Abstract
Chlamydia muridarum actively grows in murine mucosae and is a representative model of human chlamydial genital tract disease. In contrast, C. trachomatis infections in mice are limited and rarely cause disease. The factors that contribute to these differences in host adaptation and specificity remain elusive. Overall genomic similarity leads to challenges in the understanding of these significant differences in tropism. A region of major genetic divergence termed the plasticity zone (PZ) has been hypothesized to contribute to the host specificity. To evaluate this hypothesis, lateral gene transfer was used to generate multiple hetero-genomic strains that are predominately C. trachomatis but have replaced regions of the PZ with those from C. muridarum. In vitro analysis of these chimeras revealed C. trachomatis-like growth as well as poor mouse infection capabilities. Growth-independent cytotoxicity phenotypes have been ascribed to three large putative cytotoxins (LCT) encoded in the C. muridarum PZ. However, analysis of PZ chimeras supported that gene products other than the LCTs are responsible for cytopathic and cytotoxic phenotypes. Growth analysis of associated chimeras also led to the discovery of an inclusion protein, CTL0402 (CT147), and homolog TC0424, which was critical for the integrity of the inclusion and preventing apoptosis.
Keywords: chlamydia, cytopathic effect, plasticity zone
1 |. INTRODUCTION
Chlamydia is unique obligate intracellular pathogens responsible for diverse diseases in many animals including humans. Despite the wide range of hosts infected by Chlamydia, each species in the phylum shares several unique properties, including a biphasic developmental cycle (Abdelrahman & Belland, 2005). Throughout this developmental cycle, Chlamydia consistently interacts with the host cell to establish their intracellular niche, acquire nutrients, and subvert host responses. These diverse and complex molecular interactions function through chlamydial outer membrane proteins, inclusion membrane proteins, and secreted effectors (Abdelrahman & Belland, 2005; Nunes & Gomes, 2014). Because the chlamydial developmental cycle is intrinsically tied to the host, many of these chlamydial proteins play key roles in pathogenesis. Chlamydial lineages have uniquely adapted to specific hosts or tissues and the unique molecular and cellular properties therein (Nunes & Gomes, 2014). While some species-specific components and interactions have been described (Clifton et al., 2005; Damiani et al., 2014; Dehoux et al., 2011; Gauliard et al., 2015; Gomes et al., 2006), the majority of the chlamydial and cognate host factors contributing to host specificity remain elusive.
A well-established example of chlamydial host adaptation and specificity is provided by C. trachomatis and C. muridarum infections. In human genital tract infections, C. trachomatis colonizes the cervicovaginal epithelium of the lower genital tract and can ascend into the uterus and fallopian tubes of the upper genital tract (Belland et al., 2001). Once in the fallopian tubes, Chlamydia stimulates an immune response which leads to inflammation and eventual tissue damage. This can lead to a variety of symptomatic, chronic complications, including pelvic inflammatory disease, ectopic pregnancies, and infertility (Darville & Hiltke, 2010; Wiesenfeld, 2017). The mouse model of C. muridarum infection shares many key aspects of human C. trachomatis infection (Phillips et al., 2019). In female mice, C. muridarum infects and replicates at relatively high levels in the lower genital tract tissues before ascending into the uterine horns and ovarian ducts of mice, leading to hydrosalpinx and other pathologies (Campbell et al., 2014; Maza et al., 1994; Shah et al., 2005). In contrast, when immunocompetent mice are infected with C. trachomatis, replication in the vaginal vault is limited and infections are quickly cleared. Furthermore, ascension of C. trachomatis past the cervix to the upper genital tract is rarely observed in mice (De Clercq et al., 2013; Gondek et al., 2012; O’Meara et al., 2014). Similarly, there is no evidence of C. muridarum infecting humans, despite common usage in Chlamydia-research laboratories (Miller et al., 1987; Pike, 1976). These contrasting infection and pathologic capabilities of C. trachomatis and C. muridarum highlight the adaptation to their specific mammalian hosts.
Despite these differences in host-specific infectivity, C. trachomatis and C. muridarum share much of their gene content and their genomes are highly syntenous (Rajaram et al., 2015; Read et al., 2000). These Chlamydia species have condensed genomes, each comprised of an approximately 1 Mb chromosome and a 7.5 kb plasmid (Read et al., 2000; Stephens et al., 1998). C. trachomatis and C. muridarum have genome sequence identity of approximately 80% and the species share over 90% of encoded gene content (Dimond & Hefty, 2021; Read et al., 2000). Within these highly conserved genomes is a distinctive region of genetic diversity known as the plasticity zone (PZ) (Rajaram et al., 2015; Read et al., 2000). In C. trachomatis, the PZ region is ~20 kb and encodes 22 proteins including homologs to MAC/perforin (macP), multiple phospholipase D-like genes (PLD), and many proteins of unknown function (hypothetical proteins). Uniquely encoded within the PZ of C. trachomatis oculogenital pathobiovars is the tryptophan biosynthesis operon (trp operon). These gene products are a key example of species-specific adaptation as they are known to be critical for enabling C. trachomatis to survive the human interferon gamma (IFN-γ) cellular response and maintain genital tract infections (Caldwell et al., 2003; Fehlner-Gardiner et al., 2002; Rajaram et al., 2015; Roshick et al., 2006; Rottenberg et al., 2002; Xie et al., 2002). The C. muridarum PZ region is almost twice as large (~50 kb) and encodes a larger subset of PLD orthologs, three large cytotoxin homologs (LCTs), a gua operon for purine interconversion, and a different subset of species-specific hypothetical proteins (Rajaram et al., 2015; Read et al., 2000). The C. muridarum genome does not encode a tryptophan operon, reflecting the difference in mouse IFN-γ response (Dimond & Hefty, 2021; Rajaram et al., 2015; Read et al., 2000). Three LCTs are encoded by C. muridarum, whereas C. trachomatis either does not encode (e.g., LGV strains) or has a single highly truncated LCT (e.g., serovar D). As such, host-specific phenotypes have been attributed to these genes including growth-independent, immediate cytotoxicity as well as virulence in the mouse model (Belland et al., 2001; Morrison et al., 2018; Moulder et al., 1976; Nelson et al., 2007; Somboonna et al., 2011). Importantly, studies investigating the entirety of this region and multiple genes have been challenging.
As the PZ contains a substantially higher level of variability between otherwise conserved and syntenous genomes and is known to contain putative host-specific genes, this region was hypothesized to be important for host-specific pathogenesis in Chlamydia. To investigate this hypothesis, C. trachomatis and C. muridarum chimeric recombinants (Suchland et al., 2019) were generated which have the C. trachomatis PZ region replaced with full or partial regions of the C. muridarum PZ. These novel chimeras were used to evaluate relevant host-specific and gene-related phenotypes.
2 |. RESULTS
2.1 |. Generation of PZ recombinant chimera strains
A lateral gene transfer approach was used to evaluate the role of PZ genes in Chlamydia, using differently antibiotic resistant C. trachomatis and C. muridarum parental strains for generation of chimeras (Suchland et al., 2019). Briefly, dual-selection was used to cross a tetracycline-resistant C. trachomatis parent (Suchland et al., 2009) with a chloramphenicol-resistant C. muridarum transposon mutant (macP::Tn cat) (Wang et al., 2019) containing an insertion near the PZ. Recombinant progeny resistant to both selectable markers were isolated and whole-genome sequenced. From these efforts, 36 primary clones were generated with recombination within the PZ. Of these, three clones contained a majority C. trachomatis genome with recombination regions largely restricted to the PZ, replacing the C. trachomatis PZ with full or partial C. muridarum PZ (Figure 1 and Table 1). Chimera RC768 (RC–Recombinant Chlamydia) hereafter called the Left Partial PZ, contains C. muridarum genes tc0431 through tc0439, replacing the C. trachomatis MAC/perforin gene (ctl0408), the phospholipase D genes (ctl0409-411) and species-specific hypotheticals, with the C. muridarum MAC/perforin (tc0431), phospholipase D genes (tc0432-436) and the three LCTs (tc0437-439). The Left Partial PZ chimera retains the C. trachomatis trp operon (ctl0420-423), hypothetical proteins, and the oligopeptide permease 2 (oppA2). Chimera RC215, called the Full PZ, has the entire C. trachomatis PZ replaced by C. muridarum PZ. Chimera RC2043, referred to as the Right Partial PZ, contains C. muridarum genes tc0441 through tc0471 (Figure 1 and Table 1). For the Full PZ and Right Partial PZ chimeras, the recombination extends approximately 25 kb to the right of the PZ, recombining an additional 23 genes after the oppA2. Therefore, the Full PZ mutant contains the entirety of C. muridarum genes exchanged into the Left Partial and Right Partial PZ chimeras. Genome sequencing confirmed that no polymorphisms accumulated in either the C. trachomatis background or C. muridarum region of recombination in any chimera.
FIGURE 1.
