Abstract
The emergence of several zoonotic viruses in the last twenty years, especially the pandemic outbreak of SARS-CoV-2, has exposed a dearth of antiviral drug therapies for viruses with pandemic potential. Developing a diverse drug portfolio will be critical to rapidly respond to novel coronaviruses (CoVs) and other viruses with pandemic potential. Here we focus on the SARS-CoV-2 conserved macrodomain (Mac1), a small domain of non-structural protein 3 (nsp3). Mac1 is an ADP-ribosylhydrolase that cleaves mono-ADP-ribose (MAR) from target proteins, protects the virus from the anti-viral effects of host ADP-ribosyltransferases, and is critical for the replication and pathogenesis of CoVs. In this study, a luminescent-based high-throughput assay was used to screen ∼38,000 small molecules for those that could inhibit Mac1-ADP-ribose binding. We identified 5 compounds amongst 3 chemotypes that inhibit SARS-CoV-2 Mac1-ADP-ribose binding in multiple assays with IC50 values less than 100 μM, inhibit ADP-ribosylhydrolase activity, and have evidence of direct Mac1 binding. These chemotypes are strong candidates for further derivatization into highly effective Mac1 inhibitors.
Keywords: Coronavirus, SARS-CoV-2, Macrodomain, ADP-Ribose, ADP-Ribosylation, High-throughput screening
Graphical abstract
1. Introduction
COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is one of the most disruptive and deadly pandemics in modern times, with greater than 515 million cases and having led to greater than 6.2 million deaths worldwide. SARS-CoV-2 is the third CoV to emerge into the human population in the last 3 decades, following outbreaks of SARS-CoV in 2002–2003 and Middle East respiratory syndrome MERS-CoV in 2012 (Perlman, 2020). These outbreaks highlight the potential for CoVs to cross-species barriers and cause severe disease in a new host. There is a tremendous need to develop broad-spectrum antiviral therapies capable of targeting a wide range of CoVs to prevent severe disease following zoonotic outbreaks.
Coronaviruses encode for 16 highly conserved, non-structural proteins that are processed from two polyproteins, 1a and 1 ab (pp1a and pp1ab) (Fehr and Perlman, 2015). The largest non-structural protein is non-structural protein 3 (nsp3) that encodes for multiple modular protein domains. Both the SARS-CoV and the SARS-CoV-2 nsp3 proteins include three tandem macrodomains, Mac1, Mac2, and Mac3 (Srinivasan et al., 2020). Mac1 is present in all CoVs, unlike Mac2 and Mac3, and contains a conserved three-layered α/β/α fold, a common feature amongst all macrodomains. All CoV Mac1 proteins tested have mono-ADP-ribosylhydrolase (ARH) activity, though it remains unclear if they have significant poly-ARH activity (Alhammad et al., 2021; Cho et al., 2016; Egloff et al., 2006; Putics et al., 2005; Saikatendu et al., 2005; Xu et al., 2009). In contrast, Mac2 and Mac3 fail to bind ADP-ribose and instead bind to nucleic acids (Chatterjee et al., 2009; Tan et al., 2009). Mac1 homologs are also found in alphaviruses, Hepatitis E virus, and Rubella virus, indicating that ADP-ribosylation may be a potent anti-viral post-translational modification (PTM) (Makrynitsa et al., 2019; Malet et al., 2009). All viral macrodomains are members of the larger MacroD-type macrodomain family, which includes human macrodomains Mdo1 and Mdo2 (Rack et al., 2016).
ADP-ribosylation is a post-translational modification catalyzed by ADP-ribosyltransferases (ARTs, also known as PARPs) through transferring an ADP-ribose moiety from NAD+ onto target proteins or nucleic acids (Kim et al., 2020). ADP-ribose is either transferred as a single mono-ADP-ribose (MAR) unit, or it is transferred consecutively and covalently attached through glycosidic bonds to preceding ADP-ribose units to form a poly-ADP-ribose (PAR) chain. Both mono- and poly-ARTs inhibit virus replication, implicating ADP-ribosylation in the host-response to infection (Brady et al., 2019).
Several reports have addressed the role of Mac1 on the replication and pathogenesis of CoVs, mostly using the mutation of a highly conserved asparagine to alanine (N41A-SARS-CoV). This mutation abolished the MAR-hydrolase activity of SARS-CoV Mac1 (Fehr et al., 2016). This mutation has minimal effects on CoV replication in transformed cells, but reduces viral load, leads to enhanced IFN production, and strongly attenuates both murine hepatitis virus (MHV) and SARS-CoV in mouse models of infection (Eriksson et al., 2008; Fehr et al., 2015, 2016; Putics et al., 2005). More recently, we have identified mutations in the MHV-JHM Mac1 domain, predicted to abolish ADP-ribose binding, that resulted in severe replication defects in cell culture, indicating that for some CoVs Mac1 may be even more important than previously appreciated (Voth et al., 2021). Mutations in the alphavirus and HEV macrodomain also have substantial phenotypic effects on virus replication and pathogenesis (Abraham et al., 2018, 2020; Li et al., 2016; McPherson et al., 2017; Parvez, 2015).
As viral macrodomains are critical virulence factors, they are unique targets for anti-viral therapeutics (Alhammad and Fehr, 2020). Several studies have reported structures that could potentially bind to the ADP-ribose binding pocket of SARS-CoV-2 Mac1. While most of these studies were limited to in silico studies, a few have tested compound activity in biochemical assays, but have been met with minimal success (Dasovich et al., 2022; Ni et al., 2021; Russo et al., 2021; Schuller et al., 2021; Sowa et al., 2021; Virdi et al., 2020). Interestingly, the remdesivir metabolite GS-441524 bound to Mac1, though it was not tested for its ability to inhibit Mac1-ADP-ribose binding or hydrolysis activity (Ni et al., 2021). In another study, several small fragments were identified in a crystallography screen which could be good starting points for further inhibitor development (Schuller et al., 2021). However, the only compounds identified thus far that inhibit SARS-CoV-2 Mac1 with IC50 less than 100 μM are Suramin, which inhibited Mac1-ADP-ribose binding in a FRET assay with an IC50 of 8.7 μM, and Dasatanib, which inhibited Mac1 mono-ARH activity with an IC50 of ∼50 μM. Suramin targeted several divergent macrodomains and is known to have additional targets, and thus is not suitable for further evaluation (Sowa et al., 2021). Dasatinib is not a candidate for a Mac1 inhibitor as it is toxic to mammalian cells, though it may provide a scaffold for further inhibitor development. None of the identified compounds have been tested for their ability to inhibit Mac1 in cell culture or in animal models of disease.
Here, we optimized two high-throughput macrodomain-ADP-ribose binding assays, a previously described luminescent-based AlphaScreen™ (AS) assay, and a novel fluorescence polarization (FP) assay (Schuller et al., 2017, 2021). We used the AS assay to screen ∼38,000 compounds for their ability to inhibit SARS-CoV-2 Mac1-ADP-ribose binding. We identified 5 compounds from 3 chemotypes that inhibited ADP-ribose binding by the SARS-CoV-2 Mac1 protein in both assays, some with IC50 values as low as 5–10 μM. These compounds also demonstrated some inhibition of ARH activity and have evidence of direct binding to Mac1. The profiling of the most potent inhibitor against a panel of virus and human MAR binding and hydrolyzing proteins revealed remarkable selectivity for the inhibition of SARS-CoV-2 Mac1. These compounds represent several series that can be further developed into potent Mac1 inhibitors and potential therapeutics for SARS-CoV-2 and other CoVs of interest.
