Abstract
Environmental Enteric Dysfunction (EED) refers to an incompletely defined syndrome of inflammation, reduced absorptive capacity, and reduced barrier function in the small intestine. It is widespread among children and adults in low- and middle-income countries and is also associated with poor sanitation and certain gut infections possibly resulting in an abnormal gut microbiota, small intestinal bacterial overgrowth (SIBO) and stunting.
We investigated bacterial pathogen exposure in stunted and non-stunted children in Antananarivo, Madagascar by collecting fecal samples from 464 children (96 severely stunted, 104 moderately stunted and 264 non-stunted) and the prevalence of SIBO in 109 duodenal aspirates from stunted children (61 from severely stunted and 48 from moderately stunted children). SIBO assessed by both aerobic and anaerobic plating techniques was very high: 85.3% when selecting a threshold of ≥105 CFU/ml of bacteria in the upper intestinal aspirates. Moreover, 58.7% of the children showed more than 106 bacteria/ml in these aspirates. The most prevalent cultivated genera recovered were Streptococcus, Neisseria, Staphylococcus, Rothia, Haemophilus, Pantoea and Branhamella. Feces screening by qPCR showed a high prevalence of bacterial enteropathogens, especially those categorized as being enteroinvasive or causing mucosal disruption, such as Shigella spp., enterotoxigenic Escherichia coli, enteropathogenic E. coli and enteroaggregative E. coli. These pathogens were detected at a similar rate in stunted children and controls, all showing no sign of severe diarrhea the day of inclusion but both living in a highly contaminated environment (slum-dwelling). Interestingly Shigella spp. was the most prevalent enteropathogen found in this study (83.3%) without overrepresentation in stunted children.
Author summary
About 2 million children under the age of 5 suffer from stunted growth in Madagascar. Although deficient diet is the major cause of undernutrition, impaired absorption or assimilation caused by Environmental Enteric dysfunction (EED) has been proposed to play an important role in stunting. EED is widespread among children and adults in low- and middle-income countries (LMIC) and is also associated with undernutrition, poor sanitation, certain gut infections resulting in an abnormal gut microbiota and small intestinal bacterial overgrowth (SIBO) although the role of SIBO in EED remains unclear. The current study highlights the presence at high concentrations of bacterial taxa usually found in the oro-pharyngeal sphere in a high number of duodenal fluids of stunted children. This uncommon presence suggests a decompartmentalization of the gastrointestinal tract and a possible pro-inflammatory effect due to the ectopic presence of some of these bacteria in the duodenum.
The study also points to a high prevalence of enteropathogens (especially Shigella spp.) in the feces of both stunted and control children, hence preventing from proposing a direct association with stunting. This suggests that, beside combatting poverty and improving diet, environmental sanitation, quality of water sources, hygiene promotion and health education are key points to mitigate stunting and restore nutritional benefits.
Introduction
Stunting is a major public health and economic development concern in Madagascar which is the 5th most affected country in the world with 48.5% of all children under 5 (ca. 2 million) suffering from stunted growth [1]. Although deficient diet is the major cause of undernutrition, other factors such as Environmental Enteric Dysfunction (EED) has been proposed to be an associate driver of stunting in low and middle-income countries (LMICs) [2,3]. EED is a chronic inflammatory condition of the gut occurring among children living in unsanitary conditions [4], or among adults returning from deployment to LMICs [5,6]. According to estimates, greater than 75% of all children in LMICs could suffer from this syndrome at different degrees of severity [7]. Certain gut infections resulting in an abnormal gut microbiota and small intestinal bacterial overgrowth (SIBO) may also play a role in EED [5].
A study in Bangladesh found enteric infections in the first two years of life, mainly Shigella and Enterotoxigenic E. coli (ETEC), to be associated with EED and stunting [8]. Two additional studies found entero-aggregative E. coli (EAEC) and Campylobacter to be associated with markers of gut inflammation and stunting [9,10]. Even in the absence of diarrhea, these recurrent enteric infections may result in an imbalanced gut microbiota (i.e. dysbiosis) leading to growth faltering mediated through systemic inflammation [11].
SIBO is defined as the presence of excessive bacteria (greater than 105 CFU/ml) in the small intestine. It is frequently implicated as the cause of chronic diarrhea and malabsorption. SIBO can be measured noninvasively by hydrogen breath testing or by anaerobic and aerobic cultures of endoscopically aspirated upper gastrointestinal fluid [12]. In a study performed on Bangladeshi children, SIBO was associated with a decreased length-for-age Z score since birth and poor sanitation but was independent of frequent or recent diarrheal disease. It was also associated with intestinal inflammation but not with increased permeability or systemic inflammation [13].
Both repetitive gut infections and SIBO can co-exist in an intestine weakened by undernutrition and might lead themselves to micronutrient malabsorption fueling a vicious cycle with long-lasting effects.
AFRIBIOTA is a case-controlled study for stunting, conducted in children in Antananarivo, Madagascar and in Bangui, Central African Republic, in which 460 children aged 2–5 years with no overt signs of gastrointestinal disease were recruited in each country [14]. In a preliminary study (feces from 153 children and 12 duodenal aspirates) [15], we showed vast majority of the stunted children (>80%) showed SIBO dominated by bacteria normally residing in the oro-pharyngeal tract. There was also an overrepresentation of oral bacteria in fecal samples of stunted children. Escherichia coli/Shigella spp. and Campylobacter spp. were found by a metataxonomic approach to be more prevalent in stunted children, while the butyrate-producers (Clostridia) were reduced [15].
The current research aimed to assess SIBO and gut infections in a population of children (N = 464) recruited in Antananarivo, Madagascar, using a quantitative culture-based technique on duodenal aspirates from stunted children and a more sensitive and accurate molecular-based approach (qPCR) than the classical metataxonomic approach [16,17,18], targeting the main enteropathogenic bacterial species and pathobionts in the feces.
Methods
Ethics statement
The study protocol for AFRIBIOTA has been approved by the Institutional Review Board of the Institut Pasteur (2016–06/IRB) and the National Ethical Review Boards of Madagascar (55/MSANP/CE, May 19th 2015). All participants received oral and written information about the study and the legal representatives of the children provided written consent to participate in the study.
Recruitment of participants
The case control was extensively described in a preliminary study [15] and is similar to the full-study. Briefly, the study population comprises HIV-negative children aged 2 to 5 years, neither suffering from acute malnutrition, nor from any other severe diseases (such as dysenteric syndrome, severe acute respiratory infections (SARI)/influenza-like illness (ILI), meningitis, malaria, acute otitis media, varicella, measles, …) who were recruited in the community of the two districts of Ankasina or Andranomanalina Isotry, and in two hospitals (Centre Hospitalier Universitaire Joseph Ravoahangy Andrianavalona (CHU-JRA) and Centre de Santé Materno-Infantile, Tsaralalana) in Antananarivo, Madagascar. Children were recruited either in the community (community-recruited children) or directly in the hospital (hospital-recruited children). The caregivers of all children received oral and written explanations of the study and provided written consent to participate. Anthropometric measurements were collected at the community-health centers and in the hospital. All children were admitted to the hospital for sample collection. Height was measured to the nearest 0.1 cm in a standing position using collapsible height boards; weight was measured using a commercial weighing scale to the nearest 100 g. All measurements were taken at least twice and repeated if the measurements were more than 0.1 cm/100 g apart from each other. The children were classified according to the median height of the WHO reference population [16] in three different groups: severe stunting (height-for-age z-score ≤ -3SD), moderate stunting (height-for-age z-score between -3SD and -2SD) and not stunted (height-for-age z-score ≥ -2SD). The non-stunted (control subjects) were matched for living area and sampling time period and were recruited during the entire study period (December 2016—March 2018). The realized sample size was 490 children: 211 stunted children and 279 not stunted. All children (hospital- and community-recruited children) were asked to come to the hospital for sample collection for the safety of the children (see hereafter).
Collection of stools, gastric and duodenal aspirates
All those conditions have been previously described in Vonaesch et al., 2018 [15]. Briefly, stools were collected in the morning at the hospital (directly before coming to the hospital for the community-recruited children) and the time of defecation recorded. If community-recruited children were able to again emit feces in the hospital, these feces were also collected and directly snap-frozen in liquid nitrogen before being transferred to -80°C. The time spent between defecation and freezing (liquid nitrogen/-80°C) was recorded on specific tracking sheets for each sample. The time for freezing the emitted feces was comprised between 6 min and 23.66h with a mean of 3.69h.
