Summary:
Unlimited generation of chimeric antigen receptor (CAR) T cells from human induced pluripotent stem cells (iPSCs) is an attractive approach for ‘off-the-shelf’ CAR T cell immunotherapy. Approaches to efficiently differentiate iPSCs into canonical αβ T cell lineages, while maintaining CAR expression and functionality, however, have been challenging. We report that iPSCs reprogrammed from CD62L+ naïve and memory T cells followed by CD19-CAR engineering and 3D-organoid system differentiation confers products with conventional CD8αβ-positive CAR T cell characteristics. Expanded iPSC CD19-CAR T cells showed comparable antigen-specific activation, degranulation, cytotoxicity and cytokine secretion compared to conventional CD19-CAR T cells, and maintained homogenous expression of the TCR derived from the initial clone. iPSC CD19-CAR T cells also mediated potent antitumor activity in vivo, prolonging survival of mice with CD19+ human tumor xenografts. Our study establishes feasible methodologies to generate highly functional CAR T cells from iPSCs to support the development of ‘off-the-shelf’ manufacturing strategies.
eTOC blurb:
Wang Z. et al. establishes feasible methodologies to generate highly functional CAR T cells from T cell derived iPSCs using 3D-organoid culture thereby supporting the development of ‘off-the-shelf’ manufacturing strategies.
Graphical Abstract

Introduction
Chimeric Antigen Receptor (CAR) T cell immunotherapy is a revolutionary cancer treatment, which has achieved great success in treating hematopoietic malignancies and shown potential promise against solid tumors (Brown et al., 2016; June et al., 2018; Ruella et al., 2018). The current autologous strategy of CAR T production requires individualized blood apheresis and manufacture (Levine et al., 2017), which poses major hurdles for its broad application, such as high cost, inefficient and unsecured production, limited dose and intrinsic variation of T cell fitness(Kohl et al., 2018; Lin et al., 2019; Thommen and Schumacher, 2018). It has been proposed that these hurdles might be overcome by the generation of ‘off-the-shelf’ CAR T cell products from an allogenic healthy donor or from induced pluripotent stem cells (iPSCs) (Themeli et al., 2015; Yang et al., 2015).
Primary human T cells, whether autologous or allogenic, have limited ex vivo expansion potential and gradually become exhausted following cell culture (Akbar and Henson, 2011; Ghassemi et al., 2018; Long et al., 2015). In contrast, iPSCs can proliferate almost infinitely, while maintaining their pluripotency and lineage differentiation potential (Inoue et al., 2014; Murry and Keller, 2008), making them an attractive cell population from which to generate a limitless supply of CAR T cells. Further genetic editing of iPSCs has also proved to be a viable strategy to broaden their immune compatibility, thus further supporting their potential for creating an ‘off-the-shelf’ cell product (Deuse et al., 2019; Poirot et al., 2015). However, studies have also revealed the complexity of in vitro T cell development from stem cells (Kennedy et al., 2012; Minagawa et al., 2018; Montel-Hagen et al., 2019a; Nishimura et al., 2013; Timmermans et al., 2009; Vizcardo et al., 2018) and the disturbance of T cell differentiation pathways by CAR expression (Themeli et al., 2013). The reported studies establishing proof-of-concept for iPSC-differentiation to generate CAR T cells have so far demonstrated an innate γδ T cell-like profile and skewed CAR T cell functionality (Themeli et al., 2013). More recently, a 3D-organoid culture system was developed to successfully generate functional mature T cells from hematopoietic stem cells and embryonic stem cells (ESC) (Montel-Hagen et al., 2019a), however successful generation of CAR T cells by this method has yet to be reported. Interestingly, the finding that such ESC-derived T cells display unpredictable TCR repertoires, while T cells derived from either TCR-transduced ESC or T-cell derived iPSC (TiPSC) preserve the predicted TCR (Minagawa et al., 2018; Montel-Hagen et al., 2019a), suggests that the pre-existing TCR has a directional effect on T cell differentiation. Thus, we hypothesized that the use of both TiPSC and a 3D-culture system to generate CAR T cells would facilitate the production of T cells with more conventional phenotypes and improved functionality.
Results:
Generation of iPSC-derived CAR T cells with a conventional T cell phenotype
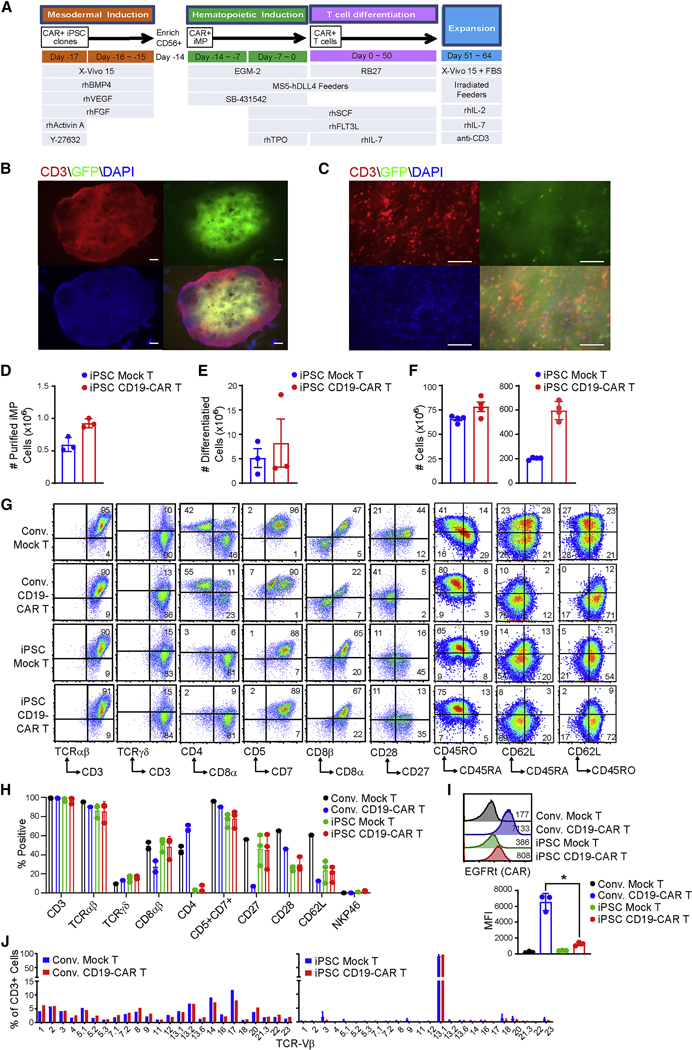
In the current study, we utilized iPSC clones derived from primary CD62L+ naïve and memory T cells (Tn/mem), a T cell population that has been proposed to have superior persistence and improve clinical outcomes in CAR T cell therapy (McLellan and Ali Hosseini Rad, 2019; Morgan and Schambach, 2018; Popplewell et al., 2018; Samer K. Khaled, 2018; Zah et al., 2020). The Tn/mem cells that had been enriched from the peripheral blood of healthy human donors were transduced and reprogrammed by episomal plasmids encoding KLF4, SoX2, OCT-4, C-MYC and LIN28, along with P53 shRNA (Okita et al., 2013), and multiple integration-free iPSC clones were screened and characterized (Figure S1). iPSC clone pluripotency was confirmed by alkaline phosphatase staining and examination of stem cell markers SSEA3, SSEA4, TRA1–60, TRA1–81 and CD30 (Figure S1A, B), with EBNA PCR demonstrating that the iPSC clones were integration free (Figure S1C). Qualified clones were transduced with clinical grade lentivirus encoding a CD19-targeting CAR (NCT02146924) (Popplewell et al., 2018; Samer K. Khaled, 2018) and CAR+ cells were single cell sorted by flow cytometry, colonized, expanded and banked. Both mock-transduced and CAR-expressing clones maintained stem cell markers expression (Figure S1D). The parental iPSC and CD19-CAR+ iPSC clones were further tested by teratoma formation assay to confirm their pluripotency potential to generate ectoderm, endoderm and mesoderm germ layers (Figure S1E). To direct the differentiation of CD19CAR expressing iPSC into CD19-CAR expressing T cells, we adapted the modified embryonic and induced pluripotent stem cells based Artificial Thymic Organoid (PSC-ATO) system of Montel-Hagen et al (Montel-Hagen et al., 2019a). First, the CD19-CAR+ iPSCs were cultured in feeder-free conditions for the first three days to induce mesodermal differentiation (Figure 1A). CD56+CD326- iPSC mesodermal progenitor cells (iMP) were then enriched by magnetic selection of CD56+ cells, and went through iPSC mesodermal organoid culture (iMO) with MS5-hDLL4 feeder cells to differentiate into hematopoietic progenitors (14 days), followed by T cell commitment and differentiation (additional 5–7 weeks) (Figure 1A). In situ staining and imaging of mature organoid cultures demonstrated a heterogeneous tissue-like architecture with CD3+ T cells and GFP+ MS5-DLL4 feeder cells (Figures 1B, 1C, and S1F). The cell yield from iPSC to iMP, and from iMP to T cells was comparable between CD19-CAR+ and mocktransduced groups (Figure 1D, 1E), and PSC-ATO differentiated iPSC T cells, both mock-transduced or CD19-CAR+, can be efficiently expanded to clinically relevant numbers using a modified rapid expansion method (REM) (Wang et al., 2011b). Approximately 75-fold expansion was achieved in 2-weeks during REM expansion and 600 million CD19-CAR T cells were generated from 1 million CAR+ iPSC cells (Figure 1F). The resulting expanded iPSC CD19-CAR T cells were then harvested for phenotypic characterization and expansion. The PSC-ATO-differentiated and expanded iPSC CD19-CAR T cells demonstrated a CD3/CD5/CD7/TCRαβ/CD8αβ-positive, NKG2A/NKP46/CD16/CD19-negative phenotype. As a benchmark for conventional CAR T cell phenotype and function we utilized PBMC-derived CD19-CAR T cells generated from the same donor using standard and clinically relevant CD3/CD28 bead stimulation and IL2/IL15 expansion procedures (Brentjens et al., 2011; Wang et al., 2016). We demonstrate that iPSC CAR T cells are phenotypically similar to the CD8+ subpopulation of conventional CD19-CAR T cells (Figures 1G, 1H, S1G, S1H and S1I). Furthermore, similar to conventional CD19-CAR T cells, the iPSC CD19-CAR T cells are composed of populations in different stages of differentiation, including stem-cell-like T cells and memory T cells based on CD62L, CD45RA and CD45RO profiles (Figure 1G, 1H). They also express similar levels of FasL, but higher levels of CD56, NKG2D and NKP44 compared to conventional CD19-CAR T cells (Figure S1H, S1I). Similar phenotypes were also seen when we generated iPSC CAR T cells using the same methods but from different colonized iPSC lines carrying other CAR constructs - specifically a 41BB-containing CD19-targeting CAR (CD19(BBz)-CAR), or another clinical grade lentivirus encoding a Chlorotoxin-targeting CAR (CLTX-CAR; NCT04214392)(Wang et al., 2020) (Figure S1K, S1L). Interestingly, iPSC CD19-CAR T cells appear to express much less CAR/transgene than conventional CD19-CAR T cells (Figure 1I, S1M). TCR repertoire analysis by flow cytometry (Figures 1J, S2A, S2B and S2D) and PCR of gDNA (Figures S2C, S2E) demonstrate that the iPSC mock-transduced and CD19-CAR+ T cells preserve their clonal TCR, while the conventional T cells, as well as cell products generated from cord blood-, fibroblast, or effector T cell-derived iPSC, are highly polyclonal.
Figure 1. Generation of iPSC-derived CD19-CAR T cells.
(A) Schematic of events (top), cell type (middle) and media conditions (bottom) during PSC-ATO culture. Reference online STAR Methods. (B, C), Seven-week organoid cultures of iPSC CD19-CAR T cells with GFP+ DLL4+ MS5 feeder cells were fixed by 2% paraformaldehyde and stained with CD3 (red) and DAPI (blue) in situ. Composites of individual images have been stitched together. White bars indicate scales of 500 μm (B) and 100 μm (C). (D) Number of differentiated mesodermal progenitor cells (iMP) derived from 1 million mock-transduced or CD19-CAR expressing Tn/mem-iPSC. Data of three separate experiments is depicted, with mean ± S.E.M as bars. (E) Number of differentiated T cells derived from 1 million mock-transduced or CD19-CAR expressing iMP. Data of three separate experiments is depicted, with mean ± S.E.M as bars. (F) Yield of iPSC-derived Mock T and CD19-CAR T cells. T cells were expanded as indicated in (A). Numbers of cells expanded from 1 × 106 differentiated T cells (Left) or from 1 × 106 iPSC cells (Right) are depicted using data of three separate experiments, with mean ± S.E.M as bars. (G) Representative flow cytometric analysis of the indicated markers on conventional (Conv.) vs. iPSC-derived mock-transduced (Mock) and CD19-CAR expressing T cells. Percentages of cells expressing each marker are indicated in the relevant quadrants, which were drawn based on isotype control staining. (H) Percentages of cells staining with the indicated markers in three separate experiments, with mean ± S.D. as bars. (I) Comparison of transgene expression levels on conventional (Conv.) vs. iPSC-derived Mock T and CD19-CAR T cells. Top, representative histograms of EGFRt staining as a marker for CAR expression, with mean fluorescence intensity (MFI) indicated. Bottom, transgene MFI data of three separate experiments is depicted, with mean ± S.D. as bars. *, P = 0.0011 using Student’s t-test. (J) TCR Vx repertoire of conventional vs. iPSC-derived Mock T and CD19-CAR T cells.
