Abstract
Viral infections are responsible for the deaths of millions of people throughout the world. Since outbreak of highly contagious and mutant viruses such as contemporary sars-cov-2 pandemic, has challenged the conventional diagnostic methods, the entity of a thoroughly sensitive, specific, rapid and inexpensive detecting technique with minimum level of false-positivity or -negativity, is desperately needed more than any time in the past decades. Biosensors as minimized devices could detect viruses in simple formats. So far, various nucleic acid, immune- and protein-based biosensors were designed and tested for recognizing the genome, antigen, or protein level of viruses, respectively; however, nucleic acid-based sensing techniques, which is the foundation of constructing genosensors, are preferred not only because of their ultra-sensitivity and applicability in the early stages of infections but also for their ability to differentiate various strains of the same virus. To date, the review articles related to genosensors are just confined to particular pathogenic diseases; In this regard, the present review covers comprehensive information of the research progress of the electrochemical, optical, and surface plasmon resonance (SPR) genosensors that applied for human viruses' diseases detection and also provides a well description of viruses' clinical importance, the conventional diagnosis approaches of viruses and their disadvantages. This review would address the limitations in the current developments as well as the future challenges involved in the successful construction of sensing approaches with the functionalized nanomaterials and also allow exploring into core-research works regarding this area.
Keywords: Genosensor, Virus detection, Electrochemical biosensors, Optical-base sensing platforms
1. Introduction
Viral infections are one of the global concerns that threaten the health, social, and economic dimensions of the life of millions of people. The well-characterized and newly-emerged viruses caused dynamic epidemics and pandemics around the world. A wide range of viruses with special structural and pathogenicity features have been detected by scientists. For example, Hepatitis B virus (HBV), Hepatitis C virus (HCV), Human immunodeficiency virus (HIV), Human papillomavirus (HPV), Influenza viruses, Arboviruses like Zika virus (ZIKV), Dengue virus (DENV), and Chikungunya are most important and studied infectious agents for human [[1], [2], [3], [4], [5], [6], [7]]. Although most viral diseases are asymptomatic or mild, however, remarkable number of viruses are caused by deadly acute or chronic illnesses. Hepatitis viruses (A to E and G) target the liver and are responsible for hepatic and extrahepatic failures such as chronic hepatitis, occult hepatitis cirrhosis, hepatocellular carcinoma, hematologic, and renal disorders [4]. According to the World Health Organization (WHO) report, 1/3 people in the world have been infected by either HBV or HCV and around 1.3 million people have died of viral hepatitis in 2015 [8]. Acquired immune deficiency syndrome (AIDS), tumors, aplastic and hemolytic anemia are fatal diseases are caused by Retroviruses [[9], [10], [11]]; for instance, 1.94 million new cases of HIV infection and 0.95 million deaths were reported in 2017 [12]. Arboviruses led to acute viral hemorrhagic fevers (VHFs) including; Dengue fever (DF), Dengue hemorrhagic fever (DHF), dengue shock syndrome (DSS), bleeding from organs, Marburg hemorrhagic fever (MHF), Ebola hemorrhagic fever (EHF), and yellow fever [[13], [14], [15]]. Different subtypes of influenza virus have resulted in acute and deadly pandemics in different nations during the 20thand earliest of the current century [16]. Meanwhile, severe acute respiratory syndrome coronavirus 2 (SARS- CoV-2) is responsible for the newly emerged viral pandemic named COVID-19 which is still killing millions of people [17]. In acute infections, clinical signs are nonspecific and time is a very important factor in the control of rapidly epidemiologically dangerous diseases such as Ebola, Dengue fever, Influenza, SARS-CoV-2, and MERS. On the other side, effective prevention of infection spread, clinical management, and successful therapeutic in chronic diseases including HIV and viral hepatitis depends on early diagnoses of the pathogen [18]. Consequently, detection tests of viral diseases should be applicable in the early stages of infection.
Generally, different ranges of old and newly-emerged diagnostic tests and toolkits including electron microscope (EM), virus isolation by cell culture, immunoassay-based tests, amplification-based approaches, next-generation sequencing (NGS), and mass spectrometry (MS) have been developed for recognition of viruses [[19], [20], [21], [22], [23], [24]](Table 1 ). Each diagnostic test has its advantages and disadvantages which restrict its application in the detection of viral pathogens (Table 1). Since 1941, electron microscope (EM) was a useful tool for the direct detection of viruses in human specimens [25]. Although, EM is one of the direct diagnostic techniques for visualization and counting of the viral particles but now is seldom considered as a detection method [26,27]. Cell culture-based techniques are direct gold standard methods for virus propagation and isolation and also are used for assessing the cytotoxicity and cytopathic effects (CPE) of viruses [28]. Since the 1960s, virus culturing in the human and animal cell line was popular in clinical virology, nevertheless, low sensitivity, long time required for virus isolation, and need for experienced personnel limit the usefulness of this diagnostic assay in the clinical [27,29]. Thus, cell culture-based methods could not be considered as a rapid test for the diagnosis of viral illnesses at the onset of infection. Antibody production is one of the immune system responses to fight against viruses. Thus, testing the level of antibodies or antigens by immunoassay-based techniques is an effective approach for the diagnosis of viral pathogens in clinical specimens [30,31]. Up to now, different types of immunoassays were designed and tested for viruses (Table 1), however, The Enzyme-Linked Immunosorbent Assay (ELISA) and Radio-immunoassay (RIA) are well-known [20,32]. The formation of the antigen-antibody complex is a general basis of them [33]. Although they have acceptable sensitivity, specificity and are easily settled but the high cost of reagents, being time-consuming, false-positive or negative results are their drawbacks and -restrictions in clinical diagnostic [34,35].
Table 1.
A summary of diagnosis testing approaches of viral infections.
| Diagnostic Test Methods Advantages Disadvantages Ref | ||||
|---|---|---|---|---|
| Electron microscope (EM) | TEMa SEMb |
Good for direct morphology information and counting of viral particles | Expensive, time-consuming, required to substantial technical skills | [71] |
| Virus isolation | Conventional cell culture shell vial technique | Acceptable specificity and sensitivity | Low sensitivity, long time required for virus isolation, need for experienced personnel | [20] |
| Immunoassay-based tests | ELISAc RIAd CAe MEIAf CLIAg FPIAh HIi |
High sensitivity and specificity, easily settled | High cost of reagents, time-consuming, false-positive or negative results, | [29], [72], [73], [74], [75], |
|
Molecular techniques . |
PCRj Real-time PCR RT-PCRk DNA Microarrays LAMPl NGSm |
cost-effectiveness, reliability and accuracy of results | Need to experienced operators, risk of contamination, need to specific primers for every target | [22], [76], [77] |
Transmission electron microscopy.
Scanning electron microscopy.
Enzyme-linked immunosorbent assay.
Radioimmunoassay.
Immunochemiluminescent Assay.
Micro-particle enzyme immunoassay.
Chemiluminescent immunoassay.
Fluorescence polarization immunoassay.
Hemagglutination Inhibition.
Polymerase chain reaction.
Reverse Transcription-PCR.
Loop-Mediated Isothermal Amplification.
Next-Generation Sequencing.
In the past years, virus testing methods have experienced significant developments and molecular procedures have been become inevitable methods for the detection of viruses, with accurate and rapid results [36,37]. Currently, gene amplification-based methods are considered as a reference method for the detection of many viruses [22]. The molecular diagnostic procedures have wide applications in virology and they are used for virus detection and co-infections, genotyping genome sequencing, measuring the viral load, and management of infection [[38], [39], [40], [41], [42], [43]]. Up to now, several modifications of PCR (polymerase chain reaction) were designed and patented to increase the capability of molecular tests (Table 1) [29,44]. However, conventional PCR, real-time PCR, and reverse transcription-PCR (RT-PCR) are the most popular and routine variants of molecular tests for the detection of the virus genome in clinical laboratories [45,46]. Besides the strengths of molecular diagnostic approaches including great sensitivity and specificity, this method is very time-wasting and creates the need for the assessment of virus genome in a faster way. The importance of this issue prompted the bio-scientists to focus on fabricating and testing the smart biosensing assays to immediate detection of viruses with the highest sensitivity, accuracy, specificity, selectivity, and simplicity, and eventually, their attempts led to designing several nucleotide and immunoassay-based sensors for the detection of wide range viral pathogens in clinical samples [3,36,[47], [48], [49], [50]].
However, there exists some challenges and direction towards developing genosensors including: sensitivity, repeatability, analysis time and non-specific signals. For example, small dimensions of the nucleic acids and the nature of the attachment of DNA/RNA probe onto the electrode surface and large amount of sequences required to sensitize the sensor, make more difficult repeatability, sensitivity and specificity of such types of sensing devices [[51], [52], [53]]. Hence in recent years, they have been partially overcome by the positioning of the reporter between the base mismatches and the electrode surface and appearance new sensors fabricated by nanotechnology. But to succeed in commercialization requires more effort [[54], [55], [56], [57]].
There are various review articles concerning the biosensor's application in health care and medical laboratory [[58], [59], [60], [61], [62]]. These articles include studies about a particular type of biosensor, e.g. electrochemical, for detecting none-human and human-infecting viruses [[63], [64], [65]] or pathogens in general [[66], [67], [68], [69]] and implementation of biosensor in diagnosis of a specific infectious disease like human papilloma virus (HPV) [70]. Hence, in this review, we have contributed comprehensive information through utilized genosensors in virus detection. First clinical approaches for recognition of various kinds of viruses will be reviewed. Afterwards, different sensing platforms based on genosensing assays for highly sensitive determination of viruses will be discussed. Finally, a summary and future outlook of the current limitations and upcoming challenges in analyzing of various kinds of viruses will be explained as broadly as possible.
2. Genosensors
Biosensors have become very well-known in the last two decades and a wide range of them have increasingly been utilized in the field of food, forensics, bioterrorism, environmental, and in the therapeutic and diagnostic area of medicine [[78], [79], [80]]. Generally, biosensors are small analytical tools used for quantification and/or detection of biological analytes. In these bio-sensing platforms, biologically active elements including DNA, RNA, enzyme, antibody, protein, etc produce a measurable signal using the transducers [[81], [82], [83]]. The surprising advances in the biosensor branch and integration of this field with medicine, and also the inability of conventional diagnostic techniques to monitor the infectious agents with ultra-sensitivity lead to the development of diagnostic methods for recognition of pathogenic microorganisms (viruses, bacteria, fungi) in the clinical samples [[84], [85], [86]]. Although, nucleic acid, immuno, and protein-based biosensors could recognize and measure the genome, antigen, or protein level of microorganisms and the triggered immune responses against a pathogen, however, nucleic acid-based sensing techniques because of having ultra-sensitivity and applicability in the early stages of infections are preferred to the others [84].
Genes are genomic distinct region that contains the information for the synthesis of proteins. Gene-based recognition approaches such as nucleotide-based sensors rapidly developed for the detection of genes illnesses, especially viral infections in recent years [87]. The nucleotide-based sensing (NABSs) or so-called “genosensors” are biological devices that could recognize the target nucleic acids (DNA or RNA) based hybridization reaction [88]. The single-strand DNA (ssDNA) sequences called probes and target nucleic acid sequence are recognition elements of genosensors which their hybridization is monitored by a direct format, but, sometimes, the ss-DNA probe-target DNA complex on the surface of the sensor could not produce desirable changes in the transduction values, so, to improve the detection limits, sandwich, and competitive formats are preferred to the direct ones [[89], [90], [91], [92]]. The principle of direct format is based on label-free detection by immobilization of ssDNA probe on a transducer surface, while in the sandwich and competitive formats, a mixture of DNA target and incubated ssDNA probe on the surface of the sensor through a specific label is used for recognition [93].
The biorecognition of oligonucleotides by genosensors is more useful and more valuable than the other types of biosensors because the complementarity of oligonucleotides is very specific, accurate, and potent. As the immobilization of single-stranded DNA sensor on the surface of recognition layer, such oligonucleotide binds to the target DNA based on base-pairing interaction. The gathering of target DNA molecules on the recognition surface gives rise to the generation of signals followed by their amplification [94]. The selection of transducer elements has a significant influence on the sensitivity and specificity of genosensors and it depends on the physio-chemical alterations of the biochemical reaction that happens in the biorecognition-layer. Genosensors have been divided into electrochemical (amperometric, voltammetric, and impedimetric), optical (surface plasmon resonance, colorimetric, fluorescent, luminescent, and interferometric), calorimetric, mass-based (magnetic, piezoelectric, quartz crystal microbalance, and acoustic-wave) based on transducer element [95],and, newly, they mostly considered for the detection of viruses genome based on hybridization ability of oligonucleotides [91,[96], [97], [98]].
Electrochemical genosensors are most popular because of their high sensitivity and specificity, rapid, and low-cost bio-sensing platforms for viral infections detection [99]. Fig. 1 represents the structure of an electrochemical genosensor developed for a DNA virus genome diagnosis; in these sensors, a hybridization event between an immobilized ssDNA probe onto the surface of the electrode and its complementary DNA produces a detectable quantitative signal, and eventually signal detection is done in two ways; i: direct detection (label-free) based on nucleotide oxidation in the DNA probe and ii: indirect detection (label-based) through using an indicator of the hybridization process [100]. Although, increasing the immobilization and DNA probe accessibility for DNA target is the main disadvantage of these genosensors but this foible is easily improvable by using form metal and oxide nanomaterials and nanoparticles including silver [101], gold [48], osmium [102], palladium [103], and platinum [104]; among them, gold nanoparticles are first choice to increase the sensitivity of electrochemical genosensors for the detection of DNA [105].
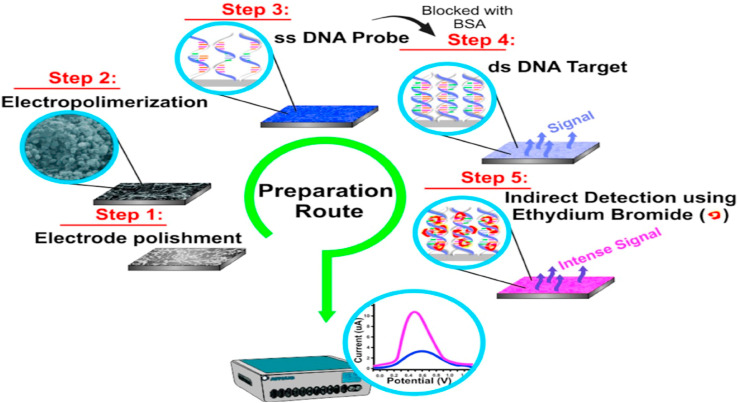
Fig. 1.
Schematic presentation of an electrochemical genosensor developed for detection of a DNA virus. Step 1) Preparation and modification of graphite electrode with poly(4-aminophenol) on the sensitive surface, Step 2) Immobilization of oligonucleotide probes (ssDNA) by dropping poly (GA) or HepB1 onto the modified graphite electrodes, Step 3)Hybridization Of negative and positive (ds DNA) serum samples with poly (CT) or HepB2, Step 4)The electrode rinsing by immersion in phosphate buffer solution for 30 s under agitation, and Step 5) Analysis and detection of transduced electrochemical signal in sera samples after upon adding ethidium bromide to the electrode surface. This figure is obtained with permission from Ref. [100].
Piezoelectric bio-sensing devices or more precisely piezoelectric crystals are label-free monitoring methods that work based on oscillations change because of a mass bound on the transducer like quartz crystal disks. Good sensitivity and low price are the benefits of these types of sensors, however they are not completely commercialized and still applied in experimental studies [106,107]. Surface plasmon resonance (SPR) sensors are other types of are optical sensors that work based on internal reflection in a glass prism. In SPR genosensors polarized plasmon between analytes and biomolecules are online detectable without any need for labeling process. Besides these advantages, in the conventional SPR method, expensive sensor chips are used in biosensing tools and protein or ligand immobilization requires complicated chemistry, as well [108,109].
The quartz crystal microbalance (QCM) is a type of platform for biosensors that working on piezoelectric principle. Even though QCM-based biosensors are not as common as other biosensing methods like optical or voltametric ones, they are very popular in the field of pathogen detection because this method is highly sensitive, cheap and able to detect any types of biomolecules via a label-free method as well as quick response [110,111]. This system provides a selective and sensitive scheme to detect viral infections, and can be applied to an early screening of epidemic situations; further, provide effective methods for outbreak management and public health management [112].
3. Investigation of the various genosensing platforms for detection of different viral pathogens
3.1. Hepatitis B virus (HBV)
The most important member of viral hepatitis causes is HBV. This virus is a small DNA virus that could expose the liver of an infected person to several horrible liver-related diseases such as liver cancer or hepatocellular carcinoma. The infection is a worldwide health challenge that involves approximately 300 million people around the world with various manifestations from simple liver failure to cancer. The thing which deteriorates this infection is to become chronic if the virus, not is cleared from the patient's body by the immune system. Furthermore, the routine molecular and immune-based detection methods used for HBV monitoring are applicable only after several times upon infection [2]; hence, to overcome this limitation, abundant types of genosensors have been designed and used to diagnose this virus genome in the early days post-infection from clinical specimens during recent years [4,48,105]. Table 2 gives an overview of different types of genosensors designed and used for HBV detection.
Table 2.
The overview of various reported genosensing platforms for the detection of HBV.
| Genosensor type Method electrode assay time linear range detection limit Ref | |
|---|---|
|
Electrochemical Piezoelectric SPR |
Impedimetric Gold−1 × 10−18 M − 1 × 10−6 M 1 × 10−18 M [126] Voltammetric Cobalt phenanthroline−281–563 × 10 −6 M 2.46 × 10−8M [127] Impedimetric Glassy carbon−0.0005–0.5 × 10 −9 M 0.082 × 10 −12 M [128] Impedimetric Gold ∼ 1 h 3.48285–15.00996 × 10 −15 M0.33,705 × 10 −15 M [129] Voltammetric Gold ∼170 min 1.10 × 103–1.21 × 105 copies/mL 1100 copies/mL [130] Voltammetric Gold−4.00 × 10−13M to 4.00 × 10−9M 3.00 × 10−13M [131] Voltammetric Gold−1.0 × 10−12–10.0 × 0–6 M 2.0 × 10−12M [132] QCM Gold 30 min - 1.6 × 10−18 M [133] QCM Gold 1 h 0.8–38.8 × 10−3M 0.8 × 10 −3M [134] QDs ∼ 4 h 106 IU/mL to 101 IU/mL. 103 IU/mL [135] LSPR Silver 70 min0.5–100 × 10−15M 5 × 1017 M [136] FRET Gold 50 min 0.045–6.0 × 10 −9 M15 × 10 −12M [137] |
QDs; Quantum Dots, LSPR; localized-SPR, FRET; Fluorescence resonance energy transfer.
Electrochemical genosensors are commonly designed bio-sensing platforms for HBV detection [99]. Until now, various types of electrochemical genosensors based on transducer elements including amperometric, voltammetric, and impedimetric [113] sensors with variable detection of limit (LOD) were developed for detecting of HBV genome [[114], [115], [116], [117], [118], [119]]. For example, in 2020, M Shariati et al., have fabricated a highly sensitive impedimetric electrochemical genosensor based on tin-doped WO3/In2O3 nanowire (WO3/indium tin oxide (ITO) NW) for HBV diagnosis. In this bio-sensing method, tin-dopedWO3/In2O3 nanowires have synthesized through physical vapor deposition and then four DNA sequences including thiolated ssDNA probe, complementary (a sequence of HBV genome), non-complimentary, and mismatch targets were used for bio-recognition aims on the surface of these nanowires. The label-free designed genosensor detected the hybridization among probe and HBV DNA through the oxidation reactions at very low concentrations of DNA with liner range at 0.1 × 10 −12 M to10−5 M. This is an electrochemical bio-detection under laser emission led to releasing of more charged carriers and their interaction with DNA on the electrode surface and achieved the detection limit of 1 fM. Applying the laser amplification, photogeneration process, and electron-hole recombination rate processes are unique features of this study, which all of them led to distinguishable detection of complementary HBV DNA from the mismatch and non-complementary target sequences with ultrahigh sensitivity, high selectivity, and 96% stability of its initial activity after six weeks [120].