2. Methods
2.1. Reagents
All plasmids and proteins used were expressed and purified as previously described (Alhammad et al., 2021; Dasovich et al., 2022; Sowa et al., 2021, 2022). All compounds were repurchased from MolPort except for compounds 6 and 10, which were repurchased from ChemDiv. After reordering once, compounds 10 and 11 became unavailable and thus were resynthesized according to the literature (Capobianco et al., 2021). ADP-ribosylated peptides were purchased from Cambridge peptides.
2.2. Differential scanning fluorimetry (DSF)
Thermal shift assay with DSF involved use of LightCycler® 480 Instrument (Roche Diagnostics). In total, a 15 μL mixture containing 8× SYPRO Orange (Invitrogen), and 10 μM macrodomain protein in buffer containing 20 mM HEPES-NaOH, pH 7.5 and various concentrations of ADP-ribose or hit compounds were mixed on ice in 384-well PCR plate (Roche). Fluorescent signals were measured from 25 to 95 °C in 0.2 °C/30/Sec steps (excitation, 470–505 nm; detection, 540–700 nm). The main measurements were carried out in triplicate. Data evaluation and Tm determination involved use of the Roche LightCycler® 480 Protein Melting Analysis software, and data fitting calculations involved the use of single site binding curve analysis on GraphPad Prism. The thermal shift (ΔTm) was calculated by subtracting the Tm values of the DMSO from the Tm values of compounds.
2.3. AlphaScreen (AS) assay
The AlphaScreen reactions were carried out in 384-well plates (Alphaplate, PerkinElmer, Waltham, MA) in a total volume of 40 μL in buffer containing 25 mM HEPES (pH 7.4), 100 mM NaCl, 0.5 mM TCEP, 0.1% BSA, and 0.05% CHAPS. All reagents were prepared as 4× stocks and 10 μL volume of each reagent was added to a final volume of 40 μL. All compounds were transferred acoustically using ECHO 555 (Beckman Inc) and preincubated after mixing with purified His-tagged macrodomain protein (250 nM) for 30 min at RT, followed by addition of a 10 amino acid biotinylated and ADP-ribosylated peptide [ARTK(Bio)QTARK (Aoa-RADP)S] (Cambridge peptides) (625 nM). After 1 h incubation at RT, streptavidin-coated donor beads (7.5 μg/mL) and nickel chelate acceptor beads (7.5 μg/mL); (PerkinElmer AlphaScreen Histidine Detection Kit) were added under low light conditions, and plates were shaken at 400 rpm for 60 min at RT protected from light. Plates were kept covered and protected from light at all steps and read on BioTek plate reader using an AlphaScreen 680 excitation/570 emission filter set. For counter screening of the compounds, 25 nM biotinylated and hexahistidine-tagged linker peptide (Bn-His6) (PerkinElmer) was added to the compounds, followed by addition of beads as described above. For data analysis, the percent inhibition was normalized to positive (DMSO + labeled peptide) and negative (DMSO + macrodomain + peptide, no ADPr) controls. The IC50 values were calculated via four-parametric non-linear regression analysis constraining bottom (=0), top (=100), & Hillslope (=1) for all curves.
2.4. Fluorescence polarization (FP) assay
The FP assay was performed in buffer containing 25 mM Tris pH7.5, 50 mM NaCl, 0.025% TritonX-100. All reagents were prepared as 2× stocks and 10 μL volume of each reagent was added to a final volume of 20 μL. Compounds were preincubated with His-Macrodomain proteins (4 μM) for 30’, RT in black 384 well plates (Corning 3575 plates), followed by addition of 50 nM of fluorescein labeled ADP-ribosylated peptide [5 Flu-ARTKQTARK (Aoa-RADP)S]. After mixing for a minute, the plate was incubated at 25 °C, protected from light and fluorescence polarization was read after 30 min, 1 h and 2 h using a plate reader. Data analysis was done as described for the AS assay.
2.5. Gel-based inhibition of mono-ADP-ribosylhydrolase activity (de-MARylation)
PARP10-CD protein was auto-MARylated through incubation for 20 min at 37 °C with 1 mM final concentration of β-Nicotinamide Adenine Dinucleotide (β NAD+) (Millipore-Sigma) in a reaction buffer (50 mM HEPES, 150 mM NaCl, 0.2 mM DTT, and 0.02% NP-40). MARylated PARP10-CD was aliquoted and stored at −80 °C. To test the ability of identified compounds for their ability to inhibit MARylation activity of Mac1, we first incubated each compound with purified SARS-CoV-2 Mac1 in the reaction buffer at 37 °C for 30 min. Then, MARylated PARP10-CD was added to this mixture solution and further incubated for 30 min at 37 °C. The reaction was stopped with addition of 2× Laemmli sample buffer containing 10% β-mercaptoethanol. Protein samples were heated at 95 °C for 5 min before loading and separated onto SDS-PAGE cassette (Thermo Fisher Scientific Bolt™ 4–12% Bis-Tris Plus Gels) in MES running buffer. For immunoblotting, the separated proteins were transferred onto polyvinylidene difluoride (PVDF) membrane using iBlot™ 2 Dry Blotting System (ThermoFisher Scientific). The blot was blocked with 5% skim milk in 1xPBS and probed with the anti-mono-ADP-ribose binding reagent/antibody MABE1076 (α-MAR), and anti-GST tag monoclonal antibody MA4-004 (ThermoFisher Scientific). The primary antibodies were detected with secondary anti-rabbit and anti-mouse antibodies (LI-COR Biosciences). All immunoblots were visualized using Odyssey® CLx Imaging System (LI-COR Biosciences). The images were quantitated using LI-COR Image Studio software.
2.6. ADP-ribosylhydrolase assay
The recently published assay, ADPr-Glo, was used to examine the impact of our top hit compounds on SARS-CoV-2 enzymatic activity (Dasovich et al., 2022). Briefly, the compounds were preincubated with SARS-CoV-2 Mac1 (2 nM) and NudF (125 nM) at ambient temperature for 30 min prior to the addition of MARylated PARP10-CD derived substrate. The substrate (20 μM) was then incubated with the SARS-CoV-2 Mac1 and NudF at ambient temperature for 30 min. The reaction products were measured with AMP-Glo. To confirm that our compounds do not inhibit NudF or interfere with AMP detection by AMP-Glo, we used a counter screen where 2 μM ADP-ribose was used instead of the ADP-ribosylated substrate and the macrodomain was omitted from the reaction. Luminescence signal was converted to AMP concentration via interpolation from an AMP standard curve. Data plotted are AMP generated by the macrodomain and NudF, subtracted by AMP generated from NudF alone. Percent inhibition was calculated and non-linear regression analysis was performed in GraphPad Prism.
2.7. A FRET based binding assay and inhibitor profiling
FRET was utilized for the profiling compound 6 with a panel of human and viral macrodomains to determine their specificity (Sowa et al., 2021, 2022). The assay is based on the site-specific introduction of cysteine-linked mono-ADP-ribose to the C-terminal Gαi peptide (GAP) by Pertussis toxin subunit 1 (PtxS1) fused to YFP. To generate the FRET signal ADP-ribosyl binders were fused to CFP. Samples were prepared in the assay buffer (for most binders; 10 mM Bis-Tris propane pH 7.0, 3% (w/v) PEG 20,000, 0.01% (v/v) Triton X-100 and 0.5 mM TCEP), (for TARG1; 10 mM Bis-Tris propane pH 7.0, 150 mM NaCl, 0.01% (v/v) Triton X-100 and 0.5 mM TCEP), (for PARG; 10 mM Bis-Tris propane pH 7.0, 25 mM NaCl, 0.01% (v/v) Triton X-100 and 0.5 mM TCEP) in a 384-well black polypropylene flat-bottom plates (Greiner, Bio-one) with 10 μL reaction volume per well. The reactions consisted of 1 μM CFP-fused binders and 5 μM MARylated YFP-GAP. Reactions were excited at 410 nm (20 nm bandwidth), while the emission signal was measured at 477 nm (10 nm bandwidth) and 527 nm (10 nm bandwidth). Afterwards, blanks were deducted from the individual values and the radiometric FRET (rFRET) was calculated by dividing the fluorescence intensities at 527 nm by 477 nm. Compound was dispensed with Echo acoustic liquid dispenser (Labcyte, Sunnyvate, CA). Dispensing of larger volumes of the solutions was carried out by using Microfluidic Liquid Handler (MANTIS®, Formulatrix, Beford, MA, USA). Measurements were taken with Tecan Infinite M1000 pro plate reader.