Transcriptional profile of iPSC-derived CAR T cells
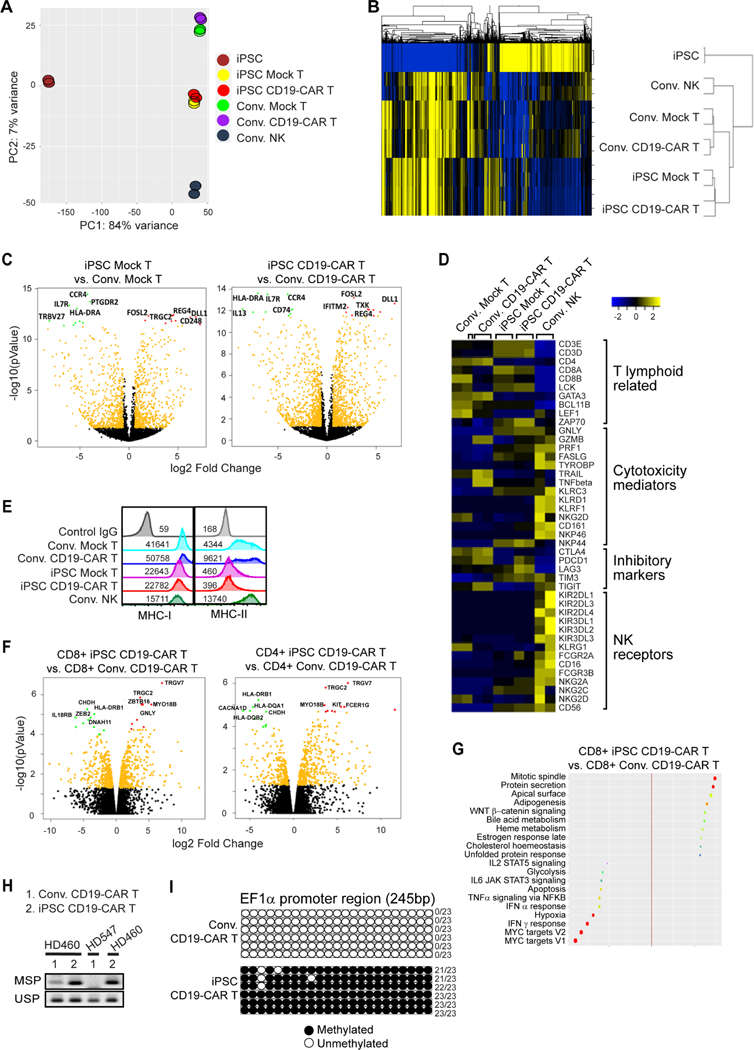
To explore differences between iPSC CD19-CAR T cells and conventional, PBMC-derived CD19-CAR T cells from the same donor, we did bulk RNA deep sequencing analysis (Supplemental Excel Table 1). NK cells from the same donor were also used for comparison. Principle components analysis (PCA) showed that iPSC Mock T and iPSC CD19-CAR T cells displayed similar transcription profiles as conventional mock-transduced T cells, CD19-CAR T cells or NK cells derived from the same donor (PC score ~7% variance), but were dramatically distinguished from iPSCs (PC score ~84% variance) (Figure 2A). Hierarchical clustering of global transcriptional profiles showed that iPSC-derived T cells were more similar to conventionally derived T cells than to NK cells (Figure 2B). Looking at the most significantly differentiated genes, it was observed that the iPSC CD19-CAR T cells expressed lower levels of IL-13, HLA-DR, IL7R, CCR4, and CD74, but higher levels of DLL1, FOSL2, TXK, REG4, and IFITM2 compared to the conventional CD19-CAR T cells (Figures 2C and S2F). Evaluation of selected functional related gene sets revealed that iPSC CD19-CAR T cells expressed higher levels of T lymphocyte genes CD3E, CD3D, CD8, LCK and ZAP70, and lower levels of CD4, GATA3, BCL11B and LEF1 genes as compared to conventional CD19-CAR T cells (Figure 2D). For cytotoxic mediator genes, iPSC CD19-CAR T cells express more GNLY and PRF1, but less GZMB compared to conventional CD19-CAR T cells. For T cell inhibitory genes, iPSC CD19-CAR T express less CTLA4, PD1, and TIGIT, but more LAG3 and TIM3 (Figure 2D). iPSC CD19-CAR T cells do not express NK cell signature genes, which is similar to conventional CD19-CAR T cells (Figure 2D). iPSC-derived T cells also demonstrated lower levels of MHC genes than conventional T cells and did not show biased gene signature towards exhaustion phenotype (Figure S3A). Further gene set enrichment analysis showed downregulated MYC target gene, IFNγ response signatures in iPSC CD19-CAR T cells versus conventional CD19-CAR T cells (Figure S3B), which may relate to the microenvironment in 3D organoid culture and indicates unique metabolic signatures representing lower activation status at steady state as compared to conventional CAR T cells (Palazon et al., 2017; Pavlacky and Polak, 2020; Wang et al., 2011a). Interestingly, iPSC CD19-CAR T cells display decreased MHC-II and MHC-I expression levels by flow cytometry (Figure 2E), which is consistent to RNA seq data (Figure S3A).
Figure 2. Gene and signaling signature of iPSC CD19-CAR T cells.
(A) Principle components analysis (PCA) and (B) hierarchical clustering of global transcriptional profiles of two samples of iPSC, conventional (Conv.) mock-transduced (Mock) or CD19-CAR T cells, iPSC-derived Mock T or CD19-CAR T cells, or conventional PBMC-derived NK cells. (C) Vocano plots of iPSC Mock T vs. Conv. Mock T cells (left), or of iPSC CD19-CAR T vs. Conv. CD19-CAR T cells (right). Top five upregulated genes in conventional cells are highlighted with green dots, while those in iPSC-derived cells are highlighted by red dots. (D) Heat map of z score value of T lymphoid related genes, cytotoxicity mediators, inhibitory markers and NK receptor genes. (E) HLA ABC (MHC-I) and HLA-DR,DP,DQ (MHC-II) expression levels on the surface of Conv. Mock T, Conv.CD19-CAR T, iPSC Mock T and iPSC CD19-CAR T cells, with mean fluorescence intensity (MFI) indicated. (F) Vocano plots of CD8+ iPSC CD19-CAR T vs. CD8+ Conv. CD19-CAR T cells (left), or of CD4+ iPSC CD19-CAR T vs. CD4+ Conv. CD19-CAR T cells (right). Top five upregulated genes in conventional cells are highlighted with green dots, while those in iPSCderived cells are highlighted by red dots. (G) Bubble plot showing top up- or down- regulated signaling pathway derived from GSEA comparison of CD8+ iPSC CD19-CAR T vs. CD8+ Conv. CD19-CAR T cells. (H) Bisulfite converted genomic DNA was used as a template for PCR analysis using methylationspecific primers (MSP) and unmethylation-specific primers (USP) within the EF1α promoter. (I) EF1α promoter methylation determination by bisulfite sequencing. Region 114–360bp of EF1α promoter was PCR amplified from bisulfite converted genomic DNA, sub-cloned, and 6 clones for each group were sequenced. Number of methylated CG sites for each clone, out of the 23 CG sites in this 245bp region, are indicated at the right of each row.
Since the percentages of CD8αβT cells were different between iPSC and conventional CAR T cells, we also compared their gene expression phenotypes following CD8+ sorting (Figures S3C–F, 2F, 2G). The differences in phenotypes observed for the bulk comparisons were confirmed when comparing sorted CD8+ iPSC derived to sorted CD8+ conventional CAR T cells, including downregulated MHCI/II, decreased INFγ response/MYC target gene pathways, and upregulated GNLY expression in iPSC derived CD8+CAR+CD19-CAR T cells.
As shown by flow cytometry that the iPSC CD19-CAR T cells expressed much lower levels of CAR transgene than conventional CAR T cells (Figure 1I, S1M). However, the CAR expression levels in CAR transduced, colonized iPSCs was quite high and clearly distinguishable from mock-transduced iPSCs (Figure S1D), indicating that subsequent CAR downregulation might be mediated by transcriptional or translational regulation during cell differentiation. In our study, as in many lentivirus-based CAR T platforms (Porter et al., 2011; Programs, 2019), CAR transgene expression was driven by the EF1α promoter which contains many CpG islands (Figure S3G). Thus, we hypothesized that the CpG enriched EF1α promoter might be methylated during the cell differentiation of CD19-CAR expressing iPSC into CD19-CAR expressing T cells, and lead to transcriptional downregulation of the CAR. Examination of the methylation status by bisulfite specific PCR using bisulfite converted genomic DNA as template, showed that the EF1α promoter methylation status was significantly enhanced in iPSC CD19-CAR T cells when compared to conventional CD19-CAR T cells derived from the same donor (Figure 2H). This hyper-methylation was confirmed upon further bisulfite sequencing analysis of a 245 bp region of the EF1α promoter containing 23 sites of CpG (Figure 2I). These data suggest that EF1α promoter hypermethylation occurs during T cell differentiation from iPSC, resulting in the downregulated CAR expression in iPSC CD19-CAR T cells.
Taken together, these studies establish that iPSC CAR T cells have an RNA expression signature that is overall similar to conventional CAR T cells, albeit with a relatively lower activation status at steady state, which is associated with lower CAR expression levels due to transgene promoter hyper-methylation during differentiation.
Functional analysis of iPSC-derived CAR T cells
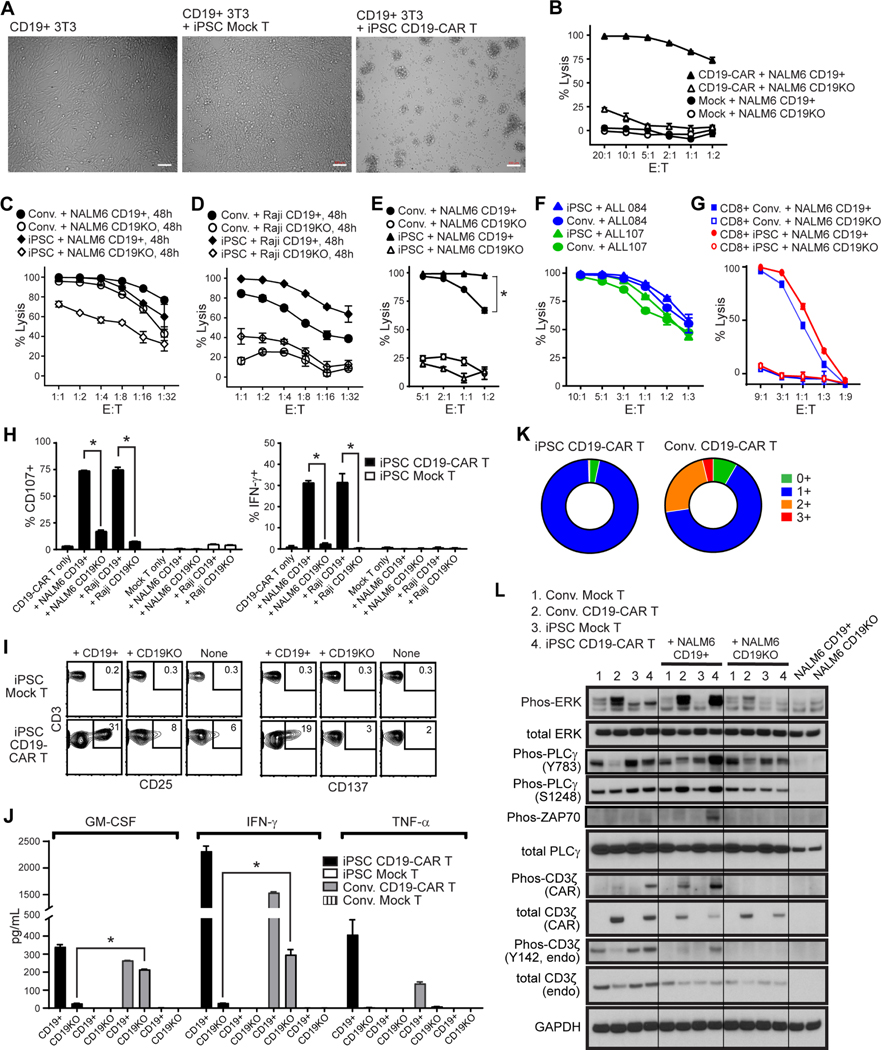
We next evaluated the effector function of REM expanded iPSC CD19-CAR T cells to lyse CD19 expressing targets in vitro. iPSC CAR T cells mediated potent CAR-directed cytolytic activity against CD19+ 3T3 cells (Figure 3A), NALM6 cells (Figure 3B, 3C), and Raji cells (Figure 3D), but not their CD19-negative counterparts (Figure S3H). We use PBMC derived conventional CD19-CAR T cells as comparison control, which was produced by clinically relevant procedure and did not go through REM expansion. Importantly, the killing activity of iPSC CD19-CAR T cells was comparable to conventional PBMC-derived CD19-CAR T cells from the same donor (Figure 3E–G, S3I). The innate-like natural killing activity of the CD8+ iPSC CD19-CAR T cells and conventional CD19-CAR T cells against K562 cells was negligible, and significantly lower than that of NK cells (Figure S3J). Upon CD19+ tumor cell stimulation, iPSC CD19-CAR T cells also demonstrated potent degranulation, expression of intracellular IFNγ, surface expression of activation markers CD25 and CD137/4–1BB, and Th1 cytokine release in an antigen-dependent manner (Figures 3H–J). Interestingly, without antigenic stimulation, the levels of GMCSF and IFN- are much lower in the supernatant from iPSC CD19-CAR T cells than that of conventional CD19-CAR T cells (Figure 3J), which is consistent with lower basal ERK protein phosphorylation level (Figure 3L) and suggests lower levels of CAR tonic signaling. Furthermore, upon serial challenge with CD19+ tumor cells, iPSC CD19-CAR T cells displayed decreased expression of PD-1, TIM-3 and LAG3 as compared to conventional CD19-CAR T cells, indicating a less exhausted phenotype. (Figure 3K).
Figure 3. Functional profile of iPSC CD19-CAR T cells.
(A), Brightfield images after 4 hour co-culture of iPSC-derived mock-transduced (Mock) or CD19-CAR T cells with CD19+ 3T3 cells at an effector-to-target (E:T) ratio of 4:1. White bars indicate scale of 100 μm. (B-E), Cytotoxic activity of iPSC CD19-CAR T cells against CD19+ or CD19-negative/knockout (CD19KO) NALM6 (B, C, E), or Raji (D) target cells when co-cultured at the indicated E:T ratios for 4h (B, E) or 48h (C, D). Lytic activity was compared to that of iPSC-derived mock transduced T cells (MOCK, B) or conventional CD19-CAR T cells (Conv., C, D, E). Mean ± S.D. values of duplicate cultures are depicted. *, P < 0.001 by two way ANOVA test in (E). (F), Cytotoxic activity of iPSC-derived (iPSC) or conventional (Conv.) CD19-CAR T cells against patient derived ALL cells when co-cultured at the indicated E:T ratios for 4h. (G) Cytotoxic activity of flow cytometry sorted CD8+ iPSC-derived (CD8+ iPSC) or conventional CD8+ CD19-CAR T cells (CD8+ Conv.) against CD19+ or CD19-negative/knockout (CD19KO) NALM6. (H) Degranulation (i.e., surface CD107, left) and intracellular IFN-g levels (right) in iPSC-derived mock-transduced (Mock) or CD19-CAR T cells was measured by flow cytometry after co-culture with the indicated stimulator cells (X-axis labels) at an E:T ratio of 1:1 for 5 hours in the presence of the Golgi Stop protein transport inhibitor. *, P < 0.01 by Students t-test. (I) Flow cytometric analysis of activation markers were compared between iPSCderived Mock T and CD19-CAR T cells that were unstimulated (None), or stimulated with CD19+ or CD19-negative/knockout (CD19KO) NALM6 at an E:T ratio of 1:1 for 24 hours. Percentages of CD3+ cells expressing CD25 or CD137/4–1BB are indicated in each contour plot, with gates drawn based on isotype control staining. (J) Cytokine production by iPSC-derived or conventional (Conv.) Mock T or CD19-CAR T cells was measured by Bio-Plex analysis of supernatants harvested 24 hours after coculture with CD19+ or CD19-negative/knockout (CD19KO) NALM6 cells at an E:T ratio of 1:1. *, P < 0.001 by Student’s t-test. (K) T cell exhaustion marker profile of iPSC-derived or conventional (Conv.) CD19-CAR T cells after being re-challenged by CD19+ NALM6 cells every 2 days for a total of 3 stimulations at an E:T ratio of 1:2. Cells were stained with anti-PD-1, anti-TIM-3, anti-LAG-3 and percentages of CD3+ cells staining for no (0+), one (1+), two (2+) or all three (3+) markers were determined by flow cytometry. (L) Western Blot analysis of ERK, phosphorylated ERK, PLCγ, PLCγ phorphorylated at Y782, PLCγ phosphorylated at Ser1248, endogenous CD3ζ, phosphorylated endogenous CD3ζ, CD3ζ within the CAR, phosphorylated CD3ζ within the CAR, or GAPDH as a loading control in the indicated T cells cultured for 60 minutes alone, or with NALM6 tumors that are either CD19+ or CD19-negative (CD19KO). Tumor cells cultured alone were also examined as controls. Composites of individual images have been stitched together.
