Abstract
Besides pandemic SARS-CoV-2, also endemic seasonal human common cold coronaviruses (hCoVs) have a significant impact on human health and economy. Studies on hCoVs and the identification of antivirals are therefore crucial to improve human well-being. However, hCoVs have long been neglected and the methodology to study virus infection, replication and inhibition warrants being updated. We here evaluated the established plaque-based assays to determine viral titers and cell-to-cell spread and developed protocols for the immunodetection of the viral nucleocapsid protein by flow cytometry and in-cell ELISA to study infection rates at early time points. The developed protocols allow detection of hCoV-229E infection after 2, and hCoV-NL63 and -OC43 infection after 3 days at a single cell level or in a 96 well microtiter format, in large sample numbers without being laborious or expensive. Both assays can be applied to assess the susceptibility of cells to hCoV infection and replication, and to determine the efficacy of antiviral compounds as well as neutralizing antibodies in a sensitive and quantitative manner. Application revealed that clinically applied SARS-CoV-2 targeting monoclonal antibodies are inactive against hCoVs, but that the viral polymerase targeting antivirals remdesivir and molnupiravir are broadly active also against all three hCoVs. Further, the in-cell ELISA provided evidence that nirmatrelvir, previously shown to broadly inhibit coronavirus proteases, also prevents replication of authentic hCoVs. Importantly, the protocols described here can be easily adapted to other coronavirus strains and species as well as viruses of other families within a short time. This will facilitate future research on known and emerging (corona)viruses, support the identification of antivirals and increase the preparedness for future virus outbreaks.
Keywords: Seasonal coronaviruses, Antiviral testing, Drug screening, In-cell ELISA
1. Introduction
The ongoing severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) pandemic has demonstrated the danger from zoonotic spillovers of coronaviruses into the human population (Dhama et al., 2020). Similar to SARS-CoV-2, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) have emerged in 2002 and 2012 as highly human pathogenic viruses (Cui et al., 2019). Studies on these coronavirus outbreaks have also drawn attention to the long neglected human coronaviruses (hCoV) (Ljubin-Sternak et al., 2021) that have entered the human population already 70–820 years ago (Forni et al., 2017) and have only been considered as harmless seasonal common cold viruses (Corman et al., 2018; Cui et al., 2019; da Costa et al., 2020; Hartenian et al., 2020). HCoV-229E and -OC43 have already been known since the 1960s, while hCoV-HKU1 and -NL63 have only recently been identified in 2004 and 2005, respectively (Corman et al., 2018; Cui et al., 2019; van der Hoek et al., 2004; Woo et al., 2005). While hCoV-NL63 and -229E belong to the genera of alphacoronaviruses, -OC43 and -HKU1 are betacoronaviruses (Corman et al., 2018). These four coronaviruses are endemic in the human population and circulate with annual peaks of infections in the winter months (Killerby et al., 2018; Li et al., 2020; Park et al., 2020), where they usually cause mild and self-limiting upper respiratory tract infections (Pyrc et al., 2007) and account for about 15–30% of common cold cases (Corman et al., 2018). Nonetheless, hCoVs are involved in 10–20% of hospitalizations due to respiratory illness (Fischer et al., 2021; Pyrc et al., 2007) and can cause severe courses of disease with bronchiolitis and pneumonia, as well as enteric and even neurological diseases, especially in young children and immunocompromised adults (Corman et al., 2018; Pyrc et al., 2007). Furthermore, the economic costs due to mild common cold infections (though not exclusively by hCoVs) are estimated with 24–40 billion USD annually in the U.S. (Bramley et al., 2002; Fendrick et al., 2003). Thus, although usually causing mild infections, seasonal hCoVs have a significant impact on human health, the economy and the health systems. Accordingly, there is an urgent need for drugs to treat or prevent hCoV infections. So far, however, there is no vaccine or antiviral drug against hCoVs available.
The progress made by SARS-CoV-2 vaccine development and antiviral drug research might also be beneficial for hCoVs (Cannalire et al., 2020; Malone et al., 2022) as coronaviruses share similarities in their spike proteins (Grifoni et al., 2020) and enzymes such as the most conserved RNA dependent RNA polymerase (Pyrc et al., 2007) or the 3C-like protease (Dai et al., 2020). Thus, methods to readily study hCoV biology, infection, inhibition or serum neutralization in a high-throughput and quantitative manner are highly warranted. Common methods for in vitro wildtype hCoV infection experiments include indirect detection of virus-induced cytopathic effects (CPE) or direct detection of viral antigens in infected cells by western blotting, fluorescence staining, or by detecting viral titers of progeny virus by qPCR or virus titrations (Carbajo-Lozoya et al., 2014; Flint and Racianello, 2015; Hirose et al., 2021; Hsu et al., 2021; Schirtzinger et al., 2022; Wang et al., 2021b). These methods can be laborious, time-consuming and lack sensitivity or specificity. Moreover, increasing sample sizes in these assays come with higher workloads and costs, making high-throughput analyses impractical. In the case of analyzing spike protein function, these drawbacks can be circumvented by working with pseudotyped lenti- or rhabdoviral particles, however, these do not recapitulate viable virus infection and replication (Li et al., 2018). Thus, we here established protocols for quantifying authentic hCoV-229E, -NL63, -OC43 infection and replication in cell lines by immunodetection of the viral nucleocapsid protein applying flow cytometry or in-cell ELISA. By specifically detecting newly synthesized nucleocapsid protein, infection can be quantified on a single cell level or in bulk cultures in a 96 well microtiter format. We validated the assays by analyzing the broadly active polymerase inhibitors remdesivir and molnupiravir (Malone et al., 2022) and the cellular protease inhibitors camostat mesylate and E-64d (Hoffmann et al., 2020). The assays allow to determine the half maximal inhibitory concentrations (IC50) of the antivirals and confirm that remdesivir and molnupiravir are active against all hCoVs. In addition, the assays prove that the viral 3C-like protease inhibitor nirmatrelvir (Cannalire et al., 2020) also inhibits all three authentic hCoVs. In contrast, monoclonal antibodies designed for and used in the therapy of SARS-CoV-2 were not active against the hCoVs. Conclusively, the described assays offer a quantitative tool to study hCoV biology and evaluate infection rates and activity of drugs or antibodies against seasonal coronaviruses.
2. Methods
2.1. Cell culture
Huh-7 cells (hepatocyte-derived carcinoma; kindly provided by Anna-Laura Kretz, Department for General and Visceral Surgery, Ulm University) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 10% heat-inactivated fetal calf serum (FCS). Caco-2 cells (human epithelial colorectal adenocarcinoma; kindly provided by Prof. Barth, Ulm University) were grown in DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 1x non-essential amino acids, and 10% FCS. Vero E6 cells (African green monkey derived epithelial kidney; ATCC® CRL-1586™) were grown in DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 1x non-essential amino acids, and 10% FCS for culture and 2.5% FCS for seeding, respectively. Calu-3 cells (human non-small-cell lung adenocarcinoma; ATCC® HTB-55™) were cultured in Minimum Essential Medium Eagle (MEM) supplemented with 100 units/ml penicillin, 100 µg/ml streptomycin, 1 mM sodium pyruvate, 1x non-essential amino acids, 10% FCS. LLC-MK-2 cells (rhesus monkey kidney) were cultured in MEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1x non-essential amino acids, and 8% FCS. HCT-8 cells (human ileocecal adenocarcinoma; ATCC® CCL-244™) were grown in RPMI-1640 medium which was supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FCS. All cells were cultivated in a ventilated 175 ml flask at 37 °C in a 5% CO2 humidified incubator. All cell lines were regularly tested for mycoplasma contamination and remained negative.
2.2. Virus strains and propagation
HCoV-229E (ATCC® VR-740™) was propagated on Huh-7 cells, -NL63 (Netherland 63) on LLC-MK2 cells (both NL63 and LLC-MK2 were kindly provided by Lia van der Hoek, Amsterdam, The Netherlands), and -OC43 (Organ Culture 43; ATCC® CR-1558 ™) on HCT-8 cells. To this end, 80% confluent cells were inoculated with a MOI of 0.1 in 10 ml medium supplemented with 2% FCS and kept at 33 °C. One day post-inoculation cells were washed with PBS before 10 ml of fresh medium supplemented with 2% FCS was added. The following days, cells were monitored daily under the light microscope until a strong cytopathic effect (CPE) was visible (5 days for hCoV-229E and -NL63 and 7 days for hCoV-OC43). Virus stocks were then harvested by centrifuging the supernatant for 5 min at 1300 rpm to remove cellular debris and storing aliquots at −80 °C.
