ABSTRACT
Sox8 is a developmentally important transcription factor that plays an important role in sex maintenance and fertility of adult mice. In the B-type spermatogonial cells, Sox8 is regulated by the long noncoding RNAs (lncRNA) Mrhl in a p68-dependant manner under the control of the Wnt signaling pathway. The downregulation of Mrhl leads to the meiotic commitment of the spermatogonial cells in a Sox8-dependant manner. While the molecular players involved in the regulation of transcription at the Sox8 promoter have been worked out, our current study points to the involvement of the architectural proteins CTCF and cohesin in mediating a chromatin loop that brings the Sox8 promoter in contact with a silencer element present within the gene body in the presence of lncRNA Mrhl concomitant with transcriptional repression. Further, lncRNA Mrhl interacts with the Sox8 locus through the formation of a DNA:DNA:RNA triplex, which is necessary for the recruitment of PRC2 to the locus. The downregulation of lncRNA Mrhl results in the promoter-silencer loop giving way to a promoter-enhancer loop. This active transcription-associated chromatin loop is mediated by YY1 and brings the promoter in contact with the enhancer present downstream of the gene.
KEYWORDS: 3D chromatin organization, epigenetics, gene regulation, lncRNA
INTRODUCTION
Sox8 is a developmentally important transcription factor that is critical for the maintenance of adult male fertility. Sox8 knockout mice become progressively infertile because of age-related degeneration of spermatogenesis (1). The Sertoli-specific deletion of Sox9 (another essential transcription factor involved in mammalian sex determination) in Sox8-null embryonic mice results in failure to achieve the first wave of spermatogenesis (2). The deletion of both Sox8 and Sox9 in adult Sertoli cells results in testis-to-ovary genetic reprogramming (3), while the deletion Sox8 alone is sufficient for ovarian-to-testicular genetic reprogramming in the absence of R-spondin1 (4).
Regulation of gene expression in mammals is a complex interplay of intricately controlled events. While decades of work have brought about some understanding of these regulatory events, including the role of transcriptional factors and the role of proximal and distal regulatory elements, long noncoding RNAs (lncRNAs) are the latest entrants to this hotbed of research. It is emerging that lncRNAs are inextricably involved in every step of gene regulation.
Mrhl (meiotic recombination hot spot locus) lncRNA is a 2.4-kb-long monoexonic transcript that is transcribed from an independent transcription unit within the 15th intron of the phkb gene in mice (5). This chromatin-bound lncRNA regulates the expression of multiple genes in the mouse spermatogonial cells in a Wnt signaling-dependent manner. Sox8 is one of the genes that the lncRNA regulates by binding to the promoter in a p68-dependent manner (6).
Our group has previously deciphered the gene regulatory mechanism in play at the promoter of Sox8 in mouse spermatogonial cells. Mrhl binds at the Sox8 promoter ∼140 bp upstream of the transcription start site (TSS), and the Mad-Max transcription factors along with the corepressors Sin3a and HDAC1 are also bound at the Sox8 promoter close to the Mrhl binding site in the presence of Mrhl. Wnt signaling-mediated transcriptional regulation of Sox8 brings about substantial changes in the chromatin dynamics of the promoter. There is a concomitant transcriptional activation of Sox8 expression with downregulation of Mrhl. Associated changes include the Mad-Max complex being replaced by the Myc-Max transcription factors and increased levels of H3K4me3 and H3K9ac histone modifications and decreased levels of H3K27me3 modification at this locus. Simultaneously, beta-catenin binds at the Wnt responsive element present at the promoter (7). Activation of the Wnt signaling cascade in Gc1-spg cells results in their meiotic commitment marked by the increase in the levels of premeiotic and meiotic markers (8), including that of the meiotic gatekeeper, Stra8, in a Sox8-dependent manner.
While chromatin-associated lncRNAs interact with DNA through protein bridges, an alternative mode of interaction is by directly binding to DNA. The potential of lncRNAs to form hybrid structures, such as DNA:DNA:RNA triplexes to directly interact with DNA has been explored in the past decade. LncRNAs, including Meg3, KHPS1, and PARTICLE, have been reported to form triplex structures at genomic regions having adenine-guanine (AG)-rich motifs (9–11). In addition to acting as tethers, triplexes have been shown to act as platforms for the recruitment of DNA methyltransferase complex DNMT3b by promoter-associated RNA (pRNA) at the ribosomal DNA (rDNA) locus or the polycomb repressive complex PRC2 by lncRNAs MEG3 and Fendrr (9, 12, 13) and thereby influence the expression of genes in the vicinity.
Recent advances indicate a role for lncRNAs in bringing together distant gene regulatory elements in three-dimensional (3D) space to regulate gene expression. CCCTC-binding factor (CTCF), the master architectural protein in mammalian cells, mediates interactions between distant genomic elements, resulting in context-dependent functional outcomes. The cohesin complex is essential in stabilizing these CTCF mediated 3D contacts while various proteins such as Ying-Yang1 (YY1), DDX5/p68, and CHD8 among others supplement CTCF’s role in genome organization (14). CTCF can bind to lncRNAs through its RNA-binding domain (15), and the deletion of the RNA-binding domain within CTCF compromises its ability to homodimerize—an essential ability to enable chromatin looping (16). The lncRNA CCAT1-L is transcribed from a locus 515 kb away from the MYC promoter. Transcribed specifically in colorectal cancers, it associates with CTCF to bring the MYC promoter in contact with the locus 515 kb away, which functions as an enhancer for MYC (17). The role of the CTCF-cohesin complex along with lncRNA SRA and the DEAD box RNA helicase DDX5/p68 in maintaining imprinting at the H19/Igf2 has been extensively studied (18).
In the present study, we show that Mrhl, in association with the nuclear organizing factors CTCF, cohesin, and YY1, mediates the formation of dynamic chromatin loops at the locus. In addition, Mrhl forms a triplex at the Sox8 promoter and contributes to the regulation of gene expression through the triplex-mediated recruitment of PRC2.
RESULTS
Mrhl forms a triplex at the Sox8 locus.
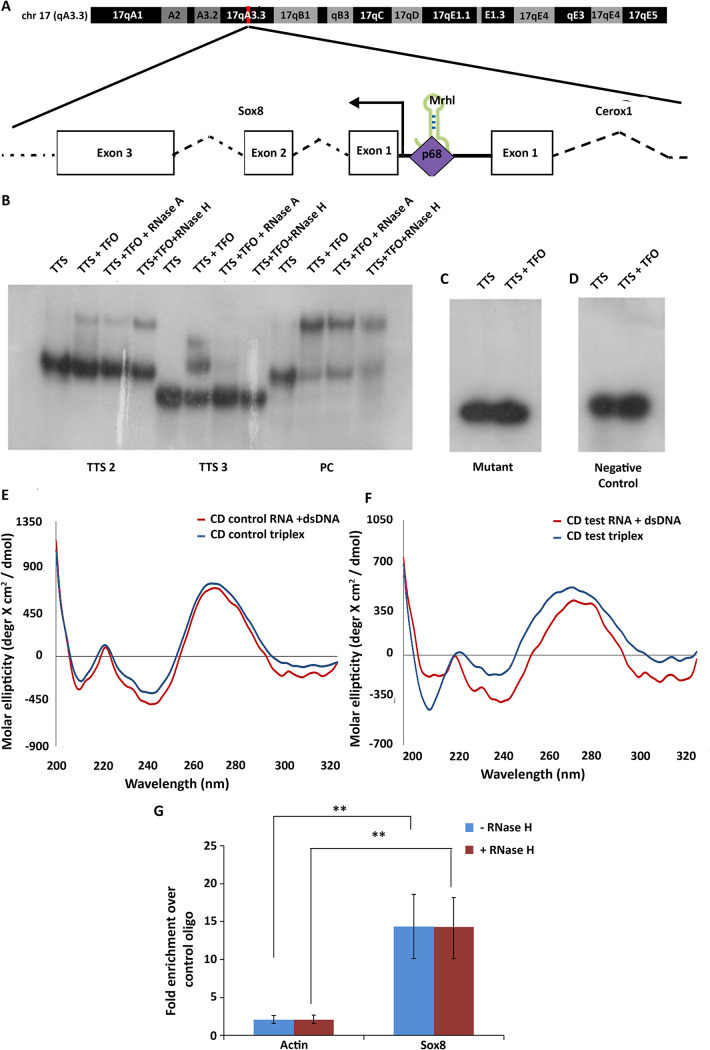
Sox8 is transcribed from a bidirectional promoter on chromosome 17. The binding of Mrhl at the promoter of Sox8, 140 bp upstream of the TSS, is dependent on DDX5/p68 (Fig. 1A). Since multiple chromatin-associated lncRNAs interact with DNA through the formation of DNA:DNA:RNA triple helix structures, it was of interest to investigate if Mrhl lncRNA too interacts with the Sox8 locus through the formation of a triplex.
FIG 1.
Mrhl interacts at the Sox8 locus. (A) The mouse Sox8 locus on chromosome 17. Mrhl lncRNA binds at the bidirectional promoter ∼140 bp upstream of the TSS in a p68-dependent manner to maintain Sox8 in the transcriptionally repressed state. (B) EMSA performed for the indicated DNA oligonucleotide incubated with RNA oligonucleotide TFO1. The lanes have the reaction components indicated above them. (C) EMSA performed with mutant TTS2 and RNA TFO1 where no shift in mobility is observed. (D) EMSA for negative control TFO/TTS pair. (E) Artificial spectrum generated by summation of individual CD spectra recorded for TTS2 only and NC TFO only (red) overlaid with CD spectrum of triplex reaction for the oligonucleotides (blue). (F) Artificial spectrum generated by summation of individual CD spectra recorded for TTS2 only and TFO1 only (red) overlaid with CD spectrum of triplex reaction for the same oligonucleotides (blue). Plots are an average of 4 independently recorded spectra. (G) Results of the in-nucleus triplex pulldown assay show significant enrichment of the Sox8 TTS region in the TFO1 oligonucleotide-associated chromatin fraction over NC TFO-associated chromatin fraction both without and with RNaseH digestion. Data in the graph have been plotted as mean ± standard deviation (SD); n = 3. ***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; NS, not significant (two-tailed Student's t test).
A majority of the lncRNA:DNA triplexes have been reported to form at genomic regions rich in AG-repeats. The region ∼1.2 kilobase pairs (kb) upstream of the Sox8 TSS harbors ∼50 AG-repeats and is a prime candidate for triplex formation. Initial in silico predictive analysis was done using the triplexator tool. Triplex-forming oligonucleotide (TFO)-triplex target site (TTS) pairs with a score of 10 and above (Table 1) were considered for further experimentation. The TTS sequences were found to be overlapping within a 33-bp DNA sequence between the regions −1224 and −1257 of the Sox8 promoter, while 2 sequences mapping to 2 distinct regions of Mrhl RNA, one mapping to the 5′ end of the lncRNA and the other to the 3′ end, were identified as TFO sequences. The 33-nucleotide (nt) region from the Sox8 locus was split between 3 DNA oligonucleotides for downstream experiments while 2 RNA oligonucleotides were generated, each mapping to the two different predicted regions.
TABLE 1.
Triplexator predictionsa
| TFO start (nucleotide position within Mrhl) | TFO end (nucleotide position within Mrhl) | TTS start (nucleotide position from TSS) | TTS end (nucleotide position from TSS) | Score |
|---|---|---|---|---|
| 391 | 404 | 1224 | 1237 | 11 |
| 389 | 400 | 1224 | 1235 | 10 |
| 389 | 404 | 1226 | 1241 | 12 |
| 389 | 404 | 1232 | 1247 | 12 |
| 389 | 401 | 1245 | 1257 | 10 |
| 2362 | 2374 | 1106 | 1118 | 10 |
| 2362 | 2374 | 1239 | 1251 | 10 |
Predictions with a score of 10 and above for regions within Mrhl lncRNA (TFO start and end) and Sox8 promoter (TTS start and end).