Next, we explored CAR T cell signaling upon co-culture with either parental CD19+ or CD19 KO NALM6 cells. iPSC CD19-CAR T cells demonstrated ERK1/2 Thr202/Thr204, and PLCγ Ser1248 phosphorylation in an antigen specific manner that was comparable to that of conventional CD19-CAR T cells (Figure 3L). However, the PLCγ Y783, ZAP70 and endogenous CD3ζ phosphorylation levels were higher in antigen stimulated iPSC CD19-CAR T cells than antigen stimulated conventional CD19-CAR T cells, which support the potent cytotoxicity activity. Interestingly, both the endogenous CD3ζ Y142 and CAR-associated CD3ζ phosphorylation in CD19-CAR T cells was suppressed by co-culture with CD19 negative NALM6 cells, indicating an immunosuppressive effect of cancer cells (Figure 3L). Western blot analysis also confirmed that the iPSC CD19-CAR T cells expressed much lower levels of CAR transgene than conventional CAR T cells (Figure 3L), which is consistent to flow cytometry data (Figure 1I). It may also explain their lower activation level in the absence of antigen as measured by ERK phosphorylation (Figure 3L) and cytokine secretion (Figure 3H), since lower CAR expression has been show to favor lower tonic signaling (Eyquem et al., 2017).
Overall, these studies establish that iPSC CD19-CAR T cells yield products with comparable in vitro effector activity as compared to conventional CAR T cell expanded using clinically relevant methodologies.
Anti-tumor efficacy of iPSC-derived CAR T cells
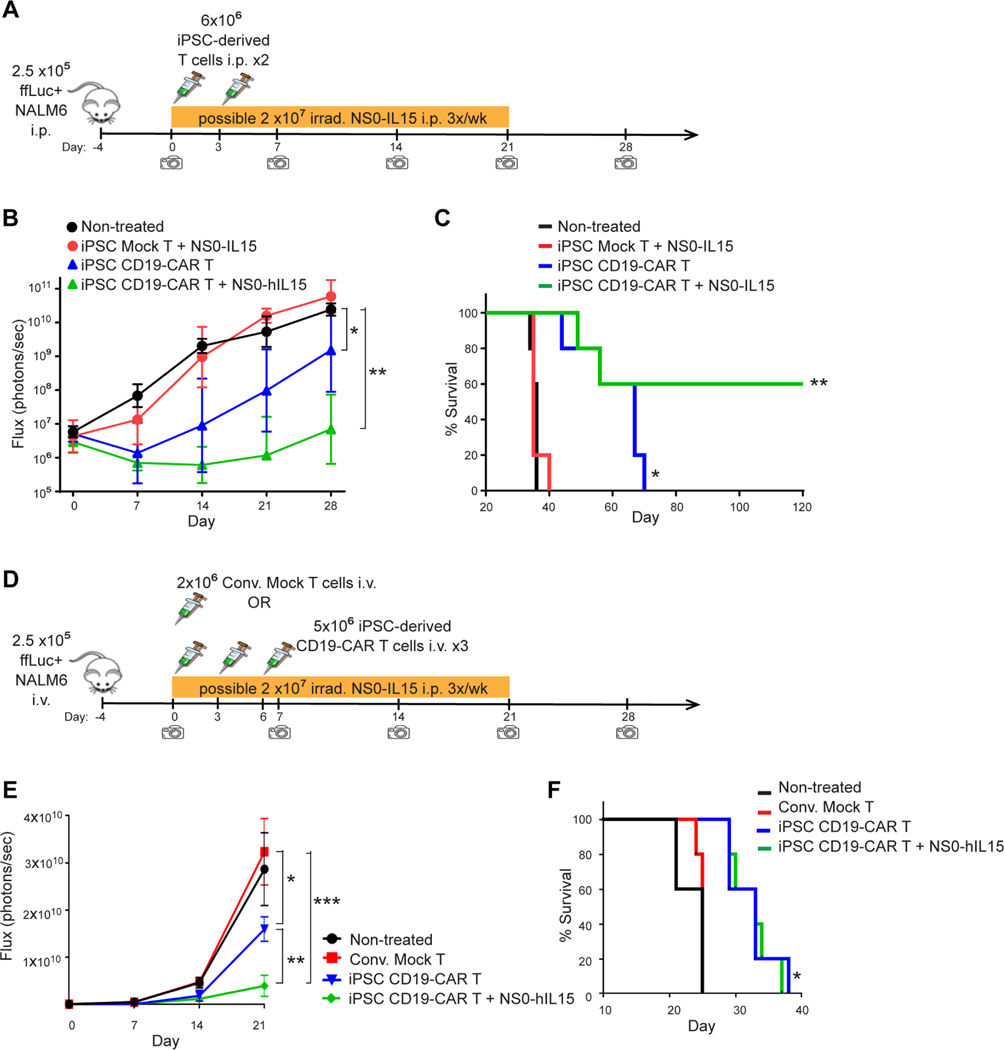
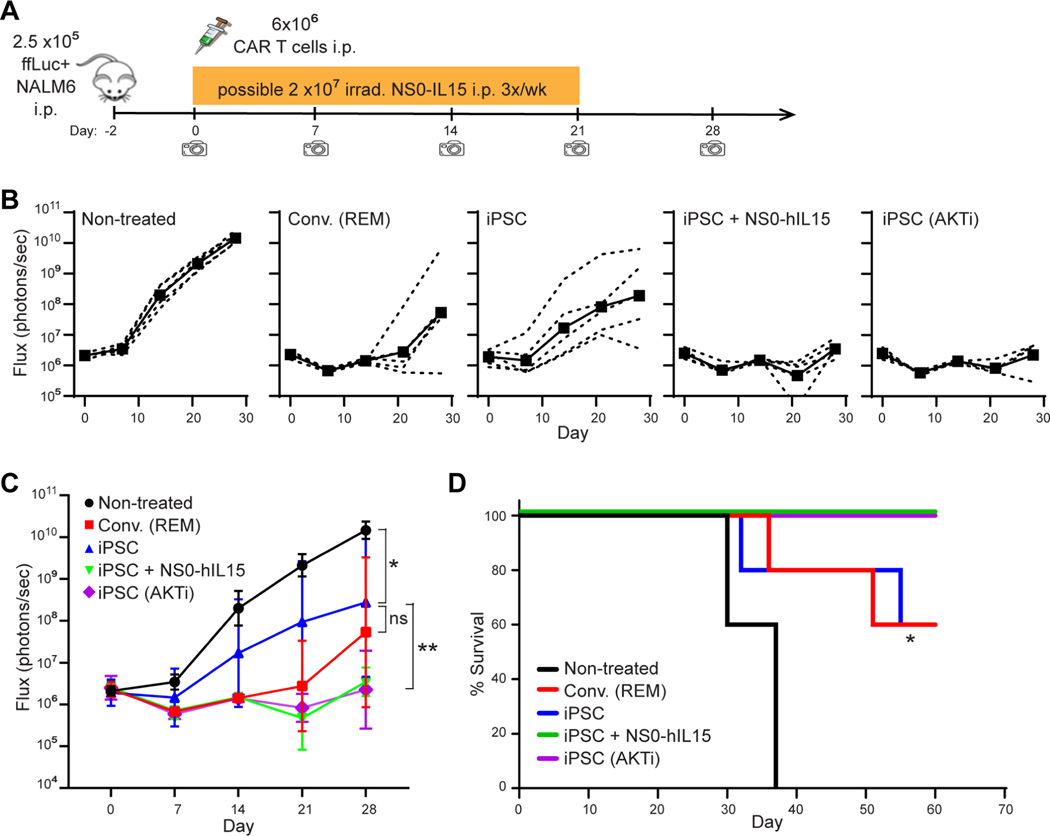
While the reduced CAR expression resulted in less activation in the absence of antigen (Figure 3J), and might account for the less exhausted phenotype in the presence of antigen challenge (Figure 3K), the iPSC CD19-CAR T cells still appeared to exhibit robust antigen-specific cytotoxic activity in vitro (Figure 3A–G). However, to better evaluate the anti-tumor activity of these T cells, we next carried out in vivo therapeutic assays in mouse xenograft models using NALM6 cells expressing firefly luciferase to allow for bioluminescent imaging (images provided in Figure S4). In the intraperitoneal (i.p.) tumor model (Figure 4A), i.p. administration of iPSC CD19-CAR T cells dramatically delayed tumor progression (Figure 4B) and significantly prolonged the mouse survival (P=0.004) (Figure 4C). Because the iPSC CAR T cells are primarily CD8+ with a minor CD4+ helper population, and extended ex vivo culture of T cells is known to have deleterious effects on in vivo persistence and activity (Akbar and Henson, 2011), we evaluated the potential of augmenting in vivo antitumor activity by supplementing with exogenous IL15 cytokine support as proposed by others (Alizadeh et al., 2019; Pilipow et al., 2015). Combination of iPSC CD19-CAR T cells with human IL15 secreting nurse cells (NS0-hIL15) did indeed enhance this therapeutic effect, leading to complete cure in 3 out 5 mice (Figure S4A). The therapeutic benefit of iPSC CD19-CAR T cells was also demonstrated in a more aggressive intravenous (i.v.) mouse tumor model (Figure 4D, 4E, 4F and S4B), again showing significantly improved mouse survival (P=0.0035). To sideby-side compare the in vivo anti-tumor potency of iPSC CAR T cells with conventional CAR T cells, we administrated the same dose of expanded iPSC CD19-CAR T cells and PBMC CD19-CAR T cells which had undergone ex vivo REM expansion similar to the iPSC CAR T cells and have a comparable percentage of CD8+ cells (Figures 5, S4C and D). The iPSC CD19-CAR T cells demonstrated comparable anti-tumor efficacy with REM expanded PBMC CD19-CAR T cells, and combination with NS0-hIL15 or pre-treatment with AKT inhibitor AZD5363 (the benefits of which have been shown in (Urak et al., 2017)) further enhanced the anti-tumor potency of the iPSC CD19-CAR T cells (Figure 5, S4D). A similar pattern was also shown in a more aggressive i.v. model using purified CD8+ populations from REM expanded PBMC-derived CAR T cells and iPSC CAR T cells (Figure S5). In summary, the iPSC CD19-CAR T cells which were produced by the PSC-ATO culture system from CAR expressing Tn/mem cells, demonstrated potent anti-tumor efficacy in vivo.
Figure 4. iPSC CD19-CAR T cells demonstrate potent anti-tumor activity in vivo.
(A) Schema of animal studies using intraperitoneal (i.p.) tumor model. On day −4, NSG mice were inoculated i.p. with 2.5×105 ffluc+ NALM6 cells. Mice were then either left untreated, or treated with 6×106 iPSC-derived mock-transduced (Mock) or CD19-CAR T cells i.p. on days 0 and 3; in one group receiving iPSC CD19-CAR T cells, 2×107 irradiated NS0-hIL15 cells were also administered 3 times a week for 3 weeks. Tumor burden was determined by weekly bioluminescent imaging. (B), Geometric mean ± 95% CI of i.p. tumor ffLuc Flux over time. Using two-way ANOVA test: *, P = 0.0008, ** P < 0.0001. (C), Kaplan-Meier survival analysis of i.p. xenografted mice. Using Mantel-Cox test: *, P = 0.0034 comparing the iPSC CD19-CAR T treated group to the non-treated group; **, P = 0.0016 comparing the iPSC CD19-CAR T + NS0-hIL15 treated group to the iPSC CD19-CAR T treated group. (D), Schema of animal studies using intravenous (i.v.) tumor model. On day −4, NSG mice were inoculated i.v. with 2.5×105 ffluc+ NALM6 cells. Mice were then either left untreated, or treated with 5×106 iPSC-derived CD19-CAR T cells i.v. on days 0, 3 and 6; where indicated, 2×107 irradiated NS0-hIL15 cells were administered 3 times a week for 3 weeks. Other control groups included mice that received 2×106 donor-matched Tn/mem-derived Mock T at day 0. Tumor burden was determined by weekly bioluminescent imaging. (E), Geometric mean ± 95% CI of i.v. tumor ffLuc Flux over time. Using two-way ANOVA test: *, P = 0.0019, **, P = 0.0002, ***, P < 0.0001. (F), Kaplan-Meier survival analysis of i.v. xenografted mice. Using Mantel-Cox test: *, P = 0.0035 comparing either iPSC CD19-CAR T treated group to the non-treated group.
Figure 5. Combination with IL15 or pre-treatment with AKT inhibitor (AKTi) improves anti-tumor activity of iPSC CD19-CAR T cells in vivo.
(A) Schema of animal studies using intraperitoneal (i.p.) tumor model. On day −2, NSG mice were inoculated i.p. with 2.5×105 ffluc+ NALM6 cells. Mice were then either left untreated, or treated i.p. on day 0 with 6×106 Conv. CD19-CAR T cells that had undergone REM, or iPSC CD19-CAR T cells with or without AKT inhibitor (AZD5363) treatment during expansion; in one group receiving iPSC CD19-CAR T cells, 2×107 irradiated NS0-hIL15 cells were also administered i.p. 3 times a week for 3 weeks. Tumor burden was determined by weekly bioluminescent imaging. (B, C), Tumor ffLuc Flux of individual mice (dashed lines) and geometric mean ± 95% CI of tumor ffLuc Flux (solid lines) over time. Using two-way ANOVA test: *, P = 0.0157, iPSC CD19-CAR T vs. Non-treated; **, P = 0.0178, comparing iPSC CD19-CAR T + NS0+hIL15 or iPSC CD19-CAR T (AKTi) treated groups to iPSC CD19-CAR T treatment alone. (D), Kaplan-Meier survival analysis of i.p. xenografted mice. Using Mantel-Cox test: *, P = 0.0008 comparing the iPSC CD19-CAR T treated group to the non-treated group; P = 0.1342 comparing the iPSC CD19-CAR T + NS0-hIL15 treated group, or iPSC CD19-CAR T (AKTi) treated group to the iPSC CD19-CAR T treated group.
