Abstract
Development of the craniofacial structures requires the precise differentiation of cranial neural crest cells into osteoblasts or chondrocytes. Here, we explore the epigenetic and non-epigenetic mechanisms that are required for the development of craniofacial chondrocytes. We previously demonstrated that the acetyltransferase activity of the highly conserved acetyltransferase GCN5, or KAT2A, is required for murine craniofacial development. To further test the potential cell autonomous function, we hypothesize that GCN5 is required for chondrocyte development following the arrival of the cranial neural crest within the pharyngeal arches. Here, we show that Gcn5 is required cell autonomously in the cranial neural crest. Using a combination of in vivo and in vitro inhibition of GCN5 acetyltransferase activity, we demonstrate that GCN5 is a potent activator of chondrocyte maturation, acting to control chondrocyte maturation and size increase during pre-hypertrophic maturation to hypertrophic chondrocytes. Rather than acting as an epigenetic regulator of histone H3K9 acetylation, our findings suggest GCN5 primarily acts as a non-histone acetyltransferase to regulate chondrocyte development. Here, we investigate the contribution of GCN5 acetylation to the activity of the mTORC1 pathway. Our findings indicate that GCN5 acetylation is required for activation of this pathway, either via direct activation of mTORC1 or through indirect mechanisms. We also investigate one possibility of how mTORC1 activity is regulated through RAPTOR acetylation, which is hypothesized to enhance mTORC1 downstream phosphorylation. This study contributes to our understanding of the specificity of acetyltransferases, and the cell type specific roles in which these enzymes function.
Keywords: Craniofacial development, neural crest, acetyltransferase, GCN5, Kat2a, chondrocyte
Introduction:
Embryonic development of the craniofacial structures is a complex process, requiring the migration and post-migratory differentiation of cranial neural crest cells (cNCC) for the formation of bones such as the mandible and maxilla. These structures are critical for vertebrate function and survival, as neurocristopathies are a prevalent birth defect worldwide (about 1 in 1000 live births [1]). Thus, there is a critical need to understand the molecular mechanisms that control craniofacial development. In mice, cNCC migrate into the pharyngeal arches, and post-migration these cells differentiate towards different terminal fates [2]. cNCC contribute to craniofacial bone via either an intramembranous or endochondral ossification pathway and also differentiate into cartilage of the head and neck, as well as glia [3]. Transcriptional regulatory mechanisms are a fundamental process of cell type specific transcriptional control, and numerous transcription factors and enhancers have been identified that specifically govern the migration and specification of cNCC, forming a gene regulatory network [4]. However, considerably less is known about the post-translational modifications that are required for post-migratory differentiation of cNCC.
We previously described that the acetyltransferase activity of GCN5, or KAT2A, is required for murine and zebrafish craniofacial development [5]. Using a catalytically inactive allele of Gcn5, named Gcn5hat [6], we identified several craniofacial defects in embryonic day E16.5 homozygous Gcn5hat/hat mutant mouse embryos, including hypoplasia of the mandible, and hypoplastic cartilage templates including Meckel’s cartilage and cartilage of the malleus and incus. Analysis of the specification and migratory marker AP2 was unchanged in Gcn5hat/hat mutants as compared to wildtype, suggesting that specification and migration of cNCC was not impaired. We thus determined that GCN5 acetyltransferase activity is not required for cNCC epithelial-to-mesenchymal transition and migration or early cNCC specification, suggesting that GCN5 acts at later stages of cNCC development.
Craniofacial cartilage that is derived from the cNCC can either become permanent cartilage structures, such as the cartilage rings of the trachea and nasal capsule cartilage, or cNCC can differentiate into the cartilage primordia that will be replaced by bone via endochondral ossification, as is the case with the middle ear bones [7]. While the mechanisms that direct the formation of permanent cartilage versus cartilage that will be converted to endochondral bone are not clear, cNCC nevertheless require the expression of the transcription factor SOX9 in order to differentiate into chondrocytes and form cartilage. cNCC-specific loss of Sox9 in mice results in a complete absence of facial cartilage and endochondral craniofacial bones [8]. Downstream targets of SOX9 that are required for skeletal growth and development have been extensively described in studies of development of the long bones, which form through endochondral ossification. Two SOX9 targets required for proper chondrocyte development are the extracellular matrix proteins encoded by the genes Acan and Col2a1 [9]. The expression of both of these extracellular matrix proteins is considered a hallmark of pre-hypertrophic chondrocytes [10]. These chondrocytes then progress to mature hypertrophic chondrocytes that express high levels of Mmp13 and Col10a1, which encode matrix metalloproteinase 13 and collagen 10. While the temporal progression of these markers has been established in chondrocyte maturation, the epigenetic and non-epigenetic regulatory mechanisms that regulate these steps are not fully elucidated.
Epigenetic and non-epigenetic protein modifications provide additional layers of transcriptional regulation, serving to turn on or off signaling pathways and RNA transcription in response to extracellular or intracellular signaling [11]. Of the many protein modifiers known, the acetyltransferase GCN5 has emerged as a prominent player in cell signaling, participating both epigenetically and non-epigenetically in pathways such as retinoic acid signaling [12]; the TGFβ pathway [13, 14]; FGF signaling [15]; and Notch signaling [16]. In addition to non-epigenetic protein targets, GCN5 also has known epigenetic targets— the best characterized being histone H3 lysine 9 acetylation (H3K9ac) [17, 18]. H3K9ac is extensively described as a mark of active transcription, and H3K9ac histone modification assists in the opening of compacted chromatin for transcriptional machinery [19]. ChIP-seq studies support GCN5 localization to loci of active transcription [20] and interaction with RNA polymerase machinery in yeast [21]. Further studies have also revealed that GCN5, as part of its protein complex ATAC, also binds with enhancers that are distinct from those bound by the closely related p300 acetyltransferase [22]. Despite the role of GCN5 in H3K9ac, our previous results indicated that loss of GCN5 activity does not significantly affect global H3K9ac levels in the embryonic craniofacial cells. Furthermore, here we present evidence that GCN5 is not required for H3K9ac at Acan enhancers specific for pre-hypertrophic chondrocytes. These results suggest a non-epigenetic role for GCN5 in craniofacial development.
Here, we turn our attention to the maturation and development of the cNCC derived chondrocytes. Despite disparate cellular origins, endochondral ossification of the craniofacial bones appears to resemble endochondral ossification of the limb bones and is directed by many of the same signaling mechanisms. The mTOR signaling pathway has been described as an important regulator of chondrocyte growth and skeletal development in long bone formation and is a putative target for GCN5 acetylation [23–26]. These studies identified downstream targets of mTOR complex 1 (mTORC1) signaling as being active in pre-hypertrophic chondrocytes and required for long bone skeletogenesis. We describe here that inhibition of GCN5 disrupts the mTORC1 pathway during cNCC chondrocyte maturation. Our findings present a non-epigenetic role for GCN5 in craniofacial bone and cartilage development, acting as an activator that is required for craniofacial chondrocyte maturation.
Results:
GCN5 is required cell autonomously in cranial neural crest cells
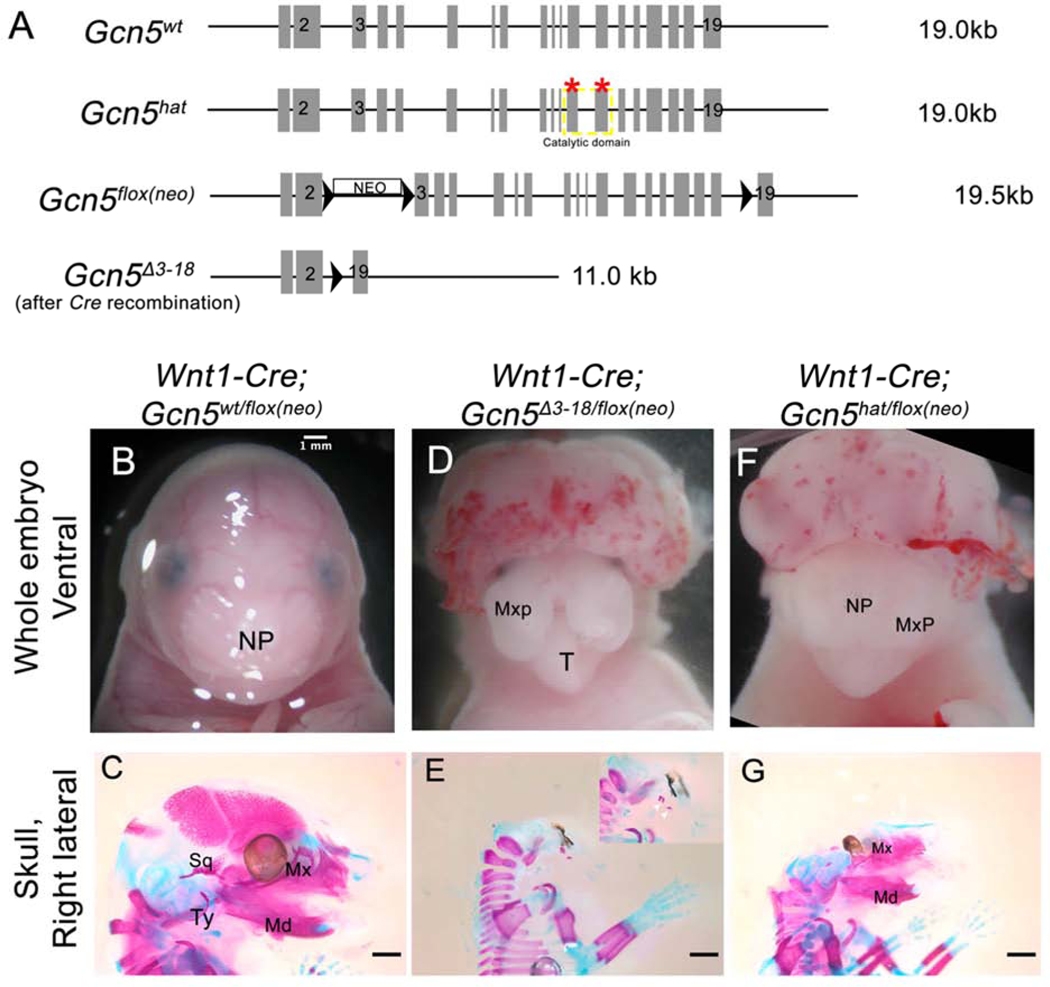
Our previous studies showed that GCN5 acetyltransferase activity is required for proper formation of the craniofacial bones and cartilage. However, as this was a ubiquitous knock-out of GCN5 acetyltransferase (HAT) activity, it is unclear in which tissue(s) GCN5 activity is needed. To test the requirement for GCN5 in cNCC, we used the Wnt1-Cre allele, which drives Cre recombinase in cNCC [27], in combination with a floxed allele of Gcn5, Gcn5flox(neo) [28], which contains three LoxP sites flanking the coding sequence of Gcn5, allowing for complete deletion of Gcn5 coding sequence from exon 3 to 18 in the presence of Cre recombinase, named here Gcn5Δ3−18 (Figure 1A). First we created a global Gcn5Δ3−18 deletion using the germline Sox2-Cre [29] because the Gcn5flox(neo) allele is hypomorphic and we were not able to raise viable homozygous Gcn5flox(neo)/flox(neo) mice [30]. Gcn5Δ3−18/+ mice were crossed to Wnt1-Cre to create compound heterozygous Wnt1-Cre; Gcn5Δ3−18/+ mice. These animals were then crossed with Gcn5flox(neo)/+ mice to generate embryos with the genotype Wnt1-Cre; Gcn5Δ3−18/flox(neo) wherein GCN5 function is lost in the neural crest. Embryos with the genotype Wnt1-Cre; Gcn5flox(neo)/+ were used as controls. Out of 30 total embryos (genotypes and phenotypes are listed in Supplemental Table 1), 3 embryos with the genotype Wnt1-Cre; Gcn5Δ3−18/flox(neo) had severe craniofacial defects, including severe neural tube closure defects, clefting of the nasal prominence as well as the maxilla and palate, and absence of the mandible (Figure 1D). Skeletal staining showed that almost all of the craniofacial skeleton was absent, including the mandible and Meckel’s cartilage, with only a few small bony elements underneath the eye (Figure 1E inset, white arrowheads). These results support our hypothesis that GCN5 is required cell autonomously in the cNCC.