Electrophoretic mobility shift assay (EMSA) was carried out to check for interaction between DNA TTS and RNA TFO oligonucleotides in vitro. A positive control TFO-TTS pair from within Meg3 lncRNA and its target TGFBR1 gene (9) and a negative control TFO-TTS pair with no AG bias taken from within the Sox8 locus and Mrhl RNA were included for EMSA. Of the 5 TFO-TTS pairs probed, 2 combinations with Mrhl TFO1 as the RNA oligonucleotide, TTS2, and TTS3, showed a mobility shift corresponding to the formation of triplex. Triplex reactions were additionally subjected to RNase H digestion, which specifically degrades DNA:RNA duplexes to rule out any shift in mobility arising due to this hybrid duplex, in addition to RNase A digestion controls. While both TTS-TFO pairs showed a decrease in the intensity of signal with RNaseA treatment, only one (Mrhl TFO1-TTS2) was resistant to RNase H digestion (Fig. 1B). Since the purine bases G and A participate in the formation of Hoogsteen base pairing involved in the formation of triplex structures, the G bases were mutated to C in TTS2 to look for the effect on triplex formation. We observed a loss in the mobility shift when the mutant oligonucleotides were used (Fig. 1C). This pair was selected as a probable triplex-forming region between Mrhl and the Sox8 locus.
To validate the results from EMSA, circular dichroism spectra for the TFO-TTS pairs TTS2-TFO1 and TTS2-NC TFO were recorded. The spectrum recorded for the test oligonucleotide showed characteristics of a triplex with two minima—a strong negative peak at 210 nm and an additional negative peak at 240 nm (Fig. 1E, blue line), whereas the spectrum recorded for the control oligonucleotide was not characteristic of a triplex spectrum (Fig. 1F, blue line). Artificial spectra were generated by plotting the sum of the individual spectra for either TTS2 and TFO1 or TTS2 and NC TFO. When the artificial spectra were overlaid with the spectra obtained from the triplex reactions, an overlap in the spectra for the control oligonucleotide was observed (Fig. 1F) but not for the test TFO1 (Fig. 1E) containing spectra, indicating that the spectrum for the reaction containing control oligonucleotide is a result of the contribution of the individual reaction components.
Further, the formation of triplex within the nucleus was investigated by using biotinylated TFO 1 or control TFO as bait to pulldown associated chromatin. Compared to the control oligonucleotide, TFO 1 pulldown fraction was significantly enriched for the Sox8 TTS2-containing locus but not for a control region from the beta-actin locus (Fig. 1G). This enrichment was not significantly affected by RNaseH digestion, thereby confirming that the TFO from within Mrhl lncRNA interacts with the Sox8 locus through the formation of a DNA:DNA:RNA triplex.
Triplex formation regulates Sox8 expression through PRC2 recruitment.
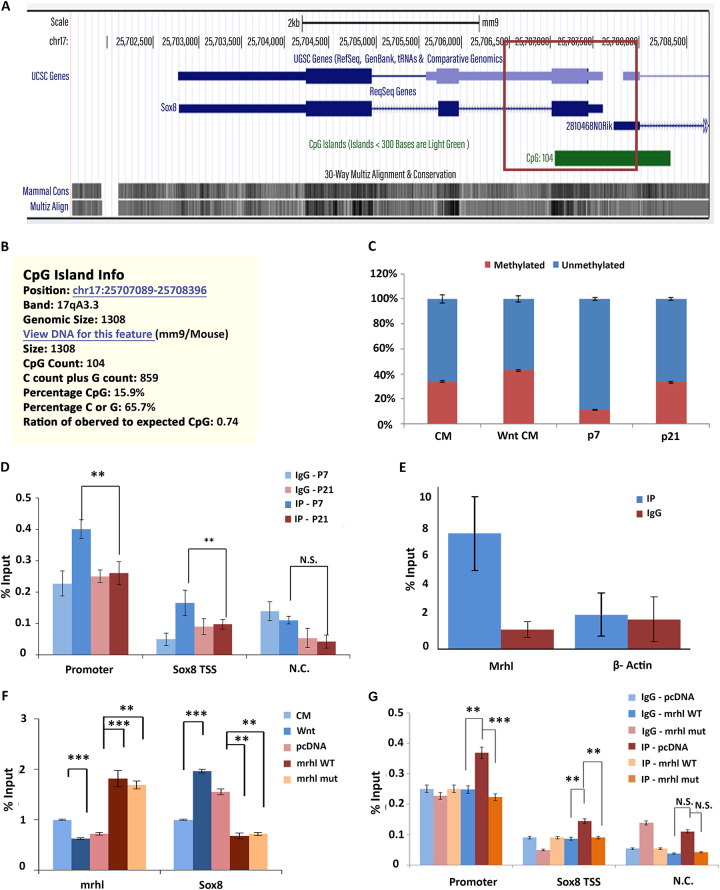
Triplex formation by lncRNA can regulate genes present in its vicinity—either by activating or repressing gene transcription. Specifically, the triplex structure could act as a platform for the recruitment of DNMT3b, which methylates the CpG island present in the vicinity, thereby leading to gene repression (12)—a mechanism that could be relevant to Sox8 as the presence of Mrhl maintains Sox8 in the transcriptionally repressed state. Since the Sox8 promoter and the TTS is located within a CpG island (Fig. 2A), we wanted to investigate if lncRNA triplex formation leads to methylation of this CpG island. The methylation status was investigated in the murine cell line Gc1-spg, derived from the B-type spermatogonia and a cell line in which the role of Mrhl lncRNA has been extensively characterized, in the absence and presence of the Wnt ligand. Under control conditions, Mrhl is actively expressed and maintains Sox8 in the transcriptionally repressed state, while activation of the Wnt signaling pathway results in the downregulation of Mrhl lncRNA, which activates Sox8 expression (6). To our surprise, no reduction in methylation levels was observed when Sox8 transcription was activated. These results were corroborated by the results of methylation levels in mice testes of 7-day-old (P7) and 21-day-old (P21) mice (Fig. 2C). Testes of P7 mice have a predominant population of spermatogonial cells (cells without activation of Wnt signaling), while testes of P21 mice have a predominant population of spermatocytes (with activation of Wnt signaling). Much of the results pertaining to Mrhl lncRNA and Sox8 expression and regulation in control and Wnt-signaling activated cells are also reflected in P7 and P21 mice testes, respectively (6–8).
FIG 2.
PRC2 occupancy at the Sox8 locus. (A) CpG island encompassing the Sox8 promoter. (B) Information about the CpG island. (C) Percent CpG methylation at the Sox8 CpG island in both cells (increase in wnt when compared to that of the control) and mice testis (increase in p21 when compared to p7). (D) ChIP-qPCR for Ezh2 subunit of PRC2 at the Sox8 locus in Gc1-spg cells under control and Wnt signaling activated conditions. (E) UV-RIP for Ezh2 shows no significant enrichment of Mrhl in IP fraction over the IgG control. (F) ChIP-qPCR for Ezh2 performed in cells transfected with either vector control, WT, or TFO mutant constructs of Mrhl. (G) The expression levels of Mrhl and Sox8 in cells grown under control or Wnt-activated conditions and cells transfected with vector only control Mrhl WT and Mrhl TFO mutant constructs with Wnt activation. Data in the graph have been plotted as mean ± SD; n = 3. ***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; NS, not significant (two-tailed Student's t test).
We have shown earlier that Sox8 repression correlates with high levels of the repressive histone modification H3K27me3 in spermatogonia (7). So, we next checked for the presence of PRC2, the enzyme complex that catalyzes methylation of H3K27, at the promoter in the presence of Mrhl by performing chromatin immunoprecipitation (ChIP) for Ezh2, a core subunit of PRC2 and also a subunit that mediates PRC2-RNA interaction. The occupancy of Ezh2, which was observed under control conditions, was found to reduce upon activation of the Wnt signaling pathway in the Gc1-spg cells (Fig. 2D).
Since multiple features, including high GC content of DNA, unique DNA conformations, presence of a guide lncRNA or the RNA:DNA:DNA triplex structure, can recruit PRC2 to the target locus, we wanted to understand the contribution of the triplex formation by Mrhl lncRNA to the occupancy of PRC2 at the Sox8 locus. To check if Mrhl lncRNA was acting as a guide to recruit PRC2 to the Sox8 locus, we performed UV-RNA Immunoprecipitation (RIP) to check for interaction between Mrhl and Ezh2. The success of IP was confirmed by Western blotting (data not shown). However, there was no significant enrichment of Mrhl in the IP fraction over isotype control (2E). So next, we wanted to check if the DNA: RNA triplex was acting as a platform to recruit PRC2. To this end, we attempted to rescue Ezh2 occupancy by expressing Mrhl lncRNA containing either the wild-type (WT) TFO or mutated TFO. While WT Mrhl lncRNA expressed in trans in the presence of the Wnt cue resulted in the rescue of Ezh2 occupancy, mutant TFO did not have a similar effect (Fig. 2F). Therefore, we conclude that the formation of triplex by Mrhl lncRNA at the Sox8 locus recruits PRC2 to the Sox8 locus. Furthermore, we also looked at the ability of TFO mutant Mrhl to rescue Sox8 transcript levels. WT Mrhl was able to rescue the expression of Sox8 to the levels comparable to control conditions (Fig. 2G). Interestingly, the TFO mutant was able to transcriptionally repress Sox8 similar to WT Mrhl. This indicates that triplex formation, while required for PRC2 recruitment, is not necessary to maintain Sox8 in the repressed state and points to an additional role for Mrhl lncRNA in repressing Sox8 transcription.