Gastric and duodenal samples were collected using a pediatric nasogastric tube with stent (Vygon, France), and were only collected for stunted children due to ethical concerns. The nasogastric tube was introduced in the infant’s nose sitting in an upright position and moved to the stomach. Proper placement in the stomach was controlled using the syringe-air test and confirmed by an acid pH (pH ≤ 4). Once at least 4 ml of liquid were aspirated, the tube was moved 5 cm forward. The child was then laid down on the right side as to facilitate the tube’s passage in the duodenum. Pyloric passage was monitored regularly and was confirmed when the pH reached a pH ≥ 5. In this case, a second aspiration was performed and identified as duodenal. The procedure was interrupted if the pH did not change to a pH ≥ 5 within 1h30 min after introducing the tube. We also ensured that the first ml of aspiration from both the stomach and the duodenum were discarded. This allowed flushing out possible contaminating bacteria and minimize the carry-over from the more proximal compartments. 100 μl of fresh duodenal sample were inoculated directly in 0.9 ml of Robertson’s Cooked Meat (RCM) medium and processed for culture (see below). The rest of the aliquots were directly snap-frozen in liquid nitrogen and then transferred the same day to a -80°C freezer.
Culture of duodenal aspirates, feces and identification of colonies
RCM-diluted duodenal aspirations were brought within a time frame of 30 min to the Institut Pasteur of Madagascar, where they were further diluted and streaked on plates according to the protocol described in Chandra et al. [17]. In brief, the duodenal samples were analyzed using plating techniques in aerobic and anaerobic culture conditions after the RCM-diluted duodenal aspirates were serially diluted (1:100, 1:1,000) in phosphate buffered saline containing 1% peptone and 20 μL of each dilution and inoculated onto 3 separate culture plates. Aerobic culture was performed on chocolate agar, 5% sheep blood agar, and MacConkey’s medium. Anaerobic cultures were performed directly from the RCM tube (without dilution) onto 5% sheep blood agar containing haemin (5 μg/ml) and vitamin K (menadione) (1 μg/mL). The plates were incubated in an anaerobic jar or aerobically at 37°C for 48 h. Total bacterial count per ml of duodenal aspirate was calculated based on the total count on the chocolate agar plates (incubated aerobically) and the anaerobic culture. The following formula was used to calculate the total CFU load: No. of CFU per 20 μL of fluid inoculated x x 1,000 X dilution on plate.
The isolated colonies were counted by morphotype and at the beginning 2–3 colonies for every morphotype were re-isolated. Re-isolated colonies were all identified by MALDI-TOF mass spectrometry (Bruker Biotyper, Bruker Daltonics, Bremen, Germany). As colonies from the same morphotype from the same plate were all identical (same identification), finally only one colony/morphotype was isolated and subcultured. Cultures were considered positive for SIBO if the total bacterial count was ≥105 CFU per mL of duodenal fluid [12,17].
To isolate Shigella spp. from the last 143 feces samples, a fresh scoop of feces taken with an inoculation loop was suspended in 5ml of physiological water and 20 μL were plated on Hektoen and XLD media. Suspected Shigella isolates (green or pink colonies on Hektoen or XLD, respectively) were purified, stored and identified with API galleries. There were sent to the French National Reference Center for E. coli, Shigella and Salmonella (FNRC-ESS), Institut Pasteur, Paris for species confirmation and serotyping. Serotyping was done by slide agglutination assays using a complete set of antisera allowing recognition of all described Shigella serotypes [18].
Data collection
To assess risk factors for acquiring pathogens, a questionnaire was developed and administered to children and their caregivers. In brief, the questionnaire contained four sections: 1. Socio-demographic data: age, gender, community setting, education and occupation of parents, family marital status; 2. Environmental factors: housing conditions (proximity of the housing to landfill, soil type, lavatories and showers) and quality of drinking water; 3. Behavior habits: type of toilet commonly used, hand washing habit, exposure to sewage and garbage; 4. Medical status/history: dental cavities, cough, stuffy nose, runny nose, delivery mode.
Statistical analysis
The data were encoded in an Excel (microbiological data) and an Access (socioeconomic data) database and analyzed using the R statistical software version 3.6.2. Comparisons between groups (controls and stunted children MS+SS) were determined using Pearson’s χ2-test or Fisher’s exact test, as appropriate. Values of p < 0.05 were considered to be statistically significant.
For risk factors analysis, bivariate logistic regression analysis was carried out between factors (S1 and S2 Tables) and the presence of SIBO using the Chi-squared test or the Fisher’s exact test. Variables having P-value of ≤0.25 were entered into multivariate logistic regression for final analysis. Additionally, we correlated the pathogen presence in stools with the measure of CFU/ml grown from duodenal aspirates in aerobic and anaerobic culture conditions using the non-parametric Wilcoxon’s rank sum test.
For the clustering analysis, the data consisted in 0s and 1s corresponding to the infection status (detection of the pathogen by qPCR) of the children: 0 = uninfected and 1 = infected. The infection profiles for the children obtained were clustered with a Hierarchical Clustering based on a binary distance coupled with Ward’s agglomeration method. No clustering was applied on the variables.
DNA extraction and real-time PCR
Samples were extracted by commercial kits using a Qiacube instrument (Cador Pathogen 96 QIAcube HT Kit, Qiagen France SAS, Courtaboeuf, France). DNA extractions were performed following the manufacturer’s recommendations with an additional bead-beating step to increase mechanical disruption. In brief, 200 mg of freshly thawed sample were mixed with 1.4 mL of ASL buffer at 4°C and vigorously vortexed for 1 min. The suspension was transferred into a Pathogen Lysis Tube S (Qiagen) containing 2 mg of sterile glass beads (100μM diameter). Samples were mechanically disrupted using a TissueLyser II (Qiagen Retsch GmbH, Hilden, Germany) for 10 min at 30Hz. The suspension was then incubated at 95°C for 5 min, vortexed for 15 sec and centrifuged at 14,000 x g for 1 min to ensure that no solid particles were transferred to the subsequent steps. A volume of 1.2 mL of the supernatant was then transferred into a new tube containing an InhibitEX tablet (Qiagen). Samples were vigorously vortexed for at least 1 min or until complete dissolution of the InhibitEX tablet. Three steps of centrifugation and supernatant transfer were performed thereafter (14,000 x g for 3 min). The remainder of the protocol was performed as recommended by the manufacturer. All samples were eluted in 150 μL AE buffer. DNA concentrations and purity were assessed via 260/280 and 260/230 absorbance ratios by spectrophotometry (Nanodrop 2000 Spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA). Samples were stored at -80°C until molecular analyses.
Amplifications were carried out in an ABI StepOne instrument (Applied Biosystems, Nairobi, Kenya). After an initial denaturation step (95°C for 10 min), 45 cycles of two-step PCR (95°C for 15 s and 50–56°C for 60 s according to the duplex reaction) were performed in 5 parallel duplex reactions, targeting a broad range of diarrheagenic bacterial agents as described in Table 1. The result for each agent was recorded as the Ct value, which is inversely related to the pathogen load in each specimen. Ct values < or equal to 37 were considered as positive. Standard curves for each target were established and the linearity ensured the Ct cutoff value of 37 was applicable.
