ABSTRACT
The Borrelia burgdorferi BB0323 protein undergoes a complex yet poorly defined proteolytic maturation event that generates N-terminal and C-terminal proteins with essential functions in cell growth and infection. Here, we report that a borrelial protease, B. burgdorferi high temperature requirement A protease (BbHtrA), cleaves BB0323 between asparagine (N) and leucine (L) at positions 236 and 237, while the replacement of these residues with alanine in the mutant protein prevents its cleavage, despite preserving its normal secondary structure. The N-terminal BB0323 protein binds BbHtrA, but its cleavage site mutant displays deficiency in such interaction. An isogenic borrelial mutant with NL-to-AA substitution in BB0323 (referred to as Bbbb0323NL) maintains normal growth yet is impaired for infection of mice or transmission from infected ticks. Notably, the BB0323 protein is still processed in Bbbb0323NL, albeit with lower levels of mature N-terminal BB0323 protein and multiple aberrantly processed polypeptides, which could result from nonspecific cleavages at other asparagine and leucine residues in the protein. The lack of infectivity of Bbbb0323NL is likely due to the impaired abundance or stoichiometry of a protein complex involving BB0238, another spirochete protein. Together, these studies highlight that a precise proteolytic event and a particular protein-protein interaction, involving multiple borrelial virulence determinants, are mutually inclusive and interconnected, playing essential roles in the infectivity of Lyme disease pathogens.
KEYWORDS: Borrelia burgdorferi, Lyme disease, protein cleavage, serine protease
INTRODUCTION
Lyme disease, caused by the spirochete Borrelia burgdorferi sensu lato, is a prevalent arthropod-borne disease in many parts of the world (1). The pathogen survives in an enzootic life cycle involving Ixodes scapularis ticks and diverse vertebrates (2). In infected mammals, the pathogen can cause various clinical complications, including arthritis, carditis, and neurological disorders (3, 4). The microbe features an unusual genome encoding many proteins of undefined functions (5–7), which can be differentially expressed in specific environments (8, 9) and can contribute to microbial persistence (2). Currently, as a vaccine to prevent Lyme disease in humans is unavailable (10, 11), a more thorough understanding of spirochete biology and pathogenesis and the discovery of novel comprehensive interventions against infection remain highly warranted research goals.
The molecular mechanisms that allow spirochetes to establish infection in the host are largely unknown. Over the past decades, many unique gene products with unknown structure or function that are essential for microbial infection have been characterized (2). One of these proteins, annotated as BB0323, was identified to be associated with cell fission (12) and infectivity (13–15). BB0323 is an outer membrane (OM)-associated periplasmic protein (14, 15) that houses a C-terminal peptidoglycan-binding motif called LysM (16, 17). Gene deletion studies (12–15) suggest that BB0323 is functionally analogous to the bacterial Tol-Pal protein complex (18–20), which supports OM stability. However, BB0323 shares no sequence homology with Tol-Pal proteins. Additionally, BB0323 is a virulence determinant, as the deletion of its C-terminal portion renders the spirochete noninfectious, while not impacting fission or OM stability or any other apparent defects in B. burgdorferi growth in culture (14). Recently, the crystal structure of the BB0323 N-terminal protein was solved (21–24), indicating a rare structure for a bacterial protein. Its overall fold belongs to the spectrin superfamily proteins (25) that function as linkers between actin and other cytoskeletal proteins. Spectrin proteins are seldom present in bacteria, except for EzrA proteins, which inhibit FtsZ polymerization and ensure proper formation of the Z-ring at mid-cell (26). Nevertheless, it remains enigmatic as to how BB0323 spectrin repeats support fission, OM stability, or infectivity.
We have previously shown that BB0323 interacts with at least two spirochete proteins that impact its maturation, stability, and biological functions (13, 14, 27). Its maturation is influenced by a member of the high-temperature requirement A (HtrA) family of serine proteases, called B. burgdorferi HtrA (BbHtrA) (14), which cleaves BB0323 into two proteins, representing N-terminal and C-terminal domains that support essential functions in spirochete fission and infectivity. The BbHtrA mutant is noninfectious, where the proteolytic processing of BB0323 is impaired (28). As BbHtrA is involved in the maturation of additional borrelial proteins required for infection (29–33), the significance of its cleavage of BB0323 remains unclear. BB0323 also interacts with another protein of unknown function, BB0238 (13, 34). The interaction of the BB0323 N-terminal protein with BB0238 contributes to the establishment of Borrelia infection and transmission from ticks to mammals (27). Notably, the cellular compartment where BB0323 and BB0238 interacts and where BbHtrA processes BB0323 remain unknown, but these proteins have been shown to be localized in the periplasm (13, 28). In the current study, we showed that BbHtrA-mediated maturation of BB0323 is essential for spirochete infectivity and optimal formation of the BB0323-BB0238 complex, highlighting the usefulness of these antigens as novel therapeutic targets against Lyme disease.
RESULTS
Identification and localization of BbHtrA protease cleavage site in BB0323.
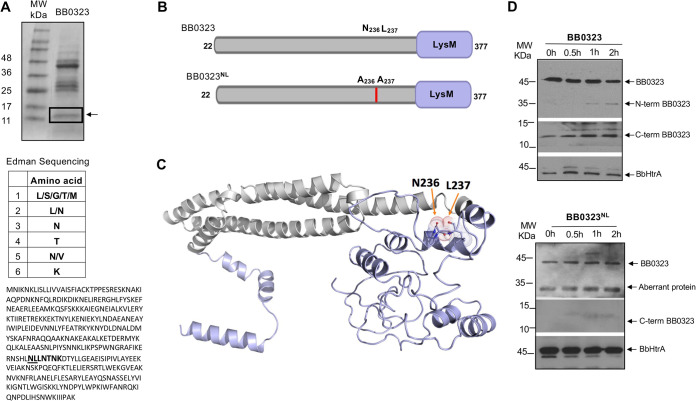
To identify the cleavage site of BbHtrA in BB0323, we performed an in vitro digestion assay using recombinant proteins. The cleaved C-terminal polypeptide, which migrated in an SDS-PAGE gel around 12 kDa (Fig. 1A, upper portion), was excised and subjected to N-terminal sequencing by Edman degradation. The first six amino acids, as obtained by sequencing (Fig. 1A, middle portion), matched BB0323 residues LLNTNK, spanning positions 237 to 242, suggesting that the N-terminal sequencing originated from the 237th residue of BB0323 (Fig. 1A, lower portion). Based on these results, we predict that the potential BbHtrA cleavage site in BB0323 encompasses the asparagine 236 (N236) and leucine 237 (L237) residues.
FIG 1.
