCN2A. (B) Treemap of confirmed pathogenic ion channel variants associated with epilepsy separated by the type of channel (Na+, K+, Ca2+, Cl−, nonselective Na+/K+, or ion channel accessory protein genes). A list of all ion channel/accessory protein genes was used to perform a batch search for pathogenic variants identified in those genes using HGMD database. Variants were then manually cross referenced for clinical phenotypes related to epilepsy or seizures (including SUDEP that was not cardiac associated). (C) Numbers of variants reported to ClinVar database within each gene SCN1A, KCNQ2 and SCN2A separated by their pathogenicity profile.
With this compiled list of ion channel/accessory protein genes, we next performed a batch search for disease-causing variants using the human gene mutation database (HGMD) and cross-referenced the results for epilepsy or seizure-related clinical phenotypes (Figure 1B). The HGMD database curates published “gene lesions responsible for human inherited disease” [28]. Figure 1B illustrates the relative number of pathogenic variants within each specific gene, with SCN1A, SCN2A, KCNQ2 and SCN8A representing the highest heterogeneity in pathogenic variants. Importantly, while in Figure 1A we present the number of patients identified with epilepsy-associated variants in specific genes from selected studies, Figure 1B represents the number of different variants per gene confirmed to be pathogenic or likely pathogenic. Some variants are recurring in different individuals or segregate in families. This is true for some of the less severe or self-limited epilepsy syndromes. Moreover, because Na+, Ca2+ and Cl− channel genes are nearly 4 times the size of most K+ channel genes, the number of possible variants would be higher by default (e.g., protein length of Nav1.1/SCN1A (NP_001159435.1) is 2009 amino acids while KCNQ2 (NP_742105.1) is only 872 amino acids).
A critical challenge is ascribing functional relevance to the many rare variants continuously being reported in patients with epilepsy that occur within ion channel genes. Figure 1C illustrates the pathogenicity classifications of rare variants found in SCN1A, KCNQ2 and SCN2A genes, based on the ClinVar database, which tracks genetic variation in relation to disease [29]. The variant classification includes the following categories: pathogenic/likely pathogenic, benign/likely benign, conflicting interpretations or uncertain significance. Most KCNQ2 variants are cast as pathogenic/likely pathogenic, while for SCN2A most of the variants are of uncertain significance. Experimentally assessing the impact of genetic variants in ion channel function is labor-intensive but recent advances in automated electrophysiology have allowed for large scale analysis, which will contribute to improving the accuracy of variant classification and facilitate clinical diagnosis and genetic counseling [30-32].
Modeling channelopathy-associated epilepsy using human iPSC technology
There are advantages and disadvantages of different model systems to study ion channel variants and their functional consequences (for a review see [33]). One of the major limitations of animal models is that orthologues of human channels have varying expression patterns among cell types and brain regions, and differences in developmental time course and subcellular localization that collectively influence how neurons mature and behave [34-37]. As an example, a recent study found that compared to rat cortical pyramidal neuronal dendrites, human neuronal dendrites exhibit lower expression of ion channels, which leads to higher input resistance and reduced AP burst firing [34,37]. These species differences can impede the discovery of human-specific disease mechanisms and development of therapeutic strategies. Therefore, to understand how genetic variants in ion channels cause human pathology, it is important to study them in the context of human neurons. Stem cell technologies and specifically induced pluripotent stem cells (iPSCs, see Glossary) have enabled the development of such human neuronal models.
As is the case with all disease modeling systems, the value of research findings from in vitro iPSC-based models depends on thoughtful experimental design. Special consideration should be given to rigorous quality controls (QC) to account for the variability and associated limitations of iPSC-technology (Figure 2, Text Box 1; [38]). The primary research question should guide the selection of the disease-relevant differentiation protocols and platforms for discerning in vitro phenotypes (discussed in Part III). Basic experimental design considerations include: 1) selecting patient donors and properly matched controls for comparisons, 2) powering assays with enough clones per genetic background and adequate biological and technical replicates, 3) determining the suitable differentiation protocol, and 4) selecting appropriate platforms to interrogate disease-relevant cellular phenotypes that can constitute accurate read-outs for drug screening and mechanistic studies.
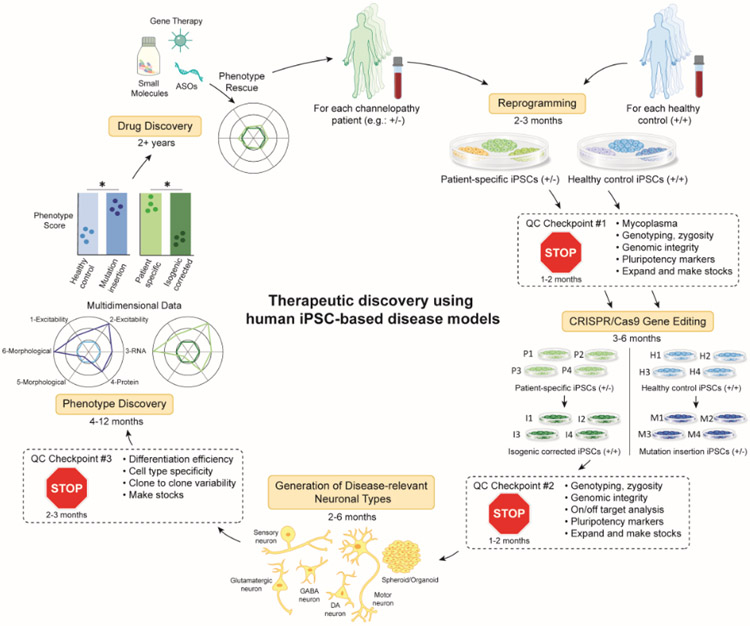
Figure 2. Flowchart of research practices for iPSC disease modeling and therapeutic discovery.
Here we present an example flowchart using a patient, heterozygous for a disease-associated mutation (+/−), although this process can be applied to patients with other types of mutations. Somatic cells obtained from a patient or alternatively a healthy control subject are reprogrammed into iPSCs (See Table S2). Several clones can be screened at QC (Quality Control) checkpoint 1 (See Text box 1). Genetic editing technology, such as CRISPR/Cas9 can be used to generate isogenic controls where a pathogenic variant is corrected in patient-derived lines or inserted in healthy control iPSCs. Several clones of each should be screened at QC checkpoint 2. iPSCs can then be differentiated into disease relevant cell types using 2D or 3D protocols (See Table S3). Each type of differentiation protocol must be optimized and screened at QC checkpoint 3. At this point, in vitro phenotype discovery is initiated in which any number of assays relevant to the disease (functional, morphological, biochemical) are performed and multi-dimensional metrics are compiled into an ultimate phenotype score. Ideally, there should be overlap in the phenotypes scores between patient lines that have been corrected (P->I) and healthy control lines that have been mutated (H->M). Once a robust, reproducible in vitro phenotype is established, drug discovery can be initiated. This workflow should be repeated ideally with multiple patients and healthy controls. The approximate timeline for completing each step is noted throughout the figure.