Table 1. Primers and probes targeting DNA of diarrheagenic agents.
| Pathogen | Duplex/annealing t° | Forward primer | Reverse primer | Probe | Fluorophores | Target gene/Access number—GenBank | Reference |
|---|---|---|---|---|---|---|---|
| Salmonella spp. | 1 / 50°C | CGGGTTGCGTTATAGGTCTGA | TGAAATACGATGCGAACAACATC | AATACTGCGCTGCCAGAT | HEX-BQ1 | Outer membrane protein, ompC / AIH09487.1 | [19] |
| Shigella spp. | 2 / 56°C | ACCGGCGCTCTGCTCTC | GCAATGTCCTCCAGAATTTCG | CTGGGCAGGGAAATGTTCCGCC | HEX-BQ1 | invasion plasmid antigen H, ipaH / DQ132807.1 | [19] |
| ETEC | 1 / 50°C | AAGCATGAATAGTAGCAATTACTGCT -> AAGCATGAATRGTAGCAATTACTGCT* | TTAATAGCACCCGGTACAAGCA | AACAACACAATTCAC -> TACAACACAATTCAC* | FAM-BQ1 | Heat-stable enterotoxin ST, estIa / M29255.1 | [19]* |
| ETEC | 2 / 56°C | TCCGGCAGAGGATGGTTACA | CCAGGGTTCTTCTCTCCAAGC | AGCAGGTTTCCCACCGGATCACC | FAM-BQ1 | Heat-labile enterotoxin LT, eltB / BAI49232.1 | [19] |
| EPEC | 3 / 50°C | CATTGATCAGGATTTTTCTGGTGATA | CTCATGCGGAAATAGCCGTTA | ATACTGGCGAGACTATTTCAA | FAM-BQ1 | Intimin, eae / CAG17538.1 | [20] |
| EPEC | 3 / 50°C | TGGTGCTTGCGCTTGCT | CGTTGCGCTCATTACTTCTG | CAGTCTGCGTCTGATTCCAA | HEX-BQ1 | bundle-forming pilus, bfpA / AB247927.1 | [20] |
| EAEC | 4 / 52°C | GAATCGTCAGCATCAGCTACA | CCTAAAGGATGCCCTGATGA | CGGACAACTGCAAGCATCTA | FAM-BQ1 | transcriptional activator of aggregative adherence fimbriae expression, aggR / AF411067.1 | [21] |
| EAEC | 4 / 52°C | CATTTCACGCTTTTTCAGGAAT | CCTGATTTAGTTGATTCCCTACG | CACATACAAGACCTTCTGGAGAA | HEX-BQ1 | part of the aai gene cluster, encoding a type VI secretion system, aaiC / FN554766.1 | [21] |
| Campylobacter jejuni -> Campylobacter jejuni/coli | 5 / 53°C | CTGCTAAACCATAGAAATAAAATTTCTCAC -> CWGCTAAACCATARAAATAAAATTTCTCAC* | CTTTGAAGGTAATTTAGATATGGATAATCG -> YTTTGAAGGTAATTTAGATATGGATAATCG* | CATTTTGACGATTTTTGGCTTGA -> CATTTTGAYGATTTTTGGCTTGA* | HEX-BQ1 | Fibronectin-binding protein, cadF / AJK71638.1 | [22]* |
| Vibrio cholerae | 5 / 53°C | CCACTTAGTGGGTCAAACTATATTGTC | ATGCCCCTAATACATCATTAACGTT | AGCCACTGCACCCAA | FAM-BQ1 | Cholera toxin A subunit, ctxA / X58785.1 | [19] |
* with modifications in the nucleotide sequence of primers and/or probe
Bacterial agents and target sequences
The targets for real-time PCR are presented in Table 1. Bacterial PCRs were developed from available publications concerning suitable target regions. All sequences were controlled and blasted in GenBank and some primers were adapted according to new retrieved sequences from GenBank. Salmonella spp. and Shigella spp. were identified by amplification of the outer-membrane protein C and the invasion plasmid antigen H (ipaH) gene (which also may be present in enteroinvasive E. coli [EIEC]), respectively.
The presence of ipaH gene present on the virulence plasmid and on the chromosome was also tested by conventional PCR with the primers described in Phantouamath et al. [23]
For enterotoxigenic Escherichia coli (ETEC), heat-labile toxin (eltB) and heat-stable toxin (estA) coding regions were targeted. For enteropathogenic E. coli (EPEC), the bundle-forming pilus (encoded by the bfpA gene) carried by the EPEC adherence factor (EAF) plasmid and the intimin (eae gene for EPEC attaching and effacing), an outer membrane adhesion essential for the intimate attachment of the EPEC or enterohemorrhagic E. coli (EHEC) to enterocytes were the targets. For enteroaggregative E. coli (EAEC), aggR and aaiC genes were amplified. The aggR gene encodes a transcriptional activator of the aggregative adherence fimbriae expression in enteroaggregative E. coli and aaiC, is part of the aai gene cluster, encoding a type VI secretion system. When one of the two targets was detected for ETEC (estIa or eltB) or EAEC (aggR or aaiC), the pathogen was considered to be present. Since eae can be present both in EPEC and EHEC, we considered only the presence of bfpA gene for EHEC detection.
Fibronectin-binding protein (cadF) gene and the cholera toxin (CT) subunit A gene (ctxA) were the targeted genes for Campylobacter jejuni/coli and Vibrio cholerae, respectively. Sufficient amplification efficiencies were documented for each realtime PCR by analyzing serial dilutions of pUC57 plasmids carrying all synthetic target inserts (GeneCust Europe, Dudelange, Luxembourg). By comparing Ct values for each target amplified alone or in duplex reactions, it was confirmed that performance was not compromised by multiplexing (5 duplex reactions). In addition to optimization, which focused on analytical sensitivity, diagnostic accuracy was evaluated by analyzing well-characterized bacterial strains from the Culture Collection, Pasteur Institute of Madagascar (Salmonella Typhimurium [BEX125], Shigella flexneri [BEX126], E. coli Diego 1120 ETEC (astA, estA, eltB, uidA) [BEX228], E. coli Tul 2322 EPEC (escV, bfpA, uidA, eae) [BEX217], E. coli Morondava 1612 EAEC (eae, astA, aggR, pic, aaiC, uidA) [BEX216], Campylobacter jejuni [BEX112], Vibrio cholerae [BEX227]). Negative and positive controls were included in each qPCR amplification run.
DNA extraction, whole genome sequencing and phylogenetic analysis
DNA was extracted from the Shigella isolates with cador Pathogen 96 QIAcube HT Extraction Kit (QIAGEN, Paris, France) on a Qiacube HT from 5 mL of liquid cultures grown overnight at 37°C in Luria-Bertani infusion medium, following the manufacturer’s protocol. Purity and DNA quantity were assessed using Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). As previously described [24], illumina sequencing libraries were prepared by using Nextera XT DNA Sample Kit (Illumina, San Diego, CA, USA) with indexed-encoded adapters from Illumina, according to the manufacturer’s instructions. The libraries were pooled for sequencing on NextSeq 500 platform (Illumina) using 2 × 150-bp runs. FqCleaner (version 3.0) was used to eliminate adaptor sequences, reduce redundant or overrepresented reads, correct sequencing errors, merge overlapping paired reads, and discard reads with Phred scores (measure of the quality of identification of nucleobases generated by automated DNA sequencing) <20. The Illumina sequence data were assembled using Spades software [25].
The average nucleotide identity (ANI) values were calculated in EzGenome (https://www.ezbiocloud.net/taxonomy) [26] and Genome-to-Genome Distance Calculator (GGDC; http://ggdc.dsmz.de) [27], respectively. Whole-genome-based taxonomic analysis was performed by Type Strain Genome Server (TYGS) (at https://tygs.dsmz.de) [28]. The phylogenomic tree was constructed using FastME [29] from the genome blast distance phylogeny (GBDP). The trees were rooted at the midpoint [30]. Branch supports were inferred from 100 pseudo-bootstrap replicates. MLST, wgMLST, rMLST and cgMLST were obtained using Enterobase and virulence factor were identified using abricate package.
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAIQVZ000000000 for Shigella flexneri HJRA178 and JAIQWA000000000 for Shigella flexneri HJRA198. The version described in this paper is version JAIQVZ010000000 and JAIQWA010000000. Raw sequence data for those strains were deposited under strain ESC_WA5556AA for Shigella flexneri HJRA198 and ESC_WA5569AA for Shigella flexneri HJRA178 in Enterobase.
Gentamicin invasion assay
The gentamicin invasion assay was performed to determine the invasion rate of viable Shigella isolates inside the HEp-2 cells. The methodology used was described in the Bio-protocol 9(13): e3292. [31]. In brief, the amount of 3.105 HEp-2 cells per well were initially seeded in a 6-well microplate. After 24 hours of incubation at 37°C in 10% CO2, the cell cultures were inoculated with Shigella isolates at multiplicity of infection (MOI) for this experiment of 5 (about 5 x 106 bacteria). The infected cells were incubated for 1 hour, washed three times with PBS and incubated for an additional hour with the gentamicin solution. The antibiotic solution was removed, the cells were washed three times with PBS and the infected cells were lysed with 1 ml of sodium deoxycholate solution. The remaining Shigella were quantified by CCU methodology and performed in duplicate. These CCU were compared with the initial values of Shigella suspensions.
