ABSTRACT
The ability to distinguish between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) is of ongoing interest due to differences in transmissibility, responses to vaccination, clinical prognosis, and therapy. Although detailed genetic characterization requires whole-genome sequencing (WGS), targeted nucleic acid amplification tests can serve a complementary role in clinical settings, as they are more rapid and accessible than sequencing in most laboratories. We designed and analytically validated a two-reaction multiplex reverse transcription-quantitative PCR (RT-qPCR) assay targeting spike protein mutations L452R, E484K, and N501Y in reaction 1 and del69–70, K417N, and T478K in reaction 2. This assay had 95 to 100% agreement with WGS for 502 upper respiratory tract swab samples collected between 26 April 2021 and 1 August 2021, consisting of 43 Alpha, 2 Beta, 20 Gamma, 378 Delta, and 59 non-VOC infections. Validation in a separate group of 230 WGS-confirmed Omicron variant samples collected in December 2021 and January 2022 demonstrated 100% agreement. This RT-qPCR-based approach can be implemented in clinical laboratories already performing SARS-CoV-2 nucleic acid amplification tests to assist in local epidemiological surveillance and clinical decision-making.
KEYWORDS: COVID-19, Omicron, SARS-CoV-2, variant
INTRODUCTION
Since the original strain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first discovered in late 2019, numerous new variants have been identified, including variants of concern (VOCs) Alpha (B.1.1.7 and sublineages), Beta (B.1.351), Gamma (P.1 and sublineages), Delta (B.1.617.2 and sublineages), and Omicron (B.1.1.529 and sublineages) (1). Importantly, these VOCs differ in their clinical prognosis, transmissibility, antibody susceptibility, and response to vaccination (2–22). Whole-genome sequencing (WGS) has played a critical role in identifying the emergence of these new variants (23–25), and millions of distinct sequences have been deposited in public repositories such as the Global Initiative on Sharing Avian Influenza Data (GISAID) database (26). However, WGS has a relatively long turnaround time, is labor-intensive, and requires instruments, bioinformatic support, and specially trained staff that may not be widely available to many clinical laboratories. Therefore, the development of reverse transcription-quantitative PCR (RT-qPCR) assays to presumptively type SARS-CoV-2 variants may be an important real-time complement to WGS epidemiological surveillance and may directly impact the clinical care of individual patients by informing selection of expensive and potentially difficult-to-source monoclonal antibody therapies (2, 7, 13–17, 20, 21, 27). It is important to note that such presumptive typing assays may provide atypical results for emerging strains due to mutations within primer and/or probe binding sites. Therefore, they must be intelligently designed, thoroughly validated, and interpreted carefully.
In this study, we report the design of a multiplex RT-qPCR assay that detects the del69–70, K417N, and T478K mutations in SARS-CoV-2 spike protein and targets the wild-type 69 to 70 (wt69–70) sequence as an internal control. We further evaluate the performance of this assay in combination with our previously described RT-qPCR assay for the detection of L452R, E484K, and N501Y (28) and demonstrate the utility of this targeted mutational analysis to accurately distinguish among VOCs.
MATERIALS AND METHODS
Assay design.
The spike protein mutations associated with each variant that are interrogated by the RT-qPCR assays are summarized in Fig. 1. In the first reaction (reaction 1), we utilized our previously described RT-qPCR assay to detect L452R, E484K, and N501Y mutations in the spike receptor binding domain (RBD) (28). The present study describes the combination of this assay with a second, newly designed reaction (reaction 2), which detects the deletion of amino acids 69 and 70 in the spike N-terminal domain (del69–70) as well as K417N and T478K mutations in the RBD. We use allele-specific RT-qPCRs with probe sequences designed to maximize the difference in annealing temperatures between mutant and wild-type sequences, allowing differential binding and amplification. The primer/probe sequences for each mutation site are summarized in Table 1, and the guidance for interpretation and reporting is described in Table 2. Additional details are provided in the supplemental material, including single-stranded DNA (ssDNA) sequences for analytical experiments (see Table S1 in the supplemental material), analytical validation data (see Table S2), and in silico analysis of primer and probe sequences (see Fig. S1).
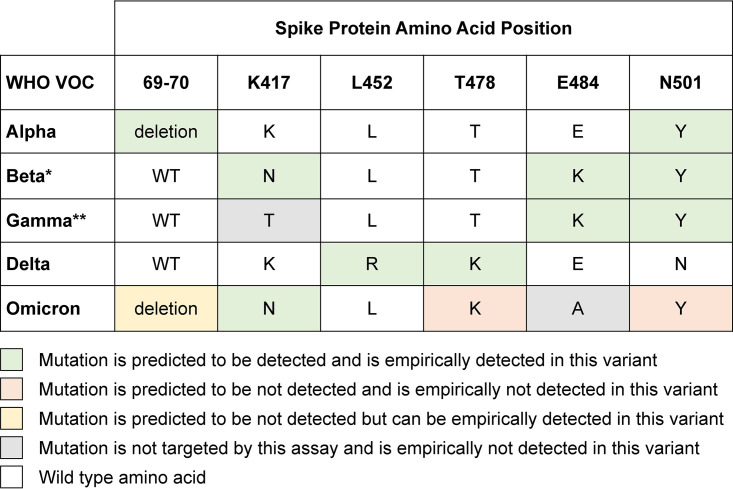
FIG 1.
Summary of current WHO-designated VOC along with their expected spike mutations at sites targeted by this two-reaction multiplex SARS-CoV-2 RT-qPCR genotyping approach. These reactions are designed to detect the following mutations: del69–70, K417N, L452R, T478K, E484K, and N501Y. Shading indicates predicted versus empirical performance of this assay for the detection and differentiation of these VOCs. The predicted detection for the VOC Omicron is based on both the sequence at the target sites and known adjacent mutations in the probe binding site. *, Known limitation of the assay in differentiating VOC Beta from the VOI Mu; **, known limitation of the assay in differentiating VOC Gamma from the VOI Mu.
TABLE 1.