2.3. Tissue culture infection dose 50 (TCID50)
Infectious titers of hCoV-229E, -NL63, or -OC43 were determined on Huh-7, Caco-2, or HCT-8 cells, respectively, in a 96 well format. To this end 25,000 (Huh-7, Caco-2) or 30,000 (HCT-8) cells were seeded per well in 100 µl medium supplemented with 2% FCS. The following day, 62 µl of medium was added before the cells were inoculated with 18 µl of a 10-fold serial dilution of the virus stock and kept at 33 °C. At 7 days post infection, CPE (for hCoV-229E and -NL63) or rounding of cells (for -OC43) was evaluated by light microscopy and TCID50 calculated according to Reed-Münch. TCID50 values were used to calculate the multiplicities of infection (MOI).
2.4. Cell viability assay
The effect of antivirals on the metabolic activity of cells was analyzed using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega #G7571). Metabolic activity was examined under conditions corresponding to the respective infection assays. The CellTiter-Glo® assay was performed according to the manufacturer's instructions. Briefly, medium was removed from the culture at the respective days of incubation and 50 µl of PBS and 50 µl of substrate reagent were added. After 10 min, luminescence of the samples was measured in an Orion II Microplate Luminometer (Titertek Berthold). Untreated controls were set to 100% viability.
2.5. Plaque assay
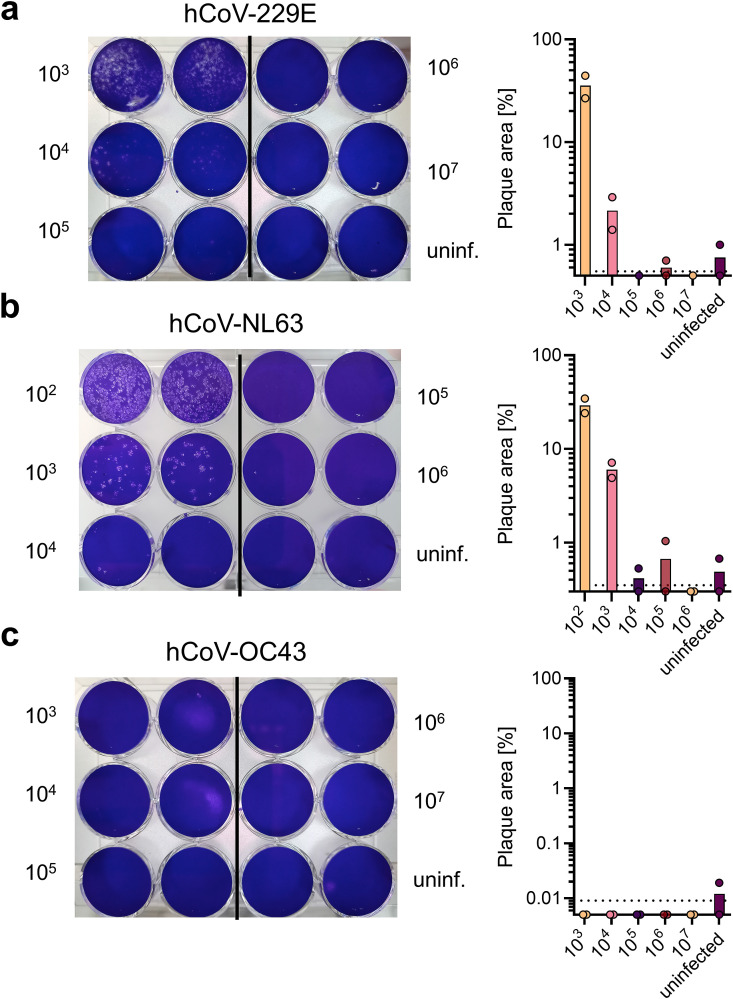
Plaque assays were performed in 12-well plates seeded with 700,000 Huh-7 cells for hCoV-229 E, 700,000 Caco-2 or 400,000 LLC-MK2 cells for -NL63, 700,000 Huh-7 cells for -OC43 to reach a 100% confluent cell monolayer. The next day, cells were washed once with PBS and 400 µl PBS added. Virus stocks were titrated 1:10 in medium and 100 µl per dilution were added in duplicates to the cells to reach a final volume of 500 µl. Cells were incubated for 2 h at 33 °C while shaking every 20 min, and subsequently overlaid with 1.5 ml 0.6% Avicel in medium. 4- (hCoV-229 E) and 5-days (hCoV-NL63, -OC43) post infection, cells were fixed by adding 1 ml of 8% PFA and incubating for 45 min at room temperature. Cells were then washed once with PBS and stained for 20 min with 0.5% crystal violet and 0.1% triton. Thereafter, cells were washed with PBS, dried, and plaques counted. Virus-induced plaques were quantified in ImageJ 1.53c by assessing number of pixels occupied by plaques.
2.6. Detection of hCoV infection by flow cytometry
To establish infection and staining conditions for flow cytometric analyses, 96-well plates were seeded with 25,000 cells (Huh-7 for hCoV-229 E and -OC43 or Caco-2 for -NL63) in 100 µl medium supplemented with 2% FCS. The next day, 62 µl medium was added and cells inoculated with 18 µl of a 5-fold titration of virus stock, to reach a final volume of 180 µl. After 2 h and 2, 3, or 4 days after incubation at 33 °C, cells were detached using 50 µl 0.05% trypsin, and fixed by adding 50 µl of 8% PFA for 30 min. Cells were then transferred into a 96 well V-plate, centrifuged for 1 min at 3,000 rpm before antibody staining. Permeabilization and staining was performed for 40 min at 4 °C with 50 µl of 1:5,000 or 1:10,000 diluted rabbit anti-nucleocapsid protein antibodies (anti-229 E (40640-T62); anti-NL63 (40641-T62); anti-OC43 (40643-T62); all Sino Biological) in buffer B (Nordic MUbio Kit). Subsequently, cells were washed two times with 200 µl FACS buffer (1% FCS in PBS) before they were stained for another 30 min with 50 µl of 1:5,000, 1:10,000 and 1:15,000 dilutions of Alexa Fluor 647 conjugated goat anti-rabbit IgG antibody (#A32733; Thermo Fisher) in FACS buffer at 4 °C. After two more washing steps in FACS buffer, cells were analyzed by flow cytometry at a sample flow rate of 120 μl/min. To evaluate the activity of remdesivir, cells in 144 µl of medium were treated with 18 µl titrated remdesivir for 10 min before inoculation with 18 µl of hCoV-229E (MOI 0.028), -NL63 (MOI 0.018) or -OC43 (MOI 0.0088). 2 (229E) or 3 (NL63, OC43) days post infection, the established protocol was applied (Table 1 ), infection rates quantified and half maximal inhibitory concentrations (IC50) determined.
Table 1.
Final SOP parameters for the optimal detection of hCoV infection by flow cytometry.