Text Box 1. Sources of variability in iPSC-based disease modeling experiments.
Variability between distinct iPSC clones generated from the same individual can arise from somatic mutations or loss of genetic integrity either already present in a subpopulation of the original parental cells (parental mosaicism) or introduced stochastically during reprogramming, cell culture, or during CRISPR/Cas9 gene editing. Such genomic events can unintentionally be sub-cloned during passaging and lead to genetic drift away from the original patient cells [110-114]. The use of non-integrating reprogramming vectors, which has now become standard practice, avoids integration of DNA into the somatic cell genome, and has also been associated with lower copy number variation [115]. Regular assessment of genomic integrity through G-band karyotyping, array-CGH (Comparative Genomic Hybridization), or by WGS can be used to identify genomic abnormalities or variations of sub-clonal iPSCs. Additionally, standardizing culture conditions and use of defined reagents to maintain and propagate iPSC lines is highly recommended, as these can have profound effects on iPSC heterogeneity. While discovery of biological phenotypes using multiple clones per each patient and isogenic control iPSC line is strongly recommended (Figure 2), as it ensures robust results, it adds significant cost and effort related to the maintenance, differentiation, and assessment of several clones in parallel. An alternative approach could be performing extensive QC on a single pair of patient and edited iPSC clones for initial experiments, followed by validation of a measurable phenotype across additional iPSC clones or lines. The choice of approach will ultimately depend both on available resources and considerations around the degree to which detection of subtle phenotypes require additional powering with larger sets of cellular reagents and proper statistical modeling.
Phenotype variability between independent differentiations and between technical replicates of the same differentiated clone can arise from differences in cellular composition (varying cell type or maturation) or may simply be inherent to the type of differentiation protocol [116,117]. For instance, a recent study compared the production efficiency and cellular composition of cells generated from 3 independent differentiations of the same 2 iPSC lines across 5 different laboratories using a well-established protocol to generate a mixture of cortical neurons and astrocytes [116]. Analysis of single cell gene expression profiles revealed significant heterogeneity of cell type composition between labs that masked genotypic effects between the two iPSC lines in the multi-site study [116]. Transcription factor-based differentiation protocols tend to produce more homogeneous and mature populations of cell types [118,119], but may suffer from potentially skipping relevant developmental steps associated with the process of neuronal progenitor differentiation [120]. Some considerations to minimize variability with any differentiation protocol include stringent differentiation QC metrics, consistency of key reagents and technique, and statistical modeling of mixed effects to remove unwanted variation associated only with technical effects [116].
Sources of variability and proper controls when using iPSC-based disease models
The derivation of iPSCs and their subsequent differentiation into disease relevant cell types is a multistep process, and small variations at each step can accumulate and have profound effects on biological function including disease-related in vitro phenotypes (for in-depth reviews see [39-43]). The inter-individual differences in genetic background of human subjects have been shown to be a key factor in iPSC differentiation potential and efficiency [44-46]. According to an extensive study, common genetic variation is the primary driver of molecular heterogeneity between individual iPSC lines and can account for up to 26% and 45% of differences in transcript and protein expression, as measured by RNA sequencing and immunostaining, respectively [44]. As a result, using iPSCs to model diseases with polygenic or unknown genetic origins can be challenging. In these cases, comparisons of multiple patient lines to multiple healthy controls are required to build confidence that differences between these two groups reflect a disease-associated mechanism.
To circumvent the effects of genetic background, CRISPR/Cas9-based gene editing can be used to generate isogenic control iPSC lines (see Glossary). This approach is warranted when modeling monogenic disorders, such as channelopathy-associated epilepsy [47]. Correcting a genetic variant within a patient-derived cell line can assess its necessity, while introducing a variant within a healthy control cell line can assess its sufficiency to cause specific phenotypes [48]. The availability of multiple channelopathy patient iPSC lines (Table S2) and the advancement of gene editing protocols [49], allow researchers to address fundamental questions about the impact of genetic variants on ion channel function and neuronal homeostasis, and can facilitate the classification of patient variants. Variability between distinct iPSC clones as well as between independent differentiations can arise from many factors and use of multiple clones and stringent differentiation QC metrics can be helpful strategies (Text Box 1).
Selection of differentiation protocol to study disease pertinent cell types
As epilepsy is a brain disorder, the most disease-relevant cell types are cortical excitatory and inhibitory neurons, and glial cells. Traditional iPSC protocols are based on the differentiation of adherent 2D cultures using small molecules or expression of key transcription factors. The development of 3D organoids allows for the differentiation of multi-cellular models that recapitulate certain spatial and temporal aspects of human brain development [50]. Table S3 includes a list of both 2D and 3D differentiation protocols that may be relevant for studying channelopathy-associated epilepsy. For in depth reviews of iPSC differentiation protocols see [51-54], for transcription factor-based protocols see [55], and organoid protocols see [56,57]. Complete understanding of disease pathogenesis requires an appreciation of its dynamic progression and of how molecular, cellular and tissue-level biological activities are altered spatially and temporally. iPSC-based models allow for the observation of disease-relevant cells over time as they mature and age in vitro, and thus can highlight the origin and progression of disease mechanisms (Figure 3, top). Emerging organoid-based differentiation protocols (Table S3) may be particularly relevant for the investigation of very early onset channelopathy-associated epilepsy as they recapitulate many spatiotemporal features of the developing early human brain [58]. Moreover, Trujillo et al., recently showed that cortical organoids exhibit activity reminiscent of late embryonic, early postnatal brain oscillations [59], suggesting that these models may be used to examine salient features of early onset seizure activity induced by ion-channel mutations.
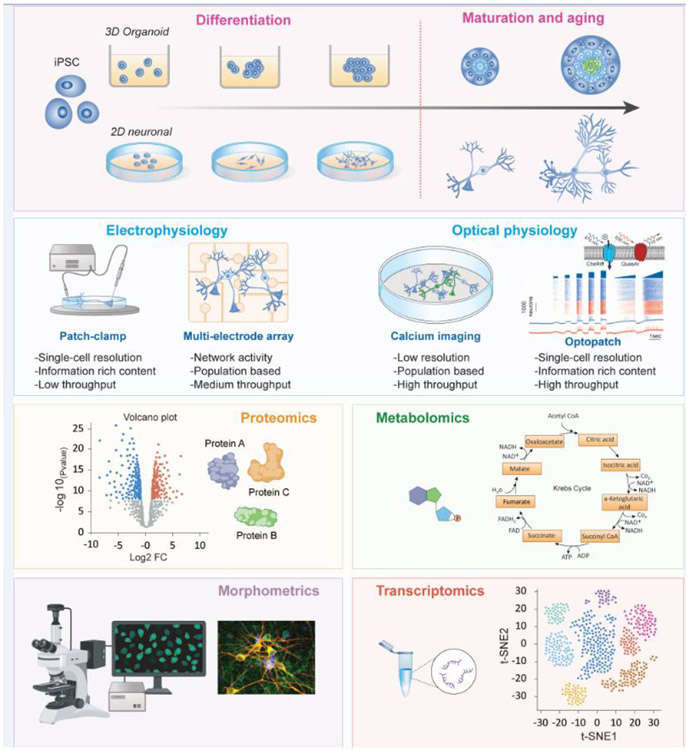
Figure 3. Longitudinal assessment of neurons on multiple phenotypic platforms.