Results
Description of the study population
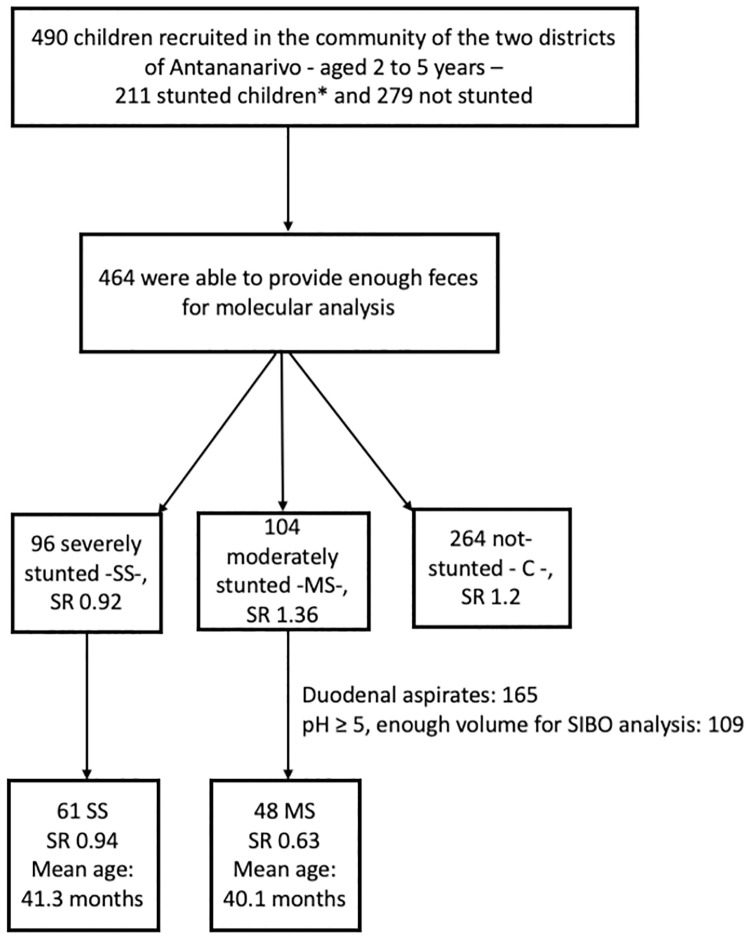
The study population of the Afribiota full-project (Madagascar) comprises 490 children recruited in the community of the two districts of Antananarivo, Madagascar, of which 464 were able to provide enough feces for molecular analysis. Among these 464 children, 96 were severely stunted -SS-, 104 moderately stunted -MS- and 264 non-stunted considered as controls—C -. Sex ratio (M/F) was distributed as follows: 0.92, 1.36 and 1.2 for SS, MS and C, respectively. Duodenal aspirates (N = 109) were analyzed using bacteriological techniques and stools were analyzed by qPCR (Fig 1).
Fig 1. Flowchart inclusion process.
HIV-negative children aged 2 to 5 years, neither suffering from acute malnutrition, nor from any other severe disease recruited in the community of the two districts of Ankasina or Andranomanalina Isotry, and in two hospitals (Centre Hospitalier Universitaire Joseph Ravoahangy Andrianavalona (CHU-JRA) and Centre de Santé Materno-Infantile, Tsaralalana) in Antananarivo, Madagascar. They were classified in three different groups*: severe stunting—SS—(height-for-age z-score ≤ -3SD), moderate stunting—MS—(height-for-age z-score between -3SD and -2SD) and not stunted—C for controls—(height-for-age z-score ≥ -2SD). SR: Sex ratio.
SIBO
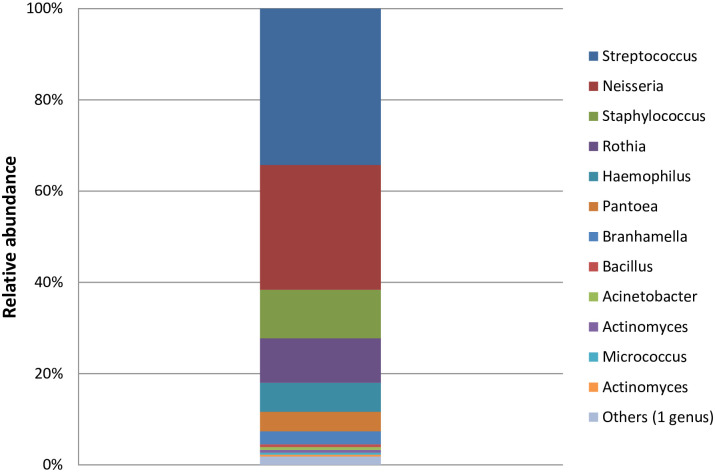
In our study, a total of 165 duodenal aspirates were collected; however, due to several limitations and constraints (i.e. pH ≥ 5, enough volume for subsequent analyses), only 109 samples were investigated, 61 from SS and 48 from MS children (Fig 1). This is a substantial extension of the initial dataset presented in ref. 15 (12 duodenal samples). The duodenal samples were analyzed using plating techniques in aerobic and anaerobic culture conditions at 37°C. The prevalence of SIBO (≥105 CFU/ml of bacteria in the upper intestinal aspirates) was 85.3% (93 children out of 109) with 58.7% hosting more than 106 bacteria/ml (Table 2). The gender distributions were similar for the two groups, with a sex ratio of 0.94 and 0.63 for SS and MS, respectively. The mean age was 41.3 and 40.1 months for SS and MS, respectively. Per aspirate, two to eleven different morphotypes were randomly chosen and identified. In total, 51 different species were identified. Streptococcus, Neisseria, Staphylococcus, Rothia, Haemophilus, Pantoea and Branhamella were the most prevalent genera cultivated (Fig 2).
Table 2. CFU values in duodenal aspirates of stunted children (N = 109).
SIBO is defined as greater than 105 CFU/ml of upper intestinal aspirate as assessed by both anaerobic and aerobic cultures [16].
| CFU values | N (%) |
|---|---|
| 0 | 7/109 (6.4) |
| 102<cfu<105 | 9/109 (8.3) |
| 105<cfu<106 | 29/109 (26.6) |
| 106<cfu<107 | 29/109 (26.6) |
| 107<cfu<108 | 28/109 (25.7) |
| cfu> = 108 | 7/109 (6.4) |
Fig 2. Number of isolates of the 12 most-abundant genera identified by culture techniques in duodenal aspirates of stunted children.
The color code for the different genera is given on the right.
SIBO and risk factors
SIBO was determined in stunted children only. Consequently, risk factors were assessed only for these children. According to the univariate analysis, age group, dental cavities, mode of delivery, lavatories, showers, household waste and soap usage for hand washing (mother) showed a p-value less than 0.25 to SIBO. A multivariable logistic regression analysis was carried out on these variables and none of these were found to be statistically associated with SIBO (p>0.05) (Tables 3 and S1 and S2). Regarding the pH values measured in the stomach samples from stunted children, they were low and there were no significant differences in a bivariate analysis for stomach pH and SIBO (S3 Table).
Table 3. Possible risk factors associated with SIBO.
Only variables having a P-value ≤0.25 in the univariate analysis (S2 Table) were entered into multivariate logistic regression for a final analysis (S3 Table). All variable were recorded by field workers (clinical research associates).
| Variables | SIBO | p-values | |
|---|---|---|---|
| positive | negative | ||
| Age group | |||
| [2–3] | 46 | 6 | 0.13 |
| [4–5] | 43 | 14 | |
| Dental cavities | |||
| yes | 34 | 11 | 0.25 |
| no | 55 | 9 | |
| Delivery mode | |||
| Vaginal delivery | 80 | 20 | 0.20 |
| Caesarean delivery | 9 | 0 | |
| Lavatories | |||
| Collective | 79 | 17 | 0.14 |
| Individual | 6 | 0 | |
| No toilets | 4 | 3 | |
| Showers | |||
| Inside house | 2 | 2 | 0.14 |
| Outside house | 44 | 7 | |
| No showers | 43 | 11 | |
| Household waste | |||
| Burn | 13 | 0 | 0.12 |
| Threw | 76 | 20 | |
| Soap usage for hand washing (mother) | |||
| Before meals | 50 | 15 | 0.07 |
| Before and after meals | 38 | 4 | |
| After meals | 1 | 0 | |
| Never | 0 | 1 | |
Pathogens detection rates
In total, 464 fecal samples (264 controls -C-, 104 MS and 96 SS children) were included in the analysis. Negative and positive (pUC57 plasmids carrying synthetic target inserts and reference strains) controls were included in each qPCR amplification run. Table 4 (see also S4 Table) presented the percentages of bacterial diarrheagenic agents found in stunted children (moderately and severely stunted) and controls (CT values < or equal to 37.0 were considered as positive). At least one bacterial pathogen was detected for 91.8% (90.9% in controls (N = 240) and 92% in stunted children (99 MS and 85 SS)).