Discussion
Generation of T cells and CAR T cells using extrathymic culture systems, whether they are single-layer or 3D-organoid co-cultures, remains a challenge (Maeda et al., 2016; Montel-Hagen et al., 2019a; Vizcardo et al., 2018; Vizcardo et al., 2013; Zhao et al., 2007). The previously reported iPSC CAR T cells generated by a mono-layer co-culture system displayed an innate-like phenotype (i.e., CD8αα+), as well as less-efficient antigen-specific cytotoxicity and cytokine secretion compared to conventional CAR T cells (Themeli et al., 2013). More recently, a 3D-organoid culture system was reported to facilitate generation of mature and functional CD3+CD8αβ+ and CD3+CD4+ conventional T cells and TCRtransgenic T cells (Montel-Hagen et al., 2019a); and in making further modifications to this organoid culture strategy, we demonstrate the successful generation of iPSC CAR T cells with a conventional T cell phenotype and CAR T cell function. Specifically, by using Tn/mem-derived iPSCs that were gene modified to express the CAR, and an PSC-ATO culture system to drive differentiation, iPSC CAR T cells were generated expressing conventional CD5+CD7+TCRαβ+TCRγδ-CD8αβ+ T cell phenotypes, and exhibited potent cytotoxic killing and Th1 cytokine secretion activity that was comparable to conventional CAR T cells derived from the same donor. Such improvements validate the potential utility of iPSCs for generating therapeutic CAR T cell products.
It is also potentially advantageous that our Tn/mem-derived iPSC CAR T cells displayed a more homogenous, monoclonal TCR repertoire, which was different from the polyclonal phenotype of iPSC reprogrammed from CD34+ cord blood, fibroblasts, or effector T cells as described here, as well as that of ESC-derived T cells (Montel-Hagen et al., 2019a; Nishimura et al., 2013). Furthermore, the use of terminally differentiated effector T cells to generate the iPSCs resulted in regenerated CD8αβ T cells that lost their clonal antigen specificity by additional TCR rearrangement, with TCR stability only being induced upon TCR transduction of the iPSCs (Minagawa et al., 2018). This suggests that starting with a less-differentiated Tn/mem population may have unique effects on TCR rearrangement during re-differentiation, which may or may not relate to the allelic exclusion effect of pre-existing TCR loci (Brady et al., 2010). Regardless, selection of Tn/mem-derived iPSC clones of a known and/or innocuous TCR to minimize potential graft-versus-host toxicities would then be relevant to the manufacture of an ‘off-the-shelf’ iPSC CAR T cell products.
The lower expression levels of MHC and dominance of CD8 on our iPSC CAR T cells may also relate to the unique effects of starting with Tn/mem-derived iPSC clones, or it may be related to the lack of thymic epithelial cells in the culture system (Vizcardo et al., 2018). While low MHC expression may be desirable for reducing T cell mediated rejection and facilitating iPSC CAR T cell persistence after adoptive transfer, it might be important to improve the balance between the CD4+ and CD8+ populations, since CD4+ CAR T cells have recently been shown to play important role in adoptive immune cell therapy (Wang et al., 2018). A more balanced CD4/CD8 lineage differentiation may be obtained by manipulating either the culture conditions during differentiation or the lineage selection pathways by gene editing (Singer et al., 2008).
The comparison of gene expression profiles of bulk, CD4 and CD8 positive populations between iPSC CD19-CAR T cells and conventional CD19-CAR T cells revealed interesting differences. The upregulation of DLL1 expression in iPSC CAR T cells could contribute to the development of CD8 and Th1 cells (Kelliher and Roderick, 2018; Maekawa et al., 2003), and expression of cytokines, such as IFNɣ/IL2/IL17/IL4 (Zhang et al., 2011), which may be advantageous for anti-tumor activity. The elevated phosphorylation level of PLC (Y783), CAR and endogenous CD3zeta is indicator of T cell activation status, which may contribute to potent anti-tumor cytotoxicity of iPSC CAR T cells, but may also lead to an activation-dependent differentiated phenotype. The low percentage of CD62L+ and diminished expression of IL7R confirmed a more differentiated phenotype and indicates that improving the less-differentiated stem-like phenotype of iPSC CAR T cells would benefit the fitness of the product. While our REM protocol drove more robust expansion than stimulation with CD3/CD28 Dynabeads™ (i.e., as typically used to expand conventional CAR T cells; data not shown), further optimization of the iPSC CAR T cell expansion methods could be beneficial. Interestingly, our study has shown that the previously reported strategies of combining CAR T cells with supportive IL15 cytokine (Alizadeh et al., 2019), or treating CAR T cells with AKT inhibitor (Urak et al., 2017) during cell expansion can benefit iPSC derived CAR T cells, which encourages us to explore more strategies to further improve the in vivo activity.
We have demonstrated that decreased CAR expression in the iPSC CAR T cells was related to the hyper-methylation of the EF1α promoter. While promoter methylation has been known to regulate gene expression (Hofmann et al., 2006) and could be acquired progressively and lead to subsequently epigenetic suppression of transgenic product in TCR adoptive therapy (Nowicki et al., 2020), the differentiation-induced hyper-methylation of the CAR transgene promoter represents a unique mechanism by which one might regulate CAR expression. The use of other PSC differentiation resistant promoters such as a ubiquitin promoter might be explored to drive higher transgene expression (Jiang et al., 2010). However, lowering CAR expression might be more desirable than previously appreciated, as it has been reported that optimal basal/low CAR expression can reduce tonic signaling and sustain CAR T cell functions (Eyquem et al., 2017). In our study, the basal pERK phosphorylation level and the basal GM-CSF/IFNγ secretion levels of iPSC CD19-CAR T cells without antigen stimulation was much lower than that seen in conventional CD19-CAR T cells. In contrast, upon antigen encounter, cell signaling by iPSC CD19-CAR T cells, as measured by pERK, PLCγ(Y783) and ZAP70 phosphorylation, as well as Th1 cytokine secretion was comparable to or higher than that of conventional CD19-CAR T cells, in spite of the lower CAR expression. Interestingly, while phosphorylation of both the endogenous and CAR-associated CD3ζ sequences was suppressed by NALM6 tumor cells (Figure 3L), a phenomenon which has not been reported with anti-CD3 or antigen coated beads (Salter et al., 2018; Sun et al., 2020), the cytotoxic activity observed against CD19+ NALM6 tumors (Figure 3B, 3C, 3E) suggest that iPSC CD19-CAR T cells overcame this suppression better than conventional CD19-CAR T cells. Together these data suggest iPSC CAR T cells may exhibit an antigen-specificity profile that is beneficial for both safety and efficacy.
Various off-the-shelf CAR T cell strategies are in early stages of clinical testing (Depil et al., 2020), and our data builds on these advances by establishing a platform for effective iPSC differentiation to generate CAR T cells with canonical T cell phenotype and CAR T function, thereby supporting further clinical translation. This would require the establishment of GMP compliant iPSC lines with designated differentiation potential of TCR repertoire, robust, scalable and GMP compliant systems for gene engineering, CAR+ iPSC directional differentiation and expansion into CAR T cell product, etc. Importantly, our study demonstrated that iPSC-ATO 3D culture can be used to generate T cells harboring different clinically relevant CAR constructs such as CD19-CAR and CLTX-CAR. Another key consideration for clinical translation is to ensure negligible graft-vs-host and/or host-vs-graft potential of the iPSC CAR T cells, and it has also been proposed that endogenous TCR and MHC-I/II genes could be targeted by gene editing technologies (Deuse et al., 2019; Poirot et al., 2015). However, how such gene manipulations might further affect functional CAR T cell development from iPSC cells is still being explored. In summary, our study presents a feasible method using Tn/mem-derived, CAR expressing iPSC cells and PSC-ATO culture to generate CAR T cells with a canonical T cell phenotype and conventional CAR T cell attributes, and supports further optimizations for clinical translation.
Limitations of the study
Despite the promise of the iPSC CAR T cell platform described here, there remain various ways in which this strategy can be improved, including the incorporating approaches to enhance antitumor potency. We showed that iPSC CD19 CAR T cells and conventional CD8+ CAR T cells had comparable antitumor activity in several tumor models, however, both mediated only modest survival benefit in aggressive i.v. NALM6 tumor models, possibly due to prolonged ex vivo expansion, which has been well established to limit therapeutic activity (Ghassemi et al., 2016). Shortening the time of expansion, optimizing the expansion conditions and/or combination with supportive cytokines may be strategies to improve the in vivo therapeutic effect of iPSC CAR T cells. While CAR constructs incorporating cytokines, such as IL15, have shown utility in enhancing CAR T cell potency (Hoyos et al., 2010; Lanitis et al., 2021), engineered expression of IL-15 is expected to negatively affect iPSC differentiation into T cells because IL15 can strongly drive differentiation of lymphoid-progenitor cells towards NK cells instead of T cells (Mrozek et al., 1996). Alternatively, combination of matured iPSC CAR T cells with IL15 administration or pretreatment with supportive cytokines or small molecular inhibitors during ex vivo expansion can be used as a strategy to further enhance the functionality of iPSC CAR T cells. In this study, we have explored such strategies by combining treatment with hIL15 in vivo, or manufacturing in the presence of AKTi, and both methods demonstrated some therapeutic improvement. It would also be interesting to examine the in vivo anti-tumor effects of unexpanded iPSC CAR T cells, or of iPSC CAR T cells that exhibit higher CAR expression levels (e.g., which may be achieved by switching the EF1a transgene promoter to another methylation resistant promoter). Such strategies are currently under investigation by our group, and overcoming the current limitations will provide a foundation for the further development of strategies aimed at producing an unlimited number of potent, allogeneic ‘off-the-shelf’ CAR T cells.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Christine Brown (CBrown@coh.org).
Materials Availability
All unique/stable reagents generated in this study will be made available on request, but we may require a payment and /or a completed Materials Transfer Agreement if there is potential for commercial application.
Data Availability
The RNA seq datasets generated in this study have been deposited at NCBI GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human SSEA-3 | BD Biosciences | (clone MC-6310) RRID:AB_1645542 |
| Anti-human SSEA-4 | BD Biosciences | (clone MC813–70) RRID:AB_2033991 |
| Anti-human Tra-1–60 | BD Biosciences | (clone TRA-1–60) RRID:AB_10565983 |
| Anti-human TRA-1–81 | BD Biosciences | (clone TRA-1–81) RRID:AB_1645493 |
| Anti-human EpCAM (CD326) | BioLegend | (clone 9C4) RRID:AB_756082 |
| Anti-human CD56 | BD Biosciences | (clone NCAM16.2) RRID:AB_400025 |
| Anti-human CD30 | BD Biosciences | (clone BerH8) RRID:AB_393541 |
| Anti-human CD3 (BV510) | BD Biosciences | (clone UCHT1) RRID:AB_2732053 |
| Anti-human CD3 (APC) | BD Biosciences | (clone SK7) RRID:AB_400513 |
| Anti-human CD4 (Per-CP) | BD Biosciences | (clone SK3) RRID:AB_400282 |
| Anti-human CD4 (PE-Cy7) | BD Biosciences | (clone SK3) RRID:AB_396897 |
| Anti-human CD8α (PE) | BD Biosciences | (clone SK1) RRID:AB_400005 |
| Anti-human CD8α (Per-CP) | BD Biosciences | (clone SK1) RRID:AB_400280 |
| Anti-human CD8α (APC-Cy7) | BD Biosciences | (clone SK1) RRID:AB_400383 |
| Anti-human CD8β | BD Biosciences | (clone 2ST8.5H7) RRID:AB_1645747 |
| Anti-human CD5 | BD Biosciences | (clone UCHT2) RRID:AB_2737700 |
| Anti-human CD7 | BD Biosciences | (clone M-T701) RRID:AB_2738544 |
| Anti-human TCR αβ | BD Biosciences | (clone T10B9.1A-31) RRID:AB_2738437 |
| Anti-human TCR γδ | BD Biosciences | (clone B1) RRID:AB_396061 |
| Anti-human TCR γδ | BD Biosciences | (clone 11F2) RRID:AB_2827402 |
| Anti-human NKG2A | R&D Systems | (clone 131411) RRID:AB_2132978 |
| Anti-human NKG2D (CD314) | BD Biosciences | (clone 1D11) RRID:AB_10896282 |
| Anti-human NKp44 (CD336) | BD Biosciences | (clone p44–8) RRID:AB_647239 |
| Anti-human NKP46 (CD335) | BioLegend | (clone 9E2) RRID:AB_2561649 |
| Anti-human CD19 | BD Biosciences | (clone SJ25C1) RRID:AB_396893 |
| Anti-human CD16 | BD Biosciences | (clone NKP15 ) RRID:AB_400317 |
| Anti-human Fas L (CD178) | BD Biosciences | (clone NOK-1) RRID:AB_2738713 |
| Anti-human CD62L (PE) | BD Biosciences | (clone SK11) RRID:AB_400205 |
| Anti-human CD62L (FITC) | BD Biosciences | (clone SK11) RRID:AB_400302 |
| Anti-human CD45RA | BD Biosciences | (clone HI100) RRID:AB_398468 |
| Anti-human CD45RO | BD Biosciences | (clone UCHL1) RRID:AB_395883 |
| Anti-human EGFR (APC) | BioLegend | (clone AY13) RRID:AB_11150410 |
| Anti-human EGFR (PE-Cy7) | BioLegend | (clone AY13) RRID:AB_2562159 |
| Anti-human CD28, InVivoMab | Bio X Cell | (clone 9.3) RRID:AB_2687729 |
| Anti- human CD27 | BD Biosciences | (clone M-T271) RRID:AB_395834 |
| Anti-human CD28 (APC-H7) | BD Biosciences | (clone CD28.2) RRID:AB_11154032 |
| Anti-human CD279 (PD-1) | ThermoFisher Scientific | (clone eBioJ105) RRID:AB_2573976 |
| Anti-human LAG3 /CD223 | Lifespan | (clone 17B4) RRID:AB_1650048 |
| Anti-human TIM-3 | R&D Systems | (clone 344823) RRID:AB_2232901 |
| Anti-human IgG Fc | Abcam | Abcam # ab98622, RRID:AB_10673586 |
| Pan p44/42 MAPK (phosphorylated Erk1/2) antibody | Cell Signaling Technology | Polyclonal, Cat# 9101, RRID:AB_331646 |
| Pan p44/42 MAPK (Erk1/2) antibody | Cell Signaling Technology | (clone 3A7) RRID:AB_10695739 |
| Anti-human, -mouse phospho-PLCγ1 (Tyr783) antibody | Cell Signaling Technology | (clone D6M9S) RRID:AB_2728690 |
| Anti-human, -mouse, -monkey phospho-PLC 1 (Ser1248) antibody | Cell Signaling Technology | (clone D25A9) RRID:AB_10890863 |
| Anti-human, -rat, -mouse PLCγ1 | Cell Signaling Technology | Polyclonal, Cat# 2822, RRID:AB_2163702 |