The use of iPSC-based 2D or 3D models allows for the characterization of cells over time (i.e., disease progression; top panel). Functional, morphometric, and biochemical assays as highlighted here can be used to define a robust phenotype in disease-relevant cells. The application of multiple assays in delineating an in vitro phenotype allows for the accumulation of rich datasets to ultimately generate complex multidimensional data analyses (bottom panels). The choice of assay depends on the predicted disease- and cell-specific mechanisms, for example choosing a single cell versus network assay. The power of any measured phenotype, however, lies in both the number of measurements acquired to account for any sample-to-sample or cell-to-cell variability, as well as including appropriate isogenic controls.
Importantly, ion channel expression is not exclusive to the brain and some ion channels associated with epilepsy are also expressed in other tissues which may explain some comorbidities. For example, Dravet syndrome can be associated with cardiac arrhythmia. Studying patient iPSC-derived cardiomyocytes along with neurons has revealed important in vitro features relevant to both epilepsy and abnormal heart rhythms [60].
Indeed, the necessity for selection of appropriate cell types for investigation is highlighted in the iPSC-modeling studies of Dravet Syndrome associated with SCN1A loss-of-function mutations. Electrophysiological experiments on iPSC-derived patient neurons have revealed a variety of phenotypes in both excitatory and inhibitory neuronal subtypes (Table S2). Early iPSC-modeling studies reported higher sodium channel currents and overall hyperexcitability in cultures with mixed populations of excitatory and inhibitory neurons [61,62]. More recent studies reveal distinct differences in the behavior of neuronal subtypes wherein iPSC-derived inhibitory neurons displayed decreased sodium channel currents and reduced excitability [63], while excitatory neurons displayed no phenotype [64], which is reminiscent of early rodent studies [65,66]. Phenotypic variability could arise from differences in patient’s genetic background and the specific mutation tested, neuronal lineage, subtype differentiation protocols (Table S3), cell maturation stage (Figure 3), and/or choice of assay (network vs single cell) [67]. Ideal phenotypic studies would utilize differentiation protocols with well-characterized cell subtypes, assess multiple time points, and use isogenic controls [48,60,68]. Additionally, inclusion of pharmacological, or genetic rescue experiments with phenotype reversal [69] will build strong confidence in the measured phenotype as described in this review.
Tools for phenotyping patient iPSC-derived neurons
There are many available tools that can be utilized to systematically examine the function of neurons derived from channelopathy-associated epilepsy iPSC lines. Given the clinical presentation of patients, most published studies have focused on assessing neuronal excitability (Table S2). In the section below we discuss several phenotypic platforms and their applications in relevant iPSC models. During selection and optimization of the assay used to establish a disease-associated phenotype, it is important to consider the required throughput, resolution, and reproducibility. A variety of functional and molecular approaches can be used separately, or in combination, to generate the source data for phenotype discovery (Figures 2-3). Principal component-based analysis of different phenotypic metrics (e.g., excitability and morphological measurements) can facilitate the robust separation between patient and isogenic control neurons. In such analytical models, disparate measures may be weighted unequally based on their ability to distinguish patient and control neurons.
Manual and automated patch-clamp electrophysiology
Patch-clamp (see Glossary) recording provides direct and precise measurements of electrical activity such as membrane potential and current density, at single cell level. These methods can generate rich datasets and offer flexibility to explore disease mechanisms and characterize pharmacological effects [70]. Several studies have used manual patch-clamp to assess the functional effects of epilepsy-associated ion channel variants using iPSC-differentiated neurons (SCN1A, SCN2A, SCN8A, KCNQ2, CACNA1C; Table S2). While these have effectively revealed mutation-associated alterations on the firing properties of iPSC-differentiated neurons, they have also demonstrated the necessity to record from a substantial number of neurons due to high inherent variability. As manual patch-clamp is very labor-intensive it is not adequate for larger scale phenotypic and drug discovery projects. Automated patch-clamp instruments can provide high quality electrophysiological data in 384-well format and are a key resource for molecular characterization of epilepsy associated ion channel gene variants in heterologous expression systems [30-32]. In a recent study Vanoye et al., reported the high-throughput evaluation of more than 80 KCNQ2 epilepsy-associated variants revealing strong functional and pharmacological heterogeneity [32]. However, automated patch-clamp recording requires dissociated cells, which disrupts the morphological integrity of mature neurons, and precludes measurements of network neuronal function.
Multielectrode arrays (MEAs)
MEAs provide a nonperturbative way to measure the electrical activity of a network of cultured neurons with high temporal resolution for extended periods. Neurons are plated on a surface with embedded electrodes that offer an extracellular, alternating capacitive (AC)-coupled measure of action potentials (APs). Spatial resolution is determined by electrode size, which is typically larger than individual neurons, such that each electrode provides a composite view of the neurons that are in close contact with its surface [71]. Transparent electrodes and concurrent viability assessment assays will facilitate the interpretation of recording data [72]. An advantage of MEAs is that it is possible to make long-term recordings from neuronal cultures within an incubator and assess dynamic changes in network activity [73]. MEAs have been used to assess the spontaneous activity of excitatory neurons derived from KCNQ2-DEE patient iPSC lines [74]. The patient-derived neurons exhibited a higher propensity to fire in bursts compared to isogenic controls, a phenotype that remarkably resembled the burst-suppression pattern typically seen on corresponding KCNQ2-DEE patient EEGs. A bursting phenotype was also found in neurons differentiated from an iPSC line with an engineered KCNT1 gain-of-function mutation [75]. Another study compared the functional profiles of iPSC-derived excitatory neurons from multiple patients with SCN8A-associated epilepsy to several healthy controls. Patient neurons were found to exhibit resurgent Na+ currents and slower AP repolarization leading to prescription of Riluzole in these patients, which blocked resurgent Na+ currents and reduced seizure frequency [69].
Fluorescent assays of intracellular calcium or membrane voltage
Changes in intracellular calcium or membrane potential can be followed using kinetic plate readers including FLIPR (Molecular Devices), FDS6000 (Hamamatsu), PanOptic (WaveFront Biosciences), Bolt (Photoswitch), Cellaxes (Cellectricon), and Kinetic Imaging Cytometer (Vala). Some of these instruments can read all wells in a plate in a single scan and assays using these platforms can be readily adapted for high-throughput screening to support phenotyping and drug discovery efforts. Calcium measurements can be performed using genetically encoded calcium indicators (GECIs, see Glossary) or calcium-sensitive fluorescent dyes, whereas membrane potentials can be measured with genetically encoded voltage sensors (GEVIs, see Glossary), or voltage-sensitive dyes. Fluorescent plate readers can provide viable means to measure pharmacological reversal of channelopathy-associated cellular phenotypes using heterologous expression systems. However, commercial kinetic plate readers do not typically afford the sensitivity required to use GEVIs in neuronal assays.