Table 4. Comparison between stunted children and controls for the presence of bacterial diarrheagenic agents in fecal samples.
| % for controls (N = 264) | % for MS = (N = 104) | % for SS = (N = 96) | P-value* | |
|---|---|---|---|---|
| Salmonella spp. | 7.95 (N = 21) | 5.21 (N = 5) | 12.5 (N = 12) | 0.13 |
| Shigella spp. | 80.68 (N = 213) | 90.38 (N = 94) | 84.38 (N = 81) | 0.07 |
| ETEC estIa | 4.17 (N = 11) | 5.77 (N = 6) | 6.25 (N = 6) | 0.66 |
| ETEC eltB | 29.92 (N = 79) | 40.38 (N = 42) | 32.29 (N = 31) | 0.15 |
| EPEC bfpA | 13.26 (N = 35) | 11.54 (N = 12) | 18.75 (N = 18) | 0.29 |
| EPEC eae | 37.50 (N = 99) | 36.54 (N = 38) | 36.46 (N = 35) | 0.97 |
| EAEC aggR | 31.06 (N = 82) | 27.88 (N = 29) | 29.17 (N = 28) | 0.82 |
| EAEC aaiC | 17.80 (N = 47) | 25.96 (N = 27) | 22.92 (N = 22) | 0.18 |
| Campylobacter jejuni/coli | 14,39 (N = 38) | 11,54 (N = 12) | 13.54 (N = 13) | 0.77 |
%: percentage; N: total number of sample analysis; MS: moderately stunted; SS: severely stunted;
Comparisons between groups (controls and stunted children MS+SS) were determined using Pearson’s χ2-test or Fisher’s exact test, as appropriate. Only values with p < 0.05 could be considered to be statistically significant*.
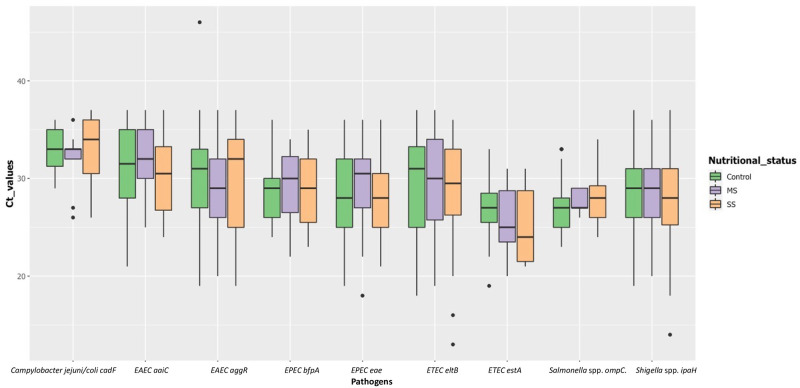
All pathogens were detected at similar prevalence in MS/SS and in C (no statistical differences). The highest prevalence was found for the ipaH gene (Shigella spp. or enteroinvasive E. coli) with values up to 90.4% in MS and equal to 83.3 for the three groups altogether (MS, SS and C). The mean, median, lowest and highest CT values for ipaH were 30, 31.5, 14.5 and 45, respectively. This suggests that the bacterial load was relatively high (Fig 2). The CT values were not significantly lower in stunted children than controls for ipaH, as for the other genes/pathogens (Fig 3).
Fig 3. Box plot showing CT values for pathogens targeted by real-time PCR among stunted (104 moderately stunted and 96 severely stunted) and controls (N = 264).
Boxes show the median (midline) and the 25th and 75th percentiles, and bars indicate the 10th and 90th percentiles. Only the positive subjects (CT values = <37) for the respective pathogen agent was considered.
When considering the presence of only one of the two genes for ETEC and EAEC, and only bfpA for EPEC since eae can be present both in EPEC and EHEC, ETEC and EAEC were present in about one-third (ETEC = 32.8 and EAEC = 29.9%) of the children (C, MS and SS children) and EPEC in 14% of the children. Campylobacter jejuni/coli and Samonella spp. were present in 13.6% and 8.2% of all children. No Vibrio cholerae were found. Infection with more than one pathogen was a common finding for both stunted children and asymptomatic controls: Out of 464 children, 173 (37.3%) carried 3 or more (until 5) pathogens, 151 (32.5%) and 107 (23%) carried two or one pathogen, respectively. No pathogens were detected in only 7.1% (N = 33) of the children.
Using the Wilcoxon’s rank sum test, we correlated the pathogen presence in stools with the CFU/ml values from duodenal aspirates in aerobic and anaerobic culture conditions. The only pathogen significantly associated with the duodenal CFU count was Campylobacter spp. (p = 0.026) (S5 Table).
The distributions of the CFU in duodenal aspirates with SIBO matching with feces contaminated or not by Campylobacter spp. are presented in S1 Fig.
Hierarchical clustering of enteropathogens according to their virulence factors and nutritional the status of children
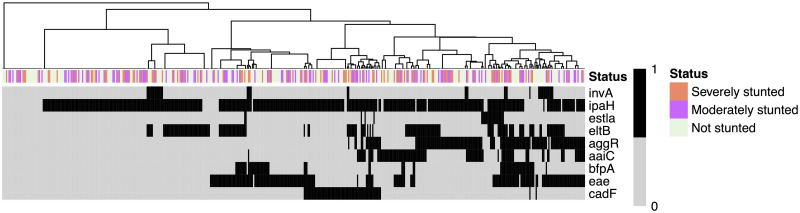
A hierarchical clustering based on a binary distance and Ward’s agglomeration method allowed us to highlight 8 different clusters of pathogen detection profiles (Fig 4). A linear model was then applied to predict if HAZ correlated with the different virulence genes (invA, ipaH, estla, eltB, aggR, aaiC, bfpA, eae and cadF). The model showed a statistically not significant and weak proportion of variance (R2 = 0.03, F(9, 450) = 1.34, p = 0.214, adj. R2 = 6.61e-03). However, within this model, the effect of aaiC was statistically significant and negative (beta = -0.31, 95% CI [-0.58, -0.03], t(450) = -2.19, p < .05).
Fig 4. Heatmap showing gene frequency detected by qPCR and height for age Z (HAZ) scores (x-axis: isolates; y-axis: targeted genes).
The dendrogram fitted on the isolates (i.e. the subjects) were computed based on an asymmetric binary distance coupled with a Ward agglomeration method based on the presence—black color—or absence of the genes—gray color. The clusters derived from the hierarchical clustering were not shown to be significantly associated to HAZ. No clustering was performed on the variables (targeted genes).
Shigella spp. carriage
During the study, we decided to isolate Shigella spp. from the feces collected (N = 143 samples) by cultivating them on Hektoen and XLD media. Very few green or pink colonies grew respectively on Hektoen or XLD [from 41 feces samples] on which 1 representative was isolated for further study. Only two isolates (from HJRA178—HJRA198 samples) were positive (qPCR) and were identified by API20E as Shigella flexneri (ID scores of 69.1 and 82.4%). They were isolated from a severely (HJRA178) and a moderated stunted child (HJRA198), both female, showing no recent clinical sign of diarrhea.
The two isolates from HJRA178—HJRA198 samples were sequenced and their ANI and dDDH values based on full genome sequence regarding the closest relatives’ strains were significantly higher than the recommended cut-off points for species boundary [27] to indicate that they belong to the genus Shigella and the species flexneri with the closest relative being Shigella flexneri ATCC 29903 (S6 Table). In addition, the phylogenomic tree constructed on the TYGS using FastME from the genome blast distance phylogeny (GBDP) [26,27] provided further evidence on the taxonomic position of these strains within the genus Shigella and the species flexneri (S2 Fig). These two isolates clustered with Shigella flexneri ATCC 29903. Genotypic differences between the two isolates were also found based on their MLST, wgMLST and cgMLST and virulence factors (S7 Table).
The two Shigella isolates from HJRA 178 and HJRA 198 stool samples were also referred to the French National Reference Center for Shigella at Institut Pasteur, Paris, France and were serotyped as S. flexneri 1b and S. flexneri 4v, respectively.