2.8. Computational modeling
Hit compounds were docked into the ADPr-bound (6WOJ), 3 unique unbound conformations (7KR0, 7KR1, 6WEY) and two small molecules bound (5RSG, 5RTT) structures of SARS-CoV-2 Mac1 (Alhammad et al., 2021; Frick et al., 2020; Schuller et al., 2021). The proteins and ligands were prepared using Schrodinger Maestro and were subsequently docked using Glide with XP precision, analog compounds 6, 7, 8, and 9 were re-docked using a core constraint to a high scoring, intuitive pose of compound 7, and high scoring poses were subjected to a Prime MM-GBSA minimization, allowing flexibility for any residue within 5 Å of the ligand (Friesner et al., 2004, 2006; Halgren et al., 2004; Jacobson et al., 2002, 2004; Sastry et al., 2013).
3. Results
3.1. Comparison of viral and human macrodomains in two high-throughput ADP-ribose binding assays
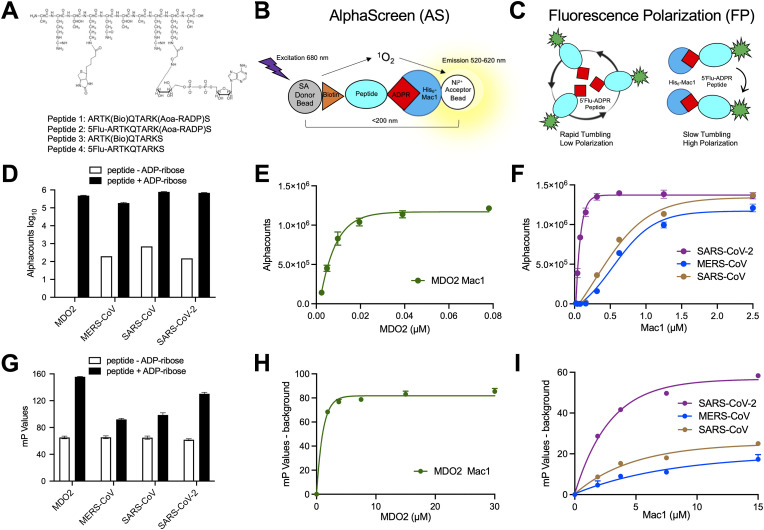
Here we established two distinct ADP-ribose binding assays for multiple macrodomain proteins (Fig. 1 A–C). First, we adopted a previously published AS assay, where a short peptide was modified at a leucine residue with ADP-ribose through an amino-oxyacetic acid linkage, and at a second leucine residue with biotin (Fig. 1A) (Schuller et al., 2017). Streptavidin donor beads and Ni2+ acceptor beads induce a light signal if the His-tagged Mac1 protein interacts with the biotinylated peptide (Fig. 1B). We also developed an FP assay as an orthogonal method to evaluate interactions of macrodomains with ADP-ribosylated peptide. This assay used the same peptide but with fluorescein attached instead of biotin and measures polarization of the fluorescent signal (Fig. 1C). We then tested 4 separate macrodomains for their ability to bind to these peptides, the human macrodomain Mdo2, and Mac1 from SARS-CoV, MERS-CoV, and SARS-CoV-2. All 4 macrodomains bound to the ADP-ribosylated control peptides better than to non-ADP-ribosylated peptides (Fig. 1D,G). The AS assay had an especially strong signal-to-background ratio, ranging from ∼0.75–2 × 103. To further study the binding of Mac1 proteins to AS and FP peptides, we evaluated binding in a dose-dependent assay. Of these four proteins, the human MDO2 demonstrated the greatest level of polarization with a peak of signal at ∼3.75 μM in the FP assay and reached a maximum signal in the AS assay at 40 nM (Fig. 1E,H). The SARS-CoV-2 Mac1 reached the next highest level of polarization in the FP assay and reached a maximum signal in the AS assay at 0.625 μM, while the SARS-CoV and MERS-CoV Mac1 both reached their maximum signal in the AS assay at ∼1.25–2.5 μM (AS) and performed similarly in the FP assay, respectively (Fig. 1F,I).
Fig. 1.
Coronavirus Mac1 binding to ADP-ribosylated peptides. (A) Illustration of the amino-oxyacetic acid modified lysine-conjugated ADP-ribosylated peptide with an additional biotin conjugated to a different lysine residue and included are the amino acid sequences and modification sites of peptides used in this study. (B–C) Cartoon diagrams depicting a bead-based AS (A) and FP (B) assays for measuring macrodomain interactions with an ADP-ribosylated peptide. (D) Macrodomain proteins were incubated with peptide #1 or peptide #3 for 1 h at RT and Alphacounts were determined as described in Methods. (E–F) Peptide #1 was incubated with indicated macrodomains at increasing concentrations and Alphacounts were measured as previously described. (G) Mac1 proteins were incubated at indicated concentrations with peptide #2 or peptide #4 and the plate was incubated at 25 °C for 1 h before polarization was determined. (H–I) Peptide #2 was incubated with indicated macrodomain proteins at increasing concentrations and polarization was determined as previously described. All data represent the means ± SD of 2 independent experiments for each protein.
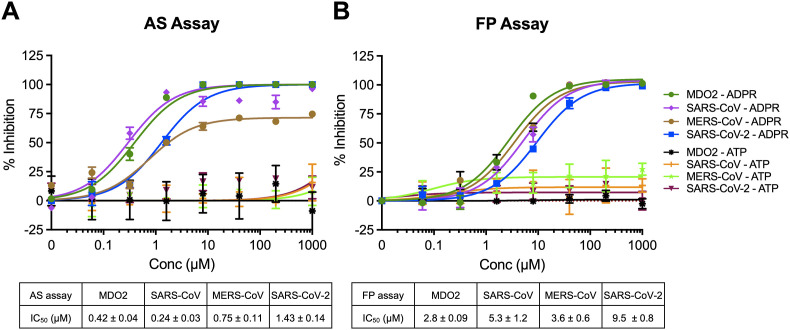
Next, we tested the ability of free ADP-ribose to inhibit the binding of Mac1 to the ADP-ribosylated peptide. For these displacement assays, the amounts of beads, peptide, and Mac1 protein to be used were optimized to obtain a robust signal while limiting the amounts of reagents used for screening purposes (see Methods). The addition of free ADP-ribose, but not ATP, into the AS and FP assays inhibited human macrodomain and CoV Mac1 binding to the ADP-ribosylated peptides, confirming that these assays can be used to identify macrodomain binding inhibitors (Fig. 2 ). IC50 values for free ADP-ribose ranged between 0.24 μM with SARS-CoV Mac1 to 1.43 μM with SARS-CoV-2 using free ADP-ribose in the AS assay (Fig. 2A). Similar results, albeit higher IC50 values, were observed in the FP assay, likely because of a higher amount of Mac1 used in this assay (4 μM vs 250 nM), with IC50 values ranging from 2.8 μM to 9.5 μM (Fig. 2B).