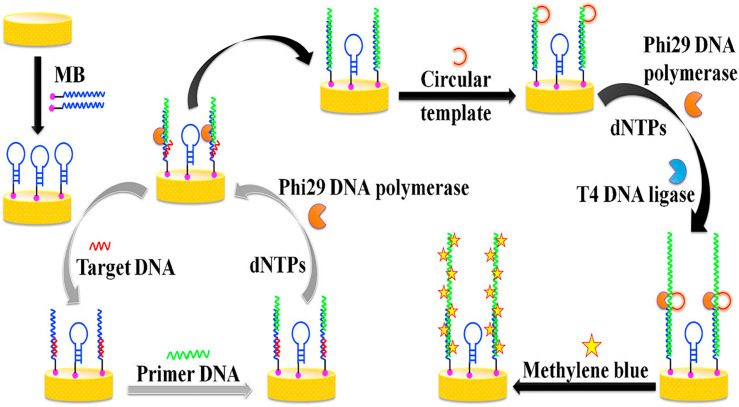
In an investigation in 2017, an electrochemical genosensor platform was constructed for the diagnosis of HBV genome in the clinical samples by the cascade signal amplification based on a molecular beacon (MB) mediated circular strand displacement (CSD) and rolling circle amplification (RCA) strategies. In this label-free genosensor, target DNA was dropped on the surface of a gold electrode modified with molecular beacon (MB/GE), then MB mediated CSD was performed and eventually viral genome bio-sensing was followed by adding the circular template onto the electrode surface. To complete the detection, methylene blue was added onto the surface of gold electrodes to increase the differential pulse voltammetric (DPV) response of their sensor (Fig. 2 ). The platform's LOD reached 2.6 × 10−18 M with a liner range of 1 × 10−17 to 7 × 10−16 M of target DNA [115].
Fig. 2.
The scheme illustrates the detection of the HBV genome via a voltammetric electrochemical genosensor preparation process. Present diagram represents the preparation and modification processes of gold electrode modified with molecular beacon (MB/GE) using bare GE and MB to bio-sensing of the ssDNA sequence derived from the genome of hepatitis B virus (HBV) by circular strand displacement (CSD) and rolling circle amplification (RCA) strategies mediated by a molecular beacon (MB). Hence, the target ssDNA hybridizes with the loop section of the MB/GE, while the rest of partial DNA sequences of MB hybridizes with primer DNA. In the following steps, the RCA initiates in the presence of dNTPs, Phi 29 DNA polymerase, and T4DNA ligase to produce a long DNA strand with multiple tandem-repeat sequences and this results led to detection of the induced voltammetric signals in the presence of electrochemical probe Methylene Blue. This figure is obtained with permission from Ref. [115].
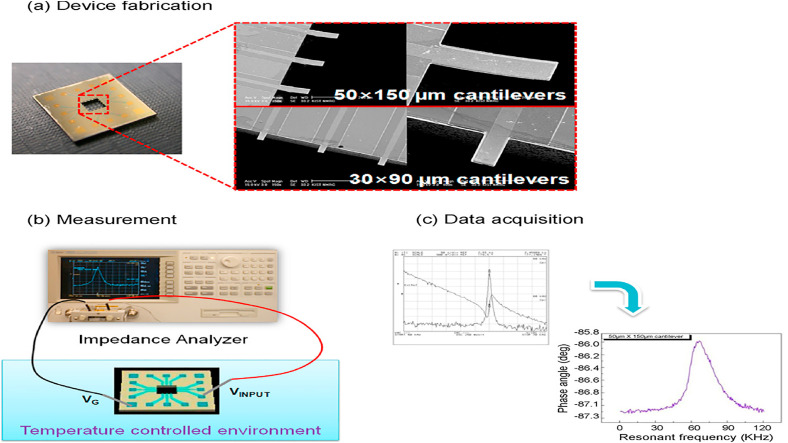
Besides the electrochemical assays, piezoelectric genosensors were designed for HBV monitoring [36,121]. In the last two decades, three types of these label-free bio-sensing platforms such as quartz crystal microbalance (QCM) [122], microcantilever biosensors [123], and surface acoustic wave (SAW) [2] were tested for detecting HBV DNA in clinical samples; some of them are listed in Table 2. For real-time detection of HBV-specific sequences genome, Ya et al., have constructed a peptide nucleic acid (PNA) piezoelectric genosensor. In the PNA QCM platform, the biotin-avidin method was applied to deposition of PNA and RecA protein-coated ssDNA probe on the surface of a crystal disk following a common traditional procedure. The data showed that hybridization was done in less than 1-h time with 19.3242 × 10−18 M limit of detection rate and 94.44% specificity in comparison with real time-PCR assay. The constructed PNA probe could able to distinguish sequences of target DNA that were different only in one base, hence, improving the efficiency sensitivity of the sensor and reducing time needs for doing the test were the main reasons for using this protein-coated probe [122]. In another piezoelectric sensing platform, a silica nanoparticle (SiNPs)-improved dynamic microcantilever platform was tested for HBV DNA detection. Conjugation of detection probe with silica nanoparticles and capture probe with the surface of microcantilever disks were conducted to detection of 243 base pair (bp) of the core region of HBV target DNA. The sequence 243 bp of target DNA was diagnosed up to pM level without using of nanoparticles (NPs) and up to fM level in silica NPs-improved microcantilever platform. As depicted in Fig. 3 , a linear correlation was seen among resonant frequency alterations and HBV target concentrations, which was due to the increased mass arising from the binding of the probe to the HBV DNA and SiNPs to the HBV DNA to improve the signal. The detection limit of 2.3 × 10−15 M and around 2–3 folds of enhancement in sensitivity were salient features of this label-free procedure [124].
Fig. 3.
Measurement of the resonant frequency and data receiving from a dynamic microcantilever: (a)is a picture of a device containing 12 microcantilevers and SEM photographs of 2 kinds of microcantilevers, (b)is a diagram of equipment utilized for estimation of the frequency resonant from the dynamic microcantilevers, which uses form an impedance analyzer, and (c) results of frequency resonant which is achieved the impedance signal. This figure is obtained with permission from Ref. [124].
Furthermore, SPR sensors were utilized for HBV genome detection [105] (Table 2). These NABSs sensors are optical sensors that work based on internal reflection in a glass prism. In SPR genosensors polarized plasmon between analytes and biomolecules are online detectable without any need for the labeling process [108,109]. In a research work, Chuang and co-workers, a simple and inexpensive SPR genosensor platform were constructed based on a loop-mediated isothermal amplification (LAMP) way to the determination of HBV DNA. Their sensor was a new SPR based LAMP geno-sensing cartridge containing a polycarbonate (PC) prism and a polymethyl methacrylate (PMMA) surface. The limit of detection 4.494 × 10−18 M and almost 0.0011 refractive indexes (RI) were achieved by the SPRLAMP genosensing system for HBV DNA templates in only 17 min [125].
3.2. Hepatitis C virus
Hepatitis C virus (HCV), a positive-sense single-stranded RNA virus from the Flaviviridae family, is responsible for blood-borne non-A and non-B hepatitis [138]. The clinical manifestations of this infectious agent are variable from hepatic findings such as chronic cirrhosis and hepatocellular carcinoma to non-specific extrahepatic signs including cryoglobulinemia, renal, and dermatologic disorders. HCV could infect ∼130–170 million individuals around the world by transmission through contact with infected blood, organs, and body fluids [139]. Generally, PCR-based methods are known as the gold standard detection tests of HCV infection in the clinical laboratories, however, they are costly, time-consuming, and are applicable by specialized equipment and technician several months post-infection [140]. On the other hand, HCV patients have a low cure chance. So, due to the aforementioned restrictions of conventional HCV monitoring techniques, different types of electrochemical, Piezoelectric, andSPR-NABSs were developed to HVC RNA detection in the early stages of infection and increase the efficiency of treatment [139,[141], [142], [143], [144]]. Although the earlier applied DNA-based sensors for HCV detection require PCR technology for cDNA production from viral RNA [139,140,143], fortunately, using nanoparticles and nanomaterials improved the PCR-based genosensors disadvantages with increased portability, accuracy, and reliability [139]. For example, a rapid, simple, and cost-effective genosensor for direct detection of unamplified HCV RNA using gold nanoparticles with the sensitivity of 92%, specificity of 88.9%, and detection limit of 50 copies/reaction within 1 min was designed for HCV genome screening in the clinical specimens [140,145]. Albeit, HCV infection is detectable by both immunosensors and genosensors, but immunosensors are unable to discriminate between current and previous infection, and also detect the pathogen in the absence of antibody during the acute phase. Hence, NABSs with the highest sensitivity and selectivity are the best choices for HCV genome screening, viral load measurement, and HCV genotyping in clinical specimens [146,147]. Table 3 represents some examples of various types of genosensors developed for HVC detection.
Table 3.
List of some nucleic acid-based biosensors (NABSs) for HCV genome detection investigated in present article.
| Genosensor type Method Electrode assay time linear range detection limit Ref | |
|---|---|
|
Electrochemical Piezoelectric SPR |
Amperometric Graphite 30 min - 50 IU/mL [158] Impedimetric Silica - 100-106 copies/mL 90 copies/mL [150] Voltammetric Pencil Graphite 30 min 0.05–0.75 × 10 −3 M 6.5 × 10−9 M [148] Voltammetric Gold 60 min 1.5–2 × 10 −6 M0.02 × 10 −6M [159] Voltammetric Gold - 0.05–4.0 × 10 −6M 0.5 ± 0.2 × 10 −9 M [160] Voltammetric Ag/AgCl 10 min 2.57 × 10−21-3.16 × 10−22M 1.82 × 10−21M [161] Voltammetric pencil graphite 60 min 0.05–0.75 × 10 −6 M 54.9 × 10 −9 M [162] Impedimetric Gold 20 min 1.36–4.53 × 10 −9 M1.36 × 10 −9 M [163] QCM Gold 10 min - 4.494 × 10 −7M [143] SPR Gold 30 min 25–2000 copies/reaction 50 copies/reaction [140] |
PGA/PGE; poly(l-glutamic acid)-modified pencil graphite electrode.
Electrochemical sensors are the most popular bio-sensing devices for HCV RNA monitoring and could detect this virus genome directly (label-free) or indirectly (label-based) [147,148]. In the direct method, electroactivity of HCV cDNA bases results in detectable electrochemical signal changes [149,150]. While in the indirect method, target cDNA is detectable after labeling with a nanoparticle or a redox-active enzyme [142,151]. An in-vitro electrochemical diagnostic platform with a highly sensitive and selective were constructed for early detection of HCV-genome in the clinical samples. The immobilization of gold nanostructures modified indium tin oxide (ITO) substrate as a probe with the untranslated regions (5′-UTR) of HCV-RNA was monitored using Raman spectra and square wave voltammetry (SWV) techniques (Fig. 4 A). The designed genosensor showed the limit of detection 264.5 IU/mL and ∼101.5 IU/mL based Raman spectroscopy and SWV results, respectively [152]. Another simple label-free genosensor for detection of HCV genotype 1a (HCV1a) was designed using poly (l-glutamic acid) (PGA) modified pencil graphite electrode (PGE) by Donmez and co-workers in 2015. Inosine-substituted 20 per probes were immobilized on the surface of PGA modified PGE surface by covalent linking. During the hybridization of probe and complementary region of HCV genome, the guanine oxidation signal was detected through SWV technique, which oxidation peak was seen at +1.05 V HCV1a RNA was detected in linear range 5 × 10 −8 M to 1.0 × 10 −9M and limit of detection 40.6 nM by this cheap and disposable PGA-modified genosensor [142].
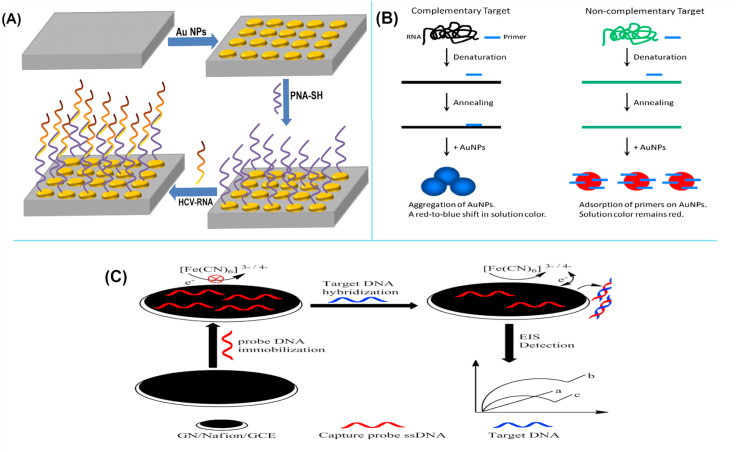
Fig. 4.
(A): A schematic picture from the development of a voltammetric electrochemical HCV-RNA genosensor, first step: Fabrication of gold (Au) nanostructures modified indium tin oxide (ITO) substrate; Second step: immobilization of peptide nucleic acid (PNA-SH) onto Au nanodots modified ITO substrate; and Third step: probe binding with the complementary regions of HCV-RNA. (B): A schematic diagram of a label-free colorimetric SPR genosensor using gold nanoparticles for detection of unamplified 5′UTR of HCV RNA. In the presence of complementary target RNA and primer, the AuNPs were aggregated and the solution color changed from red to blue, while upon introduction of non-complementary target RNA, primers were adsorbed on AuNPs and the solution color remained red. (C): Schematic shows the structure of an impedimetric biosensor and detection of the target through the electrochemical impedance spectroscopy (EIS) strategy. The figures are obtained with permission from Refs. [140,152], and [176].
Despite recent advances in the production of antiviral therapeutics, there is no effective prophylactic vaccine for HCV infection due to the high rate of genetic variation of the HCV virus genome during replication in human host cells. HCV isolates are divided into six main genotypes (from 1 to 6) which subtype 3a is a more risky and dominant subtype globally [153]. Accordingly, the development of a reliable and sensitive HCV genotyping platform is very important to choose proper antiviral therapy and predicting of responses to treatment. Hence, many electrochemical genosensors were designed for HCV genotyping [[153], [154], [155]], and, in 2012, a commercial HCV genotyping electrochemical sensor with high sensitivity (97%) was enrolled in worldwide markets, called eSensor HCV Genotyping Test [156].
Publishes demonstrated that piezoelectric genosensor due to the real time monitoring of hybridization between the immobilized oligonucleotide probe and the complementary oligonucleotides of HCV genome in simple format are better than other types of NABSs. Also, it has been suggested that they are best choices for HCV RNA detection in real samples, from the economic point of view; however, only a few studies exist with HCV genome detection via these types of NABSs. An oligonucleotide probe modified piezoelectric quartz crystal was constructed for detection of RT-PCR amplification of HCV genome from sera of infected patients; the Amplicor commercial kit was used for production of HCV amplicons. For sensor designing, monolayer of cystamine was formed on the gold electrodes to generate the smooth and rough sensing surface and then immobilization of different biotinylated, avidin (streptavidin)-modified oligonucleotide probes for detection of HCV 1, HCV2A/C, HCV2B, and HCV3 was applied through the biotin/avidin interaction. In this real-time monitoring QCM contribution, the total length 244 bases of HCV genome were diagnosed during only 10 min. Meanwhile, they pointed out that the reproducibility and performance of smooth crystals for HCV genome monitoring were better than rough sensors; this might be because of the less reproducible surface finish affecting also subsequently attached biolayers [143].
Also, for having the possibility of a high sensitive and label-free detection of HCV genome, Hwang et al. designed a nanomechanical microcantilever genosensor based on vibration modes for label-free detection of HCV helicase using RNA aptamer as receptor molecules. Production of detectable dynamic response on the microcantilever surface during the receptor-ligand binding event was the detection principle of their nanomechanical piezoelectric genosensor. Interaction between HCV helicase and RNA aptamer led to the detection of a low concentration of HCV genome (as much as 2.247 × 10−13 M) in the real samples. Furthermore, they suggested that using RNA aptamers as receptor molecules for the oscillating microcantilevers might be useful to increase the sensitivity of label-free detection [157].
It has been suggested that gold nanoparticles due to the showing of the unique phenomenon known as SPR are the best nanomaterials for use in the construction of colorimetric assays. Therefore, Sherif et al. designed the first rapid, simple, cheap, and label-free SPR genosensor using unmodified gold nanoparticles for detection of unamplified 5′UTR of HCV RNA obtained from infected sufferings. As depicted in Fig. 4B, the color changes from red to blue were detected in HCV positive samples within 1 min in the constructed colorimetric assay. The sensitivity, specificity, and detection limits of the colorimetric assay were 92%, 88.9%, and 50 copies/reaction, respectively. Detection of unamplified HCV RNA in clinical samples and elimination of the thermal cycling and detection instruments for cDNA synthesis are the main advantages of this SPR assay [140].
3.3. Human immunodeficiency virus (HIV)
Human immunodeficiency virus (HIV), a lentivirus, causes one of the great worldwide health challenges named acquired immunodeficiency syndrome (AIDS) [164]. The existed statistics demonstrated that around 36.9 million individuals of mankind were positive for HIV infection with a 0.8% prevalence rate at the end of 2017 [165]. Two types of HIV viruses including HIV-1 and HIV-2 are known as causative agents for AIDS, which have been classified according to their origin. Scientists suggested that HIV-1 was originated from chimpanzees, while HIV-2 originally transmitted from monkey species. However, HIV-1 is more viral and pandemic than HIV-2 and is responsible for most HIV infection cases [166]. The virus transforms from direct contact with body fluids and infects immune system cells which eventually led to immune system suppression of infected persons and susceptible them to opportunistic infections [167]. HIV infection is divided into three phases including acute, asymptomatic (window period), and AIDS phases [168]. Generally, antiretroviral therapy (ART) is a globally successful method to suppress HIV infection. But a remarkable ratio of HIV-infected individuals in low incoming and developing nations do not receive any effective ARTs because of the limited availability of rapid, sensitive, cheap diagnostic methods of HIV infection. For example, around 46% of HIV patients who needed ART worldwide did not receive medicine by the end of 2011 [169]. Therefore, HIV detection as soon as it enters the human body is a very important point to get rid of CD4 counting with flow cytometry and viral load measurement with real-time PRC gaps and also prevention of viral transmission during acute and window period phases of infection and increasing the efficiency of ART [164,166,170].
In the past two recent decades, the integration of virology with nanotechnology and bio-sensing platforms has opened helpful avenues to the designing of new detection methods for the HIV. To date, scientists tested and designed both types of immunosensors and genosensors to detection of HIV in clinical and research studies, but genosensors are preferred more than immunosensors due to having high sensitivity and applicability in the early days upon infection especially window period which antibody against the virus is not detectable [166,[171], [172], [173], [174], [175], [176]]. Table 4 is a list of various genosensors fabricated for HIV monitoring.
Table 4.
The various genosensing assays utilized for HIV genome detection.
| Genosensor type Method electrode assay time linear range detection limit Ref | |
|---|---|
|
Electrochemical Piezoelectric SPR |
Voltammetric GR/AuNCs/GCE - 10 −16 -10 −7M 3 × 10 −17 M [186] Voltammetric SPE - 5 × 10 −11 – 3 × 10 −10 M 5 × 10 −11 M [187] Amperometric gold 2 h 10 −10-10 −5M 7.05 × 10 −12 M [188] Impedimetric platinum 3 min 1 × 10−9 M − 1 × 10−6 M 5 × 10−10 M [189] Impedimetric GC 2.30 h 1 × 10−12 M − 1 × 10−9 M 3 × 10−13 M [190] - CPE 30 min 1 × 10−7 M −1.2 × 10−6 M 4 × 10−9 M [191] Microcantilever - 25 min 10 −6 – 2 × 10 −19M 400 ng/mL [175] SERS gold - 0-10−13M 10−19M [192] |
GR/AuNCs/GCE; GR-AuNCs modified glass carbon electrode, GC; Glassy carbon, CPE; carbon paste electrode, SERS; surface-enhanced Raman spectroscopy, SPE; Screen printed electrode.
The electrochemical genosensor technology is one of the most studied and interesting NABSs devices for monitoring HIV genome sequences due to their inherent advantages mentioned in the previous sections of this review [[177], [178], [179]]. Forasmuch as, HIV-1 is responsible for most of the HIV infection around the world [180]; hence, most electrochemical constructions were designed for this type of virus [[181], [182], [183]]. Nevertheless, there is a few reports from HIV-2 genome monitoring by this type of genosensor [184]. Fig. 4C shows an impedimetric DNA biosensor that were fabricated by Qiaojuan and co-workers for HIV genome. In this report, the ssDNA probe was adsorbed on the graphene-Nafion modified on the surface of glassy carbon electrode through the π–π∗ stacking interactions. In the presence of HIV genome, the ssDNA probe hybridization with the target DNA formed a double-stranded DNA (dsDNA) helix, which formation of this helix induced releasing of ds-DNA from the biosensor surface. HIV DNA Hybridization of graphene-Nafion/GCE led to induction of changes in negative charge and conformational transition; which these changes were detected by electrochemical impedance spectroscopy (EIS) strategy. The fabricated biosensor was capable to detect the HIV genome in linear range 1 × 10−13 to 1 × 10−10 M and limit of detection 2.3 × 10−14. Furthermore, it was point out that Nafion could stabilize graphene and also induce graphene dispersion [176].