Figure 1. Conditional deletion of Gcn5 using Wnt1-Cre results in craniofacial defects.
(A) Representation of Gcn5 alleles (adapted from [6]: red stars indicate two point mutations that abolish GCN5 acetyltransferase activity in Gcn5hat, the catalytic domain of GCN5 is outlined with dashed yellow box; Gcn5flox(neo) conditional allele has three LoxP sites (black arrowheads) and a neomycin cassette between exons 2 and 3; Gcn5Δ3−18 is the deletion allele following Cre recombinase. (B, D, F) Whole mount images of E16.5 wild type (B) and Wnt1-Cre neural crest conditional mutants (D, F). (C, E, G) Right lateral views of craniofacial skeleton of E16.5 wild type (C) and Gcn5 conditional mutants (E, G) stained with alcian blue (cartilage) and alizarin red (bone). Embryos in (C-G) represent separate embryos from (B-F). Inset in (E) shows higher magnification, white arrowheads point to hypoplastic bone. NP, Nasal Prominence; Mxp, Maxillary prominence; T, Tongue; Md, Mandible; Mx, Maxilla; Sq, Squamosal; Ty, Tympanic ring. White and black scale bars represent 1mm.
To determine if the acetyltransferase activity of GCN5 is required cell autonomously in cNCC for craniofacial development, we crossed the HAT mutation, that results in full length GCN5 protein which is enzymatically inactive and cannot acetylate both histone and non-histone proteins, with Wnt1-Cre. We identified embryos of the genotype Wnt1-Cre; Gcn5hat/flox(neo) as conditional mutants with a loss of GCN5 protein and GCN5 HAT activity in the cNCC. Out of 19 total embryos, we identified 3 embryos with the genotype Wnt1-Cre; Gcn5hat/flox(neo) at the expected frequency (Supplemental Table 2). Wnt1-Cre; Gcn5hat/flox(neo) embryos had a neural tube closure defect and a less severe craniofacial defect than Wnt1-Cre; Gcn5Δ3−18/flox(neo) mutant embryos, which we expected as the HAT allele still generates full-length protein [6]. Gross observation of Wnt1-Cre; Gcn5hat/flox(neo) embryos did not show obvious craniofacial defects nor cleft of the nasal prominence or the palatal shelves (Figure 1F). However, alizarin red (bone) and alcian blue (cartilage) staining of Wnt1-Cre; Gcn5hat/flox(neo) embryos did reveal several defects in the mandibular portion of the craniofacial skeleton (Figure 1G). The angular, condylar, and coronoid processes were consistently diminished on both sides of the mandible. The squamosal and sphenoid bones were also very hypoplastic. In addition, the malleus and incus cartilage templates in mutants were also hypoplastic compared to controls (Figure 2A, B). In contrast, the premaxilla and maxilla bones appeared to be normal in shape and size. Lastly, it should be noted that exencephaly was observed in mutants that carried alleles of both genotypes, Gcn5hat/flox(neo) or Gcn5Δ3−18/flox(neo), although exencephaly was incompletely penetrant and craniofacial defects were observed in embryos with normal neural tube closure. Previous reports from the Dent lab also reported weakly penetrant exencephaly with these allelic combinations [30]. Taken together, these observations support our hypothesis that GCN5 enzymatic activity is required cell autonomously during cNCC development.
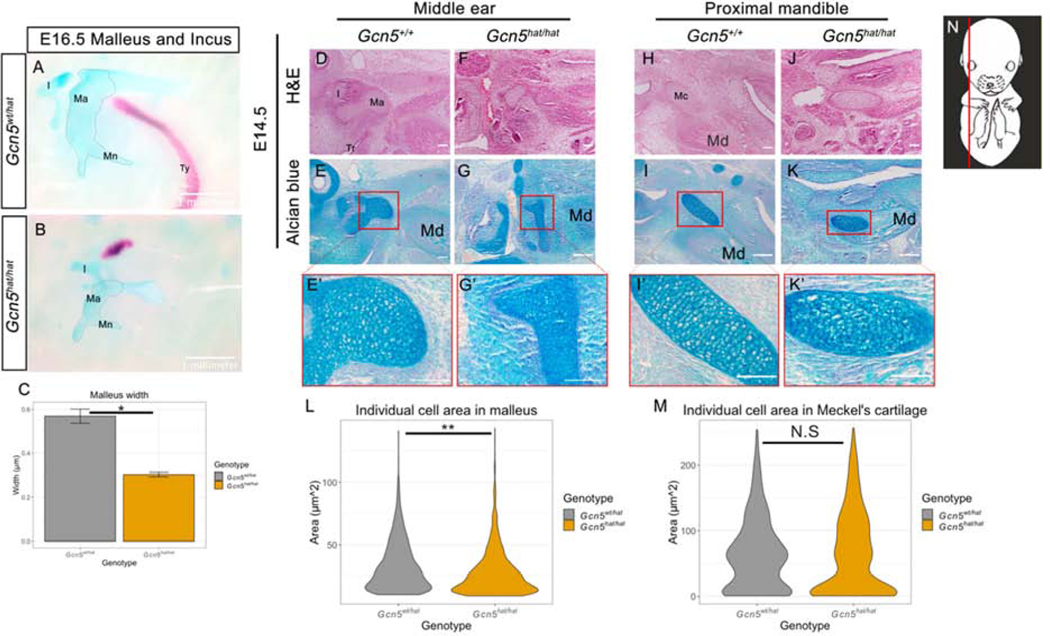
Figure 2. Histological staining reveals hypoplastic craniofacial cartilage primordia in Gcn5hat/hat mutants.
(A, B) Whole mount malleus and incus stained with alcian blue and alizarin red dissected from E16.5 Gcn5wt/hat (A) and Gcn5hat/hat (B) embryos. (C) Quantification of malleus width, multiple measurements taken from right and left ear ossicles from one embryo per genotype. (D, F) Hematoxylin and Eosin (H&E) staining of middle ear sagittal sections from E14.5 control (D) and Gcn5hat/hat embryos (F). (E, G) Alcian blue (AB) staining of E14.5 control (E) and Gcn5hat/hat middle ear sagittal sections (G). (E’, G’) Higher magnification of red box shown in (E, G). (H, J) H&E staining of mandible sagittal sections from E14.5 control (H) and Gcn5hat/hat mutant (J). (I, K) AB staining of E14.5 control (I) and Gcn5hat/hat (K) mandible sagittal sections. (I’, K’) Higher magnification of red box shown in (I, K). (L) Quantification of individual chondrocyte cell area within malleus for wild type and Gcn5hat/hat mutants. Y axis shows area in μm2. (M) Quantification of individual cell area within Meckel’s cartilage for wild type and Gcn5hat/hat mutants. Y axis shows area in μm2. (N) Representation of plane of sagittal sections used for histological analyses, adapted from EMAP eMouse Atlas Project [60]. Ma, Malleus; I, Incus; Sc, Semicircular canal; Tr, Tubo tympanic recess; Ty, Tympanic ring; Mc, Meckel’s Cartilage; Md, Mandible. Scale bars represent 100μm. N = 3 biological replicates for measurements in (L) and (M). P < 0.05 considered significant. * represents P < 0.05; ** represents P < 0.005. Error bars represent standard error of the mean.