Fig. 2.
Free ADP-ribose inhibits macrodomain binding to ADP-ribosylated peptides. ADP-ribose (ADPr) competition assays were used to block the interaction between macrodomain proteins and ADP-ribosylation peptides in the AS (A) or FP (B) assays. ATP was used as a negative control. Data was analyzed as described in Methods. The data represent the means ± SD of 2 independent experiments for each protein.
3.2. High-throughput screening (HTS) for SARS-CoV-2 Mac1 inhibitors
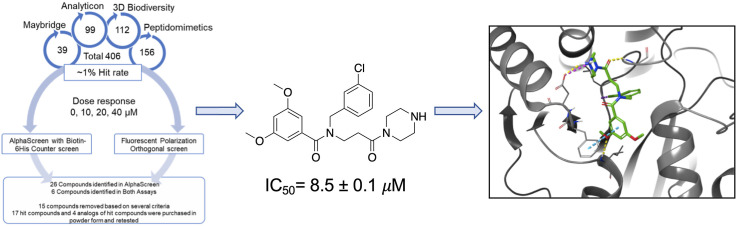
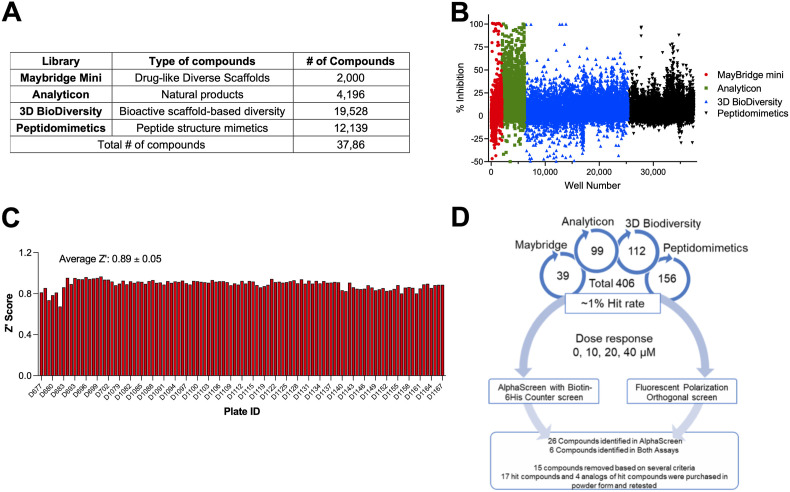
We next performed a small pilot screen of ∼2000 compounds from the Maybridge Mini Library of drug-like scaffolds at 10 μM using both AS and FP assays (Fig. 3 A–B). We identified 39 compounds that significantly inhibited Mac1-ADP-ribose binding at >3 standard deviations (3SD) plus the plate median (Fig. 3A–B). After performing dose-response curves we found that 2 compounds inhibited binding in both assays (Fig. 4 A-B). We then tested these compounds in a counter screen, which is also an AS assay that utilizes a biotinylated-His peptide that gives off a strong signal with the addition of streptavidin donor and nickel acceptor beads. These two compounds did not affect the signal from our counter screen indicating that they do not intrinsically inhibit the assay. After this initial validation of our screen, three additional libraries were chosen to include a total number of 35,863 compounds from the Analyticon, 3D BioDiversity, and Peptidomimetics libraries (Fig. 3A). We chose the AS assay as our primary HTS assay, as the average Z′ score for the AS was higher than the Z′ score from the FP assay in our original screen (0.82 vs 0.67). In this larger screen, the average Z’ was 0.89 ± 0.05, indicating a strong separation between positive and negative controls (Fig. 3C). Using the same hit criteria described above for each individual library, we identified 406 hits resulting in a 1% hit rate (Fig. 3D). Of note, the Analyticon library produced a lot of non-specific inhibitors, indicating a lot of these compounds likely inhibit the assays themselves (Fig. 3B). We next performed dose-response (10–40 μM) curves of these 406 compounds in our primary (AS), orthogonal (FP), and counter screen (Bn-His6) assays (Fig. 3D). From the 406 original hits, 26 compounds were identified that inhibited SARS-CoV-2 Mac1-ADP-ribose binding in the AS assay in a dose-dependent fashion, and 6 compounds were identified that inhibited Mac1 binding in both AS and FP assays (Fig. 3D). Of these 32 hit compounds, we re-purchased 17 of them, excluding 15 based on several selection criteria, including substantial inhibition of the counter screen, high IC50 values in the AlphaScreen, pan-assay interference compounds, and compound availability (Fig. 3D). The remaining 17 compounds along with 4 analogs were repurchased or resynthesized (see Methods).
Fig. 3.
High-throughput screen for SARS-CoV-2 Mac1 inhibitors. (A) List of libraries that were screened, the number of compounds from each library, and the type of compounds each library contains. (B) Scatterplot showing the % inhibition of each compound in the screen. The cutoff for a hit was the plate median +3 standard deviations. (C) Z′ scores were determined for each plate in the screen. The average Z′ score was 0.89 ± 0.05. (D) Dose response confirmation. From the original screen, we identified 406 potential hits, these hits were retested in a dose-response assay on both the AS and FP assays and were also counterscreened against a biotinylated 6 His peptide. After these assays and other exclusion criteria, 17 hit compounds and 4 analogs were repurchased or resynthesized.
Fig. 4.
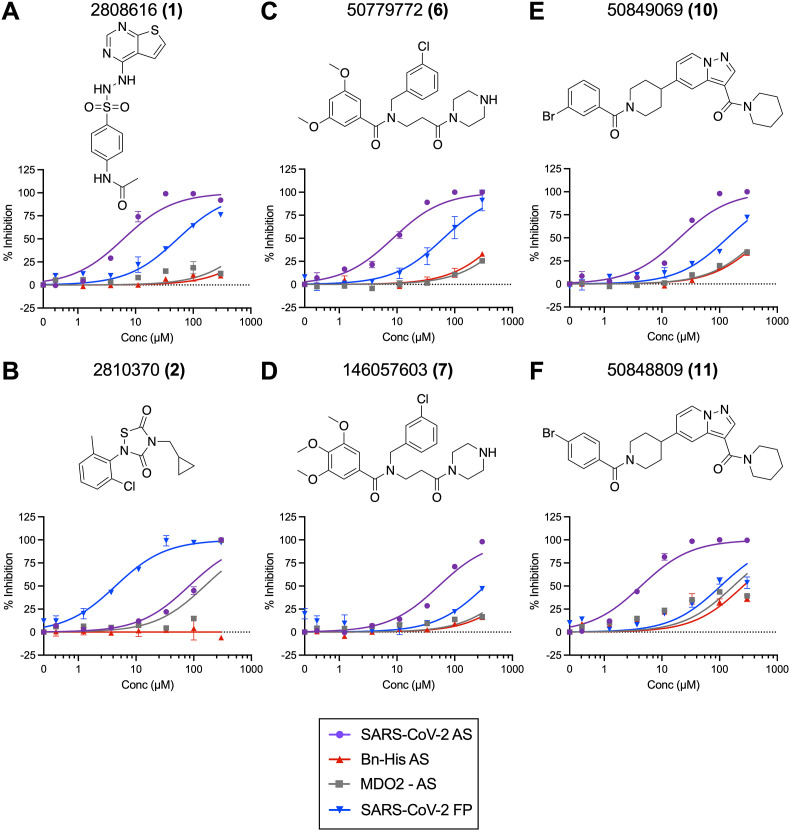
Identification of chemical compounds that inhibit SARS-CoV-2 Mac1 ADP-ribose binding. Dose-response curves representing hit compounds identified in the HTS that inhibit both AS and FP assays. (A–B) Maybridge Mini Library compounds 1 (A), and 2 (B). (C–D) Compound 6 (C) and its analog, 7 (D). (E–F) Compound 10 (E) and its analog 11 (F). Data was analyzed as described in Methods. Data represent the means ± SD of at least 2 independent experiments for each compound with each protein. Structures were created using ChemDraw.