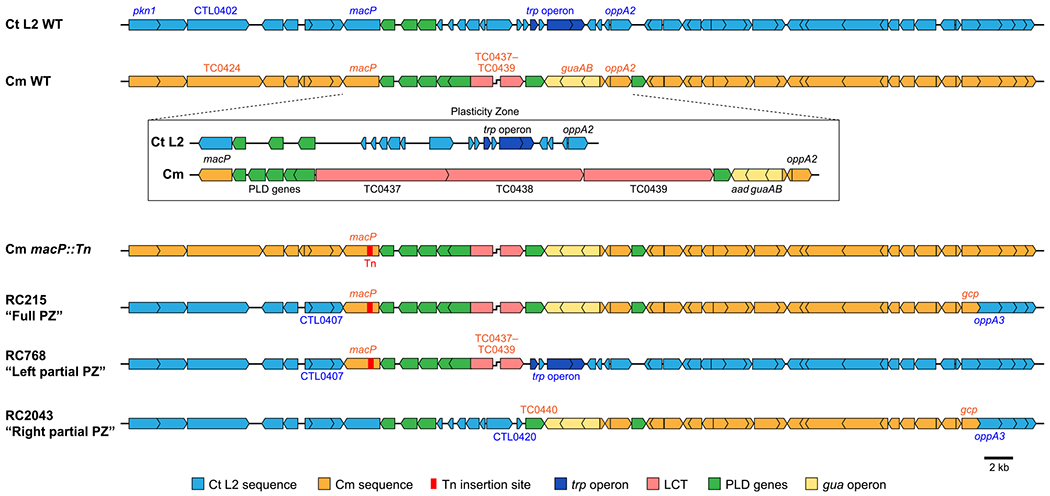
Schematic representations of Chlamydia and plasticity zome chimeras. Gene organization for the region surrounding the plasticity zone and detailed inset for the plasticity zone of the parental strains C. trachomatis (LGV/Bu/434) and C. muridarum (VR-123). Gene organization and location of the transposon insertion (red) for the C. muridarum mutant strain (Cm macP::Tn) used to generate C. trachomatis chimera RC215 (Full PZ), RC768 (Left Partial PZ), and RC2043 (Right Partial PZ). Genes in blue are C. trachomatis and orange genes represent those recombined from C. muridarum. Related plasticity zone genes share colors including: trp operon (dark blue), LCTs (pink), phospholipase D genes (green), and the gua operon (yellow)
TABLE 1.
C. muridarum genes in full plasticity zone chimera strain
| Partial chimera | Gene | Name | Function | ID/Sim %A | E(e-) |
|---|---|---|---|---|---|
| L | tc0431 | macP | MAC/perforin family protein | 71/82 | 0 |
|
|
|||||
| L | tc0432 | PLD | Phospholipase D family protein | 32/51B | 39 |
| L | tc0433 | PLD | Phospholipase D family protein | 33/50B | 37 |
| L | tc0434 | PLD | Phospholipase D family protein | 36/53B | 34 |
| L | tc0435 | PLD | Phospholipase D family protein | 29/52B | 24 |
| L | tc0436 | PLD | Phospholipase D family protein | 34/51B | 31 |
| L | tc0437 | Large cytotoxin (LCT) | Unique | ||
| L | tc0438 | Large cytotoxin (LCT) | Unique | ||
| L | tc0439 | Large cytotoxin (LCT) | Unique | ||
| R | tc0440 | PLD | Phospholipase D family protein | 36/54B | 41 |
| R | tc0441 | Hypothetical protein | Unique | ||
| R | tc0442 | guaA | GMP synthase | Unique | |
| R | tc0443 | guaB | Inosine-5′-monophosphate dehydrogenase, putative | Unique | |
| R | tc0444 | Hypothetical protein | Unique | ||
| R | tc0445 | Hypothetical protein | Unique | ||
|
|
|||||
| R | tc0446 | oppA2 | Peptide ABC transporter substrate-binding protein | 73/86 | 0 |
| R | tc0447 | PLD | Phospholipase D family protein | 36/52B | 29 |
| R | tc0448 | dsbB | Disulfide formation protein | 86/92 | 75 |
| R | tc0449 | dsbG | Putative disulfide bond chaperone | 89/94 | 163 |
| R | tc0450 | Conserved hypothetical protein | 82/91 | 0 | |
| R | tc0451 | Conserved hypothetical protein | 66/77 | 67 | |
| R | tc0452 | ABC transporter, ATP-binding protein | 77/89 | 134 | |
| R | tc0453 | Conserved hypothetical protein | 84/92 | 134 | |
| R | tc0454 | kdsB | 3-deoxy-manno-octulosonate cytidylyltransferase | 87/93 | 169 |
| R | tc0455 | pyrG | CTP synthase | 86/93 | 0 |
| R | tc0456 | ruvX | Holliday junction resolvase | 84/94 | 92 |
| R | tc0457 | zwf | Glucose-6-phosphate 1-dehydrogenase | 92/97 | 0 |
| R | tc0458 | devB | 6-phosphogluconolactonase | 88/92 | 172 |
| R | tc0459 | dnaX | DNA polymerase III, tau subunit | 82/92 | 0 |
| R | tc0460 | tmk | Thymidylate kinase | 81/92 | 0 |
| R | tc0461 | gyrA | DNA gyrase, subunit A | 96/97 | 0 |
| R | tc0462 | gyrB | DNA gyrase, subunit B | 96/97 | 0 |
| R | tc0463 | Conserved hypothetical protein | 80/98 | 78 | |
| R | tc0464 | inc | Inclusion membrane protein | 46/62 | 59 |
| R | tc0465 | tgt | Queuine tRNA-ribosyltransferase | 91/94 | 0 |
| R | tc0466 | mgtE | Magnesium transporter | 99/99 | 0 |
| R | tc0468 | Conserved hypothetical protein | 71/84 | 0 | |
| R | tc0469 | inc | Inclusion membrane protein | 57/73 | 32 |
| R | tc0470 | tsaD | tRNA (adenosine(37)-N6)-threonylcarbamoyltransferase | 89/94 | 0 |
Note:
Protein sequences compared to C. trachomatis LGV/434/Bu (GenBank AM884176.1).
Protein identity and similarity to the closest PLD homolog in C. trachomatis LGV/434/Bu (GenBank AM884176.1).
Gray indicates plasticity zone genes.
E is E-value calculated by NCBI Blast (Altschul et al., 2005).
2.2 |. Transcriptional analysis supports expression of C. muridarum PZ genes in a C. trachomatis background
To confirm expression of C. muridarum PZ genes in a predominantly C. trachomatis background, RNA sequencing was performed on parental and mutant-infected cells at 24 hr post-infection (hpi). Sequencing reads aligning to the C. muridarum PZ were present for all genes in the Full PZ mutant and at levels equal to those of C. muridarum, with the exception of tc0452. Quantitative PCR was performed on tc0452 and tc0451 revealing that these genes are transcribed at very low levels (data not shown), which could contribute to differences in the RNAseq. Using this method, the Full PZ transcript levels for tc0452 and tc0451 were not significantly different from the C. muridarum parental strain. These results support that transcription of C. muridarum PZ genes still occurred in C. trachomatis and at equal levels to parental C. muridarum. Analysis of transcript levels for the remainder of the genome revealed limited differential expression (Figure S1). Ten genes in the Full PZ clone exhibited significant differential expression compared to parental C. trachomatis and the levels were two-fold or less for all but one of these genes (Table S1). The affected genes are dispersed across the genome, with four being up-regulated in the Full PZ mutant and six being down-regulated.
2.3 |. C. muridarum PZ genes are partially involved in in vitro growth and developmental cycle regulation
Chlamydia undergoes a well-described developmental cycle (Abdelrahman & Belland, 2005; Gitsels et al., 2019), although the timing of key events varies by species. C. muridarum undergoes development within cells that is accelerated by about 6 hr relative to C. trachomatis (Messinger et al., 2015). To investigate whether PZ recombination affected the in vitro developmental cycle, temporal analysis of progeny production was evaluated. Consistent with prior observations, wildtype C. muridarum showed peak progeny production from 18–24 hr while C. trachomatis progeny peaked after 36 hpi (Figure 2a). The C. muridarum transposon parent (Cm macP::Tn) that was used to generate chimeras, exhibited growth features that matched wildtype C. muridarum. Each of the PZ chimeras displayed progeny production patterns that closely correlated to C. trachomatis. The Left Partial PZ chimeras closely mirrored wildtype C. trachomatis progeny production, whereas the Full PZ and Right Partial PZ chimeras matched the C. trachomatis parental production at 24 hpi but displayed a log lower progeny production at 36 and 42 hpi.
FIGURE 2.
Growth and morphology of Chlamydia PZ chimeras, (a) Progeny production was assessed from 0 to 42 hr post-infection. The blue dot on y-axis represents normalized initial infection concentration for all strains. Statistical analysis was performed by a two-tailed unpaired student’s t-test between samples at each time point. * indicates significant difference (P-value <0.05) for Full PZ and Right Partial PZ relative to C. trachomatis. (b) Inclusion sizes were measured at 20X magnification at 24 hr post-infection (n > 70) and two-tailed unpaired students t-tests performed to compare all data sets (*** indicates P-value <0.0001). (c) Immunofluorescent confocal microscopy of L929 mouse fibroblast cells infected with respective Chlamydia strains 24 hr post-infection. Green (Chlamydia MOMP), red (Evans Blue – host cytosol), blue (DAPI – DNA). Representative images of at least 5 fields of view. Scare bar = 5μm
As a complementary approach, the development and morphologies of primary inclusions for each chimera at 24 hpi was analyzed (Figure 2c). C. muridarum and C. muridarum macP::Tn inclusions were much larger and filled with EBs at a higher density than C. trachomatis at 24 hpi, consistent with growth rate differences. The inclusion morphology of the Left Partial PZ chimera was similar to C. trachomatis (Figure 2c), matching the levels and pattern of progeny production (Figure 2a). The Full PZ and Right Partial PZ mutants produced inclusions that were smaller and appeared to be less densely packed as the wild type C. trachomatis parent (Figure 2c). Inclusions were also imaged and mean area was quantified (Figure 2b). C. muridarum and C. muridarum macP::Tn had significantly larger inclusions than C. trachomatis, consistent with their more rapid developmental cycle. The mean size of the Left Partial PZ chimera was similar to C. trachomatis, also matching observations with progeny production (Figure 2a) and imaging (Figure 2c). In contrast, the Full PZ and the Right Partial PZ inclusions were significantly smaller compared to both C. trachomatis and C. muridarum. These observations support that PZ chimeras exhibited growth phenotypes more similar to C. trachomatis, with the Left Partial PZ chimera closely matching those of C. trachomatis. These data also support that the 34 kb region of C. muridarum which crossed into the Full PZ and Right Partial PZ may have removed C. trachomatis genes that contribute to optimal growth, or have added C. muridarum PZ gene products that disrupt optimal development.