| Virus | Cell line | Days post infection | MOI | Primary antibody | Secondary antibody |
|---|---|---|---|---|---|
| HCoV-229E | Huh-7 | 2 | 0.028 | 1:5,000 | 1:5,000 |
| HCoV-NL63 | Caco-2 | 3 | 0.018 | 1:5,000 | 1:5,000 |
| HCoV-OC43 | Huh-7 | 3 | 0.008 | 1:5,000 | 1:5,000 |
2.7. Detection of hCoV infection by in-cell ELISA
For in-cell ELISA, 25,000 cells (Huh-7 for hCoV-229E or -OC43 and Caco-2 for -NL63) were seeded in 100 µl of medium supplemented with 2% FCS in 96 well plates. The following day, 62 µl fresh medium was added and the cells inoculated with 18 µl of a 2- or 10-fold serial dilution of virus stocks and cells incubated at 33 °C. 2 h, 2, 3, 4 or 6 days post infection, cells were fixed by adding 180 µl of 8% PFA for 30 min at room temperature and permeabilized for 5 min using 0.1% triton. Subsequently, cells were washed with PBS once and stained with 50 µl of 1:5,000, 1:7,500, 1:10,000 or 1:15,000 diluted rabbit anti-nucleocapsid protein antibodies (anti-229E (40640-T62); anti-NL63 (40641-T62); anti-OC43 (40643-T62); all Sino Biological) in antibody dilution buffer (10% FCS and 0.3% tween 20 in PBS) for 1 h at 37 °C. Subsequently, cells were washed twice with washing buffer (0.3% tween 20 in PBS) and stained using 50 µl of an HRP-coupled anti-rabbit secondary antibody (#31460, Thermo Fisher) at dilutions of 1:7,500, 1:10,000 and 1:15,000 for 1 h at 37 °C. After 3 washes, 50 µl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added for 5 min under light protection before the enzymatic reaction was stopped using 0.5 M H2SO4. Optical density was measured at 450 nm and baseline corrected at 620 nm using an ELISA microplate reader. Susceptibility to infection of Huh-7 (25,000 seeded per well), Caco-2 (25,000 seeded per well), Calu-3 (40,000 seeded per well), or Vero E6 (20,000 seeded per well) cells was analyzed by inoculation with titrated hCoV virus stocks (maximum MOI of 0.004 for hCoV-229E, 0.016 for -NL63, and 0.012 for -OC43). Antivirals were evaluated by pretreating cells with increasing concentrations of remdesivir (S8932, Selleckchem), molnupiravir (S8969, Selleckchem), nirmatrelvir (S9866, Selleckchem), camostat mesylate (SML0057, Sigma), or E-64d (E8640, Sigma) before in-cell ELISA was performed according to the optimal determined conditions (Table 2 ) and half maximal inhibitory concentrations (IC50) determined. For analysis of monoclonal anti-SARS-CoV-2 antibodies imdevimab, casirivimab (Roche) or bamlanivimab (NDC 0002–7910, Eli Lilly), virus was preincubated with titrated antibodies for 30 min before inoculation of target cells at the optimal conditions (Table 2).
Table 2.
Final SOP parameters for the optimal detection of hCoV infection by in-cell ELISA.
| Virus | Cell line | Days post infection | MOI | Primary antibody | Secondary antibody | Z′ factor | S/N ratio |
|---|---|---|---|---|---|---|---|
| HCoV-229E | Huh-7 | 2 | 0.002 | 1:5,000 | 1:15,000 | 0.88 | 22.56 |
| HCoV-NL63 | Caco-2 | 6 | 0.01 | 1:5,000 | 1:10,000 | 0.83 | 6.81 |
| HCoV-OC43 | Huh-7 | 3 | 0.006 | 1:5,000 | 1:10,000 | 0.92 | 21.75 |
2.8. Data and statistical analysis
Data analysis was performed using Microsoft Excel 2019 and GraphPad Prism version 9.2.0 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com, R (version 4.0.1) and SAS (version 9.4). Plaque assays were quantified using ImageJ. Error bars were plotted as SD (standard deviation) for experiments with technical replicates and as SEM (standard error of mean) for experiments with more than one biological replicate. Compound dilutions that caused a 50% reduction of infection (IC50) were calculated using a non-linear regression model (inhibitor vs. normalized response, variable slope). Z’ factors were determined as described (Iversen et al., 2006). Correlations were assessed by non-parametric spearman correlation.
Role of funding source
Funders had no role in study design, data collection, data analyses, interpretation, or writing of the report.
3. Results
A classic method in virology for determining infectious titers is the visualization of virus-induced plaques in a cell monolayer, which results from viral cell-to-cell spread and cytopathic effects (CPE) (Flint and Racianello, 2015). We applied this method to hCoV-229E, -NL63, and -OC43 in different cell lines that were reported to be permissive for hCoVs (Bracci et al., 2020; Gerna et al., 2006; Herzog et al., 2008). We observed plaques after different times of viral infection (Fig. 1 ) and found that plaque formation in Huh-7 or Caco-2 cells was optimal after 4 days of infection for hCoV-229E and 5 days for -NL63 (Fig. 1a and b). Titers were determined as 2.6 × 106 plaque forming units (PFU)/ml for hCoV-229E and 3.1 × 106 PFU/ml for -NL63. LLC-MK2 cells are also described to support hCoV-NL63 replication, however, failed in our plaque assays due to cell detachment in the presence of virus (Supplemental Fig. 1) (Herzog et al., 2008). Counting plaques by software analysis gave similar results as by eye (1.8 × 106 PFU/ml for hCoV-229E and 4.5 × 106 PFU/ml for -NL63). Depending on viral replication time, spread and degree of CPE, plaque sizes may differ between viral species and variants (Jeong et al., 2021; Kato et al., 2017). Thus, plaque area analysis can also provide information on the rate of viral replication and cell-to-cell-spread. We determined plaque areas by software analysis and found that when the plaque area was less than 10% counting of plaques was possible, while when the area was greater than 30% counting was not possible (Fig. 1a and b). Furthermore, as viral titers decreased, the total area of plaques also decreased, making this measurement tool suitable for quantifying the efficiency of agents that act antivirally or virucidally or affect viral replication or cell-to-cell spread. In our experiments, hCoV-OC43 infection of Huh-7 cells did not lead to plaque formation (Fig. 1c). This is consistent with the published finding that hCoV-OC43 only induces morphological changes but no CPE (Hirose et al., 2021) and rules out the plaque assay as a general method for quantifying -OC43 infection in all permissive cell lines (Bracci et al., 2020). Conclusively, measuring plaque numbers and areas is suitable for determining titers and viral spread of hCoV-229E or -NL63 but not -OC43. However, this test does not allow to assess early virus infection and is unsuitable for high-throughput applications.
Fig. 1.
Plaque assays for quantifying infectious hCoVs −229E, -NL63 and -OC43 titers. Plaque assays were performed by inoculation of target cells in duplicates with a 10-fold serial dilution of respective virus stock. Cells were incubated for 2 h and subsequently overlaid with Avicel. To visualize plaques, cells were stained with crystal violet (left panels). Plaques were counted by eye and plaque area was quantified using ImageJ (right panels). Indicated are means from a single experiment carried out in duplicates. a) hCoV-229E titration was performed on Huh-7 cells and evaluated 4 days post infection, b) hCoV-NL63 titration was performed on Caco-2 cells and evaluated 5 days post infection, and c) hCoV-OC43 titration was performed on Huh-7 cells and evaluated 5 days post infection. Dashed line indicates limit of detection.
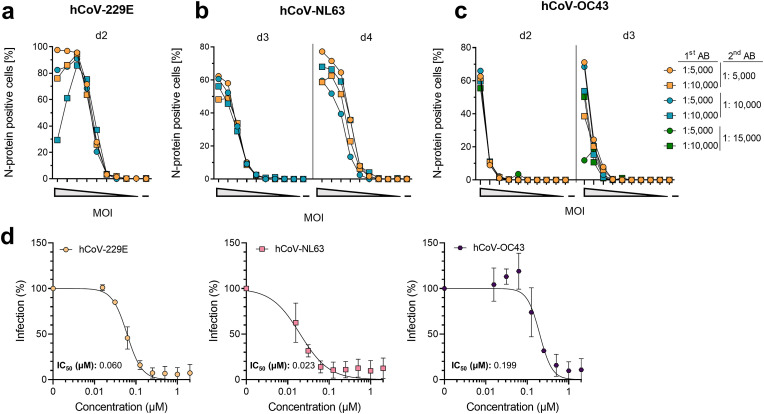
For early detection of hCoV infection at the single cell level, we stained for newly translated hCoV nucleocapsid proteins and detected them by flow cytometry. To this end, hCoV permissive Huh-7 and Caco-2 cells (Gerna et al., 2006; Herzog et al., 2008) (Fig. 1) were inoculated in a 96-well format with decreasing multiplicities of infection (MOI). MOIs were inferred from tissue culture infectious dose 50 (TCID50) titrations to also include hCoV-OC43 that did not allow PFU determination (Fig. 1c). Viral replication was stopped at various time points post infection by fixing the cells in paraformaldehyde and staining for hCoV nucleocapsid proteins with appropriate primary anti-nucleocapsid antibodies and fluorescently labelled detection antibodies at various dilutions. Cells were then analyzed by flow cytometry for the mean fluorescence intensities (MFI) and the percentage of nucleocapsid positive cells (Supplemental Fig. 2 and 2a–c ). To ensure that only newly translated nucleocapsid protein is detected, we also determined the nucleocapsid protein from the viral inoculum after 2 h of infection (input control) and subtracted this value from all other conditions. The experiment resulted in a maximum of 97% nucleocapsid-positive cells already 2 days post hCoV-229E infection at a maximum MOI of 0.7 (Fig. 2a). At an MOI of 0.45, 62% hCoV-NL63-infected cells were detected after day 3, increasing to 77% after day 4 (Fig. 2b). Similarly, after 2 days, at an MOI of 0.044, 62% hCoV-OC43-infected cells were detected, and the proportion did not further increase until the third day (Fig. 2c). At high MOIs, infection plateaued at around 100% for hCoV-229E, but decreased similarly to the other hCoVs with lower MOIs. MFI measurements showed highest fluorescence signals even at the lowest antibody dilutions and were consistent with the percentage of nucleocapsid positive cells (Supplemental Fig. 3 a-c). Using the highest concentration of primary and detection antibody gave the best results in terms of sensitivity and MOI-dependent infection and a standard operating procedure (SOP) was established for each virus (Table 1).