Reaction 2 primer and probe oligonucleotide sequences
| Namea | Sequence (5′→3′) | Final concn (nM) |
|---|---|---|
| Primers | ||
| del69–70_FWD | CATTAAATGGTAGGACAGGGTTA | 300 |
| del69–70_REV | ACATTCAACTCAGGACTTGTT | 300 |
| K417N_FWD | GCAGCCTGTAAAATCATCTG | 300 |
| K417N_REV | CATTTGTAATTAGAGGTGATGAAGTC | 300 |
| T478K_FWD | AAAGGAAAGTAACAATTAAAACCT | 300 |
| T478K_REV | AGGAAGTCTAATCTCAAACCT | 300 |
| Probes | ||
| del69–70_MT_HEX | HEX-TTGGTCCCAGAGATAGCATG-BHQ1 | 50 |
| wt69–70_WT_CY3.5b | CY3.5-GGTCCCAGAGACATGTATAG-BHQ2 | 50 |
| K417N_MT_CY5 | CY5-TAATCAGCAATATTTCCAGT-BHQ2 | 50 |
| T478K_MT_FAM | FAM-ACCATTACAAGGTTTGCTAC-BHQ1 | 50 |
FWD, forward; REV, reverse; WT, wild-type; MT, mutant; CY3.5, cyanine 3.5; BHQ, black hole quencher.
Included as an internal amplification control for samples without the del69–70 mutation.
TABLE 2.
Interpretation and reporting of the two-reaction multiplex genotyping RT-qPCR results
| Reaction 1 resultsa |
Reaction 2 results |
Action (see comments)b | ||||||
|---|---|---|---|---|---|---|---|---|
| N501WT (ROX/Cy3.5) | N501Y (FAM) | E484K (CY5) | L452R (HEX) | 69_70WT (ROX/Cy3.5) | T478K (FAM) | K417N (CY5) | Del69_70 (HEX) | |
| DTD ≤40 | NDET | NDET | DTD ≤40 | DTD ≤40 | DTD ≤40 | DTD ≤40 | NDET | Report as 1 |
| DTD ≤40 | NDET | NDET | DTD ≤40 | DTD ≤40 | DTD ≤40 | NDET | NDET | Report as 1 |
| DTD ≤40 | NDET | NDET | DTD ≤40 | NDET | DTD ≤40 | DTD ≤40 | NDET | Report as 1 |
| NDET | DTD ≤40 | DTD ≤40 | NDET | DTD ≤40 | NDET | DTD ≤40 | NDET | Report as 2 |
| NDET | DTD ≤40 | DTD ≤40 | NDET | DTD ≤40 | NDET | NDET | NDET | Report as 3 |
| NDET | DTD ≤40 | DTD ≤40 | NDET | NDET | NDET | NDET | DTD ≤40 | Report as 4 |
| NDET | DTD ≤40 | NDET | NDET | NDET | NDET | NDET | DTD ≤40 | Report as 4 |
| NDET | NDET | NDET | NDET | NDET | NDET | DTD ≤40 | DTD ≤40 | Report as 5 |
| NDET | NDET | NDET | NDET | NDET | NDET | DTD ≤40 | NDET | Report as 5 |
| NDET | NDET | NDET | NDET | NDET | NDET | NDET | NDET | Unable to genotype (refer to 6) |
| Any target with CT value of >40 or abnormal/inconclusive amplification curves | Review (refer to 7) | |||||||
| Any scenarios not designated above | Review (refer to 7) | |||||||
DTD ≤40, detected with a CT value of ≤40; NDET, not detected.
Result interpretation: 1. Probable VOC Delta; increased transmissibility; decreased susceptibility to bamlanivimab (LY-CoV555). 2. Possible VOC Beta versus VOI Mu; decreased susceptibility to bamlanivimab, etesevimab (LY-CoV016, JS016, CB6ETE), bamlanivimab plus etesevimab, casirivimab (REGN10933), and regdanvimab (CT-P59). 3. Possible VOC Gamma versus VOI Mu; decreased susceptibility to bamlanivimab, etesevimab, bamlanivimab plus etesevimab, casirivimab, and regdanvimab. 4. Probable VOC Alpha; decreased susceptibility to etesevimab. 5. Probable VOC Omicron; increased transmissibility; decreased susceptibility to bamlanivimab, etesevimab, bamlanivimab plus etesevimab, casirivimab, imdevimab (REGN10987), casirivimab plus imdevimab (REGN-COV2), cilgavimab (COV2-2130, AZD1061), tixagevimab (COV2-2196, AZD8895), cilgavimab plus tixagevimab, and regdanvimab; Decreased susceptibility to convalescent plasma. 6. Unable to interpret results due to low level of viral RNA. 7. Triage for medical director review (mutations uncommonly seen together, which could indicate possible mixed infection, contamination, or nonspecific amplification; analyze additional information, including previous results and curve shape; potential new variant or novel mutation in primer/probe sites causing dropout); consider sequencing if the level of viral RNA is sufficient. Interpretations regarding monoclonal antibody susceptibility and plasma susceptibility were based on data from the Stanford Coronavirus Antiviral and Resistance Database (https://covdb.stanford.edu). Decreased susceptibility to monoclonal antibodies and convalescent plasma was defined as a >10-fold reduction in neutralization.
Clinical specimens.
The samples included in the initial phase of this study were upper respiratory tract swab specimens collected from patients as part of routine clinical care between 26 April 2021 and 1 August 2021. Testing was performed at the Stanford Clinical Virology Laboratory, which provides virological testing for all Stanford-affiliated hospitals and outpatient centers in the San Francisco Bay Area. These initial SARS-CoV-2 nucleic acid amplification tests (NAATs) prior to genotyping were conducted according to manufacturer and emergency authorization instructions, as described previously (28) and in the supplemental material. All samples that tested positive for SARS-CoV-2 RNA were subjected to genotyping. We then excluded samples that were initially tested by laboratory-based methods with cycle threshold (CT) values of ≥35 or relative light unit (RLU) values of ≤1,100. We included all available samples that were initially tested at or near the point of care, because CT data were not readily available for real-time specimen triage for these samples. We also excluded follow-up specimens to eliminate patient-level duplicates. Subsequent validation of this assay for Omicron variant detection was conducted using a convenience set of 230 Omicron variant samples with available WGS data that were collected between 2 December 2021 and 5 January 2022. This study was conducted with Stanford University institutional review board approval (protocol 57519), and the requirement for individual consent was waived.