2.4 |. PZ chimeras exhibit infection capabilities similar to C. trachomatis in the mouse model
Ascension from the vaginal vault to the upper genital tract is a critical pathogenic capability and distinguishing the difference between C. trachomatis and C. muridarum in mice. C. muridarum reliably ascends in mice whereas C. trachomatis rarely is observed in the upper genital tract following vaginal infection. To evaluate the hypothesis that genes encoded in the C. muridarum PZ contribute to this capability and reflect species-specific adaptations, PZ chimeras and parental strains were intravaginally infected into mice. After seven days post-infection, both vaginal vaults and uterine horns were harvested from the mice and bacterial burdens were assessed (Figure 3a). C. muridarum and C. muridarum macP::Tn infected the vaginal vault and the upper genital tract at similar mean bacterial burdens. In contrast, C. trachomatis infected the lower genital tract with over a two-log lower bacterial burden than wildtype C. muridarum (p <.05) and was not detected in the uterine horns. All PZ chimeras had reduced bacterial burdens in the lower genital tract when compared to C. trachomatis, although these differences were not statistically significant. Similar to C. trachomatis, PZ chimeras were not detected in the uterine horns following vaginal infections. Based on previous studies suggesting that the C57Bl/6 mouse model is more robust against chlamydial infection (Chen et al., 2014), the same experiment was conducted in the more susceptible C3He/J mouse model (Figure S2a) with observations similar to those with the C57Bl/6 model (Figure 3).
FIGURE 3.
Mouse infections with PZ chimeras. C57Bl/6 mice were infected with 105 IFU either (a) intravaginally or (b) transcervically. Lower genital tracts (vaginal vault) and upper genital tracts (uterine horns) were harvested seven days post-infection. Bacterial burdens were assessed using ddPCR to compare genome copies of Chlamydia to host in each tissue. ND indicates that organisms were not detectable when compared to mock infection threshold levels, (c) Bacterial shedding was measured by vaginal swab at 3, 7, 14, and 21 days post-intravaginal infection. Student’s t-tests were performed between C. trachomatis and PZ chimeras, C. muridarum and C. muridarum macP::Tn, and C. muridarum and C. trachomatis. For panels (a) and (b) * indicates p <.05 between C. trachomatis and C. muridarum. For panel (c) * indicates p <.05 between C. muridarum and C. trachomatis or C. muridarum macP::Tn. P values for all other comparisons were >0.05
The inability to ascend in the mouse female reproductive tract could indicate an inability to replicate or establish a niche within the endometrial tissues. Therefore, transcervical infection was conducted by infecting the bacteria directly into the uterine horns using an implantation device to bypass the cervix (Helble et al., 2019). Both vaginal vaults and uterine horns were harvested after seven days post-infection (Figure 3b). C. muridarum infection levels in the uterine horns were similar to those observed following vaginal infections (Figure 3a). C. trachomatis was able to colonize the uterine horns with approximately two-log fold lower bacterial burdens than C. muridarum (p <.05), similar to what is seen in the lower genital tract in an intravaginal infection (Figure 3a). Both the Left and Full PZ chimeras had bacterial burdens slightly higher than C. trachomatis in the uterine horns, though these differences were not statistically significant (p >.05). These data suggest that the cost associated with the recombined PZ may be specific to colonization of the lower genital tract. Along with the upper reproductive tract, bacterial burdens in the lower reproductive tract were analyzed after transcervical infections. Interestingly, only C. muridarum strains were able to disseminate downward and colonize the lower reproductive tract while C. trachomatis and the PZ chimeras were not detected in lower genital tracts, which indicates that the bacterial dissemination restriction may occur in both directions and that the cervix may serve as a key barrier.
The reduced infection levels may suggest that the absence or presence of PZ genes was a burden to the chimeric strains during murine infection. To further investigate this, vaginal shedding was examined (Figure 3c). C. muridarum shedding was detected until 21 days post-infection, with a peak at 7 days post-infection, while wildtype C. trachomatis was quickly cleared with organisms undetectable after 14 days post-infection. In parallel with the bacterial burdens observed in the vaginal vault (Figure 3a), all of the PZ chimeras were shed at levels lower than C. trachomatis. This was a general trend, though not statistically significant (p >.05). This analysis also revealed that the C. muridarum macP::Tn strain had significantly lower shedding levels compared to parental C. muridarum at day 7 (p <.05).
Recent research by Morrison et al. (2018) investigated the role of individual LCTs in chemical mutagenized clones containing multiple SNPs, including non-sense mutations in LCTs. As the Full and Left Partial PZ chimeras encoded all three LCTs, it provided an opportunity to investigate the role of these and other C. muridarum PZ genes in conferring a gastrointestinal advantage. Mice were infected intrarectally and swabs were collected at peak C. muridarum rectal shedding (Figure S2b). C. muridarum exhibited high bacterial burdens; however, the C. muridarum macP::Tn strain showed severe attenuation in the rectal model with approximately three-log lower detectable IFUs (p <.005). This attenuation appears to be tissue-specific, as this mutant shared similar infection levels as parental C. muridarum in the genital tract model (Figure 3a,b), suggesting a possible role for macP in gastrointestinal infections. Importantly, C. trachomatis parental strain and the Full or Left PZ chimeras were not detectable in the rectal swabs or tissue at either day 3 or day 7 post-infection (Figure S2b and data not shown). As the Full and Left Partial PZ encode all three LCT, this supports that the LCTs and associated PZ genes are not independently sufficient to confer the ability for C. trachomatis to infect rectal tissues, even as the lower infection levels observed with C. muridarum macP::Tn are considered.
2.4.1 |. Cytotoxin genes (LCT) from C. muridarum are insufficient for immediate host-cell toxicity
Another phenotype that is distinct between C. trachomatis L2 and C. muridarum is the growth-independent and immediate cellular toxicity observed following exposure to high multiplicity of infection (MOI) with C. muridarum (Belland et al., 2001). This immediate cytotoxicity phenotype has been associated with the 3 LCTs encoded within the PZ and has been characterized by an early, approximately 4 hpi, breakdown of the host actin cytoskeleton and eventual host-cell death (Belland et al., 2001; Bothe et al., 2015). Belland et al investigated this phenotype both in C. muridarum and in C. trachomatis serovar L2, which did not have the cytotoxic phenotype. It was posited that the LCT genes, present in C. muridarum but absent or severely truncated in C. trachomatis, were responsible for producing this phenotype. The chimeras containing the three LCTs (Full and Left Partial) compared to the chimera without LCTs (Right Partial), along with parental C. trachomatis and C. muridarum, provided an opportunity to further investigate this phenotype and the contribution of the LCTs.
Each PZ chimeric strain, along with the wildtype parent strains, was infected onto a cellular monolayer at a high MOI (~100), as was performed by Belland et al (Belland et al., 2001). After 4 hr of incubation, actin arrangement was analyzed by immunofluorescence microscopy (Figure 4a). Cells infected with C. trachomatis L2 had an elongated fibroblast shape with actin filaments visible within the cytosol and concentrated around the membrane, which were structurally not discernible from uninfected cells. Moreover, the confluency and attachment of the monolayer was maintained. In contrast, high MOI C. muridarum-infected cells lost fibroblastic shape and instead had a distinct cell rounding with puncta of concentrated actin in the center of the cell, indicating a breakdown of the actin filaments. Additionally, cellular adherence appeared to be severely disrupted as limited cells were retained in the samples. When the PZ chimeras were analyzed for cellular effects, the Full PZ chimera exhibited cell rounding and loss of attachment, similar to C. muridarum. Unexpectedly, the Left Partial PZ chimera, which also encoded all three LCTs, did not cause cell rounding nor disruption of cellular monolayer and adherence. In contrast, the Right Partial PZ chimera caused cell rounding and loss of adherence, despite not encoding any of the LCTs.
FIGURE 4.
Immediate cytopathic and cytotoxicity effects of PZ Chimeras. Cellular imaging and LDH release experiments were performed on the same samples infected for 4 hr with 100 MOI of parental or PZ chimera Chlamydia strains, (a) Representative images of L929 cells infected with indicated Chlamydia strains with host nuclei stained with DAPI (blue) and host actin is stained with FITC labeled phalloidin (green), (b) Average levels of LDH released from cells and compared to levels released in a positive control sample (% cytotoxicity). Values are from triplicate biologic experiments. *P-value <0.005
To further evaluate cellular effects associated with the PZ genes, cell membrane integrity and LDH release were evaluated (Figure 4b) using supernatant from the same samples used for imaging (Figure 4a). C. muridarum elicited relatively high levels of LDH release, whereas C. trachomatis and the Left PZ chimera caused the limited release. Interestingly, while both the Full and Right PZ chimeras caused substantial cell rounding and loss of adherence, LDH release levels were very low and similar to those observed with C. trachomatis. To evaluate if these observations were shared between other cell types, Hela cells were also tested for cytopathic and cytotoxic effects revealing very similar results (data not shown). Expression of the PZ genes was previously evaluated in the Full PZ chimera by RNAseq showing gene expression levels equal to parental C. muridarum (Figure S1); however, the absence of cytopathic and cytotoxic effects with the Left PZ chimera could be due to a loss of LCT expression. To validate that the LCTs were expressed similarly in the Left Partial PZ strain, ddPCR was used to measure transcript levels relative to parental C. muridarum. At 24 hpi, transcripts were detected for all three LCTs with levels similar to wildtype C. muridarum and the Full PZ chimera (data not shown).
Overall, these results suggest that there is a genetic disconnect between the observed cytopathic (cell rounding and adherence loss) and cytotoxic (LDH release) effects. These data support that one or more of the 30 genes encoded to the right of the LCTs (Table 1 and Figure 1) may play an important role in this cytopathic effect. Based upon gene products that are more unique to C. muridarum, candidate genes within this region include two phospholipases (tc0440 and tc0447), two inclusion membrane proteins (tc0464 and tc0469), and three hypothetical proteins specific to C. muridarum (tc0444, tc0445 and tc0467). While it is less likely that highly conserved bacterial proteins, particularly those involved in known pathways like RuvX (tc0456, Table 1), are responsible for this phenotype, this possibility cannot be excluded. The absence of LDH release with Right or Full PZ chimeras (Figure 4b) also supports that LCTs were not solely responsible for this C. muridarum cytotoxic phenotype, suggesting that non-PZ genes likely contribute to this effect.