Craniofacial cartilage primordia are hypoplastic in Gcn5hat mutants
To explore the molecular mechanisms of GCN5 enzymatic activity in cNCC derivatives, the following studies use in vivo and in vitro approaches. Gcn5hat/hat homozygous mutants have multiple hypoplastic cartilage structures, while the intramembranous bones are affected with less severity [5]. We therefore hypothesized that GCN5 HAT activity is required for chondrocyte maturation and growth during endochondral bone formation. Chondrocyte development in both craniofacial skeleton and long bone formation initiates by condensation of the mesenchymal cells and then proceeds through multiple steps of differentiation. Therefore, we turned our attention to early stages of craniofacial endochondral bone development in the Gcn5hat murine model. Craniofacial bones that form via endochondral ossification include the malleus, incus, and stapes. During development, the malleus forms at the proximal end of Meckel’s cartilage, and is initially attached until post-natal development [31]. Analysis of E16.5 Gcn5hat/hat skeletal preparations revealed that the malleus is hypoplastic at later stages, with the width of the malleus reduced to half the size of control (Figure 2A, B, C). Despite the hypoplasia of the malleus, the overall identity of the ossicle did not appear to be disrupted.
Our observations of skeletal defects in the E16.5 Gcn5hat/hat embryo turned our attention to the role of GCN5 during earlier stages of chondrocyte development. During the timeline of chondrocyte development, cells change their shape and size in a well characterized manner that can be observed histologically [32]. Serial sagittal sections from the mandibular and middle ear regions of E14.5 control and Gcn5hat/hat mutant embryos were stained with either hematoxylin and eosin (H&E) or alcian blue (AB) to stain the chondrocyte extracellular matrix (Figure 2). Sections were matched by the location of the eye and semi-circular canals. By at least E14.5 in control embryos, the cartilage primordium that will form the middle ear bones are visible in the proximal region of the mandibular prominence, with hypertrophic chondrocytes observed as AB positive cells, with large cytoplasm and cell size (Figure 2D, E, E’). Hypertrophic chondrocytes are also visible in E14.5 control embryos in Meckel’s cartilage, which forms proximal to distal within the mandible (Figure 2H, I, I’). In E14.5 Gcn5hat/hat mutants, the cartilage primordium of two middle ear ossicles, the malleus and incus, was hypoplastic (Figure 2G, G’). In control E14.5 embryos, the body of the malleus is large and rounded, and can easily be followed from Meckel’s cartilage, with the manubrium visibly extending ventrally and the head of the malleus extending towards the incus (Figure 2E). In Gcn5hat/hat embryos, the body of the malleus is visibly narrower and much less rounded than that of control embryos (Figure 2F, G, G’). Despite the narrow body of the malleus in Gcn5hat/hat embryos, patterning of the ossicle did not appear to be disrupted, as the ventrally extending manubrium and extending head of the malleus were visible (Figure 2G, G’). However, the incus in Gcn5hat/hat embryos could not be easily identified. Quantification of individual cell area within the malleus at E14.5 was also significantly reduced in Gcn5hat/hat mutants compared to controls (Figure 2L). However, quantification of cell area within Meckel’s cartilage did not show a significant change in Gcn5hat/hat mutants compared to controls (Figure 2J, K, K’, M and Supplemental Figure 1). Overall, the histological defects in mutant craniofacial chondrocytes, in particular the middle ear bones, suggest that cartilage primordium shape and cell size increase is impaired, parameters that reflect a defect in chondrocyte maturation to hypertrophy.
GCN5 acetylation is required for the maturation of cNCC derived chondrocytes
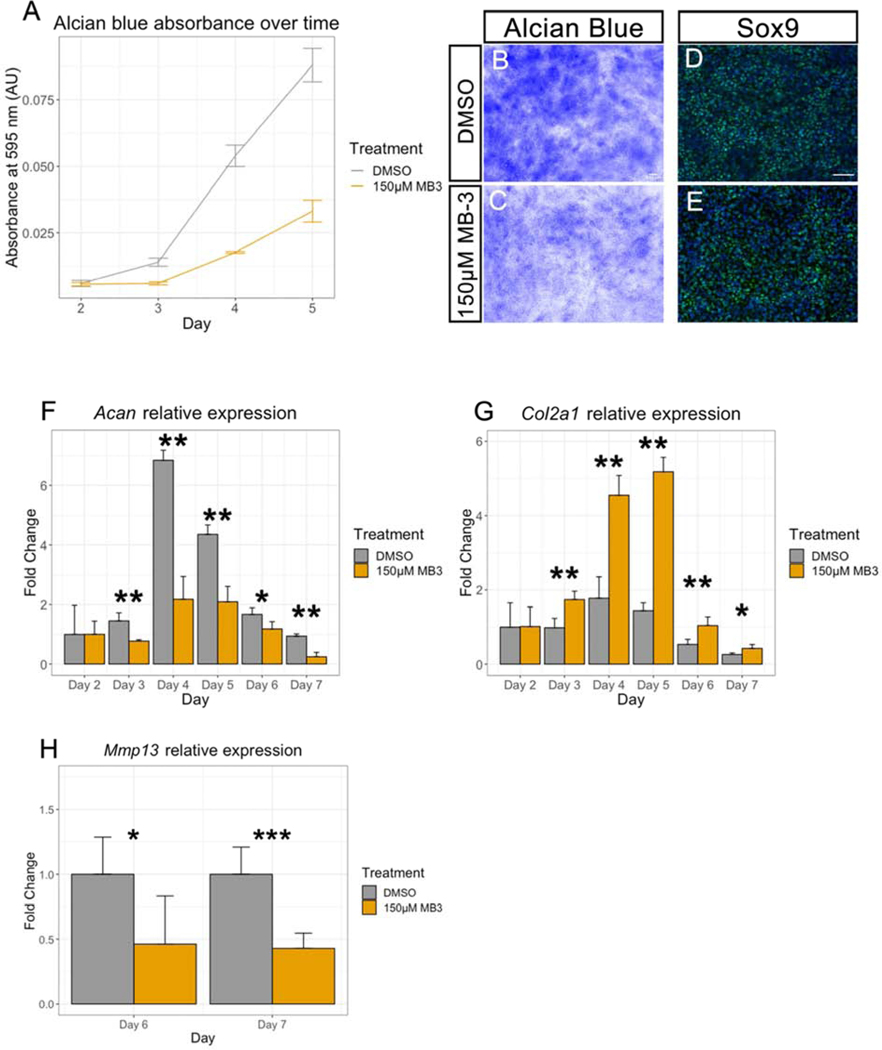
Our in vivo observations suggest that GCN5 HAT activity is required for the growth and size increase of cNCC derived chondrocytes as pre-hypertrophic chondrocytes mature to hypertrophy and ultimately form the endochondral bones. To test this hypothesis further, we turned to an in vitro micromass culture method to recapitulate chondrocyte development and identify which stage of chondrocyte development is affected. During chondrocyte development, chondrocytes will change their transcriptional profile as they reach maturation, allowing us to assess the different stages of chondrocyte maturation. Micromass cultures were performed using the cNCC cell line O9–1 [33]. As we are not aware of a timeline of cNCC micromass differentiation relative to markers of pre-hypertrophic and hypertrophic differentiation, we first established a baseline of in vitro maturation. Beginning with plating on day 0, there was a visible condensation of the mesenchymal cells by day 1, an increase in AB staining between days 3–4 with further increase to day 5, the expression of pre-hypertrophic markers Col2a1 and Acan by day 4, and the mature Mmp13 hypertrophic chondrocyte marker detectable by day 6 (Figure 3; Supplemental Figure 2A). The progression and timing are similar to in vivo chondrocyte development [10], as shown for example by the timeline of both Acan and Col2a1 expression where cells initially expressed low levels of both genes, peaking by day 4 of in vitro development, and sharply decreasing as cells proceeded to maturation (Supplemental Figure 2B). We then treated the O9–1 micromass cultures with the GCN5 inhibitor MB-3 [34] or DMSO as a vehicle control. MB-3 binds with the Acetyl-CoA pocket of GCN5 and inhibits the acetyltransferase activity of GCN5. We first performed a dose response of micromass cells to MB-3, with AB absorbance at 595nm wavelength used as a read out for chondrocyte function and maturation (Supplemental Figure 2C). From this we chose an intermediate concentration of 150μM to continue with our analyses. MB-3 has specificity for GCN5 (GCN5 IC50=100μM, CBP IC50=500μM) [34] and we have previously used a concentration of 100–200μM for cell culture experiments [12]. At 150μM MB-3, a significant decrease in AB absorbance was detected in day 3 micromass cultures. The same trend was observed at all time points tested following day 3 (Figure 3A–C), suggesting deficiencies in chondrogenesis. Delaying the addition of MB-3 by 48 hours was also sufficient to inhibit chondrogenesis (Supplemental Figure 2A, D), suggesting GCN5 enzymatic activity is required for cartilage development, but after the initial condensation and initiation of chondrogenesis. Similarly, washing out MB-3 treatment after 48 hours in culture restored AB absorbance levels to that of controls, supporting a later role for GCN5 in pre-hypertrophic and/or hypertrophic chondrogenesis (Supplemental Figure 2A, E).
Figure 3. Early and late chondrocyte markers are dysregulated in MB-3 treated micromass cultures.