| Anti-human, -mouse phospho-Zap-70 (Tyr319)/Syk (Tyr352) | Cell Signaling Technology | (clone 65E4) RRID:AB_2218658 |
| Anti-human, -mouse, anti-CD3ζ (pY142) | BD Biosciences | (clone K25–407.69) RRID: AB_647307 |
| Bacterial and virus strains | ||
| NEB 5-alpha Competent E. coli | NEB | C2987 |
| Biological samples | ||
| PBMC from healthy donor | This paper | N/A |
| Primary B-ALL patient samples | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Proleukin (aldesleukin), rhIL-2 | Novartis Oncology | NCD 65483–116-07 |
| rhIL-15 | CellGenix, US Operations | 1413–050 |
| Activin A | R&D Systems | 338-AC-010 |
| rhBMP-4 | R&D Systems | 314-BP-010 |
| rhVEGF | R&D Systems | 293-VE-010 |
| rhFGF-basic | Peprotech | AF-100–18B |
| ROCK inhibitor, Y-27632 dihydrochloride | StemCell Technologies | 72304 |
| TGFβ-RI inhibitor, SB-431542 | StemCell Technologies | 72234 |
| rhFlt3-Ligand | Peprotech | AF-300–19 |
| rhIL-7 | Peprotech | AF-200–07 |
| rhTPO | Peprotech | 300–18 |
| rhSCF | Peprotech | AF-300–07 |
| MEM α, with nucleosides | GIBCO | 12571063 |
| Fetal bovine serum, defined, heat-inactivated | HyClone | SH30070.03IH |
| X-VIVO 15 Serum-free Hematopoietic Cell Medium | Lonza | BEBP04–744Q |
| EGM-2, Endothelial Cell Growth Medium-2 Bullet Kit | Lonza | CC-3162 |
| RPMI 1640 Medium with L-Glutamine and HEPES | Lonza | 12–115F |
| GLutaMAX Supplement | ThermoFisher Scientific | 35050061 |
| 2-Mercaptoethanol, 55 mM | ThermoFisher Scientific | 21985023 |
| MEM Non-Essential Amino Acids Solution (100X) | ThermoFisher Scientific | 11140050 |
| L-Ascorbic acid 2-phosphate sesquimagnesium salt hydrate | Sigma | A8960 |
| Phosphate-buffered saline | Irvine Scientific | 9240 |
| Penicillin-Streptomycin (100X) | Lonza | 17602E |
| mTeSR1 | StemCell Technologies | 85850 |
| DMEM/F12, with HEPES | ThermoFisher Scientific | 11330057 |
| Matrigel hESC-Qualified Matrix, LDEV-free | Corning | 354277 |
| ReLeSR | StemCell Technologies | 5872 |
| CloneR | StemCell Technologies | 5889 |
| StemPro Accutase Cell Dissociation Reagent | ThermoFisher Scientific | A1110501 |
| DNase I | Zymo Research | E1010 |
| B-27 Supplement (50X) | ThermoFisher Scientific | 17504–044 |
| TrypLE Express Enzyme (1X) | ThermoFisher Scientific | 12605010 |
| Protamine sulfate | APP Pharmaceuticals | NDC 63323–229-05 |
| XenoLight D-luciferin potassium salt | PerkinElmer | 122799 |
| Hanks’ Balanced Salt Solution, w/o Ca++, Mg++ | GIBCO | 14175103 |
| 0.05% Trypsin/0.53mM EDTA in HBSS w/o Ca++, Mg++ | Corning | MT25051CI |
| Brefeldin A Golgi Plug | BD Biosciences | 555029 |
| Critical commercial assays | ||
| Leukocyte Alkaline Phosphatase Kit | Sigma | 86R-1KT |
| IOTest beta Mark TCR Vβ Repertoire Kit | Beckman Coulter | IM3497 |
| TCRB T-Cell Clonality Assay Kit | Invivoscribe | 12000011 |
| CD34 MicroBead Kit UltraPure | Miltenyi Biotec | 130–100-453 |
| EasySep Human CD56 Positive Selection Kit II | Stem Cell Technologies | 17855 |
| EasySep Human EpCAM Positive Selection Kit II | Stem Cell Technologies | 17846 |
| Dynabeads™ Human T-Expander CD3/CD28 | ThermoFisher Scientific | 11141D |
| RNeasy Micro Kit | Qiagen | 74004 |
| Quick-RNA Microprep Kit | Zymo Research | R1050 |
| EZ DNA Methylation-Lightning Kit | Zymo Research | D5030 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | 554714 |
| Luminex Cytokine Mouse Magnetic 10-Plex Panel | ThermoFisher Scientific | LMC0001M |
| Millicell Cell Culture Insert, 30 mm, hydrophilic PTFE, 0.4 μm | Millipore Sigma | PICM0RG50 |
| Deposited data | ||
| RNA expression data by high throughput sequencing | This paper | NCBI GEO: GSE193422 |
| Experimental models: Cell lines | ||
| T cell derived iPSC lines | this paper | N/A |
| Fibroblast derived iPSC line | ATCC | DYS0100, ACS-1019 |
| MS5-hDLL4-eGFP | Gay Crooks’ lab | N/A |
| NALM6.ffluc.eGFP | this paper | N/A |
| NALM6.ffluc.eGFP_CD19KO | this paper | N/A |
| Raji.ffluc.eGFP | this paper | N/A |
| Raji.ffluc.eGFP_CD19KO | this paper | N/A |
| 3T3_CD19 | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) | Jackson Laboratory | Strain #005557 |
| Oligonucleotides | ||
| Primers for EF1α promoter PCR and sequencing, see Extended Table 2 | This paper | N/A |
| Recombinant DNA | ||
| CD19-CAR_pHIV (CD19-CAR) | Forman lab | N/A |
| CLTX-CAR_pHIV (CLTX-CAR) | Forman lab | N/A |
| hOCT3/4-shp53_pCXLE | addgene, gift from Shinya Yamanaka | RRID:Addgene_27077 |
| hSK_pCXLE (SOX2, KLF4) | addgene, gift from Shinya Yamanaka | RRID:Addgene_27078 |
| hUL_pCXLE (L-MYC, LIN28) | addgene, gift from Shinya Yamanaka | RRID:Addgene_27080 |
| EBNA_pCXLE | addgene, gift from Shinya Yamanaka | RRID:Addgene_37624 |
| Software and algorithms | ||
| GraphPad Prism GraphPad Software | GraphPad Software | https://www.graphpad.com/ |
| Flowjo | Tree Star Inc. | https://www.flowjo.com/solutions/flowjo |
| Zen | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| BZ-X800 Analyzer | Keyence | https://www.keyence.com/landing/microscope/lp_fluorescence.jsp |
| R package “DESeq2” (v.3.10) |
R | https://www.r-project.org |
| Cluster (v.3.0) (clustering algorithms) |
Human Genome Center Institute of Medical Science, University of Tokyo | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| JavaTreeView (v.1.1.6r4) | Stanford University School of Medicine | http://jtreeview.sourceforge.net/ |
| “edgeR” (v.3.28.0) | R | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| “ggplot2” (v.3.2.1) | R | https://cran.r-project.org/web/packages/ggplot2/index.html |
| GSEA (v.4.0.3). (gene set enrichment analysis) | Broad Institute | https://www.gsea-msigdb.org/gsea/index.jsp |
| BioRender | BioRender.Inc | https://app.biorender.com/ |
| Other | ||
| N/A | ||
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Cell lines
Details in Key Resources Table. All experiments were conducted under protocols approved by IBC and SCRO of City of Hope.
Mice
All mouse experiments were conducted with protocols approved by City of Hope Institutional Animal Care and Use Committee. Tumor xenograft models were generated using 6 to 8 week-old NOD/SCID/IL2Rγ– /– (NSG) mice as previously described (Jackson Laboratory)(Urak et al., 2017). Briefly, on day 0, ffLuc+ NALM6 cells (2.5 × 105) were injected either intraperitoneally (i.p.) or intravenously (i.v.) into the NSG mice. After 2 or 4 days, mice were then treated with iPSC-derived or conventional CAR T or Mock T cells as described for each experiment. Mice in the indicated groups were injected i.p. three times per week with 20 × 106 irradiated (80 Gy) human hIL-15-secreting nurse cells (IL15-NS0) (Wang et al., 2011b). Reference schematics of Figure 4A, 4D, Figure 5A and S7B for injections of T cells in each tumor model. Tumor growth was determined weekly by in vivo biophotonic imaging using a Xenogen IVIS 100. Mice were also monitored for survival, with euthanasia applied according to the American Veterinary Medical Association Guidelines. Composites of individual mouse images are depicted in each group on day 0 in Figure S4 and S5.
Methods details
DNA constructs
The CD19-targeted chimeric antigen receptor (CD19-CAR) construct is the same as currently used in our clinical studies targeting B cell leukemia/lymphoma (NCT02146924) (Popplewell et al., 2018; Samer K. Khaled, 2018). The CD19-CAR contains an anti-CD19 scFv domain derived from the FMC63 mAb (Nicholson et al., 1997), an IgG4 Fc spacer with two point mutations (L235E and N297Q) within the CH2 region, a CD28 transmembrane domain, a CD28ζ costimulatory domain, and a CD3ζ signaling domain. A T2A ribosome skip sequence (Donnelly et al., 2001) then separates this CAR sequence from a truncated human epidermal growth factor receptor sequence (huEGFRt) which can be used as a selection marker and safety switch(Jonnalagadda et al., 2015; Urak et al., 2017; Wang et al., 2011c). The CD19(BBz)-CAR construct has the same anti-CD19 SCFV sequence and CD3ζ signaling domain, with CD4 transmembrane domain and 41BB co-activation domain. The Chlorotoxin-targeting CAR (CLTX-CAR) is a chlorotoxin peptide based CAR construct targeting glioblastoma (GBM), which is in clinical trial stage (clinicaltrials.gov # NCT04214392)(Wang et al., 2020). The episomal plasmids encoding OCT3/4/shp53, SOX2/KLF4, L-MYC/LIN28, and EBNA were gifts from Dr. Shinya Yamanaka(Okita et al., 2013).
Tn/mem isolation
Blood products were obtained from healthy donors under protocols approved by the COH IRB, and naïve and memory T (Tn/mem) cells were isolated following similar procedures described in previous studies(Wang et al., 2012). In brief, human peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation over Ficoll-Paque (GE Healthcare) and then underwent sequential rounds of CliniMACS/AutoMACS (Miltenyi Biotec) depletion to remove CD14- and CD25-expressing cells, followed by a CD62L-positive selection for Tn/mem cells.
Generation of iPSCs from Tn/mem
Tn/mem cells were reprogrammed into pluripotent stem cells (iPSCs) by an integration-free method modified from a published protocol (Okita et al., 2013). In brief, one million Tn/mem cells were electroporated with 3 μg of plasmid mixture using the Human T Cell Nucleofector Kit and the Nucleofector 4D electroporation device (Lonza). The plasmid mixture was composed of episomal plasmids encoding OCT3/4, SOX2, KLF4, L-MYC, LIN28, and shRNA for TP53 (Okita et al., 2013). The transfected cells were cultured in X-VIVO 15 medium (Lonza) supplemented with 10% FBS (HyClone), 50 U/mL rhIL-2 (Novartis Oncology), 0.5 ng/mL rhIL-15 (CellGenix) and Dynabeads Human T-Expander CD3/CD28 (ThermoFisher Scientific) (bead to cell ratio of 1:1). Two days after the transfection, an equal volume of pluripotent stem cell (PSC) medium containing rhFGF-basic and 10 μM Y27632 was added (Okita et al., 2013). The medium was then completely changed to PSC medium 4 days after transfection. iPSC colonies were visible at day 20–30 and individual colonies were picked under a microscope for further culture/expansion in cGMP-grade mTeSR1 medium (StemCell Technologies) in Matrigel-coated (Corning) plates.
Generation of CAR-positive, clonal iPSC lines
Before lentivirus transduction, iPSC cultures were dissociated with Accutase (ThermoFisher Scientific) treatment and the cells were seeded at a density of 105 per well in 12-well plates in mTeSR1 medium supplemented with 1X CloneR and 10 μM ROCK inhibitor Y-27632 dihydrochloride (StemCell Technologies). After overnight culture, cGMP lentivirus encoding CD19-CAR was added to the culture with 10 μg/mL protamine sulfate (APP Pharmaceuticals) to transduce the iPSCs (multiplicity of infection [MOI] = 1). The transduced cells were cultured for at least two passages before single cell sorting by flow cytometry and iPSC colonization. Clonal CAR-positive cells were again expanded in mTeSR1 medium on Matrigel-coated plates, and banked for subsequent differentiation.
Integration detection by PCR
EBNA1 is a common component of all episomal vectors (Black and Vos, 2002). To detect genomic integration of episomal plasmids used for iPSC reprogramming from T cells, PCR was performed to amplify integrated EBNA components from genomic DNA using primers as follows: EBNA1_For: ATCAGGGCCAAGACATAGAGATG, EBNA1_Rev: GCCAATGCAACTTGGACGTT. Plasmid integration free iPSC clones did not show EBNA1 signal. FBX15, which was expressed on pluripotent stem cells, was used as house-keeping gene here and was amplified by the following primers: FBX15_For: GCCAGGAGGTCTTCGCTGTA; FBX15_Rev: AATGCACGGCTAGGGTCAAA.
Teratoma formation assay
Two million dissociated iPSCs were suspended in 200 uL medium (100 uL PBS (Irvine Scientific) and 100 uL Matrigel) and injected subcutaneously into NSG mice. After 5–8 weeks, teratomas were harvested in PBS, fixed overnight in 4% paraformaldehyde (Boston BioProducts) at room temperature, and maintained thereafter in 70% ethanol for processing. Samples were submitted to the City of Hope Histology Core Facility for sectioning and hematoxylin and eosin staining. Sections were examined, interpreted, and photographed microscopically.
Differentiation of Tn/mem-derived, CAR+ iPSC into CAR+ T cells by PSC-ATO culture
The schema of the sequential differentiation protocol is outlined in Fig. 1A. First, mesoderm commitment was induced as previously described (Chin et al., 2016; Evseenko et al., 2010; Montel-Hagen et al., 2019b). Briefly, iPSCs were harvested as a single cell suspension after Accutase treatment, resuspended at 1 × 106 cells/mL in X-VIVO 15 medium containing 10 ng/mL rhActivin A (R&D Systems), 10 ng/mL rhBMP4 (R&D Systems), 10 ng/mL rhVEGF (R&D Systems), 10 ng/mL rhFGF (Peprotech), and 10 μM ROCK inhibitor Y-27632 dihydrochloride (StemCell Technologies). Three million cells per well were plated in Matrigel-coated 6-well plates. Medium was then changed daily with X-VIVO 15 containing 10 ng/mL rhBMP4, 10 ng/mL rhVEGF, and 10 ng/mL rhFGF. Three days later (Day −14 in Fig. 1A), cells were washed 3 times with PBS (Irvine Scientific) and incubated with 1 mL per well Accutase for 5–7 minutes at 37°C. Cells were harvested, washed in PBS containing 1 mM EDTA and 2% FBS, and CD56+CD326- human iPSC mesodermal progenitors (iMP) were isolated by CD56 enrichment using EasySep Positive Selection kits (StemCell Technologies). Flow cytometry was performed to confirm CD56+CD326- phenotype of the iMP.
iPSC mesodermal organoids (iMOs) were generated by aggregating iMP cells and MS5-hDLL4 feeder cells. On day −14, MS5-hDLL4 cells were harvested with trypsin and washed into hematopoietic induction medium composed of EGM-2 (Lonza) with 10 μM Y-27632 and 10 μM TGF-βRI inhibitor SB-431542 (StemCell Technologies). After using a 40 μm nylon mesh strainer to remove aggregates, 5 × 105 MS5-hDLL4 cells were combined with 0.5–1 × 104 purified iMP cells in 1.5 mL microfuge tubes and centrifuged at 300 × g for 5 min at 4°C in a swinging bucket centrifuge. Up to 12 iMOs were prepared in each tube. After carefully removing the supernatant, the MS5-hDLL4/iMP cell pellet was resuspended by brief pulse vortexing in hematopoietic induction medium (i.e., EGM-2 with 10 μM SB-431542) at 6 μl per iMO. Two 6 μL aliquots of cells were plated as iMOs on one Millicell transwell insert (Millipore Sigma) per well in 6well plates containing 1.5 mL hematopoietic induction medium. Medium was changed completely every 2–3 days for one week. On day −7, medium was changed to EGM-2 with 10 μM SB-431542 plus 5 ng/mL rhTPO (Peprotech), 5 ng/mL rhFLT3L (Peprotech), and 50 ng/mL rhSCF (Peprotech).