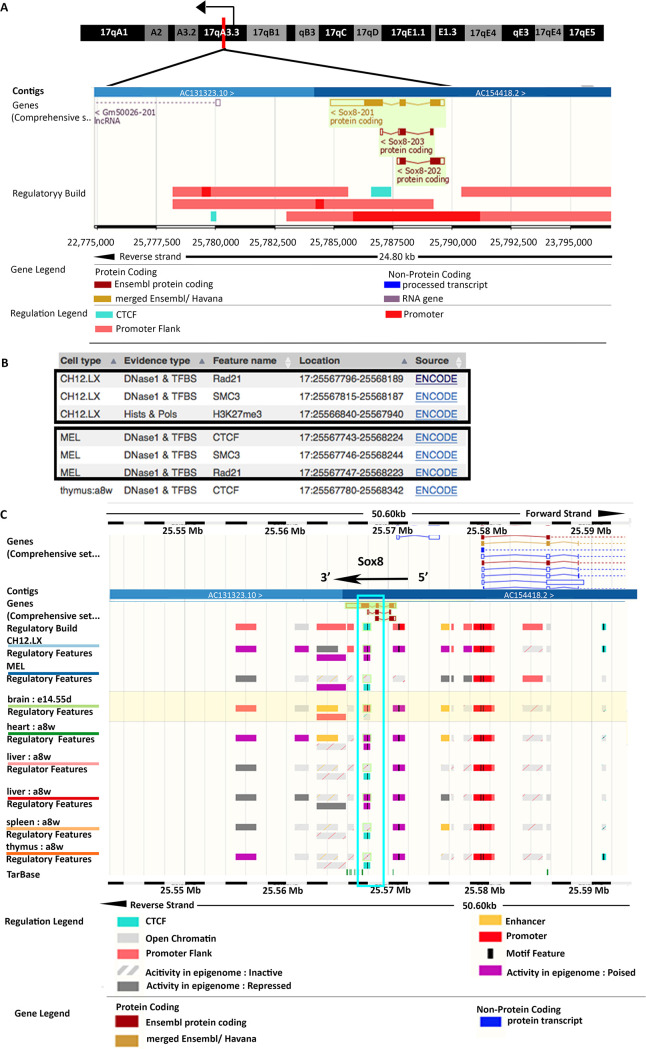
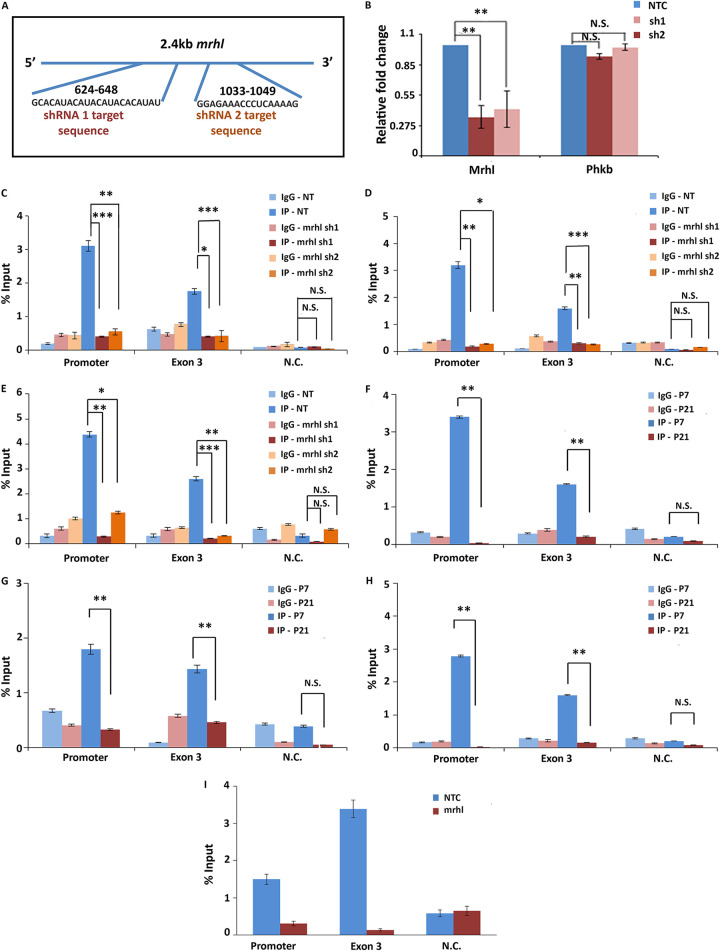
CTCF and cohesin bind at the Sox8 locus in the presence of lncRNA Mrhl.
The p68-dependent gene repression of Sox8 by Mrhl lncRNA is reminiscent of lncRNA SRA and p68 at the imprinted H19/Igf2 locus where the two molecules associate with architectural proteins CTCF and cohesin to silence the Igf2 gene on the maternal allele (18). We looked for potential CTCF and cohesin binding sites at the Sox8 locus to check if a similar mechanism of gene repression was operating here. The ENSEMBL database showed that the transcription factor CTCF binds at a binding site present in exon 3 of the Sox8 gene (Fig. 3A). An inverse correlation between Sox8 expression pattern and CTCF binding was observed across various tissues (Fig. 3C) (activity at the Sox8 gene locus is considered as an indication of Sox8 expression), indicating that CTCF binding corresponds to the transcriptionally repressed state of the gene. The cohesin complex, too, binds at this region along with CTCF (Fig. 3B). CTCF binding is observed in those tissues in which Mrhl is expressed, suggestive of cooperation between Mrhl and CTCF in the regulation of Sox8 in multiple tissues (Fig. 3C).
FIG 3.
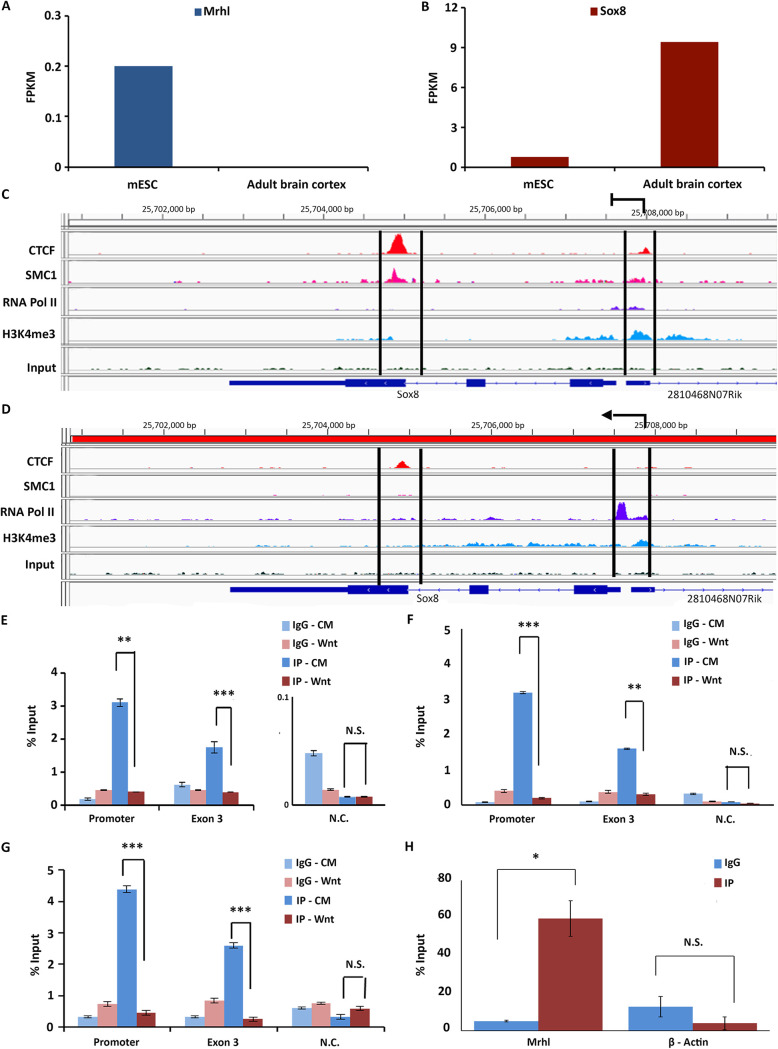
Occupancy of architectural proteins at the Sox8 locus in spermatogonia. (A) Sox8 locus harboring CTCF binding site within exon 3. (B) Cohesin subunit SMC3 and Rad21 are found to bind close to the CTCF binding site in exon 3 of Sox8. (C) CTCF appears to be bound at the binding site within Sox8 in those tissue in which Mrhl is expressed but not in some other in which Mrhl is not expressed. Mrhl expression levels in mESC and adult brain cortex.
We then analyzed publicly available ChIP-sequencing (ChIP-seq) data sets to visualize the binding of the 2 architectural proteins at the Sox8 locus in more detail. Due to the unavailability of required ChIP-seq data sets in spermatogonial cells, mouse embryonic stem cells (mESC) and adult brain cortex were used as surrogate systems. Mrhl lncRNA is expressed in mESC while it is not expressed in the adult brain cortex (19). The inverse relationship between Mrhl and Sox8 expression in these two tissues was confirmed by analyzing publicly available RNA-seq data sets (Fig. 4A and B). ChIP-seq analysis for CTCF, SMC1 subunit of cohesin, RNA polymerase II (RNA PolII), and H3K4me3 to indicate transcriptional activity at the locus was performed in these two surrogate systems. This confirmed the occupancy of CTCF within exon 3 of Sox8 in mESC, in which Sox8 is not actively transcribed (low transcript levels in RNA-seq and low relative levels of RNA PolII at promoter and H3K4me3 at the locus).
FIG 4.
(A) Mrhl expression in mESC and adult brain cortex. (B) Sox8 expression in mESC and adult brain cortex. (C) Analysis of ChIP-seq data sets in mESC showing the presence of CTCF and cohesin (SMC1) at both exon 3 and the promoter. (D) Analysis of ChIP-seq data sets in adult brain cortex showing reduced occupancy of CTCF and cohesin (SMC1) at exon 3 and promoter. Results for ChIP-qPCR for CTCF (E), Rad21 (F), and p68 (G) showing their occupancy patterns at the promoter and exon 3 of Sox8 gene in cells without and with Wnt activation. (H) UV-RIP for CTCF shows enrichment of Mrhl in IP fraction over the IgG fraction but not for beta-actin. Data in the graph have been plotted as mean ± SD; n = 3. ***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; NS, not significant (two-tailed Student's t test).
The same was found to be true for cohesin (the presence of SMC1 has been considered as evidence of cohesin occupancy). Interestingly, an additional peak for both CTCF and cohesin was observed at the promoter of Sox8 very close to the Mrhl binding site (Fig. 4C). Upon transcriptional activation (adult brain cortex) (higher transcript levels and relative occupancy of RNA PolII at promoter and H3K4me3 at the locus), CTCF and cohesin occupancy were not observed at the promoter and the occupancy at exon 3 was significantly reduced (Fig. 4D). This occupancy pattern was experimentally validated by performing ChIP for CTCF and Rad21 subunit of the cohesin complex in Gc1-spg cells without and with activation of the Wnt signaling pathway. A gene desert locus in chromosome 3 was chosen as a negative control for the ChIP quantitative PCR (qPCR). To confirm that ChIP experiments were working, Western blotting was carried out for the proteins (data not shown). The results from these experiments showed protein occupancy for both CTCF and Rad21 at both exon 3 and Sox8 promoter regions under control conditions. In the Wnt-activated cells, CTCF and Rad21 binding at both positions in the Sox8 locus was significantly reduced (Fig. 4E and F). We knew from previous p68 ChIP experiments that it binds at the Sox8 promoter at the Mrhl binding site. Our current p68 ChIP experiment showed significant enrichment of p68 at exon 3 of Sox8 in the Gc1-spg cells under Wnt-inactivated conditions. This occupancy too reduced upon Mrhl downregulation by Wnt signaling activation (Fig. 4G). Since lncRNAs have been reported to bind to and guide CTCF to their target sites, we wanted to see if Mrhl lncRNA too binds to CTCF, and to check for this interaction, we performed CTCF UV-RNA IP. We could see a significant enrichment of Mrhl in the IP fraction compared to the isotype control (Fig. 4H), indicative of the possibility that Mrhl recruits CTCF to the Sox8 locus.
To understand if the differential binding of these proteins at the Sox8 locus was dependent upon Mrhl or was due to an indirect, downstream effect of the activation of the Wnt pathway, their occupancy was investigated in cells in which Mrhl was depleted through RNA interference (RNAi). ChIP was carried out in cells in which two different inducible lentiviral short hairpin RNA (shRNA) constructs targeting Mrhl or a nontarget control construct were integrated (Fig. 5A). Similar to what was observed under Wnt-induced conditions, occupancy of CTCF, Rad21, and p68, observed at both exon 3 and promoter of Sox8 in cells with nontarget control, was depleted upon RNAi-mediated Mrhl downregulation indicative of their Mrhl lncRNA-dependent occupancy at the Sox8 locus (Fig. 5C to E).
FIG 5.