(A) Alcian blue absorbance at 595nm plotted over time in absorbance units (AU). Grey line shows DMSO control, gold lines shows 150μM MB-3 treatment. Measurements were taken for day 2, 3, 4, and 5. (B, C) AB staining of day 4 micromass cultures treated with DMSO vehicle control (B) or 150μM MB-3 (C). (D, E) SOX9 immunofluorescence staining on day 4 micromass cultures treated with DMSO vehicle control (D) or 150μM MB-3 (E). (F, G, H) qPCR analysis of pre-hypertrophic and hypertrophic chondrocyte markers. Acan is shown in (F), Col2a1 is shown in (G), and Mmp13 is shown in (H). RNA was extracted from days 2 – 7 micromass cultures treated with DMSO vehicle control (grey) or 150μM MB-3 (gold). Cp values from (F) and (G) were normalized first to GusB, and then all days were normalized to day 2 micromass. Y axis shows Log2 Fold change from day 2 expression. Cp values from (H) were normalized to GusB. *** and ** represents p < 0.0005; * represents p < 0.05, n =3 for all experiments. Scale bars shows 100μm. Error bars represent standard error of the mean.
In order to narrow the time frame in which GCN5 activity is required, we investigated the transcriptional regulation of differentiating chondrocytes in response to inhibition of GCN5 acetylation activity. Examination of expression of SOX9, which is often considered to be a master transcription factor required for chondrocyte specification [9], in day 4 micromass treated with MB-3 showed no obvious difference in immunofluorescence staining intensity compared to controls (Figure 3D, E). This is consistent with our above observations that GCN5 activity is not required for the initiation of chondrogenesis (Supplemental Figure 2E). We therefore turned to later markers of chondrocyte development. Analysis of Acan, Col2a1, and Mmp13 expression over 7 days revealed that in the presence of the GCN5 inhibitor MB-3, the expression of these markers became severely dysregulated compared to DMSO controls. Expression of both Acan and Mmp13 was significantly decreased with MB-3 treatment starting at day 3 for Acan and day 6 for Mmp13, suggesting that the cartilage cells do not proceed toward maturation (Figure 3F, H; Supplemental Figure 3A, C). Consistent with this idea, day 6 MB-3 treated micromass cultures did not show a recovery of Acan expression, indicating that cells are not just delayed in their maturation. On the other hand, expression of Col2a1 was significantly increased starting at day 3 and continuing through day 7 (Figure 3G; Supplemental Figure 3B). The dysregulation observed with inhibition of GCN5 suggests an uncoupling of the normal transition from immature chondrocytes to mature, hypertrophic chondrocytes. Similar results have been observed by others with Col2a1-Cre deletion of the tumor suppressor Lkb1, which resulted in an increase in Col2a1 expression and a decrease in Mmp13 and Col10a1 expression in maturing chondrocytes [35]. Even though Acan and Col2a1 are considered equivalent markers of immature chondrocytes, our results suggest that Acan and Col2a1 are regulated by separate mechanisms. Acan null mice do not show a loss of Col2a1 expression [36], consistent with our observations. Taken together, these data suggest that chondrocytes are stalled and fail to reach a mature, hypertrophic stage when GCN5 acetylation is inhibited.
H3K9 acetylation is not altered by GCN5 inhibition during cNCC chondrogenesis
The significant decrease in Acan expression raised the hypothesis that GCN5 is directly regulating expression at the histone level. GCN5 was first described in Tetrahymena and yeast as a histone acetyltransferase, participating in active transcription [37, 38]. GCN5 has been shown to preferentially acetylate H3K9 [6, 18] and acetylation of H3K9 on histone tails is considered an active transcription mark and is necessary for transcriptional machinery to access DNA during transcription [39]. Therefore, we hypothesized that GCN5 acetylates H3K9 at the Acan locus to turn on Acan expression during chondrocyte maturation.
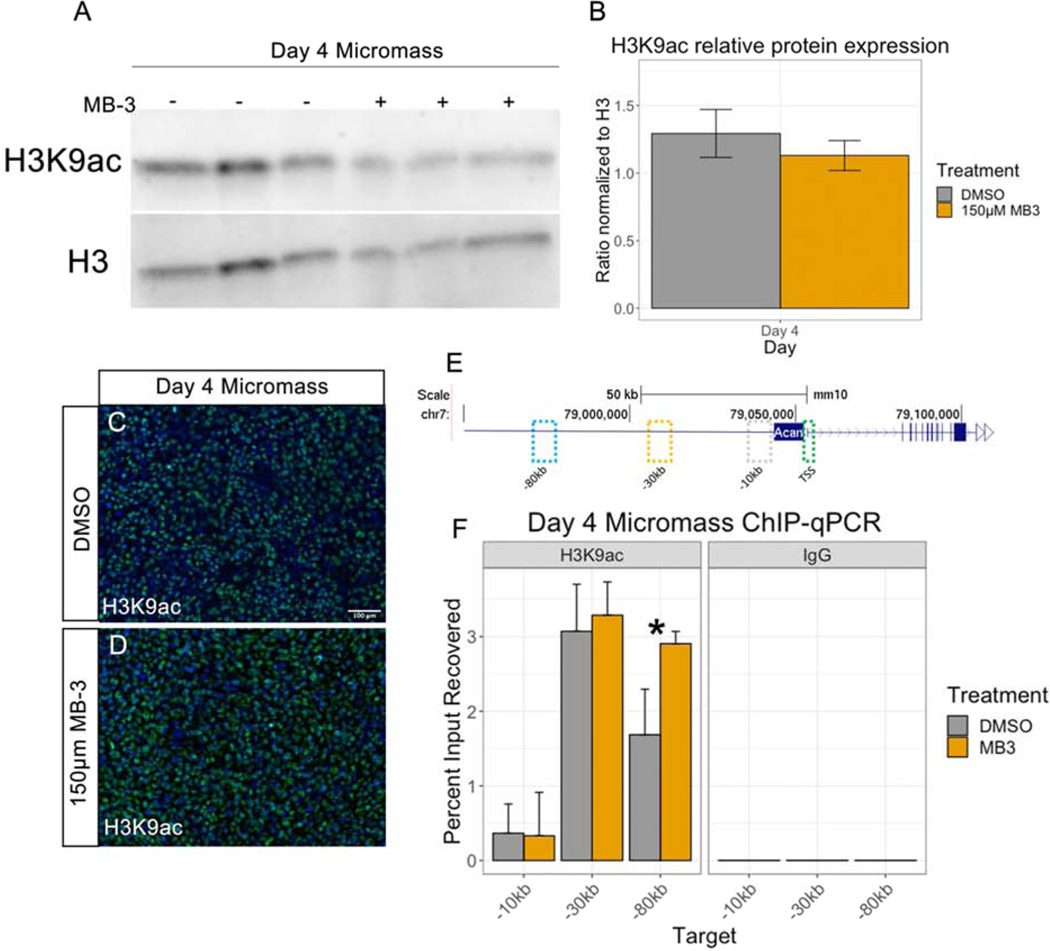
We first determined if total H3K9ac is decreased in micromass cultures treated with GCN5 inhibitor MB-3 compared to DMSO controls. Both western blot and immunofluorescence of day 4 micromass cultures did not show a significant decrease in H3K9ac in MB-3 treated micromass cultures compared to DMSO control (Figure 4A–D). This is similar to our in vivo findings of E12.5 Gcn5hat/hat frontonasal prominence where there was no significant change in H3K9ac immunofluorescence [5]. As acetylation of H3K9 could be altered at specific genes, we specifically focused on the Acan locus. Multiple enhancers for Acan exist both upstream and downstream of the transcription start site (TSS), and others have shown that these enhancers are specific to pre-hypertrophic chondrocytes and may act redundantly [40, 41]. Therefore, we designed primers around three of these enhancer sites at −10kb, −32kb, and −80kb upstream of the TSS (Figure 4E). Using chromatin from day 4 micromass cultures treated with MB-3 or DMSO followed by H3K9ac ChIP-qPCR, there was no significant change in the percent input recovered with H3K9ac pulldown at two of the three genomic regions tested (Figure 4F). While we did observe a significant increase at the −80kb region, this increase does not reflect an increase in transcription of Acan (Figure 3F). Taken together with our previous in vivo finding of no change in global H3K9ac [5], with only a slight change in Acan at one locus, suggest that GCN5 acetylation of H3K9 is not the major driver of craniofacial chondrocyte development.
Figure 4. H3K9ac is unchanged in MB-3 treated micromass cultures.
(A) Western blot analysis of H3K9ac in day 4 micromass treated with DMSO vehicle control or 150μM MB-3. (B) Quantification of western blots, y axis shows ratio of H3K9ac over H3 loading control. (C, D) Immunofluorescence staining with anti-H3K9ac antibody on day 4 micromass cultures treated with DMSO vehicle control (C) or 150μM MB-3 (D) and counterstained with DAPI. (E) Enhancer targets used for qPCR on chromosome 7, upstream of Acan TSS. (F) ChIP-qPCR analysis of H3K9ac status around Acan enhancer elements in DMSO and 150μM MB-3 treated day 4 micromass. IgG was used as a negative control. Y axis shows percent input of DNA recovered (3% of input used), n = 2 experiments performed on separate days. Scale bar represents 100μm. Error bars represent standard error of the mean. * represents P > 0.05.