As the two Shigella isolates were isolated from feces of asymptomatic stunted children, we investigated whether the two isolates of S. flexneri were able to invade Hep2 cells by using the classical gentamicin protection assay [27]. After incubation and treatment with gentamicin the total number of Hep2 cell- internalized bacteria was calculated and compared to a wild-type (M90T) and an invasion-deficient (mixD) strain. The gentamicin assay was realized in duplicate. Invasion was defined as the total number of intracellular bacteria in cells (extracellular bacteria were killed by gentamicin, a cell-impermeable antibiotic). The HJRA178 isolate (S. flexneri 1b = 3109 in S3 Fig) displayed the same behavior than the invasion-deficient (mixD) mutant and the HJRA198 isolate (S. flexneri 4v = 3110 in S3 Fig) the same behavior than the wild-type (M90T) strain. This confirmed that one of the two strains had intact invasiveness properties while the other one did not. The HJRA178 isolate displayed the Congo red binding (Crb+) phenotype associated the infectivity properties of S. flexneri whereas the HJRA198 isolate did not (Crb-) confirming the data obtained in the gentamicin protection assay.
Discussion
In this study, duodenal aspirates from stunted children (N = 109) were analyzed to characterize the bacterial population in the duodenum of stunted children and to investigate SIBO. Unfortunately, there is currently no single valid test for SIBO, and the accuracy of all current tests remains limited due to the failure of culture to be a gold standard. Even the value of 105 CFU/ml has begun to be questioned by consensus and review of the literature. Some studies suggest normal subjects rarely exceed 103 CFU/ml and that this should be the defining threshold for SIBO [32].
However, if we consider SIBO defined as greater than 105 CFU/ml upper intestinal aspirate as assessed by both anaerobic and aerobic cultures, this study confirms the preliminary results obtained in the Afribiota study [15] showing a very high prevalence of SIBO in stunted children (85.3%) with 58.7% having more than 106 bacteria/ml of duodenal fluid. Studies of children living in shantytowns in South America and Asia have detected SIBO at lower prevalence; from 16% (Bangladesh) to 61% (São Paulo, Brazil) [13,33,34,35]. These high SIBO positivity rates in our study also reflect the fact that the cultivation approach is more sensitive than the breath tests utilized in other studies. The presence of excessive bacteria in the small intestine is typically associated with a malabsorptive syndrome occurring in the context of gut stasis syndromes [36]. It was also suggested that SIBO is associated with intestinal inflammation [13], especially in resource-poor communities where an extreme poverty and unhygienic conditions prevail.
The duodenum and proximal jejunum normally contain small numbers of bacteria (< 104 organisms per mL), usually lactobacilli, enterococci, streptococci (from the Lactobacillales order), gram-positive aerobes such as Veillonella (Firmicutes) or facultative anaerobes from Moraxellaceae and transiently non-oral genera from Enterobacteriaceae [37]. In this study, the most prevalent genera cultivated from the small intestine were Streptococcus, Neisseria, Staphylococcus, Rothia, Haemophilus, Pantoea and Branhamella. Those genera are generally oral taxa [38] and their presence in high numbers in duodenal fluids is uncommon. Our previous study on a relatively small number of stunted children in Antananarivo (Madagascar) and Bangui (CAR) showed a similar microbiota in both the stomach and duodenum of stunted children and a clear over-representation of pathobionts and of oropharyngeal species in the stools suggesting a decompartmentalization of the gastrointestinal tract [15]. We speculate that in these stunted children, gut maintenance declines and microbes can stray from traditional zones, negatively impacting intestinal homeostasis, host health and altered nutriment absorption [15].
In a recent study conducted on stunted undernourished children with enteropathy living in Bangladesh, the authors showed after esophagogastroduodenoscopy and duodenal biopsies a shared group of 14 taxa (not typically classified as enteropathogens) negatively correlated with linear growth and positively correlated with duodenal proteins involved in inflammatory responses [39]. The three most strongly correlated bacteria with the duodenal inflammatory markers were a Veillonella species, a Streptococcus species and Rothia mucilaginosa. Dysbiosis in the oral microbiota, and the significant increase of Veillonella sp. also positively correlated with elevated levels of inflammatory cytokines (IL-1ß, IFN-ɣ, TNF-α, IL-8) and immunoglobulin A in the saliva in patients with autoimmune liver disease [40] and in patients with inflammatory bowel disease (dominant genera: Streptococcus, Prevotella, Neisseria, Haemophilus, Veillonella and Gemella correlating to increased IL-1ß and lysozyme levels) [41]. The collected data emphasizes the possible role of the ectopic colonization in the upper part of the small intestine by some oral bacteria with pro- inflammatory features that could be tested in animal models with our isolated bacteria from the duodenum. Another explanation might be lack or the reduced concentration of gastric acid in the stunted children in LMICs (i.e. due to Helicobacter pylori contamination) which may lead to this bacterial overgrowth since gastric secretions is one of the normal defense mechanisms controlling the intestinal microflora [42]. However, the pH values measured in the stomach samples of our stunted children were low and there were no significant differences in a bivariate analysis for pH and SIBO (S4 Table).
This study also assessed the possible association of SIBO with potential risk factors among the participants (education, possible exposure to contaminants, hygiene, …). Although some studies on SIBO have revealed an association with some risk factors such as open sewer outside the home (OR, 4.78; 95% CI, 1.06 to 21.62) [13], the present study did not find any association between SIBO and the variables assessed. The absence of association could be due to the high prevalence of SIBO in this study (85.3% vs 16.7% and 13.5% for the studies referred to in 34 and 35).
The current research also aimed to assess by a molecular approach the carriage of intestinal pathogens in the feces, one of the important etiological causes which may be associated to EED in the children population. Despite the disadvantages of molecular approaches, such as being unable to determine whether bacteria are still alive or metabolically active [43], they offer a high level of sensitivity. For example, it has been shown that systematic application of multiplex qPCR enhances from 18 to 30% the detection of bacteria, parasites, and viruses in stool samples [44,45]. The current study shows a high prevalence of bacterial enteropathogens, especially those categorized as “enteroinvasive” or causing mucosal disruption such as Shigella spp., ETEC, EPEC and EAEC. Unexpectedly, all these pathogens were detected at similar rate in MS/SS and in C (no statistical differences) and all the children showed no sign of severe diarrhea 15 days before the inclusion. Particularly, Shigella spp. was highly prevalent in the qPCR approach, also confirmed by a conventional PCR approach on a limited number of samples. To assert the presence of living Shigella spp. in the feces samples, isolation was performed at the end of the cross-sectional study directly from stool streaked on XLD and Hektoen plates and only two samples gave rise to two S. flexneri isolates confirmed by whole genome sequence analysis and serotyping. This very low recovery of Shigella isolates could be explained by the fact that no specific enrichment broth exists for Shigella (except Selenite-F [46]) or that there were no-metabolically active Shigella in the samples.
Subclinical infections (an infection in which symptoms are either absent or sufficiently mild to escape diagnosis [47]) or asymptomatic carriage (one who harbors pathogenic organisms without clinically recognizable symptoms and may infect others [48]) of intestinal pathogens seems common in low- and middle-income countries suffering from poor sanitary conditions. For instance, in Rwanda, high proportions of asymptomatic carriage (young healthy controls) were observed for adenovirus (50%), ETEC—based on eltB detection—(47%), EPEC—eae—(23%), Campylobacter (22%) and Shigella—ipaH—(17%) [49]. In children living in the Peruvian amazon, Shigella isolates (S. flexneri accounted for 67.1% of isolates) were also obtained by bacteriological cultures from 3.2% of surveillance stool cultures in the absence of diarrheal illness [50].
Asymptomatic carriage in young children has been attributed to several factors, including breastfeeding and maternal immunity, immunity gained from previous clinical or subclinical infections, and the intestinal microbiome acting as a gut barrier (which affects the likelihood that an enteric pathogen induces disease) [51]. In the case of Shigella it can be also interpreted as a prolonged pathogen excretion after illness (up to 17 months) [50] or a reinfection with the same strain from a very contaminated area with short- to medium term and serotype-specific immunity in children provided following clearance [52]. The carriage of a microorganism with limited pathogenicity could be also considered what could be the case for one of the two Shigella isolates (from the HJRA178 sample) as shown by the genomic analysis of the virulence factors and the inability to invade Hep2 cells. The limited pathogenicity could be explained by the loss of the virulence plasmid harboring the Mxi/Spa secretory apparatus encoded by two operons comprising about 25 genes. However, the other Shigella isolate (from the HJRA198 sample) was fully virulent and invasive suggesting different patterns of infection.