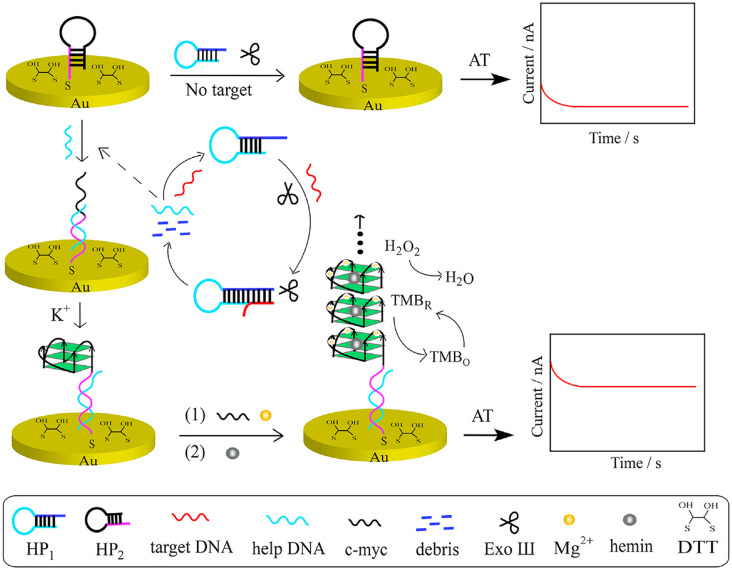
In a report, an electrochemical biosensor consisting of exonuclease III-assisted target recycling and guanine nanowire amplification (GWA) techniques were utilized for HIV genome detection. The Hairpin probe 1 (HP1), Hairpin probe 2 (HP2), Exonuclease III (Exo III), c-myc, Hemin/G-quadruplex and gold electrode were utilized for the construction of the genosensor and improvement of its sensitivity and selectivity (Fig. 5 ). The presence of target HIV DNA caused structural conformations in the HP1 and also induced the Exo Ш cleavage activity for gaining a digestion product named help DNA towards double-strand DNA. The obtained help DNA hybridized with the G-quadruplex sequence-locked HP1, eventually, formation of G-quadruplex structure with help of K+ and Mg2+ triggered guanine nanowire amplification (GWA). The applicability of the developed genosensor was evaluated by doing recovery experiments in the serum of healthy individuals. In the standard addition test, 10−10 M, 10 −9 M, and 25 × 10 −9M of HIV genome were added to the diluted human serum specimens. The developed biosensors showed good accuracy and potential for HIV genome detection in the clinical samples by exhibiting 99.5%, 97.8%, and 94.1% recoveries, respectively. The proposed bio-sensing strategy had good performance for diagnosis of target DNA at concentrations ranging from 10−11 M to 10−7 M and LOD 3.6 × 10 −12M, which was lower than reported genosensors [185].
Fig. 5.
Schematic exhibits the preparation process of the fabricated biosensor fabrication HIV diagnosis.
Besides the electrochemical strategies, a potential microcantilever genosensor as a HIV detector was employed by Abdullah and coworkers to detection of HIV ssDNA and ssRNA at conserved regions. In this label-free manner, several HIV genome sequences at different lengths and concentrations were tested for a deeper understanding of DNA–DNA and DNA-RNA hybridization. The ssDNA probe hybridization with ssDNA complementary target formed a dsDNA that induced comprehensive stress on the microcantilever surface, while there was no deflection in active microcantilever and hybridization event when non-complementary sequences were introduced with ssDNA probe. It found that the variation in the microcantilever deflection upon the introduction of different concentrations of target DNA is the reason for the high sensitivity of this piezoelectric genosensor. The detection limit of 8.988 × 10 −10 M was reported for the HIV genome by the fabricated microcantilever genosensor [175].
An innovative label-free SPR bio-sensing strategy with maximum sensitivity and duplicability was proposed for online monitoring of HIV DNA using the hairpin capture probes for immobilization on the gold sensing chip through Au–S bond and generation of a double-stranded complex. The sensor worked based on entropy-driven strand displacement reactions (ESDRs) and double-layer DNA tetrahedrons (DDTs). The advanced SPR system could efficiently detect virus genome in a linear range from 10 −12 M to 15 × 10 −8 M and limit of detection 48 × 10 −15M during window period in absence of antibody. In comparison with other biological sensors, this simple and low-cost sensor could be done without any need for chemical complexes and enzymes for only 1 h [174].
3.4. Human papilloma virus
Human papillomavirus (HPV) has a circular, double-stranded DNA genome that encodes early (E), and late (L) genes, which among them, E6 and E7 are necessary for transformation. According to the risk of cancer, infections are classified into two high risk and low-risk groups [193]. High-risk subtypes of the HPVs are HPV-16 and HPV-18 that are responsible for carcinogenesis and cervical cancers [194]. Pap smear is a traditional method for the detection of cervical cancer. PAP test has a sensitivity ranging from 30 to 87% [195]. So high rate of false-negative, poor specificity, and sensitivity are these methods' limitations. However, by developing molecular methods, novel strategies based on the use of HPV DNA assays expanded for the detection and identification of the HPV types [196,197].
The Benefit of molecular methods is their high sensitivity for HPV detection. Nevertheless, the methods mentioned have drawbacks such as high cost, the requirement of the trained operator, and advanced equipment [193]. Real-time PCR Detection range is 12.8 DNA copies [198]. Therefore, Due to the disadvantages of the existing methods, in recent years, DNA sensor platforms for HPV-DNA detection are expanding as a simple, rapid, economical, sensitive, and specific diagnostic method [199].
Avelino et al. developed a biosensor composed of polypyrrole (PPy) films and gold nanoparticles (AuNPs) for specific detection of HPV genotypes in cervical specimens [200]. The biosensor was fabricated by using flexible electrodes based on polyethylene terephthalate coated with indium tin oxide (PET/ITO electrodes). In this study, AuNP loaded polymer films were obtained by the electrosynthesis method, and then modified oligonucleotides were designed to recognize HPV families were chemically immobilized on the nanostructured platform. CV and EIS were used to measure the electrode modification and monitoring of molecular hybridization. They obtained a linear performance in a range of 2.247 × 10−10 M to 2.247 × 10 −15 M, LOD of 199.983 × 10 −14 M, and LOQ of 6.0669 × 10 −12 M with a regression coefficient of 220.206 × 10 −14. Screening tests on cervical specimens showed high specificity, selectivity, and sensitivity for HPV families.
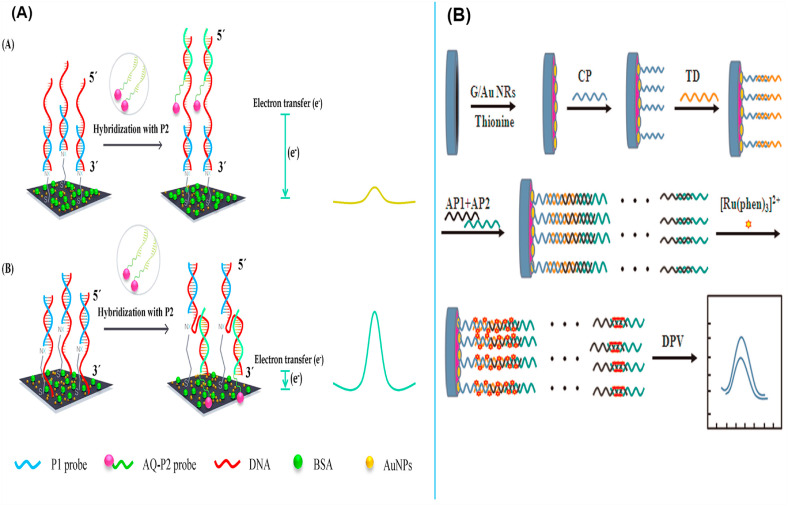
As shown in Fig. 6 A, an electrochemical DNA sensors based on a sandwich-hybridization using pyrrolidinyl peptide nucleic acid probes was fabricated to detect high-risk human HPV DNA sequences [201]. They first immobilized a capture PNA probe on a gold-deposited SPCE. Second, to obtain a higher signal response, an anthraquinone-labeled signaling probe (AQ P2) was designed to bind to the target DNA sequence at upstream (ASU) or downstream (ASD) positions. Compare to the ASU sensor, the ASD sensor showed a higher signal response. This biosensor detected the target DNA selectively in a detection range of 5 × 10 −10 -10 −7 M, and the limits of detection of 150 × 10 −12 and 153 × 10 −12M were acquired for HPV type 16 and 18 sequences respectively. Furthermore, to specific detection of HPV DNA in clinical samples, Huang et al. introduced an ultrasensitive electrochemical DNA biosensor platform based on graphene/Au nanorod/polythionine (G/Au NR/PT) by using electrochemical impedance spectroscopy and differential pulse voltammetry techniques [202] (Fig. 6B). In this biosensor, two auxiliary probes were used to long-range self-assemble DNA nanostructure, in which the target DNA triggered the connection of the DNA structure to the capture probe immobilized onto the electrode surface. [Ru(phen)3]2+ was utilized as a redox indicator and enhanced significantly electrochemical signals. The DNA biosensor displayed a detection range of 1.0 × 10−13 to 1.0 × 10−10 M and a detection limit value of 4.03 × 10−14 M.
Fig. 6.
(A): The fabricated electrochemical DNA sensors based on a sandwich-hybridization using pyrrolidinyl peptide nucleic acid probes to detect high-risk human HPV DNA sequences. (B): A schematic diagram of the constructed long-range self-assembled DNA electrochemical sensor for HPV detection. These figures are obtained with permission from ref [201,202].
In a research, a highly sensitive label-free electrochemical DNA hybridization biosensor was designed for the detection of specific DNA sequences of HPV [203]. In this genosensing platform, the thiol-modified single-stranded DNA probe (HS ssDNA) coated onto the sensing surface of a screen-printed gold electrode (SPGE). Hybridization among HS ssDNA with its target DNA (complementary DNA) in solution to form double-stranded DNA (dsDNA) onto the surface of SPGE was performed. Under optimal conditions, the detection limit value of this biosensor is lower than 4.494 × 10 −15M.Karimizefreh et al. introduced an impedimetric biosensor for the specific detection of HPV DNA type16based on glassy carbon-gold nanosheet (GC-GNS) electrodes [204]. They synthesized a glassy carbon electrode modified with gold nanosheets. A single-stranded oligonucleotide (ssDNA) acting as the DNA probe was placed on the surface of the GC GNS electrode. They have demonstrated that the use of gold nanosheet can increase the active area of the gold nanosheet modified electrode, furthermore, this sensor can distinguish between single base pair mismatches, complementary and non-complementary HPV ssDNA, and as a result improve the sensitivity of the biosensor. The suggested biosensor had a good selectivity with a remarkable detection limit (15 × 10 −14 M).
Besides the discussed genosensing approaches, Bartolome et al. synthesized carbon nano onions (CNOs) modified Glassy carbon electrodes as a biosensing platform for the detection of oncogenic human papillomaviruses [205]. These GCE/CNO electrodes were used for the amperometric detection of oncogene sequences of HPV DNA by immobilizing short biotinylated or thiolated DNA probes. They showed that this biosensor had a better sensitivity and a low limit of detection (54 × 10 −11 M).
3.5. Influenza viruses
Influenza is a viral infectious disease that has threatened human health and life as well as caused numerous medical issues and considerable financial burdens. The influenza viruses are negative-stranded enveloped RNA viruses that belong to the Orthomyxoviridae family andare divided into A, B, and C types [206].
In recent years, significant advances have been achieved in laboratory diagnostics for the detection of influenza virus infection. Virus isolation via cell culturing, shell vial culturing, Serological assay, and polymerase chain reaction (RT-PCR), are the methods currently used for the laboratory diagnosis of influenza viruses [207,208]. Routinely real-time PCR technique has been developed for the specific detection of influenza viruses and also have been for typing and subtyping. However, it has its limitations: relatively long detection time (∼2 h), expensive instruments, sample handling, post-PCR analysis, and requirement of well-trained technicians [208,209]. Nevertheless, rapid and accurate detection of influenza viruses to prevent the occurrence of serious influenza outbreaks is instantly necessary. Recent advances in biosensors for virus detection have led to the development of simple, rapid, and sensitive biosensors for the detection of influenza viruses, including Electrochemical DNA-Biosensor and impedance biosensors [206,[210], [211], [212]].
Tam et al. presented a direct and label-free detection method for the type A influenza virus by using carbon multi-walled nanotubes (MWCNTs) [213]. They have been used DNA-functionalized MWCNTs to determine the concentration of the target DNA sequence on the surface of sensors. The results show that this biosensor can reduce the response time required for influenza virus type A to 4 min. they also obtained detection limit values as low as 5 × 10 −10M of the target DNA samples.
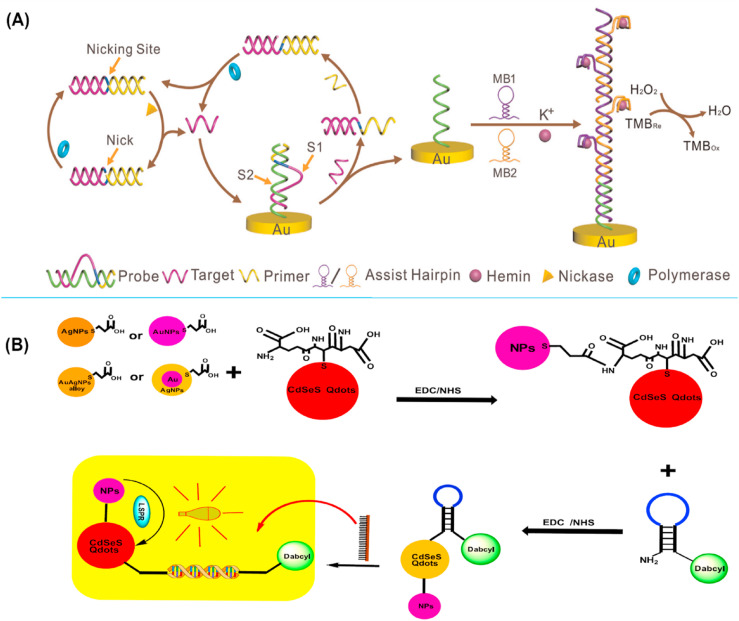
Additionally, DNA based label-free electrochemical biosensor with a duel amplification strategy was developed for the detection of the avian influenza A virus [214]. The isothermal exponential amplification reaction (EXPAR) coupled with hybridization chain reaction (HCR) of DNAzymes nanowires has been developed for the ultra-sensitive detection of low-abundance DNA of Avian influenza A(H7N9) in the clinical samples (Fig. 7 A). This method has low cost, high efficiency, and rapid amplification detection under isothermal conditions. They demonstrated that the detection limit of this system were 9.4 × 10 −15M.
Fig. 7.
(A); Schematic picture of dual amplification strategy based on isothermal exponential amplification reaction (EXPAR) coupled with hybridization chain reaction (HCR) of DNAzymes nanowires for electrochemical genosensor technique. (B); Conjugation of plasmonic NPs with l-glutathione-capped CdSeS alloyed Qdots makes NP-Qdots hybrids that further conjugate to MB. Finally, an LSRP-mediated fluorescent signal is detected as a result of the hybridization of target RNA and MB. The figures are obtained from ref [214,225].
On the other study, an electrochemical biosensor by using DNA tetrahedral nanostructural probe was immobilized onto the gold surface was developed to detect the hemagglutinin (HA) gene of influenza A (H7N9) virus from clinical throat-swab samples by amperometric measurements by Dong et al. [215]. This biosensor can specifically detect the target DNA of influenza A virus (H7N9) from other types of influenza A viruses (H1N1 and H3N2) and even could detect single-stranded mismatch oligonucleotides. They showed that through the combined use of the DNA tetrahedral structure as a probe and avidin HRP as the signal amplifier the detection limit of the biosensor could reach 10−13 M.
3.6. Arboviruses
Arboviruses are known as viruses that are transmitted by blood-feeding arthropods [216]. Arthropod-borne viruses (arboviruses) are a unique, diverse, and fascinating group of RNA viruses that belong to different families, including Bunyaviridae, Flaviviridae, and Togaviridae, which have the same characteristic of being transmitted to humans or other hosts by arthropods [217]. Some of the most well-known arboviruses include the zika virus, dengue virus, and chikungunya virus. This group of viruses is associated with worldwide outbreaks and a serious public health problem [7]. Dengue virus (DENV) and zika virus (ZIKV) are enveloped positive-sense single-stranded RNA viruses, members of the Flaviviridae family, genus Flavivirus. Chikungunya virus (CHIKV) is a single-stranded RNA virus, belonging to the Togaviridae family and genus Alphavirus [218]. Similarities in some clinical manifestations of the disease caused by the CHIKV to the DENV and the ZIKV, make the diagnosis of CHIKV difficult, especially in endemic areas with DENV and ZIKV circulation. Conventional diagnostic assays that are exploited for Zika, DENV-1–4, and CHIKV detection are RT-PCR and IgM capture ELISA and as well as MAC-ELISA. However, the RT-PCR detection method is highly sensitive and specific for ZIKV, considered the gold standard test for ZIKV [[219], [220], [221]]. However, the RT-PCR method is cumbersome, requires centralized laboratory facilities, skilled individuals to operate, and as well as MAC-ELISA has reduced sensitivity due to false-negative results associated with cross-reaction caused by arboviruses [[222], [223], [224]].
By identification of ZIKV genome in only one drop of sample, an impedimetric electrochemical DNA biosensor, using disposable, three-contact electrodes, has shown great selectivity with a 25.0 ± 1.7 × 10 −9 M detection limit. Being a label-free genosensor, allows EIS to easily measure electrical changes of the electrode surface, caused by hybridization of capture probes and target sequences [89]. The detection of ZIKV RNA has been also successfully conducted by using fluorescence signals from semiconductor quantum dot (Qdot) nanocrystals, mediated by localized surface plasmon resonance (LSPR) from four different plasmonic nanoparticles (NPs). The NP-Qdot conjugates to DNA loop sequence of molecular beacon (MB) probes, which hybridization of them with target RNA, makes alterations in optical transduction in NP-Qdot reporter fluorophore (Fig. 7B) [225]. Moreover, genosensors are capable of differentiating the ZIKV genome from its homologous arboviruses, DENV and CHIKV. To achieve this goal, the target genome binds to capture probe and signal probe, from the 3′ and 5′-ends, then, signal probe is recognized by specific antibodies and signals are measured by both electrochemical and calorimetric formats [91].
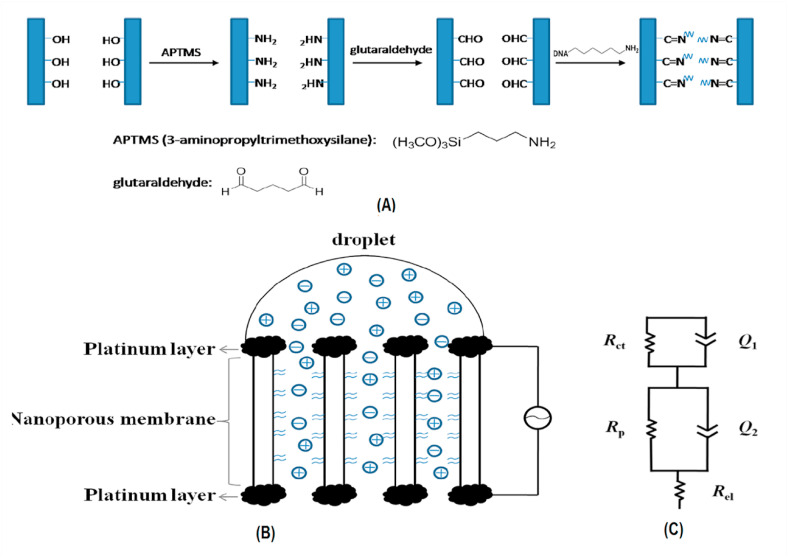
Detection of DENV has been experimented by several silicon nanowires (SiNW)-based genosensors including the one, accomplished by G Zhang et al. In this sensing method, SiNW is functionalized with the attachment of peptide nucleic acid (PNA) on its surface and hybridization of the complementary target sequence of DENV, which is acquired by RT-PCR, to PNA makes alterations in SiNW resistance, measured by electric sensing technique with LOD of 10−14 M [226]. Piezoelectric biosensors, also called quartz crystal microbalance (QCM), are considered sensitive and specific in identifying the DENV genome as well as other sensing methods. To be more detailed, first, the target DNA sequence, obtained from the DENV genome, hybridize into two different probes from 3′ and 5' ends in a sandwich format, second, this complex binds to another probe via by excess target, that acts as a bridge in this layered probe complex. Ultimately the frequency changes (Δf), amplified by probes for enhancing detection sensitivity, are being measured by QCM system with LOD of 2 pfu/mL [227]. Another procedure, in which DENV DNA is detected with high sensitivity and selectivity, is a label-free impedimetric sensor based on nanoporous alumina membrane (Fig. 8 ). Both sides of the alumina membrane are coated by thin platinum electrodes and DNA probes are linked into the pores of the alumina membrane. The impedance changes, induced by binding of target DNA and probes, are measured by impedance spectroscopy and show a 2.7 × 10−12 M limit of detection [228]. Other DENV and ZIKV-specific genosensors are shown in Table 5. Unless two other arboviruses, chikungunya virus detection has been little investigated by genosensors, however, the studies whose summaries are displayed in Table 5, have been demonstrated to be efficient and reliable in detecting the virus.