Architectural proteins at the Sox8 locus in Mrhl-silenced cells and mice testes. (A) The two different regions within Mrhl targeted by the two shRNA. (B) Mrhl silencing efficiency of the two shRNA-silencing efficiencies of ∼65% and ∼55%, respectively, were observed for Mrhl while the transcript levels of the host phkb gene were not perturbed significantly. Results for ChIP-qPCR for CTCF (C), Rad21 of cohesin (D), and p68 (E) show significant reduction in occupancy of CTCF at both the promoter and exon 3 of Sox8 upon induction of silencing of Mrhl with both shRNA construct 1 and shRNA construct 2. Occupancy of CTCF (F), Rad21 (G), and p68 (H) is observed at the promoter and exon 3 of the Sox8 locus in P7 mice testes, and a significantly reduced occupancy is observed in P21 mice testes. (I) ChIP-qPCR performed with CTCF-specific antibody indicates that CTCF and not CTCFL is bound at the Sox8 locus. Data in the graph have been plotted as mean ± SD; n = 3. ***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; NS, not significant (two-tailed Student's t test).
We also investigated the status of occupancy of these proteins in the testes of mice in an attempt to address the biological relevance of the participation of CTCF and cohesin in Sox8 gene regulation. Results of ChIP carried out for CTCF, Rad21, and p68 in P7 and P21 mouse testes corroborated with the results from control Gc1-spg and Wnt-induced/RNAi-induced Mrhl-downregulated cells, respectively (Fig. 5F to H). The 3 proteins were found to be bound at the Sox8 locus in the testes of 7-day-old mice, and a significant reduction in the occupancy was observed in the testes of 21-day-old mice.
YY1 binds at the Sox8 locus upon Mrhl downregulation.
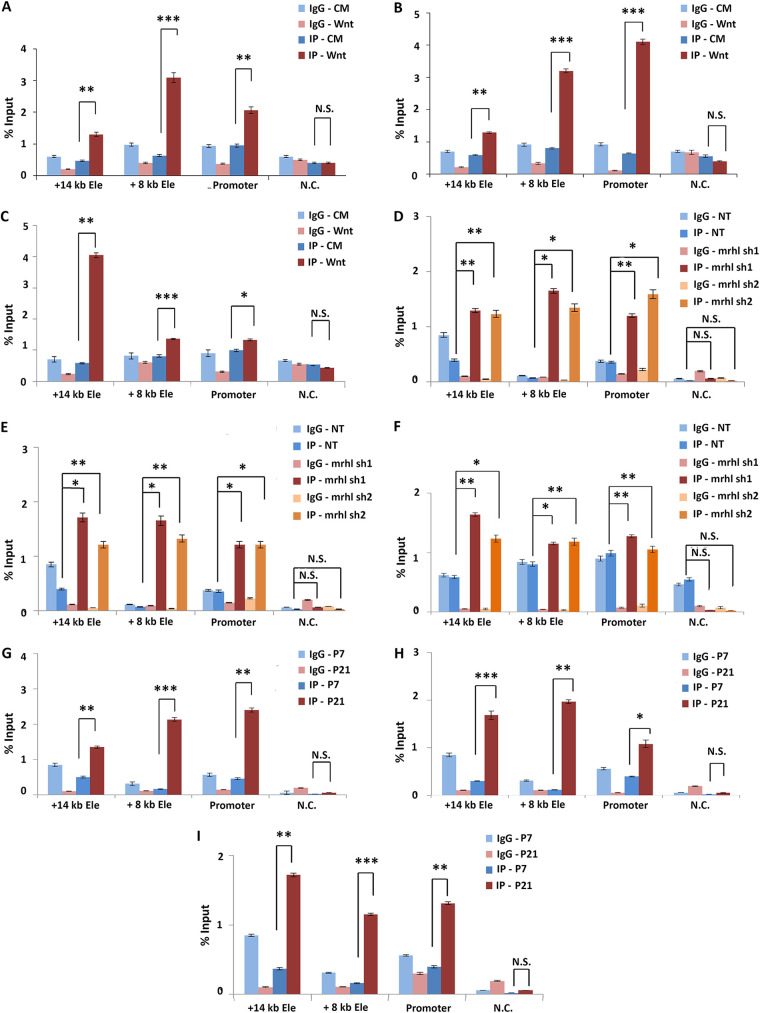
CTCF and cohesin occupancy at the Sox8 promoter was not seen in the ENSEMBL database. In silico analysis using the Gene Promoter, Miner tool was performed to look for probable interacting partners of CTCF that could be present at the Sox8 promoter. The analysis of the promoter for transcription factor binding sites revealed the presence of a binding site for YY1, a CTCF interacting transcription factor. Exon 3 has characteristics of an enhancer-blocker or a silencer element, a type of cis-regulatory element of a gene that contributes to transcriptional silencing by contacting and recruiting repressive transcription factors, such as CTCF to the promoter. If so, when not in the transcriptionally repressed state, the Sox8 promoter is free to interact with an enhancer element. YY1 has been reported to bind at enhancers of genes and facilitate active transcription by enabling enhancer-promoter contact. As per the ENSEMBL database, two enhancer elements are present in the immediate vicinity of the Sox8 gene, one upstream and one downstream of Sox8, and activity in both of these elements correlated with transcriptional activity of Sox8 (Fig. 3C). In an attempt to identify if YY1 enables the active transcription of Sox8 by binding to its promoter and Sox8-specific enhancer, we set out to identify potential enhancers for Sox8 in the mouse spermatogonial cells. Multiple tissue-specific enhancers have been reported to regulate the expression of the members of the SoxE group of transcription factors. Specifically, the expression of Sox9, another essential transcription factor for sex determination and maintenance of mammalian testis, is regulated by multiple testis-specific enhancers, either synergistically or redundantly (20). An early attempt to identify enhancers for Sox8 identified 7 evolutionarily conserved regions in the vicinity of the gene with the potential to act as enhancers. However, none of these elements acted as an enhancer in the embryonic gonad (21). Another study hinted at the presence of two regulatory elements downstream of the gene, specifically in cells of gonadal origin (22). Collating information from these different sources, we identified two regions downstream of Sox8, one 8 kb downstream and another on 14 kb downstream, as putative enhancers. Of these two regions, the element proximal to Sox8 harbors one of the 7 evolutionarily conserved elements (E6).
We performed ChIP for the enhancer-specific histone marks and for YY1 in Gc1-spg cells with and without activation of the Wnt signaling pathway to explore if (i) either one of the two putative elements gained enhancer-specific histone modifications concomitant with Sox8 transcriptional activation and (ii) if YY1 could potentially be regulating Sox8 expression by binding to the enhancer. Both the putative enhancer as well as the gene promoter regions had a significant increase in the levels of H3K4me1 and H3K27ac marks upon Wnt induction (Fig. 6A and B). Additionally, increased occupancy of YY1 at both of these enhancers and the gene promoter was observed with Wnt activation (Fig. 6C). The same trend was also observed in the RNAi-mediated Mrhl knockdown cells. While the levels of H3K4me1 and H3K27ac at both putative enhancer elements were low in cells with nontarget control shRNA, the levels increase significantly in cells containing both Mrhl-targeting shRNA (6D and E). Similarly, the occupancy of YY1 increased significantly at both the elements as well as the promoter upon Mrhl depletion (Fig. 6F). Finally, these observations were found to be biologically relevant since the testes of 21-day-old mice showed significantly higher levels of H3K4me1 and H3K27ac marks (Fig. 6G and H) as well as the occupancy of YY1 (Fig. 6I) at both the enhancer regions as well as the promoter of Sox8 when compared to testes of 7-day-old mice.
FIG 6.
Enhancers at the Sox8 locus. Results from ChIP-qPCR experiment for H3K27ac (A), H3K4me1 (B), and YY1 (C) show a significant increase in the levels of this modification with Wnt signaling-induced Sox8 transcriptional activation at the promoter and both the enhancer elements. Results from ChIP-qPCR experiment for H3K27ac (D), H3K4me1 (E), and YY1 (F) show a significant increase in the levels of this modification with Sox8 transcriptional activation at the promoter and both the enhancer elements in Mrhl knockdown cells when compared to cells expressing nontarget control shRNA. Results from ChIP-qPCR experiment for H3K27ac (G), H3K4me1 (H), and YY1 (I) show a significant increase in the levels of this modification in P21 mice testes when compared to those in P7 mice testes at the promoter and both the enhancer elements. Data in the graph have been plotted as mean ± SD; n = 3. ***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; NS, not significant (two-tailed Student's t test).
Mrhl mediates a looping switch at the Sox8 locus.
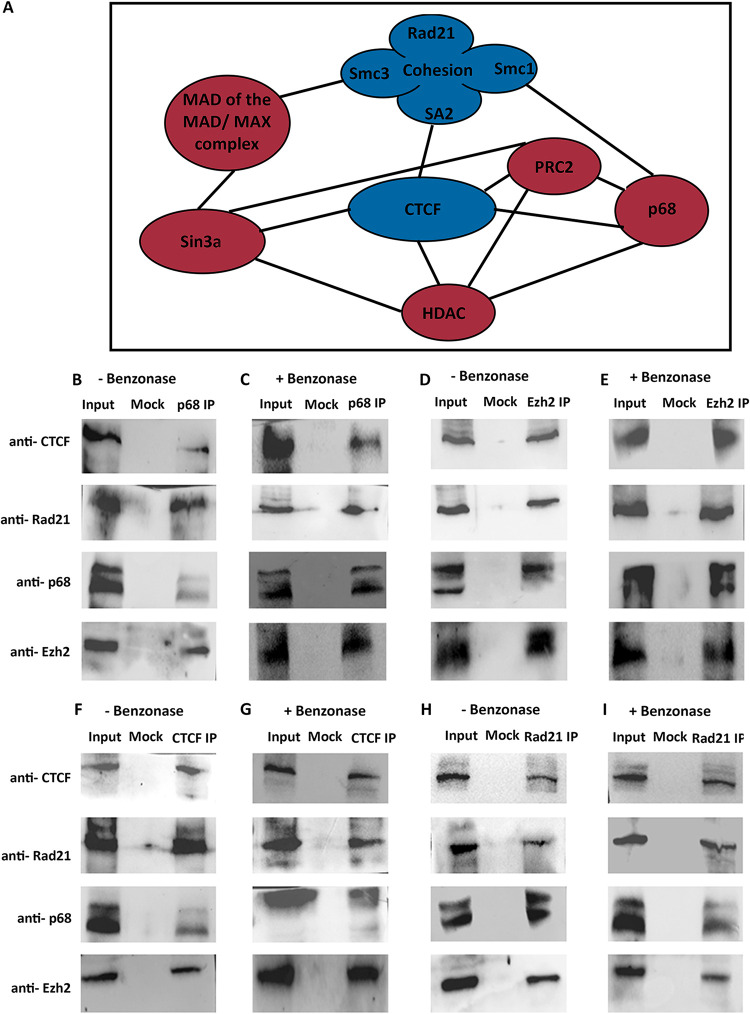
Many of the proteins bound at the promoter of Sox8, including PRC2, p68, HDAC, Sin3a, and MAD-Max, have been reported to interact with the proteins identified as bound within exon 3 of Sox8 including CTCF and cohesin (Fig. 7A). We confirmed that these interactions were observed in the mouse spermatogonial cells as well by performing co-immunoprecipitation (Co-IP) for the proteins CTCF, Rad21, p68, and Ezh2, both with and without Benzonase treatment to understand the role of nucleic acids in mediating these interactions. We observed that each one of these proteins was interacting with all of the others in a non-nucleic acid-dependent manner (Fig. 7F to I). We reasoned that occupancy of proteins such as p68 and Rad21 was detected at both the promoter and exon 3 because this protein complex was bringing the promoter and exon 3 in contact with each other through looping of chromatin at the Sox8 locus. Silencer elements have been reported to repress gene transcription in precisely this manner—by binding to transcriptional repressors such as CTCF, contacting the gene promoter, and preventing promoter-enhancer interaction (23). Additionally, we hypothesized that Mrhl downregulation resulted in the dissociation of the promoter-silencer contact, freeing the promoter to come in contact with the downstream enhancer element.