Re-purchased compounds were evaluated in dose-response assays against both SARS-CoV-2 Mac1 and human MDO2 protein. Our cutoff criteria included: i) compound must inhibit both primary and orthogonal assays with at least 75% inhibition in AS assay and at or near 50% inhibition in the FP assay, and ii) less than 30% inhibition of the Bn-His6 counter screen. Among the 17 selected and the 4 analogs compounds, six compounds inhibited ADP-ribose binding of SARS-CoV-2 Mac1 in both AS and FP assays with no substantial inhibition of the Bn-His6 counter screen. These were compounds 1,2,6,7,10, and 11 (Table 1 ). IC50 values ranged from 6.2 μM to 83.0 μM in AS assay and 4.4 μM–338 μM in FP assay (Table 1, Fig. 4). Compounds 1, 10, and 11 also had some inhibitory activity against the MERS-CoV Mac1 protein, though the inhibition of MERS-CoV Mac1 was lower than the inhibition demonstrated against SARS-CoV-2 (Table 1). In addition, only compound 2 inhibited MDO2, indicating that these compounds were broadly specific for viral macrodomains. Compounds 4, 8, 9, and 14 showed inhibition in the alphascreen assay, no inhibition in the counterscreen, but did not meet the benchmark in the FP assay (Table S1, Fig. S1). Compounds 8 and 9 represent analogs of compounds 6 and 7.
Table 1.
IC50 of the selected compounds in AS, FP, and FRET assays.
| Compound Identification number (CID) | Vendor ID | IC50 AS SARS-CoV-2 (μM) | IC50 FP SARS-CoV-2 (μM) |
IC50 AS MDO2 (μM) | IC50 AS MERS (μM) | IC50 FRET SARS-CoV-2 (μM) |
|---|---|---|---|---|---|---|
| ADP-ribose | 1.5 ± 0.05 | 9.74 ± 0.75 | 0.42 ± 0.05 | 1.25 ± 0.23 | N.D. | |
| Compounds inhibit both AS and FP | ||||||
| 2808616 (1) | GK02919 | 6.2 ± 0.7 | 53.6 ± 7.4 | >300 | 39.6 ± 4.0 | N.D. |
| 2810370 (2) | HTS01833 | 83.0 ± 5.1 | 4.4 ± 0.3 | >300 | >300 | N.D. |
| 50779772 (6) | F594-1001 | 8.5 ± 0.1 | 68.0 ± 29.8 | >300 | >300 | 45 ± 10.9 |
| 146057603 (7) | F594-1011 | 52.9 ± 0.1 | 338 ± 4.9 | >300 | >300 | N.D. |
| 50849069 (10) | Z269-0281 | 26.7 ± 6.5 | 75.2 ± 10.6 | >300 | 181.3 ± 14.4 | N.D. |
| 50848809 (11) | Z269-0215 | 56.9 ± 33.7 | 27.6 ± 11.1 | 174.15 ± 6.6 | 181.8 ± 4.7 | N.D. |
(N.D.): not determined.
3.3. Selected compounds demonstrate evidence of SARS-CoV-2 Mac 1 binding
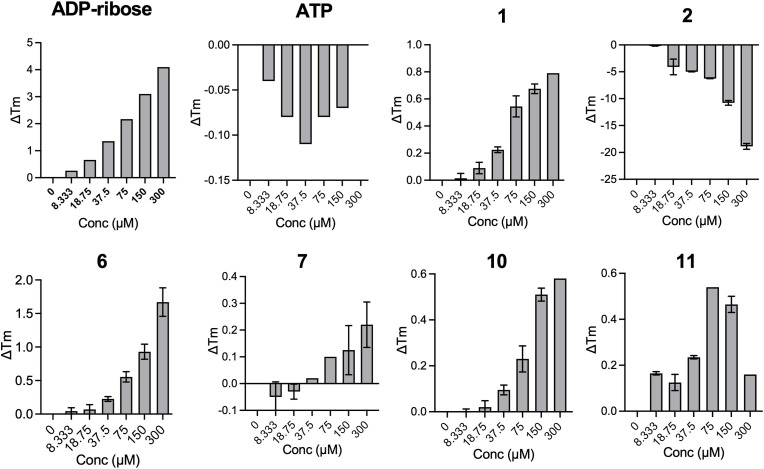
Next, we set out to test the hypothesis that these compounds inhibit the Mac1-ADP-ribose interaction by binding to Mac1 and not other components of the assay, such as the peptide. To test for Mac1 binding, we used a differential scanning fluorimetry (DSF) assay as previously described (Alhammad et al., 2021) and tested our top 6 hit compounds (Fig. 5 , Fig. S2) and compounds 8 and 9, as they are analogs of 6 and 7 (Fig. S3). In this assay, compound binding to Mac1 should increase the melting temperature of Mac1. The addition of free ADP-ribose, which binds to Mac1, showed a dose-dependent increase of approximately 4 °C in the melting temperature of Mac1, while the negative control, ATP, had no effect, as previously demonstrated (Alhammad et al., 2021). Compounds 1, 6, 7, 10, and 11 showed dose-dependent shifts in the melting temperature of Mac1 with peaks ranging from 0.22 to 1.67 °C (Table 2 ), providing strong evidence that these compounds bind to Mac1, albeit not with the same affinity as ADP-ribose. On the other hand, compound 2 resulted in highly irregular thermal shift curves, and compounds 8 and 9 showed minimal activity, indicating that these compounds may not bind to Mac1 (Fig. 5, Figs. S2–S3). These results provide evidence that 5 of our 6 hit compounds (1, 6, 7, 10, and 11) directly bind to SARS-CoV-2 Mac1.
Fig. 5.
Thermal stability of SARS-CoV-2 Mac1 after incubation with hit compounds. The top 6 hit compounds were tested for their ability to increase the thermal stability of SARS-CoV-2 Mac1 in a differential scanning fluorimetry assay (DSF). The data represent the means ± SD of the ΔTm from two independent experiments.
Table 2.
Peak thermal shifts for DSF assay.