Fig. 8.
(A) A picture of DNA immobilization procedure; (B) Schematic representation of nanoporous alumina membrane for impedimetric biosensing of DENV DNA; (C) The equivalent circuit model of the impedimetric experimental results. The figure is obtained from ref [228].
Table 5.
Summary of the various genosensing studies regrading arbovirus virus.
| Virus | Genosensor type | Method | Electrode | Assay time | Linear rang | Detection limit | Ref |
|---|---|---|---|---|---|---|---|
| ZIKV | Electrochemical | Impedimetric | Gold | 50 min | 1.0 × 10−12 _ 1.0 × 10−6M | 82 × 10−14M | [229] |
| ZIKV | Electrochemical | Voltammetric | 1.SPAuE 2.SPCE/Au | _ | 1.1 × 10−15_6 × 10−13 M 2.5 × 10−15_10−11M | 0.2–33 × 10 −15M | [230] |
| ZIKV | Optical | Fluorescence | MOF | _ | 0–6 × 10−7 M | 23 × 10 −11M | [231] |
| DENV | Electrochemical | Voltammetric | Alumina | 45 min | 10−12-10−6 M | 9.55 × 10−12 M | [232] |
| DENV | Electrochemical | Voltammetric | ZnO/Pt–Pd | _ | 1 × 10−6_100 × 10−6 M | 4.3 × 10−5 M | [97] |
| DENV | Optical | Calorimetric | PSiNs | 90 min | 1.0 × 10−16-1.0 × 10−10 M | 2 × 10 −19 M | [233] |
| DENV | Electrical | Electrical | Silicon | _ | 10−14-10−5 M | 2.0 × 10 −15M | [234] |
| DENV | Electrical | Electrical | SiNW | _ | 1.0 × 10−11-1.0 × 10−7 M | 1.63 × 10−12 M | [235] |
| DENV | Electrical | Electrical | SiNW | _ | 1.0 × 10−9-1.0 × 10−13 M | 1.985 × 10−14 M | [236] |
| CHIKV | Electrochemical | Voltammetric | Gold | _ | 10−10 -10−4 M | 3.4 × 10 −9 M | [237] |
| CHIKV | Electrochemical | Voltammetric | Gold | _ | 10−10 -10−4 M | 1.0 × 10−9 M | [238] |
SPAuE; screen-printed gold electrode, SPCE/Au; screen-printed carbon electrode decorated with gold nanostructure, MOF; metal-organic framework, ZnO/Pt–Pd; Zinc oxide/platinum-palladium, PSiNs; porous silica nanospheres, SiNW; silicon nanowire.
3.7. Other viruses
Aside from the viruses discussed earlier, genosensors have been utilized to detect other viruses like adeno and herpes virus and recently discovered viruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that will be reviewed in this section.
3.7.1. Coronaviruses
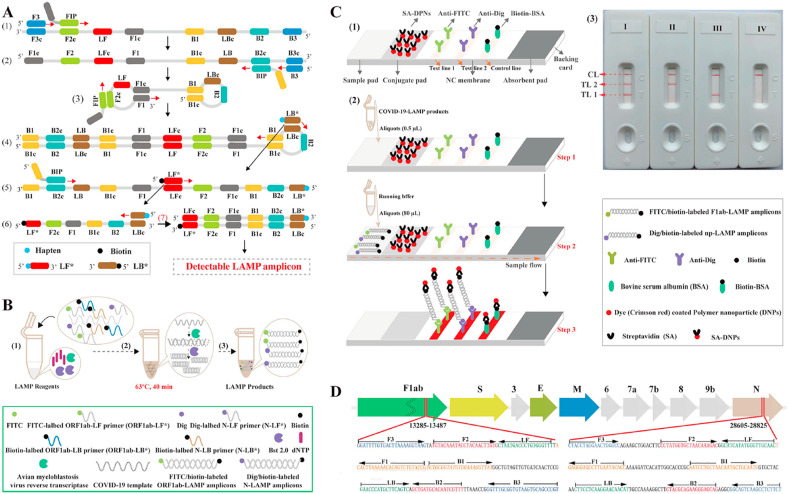
Coronaviruses in general have been detected by different genosensors as well as other respiratory viruses. However, since some of the species of this family are related to severe diseases including middle eastern respiratory syndrome (MERS) and SARS-CoV-2, the necessity of a sensitive, selective, and rapid detecting method based on biosensor is more important [239,240]. With so many deaths and hospitalized cases around the world, the coronavirus disease (COVID-19) caused by SARS-CoV-2 is known to be the third widespread pandemic of coronaviruses in the last 2 decades. Despite several false-positive and false-negative cases reported in laboratory diagnosis, RT-PCR has been regularly used for SARS-CoV-2 detection. Thus, biosensors especially the ones that detect the viral genome can compensate for the deficiencies of conventional methods. To develop an efficient genosensor, the plasmonic photothermal (PPT) effect combined with LSRP sensing transduction, have made a double-function biosensor in which the hybridization of the target sequence with complementary DNA receptor-functionalized gold nanoislands (AuNIs), can produce two different wavelengths that contribute to high sensitivity and reliability of this approach [241]. The combination of a lateral flow biosensor (LFB) with a multiplex reverse transcription loop-mediated isothermal amplification (mRT-LAMP)had also led to the development of an assay for SARS-CoV-2 detection with an LOD of 12 copies of detection target per reaction (Fig. 9 ). By utilizing two LAMP primer sets, two different sequences of the virus are amplified at the same time then labeled to be detected. The interpretation of the results accomplished by LFB becomes appears on the LFB test line with three lines that two of them are indicators of 2 different SARS-CoV-2 genes and one for control [242]. To provide more feasibility in result interpretation, an AuNPs-based colorimetric biosensor with the visual naked-eye response is fabricated to identify SARS-CoV-2 nucleocapsid protein gene from extracted RNA sample and shows LOD of 4 × 10 −16 M. In this system, shown in picture 15, when antisense oligonucleotide (ASO)-capped AuNPs hybridize with target RNA, they become agglomerated and change their surface plasmon resonance. After adding RNase H to this solution, target RNAs will be removed from AuNPs which results in visible precipitation from the solution [243]. In COVID-19 pandemic, some evidence supporting the possibility of transmitting SARS-CoV-2 through food products. So, monitoring and assessment food products for hazardous agents are important to progress in healthcare. Thus, advances in the producing mobile sensor devices to screening food products for fecal–oral transmission of viruses offer a significant outlook in-home healthcare [244]. Other approaches for genome detection of SARS-CoV-2 are summarized in Table 6 .
Fig. 9.
The combination of a lateral flow biosensor (LFB) with a multiplex reverse transcription loop-mediated isothermal amplification (mRT-LAMP) biosensor for detection of SARS-CoV-2 RNA. A) Is a schematic picture of LAMP assay with LF∗ and LB∗. Top row is an outline of LAMP with LF∗ and LB∗, while bottom row, schematically represents the new forward/backward loop primers (LF∗/LB∗). LF∗ was labeled with a hapten and LB∗ was labeled with biotin. B) Mechanistic representation of the SARS-CoV-2 RNA detection by RT-LAMP-LFB technique; this assay steps are amplification mixtures preparation (1), RT-LAMP reaction (2), and detectable SARS-CoV-2 RT-LAMP products (3). C) The principle of LFB for visualization of SARS-CoV-2 RT-LAMP products; the details of LFB (1), the principle of LFB for SARS-CoV-2 RT-LAMP products (2), Interpretation of the SARS-CoV-2 RT-LAMP results (3). D) Primer design of SARS-CoV-2 mRT-MCDA-LFB assay; up row shows SARS-CoV-2 genome organization and bottom row indicates nucleotide sequence and location of ORF1ab and N gene used to primer designing. The figure is obtained with permission from ref [242].
Table 6.
Summary of genosensors of SARS-CoV-2 detection.
| Genosensor type | Method | Electrode | Assay time | Linear rang | Detection limit | Ref |
|---|---|---|---|---|---|---|
| Electrochemical | EIS | Carbon | _ | 10−17-10−12 M | 200 copies/mL | [245] |
| Voltammetry | Gold | 5 min | 585.4 × 103_5.854 × 1010copies/mL | 6.9 × 10 3copies/mL | [246] | |
| Voltammetry | Carbone | >2 h | 1–1 × 1012 copies/mL | 103 copies/mL | [247] | |
| Voltammetry | PANI nanowire | _ | 10−14- 10−9 M | 3.5 × 10−15 M | [248] | |
| Electrochemiluminescence | EIS | Gold | _ | 10−15 _ 10−10M | 2.67 × 10 −15M | [249] |
| Optomagnetic | Volumetric | MNP | 100 min | 10 −16 -10 −14M | 4 × 10 −16M | [250] |
PANI; electropolymerized polyaniline, MNP; magnetic nanoparticle.
3.7.2. Herpes viruses
From transient erythematous vesicles to various cutaneous manifestations including malignancies, all eight members of the Herpesviridae family are capable of developing an asymptomatic infection in humans. What makes different species of this family to be important from a diagnostic point of view, is the characteristic of the latent phase in the virus infection cycle, especially in immunocompromised individuals [251]. Here we refer to some of the studies that have employed genosensors for the detection of herpes viruses.
One of the efficient electrochemical genosensors for herpes virus detection is developed by immobilization of oligonucleotides on graphite electrodes which hybridization of the target sequence with complementary probes, making voltammetric changes measured by voltammogram with LOD of approximately 14.77 × 10 −12 M. Moreover, this DNA biosensor can distinguish between herpes simplex virus I (HSV I) and Herpes simplex virus II (HSV II), which are responsible for facial and genital sores respectively [252]. Similar to other members of the Herpesviridae family, cytomegalovirus has double-stranded DNA and is related to several diseases particularly to immunosuppressed patients like organ transplant recipients [251]. By utilizing two different methods for the preparation of gold electrode, a direct capacitive DNA biosensor demonstrates to be efficient when CMV DNA specifically hybridize to oligonucleotides that are immobilized on the surface of electrodes followed by displacement of water and ions from electrode surface which results in capacitance alteration with LOD of 102 molecules/mL [253]. This was one of the first DNA biosensors applied for herpesvirus detection, however, subsequent researches are greater in complexity and specificity. Table 7 shows DNA biosensors for HSV, CMV, Epstein - Barr virus (EBV), human herpesvirus 5 (HHV-5) and Kaposi's sarcoma-associated herpesvirus (KSHV).
Table 7.
Summary of herpes viruses' DNA biosensor.
| Virus | Genosensor type | Method | Electrode | Assay time | Linear rang | Detection limit | Ref |
|---|---|---|---|---|---|---|---|
| HSV I | Piezoelectric | Acoustic | QCM | 3 min | 5.2 × 10−11_1.3 × 10−7 M | 10−18 M | [254] |
| HSV I | Piezoelectric | Acoustic | Gold | 6 min | _ | 5.2 × 10−12 M | [255] |
| HSV I | Capacitive | Impedimetric | Aluminum | 30 s | _ | 21 × 10 −17 M | [256] |
| CMV | Electrochemical | Voltammetric | SPE | _ | 1 × 10−10M | 6 × 10 −16M | [257] |
| CMV | Electrochemical | Amperometric | SPE | _ | 3 × 10 −8_ 2 × 10−7 M | 3 × 10 −11 M | [258] |
| CMV | Optical | Fluorometric | GO | _ | _ | 4.15 × 105IFU/ml | [259] |
| CMV | Optical | SPR | Gold | <1 h | _ | 24_108 × 10 −15M | [260] |
| CMV | Piezoelectric | PCR | QCM | _ | 0_500 × 10 −9 M | 25 × 10 −9 M | [261] |
| CMV | Piezoelectric | SDA | Gold | >3 h | _ | _ | [262] |
| EBV | Electrochemical | Voltammetric | Graphite | _ | 3.78_756 × 10 −6 M | 17.32 × 10 −9 M | [263] |
| EBV | Electrochemical | Microfluidic | Gold | 10 min | 1.0 × 103_1.0 × 10−1 copies/mL | 1.1 × 103 copies/mL | [264] |
| EBV | Electrochemical | EIS | Gold | _ | 1 × 10−15_1 × 10−9 M | 38 × 10−17 M | [265] |
| EBV | LF | LF | Gold | _ | _ | _ | [266] |
| EBV | Electrochemical | Signal amplification | QCM-D | _ | 11.235 × 10−11 _2.247 × 10−9 M | 112.35 × 10−12M | [267] |
| HHV-5 | Electrochemical | Voltammetric | Zn–Ag | 5 s | 113_103 and 3 × 105_106copies/mL | 97 copies/mL | [268] |
| KSHV | LF | SERS | Gold | 20 min | 0_100 × 10 −12 M | 0.043 × 10 −12 M | [269] |
SPE; screen-printed carbon electrode, IFU; immunofluorescence focus unit, SPR; surface plasmon resonance, GO; graphene oxide, SDA; strand displacement amplification, EIS; electrochemical impedance spectroscopy, LF; lateral flow, QCM-D; quartz crystal microbalance with a dissipation monitoring, Zn–Ag; zinc-silver, SERS; surface-enhanced Raman scattering.
3.7.3. Adenovirus
Adenoviruses are double-stranded DNA viruses that are transmitted via either the respiratory tract or the fecal-oral route. Human adenoviruses are capable of establishing severe disease in children, the elderly, and immunocompromised patients, however, they are mostly asymptomatic in healthy individuals [270]. Despite the prosperity of most genosensors in virus detection, eSensor respiratory viral panel (eSensor RVP) shows some cross-reactivity in the detection of different adenovirus serotypes when compared to real-time PCR. eSensor RVP is a commercial electrochemical biosensor with gold electrodes using voltammetry for the detection of the genome of respiratory viruses such as influenza and rhinoviruses [271]. While commercial biosensors might not be highly selective and sensitive in the diagnosis of some virus serotypes, the ones developed in laboratories indicate more success compared to conventional methods like PCR. Table 8 summarizes some of the genosensors, applied for adenovirus and also picornaviruses detection. In addition to before mentioned viruses, other viruses are applied for genome detection by biosensors. In Table 9 we have displayed the information about related researches for paramyxovirus, rhabdoviruses, and reoviruses diagnosis.
Table 8.
Information about some of the genosensors applied for adenovirus and picornaviruses detection.
| Virus | Genosensor type | Method | Electrode | Assay time | Linear rang | Detection limit | Ref | |
|---|---|---|---|---|---|---|---|---|
| Adenovirus | Adenovirus | Electrochemical | Amperometric | Carbone | 3_5 h | _ | _ | [272] |
| Electrochemical | Conductometric | Ag/AgCl | _ | 0.4_1.0 μM/tDNA | _ | [273] | ||
| Optical | Fluorescent | Silver | _ | _ | 1 × 10−9 M | [274] | ||
| Picornaviruses | HAV | Electrochemical | Voltammetry | Gold | Few minutes | _ | 13.693 × 10−9M | [275] |
| HAV | Optical | Ratiometric | Silver | _ | _ | 0.5 × 10−9 M | [276] | |
| HAV | Optical | FRET | QD | 30 min | 2 × 10−10-5 × 10−8M | 1.3 × 10−11M | [277] | |
| HAV | Optical | Chemiluminescent | Silica Fiber | _ | 11.23 × 10−6_2.247 × 10-5 M | 11.23 × 10−6M | [278] | |
| Coxsackievirus | Electrochemical | EIS | GO | _ | 0.01_20 × 10−6M | 2.5 × 10−9 M | [279] | |
| Enterovirus | LF | LF | Gold | <60 s | _ | 1 × 10−7 M | [280] | |
Ag/AgCl, silver chloride; tDNA, target DNA; HAV, hepatitis A virus; FRET, fluorescence resonance energy transfer; QD, quantum dot; EIS, electrochemical impedance spectroscopy; GO, graphene oxide; LF, lateral flow.
Table 9.
The list of genosensors for applied for paramyxovirus, rhabdoviruses, and reoviruses diagnosis.
| Virus | Genosensor type | Method | Electrode | Assay time | Linear rang | Detection limit | Ref | |
|---|---|---|---|---|---|---|---|---|
| Paramyxovirus | SV40 | Optical | Fluorescence | GO | _ | 40.0_260 × 10 −9 M | 14.3 × 10 −9M | [281] |
| SV40 | Optical | Fluorescence | MB | _ | 1_600 × 10 −9 M | 0.4 × 10 −9 M | [282] | |
| RSV | Electrochemical | Voltammetric | Gold | _ | _ | 13.6 × 10−18 mol | [283] | |
| NDV | Electrochemical | Potentiometric | Nitrocellulose | 45_60 min | _ | 0.30 × 10−15 mol | [284] | |
| Rhabdoviruses | Rabies | Optical | 1.fluorescence 2.colorimetric | _ | _ | 1.0.1_6 × 10 −9M 2.0.5_60 × 10 −9 M |
1.15 × 10−12M 2.60 × 10 −12 M |
[285] |
| Lyssaviruses | Optical | Fluorescent | _ | _ | _ | _ | [286] | |
| Reovirus | Ibaraki virus | Magnetic | FRET | Magnetic | 18 min | _ | 1.9 × 10−12 M | [287] |
SV40, Simian virus 40; GO, graphene oxide; MB, molecular beacon; RSV, respiratory syncytial virus; NDV, Newcastle disease virus; FRET, fluorescent resonance energy transfer.
4. Simultaneous multi-target detection of viruses
Besides the mentioned mono-target detection genosensor platforms that were constructed for the determination of one viral infection, some scientists are focused to design biosensors with capability to simultaneously diagnose different viruses from clinical specimens at the same time. Because this strategy provides multiplex virus genome detection systems with the lowest price and time-wasting which is an important health issue for all nations especially developing ones [286,[288], [289], [290]]. A multi-target SPR genosensor was combined with gene chip technology by Lei Shi et al. for simultaneous detection of nine respiratory viruses' genome including influenza A and B, H1N1, parainfluenza virus 1–3 (PIV-1, 2, 3), respiratory syncytial virus (RSV), adenovirus (ADV), and severe acute respiratory syndrome coronavirus (SARS) from clinical swaps within 30 min. In this strategy, specific oligonucleotides of the mentioned respiratory viruses' genome have been immobilized in a chip. In the present SPR sensor, PCR primers were labeled with biotin and also streptavidin used for further amplifying the signal to increase the sensitivity of the sensor. According to this report, the complementary probes are specifically hybridized with the genome of the target virus without any nonspecific and cross-hybridization between the viruses' genome and probes. This type of multi-target genosensor is enabling to concurrent identifying of respiratory viruses with high sensitivity, specificity, and reproducibility at the same time with LOD: 5, 1, 1, 2.5, 3.5, 3, 0.5, 2, and 3 × 10 −9 M for Influ A, Influ B, PIV1, PIV2, PIV3, RSV, ADV, SARA, H1N1, respectively [291].
In 2019, for the first time, a multifunctional molecularly imprinted sensor for simultaneous detection of hepatitis viruses including HAV and HBV was provided. As simultaneously detection of HAV and HBV viruses is very difficult because of their similarity of the outer structure therefore some modifications such as hydrophilic monomers and metal chelation are used to reduce the cross-hybridization and non-specific binding as well as to increase the specificity of the biosensor. The acceptable selectivity and sensitivity were achieved for the detection of the two viruses with the limits of detection of 3.4 × 10−12 M and 5.3 × 10−12 M for HAV and HBV, respectively, less than 20 min [289]. Additionally, an innovative electrochemical genosensor array with high sensitivity was fabricated for real-time multiplexed monitoring of three of the most common high-risk HPV types (including 16, 18 and 45) DNA sequences from the clinical samples using a co-immobilization strategy of a thiolated probe and bipodal alkanethiol for the modification of the gold electrodes and sandwich assay with a horseradish peroxidase (HRP) labeled reporter probe for the diagnosis of the three HPV types. The proposed electrochemical genosensor array detected HPV16, 18, and 45 DNA with limits of detection of 220 × 10−12 M, 170 × 10−12 M and 110 × 10−12 M, respectively at the same time. Also, there was insignificant cross-reactivity and non-specific binding between all target viruses and HRP-labeled probes [288].
On the other study, a simple and very cheap label-free electrochemical multiplexed genosensor by using the six gold working electrodes, a gold auxiliary electrode, and a printed Ag/AgCl reference electrode to simultaneously detect the specific oligonucleotide sequences of HIV-1 and HIV-2 genome. The voltammetric system could detect the DNA of HIV-1 and HIV-2 with good sensitivity and specificity at the detection limits of 0.1 × 10−9 M without the obvious cross-interference under the optimized conditions. Meanwhile, single-base mutation oligonucleotides, random oligonucleotides, and complementary target genome easily discriminated from each other. This research demonstrates that using different hairpin-DNA probes can be helpful to design the sensitive label-free electrochemical genosensor for simultaneous diagnosis of DNA sequences for HIV-1 and HIV-2 as well as various viral pathogens [184].