On day 0, the artificial thymic organoid (ATO) T cell differentiation phase was initiated with a switch to serum-free ATO culture medium containing 10 ng/mL rhSCF, 5 ng/mL rhFLT3L, and 5 ng/mL rhIL-7 in RB27 medium that consisted of RPMI 1640 (Lonza), with 4% B27 Supplement (ThermoFisher Scientific), 30 μM L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma), 1% GlutaMAX (ThermoFisher Scientific), 1% Penicillin-Streptomycin (Lonza), 55 uM 2-mercaptoethanol (ThermoFisher Scientific), and 1% MEM Non-essential Amino Acids (ThermoFisher Scientific). Medium was changed completely every 2–3 days. After 5–7 weeks of differentiation, iMO-ATO-derived CAR+ T cells were harvested by pipetting 1–2 mL of X-VIVO 15 with 10% FBS onto the surface of each transwell insert and disaggregating the iMO-ATO by repeated aspiration with a P1000 pipettor. Single cells were isolated by passing the disaggregated cell suspension through a 40 μm nylon mesh strainer. An aliquot of the recovered cells was stained with the indicated antibodies for phenotyping by flow cytometry and the remaining cells were cultured in a previously described rapid expansion method (REM) (Wang et al., 2011b; Wang et al., 2015), with minor modifications. Briefly, 1 × 106 iMO-ATO-derived T cells were combined with 50 × 106 ɣ-irradiated (35 Gy) PBMCs and 10 × 106 ɣ-irradiated LCL cells (80 Gy) in 50 mL X-VIVO 15 medium containing 10% FBS, 20 ng/mL anti-CD3 (Miltenyi Biotec), 50 U/mL rhIL-2 and 10 ng/mL rhIL-7. REM cultures were maintained for 14 days, with half-volume medium changes every 48 hours. In the indicated experiment, 0.1 μM AKT inhibitor AZD5363 was added during this expansion of iPSC CAR T cells.
Immunohistochemistry
The PSC-ATO organoids were fixed and permeabilized with the Fixation/Permeabilization Solution Kit (BD Biosciences), stained with PE-anti-CD3 and DAPI in permeabilization buffer for 15 minutes and then rinsed with wash buffer three times. In situ images were taken with a BZ-X810 fluorescence microscope (Keyence).
Generation of conventional CD19-CAR T cells
PBMC were stimulated with Dynabeads Human T-Expander CD3/CD28 at a ratio of 1:2 (cells:beads) in X-VIVO 15 medium containing 10% FBS, 50 U/mL rhIL-2, and 0.5 ng/mL rhIL-15. Cells were transduced with clinical grade lentivirus to express CD19CAR with 25 μg/mL protamine sulfate (APP Pharmaceuticals). Cultures were then maintained at 37°C, 5% CO2 under the same medium and cytokine conditions. Fresh cytokines were supplied every other day. On day 7 after transduction, the CD3/CD28 Dynabeads were removed from cultures using the DynaMag-50 magnet (ThermoFisher Scientific). The cells were expanded in culture until harvest at day 17 or as indicated.
The PBMC-derived, CAR+ T cells were enriched by EasySep kit with anti-EGFRt antibody (StemCell Technologies) and used for phenotype characterization and functional assays; PBMC-derived CAR+ T cells used in the in vivo assays were not enriched, but dosed based on CAR+. The REM expanded PBMC-derived CAR T cells which were used in Figure 5, were expanded using the same conditions as iPSC CD19-CAR T cells. Briefly, 1 × 106 conventional CD19-CAR T cells were combined with 50 × 106 ɣ-irradiated (35 Gy) PBMCs and 10 × 106 ɣ-irradiated LCL cells (80 Gy) in 50 mL X-VIVO 15 medium containing 10% FBS, 20 ng/mL anti-CD3 (Miltenyi Biotec), 50 U/mL rhIL-2 and 10 ng/mL rhIL-7. REM cultures were maintained for 14 days, with half-volume medium changes every 2 days.
Isolation and culture of conventional NK cells
Primary NK cells were isolated from healthy donor PBMC using the EasySep Human CD56 Positive Selection Kit II (Stemcell technologies) per manufacture’s protocol. Enriched CD56+ NK cells were cultured with 100 Gy irradiated K562 aAPC cells which were genetically modified with mbIL21 in a ratio of 1:2. The cells were cultured in X-VIVO 15 medium containing 10% FBS and 100 U/mL rhIL-2(Denman et al., 2012). The medium was refreshed every 2~3 days and the expanded conventional NK cells were then frozen down or examined for surface markers expression after 14 days of expansion.
Flow cytometry
iPSCs were dissociated with Accutase (ThermoFisher Scientific) and resuspended in mTeSR1 medium with 1X CloneR supplement (StemCell Technologies). iPSC phenotype was examined using fluorochrome-conjugated antibodies against EGFR (to detect transgene), SSEA3, SSEA4, TRA1–60, TRA1–81, and CD30. T cells were harvested and stained as described previously(Jonnalagadda et al., 2015). T cell phenotype was examined using fluorochrome-conjugated antibodies against CD3, CD4, CD8α, CD8β, CD5, CD7, TCRαβ, TCRɣδ, CD16, CD56, CD27, CD28, NKP44, NKP46, NKG2A, NKG2D, CD178 (FasL), and CD19. CAR expression was determined by staining for the truncated EGFR. Memory-associated phenotypes were evaluated with fluorochrome-conjugated antibodies against CD45RO, CD45RA, and CD62L. To compare the phenotypes of CD8 single positive populations, cells were stained with anti-EGFRt (CAR) and anti-CD8/anti-CD4 antibodies and CD8+CAR+ or CD4+CAR+ T cells were sorted by FACS Aria SORP (BD).
T cell receptor Vβ staining was performed with the IOTest Beta Mark TCR Repertoire Kit (Beckman Coulter) which consists of monoclonal antibodies (mAbs) designed to identify 24 distinct TCR Vβ families. Each set consisted of three distinct anti-Vβ family-specific mAbs labelled with fluorescein isothiocyanate (FITC), phycoerythrin (PE) or doubly labelled with FITC and PE. The T cell population was also co-stained with APC-anti-CD3 antibody and the CD3-positive population was gated on for analysis.
Data were acquired on MacsQuant Analyzer 10 (Miltenyi Biotec) or Fortessa (Becton Dickinson) flow cytometers and analyzed with FlowJo (v10.6.1).
PCR based TCR β clonality assay
Genomic DNA was extracted by DNeasy kit (Qiagen) and used as PCR template. The PCR assay was set up according to the protocol of IdentiClone TCRB+TCRG T-Cell Clonality Assay Kit (Invivoscribe) (Langerak et al., 2012; van Dongen et al., 2003), TCRB tube A and B primer master mix target framework regions within the variable region and joining region of the TCR beta chain locus. TCRB Tube C targets the diversity and joining regions of the TCR beta chain locus. The specimen control size ladder master mix targets multiple genes and generates a series of amplicons to serve as quality control of input DNA. The primers are fluorescence labelled and fragment analysis was performed to detect the fragment size of PCR products concomitantly with regular DNA agarose gel examination.
In vitro T cell functional assays
Effector cells (iPSC CD19-CAR T, iPSC Mock T, conventional CD19-CAR T or conventional Mock T cells) were washed, resuspended in fresh medium containing 50 U/mL rhIL-2 and 0.5 ng/mL rhIL-15 and co-cultured in 96-well U-bottom plates with the indicated tumor cells at the indicated effector-to-target (E:T) ratios for 4 hours or 48 hours. Cytotoxic activity was then routinely evaluated by flow cytometry by enumerating viable (i.e., DAPI-negative) GFP-expressing tumor cells; for primary ALL cells, DAPI/CD19+ cells were enumerated. Alternatively, for luciferase based cytotoxicity assays, at each timepoint, D-luciferin potassium salt (PerkinElmer) was added to each well at a final concentration of 0.14 mg/mL and plates were incubated at 37°C for 10 minutes. Following the incubation with luciferin, the contents of each culture plate were mixed carefully and transferred to an opaque 96-well U-bottom plate with a multichannel pipettor. Bioluminescent flux was read with a Cytation 3 plate reader (Biotek). For each tumor line, replicate wells of tumor cells alone were used to generate internal MIN (0% viability) and MAX (100% viability) references for the calculation of percent lysis; the MIN was obtained by the addition of SDS to a final concentration of 1% ten minutes before the addition of luciferin (Brown et al., 2005).
To evaluate T cell activation, iPSC-derived or conventional CAR T or Mock T cells were incubated with the indicated tumor cells at an E:T ratio of 1:1 for five hours in the presence of CD107a antibody and GolgiStop protein transport inhibitor (BD Biosciences). Cells were then harvested, fixed, permeabilized, and stained for intracellular cytokines. Degranulation (CD107a staining) and intracellular cytokine staining (e.g. IFNγ) on CD3-gated cells was then examined by flow cytometry. Similar co-cultures without GolgiStop were harvested for staining of surface activation markers CD25 and CD137/4–1BB on CD3-gated cells was evaluated by flow cytometry.
To further characterize cytokine production, iPSC-derived or conventional CAR T or Mock T cells were co-incubated for 24 hours with the indicated NALM6 tumor cells at an E:T ratio of 1:1 in medium without added cytokines. Supernatants were collected and cytokine levels were quantified with the Cytokine 10-Plex Human Panel Kit (ThermoFisher Scientific) by a Bio-Plex reader (Bio-Rad). Similar co-cultures were harvested for flow cytometric analysis of surface activation markers CD25 and CD137/4–1BB on CD3-gated cells.
For In vitro repetative challenge assay, 105 CAR T cells were co-cultured with 4X105 CD19+ NALM6 cells at E:T ratio of 1:4, and re-challenged every other day with 4X105 NALM6 cells for 3 times. The cells were then stained with surface exhaustion markers PD-1, TIM-3 and LAG-3 together with T cells markers. Each of exhaustion marks was evaluated on CD3-gated cells by flow cytometry(Wang et al., 2019).
RNA and protein analysis
RNA was extracted with the Quick-RNA Microprep kit (Zymo Research) and treated with DNase I. RNA deep sequencing was performed by the City of Hope Integrative Genomics Core Facility. Briefly, stranded RNA-seq libraries were prepared using the KAPA mRNA HyperPrep kit (Roche), according to the manufacturer’s recommended protocol. Libraries were quantified using Qubit quantification kit (Thermofisher Scientific) and loaded onto the HiSeq 2500 sequencing platform (Illumina) for single-end 51-bp sequencing. Base calling was done using Illumina Real Time Analysis (RTA) v1.18.64.
For protein analysis by western blot, the harvested cells were lysed in RIPA buffer (ThermoFisher Scientific) and protein extraction was quantified with a BCA protein assay kit (ThermoFisher Scientific). The Bolt Mini Gel System (ThermoFisher Scientific) was used for gel electrophoresis and protein transfer. Anti-p44/42 MAPK (Erk1/2) and anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204); anti-PLCγ1, antiphospho-PLCγ1 (Tyr783) and anti-phospho-PLCγ1(Ser1248); anti-CD3ζ, and anti-phosphoCD3ζ(Y142); and anti-phospho-ZAP70 were used to interrogate CAR T and T cell signaling pathways (see Resource Table for antibody details).
Bioinformatics analysis of RNA seq data
To analyze the RNA seq data, the 2-D visualization of PCA was implemented using R package “DESeq2” (v.3.10) based on the PCA algorithm. Heatmaps of z-scores were generated by Cluster (v.3.0) and JavaTreeView (v.1.1.6r4) using a hierarchical clustering approach. Differentially expressed gene (DEG) analysis was performed with R package “edgeR” (v.3.28.0) (Robinson et al., 2010). The pipelines of deriving DEG involved the quantile-adjusted conditional maximum likelihood (qCML), and the quasilikelihood (QL) F-test. Bubble plots were acquired with R package “ggplot2” (v.3.2.1). The Gene Set Enrichment Analysis (GSEA) algorithm was run on GSEA (v.4.0.3) (Mootha et al., 2003; Subramanian et al., 2005). The resources of bioinformatic software packages are listed in the table of ‘Key resources’.
Bisulfite conversion, PCR, and sequencing
Genomic DNA was prepared by DNeasy kit (Qiagen). 500 ng genomic DNA was treated with sodium bisulfite to convert unmethylated cytosines using the EZ DNA Methylation-Lightning Kit (Zymo Research). Reactions were carried out per manufacture’s protocol. Methylation-specific PCR was performed using primers described in Supplemental Table 2. 245 bp PCR fragments of EF1a promoter from bisulfite converted gDNA of iPSC CD19-CAR T cells and conventional CD19-CAR T cells were amplified using primers described in Supplemental Table 2. The PCR fragments were subcloned into a pCR4-TOPO vector (Thermo Fisher Scientific) and six clones of each group were sequenced by Sanger Sequencing. The sequencing results were aligned to original and putative methylated sequences to determine the methylation status of CG sites.
QUANTIFICATION AND STATISTICAL ANALYSIS
For animal studies, Kaplan-Meier survival analysis was performed and statistical significance was calculated using log-rank (Mantel-Cox) tests. For other data analysis, Student’s t-tests were performed. Two-tailed analysis was used in all cases and P < 0.05 was considered statically significant. The statistical details and results were described in the figure legend.
Supplementary Material
Supplemental excel table 1(Table S1): RNA seq data of iPSC derived and conventional CD19-CAR T cells. Related to Figure 2.