| Peak ΔTm |
||||||
|---|---|---|---|---|---|---|
| Compound | 1 | 2 | 6 | 7 | 10 | 11 |
| Average ± SD | 0.68 ± 0.04 | −18.88 ± 0.53 | 1.67 ± 0.21 | 0.22 ± 0.08 | 0.51 ± 0.03 | 0.47 ± 0.04 |
| Conc (μM) | 150 | 300 | 300 | 300 | 150 | 150 |
3.4. Hit compounds inhibit ADP-ribosylhydrolase activity in vitro
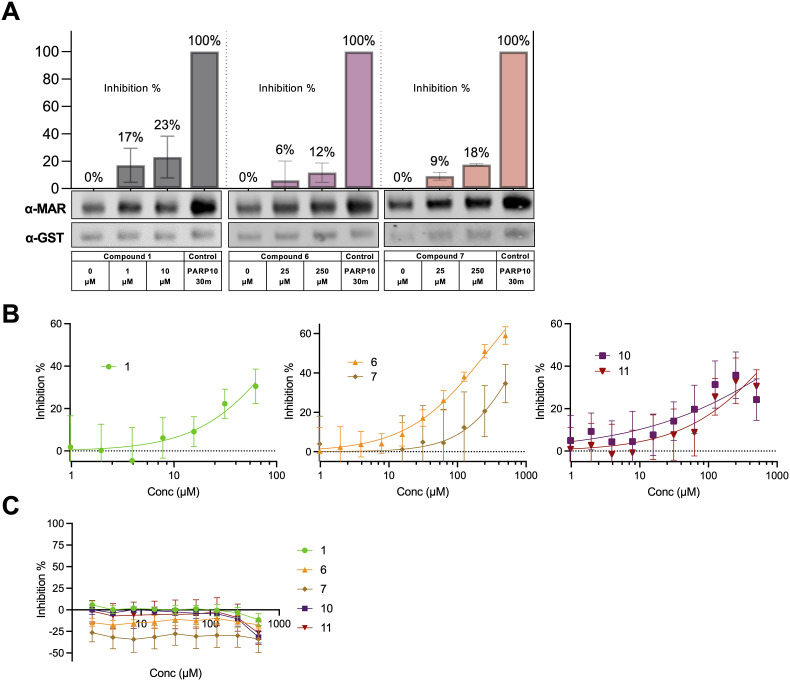
SARS-CoV-2 Mac1 is a mono-ADP-ribosylhydrolase that removes mono-ADP-ribose from target proteins (Alhammad et al., 2021; Dasovich et al., 2022; Li et al., 2016). Next, we examined the ability of our top 5 hit compounds to inhibit the enzymatic activity of SARS-CoV-2 Mac1 using two distinct assays. The first approach was a gel-based Mac1 ADP-ribosylhydrolase assay where we tested each compound against the SARS-CoV-2 Mac1 protein. Compound 1 tended to precipitate in these assays at higher concentrations, and so we used lower concentrations for this compound than others. Compounds 1, 6, and 7 exhibited a dose-dependent inhibition of Mac1 ADP-ribosylhydrolase activity (Fig. 6 A). We were unable to detect any significant inhibition with 10 and 11 in this assay.
Fig. 6.
Impact of hit compounds on SARS-CoV-2 ADP-ribosylhydrolase activity. (A) Top hit compounds were tested in the gel-based deMARylation assay as described in Methods. Proteins were analyzed by Immunoblotting with anti-GST (PARP10) and anti-MAR binding reagent (MABE1076). Gels were quantitated using Image Studio software. The bar graph above each immunoblot represents the mean inhibition ± SD from at least two independent experiments. (B) Top hit compounds were tested in the ADPr-Glo assay as described in Methods. Data represent the means ± SD of 2 independent experiments with 4 technical replicates for each compound. (C) The top hit compounds were also tested in the counter screen to the ADPr-Glo assay that omits Mac1 and uses free ADP-ribose instead of MARylated PARP10. Data represent the means ± SD of 4 technical replicates for each compound.
Next, we utilized a recently published high-throughput luminescence-based ADP-ribosylhydrolyase assay (Dasovich et al., 2022). Here we found that 1, 6, 7, 10 and 11 all showed dose-dependent inhibition of ADP-ribosylhydrolase activity (Fig. 6B) without impacting the counter screen (Fig. 6C). Compound 6 was clearly the most efficient inhibitor, as it had a peak of ∼60% inhibition, similar to Dasatinib, which we previously identified in a separate HTS (Dasovich et al., 2022). These results indicate that the identified Mac1 inhibitors block Mac1 binding and Mac1 enzymatic activity.
3.5. Selectivity profiling
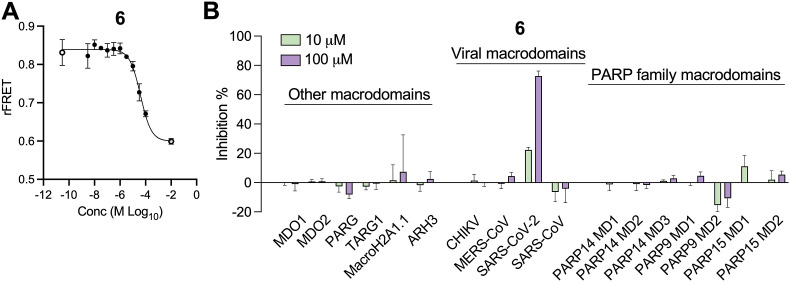
As compound 6 inhibited both Mac1 ADP-ribose binding and hydrolysis activity, and showed the strongest evidence of direct Mac1 binding, we tested its ability to inhibit 16 different macrodomains using a recently developed FRET-based assay (Sowa et al., 2021). Again, 6 demonstrated dose-dependent inhibition of Mac1-ADP-ribose binding in this assay, consistent with our AS results but with a slightly higher IC50 of 45.0 ± 10.9 μM (Fig. 7 A). Remarkably, when tested against 16 different human and viral macrodomains in this assay, 6 only inhibited SARS-CoV-2 Mac1, having only minimal levels of inhibition of all other macrodomain proteins, including other CoV macrodomains (Fig. 7B). These results indicate that this compound is highly selective for the SARS-CoV-2 Mac1 protein.
Fig. 7.
Compound 6 is highly selective for the SARS-CoV-2 Mac1 protein. (A–B) Compound 6 was tested in a FRET-based assay for its ability to inhibit SARS-CoV-2 Mac1 protein in a dose-dependent manner (A) and for its ability to inhibit a panel of 17 macrodomain containing proteins (B). The data in means ± SD are shown as a single experiment representative of 3 independent experiments.
3.6. Structure activity relationship (SAR)
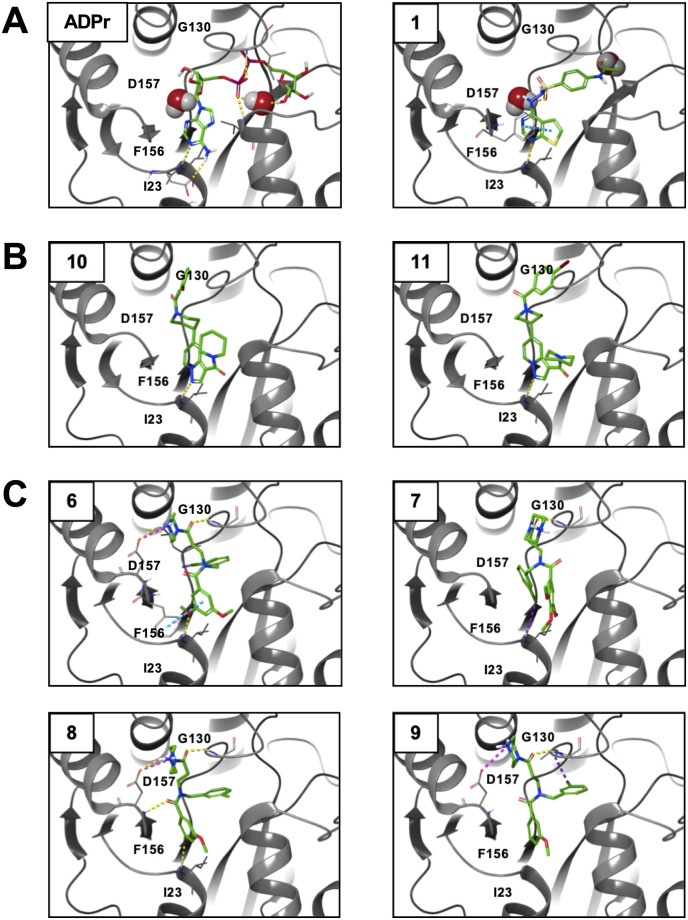
The top 5 compounds could be separated into 3 chemotypes based on their structures. To analyze the involved residues and type of connection between selected hit compound and Mac1, we used computational docking analysis to get an initial structure activity relationship (SAR) by predicting poses of compounds in Mac1 structures (Fig. 8 ). In addition to our 5 hit compounds, we also docked compounds 8 and 9 as they are analogs of 6 and 7 and could give further insight into SAR, even though we either detected minimal or no direct Mac1 binding by these compounds. These seven compounds were docked against the ADP-ribose bound structure of SARS-CoV-2 Mac1 (PDB 6WOJ) as well as three apo structures of Mac1 (PDB 7KR0, 7KR1, 6WEY). Docking and glide emodel scores were calculated for each compound against all four structures and the best structure was chosen based on these scores (Table S2). Analog compounds 6, 7, 8, and 9 were assessed both based on score and visual inspection and were re-docked using a core constraint to a high scoring, intuitive pose of compound 7. All top scoring poses were subsequently minimized using Prime, allowing flexibility within 5 Å of the ligand. Compound 1 was its own chemotype but has a sulfonohydrazide that is also found in a compound identified in a previous screen for macrodomain inhibitors (Ekblad et al., 2018). It also has a thienopyrimidine that is similar to the pyrrolopyrimidine found in the compounds identified in the fragment screen by Schuller et al. (2021). It makes a hydrogen bond with a backbone amine of D22, pie-stacking interactions with F156, and extends with a benzene ring into the distal ribose pocket inserting in between the GGG and GIF loops (Fig. 8A). Compounds 10 and 11 are close analogs with a single difference of positioning in the bromobenzoyl moiety on the piperidine ring (Fig. 4E–F). These compounds had similar activity across the board in our assays, making it difficult to analyze their SAR. While they docked into the binding pocket, these docking poses only indicate a single hydrogen bond with the backbone amine of D22 (Fig. 8B). In contrast, compounds 6, 7, 8, and 9 are close analogs of each other and have a wide range of inhibitory and binding activity. IC50 values for these compounds range from 8.5 to several hundred μM (Table 1). Direct binding also varied substantially, with Tm's ranging from ∼1.7 °C (6) to undetectable binding (8). These compounds all have the same base structure, including a beta-alanine core substituted with a N-benzyl or N-chlorobenzyl group, a methoxy benzoyl group and a piperazine amide (Fig. 4C–D, Figs. S1A–B). The main difference between 6 and its analogs are the addition of a methoxy group on the benzoyl group (7), the loss of a chlorine (8), and a missing methoxy group (9). Each of these changes reduces the activity of this series indicating that i) the orientation of the methoxy groups on 6 is likely important for its increased activity, ii) reorienting 7 to accommodate the 4-methoxy group likely decreases activity due to the disruption of multiple interactions, and iii) the chlorine likely makes a critical halogen bond with a backbone amino group of L126 in the binding pocket (Fig. 8C). Looking at hit compounds we noted that both compounds 1 and 6 are predicted to have multiple interactions at the base of the pocket. All other compounds only had a single interaction in this region, indicating that securing the compounds to this region of the pocket is likely critical for inhibiting Mac1-ADP-ribose binding. This is consistent with prior studies that have shown that the aspartic acid at the base of the binding pocket is critical for ADP-ribose binding by both human and viral macrodomains (Karras et al., 2005; McPherson et al., 2017).