Identification of BbHtrA cleavage site in BB0323. (A) In vitro cleavage of full-length BB0323. Recombinant full-length BB0323 was incubated with BbHtrA at 37°C for 2.5 h. The products were subjected to 15% SDS-PAGE and transferred to a nylon membrane (upper portion). The area encompassing the C-terminal fragment, as indicated by a box (arrow), was dissected and subjected to N-terminal sequencing. The identities of the first six residues, as revealed by N-terminal sequencing (middle portion), and their localization (highlighted in bold letters) in the full-length BB0323 protein sequence (lower portion) are shown. Residues encompassing the cleavage site are underlined. (B) Schematic representation of full-length BB0323 proteins. The full-length BB0323 without the signal peptide and the mutated protein (BB0323NL) with amino acid replacement of asparagine (N) and leucine (L) at positions 236 and 237 with two alanine residues are shown. (C) A homology-guided structure of full-length BB0323. The five N-terminal α-helices and the location of BbHtrA cleavage site in the predicted model with positions of cleavage site residues, N236 and L237, are indicated. (D) Mutation in the cleavage site impairs proteolytic processing of BB0323. Recombinant BbHtrA was incubated with full-length BB0323 (arrow, upper portion) or BB0323NL (arrow, lower portion) at 37°C for the indicated times. Samples were resolved on SDS-PAGE and subjected to immunoblotting with specific antibodies to detect cleaved N-terminal (arrow, upper portion) or cleaved C-terminal product (arrow, middle portion). The levels of protease were detected using BbHtrA antiserum (arrow, bottom portion). In the case of BB0323NL digestion, no N-terminal BB0323 or C-terminal BB0323 was detectable, except for a nonspecific aberrantly reactive protein present at all tested time points.
Site-directed mutagenesis of BbHtrA cleavage site in BB0323 impairs proteolysis and impacts protease interaction.
To further study the cleavage site involving the N236 and L237 residues, a mutated version of full-length BB0323, with replacement of both residues with alanine (A), was generated and named as BB0323NL (Fig. 1B). A homology-guided structure of full-length BB0323, based on the available crystallized portion of N-terminal BB0323 (PDB code 6RJX) and iterative template-based fragment assembly simulations of the rest of the protein region using I-TASSER on high-performance computing systems available at the National Institutes of Health, suggested possible surface accessibility of the N236 and L237 residues (Fig. 1C). To test whether the mutation affected the enzymatic processing of the mutated full-length protein, BB0323NL, by BbHtrA, a proteolytic assay was performed, as developed earlier (14). The wild-type recombinant BB0323 and BB0323NL were separately incubated with recombinant BbHtrA, and the cleavage was tested at various time points. We observed the appearance of a cleaved N-terminal BB0323 fragment using an anti-BB0323 antibody, with increased intensity over the course of incubation with BbHtrA (Fig. 1D, upper portion). Consistently, we also observed a reduction in the full-length BB0323 protein (Fig. 1D, upper portion). The levels of cleaved C-terminal BB0323 protein also increased with prolonged incubation, as detected with far-Western blotting (Fig. 1D, upper portion). In contrast, neither an optimal level of cleaved N-terminal polypeptide nor an optimal level of C-terminal polypeptide could be detected when BB0323NL was incubated with BbHtrA (Fig. 1D, lower portion); instead, we detected an aberrant nonspecific protein (presumably copurified glutathione S-transferase [GST] used as a fusion tag) that was present at all time points (Fig. 1D, lower portion) and also when a previously described (14) catalytically inactive form of BbHtrA protein was used in the digestion assay (data not shown). Thus, the unaltered intensity of the full-length protein, together with the undetectable C terminus, confirmed that the point mutation impairs BB0323 proteolytic processing by BbHtrA.
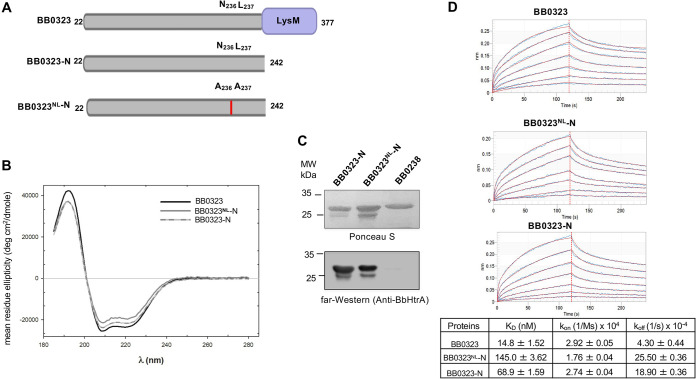
Next, we assessed whether the cleavage site mutation affects the BB0323-BbHtrA interaction using a far-Western assay. As full-length BB0323 is difficult to purify in larger quantities, an N-terminal BB0323 protein including the BbHtrA cleavage site, spanning residues 22 to 242 (termed BB0323-N), and a mutated version with similar change of N236 and L237 to alanine residues, designated BB0323NL-N, were also generated (Fig. 2A). A far-UV circular-dichroism (CD) study confirmed that the secondary structure of the N-terminal BB0323 or the mutated recombinant protein was not altered, compared to the full-length protein, as reflected by comparable percentages of structural features, such as α-helicity and β-turn of full-length BB0323 (72% and 11%) and the BB0323NL-N (62% and 13%) and BB0323-N (66% and 12%) proteins with similar CD profiles (Fig. 2B). A far-Western analysis suggested a similar interaction of the recombinant BB0323 N-terminal proteins and BbHtrA, which did not occur in the control borrelial protein, BB0238, a known BB0323 interaction partner (Fig. 2C). Biolayer interferometry (BLI) was applied to obtain further insights into the interaction kinetics. The analysis of the association and dissociation sensorgrams showed that full-length BB0323 binds to BbHtrA with an extremely tight affinity (KD [equilibrium dissociation constant] = 1.48 × 10−8 M [∼15 nM]), while the KD of the N-terminal BB0323 protein is 6.89 × 10−8 M (∼70 nM). The KD of the cleavage site mutant protein BB0323NL-N is 1.45 × 10−7 M (145 nM), which is substantially higher than for the wild type full-length or N-terminal protein (Fig. 2D), suggesting substantially reduced affinity for binding to the protease.
FIG 2.