All-optical electrophysiology
Optical stimulation and recording methods avoid the use of electrodes and can be performed on many neurons in parallel, and with the required spatial resolution to resolve signal from individual cells and temporal resolution to resolve individual APs. An all-optical approach (termed Optopatch™, see Glossary) was developed to address the throughput limitations associated with manual electrophysiology while maintaining the information-rich content [76,77]. This approach enables high throughput interrogation of cellular phenotypes (recording from thousands of neurons per condition in 96- to 384-well plates) and a platform for drug screening. Implementation of this approach requires optogenetic actuators and voltage reporters with distinct spectral properties. As an example, channelrhodopsin can be stimulated with blue light to excite neurons and archaerhodopsin with red light to measure changes in membrane potential. The optogenetics “toolbox” of actuators and reporters is continually evolving to meet experimental requirements [78]. In addition, blue light can be delivered in arbitrarily defined sequences to enable flexibility to probe neuronal function and can be spatially patterned to deliver stimulation to individual neurons or groups of neurons to evaluate network function and synaptic transmission. Widefield imaging can be used to detect hundreds of neurons at once with sequential well-by-well scanning across a multi-well plate. An automated analysis pipeline identifies individual spiking neurons and generates fluorescence versus time signals that are equivalent to standard electrophysiological records (Figure 3) [77,79]. This platform can generate rich datasets containing >300 functional parameters per neuron, providing a unique substrate for generation of complex, multiparameter cellular phenotypes using machine learning approaches. Measurements can also be performed using other GEVIs, voltage-sensitive dyes, and GECIs [80]. Optopatch measurements have been applied in iPSC-models of neurodegenerative diseases [77,79,81], while efforts focusing on the characterization of epilepsy-associated variants are ongoing.
Live cell imaging
Morphometric measures of neuronal properties can provide insight into long-term structural manifestations of ion channel variants [82]. High content analysis of endpoint measurements along with kinetic readouts of neuronal morphology and protein trafficking can reveal cellular defects associated with channelopathies and provide orthogonal measurements to supplement functional readouts generated using optical or electrophysiological measurements. Using this method, neurons derived from Timothy syndrome patient-specific iPSCs with mutations in CACNA1C were found to exhibit dendritic retraction in cortical neurons [83], and abnormal interneuron migration in 3D assembloids when compared to mutation corrected isogenic controls [84].
Omics analyses
Comparative large-scale expression profiles (at the RNA and protein level) provide an unbiased view of responses to genetic variants and can facilitate the interpretation of functional changes identified in patient neurons. Transcriptomic analysis can provide insights into homeostatic responses of neurons to pathogenic ion channel variants [85], while proteomic studies of iPSC-derived neurons can reveal key pathways associated with early-onset epileptic encephalopathy [86]. Current technologies may not support large-scale screening to identify potential therapies but can provide a candidate list of genes for focused higher-throughput analysis. Single cell-based transcriptomic methods can assess changes in transcript levels, as well as identify effects on specific sub-populations of cells, and track changes in cellular lineage [87]. For example, RNA sequencing was successfully used in a recent study to discover new disease-related mechanisms involving dysregulated transcriptomic pathways for chromatin remodeling and neurodevelopment in mutant SCN1A patient iPSC-derived GABAergic neurons [88]. The accurate measurement of cellular metabolites that are critical for energy production, respiration and transmethylation is another emerging analysis tool [89], that can provide an insightful view of the effects of ion-channel variants and excitability to neuronal homeostasis.
Development of therapeutics using iPSC-based models of channelopathy-associated epilepsy
A robust phenotype established using the assays described in the previous section forms the basis for discovering therapeutics. In both de novo and repurposed drug screening, human iPSC-based neuronal models of channelopathy-associated epilepsy can serve either as primary screening tools or as secondary assays for candidate therapeutic platforms as discussed below.
Small molecules
Screens of diverse small molecule and biological libraries containing tens of thousands of candidates provide a promising path to discovery of therapeutics. Currently, the cost and effort required to produce and screen large batches of iPSC-derived neurons can constrain ultra-high-throughput screening (uHTS) projects in which a million or more agents are tested, though improvements and automation of neuronal production methods will enable broader adoption of this approach. There are ~28 FDA-approved antiseizure medications (ASMs) available on the US market [90] with variable efficacies in the treatment of epilepsy. Importantly, only a few of ASMs are designed to address the underlying pathophysiology of channelopathy-associated epilepsy. ASMs mainly act to reduce neuronal excitability through several mechanisms, for example by blocking sodium channels (e.g., carbamazepine, lacosamide, phenytoin, etc.) or boosting the effect of GABA (e.g., clonazepam, phenobarbital) the primary inhibitory neurotransmitter in the brain (for review see [91]). The research, development, and approval process for small molecules from bench to bedside can take up to 15 years [92]. Drug repurposing provides an alternative and potentially shorter avenue for therapeutic discovery of ASMs [93]. This is especially attractive in the case of rare, severe diseases (such as DEEs) in which the patient population is often not large enough to justify the development costs and time required for pharmaceutical development. An example of drug repurposing is use of quinidine, a class 1 antiarrhythmic agent and broad-spectrum sodium and potassium channel blocker for treating KCNT1 (encoding a sodium-activated potassium channel)-related epilepsies [94]. The functional classification of certain KCNT1-associated epilepsy mutations as gain-of-function, suggested that quinidine may be a rational treatment for this severe infantile epilepsy, which is refractory to traditional ASMs. Several studies exploring the use of quinidine in this condition showed positive responses in children with specific seizure subtypes and location of KCNT1 mutation [95], although potential off-target cardiac effects and lower efficacy in adults with other seizure subtypes in other studies limited its usefulness [96].
Oligonucleotide-based therapeutics
Oligonucleotide-based therapeutics that modulate gene expression, including short interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs), have emerged as potentially transformative approaches in the treatment of disorders of the nervous system. siRNAs are small, highly specific, double-stranded complexes that trigger the RNA interference (RNAi) pathway to degrade a target mRNA [97]. More than 30 siRNA-based therapeutics have entered clinical trials [97]. The sequence complementarity of ASOs allows precise binding to and modulation of levels of an RNA target [98]. Specific chemical modifications stabilize ASOs and allow them to downregulate target gene expression by RNase H-mediated decay (ASOs synthesized with “gapmer” chemistry), or by modulating mRNA splicing, stability, or downstream translation (ASOs synthesized with RNA-like “steric blocking” chemistry). Consequently, gain-of-function mutations may be rescued with “gapmer” ASOs and loss-of-function mutations with “steric blocking” ASOs. Many genetic diseases including DEEs can therefore be addressed by ASOs designed to correct dysregulated expression levels at their root cause. ASOs have demonstrated clinical success in the treatment of Spinal Muscular Atrophy (Clinical Trials identifier: NCT01839656, NCT01703988) [99,100], and there is a pipeline of compounds in development and testing for treating epileptic channelopathies including molecules that boost the expression of SCN1A for Dravet Syndrome (Clinical Trials identifier: NCT04442295, NCT04740476) [101]. The timeline from project inception to clinical trials for ASO-based therapeutics is typically shorter than the timeline for more traditional small molecule-based therapeutics [102], while iPSC-based models have been an invaluable platform for assessing the specificity, selectivity, and efficacy of candidate ASOs in vitro.