WGS.
To validate the genotyping RT-qPCRs, we tested their performance against WGS for a subset of the samples in the initial cohort from 26 April 2021 to 1 August 2021 with CT values of <30. Samples with nondominant variant typing by RT-qPCR were prioritized for sequencing, with the remaining isolates chosen randomly to fill a sequencing run. WGS was conducted as described previously, using a laboratory-developed pipeline consisting of long-range PCR followed by fragmentation and then single-end 150-cycle sequencing using the MiSeq reagent kit v3 (Illumina, San Diego, CA) (28). Genomes were assembled via a custom assembly and bioinformatics pipeline using NCBI GenBank accession number NC_045512.2 as the reference. Whole-genome sequences with ≥75% genome coverage to a depth of at least 10 reads were accepted for interpretation. Mutation calling required a depth of at least 12 reads with a minimum variant frequency of 20%. PANGO lineage assignment was performed using the Pangolin Coronavirus Disease 2019 (COVID-19) Lineage Assigner (https://pangolin.cog-uk.io) running Pangolin v3.1.17, while Nextclade Web v1.13.1 and auspice.us v0.8.0 were used to perform phylogenetic placement (3, 29, 30). Both lineage and clade assignments were performed on 1 February 2022. WGS data were deposited in GISAID (see Table S3).
Statistical analysis.
Positive percent agreement (PPA) and negative percent agreement (NPA) were reported with Clopper-Pearson score 95% binomial confidence intervals (CIs) using WGS as the reference method. Analyses were conducted using the R statistical software package. This study was reported in accordance with Standards for Reporting of Diagnostic Accuracy Studies (STARD) guidelines.
RESULTS
During the initial study period of 26 April 2021 to 1 August 2021, the Stanford Clinical Virology Laboratory received 102,158 specimens from 70,544 unique individuals. A total of 1,657 samples from unique individuals tested positive for SARS-CoV-2, of which 1,093 (66%) had genotyping RT-qPCR 1 and 2 performed and 502 (30.3%) had successful WGS performed (see Fig. S2 in the supplemental material). The lower limits of detection for the mutation site probes in reaction 2 were 14.8 copies/μL template for del69–70 (hexachlorofluorescein [HEX]), 16.4 copies/μL template for K417N (cyanine 5 [CY5]), and 2.1 copies/μL template for T478K [5(6)-carboxyfluorescein [FAM]]. The median number of aligned reads for WGS was 485,870 (interquartile range [IQR], 289,363 to 655,481 reads), while the median genome coverage to a depth of at least 10 reads was 99.3% (IQR, 97.1 to 99.3%). Of note, reaction 1 was performed in near real time, while reaction 2 was performed retrospectively. Overall, this subset of sequenced samples had patient and testing characteristics that closely resembled those of the larger cohorts (see Table S4).
The assay yielded “unable to genotype” results for 152 of 1,093 samples (14%) due to lack of amplification of any target in either or both reactions. Assay failure occurred predominantly in samples that were originally tested at or near the point of care (119/341 samples [35%]), where all positive samples were triaged for genotyping without any filter. In contrast, assay failure occurred much less frequently in samples that were originally tested in the moderate-to-high-complexity virology laboratory (33/752 [4%]), where samples with lower viral RNA levels (CT values of ≥35) were not triaged for genotyping. In the group of 752 samples tested in the virology laboratory, 601 had known CT values. Among those 601 samples, 68 had CT values of >30, of which 11/68 (16%) failed amplification.
PPA and NPA values for the six individual mutations targeted by the genotyping RT-qPCRs, compared to WGS, were calculated for the 502 samples for which both RT-qPCR and WGS were performed (see Fig. S2). The number of samples that tested positive for each mutation reflects the natural prevalence of each mutation during that time period. For the combination of reactions 1 and 2, the PPAs for del69–70, L452R, T478K, E484K, and N501Y were 100% (Table 3). Across all six loci, only K417N had a false-negative result, resulting in a PPA of 96% (27/28 samples); in that sample, WGS showed a synonymous T-to-C mutation at position 1254 of the spike gene, corresponding to amino acid position 418, changing the codon from ATT to ATC. This single-base pair substitution likely decreased the annealing temperature, causing probe dropout and a false-negative result.
TABLE 3.
Comparison of RT-qPCR and WGS results for SARS-CoV-2 spike gene mutation detection in the initial cohort (n = 502)
| Spike mutation and RT-qPCR result | No. with positive WGS result | No. with negative WGS result | PPA (95% CI) (%) | NPA (95% CI) (%) |
|---|---|---|---|---|
| Del69–70 | ||||
| Positive | 43 | 0 | 100 (92–100) | 100 (99–100) |
| Negative | 0 | 459 | ||
| K417N | ||||
| Positive | 27 | 0 | 96 (82–100) | 100 (99–100) |
| Negative | 1a | 474 | ||
| L452R | ||||
| Positive | 403 | 5b | 100 (99–100) | 95 (89–98) |
| Negative | 0 | 94 | ||
| T478K | ||||
| Positive | 379 | 0 | 100 (99–100) | 100 (97–100) |
| Negative | 0 | 123 | ||
| E484K | ||||
| Positive | 35 | 3c | 100 (90–100) | 99 (98–100) |
| Negative | 0 | 464 | ||
| N501Y | ||||
| Positive | 70 | 0 | 100 (95–100) | 100 (99–100) |
| Negative | 0 | 432 |
False-negative RT-qPCR result due to a synonymous mutation at spike amino acid position 418 (codon ATT to ATC), causing probe dropout.
False-negative WGS results due to insufficient read counts (<12 reads) at this codon. Manual review of the sequences revealed 3 to 9 mutant reads in each sample.
These 3 samples were found by WGS to be positive for E484Q. While positive for the E484K target by RT-qPCR, these samples had a distinct blunted amplification curve associated with E484Q, as described previously (31).
The NPA values for del69–70, K417N, T478K, and N501Y were 100% (Table 3). L452R had an NPA value of 95% (94/99 samples), and E484K had an NPA value of 99% (464/467 samples). At the L452 locus, 5 samples that were positive for the L452R mutation by RT-qPCR were negative by WGS. Manual review of the WGS data showed that these were likely false-negative WGS results due to insufficient coverage (<12 reads) at this codon. Three to 9 reads containing the L452R mutation were identified in the WGS primary data for each of these five samples. These 5 samples were all in the Delta lineage, based on mutations found at other positions by sequencing.