The mutated N-terminal BB0323 binds to BbHtrA with a lower affinity than ones carrying the native cleavage site. (A) Schematic representation of N-terminal BB0323 proteins. The full-length BB0323, N-terminal BB0323 protein carrying the BbHtrA cleavage site, spanning amino acid residues 22 to 242 (BB0323-N), and its mutated version (BB0323NL-N) with changes of N and L residues to AA at positions 236 and 237 are indicated. (B) Amino acid alterations in BB0323 do not affect protein secondary structure. The recombinant proteins were analyzed by circular-dichroism spectroscopy. (C) Far-Western analysis of BB0323-BbHtrA interaction. The recombinant N-terminal BB0323 protein carrying the native BbHtrA cleavage site (BB0323-N), its mutated version BB0323NL-N, and a control protein (BB0238) that binds BB0323 but fails to interact with BbHtrA were separated by SDS-PAGE and transferred to a nitrocellulose membrane (upper portion), incubated with recombinant BbHtrA, and probed with specific anti-BbHtrA antibodies (lower portion). (D) Biolayer Interferometry (BLI) sensorgrams of BB0323-BbHtrA interaction. The BLI results showed that compared to BB0323NL-N showed weaker interaction with BbHtrA than did the full-length BB0323 and BB0323-N proteins. The kinetic and thermodynamic parameters of the interaction between BB0323 proteins and BbHtrA are summarized in the table. Blue jagged lines indicate the experimental data, while the smooth red line represents the fitted model, as plotted for each protein and each concentration of ligand analyzed. The model was selected for the best fit of all of the data for all of the curves rather than an individual curve. The red dotted vertical line shows the breakpoint between the 120-s association and the 120-s dissociation as described in Materials and Methods.
BbHtrA-mediated cleavage of BB0323 does not impact spirochete fission or growth in vitro.
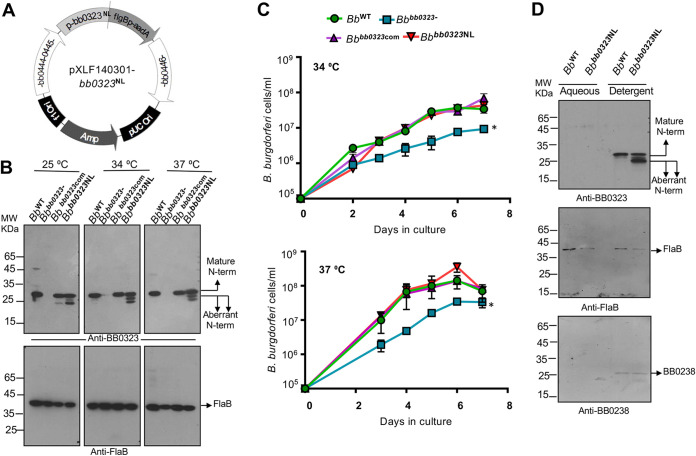
To examine the importance of BbHtrA-mediated cleavage of BB0323 in spirochete biology, we generated a B. burgdorferi mutant that produces BB0323NL instead of the wild-type protein, designated Bbbb0323NL (Fig. 3A and B). Interestingly, in mutant spirochetes, in addition to the mature 27-kDa N-terminal BB0323, two aberrantly processed polypeptides of lower molecular weights were also present when cells were grown at various temperatures ranging from 25 to 37°C (Fig. 3B). While the exact nature of origin of these aberrant polypeptides remains uncertain, the nonspecific cleavage of BB0323 at two other cognate asparagine lysine pairs, such as in positions 147 to 148 and 207 to 208, cannot be ruled out. However, Bbbb0323NL displayed no growth defects (Fig. 3C) and no obvious cell fission defects or cell clumps, as noted for bb0323-deficient mutants (15), indicating that the optimal cleavage of BB0323 by BbHtrA is not required for maintaining normal cell growth and morphology. The cleavage site mutation also did not alter the membrane association of BB0323 or its interaction partner BB0238 (13), as analyzed by a Triton X-114 phase partitioning assay (Fig. 3D).
FIG 3.
Alteration of BbHtrA cleavage site in BB0323 impairs processing of N-terminal domains but does not affect spirochete growth and biology in vitro. (A) Diagram of the vector construct (pXLF14301-bb0323NL) for introducing mutants in B. burgdorferi. The construct was generated via the chromosomal insertion of bb0323N236AL237A into the bb0323 deletion mutant, in order to produce bb0323NL complemented B. burgdorferi isolates (designated Bbbb0323NL). (B) Production of mature N-terminal BB0323 domains in B. burgdorferi isolates. The wild type (WT), bb0323 mutant (bb0323-), bb0323 complement (bb0323com), and cleavage site mutant (Bbbb0323NL) were grown at different temperatures (25 to 37°C). The spirochetes were harvested and lysates were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and blotted with antiserum against BB0323-N (upper portion) and FlaB (lower portion). Mature N-terminal proteins with expected molecular weights of ∼27 kDa (larger arrows), as well as polypeptides of slightly lower molecular weights (small arrows), are indicated. (C) The Bbbb0323NL cleavage site mutants display normal growth in culture compared to wild-type cells. B. burgdorferi cells were diluted to a density of 105/mL and grown at 34°C and 37°C in BSK-H medium. Samples were counted under a dark-field microscope every 24 h using a Petroff-Hausser cell counter. The bb0323 deletion mutant (Bbbb0323-) display significant growth defects compared to other isolates (*, P < 0.05). (D) N-terminal BB0323 proteins are associated with spirochete membrane fractions. Spirochete lysates were subjected to Triton X-114 phase partitioning of aqueous and detergent phases and immunoblotted using antibodies against BB0323, FlaB, and BB0238.
Precise BbHtrA-mediated cleavage of BB0323 is required for spirochete infectivity.
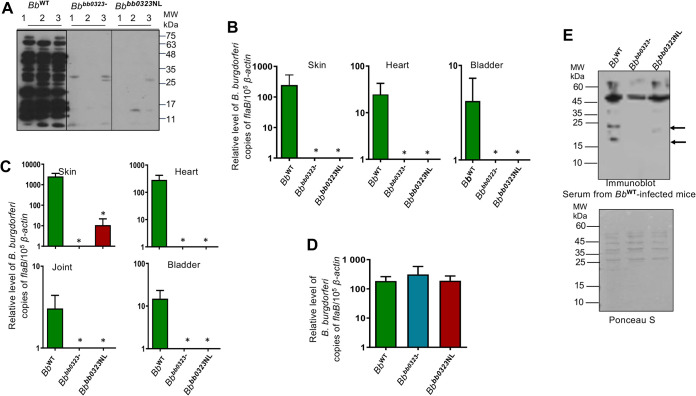
To assess whether BbHtrA-mediated cleavage of BB0323 is required for spirochete infectivity, mice were inoculated intradermally with either wild-type, bb0323 mutant, or Bbbb0323NL spirochetes. After 14 days of infection, antibody responses against B. burgdorferi were detectable in mice infected with the wild type but not in mice infected with the bb0323 mutant or Bbbb0323NL isolate, suggesting a loss of infectivity in both mutants (Fig. 4A). Unlike for the wild type, mutant spirochete burdens in tested murine tissues were mostly indiscernible (Fig. 4B). Furthermore, when murine tissues were cultured, no mutant spirochetes were recovered, whereas the wild type could be readily isolated (see Table S1 in the supplemental material). These results suggest that the precise cleavage of BB0323 at N236 and L237 is essential for B. burgdorferi persistence in mammalian hosts.