FIG 7.
(A) Protein-protein interactions that have been reported for the molecules occupying the Sox8 locus. Co-IP performed in Gc1-spg cells without Benzonase for p68 (B), Ezh2 (D), CTCF (F) and p68 (H), and WB performed with antibody indicated on the left side show their interaction with each other. Co-IP performed with Benzonase for p68 (C), Ezh2 (E), CTCF (G) and p68 (I) and WB with antibodies as indicated on the left show their interaction with each other even in the absence of nucleic acid intermediates.
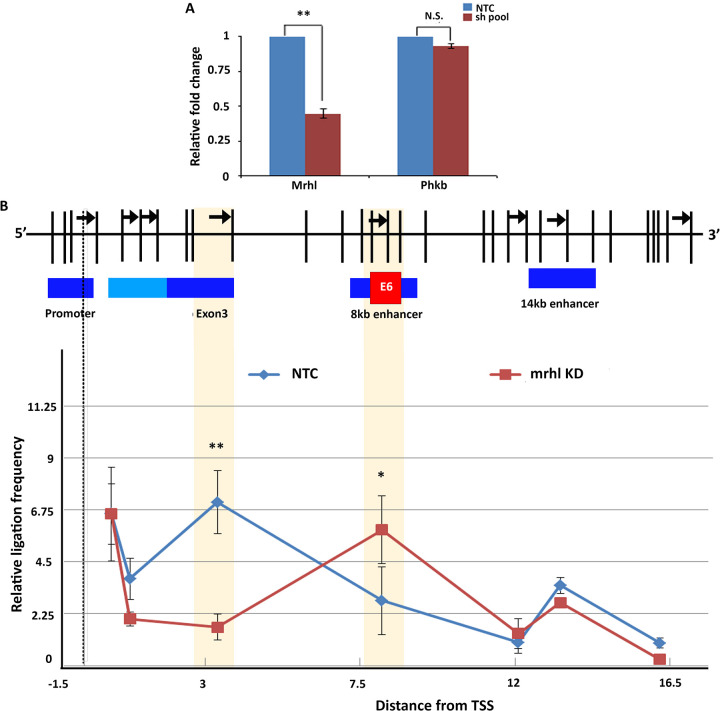
To validate the hypothesis, we looked at the chromatin looping status in the presence and absence of lncRNA Mrhl through chromosome conformation capture (3C). From 3C performed in Gc1-spg cells with and without RNAi, we observed that the Sox8 promoter was indeed brought in contact with exon 3 in a Mrhl-dependent manner supporting our hypothesis. Interestingly, of the two enhancers identified downstream of Sox8, the interaction frequency between the promoter and the proximal enhancer (present 8 kb downstream of TSS) was found to be higher in Mrhl knockdown cells, while there was no significant difference in the interaction frequency between the promoter and more distal enhancer (14 kb downstream of TSS) upon Mrhl depletion, suggesting an enhancer function for the proximal element in the mouse spermatogonial cells (Fig. 8B).
FIG 8.
(A) Mrhl was targeted for silencing through RNAi using a pool of both sh1 and sh2 shRNA constructs, and a silencing efficiency of 65% was observed. The transcript levels of the host phkb gene were not significantly perturbed. (B) Chromosome conformation capture performed in control and Mrhl knockdown Gc1-spg cells indicates that the promoter is brought in contact with exon 3 in the presence of Mrhl lncRNA. Upon downregulation of Mrhl, the promoter-exon 3 contact gives way to a promoter-enhancer contact, whereby the enhancer element present 8 kb downstream. Data in the graph have been plotted as mean ± SD; n = 3. ***, P ≤ 0.0005; **, P ≤ 0.005; *, P ≤ 0.05; NS, not significant (two-tailed Student's t test).
DISCUSSION
A member of the SoxE group, Sox8 is essential for the maintenance of male fertility as Sox8-null mice show progressive degeneration of spermatogenesis resulting in infertility (1). Specifically, Sox8 expression in Sertoli cells is essential for germ cell differentiation (24). Most of the previous studies have focused on understanding the role of Sox8 in Sertoli cells in mammalian testes. Studies from our group were the first to not only report the expression of Sox8 in spermatogenic cells but also explore the potential role of this transcription factor in meiotic commitment, likely via the master regulator of meiosis in spermatogenesis, Stra8 (25). In this context, it was of importance to study the detailed molecular events during the regulation of Sox8 by Mrhl lncRNA.
The formation of a DNA:DNA:RNA triplex is a mechanism of interaction that is common to many chromatin-bound lncRNAs, such as Meg3, PARTICLE, HOTAIR, and KHPS1 (9, 11, 26, 27). In the current study, we show that Mrhl too interacts at the Sox8 locus directly with the chromatin through the formation of DNA:DNA:RNA triplex. Mrhl lncRNA harbors multiple potential triplex-forming regions within it. The in silico predictions for the Sox8 locus suggested that two different regions seemed to have triplex-forming potential—one mapping to the 5′ end and another toward the 3′ end. While the sequence toward the 5′ end participates in triplex formation at the Sox8 locus in the Gc1-spg cells, it is possible that the sequence toward the 3′ end forms triplex in a different context. The predictions from Triplexator using different genomic regions, such as Pou3f2, Runx2, or FoxP2 (19), show that other regions within Mrhl lncRNA, too, have the potential to form triplex. Mrhl likely interacts at multiple other loci through the formation of DNA:DNA:RNA triplex through different TFOs present within it, in a context-dependent manner.
PRC2 in Sox8 gene regulation.
Many genes are repressed through methylation of CpG-rich DNA at their promoters. Further, triplex formation by the ncRNA pRNA acts as a platform for the recruitment of the DNA methyltransferase DNMT3b at the rDNA promoter which goes on to methylate the DNA at the gene promoter, thereby repressing transcription (12). The presence of a 1.3-kb-long CpG island encompassing the promoter of Sox8 suggested a probable mechanism of gene repression through the methylation of this CpG island. However, no reduction in methylation levels was observed experimentally corresponding to Sox8 activation in either the mouse spermatogonial cell line Gc1-spg upon Wnt induction or in 21-day-old mouse testes, suggesting that DNA methylation is not the mechanism of epigenetic repression of Sox8.
The presence of high levels of H3K27me3 repressive histone mark in the Sox8 transcriptional repressed state (7) was indicative of the presence of PRC2 at the Sox8 locus. PRC2 is the multiprotein complex responsible for catalyzing the methylation of H3K27. A common feature of the mammalian PRC2-binding region is the presence of CpG islands (CGIs) and more specifically, CpG-rich regions that are adjacent to the TSS of silenced genes. Multiple studies indicate that high-density DNA methylation seems to be mutually exclusive with PRC2 since most of the CGIs or CG-rich regions occupied by PRC2 are hypomethylated (28). In agreement with these studies, the levels of methylation are lower at the promoter of Sox8, which is situated within the CpG island, in the Sox8 transcriptionally repressed state than in the active state.
Another factor influencing PRC2 binding to target loci is its interaction with RNA molecules. It is now believed that lncRNA interaction, including those with PARTICLE and Meg3, could be one of the mechanisms by which PRC2 gains target specificity (9, 11). Different subunits of PRC2 recognize and bind to different secondary structures/DNA sequences through which they get targeted specifically to genomic loci. For instance, an unmethylated GCG trinucleotide motif showing an unwound DNA helix can specifically recruit PRC2-MTF2 while the Suz12 subunit has been reported to bind to the two-hairpin motif present in RNA molecules. PRC2 subunit JARID2 preferentially binds to GC-rich DNA sequences (28). At the Sox8 locus, multiple possible modes of recruitment of PRC2 exist, namely, the presence of a hypomethylated CpG island, the presence of Mrhl lncRNA, and also the formation of triplex by Mrhl lncRNA. While we see that Mrhl does not directly interact with the Ezh2 subunit of PRC2, through functional rescue with wild-type and TFO mutant Mrhl, we have shown that triplex formation indeed recruits PRC2 to the Sox8 locus.
CTCF and cohesin in Sox8 gene regulation.
CTCF in mammals is the master architectural protein and, along with cohesin, has been implicated in organizing chromatin architecture at different genomic scales from chromatin loops at the scale of a single locus to the organization of TADs. Using a combinatorial approach, we show the Mrhl-dependent occupancy of CTCF and cohesin at the Sox8 locus along with the DEAD box RNA helicase p68. The popular loop-extrusion model of chromatin loop formation proposes that the cohesin protein complex slides along chromatin forming a growing loop until it meets two CTCF molecules bound with convergent orientation. This prevents cohesin from sliding further. Preliminary in silico analysis suggests the presence of CTCF binding sites both at the promoter and within exon 3 of Sox8 (Table 2). A limitation of the prediction software utilized for this study is that it does not exhaustively predict the presence of all CTCF binding sites present within the sequence but only the sequence with the highest score. Heterogeneity is observed in CTCF binding motifs. In each species, the CTCF binding profile is composed of substantial numbers of both deeply conserved and evolutionarily recent sites. CTCF binding sites at TAD boundaries are highly conserved across species, while evolutionarily recent sites play a role in modulating gene regulation (29). Further, cell-type-specific CTCF bound sites have also been reported to have a varied binding motif compared to that of constitutively bound sites (30).
TABLE 2.
Predicted CTCF binding site from CTCFBSDB 2.0 database based in the DNA sequences of Sox8 promoter and exon 3a
| Motif PWM | Motif sequence | Input sequence name | Motif length | Motif orientation | Score |
|---|---|---|---|---|---|
| EMBL_M1 | CGCCGCCTAGTGGA | Exon 3 | 14 | − | 12.427 |
| EMBL_M1 | GGTCACCTGGTGGC | Promoter | 14 | − | 10.1556 |
| EMBL_M2 | GGAACAGCA | Exon 3 | 9 | + | 11.4118 |
| EMBL_M2 | GTCACTGCC | Promoter | 9 | − | 6.0656 |
| MIT_LM2 | TGTCCACTAGGCGGCGCCC | Exon 3 | 19 | + | 7.22188 |
| MIT_LM2 | GAGCCACCAGGTGACCCTG | Promoter | 19 | + | 5.51758 |
| MIT_LM7 | TGTCCACTAGGCGGCGCCCT | Exon 3 | 20 | + | 11.2928 |
| MIT_LM7 | GAGCCACCAGGTGACCCTGG | Promoter | 20 | + | 9.84716 |
All results with a score higher than 3 have been listed in the table. The “+” and “−” corresponds to the sense and antisense strands respectively.
CTCFL is a testis-specific paralog that is expressed only transiently in premeiotic male germ cells together with CTCF, and the two paralogs compete for binding at a subset of the CTCF binding sites (31). CTCFL functions as a transcription factor and does not have a role in chromatin organization since it cannot anchor cohesin to chromatin like CTCF can (32). ChIP qPCR using CTCF-specific antibody in the spermatogonial cells performed by us (Fig. 5I) further confirms that CTCF and not CTCFL is bound at the Sox8 locus.
DNA methylation at the DMR regulating the imprinting at the H19/Igf2 locus prevents the binding of CTCF to its cognate binding site within the DMR. The slight increase in the methylation at the CpG island of the Sox8 promoter (Fig. 2C) upon its transcriptional activation possibly may serve the same purpose.
Most of the molecular players involved in transcriptional repression of Sox8 have been reported to interact with each other. The CTCF-cohesin interaction has been extensively explored, while p68 interacts directly with cohesin and this interaction stabilizes CTCF-cohesin interaction (18). However, a recent study identified p68 to be a protein-interacting partner of CTCF through mass spectrometry (33). Members of the cohesin complex interact with various subunits of PRC2 (34), while CTCF interacts with the Suz12 subunit (35). We have confirmed many of these interactions to be independent of nucleic acids in the mouse spermatogonial cells. While it is difficult to postulate the order in which these molecules get recruited to the Sox8 locus, we know that the binding of p68, CTCF, and Ezh2 are dependent on the presence of Mrhl lncRNA.