2.4.2 |. CT147 is an inclusion membrane protein that plays a role in preventing host-cell death
During the generation of PZ chimeras, a strain termed RC826 was difficult to propagate. Fluorescent imaging of this strain at 26 hpi revealed that the integrity of the inclusion appeared to be compromised early in the development cycle and dispersal of Chlamydia within the host cell was noticeable due to inclusion lysis (Figure 5b). In order to determine which gene(s) may be responsible for this phenotype, additional recombination was performed and certain sequenced chimeras were selected for investigation of this phenotype. Most importantly, RC1323 was generated, which has an identical genotype to RC826 except for a single gene difference: RC1323 retains the C. trachomatis gene ctl0402 and RC826 has incorporated the C. muridarum ortholog tc0424 (Figure 5a). RC1323 displayed inclusion morphology matching that of parental C. trachomatis (Figure 5b). Additional chimeras (Figure S3) were analyzed and recombinant strains that contained the native ctl0402 appeared to have wildtype growth, while those with a recombined tc0424, exhibited a disrupted developmental cycle with early inclusion lysis (data not shown). To confirm that the replacement of ctl0402 with tc0424 was responsible for the abnormal growth, a backcross was performed to replace tc0424 in RC826 with the native ctl0402. The resulting chimera, RC3254, was able to complete the developmental cycle and did not cause early inclusion lysis and dispersal of EBs (Figure 5b).
FIGURE 5.
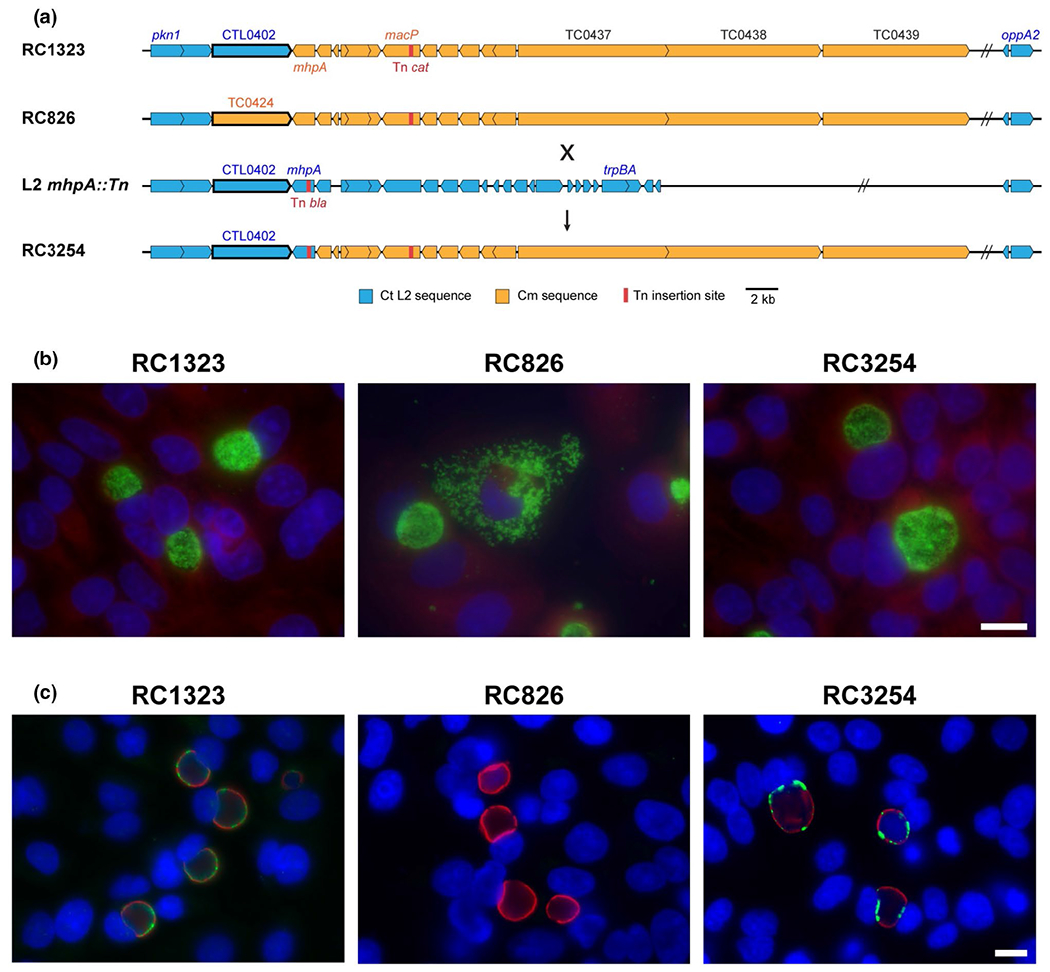
Genetic diagram of CTL0402 (CT147) associated chimeras and immunofluorescence microscopy of early inclusion lysis phenotype. (a) Open reading frame maps of chimeric strains carrying the CTL0402 sequence (RC1323), the C. muridarum homolog TCO424 (RC826) and a backcrossed recombinant that re-introduced the C. trachomatis homolog (RC3254). The transposon mutant L2 mhpA:Tn was used to generate the backcross. (b) Immunofluorescence microscopy at 26 hpi showing wildtype inclusion development in RC1323 but an early inclusion lysis phenotype in RC826, followed by chlamydial dispersal. Chlamydial developmental forms are labeled green with monoclonal antibody L2-I45, host nuclei are stained with DAPI (blue) and host cytosol is stained with Evan’s Blue (red). (c) Infected samples fixed and stained at 18 hpi with showing that CTL0402 protein (green) is found in the inclusion membrane in RC1323 and the backcrossed strain (RC3254), while it is not detected in RC826. The inclusion membrane protein IncG (red) is present in each strain
Both CTL0402 and TC0424 are putative inclusion membrane proteins, as they contain the characteristic bilobed hydrophobic domains (Bannantine et al., 2000). The presence of CTL0402 on the inclusion membrane in the chimeras was verified with fluorescent labeling (Figure 5c). CTL0402 was detected at the inclusion membrane for both RC1323 and RC3254. In contrast, antibodies to CTL0402 did not label the inclusion of RC826, which encodes for the C. muridarum homolog TC0424. The early inclusion lysis in strains with TC0424, rather than CTL0402, could be consistent with improper inclusion maintenance, a possible function of these inclusion membrane proteins. To identify what was occurring mechanistically and temporally to cause the early lysis of the inclusions, live time-lapse microscopy was performed on host cells infected with recombinant strains as well as C. muridarum and C. trachomatis L2 controls. Inclusions appear to develop normally in C. trachomatis, C. muridarum, and RC1323-infected cells. However, RC826-infected cells displayed a membrane-blebbing phenotype characteristic of apoptosis occurring as early as 16 hr post-infection, and the majority of infected cells exhibited this phenotype by 22 hr post-infection (Figure 6a and Figure S4). In wildtype C. muridarum-infected cells, occasional apoptosis is known, though this typically does not occur until around 20–22 hr.
FIGURE 6.
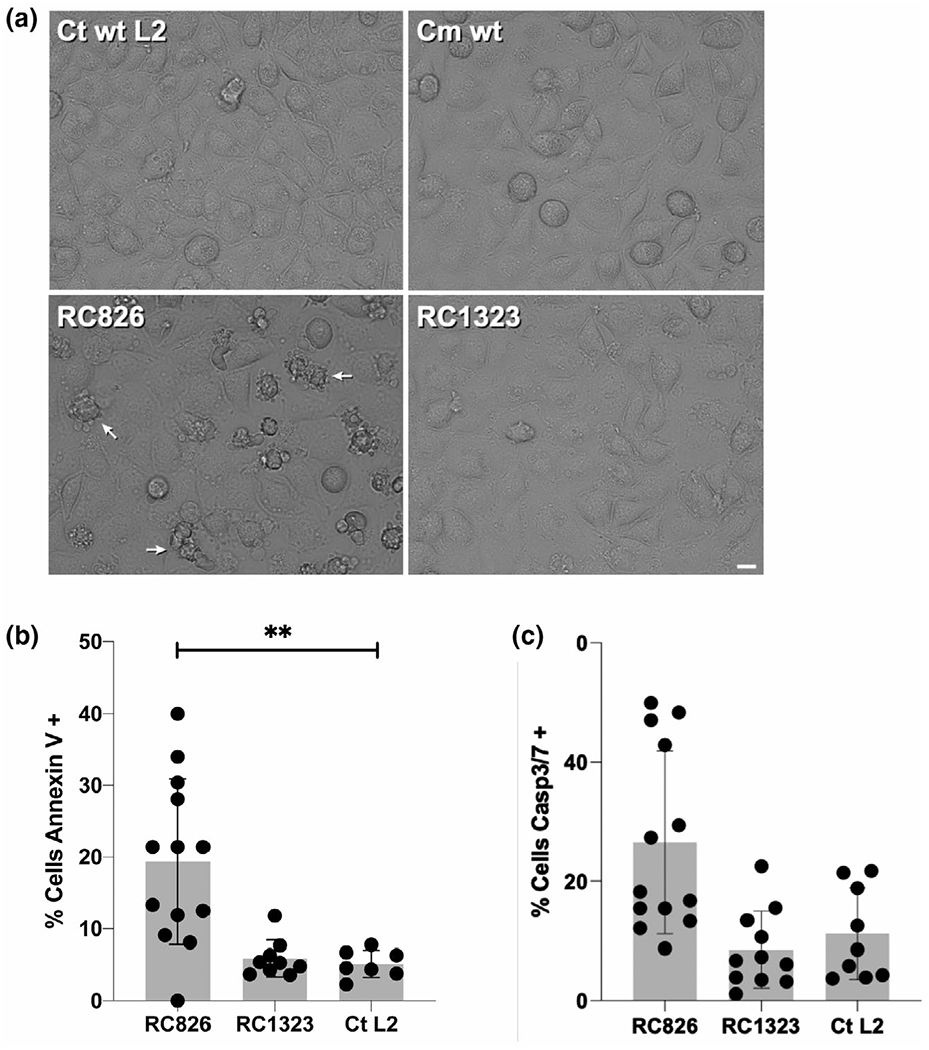
Qualitative and quantitative assays demonstrating host-cell apoptosis of cells infected with RC826. (a) Images taken from live infection videos (Figure S4) at 16 hr post-infection. Arrows point to cells undergoing apoptosis. (b) Host cells infected with wildtype C. trachomatis or chimeric strains were stained with Annexin V AlexaFluor 594. Percent cells positive for external plasma membrane annexin V staining in each sample compared with an uninfected control were plotted. Significance was calculated by one-way ANOVA (adjusted P-value = 0.0006). (c) Host cells infected as in (b) and stained at 28 hr post-infection with Image-iT live caspase 3/7 kit and plotted as percent stained compared with uninfected controls
Apoptosis can be assessed by examination of a variety of cellular markers, including the presence of phosphatidylserine in the outer leaflet of the plasma membrane and the detection of activated effector caspases-3/7 (Segawa & Nagata, 2015). To determine if the membrane-blebbing phenotype observed in the time-lapse microscopy was apoptosis, infected host cells were labeled with Annexin V, to detect phosphatidylserine in the outer membrane and FLICA labeling of the activated caspases 3/7. In both experiments, apoptosis markers were detected in a significantly higher percentage of cells when infected with RC826 over RC1323 or wildtype C. trachomatis (Figure 6b,c). Together, these data support that C. muridarum TC0424 expression in C. trachomatis as a replacement for CTL0402 is responsible for the early lysis phenotype which is also associated with the induction of cellular apoptosis.