FIG 4.
BbHtrA-mediated cleavage of BB0323 is essential to spirochete infectivity in murine hosts. (A) Serological response in infected mice. Mice were infected with wild-type (BbWT), bb0323 mutant (Bbbb0323-), and cleavage site mutant (Bbbb0323NL) spirochetes via syringe inoculation. Sera were collected 2 weeks after inoculation and used for immunoblotting against B. burgdorferi cell lysates. (B) Spirochete levels in the murine skin, heart, and bladder were assessed by measuring copies of B. burgdorferi flaB cDNA using qPCR and normalizing against mouse β-actin. The pathogen burden was significantly reduced in all mouse tissues infected with Bbbb0323- and Bbbb0323NL compared to those infected with BbWT (*, P < 0.05). Data represent the means ± SEM of four qPCR analyses of B. burgdorferi levels derived from two independent animal experiments. (C) Transmission of spirochetes by ticks to naive hosts. Ticks were microinjected with equal numbers of either BbWT, Bbbb0323-, and Bbbb0323NL spirochetes and allowed to engorge on naive mice. Two weeks after tick feeding, samples of the skin, heart, joint, and bladder tissues were collected from mice, and the isolated cDNA preparations were assessed for spirochete levels using qPCR. The B. burgdorferi burden was significantly reduced in all mouse tissues parasitized by ticks infected with Bbbb0323- and Bbbb0323NL isolates compared to the WT (*, P < 0.05). (D) Spirochete burdens in ticks. The ticks, which were microinjected with different groups of spirochetes as detailed for panel C, were collected after blood meal engorgement on mice and subjected to analysis of pathogen burden by measuring cDNA copies of the B. burgdorferi flaB gene using qPCR and normalizing against tick β-actin. Data represent the means ± SEM of four qPCR analyses of B. burgdorferi levels derived from two independent animal experiments. (E) The bb0323 mutants display altered immunogenic protein profiles compared to the wild type. Cell lysates from BbWT, Bbbb0323-, and Bbbb0323NL spirochetes immunoblotted with serum samples collected from mice were infected with wild-type spirochetes (as detailed for panel A). The protein bands absent from bb0323 mutants are indicated (arrows).
In our examination of whether Bbbb0323NL is able to transmit via tick bite, groups of nymphs were microinjected with spirochetes and allowed to parasitize naive mice. Twelve days after tick feeding, serological analysis of the antibody responses against B. burgdorferi in mice (data not shown), as well as the measurement of spirochete levels in various tissues using quantitative PCR (qPCR), showed that both the bb0323 deletion mutants and the Bbbb0323NL isolate were severely impaired in establishing infection in mice (Fig. 4C), although their levels were comparable with those of the wild-type isolate in replete ticks (Fig. 4D). Notably, immunoblot analysis using serum samples collected from wild-type-infected mice showed that both the bb0323 mutant and Bbbb0323NL spirochetes display an altered immunogenic protein profile compared to that of the wild-type spirochetes (Fig. 4E). Together, these data indicate that the precise cleavage of BB0323 is required for B. burgdorferi persistence in mice and transmission via tick-borne infection but is not necessary for spirochete survival in ticks.
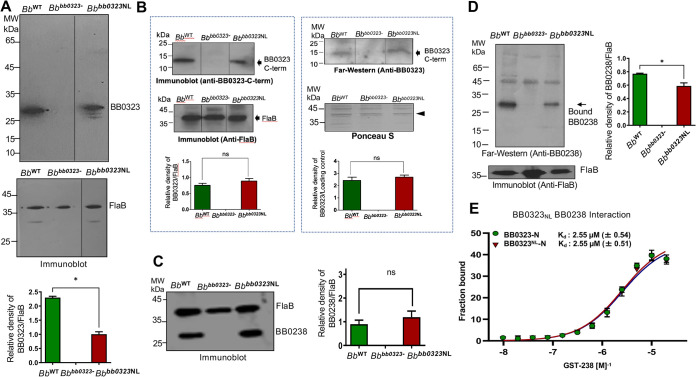
Optimal level of mature BB0323 N-terminal polypeptides is essential for borrelial infectivity.
We next sought to determine the potential mechanism behind the Bbbb0323NL isolate’s inability to establish infection in mice. A densitometric analysis showed that the relative level of mature 27-kDa N-terminal BB0323 polypeptides was significantly lower in the Bbbb0323NL isolate (Fig. 5A) than in the wild-type strain. We have previously shown that the C-terminal BB0323 protein is essential for B. burgdorferi survival in mice and interacts with the N-terminal protein (14), which remains unaltered in the Bbbb0323NL isolate (Fig. 5B). Although bb0323 deletion mutants display severe defects in cell growth and fission (12, 15), like wild-type spirochetes, Bbbb0323NL isolates showed normal growth pattern (Fig. 3C), including atypical zonal cellular distribution of peptidoglycan (35), as detected by labeling with the fluorescent d-alanine analog 7-hydroxycoumarin-amino-d-alanine (HADA) (Fig. S2). Previously, it has also been demonstrated that the interaction between the N-terminal BB0323 protein and BB0238 is essential for the posttranslational stability of both proteins and for B. burgdorferi infectivity in mammals (13). However, despite the observation that BB0238 expression levels were not altered in the Bbbb0323NL isolate (Fig. 5C), it interacted only with the mature 27-kDa N-terminal BB0323 protein and not with the two N-terminal polypeptides of aberrantly lower molecular weights (Fig. 5D). The microscale thermophoresis (Fig. 5E) and biolayer interferometry (data not shown) analyses confirmed that the cleavage site mutations in the recombinant BB0323 N-terminal protein did not alter its specificity and affinity toward BB0238. These results collectively highlight that a precise level of mature N-terminal BB0323 polypeptides is required to maintain normal levels (and possibly the stoichiometry) of the BB0323-BB0238 complex, which is shown to be essential for mammalian infection by spirochetes (13, 27).
FIG 5.