Gene therapy
In adeno-associated virus (AAV)-mediated gene delivery a non-replicating viral capsid is used to supply a functional copy of a gene that may be missing or mutated in a loss-of-function state (e.g., SCN1A in Dravet Syndrome) [103]. To date, ~150 clinical trials involving AAVs have commenced and have shown acceptable safety profiles and clinical benefits in genetic diseases [104]. Advantages of AAV gene therapy include broad tissue tropism, long-term expression profiles, and the ability to transduce non-dividing cells [105]. Several serotypes show specificity to the CNS [106]. Ongoing challenges with gene delivery include appropriate biodistribution, cell-to-cell heterogeneity in expression levels, and limitations in cargo size [107,108]. While the primary application for gene therapy will be for loss-of-function channelopathy-associated epilepsy [109], AAVs can also be used to downregulate the expression of a target by delivering regulatory RNA (miRNA or shRNA) to counteract a gain-of-function disease state. Human iPSC-derived neuronal and multicellular organoid channelopathy models can be effectively used to assess target engagement, cell-type specificity, and efficacy of AAV-based gene modulation strategies in the context of a human genetic background.
Concluding remarks and future perspectives
Channelopathy-associated epilepsies represent a major therapeutic challenge. The advent of iPSC technology has enabled the development of patient-specific neuronal and multicellular models of human disease. This review serves as a resource for researchers and clinicians that are interested in adopting iPSCs as a model system for these devastating diseases. The first wave of published iPSC studies has showcased the impact of different ion-channel mutations on neuronal physiology, revealing several disease-associated phenotypes including irregular excitability patterns, defective interneuronal migration and dyshomeostatic plasticity. We anticipate that over the next decade several outstanding challenges particularly in relation to establishing the association of iPSC-based in vitro findings with clinical relevance (e.g., prediction of drug treatment responses or disease severity) will be resolved (see Outstanding Questions). As iPSC models and phenotyping platforms will continue to advance, their utility in personalized therapeutic projects as well as in broad drug screening campaigns will increase and affect how we diagnose, investigate, and treat epilepsy.
Outstanding questions.
What is the relationship between iPSC-derived cellular phenotypes and the parental patient clinical phenotypes?
How is disease severity and onset linked to the functional effects of a given pathogenic variant?
Can iPSC-based models of channelopathy-associated epilepsy predict drug treatment responses in patients?
How do mutations in ion channel genes impact the development and plasticity of human neurons?
Supplementary Material
Highlights.
Mutations in ion channel genes account for approximately 45% of all cases of genetic epilepsy
Modeling channelopathy-associated epilepsy using iPSC technology provides access to human neurons and requires strict quality control measures
A range of technologies can be effectively used for physiological and pharmacological assessment of iPSC-derived patient neurons in cell culture models including advanced functional measurements and -omics based tools
iPSC-based models for channelopathy-associated epilepsy can be used to develop and assess potential therapeutics such as small molecules, antisense oligonucleotides and gene-therapy approaches
Acknowledgments
This work was supported by the US National Institutes of Health (NIH) National Institute on Neurological Disorders and Stroke (NINDS) U54NS10887, R21NS125503 and the New York Stem Cell Foundation. E.K. is a Les Turner ALS Center Investigator and a New York Stem Cell Foundation – Robertson Investigator.
Glossary
- WES and WGS
sequencing of whole exome or whole genome respectively.
- iPSCs
induced pluripotent stem cells that are generated by induced reprogramming of somatic cells through the forced expression of transcription factors.
- Isogenic control iPSC lines
cell lines engineered by gene editing methods such as CRISPR/Cas9 to introduce or correct a specific mutation and are otherwise genetically identical to the parental control line.
- Patch-clamp
a classic electrophysiology technique based on the use of electrodes to study ionic currents in individual cells.
- GEVIs and GECIs
genetically encoded voltage indicator and genetically encoded calcium indicators are protein sensors of voltage and calcium changes respectively. They are incorporated into cells by translation and are used to acquire excitability information.
- Optopatch™
an all-optical system in which an optogenetic actuator and reporter allow for high-throughput, light-based stimulation and recording of action potentials in individual neurons.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
CMA, LAW, GTD and OBM are employees of Q-State Biosciences and may own stock or stock options in the company. EK has been a paid consultant for Q-State Biosciences in the past and owns stock options in the company.