3 |. DISCUSSION
In this study, inter-species recombination was used to evaluate the role of the PZ in C. trachomatis and C. muridarum. Key recombinants around the plasticity zone were generated which resulted in the first C. trachomatis strain that contains the entire PZ from C. muridarum. Chimeras were also generated that contained Left and Right partial PZ regions. These chimeras allowed for the collective assessment of this multi-gene locus and contributions to various phenotypes, including those associated with mammalian infection. Among the primary observations was the limited ability of the PZ locus from C. muridarum to solely confer enhanced infection capabilities for C. trachomatis in mice. The absence of detectable enhancement in infection capabilities is not overly surprising as these processes are complex and, very likely, incorporate a suite of diverse molecular interactions. However, it would have been a powerful observation relative to the importance of PZ contents had enhanced infection phenotypes been observed. Another major observation was the contrasting cytotoxic and cytopathic phenotypes by the PZ chimeras that support that gene products, excluding the LCTs, are likely responsible for these phenotypes. Additionally, an inclusion membrane protein (ctl0402 - tc0424) that displayed a species-specific incompatibility led to a loss of inclusion membrane integrity and cellular apoptosis was discovered. Overall, this inter-species LGT approach focused on a single locus has enabled multiple discoveries that are expected to serve as an example for future efforts as well as studies on specific gene contributions and mechanisms associated with these key phenotypes.
A central hypothesis for this study was that the C. muridarum plasticity zone contains gene products that contribute to host specificity which could provide enhanced infection capabilities to C. trachomatis in lower and upper reproductive tissues of mice. Despite the presence of C. muridarum PZ genes, these chimeras displayed very poor replication in the vaginal vault and no ascension into the upper genital tract similar to C. muridarum in this mouse model (Figure 3a). These observations should not be interpreted as PZ genes are not important for host specificity or infection processes, merely that this locus alone was unable to sufficiently complement C. trachomatis and enable a detectable ‘gain-of-infection function’ phenotype in murine vaginal infection. Moreover, PZ expression analysis supported transcription within chimeras but it is possible that post-transcriptional processes are not similar between species and thus affecting interpretations. Nevertheless, the inability of the PZ to enable gain-of-infection phenotype may indicate that PZ gene products require contributions from specific C. muridarum gene products encoded elsewhere in the genome for vaginal infection in mice. Interestingly, the Full PZ chimera strain did trend toward higher bacterial burdens relative to C. trachomatis when upper genital tracts were infected directly (Figure 3b) suggesting that contents of the PZ may play a more critical role in uterine horn tissues or perhaps that the adverse effects of containing these C. muridarum genes are lessened in the upper reproductive tract. Generally, the lower reproductive tract is lined with stratified, squamous epithelial cells which form a more protective barrier than the columnar epithelium of the upper reproductive tissues (Lee et al., 2015). Additionally, there are differences in the immune cells of the upper and lower tracts; particularly, the vagina-facing ectocervix contains dendritic cells, which are not found in the upper reproductive tract. The endocervix, which is part of the upper reproductive tract, contains mucus-producing glands to prevent microbial pathogen access to the uterus (Trifonova et al., 2014).
Along these lines, prior studies have evaluated C. muridarum mutant strains that contain gene disruptions in certain PZ genes (Morrison et al., 2018; Rajaram et al., 2015), revealing no effect on vaginal infections and providing support for a less critical role for these gene products in the lower genital tract environment. Interestingly, disruptions to individual LCT genes have rendered strains defective in gastrointestinal infections, although these were associated with other mutations (e.g., tc0600) providing support that certain PZ genes may be critical in specific tissue environments (Morrison et al., 2018). As multiple PZ gene products may contribute collectively to successful lower and upper genital tract infections, possibly in combination with other C. muridarum gene products, it will be useful to evaluate C. muridarum strains with a fully disrupted PZ locus. Based upon the advances described herein, the application of LGT with selected C. trachomatis transposon clones (LaBrie et al., 2019; Wang et al., 2019) are expected to enable the generation of reciprocal PZ chimeras and assessment of these gene products in the context of C. muridarum.
One of the more unexpected observations was the apparent genetic discordance between cytotoxic and cytopathic phenotypes. As highlighted, immediate cytotoxicity has been established with C. muridarum, although some strains of C. trachomatis (excluding LGV) have displayed this activity to a substantially reduced degree (Belland et al., 2001). This immediate cytotoxicity has been hypothesized to be correlated with three large LCT gene products with domains similar to Clostridium difficile A/B toxin. The combinatorial analysis of shared and unique loci revealed that the Left Partial PZ chimera, which encodes three LCTs, did not exhibit cytotoxicity or cytopathic phenotypes (Figure 4a,b). The absence of these phenotypes in the Left Partial PZ chimera is unlikely to be due to expression issues as equal LCT transcription was confirmed by the Left Partial and parental C. muridarum. Moreover, the Right Partial PZ chimera that does not encode the LCTs did induce cytopathic effects similar to that of parental C. muridarum and the Full PZ chimera. These observations call into question the genes that contribute to immediate cytotoxicity. There have been few direct analyses on LCTs and cytotoxicity to support causation, largely due to difficulty in cloning these genes and proteins. The most compelling observation in support of LCT and cytotoxicity was from a recent study on C. muridarum strains that contain non-sense mutations in each of the LCTs (Rajaram et al., 2015). This study reported no loss of cell rounding for any of the LCT mutant strains; however, two mutant strains did exhibit a decrease in LDH release. As these mutant strains had other mutations, contributions by other mutations to this loss of LDH release must be considered. Additional important studies on cytotoxicity include those by Thalmann et al. and Bothe et al. which focus on C. trachomatis serovar D protein ct166, which contains similarity to the C. muridarum cytotoxins (Lee et al., 2015; Segawa & Nagata, 2015). C. trachomatis D is known to exhibit a version of cytotoxicity and these studies demonstrated that the cytotoxicity was attributed to actin rearrangement and was dependent on glucosylation of Rac1 (Thalmann et al., 2010). One critical aspect of this study was the use of CT166-expressing HeLa cells which demonstrated that CT166 was sufficient to cause reduced cell proliferation. This study appears to present some contradicting findings, in that the LCTs do not appear to be necessary for cytotoxicity (LDH release) or cytopathic effect (cell rounding and loss of adherence) in the context of chimeras. One potential explanation for this difference is that C. muridarum cytotoxicity is mechanistically distinct from the cytotoxicity induced by endogenously expressed CT166. Additional studies will be required to compare the mechanism of actin reorganization, determine the role of Rho GTPases in C. muridarum cytotoxicity, and define the genetic components involved in this process.
There are 30 genes encoded in the Right Partial PZ region that could contribute to the observed immediate cytotoxicity (Table 1). The vast majority of these genes are highly conserved with C. trachomatis and are less likely to contribute to this phenotype; however, there are some unique and conserved genes to consider. There are three hypothetical proteins that are unique to C. muridarum (tc0441, tc0444, and tc0445) and two phospholipase D proteins (tc0440 and tc0447) that share close to 50% similarity with their C. trachomatis orthologs. Phospholipase D proteins typically hydrolyze phosphatidylcholine and phosphatidylethanolamine to generate phosphatidic acid, which can serve as a signaling molecule (Selvy et al., 2011). They can also serve as secreted effector proteins, cause cytotoxicity, and are documented virulence factors in other bacteria (Flores-Diaz et al., 2016). There are two inclusion membrane proteins also encoded in this region (tc0464 and tc0469) which are also expected to be secreted effector proteins. Future studies focused on disruptions to these gene candidates, as well as obtaining and investigating Right Partial PZ chimera sub-clones that contain fewer genes, will be instrumental in determining more directly the genetic contributions to immediate cytotoxicity.