The cleavage site mutant Bbbb0323NL isolates generate lesser amounts of functional N-terminal proteins, resulting in suboptimal recruitment of the interaction partner BB0238. (A) The Bbbb0323NL cleavage site mutant isolates produce significantly lower levels of mature 27-kDa N-terminal BB0323 protein. Comparable amounts of spirochete lysates from the WT, bb0323 deletion mutant (bb0323-), and the cleavage site mutant (Bbbb0323NL) isolates were separated by SDS-PAGE. The gel was then transferred to a nitrocellulose membrane and immunoblotted with antibodies against BB0323 (upper portion). The loading control was measured with anti-FlaB (lower portion). The densities of N-terminal protein were quantified by ImageJ software, with the values normalized against the corresponding density of FlaB. The amount of the upper band of BB0323NL protein was significantly decreased compared to the wild type (*, P < 0.05). (B) Analysis of C-terminal BB0323 fragments in Bbbb0323NL isolates. The Bbbb0323NL point mutation did not affect cleavage of the BB0323 C terminus (arrow, left portion). The proteins of WT, bb0323 mutant (bb0323), and Bbbb0323NL complemented spirochetes were separated using SDS-PAGE and transferred to a nitrocellulose membrane, which was probed with antibodies against the BB0323 C terminus (left upper portion), while the equal protein loading was shown by immunoblotting with anti-FlaB antibodies (arrow, left lower portion). The BB0323 C-terminal protein binds to the N-terminal BB0323, as assessed by far-Western analysis (arrow, right upper portion). The proteins of WT, bb0323 mutant (bb0323-), and Bbbb0323NL complemented spirochetes were separated using SDS-PAGE and transferred to a nitrocellulose membrane, which was incubated with recombinant BB0323-N protein purified from E. coli and probed with antiserum against the N terminus of BB0323 (right upper portion), while the equal protein loading is shown by Ponceau S stain (right lower portion). The bottom graphs represent the densities of the protein bands of corresponding BB0323 immunoblots, normalized against controls, either FlaB (left portion) or a protein stained with Ponceau S (arrowhead, right portion). (C) The cleavage site mutation did not reduce BB0238 protein levels in Bbbb0323NL. The protein lysates of WT, bb0323 mutant (bb0323-), and Bbbb0323NL isolates were separated by SDS-PAGE. The BB0238 protein level was detected with specific antibodies against BB0238 using Western blotting, with similar levels observed in Bbbb0323NL and the wild type. (D) Only mature 27-kDa N-terminal BB0323 protein is capable of interaction with BB0238. The protein lysates of WT, bb0323-, and Bbbb0323NL isolates were separated by SDS-PAGE and transferred to a nitrocellulose membrane, incubated with BB0238 recombinant protein, and probed with specific anti-BB0238 (upper portion). Only the mature 27-kDa N-terminal BB0323 peptides, not the lower BB0323 polypeptides, displayed binding. The density of the protein bands was scanned and normalized with FlaB (lower portion). The level of recruited BB0238 in the protein complex was significantly decreased compared to the wild-type protein (*, P < 0.05). (E) The binding affinity of recombinant BB0323 N-terminal proteins with BB0238 is not altered by cleavage site mutation. A microscale thermophoresis assay was performed to measure binding kinetics of BB0323-N and BB0323NL-N, which were essentially similar.
DISCUSSION
BB0323 undergoes multiple, and potentially complex, series of poorly defined proteolytic events in which the first cleavage is likely mediated by BbHtrA (14), which is followed by additional cleavage by another protease, a C-terminal protease called CtpA (36). The proteolytically processed BB0323 N- and C-terminal proteins independently and/or cooperatively support multiple aspects of borrelial biology and infection (14). Although the mechanism of BB0323 maturation awaits future studies, we show that at least one of the proteolytic cleavage events is mediated by BbHtrA (BB0104) (28, 31, 33) and occurs between amino acid positions 236 and 237. Despite the critical role of such BB0323 cleavage in spirochete infectivity, it is dispensable for other known BB0323 functions, including fission (12, 15), OM stability (15), and support of the posttranslational stability of BB0238 (13), which are otherwise affected by the deletion of the bb0323 gene. Nevertheless, as the BbHtrA-mediated cleavage of BB0323 is essential for spirochete infection, these proteins and their interaction with each other constitute novel therapeutic targets for the prevention of Lyme borreliosis.
The present study provided new evidence for another critical role of the HtrA family of proteases (37–39), particularly in the controlled processing of an essential microbial virulence determinant. Besides BB0323, the protease influences the degradation of additional borrelial virulence determinants (29–32), such as BmpD, CheX, P66, and FlaB, as well as host aggrecan (33). Given its pleiotropic roles in the degradation of multiple proteins, it is difficult to tease out its role in the processing and/or stability of a single target, specifically BB0323. Although the HtrA proteins can transition from chaperone to protease functions, our study clearly shows that only the latter function of BbHtrA is required to produce an optimal level of N-terminal BB0323 proteins, as required for spirochete infectivity in mammals. Note that the impaired production or stability of N-terminal BB0323 polypeptides (such as in Bbbb0323NL) also occurs in the BbHtrA (28) or bb0238 mutants (13). These mutants still maintain a lower level of the N-terminal BB0323 protein that is sufficient for supporting normal fission and produce the protein independently of BbHtrA protease activity (28) or BB0238-mediated posttranslational stability (13). As the bb0323 gene deletion completely removes N-terminal BB0323 polypeptides, prevents normal fission, and destabilizes the outer membrane (15), the identification of additional BB0323 cleavage mechanisms would lead to new avenues for therapeutic drugs or biologics that prevent borrelial fission and biology.
The crystal structure of BB0323 N-terminal protein has recently been reported (21); it simulates those of spectrins (40), which are rarely present in bacteria, except for EzrA proteins, which regulate the formation of septation rings (41, 42). As speculated (21) and supported by studies (13–15), the BB0323 complex, via its N-terminal spectrin repeats and C-terminal LysM domain, may function to stabilize the OM by bridging this structure to the underlying peptidoglycan layer or unidentified membrane components. However, the existence of such a BB0323-supported membrane bridge remains paradoxical, particularly given the existence of intervening periplasmic flagellar filaments that are present throughout spirochete cell growth, rotate around the protoplasmic cylinder (43, 44), and even pass through the cell division sites (45).