Supplemental Information
Supplemental Information associated with this article can be found at doi:XXXXXXX’
References
- 1.Zack MM and Kobau R (2017) National and State Estimates of the Numbers of Adults and Children with Active Epilepsy - United States, 2015. MMWR Morb Mortal Wkly Rep 66, 821–825. 10.15585/mmwr.mm6631a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiest KM et al. (2017) Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology 88, 296–303. 10.1212/WNL.0000000000003509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J et al. (2017) Epilepsy-associated genes. Seizure 44, 11–20. 10.1016/j.seizure.2016.11.030 [DOI] [PubMed] [Google Scholar]
- 4.Martinez LA et al. (2021) Genetics in Epilepsy. Neurol Clin 39, 743–777. 10.1016/j.ncl.2021.05.005 [DOI] [PubMed] [Google Scholar]
- 5.Fisher RS et al. (2017) Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 58, 522–530. 10.1111/epi.13670 [DOI] [PubMed] [Google Scholar]
- 6.Katyayan A and Diaz-Medina G (2021) Epilepsy: Epileptic Syndromes and Treatment. Neurol Clin 39, 779–795. 10.1016/j.ncl.2021.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Yang L et al. (2019) Clinical and genetic spectrum of a large cohort of children with epilepsy in China. Genet Med 21, 564–571. 10.1038/s41436-018-0091-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Marmiesse A et al. (2019) Rare Variants in 48 Genes Account for 42% of Cases of Epilepsy With or Without Neurodevelopmental Delay in 246 Pediatric Patients. Front Neurosci 13, 1135. 10.3389/fnins.2019.01135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganesan S et al. (2020) A longitudinal footprint of genetic epilepsies using automated electronic medical record interpretation. Genet Med 22, 2060–2070. 10.1038/s41436-020-0923-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler KM et al. (2017) Diagnostic Yield From 339 Epilepsy Patients Screened on a Clinical Gene Panel. Pediatr Neurol 77, 61–66. 10.1016/j.pediatrneurol.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costain G et al. (2019) Clinical Application of Targeted Next-Generation Sequencing Panels and Whole Exome Sequencing in Childhood Epilepsy. Neuroscience 418, 291–310. 10.1016/j.neuroscience.2019.08.016 [DOI] [PubMed] [Google Scholar]
- 12.Lee J et al. (2020) Determining the best candidates for next-generation sequencing-based gene panel for evaluation of early-onset epilepsy. Mol Genet Genomic Med 8, e1376. 10.1002/mgg3.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang SS et al. (2019) Diagnostic Yield of Epilepsy Panel Testing in Patients With Seizure Onset Within the First Year of Life. Front Neurol 10, 988. 10.3389/fneur.2019.00988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Symonds JD et al. (2019) Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain 142, 2303–2318. 10.1093/brain/awz195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindy AS et al. (2018) Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 59, 1062–1071. 10.1111/epi.14074 [DOI] [PubMed] [Google Scholar]
- 16.Na JH et al. (2020) Targeted gene panel sequencing in early infantile onset developmental and epileptic encephalopathy. Brain Dev 42, 438–448. 10.1016/j.braindev.2020.02.004 [DOI] [PubMed] [Google Scholar]
- 17.Ko A et al. (2018) Targeted gene panel and genotype-phenotype correlation in children with developmental and epileptic encephalopathy. Epilepsy research 141, 48–55. 10.1016/j.eplepsyres.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 18.Nashabat M et al. (2019) The landscape of early infantile epileptic encephalopathy in a consanguineous population. Seizure 69, 154–172. 10.1016/j.seizure.2019.04.018 [DOI] [PubMed] [Google Scholar]
- 19.Zhou P et al. (2018) Novel mutations and phenotypes of epilepsy-associated genes in epileptic encephalopathies. Genes Brain Behav 17, e12456. 10.1111/gbb.12456 [DOI] [PubMed] [Google Scholar]
- 20.Shellhaas RA et al. (2017) Profile of neonatal epilepsies: Characteristics of a prospective US cohort. Neurology 89, 893–899. 10.1212/WNL.0000000000004284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trump N et al. (2016) Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. Journal of medical genetics 53, 310–317. 10.1136/jmedgenet-2015-103263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrini E et al. (2017) Diagnostic Targeted Resequencing in 349 Patients with Drug-Resistant Pediatric Epilepsies Identifies Causative Mutations in 30 Different Genes. Human mutation 38, 216–225. 10.1002/humu.23149 [DOI] [PubMed] [Google Scholar]
- 23.Balciuniene J et al. (2019) Use of a Dynamic Genetic Testing Approach for Childhood-Onset Epilepsy. JAMA Netw Open 2, e192129. 10.1001/jamanetworkopen.2019.2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snoeijen-Schouwenaars FM et al. (2019) Diagnostic exome sequencing in 100 consecutive patients with both epilepsy and intellectual disability. Epilepsia 60, 155–164. 10.1111/epi.14618 [DOI] [PubMed] [Google Scholar]
- 25.Rochtus A et al. (2020) Genetic diagnoses in epilepsy: The impact of dynamic exome analysis in a pediatric cohort. Epilepsia 61, 249–258. 10.1111/epi.16427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh R et al. (2017) Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol 18, 225. 10.1186/s13059-017-1353-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards S et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenson PD et al. (2020) The Human Gene Mutation Database (HGMD((R))): optimizing its use in a clinical diagnostic or research setting. Human genetics 139, 1197–1207. 10.1007/s00439-020-02199-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ et al. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42, D980–985. 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng CA et al. (2020) High-throughput phenotyping of heteromeric human ether-a-go-go-related gene potassium channel variants can discriminate pathogenic from rare benign variants. Heart Rhythm 17, 492–500. 10.1016/j.hrthm.2019.09.020 [DOI] [PubMed] [Google Scholar]
- 31.Kang SK et al. (2019) Spectrum of KV 2.1 Dysfunction in KCNB1-Associated Neurodevelopmental Disorders. Annals of neurology 86, 899–912. 10.1002/ana.25607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanoye CG et al. (2022) High-throughput evaluation of epilepsy-associated KCNQ2 variants reveals functional and pharmacological heterogeneity. JCI Insight. 10.1172/jci.insight.156314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall GF et al. (2021) Modelling epilepsy in the mouse: challenges and solutions. Dis Model Mech 14. 10.1242/dmm.047449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaulieu-Laroche L et al. (2018) Enhanced Dendritic Compartmentalization in Human Cortical Neurons. Cell 175, 643–651 e614. 10.1016/j.cell.2018.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardoso-Moreira M et al. (2020) Developmental Gene Expression Differences between Humans and Mammalian Models. Cell Rep 33, 108308. 10.1016/j.celrep.2020.108308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S et al. (2014) Comparison of the transcriptional landscapes between human and mouse tissues. Proceedings of the National Academy of Sciences of the United States of America 111, 17224–17229. 10.1073/pnas.1413624111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaulieu-Laroche L et al. (2021) Allometric rules for mammalian cortical layer 5 neuron biophysics. Nature 600, 274–278. 10.1038/s41586-021-04072-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson NC et al. (2021) Balancing serendipity and reproducibility: Pluripotent stem cells as experimental systems for intellectual and developmental disorders. Stem Cell Reports 16, 1446–1457. 10.1016/j.stemcr.2021.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engle SJ et al. (2018) Best Practices for Translational Disease Modeling Using Human iPSC-Derived Neurons. Neuron 100, 783–797. 10.1016/j.neuron.2018.10.033 [DOI] [PubMed] [Google Scholar]