Fig. 8.
Computational modeling of identified compounds with SARS-CoV-2 Mac1 structures. Indicated compounds were docked and modeled with SARS-CoV-2 Mac1 structures using Maestro Schrödinger software and separated into 3 groups. (A) – Compound 1; (B) Compounds 6, 7, 8, 9; (C) Compounds 10, 11. Yellow lines - hydrogen bonds; Cyan lines - pi-pi interactions; magenta lines – weak hydrogen bonds; and purple lines – halogen bond. Water molecules are indicated where the structure used to model the compound also had a water-mediated interaction with the solved ligand and amino acid side chains were shown if an interaction between the indicated compound and the side chain were predicted from the modeling results.
4. Discussion
The conserved CoV macrodomain or Mac1 is critical for viral pathogenesis, and thus represents a potential therapeutic target (Leung et al., 2022). However, prior to COVID-19, there were no available inhibitors for viral macrodomains. Mac1 represents a very unique viral target, distinct from the protease and polymerase proteins. It impacts both virus replication and blocks IFN responses independently. Thus, inhibitors may not only decrease viral replication but also boost anti-viral immunity in vivo. To facilitate drug discovery, there are more than 500 crystal structures of viral macrodomains deposited into the PDB which have revealed 3 druggable pockets, and there are now several assays available to identify novel inhibitors. With the outset of COVID-19, several groups have started to screen for inhibitors of the SARS-CoV-2 Mac1. The compounds identified here represent some of the more potent inhibitors of the SARS-CoV-2 Mac1 domain to date, though it's hard to compare with screens that utilized different assays (Dasovich et al., 2022; Sowa et al., 2021). They showed activity in multiple assays and had strong specificity towards the SARS-CoV-2 Mac1 protein (Fig. 7B), an important feature considering that all other species, including humans, contain macrodomains. Despite these advances, the IC50 values of these inhibitors will likely need to be improved before these compounds could be considered for cell culture and animal testing. It will also be important to address issues of solubility, stability, and permeability of these compounds in cell-based assays. Despite these challenges it seems likely that multiple Mac1 inhibitors capable of targeting this domain during a coronavirus infection will be developed in the near future, due to the number of researchers currently developing Mac1 inhibitors. These inhibitors could be used both to better understand the molecular function of the macrodomain during infection and ultimately tested as a novel therapy for COVID-19 or other emerging CoVs.
Declaration of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
ARF would like to the KU Synthetic Chemistry Core facility for help in ordering and confirming compounds. DVF would like to acknowledge McDaniel College Student-Faculty Summer Research Fund, the Jean Richards Fund, the Schofield fund, and the Scott and Natalie Dahne fund. DVF would also like to acknowledge Mr. Kristopher Mason and Vaccitech for the use of their mass spectrometer. LL would like to acknowledge the use of the facilities of the Biocenter Oulu Structural Biology core facility, a member of Biocenter Finland, Instruct-ERIC Centre Finland and FINStruct. This research was funded by National Institutes of Health (NIH) grants P20 GM113117, P30GM110761, a CTSA grant from NCATS awarded to the University of Kansas for Frontiers: University of Kansas Clinical and Translational Science Institute (#UL1TR002366), and University of Kansas start-up funds to ARF, and by Sidrid Jusélius foundation grant to LL, and Johns Hopkins Bloomberg School of Public Health Discretionary Fund to AKLL. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or NCATS.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105344.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abraham R., Hauer D., McPherson R.L., Utt A., Kirby I.T., Cohen M.S., Merits A., Leung A.K.L., Griffin D.E. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. U. S. A. 2018;115(44) doi: 10.1073/pnas.1812130115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham R., McPherson R.L., Dasovich M., Badiee M., Leung A.K.L., Griffin D.E. Both ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain affect neurovirulence in mice. mBio. 2020;11(1) doi: 10.1128/mBio.03253-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y.M.O., Fehr A.R. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses. 2020;12(4):384. doi: 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y.M.O., Kashipathy M.M., Roy A., Gagne J.P., McDonald P., Gao P., Nonfoux L., Battaile K.P., Johnson D.K., Holmstrom E.D., Poirier G.G., Lovell S., Fehr A.R. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J. Virol. 2021;95(3) doi: 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady P.N., Goel A., Johnson M.A. Poly(ADP-Ribose) polymerases in host-pathogen interactions, inflammation, and immunity. Microbiol. Mol. Biol. Rev. 2019;83(1) doi: 10.1128/MMBR.00038-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianco A.J., Schürer S.C., Zhu X., Kelley T.T. Google Patents; 2021. Inhibitors of the Notch Transcriptional Activation Complex Kinase (" Nack") and Methods for Use of the Same. [Google Scholar]
- Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., Saikatendu K., Buchmeier M.J., Kuhn P., Wüthrich K. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J. Virol. 2009;83(4):1823–1836. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C.C., Lin M.H., Chuang C.Y., Hsu C.H. Macro domain from Middle East respiratory syndrome coronavirus (MERS-CoV) is an efficient ADP-ribose binding module: crystal structure and biochemical studies. J. Biol. Chem. 2016;291(10):4894–4902. doi: 10.1074/jbc.M115.700542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasovich M., Zhuo J., Goodman J.A., Thomas A., McPherson R.L., Jayabalan A.K., Busa V.F., Cheng S.J., Murphy B.A., Redinger K.R., Alhammad Y.M.O., Fehr A.R., Tsukamoto T., Slusher B.S., Bosch J., Wei H., Leung A.K.L. High-throughput activity assay for screening inhibitors of the SARS-CoV-2 Mac1 macrodomain. ACS Chem. Biol. 2022;17(1):17–23. doi: 10.1021/acschembio.1c00721. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80(17):8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblad T., Verheugd P., Lindgren A.E., Nyman T., Elofsson M., Schuler H. Identification of poly(ADP-ribose) polymerase macrodomain inhibitors using an AlphaScreen protocol. SLAS Discov. 2018;23(4):353–362. doi: 10.1177/2472555217750870. [DOI] [PubMed] [Google Scholar]