Our study shows that the function of BB0323, particularly for the mature N-terminal protein, requires multifaceted protein-protein interaction (PPI) events involving two borrelial proteins, BbHtrA (14, 28) and BB0238 (13). Notably, these events are mutually inclusive, as the prevention of BbHtrA-mediated proteolytic processing also affected the BB0323-BB0238 PPI event essential for infection (27). A precisely regulated proteolysis of transcription factors in Vibrio cholerae was reported to influence pathogen adaptation under stress conditions (46). Likewise, Streptococcus pyogenes virulence factors, such as SpeB and streptolysin S, require extensive processing for the generation of their biologically active forms (47, 48). In addition to PPI, a regulated proteolysis of BB0323 also supports existence and/or functions of additional unidentified spirochete proteins, especially ones that are expressed during murine infection. While the proteolytic maturation pathways of borrelial proteins, including BB0323, that dictate spirochete infection and virulence remain enigmatic, a further understanding of the proteolysis and PPIs involving three functionally interdependent borrelial proteins—BB0323, BB0238, and BbHtrA—would enrich our knowledge of atypical spirochete biology and support the development of new interventions against Lyme borreliosis.
MATERIALS AND METHODS
Bacteria, mice, and ticks.
The B. burgdorferi infectious isolate B31-A3 was grown in complete Barbour-Stoenner-Kelly H (BSK-H) medium at 34°C (15). Four- to 6-week-old female C3H/HeN mice were purchased from the National Institutes of Health. All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee and Institutional Biosafety Committee of the University of Maryland, College Park, MD. Ixodes scapularis ticks used in the study were raised in the laboratory (13, 15); the tick egg masses were purchased from the Oklahoma State University Tick Rearing Facility. The hatched larvae were fed on mice and reared in the laboratory until molt into nymphs.
Quantitative PCR.
The primers used in quantitative PCR (qPCR) are listed in Table S2. For qPCR, total RNA was isolated from ticks and murine tissues using TRIzol reagent (Invitrogen), treated with DNase I (New England BioLabs [NEB]), reverse transcribed to cDNA (Thermo), and analyzed by qPCR, as detailed previously (13, 15). For the quantification of spirochetes, flaB cDNA copies were measured using qPCR and normalized to tick or mouse β-actin levels (13, 15).
Site-directed mutagenesis, generation of recombinant proteins, and generation of antisera.
The recombinant full-length BB0323 and an N-terminal BB0323 protein spanning residues 22 to 242 (termed BB0323-N) were produced as detailed previously (13, 14). A mutated full-length BB0323 protein (designated BB0323NL) and an N-terminal BB0323 protein (designated BB0323NL-N) were also generated by replacing two residues at the site where BbHtrA cleaves the protein—asparagine at position 236 (N236) and leucine at position 237 (L237)—with two alanine (AA) residues, using oligonucleotide primers (Table S2) and a Q5 site-directed mutagenesis kit (NEB). The clones were sequenced to ensure authenticity, and the recombinant protein was produced in Escherichia coli, as described previously (13, 15). The generation of anti-BB0323 murine antisera used to detect recombinant full-length BB0323 and processed BB0323 in Borrelia extracts and far-Western blotting, was described earlier (13, 14). An aliquot of antibody against BB0323 C terminus was obtained from Philip Stewart and Patricia Rosa at the National Institutes of Health.
Protease cleavage assay and N-terminal sequencing.
The in vitro cleavage assay of BB0323 proteins by BbHtrA was performed as described previously (14). Briefly, recombinant proteins were incubated with BbHtrA at 37°C and analyzed by Western blotting using respective antisera. For N-terminal sequencing, the digested products were transferred to a nylon membrane, stained with amido black, and modified with phenyl isothiocyanate (PITC). Subsequently, the derivatized terminal amino acid was removed by acid cleavage in a form of phenylthiohydantoin (PTH) derivative, thus leaving a new α-amino group on the next amino acid available for the next round of PITC reaction. The protein sequence was thereby analyzed through a series of reactions with PITC and the cleavage of one PTH at a time.
Triton X-114 phase partitioning assay, far-Western blotting, and Western blotting.
The spirochetes were resuspended in phosphate-buffered saline (PBS; pH 7.4), sonicated, and extracted with 2% Triton X-114. The aqueous and detergent phases were separated by centrifugation, washed with ice-cold acetone, and centrifuged at 13,000 × g for 15 min at 4°C. The protein pellets were resuspended in PBS and subjected to SDS-PAGE and immunoblot analysis as previously (49, 50). Far-Western blotting and Western blotting analyses were performed as detailed previously (14). For detection of the C terminus by far-Western blotting, the nitrocellulose membrane was incubated with recombinant BB0323-N and probed with antiserum against the N terminus of BB0323. The densities of protein bands were measured by ImageJ and normalized by FlaB levels in each sample.
Circular dichroism.
The proteins were dialyzed into 10 mM KPO4 buffer at pH 7.6. Concentrations were determined by A280 and extinction coefficients of 65,320 M−1cm−1 for BB0323 and 27,390 M−1cm−1 for BB0323NL-N and BB0323-N proteins. Circular-dichroism (CD) spectra were collected using a Chirascan CD spectrometer (Applied Photophysics LTD, Surrey, United Kingdom) using a 0.1-mm demountable quartz cell over a wavelength range of 188 to 280 nm. All CD data were collected at 25°C using 1-nm steps and 3-s averaging per data point. Repeated scans resulted in very similar data. Data were converted to mean residue molar ellipticity (degrees per square centimeter per decimole) after baseline subtraction of the buffer. Secondary structure deconvolution of the CD data was estimated using the CDNN deconvolution program (CDNN courtesy of Gerald Bohn, 1997, at the Institut für Biotechnologie, Martin-Luther Universität Halle-Wittenberg, through distribution with Applied Photophysics software).
Biolayer interferometry (BLI).
Anti-penta-HIS (HIS1K) biosensors (ForteBio) were hydrated in 200 μL of kinetics buffer (KB) and proteins were diluted in KB to final concentrations of 5.5 nM to 0.7 μM and analyzed as follows: baseline (KB), 60 s; load (BB0323), 300 s; association (BbHtrA), 120 s; and dissociation (KB), 120 s. The results were analyzed with a global fit and 2:1 heterogeneous ligand-binding model according to the manufacturer’s software platform. We calculated the full R2 coefficient of determination for the goodness of the curve fit for the 2:1 model; values were 0.9975 for BB0323, 0.9987 for BB0323NL-N, and 0.9988 for BB0323-N.
Microscale thermophoresis.
Microscale thermophoresis (MST) analysis was performed with a Monolith NT instrument according to the manufacturer’s instructions (NanoTemper Technologies), using dilutions of BB0323 proteins and NanoTemper Red-Tris-nitrilotriacetic acid (NTA) dye in PBS-T buffer (PBS [pH 7.4] with 0.05% Tween 20). A 12-point 1:1 serially diluted mixture of 20 μM GST-BB0238 and 10 nM labeled His-BB0323 protein was incubated and analyzed at 40% MST power and 10% light emitting diode (LED) power for 10 s. Data from 3 biological replicates were analyzed using MO.Affinity analysis software and evaluated in a KD fit model that describes a molecular interaction with 1:1 stoichiometry, according to the law of mass action.