Although cross-hybridization and interference are the main concerns of genosensors application for simultaneous detection of viruses at the same time. But the cross-interference assays that have been conducted for the multiplexed genosensing systems showed that there is no report of being obvious cross-interference and non-specific binding between the genome of the tested pathogens and used specific probes in all fabricated and reported genosensors [201,286,[288], [289], [290]]; however further investigations especially proteomics research are needed for more confirmations.
5. Commercial genosensors
As previously mentioned, low cost, high sensitivity and Low limit of detection of biomolecules have resulted in the development of biosensors. Although, only small types of biosensors are commercially available, Innovations in this technology have enabled many biosensors to be commercialized. At-home testing kits give rapid results, Self-testing is safe and do not require you to send samples to a laboratory for analysis. Commercial biosensors are available for viral infections specifically detecting the viral antigens, Nucleic acid, and antibodies in biological specimens [292]. The commercially available gene sensors and their characteristics are shown in Table 10 .
Table 10.
Commercially available clinical genosensors and their characteristics.
|
Company |
Model | Sensitivity | Specificity | Sample | Virus | Reference |
|---|---|---|---|---|---|---|
| Quidel, USA |
Solana Influenza A + B Assay | Influenza A: 97.2% (95% CI 95.0%–98.4%) Influenza B: 100.0% (95% CI 95.4%–100%) |
Influenza A: 96.7% (95% CI 95.4%–97.7%) Influenza B: 98.9% (95% CI 98.2%–99.4%) |
nasal and nasopharyngeal swabs | Influenza A&B | [292] |
| Lyra Influenza A + B Assay | A: 100% B: 98.4% |
A: 98.7% B: 95.5% |
nasal swabs | Influenza A (HIN1,H3N2,H7N9) &B | ||
| Solana SARS-CoV-2 Assay | Nasal: 97.2% Nasopharyngeal: 100% |
Nasal: 100% Nasopharyngeal: 96.9% |
Nasal swabs and nasopharyngeal swabs | SARS-CoV-2 | ||
| Lyra RSV + hMPV Assay | RSV 96% hMPV 95% | RSV 98% hMPV 98.9% | Nasal swabs And nasopharyngeal swab |
Respiratory syncytial virus (RSV) and Human metapneumovirus (hMPV) | ||
| Alere, USA |
Alere i Influenza A & B assay | A: 55.2–100% B: 45.2–100% |
A: 62.5–100% B: 53.6–100% |
Nasal swabs And nasopharyngeal swab |
Influenza A & B | [293] |
6. Conclusion and future perspectives
Among all infectious agents, viruses, in general, are considered to be the most life-threatening pathogens not only for humans but for another alive creature including bacteria. The virus's unique non cellular structure makes it resistant to antimicrobial therapies and calls for specific antiviral medicines which target the specific components of the virus, with precise diagnosis as a prerequisite for antiviral treatment. Another challenging issue in defeating these vicious foes is their ability to trick the immune system and generate an anergy phase in which immune cells cannot respond to antigens. This condition impedes antibody-based detection of the virus because the immune cells do not recognize them as pathogens and thus the production of specific antibodies will not take place as the window period of HIV. In the case of hepatitis B window period, nor HBV-specific antibody nor it's a surface antigen (HBsAg) is detectable in patient's serum [294]. Moreover, some viruses like the Herpesviridae family have a latent phase in the host cells in which they only transcribe some of their vital genes and hide from the immune system.
Among molecular conventional methods, PCR an example of a precise one, has been vastly utilized in the recent pandemic of COVID-19. With regards to its approximate accuracy in SARS-CoV-2 genome detection, the number of false positivity or false negativity necessitates an alternative method to prevent prevailing errors. Noted obstacles in rapid and accurate detection of viruses and their infectious phase, leads to the development of biosensor that does not possess the deficiencies of conventional detecting methods and is even capable of discriminating virus serotypes that have cross-reactivity. While detection of antibody, antigen, or whole virus might undergo inaccuracy due to delayed immune response or latency of virus life cycle, sensing techniques that are designed to detect virus genome can specifically be applied for different stages of infection by targeting particular genes. Despite conventional methods, genosensors can be operated in a selective, specific, time-saving, and effortless manner even by inexpert individuals and the difficulties in early stages of manufacturing different genosensors including costly instruments and materials, suitable DNA probe availability in label-based sensors, and other complications in small nucleic acid detection like sensitivity and specificity, have been resolved over time in respect of researcher's efforts.
From various types of fabricated genosensors, the majority of them have been used electrochemical sensing techniques and the most utilized electrodes have been the gold ones. Sensing methods especially, optical ones by which the results can be observed with naked-eye are more feasible, besides, label-free genosensors have provided a less-laborious detecting procedure. The limit of detection represented by different studies has been mostly comparable or lower to the PCR method indicating that genosensors are promising for future laboratory diagnostic approaches and can be a potential alternate for current methods. In this review we attempt to bring up the application of genosensors for virus detection, however many viruses remained unassessed because they were less of a consideration in terms of epidemiology or severity of the diseases they cause. Unlike plenty of researches accomplished about virus detection by genosensor, this advanced device needs more investigation to be proved for routine application.
Compliance with ethical standards
The authors declare that they have no competing interests.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are also very much grateful for financial support from the Immunology Research Center, Tabriz University of Medical Sciences.
References
- 1.Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao C.-Y., Fu W.-L. Biosensors for hepatitis B virus detection. World J. Gastroenterol.: WJG. 2014;20 doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farzin L., Shamsipur M., Samandari L., Sheibani S. HIV biosensors for early diagnosis of infection: the intertwine of nanotechnology with sensing strategies. Talanta. 2020;206 doi: 10.1016/j.talanta.2019.120201. [DOI] [PubMed] [Google Scholar]
- 4.Hassanpour S., Baradaran B., de la Guardia M., Baghbanzadeh A., Mosafer J., Hejazi M., Mokhtarzadeh A., Hasanzadeh M. Diagnosis of hepatitis via nanomaterial-based electrochemical, optical or piezoelectrical biosensors: a review on recent advancements. Microchim. Acta. 2018;185:1–24. doi: 10.1007/s00604-018-3088-8. [DOI] [PubMed] [Google Scholar]
- 5.McQuillan G.M., Kruszon-Moran D., Markowitz L.E., Unger E.R., Paulose-Ram R. US Department of Health and Human Services, Centers for Disease Control and …2017; United States: 2011-2014. Prevalence of HPV in Adults Aged 18-69. [Google Scholar]
- 6.Bruno J.G., Carrillo M.P., Richarte A.M., Phillips T., Andrews C., Lee J.S. Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes. 2012;5:1–12. doi: 10.1186/1756-0500-5-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva J.V., Jr., Lopes T.R., de Oliveira-Filho E.F., Oliveira R.A., Durães-Carvalho R., Gil L.H. Current status, challenges and perspectives in the development of vaccines against yellow fever, dengue, Zika and chikungunya viruses. Acta Trop. 2018;182:257–263. doi: 10.1016/j.actatropica.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Jefferies M., Rauff B., Rashid H., Lam T., Rafiq S. Update on global epidemiology of viral hepatitis and preventive strategies. World J. Clin. Cases. 2018;6:589. doi: 10.12998/wjcc.v6.i13.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesarman E., Damania B., Krown S.E., Martin J., Bower M., Whitby D. Kaposi sarcoma. Nat. Rev. Dis. Prim. 2019;5:1–21. doi: 10.1038/s41572-019-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portillo C.J., Holzemer W.L., Chou F.-Y. HIV symptoms. Annu. Rev. Nurs. Res. 2007;25:259–291. [PubMed] [Google Scholar]
- 11.Gessain A., Cassar O. Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 2012;3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank T.D., Carter A., Jahagirdar D., Biehl M.H., Douwes-Schultz D., Larson S.L., Arora M., Dwyer-Lindgren L., Steuben K.M., Abbastabar H. Global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2017, and forecasts to 2030, for 195 countries and territories: a systematic analysis for the Global Burden of Diseases, Injuries, and Risk Factors Study 2017. Lancet. 2019;6:e831–e859. doi: 10.1016/S2352-3018(19)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratageri V.H., Shepur T., Wari P., Chavan S., Mujahid I., Yergolkar P. Clinical profile and outcome of dengue fever cases. Indian J. Pediatr. 2005;72:705–706. doi: 10.1007/BF02724083. [DOI] [PubMed] [Google Scholar]
- 14.Hartman A.L., Towner J.S., Nichol S.T. Ebola and marburg hemorrhagic fever. Clin. Lab. Med. 2010;30:161–177. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Monath T.P., Vasconcelos P.F. Yellow fever. J. Clin. Virol. 2015;64:160–173. doi: 10.1016/j.jcv.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 16.Cox N.J., Subbarao K. Global epidemiology of influenza: past and present. Annu. Rev. Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 17.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. International journal of antimicrobial agents; 2020. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Corona Virus Disease-2019 (COVID-19): the Epidemic and the Challenges. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wandtke T., Woźniak J., Kopiński P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7:751–780. doi: 10.3390/v7020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roingeard P. Viral detection by electron microscopy: past, present and future. Biol. Cell. 2008;100:491–501. doi: 10.1042/BC20070173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souf S. Recent advances in diagnostic testing for viral infections. Biosci. Horiz.: Int. J. Stud. Res. 2016;9 [Google Scholar]
- 21.Zhang N., Wang L., Deng X., Liang R., Su M., He C., Hu L., Su Y., Ren J., Yu F. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92:408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cobo F. Suppl 1: application of molecular diagnostic techniques for viral testing. Open Virol. J. 2012;6:104. doi: 10.2174/1874357901206010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barzon L., Lavezzo E., Militello V., Toppo S., Palù G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12:7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singhal N., Kumar M., Kanaujia P.K., Virdi J.S. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015;6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazelton P.R., Gelderblom H.R. Electron microscopy for rapid diagnosis of emerging infectious agents. Emerg. Infect. Dis. 2003;9:294. doi: 10.3201/eid0903.020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golding C.G., Lamboo L.L., Beniac D.R., Booth T.F. The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci. Rep. 2016;6:26516. doi: 10.1038/srep26516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cella L.N., Blackstock D., Yates M.A., Mulchandani A., Chen W. Critical Reviews™ in Eukaryotic Gene Expression; 2013. Detection of RNA Viruses: Current Technologies and Future Perspectives; p. 23. [DOI] [PubMed] [Google Scholar]
- 28.Landry M.L., Tang Y.-W. Manual of Molecular and Clinical Laboratory Immunology; 2016. Immunologic and Molecular Methods for Viral Diagnosis; pp. 538–549. [Google Scholar]
- 29.Reta D.H., Tessema T.S., Ashenef A.S., Desta A.F., Labisso W.L., Gizaw S.T., Abay S.M., Melka D.S., Reta F.A. Molecular and immunological diagnostic techniques of medical viruses. Int. J. Microbiol. 2020:2020. doi: 10.1155/2020/8832728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R.L. Atmar, Immunological Detection and Characterization, Viral Infections of Humans, Springer2014, pp. 47-62.
- 31.Kumar P. Methods for rapid virus identification and quantification. Mater. Methods. 2013;3:207. [Google Scholar]
- 32.Lequin R.M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA) Clin. Chem. 2005;51:2415–2418. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- 33.Gan S.D., Patel K.R. Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Invest. Dermatol. 2013;133:e12. doi: 10.1038/jid.2013.287. [DOI] [PubMed] [Google Scholar]
- 34.S. Hosseini, P. Vázquez-Villegas, M. Rito-Palomares, S.O. Martinez-Chapa, Advantages, Disadvantages and Modifications of Conventional ELISA, Enzyme-linked Immunosorbent Assay (ELISA), Springer2018, pp. 67-115.
- 35.Behzadi P., Ranjbar R., Alavian S.M. Nucleic acid-based approaches for detection of viral hepatitis. Jundishapur J. Microbiol. 2015;8 doi: 10.5812/jjm.17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassanpour S., Baradaran B., de la Guardia M., Baghbanzadeh A., Mosafer J., Hejazi M., Mokhtarzadeh A., Hasanzadeh M. Diagnosis of hepatitis via nanomaterial-based electrochemical, optical or piezoelectrical biosensors: a review on recent advancements. Microchim. Acta. 2018;185:568. doi: 10.1007/s00604-018-3088-8. [DOI] [PubMed] [Google Scholar]
- 37.Ghaffari S.H., Obeidi N., Dehghan M., Alimoghaddam K., Gharehbaghian A., Ghavamzadeh A. Monitoring of cytomegalovirus reactivation in bone marrow transplant recipients by real-time PCR. Pathol. Oncol. Res. 2008;14:399–409. doi: 10.1007/s12253-008-9030-3. [DOI] [PubMed] [Google Scholar]
- 38.Prabhakar P.K., Lakhanpal J. Molecular Biology Reports; 2020. Recent Advances in the Nucleic Acid-Based Diagnostic Tool for Coronavirus; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Versalovic J., Lupski J.R. Molecular detection and genotyping of pathogens: more accurate and rapid answers. Trends Microbiol. 2002;10:s15–s21. doi: 10.1016/s0966-842x(02)02438-1. [DOI] [PubMed] [Google Scholar]
- 40.Stals A., Mathijs E., Baert L., Botteldoorn N., Denayer S., Mauroy A., Scipioni A., Daube G., Dierick K., Herman L. Molecular detection and genotyping of noroviruses. Food Environ. Virol. 2012;4:153–167. doi: 10.1007/s12560-012-9092-y. [DOI] [PubMed] [Google Scholar]
- 41.Park Y., Kim B.S., Choi K.H., Shin D.H., Lee M.J., Cho Y., Kim H.-S. A novel multiplex real-time PCR assay for the concurrent detection of hepatitis A, B and C viruses in patients with acute hepatitis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syrmis M.W., Whiley D.M., Thomas M., Mackay I.M., Williamson J., Siebert D.J., Nissen M.D., Sloots T.P. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J. Mol. Diagn. 2004;6:125–131. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noorbazargan H., Nadji S.A., Samiee S.M., Paryan M., Mohammadi-Yeganeh S. New design, development, and optimization of an in-house quantitative TaqMan Real-time PCR assay for HIV-1 viral load measurement. HIV Clin. Trials. 2018;19:61–68. doi: 10.1080/15284336.2018.1440991. [DOI] [PubMed] [Google Scholar]
- 44.Mumford R., Boonham N., Tomlinson J., Barker I. Advances in molecular phytodiagnostics–new solutions for old problems. Eur. J. Plant Pathol. 2006;116:1–19. doi: 10.1007/s10658-006-9037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobo F., Talavera P., Concha A. Diagnostic approaches for viruses and prions in stem cell banks. Virology. 2006;347:1–10. doi: 10.1016/j.virol.2005.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gruber F., Falkner F.G., Dorner F., Hämmerle T. Quantitation of viral DNA by real-time PCR applying duplex amplification, internal standardization, and two-color fluorescence detection. Appl. Environ. Microbiol. 2001;67:2837–2839. doi: 10.1128/AEM.67.6.2837-2839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eivazzadeh-Keihan R., Pashazadeh-Panahi P., Mahmoudi T., Chenab K.K., Baradaran B., Hashemzaei M., Radinekiyan F., Mokhtarzadeh A., Maleki A. Dengue virus: a review on advances in detection and trends–from conventional methods to novel biosensors. Microchim. Acta. 2019;186:329. doi: 10.1007/s00604-019-3420-y. [DOI] [PubMed] [Google Scholar]
- 48.Ahangar L.E., Mehrgardi M.A. Amplified detection of hepatitis B virus using an electrochemical DNA biosensor on a nanoporous gold platform. Bioelectrochemistry. 2017;117:83–88. doi: 10.1016/j.bioelechem.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Tan Z., Dong H., Liu Q., Liu H., Zhao P., Wang P., Li Y., Zhang D., Zhao Z., Dong Y. A label-free immunosensor based on PtPd NCs@ MoS2 nanoenzymes for hepatitis B surface antigen detection. Biosens. Bioelectron. 2019;142 doi: 10.1016/j.bios.2019.111556. [DOI] [PubMed] [Google Scholar]
- 50.Hassanpour S., Baradaran B., Hejazi M., Hasanzadeh M., Mokhtarzadeh A., de la Guardia M. Recent trends in rapid detection of influenza infections by bio and nanobiosensor. Trac. Trends Anal. Chem. 2018;98:201–215. [Google Scholar]
- 51.Sohrabi H., Majidi M.R., Asadpour-Zeynali K., Khataee A., Mokhtarzadeh A. Bimetallic Fe/Mn MOFs/MβCD/AuNPs stabilized on MWCNTs for developing a label-free DNA-based genosensing bio-assay applied in the determination of Salmonella typhimurium in milk samples. Chemosphere. 2022;287 doi: 10.1016/j.chemosphere.2021.132373. [DOI] [PubMed] [Google Scholar]
- 52.Sohrabi H., Majidi M.R., Asadpour-Zeynali K., Khataee A., Dastborhan M., Mokhtarzadeh A. A PCR-free genosensing platform for detection of Shigella dysenteriae in human plasma samples by porous and honeycomb-like biochar decorated with ultrathin flower-like MoS2 nanosheets incorporated with Au nanoparticles. Chemosphere. 2022;288 doi: 10.1016/j.chemosphere.2021.132531. [DOI] [PubMed] [Google Scholar]
- 53.Sohrabi H., Khataee A., Ghasemzadeh S., Majidi M.R., Orooji Y. Layer double hydroxides (LDHs)-based electrochemical and optical sensing assessments for quantification and identification of heavy metals in water and environment samples: a review of status and prospects. Trends Environ. Anal. Chem. 2021;31 [Google Scholar]
- 54.Mir M., Martínez-Rodríguez S., Castillo-Fernández O., Homs-Corbera A., Samitier J. Electrokinetic techniques applied to electrochemical DNA biosensors. Electrophoresis. 2011;32:811–821. doi: 10.1002/elps.201000487. [DOI] [PubMed] [Google Scholar]
- 55.Subak H., Ozkan-Ariksoysal D. Label-free electrochemical biosensor for the detection of Influenza genes and the solution of guanine-based displaying problem of DNA hybridization. Sensor. Actuator. B Chem. 2018;263:196–207. [Google Scholar]
- 56.Sohrabi H., Hemmati A., Majidi M.R., Eyvazi S., Jahanban-Esfahlan A., Baradaran B., Adlpour-Azar R., Mokhtarzadeh A., de la Guardia M. Recent advances on portable sensing and biosensing assays applied for detection of main chemical and biological pollutant agents in water samples: a critical review. Trac. Trends Anal. Chem. 2021;143 [Google Scholar]
- 57.Pashazadeh-Panahi P., Belali S., Sohrabi H., Oroojalian F., Hashemzaei M., Mokhtarzadeh A., de la Guardia M. Metal-organic frameworks conjugated with biomolecules as efficient platforms for development of biosensors. Trac. Trends Anal. Chem. 2021;141 [Google Scholar]