Inhibition of GCN5 disrupts the mTORC1 signaling pathway
The lack of evidence for an epigenetic change related to craniofacial chondrocyte development refocused our attention to non-epigenetic pathways that are required for chondrocyte growth and maturation. The mTORC1 signaling pathway has been described as a regulator of skeletal growth and development in long bone formation. The mTORC1 protein complex contains the proteins mTOR and RAPTOR [42]. Together, this complex phosphorylates and activates downstream pathways. These include the ribosome kinase S6K1 (p70S6K1), which phosphorylates the ribosomal protein rpS6 to facilitate protein synthesis. mTORC1 also phosphorylates binding protein 4E-BP1, which is required to release 4E-BP1 from eIF4E in order to activate cap dependent translation [42]. Both phosphorylated p-S6K1 and p-4E-BP1 have been reported to be active in pre-hypertrophic chondrocytes, but not hypertrophic chondrocytes of the limb bones [23, 35] and that deletion of the mTORC1 suppressor Lkb1 results in chondrocyte stalling, similar to our results presented here [35]. Others have also suggested that GCN5 is required for inhibition of the mTORC1 pathway [43, 44]. Therefore, we hypothesized that GCN5 is involved in mTORC1 signaling during chondrocyte maturation.
To determine if mTORC1 signaling is active in the cNCC in vitro system, we analyzed mTORC1 signaling over the time course of O9–1 micromass differentiation. Consistent with reports by others in long bone development, we observed no active signaling in undifferentiated cells and the strongest signal in day 4 pre-hypertrophic chondrocytes as assessed by p-mTOR and p-rpS6 (Supplemental Figure 4A). Active mTORC1 signaling dramatically decreased following day 4, consistent with reports that mTORC1 signaling is not active in hypertrophic chondrocytes. We next treated micromass cultures with the mTORC1 inhibitor rapamycin to determine whether mTORC1 activity is required for cNCC differentiation. Consistent with previous publications of long bone development, rapamycin treatment inhibited chondrogenesis in a dose dependent manner, determined by a significant reduction in AB absorbance compared to DMSO controls (Supplemental Figure 4B). To determine if rapamycin and MB-3 act synergistically, micromass cultures were treated with varying doses of rapamycin or MB-3 individually or in combination and AB absorbance was measured at day 4 of differentiation (Supplemental Figure 4C). In none of the treatments did we observe a synergistic effect of combined rapamycin and MB-3 treatment relative to either drug alone. These results suggest that inhibition of GCN5 with MB-3 and mTORC1 with rapamycin do not result in a combined effect on chondrocyte development.
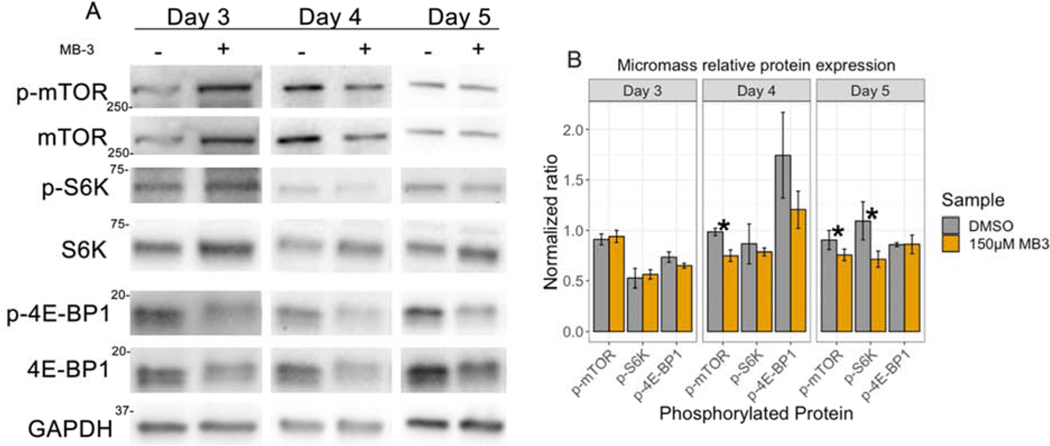
To asses if GCN5 acetylation is required for normal mTORC1 signaling, we tested if the phosphorylation levels of mTOR and its downstream targets were changed in micromass cultures treated with MB-3 compared to DMSO controls. Total protein lysates from day 3, day 4, and day 5 micromass cultures were analyzed with western blot and normalized to non-phosphorylated protein levels. p-mTOR was significantly decreased with MB-3 treatment at both days 4 and day 5 (Figure 5A, B). The phosphorylation levels of p-S6K1 were not significantly changed at day 3 and 4 but were decreased at day 5. p-4E-BP1 levels were decreased in MB-3 treated micromass at day 4, however not significantly, and phosphorylation levels were not changed in day 3 and 5 MB-3 treated micromass (Figure 5A, B). Total protein levels of 4E-BP1 were decreased with MB-3 treatment when compared to GAPDH; however, the ratio of phosphorylated to non-phosphorylated signal was not significantly changed between control and MB-3 treated micromass (Figure 5B). Together, these results show that in the presence of the GCN5 inhibitor MB-3, some components of mTORC1 signaling are decreased in MB-3 treated micromass during chondrocyte development.
Figure 5. mTORC1 signaling is dysregulated in MB-3 treated micromass cultures.
(A) Western blot analysis of mTORC1 pathway. Phosphorylated and un-phosphorylated proteins were evaluated in day 3, 4, and 5 micromass cultures treated with 150μM MB-3. Blots were first immunoblotted for phosphorylated proteins, then stripped, and re-probed for non-phosphorylated proteins. (B) Quantification of western blot densitometry. Phosphorylated proteins were normalized to non-phosphorylated protein. Y axis shows ratio of phosphorylated to non-phosphorylated protein. N = 4 experiments performed on separate days. P < 0.05 considered significant. * represents P < 0.05. Error bars represent standard error of the mean.
The decrease observed in mTORC1 signaling suggested that GCN5 acetylation could function in activation of this pathway, and that GCN5 may act mechanistically through mTORC1 to regulate protein synthesis and mRNA translation during chondrocyte maturation. Recent studies indicate that the mTORC1 protein component RAPTOR contains a lysine residue on the long isoform that is acetylated by the lysine acetyltransferase p300 and RAPTOR acetylation is required for mTORC1 activity and enhanced downstream phosphorylation [45]. RAPTOR is also required for skeletal development in the limb buds [26]. To test if RAPTOR acetylation is decreased upon GCN5 inhibition, we used protein lysate from day 4 micromass cultures treated with MB-3 or DMSO control, and immunoprecipitation for acetylated-lysine, followed by western blot analysis with anti-RAPTOR antibody (Supplemental Figure 5A). Analysis and quantification of RAPTOR from immunoprecipitations did not show a significant decrease in RAPTOR acetylation of the long isoform (Supplemental Figure 5B). These results suggest that GCN5 mediated acetylation of RAPTOR is not a significant aspect of cNCC chondrocyte development.
Discussion:
GCN5 has cell autonomous roles in the cNCC both enzymatically and non-enzymatically
We previously reported that GCN5 HAT activity is necessary for craniofacial development, but it was unclear in which cell type GCN5 function is required. Here, we present evidence that GCN5 is required cell autonomously in cNCC. Using the Wnt1-Cre recombinase driver to conditionally delete Gcn5 in cNCC, the resulting mouse embryos show severe craniofacial defects, with most of the bone and cartilage absent from the craniofacial region (Figure 1E). In contrast, deletion of one allele of Gcn5 in the cNCC in combination with one allele that generates catalytically dead, full length GCN5 results in less severe craniofacial defects (Figure 1F). The differences in phenotype suggests that GCN5 acetyltransferase activity as well as non-HAT related functions are required for craniofacial development. This is perhaps not surprising as GCN5 has been described to participate in many activities within the cell, including interacting with RNA Polymerase II [21], and as a protein component of the large complexes SAGA and ATAC [46], raising the possibility that GCN5 also has critical structural roles as part of these complexes. Lastly, we also demonstrate that the enzymatic activity of GCN5 is required for cNCC chondrocyte maturation in vitro. In the absence of GCN5 activity, cNCC pre-hypertrophic chondrocytes become stalled in an immature stage, and do not develop into mature chondrocytes (Figure 6). These results are also reflected in vivo, as the cell size of individual chondrocytes from Gcn5hat/hat are smaller compared to control, reflecting a failure of immature chondrocytes to increase cell size as they proceed to hypertrophy.