- Eriksson K.K., Cervantes-Barragan L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1''-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82(24):12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89(3):1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7(6) doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. In: Maier H.J., Bickerton E., Britton P., editors. vol. 1282. Springer; New York: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. (Coronaviruses). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D.N., Virdi R.S., Vuksanovic N., Dahal N., Silvaggi N.R. Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59(28):2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Jacobson M.P., Friesner R.A., Xiang Z., Honig B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002;320(3):597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Jacobson M.P., Pincus D.L., Rapp C.S., Day T.J., Honig B., Shaw D.E., Friesner R.A. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55(2):351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.S., Challa S., Jones A., Kraus W.L. PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis. Genes Dev. 2020;34(5–6):302–320. doi: 10.1101/gad.334433.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K.L., Griffin D.E., Bosch J., Fehr A.R. The conserved macrodomain is a potential therapeutic target for coronaviruses and alphaviruses. Pathogens. 2022;11(1) doi: 10.3390/pathogens11010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Debing Y., Jankevicius G., Neyts J., Ahel I., Coutard B., Canard B. Viral macro domains reverse protein ADP-ribosylation. J. Virol. 2016;90(19):8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrynitsa G.I., Ntonti D., Marousis K.D., Birkou M., Matsoukas M.T., Asami S., Bentrop D., Papageorgiou N., Canard B., Coutard B., Spyroulias G.A. Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose. J. Struct. Biol. 2019;206(1):119–127. doi: 10.1016/j.jsb.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet H., Coutard B., Jamal S., Dutartre H., Papageorgiou N., Neuvonen M., Ahola T., Forrester N., Gould E.A., Lafitte D., Ferron F., Lescar J., Gorbalenya A.E., de Lamballerie X., Canard B. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83(13):6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R.L., Abraham R., Sreekumar E., Ong S.E., Cheng S.J., Baxter V.K., Kistemaker H.A., Filippov D.V., Griffin D.E., Leung A.K. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. U. S. A. 2017;114(7):1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X., Schroder M., Olieric V., Sharpe M.E., Hernandez-Olmos V., Proschak E., Merk D., Knapp S., Chaikuad A. Structural insights into plasticity and discovery of remdesivir metabolite GS-441524 binding in SARS-CoV-2 macrodomain. ACS Med. Chem. Lett. 2021;12(4):603–609. doi: 10.1021/acsmedchemlett.0c00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez M.K. The hepatitis E virus ORF1 'X-domain' residues form a putative macrodomain protein/Appr-1''-pase catalytic-site, critical for viral RNA replication. Gene. 2015;566(1):47–53. doi: 10.1016/j.gene.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020;382(8):760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79(20):12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack J.G., Perina D., Ahel I. Macrodomains: structure, function, evolution, and catalytic activities. Annu. Rev. Biochem. 2016;85:431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- Russo L.C., Tomasin R., Matos I.A., Manucci A.C., Sowa S.T., Dale K., Caldecott K.W., Lehtiö L., Schechtman D., Meotti F.C. bioRxiv; 2021. The SARS-CoV-2 Nsp3 Macrodomain Reverses PARP9/DTX3L-dependent ADP-Ribosylation Induced by Interferon Signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13(11):1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Schuller M., Correy G.J., Gahbauer S., Fearon D., Wu T., Diaz R.E., Young I.D., Carvalho Martins L., Smith D.H., Schulze-Gahmen U., Owens T.W., Deshpande I., Merz G.E., Thwin A.C., Biel J.T., Peters J.K., Moritz M., Herrera N., Kratochvil H.T., Consortium Q.S.B., Aimon A., Bennett J.M., Brandao Neto J., Cohen A.E., Dias A., Douangamath A., Dunnett L., Fedorov O., Ferla M.P., Fuchs M.R., Gorrie-Stone T.J., Holton J.M., Johnson M.G., Krojer T., Meigs G., Powell A.J., Rack J.G.M., Rangel V.L., Russi S., Skyner R.E., Smith C.A., Soares A.S., Wierman J.L., Zhu K., O'Brien P., Jura N., Ashworth A., Irwin J.J., Thompson M.C., Gestwicki J.E., von Delft F., Shoichet B.K., Fraser J.S., Ahel I. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 2021;7(16) doi: 10.1126/sciadv.abf8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller M., Riedel K., Gibbs-Seymour I., Uth K., Sieg C., Gehring A.P., Ahel I., Bracher F., Kessler B.M., Elkins J.M., Knapp S. Discovery of a selective allosteric inhibitor targeting macrodomain 2 of polyadenosine-diphosphate-ribose polymerase 14. ACS Chem. Biol. 2017;12(11):2866–2874. doi: 10.1021/acschembio.7b00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa S.T., Galera-Prat A., Wazir S., Alanen H.I., Maksimainen M.M., Lehtio L. A molecular toolbox for ADP-ribosyl binding proteins. Cell Rep Methods. 2021 doi: 10.1016/j.crmeth.2021.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa S.T., Galera-Prat A., Wazir S., Alanen H.I., Maksimainen M.M., Lehtio L. Preparation of screening assays for ADP-ribosyl readers and erasers using the GAP-tag as a binding probe. STAR Protoc. 2022;3(1) doi: 10.1016/j.xpro.2022.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdi R.S., Bavisotto R.V., Hopper N.C., Vuksanovic N., Melkonian T.R., Silvaggi N.R., Frick D.N. Discovery of drug-like ligands for the Mac1 domain of SARS-CoV-2 Nsp3. SLAS DISCOVERY: Advancing the Science of Drug Discovery. 2020;25(10):1162–1170. doi: 10.1177/2472555220960428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth L.S., O’Connor J.J., Kerr C.M., Doerger E., Schwarting N., Sperstad P., Johnson D.K., Fehr A.R. Unique mutations in the MHV macrodomain differentially attenuate virus replication, indicating multiple roles for the macrodomain in coronavirus replication. J. Virol. 2021;95(15) doi: 10.1128/JVI.00766-21. JVI. 00766-00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.Y., Cong L., Chen C., Wei L., Zhao Q., Xu X.L., Ma Y.L., Bartlam M., Rao Z.H. Crystal structures of two coronavirus ADP-ribose-1 ''-monophosphatases and their complexes with ADP-ribose: a systematic structural analysis of the viral ADRP domain. J. Virol. 2009;83(2):1083–1092. doi: 10.1128/JVI.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.