A powerful demonstration of the utility of chlamydial LGT studies, as well as significant biologic discovery, were the observations related to a single gene (ctl0402/tc0424) and early lysing of Chlamydia inclusions (Figure 5). The initial observation was of a chimera, RC826, that was difficult to propagate and genomic comparison to similar chimeras that did not exhibit this growth defect (Figure S3). The comparison to RC1323 enabled a single gene to be identified in association with the early lysis phenotype and genetic complementation was achieved using a Tn mutant strain (L2 mhpA::Tn) and backcrossing with RC826. This gene product is an inclusion membrane protein and these observations support that TC0424 in a C. trachomatis background leads to the integrity of the inclusion being compromised and lysing relatively early in the developmental cycle. This uncontrolled spilling of the inclusion contents is expected to induce apoptosis that was observed by time-lapse microscopy (Figure S4) and membrane markers (Figure 6). These Inc orthologs share 59% identity and 76% similarity with almost complete sequence coverage and limited gaps (Table 1). As such, it is expected that the differences that lead to the incompatibility are subtle molecular aspects that likely affect stable protein-protein interactions. These interactions could be with other inclusion membrane proteins likely in a complex, as is seen with many inclusion membrane proteins (Rockey et al., 2002). This inability to form a stable heterocomplex could disrupt host-cell interactions that lead to loss of inclusion integrity. Additionally, interacting partners could be host proteins, including those involved in apoptotic signaling pathways, as is the case for Chlamydia pneumoniae inclusion membrane protein Cpn1027 (Flores & Zhong, 2015). This inclusion membrane protein (CT147) has been investigated in C. trachomatis serovar D and was annotated as early endosomal antigen-1 (Belland et al., 2003). It was transcribed very early (1 hpi) with protein detected by 8 hpi and eventually localized to the inclusion membrane. Protein partners have not been identified and future studies investigating this hypothesis will require these efforts. Additionally, it may be expected that investigations of other chimeras with greater genomic representation could reveal strains that phenocopy RC826 and lead to protein partner candidates. This direction seems promising given the success of these efforts to identify a single gene associated with inclusion maintenance process.
There have been other reported cases of inclusion membrane protein disruption leading to apoptosis of the host cell (Giebel et al., 2019; Weber et al., 2017). In these studies, random chemical (C. muridarum) or TargeTron directed (C. trachomatis) mutations in inclusion membrane proteins resulted in inclusion lysis and apoptosis. A study performed in C. muridarum showed inclusion lysis of a TC0574 mutant was mediated by interferon gamma (Giebel et al., 2019). In contrast, the observations in this study were performed in cells untreated with interferon, suggesting that the mechanism of action for the TC0424/CTL0402 inclusion lysis is distinct from those observed in TC0574. In C. trachomatis, TargeTron mutations in ct229, incC, and ct383 each resulted in inclusion membrane lysis, similar to what was observed in this work (Weber et al., 2017). In those mutants, it was demonstrated that premature inclusion lysis led to host-cell apoptosis. Similar correlation is expected here, as well although additional studies can be performed to determine the timing of these events, along with the mechanism for premature lysis. From the Weber et al. study, it appears that premature inclusion lysis was not caused by the same mechanism in all three mutants investigated (Weber et al., 2017), and it will become important to define these mechanisms in the TC0424/CTL0402 recombinant in subsequent studies. While it is expected that the early lysis phenotype is resulting in host-cell apoptosis, as was seen with previous inclusion membrane protein mutants (Weber et al., 2017), the possibility that necroptosis is instead occurring should be considered. It is possible that necroptosis is resulting from the inclusion disruption, but in either case, it is clear that the inclusion instability results in host-cell death.
Overall, this study demonstrates the utility of combining LGT and selected transposon insertion clones to investigate a multi-gene locus with capabilities to identify a single gene associated with important phenotypes. As this study focused on the PZ locus, the transposon libraries in C. muridarum (Wang et al., 2019) and C. trachomatis (LaBrie et al., 2019) with complementary selection markers provide for extensive possibilities to expand similar studies to other candidate loci. Similarly, these will enable larger scale screens to discover regions of interest associated with critical cellular processes and virulence-related phenotypes through mouse models. These should also enable the reciprocal exchange of the C. trachomatis PZ into a C. muridarum background and evaluation of these genetic modifications on vaginal and gastrointestinal infections in mice. Together, this and similar recent studies (Jeffrey et al., 2013; Suchland et al., 2009, 2019) provide a growing foundation for new genetic tools and approaches within Chlamydia to better decipher the basic biology and pathogenesis for this important intracellular pathogen.
4 |. MATERIALS AND METHODS
4.1 |. Chlamydial strains
C. trachomatis serovar L2/tet (Suchland et al., 2009), C. muridarum (ATCC VR-123), C. muridarum macP::Tn (UWCM007) (Wang et al., 2019), and chimeras were propagated in L929 mouse fibroblast cells (ATCC CCL-1) using RPMI 1640 medium (Invitrogen, Grand Island, NY) supplemented with 5% heat-inactivated fetal bovine serum (FBS) plus 10 μg/ml gentamycin (Fisher Scientific, Pittsburgh, PA) at 37°C and 5% CO2. All EBs were stored in sucrose-phosphate-glutamic acid (SPG) media at −80°C. Chimeras used in this study (GeneBank accession numbers): RC768 (Left Partial PZ; CP042748), RC215 (Full PZ; CP042772), RC2043 (Right Partial PZ; CP042717), RC826 (CP042747), RC1323 (CP042791) and RC3254.
4.2 |. RNAseq transcriptional analysis
PZ chimeric strains and their parents were grown in L929 cells at a MOI of 5. At 20 hpi, the infections were harvested for RNA using TRIzol (Invitrogen). RNA was purified by phenol/chloroform extraction followed by DNase treatment with TURBO DNase (Invitrogen). Residual DNA contamination was assessed by converting a portion of the RNA into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher), with control reactions containing no reverse transcriptase, and performing PCR on cDNA reactions. Ribosomal RNA was then depleted using NEBNext rRNA Depletion (Human/Mouse/Rat) (New England Biosciences) and RiboMinus Transcriptome Isolation bacterial (Invitrogen) kits. A final purification step was performed using the RNeasy Mini kit (Qiagen). rRNA depletion was assessed by running samples on a 2% agarose gel. RNA quality was assessed using Qubit quantification and TapeStation gel analysis. Library prep for RNAseq was performed using NEBNext Standard mRNA library kit without the polyA selection step. Sequencing was performed on the NextSeq550 with single read 50bp read length. Geneious Prime (Version 2019.1.1) was used for the data analysis. Reads were aligned to Chlamydia muridarum (NC002620) and/or Chlamydia trachomatis L2/tet9 (CP035484.1). An average of approximately 75 million reads were sequenced for each sample with approximately 3.3 million reads mapping to each Chlamydia genome. Transcript levels were normalized to reads per million. RNAseq analysis was done in duplicate with RNA harvested from two independent infections. Significant differential expression ratios were determined using two standard deviations from the average expression ratio for all RNAseq runs (Figure S1). To verify RNAseq, cDNA was generated and used in a PCR to confirm presence or absence of genes. Using the above RNA extraction protocol, RNA was extracted from the PZ chimeric strains and wildtype parents at 20 hpi. cDNA was prepared using the Applied Biosystems High-Capacity cDNA reverse Transcription Kit using the manufacturer’s instructions. PCR was performed to detect transcripts. To quantify transcript levels, ddPCR was performed with total cDNA using EvaGreen Supermix (Bio-rad) protocols (Hindson et al., 2011). Positive droplets were normalized to secY levels using dual C. trachomatis and C. muridarum secY primers (Table S2).
4.3 |. Progeny production assay and inclusion size measurement
Mouse fibroblast L929 cells were infected (550xg; 30min) with C. trachomatis L2 (LGV/434/Bu), C. muridarum (ATCC VR-123), the transposon mutant C. muridarum (macP::Tn cat) or one of the three PZ recombinants (MOI = 0.3–0.5). At 0, 12, 18, 24, 36 or 42-hr post-infection, cells were washed with 1X Hanks’ balanced salt solution (HBSS) chlamydial EBs were released through water lysis (Weber et al., 2017). In order to determine titers at each time point, samples were infected with serial dilution into 96-well plates and allowed to grow for 24 hr before methanol fixation and subsequent staining. Chlamydial inclusions were stained using the PathoDX Chlamydia Culture Confirmation Kit (ThermoFisher Scientific). In brief, the Chlamydia infected cellular monolayer was washed twice with 1X phenol-free HBSS and fixed in 100% methanol for 10 min before adding a 1:40 dilution of stain. Total DNA was stained using DAPI. Inclusions were visualized under 10X magnification using the EVOS FL Auto 2 microscope (Thermo Scientific, Waltham, MA) and were enumerated manually and/or on MIPAR software using similar parameters as mentioned below. Final progeny production was normalized to C. trachomatis wildtype MOI at 0 hr post-infection and normalized values were plotted as mean and standard deviation followed by two-tailed unpaired t-test at 36 hr to see if the chimeras were significantly different with a p-value <0.05. Inclusion sizes were measured at 24 hpi by capturing images from the center of the field under 20X magnification using the EVOS FL Auto 2 microscope (Thermo Scientific, Waltham, MA). Quantification of inclusion area was done using MIPAR software (Sosa et al., 2014) with a contrast range between 90 and 255 units, expected inclusion area of 30 μm2 −1600 μm2 and a roundness threshold of ≤0.7. Two-tailed unpaired t-test was run between the chlamydial parents and chimeras to see if there was any significant difference in the inclusion sizes with a p-value <0.0001.
4.4 |. Cytotoxicity imaging and LDH assay
Methods used by Belland et al. were followed with minor alterations (Belland et al., 2001). 96-well plates were seeded to 80% confluency with L929 mouse fibroblast and HeLa cells. Each plate was mock infected with SPG or infected with wild type C. trachomatis L2, C. muridarum, or indicated chimera mutant strain at an MOI of 100. Infected cells were centrifuged at 550 x g for 30 min. Inoculum was then aspirated and RPMI growth media added to wells. At 4 hpi, 50 μl of supernatant from 96-well plates infected at an MOI of 100 was transferred from infected and mock-infected wells to a new 96-well plate and assayed for LDH release using Invitrogen CyQUANT LDH Cytotoxicity Assay (Life Technologies Corporation, Eugene, OR). The remaining supernatant within the 96-well plates was aspirated and cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Wells were aspirated and washed with HBSS. Wells were then stained with a 1:250 dilution of FITC conjugated phalloidin stain for 2 hr at room temperature. Wells were aspirated, and overlayed with 90% glycerol tris solution, and imaged at 40X magnification on an EVOS FL Auto 2. To directly quantify MOI being utilized for these experiments, 96-well plates with confluent layers of L929 mouse fibroblast were infected with serial dilutions of each inoculum. At 24 hpi, wells from the serial dilution plate were fixed in methanol for 10 min and stained with Chlamydia trachomatis detection kit for 2 hr at room temperature. Wells were aspirated, overlayed with 90% glycerol tris solution and IFUs quantified. Simultaneous determination of individual samples of Chlamydia being experimentally evaluated revealed MOI ranging from 90 to 110. All experiments were repeated on three separate days with new monolayers.