Silencer and enhancer elements in Sox8 gene regulation.
In addition to promoters, silencers/insulators and enhancers together make up cis-regulatory elements (CREs) of a gene. H3K27me3 mark enrichment has been found to be enriched within silencer elements. Most H3K27me3+ silencer elements are also DNase I hypersensitive and have binding sites for ubiquitous repressors such as CTCF, SMAD group of proteins, and tissue-specific TFBS (36). Additionally, many H3K27me3-DHS coincided with active histone modifications, such as H3K4me1 and H3K27ac. The element within exon 3 has many of these characteristics. In addition to the occupancy of CTCF and cohesin within this genomic region, the ENSEMBL database suggests that this element is DNase hypersensitive and shows the presence of both H3K4me1 and H3K27me3. The results of the 3C experiment further indicate that this element contacts the gene promoter in the transcriptionally repressed state. Taken together, this evidence supports the “silencer” function of exon 3 of Sox8.
We have identified two putative enhancers for Sox8 in spermatogonial cells located downstream of the gene. We observe activity at both of these enhancer elements upon Mrhl knockdown as evidenced from enhancer-specific histone modification ChIP experiments. Only one of these two regions, the enhancer present 8 kb downstream, contacts the promoter of Sox8 as observed from the 3C experiment and is likely to drive the expression of Sox8 in meiotically committed spermatogonia. This enhancer harbors within it one of the evolutionarily conserved elements, E6 (21), as a putative enhancer. However, this does not mean that the other enhancer element has no role to play in regulating Sox8 expression or that the E6-harboring enhancer is the sole enhancer regulating the expression of Sox8. Further, this study has been performed in embryonic gonads and gives us no information on the postnatal activity of this regulatory element. The possibility that the E6 harboring enhancer may not be a testis-specific enhancer exists.
Sox9 is regulated by multiple tissue-specific enhancers and, in the testis alone, is regulated by multiple enhancers acting either redundantly or synergistically (20, 37). Our study has focused on characterizing the regulatory elements of Sox8 in a genomic region of 25 to 30 kb only. Extensive characterization, including genomic deletion of the regulatory elements in a larger region, is required to identify and better understand the possible interplay between various enhancers in regulating Sox8 expression. Such characterization is beyond the scope of the current study. Further, the possibility of trans-interactions regulating Sox8 expression has not been explored.
YY1 in Sox8 gene regulation.
Of all the regulatory proteins identified at the Sox8 locus, YY1 is the only one with contradictory functional roles. YY1 can act both as a transcriptional activator or transcriptional repressor in a context-dependent manner. As an architectural protein too, YY1 can mediate the formation of chromatin loops, which can either have gene repressive or activating outcomes.YY1 dimerizes with CTCF to mediate chromatin loop formation to repress E6 and E7 oncogenes of the human papillomavirus genome in infected cells (38). At the same time, YY1 binds to promoter-proximal elements and active enhancers and forms dimers that facilitate the interaction of these DNA elements (39). Thus, YY1 at the Sox8 locus was a wildcard that could be involved in either function. However, the results from the ChIP experiments clearly indicated the association of YY1 at the regulatory elements only in the Sox8 transcriptionally active state. Further, the occupancy of YY1 at the promoter and enhancer elements suggested a role for it in facilitating the interaction of the enhancer with the promoter, and this has been validated by chromosome conformation capture.
Mrhl lncRNA possibly possesses multiple functional domains within it. At the Sox8 locus, a region from the 5′ end of the lncRNA participates in triplex formation. Results from previous work suggest that a region toward the 3′ end of Mrhl is involved in its interaction with p68 (unpublished data). The gene regulatory function is an outcome of the combinatorial function of all the different domains of Mrhl.
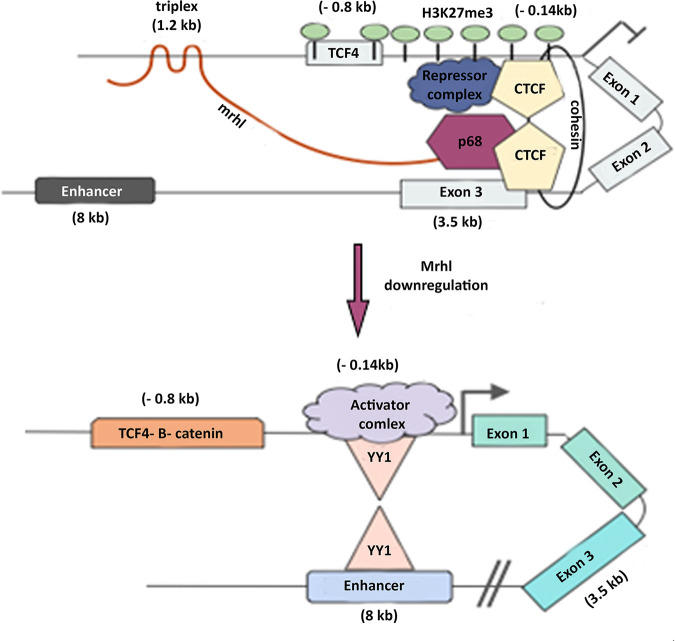
In the current study, we have demonstrated that the lncRNA Mrhl is involved in mediating chromatin looping at the Sox8 locus in association with the architectural proteins CTCF and cohesin to maintain the gene in the transcriptionally repressed state. The downregulation of Mrhl lncRNA results in a rearrangement of the looping interaction at the locus, whereby the promoter-silencer contact gives way to a promoter-enhancer contact mediated by YY1. Further, Mrhl forms a DNA:DNA:RNA triplex at the distal promoter of Sox8 that is required for the recruitment of PRC2, which then trimethylates H3K27 at the Sox8 gene locus (summarized in Fig. 9).
FIG 9.
Figure summarizing the regulation of Sox8. Sox8 is maintained in the transcriptionally repressed state when Mrhl lncRNA is bound at the promoter.
The mechanism of silencing at the Sox8 locus by Mrhl lncRNA via triplex formation, PRC2 recruitment, and the involvement of CTCF, cohesin, and p68 fits into the growing theme of gene silencing mechanism by lncRNAs. Stating that Mrhl associates with this protein complex to mediate the formation of a repressive loop is a simplistic view of events. Taking into account the very large size of the repressive complex made up of CTCF, cohesin complex, Sin3a, HDAC1, Mad-Max transcription factor dimer, p68, PRC2, and Mrhl, it would be more realistic to state that Mrhl creates a repressive environment around the Sox8 locus.
In the recent update of ENSEMBL, the human Sox8 locus can be seen to not only contain a conserved CTCF binding site in exon 3 but also has binding of many of the regulatory proteins observed at the mouse Sox8 locus. While the data are indicative of a conserved regulatory mechanism, the involvement of an lncRNA in the regulation of Sox8 in humans remains to be seen.
In summary, we have delineated the detailed molecular mechanism of regulation of Sox8 gene expression by Mrhl lncRNA, which has significant implications toward our understanding of the role of Sox8 in male germ cell differentiation, particularly the meiotic commitment of B-type spermatogonia.
MATERIALS AND METHODS
Cell lines and reagents.
Gc1-spg cell line (CRL-2053) was obtained from ATCC. L-control cell line (ATCC CRL-2648) and L-Wnt3A cell line (ATCC CRL-2647) were kind gifts from Jomon Joseph (NCCS, India).
All chemical reagents were purchased from Sigma-Aldrich and were of analytical reagent (AR) grade. Lipofectamine 2000 (11668027) and Dynabeads protein A (10002D) were purchased from Invitrogen. cDNA synthesis reagents were procured from Thermo Fisher Scientific. DpnII restriction enzyme (R0543), DNase I (MO303), and T4 DNA ligase (M0202S) were purchased from New England BioLabs. Mrhl shRNAs (custom synthesized) and nontarget control (SHC332) were procured from Sigma-Aldrich. qPCR was performed using Bio-Rad’s CFX96 machine. DNeasy blood and tissue kit (69504), EpiTect II DNA methylation enzyme kit (335452), and EpiTect methyl II PCR system (EPMM104707-1A) for CpG methylation analysis were procured from Qiagen. [γ-32P]ATP was sourced from BRIT, Hyderabad, India.
A list of antibodies that were used is as follows with the manufacturer and catalog number in parenthesis: CTCF (AbCam; ab188408), CTCF (Cell Signaling Technology; 3418), Rad21 (AbCam; ad9920), p68/DDX5 (Cusabio; CSB-PA003685), YY1 (Diagenode; C15410345), H3K4me1 (Diagenode; C15410037), H3K27ac (Diagenode; C15410174), and Ezh2 (Diagenode; C15410039).
Cell culture and preparation of control and Wnt3A-conditioned media.
Gc1-spg cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin.
Preparation of control and Wnt3A-conditioned media was done as per the manufacturer's (ATCC) instructions. The collected medium was centrifuged at 500 × g for 5 min and used after filter sterilization.
Gc1-spg cells were grown in control or Wnt-conditioned media for 72 h for Wnt induction. All cell lines were checked for Mycoplasma contamination every 2 months.
Generation of stable knockdown lines.
All shRNA plasmids were transfected at a concentration of 1.5 μg/mL Lipofectamine 2000 at 70% cell confluence. To select the positive transfectants, cells were grown in selection medium containing puromycin at a final concentration of 3 μg/mL for 72 h. To induce shRNA expression, cells were grown in complete medium in the presence of 0.5 mM IPTG and 2.5 μg/mL puromycin for 96 h.
Cloning.
Full-length WT Mrhl and TFO mutant Mrhl were cloned into the pCDNA3.1 vector between the HindIII and BamHI sites. Clones were confirmed by Sanger sequencing.
Preparation of the mice testicular samples.
The testicular samples prepared were harvested from BALB/c mice in the age groups of 7 days post partum (dpp) and 21 dpp. The dissected testis was decapsulated in phosphate-buffered saline (PBS) (pH 7.4) on ice by removing the tunica albuginea. The seminiferous tubules were released into PBS (pH 7.4) and subjected to homogenization to procure a single-cell suspension.
Chromatin immunoprecipitation.
ChIP was performed as previously described (7). Briefly, cross-linked cells were resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl). This was followed by sonication of the lysate to enrich for chromatin in the size range of 200 to 600 bp. After removal of debris by centrifugation, the lysate was incubated with either a 3- to 5-μg specific antibody or a corresponding amount of isotype control for immunoprecipitation overnight. The immune complexes were allowed to bind to the protein A Dynabeads, and the beads bound by immune complexes were subjected to washes with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl), LiCl wash buffer (1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-Cl pH 8.0), and Tris-EDTA (TE) (10 mM Tris-Cl pH 8.0, 1 mM EDTA). The beads were then either processed directly for Western blotting, or the immunoprecipitated material was eluted from the beads by adding elution buffer (0.1 M NaHCO3, 1% SDS). The supernatant was treated with proteinase K (Life Technologies), and cross-links were reversed. Eluted DNA was used for quantitative PCR. All primers used in the study have been listed in Table 3.
TABLE 3.