For the E484K target, there were 3 samples that tested positive for the E-to-K mutation but in fact had a E484Q mutation, as determined by WGS. In both the E-to-K mutation (GAA to AAA) and the E-to-Q mutation (GAA to CAA), there was a single base substitution at the first position of the codon, resulting in nonspecific probe binding. These three samples had a distinct blunted amplification curve with high CT values associated with E484Q, as described previously (31). While this cross-reactivity is a limitation of the E484K probe design, the unusually shaped amplification curves were identified and flagged for medical director review as part of the assay interpretation protocol (Table 2). Presumptive typing for such cases would need to be deferred until WGS confirmation.
Of note, there was a subset of variant AY.2, involving 4 specimens in our cohort, that had a V70F mutation causing both del69–70 and wt69–70 probes not to bind. However, because this variant would have T478K and K417N detected, the wt69–70 signal was not needed as an amplification control. This subset of the AY.2 genotype is expected to be positive for L452R and N501 wild-type internal controls while negative for N501Y and E484K in reaction 1 and positive for K417N and T4748K while negative for del69–70 and wt69–70 internal controls in reaction 2. This scenario has been reflected in the clinical interpretation table (Table 2).
SARS-CoV-2-positive specimens collected starting on 2 December 2021 began to show an unusual combination of mutations, i.e., the presence of K417N and del69–70 only in reaction 2, with all targets, including the internal control N501, not detected in reaction 1. Based on in silico analysis, we determined that these cases likely represented the Omicron variant. While most Omicron variant strains possess del69–70, K417N, T478K, and N501Y mutations, they also have A67V, S477N, and Q498R mutations, which would be predicted to interfere with binding of the del69–70/wt69–70, T478K, and N501Y/N501 probes, respectively. Although the E484K probe demonstrated cross-reactivity with strains containing the E484Q mutation, as described above, the E484K probe did not detect the E484A mutation in the Omicron variant since it differed by ≥2 bases. The del69–70 probe likely was able to retain some degree of binding due to the wider melting temperature differential of a 6-nucleotide deletion, compared to a point mutation. Therefore, we validated this assay for Omicron detection using a set of 230 SARS-CoV-2-positive samples confirmed to be Omicron by WGS. We found that the unique pattern of K417N and del69–70 in reaction 2, along with the failure to amplify any target, including the internal control, in reaction 1, was present in 230/230 (100% [95% CI, 98 to 100%]) Omicron samples tested. This pattern was not seen in any of the 1,093 non-Omicron samples previously genotyped.
We next predicted the World Health Organization (WHO) variant designation of samples using RT-qPCR results and correlated them with the PANGO lineage assignments based on WGS data (Table 4). Mapping the genotyping results for the cohort based on RT-qPCR mutation analysis onto the Nextclade phylogenetic tree demonstrated close correlation with their WHO variant designations (Fig. 2). Among the 732 clinical samples that were tested by both RT-qPCR and WGS, 43 samples (5.9%) were Alpha (B.1.1.7 or Q.3), 2 samples (0.3%) were Beta (B.1.351), 20 samples (2.7%) were Gamma (P.1 and sublineages), 378 samples (51.6%) were Delta (B.1.617.2 or AY sublineages), and 230 samples (31.4%) were Omicron (B.1.1.529 or BA sublineages). There were no RT-qPCR false-negative results in assigning samples to these lineages. In addition, 59 samples (8.1%) that were tested by WGS that did not correspond to a WHO VOC as of 2 February 2022. Within this subset, there were 4 samples that were erroneously assigned as Gamma and 1 that was assigned as Beta by RT-qPCR. By WGS, these samples were variant of interest (VOI) Mu (B.1.621 or BB.2). This variant shares mutations E484K and N501Y with both the Beta and Gamma variants. A subset of Mu also includes the K417N mutation, which is seen in the Beta variant. Thus, our PCR assay could not distinguish VOI Mu from VOCs Beta and Gamma. Our interpretation table information reflects this limitation (Table 2). The remaining 54 samples did not contain mutation patterns associated with VOCs by either RT-qPCR or WGS.
TABLE 4.
Comparison of RT-qPCR and WGS results for SARS-CoV-2 VOC detection (n = 732)
| WHO VOC and PANGO lineage by WGS | No. with RT-qPCR result indicating:b |
||||||