Labeling of peptidoglycan in B. burgdorferi.
B. burgdorferi peptidoglycan labeling with 3-[[(7-hydroxy-2-oxo-2H-1-benzopyran-3-yl)carbonyl]amino]-d-alanine hydrocholoride (HADA) was performed as detailed previously (35). Briefly, the wild or mutant spirochetes were grown at 37°C in BSK medium until mid-log stage and incubated with 0.2 mM HADA resuspended in 1% dimethyl sulfoxide (DMSO). The Borrelia cells treated with only DMSO were used as a control. The samples were spotted on 1% (wt/vol) agarose/PBS pads and imaged using an Observer Z1 motorized epifluorescence microscope (Zeiss Inc.) at room temperature.
Genetic manipulation of B. burgdorferi.
To generate borrelial mutants with amino acid replacements as in BB0323NL, a DNA insert representing NL-to-AA replacement in BB0323 was inserted into the genome of the bb0323 deletion mutant B. burgdorferi, as detailed earlier (14). The plasmid pXLF14301-bb0323com (14, 15) was used as parental plasmid. The primers 5′-CAGTCACTTAGCCGCTTTAAATACTAATAAAGACACTTATC-3′ and 5′-TTTCTTTCTTTAATGAATGCTCTAC-3′ were used to generate point mutations using the Q5 site-directed mutagenesis kit (NEB) according to the manufacturer’s instructions. The construct was sequenced to confirm the mutation and electroporated into the bb0323 mutant. Although two transformant clones showed aberrant processing of BB0323 (Fig. S1), the first clone, designated Bbbb0323NL, with similar endogenous plasmid profiles, protein expression, and growth analysis, was selected for further studies, as detailed earlier (14).
Infection studies using mice and ticks.
To assess spirochete infectivity, C3H/HeN mice were injected intradermally with Borrelia (105 cells/mouse; 3 mice/group). Fourteen days after infection, serum was collected to assess the antibody responses against B. burgdorferi. Samples of the skin, heart, and bladder were collected to quantify Borrelia levels using qPCR analysis of cDNA copies of the flaB gene and normalizing against murine β-actin, as described previously (14, 15).
To study pathogen transmission from ticks, naive nymphs were microinjected with spirochetes, and after overnight incubation, they were allowed to parasitize mice (5 ticks/mouse; 3 mice/group). Spirochete burdens in engorged ticks were analyzed using qPCR analysis of copies of the flaB gene and normalizing against tick β-actin, as described previously (14, 15). Twelve days following tick feeding, murine tissues (skin, joints, heart, and bladder) were tested for spirochete burden via qPCR, as described previously (14, 15).
Statistical analysis.
All numerical results are expressed as means ± standard errors of the means (SEM). The significance of differing mean values between groups was evaluated using the Student t test or analysis of variance (ANOVA) using GraphPad Prism software, version 4.0 and version 9.0 (GraphPad software, CA).
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
ACKNOWLEDGMENTS
We are thankful to Kathryn Nassar for assistance with the preparation of the manuscript. We are thankful to Philip Stewart and Patricia Rosa for providing an antibody against BB0323 C terminus.
This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (award numbers R01AI080615, R01AI116620, and P01AI138949 to U.P.) and the Intramural Research Program of the National Center for Advancing Translational Sciences, NIH (1ZIATR000355-04).
We declare that there are no potential conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Utpal Pal, Email: upal@umd.edu.
Guy H. Palmer, Washington State University
REFERENCES
- 1.Mead PS. 2015. Epidemiology of Lyme disease. Infect Dis Clin North Am 29:187–210. 10.1016/j.idc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J Clin Invest 113:1093–1101. 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadelman RB, Wormser GP. 1998. Lyme borreliosis. Lancet 352:557–565. 10.1016/S0140-6736(98)01146-5. [DOI] [PubMed] [Google Scholar]
- 5.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 6.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 7.Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, Huang WM, Vujadinovic M, Aron JK, Vargas LC, Freeman S, Radune D, Weidman JF, Dimitrov GI, Khouri HM, Sosa JE, Halpin RA, Dunn JJ, Fraser CM. 2012. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One 7:e33280. 10.1371/journal.pone.0033280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer R, Caimano MJ, Luthra A, Axline D, Jr, Corona A, Iacobas DA, Radolf JD, Schwartz I. 2015. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol 95:509–538. 10.1111/mmi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revel AT, Talaat AM, Norgard MV. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA 99:1562–1567. 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marconi RT, Earnhart CG. 2010. Lyme disease vaccines, p 467–486. In Samuels DS, Radolf JD (ed), Borrelia, molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 11.Gomes-Solecki M, Arnaboldi PM, Backenson PB, Benach JL, Cooper CL, Dattwyler RJ, Diuk-Wasser M, Fikrig E, Hovius JW, Laegreid W, Lundberg U, Marconi RT, Marques AR, Molloy P, Narasimhan S, Pal U, Pedra JHF, Plotkin S, Rock DL, Rosa P, Telford SR, Tsao J, Yang XF, Schutzer SE. 2020. Protective immunity and new vaccines for Lyme disease. Clin Infect Dis 70:1768–1773. 10.1093/cid/ciz872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart PE, Hoff J, Fischer E, Krum JG, Rosa PA. 2004. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl Environ Microbiol 70:5973–5979. 10.1128/AEM.70.10.5973-5979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kariu T, Sharma K, Singh P, Smith AA, Backstedt B, Buyuktanir O, Pal U. 2015. BB0323 and novel virulence determinant BB0238: Borrelia burgdorferi proteins that interact with and stabilize each other and are critical for infectivity. J Infect Dis 211:462–471. 10.1093/infdis/jiu460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kariu T, Yang X, Marks CB, Zhang X, Pal U. 2013. Proteolysis of BB0323 results in two polypeptides that impact physiologic and infectious phenotypes in Borrelia burgdorferi. Mol Microbiol 88:510–522. 10.1111/mmi.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Yang X, Kumar M, Pal U. 2009. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis 200:1318–1330. 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 17.Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, Fujimoto Y, Hounslow AM, Lacroix-Desmazes S, Fukase K, Foster SJ, Williamson MP. 2014. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun 5:4269. 10.1038/ncomms5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 180:4872–4878. 10.1128/JB.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llamas MA, Ramos JL, Rodriguez-Herva JJ. 2000. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol 182:4764–4772. 10.1128/JB.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. 2010. The Caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol 192:4847–4858. 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brangulis K, Akopjana I, Kazaks A, Tars K. 2019. Crystal structure of the N-terminal domain of the major virulence factor BB0323 from the Lyme disease agent Borrelia burgdorferi. Acta Crystallogr D Struct Biol 75:825–830. 10.1107/S2059798319010751. [DOI] [PubMed] [Google Scholar]