List of all primers and oligonucleotides used for the current study
| Primer or oligonucleotide | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| Primer for 3C | ||
| Sox C | CCAAGTGCAGCTAGGAGTCTCTC | |
| Sox test 1 | AGCACCTGCGACACGGCATC | |
| Sox test 2 | CTGGGAGCAGTACCTGCCAGAGG | |
| Sox test 3 | GGCAGAAGTTTGGATATCCAGAAGC | |
| Sox test 4 | GCCTGCCTCTGTCTACGCTTGG | |
| Sox test 5 | CCAGTGCTTGAAACTCAATGGATGG | |
| Sox test 6 | TCTCTCTGCTCGCCCTCATCC | |
| Sox test 7 | CTGCAATCCCAGCACTGGAG | |
| Ercc3 1 | CTGACCCTCAGCCTGTTAGAGC | |
| Ercc3 2 | ACCAGTCTTGCCTTGTGTCAGC | |
| ChIP RT primer | ||
| Sox8 promoter | AGAGGGCTAAGGGTGACTGACT | GTTTGGTTGCAATAGCGGATTC |
| Sox8 exon 3 | GATAACCTCGCTGCTGAGCTCGG | CTGGTGTCACCCACCAGCTCC |
| Sox8 enhancer 8 kb | CCGCTATCCAGATCACCAGG | CTGCTGAGTGACCGATGAGAC |
| Sox8 enhancer 16 kb | GCCTCAGGACTCACATCTGGC | TGTGGGTCCTTGCCAGGAGC |
| Sox8 triplex region | CCTTAATGGTGACCTTATTCTATTCTAG | CCTTTCTTGGCAGGTAATGG |
| Actin promoter N.C. | CGCTCACTCACCGGCCTC | GTCCGGGCCTCGATGCTG |
| Gene desert N.C. | TGGCTGTCCTGGCCTGC | GGCAGCCTATGCAGCATTCAATG |
| Quantification primers | ||
| Mrhl | TGAGGACCATGGCTGGACTCT | AGATGCAGTTTCCAATGTCCAAAT |
| Beta-actin | AGGTCATCACTATTGGCAACG | TACTCCTGCTTGCTGATCCAC |
| Phkb | AAGCCCAGCAATGAGGACTC | AGCACCCACCACACTAACAC |
| Sox8 | TGCTGAGCTCTGCGTTATGGAG | GTCTGGTGCCTATGCCTGTGC |
| Cloning primers | ||
| Mrhl FP | ATGCAAGCTTTGACTTGCTCTTCATTAGATC | |
| Mrhl TFO mut FP | CATTGAAACTCACACACACACATGGCATCTCTCAGGTCAC | |
| Mrhl TFO mut RP | TGTGACCTGAGAGATGCCATGTGTGTGTGTGAGTTTCAATG | |
| Mrhl RP | ATGCGGATCCAGGAGGAATGAAGTATCCAC | |
| EMSA oligonucleotides | ||
| Positive control DNA | AGAGAGAGGGAGAGAG | CTCTCTCCCTCTCTCT |
| TTS1 | GGGAGGGAGACAGAGAGG | CCTCTCTGTCTCCCTCCC |
| TTS2 | GGAAGAGGGAGGGAGA | TCTCCCTCCCTTCTTCC |
| TTS3 | AGACAGAGAGGGA | TCCCTCTCTGTCT |
| TTS4 | AGCAGGAAGCAGG | CCTGCTTCCTGCT |
| TTS5 | AGAGGGAAGAGGG | CCCTCTTCCCTCT |
| TTS negative control | ACCACGTGGGCCAGGCGC | GCGCCTGGCCCACGTGGT |
| TTS mutant | GGAACACCCACCCAGA | TCTGGGTGGGTGTTCC |
| Positive control RNA | CGGAGAGCAGAGAGGGAGCG | |
| TFO1 | UGAGAGAGAGAGAUGG | |
| TFO2 | AGAAGAAGGAAGAC | |
| Negative control RNA | CUUAUACUGCAUAAAU | |
| In nucleus pulldown oligonucleotide | ||
| TFO1 | psoralen-UGAGAGAGAGAGAUGG-biotin | |
| NC TFO | psoralen-CUUAUACUGCAUAAAU-biotin |
UV-RNA immunoprecipitation.
UV RIP was performed as described by Schaukowitch et al. (40). Briefly, cells were cross-linked under UV at 400 mJ/cm2 and resuspended in low-salt lysis buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 150 mM NaCl). Nuclei were pelleted and lysed in high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl) and diluted with IP buffer (1 mM EDTA, pH 8.0, 0.5 mM EGTA, pH 8.0, 10 mM Tris-HCl, pH 8.0, 1% Triton X-100, 0.1% deoxycholate [DOC]), protease inhibitors, and RNasin plus (50 U/mL). After removal of debris through centrifugation, the sample was split and either specific antibody or isotype control was added and incubated overnight. The immune complexes were allowed to bind to protein A Dynabeads, and the beads bound by immune complexes were subjected to washes with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl), LiCl wash buffer (1%NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-Cl pH 8.0), and TE (10 mM Tris-Cl pH 8.0, 1 mM EDTA). RNA was extracted from the beads in elution buffer (10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0, 1% SDS) and treated with proteinase K. RNA isolation was further done using TRIzol as per the manufacturer’s instructions. cDNA was synthesized and used as a template for qPCR.
Western blotting.
After SDS-PAGE and transfer, the membranes were blocked in 5% skimmed milk for 1 h at room temperature and then incubated with the respective primary antibody dissolved in 1% skimmed milk prepared in 1× PBS with 0.1% Tween 20 (0.1% PBST). The membrane was then washed with 0.1% PBST and incubated with the respective secondary antibody dissolved in 1% skimmed milk for an hour at room temperature. Washes were then given using 0.1% PBST, and the blot was developed using Millipore’s Immobilon Forte Western horseradish peroxidase (HRP) substrate, and the image was captured on Bio-Rad’s ChemiDoc.
Coimmunoprecipitation.
Coimmunoprecipitation was performed as described by Spruce et al. (41). Cells were resuspended in protein extraction buffer (50 mM HEPES-KOH pH 7.4, 137 mM NaCl, 10% glycerol, 0.4% NP-40) containing protease inhibitors either with or without Benzonase nuclease (250 U), and debris was removed. Protein A Dynabeads were conjugated to either specific antibody or isotype control and added to the cell lysate for overnight binding at 4°C. The beads were washed with wash buffer (50 mM HEPES-KOH pH 7.4, 150 mM NaCl, 0.4% NP-40) the following day and were boiled in SDS loading dye for immunoblotting.
Circular dichroism spectroscopy.
Circular dichroism (CD)-spectra were recorded on a Jasco 500A spectropolarimeter. Each spectrum is the average of 4 consecutive spectra (independent replicates) and baseline corrected with a spectrum of pure buffer. CD-spectra were recorded on Mrhl single-stranded RNA (ssRNA) TFO, NC ssRNA TFO, and the double-stranded DNA (dsDNA) oligonucleotides separately as well as on a 1:1 mix of the two in 1× triplex-forming buffer (10 mM Tris pH 7.5, 25 mM NaCl, and 10 mM MgCl2). The mixed samples or individual RNAs and dsDNAs were heated to 70°C for 10 min and slowly cooled to room temperature and incubated at room temperature for 2 h. The samples were incubated overnight at 4°C. The measurements were performed at 5°C The data presented in the spectra is the molar ellipticity given based on the concentration of nucleotides in the samples.
Triplex electrophoretic mobility shift assay.
Electrophoretic mobility shift assay was performed as per the protocol of Mondal et al. (9). The double-stranded oligonucleotides were end-labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP and purified using G-25 columns (GE Healthcare). To remove secondary structures present in them, RNA oligonucleotides were heated at 70°C and incubated on ice. The binding reaction was carried out using labeled dsDNA oligonucleotides (corresponding to 10,000 cpm), 1× triplex-forming buffer, and 50× molar excess of RNA oligonucleotides and incubated for 6 h at room temperature. In the control assay, the triplex reaction was treated with either 5 units of RNase H (NEB) or 10 μg of RNase A (Life Technologies) for 20 min. The triplex formation was monitored on 20% polyacrylamide Tris-borate-EDTA (TBE) gel in 1× TBE buffer supplemented with 8 mM MgCl2. The gel was dried and exposed to X-ray films.
In vitro triplex pulldown assay.
Nuclei from Gc1-spg cells were prepared by resuspending the cells in 1× nuclei isolation buffer (40 mM Tris-HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100, 1.28 M sucrose). Ten micromolar psoralen-biotinylated TFO1 or NC TFO RNA oligonucleotides (purchased from Sigma-Aldrich) were incubated with the Gc1-spg nuclei for 2.5 h at 30°C in 1× triplex-forming buffer followed by 10 min of UV treatment to induce photo-adduct formation. Nuclei were lysed using Bioruptor (10 cycles, 30 s on and 30 s off; Diagenode). The supernatants were incubated with 50 μL streptavidin-magnetic beads at 30°C. In case of the RNase H control reaction, the supernatants were treated with 15 units of RNase H for 20 min before streptavidin-magnetic beads were added. Following beads capture, the beads were washed in 1× triplex-forming buffer to remove the nonspecifically bound DNA fragments, and then beads were resuspended in DNA isolation buffer. Resuspended beads bound to DNA-RNA triplex were treated with RNase A (20 μg) for 30 min followed by proteinase K treatment. Precipitated DNA was used as the template for qPCR.
Chromosome conformation capture assay.
Bacterial artificial chromosome (BAC) plasmids containing the Sox8 locus (RP23-70O24) and the control Ercc3 locus (RP23-148C24) (BACPAC resources, Emeryville, CA) were purified by the alkaline lysis method. The plasmids were mixed in equimolar ratio, and 20 μg of mixed plasmid was subjected to restriction digestion with the enzyme Sau3AI (NEB; catalog number R0169) (isoschizomer of DpnII) overnight at 37°C. DNA was precipitated and resuspended in ligation master mix (1× NEB ligation buffer, 0.8% Triton X-100, 0.5× bovine serum albumin [BSA], 1,600 U of T4 DNA ligase) (NEB; catalog number M0202). The reaction was incubated at 21°C for 4 h. Ligated DNA was precipitated and used as a template for PCR.
Contact library was generated as described by Mumbach et al. (42) with modifications. Briefly, nuclei were isolated from cross-linked cells by resuspending cells in cell lysis buffer (10 mM Tris-Cl pH 8.0, 1.5 mM MgCl2, 10 mM KCl,0.5 mM dithiothreitol [DTT], 0.05% NP-40, 1× mPIC, and 0.2 μM phenylmethylsulfonyl fluoride [PMSF]) and incubating on ice for 30 min. Pelleted nuclei were washed once with cell lysis buffer and permeabilized using 0.7% SDS by incubating at 62°C for 15 min. Triton X-100 was added to a final concentration of 10%. Fifty microliters of 10× DpnII buffer and 375 U of DpnII (NEB; catalog number R0543) restriction enzyme were added, and the reaction was incubated overnight at 37°C with shaking at 900 rpm. The reaction was heat-inactivated at 62°C for 20 min. In situ ligation was carried out by adding a ligation master mix and incubating at 21°C for 4 h with shaking. Nuclei were pelleted and resuspended in SDS lysis buffer. The lysate was subjected to proteinase K treatment and reverse cross-linking overnight at 65°C. DNA precipitated was then used as template for PCR. The relative frequency of interaction was calculated as described by Naumova et al. (43) from agarose gel images.
CpG methylation assay.
The methylation status at the Sox8 promoter was scored for using the EpiTect methyl II PCR kit from Qiagen according to the manufacturer's instructions.
Systems analysis. (i) Triplexator prediction.
To identify all putative triplexes that can form between Mrhl lncRNA and the Sox8 promoter, analysis was run with default parameters (44) to identify TFO-TTS pairs in single-strand and duplex sequences.
(ii) ChIP-sequencing data analysis.