Fig. 2.
Detection of hCoV-229E, -NL63 and -OC43 infection by flow cytometry. a) 5-fold titrated hCoV-229E at a maximum MOI of 0.7 was inoculated onto Huh-7 cells, b) 5-fold titrated hCoV-NL63 at a maximum MOI of 0.45 was inoculated onto Caco-2 cells, and c) 5-fold titrated hCoV-OC43 at a maximum MOI of 0.004 was inoculated onto Huh-7. 2 h (input control) and 2, 3 or 4 days post infection, cells were fixed. Immunostaining was performed using indicated concentrations of a virus-specific anti-nucleocapsid first antibody (1st AB) and an Alexa Fluor 647 coupled secondary detection antibody (2nd AB). Data are from a single experiment conducted in pooled duplicates. d) For applying flow cytometry to evaluate antiviral activities, respective target cells were seeded and the next day pre-treated with a 2-fold serial dilution of remdesivir for 10 min. Next, cells were inoculated with hCoV-229E (MOI 0.028, left panel), -NL63 (MOI 0.018, center panel), or -OC43 (MOI 0.008, right panel). At 2 (229E) or 3 (NL63, OC43) days post infection, cells were fixed and stained. Data were normalized to untreated virus control (100%) and are presented as mean ± SD of two independent experiments conducted in triplicates. Half-maximal inhibition values (IC50) were calculated using a non-linear regression model.
Applying the SOPs, we investigated the feasibility of this assay for antiviral drug testing. For this, cells were incubated with the nucleoside analogue remdesivir before infection. The adenosine analogue remdesivir is a broadly active coronavirus RNA-dependent RNA polymerase inhibitor (Malone et al., 2022) that has already been shown to also be active against hCoV-229E, -NL63, and -OC43 (Brown et al., 2019; Hsu et al., 2021; Parang et al., 2020; Wang et al., 2021a). Our experiments confirm that all tested hCoVs are inhibited by remdesivir, with respective half maximal inhibitory concentrations (IC50) of 0.06 µM for −229E, 0.023 µM for -NL63, and 0.19 µM for -OC43 (Fig. 2d). Calculation of IC50 values from the MFIs gave similar results (Supplemental Fig. 3d). Thus, nucleocapsid staining and flow cytometric analysis allow early and reliable quantification of hCoV infection rates and determination of the antiviral activity of drugs.
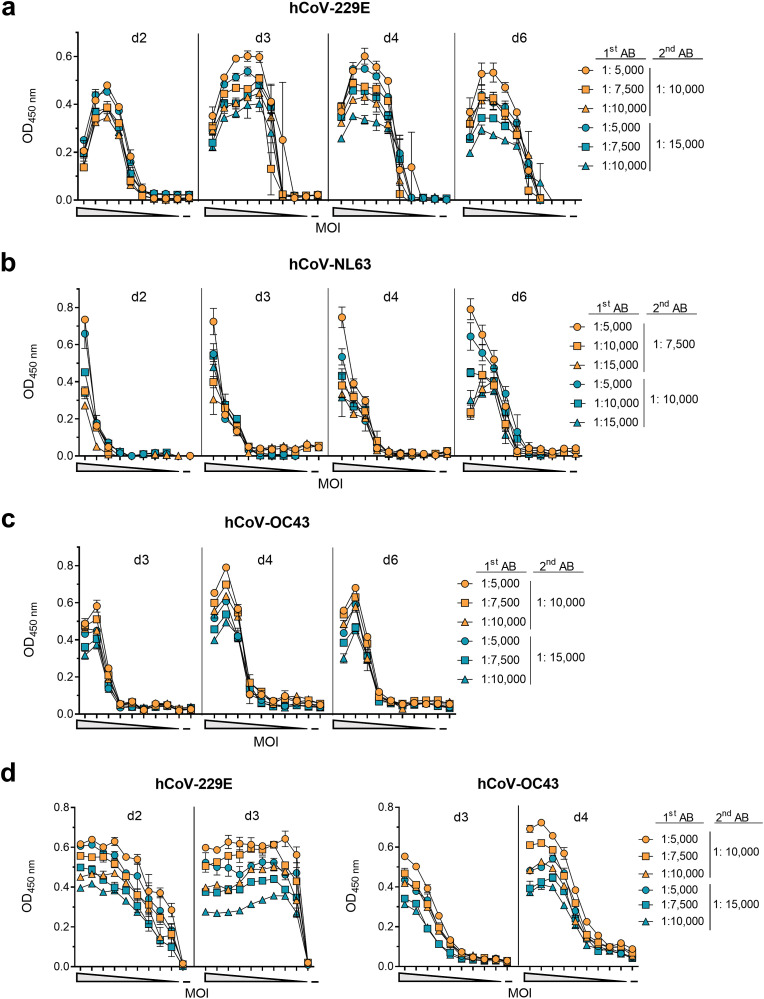
Determination of infection rates by detection of viral proteins using in-cell ELISAs has proven very useful in the investigation of Zika virus (Aubry et al., 2016; Müller et al., 2017), and SARS-CoV-2 (Conzelmann et al., 2020; Schöler et al., 2020). To adapt this assay for hCoVs, we inoculated susceptible cells seeded in microtiter plates with decreasing MOIs of the hCoVs. At various time points, cells were fixed, permeabilized and stained with primary antibodies detecting the viral nucleocapsid proteins followed by an HRP-coupled secondary antibody (Fig. 3 ). High MOIs resulted in strong CPEs with massive cell death and detachment, resulting in low OD values (Fig. 3a). Lower MOIs resulted in higher ODs, which then decreased with further MOI dilutions. This MOI-dependent correlation with OD values was seen for all tested hCoVs (Fig. 3b and c; Supplemental Table 1). For all viruses, nucleocapsid could already be detected 2–3 days after inoculation, and at later time points also at low MOIs. However, hCoV-NL63 replicated the slowest and an optimal MOI correlation (r = 0.98, p < 0.001) was observed after 6 days (Fig. 3b; Table 1; Supplemental Fig. 4). For hCoV-229E and -OC43 optimal MOI correlations (r = 0.98, p < 0.001; r = 0.98, p < 0.001) were determined after 2 or 3 days, respectively (Fig. 3d; Table 1; Supplemental Fig. 4). Correspondingly, optimal assay performance was observed upon minimal dilution of the primary antibody but high dilution of the detection antibody (Table 2). For SOPs resulting in optimal and early assay evaluation, we chose MOIs of 0.002, 0.01, 0.006 for hCoV-229E, -NL63, and -OC43, respectively, which results in high ODs >0.55 and signal to noise ratios >6.8 (Table 2). At these conditions, the resulting Z’ factors of 0.83, 0.88, and 0.92 indicate low intra-assay variation, showing that this in-cell ELISA is suitable for high-throughput applications (Iversen et al., 2006) (Table 2).
Fig. 3.