The MAL-ED Network Investigators [11] study showed an association between enteropathogens (mainly Campylobacter and EAEC) in non-diarrheal stools and reduced linear growth (length and weight). The high proportion of enteropathogens in this study, especially Shigella spp. but also ETEC and EAEC, in both populations (stunted and non-stunted), supports the fact that carriage of enteropathogens could not be only associated with the population of stunted children and could prevented us to propose a direct association with stunting. However, in our previous study conducted on a more restricted number of samples by 16S rDNA sequencing, members of Escherichia coli/Shigella and Campylobacter sp. were more prevalent in stunted children compared with non-stunted controls [15]. Our data suggest that, beside combatting poverty and improving diet, environmental sanitation, quality of water sources, hygiene promotion and health education are key points to mitigate stunting and restore nutritional benefits even if a recent study provided evidence for the important role of timing of stunting on the recovery from the phenomenon rather than WASH practices [53].
Limitations
This study has however some limitations such as a single run of qPCR amplification on individual samples. Moreover, the integration of qPCR amplification results for enteric parasites [43] and viral infections was not considered either.
Regarding the duodenal sampling technique via nasogastric tube, we ensured the first ml of aspiration from both stomach and duodenum were discarded in order to flush out possible contaminating bacteria and carry-over from the more proximal compartments.
This study was not intended to directly measure evidence of EED but two investigate two major–possibly combined—etiologies, which may account for EED: SIBO and exposure to enteropathogens.
Supporting information
Distributions of the CFU in duodenal aspirates with SIBO matching with feces not contaminated by Campylobacter spp. [0 (in blue)]; and in duodenal aspirates with SIBO matching with feces contaminated by Campylobacter spp. [1 (in yellow)].
(TIF)
The phylogenomic tree includes the two Shigella isolates from HJRA178—HJRA198 samples constructed on the Type Strain Genome Server (TYGS) (at https://tygs.dsmz.de) using FastME from the genome blast distance phylogeny (GBDP). [26,27].
(TIF)
(TIF)
(XLSX)
(XLSX)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
Acknowledgments
We wish to thank all participating families, the AFRIBIOTA Consortium, the participating hospitals in Antananarivo, as well as the Institut Pasteur, the Institut Pasteur de Madagascar, and members of the scientific advisory board for their continuous support; Prof. Jean-Louis Demarquez for training sessions to teach the local health professionals the methods used for duodenal aspirations; Aurélie Etienne for precious help with the clinical procedures and first aspirations performed; the field workers Tseheno Harisoa and Rado Andrianantenaina, as well as all implicated community health workers, for countless hours spent in the field; the Centre de Recherche Translationelle and the Direction Internationale of the Institut Pasteur, and especially Paméla Palvadeau, Jane Lynda Deuve, Marc Rovatiana Ranarijesy, Kanto Liantsoa Razanakolona, Cécile Artaud, Nathalie Jolly, Sophie Jarrijon, Mamy Ratsialonina, Jean-François Damaras, Marie-Noelle Ungeheuer, and Laurence Arowas for precious help in setting up and steering the AFRIBIOTA project and managing the funds and the biobank. We would like to thank the staff of the “Plateforme de Microbiologie Mutualisée (P2M)” at Institut Pasteur Paris where the whole genome sequencing was performed, Sophie Lefevre from the Centre National de Référence des Escherichia coli, Shigella et Salmonella in Paris, France for Shigella serotyping, and Claude Parsot and Alexandre Grassart for the interesting discussions on Shigella.
Thanks are also due to Daniel Falush and Khashayar Shahin for carefully reading and commenting the manuscript.
Data Availability
All relevant data used for the analyses are within the manuscript and Supporting information files.
Funding Statement
This project was funded by the Total and Petram Foundations and by Institut Pasteur. LA, PA and RR wages were supported by the Total Foundation and MR wage by the Petram Foundation. PV was supported by an Early Postdoctoral Fellowship (P2EZP3_152159), an Advanced Postdoctoral Fellowship (P300PA_177876) as well as a Return Grant (P3P3PA_17877) from the Swiss National Science Foundation, a Roux-Cantarini Fellowship (2016), a L'Oréal-UNESCO for Women in Science France Fellowship (2017) and an Excellence Scholarship from the University of Basel (Forschungsfonds, 2019). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.UNICEF, Madagascar, Programme Nutrition. https://www.unicef.org/madagascar/en/programme/nutrition. Last accession February 6th, 2020.
- 2.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–21. doi: 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keusch GT, Rosenberg IH, Denno DM, Duggan C, Guerrant RL, Lavery JV, et al. Implications of acquired environmental enteric dysfunction for growth and stunting in infants and children living in low- and middle-income countries. Food Nutr Bull. 2013;34(3):357–64. doi: 10.1177/156482651303400308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keusch GT, Denno DM, Black RE, Duggan C, Guerrant RL, Lavery JV, et al. Environmental enteric dysfunction: pathogenesis, diagnosis, and clinical consequences. Clin Infect Dis. 2014;59 Suppl 4:S207–12. doi: 10.1093/cid/ciu485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane RJ, Jones KD, Berkley JA. Environmental enteric dysfunction: an overview. Food Nutr Bull. 2015. Mar;36(1 Suppl):S76–87. doi: 10.1177/15648265150361S113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tickell KD, Atlas HE, Walson JL. Environmental enteric dysfunction: a review of potential mechanisms, consequences and management strategies. BMC Med. 2019. Nov 25;17(1):181. doi: 10.1186/s12916-019-1417-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay S, Gaudier E, Campbell DI, Prentice AM, Albers R. Environmental enteropathy: new targets for nutritional interventions. Int Health. 2010;2(3):172–80. doi: 10.1016/j.inhe.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 8.George CM, Burrowes V, Perin J, Oldja L, Biswas S, Sack D, et al. Enteric Infections in Young Children are Associated with Environmental Enteropathy and Impaired Growth. Trop Med Int Health. 2018;23(1):26–33. doi: 10.1111/tmi.13002 [DOI] [PubMed] [Google Scholar]
- 9.Rogawski ET, Guerrant RL, Havt A, Lima IFN, Medeiros PHQS, Seidman JC, et al. Epidemiology of enteroaggregative Escherichia coli infections and associated outcomes in the MAL-ED birth cohort. PLoS Negl Trop Dis. 2017;11(7):e0005798. doi: 10.1371/journal.pntd.0005798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MAL-ED Network Investigators. Relationship between growth and illness, enteropathogens and dietary intakes in the first 2 years of life: findings from the MAL-ED birth cohort study. BMJ Glob Health. 2017;2(4):e000370. doi: 10.1136/bmjgh-2017-000370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosek MN; MAL-ED Network Investigators. Causal Pathways from Enteropathogens to Environmental Enteropathy: Findings from the MAL-ED Birth Cohort Study. EBioMedicine. 2017. Apr;18:109–117. Epub 2017 Mar 8. doi: 10.1016/j.ebiom.2017.02.024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dukowicz AC, Lacy BE, Levine GM. Small intestinal bacterial overgrowth: a comprehensive review. Gastroenterol Hepatol (N Y). 2007. Feb;3(2):112–22. . [PMC free article] [PubMed] [Google Scholar]
- 13.Donowitz JR, Haque R, Kirkpatrick BD, Alam M, Lu M, Kabir M et al. Small Intestine Bacterial Overgrowth and Environmental Enteropathy in Bangladeshi Children. mBio. 2016;7(1):e02102–e2115. doi: 10.1128/mBio.02102-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vonaesch P, Randremanana R, Gody JC, Collard JM, Giles-Vernick T, Doria M et al. Identifying the etiology and pathophysiology underlying stunting and environmental enteropathy: study protocol of the AFRIBIOTA project. BMC Pediatr. 2018;18(1):236. doi: 10.1186/s12887-018-1189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vonaesch P, Morien E, Andrianonimiadana L, Sanke H, Mbecko JR, Huus KE et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc Natl Acad Sci U S A. 2018;115(36):E8489–E8498. doi: 10.1073/pnas.1806573115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO Multicentre Growth Reference Study Group (2021) WHO Child Growth Standards, World Health Organization, Geneva. https://www.who.int/tools/child-growth-standards/who-multicentre-growth-reference-study
- 17.Chandra S, Dutta U, Noor MT, Taneja N, Kochhar R, Sharma M et al. Endoscopic jejunal biopsy culture: a simple and effective method to study jejunal microflora. Indian J Gastroenterol. 2010;29(6):226–230. doi: 10.1007/s12664-010-0072-6 [DOI] [PubMed] [Google Scholar]
- 18.Langendorf C, Le Hello S, Moumouni A, Gouali M, Mamaty AA, Grais RF et al. Enteric bacterial pathogens in children with diarrhea in Niger: diversity and antimicrobial resistance. PLoS One. 2015. Mar 23;10(3):e0120275. doi: 10.1371/journal.pone.0120275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elfving K, Andersson M, Msellem MI, Welinder-Olsson C, Petzold M, Björkman A, et al. Real-time PCR threshold cycle cutoffs help to identify agents causing acute childhood diarrhea in Zanzibar. J Clin Microbiol. 2014;52(3):916–23. doi: 10.1128/JCM.02697-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, et al. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51(2):472–80. doi: 10.1128/JCM.02658-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.EU protocol for outbreak investigation. Detection of Enteroaggregative Escherichia coli in food by Real Time PCR amplification of the aggR and aaiC genes. EU RL_Method_05_Rev 1. 05/10/2013. EU Reference Laboratory for E. coli. Istituto Superiore di Sanità.