- 58.H. Sohrabi, M.R. Majidi, P. Khaki, A. Jahanban-Esfahlan, M. de la Guardia, A. Mokhtarzadeh, State of the Art: Lateral Flow Assays toward the Point-of-care Foodborne Pathogenic Bacteria Detection in Food Samples, Comprehensive Reviews in Food Science and Food Safety, DOI. [DOI] [PubMed]
- 59.Sohrabi H., Bolandi N., Hemmati A., Eyvazi S., Ghasemzadeh S., Baradaran B., Oroojalian F., Majidi M.R., de la Guardia M., Mokhtarzadeh A. State-of-the-art cancer biomarker detection by portable (bio) sensing technology: a critical review. Microchem. J. 2022 [Google Scholar]
- 60.Sohrabi H., Javanbakht S., Oroojalian F., Rouhani F., Shaabani A., Majidi M.R., Hashemzaei M., Hanifehpour Y., Mokhtarzadeh A., Morsali A. Nanoscale Metal-Organic Frameworks: recent developments in synthesis, modifications and bioimaging applications. Chemosphere. 2021;281 doi: 10.1016/j.chemosphere.2021.130717. [DOI] [PubMed] [Google Scholar]
- 61.Sohrabi H., Majidi M.R., Arbabzadeh O., Khaaki P., Pourmohammad S., Khataee A., Orooji Y. Recent advances in the highly sensitive determination of zearalenone residues in water and environmental resources with electrochemical biosensors. Environ. Res. 2022;204 doi: 10.1016/j.envres.2021.112082. [DOI] [PubMed] [Google Scholar]
- 62.Sohrabi H., Majidi M.R., Fakhraei M., Jahanban-Esfahlan A., Hejazi M., Oroojalian F., Baradaran B., Tohidast M., de la Guardia M., Mokhtarzadeh A. Talanta; 2022. Lateral Flow Assays (LFA) for Detection of Pathogenic Bacteria: A Small Point-Of-Care Platform for Diagnosis of Human Infectious Diseases. [DOI] [PubMed] [Google Scholar]
- 63.Brazaca L.C., Dos Santos P.L., de Oliveira P.R., Rocha D.P., Stefano J.S., Kalinke C., Munoz R.A.A., Bonacin J.A., Janegitz B.C., Carrilho E. Biosensing strategies for the electrochemical detection of viruses and viral diseases–A review. Anal. Chim. Acta. 2021;1159 doi: 10.1016/j.aca.2021.338384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong S., Lee C. The current status and future outlook of quantum dot-based biosensors for plant virus detection. Plant Pathol. J. 2018;34:85. doi: 10.5423/PPJ.RW.08.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orooji Y., Sohrabi H., Hemmat N., Oroojalian F., Baradaran B., Mokhtarzadeh A., Mohaghegh M., Karimi-Maleh H. An overview on SARS-CoV-2 (COVID-19) and other human coronaviruses and their detection capability via amplification assay, chemical sensing, biosensing, immunosensing, and clinical assays. Nano-Micro Lett. 2021;13:1–30. doi: 10.1007/s40820-020-00533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cesewski E., Johnson B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020;159 doi: 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.da Silva A.D., Paschoalino W.J., Neto R.C., Kubota L.T. Case Studies in Chemical and Environmental Engineering; 2022. Electrochemical Point-Of-Care Devices for Monitoring Waterborne Pathogens: Protozoa, Bacteria, and Viruses–An Overview. [Google Scholar]
- 68.Rosario R., Mutharasan R. Nucleic acid electrochemical and electromechanical biosensors: a review of techniques and developments. Rev. Anal. Chem. 2014;33:213–230. [Google Scholar]
- 69.Tosar J., Branas G., Laíz J. Electrochemical DNA hybridization sensors applied to real and complex biological samples. Biosens. Bioelectron. 2010;26:1205–1217. doi: 10.1016/j.bios.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 70.Mahmoodi P., Fani M., Rezayi M., Avan A., Pasdar Z., Karimi E., Amiri I.S., Ghayour-Mobarhan M. Early detection of cervical cancer based on high-risk HPV DNA-based genosensors: a systematic review. Biofactors. 2019;45:101–117. doi: 10.1002/biof.1465. [DOI] [PubMed] [Google Scholar]
- 71.Blancett C.D., Fetterer D.P., Koistinen K.A., Morazzani E.M., Monninger M.K., Piper A.E., Kuehl K.A., Kearney B.J., Norris S.L., Rossi C.A. Accurate virus quantitation using a Scanning Transmission Electron Microscopy (STEM) detector in a scanning electron microscope. J. Virol Methods. 2017;248:136–144. doi: 10.1016/j.jviromet.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 72.Mixson-Hayden T., Dawson G.J., Teshale E., Le T., Cheng K., Drobeniuc J., Ward J., Kamili S. Performance of ARCHITECT HCV core antigen test with specimens from US plasma donors and injecting drug users. J. Clin. Virol. 2015;66:15–18. doi: 10.1016/j.jcv.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 73.Gupta E., Pandey P., Kumar A., Sharma M., Sarin S. Correlation between two chemiluminescence based assays for quantification of hepatitis B surface antigen in patients with chronic hepatitis B infection. Indian J. Med. Microbiol. 2015;33:96. doi: 10.4103/0255-0857.148400. [DOI] [PubMed] [Google Scholar]
- 74.Trombetta C.M., Remarque E.J., Mortier D., Montomoli E. Comparison of hemagglutination inhibition, single radial hemolysis, virus neutralization assays, and ELISA to detect antibody levels against seasonal influenza viruses. Influenza Respir. Viruses. 2018;12:675–686. doi: 10.1111/irv.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morota K., Fujinami R., Kinukawa H., Machida T., Ohno K., Saegusa H., Takeda K. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J. Virol Methods. 2009;157:8–14. doi: 10.1016/j.jviromet.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 76.Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barzon L., Lavezzo E., Costanzi G., Franchin E., Toppo S., Palù G. Next-generation sequencing technologies in diagnostic virology. J. Clin. Virol. 2013;58:346–350. doi: 10.1016/j.jcv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 78.Kavita V. DNA biosensors-a review. J. Bioeng Biomed. Sci. 2017;7:222. [Google Scholar]
- 79.Hasanzadeh M., Sadeghi S., Bageri L., Mokhtarzadeh A., Shadjou N., Mahboob S. Poly-dopamine-beta-cyclodextrin: a novel nanobiopolymer towards sensing of some amino acids at physiological pH. Mater. Sci. Eng. C. 2016;69:343–357. doi: 10.1016/j.msec.2016.06.081. [DOI] [PubMed] [Google Scholar]
- 80.Hasanzadeh M., Tagi S., Solhi E., Shadjou N., Jouyban A., Mokhtarzadeh A. Immunosensing of breast cancer prognostic marker in adenocarcinoma cell lysates and unprocessed human plasma samples using gold nanostructure coated on organic substrate. Int. J. Biol. Macromol. 2018;118:1082–1089. doi: 10.1016/j.ijbiomac.2018.06.091. [DOI] [PubMed] [Google Scholar]
- 81.Touhami A. Biosensors and nanobiosensors: design and applications. Nanomedicine. 2014;15:374–403. [Google Scholar]
- 82.Rafique B., Iqbal M., Mehmood T., Shaheen M.A. Sensor Review; 2019. Electrochemical DNA Biosensors: A Review. [Google Scholar]
- 83.Hasanzadeh M., Mokhtarzadeh A., Shadjou N., Mahboob S. An innovative immunosensor for detection of tumor suppressor protein p53 in unprocessed human plasma and cancer cell lysates. Int. J. Biol. Macromol. 2017;105:1337–1348. doi: 10.1016/j.ijbiomac.2017.07.165. [DOI] [PubMed] [Google Scholar]
- 84.Abu-Salah K.M., Zourob M.M., Mouffouk F., Alrokayan S.A., Alaamery M.A., Ansari A.A. DNA-based nanobiosensors as an emerging platform for detection of disease. Sensors. 2015;15:14539–14568. doi: 10.3390/s150614539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eivazzadeh-Keihan R., Pashazadeh-Panahi P., Baradaran B., de la Guardia M., Hejazi M., Sohrabi H., Mokhtarzadeh A., Maleki A. Recent progress in optical and electrochemical biosensors for sensing of Clostridium botulinum neurotoxin. Trac. Trends Anal. Chem. 2018;103:184–197. [Google Scholar]
- 86.Mobed A., Baradaran B., de la Guardia M., Agazadeh M., Hasanzadeh M., Rezaee M.A., Mosafer J., Mokhtarzadeh A., Hamblin M.R. Advances in detection of fastidious bacteria: from microscopic observation to molecular biosensors. Trac. Trends Anal. Chem. 2019;113:157–171. [Google Scholar]
- 87.Ju H.-X., Ye Y.-K., Zhao J.-H., Zhu Y.-L. Hybridization biosensor using di (2, 2′-bipyridine) osmium (III) as electrochemical indicator for detection of polymerase chain reaction product of hepatitis B virus DNA. Anal. Biochem. 2003;313:255–261. doi: 10.1016/s0003-2697(02)00625-5. [DOI] [PubMed] [Google Scholar]
- 88.Manzanares-Palenzuela C.L., Martín-Fernández B., López M.S.-P., López-Ruiz B. Electrochemical genosensors as innovative tools for detection of genetically modified organisms. Trac. Trends Anal. Chem. 2015;66:19–31. [Google Scholar]
- 89.Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 90.Wu C.-C., Yen H.-Y., Lai L.-T., Perng G.-C., Lee C.-R., Wu S.-J. A label-free impedimetric genosensor for the nucleic acid amplification-free detection of extracted RNA of dengue virus. Sensors. 2020;20:3728. doi: 10.3390/s20133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alzate D., Cajigas S., Robledo S., Muskus C., Orozco J. Genosensors for differential detection of Zika virus. Talanta. 2020;210 doi: 10.1016/j.talanta.2019.120648. [DOI] [PubMed] [Google Scholar]
- 92.Tu J., Torrente-Rodríguez R.M., Wang M., Gao W. The era of digital health: a review of portable and wearable affinity biosensors. Adv. Funct. Mater. 2020;30 [Google Scholar]
- 93.Paniel N., Baudart J., Hayat A., Barthelmebs L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods. 2013;64:229–240. doi: 10.1016/j.ymeth.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 94.Drummond T.G., Hill M.G., Barton J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 95.Perumal V., Hashim U. Advances in biosensors: principle, architecture and applications. J. Appl. Biomed. 2014;12:1–15. [Google Scholar]
- 96.Malecka K., Stachyra A., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecka H., Radecki J. Electrochemical genosensor based on disc and screen printed gold electrodes for detection of specific DNA and RNA sequences derived from Avian Influenza Virus H5N1. Sensor. Actuator. B Chem. 2016;224:290–297. [Google Scholar]
- 97.Singhal C., Pundir C., Narang J. A genosensor for detection of consensus DNA sequence of Dengue virus using ZnO/Pt-Pd nanocomposites. Biosens. Bioelectron. 2017;97:75–82. doi: 10.1016/j.bios.2017.05.047. [DOI] [PubMed] [Google Scholar]
- 98.Singhal C., Ingle A., Chakraborty D., Pundir C., Narang J. Impedimetric genosensor for detection of hepatitis C virus (HCV1) DNA using viral probe on methylene blue doped silica nanoparticles. Int. J. Biol. Macromol. 2017;98:84–93. doi: 10.1016/j.ijbiomac.2017.01.093. [DOI] [PubMed] [Google Scholar]
- 99.Pänke O., Kirbs A., Lisdat F. Voltammetric detection of single base-pair mismatches and quantification of label-free target ssDNA using a competitive binding assay. Biosens. Bioelectron. 2007;22:2656–2662. doi: 10.1016/j.bios.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 100.Castro A.C.H., França E.G., de Paula L.F., Soares M.M., Goulart L.R., Madurro J.M., Brito-Madurro A.G. Preparation of genosensor for detection of specific DNA sequence of the hepatitis B virus. Appl. Surf. Sci. 2014;314:273–279. [Google Scholar]
- 101.Li Y. 2018. RESEARCH ARTICLE A Sensitive Colorimetric DNA Biosensor for Specific Detection of the HBV Gene Based on Silver-Coated Glass Slide and G-Quadruplex-Hemin DNAzyme. [DOI] [PubMed] [Google Scholar]
- 102.Pourakbari R., Shadjou N., Yousefi H., Isildak I., Yousefi M., Rashidi M.-R., Khalilzadeh B. Recent progress in nanomaterial-based electrochemical biosensors for pathogenic bacteria. Microchim. Acta. 2019;186:820. doi: 10.1007/s00604-019-3966-8. [DOI] [PubMed] [Google Scholar]
- 103.Pei F., Wang P., Ma E., Yang Q., Yu H., Gao C., Li Y., Liu Q., Dong Y. A sandwich-type electrochemical immunosensor based on RhPt NDs/NH2-GS and Au NPs/PPy NS for quantitative detection hepatitis B surface antigen. Bioelectrochemistry. 2019;126:92–98. doi: 10.1016/j.bioelechem.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 104.Cabral D.G., Lima E.C., Moura P., Dutra R.F. A label-free electrochemical immunosensor for hepatitis B based on hyaluronic acid–carbon nanotube hybrid film. Talanta. 2016;148:209–215. doi: 10.1016/j.talanta.2015.10.083. [DOI] [PubMed] [Google Scholar]
- 105.Wang X., Li Y., Wang H., Fu Q., Peng J., Wang Y., Du J., Zhou Y., Zhan L. Gold nanorod-based localized surface plasmon resonance biosensor for sensitive detection of hepatitis B virus in buffer, blood serum and plasma. Biosens. Bioelectron. 2010;26:404–410. doi: 10.1016/j.bios.2010.07.121. [DOI] [PubMed] [Google Scholar]
- 106.Pohanka M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials. 2018;11:448. doi: 10.3390/ma11030448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pohanka M. The piezoelectric biosensors: principles and applications. Int. J. Electrochem. Sci. 2017;12:496–506. [Google Scholar]
- 108.Liu Y.-P., Yao C.-Y. Rapid and quantitative detection of hepatitis B virus. World J. Gastroenterol. 2015;21:11954. doi: 10.3748/wjg.v21.i42.11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Englebienne P., Van Hoonacker A., Verhas M. Surface plasmon resonance: principles, methods and applications in biomedical sciences. Spectroscopy. 2003;17:255–273. [Google Scholar]
- 110.Fulgione A., Cimafonte M., Della Ventura B., Iannaccone M., Ambrosino C., Capuano F., Proroga Y.T.R., Velotta R., Capparelli R. QCM-based immunosensor for rapid detection of Salmonella Typhimurium in food. Sci. Rep. 2018;8:1–8. doi: 10.1038/s41598-018-34285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pohanka M. Quartz crystal microbalance (QCM) sensing materials in biosensors development. Int. J. Electrochem. Sci. 2021;16:2. [Google Scholar]
- 112.Ni Y., Yan Y., Xie Y., Liu C., Li X., Zhu J. IEEE; 2021. Design of QCM Frequency Acquisition Module for the Detection of RNA Viruses, 2021 40th Chinese Control Conference (CCC) pp. 5721–5727. [Google Scholar]
- 113.Khosravi-Nejad F., Teimouri M., Marandi S.J., Shariati M. The highly sensitive impedimetric biosensor in label free approach for hepatitis B virus DNA detection based on tellurium doped ZnO nanowires. Appl. Phys. A. 2019;125:616. [Google Scholar]
- 114.Shariati M. The field effect transistor DNA biosensor based on ITO nanowires in label-free hepatitis B virus detecting compatible with CMOS technology. Biosens. Bioelectron. 2018;105:58–64. doi: 10.1016/j.bios.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 115.Huang S., Feng M., Li J., Liu Y., Xiao Q. Voltammetric determination of attomolar levels of a sequence derived from the genom of hepatitis B virus by using molecular beacon mediated circular strand displacement and rolling circle amplification. Microchim. Acta. 2018;185:1–9. doi: 10.1007/s00604-018-2744-3. [DOI] [PubMed] [Google Scholar]
- 116.Khosravi-Nejad F., Teimouri M., Marandi S.J., Shariati M. The highly sensitive impedimetric biosensor in label free approach for hepatitis B virus DNA detection based on tellurium doped ZnO nanowires. Appl. Phys. A. 2019;125:1–8. [Google Scholar]
- 117.Ariksoysal D.O., Karadeniz H., Erdem A., Sengonul A., Sayiner A.A., Ozsoz M. Label-free electrochemical hybridization genosensor for the detection of hepatitis B virus genotype on the development of lamivudine resistance. Anal. Chem. 2005;77:4908–4917. doi: 10.1021/ac050022+. [DOI] [PubMed] [Google Scholar]
- 118.Narang J., Singhal C., Malhotra N., Narang S., Pn A.K., Gupta R., Kansal R., Pundir C. Impedimetric genosensor for ultratrace detection of hepatitis B virus DNA in patient samples assisted by zeolites and MWCNT nano-composites. Biosens. Bioelectron. 2016;86:566–574. doi: 10.1016/j.bios.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 119.Erdem A., Ariksoysal D.O., Karadeniz H., Kara P., Sengonul A., Sayiner A.A., Ozsoz M. Electrochemical genomagnetic assay for the detection of hepatitis B virus DNA in polymerase chain reaction amplicons by using disposable sensor technology. Electrochem. Commun. 2005;7:815–820. [Google Scholar]
- 120.Shariati M., Sadeghi M. Ultrasensitive DNA biosensor for hepatitis B virus detection based on tin-doped WO 3/In 2 O 3 heterojunction nanowire photoelectrode under laser amplification. Anal. Bioanal. Chem. 2020;412:5367–5377. doi: 10.1007/s00216-020-02752-z. [DOI] [PubMed] [Google Scholar]
- 121.Zhou X., Liu L., Hu M., Wang L., Hu J. Detection of hepatitis B virus by piezoelectric biosensor. J. Pharmaceut. Biomed. Anal. 2002;27:341–345. doi: 10.1016/s0731-7085(01)00538-6. [DOI] [PubMed] [Google Scholar]
- 122.Yao C., Zhu T., Tang J., Wu R., Chen Q., Chen M., Zhang B., Huang J., Fu W. Hybridization assay of hepatitis B virus by QCM peptide nucleic acid biosensor. Biosens. Bioelectron. 2008;23:879–885. doi: 10.1016/j.bios.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 123.Huang C.-W., Hsueh H.-T., Huang Y.-J., Liao H.-H., Tsai H.-H., Juang Y.-Z., Lin T.-H., Lu S.-S., Lin C.-T. A fully integrated wireless CMOS microcantilever lab chip for detection of DNA from Hepatitis B virus (HBV) Sensor. Actuator. B Chem. 2013;181:867–873. [Google Scholar]
- 124.Cha B.H., Lee S.-M., Park J.C., Hwang K.S., Kim S.K., Lee Y.-S., Ju B.-K., Kim T.S. Detection of Hepatitis B Virus (HBV) DNA at femtomolar concentrations using a silica nanoparticle-enhanced microcantilever sensor. Biosens. Bioelectron. 2009;25:130–135. doi: 10.1016/j.bios.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 125.Chuang T.-L., Wei S.-C., Lee S.-Y., Lin C.-W. A polycarbonate based surface plasmon resonance sensing cartridge for high sensitivity HBV loop-mediated isothermal amplification. Biosens. Bioelectron. 2012;32:89–95. doi: 10.1016/j.bios.2011.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang S., Li L., Jin H., Yang T., Bao W., Huang S., Wang J. Electrochemical detection of hepatitis B and papilloma virus DNAs using SWCNT array coated with gold nanoparticles. Biosens. Bioelectron. 2013;41:205–210. doi: 10.1016/j.bios.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 127.Erdem A., Kerman K., Meric B., Akarca U.S., Ozsoz M. DNA electrochemical biosensor for the detection of short DNA sequences related to the hepatitis B virus. Electroanalysis: Int. J. Devoted Fund. Practical Aspect. of Electroanalysis. 1999;11:586–587. [Google Scholar]
- 128.Liu L., Wang X., Ma Q., Lin Z., Chen S., Li Y., Lu L., Qu H., Su X. Multiplex electrochemiluminescence DNA sensor for determination of hepatitis B virus and hepatitis C virus based on multicolor quantum dots and Au nanoparticles. Anal. Chim. Acta. 2016;916:92–101. doi: 10.1016/j.aca.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 129.Oliveira D.A., Silva J.V., Flauzino J.M., Castro A.C., Moço A.C., Soares M.M., Madurro J.M., Brito-Madurro A.G. Application of nanomaterials for the electrical and optical detection of the hepatitis B virus. Anal. Biochem. 2018;549:157–163. doi: 10.1016/j.ab.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 130.Zhao F., Bai Y., Cao L., Han G., Fang C., Wei S., Chen Z. New electrochemical DNA sensor based on nanoflowers of Cu3 (PO4) 2-BSA-GO for hepatitis B virus DNA detection. J. Electroanal. Chem. 2020;867 [Google Scholar]
- 131.Zheng J., Chen C., Wang X., Zhang F., He P. A sequence-specific DNA sensor for Hepatitis B virus diagnostics based on the host–guest recognition. Sensor. Actuator. B Chem. 2014;199:168–174. [Google Scholar]
- 132.Shakoori Z., Salimian S., Kharrazi S., Adabi M., Saber R. Electrochemical DNA biosensor based on gold nanorods for detecting hepatitis B virus. Anal. Bioanal. Chem. 2015;407:455–461. doi: 10.1007/s00216-014-8303-9. [DOI] [PubMed] [Google Scholar]
- 133.Wu W., Kirimli C.E., Shih W.-H., Shih W.Y. Real-time, in situ DNA hybridization detection with attomolar sensitivity without amplification using (pb (Mg1/3Nb2/3) O3) 0.65–(PbTiO3) 0.35 piezoelectric plate sensors. Biosens. Bioelectron. 2013;43:391–399. doi: 10.1016/j.bios.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 134.Giamblanco N., Conoci S., Russo D., Marletta G. Single-step label-free hepatitis B virus detection by a piezoelectric biosensor. RSC Adv. 2015;5:38152–38158. [Google Scholar]