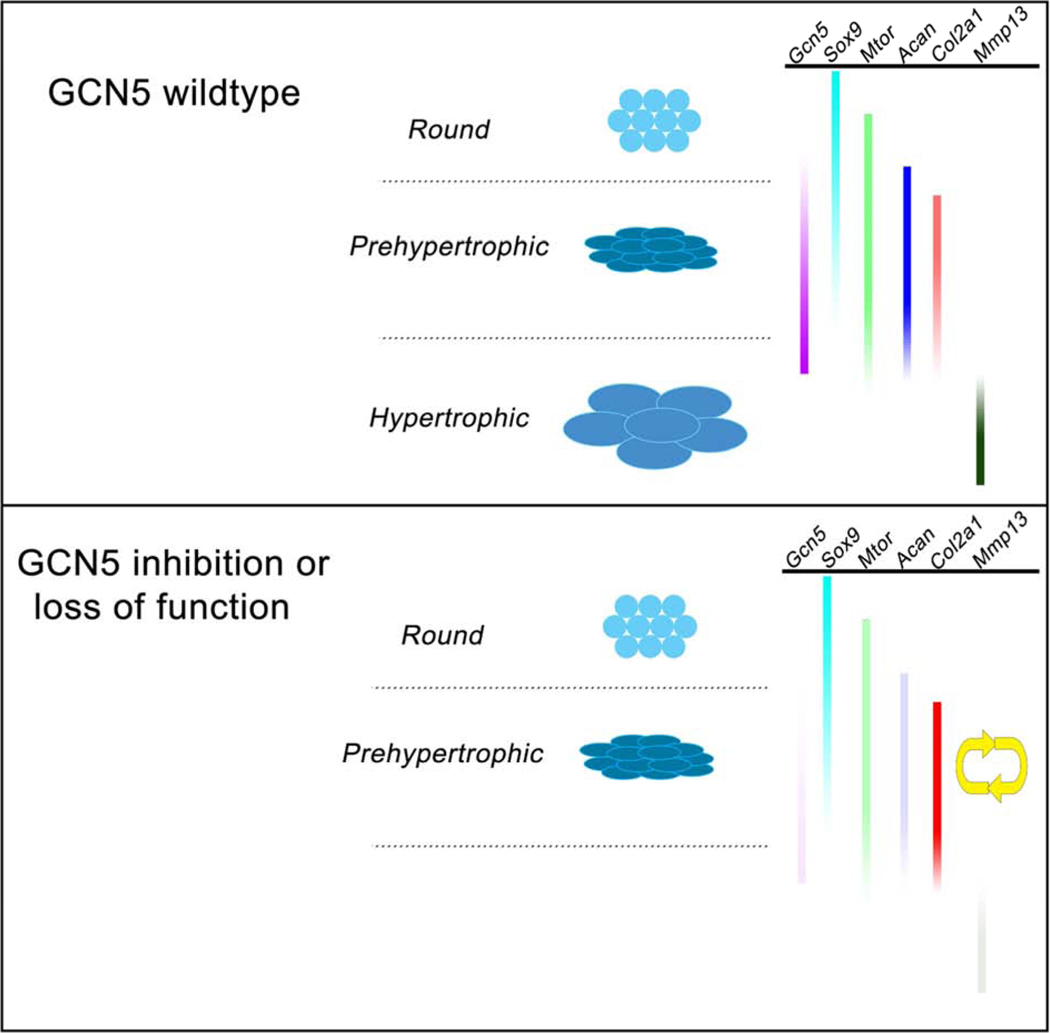
Figure 6. GCN5 acetyltransferase activity is required for chondrocyte maturation.
Diagrams representing stages of chondrocyte development when GCN5 acetylation is required. Top diagram represents the wild type condition. Chondrocytes progress normally to a hypertrophic stage in the presence of GCN5 activity (purple bar). Bottom diagram represents the mutant condition. Chondrocytes that have lost or reduced GCN5 acetylase activity become stalled and do not progress to maturation. Genes representative of chondrocyte stages are shown and their changes in expression in Gcn5hat/hat mutants is indicated by change in shading relative to wild type.
While the SAGA and ATAC complexes can also recruit the GCN5 family member PCAF (Kat2b), it is possible that during cNCC development, PCAF is not able to compensate for a loss of GCN5. Previous reports identified that expression of Pcaf is much lower than Gcn5 during early murine development, and that these family members have temporal and tissue specific functions [47, 48]. It also remains to be determined whether GCN5 and p300 have overlapping functions in craniofacial development as p300 homozygous null mice die early in embryonic development and conditional deletion of p300 with Wnt1-Cre has not been performed. More broadly, Gcn5hat/hat mutants do not display defects in bone and cartilage of the ribs or limbs despite Gcn5 being ubiquitously expressed, which could suggest that p300 activity is more prominent in these skeletal elements. More work will be needed to determine the relationship between GCN5 and p300 in skeletal development. Nonetheless, our studies indicate that GCN5 has both enzymatic and non-enzymatic roles that are required cell autonomously in cNCC.
GCN5 mediated histone acetylation is not required for chondrocyte development
GCN5 has been extensively described as a histone acetyltransferase, targeting histone 3 lysine residue K9, and to some extent K14, K18, and K36 [49]. In yeast, GCN5 acts to globally acetylate histones across the entire genome [50]. The results from mammalian studies yield discrepancies in the requirement for GCN5 to globally acetylate histones. For instance, in murine development, knock-out of GCN5 HAT activity does not cause a global loss of H3K9ac [5, 6]. In fact, knock-out of both Gcn5 and Pcaf is required to globally ablate H3K9 acetylation [51]. Therefore, an important question that remains is how much can PCAF compensate for a loss of GCN5 histone acetyltransferase activity? Nevertheless, for GCN5, it is obvious that what was classically described as a histone acetyltransferase, instead has many roles that deviate from its canonical function. We show here that GCN5 mediated histone acetylation does not appear to be required for cNCC pre- and hypertrophic chondrocyte development, and that GCN5 is instead dispensable for H3K9ac during chondrocyte development. Instead, our results indicate that GCN5 enzymatic activity participates in non-epigenetic mechanisms, consistent with other non-epigenetic pathways described for GCN5 [12, 43, 52, 53]. Furthermore, it is unclear from the literature at what temporal and spatial regions GCN5 histone acetylation is required during development. Our results suggest that craniofacial chondrocyte development is not directly dependent on GCN5 histone acetylation.
GCN5 has cell type specific activities
Numerous studies have identified specific roles for GCN5 in disparate cell types and stages of development. While GCN5 is ubiquitously expressed and highly conserved, the phenotypes observed in Gcn5hat/hat mutants severely affect the neural and cranial regions with little effect on other tissues, as we have described in this study and others have noted previously [6]. Therefore, some tissue specificity is achieved through GCN5 enzymatic activity. However, the pathways previously described for GCN5 are not necessarily required within the context of cNCC chondrocyte maturation. For example, work from our group identified an interaction between GCN5 acetylation and retinoic acid signaling [12]. However, during chondrocyte maturation, retinoic acid negatively regulates chondrocyte maturation and causes severe cell death [32]. Other reports have also identified that GCN5 acetylation is required for the inhibition of the mTORC1 pathway during fasting in skeletal muscle [43]. Our results do not suggest that GCN5 is required for inhibition of mTORC1 in cNCC derived chondrocytes (Figure 5A). Instead, our results indicate that GCN5 acetylation is required for activation of the mTORC1 pathway in cNCC chondrocyte maturation, thus raising the question about how GCN5 acetylation could be activating the pathway? As we did not observe a change in RAPTOR acetylation, this suggests that upstream activators of mTORC1 could be direct targets of GCN5 acetylation. Upstream activators of mTORC1 include AKT. However, studies identified that AKT acetylation inhibited its activity through hinderance of AKT binding with PIP3 and a reduction in downstream AKT phosphorylation [54]. In consideration of additional proteins that modulate mTORC1 activity, one potential target of GCN5 acetylation could be TSC2, which when complexed with TSC1, the TSC1/2 complex inhibits mTORC1 activity. One study investigating TSC2 acetylation identified that TSC2 acetylation was required for TSC2 protein degradation, thus preventing TSC1/2 inhibition of mTORC1 activity [55]. In the present study we did not test TSC2 acetylation nor the acetylation of other mTORC1 regulators, however, our results indicate that it is highly likely that a novel direct target of GCN5 acetylation that has not previously been described is required for mTORC1 activation. Taken together, these results suggest that GCN5 has disparate functions depending on cell type and unknown protein targets that in the future can be uncovered through unbiased sequencing approaches.
Craniofacial chondrocyte maturation is similar to endochondral ossification of the limbs
Endochondral ossification within the limb skeleton has been well studied in mouse, identifying many of the regulatory mechanisms that are required for long bone development. In regards to the craniofacial skeleton, some of the mechanisms that are required for endochondral bone development in the limb have also been studied during craniofacial development [56]. Our results here and work from others strongly supports the hypothesis that mechanisms that regulate endochondral bone formation in the craniofacial skeleton are similar to endochondral bone formation of the limbs. This includes the temporal progression of molecular markers of chondrocyte differentiation and mTORC1 signaling [24, 25, 57, 58]. Our results also indicate that mTORC1 signaling is required for chondrocyte hypertrophy in the craniofacial skeleton (Supplemental Figure 3A,B), consistent with a recent report that conditional deletion of mTOR with Wnt1-Cre results in craniofacial defects [59]. While we do not observe long bone defects in homozygous Gcn5hat/hat mutants, it is possible that there are compensatory mechanisms and/or cell type specific mechanisms for GCN5 in the limb versus the cNCC. Our results here describe a cell-autonomous function for GCN5 in the progression of cNCC derived chondrocyte maturation.
Materials and Methods
Mice and Genotyping:
Mice were bred and utilized according to protocol #2590 set by the Institution for Animal Care and Use at the University of Colorado Boulder. Mice were kept on a C57/Bl6J background under standard light and ambient conditions. Vaginal plugs were checked in mornings for timed mating, with positive identification of a plug considered to be embryonic day 0.5. Dissections were carried out according to NIH guidelines and standards. Genotyping of Wnt1-Cre (H2az2Tg(Wnt1-cre_1Rth)) and all Gcn5 alleles was carried out as previously described [6, 27, 30].
Cell culture and micromass assays:
The cell line O9–1 was purchased from Millipore-Sigma (cat. #SCC049), early passage aliquots frozen, and single aliquots passaged up to 12 times. Cells were periodically checked for mycoplasma contamination with MycoAlert (Lonza cat. LT07–218). Cells were maintained as previously described [33]. Briefly, cells were passaged onto Geltrex (ThermoFisher cat. #A1413201) coated plates, in a 50:50 mixture of conditioned O9–1 media (conditioned by STO cells) and unconditioned O9–1 media. O9–1 media consisted of DMEM, 15% FBS, 1% NEAA, 2% L-Glutamine, .01% Beta-mercaptoethanol, 25 ng/mL FGF, and 1000 U/mL LIF. Cells were maintained in standard incubator conditions (5% CO2, 37°C). For micromass cultures, cells were expanded to confluency and then harvested and resuspended at a density of 2*107 cells/mL in DMEM/10% FBS. The high-density cell suspension was then plated in 15 μL drops into 12-well (1 drop per well) or 6-well plates (5 drops per well) and cells were allowed to attach for 2 hours, followed by flooding wells with media. Media was changed every day. Micromass differentiation media for chondrogenesis consisted of DMEM, 10% FBS, 1% ITS-A (ThermoFisher Cat. #51300044), 50 μg/mL Ascorbic Acid, and 100 nM Dexamethasone. MB-3 (Millipore-Sigma cat. #M2449) was reconstituted in DMSO at a stock concentration of 200mM and added fresh to media every day. DMSO as a vehicle control was added to media at an equal amount to MB-3. Rapamycin (Millipore-Sigma cat. #553210) was reconstituted in DMSO at a stock concentration of 1μM.