|---|---|---|---|---|---|---|---|
| Alpha | Beta | Gamma | Delta | Omicron | Non-VOC | Total no. | |
| All Alpha | 43 | - | - | - | - | - | 43 |
| B.1.1.7 | 37 | - | - | - | - | - | 37 |
| Q.3 | 6 | - | - | - | - | - | 6 |
| All Beta | - | 2 | - | - | - | - | 2 |
| B.1.351 | - | 2 | - | - | - | - | 2 |
| All Gamma | - | - | 20 | - | - | - | 20 |
| P.1 | - | - | 13 | - | - | - | 13 |
| P.1.10 | - | - | 5 | - | - | - | 5 |
| P.1.17 | - | - | 2 | - | - | - | 2 |
| All Delta | - | - | - | 378 | - | - | 378 |
| B.1.617.2 | - | - | - | 29 | - | - | 29 |
| AY.1 | - | - | - | 20 | - | - | 20 |
| AY.2 | - | - | - | 5 | - | - | 5 |
| AY.3 | - | - | - | 5 | - | - | 5 |
| AY.4 | - | - | - | 1 | - | - | 1 |
| AY.13 | - | - | - | 32 | - | - | 32 |
| AY.14 | - | - | - | 59 | - | - | 59 |
| AY.19 | - | - | - | 1 | - | - | 1 |
| AY.20 | - | - | - | 5 | - | - | 5 |
| AY.23 | - | - | - | 1 | - | - | 1 |
| AY.25 | - | - | - | 7 | - | - | 7 |
| AY.25.1 | - | - | - | 25 | - | - | 25 |
| AY.26 | - | - | - | 15 | - | - | 15 |
| AY.35 | - | - | - | 2 | - | - | 2 |
| AY.43 | - | - | - | 2 | - | - | 2 |
| AY.44 | - | - | - | 77 | - | - | 77 |
| AY.46.2 | - | - | - | 1 | - | - | 1 |
| AY.47 | - | - | - | 8 | - | - | 8 |
| AY.48 | - | - | - | 1 | - | - | 1 |
| AY.52 | - | - | - | 1 | - | - | 1 |
| AY.54 | - | - | - | 3 | - | - | 3 |
| AY.59 | - | - | - | 1 | - | - | 1 |
| AY.62 | - | - | - | 1 | - | - | 1 |
| AY.67 | - | - | - | 3 | - | - | 3 |
| AY.74 | - | - | - | 1 | - | - | 1 |
| AY.75 | - | - | - | 10 | - | - | 10 |
| AY.98.1 | - | - | - | 1 | - | - | 1 |
| AY.100 | - | - | - | 3 | - | - | 3 |
| AY.103 | - | - | - | 26 | - | - | 26 |
| AY.110 | - | - | - | 9 | - | - | 9 |
| AY.114 | - | - | - | 1 | - | - | 1 |
| AY.116.1 | - | - | - | 2 | - | - | 2 |
| AY.118 | - | - | - | 5 | - | - | 5 |
| AY.119 | - | - | - | 4 | - | - | 4 |
| AY.120.1 | - | - | - | 1 | - | - | 1 |
| AY.121 | - | - | - | 3 | - | - | 3 |
| AY.122 | - | - | - | 5 | - | - | 5 |
| AY.126 | - | - | - | 2 | - | - | 2 |
| All Omicron | - | - | - | - | 230 | - | 230 |
| B.1.1.529/BA.1 | - | - | - | - | 123 | - | 123 |
| BA.1.1 | - | - | - | - | 107 | - | 107 |
| All non-VOC | - | 1 | 4 | - | - | 54 | 59 |
| A.2.5 | - | - | - | - | - | 6 | 6 |
| B.1 | - | - | - | - | - | 3 | 3 |
| B.1.1.318 | - | - | - | - | - | 1 | 1 |
| B.1.1.519 | - | - | - | - | - | 1 | 1 |
| B.1.311 | - | - | - | - | - | 1 | 1 |
| B.1.427 | - | - | - | - | - | 3 | 3 |
| B.1.429 | - | - | - | - | - | 8 | 8 |
| B.1.526 | - | - | - | - | - | 10 | 10 |
| B.1.621a | - | 1 | 2 | - | - | - | 3 |
| BB.2a | - | - | 2 | - | - | - | 2 |
| B.1.627 | - | - | - | - | - | 1 | 1 |
| B.1.637 | - | - | - | - | - | 11 | 11 |
| XB | - | - | - | - | - | 9 | 9 |
| All | 43 | 3 | 24 | 378 | 230 | 54 | 732 |
VOI Mu with E484K and N501Y mutations and a subset with K417N, which overlaps with VOCs Beta and Gamma.
Dashes indicate none detected.
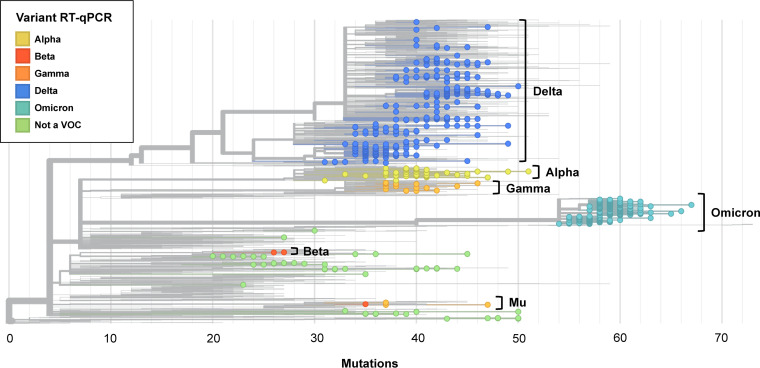
FIG 2.
Nextclade phylogenetic tree of 3,097 SARS-CoV-2 genomes, including all 732 of the sequenced genomes from this study and 2,365 genomes from the Nextstrain global reference tree as of 2 February 2022. The 732 included genomes are colored according to RT-qPCR-genotyping-predicted variant type, with each circle representing a sequenced genome. Branch lengths correspond to nucleotide divergence. Sequenced genomes span the breadth of the reference tree. Annotation to the right of the tree indicates the variant type based on WGS. Variant determination by RT-qPCR matched that by WGS except for 1 sequence typed as Beta and 4 sequences typed as Gamma by RT-qPCR, which clustered with VOI Mu by WGS.
DISCUSSION
The ability to distinguish between SARS-CoV-2 VOCs is important for epidemiological surveillance and, in certain circumstances, the care of individual COVID-19 patients. In this study, we describe a two-reaction, multiplex RT-qPCR genotyping approach that examines the spike mutations del69–70, K417N, L452R, T478K, E484K, and N501Y. This targeted mutational analysis can be used to differentiate between the WHO VOCs Alpha (B.1.1.7 and sublineages), Beta (B.1.351), Gamma (P.1 and sublineages), Delta (B.1.617.2 and sublineages), and Omicron (B.1.1.529 and sublineages), as well as to identify samples that cannot be categorized as a known VOC or VOI. Because the first part of this approach, reaction 1, was described previously, this study focuses on reaction 2 and the integrated results of the two-reaction test (28). Overall, these reactions showed high levels of concordance with WGS, demonstrating PPA values of 96 to 100% and NPA values of 95 to 100% for all targeted mutations.