- 135.Zhang C., Chen Y., Liang X., Zhang G., Ma H., Nie L., Wang Y. Detection of hepatitis B virus M204I mutation by quantum dot-labeled DNA probe. Sensors. 2017;17:961. doi: 10.3390/s17050961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li M., Cushing S.K., Liang H., Suri S., Ma D., Wu N. Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA. Anal. Chem. 2013;85:2072–2078. doi: 10.1021/ac303387a. [DOI] [PubMed] [Google Scholar]
- 137.Lu X., Dong X., Zhang K., Han X., Fang X., Zhang Y. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst. 2013;138:642–650. doi: 10.1039/c2an36099c. [DOI] [PubMed] [Google Scholar]
- 138.Al Olaby R.R., Azzazy H.M. Hepatitis C virus RNA assays: current and emerging technologies and their clinical applications. Expert Rev. Mol. Diagn. 2011;11:53–64. doi: 10.1586/erm.10.101. [DOI] [PubMed] [Google Scholar]
- 139.Uliana C.V., Riccardi C.S., Yamanaka H. Diagnostic tests for hepatitis C: recent trends in electrochemical immunosensor and genosensor analysis. World J. Gastroenterol.: WJG. 2014;20 doi: 10.3748/wjg.v20.i42.15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shawky S.M., Bald D., Azzazy H.M. Direct detection of unamplified hepatitis C virus RNA using unmodified gold nanoparticles. Clin. Biochem. 2010;43:1163–1168. doi: 10.1016/j.clinbiochem.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 141.Warkad S.D., Nimse S.B., Song K.-S., Kim T. HCV detection, discrimination, and genotyping technologies. Sensors. 2018;18:3423. doi: 10.3390/s18103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Donmez S., Arslan F., Arslan H. A nucleic acid biosensor for detection of hepatitis C virus genotype 1a using poly (l-glutamic acid)-modified electrode. Appl. Biochem. Biotechnol. 2015;176:1431–1444. doi: 10.1007/s12010-015-1655-6. [DOI] [PubMed] [Google Scholar]
- 143.Skládal P., dos Santos Riccardi C., Yamanaka H., da Costa P.I. Piezoelectric biosensors for real-time monitoring of hybridization and detection of hepatitis C virus. J. Virol Methods. 2004;117:145–151. doi: 10.1016/j.jviromet.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 144.Shawky S.M., Awad A.M., Allam W., Alkordi M.H., El-Khamisy S.F. Gold aggregating gold: a novel nanoparticle biosensor approach for the direct quantification of hepatitis C virus RNA in clinical samples. Biosens. Bioelectron. 2017;92:349–356. doi: 10.1016/j.bios.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abduo S.M.S., El H.M.E.-S.A. Google Patents; 2016. Direct Detection of Unamplified Hepatitis C Virus RNA Using Unmodified Gold Nanoparticles. [Google Scholar]
- 146.Singh R., Mukherjee M.D., Sumana G., Gupta R.K., Sood S., Malhotra B. Biosensors for pathogen detection: a smart approach towards clinical diagnosis. Sensor. Actuator. B Chem. 2014;197:385–404. [Google Scholar]
- 147.Hejazi M., Pournaghi-Azar M., Ahour F. Electrochemical detection of short sequences of hepatitis C 3a virus using a peptide nucleic acid-assembled gold electrode. Anal. Biochem. 2010;399:118–124. doi: 10.1016/j.ab.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 148.Pournaghi-Azar M.e.H., Ahour F., Hejazi M.e.S. Differential pulse voltammetric detection of hepatitis C virus 1a oligonucleotide chain by a label-free electrochemical DNA hybridization biosensor using consensus sequence of hepatitis C virus 1a probe on the pencil graphite electrode. Electroanalysis: Int. J. Devoted Fund. Practical Aspect. of Electroanalysis. 2009;21:1822–1828. [Google Scholar]
- 149.Park J.-Y., Lee Y.-s., Chang B.-Y., Kim B.H., Jeon S., Park S.-M. Label-free impedimetric sensor for a ribonucleic acid oligomer specific to hepatitis C virus at a self-assembled monolayer-covered electrode. Anal. Chem. 2010;82:8342–8348. doi: 10.1021/ac1019232. [DOI] [PubMed] [Google Scholar]
- 150.Singhal C., Ingle A., Chakraborty D., Pn A.K., Pundir C., Narang J. Impedimetric genosensor for detection of hepatitis C virus (HCV1) DNA using viral probe on methylene blue doped silica nanoparticles. Int. J. Biol. Macromol. 2017;98:84–93. doi: 10.1016/j.ijbiomac.2017.01.093. [DOI] [PubMed] [Google Scholar]
- 151.dos Santos Riccardi C., Dahmouche K., Santilli C.V., da Costa P.I., Yamanaka H. Immobilization of streptavidin in sol–gel films: application on the diagnosis of hepatitis C virus. Talanta. 2006;70:637–643. doi: 10.1016/j.talanta.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 152.El-Said W.A., Choi J.-w. High selective spectroelectrochemical biosensor for HCV-RNA detection based on a specific peptide nucleic acid. Spectrochim. Acta Mol. Biomol. Spectrosc. 2019;217:288–293. doi: 10.1016/j.saa.2019.03.115. [DOI] [PubMed] [Google Scholar]
- 153.Lereau M., Fournier-Wirth C., Mayen J., Farre C., Meyer A., Dugas V., Cantaloube J.-F.o., Chaix C., Vasseur J.-J., Morvan F.o. Development of innovative and versatile polythiol probes for use on ELOSA or electrochemical biosensors: application in hepatitis C virus genotyping. Anal. Chem. 2013;85:9204–9212. doi: 10.1021/ac401941x. [DOI] [PubMed] [Google Scholar]
- 154.Liu S., Wu P., Li W., Zhang H., Cai C. Ultrasensitive and selective electrochemical identification of hepatitis C virus genotype 1b based on specific endonuclease combined with gold nanoparticles signal amplification. Anal. Chem. 2011;83:4752–4758. doi: 10.1021/ac200624f. [DOI] [PubMed] [Google Scholar]
- 155.Sam S.S., Steinmetz H.B., Tsongalis G.J., Tafe L.J., Lefferts J.A. Validation of a solid-phase electrochemical array for genotyping hepatitis C virus. Exp. Mol. Pathol. 2013;95:18–22. doi: 10.1016/j.yexmp.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 156.Zhang Z., Li X., Wang C., Xiong Y., Liu P., Zhang C. Cobalt complex plays a dual role in the construction of E-DNA sensor. J. Electroanal. Chem. 2013;690:117–120. [Google Scholar]
- 157.Hwang K.S., Lee S.-M., Eom K., Lee J.H., Lee Y.-S., Park J.H., Yoon D.S., Kim T.S. Nanomechanical microcantilever operated in vibration modes with use of RNA aptamer as receptor molecules for label-free detection of HCV helicase. Biosens. Bioelectron. 2007;23:459–465. doi: 10.1016/j.bios.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 158.Uliana C.V., Riccardi C.S., Tognolli J.O., Yamanaka H. Optimization of an amperometric biosensor for the detection of hepatitis C virus using fractional factorial designs. J. Braz. Chem. Soc. 2008;19:782–787. [Google Scholar]
- 159.Liu S., Hu Y., Jin J., Zhang H., Cai C. Electrochemical detection of hepatitis C virus based on site-specific DNA cleavage of Bam HI endonuclease. Chem. Commun. 2009:1635–1637. doi: 10.1039/b900690g. [DOI] [PubMed] [Google Scholar]
- 160.Liu S., Wang Q., Chen D., Jin J., Hu Y., Wu P., Zhang H., Cai C. Electrochemical approach for the specific detection of hepatitis C virus based on site-specific DNA cleavage of Bam HI endonuclease. Anal. Methods. 2010;2:135–142. [Google Scholar]
- 161.Riccardi C.d.S., Kranz C., Kowalik J., Yamanaka H., Mizaikoff B., Josowicz M. Label-free DNA detection of hepatitis C virus based on modified conducting polypyrrole films at microelectrodes and atomic force microscopy tip-integrated electrodes. Anal. Chem. 2008;80:237–245. doi: 10.1021/ac701613t. [DOI] [PubMed] [Google Scholar]
- 162.Donmez S., Arslan F., Arslan H. Sequence-specific label-free nucleic acid biosensor for the detection of the hepatitis C virus genotype 1a using a disposable pencil graphite electrode. Artif. Cell Nanomed. Biotechnol. 2016;44:912–917. doi: 10.3109/21691401.2014.998831. [DOI] [PubMed] [Google Scholar]
- 163.Oliveira D.A., Silva J.V., Flauzino J.M., Sousa H.S., Castro A.C., Moço A.C., Soares M.M., Madurro J.M., Brito-Madurro A.G. Carbon nanomaterial as platform for electrochemical genosensor: a system for the diagnosis of the hepatitis C in real sample. J. Electroanal. Chem. 2019;844:6–13. [Google Scholar]
- 164.Lifson M.A., Ozen M.O., Inci F., Wang S., Inan H., Baday M., Henrich T.J., Demirci U. Advances in biosensing strategies for HIV-1 detection, diagnosis, and therapeutic monitoring. Adv. Drug Deliv. Rev. 2016;103:90–104. doi: 10.1016/j.addr.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Farzin L., Shamsipur M., Samandari L., Sheibani S. Talanta; 2019. HIV Biosensors for Early Diagnosis of Infection: the Intertwine of Nanotechnology with Sensing Strategies. [DOI] [PubMed] [Google Scholar]
- 166.Fatin M., Ruslinda A., Arshad M.M., Tee K., Ayub R., Hashim U., Kamarulzaman A., Gopinath S.C. HIV-1 Tat biosensor: current development and trends for early detection strategies. Biosens. Bioelectron. 2016;78:358–366. doi: 10.1016/j.bios.2015.11.067. [DOI] [PubMed] [Google Scholar]
- 167.Cooper A., García M., Petrovas C., Yamamoto T., Koup R.A., Nabel G.J. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 168.Rodrigo M.A.M., Heger Z., Cernei N., Jinemez A.M.J., Zitka O., Adam V., Kizek R. HIV biosensors–the potential of the electrochemical way. Int. J. Electrochem. Sci. 2014;9:3449–3457. [Google Scholar]
- 169.Shafiee H., Lidstone E.A., Jahangir M., Inci F., Hanhauser E., Henrich T.J., Kuritzkes D.R., Cunningham B.T., Demirci U. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci. Rep. 2014;4:1–7. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Conway J.M., Ribeiro R.M. Modeling the immune response to HIV infection. Curr. Opin. Struct. Biol. 2018;12:61–69. doi: 10.1016/j.coisb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhou J., Gan N., Li T., Hu F., Li X., Wang L., Zheng L. A cost-effective sandwich electrochemiluminescence immunosensor for ultrasensitive detection of HIV-1 antibody using magnetic molecularly imprinted polymers as capture probes. Biosens. Bioelectron. 2014;54:199–206. doi: 10.1016/j.bios.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 172.Kheiri F., Sabzi R., Jannatdoust E., Shojaeefar E., Sedghi H. A novel amperometric immunosensor based on acetone-extracted propolis for the detection of the HIV-1 p24 antigen. Biosens. Bioelectron. 2011;26:4457–4463. doi: 10.1016/j.bios.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 173.Giannetto M., Costantini M., Mattarozzi M., Careri M. Innovative gold-free carbon nanotube/chitosan-based competitive immunosensor for determination of HIV-related p24 capsid protein in serum. RSC Adv. 2017;7:39970–39976. [Google Scholar]
- 174.Diao W., Tang M., Ding S., Li X., Cheng W., Mo F., Yan X., Ma H., Yan Y. Highly sensitive surface plasmon resonance biosensor for the detection of HIV-related DNA based on dynamic and structural DNA nanodevices. Biosens. Bioelectron. 2018;100:228–234. doi: 10.1016/j.bios.2017.08.042. [DOI] [PubMed] [Google Scholar]
- 175.Alodhayb A., Brown N., Saydur Rahman S., Harrigan R., Beaulieu L. Towards detecting the human immunodeficiency virus using microcantilever sensors. Appl. Phys. Lett. 2013;102 [Google Scholar]
- 176.Gong Q., Wang Y., Yang H. A sensitive impedimetric DNA biosensor for the determination of the HIV gene based on graphene-Nafion composite film. Biosens. Bioelectron. 2017;89:565–569. doi: 10.1016/j.bios.2016.02.045. [DOI] [PubMed] [Google Scholar]
- 177.Zhang J., Wu X., Yang W., Chen J., Fu F. Ultra-sensitive electrochemical detection of single nucleotide polymorphisms based on an electrically controllable magnetic gold electrode. Chem. Commun. 2013;49:996–998. doi: 10.1039/c2cc37783g. [DOI] [PubMed] [Google Scholar]
- 178.Odenthal K.J., Gooding J.J. An introduction to electrochemical DNA biosensors. Analyst. 2007;132:603–610. doi: 10.1039/b701816a. [DOI] [PubMed] [Google Scholar]
- 179.Valizadeh A., Sohrabi N., Badrzadeh F. Electrochemical detection of HIV-1 by nanomaterials. Artif. Cell Nanomed. Biotechnol. 2017;45:1467–1477. doi: 10.1080/21691401.2017.1282494. [DOI] [PubMed] [Google Scholar]
- 180.Zheng L., Jia L., Li B., Situ B., Liu Q., Wang Q., Gan N. A sandwich HIV p24 amperometric immunosensor based on a direct gold electroplating-modified electrode. Molecules. 2012;17:5988–6000. doi: 10.3390/molecules17055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang Y., Chen X., Roozbahani G.M., Guan X. Graphene oxide-based biosensing platform for rapid and sensitive detection of HIV-1 protease. Anal. Bioanal. Chem. 2018;410:6177–6185. doi: 10.1007/s00216-018-1224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liang Y., Zhang Z., Wei H., Hu Q., Deng J., Guo D., Cui Z., Zhang X.-E. Aptamer beacons for visualization of endogenous protein HIV-1 reverse transcriptase in living cells. Biosens. Bioelectron. 2011;28:270–276. doi: 10.1016/j.bios.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 183.Babamiri B., Salimi A., Hallaj R. A molecularly imprinted electrochemiluminescence sensor for ultrasensitive HIV-1 gene detection using EuS nanocrystals as luminophore. Biosens. Bioelectron. 2018;117:332–339. doi: 10.1016/j.bios.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 184.Zhang D., Peng Y., Qi H., Gao Q., Zhang C. Label-free electrochemical DNA biosensor array for simultaneous detection of the HIV-1 and HIV-2 oligonucleotides incorporating different hairpin-DNA probes and redox indicator. Biosens. Bioelectron. 2010;25:1088–1094. doi: 10.1016/j.bios.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 185.Huang Y.L., Gao Z.F., Luo H.Q., Li N.B. Sensitive detection of HIV gene by coupling exonuclease III-assisted target recycling and guanine nanowire amplification. Sensor. Actuator. B Chem. 2017;238:1017–1023. [Google Scholar]
- 186.Wang Y., Bai X., Wen W., Zhang X., Wang S. Ultrasensitive electrochemical biosensor for HIV gene detection based on graphene stabilized gold nanoclusters with exonuclease amplification. ACS Appl. Mater. Interfaces. 2015;7:18872–18879. doi: 10.1021/acsami.5b05857. [DOI] [PubMed] [Google Scholar]
- 187.Dai Tran L., Nguyen B.H., Van Hieu N., Tran H.V., Le Nguyen H., Nguyen P.X. Electrochemical detection of short HIV sequences on chitosan/Fe3O4 nanoparticle based screen printed electrodes. Mater. Sci. Eng. C. 2011;31:477–485. [Google Scholar]
- 188.Guo Y., Chen J., Chen G. A label-free electrochemical biosensor for detection of HIV related gene based on interaction between DNA and protein. Sensor. Actuator. B Chem. 2013;184:113–117. [Google Scholar]
- 189.Fu Y., Yuan R., Chai Y., Zhou L., Zhang Y. Coupling of a reagentless electrochemical DNA biosensor with conducting polymer film and nanocomposite as matrices for the detection of the HIV DNA sequences. Anal. Lett. 2006;39:467–482. [Google Scholar]
- 190.Gong Q., Yang H., Dong Y., Zhang W. A sensitive impedimetric DNA biosensor for the determination of the HIV gene based on electrochemically reduced graphene oxide. Anal. Methods. 2015;7:2554–2562. [Google Scholar]
- 191.Wang J., Cai X., Rivas G., Shiraishi H., Farias P.A., Dontha N. DNA electrochemical biosensor for the detection of short DNA sequences related to the human immunodeficiency virus. Anal. Chem. 1996;68:2629–2634. doi: 10.1021/ac9602433. [DOI] [PubMed] [Google Scholar]
- 192.Hu J., Zheng P.-C., Jiang J.-H., Shen G.-L., Yu R.-Q., Liu G.-K. Sub-attomolar HIV-1 DNA detection using surface-enhanced Raman spectroscopy. Analyst. 2010;135:1084–1089. doi: 10.1039/b920358c. [DOI] [PubMed] [Google Scholar]
- 193.Mahmoodi P., Rezayi M., Rasouli E., Avan A., Gholami M., Mobarhan M.G., Karimi E., Alias Y. Early-stage cervical cancer diagnosis based on an ultra-sensitive electrochemical DNA nanobiosensor for HPV-18 detection in real samples. J. Nanobiotechnol. 2020;18:1–12. doi: 10.1186/s12951-020-0577-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cohen P.A., Jhingran A., Oaknin A., Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 195.Lie A., Risberg B., Borge B., Sandstad B., Delabie J., Rimala R., Onsrud M., Thoresen S. DNA-versus RNA-based methods for human papillomavirus detection in cervical neoplasia. Gynecol. Oncol. 2005;97:908–915. doi: 10.1016/j.ygyno.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 196.Abreu A.L., Souza R.P., Gimenes F., Consolaro M.E. A review of methods for detect human Papillomavirus infection. Virol. J. 2012;9:262. doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shah S.S., Senapati S., Klacsmann F., Miller D.L., Johnson J.J., Chang H.-C., Stack M.S. Current technologies and recent developments for screening of HPV-associated cervical and oropharyngeal cancers. Cancers. 2016;8:85. doi: 10.3390/cancers8090085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Parmin N.A., Hashim U., Gopinath S.C., Nadzirah S., Rejali Z., Afzan A., Uda M. Human Papillomavirus E6 biosensing: current progression on early detection strategies for cervical Cancer. Int. J. Biol. Macromol. 2019;126:877–890. doi: 10.1016/j.ijbiomac.2018.12.235. [DOI] [PubMed] [Google Scholar]
- 199.Baek S., Ahn J.K., Won B.Y., Park K.S., Park H.G. A one-step and label-free, electrochemical DNA detection using metal ion-mediated molecular beacon probe. Electrochem. Commun. 2019;100:64–69. [Google Scholar]
- 200.Avelino K.Y., Oliveira L.S., Lucena-Silva N., Andrade C.A., Oliveira M.D. Flexible sensor based on conducting polymer and gold nanoparticles for electrochemical screening of HPV families in cervical specimens. Talanta. 2021;226 doi: 10.1016/j.talanta.2021.122118. [DOI] [PubMed] [Google Scholar]
- 201.Jampasa S., Siangproh W., Laocharoensuk R., Yanatatsaneejit P., Vilaivan T., Chailapakul O. A new DNA sensor design for the simultaneous detection of HPV type 16 and 18 DNA. Sensor. Actuator. B Chem. 2018;265:514–521. [Google Scholar]
- 202.Huang H., Bai W., Dong C., Guo R., Liu Z. An ultrasensitive electrochemical DNA biosensor based on graphene/Au nanorod/polythionine for human papillomavirus DNA detection. Biosens. Bioelectron. 2015;68:442–446. doi: 10.1016/j.bios.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 203.Zari N., Amine A., Ennaji M. Label-free DNA biosensor for electrochemical detection of short DNA sequences related to human papilloma virus. Anal. Lett. 2009;42:519–535. [Google Scholar]
- 204.Karimizefreh A., Mahyari F., VaezJalali M., Mohammadpour R., Sasanpour P. Impedimetic biosensor for the DNA of the human papilloma virus based on the use of gold nanosheets. Microchim. Acta. 2017;184 [Google Scholar]
- 205.Bartolome J.P., Echegoyen L., Fragoso A. Reactive carbon nano-onion modified glassy carbon surfaces as DNA sensors for human papillomavirus oncogene detection with enhanced sensitivity. Anal. Chem. 2015;87:6744–6751. doi: 10.1021/acs.analchem.5b00924. [DOI] [PubMed] [Google Scholar]
- 206.Yang J.M., Kim K.R., Kim C.S. Biosensor for rapid and sensitive detection of influenza virus. Biotechnol. Bioproc. Eng. 2018;23:371–382. [Google Scholar]
- 207.Henrickson K.J. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr. Infect. Dis. J. 2004;23:S6–S10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 208.Van Elden L., Nijhuis M., Schipper P., Schuurman R., Van Loon A. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 2001;39:196–200. doi: 10.1128/JCM.39.1.196-200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee M.-S., Chang P.-C., Shien J.-H., Cheng M.-C., Shieh H.K. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J. Virol Methods. 2001;97:13–22. doi: 10.1016/s0166-0934(01)00301-9. [DOI] [PubMed] [Google Scholar]
- 210.Hushegyi A., Pihíková D., Bertok T., Adam V., Kizek R., Tkac J. Ultrasensitive detection of influenza viruses with a glycan-based impedimetric biosensor. Biosens. Bioelectron. 2016;79:644–649. doi: 10.1016/j.bios.2015.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhang H., Klose A.M., Miller B.L. Label-free, multiplex glycan microarray biosensor for influenza virus detection. Bioconjugate Chem. 2021;32:533–540. doi: 10.1021/acs.bioconjchem.0c00718. [DOI] [PubMed] [Google Scholar]