Immunofluorescence:
Micromass cultures for immunofluorescence were fixed for 30 minutes at room temperature with 4% paraformaldehyde. Cells were then permeabilized with 0.1% Triton-X100 in PBS for 10 minutes at room temperature, followed by 1-hour blocking step at room temperature with 5%BSA/1% normal goat-serum in PBS-Tween 20. The following primary antibodies were used: Rabbit anti-Sox9 conjugated with Alexa 488 (1:500, Millipore Sigma #AB5535-AF488), rabbit anti-H3K9ac (1:500, CST #9649). The following secondary antibody was used: Goat anti-rabbit Alexa 488 (1:500, ThermoFisher #A27034). Cells were counterstained with Hoechst (1:5000, ThermoFisher #62249). Cells were imaged using a Nikon E600 Upright Widefield microscope.
Histological and skeletal staining:
Embryos for histological staining were fixed overnight in 4% PFA at 4ºC, and then processed into paraffin wax as previously described [61]. Wax blocks were sectioned using a Microm HM 355S Rotary Microtome at a thickness of 10 μm onto Superfrost Plus slides. Paraffin sections were de-waxed and re-hydrated then stained with hematoxylin and eosin or alcian blue under standard conditions. Sections stained with alcian blue were counter stained with nuclear fast red (Sigma cat. #N3020) according to manufacturer’s instructions. Stained sections were mounted with Permount (ThermoFisher cat. #SP15) and imaged using an Olympus IX83 inverted microscope. Micromass cultures stained with alcian blue as previously described [62] with the exception that cells were fixed with 50% ethanol for 30 minutes at room temperature, and alcian blue absorbance was measured at 595nm. Skeletal staining was carried out as previously described [63].
RNA extraction and qPCR:
Micromass cultures for RNA extraction were harvested from 6 well plates seeded with 5 micromass drops per well. Cells were washed once with ice-cold PBS, then harvested in TRIzol Reagent (ThermoFisher cat. #15596026). Micromass were homogenized in TRIzol using a hand-held micro pestle (Kimble & Chase cat. #749520). RNA extraction from TRIzol was carried out according to manufacturer’s instructions. 1 μg of RNA was used for cDNA preparation. cDNA was prepared using First-Strand cDNA reverse transcriptase kit with Superscript III (ThermoFisher cat. #18080051) using Oligo-dT primers according to manufacturer’s instructions. cDNA was amplified by qPCR using Roche LC 480 2X Master mix with the Roche Light Cycler II and universal probes. Primers for qPCR were designed using the Roche Universal Probe Library design software. Fold change for qPCR was determined using the 2−ΔΔCp method. All target Cp values were normalized to GusB reference. Primers used are: Col2a1 Fw: 5’-aagacccagactgcctcaac-3’, Rv: 5’-cctttggccctaattttcg-3’; Acan Fw: 5’-ccagcctacaccccagtg-3’, Rv: 5’-gagggtgggaagccatgt-3’; Mmp13 Fw: 5’- cagtctccgaggagaaactatgat-3’, Rv: 5’-ggactttgtcaaaaagagctcag-3’; Fw: GusB 5’aaaatcaccctgcggttgt-3’, Rv: 5’-tgtgggtgatcagcgtctt-3’; Fw: Acan −10kb 5’-ggataaggcccaccactgt3’, Rv: 5’-ccccagaactcacaggaacta-3’; Fw: Acan −30kb 5’-gcagctgacctggtgtagact-3’, Rv: 5’ccccacatcacaagatacctg-3’; Fw: Acan −80kb 5’-agcaatttctccacattggtc-3’, Rv: 5’gggatgtctgtgtagccaatc-3’.
Protein lysate preparation and western blot:
Micromass cultures for protein extraction were washed once with ice-cold PBS and then harvested in a Tris-Triton extraction buffer containing 10mM Tris-HCl pH 7.4, 5mM EDTA, 5mM EGTA, 50mM NaCl, 1% Triton X-100 with protease inhibitor cocktail (Sigma cat. #4693159001) added fresh. Tissues were homogenized by passing through a 21-gauge syringe, and then total protein was extracted by rocking at 4ºC for 30 minutes. Lysates were cleared by centrifugation for 20 minutes at 14,000 RPM. Protein concentration was determined using Bradford reagent (BioRad cat. #5000006) and 30 μg of total protein was loaded per sample for protein separation using SDS-PAGE. Gels were transferred onto 0.22 μM PVDF membranes using wet transfer methods with either 10% methanol (MeOH) in transfer buffer for proteins less than 20 kDa (4E-BP1 and H3K9ac), or 20% MeOH in transfer buffer for all others. Membranes were blocked with either 5% non-fat milk in TBST or 5% BSA for phosphorylated antibodies. The following primary antibodies were used: Rabbit anti-p-mTOR (1:1000, CST #2974), Rabbit anti-mTOR (1:1000, CST #2983), Rabbit antip-p70S6K (1:500, CST #9234), Rabbit anti-p70S6K (1:1000, CST #2708), Rabbit anti-p-4E-BP1 (1:1000, CST #2855), Rabbit anti-4EBP1 (1:1000, CST #9644), Rabbit anti-p-rpS6 (1:1000, CST #5364), Rabbit anti-GAPDH (1:5000, Sigma cat. #G9545), Rabbit anti-H3K9ac (1:500, CST #9649), Rabbit anti-Raptor (1:500, ThermoFisher #42–4000), Rabbit anti-H3 (1:500, Abcam #ab1791). The following secondary antibodies were used: Goat anti-Rabbit HRP (1:2000, CST #7074), Goat anti-Mouse HRP (1:2000, Biorad #STAR207P). HRP was visualized using SuperSignal West Pico Plus Chemiluminescent Substrate (ThermoFisher cat. #34577). H3K9ac and H3 were visualized using SuperSignal West Femto Maximum sensitivity Chemiluminescent Substrate (ThermoFisher cat. #34095). Membranes were exposed on a BioRad Gel Doc imager. Blots were stripped with stripping buffer (ThermoFisher cat. #46428) for 15 minutes and then re-probed.
Immunoprecipitation:
Micromass cultures for immunoprecipitation were lysed with triton buffer containing 1% Triton X-100, 5mM NaCl, 5mM EDTA, 5mM EGTA, 10mM Tris-HCl, 10mM Sodium Butyrate, 1mM Trichostatin A, and protease inhibitors. Lysates were homogenized by passing lysate through a 21-gauge syringe, and then total protein was extracted by incubation for 30 minutes at 4°C. Lysates were cleared by centrifugation at 14,000 RPM at 4°C for 20 minutes. Total protein was measured using Bradford reagent. 300μg/mL of total protein was used per replicate. Lysates were pre-cleared using Dynabeads Protein G (ThermoFisher #10003D), then incubated overnight at 4°C with primary antibody. The immunocomplex was immunoprecipitated with Dynabeads Protein G for 1 hour at 4°C. Immunoprecipitates were washed using lysis buffer, then eluted with Laemmli sample buffer by boiling for 5 minutes at 95°C. Eluted samples were then used for western blot analysis. The following primary antibody was used for immunoprecipitation: Mouse anti-acetylated lysine (1:100, Santa Cruz Biotech. #sc-32268).
Chromatin Immunoprecipitation (ChIP):
Micromass cultures for ChIP were crosslinked for 14 minutes with 11% Formaldehyde and ChIP protocol was followed as previously described [64]. Chromatin was sheared in lysis buffer on ice using a sonicator probe (Heat Systems Ultrasonics, model W-185-F) until an average size of 500 – 1000 bp was achieved. Anti-H3K9ac antibody (1:50, CST #9694) or Anti-Rabbit IgG (5 μg/mL, ThermoFisher cat. #02–6102) were used for ChIP. Chromatin was immunoprecipitated using a 1:1 mixture of Dynabeads Protein A and Dynabeads Protein G (ThermoFisher cat. #10001D & 10003D). Reverse crosslinked DNA was then used for qPCR. qPCR was carried out as described in the qPCR methods above. Primers used for ChIP-qPCR are listed above.
Quantification, graphing, and statistical analysis:
Quantification for cell area (Figure 2L, M) was carried out using ImageJ. Briefly, a masked-image of the cartilage structure was used as an 8-Bit tiff image to create a threshold for the maximum intensity (Supplemental Figure 1). A range of 130–255 was used for threshold. Images after threshold cutoff were then automatically counted or measured using particle analysis. Measurement for Figure 2C was also done using ImageJ. Graphing and statistical analyses were done using R-Studio, statistical test was done with a Welch two-sample unpaired t-test. Alcian blue absorbance, western blot, and qPCR values were all analyzed and graphed using R-Studio. Statistical tests were done using Welch two-sample unpaired t-test. A P value less than 0.05 was considered significant for all experiments.
Supplementary Material
Highlights.
Gcn5 is required cell autonomously in cranial neural crest.
Craniofacial chondrocyte size and maturation are decreased upon loss of GCN5.
GCN5 histone acetylation is not required for craniofacial chondrocyte maturation.
GCN5 acts primarily through non-epigenetic mechanisms in chondrocyte maturation.