Highlights:
3D-organoid culture supports differentiation of CAR+ iPSCs into functional CAR T cells
iPSC-derived CAR T cells demonstrate conventional αβ T cell phenotypes
iPSC-derived CAR T cells show lower CAR expression and a homogeneous TCR repertoire
iPSC-derived CAR T cells demonstrate potent anti-tumor activity in vitro and in vivo
Acknowledgments
This study was funded by Mustang Research Foundation (P.I. Stephen Forman) and City of Hope IDDV Foundation (P.I Christine Brown and Zhiqiang Wang). Research reported in this publication performed by City of Hope Core facilities was supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572.
Footnotes
Declaration of interests
C.B., S.J.F., and Z.W. are listed on patent(s) relating to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- Akbar AN, and Henson SM (2011). Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nature reviews Immunology 11, 289–295. [DOI] [PubMed] [Google Scholar]
- Alizadeh D, Wong RA, Yang X, Wang D, Pecoraro JR, Kuo CF, Aguilar B, Qi Y, Ann DK, Starr R, et al. (2019). IL15 Enhances CAR-T Cell Antitumor Activity by Reducing mTORC1 Activity and Preserving Their Stem Cell Memory Phenotype. Cancer Immunol Res 7, 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J, and Vos JM (2002). Establishment of an oriP/EBNA1-based episomal vector transcribing human genomic beta-globin in cultured murine fibroblasts. Gene Ther 9, 1447–1454. [DOI] [PubMed] [Google Scholar]
- Brady BL, Steinel NC, and Bassing CH (2010). Antigen receptor allelic exclusion: an update and reappraisal. Journal of immunology 185, 3801–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, Taylor C, Yeh R, Bartido S, BorquezOjeda O, et al. (2011). Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118, 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, Ostberg JR, Blanchard MS, Kilpatrick J, Simpson J, et al. (2016). Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. The New England journal of medicine 375, 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Wright CL, Naranjo A, Vishwanath RP, Chang WC, Olivares S, Wagner JR, Bruins L, Raubitschek A, Cooper LJ, et al. (2005). Biophotonic cytotoxicity assay for high-throughput screening of cytolytic killing. J Immunol Methods 297, 39–52. [DOI] [PubMed] [Google Scholar]
- Chin CJ, Cooper AR, Lill GR, Evseenko D, Zhu Y, He CB, Casero D, Pellegrini M, Kohn DB, and Crooks GM (2016). Genetic Tagging During Human Mesoderm Differentiation Reveals Tripotent Lateral Plate Mesodermal Progenitors. Stem cells 34, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman CJ, Senyukov VV, Somanchi SS, Phatarpekar PV, Kopp LM, Johnson JL, Singh H, Hurton L, Maiti SN, Huls MH, et al. (2012). Membrane-bound IL-21 promotes sustained ex vivo proliferation of human natural killer cells. PLoS One 7, e30264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depil S, Duchateau P, Grupp SA, Mufti G, and Poirot L. (2020). ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat Rev Drug Discov 19, 185–199. [DOI] [PubMed] [Google Scholar]
- Deuse T., Hu X., Gravina A., Wang D., Tediashvili G., De C., Thayer WO., Wahl A., Garcia JV., Reichenspurner H., et al. (2019). Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nature biotechnology 37, 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MLL, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, and Ryan MD (2001). Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol 82, 1013–1025. [DOI] [PubMed] [Google Scholar]
- Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR, et al. (2010). Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 107, 13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, and Sadelain M. (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemi S, Bedoya F, Nunez-Cruz S, June C, Melenhorst J, and Milone M. (2016). Shortened T Cell Culture with IL-7 and IL-15 Provides the Most Potent Chimeric Antigen Receptor (CAR)-Modified T Cells for Adoptive Immunotherapy. J Immunol 1, 23. [Google Scholar]
- Ghassemi S, Nunez-Cruz S, O’Connor RS, Fraietta JA, Patel PR, Scholler J, Barrett DM, Lundh SM, Davis MM, Bedoya F, et al. (2018). Reducing Ex Vivo Culture Improves the Antileukemic Activity of Chimeric Antigen Receptor (CAR) T Cells. Cancer Immunol Res 6, 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E, and Pfeifer A. (2006). Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol Ther 13, 59–66. [DOI] [PubMed] [Google Scholar]
- Hoyos V, Savoldo B, Quintarelli C, Mahendravada A, Zhang M, Vera J, Heslop HE, Rooney CM, Brenner MK, and Dotti G. (2010). Engineering CD19-specific T lymphocytes with interleukin-15 and a suicide gene to enhance their anti-lymphoma/leukemia effects and safety. Leukemia 24, 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nagata N, Kurokawa H, and Yamanaka S. (2014). iPS cells: a game changer for future medicine. The EMBO journal 33, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lin X, Feng Y, Xie Y, Han J, Zhang Y, Wang ZZ, and Chen T. (2010). Hemato-endothelial differentiation from lentiviral-transduced human embryonic stem cells retains durable reporter gene expression under the control of ubiquitin promoter. Cytotechnology 62, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonnalagadda M, Mardiros A, Urak R, Wang X, Hoffman LJ, Bernanke A, Chang WC, Bretzlaff W, Starr R, Priceman S, et al. (2015). Chimeric antigen receptors with mutated IgG4 Fc spacer avoid fc receptor binding and improve T cell persistence and antitumor efficacy. Molecular therapy : the journal of the American Society of Gene Therapy 23, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, and Milone MC (2018). CAR T cell immunotherapy for human cancer. Science 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, and Roderick JE (2018). NOTCH Signaling in T-Cell-Mediated Anti-Tumor Immunity and T-CellBased Immunotherapies. Frontiers in immunology 9, 1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zuniga-Pflucker JC, and Keller G. (2012). T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell reports 2, 1722–1735. [DOI] [PubMed] [Google Scholar]
- Kohl U, Arsenieva S, Holzinger A, and Abken H. (2018). CAR T Cells in Trials: Recent Achievements and Challenges that Remain in the Production of Modified T Cells for Clinical Applications. Hum Gene Ther 29, 559–568. [DOI] [PubMed] [Google Scholar]
- Langerak AW., Groenen PJ., Bruggemann M., Beldjord K., Bellan C., Bonello L., Boone E., Carter GI., Catherwood M., Davi F., et al. (2012). EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia 26, 2159–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanitis E, Rota G, Kosti P, Ronet C, Spill A, Seijo B, Romero P, Dangaj D, Coukos G, and Irving M. (2021). Optimized gene engineering of murine CAR-T cells reveals the beneficial effects of IL-15 coexpression. J Exp Med 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine BL, Miskin J, Wonnacott K, and Keir C. (2017). Global Manufacturing of CAR T Cell Therapy. Molecular therapy Methods & clinical development 4, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JK, Muffly LS, Spinner MA, Barnes JI, Owens DK, and Goldhaber-Fiebert JD (2019). Cost Effectiveness of Chimeric Antigen Receptor T-Cell Therapy in Multiply Relapsed or Refractory Adult Large B-Cell Lymphoma. J Clin Oncol 37, 2105–2119. [DOI] [PubMed] [Google Scholar]
- Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, Smith JP, Walker AJ, Kohler ME, Venkateshwara VR, et al. (2015). 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nature medicine 21, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Nagano S, Ichise H, Kataoka K, Yamada D, Ogawa S, Koseki H, Kitawaki T, Kadowaki N, Takaori-Kondo A, et al. (2016). Regeneration of CD8alphabeta T Cells from T-cell-Derived iPSC Imparts Potent Tumor Antigen-Specific Cytotoxicity. Cancer research 76, 6839–6850. [DOI] [PubMed] [Google Scholar]
- Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, Kishihara K, and Yasutomo K. (2003). Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity 19, 549–559. [DOI] [PubMed] [Google Scholar]
- McLellan AD, and Ali Hosseini Rad SM (2019). Chimeric antigen receptor T cell persistence and memory cell formation. Immunology and cell biology 97, 664–674. [DOI] [PubMed] [Google Scholar]
- Minagawa A, Yoshikawa T, Yasukawa M, Hotta A, Kunitomo M, Iriguchi S, Takiguchi M, Kassai Y, Imai E, Yasui Y, et al. (2018). Enhancing T Cell Receptor Stability in Rejuvenated iPSC-Derived T Cells Improves Their Use in Cancer Immunotherapy. Cell stem cell 23, 850–858.e854. [DOI] [PubMed] [Google Scholar]
- Montel-Hagen A, Seet CS, Li S, Chick B, Zhu Y, Chang P, Tsai S, Sun V, Lopez S, Chen HC, et al. (2019a). Organoid-Induced Differentiation of Conventional T Cells from Human Pluripotent Stem Cells. Cell stem cell 24, 376–389.e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montel-Hagen A, Seet CS, Li S, Chick B, Zhu Y, Chang P, Tsai S, Sun V, Lopez S, Chen HC, et al. (2019b). Organoid-Induced Differentiation of Conventional T Cells from Human Pluripotent Stem Cells. Cell stem cell 24, 376–389 e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Morgan MA, and Schambach A. (2018). Engineering CAR-T Cells for Improved Function Against Solid Tumors. Frontiers in immunology 9, 2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrozek E, Anderson P, and Caligiuri MA (1996). Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood 87, 2632–2640. [PubMed] [Google Scholar]
- Murry CE, and Keller G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680. [DOI] [PubMed] [Google Scholar]
- Nicholson IC, Lenton KA, Little DJ, Decorso T, Lee FT, Scott AM, Zola H, and Hohmann AW (1997). Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol Immunol 34, 1157–1165. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kaneko S, Kawana-Tachikawa A, Tajima Y, Goto H, Zhu D, Nakayama-Hosoya K, Iriguchi S, Uemura Y, Shimizu T, et al. (2013). Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell stem cell 12, 114–126. [DOI] [PubMed] [Google Scholar]
- Nowicki TS, Farrell C, Morselli M, Rubbi L, Campbell KM, Macabali MH, Berent-Maoz B, Comin-Anduix B, Pellegrini M, and Ribas A. (2020). Epigenetic Suppression of Transgenic T-cell Receptor Expression via Gamma-Retroviral Vector Methylation in Adoptive Cell Transfer Therapy. Cancer Discov 10, 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., and Yamanaka S. (2013). An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem cells 31, 458–466. [DOI] [PubMed] [Google Scholar]
- Palazon A, Tyrakis PA, Macias D, Velica P, Rundqvist H, Fitzpatrick S, Vojnovic N, Phan AT, Loman N, Hedenfalk I, et al. (2017). An HIF-1alpha/VEGF-A Axis in Cytotoxic T Cells Regulates Tumor Progression. Cancer cell 32, 669–683 e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlacky J, and Polak J. (2020). Technical Feasibility and Physiological Relevance of Hypoxic Cell Culture Models. Front Endocrinol (Lausanne) 11, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot L, Philip B, Schiffer-Mannioui C, Le Clerre D, Chion-Sotinel I, Derniame S, Potrel P, Bas C, Lemaire L, Galetto R, et al. (2015). Multiplex Genome-Edited T-cell Manufacturing Platform for “Off-the-Shelf” Adoptive T-cell Immunotherapies. Cancer research 75, 3853–3864. [DOI] [PubMed] [Google Scholar]
- Popplewell L, X W, S. B, J W, Naranjo A, A. A, J P, L L, W C, E B, et al. (2018). CD19-CAR Therapy Using Naive/Memory or Central Memory T Cells Integrated into the Autologous Stem Cell Transplant Regimen for Patients with B-NHL. Blood 132, 610. [Google Scholar]
- Porter DL, Levine BL, Kalos M, Bagg A, and June CH (2011). Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. The New England journal of medicine 365, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Programs I. o. (2019). Programmed Cellular Immunotherapies Overview of Immuno-Oncology Programs Forward-Looking Statements. [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruella M, Xu J, Barrett DM, Fraietta JA, Reich TJ, Ambrose DE, Klichinsky M, Shestova O, Patel PR, Kulikovskaya I, et al. (2018). Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nature medicine 24, 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter AI, Ivey RG, Kennedy JJ, Voillet V, Rajan A, Alderman EJ, Voytovich UJ, Lin C, Sommermeyer D, Liu L, et al. (2018). Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Science signaling 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samer K, Khaled M, Suzette Blanchard, Xiuli Wang, Jamie Wagner, Araceli Naranjo, Jennifer Simpson, Sandra Thomas, Julie Ostberg, Christine Brown, Forman Stephen J (2018). Adult Patients with ALL Treated with CD62L+ T Naïve/Memory-Enriched T Cells Expressing a CD19-CAR Mediate Potent Antitumor Activity with a Low Toxicity Profile. Blood 132 [Google Scholar]
- Singer A, Adoro S, and Park JH (2008). Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nature reviews Immunology 8, 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Shou P, Du H, Hirabayashi K, Chen Y, Herring LE, Ahn S, Xu Y, Suzuki K, Li G, et al. (2020). THEMIS-SHP1 Recruitment by 4–1BB Tunes LCK-Mediated Priming of Chimeric Antigen Receptor-Redirected T Cells. Cancer cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themeli M, Kloss CC, Ciriello G, Fedorov VD, Perna F, Gonen M, and Sadelain M. (2013). Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nature biotechnology 31, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themeli M, Riviere I, and Sadelain M. (2015). New cell sources for T cell engineering and adoptive immunotherapy. Cell stem cell 16, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen DS, and Schumacher TN (2018). T Cell Dysfunction in Cancer. Cancer cell 33, 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans F, Velghe I, Vanwalleghem L, De Smedt M, Van Coppernolle S, Taghon T, Moore HD, Leclercq G, Langerak AW, Kerre T, et al. (2009). Generation of T cells from human embryonic stem cellderived hematopoietic zones. Journal of immunology 182, 6879–6888. [DOI] [PubMed] [Google Scholar]
- Urak R, Walter M, Lim L, Wong CW, Budde LE, Thomas S, Forman SJ, and Wang X. (2017). Ex vivo Akt inhibition promotes the generation of potent CD19CAR T cells for adoptive immunotherapy. J Immunother Cancer 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JJ., Langerak AW., Bruggemann M., Evans PA., Hummel M., Lavender FL., Delabesse E., Davi F., Schuuring E., Garcia-Sanz R., et al. (2003). Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 17, 2257–2317. [DOI] [PubMed] [Google Scholar]
- Vizcardo R, Klemen ND, Islam SMR, Gurusamy D, Tamaoki N, Yamada D, Koseki H, Kidder BL, Yu Z, Jia L, et al. (2018). Generation of Tumor Antigen-Specific iPSC-Derived Thymic Emigrants Using a 3D Thymic Culture System. Cell reports 22, 3175–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcardo R, Masuda K, Yamada D, Ikawa T, Shimizu K, Fujii S, Koseki H, and Kawamoto H. (2013). Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8(+) T cells. Cell stem cell 12, 31–36. [DOI] [PubMed] [Google Scholar]
- Wang D, Aguilar B, Starr R, Alizadeh D, Brito A, Sarkissian A, Ostberg JR, Forman SJ, and Brown CE (2018). Glioblastoma-targeted CD4+ CAR T cells mediate superior antitumor activity. JCI insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Starr R, Alizadeh D, Yang X, Forman SJ, and Brown CE (2019). In Vitro Tumor Cell Rechallenge For Predictive Evaluation of Chimeric Antigen Receptor T Cell Antitumor Function. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Starr R, Chang WC, Aguilar B, Alizadeh D, Wright SL, Yang X, Brito A, Sarkissian A, Ostberg JR, et al. (2020). Chlorotoxin-directed CAR T cells for specific and effective targeting of glioblastoma. Sci Transl Med 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. (2011a). The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Berger C, Wong CW, Forman SJ, Riddell SR, and Jensen MC (2011b). Engraftment of human central memory-derived effector CD8+ T cells in immunodeficient mice. Blood 117, 1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chang WC, Wong CW, Colcher D, Sherman M, Ostberg JR, Forman SJ, Riddell SR, and Jensen MC (2011c). A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118, 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Naranjo A, Brown CE, Bautista C, Wong CW, Chang WC, Aguilar B, Ostberg JR, Riddell SR, Forman SJ, et al. (2012). Phenotypic and functional attributes of lentivirus-modified CD19-specific human CD8+ central memory T cells manufactured at clinical scale. Journal of immunotherapy 35, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Popplewell LL, Wagner JR, Naranjo A, Blanchard MS, Mott MR, Norris AP, Wong CW, Urak RZ, Chang WC, et al. (2016). Phase 1 studies of central memory-derived CD19 CAR T-cell therapy following autologous HSCT in patients with B-cell NHL. Blood 127, 2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wong CW, Urak R, Mardiros A, Budde LE, Chang WC, Thomas SH, Brown CE, La Rosa C, Diamond DJ, et al. (2015). CMVpp65 Vaccine Enhances the Antitumor Efficacy of Adoptively Transferred CD19Redirected CMV-Specific T Cells. Clin Cancer Res 21, 2993–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jacoby E, and Fry TJ (2015). Challenges and opportunities of allogeneic donor-derived CAR T cells. Current opinion in hematology 22, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zah E, Nam E, Bhuvan V, Tran U, Ji BY, Gosliner SB, Wang X, Brown CE, and Chen YY (2020). Systematically optimized BCMA/CS1 bispecific CAR-T cells robustly control heterogeneous multiple myeloma. Nat Commun 11, 2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sandy AR., Wang J., Radojcic V., Shan GT., Tran IT., Friedman A., Kato K., He S., Cui S., et al. (2011). Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood 117, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Parkhurst MR, Zheng Z, Cohen CJ, Riley JP, Gattinoni L, Restifo NP, Rosenberg SA, and Morgan RA (2007). Extrathymic generation of tumor-specific T cells from genetically engineered human hematopoietic stem cells via Notch signaling. Cancer research 67, 2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental excel table 1(Table S1): RNA seq data of iPSC derived and conventional CD19-CAR T cells. Related to Figure 2.