Detection of hCoV-229E, -NL63 and -OC43 infection by in-cell ELISA. For establishing optimal conditions for the in-cell ELISA, target cells were seeded and the following day inoculated with a 10-fold or 2-fold serial dilutions of the respective virus. a) hCoV-229E was inoculated onto Huh-7 cells at a maximum MOI of 0.44, b) -NL63 onto Caco-2 cells at a maximum MOI of 1.6, and c) -OC43 onto Huh-7 at a maximum MOI 0.35. d) hCoV-229E and -OC43 were titrated again 2-fold at maximum MOI of 0.004 or 0.006, respectively. At the indicated time points post infection, cells were fixed and permeabilized before cells were treated with different concentrations of anti-nucleocapsid primary antibody (1st AB) and subsequently with different concentrations of HRP-coupled secondary detection antibody (2nd AB). Optical density (OD) was measured at 450 nm and baseline corrected at 620 nm. Signals obtained 2 h post infection (input control) were subtracted from later time points. Data are shown as mean of duplicates ± range, performed once.
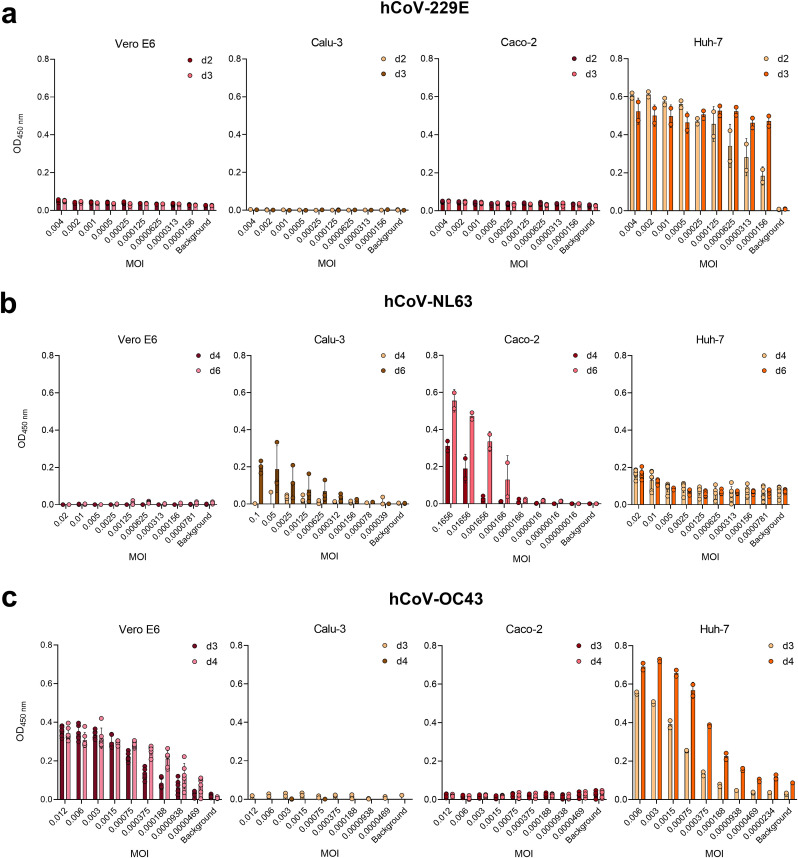
We then tested whether the in-cell ELISA can determine hCoV infection and replication in different cell lines. Vero E6 (epithelial monkey kidney), Calu-3 (epithelial human lung), Caco-2 (epithelial human colon) as well as Huh-7 (epithelial human liver) cells were inoculated with serially diluted hCoVs and infection rates were determined by in-cell ELISA (Table 2). In order not to miss weak infection or slow replication rates, we applied higher MOIs and performed the readout after additional days of cell culture. We found MOI-dependent signals of hCoV-229E infection only in Huh-7, but only marginal signals above background in Calu-3, Caco-2 and Vero E6 cells (Fig. 4 a). The cell line showing the highest OD and thus most susceptible to hCoV-NL63 infection was Caco-2, whereas modest infections signals were detected in Huh-7 and in Calu-3 cells upon high MOIs, and no infection in Vero E6 cells (Fig. 4b). hCoV-OC43 was the only virus capable to infect Vero E6 cells, and also infected Huh-7, but not Caco-2 or Calu-3 cells (Fig. 4c). These findings are in agreement with literature (Gerna et al., 2006; Herzog et al., 2008; Schirtzinger et al., 2022; Tang et al., 2005; von Brunn et al., 2015; Yoshikawa et al., 2010) and show the different tropism of hCoVs, and that the in-cell ELISA can be adapted to different cell lines, and can be used to determine cell susceptibility to infection.
Fig. 4.
Determination of cell line susceptibility to hCoV infection by in-cell ELISA. Vero E6, Calu-3, Caco-2 or Huh-7 cells were seeded and on the following day inoculated with a 2-fold serial dilution of a) hCoV-229E, b) -NL63, or c) -OC43 at indicated MOIs. According to the established in-cell ELISA, cells were fixed and stained at 2–6 days post infection (d2-d6). Input control measured 2 h post infection was subtracted from the corresponding values on later time points. Data are presented as mean ± SD of one experiment conducted in 2–6 replicates.
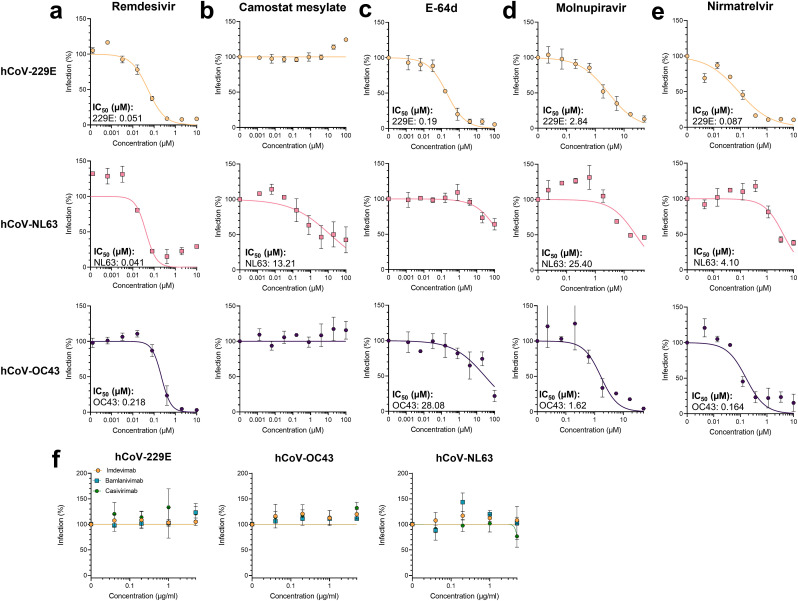
We next evaluated the in-cell ELISA SOPs for determining the activity of antivirals. Remdesivir inhibited all tested viruses with IC50 values of 0.051, 0.041, and 0.218 µM for hCoV-229E, -NL63, and -OC43, respectively (Fig. 5 a) without being cytotoxic (Supplemental Fig. 5a). These values agree with those determined by flow cytometry, and show that hCoV-NL63 is most and -OC43 least sensitive to remdesivir. We next assessed the protease inhibitors camostat mesylate and E-64d (Hoffmann et al., 2020), targeting cell surface associated TMPRSS2 or endosomal cathepsin B/L. Of note, activity of inhibitors targeting cellular factors might depend on the tested target cell, which in this experiment were Huh-7 cells for hCoV-229E and -OC43, and Caco-2 cells for -NL63. Camostat mesylate was only active against hCoV-NL63 and inhibited infection with an IC50 of 13.21 µM (Fig. 5b), which is in line with the fact that -NL63 is susceptible to TMPRSS2 cleavage (Kawase et al., 2012). E-64d was slightly cytotoxic at the highest concentration (Supplemental Fig. 5a) but to some degree inhibited all hCoVs with −229E (IC50 0.19 µM) being most sensitive (Fig. 5c). This shows that all viruses can use the endosomal entry pathway, and that hCoV-229E is most dependent on it (Shirato et al., 2017). In addition, we analyzed the novel anti-SARS-CoV-2 drugs molnupiravir (Malone et al., 2022), that received emergency use authorization by the Food and Drug Administration in the USA (U.S. Food & Drug Administration, 2021a), and nirmatrelvir (Owen et al., 2021) that is approved in the USA and the European Union (European Medicines Agency, 2022a; U.S. Food & Drug Administration, 2021b). Our assays show that all viruses are potently inhibited by the nucleoside analogue (Fig. 5d) as well as the 3C-like protease inhibitor (Fig. 5e). IC50 values are ranging from 1.62 µM to 25.40 µM for molnupiravir which is similar to the IC50 value for SARS-CoV-2 (European Medicines Agency, 2022b). Nirmatrelvir was described to have IC50 values of 0.078–0.215 µM against SARS-CoV-2 (Owen et al., 2021) and we determined values from 0.08 µM to 4.1 µM against the hCoVs. Broad activity has only been suggested due to the 3C-like protease inhibitor activity, and we could now confirm this using replicating viral isolates. This highlights the broad activity of the coronavirus-enzyme specific inhibitors, and underlines the feasibility of the in-cell ELISA for high-throughput antiviral drug testing. In a recent study, the in-cell ELISA has already been applied to study neutralizing serum titers against genuine hCoVs (Lawrenz et al., 2022). Additionally, there are several monoclonal antibodies targeting the SARS-CoV-2 spike protein applied in clinics to treat COVID-19 patients (Taylor et al., 2021). We here also tested whether the pharmaceutical antibodies imdevimab, bamlanivimab, or casirivimab also share neutralizing activities against the hCoVs. To this end, we pre-treated the hCoVs with antibodies and performed infections. As expected, the SARS-CoV-2 specific monoclonal antibodies did not cross-neutralize hCoVs (Fig. 5f, Supplemental Fig. 5b). In summary, the in-cell ELISA can be used to determine hCoV infection rates, cell susceptibility, antiviral activity of compounds as well as neutralization activities of antibodies.