Acknowledgements:
We would like to acknowledge members of the Niswander and Artinger labs for their help and feedback during the course of these experiments. We would also like to thank Lori Bulwith and the CU Boulder Office for Animal Resources staff for their support and help with mouse husbandry. Lastly, we would like to thank the CU Boulder Light Microscopy Core and Stem Cell Research and Technology Resource Center for their help with imaging.
Funding: This work is supported by NIH NIDCR R01 DE024034 to K.B.A. and L.A.N., and R01 DE024034-A1S1 to S.A.P.
Footnotes
Conflicts of interest: We have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kirby RS, The prevalence of selected major birth defects in the United States. Seminars in Perinatology, 2017. 41(6): p. 338–344. [DOI] [PubMed] [Google Scholar]
- 2.Abe M, Ruest LB, and Clouthier DE, Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor-deficient mice. Int J Dev Biol, 2007. 51(2): p. 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordero DR, et al. , Cranial neural crest cells on the move: their roles in craniofacial development. Am J Med Genet A, 2011. 155A(2): p. 270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simoes-Costa M. and Bronner ME, Establishing neural crest identity: a gene regulatory recipe. Development, 2015. 142(2): p. 242–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen R, et al. , Kat2a and Kat2b Acetyltransferase Activity Regulates Craniofacial Cartilage and Bone Differentiation in Zebrafish and Mice. J Dev Biol, 2018. 6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bu P, et al. , Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol, 2007. 27(9): p. 3405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai Y. and Maxson RE Jr., Recent advances in craniofacial morphogenesis. Dev Dyn, 2006. 235(9): p. 2353–75. [DOI] [PubMed] [Google Scholar]
- 8.Yuko Mori-Akiyama HA, David H. Rowitch, and Benoit de Crombrugghe, Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. PNAS, 2003. 100(16): p. 9360–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefebvre V. and Dvir-Ginzberg M, SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res, 2017. 58(1): p. 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozhemyakina E, Lassar AB, and Zelzer E, A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development, 2015. 142(5): p. 817–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T, Chromatin modifications and their function. Cell, 2007. 128(4): p. 693–705. [DOI] [PubMed] [Google Scholar]
- 12.Wilde JJ, et al. , Diencephalic Size Is Restricted by a Novel Interplay Between GCN5 Acetyltransferase Activity and Retinoic Acid Signaling. J Neurosci, 2017. 37(10): p. 2565–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L, Pang A, and Li Y, Function of GCN5 in the TGF-beta1-induced epithelial-tomesenchymal transition in breast cancer. Oncol Lett, 2018. 16(3): p. 3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahata K, et al. , Regulation of transforming growth factor-β and bone morphogenetic protein signalling by transcriptional coactivator GCN5. Genes to Cells, 2004. 9(2): p. 143–151. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, et al. , GCN5 Regulates FGF Signaling and Activates Selective MYC Target Genes during Early Embryoid Body Differentiation. Stem Cell Reports, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurooka H. and Honjo T, Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem, 2000. 275(22): p. 17211–20. [DOI] [PubMed] [Google Scholar]
- 17.Jin Q, et al. , Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J, 2011. 30(2): p. 249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, Edmondson DG, and Roth SY, Mammalian GCN5 and P/CAF have homologous amino terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol, 1998. 18(10): p. 5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Carey M, and Workman JL, The role of chromatin during transcription. Cell, 2007. 128(4): p. 707–19. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y, et al. , Principles of regulatory information conservation between mouse and human. Nature, 2014. 515(7527): p. 371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnet J, et al. , The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev, 2014. 28(18): p. 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krebs AR, et al. , SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell, 2011. 44(3): p. 410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan B, et al. , mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat Commun, 2016. 7: p. 11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang B, et al. , mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway. PLoS Genet, 2015. 11(8): p. e1005426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Q, et al. , mTOR/Raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell Death Differ, 2017. 24(11): p. 1886–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iezaki T, et al. , Translational Control of Sox9 RNA by mTORC1 Contributes to Skeletogenesis. Stem Cell Reports, 2018. 11(1): p. 228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chai Y, et al. , Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development, 2000. 127(8): p. 1671–9. [DOI] [PubMed] [Google Scholar]
- 28.Lin W, et al. , Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev Dyn, 2007. 236(6): p. 1547–57. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi S, et al. , Efficient gene modulation in mouse epiblast using a Sox2cre transgenic mouse strain Mechanisms of Dev., 2002. 119S: p. 97–101. [DOI] [PubMed] [Google Scholar]
- 30.Lin W, et al. , Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev Dyn, 2008. 237(4): p. 928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthwal N, Joshi L, and Tucker AS, Evolution of the mammalian middle ear and jaw: adaptations and novel structures. J Anat, 2013. 222(1): p. 147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J. and Dong S, The Signaling Pathways Involved in Chondrocyte Differentiation and Hypertrophic Differentiation. Stem Cells Int, 2016. 2016: p. 2470351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii M, et al. , A stable cranial neural crest cell line from mouse. Stem Cells Dev, 2012. 21(17): p. 3069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biel M, et al. , Design, synthesis, and biological evaluation of a small-molecule inhibitor of the histone acetyltransferase Gcn5. Angew Chem Int Ed Engl, 2004. 43(30): p. 3974–6. [DOI] [PubMed] [Google Scholar]
- 35.Lai LP, et al. , Lkb1/Stk11 regulation of mTOR signaling controls the transition of chondrocyte fates and suppresses skeletal tumor formation. Proc Natl Acad Sci U S A, 2013. 110(48): p. 19450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domowicz MS, et al. , Aggrecan modulation of growth plate morphogenesis. Dev Biol, 2009. 329(2): p. 242–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brownell JE and Allis CD, An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA, 1995. 92: p. 6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brownell JE, et al. , Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 1996. 84: p. 843–851. [DOI] [PubMed] [Google Scholar]
- 39.Venters BJ and Pugh BF, How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol, 2009. 44(2–3): p. 117–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu G, Codina M, and Fisher S, Multiple enhancers associated with ACAN suggest highly redundant transcriptional regulation in cartilage. Matrix Biol, 2012. 31(6): p. 328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda Y, et al. , A candidate enhancer element responsible for high-level expression of the aggrecan gene in chondrocytes. J. Biochem, 2013. 156(1): p. 21–28. [DOI] [PubMed] [Google Scholar]
- 42.Laplante M. and Sabatini DM, mTOR signaling at a glance. J Cell Sci, 2009. 122: p. 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee D. and Goldberg AL, Muscle wasting in fasting requires activation of NF-kB and inhibition of AKT/Mechanistic target of rapamycin (mTOR) by the protein acetylase, GCN5. J. Biogical Chem, 2015. 290(51): p. 30269–30279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lerin C, et al. , GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab, 2006. 3(6): p. 429–38. [DOI] [PubMed] [Google Scholar]
- 45.Son SM, et al. , Leucine Signals to mTORC1 via Its Metabolite Acetyl-Coenzyme A. Cell Metab, 2019. 29(1): p. 192–201 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker SP and Grant PA, The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene, 2007. 26(37): p. 5329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W, et al. , Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nature Genetics, 2000. 26: p. 229–232. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi T, et al. , Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. PNAS, 2000. 97(21): p. 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riss A, et al. , Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. J Biol Chem, 2015. 290(48): p. 28997–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imoberdorf RM, Topalidou I, and Strubin M, A role for gcn5-mediated global histone acetylation in transcriptional regulation. Mol Cell Biol, 2006. 26(5): p. 1610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Q, et al. , Gcn5 and PCAF Regulate PPARy and Prdm16 Expression To Facilitate Brown Adipogenesis. Mol Cell Biol, 2014. 34(19): p. 3746–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh TK, et al. , Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb development. J Mol Cell Cardiol, 2018. 114: p. 185–198. [DOI] [PubMed] [Google Scholar]
- 53.Wiper-Bergeron N, et al. , Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5. Proc Natl Acad Sci U S A, 2007. 104(8): p. 2703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iaconelli J, et al. , Lysine Deacetylation by HDAC6 Regulates the Kinase Activity of AKT in Human Neural Progenitor Cells. ACS Chem Biol, 2017. 12(8): p. 2139–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Aguilar A, et al. , TSC2 N-terminal lysine acetylation status affects to its stability modulating mTORC1 signaling and autophagy. Biochim Biophys Acta, 2016. 1863(11): p. 2658–2667. [DOI] [PubMed] [Google Scholar]
- 56.Abzhanov A, et al. , Regulation of skeletogenic differentiation in cranial dermal bone. Development, 2007. 134(17): p. 3133–44. [DOI] [PubMed] [Google Scholar]
- 57.Chen J. and Long F, mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development, 2014. 141: p. 2848–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newton PT, et al. , Activation of mTORC1 in chondrocytes does not affect proliferation or differentiation, but causes the resting zone of the growth plate to become disordered. Bone Rep, 2018. 8: p. 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nie X, et al. , mTOR acts as a pivotal signaling hub for neural crest cells during craniofacial development. PLoS Genet, 2018. 14(7): p. e1007491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richardson L, et al. , EMAGE mouse embryo spatial gene expression database: (2014 update). Nucleic Acids Res, 2014. 42(1): p. D835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fisher A, et al. , Paraffin embedding tissue samples for sectioning. CSH Protoc, 2008. [DOI] [PubMed] [Google Scholar]
- 62.Fantauzzo KA and Soriano P, PI3K-mediated PDGFRalpha signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes Dev, 2014. 28(9): p. 1005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rigeur D. and Lyons KM, Whole-Mount Skeletal Staining. Methods Mol Biol, 2014. 1130: p. 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rada-Iglesias A, et al. , A unique chromatin signature uncovers early developmental enhancers in humans. Nature, 2011. 470(7333): p. 279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.