Several groups previously described similar approaches to SARS-CoV-2 variant determination by RT-qPCR and digital droplet RT-PCR, particularly for the spike del69–70, E484K, and N501Y mutations (32–39). Some of those assays included additional mutation sites that were not in our study, such as spike del144 or open reading frame 1a (ORF1a) Δ3675–3677 (32, 38). Those earlier assays, published prior to the rise of Delta, primarily targeted VOCs Alpha, Beta, and Gamma. They were then followed by a surge of reports on the detection of the Delta variant. Garson et al. utilized double-mismatch allele-specific RT-PCR at L452R and T478K to differentiate the Delta variant from other VOCs among 42 U.K. patient samples (40). Aoki et al. described an approach that combined nested PCR along with high-resolution melting analysis at the same mutations, which was validated in a small Japanese patient cohort (41). Barua et al. used a slightly different approach, taking advantage of the difference in melting temperature for a probe targeted to the Delta spike mutation T478K, compared to other variants, for a Delta-specific RT-fluorescence resonance energy transfer (FRET)-PCR assay (42). Another defining feature of VOC Delta is spike del156–157, which was the target of a Delta variant PCR test developed by Hamill et al. (43). To our knowledge, the two-reaction multiplex RT-qPCR approach outlined in this study examining six different mutation sites is the most comprehensive variant genotyping test described that can identify Alpha, Beta, Gamma, Delta, and Omicron variants.
Multiplex RT-qPCR SARS-CoV-2 genotyping takes advantage of a commonly used molecular technique that can be implemented by laboratories using existing equipment, materials, and personnel. Because this assay is more accessible and has a shorter turnaround time than WGS, we envision it serving as a complement to sequencing. The genotyping RT-qPCR can provide more detailed and up-to-date epidemiological information by increasing the sample size of categorized variants in each geographic region and can be essential in tracking local outbreaks in areas without direct access to WGS. For individual patients, the turnaround time of several hours also allows the assay to directly impact clinical care. For example, VOCs show different susceptibilities to monoclonal antibody treatments, and variant reporting could include this information (Table 2) (2). Furthermore, current ongoing trials for small-molecule drugs and other treatments may yield more information about variant-specific treatment strategies.
Sequencing is still needed, however, for the identification and confirmation of novel variants. This is evidenced by the 5 VOI Mu samples originally misidentified by RT-qPCR as Gamma and Beta. The two approaches are complementary in nature, and RT-qPCR genotyping can help triage or prioritize samples for sequencing. For example, prescreening by RT-qPCR can enrich for samples with atypical mutation patterns, leading to more efficient use of sequencing resources and, potentially, more rapid identification of new variants.
This RT-qPCR approach has several limitations, as evidenced by its assay failure rate of 14% across all tested samples in our initial cohort. Because multiplex RT-qPCRs involve a mixture of multiple sets of primers and probes, they are inherently less analytically sensitive than single-target assays. For samples with RNA concentrations near the lower limits of detection, freeze-thaw cycles could impact RNA stability and may not yield consistent results due to stochastic variation. This issue could be alleviated by implementing a CT/RLU filter to genotype only samples most likely to yield interpretable results. Within our 1,093-sample cohort, the lower assay failure rate for samples tested in our clinical virology laboratory (4%), compared to near care settings (35%), is likely attributable to genotyping only specimens with higher viral RNA levels. Note, however, that, even with such filtering, mutation analysis by RT-qPCR remains more analytically sensitive than WGS. This study is also limited by the absence of VOC coinfections, such as coinfections with Delta and Omicron, although we anticipate that this RT-qPCR approach would be able to detect such cases. Future experiments will be required to confirm detection of VOC coinfections, including cases with different viral RNA levels and variant proportions.
Another limitation to this approach is the continuously evolving variant landscape, which may render such a targeted assay obsolete in a relatively short period of time. First, it is important to consider that the loss of expected internal control signal for the N501 and/or wt69–70 targets in known SARS-CoV-2 RNA-positive samples is itself useful information, analogous to the widely used spike gene target failure (36). Furthermore, the inclusion of multiple mutations in key residues that influence viral fitness and antibody escape helps guard against rapid obsolescence, as evidenced by the ability of the RT-qPCR approach to rapidly detect the emergence of the Omicron variant in our population, as well as all major variant replacements that occurred in 2021. Notably, this approach also revealed emerging community transmission of BA.2 in early 2022, with K417N and wt69–70 detected in reaction 2 and all targets, including N501, not detected in reaction 1. Nevertheless, flexibility and vigilance are required should redesign and revalidation be required as novel variants emerge.
In summary, we developed and validated a two-reaction multiplex RT-qPCR genotyping strategy that interrogates six clinically relevant mutations within the SARS-CoV-2 spike, namely, del69–70, K417N, L452R, T478K, E484K, and N501Y. This approach allows identification of WHO VOCs Alpha, Beta, Gamma, Delta, and Omicron, with excellent concordance to WGS results. Overall, this method complements WGS and is suitable for clinical decision-making, near-real-time variant surveillance, and the triage of samples for sequencing.
ACKNOWLEDGMENTS
We thank the Stanford Clinical Virology Laboratory staff members for their excellence and perseverance in caring for our community while facing the unique challenges of this pandemic. We additionally are grateful to the Stanford Protein and Nucleic Acid Facility for the synthesis of reagents on short notice.
Footnotes
Supplemental material is available online only.
Contributor Information
Benjamin A. Pinsky, Email: bpinsky@stanford.edu.
Angela M. Caliendo, Rhode Island Hospital
REFERENCES
- 1.World Health Organization. 2022. Tracking SARS-CoV2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants. Accessed 28 February 2022.