Raw FASTQ files were downloaded from the NCBI GEO repository and were reanalyzed to generate the aligned files for the visualization of regions of interest. All aligned files were aligned to the mouse genome (mm10) using Bowtie2 (45) and then sorted, indexed, and made free from PCR duplicates using Samtools (46). Aligned files were loaded in the IGV genome browser to visualize the enrichment of peaks at the regions of interest. Peak calling was done with the MACS2 pipeline (47).
(iii) RNA-sequencing data analysis.
Raw FASTQ files were downloaded from the NCBI GEO repository and were reanalyzed with the TopHat (48) and Cufflinks (49) pipeline. Aligned files were loaded on the IGV genome browser (50) to visualize the gene expression enrichment. All data sets analyzed have been listed in Table 4.
TABLE 4.
List of data sets analyzed for the present study
| Dataset | GEO accession no. |
|---|---|
| ChIP-seq dataset | |
| mESC CTCF | GSM723015 |
| mESC SMC1 | GSM560341 |
| mESC RNA Pol II | GSM723019 |
| mESC H3K4me3 | GSM723017 |
| mESC Input | GSM723020 |
| Mouse brain cortex CTCF | GSM722631 |
| Mouse brain cortex SMC1 | GSM1838869 |
| Mouse brain cortex RNA PolII | GSM722634 |
| Mouse brain cortex H3K4me3 | GSM722633 |
| Mouse brain cortex input | GSM722635 |
| RNA-seq dataset | |
| mESC | GSM723776 |
| Adult brain cortex | GSE96684 |
IACUC approval.
Experiments were performed using mice testes. The institution (JNCASR) has obtained IACUC approval for research involving the use of mice.
ACKNOWLEDGMENTS
This work was supported by the Department of Biotechnology, India (BT/01/COE/07/09 and DBT/INF/22/SP27679/2018).
M.R.S.R. acknowledges Department of Science and Technology for J. C. Bose and S.E.R.B. Distinguished fellowships and The Year of Science Chair professorship.
We declare no competing interests.
REFERENCES
- 1.O'Bryan M, Takada S, Kennedy C, Scott G, Harada S, Ray M, Dai Q, Wilhelm D, de Kretser D, Eddy E, Koopman P, Mishina Y. 2008. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol 316:359–370. doi: 10.1016/j.ydbio.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrionuevo F, Scherer G. 2010. SOX E genes: Sox9 and Sox8 in mammalian testis development. Int J Biochem Cell Biol 42:433–436. doi: 10.1016/j.biocel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Barrionuevo F, Hurtado A, Kim G, Real F, Bakkali M, Kopp J, Sander M, Scherer G, Burgos M, Jiménez R. 2016. Sox9 and Sox8 protect the adult testis from male-to-female genetic reprogramming and complete degeneration. Elife 5:e15635. doi: 10.7554/eLife.15635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson N, Gillot I, Gregoire E, Youssef S, de Rooij D, de Bruin A, De Cian M, Chaboissier M. 2020. Sox8 and Sox9 act redundantly for ovarian-to-testicular fate reprogramming in the absence of R-spondin1 in mouse sex reversals. Elife 9:e53972. doi: 10.7554/eLife.53972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishant K, Ravishankar H, Rao M. 2004. Characterization of a mouse recombination hot spot locus encoding a novel non-protein-coding RNA. Mol Cell Biol 24:5620–5634. doi: 10.1128/MCB.24.12.5620-5634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akhade V, Arun G, Donakonda S, Satyanarayana Rao M. 2014. Genome wide chromatin occupancy of Mrhl RNA and its role in gene regulation in mouse spermatogonial cells. RNA Biol 11:1262–1279. doi: 10.1080/15476286.2014.996070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kataruka S, Akhade V, Kayyar B, Rao M. 2017. Mrhl long noncoding RNA mediates meiotic commitment of mouse spermatogonial cells by regulating Sox8 expression. Mol Cell Biol 37:e00632-16. doi: 10.1128/MCB.00632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhade V, Dighe S, Kataruka S, Rao M. 2016. Mechanism of Wnt signaling induced down regulation of Mrhl long non-coding RNA in mouse spermatogonial cells. Nucleic Acids Res 44:387–401. doi: 10.1093/nar/gkv1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James A, Hoberg E, Moustakas A, Gyllensten U, Jones S, Gustafsson C, Sims A, Westerlund F, Gorab E, Kanduri C. 2015. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat Commun 6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. 2015. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell 60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 11.O'Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson MJ, Anastasov N. 2015. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep 11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz K, Mayer C, Postepska A, Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grote P, Herrmann B. 2013. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol 10:1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong C, Corces V. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saldaña-Meyer R, González-Buendía E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F, Reinberg D. 2014. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev 28:723–734. doi: 10.1101/gad.236869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldaña-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jácome-López K, Nora E, Bruneau B, Tsirigos A, Furlan-Magaril M, Skok J, Reinberg D. 2019. RNA interactions are essential for CTCF-mediated genome organization. Mol Cell 76:412–422. doi: 10.1016/j.molcel.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang J, Yin Q, Chen T, Zhang Y, Zhang X, Wu Z, Zhang S, Wang H, Ge J, Lu X, Yang L, Chen L. 2014. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero R, Felsenfeld G. 2010. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pal D, Neha C, Bhaduri U, Zenia Z, Dutta S, Chidambaram S, Rao M. 2021. LncRNA Mrhl orchestrates differentiation programs in mouse embryonic stem cells through chromatin mediated regulation. Stem Cell Res 53:102250. doi: 10.1016/j.scr.2021.102250. [DOI] [PubMed] [Google Scholar]
- 20.Gonen N, Futtner C, Wood S, Garcia-Moreno S, Salamone I, Samson S, Sekido R, Poulat F, Maatouk D, Lovell-Badge R. 2018. Sex reversal following deletion of a single distal enhancer of Sox9. Science 360:1469–1473. doi: 10.1126/science.aas9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guth S, Bösl M, Sock E, Wegner M. 2010. Evolutionary conserved sequence elements with embryonic enhancer activity in the vicinity of the mammalian Sox8 gene. Int J Biochem Cell Biol 42:465–471. doi: 10.1016/j.biocel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Moreno S, Futtner C, Salamone I, Gonen N, Lovell-Badge R, Maatouk D. 2019. Gonadal supporting cells acquire sex-specific chromatin landscapes during mammalian sex determination. Dev Biol 446:168–179. doi: 10.1016/j.ydbio.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogbourne S, Antalis T. 1998. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J 331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh AP, Harada S, Mishina Y. 2009. Downstream genes of Sox8 that would affect adult male fertility. Sex Dev 3:16–25. doi: 10.1159/000200078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima M, de Rooij D, Page D. 2019. Amplification of a broad transcriptional program by a common factor triggers the meiotic cell cycle in mice. Elife 8:e43738. doi: 10.7554/eLife.43738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalwa M, Hänzelmann S, Otto S, Kuo C, Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann A, Lee S, Teschendorff A, Denecke B, Lin Q, Widschwendter M, Weinhold E, Costa I, Wagner W. 2016. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res 44:10631–10643. doi: 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blank-Giwojna A, Postepska-Igielska A, Grummt I. 2019. lncRNA KHPS1 activates a poised enhancer by triplex-dependent recruitment of epigenomic regulators. Cell Rep 26:2904–2915. doi: 10.1016/j.celrep.2019.02.059. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Li G. 2020. Post-translational modifications of PRC2: signals directing its activity. Epigenetics Chromatin 13:47. doi: 10.1186/s13072-020-00369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kentepozidou E, Aitken S, Feig C, Stefflova K, Ibarra-Soria X, Odom D, Roller M, Flicek P. 2020. Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol 21:5. doi: 10.1186/s13059-019-1894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Essien K, Vigneau S, Apreleva S, Singh L, Bartolomei M, Hannenhalli S. 2009. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol 10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishana M, Ha C, Rodriguez-Hernaez J, Ranjbaran A, Chio E, Nora E, Badri S, Kloetgen A, Bruneau B, Tsirigos A, Skok J. 2020. Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation. Genome Biol 21:108. doi: 10.1186/s13059-020-02024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugacheva E, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk A, Strunnikov A, Zentner G, Ren B, Lobanenkov V. 2020. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc Natl Acad Sci USA 117:2020–2031. doi: 10.1073/pnas.1911708117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marino M, Rega C, Russo R, Valletta M, Gentile M, Esposito S, Baglivo I, De Feis I, Angelini C, Xiao T, Felsenfeld G, Chambery A, Pedone P. 2019. Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF. J Biol Chem 294:861–873. doi: 10.1074/jbc.RA118.004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher J, Peterson J, Reimer M, Stelloh C, Pulakanti K, Gerbec Z, Abel A, Strouse J, Strouse C, McNulty M, Malarkannan S, Crispino J, Milanovich S, Rao S. 2016. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of HoxA7 and HoxA9. Blood 128:1708. doi: 10.1182/blood.V128.22.1708.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Wang J, Yang L, Zhao C, Wu L, Xu L, Zhang F, Weng Q, Wegner M, Lu Q. 2020. CTCF-mediated chromatin looping in EGR2 regulation and SUZ12 recruitment critical for peripheral myelination and repair. Nat Commun 11:4133. doi: 10.1038/s41467-020-17955-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang D, Petrykowska H, Miller B, Elnitski L, Ovcharenko I. 2019. Identification of human silencers by correlating cross-tissue epigenetic profiles and gene expression. Genome Res 29:657–667. doi: 10.1101/gr.247007.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croft B, Ohnesorg T, Hewitt J, Bowles J, Quinn A, Tan J, Corbin V, Pelosi E, van den Bergen J, Sreenivasan R, Knarston I, Robevska G, Vu D, Hutson J, Harley V, Ayers K, Koopman P, Sinclair A. 2019. Human sex reversal is caused by duplication or deletion of core enhancers upstream of Sox9. Nat Commun 10:3351. doi: 10.1038/s41467-019-11310-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pentland I, Campos-León K, Cotic M, Davies K, Wood C, Groves I, Burley M, Coleman N, Stockton J, Noyvert B, Beggs A, West M, Roberts S, Parish J. 2018. Disruption of CTCF-YY1–dependent looping of the human papillomavirus genome activates differentiation-induced viral oncogene transcription. PLoS Biol 16:e2005752. doi: 10.1371/journal.pbio.2005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weintraub A, Li C, Zamudio A, Sigova A, Hannett N, Day D, Abraham B, Cohen M, Nabet B, Buckley D, Guo Y, Hnisz D, Jaenisch R, Bradner J, Gray N, Young R. 2017. YY1 is a structural regulator of enhancer-promoter loops. Cell 171:1573–1588. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaukowitch K, Joo J-Y, Kim T-K. 2017. UV-RNA immunoprecipitation (UV-RIP) protocol in neurons. Methods Mol Biol 1468:33–38. doi: 10.1007/978-1-4939-4035-6_4. [DOI] [PubMed] [Google Scholar]
- 41.Spruce C, Dlamini S, Ananda G, Bronkema N, Tian H, Paigen K, Carter G, Baker C. 2020. HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev 34:398–412. doi: 10.1101/gad.333542.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mumbach M, Rubin A, Flynn R, Dai C, Khavari P, Greenleaf W, Chang H. 2016. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naumova N, Smith E, Zhan Y, Dekker J. 2012. Analysis of long-range chromatin interactions using chromosome conformation capture. Methods 58:192–203. doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buske F, Bauer D, Mattick J, Bailey T. 2012. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res 22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng J, Liu T, Qin B, Zhang Y, Liu X. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, van Baren M, Salzberg S, Wold B, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson J, Thorvaldsdóttir H, Winckler W, Guttman M, Lander E, Getz G, Mesirov J. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]