- 40.Volpato V and Webber C (2020) Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis Model Mech 13. 10.1242/dmm.042317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollingsworth EW et al. (2017) iPhemap: an atlas of phenotype to genotype relationships of human iPSC models of neurological diseases. EMbO Mol Med 9, 1742–1762. 10.15252/emmm.201708191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young-Pearse TL and Morrow EM (2016) Modeling developmental neuropsychiatric disorders with iPSC technology: challenges and opportunities. Current opinion in neurobiology 36, 66–73. 10.1016/j.conb.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pamies D et al. (2017) Good Cell Culture Practice for stem cells and stem-cell-derived models. Altex-Altern Anim Ex 34, 95–132. 10.14573/altex.1607121 [DOI] [PubMed] [Google Scholar]
- 44.Kilpinen H et al. (2017) Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. 10.1038/nature22403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyttala A et al. (2016) Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Reports 6, 200–212. 10.1016/j.stemcr.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burrows CK et al. (2016) Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs. PLoS Genet 12, e1005793. 10.1371/journal.pgen.1005793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Germain PL and Testa G (2017) Taming Human Genetic Variability: Transcriptomic Meta-Analysis Guides the Experimental Design and Interpretation of iPSC-Based Disease Modeling. Stem Cell Reports 8, 1784–1796. 10.1016/j.stemcr.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Y et al. (2020) Comparisons of dual isogenic human iPSC pairs identify functional alterations directly caused by an epilepsy associated SCN1A mutation. Neurobiol Dis 134, 104627. 10.1016/j.nbd.2019.104627 [DOI] [PubMed] [Google Scholar]
- 49.De Masi C et al. (2020) Application of CRISPR/Cas9 to human-induced pluripotent stem cells: from gene editing to drug discovery. Hum Genomics 14, 25. 10.1186/s40246-020-00276-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon A et al. (2021) Long-term maturation of human cortical organoids matches key early postnatal transitions. Nature neuroscience 24, 331–342. 10.1038/s41593-021-00802-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galiakberova AA and Dashinimaev EB (2020) Neural Stem Cells and Methods for Their Generation From Induced Pluripotent Stem Cells in vitro. Front Cell Dev Biol 8, 815. 10.3389/fcell.2020.00815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang M et al. (2020) Modeling neuropsychiatric disorders using human induced pluripotent stem cells. Protein Cell 11, 45–59. 10.1007/s13238-019-0638-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niu W and Parent JM (2020) Modeling genetic epilepsies in a dish. Dev Dyn 249, 56–75. 10.1002/dvdy.79 [DOI] [PubMed] [Google Scholar]
- 54.Pacitti D et al. (2019) Organs to Cells and Cells to Organoids: The Evolution of in vitro Central Nervous System Modelling. Frontiers in cellular neuroscience 13, 129. 10.3389/fncel.2019.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flitsch LJ et al. (2020) Transcription Factor-Based Fate Specification and Forward Programming for Neural Regeneration. Frontiers in cellular neuroscience 14, 121. 10.3389/fncel.2020.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Velasco S et al. (2020) 3D Brain Organoids: Studying Brain Development and Disease Outside the Embryo. Annual review of neuroscience 43, 375–389. 10.1146/annurev-neuro-070918-050154 [DOI] [PubMed] [Google Scholar]
- 57.Sidhaye J and Knoblich JA (2021) Brain organoids: an ensemble of bioassays to investigate human neurodevelopment and disease. Cell death and differentiation 28, 52–67. 10.1038/s41418-020-0566-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley KW and Pasca SP (2022) Human brain organogenesis: Toward a cellular understanding of development and disease. Cell 185, 42–61. 10.1016/j.cell.2021.10.003 [DOI] [PubMed] [Google Scholar]
- 59.Trujillo CA et al. (2019) Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell 25, 558–569 e557. 10.1016/j.stem.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frasier CR et al. (2018) Channelopathy as a SUDEP Biomarker in Dravet Syndrome Patient-Derived Cardiac Myocytes. Stem Cell Reports 11, 626–634. 10.1016/j.stemcr.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y et al. (2013) Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Annals of neurology 74, 128–139. 10.1002/ana.23897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiao J et al. (2013) Modeling Dravet syndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Human molecular genetics 22, 4241–4252. 10.1093/hmg/ddt275 [DOI] [PubMed] [Google Scholar]
- 63.Higurashi N et al. (2013) A human Dravet syndrome model from patient induced pluripotent stem cells. Mol Brain 6, 19. 10.1186/1756-6606-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Y et al. (2016) A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet Syndrome patients. Elife 5. 10.7554/eLife.13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu FH et al. (2006) Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nature neuroscience 9, 1142–1149. 10.1038/nn1754 [DOI] [PubMed] [Google Scholar]
- 66.Cheah CS et al. (2012) Specific deletion of NaV1.1 sodium channels in inhibitory interneurons causes seizures and premature death in a mouse model of Dravet syndrome. Proceedings of the National Academy of Sciences of the United States of America 109, 14646–14651. 10.1073/pnas.1211591109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isom LL (2017) Opposing Phenotypes in Dravet Syndrome Patient-Derived Induced Pluripotent Stem Cell Neurons: Can Everyone Be Right? Epilepsy Curr 17, 244–247. 10.5698/1535-7597.17.4.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J et al. (2016) CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry 6, e703. 10.1038/tp.2015.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tidball AM et al. (2020) Variant-specific changes in persistent or resurgent sodium current in SCN8A-related epilepsy patient-derived neurons. Brain 143, 3025–3040. 10.1093/brain/awaa247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noebels JL and Jasper HH (2012) Jasper's basic mechanisms of the epilepsies (Contemporary neurology series, Vol. 80) 4th edn), Oxford University Press [Google Scholar]
- 71.Obien ME et al. (2014) Revealing neuronal function through microelectrode array recordings. Front Neurosci 8, 423. 10.3389/fnins.2014.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiang Y et al. (2018) Transparent arrays of bilayer-nanomesh microelectrodes for simultaneous electrophysiology and two-photon imaging in the brain. Sci Adv 4, eaat0626. 10.1126/sciadv.aat0626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mossink B et al. (2021) Human neuronal networks on micro-electrode arrays are a highly robust tool to study disease-specific genotype-phenotype correlations in vitro. Stem Cell Reports 16, 2182–2196. 10.1016/j.stemcr.2021.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simkin D et al. (2021) Dyshomeostatic modulation of Ca(2+)-activated K(+) channels in a human neuronal model of kCnQ2 encephalopathy. Elife 10, e64434. 10.7554/eLife.64434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quraishi IH et al. (2019) An Epilepsy-Associated KCNT1 Mutation Enhances Excitability of Human iPSC-Derived Neurons by Increasing Slack KNa Currents. J Neurosci 39, 7438–7449. 10.1523/JNEUROSCI.1628-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hochbaum DR et al. (2014) All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nature methods 11, 825–833. 10.1038/nmeth.3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams LA et al. (2019) Scalable Measurements of Intrinsic Excitability in Human iPS Cell-Derived Excitatory Neurons Using All-Optical Electrophysiology. Neurochemical research 44, 714–725. 10.1007/s11064-018-2694-5 [DOI] [PubMed] [Google Scholar]