- 2.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, Fera D, Shafer RW. 2021. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22:757–773. 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day T, Gandon S, Lion S, Otto SP. 2020. On the evolutionary epidemiology of SARS-CoV-2. Curr Biol 30:R849–R857. 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, Group CC-W, Consortium C-GU, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ. 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muik A, Wallisch A-K, Sänger B, Swanson KA, Mühl J, Chen W, Cai H, Maurus D, Sarkar R, Türeci Ö, Dormitzer PR, Şahin U. 2021. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science 371:1152–1153. 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, Liu L, Kwong PD, Huang Y, Shapiro L, Ho DD. 2021. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 29:747–751.e4. 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collier DA, De Marco A, Ferreira I, Meng B, Datir RP, Walls AC, Kemp SA, Bassi J, Pinto D, Silacci-Fregni C, Bianchi S, Tortorici MA, Bowen J, Culap K, Jaconi S, Cameroni E, Snell G, Pizzuto MS, Pellanda AF, Garzoni C, Riva A, CITIID-NIHR BioResource COVID-19 Collaboration, - Elmer A, Kingston N, Graves B, McCoy LE, Smith KGC, Bradley JR, Temperton N, Ceron-Gutierrez L, Barcenas-Morales G, COVID-19 Genomics UK (COG-UK) Consortium, Harvey W, Virgin HW, Lanzavecchia A, Piccoli L, Doffinger R, Wills M, Veesler D, Corti D, Gupta RK. 2021. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 593:136–141. 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, Yoon H, Li D, Haynes BF, Sanders KO, Gnanakaran S, Hengartner N, Pajon R, Smith G, Dubovsky F, Glenn GM, Korber B, Montefiori DC. 2021. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral Spike vaccines. Cell Host Microbe 29:529–539.e3. 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karim SSA. 2021. Vaccines and SARS-CoV-2 variants: the urgent need for a correlate of protection. Lancet 397:1263–1264. 10.1016/S0140-6736(21)00468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolze A, Cirulli ET, Luo S, White S, Wyman D, Rossi AD, Machado H, Cassens T, Jacobs S, Schiabor Barrett KM, Tsan K, Nguyen J, Ramirez JM, Sandoval E, Wang X, Wong D, Becker D, Laurent M, Lu JT, Isaksson M, Washington NL, Lee W. 2021. SARS-CoV-2 variant Delta rapidly displaced variant Alpha in the United States and led to higher viral loads. medRxiv. 10.1101/2021.06.20.21259195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, Larsen BB, Anderson C, White S, Cassens T, Jacobs S, Levan G, Nguyen J, Ramirez JM, Rivera-Garcia C, Sandoval E, Wang X, Wong D, Spencer E, Robles-Sikisaka R, Kurzban E, Hughes LD, Deng X, Wang C, Servellita V, Valentine H, De Hoff P, Seaver P, Sathe S, Gietzen K, Sickler B, Antico J, Hoon K, Liu J, Harding A, Bakhtar O, Basler T, Austin B, Isaksson M, Febbo P, Becker D, Laurent M, McDonald E, Yeo GW, Knight R, Laurent LC, de Feo E, Worobey M, Chiu C, Suchard MA, Lu JT, Lee W, Andersen KG. 2021. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv. 10.1101/2021.02.06.21251159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Chen Z, Azman AS, Sun R, Lu W, Zheng N, Zhou J, Wu Q, Deng X, Zhao Z, Chen X, Ge S, Yang J, Leung DT, Yu H. 2022. Neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants Iinduced by natural infection or vaccination: A systematic review and pooled analysis. Clin Infect Dis 74:734–742. 10.1093/cid/ciab646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supasa P, Zhou D, Dejnirattisai W, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Nutalai R, Tuekprakhon A, Wang B, Paesen GC, Slon-Campos J, Lopez-Camacho C, Hallis B, Coombes N, Bewley KR, Charlton S, Walter TS, Barnes E, Dunachie SJ, Skelly D, Lumley SF, Baker N, Shaik I, Humphries HE, Godwin K, Gent N, Sienkiewicz A, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Klenerman P, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, Hall DR, Williams MA, Paterson NG, James W, Carroll MW, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 184:2201–2211.e7. 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jäck H-M, Jahrsdörfer B, Schrezenmeier H, Müller M, Kleger A, Münch J, Pöhlmann S. 2021. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184:2384–2393.e12. 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, Nutalai R, Zhou D, Mentzer AJ, Zhao Y, Duyvesteyn HME, López-Camacho C, Slon-Campos J, Walter TS, Skelly D, Johnson SA, Ritter TG, Mason C, Costa Clemens SA, Gomes Naveca F, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Dold C, Temperton N, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert SC, Malik T, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Baillie V, Serafin N, Ditse Z, Da Silva K, Paterson NG, Williams MA, Hall DR, Madhi S, Nunes MC, Goulder P, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 184:4220–4236.e13. 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Pere H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Loriere E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Liu Y, Xia H, Zou J, Weaver SC, Swanson KA, Cai H, Cutler M, Cooper D, Muik A, Jansen KU, Sahin U, Xie X, Dormitzer PR, Shi PY. 2021. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 596:273–275. 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- 19.Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, Dushoff J, Mlisana K, Moultrie H. 2021. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 10.1101/2021.11.11.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, Metzler M, Kohmer N, Hoehl S, Helfritz FA, Wolf T, Goetsch U, Ciesek S. 2021. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 10.1101/2021.12.07.21267432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, Pere H, Veyer D, Puech J, Rodary J, Baele G, Dellicour S, Raymenants J, Gorissen S, Geenen C, Vanmechelen B, Wawina-Bokalanga T, Marti-Carreras J, Cuypers L, Seve A, Hocqueloux L, Prazuck T, Rey F, Simon-Loriere E, Bruel T, Mouquet H, Andre E, Schwartz O. 2022. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602:671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 22.Salvatore M, Bhattacharyya R, Purkayastha S, Zimmermann L, Ray D, Hazra A, Kleinsasser M, Mellan T, Whittaker C, Flaxman S, Bhatt S, Mishra S, Mukherjee B. 2021. Resurgence of SARS-CoV-2 in India: potential role of the B.1.617.2 (Delta) variant and delayed interventions. medRxiv. 10.1101/2021.06.23.21259405. [DOI] [Google Scholar]
- 23.Wang L, Cheng G. 2022. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J Med Virol 94:1728–1733. 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. 2021. Emergence of a novel SARS-CoV-2 variant in southern California. JAMA 325:1324–1326. 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, Scott R, Sconza R, Price J, Margaritis M, Bergstrom M, Spyer MJ, Miralhes PB, Grant P, Kirk S, Valerio C, Mangera Z, Prabhahar T, Moreno-Cuesta J, Arulkumaran N, Singer M, Shin GY, Sanchez E, Paraskevopoulou SM, Pillay D, McKendry RA, Mirfenderesky M, Houlihan CF, Nastouli E. 2021. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis 21:1246–1256. 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GISAID. 2022. Tracking of variants. https://www.gisaid.org. Accessed 1 February 2022.