Fig. 5.
Application of in-cell ELISA to evaluate antivirals against hCoVs. To analyze compounds, target cells were treated with 5-fold serial diluted a) remdesivir, b) camostat mesylate, c) E-64d, d) molnupiravir or e) nirmatrelvir before inoculation with hCoVs. f) To analyze antibodies, virus was pre-incubated for 30 min with imdevimab, bamlanivimab, or casirivimab before inoculating target cells. hCoV-229E experiments were performed on Huh-7 cells at an MOI of 0.002 and infection determined by in-cell ELISA 2 days later. hCoV-NL63 experiments were performed on Caco-2 cells at an MOI of 0.01 and infection determined by in-cell ELISA 6 days later. hCoV-OC43 experiments were performed on Huh-7 cells at an MOI of 0.006 and infection determined by in-cell ELISA 3 days later. Data were normalized to untreated virus control (100%) and are presented as mean ± SEM of triplicate infections (performed twice for remdesivir, molnupiravir and nirmatrelvir and three times for camostat mesylate and E-64d). Half-maximal inhibition concentrations (IC50) were calculated using a non-linear regression model.
4. Discussion
The emergence and rapid spread of SARS-CoV-2 once again illustrated the threat coronaviruses pose for public health. Coronaviruses not only burden humanity in the form of epidemics with high fatality rates but also through endemic and seasonal waves of infection that impact individual well-being (Corman et al., 2018; Fischer et al., 2021; Pyrc et al., 2007) and the economy (Bramley et al., 2002; Fendrick et al., 2003). Compared to the highly pathogenic SARS-CoV, MERS-CoV and SARS-CoV-2, studies on hCoVs are rare (Ljubin-Sternak et al., 2021) and the methodology for quantification of hCoV infection outdated, especially when it comes to the suitability for high-throughput testing. Here, we have established immunostaining-based flow cytometry and in-cell ELISA methods that allow quantification of hCoV infection in vitro. While the in-cell ELISA allows detection of bulk infected cells in a high-throughput manner, flow cytometry can be used to detect individual infected cells. Both assays can be applied to assess the susceptibility of cells to hCoV infection and replication, and to determine the efficacy of antiviral compounds and neutralizing antibodies. In addition, both methods have the advantage of providing a highly sensitive and virus-specific readout within a few days, which contrasts with CPE-based methods where infection rates are determined indirectly by virus-induced cell death that occurs with longer incubation times (Bracci et al., 2020; Montazeri Aliabadi et al., 2021). Due to the use of antibodies, material costs are slightly higher than for CPE-based assays, but very cost effective at ∼ US$ 4 for one 96 well in-cell ELISA plate. At ∼ US$ 26 per 96 well flow cytometry plate, this assay is more expensive but offers the advantage of single cell analysis, detection of additional proteins, and high sensitivity.
Viral plaque assays allow visualization of viral titers with high accuracy (Flint and Racianello, 2015) and small-scale compound testing. By determining not only plaque number but also area, the rate of viral replication and cell-to-cell spread can be quantified. However, plaque assays have the main disadvantage of being laborious and time consuming to perform (about 5 days to result) and lacking high-throughput capability. As shown for SARS-CoV-2, plaque assays can be used for screening approaches (Amarilla et al., 2021), but cannot compete with high-throughput assays. In contrast, the in-cell ELISA we developed allows quantification of hCoV infection by hCoV-229E, -NL63 and -OC43 in high sample numbers within a few days, without being laborious or expensive. Alternative qRT-PCR approaches have the disadvantage of being time consuming and expensive. We have already established in-cell ELISAs based on a similar approach for SARS-CoV-2 (Conzelmann et al., 2020) as well as for Zika virus (Müller et al., 2017). Thus, the protocols described here can be easily adapted to emerging viruses within a short time. Flow cytometry is more laborious than the in-cell ELISA, but offers higher sensitivity, allowing the analysis of single cells (Adan et al., 2017), and optionally additional expression of cellular proteins that might be affected.
For all hCoVs examined, the in-cell ELISA showed nucleocapsid expression that correlated with viral input and reached high Z’ factors over 0.8, indicating suitability for high-throughput applications (Zhang et al., 1999). In general, we aimed to measure infection as early as possible with ideally high OD and S/N-ratios. For hCoV-229E high signals were obtained already 2 days post infection and for -OC43 3–4 days post infection. As previously described, hCoV-NL63 replication was slow (Herzog et al., 2008), resulting in best OD and S/N values 6 days post infection. In this case, the higher sensitivity of the flow cytometry assay allows detection of hCoV-NL63 infection as early as 3 days post infection and may thus be the more appropriate assay. As stated above, assay results might also be affected by the choice of target cells, which may show various degrees of CPE or have high or low permissiveness for infection.
When evaluating in-cell ELISA data, we found that the assay can be adapted to different target cells but differs in performance. Vice versa, the assay is also useful in determining the susceptibility of cells to infection. In agreement with literature, hCoV-229E only infects Huh-7 cells (Gerna et al., 2006; Tang et al., 2005; von Brunn et al., 2015) but not Vero E6 (Tang et al., 2005) or Calu-3 cells (Yoshikawa et al., 2010), and -OC43 was able to infect Huh-7 (Gerna et al., 2006) and Vero E6 (Schirtzinger et al., 2022) but not Calu-3 cells (Yoshikawa et al., 2010). HCoV-NL63 was known to hardly infect Vero E6 (Gerna et al., 2006; Herzog et al., 2008) but replicated best in Caco-2 cells (Carbajo-Lozoya et al., 2014; Herzog et al., 2008) which was confirmed by our in-cell ELISA. Furthermore, our assay revealed that also Huh-7 or Calu-3 cells are suitable target cells for this coronavirus. This is in agreement with the fact that these cells express the hCoV-NL63 receptor ACE2 (Kawase et al., 2012; Zhang et al., 2021).
The choice of assay also depends on viral properties. Even if cells are permissive to infection, infection may not lead to CPE or plaque formation (Bracci et al., 2020). In the plaque assay, we observed that Huh-7 plaques appeared blurred and faint upon infection with hCoV-229E, making counting difficult but still suitable for quantification of plaque area using a software. Similarly, LLC-MK2 cells infected with hCoV-NL63 detached after infection, making plaque detection impossible (Herzog et al., 2008). None of the cell lines or conditions we studied resulted in visible plaques when infected with hCoV-OC43. Nevertheless, working with different lab-adapted strains, clinical isolates or cell lines may show different plaque morphology (Kato et al., 2017) (Bracci et al., 2020). These virus-dependent differences are most evident for hCoV-HKU1, which we did not analyze because none of the cell lines tested supported replication of this virus, that has only been cultured in primary human ciliated epithelial cells (Pyrc et al., 2010).