4.5 |. CT147-associated confocal microscopy
L929 cells were seeded at 50% confluency 24 hr prior to infection in an 8-well ibiTreat μ-Slide (Ibidi, Martinsried, Germany) and were infected with respective C. trachomatis, C. muridarum, or recombinant mutant. At 24 hpi, infected cells were fixed with 100% methanol for 10 min at room temperature. Cells were washed once with HBSS and again with PBS then stained using 180 μl of the PathoDx Chlamydia Culture Confirmation Kit (Remel Europe Ltd., Dartford, UK), mouse anti-CT147 (kind gift from Dr. Guangming Zhong, UTHSCSA), or rabbit anti-IncG diluted in PBS 1 hr and 50 min room temperature in the dark or overnight at 4°C. 20μl of 1μM 4’, 6-diamidino-2-phenylindole (DAPI) diluted 1:100 in PBS was then added to wells and allowed to stain for 10 min, room temperature in the dark. Stain was then removed, and the cells were washed with PBS. A final overlay of Vectashield antifade mounting medium (Burlingame, CA) was added and slides were stored at 4°C in the dark until imaged. Cells were visualized on an Olympus IX81/3I spinning disk confocal inverted microscope at 150X magnification and captured on an Andor Zyla 4.2 sCMOS camera (Belfast, Northern Ireland). Microscope and camera were operated using SlideBook 6 software (Intelligent Imaging Innovations, Denver, USA). Exposure time remained consistent for all fields captured, with exposure for DAPI at 2 s, GFP (MOMP/LOS) 3 s, and Evans Blue (host cytoplasm) 3 s. 5–10 images were taken per strain. 5 Z stack images at 0.35μm apart were taken per field imaged. Images were processed in SlideBook 6 and a No Neighbors Deconvolution with a subtraction constant of 0.4 was applied to all images.
4.6 |. Mouse infections
Female C57BL/6 mice or C3He/J mice (6 to 8 weeks old) were purchased from Jackson Laboratories and housed in accordance with the requirements specified by the University of Kansas Institutional Care and Use Committee. Mice were treated subcutaneously with 2.5 mg medroxyprogesterone acetate (Depo-Provera, Pfizer, NY) upon arrival (day −7) to synchronize menstrual cycles. Infectious doses of parental or chimeric Chlamydia were diluted in sucrose-phosphate-glutamic acid (SPG) buffer along with an SPG-only mock control dose. Mice were inoculated intravaginally with 5 μl of infectious dose, for a final concentration around 1 × 105 IFU/mouse, by deposition of the dose into the vagina. For transcervical infections, a non-surgical embryo transfer device (NSET, Lexington, KY) was used to inoculate mice with 10 μl diluted stock (final concentration 5 x 105 IFU/mouse). Device was inserted into the genital tract beyond the opening of the cervix. Seven days post-infection for C57BL/6 or five days post-infection for C3He/J, mice were humanely euthanized and the genital tracts including vaginal vault and uterine horns were collected in SPG. Organs were homogenized using a rotor/stator homogenizer (Biospec, Bartlesville, OK). DNA isolation was performed using a DNeasy Blood and Tissue Kit (Qiagen) to the manufacturer’s instructions. Isolated DNA was then used to determine bacterial burden by droplet digital PCR (ddPCR). Primers and probes for Chlamydial rpsR and murine rpp30 were used with ddPCR Supermix for Probes (Bio-Rad, Hercules, CA). ddPCRs were performed as previously reported (LaBrie et al., 2019) Bacterial burdens of each tissue were analyzed using QuantaSoft Software (Bio-Rad), and the results are reported as log10 ratios of Chlamydia DNA to host DNA (rpsR/rpp30 copies). Box and whisker scatter plots were generated in GraphPad Prism 8. For the vaginal shedding, C57BL/6 mice were infected as above. At days 3, 7, 14, 21 and 28 post-infection, vaginal swabs were collected by inserting a calgiswab (Puritan Medical Products, ME USA) 5 mm into the vagina and turning five times in each direction. Swabs were then placed in SPG and bead beating was used to release Chlamydia from the swab. Titers were measured identically to the progeny production assessment.
For the rectal models, female C57BL/6 were inoculated intrarectally with 5 μl of infectious dose, for a final concentration around 1 × 105 IFU/mouse, by deposition of the dose into the rectum. Rectal swabs were taken at 3 and 7 days post-infection. After 7 days post-infection, mice were humanely euthanized and GI tracts including rectum, small intestine, and spleen were collected in SPG. Rectal swabs were performed as described for vaginal swabs above. Box and whisker scatter plots were generated in GraphPad Prism 7 with statistical comparisons using unpaired, multiple t-tests with no correction for multiple comparisons.
4.7 |. Apoptotic assays
HeLa and McCoy cells were infected with wild type C. trachomatis L2 or chimera strains and analyzed for phosphatidylserine externalization and caspase 3/7 activation using the Annexin V Alexa Fluor 594 and Image-iT live caspase 3/7 assay kits (Molecular Probes), respectively, using established procedures (Sherrid & Hybiske, 2017). Infected cells were fixed and stained according to the manufacturer’s instructions, and cells were subsequently imaged on a Nikon Eclipse Ti-E inverted microscope equipped with a 60X objective and a cooled CCD camera. Images were acquired and processed using the Volocity software package (Perkin Elmer). Cells positive for external plasma membrane annexin V were scored using a software-based calculation of fluorescence intensity and distribution, along with positive and negative controls to train the software for accuracy.
4.8 |. Live in vitro imaging
L929 mouse fibroblast cells were infected in a 96-well plate at an MOI of ~5. At 16 hr post-infection, the infected plate was placed in an EVOS FL Auto 2 microscope with 5% CO2, 37°C, and humidity. One field of view for each strain was imaged by bright field using a 40X objective every 20 min for 12 hr (16 hpi-28 hpi). Images for each strain were then compiled in ImageJ (NIH, Bethesda, MD) and processed into.mp4 format using Adobe Media Encoder 2019 (Adobe, San Jose CA).
Supplementary Material
ACKNOWLEDGMENTS
These studies and associated personnel were supported by NIH (AI126785). KSH was supported by NIH T32 GM008545 (Dynamic Aspects of Chemical Biology). PSH was also supported by P20GM113117 (Center for Chemical Biology of Infectious Disease) and RNAseq was supplemented by P20GM103638 (Center for Molecular Analysis of Disease Pathways). JS was supported by the American Society for Microbiology Undergraduate Fellowship. We are very appreciative of the anti-CT147 antibodies provided by Dr. Guangming Zhong (UTHSCSA) and Dr. Richard Morrison (UAMS) for protocol and guidance with rectal mouse infection studies. We are grateful for the efforts of Nancy Schwarting with respect to mouse infections and organ isolations.
Funding information
National Institute of Allergy and Infectious Diseases, Grant/Award Number: AI126785; National Institute of General Medical Sciences, Grant/Award Number: GM008545, GM103638 and GM113117; American Society for Microbiology Undergraduate Fellowship
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
Data openly available in a public repository that issues datasets with DOIs and available on request from the authors.