- 22.Sjöling Å, Sadeghipoorjahromi L, Novak D, Tobias J. Detection of major diarrheagenic bacterial pathogens by multiplex PCR panels. Microbiol Res. 2015;172:34–40. doi: 10.1016/j.micres.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 23.Phantouamath B, Sithivong N, Insisiengmay S, et al. Pathogenicity of Shigella in healthy carriers: a study in Vientiane, Lao People’s Democratic Republic. Jpn J Infect Dis. 2005;58(4):232–234. [PubMed] [Google Scholar]
- 24.Simo Tchuinte PL, Rabenandrasana MAN, Kowalewicz C, Andrianoelina VH, Rakotondrasoa A, Andrianirina ZZ et al. Phenotypic and molecular characterisations of carbapenem-resistant Acinetobacter baumannii strains isolated in Madagascar. Antimicrob Resist Infect Control. 2019;8:31. doi: 10.1186/s13756-019-0491-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS et al. A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 2007;57:81–91. doi: 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- 27.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2016;10:2182. doi: 10.1038/s41467-019-10210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefort V, Desper R, Gascuel O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farris JS. Estimating phylogenetic trees from distance matrices. Am Nat. 1972; 106:645–668. [Google Scholar]
- 31.Sharma A, Puhar A. Gentamicin Protection Assay to Determine the Number of Intracellular Bacteria during Infection of Human TC7 Intestinal Epithelial Cells by Shigella flexneri. Bio Protoc. 2019;9(13):e3292. doi: 10.21769/BioProtoc.3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachdev AH, Pimentel M. Gastrointestinal bacterial overgrowth: pathogenesis and clinical significance. Ther Adv Chronic Dis. 2013. Sep;4(5):223–31. doi: 10.1177/2040622313496126 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khin-Maung-U, Bolin TD, Duncombe VM, Myo-Khin, Nyunt-Nuynt-Wai, Pereira SP et al. Epidemiology of small bowel bacterial overgrowth and rice carbohydrate malabsorption in Burmese (Myanmar) village children. Am J Trop Med Hyg. 1992;47(3):298–304. doi: 10.4269/ajtmh.1992.47.298 [DOI] [PubMed] [Google Scholar]
- 34.dos Reis JC, de Morais MB, Oliva CA, Fagundes-Neto U. Breath hydrogen test in the diagnosis of environmental enteropathy in children living in an urban slum. Dig Dis Sci. 2007;52(5):1253–1258. doi: 10.1007/s10620-006-9288-9 [DOI] [PubMed] [Google Scholar]
- 35.Mello CS, Rodrigues MSDC, Filho HBA, Melli LCFL, Tahan S, Pignatari ACC et al. Fecal microbiota analysis of children with small intestinal bacterial overgrowth among residents of an urban slum in Brazil. J Pediatr (Rio J). 2018;94(5):483–490. doi: 10.1016/j.jped.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 36.Adike A, DiBaise JK. Small Intestinal Bacterial Overgrowth: Nutritional Implications, Diagnosis, and Management. Gastroenterol Clin North Am. 2018. Mar;47(1):193–208. Epub 2017 Dec 7. doi: 10.1016/j.gtc.2017.09.008 . [DOI] [PubMed] [Google Scholar]
- 37.Kastl AJ Jr, Terry NA, Wu GD, Albenberg LG. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell Mol Gastroenterol Hepatol. 2020;9(1):33–45. doi: 10.1016/j.jcmgh.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caselli E, Fabbri C, D’Accolti M, Soffritti I, Bassi C, Mazzacane S, Franchi M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 2020. May 18;20(1):120. doi: 10.1186/s12866-020-01801-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen RY, Kung VL, Das S, Hossain MS, Hibberd MC, Guruge J, et al. Duodenal Microbiota in Stunted Undernourished Children with Enteropathy. N Engl J Med. 2020;383(4):321–333. doi: 10.1056/NEJMoa1916004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe K, Takahashi A, Fujita M, Imaizumi H, Hayashi M, Okai K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS One. 2018;13(7):e0198757. doi: 10.1371/journal.pone.0198757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014;21(1):15–25. doi: 10.1093/dnares/dst037 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gracey M, Cullity GJ, Suharjono, Sunoto. The stomach in malnutrition. Arch Dis Child. 1977. Apr;52(4):325–7. doi: 10.1136/adc.52.4.325 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platts-Mills JA, Liu J, Houpt ER. New concepts in diagnostics for infectious diarrhea. Mucosal Immunol. 2013;6(5):876–85. doi: 10.1038/mi.2013.50 [DOI] [PubMed] [Google Scholar]
- 44.McAuliffe GN, Anderson TP, Stevens M, Adams J, Coleman R, Mahagamasekera P et al. Systematic application of multiplex PCR enhances the detection of bacteria, parasites, and viruses in stool samples. J Infect. 2013;67(2):122–9. doi: 10.1016/j.jinf.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 45.Enserink R, Scholts R, Bruijning-Verhagen P, Duizer E, Vennema H, de Boer R, et al. High detection rates of enteropathogens in asymptomatic children attending day care. PLoS One. 2014;9(2):e89496. doi: 10.1371/journal.pone.0089496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chain Network. CHN56. SOP: Identification of E. coli, Salmonella and Shigella isolates from stool samples. https://chainnetwork.org/wp-content/uploads/2018/11/CHAIN-Ecoli-Salmonella-Shigella-Isolation-SOP.pdf Last access 2021.09.06.
- 47.Quilliam RS, Cross P, Williams AP, Edwards-Jones G, Salmon RL, Rigby D, et al. Subclinical infection and asymptomatic carriage of gastrointestinal zoonoses: occupational exposure, environmental pathways, and the anonymous spread of disease. Epidemiol Infect. 2013;141(10):2011–21. doi: 10.1017/S0950268813001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stedman’s Medical Dictionary for the Dental Professions, 2011, Second Edition, Ed. Wolters Kluver & Lippincott, Williams & Wilkins, pp. 704
- 49.Kabayiza JC, Andersson ME, Welinder-Olsson C, Bergström T, Muhirwa G, Lindh M. Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infect Dis. 2013;13:447. doi: 10.1186/1471-2334-13-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosek M, Yori PP, Pan WK, Olortegui MP, Gilman RH, Perez J, et al. Epidemiology of highly endemic multiply antibiotic-resistant shigellosis in children in the Peruvian Amazon. Pediatrics. 2008;122(3):e541–9. doi: 10.1542/peds.2008-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogt SL, Finlay BB. Gut microbiota-mediated protection against diarrheal infections. J Travel Med 2017;24 (suppl 1): S39–43. doi: 10.1093/jtm/taw086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine MM, DuPont HL, Khodabandelou M, Hornick RB. Long-term Shigella-carrier state. N Engl J Med. 1973;288(22):1169–71. doi: 10.1056/NEJM197305312882207 [DOI] [PubMed] [Google Scholar]
- 53.Das S, Fahim SM, Alam MA, Mahfuz M, Bessong P, Mduma E, et al. Not water, sanitation and hygiene practice, but timing of stunting is associated with recovery from stunting at 24 months: results from a multi-country birth cohort study. Public Health Nutr. 2021;24(6):1428–1437. doi: 10.1017/S136898002000004X [DOI] [PMC free article] [PubMed] [Google Scholar]