Both flow cytometry and in-cell ELISA were suitable to evaluate anti-coronaviral compounds. The IC50 values obtained for broad-acting remdesivir were 0.06 and 0.051 µM for hCoV-229E, 0.023 and 0.041 µM for -NL63, and 0.19 and 0.21 µM for -OC43, respectively by flow cytometry and in-cell ELISA, which is consistent between assays and within the ranges reported in the literature (Brown et al., 2019; Hsu et al., 2021; Parang et al., 2020). Only the hCoV-NL63 values differed from a study using qPCR of released viral genomes as an indicator of infection (Wang et al., 2021a). This highlights the caveats of quantifying antiviral activities by determining viral progeny after replication and supports our assay that directly measures infection rates. The described IC50 values of nucleoside analogue molnupiravir against the hCoVs (Wang et al., 2021b) are consistent with our findings, as is the described IC50 value of the SARS-CoV-2 protease inhibitor nirmatrelvir (PF-07321332) against hCoV-229E with an IC50 of 0.62 µM (Owen et al., 2021). Our findings now add evidence that nirmatrelvir is not only active against the coronavirus proteases but also against replicating hCoV-NL63 and -OC43. Thus, our in-cell ELISA showed that this orally administered drug is a broad-spectrum coronavirus inhibitor, with prospects for therapy of not only SARS-CoV-2 but seasonal coronaviruses. Furthermore, the in-cell ELISA has successfully been applied to determine neutralizing antibody titers against hCoVs (Lawrenz et al., 2022), and revealed herein that the clinically applied anti-SARS-CoV-2 monoclonal antibodies do not have broad activity against the hCoVs. This is to be expected as the RBD of the SARS-CoV-2 spike protein, which is the target of the monoclonal antibodies, is different from the hCoV RBDs (Hicks et al., 2021; Huang et al., 2020; Taylor et al., 2021). Overall, this highlights the applicability and advantage of the in-cell ELISA for antiviral drug testing.
The study of cellular protease inhibitors provides insight into viral processing and entry. Camostat mesylate inhibits TMPRSS2, which cleaves the spike proteins of SARS-CoV-2 and hCoV-229E and enables them to fuse at the plasma membrane (Hoffmann et al., 2020; Shirato et al., 2017). In contrast, the cathepsin B/L inhibitor E-64d interferes with spike processing in the endosome after viral uptake (Hoffmann et al., 2020; Shirato et al., 2017). Using in-cell ELISA, we found that hCoV-NL63 infection is more sensitive to TMPRSS2 than to inhibition of cathepsins, consistent with the fact that -NL63 appears to prefer TMPRSS2 over cathepsins (Kawase et al., 2012) which is expressed on the used Caco-2 cells (Hoffmann et al., 2020). Vice versa, hCoV-229E was strongly affected by E-64d and has previously been described to be dependent on cathepsin L (Shirato et al., 2017). However, the Huh-7 cells used do not express TMPRSS2 (Esumi et al., 2015; Zhang et al., 2021), thus inhibition by camostat mesylate was not expected and requires another cell line for evaluation of hCoV-229E and -OC43 TMPRSS2 dependency. There is a study that describes an inhibitory effect of camostat against hCoV-229E on TMPRSS2 positive cells, but this is also dependent on viral strain, isolate and passage (Shirato et al., 2017). Similar effects were described for hCoV-OC43, where the laboratory-adapted strain (VIR-1558, which was also used in our study) was not affected, but clinical isolates were affected by camostat mesylate (Shirato et al., 2018). This again highlights how viral strains and cell lines impact the experimental outcome, which needs to be considered when working on receptor or entry preferences and screening for antivirals. In particular, it is important to work with more recent hCoV isolates to investigate viral tropism and replication. Especially, strong antigenic drift has been described for hCoV-229E (Eguia et al., 2021) but laboratories are still experimenting with a 50-year-old laboratory-adapted isolate (Hamre and Procknow, 1966).
The ELISA, flow cytometry, and plaque protocols developed in this study for quantification of hCoV infections are easily adaptable to new strains and will facilitate future research on hCoVs. They have been shown to be reliable, widely applicable and suitable for high-throughput screening. Their application will enable the identification of hCoV inhibitors and screening for broad-acting anti-coronaviral compounds against highly pathogenic coronaviruses that may emerge in future. Thus, these new assays will facilitate coronavirus research and increase preparedness for future coronavirus outbreaks.
Contributors
Conceptualization, J.A.M., J.M., T.W.; Funding acquisition, J.A.M., J.M.; Investigation, J.L., T.W., A.S.; Essential resources, J.M., J.A.M.; Writing, J.A.M., T.W.; Review and editing, all authors. J.L., T.W. contributed equally. J.L., T.W., J.A.M. verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
Raw data are available from the corresponding author upon request.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 101003555 (Fight-nCoV) to J.M., the German Research Foundation (CRC1279) to J.M., and an individual research grant by the German Research Foundation (MU 4485/1-1) to J.A.M.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Daniela Krnavek and Nicola Schrott for skillful laboratory assistance. T.W. and A.S. are part of the International Graduate School in Molecular Medicine Ulm.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105343.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adan A., Alizada G., Kiraz Y., Baran Y., Nalbant A. Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- Amarilla A.A., Modhiran N., Setoh Y.X., Peng N.Y.G., Sng J.D.J., Liang B., McMillan C.L.D., Freney M.E., Cheung S.T.M., Chappell K.J., Khromykh A.A., Young P.R., Watterson D. An optimized high-throughput immuno-plaque assay for SARS-CoV-2. Front. Microbiol. 2021;12:75. doi: 10.3389/fmicb.2021.625136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry M., Richard V., Green J., Broult J., Musso D. Inactivation of Zika virus in plasma with amotosalen and ultraviolet A illumination. Transfusion. 2016;56:33–40. doi: 10.1111/trf.13271. [DOI] [PubMed] [Google Scholar]
- Bracci N., Pan H.-C., Lehman C., Kehn-Hall K., Lin S.-C. Improved plaque assay for human coronaviruses 229E and OC43. PeerJ. 2020;8 doi: 10.7717/peerj.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley T.J., Lerner D., Sames M. Productivity losses related to the common cold. J. Occup. Environ. Med. 2002;44:822–829. doi: 10.1097/00043764-200209000-00004. [DOI] [PubMed] [Google Scholar]
- Brown A.J., Won J.J., Graham R.L., Dinnon K.H., Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 2020;65:2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo-Lozoya J., Ma-Lauer Y., Malešević M., Theuerkorn M., Kahlert V., Prell E., von Brunn B., Muth D., Baumert T.F., Drosten C., Fischer G., von Brunn A. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184:44–53. doi: 10.1016/j.virusres.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann C., Gilg A., Groß R., Schütz D., Preising N., Ständker L., Jahrsdörfer B., Schrezenmeier H., Sparrer K.M.J., Stamminger T., Stenger S., Münch J., Müller J.A. An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antivir. Res. 2020;181 doi: 10.1016/j.antiviral.2020.104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa V.G., Moreli M.L., Saivish M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020;165:1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I., Malik Y.S., Sah R., Rabaan A.A., Panwar P.K., Singh K.P., Michalak I., Chaicumpa W., Martinez-Pulgarin D.F., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2 jumping the species barrier: zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Trav. Med. Infect. Dis. 2020;37:101830. doi: 10.1016/j.tmaid.2020.101830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguia R.T., Crawford K.H.D., Stevens-Ayers T., Kelnhofer-Millevolte L., Greninger A.L., Englund J.A., Boeckh M.J., Bloom J.D. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17:1–21. doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi M., Ishibashi M., Yamaguchi H., Nakajima S., Tai Y., Kikuta S., Sugitani M., Takayama T., Tahara M., Takeda M., Wakita T. Transmembrane serine protease TMPRSS2 activates hepatitis C virus infection. Hepatology. 2015;61:437–446. doi: 10.1002/hep.27426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency 2022. https://www.ema.europa.eu/en/medicines/human/EPAR/paxlovid Paxlovid [WWW Document]. URL.