- 212.Zhang H., Miller B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: state of the art. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tam P.D., Van Hieu N., Chien N.D., Le A.-T., Tuan M.A. DNA sensor development based on multi-wall carbon nanotubes for label-free influenza virus (type A) detection. J. Immunol. Methods. 2009;350:118–124. doi: 10.1016/j.jim.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 214.Yu Y., Chen Z., Jian W., Sun D., Zhang B., Li X., Yao M. Ultrasensitive electrochemical detection of avian influenza A (H7N9) virus DNA based on isothermal exponential amplification coupled with hybridization chain reaction of DNAzyme nanowires. Biosens. Bioelectron. 2015;64:566–571. doi: 10.1016/j.bios.2014.09.080. [DOI] [PubMed] [Google Scholar]
- 215.Dong S., Zhao R., Zhu J., Lu X., Li Y., Qiu S., Jia L., Jiao X., Song S., Fan C. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. ACS Appl. Mater. Interfaces. 2015;7:8834–8842. doi: 10.1021/acsami.5b01438. [DOI] [PubMed] [Google Scholar]
- 216.Gubler D.J. Human arbovirus infections worldwide. Ann. N. Y. Acad. Sci. 2001;951:13–24. doi: 10.1111/j.1749-6632.2001.tb02681.x. [DOI] [PubMed] [Google Scholar]
- 217.Bruno J.G., Carrillo M.P., Richarte A.M., Phillips T., Andrews C., Lee J.S. Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes. 2012;5:633. doi: 10.1186/1756-0500-5-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Simão E.P., Silva D.B., Cordeiro M.T., Gil L.H., Andrade C.A., Oliveira M.D. Nanostructured impedimetric lectin-based biosensor for arboviruses detection. Talanta. 2020;208 doi: 10.1016/j.talanta.2019.120338. [DOI] [PubMed] [Google Scholar]
- 219.Afsahi S., Lerner M.B., Goldstein J.M., Lee J., Tang X., Bagarozzi D.A., Jr., Pan D., Locascio L., Walker A., Barron F. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018;100:85–88. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- 220.Song J., Mauk M.G., Hackett B.A., Cherry S., Bau H.H., Liu C. Instrument-free point-of-care molecular detection of Zika virus. Anal. Chem. 2016;88:7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mahmood H.Z. 2019. A Study of Metallic and Polymeric Nanostructure-Solution Based Platforms for Optical and Chemical Arbovirus Biosensing. [Google Scholar]
- 222.Santiago G.A., Vergne E., Quiles Y., Cosme J., Vazquez J., Medina J.F., Medina F., Colón C., Margolis H., Muñoz-Jordán J.L. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Neglected Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dussart P., Labeau B., Lagathu G., Louis P., Nunes M.R., Rodrigues S.G., Storck-Herrmann C., Cesaire R., Morvan J., Flamand M. Evaluation of an enzyme immunoassay for detection of dengue virus NS1 antigen in human serum. Clin. Vaccine Immunol. 2006;13:1185–1189. doi: 10.1128/CVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Saylan Y., Erdem Ö., Ünal S., Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9:65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Adegoke O., Morita M., Kato T., Ito M., Suzuki T., Park E.Y. Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays. Biosens. Bioelectron. 2017;94:513–522. doi: 10.1016/j.bios.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 226.Zhang G.-J., Zhang L., Huang M.J., Luo Z.H.H., Tay G.K.I., Lim E.-J.A., Kang T.G., Chen Y. Silicon nanowire biosensor for highly sensitive and rapid detection of Dengue virus. Sensor. Actuator. B Chem. 2010;146:138–144. [Google Scholar]
- 227.Chen S.-H., Chuang Y.-C., Lu Y.-C., Lin H.-C., Yang Y.-L., Lin C.-S. A method of layer-by-layer gold nanoparticle hybridization in a quartz crystal microbalance DNA sensing system used to detect dengue virus. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/21/215501. [DOI] [PubMed] [Google Scholar]
- 228.Deng J., Toh C.-S. Impedimetric DNA biosensor based on a nanoporous alumina membrane for the detection of the specific oligonucleotide sequence of dengue virus. Sensors. 2013;13:7774–7785. doi: 10.3390/s130607774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Steinmetz M., Lima D., Viana A.G., Fujiwara S.T., Pessôa C.A., Etto R.M., Wohnrath K. A sensitive label-free impedimetric DNA biosensor based on silsesquioxane-functionalized gold nanoparticles for Zika Virus detection. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111351. [DOI] [PubMed] [Google Scholar]
- 230.Cajigas S., Alzate D., Orozco J. Gold nanoparticle/DNA-based nanobioconjugate for electrochemical detection of Zika virus. Microchim. Acta. 2020;187:1–10. doi: 10.1007/s00604-020-04568-1. [DOI] [PubMed] [Google Scholar]
- 231.Li J., Yang K., Wu Z., Li X., Duan Q. Nitrogen-doped porous carbon-based fluorescence sensor for the detection of ZIKV RNA sequences: fluorescence image analysis. Talanta. 2019;205 doi: 10.1016/j.talanta.2019.06.091. [DOI] [PubMed] [Google Scholar]
- 232.Rai V., Hapuarachchi H.C., Ng L.C., Soh S.H., Leo Y.S., Toh C.-S. 2012. Ultrasensitive C DNA Detection of Dengue Virus RNA Using Electrochemical Nanoporous Membrane-Based Biosensor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ariffin E.Y., Tan L.L., Yook Heng L. Optical DNA biosensor based on square-planar ethyl piperidine substituted nickel (II) salphen complex for dengue virus detection. Sensors. 2018;18:1173. doi: 10.3390/s18041173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Nuzaihan M., Hashim U., Arshad M.M., Kasjoo S., Rahman S., Ruslinda A., Fathil M., Adzhri R., Shahimin M. Electrical detection of dengue virus (DENV) DNA oligomer using silicon nanowire biosensor with novel molecular gate control. Biosens. Bioelectron. 2016;83:106–114. doi: 10.1016/j.bios.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 235.Abd Rashid J.I., Yusof N.A., Abdullah J., Hashim U., Hajian R. A novel disposable biosensor based on SiNWs/AuNPs modified-screen printed electrode for dengue virus DNA oligomer detection. IEEE Sensor. J. 2015;15:4420–4427. [Google Scholar]
- 236.Rahman S., Yusof N., Hashim U., Hushiarian R., Nuzaihan M., Hamidon M., Zawawi R., Fathil M. Enhanced sensing of dengue virus DNA detection using O2 plasma treated-silicon nanowire based electrical biosensor. Anal. Chim. Acta. 2016;942:74–85. doi: 10.1016/j.aca.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 237.Singhal C., Khanuja M., Chaudhary N., Pundir C., Narang J. Detection of chikungunya virus DNA using two-dimensional MoS 2 nanosheets based disposable biosensor. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-25824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Singhal C., Dubey A., Mathur A., Pundir C., Narang J. Paper based DNA biosensor for detection of chikungunya virus using gold shells coated magnetic nanocubes. Process Biochem. 2018;74:35–42. [Google Scholar]
- 239.Abad-Valle P., Fernández-Abedul M.T., Costa-García A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosens. Bioelectron. 2005;20:2251–2260. doi: 10.1016/j.bios.2004.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Koo B., Jin C.E., Lee T.Y., Lee J.H., Park M.K., Sung H., Park S.Y., Lee H.J., Kim S.M., Kim J.Y. An isothermal, label-free, and rapid one-step RNA amplification/detection assay for diagnosis of respiratory viral infections. Biosens. Bioelectron. 2017;90:187–194. doi: 10.1016/j.bios.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 242.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shenashen M.A., Emran M.Y., El Sabagh A., Selim M.M., Elmarakbi A., El-Safty S.A. Progress in sensory devices of pesticides, pathogens, coronavirus, and chemical additives and hazards in food assessment: food safety concerns. Prog. Mater. Sci. 2022;124 [Google Scholar]
- 245.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Song Z., Ma Y., Chen M., Ambrosi A., Ding C., Luo X. Electrochemical biosensor with enhanced antifouling capability for COVID-19 nucleic acid detection in complex biological media. Anal. Chem. 2021;93:5963–5971. doi: 10.1021/acs.analchem.1c00724. [DOI] [PubMed] [Google Scholar]
- 249.Fan Z., Yao B., Ding Y., Zhao J., Xie M., Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 251.Chisholm C., Lopez L. Cutaneous infections caused by Herpesviridae: a review. Arch. Pathol. Lab Med. 2011;135:1357–1362. doi: 10.5858/arpa.2010-0156-RS. [DOI] [PubMed] [Google Scholar]
- 252.Kara P., Meric B., Zeytinoglu A., Ozsoz M. Electrochemical DNA biosensor for the detection and discrimination of herpes simplex Type I and Type II viruses from PCR amplified real samples. Anal. Chim. Acta. 2004;518:69–76. [Google Scholar]
- 253.Berggren C., Stålhandske P., Brundell J., Johansson G. A feasibility study of a capacitive biosensor for direct detection of DNA hybridization. Electroanalysis: Int. J. Devoted Fund. Practical Aspect. of Electroanalysis. 1999;11:156–160. [Google Scholar]
- 254.Uludağ Y., Li X., Coleman H., Efstathiou S., Cooper M.A. Direct acoustic profiling of DNA hybridisation using HSV type 1 viral sequences. Analyst. 2008;133:52–57. doi: 10.1039/b711850c. [DOI] [PubMed] [Google Scholar]
- 255.Uludağ Y., Hammond R., Cooper M.A. A signal amplification assay for HSV type 1 viral DNA detection using nanoparticles and direct acoustic profiling. J. Nanobiotechnol. 2010;8:1–12. [Google Scholar]
- 256.Cheng C., Oueslati R., Wu J., Chen J., Eda S. Capacitive DNA sensor for rapid and sensitive detection of whole genome human herpesvirus-1 dsDNA in serum. Electrophoresis. 2017;38:1617–1623. doi: 10.1002/elps.201700043. [DOI] [PubMed] [Google Scholar]
- 257.Azek F., Grossiord C., Joannes M., Limoges B., Brossier P. Hybridization assay at a disposable electrochemical biosensor for the attomole detection of amplified human cytomegalovirus DNA. Anal. Biochem. 2000;284:107–113. doi: 10.1006/abio.2000.4692. [DOI] [PubMed] [Google Scholar]
- 258.Djellouli N., Rochelet-Dequaire M., Limoges B., Druet M., Brossier P. Evaluation of the analytical performances of avidin-modified carbon sensors based on a mediated horseradish peroxidase enzyme label and their application to the amperometric detection of nucleic acids. Biosens. Bioelectron. 2007;22:2906–2913. doi: 10.1016/j.bios.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 259.Lee J.-S., Kim S., Kim S., Ahn K., Min D.-H. Fluorometric viral miRNA nanosensor for diagnosis of productive (lytic) human cytomegalovirus infection in living cells. ACS Sens. 2021;6:815–822. doi: 10.1021/acssensors.0c01843. [DOI] [PubMed] [Google Scholar]
- 260.Chang Y.-F., Chou Y.-T., Cheng C.-Y., Hsu J.-F., Su L.-C., Ho J.-a.A. Analytical Chemistry; 2021. Amplification-free Detection of Cytomegalovirus miRNA Using a Modification-free Surface Plasmon Resonance Biosensor. [DOI] [PubMed] [Google Scholar]
- 261.Scarano S., Spiriti M.M., Tigli G., Bogani P., Buiatti M., Mascini M., Minunni M. Affinity sensing for transgenes detection in antidoping control. Anal. Chem. 2009;81:9571–9577. doi: 10.1021/ac901445b. [DOI] [PubMed] [Google Scholar]
- 262.Chen Q., Bian Z., Chen M., Hua X., Yao C., Xia H., Kuang H., Zhang X., Huang J., Cai G. Real-time monitoring of the strand displacement amplification (SDA) of human cytomegalovirus by a new SDA-piezoelectric DNA sensor system. Biosens. Bioelectron. 2009;24:3412–3418. doi: 10.1016/j.bios.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 263.Balvedi R., Castro A.C., Madurro J.M., Brito-Madurro A.G. Detection of a specific biomarker for Epstein-Barr virus using a polymer-based genosensor. Int. J. Mol. Sci. 2014;15:9051–9066. doi: 10.3390/ijms15059051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yang B., Fang X., Kong J. Engineered microneedles for interstitial fluid cell-free DNA capture and sensing using iontophoretic dual-extraction wearable patch. Adv. Funct. Mater. 2020;30 [Google Scholar]
- 265.Que H., Zhang D., Guo B., Wang T., Wu H., Han D., Yan Y. Label-free and ultrasensitive electrochemical biosensor for the detection of EBV-related DNA based on AgDNCs@ DNA/AgNCs nanocomposites and lambda exonuclease-assisted target recycling. Biosens. Bioelectron. 2019;143 doi: 10.1016/j.bios.2019.111610. [DOI] [PubMed] [Google Scholar]
- 266.Yuan T., Mukama O., Li Z., Chen W., Zhang Y., de Dieu Habimana J., Zhang Y., Zeng R., Nie C., He Z. A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein–Barr virus. Analyst. 2020;145:6388–6394. doi: 10.1039/d0an00663g. [DOI] [PubMed] [Google Scholar]
- 267.Garai-Ibabe G., Grinyte R., Golub E.I., Canaan A., de la Chapelle M.L., Marks R.S., Pavlov V. Label free and amplified detection of cancer marker EBNA-1 by DNA probe based biosensors. Biosens. Bioelectron. 2011;30:272–275. doi: 10.1016/j.bios.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 268.Narang J., Singhal C., Mathur A., Sharma S., Singla V., Pundir C. Portable bioactive paper based genosensor incorporated with Zn-Ag nanoblooms for herpes detection at the point-of-care. Int. J. Biol. Macromol. 2018;107:2559–2565. doi: 10.1016/j.ijbiomac.2017.10.146. [DOI] [PubMed] [Google Scholar]
- 269.Wang X., Choi N., Cheng Z., Ko J., Chen L., Choo J. Simultaneous detection of dual nucleic acids using a SERS-based lateral flow assay biosensor. Anal. Chem. 2017;89:1163–1169. doi: 10.1021/acs.analchem.6b03536. [DOI] [PubMed] [Google Scholar]
- 270.Borkenhagen L.K., Fieldhouse J.K., Seto D., Gray G.C. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg. Microb. Infect. 2019;8:1679–1687. doi: 10.1080/22221751.2019.1690953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Pierce V.M., Hodinka R.L. Comparison of the GenMark Diagnostics eSensor respiratory viral panel to real-time PCR for detection of respiratory viruses in children. J. Clin. Microbiol. 2012;50:3458–3465. doi: 10.1128/JCM.01384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.LaGier M.J., Fell J.W., Goodwin K.D. Electrochemical detection of harmful algae and other microbial contaminants in coastal waters using hand-held biosensors. Mar. Pollut. Bull. 2007;54:757–770. doi: 10.1016/j.marpolbul.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Song M.-J., Jin J.-H. Label-free electrochemical detection of the human adenovirus 40/41 fiber gene. Anal. Sci. 2015;31:159–163. doi: 10.2116/analsci.31.159. [DOI] [PubMed] [Google Scholar]
- 274.Balsam J., Rasooly R., Bruck H.A., Rasooly A. Thousand-fold fluorescent signal amplification for mHealth diagnostics. Biosens. Bioelectron. 2014;51:1–7. doi: 10.1016/j.bios.2013.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 276.Ge L., Sun X., Hong Q., Li F. Ratiometric NanoCluster Beacon: a label-free and sensitive fluorescent DNA detection platform. ACS Appl. Mater. Interfaces. 2017;9:13102–13110. doi: 10.1021/acsami.7b03198. [DOI] [PubMed] [Google Scholar]
- 277.Xie J., Zhou P., Wen M., Bai J., Yang F., Duan C. A highly selective fluorescent probe for sensitive detection of HAV in water. Anal. Methods. 2019;11:3350–3357. [Google Scholar]
- 278.Ye K., Manzano M., Muzzi R., Gin K.Y.-H., Saeidi N., Goh S.G., Tok A.I.Y., Marks R.S. Development of a chemiluminescent DNA fibre optic genosensor to Hepatitis. A Virus (HAV), Talanta. 2017;174:401–408. doi: 10.1016/j.talanta.2017.06.036. [DOI] [PubMed] [Google Scholar]
- 279.Nagar B., Balsells M., De La Escosura-Muñiz A., Gomez-Romero P., Merkoçi A. Fully printed one-step biosensing device using graphene/AuNPs composite. Biosens. Bioelectron. 2019;129:238–244. doi: 10.1016/j.bios.2018.09.073. [DOI] [PubMed] [Google Scholar]
- 280.Chauhan V.M., Elsutohy M.M., McClure C.P., Irving W.L., Roddis N., Aylott J.W. Gold–oligonucleotide nanoconstructs engineered to detect conserved enteroviral nucleic acid sequences. Biosensors. 2021;11:238. doi: 10.3390/bios11070238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wu C., Zhou Y., Miao X., Ling L. A novel fluorescent biosensor for sequence-specific recognition of double-stranded DNA with the platform of graphene oxide. Analyst. 2011;136:2106–2110. doi: 10.1039/c1an15061h. [DOI] [PubMed] [Google Scholar]
- 282.Wang M., Zhao L., Wen Y., Wu Y., Li Y. Label-free fluorescent determination of simian virus 40 using triplex DNA and G-quadruplex/N-methyl mesoporphyrin IX. Anal. Lett. 2021:1–12. [Google Scholar]
- 283.Cai Z., Song Y., Wu Y., Zhu Z., Yang C.J., Chen X. An electrochemical sensor based on label-free functional allosteric molecular beacons for detection target DNA/miRNA. Biosens. Bioelectron. 2013;41:783–788. doi: 10.1016/j.bios.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 284.Lee W.E., Thompson H.G., Hall J.G., Bader D.E. Rapid detection and identification of biological and chemical agents by immunoassay, gene probe assay and enzyme inhibition using a silicon-based biosensor. Biosens. Bioelectron. 2000;14:795–804. doi: 10.1016/s0956-5663(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 285.Zhou J., Wang W., Li S., Nie A., Song Z., Foda M.F., Lu Z., Zhao L., Han H. Dual-mode amplified detection of rabies virus oligonucleotide via Y-shaped DNA assembly. Sensor. Actuator. B Chem. 2020;304 [Google Scholar]
- 286.Vijayakumar P., Macdonald J. A DNA logic gate automaton for detection of rabies and other lyssaviruses. ChemPhysChem. 2017;18:1735–1741. doi: 10.1002/cphc.201700072. [DOI] [PubMed] [Google Scholar]
- 287.Danielli A., Porat N., Arie A., Ehrlich M. Rapid homogenous detection of the Ibaraki virus NS3 cDNA at picomolar concentrations by magnetic modulation. Biosens. Bioelectron. 2009;25:858–863. doi: 10.1016/j.bios.2009.08.047. [DOI] [PubMed] [Google Scholar]
- 288.Civit L., Fragoso A., Hölters S., Dürst M., O'Sullivan C.K. Electrochemical genosensor array for the simultaneous detection of multiple high-risk human papillomavirus sequences in clinical samples. Anal. Chim. Acta. 2012;715:93–98. doi: 10.1016/j.aca.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 289.Luo L., Zhang F., Chen C., Cai C. Visual simultaneous detection of hepatitis A and B viruses based on a multifunctional molecularly imprinted fluorescence sensor. Anal. Chem. 2019;91:15748–15756. doi: 10.1021/acs.analchem.9b04001. [DOI] [PubMed] [Google Scholar]
- 290.Grabowska I., Malecka K., Stachyra A., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecka H., Radecki J. Single electrode genosensor for simultaneous determination of sequences encoding hemagglutinin and neuraminidase of avian influenza virus type H5N1. Anal. Chem. 2013;85:10167–10173. doi: 10.1021/ac401547h. [DOI] [PubMed] [Google Scholar]
- 291.Shi L., Sun Q., He J.a., Xu H., Liu C., Zhao C., Xu Y., Wu C., Xiang J., Gu D. Development of SPR biosensor for simultaneous detection of multiplex respiratory viruses. Bio Med. Mater. Eng. 2015;26:S2207–S2216. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- 292.Bahadır E.B., Sezgintürk M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015;478:107–120. doi: 10.1016/j.ab.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 293.Wang H., Deng J., Tang Y.-W. Profile of the Alere i Influenza A & B assay: a pioneering molecular point-of-care test. Expert Rev. Mol. Diagn. 2018;18:403–409. doi: 10.1080/14737159.2018.1466703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Keechilot C.S., Shenoy V., Kumar A., Biswas L., Vijayrajratnam S., Dinesh K., Nair P. Detection of occult hepatitis B and window period infection among blood donors by individual donation nucleic acid testing in a tertiary care center in South India. Pathog. Glob. Health. 2016;110:287–291. doi: 10.1080/20477724.2016.1248171. [DOI] [PMC free article] [PubMed] [Google Scholar]