- 78.Christenson Wick Z and Krook-Magnuson E (2018) Specificity, Versatility, and Continual Development: The Power of Optogenetics for Epilepsy Research. Frontiers in cellular neuroscience 12, 151. 10.3389/fncel.2018.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Werley CA et al. (2017) All-Optical Electrophysiology for Disease Modeling and Pharmacological Characterization of Neurons. Curr Protoc Pharmacol 78, 11 20 11–11 20 24. 10.1002/cpph.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shcherbakova DM (2021) Near-infrared and far-red genetically encoded indicators of neuronal activity. J Neurosci Methods 362, 109314. 10.1016/j.jneumeth.2021.109314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kiskinis E et al. (2018) All-Optical Electrophysiology for High-Throughput Functional Characterization of a Human iPSC-Derived Motor Neuron Model of ALS. Stem Cell Reports 10, 1991–2004. 10.1016/j.stemcr.2018.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song Y et al. (2019) The Mechanosensitive Ion Channel Piezo Inhibits Axon Regeneration. Neuron 102, 373–389 e376. 10.1016/j.neuron.2019.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krey JF et al. (2013) Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nature neuroscience 16, 201–209. 10.1038/nn.3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Birey F et al. (2017) Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. 10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Springer K et al. (2021) Flexible Stoichiometry: Implications for KCNQ2- and KCNQ3-Associated Neurodevelopmental Disorders. Developmental neuroscience 43, 191–200. 10.1159/000515495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Negraes PD et al. (2021) Altered network and rescue of human neurons derived from individuals with early-onset genetic epilepsy. Molecular psychiatry 26, 7047–7068. 10.1038/s41380-021-01104-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfisterer U et al. (2020) Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat Commun 11, 5038. 10.1038/s41467-020-18752-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schuster J et al. (2019) Transcriptomes of Dravet syndrome iPSC derived GABAergic cells reveal dysregulated pathways for chromatin remodeling and neurodevelopment. Neurobiol Dis 132, 104583. 10.1016/j.nbd.2019.104583 [DOI] [PubMed] [Google Scholar]
- 89.Miljanovic N et al. (2021) Metabolomic signature of the Dravet syndrome: A genetic mouse model study. Epilepsia 62, 2000–2014. 10.1111/epi.16976 [DOI] [PubMed] [Google Scholar]
- 90.Vossler DG et al. (2018) Summary of Antiepileptic Drugs Available in the United States of America: WORKING TOWARD A WORLD WITHOUT EPILEPSY. Epilepsy Curr 18, 1–26. 10.5698/1535-7597.18.4s1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sills GJ and Rogawski MA (2020) Mechanisms of action of currently used antiseizure drugs. Neuropharmacology 168, 107966. 10.1016/j.neuropharm.2020.107966 [DOI] [PubMed] [Google Scholar]
- 92.DiMasi JA et al. (2010) Trends in risks associated with new drug development: success rates for investigational drugs. Clinical pharmacology and therapeutics 87, 272–277. 10.1038/clpt.2009.295 [DOI] [PubMed] [Google Scholar]
- 93.Brueggeman L et al. (2019) Drug repositioning in epilepsy reveals novel antiseizure candidates. Ann Clin Transl Neurol 6, 295–309. 10.1002/acn3.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Milligan CJ et al. (2014) KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Annals of neurology 75, 581–590. 10.1002/ana.24128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fitzgerald MP et al. (2019) Treatment Responsiveness in KCNT1-Related Epilepsy. Neurotherapeutics 16, 848–857. 10.1007/s13311-019-00739-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mullen SA et al. (2018) Precision therapy for epilepsy due to KCNT1 mutations: A randomized trial of oral quinidine. Neurology 90, e67–e72. 10.1212/WNL.0000000000004769 [DOI] [PubMed] [Google Scholar]
- 97.Dammes N and Peer D (2020) Paving the Road for RNA Therapeutics. Trends in pharmacological sciences 41, 755–775. 10.1016/j.tips.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hagedorn PH et al. (2017) Managing the sequence-specificity of antisense oligonucleotides in drug discovery. Nucleic Acids Res 45, 2262–2282. 10.1093/nar/gkx056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Finkel RS et al. (2021) Treatment of infantile-onset spinal muscular atrophy with nusinersen: final report of a phase 2, open-label, multicentre, dose-escalation study. Lancet Child Adolesc Health 5, 491–500. 10.1016/S2352-4642(21)00100-0 [DOI] [PubMed] [Google Scholar]
- 100.Darras BT et al. (2019) Nusinersen in later-onset spinal muscular atrophy: Long-term results from the phase 1/2 studies. Neurology 92, e2492–e2506. 10.1212/WNL.0000000000007527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Han Z et al. (2020) Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med 12. 10.1126/scitranslmed.aaz6100 [DOI] [PubMed] [Google Scholar]
- 102.Dhuri K et al. (2020) Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J Clin Med 9. 10.3390/jcm9062004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Snowball A et al. (2019) Epilepsy Gene Therapy Using an Engineered Potassium Channel. J Neurosci 39, 3159–3169. 10.1523/JNEUROSCI.1143-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuzmin DA et al. (2021) The clinical landscape for AAV gene therapies. Nat Rev Drug Discov 20, 173–174. 10.1038/d41573-021-00017-7 [DOI] [PubMed] [Google Scholar]
- 105.Wang D et al. (2019) Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 18, 358–378. 10.1038/s41573-019-0012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gray SJ et al. (2011) Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Molecular therapy: the journal of the American Society of Gene Therapy 19, 1058–1069. 10.1038/mt.2011.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kodippili K et al. (2018) Dual AAV Gene Therapy for Duchenne Muscular Dystrophy with a 7-kb Mini-Dystrophin Gene in the Canine Model. Human gene therapy 29, 299–311. 10.1089/hum.2017.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maddalena A et al. (2018) Triple Vectors Expand AAV Transfer Capacity in the Retina. Molecular therapy : the journal of the American Society of Gene Therapy 26, 524–541. 10.1016/j.ymthe.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mendell JR et al. (2017) Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. The New England journal of medicine 377, 1713–1722. 10.1056/NEjMoa1706198 [DOI] [PubMed] [Google Scholar]
- 110.Popp B et al. (2018) Need for high-resolution Genetic Analysis in iPSC: Results and Lessons from the ForlPS Consortium. Sci Rep 8, 17201. 10.1038/s41598-018-35506-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laurent LC et al. (2011) Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8, 106–118. 10.1016/j.stem.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kwon EM et al. (2017) iPSCs and fibroblast subclones from the same fibroblast population contain comparable levels of sequence variations. Proceedings of the National Academy of Sciences of the United States of America 114, 1964–1969. 10.1073/pnas.1616035114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lo Sardo V et al. (2017) Influence of donor age on induced pluripotent stem cells. Nat Biotechnol 35, 69–74. 10.1038/nbt.3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Panopoulos AD et al. (2017) iPSCORE: A Resource of 222 iPSC Lines Enabling Functional Characterization of Genetic Variation across a Variety of Cell Types. Stem Cell Reports 8, 1086–1100. 10.1016/j.stemcr.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kang X et al. (2015) Effects of Integrating and Non-Integrating Reprogramming Methods on Copy Number Variation and Genomic Stability of Human Induced Pluripotent Stem Cells. PLoS ONE 10, e0131128. 10.1371/journal.pone.0131128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Volpato V et al. (2018) Reproducibility of Molecular Phenotypes after Long-Term Differentiation to Human iPSC-Derived Neurons: A Multi-Site Omics Study. Stem Cell Reports 11, 897–911. 10.1016/j.stemcr.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lancaster MA et al. (2013) Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nehme R et al. (2018) Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep 23, 2509–2523. 10.1016/j.celrep.2018.04.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang N et al. (2017) Generation of pure GABAergic neurons by transcription factor programming. Nature methods 14, 621–628. 10.1038/nmeth.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schafer ST et al. (2019) Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nature neuroscience 22, 243–255. 10.1038/s41593-018-0295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.