- European Medicines Agency . 2022. Assessment Report EMA/719664. [Google Scholar]
- Fendrick A.M., Monto A.S., Nightengale B., Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 2003;163:487–494. doi: 10.1001/archinte.163.4.487. [DOI] [PubMed] [Google Scholar]
- Fischer N., Dauby N., Bossuyt N., Reynders M., Gérard M., Lacor P., Daelemans S., Lissoir B., Holemans X., Magerman K., Jouck D., Bourgeois M., Delaere B., Quoilin S., Van Gucht S., Thomas I., Barbezange C., Subissi L. Monitoring of human coronaviruses in Belgian primary care and hospitals, 2015–20: a surveillance study. The Lancet Microbe. 2021;2:e105–e114. doi: 10.1016/S2666-5247(20)30221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Racianello V. fourth ed. ASM Press; Washington: 2015. Principles of Virology. [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A., Baldanti F. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J. Med. Virol. 2006;78:938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog P., Drosten C., Müller M.A. Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol. J. 2008;5:138. doi: 10.1186/1743-422X-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.-P., Snead K.R., Drew M., Corbett K.S., Graham B.S., Hall M.D., Memoli M.J., Esposito D., Sadtler K. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. J. Clin. Immunol. 2021;41:906–913. doi: 10.1007/s10875-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose R., Watanabe N., Bandou R., Yoshida T., Daidoji T., Naito Y., Itoh Y., Nakaya T. In: Lee B., editor. Vol. 6. American Society for Microbiology; 2021. A Cytopathic Effect-Based Tissue Culture Method for HCoV-OC43 Titration Using TMPRSS2-Expressing VeroE6 Cells. (mSphere). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.Y., Yang C.W., Lee Y.Z., Lin Y.L., Chang S.Y., Yang R.B., Liang J.J., Chao T.L., Liao C.C., Kao H.C., Wu S.H., Chang J.Y., Sytwu H.K., Chen C.T., Lee S.J. Remdesivir and cyclosporine synergistically inhibit the human coronaviruses OC43 and SARS-CoV-2. Front. Pharmacol. 2021;12:1–11. doi: 10.3389/fphar.2021.706901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X., Xu W., Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen P.W., Eastwood B.J., Sittampalam G.S., Cox K.L. A comparison of assay performance measures in screening assays: signal window, Z’ factor, and assay variability ratio. J. Biomol. Screen. 2006;11:247–252. doi: 10.1177/1087057105285610. [DOI] [PubMed] [Google Scholar]
- Jeong G.U., Yoon G.Y., Moon H.W., Lee W., Hwang I., Kim H., Kim K.-D., Kim C., Ahn D.-G., Kim B.-T., Kim S.-J., Kwon Y.-C. Comparison of plaque size, thermal stability, and replication rate among SARS-CoV-2 variants of concern. Viruses. 2021;14:55. doi: 10.3390/v14010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato F., Tajima S., Nakayama E., Kawai Y., Taniguchi S., Shibasaki K., Taira M., Maeki T., Lim C.K., Takasaki T., Saijo M. Characterization of large and small-plaque variants in the Zika virus clinical isolate ZIKV/Hu/S36/Chiba/2016. Sci. Rep. 2017;7:16160. doi: 10.1038/s41598-017-16475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014–2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenz J., Xie Q., Zech F., Weil T., Seidel A., Krnavek D., van der Hoek L., Münch J., Müller J.A., Kirchhoff F. SARS-CoV-2 vaccination boosts neutralizing activity against seasonal human coronaviruses. Clin. Infect. Dis. 2022;37 doi: 10.1093/cid/ciac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Liu Q., Huang W., Li X., Wang Y. Current status on the development of pseudoviruses for enveloped viruses. Rev. Med. Virol. 2018;28 doi: 10.1002/rmv.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang X., Nair H. Global seasonality of human seasonal coronaviruses: a clue for postpandemic circulating season of severe acute respiratory syndrome coronavirus 2? J. Infect. Dis. 2020;222:1090–1097. doi: 10.1093/infdis/jiaa436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubin-Sternak S., Meštrović T., Lukšić I., Mijač M., Vraneš J. Seasonal coronaviruses and other neglected respiratory viruses: a global perspective and a local snapshot. Front. Public Health. 2021;9:1–10. doi: 10.3389/fpubh.2021.691163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazeri Aliabadi H., Totonchy J., Mahdipoor P., Parang K., Uludağ H. Suppression of human coronavirus 229E infection in lung fibroblast cells via RNA interference. Front. Nanotechnol. 2021;3 doi: 10.3389/fnano.2021.670543. [DOI] [Google Scholar]
- Müller J.A., Harms M., Schubert A., Mayer B., Jansen S., Herbeuval J.-P., Michel D., Mertens T., Vapalahti O., Schmidt-Chanasit J., Münch J. Development of a high-throughput colorimetric Zika virus infection assay. Med. Microbiol. Immunol. 2017;206:175–185. doi: 10.1007/s00430-017-0493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. (80-) [DOI] [PubMed] [Google Scholar]
- Parang K., El-Sayed N.S., Kazeminy A.J., Tiwari R.K. Comparative antiviral activity of remdesivir and anti-HIV nucleoside analogs against human coronavirus 229E (HCoV-229E) Molecules. 2020;25:2343. doi: 10.3390/molecules25102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee Y., Michelow I.C., Choe Y.J. Global seasonality of human coronaviruses: a systematic Review. Open Forum Infect. Dis. 2020;7 doi: 10.1093/ofid/ofaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Berkhout B., van der Hoek L. Antiviral strategies against human coronaviruses. Infect. Disord. - Drug Targets. 2007;7:59–66. doi: 10.2174/187152607780090757. [DOI] [PubMed] [Google Scholar]
- Pyrc K., Sims A.C., Dijkman R., Jebbink M., Long C., Deming D., Donaldson E., Vabret A., Baric R., Hoek L. van der, Pickles R. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 2010;84 doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirtzinger E.E., Kim Y., Davis A.S. Improving human coronavirus OC43 (HCoV-OC43) research comparability in studies using HCoV-OC43 as a surrogate for SARS-CoV-2. J. Virol. Methods. 2022;299:114317. doi: 10.1016/j.jviromet.2021.114317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler L., Le-Trilling V.T.K., Eilbrecht M., Mennerich D., Anastasiou O.E., Krawczyk A., Herrmann A., Dittmer U., Trilling M. A novel in-cell ELISA assay allows rapid and automated quantification of SARS-CoV-2 to analyze neutralizing antibodies and antiviral compounds. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.573526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 2017;91 doi: 10.1128/jvi.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B.S.F., Chan K., Cheng V.C.C., Woo P.C.Y., Lau S.K.P., Lam C.C.K., Chan T., Wu A.K.L., Hung I.F.N., Leung S., Yuen K. Comparative host gene transcription by microarray analysis early after infection of the Huh 7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 2005;79:6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food & Drug Administration . Coronavirus (COVID-19) Update: FDA Authorizes Additional Oral Antiviral for Treatment of COVID-19 in Certain Adults. 2021. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain [Google Scholar]
- U.S. Food & Drug Administration 2021. https://www.ema.europa.eu/en/medicines/human/EPAR/paxlovid Emergency Use Authorization [WWW Document]. URL.
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Ciesek S., von Brunn B., Carbajo-Lozoya J. Genetic deficiency and polymorphisms of cyclophilin A reveal its essential role for Human Coronavirus 229E replication. Curr. Opin. Virol. 2015;14:56–61. doi: 10.1016/j.coviro.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li P., Rajpoot S., Saqib U., Yu P., Li Yunlong, Li Yang, Ma Z., Baig M.S., Pan Q. Comparative assessment of favipiravir and remdesivir against human coronavirus NL63 in molecular docking and cell culture models. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-02972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li P., Solanki K., Li Y., Ma Z., Peppelenbosch M.P., Baig M.S., Pan Q. Viral polymerase binding and broad-spectrum antiviral activity of molnupiravir against human seasonal coronaviruses. Virology. 2021;564:33–38. doi: 10.1016/j.virol.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C., Chan K., Tsoi H., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Hill T.E., Yoshikawa N., Popov V.L., Galindo C.L., Garner H.R., Peters C.J., Tseng C.-T. (Kent) Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Li W., Feng J., Ramos da Silva S., Ju E., Zhang H., Chang Y., Moore P.S., Guo H., Gao S.J. SARS-CoV-2 pseudovirus infectivity and expression of viral entry-related factors ACE2, TMPRSS2, Kim-1, and NRP-1 in human cells from the respiratory, urinary, digestive, reproductive, and immune systems. J. Med. Virol. 2021;93:6671–6685. doi: 10.1002/jmv.27244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-H., Chung T.D.Y., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.