- 27.Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, Pandey R, Brown J, Zhou J, Goonawardane N, Mishra S, Whittaker C, Mellan T, Marwal R, Datta M, Sengupta S, Ponnusamy K, Radhakrishnan VS, Abdullahi A, Charles O, Chattopadhyay P, Devi P, Caputo D, Peacock T, Wattal C, Goel N, Satwik A, Vaishya R, Agarwal M, Mavousian A, Lee JH, Bassi J, Silacci-Fegni C, Saliba C, Pinto D, Irie T, Yoshida I, Hamilton WL, Sato K, Bhatt S, Flaxman S, James LC, Corti D, Piccoli L, Barclay WS, Rakshit P, et al. 2021. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599:114–119. 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Miller JA, Verghese M, Sibai M, Solis D, Mfuh KO, Jiang B, Iwai N, Mar M, Huang C, Yamamoto F, Sahoo MK, Zehnder J, Pinsky BA. 2021. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J Clin Microbiol 59:e0085921. 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagulenko P, Puller V, Neher RA. 2018. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol 4:vex042. 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HC, Kressel HY, Rifai N, Golub RM, Altman DG, Hooft L, Korevaar DA, Cohen JF, STARD Group. 2015. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 351:h5527. 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verghese M, Jiang B, Iwai N, Mar M, Sahoo MK, Yamamoto F, Mfuh KO, Miller J, Wang H, Zehnder J, Pinsky BA. 2021. A SARS-CoV-2 variant with L452R and E484Q neutralization resistance mutations. J Clin Microbiol 59:e0074121. 10.1128/JCM.00741-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boršová K, Paul ED, Kováčová V, Radvánszka M, Hajdu R, Čabanová V, Sláviková M, Ličková M, Lukáčiková Ľ, Belák A, Roussier L, Kostičová M, Líšková A, Maďarová L, Štefkovičová M, Reizigová L, Nováková E, Sabaka P, Koščálová A, Brejová B, Staroňová E, Mišík M, Vinař T, Nosek J, Čekan P, Klempa B. 2021. Surveillance of SARS-CoV-2 lineage B.1.1.7 in Slovakia using a novel, multiplexed RT-qPCR assay. Sci Rep 11:20494. 10.1038/s41598-021-99661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korukluoglu G, Kolukirik M, Bayrakdar F, Ozgumus GG, Altas AB, Cosgun Y, Ketre Kolukirik CZ. 2021. 40 minutes RT-qPCR assay for screening spike N501Y and HV69–70del mutations. bioRxiv. 10.1101/2021.01.26.428302. [DOI]
- 34.Perchetti GA, Zhu H, Mills MG, Shrestha L, Wagner C, Bakhash SM, Lin MJ, Xie H, Huang ML, Mathias P, Bedford T, Jerome KR, Greninger AL, Roychoudhury P. 2021. Specific allelic discrimination of N501Y and other SARS-CoV-2 mutations by ddPCR detects B.1.1.7 lineage in Washington State. J Med Virol 93:5931–5941. 10.1002/jmv.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banada P, Green R, Banik S, Chopoorian A, Streck D, Jones R, Chakravorty S, Alland D. 2021. A simple reverse transcriptase PCR melting-temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. J Clin Microbiol 59:e0084521. 10.1128/JCM.00845-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bal A, Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, Regue H, Semanas Q, d'Aubarede C, Billaud G, Laurent F, Gonzalez C, Mekki Y, Valette M, Bouscambert M, Gaudy-Graffin C, Lina B, Morfin F, Josset L, COVID-Diagnosis HCL Study Group. 2021. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill 26:2100008. 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neopane P, Nypaver J, Shrestha R, Beqaj SS. 2021. SARS-CoV-2 variants detection using TaqMan SARS-CoV-2 mutation panel molecular genotyping assays. Infect Drug Resist 14:4471–4479. 10.2147/IDR.S335583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogels CBF, Breban MI, Ott IM, Alpert T, Petrone ME, Watkins AE, Kalinich CC, Earnest R, Rothman JE, Goes de Jesus J, Morales Claro I, Magalhães Ferreira G, Crispim MAE, Singh L, Tegally H, Anyaneji UJ, Hodcroft EB, Mason CE, Khullar G, Metti J, Dudley JT, MacKay MJ, Nash M, Wang J, Liu C, Hui P, Murphy S, Neal C, Laszlo E, Landry ML, Muyombwe A, Downing R, Razeq J, de Oliveira T, Faria NR, Sabino EC, Neher RA, Fauver JR, Grubaugh ND, Network for Genomic Surveillance in South Africa. 2021. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol 19:e3001236. 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hale R, Crowley P, Dervisevic S, Coupland L, Cliff PR, Ebie S, Snell LB, Paul J, Williams C, Randell P, Pond M, Stanley K. 2021. Development of a multiplex tandem PCR (MT-PCR) assay for the detection of emerging SARS-CoV-2 variants. Viruses 13:2028. 10.3390/v13102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garson JA, Badru S, Parker E, Tedder RS, McClure MO. 2022. Highly sensitive and specific detection of the SARS-CoV-2 Delta variant by double-mismatch allele-specific real time reverse transcription PCR. J Clin Virol 146:105049. 10.1016/j.jcv.2021.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aoki A, Adachi H, Mori Y, Ito M, Sato K, Okuda K, Sakakibara T, Okamoto Y, Jinno H. 2021. A rapid screening assay for L452R and T478K spike mutations in SARS-CoV-2 Delta variant using high-resolution melting analysis. J Toxicol Sci 46:471–476. 10.2131/jts.46.471. [DOI] [PubMed] [Google Scholar]
- 42.Barua S, Hoque M, Kelly PJ, Bai J, Hanzlicek G, Noll L, Walz H, Johnson C, Kyriakis C, Wang C. 2021. High-resolution melting curve FRET-PCR rapidly identifies SARS-CoV-2 mutations. J Med Virol 93:5588–5593. 10.1002/jmv.27139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamill V, Noll L, Lu N, Tsui WNT, Porter EP, Gray M, Sebhatu T, Goerl K, Brown S, Palinski R, Thomason S, Almes K, Retallick J, Bai J. 2021. Molecular detection of SARS-CoV-2 strains and differentiation of Delta variant strains. Transbound Emerg Dis. 10.1111/tbed.14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, Fig. S1 and S2, and Tables S1 to S4. Download jcm.00178-22-s0001.pdf, PDF file, 0.2 MB (237.6KB, pdf)