- 22.Brangulis K, Akopjana I, Petrovskis I, Kazaks A, Jekabsons A, Jaudzems K, Viksna A, Bertins M, Tars K. 2020. Structural analysis of Borrelia burgdorferi periplasmic lipoprotein BB0365 involved in Lyme disease infection. FEBS Lett 594:317–326. 10.1002/1873-3468.13594. [DOI] [PubMed] [Google Scholar]
- 23.Brangulis K, Akopjana I, Petrovskis I, Kazaks A, Tars K. 2019. Crystal structure of Borrelia burgdorferi outer surface protein BBA69 in comparison to the paralogous protein CspA. Ticks Tick Borne Dis 10:1135–1141. 10.1016/j.ttbdis.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Brangulis K, Akopjana I, Petrovskis I, Kazaks A, Zelencova D, Jekabsons A, Jaudzems K, Tars K. 2020. BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione. Biochim Biophys Acta Gen Subj 1864:129499. 10.1016/j.bbagen.2019.129499. [DOI] [PubMed] [Google Scholar]
- 25.Liem RK. 2016. Cytoskeletal integrators: the spectrin superfamily. Cold Spring Harb Perspect Biol 8:a018259. 10.1101/cshperspect.a018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. 2004. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol 52:801–814. 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakur M, Sharma K, Chao K, Smith AA, Herzberg O, Pal U. 2017. A protein-protein interaction dictates borrelial infectivity. Sci Rep 7:2932. 10.1038/s41598-017-03279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye M, Sharma K, Thakur M, Smith AA, Buyuktanir O, Xiang X, Yang X, Promnares K, Lou Y, Yang XF, Pal U. 2016. HtrA, a temperature- and stationary phase-activated protease involved in maturation of a key microbial virulence determinant, facilitates Borrelia burgdorferi infection in mammalian hosts. Infect Immun 84:2372–2381. 10.1128/IAI.00360-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coleman JL, Toledo A, Benach JL. 2016. Borrelia burgdorferi HtrA: evidence for twofold proteolysis of outer membrane protein p66. Mol Microbiol 99:135–150. 10.1111/mmi.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coleman JL, Toledo A, Benach JL. 2018. HtrA of Borrelia burgdorferi leads to decreased swarm motility and decreased production of pyruvate. mBio 9:e01136-18. 10.1128/mBio.01136-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman JL, Crowley JT, Toledo AM, Benach JL. 2013. The HtrA protease of Borrelia burgdorferi degrades outer membrane protein BmpD and chemotaxis phosphatase CheX. Mol Microbiol 88:619–633. 10.1111/mmi.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, Qin Z, Chang Y, Liu J, Malkowski MG, Shipa S, Li L, Qiu W, Zhang JR, Li C. 2019. Analysis of a flagellar filament cap mutant reveals that HtrA serine protease degrades unfolded flagellin protein in the periplasm of Borrelia burgdorferi. Mol Microbiol 111:1652–1670. 10.1111/mmi.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell TM, Delorey MJ, Johnson BJ. 2013. Borrelia burgdorferi BbHtrA degrades host ECM proteins and stimulates release of inflammatory cytokines in vitro. Mol Microbiol 90:241–251. 10.1111/mmi.12377. [DOI] [PubMed] [Google Scholar]
- 34.Groshong AM, Fortune DE, Moore BP, Spencer HJ, Skinner RA, Bellamy WT, Blevins JS. 2014. BB0238, a presumed tetratricopeptide repeat-containing protein, is required during Borrelia burgdorferi mammalian infection. Infect Immun 82:4292–4306. 10.1128/IAI.01977-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jutras BL, Scott M, Parry B, Biboy J, Gray J, Vollmer W, Jacobs-Wagner C. 2016. Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc Natl Acad Sci USA 113:9162–9170. 10.1073/pnas.1610805113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostberg Y, Carroll JA, Pinne M, Krum JG, Rosa P, Bergstrom S. 2004. Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J Bacteriol 186:2074–2084. 10.1128/JB.186.7.2074-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 12:152–162. 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- 38.Clausen T, Southan C, Ehrmann M. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10:443–455. 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 39.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455–459. 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 40.Nicolas A, Delalande O, Hubert JF, Le Rumeur E. 2014. The spectrin family of proteins: a unique coiled-coil fold for various molecular surface properties. J Struct Biol 186:392–401. 10.1016/j.jsb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Levin PA, Kurtser IG, Grossman AD. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA 96:9642–9647. 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele VR, Bottomley AL, Garcia-Lara J, Kasturiarachchi J, Foster SJ. 2011. Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Mol Microbiol 80:542–555. 10.1111/j.1365-2958.2011.07591.x. [DOI] [PubMed] [Google Scholar]
- 43.Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ, Rowe N. 2009. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol 191:600–607. 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA 97:10899–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudryashev M, Cyrklaff M, Baumeister W, Simon MM, Wallich R, Frischknecht F. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol Microbiol 71:1415–1434. 10.1111/j.1365-2958.2009.06613.x. [DOI] [PubMed] [Google Scholar]
- 46.Pennetzdorfer N, Lembke M, Pressler K, Matson JS, Reidl J, Schild S. 2019. Regulated proteolysis in Vibrio cholerae allowing rapid adaptation to stress conditions. Front Cell Infect Microbiol 9:214. 10.3389/fcimb.2019.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu TY, Elliott SD. 1965. Streptococcal proteinase: the zymogen to enzyme transformation. J Biol Chem 240:1138–1142. 10.1016/S0021-9258(18)97551-4. [DOI] [PubMed] [Google Scholar]
- 48.Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JC. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun 68:4245–4254. 10.1128/IAI.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Lenhart TR, Kariu T, Anguita J, Akins DR, Pal U. 2010. Characterization of unique regions of Borrelia burgdorferi surface-located membrane protein 1. Infect Immun 78:4477–4487. 10.1128/IAI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang X, Lin YP, Heselpoth RD, Buyuktanir O, Qin J, Kung F, Nelson DC, Leong JM, Pal U. 2016. Middle region of the Borrelia burgdorferi surface-located protein 1 (Lmp1) interacts with host chondroitin-6-sulfate and independently facilitates infection. Cell Microbiol 18:97–110. 10.1111/cmi.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1-S2 and Tables S1-S2. Download iai.00059-22-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.