Data Availability Statement
The RNA seq datasets generated in this study have been deposited at NCBI GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human SSEA-3 | BD Biosciences | (clone MC-6310) RRID:AB_1645542 |
| Anti-human SSEA-4 | BD Biosciences | (clone MC813–70) RRID:AB_2033991 |
| Anti-human Tra-1–60 | BD Biosciences | (clone TRA-1–60) RRID:AB_10565983 |
| Anti-human TRA-1–81 | BD Biosciences | (clone TRA-1–81) RRID:AB_1645493 |
| Anti-human EpCAM (CD326) | BioLegend | (clone 9C4) RRID:AB_756082 |
| Anti-human CD56 | BD Biosciences | (clone NCAM16.2) RRID:AB_400025 |
| Anti-human CD30 | BD Biosciences | (clone BerH8) RRID:AB_393541 |
| Anti-human CD3 (BV510) | BD Biosciences | (clone UCHT1) RRID:AB_2732053 |
| Anti-human CD3 (APC) | BD Biosciences | (clone SK7) RRID:AB_400513 |
| Anti-human CD4 (Per-CP) | BD Biosciences | (clone SK3) RRID:AB_400282 |
| Anti-human CD4 (PE-Cy7) | BD Biosciences | (clone SK3) RRID:AB_396897 |
| Anti-human CD8α (PE) | BD Biosciences | (clone SK1) RRID:AB_400005 |
| Anti-human CD8α (Per-CP) | BD Biosciences | (clone SK1) RRID:AB_400280 |
| Anti-human CD8α (APC-Cy7) | BD Biosciences | (clone SK1) RRID:AB_400383 |
| Anti-human CD8β | BD Biosciences | (clone 2ST8.5H7) RRID:AB_1645747 |
| Anti-human CD5 | BD Biosciences | (clone UCHT2) RRID:AB_2737700 |
| Anti-human CD7 | BD Biosciences | (clone M-T701) RRID:AB_2738544 |
| Anti-human TCR αβ | BD Biosciences | (clone T10B9.1A-31) RRID:AB_2738437 |
| Anti-human TCR γδ | BD Biosciences | (clone B1) RRID:AB_396061 |
| Anti-human TCR γδ | BD Biosciences | (clone 11F2) RRID:AB_2827402 |
| Anti-human NKG2A | R&D Systems | (clone 131411) RRID:AB_2132978 |
| Anti-human NKG2D (CD314) | BD Biosciences | (clone 1D11) RRID:AB_10896282 |
| Anti-human NKp44 (CD336) | BD Biosciences | (clone p44–8) RRID:AB_647239 |
| Anti-human NKP46 (CD335) | BioLegend | (clone 9E2) RRID:AB_2561649 |
| Anti-human CD19 | BD Biosciences | (clone SJ25C1) RRID:AB_396893 |
| Anti-human CD16 | BD Biosciences | (clone NKP15 ) RRID:AB_400317 |
| Anti-human Fas L (CD178) | BD Biosciences | (clone NOK-1) RRID:AB_2738713 |
| Anti-human CD62L (PE) | BD Biosciences | (clone SK11) RRID:AB_400205 |
| Anti-human CD62L (FITC) | BD Biosciences | (clone SK11) RRID:AB_400302 |
| Anti-human CD45RA | BD Biosciences | (clone HI100) RRID:AB_398468 |
| Anti-human CD45RO | BD Biosciences | (clone UCHL1) RRID:AB_395883 |
| Anti-human EGFR (APC) | BioLegend | (clone AY13) RRID:AB_11150410 |
| Anti-human EGFR (PE-Cy7) | BioLegend | (clone AY13) RRID:AB_2562159 |
| Anti-human CD28, InVivoMab | Bio X Cell | (clone 9.3) RRID:AB_2687729 |
| Anti- human CD27 | BD Biosciences | (clone M-T271) RRID:AB_395834 |
| Anti-human CD28 (APC-H7) | BD Biosciences | (clone CD28.2) RRID:AB_11154032 |
| Anti-human CD279 (PD-1) | ThermoFisher Scientific | (clone eBioJ105) RRID:AB_2573976 |
| Anti-human LAG3 /CD223 | Lifespan | (clone 17B4) RRID:AB_1650048 |
| Anti-human TIM-3 | R&D Systems | (clone 344823) RRID:AB_2232901 |
| Anti-human IgG Fc | Abcam | Abcam # ab98622, RRID:AB_10673586 |
| Pan p44/42 MAPK (phosphorylated Erk1/2) antibody | Cell Signaling Technology | Polyclonal, Cat# 9101, RRID:AB_331646 |
| Pan p44/42 MAPK (Erk1/2) antibody | Cell Signaling Technology | (clone 3A7) RRID:AB_10695739 |
| Anti-human, -mouse phospho-PLCγ1 (Tyr783) antibody | Cell Signaling Technology | (clone D6M9S) RRID:AB_2728690 |
| Anti-human, -mouse, -monkey phospho-PLC 1 (Ser1248) antibody | Cell Signaling Technology | (clone D25A9) RRID:AB_10890863 |
| Anti-human, -rat, -mouse PLCγ1 | Cell Signaling Technology | Polyclonal, Cat# 2822, RRID:AB_2163702 |
| Anti-human, -mouse phospho-Zap-70 (Tyr319)/Syk (Tyr352) | Cell Signaling Technology | (clone 65E4) RRID:AB_2218658 |
| Anti-human, -mouse, anti-CD3ζ (pY142) | BD Biosciences | (clone K25–407.69) RRID: AB_647307 |
| Bacterial and virus strains | ||
| NEB 5-alpha Competent E. coli | NEB | C2987 |
| Biological samples | ||
| PBMC from healthy donor | This paper | N/A |
| Primary B-ALL patient samples | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Proleukin (aldesleukin), rhIL-2 | Novartis Oncology | NCD 65483–116-07 |
| rhIL-15 | CellGenix, US Operations | 1413–050 |
| Activin A | R&D Systems | 338-AC-010 |
| rhBMP-4 | R&D Systems | 314-BP-010 |
| rhVEGF | R&D Systems | 293-VE-010 |
| rhFGF-basic | Peprotech | AF-100–18B |
| ROCK inhibitor, Y-27632 dihydrochloride | StemCell Technologies | 72304 |
| TGFβ-RI inhibitor, SB-431542 | StemCell Technologies | 72234 |
| rhFlt3-Ligand | Peprotech | AF-300–19 |
| rhIL-7 | Peprotech | AF-200–07 |
| rhTPO | Peprotech | 300–18 |
| rhSCF | Peprotech | AF-300–07 |
| MEM α, with nucleosides | GIBCO | 12571063 |
| Fetal bovine serum, defined, heat-inactivated | HyClone | SH30070.03IH |
| X-VIVO 15 Serum-free Hematopoietic Cell Medium | Lonza | BEBP04–744Q |
| EGM-2, Endothelial Cell Growth Medium-2 Bullet Kit | Lonza | CC-3162 |
| RPMI 1640 Medium with L-Glutamine and HEPES | Lonza | 12–115F |
| GLutaMAX Supplement | ThermoFisher Scientific | 35050061 |
| 2-Mercaptoethanol, 55 mM | ThermoFisher Scientific | 21985023 |
| MEM Non-Essential Amino Acids Solution (100X) | ThermoFisher Scientific | 11140050 |
| L-Ascorbic acid 2-phosphate sesquimagnesium salt hydrate | Sigma | A8960 |
| Phosphate-buffered saline | Irvine Scientific | 9240 |
| Penicillin-Streptomycin (100X) | Lonza | 17602E |
| mTeSR1 | StemCell Technologies | 85850 |
| DMEM/F12, with HEPES | ThermoFisher Scientific | 11330057 |
| Matrigel hESC-Qualified Matrix, LDEV-free | Corning | 354277 |
| ReLeSR | StemCell Technologies | 5872 |
| CloneR | StemCell Technologies | 5889 |
| StemPro Accutase Cell Dissociation Reagent | ThermoFisher Scientific | A1110501 |
| DNase I | Zymo Research | E1010 |
| B-27 Supplement (50X) | ThermoFisher Scientific | 17504–044 |
| TrypLE Express Enzyme (1X) | ThermoFisher Scientific | 12605010 |
| Protamine sulfate | APP Pharmaceuticals | NDC 63323–229-05 |
| XenoLight D-luciferin potassium salt | PerkinElmer | 122799 |
| Hanks’ Balanced Salt Solution, w/o Ca++, Mg++ | GIBCO | 14175103 |
| 0.05% Trypsin/0.53mM EDTA in HBSS w/o Ca++, Mg++ | Corning | MT25051CI |
| Brefeldin A Golgi Plug | BD Biosciences | 555029 |
| Critical commercial assays | ||
| Leukocyte Alkaline Phosphatase Kit | Sigma | 86R-1KT |
| IOTest beta Mark TCR Vβ Repertoire Kit | Beckman Coulter | IM3497 |
| TCRB T-Cell Clonality Assay Kit | Invivoscribe | 12000011 |
| CD34 MicroBead Kit UltraPure | Miltenyi Biotec | 130–100-453 |
| EasySep Human CD56 Positive Selection Kit II | Stem Cell Technologies | 17855 |
| EasySep Human EpCAM Positive Selection Kit II | Stem Cell Technologies | 17846 |
| Dynabeads™ Human T-Expander CD3/CD28 | ThermoFisher Scientific | 11141D |
| RNeasy Micro Kit | Qiagen | 74004 |
| Quick-RNA Microprep Kit | Zymo Research | R1050 |
| EZ DNA Methylation-Lightning Kit | Zymo Research | D5030 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | 554714 |
| Luminex Cytokine Mouse Magnetic 10-Plex Panel | ThermoFisher Scientific | LMC0001M |
| Millicell Cell Culture Insert, 30 mm, hydrophilic PTFE, 0.4 μm | Millipore Sigma | PICM0RG50 |
| Deposited data | ||
| RNA expression data by high throughput sequencing | This paper | NCBI GEO: GSE193422 |
| Experimental models: Cell lines | ||
| T cell derived iPSC lines | this paper | N/A |
| Fibroblast derived iPSC line | ATCC | DYS0100, ACS-1019 |
| MS5-hDLL4-eGFP | Gay Crooks’ lab | N/A |
| NALM6.ffluc.eGFP | this paper | N/A |
| NALM6.ffluc.eGFP_CD19KO | this paper | N/A |
| Raji.ffluc.eGFP | this paper | N/A |
| Raji.ffluc.eGFP_CD19KO | this paper | N/A |
| 3T3_CD19 | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) | Jackson Laboratory | Strain #005557 |
| Oligonucleotides | ||
| Primers for EF1α promoter PCR and sequencing, see Extended Table 2 | This paper | N/A |
| Recombinant DNA | ||
| CD19-CAR_pHIV (CD19-CAR) | Forman lab | N/A |
| CLTX-CAR_pHIV (CLTX-CAR) | Forman lab | N/A |
| hOCT3/4-shp53_pCXLE | addgene, gift from Shinya Yamanaka | RRID:Addgene_27077 |
| hSK_pCXLE (SOX2, KLF4) | addgene, gift from Shinya Yamanaka | RRID:Addgene_27078 |
| hUL_pCXLE (L-MYC, LIN28) | addgene, gift from Shinya Yamanaka | RRID:Addgene_27080 |
| EBNA_pCXLE | addgene, gift from Shinya Yamanaka | RRID:Addgene_37624 |
| Software and algorithms | ||
| GraphPad Prism GraphPad Software | GraphPad Software | https://www.graphpad.com/ |
| Flowjo | Tree Star Inc. | https://www.flowjo.com/solutions/flowjo |
| Zen | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| BZ-X800 Analyzer | Keyence | https://www.keyence.com/landing/microscope/lp_fluorescence.jsp |
| R package “DESeq2” (v.3.10) |
R | https://www.r-project.org |
| Cluster (v.3.0) (clustering algorithms) |
Human Genome Center Institute of Medical Science, University of Tokyo | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| JavaTreeView (v.1.1.6r4) | Stanford University School of Medicine | http://jtreeview.sourceforge.net/ |
| “edgeR” (v.3.28.0) | R | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| “ggplot2” (v.3.2.1) | R | https://cran.r-project.org/web/packages/ggplot2/index.html |
| GSEA (v.4.0.3). (gene set enrichment analysis) | Broad Institute | https://www.gsea-msigdb.org/gsea/index.jsp |
| BioRender | BioRender.Inc | https://app.biorender.com/ |
| Other | ||
| N/A | ||
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.