REFERENCES
- Abdelrahman YM & Belland RJ (2005) The chlamydial developmental cycle. FEMS Microbiology Reviews, 29(5), 949–959. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA et al. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS Journal, 272(20), 5101–5109. 10.1111/j.1742-4658.2005.04945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Griffiths R, Viratyosin W, Brown WJ & Rockey DD (2000) A secondary structure motif predictive of proteinlocalization to the chlamydial inclusion membrane. Cellular Microbiology, 2(1), 35–47. 10.1046/j.1462-5822.2000.00029.x [DOI] [PubMed] [Google Scholar]
- Belland RJ, Scidmore MA, Crane DD, Hogan DM, Whitmire W, McClarty G et al. (2001) Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proceedings of the National Academy of Sciences of the United States of America, 98(24), 13984–13989. 10.1073/pnas.241377698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J et al. (2003) Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proceedings of the National Academy of Sciences of the United States of America, 100(14), 8478–8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe M, Dutow P, Pich A, Genth H & Klos A (2015) DXD motif-dependent and -independent effects of the chlamydia trachomatis cytotoxin CT166. Toxins, 7(2), 621–637. 10.3390/toxins7020621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D et al. (2003) Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. Journal of Clinical Investigation, 111(11), 1757–1769. 10.1172/JCI17993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J, Huang Y, Liu Y, Schenken R, Arulanandam B & Zhong G (2014) Bioluminescence imaging of Chlamydia muridarum ascending infection in mice. PLoS One, 9(7), e101634. 10.1371/journal.pone.0101634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang H, Zhou Z, Yang Z, Ding Y, Zhou Z et al. (2014) Chlamydial induction of hydrosalpinx in 11 strains of mice reveals multiple host mechanisms for preventing upper genital tract pathology. PLoS One, 9(4), e95076. 10.1371/journal.pone.0095076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Dooley CA, Grieshaber SS, Carabeo RA, Fields KA & Hackstadt T (2005) Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infection and Immunity, 73(7), 3860–3868. 10.1128/IAI.73.7.3860-3868.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani MT, Gambarte Tudela J & Capmany A (2014) Targeting eukaryotic Rab proteins: a smart strategy for chlamydial survival and replication. Cellular Microbiology, 16(9), 1329–1338. 10.1111/cmi.12325 [DOI] [PubMed] [Google Scholar]
- Darville T & Hiltke TJ (2010) Pathogenesis of genital tract disease due to Chlamydia trachomatis. Journal of Infectious Diseases, 201(Suppl 2), S114–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E, Kalmar I & Vanrompay D (2013) Animal models for studying female genital tract infection with Chlamydia trachomatis. Infection and Immunity, 81(9), 3060–3067. 10.1128/IAI.00357-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Maza LM, Pal S, Khamesipour A & Peterson EM (1994) Intravaginal inoculation of mice with the Chlamydia trachomatis mouse pneumonitis biovar results in infertility. Infection and Immunity, 62(5), 2094–2097. 10.1128/iai.62.5.2094-2097.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehoux P, Flores R, Dauga C, Zhong G & Subtil A (2011) Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics, 12, 109. 10.1186/1471-2164-12-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimond ZE & Hefty PS (2021) Comprehensive genome analysis and comparisons of the swine pathogen, Chlamydia suis reveals unique ORFs and candidate host-specificity factors. Pathogens and Disease, 79(2). 10.1093/femspd/ftaa035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD et al. (2002) Molecular basis defining human Chlamydia trachomatis tissue tropism. Journal of Biological Chemistry, 277(30), 26893–26903. 10.1074/jbc.M203937200 [DOI] [PubMed] [Google Scholar]
- Flores R & Zhong G (2015) The Chlamydia pneumoniae inclusion membrane protein Cpn1027 interacts with host cell Wnt signaling pathway regulator cytoplasmic activation/proliferation-associated protein 2 (Caprin2). PLoS One, 10(5), e0127909. 10.1371/journal.pone.0127909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Diaz M, Monturiol-Gross L, Naylor C, Alape-Giron A & Flieger A (2016) Bacterial sphingomyelinases and phospholipases as virulence factors. Microbiology and Molecular Biology Reviews, 80(3), 597–628. 10.1128/MMBR.00082-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauliard EOS, Rueden KJ & Ladant D (2015) Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis. Frontiers in Cellular and Infection Microbiology, 5(13), 13. 10.3389/fcimb.2015.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel AM, Hu S, Rajaram K, Finethy R, Toh E, Brothwell JA et al. (2019) Genetic screen in Chlamydia muridarum reveals role for an interferon-induced host cell death program in antimicrobial inclusion rupture. MBio, 10(2), e00385–19. 10.1128/mBio.00385-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitsels A, Sanders N & Vanrompay D (2019) Chlamydial infection from outside to inside. Frontiers in Microbiology, 10, 2329. 10.3389/fmicb.2019.02329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes JPNA, Bruno WJ, Borrego MJ, Florindo C & Dean D (2006) Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. Journal of Bacteriology, 118(1), 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondek DC, Olive AJ, Stary G & Starnbach MN (2012) CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. The Journal of Immunology, 189(5), 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helble JD, Reinhold-Larsson NV & Starnbach MN (2019) Early colonization of the upper genital tract by Chlamydia muridarum is associated with enhanced inflammation later in infection. Infection and Immunity, 87(9), e00405–19. 10.1128/IAI.00405-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ et al. (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry, 83(22), 8604–8610. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM & Rockey DD (2013) Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiology, 13, 142. 10.1186/1471-2180-13-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBrie SD, Dimond ZE, Harrison KS, Baid S, Wickstrum J, Suchland RJ et al. (2019) Transposon mutagenesis in Chlamydia trachomatis identifies CT339 as a ComEC homolog important for DNA uptake and lateral gene transfer. MBio, 10(4), e01343–19. 10.1128/mBio.01343-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Kim CJ, Kim DJ & Kang JH (2015) Immune cells in the female reproductive tract. Immune Network, 15(1), 16–26. 10.4110/in.2015.15.1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinger JE, Nelton E, Feeney C & Gondek DC (2015) Chlamydia infection across host species boundaries promotes distinct sets of transcribed anti-apoptotic factors. Frontiers in Cellular and Infection Microbiology, 5, 96. 10.3389/fcimb.2015.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CD, Songer JR & Sullivan JF (1987) A twenty-five year review of laboratory-acquired human infections at the National Animal Disease Center. American Industrial Hygiene Association Journal, 48(3), 271–275. 10.1080/15298668791384733 [DOI] [PubMed] [Google Scholar]
- Morrison SG, Giebel AM, Toh EC, Spencer HJ, Nelson DE & Morrison RP (2018) Chlamydia muridarum genital and gastrointestinal infection tropism is mediated by distinct chromosomal factors. Infection and Immunity, 86(7), e00141–18. 10.1128/IAI.00141-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW, Hatch TP, Byrne GI & Kellogg KR (1976) Immediate toxicity of high multiplicities of Chlamydia psittaci for mouse fibroblasts (L cells). Infection and Immunity, 14(1), 277–289. 10.1128/iai.14.1.277-289.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Taylor LD, Shannon JG, Whitmire WM, Crane DD, McClarty G et al. (2007) Phenotypic rescue of Chlamydia trachomatis growth in IFN-gamma treated mouse cells by irradiated Chlamydia muridarum. Cellular Microbiology, 9(9), 2289–2298. [DOI] [PubMed] [Google Scholar]
- Nunes A & Gomes JP (2014) Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infection, Genetics and Evolution, 23, 49–64. 10.1016/j.meegid.2014.01.029 [DOI] [PubMed] [Google Scholar]
- O’Meara CP, Andrew DW & Beagley KW (2014) The mouse model of Chlamydia genital tract infection: a review of infection, disease, immunity and vaccine development. Current Molecular Medicine, 14(3), 396–421. [DOI] [PubMed] [Google Scholar]
- Phillips S, Quigley BL & Timms P (2019) Seventy years of Chlamydia vaccine research - limitations of the past and directions for the future. Frontiers in Microbiology, 10, 70. 10.3389/fmicb.2019.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike RM (1976) Laboratory-associated infections: summary and analysis of 3921 cases. Health Laboratory Science, 13(2), 105–114. [PubMed] [Google Scholar]
- Rajaram K, Giebel AM, Toh E, Hu S, Newman JH, Morrison SG et al. (2015) Mutational analysis of the Chlamydia muridarum plasticity zone. Infection and Immunity, 83(7), 2870–2881. 10.1128/IAI.00106-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O et al. (2000) Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Research, 28(6), 1397–1406. 10.1093/nar/28.6.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DD, Scidmore MA, Bannantine JP & Brown WJ (2002) Proteins in the chlamydial inclusion membrane. Microbes and Infection, 4(3), 333–340. 10.1016/S1286-4579(02)01546-0 [DOI] [PubMed] [Google Scholar]
- Roshick C, Wood H, Caldwell HD & McClarty G (2006) Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infection and Immunity, 74(1), 225–238. 10.1128/IAI.74.1.225-238.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg ME, Gigliotti-Rothfuchs A & Wigzell H (2002) The role of IFN-gamma in the outcome of chlamydial infection. Current Opinion in Immunology, 14(4), 444–451. [DOI] [PubMed] [Google Scholar]
- Segawa K & Nagata S (2015) An apoptotic ‘eat me’ signal: phosphatidylserine exposure. Trends in Cell Biology, 25(11), 639–650. 10.1016/j.tcb.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Selvy PE, Lavieri RR, Lindsley CW& Brown HA (2011) Phospholipase D: enzymology, functionality, and chemical modulation. Chemical Reviews, 111(10), 6064–6119. 10.1021/cr200296t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AA, Schripsema JH, Imtiaz MT, Sigar IM, Kasimos J, Matos PG et al. (2005) Histopathologic changes related to fibrotic oviduct occlusion after genital tract infection of mice with Chlamydia muridarum. Sexually Transmitted Diseases, 32(1), 49–56. 10.1097/01.olq.0000148299.14513.11 [DOI] [PubMed] [Google Scholar]
- Sherrid AM & Hybiske K (2017) Chlamydia trachomatis cellular exit alters interactions with host dendritic cells. Infection and Immunity, 85(5), e00046–17. 10.1128/IAI.00046-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somboonna N, Wan R, Ojcius DM, Pettengill MA, Joseph SJ, Chang A et al. (2011) Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranuloma venereum (L(2)) and D lineages. MBio, 2(3), e00045–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa JM, Huber DE, Welk B & Fraser HL (2014) Development and application of MIPAR™: a novel software package for two- and three-dimensional microstructural characterization. Integrating Materials and Manufacturing Innovation, 3(1), 123–140. 10.1186/2193-9772-3-10 [DOI] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L et al. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science, 282(5389), 754–759. [DOI] [PubMed] [Google Scholar]
- Suchland RJ, Carrell SJ, Wang Y, Hybiske K, Kim DB, Dimond ZE et al. (2019) Chromosomal recombination targets in Chlamydia interspecies lateral gene transfer. Journal of Bacteriology, 201(23), e00365–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE & Rockey DD (2009) Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrobial Agents and Chemotherapy, 53(11), 4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann J, Janik K, May M, Sommer K, Ebeling J, Hofmann F et al. (2010) Actin re-organization induced by Chlamydia trachomatis serovar D-evidence for a critical role of the effector protein CT166 targeting Rac. PLoS One, 5(3), e9887. 10.1371/journal.pone.0009887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonova RT, Lieberman J & van Baarle D (2014) Distribution of immune cells in the human cervix and implications for HIV transmission. American Journal of Reproductive Immunology, 71(3), 252–264. 10.1111/aji.12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, LaBrie SD, Carrell SJ, Suchland RJ, Dimond ZE, Kwong F et al. (2019) Development of transposon mutagenesis for Chlamydia muridarum. Journal of Bacteriology, 201(23), e00366–19. 10.1128/JB.00366-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM, Lam JL, Dooley CA, Noriea NF, Hansen BT, Hoyt FH et al. (2017) Absence of specific Chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Reports, 19(7), 1406–1417. 10.1016/j.celrep.2017.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfeld H (2017) Screening for Chlamydia trachomatis infections in women. New England Journal of Medicine, 376(22), 2197–2198. [DOI] [PubMed] [Google Scholar]
- Xie G, Bonner CA & Jensen RA (2002) Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biology, 3(9), research0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data openly available in a public repository that issues datasets with DOIs and available on request from the authors.


