Abstract
We investigated metabolic engineering of fermentation pathways in Escherichia coli for production of optically pure d- or l-lactate. Several pta mutant strains were examined, and a pta mutant of E. coli RR1 which was deficient in the phosphotransacetylase of the Pta-AckA pathway was found to metabolize glucose to d-lactate and to produce a small amount of succinate by-product under anaerobic conditions. An additional mutation in ppc made the mutant produce d-lactate like a homofermentative lactic acid bacterium. When the pta ppc double mutant was grown to higher biomass concentrations under aerobic conditions before it shifted to the anaerobic phase of d-lactate production, more than 62.2 g of d-lactate per liter was produced in 60 h, and the volumetric productivity was 1.04 g/liter/h. To examine whether the blocked acetate flux could be reoriented to a nonindigenous l-lactate pathway, an l-lactate dehydrogenase gene from Lactobacillus casei was introduced into a pta ldhA strain which lacked phosphotransacetylase and d-lactate dehydrogenase. This recombinant strain was able to metabolize glucose to l-lactate as the major fermentation product, and up to 45 g of l-lactate per liter was produced in 67 h. These results demonstrate that the central fermentation metabolism of E. coli can be reoriented to the production of d-lactate, an indigenous fermentation product, or to the production of l-lactate, a nonindigenous fermentation product.
Lactate and its derivatives have been used in a wide range of food-processing and industrial applications (8, 27). Because lactate can be easily converted to strong, highly transparent, and readily biodegradable polyesters, it is emerging as a potential material for environmentally friendly plastics. As the physical properties of polylactate depend on the isomeric composition of lactate (28), production of optically pure lactate is a prerequisite for polymer synthesis in which lactate is used.
Lactate has been produced commercially either by chemical synthesis or by fermentation (8). In contrast to chemical processes, the fermentation process is able to produce the desired stereoisomer. Many microorganisms produce d-lactate, and some lactic acid bacteria, such as Lactobacillus bulgaricus, produce highly pure d-lactate (2). l-Lactate also has been produced by using lactic acid bacteria, such as Lactobacillus helveticus, Lactobacillus amylophilus, and Lactobacillus delbruekii (27). It has also been proposed that a mutant of the racemic mixture producer L. helveticus defective in d-lactate dehydrogenase (d-LDH) could be used for production of optically pure l-lactate (3). As lactic acid bacteria have complex nutritional requirements and very low growth rates (24), Rhizopus oryzae and Bacillus laevolacticus have been proposed as alternative producers (9, 25).
Escherichia coli has many advantageous characteristics as a production host, such as rapid growth under aerobic and anaerobic conditions and simple nutritional requirements. Moreover, well-established protocols for genetic manipulation and a large physiological knowledge base should enable the development of E. coli as a host for production of optically pure d- or l-lactate by metabolic engineering.
E. coli, a facultative anaerobe, carries out mixed-acid fermentation of glucose in which the principal products are formate (or CO2 and H2), acetate, d-lactate, succinate, and ethanol (4) (Fig. 1). Mutations in a specific fermentation pathway(s) significantly affect the overall fermentation balance or by-product pattern (6). It has been reported that pta mutants, which are not able to synthesize phosphotransacetylase (Pta), neither grow nor synthesize acetate anaerobically on glucose minimal medium (13). Similarly, an alcohol dehydrogenase (ADH)-negative adh mutant was not able to grow anaerobically on glucose (7). However, adh pta double mutants were able to grow anaerobically on glucose by lactate fermentation (14). Therefore, the acetate pathway appears to be one of the target pathways which can be manipulated to redirect the fermentation metabolic flux of E. coli to lactate production.
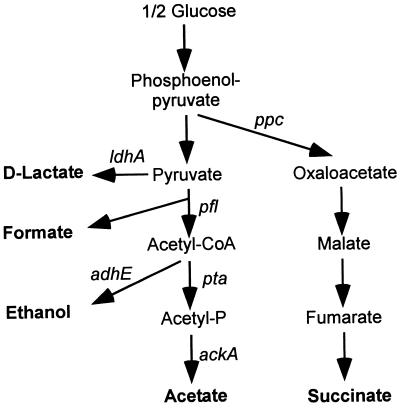
FIG. 1.
Schematic diagram of the fermentation pathways in E. coli. Gene designations: ackA, acetate kinase gene; adhE, ADH gene; ldhA, d-LDH gene; pfl, pyruvate-formate lyase gene; ppc, phosphoenolpyruvate carboxylase gene; pta, Pta gene.
In this work, using pta mutants defective in the Pta-AckA acetate production pathway, we investigated redirection of the metabolic flux from the acetate pathway to d- or l-lactate production. During anaerobic cultivation of the E. coli RR1 pta ppc mutant, fermentative metabolism was redirected to d-lactate production for recycling of NADH produced by glycolysis. When l-LDH from Lactobacillus casei was introduced into a pta ldhA mutant lacking enzymes leading to the production of acetate and d-lactate, optically pure l-lactate was produced as the major fermentation product. The lactate productivity was increased by growing the cells first under aerobic conditions before shifting them to anaerobic conditions, which are favorable for the production of lactate.
MATERIALS AND METHODS
Bacterial strains and plasmid.
The bacterial strains and plasmid used in this study are listed in Table 1. All of the cultivations were carried out with E. coli RR1 and its derivatives. Each mutation in the pta, ppc, or ldhA gene was introduced into E. coli RR1 by P1 transduction by using lysates of the appropriate strain. The mutants constructed had stable phenotypes during cultivation, as confirmed by resistance to antibiotics and by by-product patterns on M9 medium containing glucose. To construct a ppc strain, the structural gene of phosphoenolpyruvate carboxylase was cloned by PCR performed with primers designed on the basis of the previously published sequence (12). The resulting PCR product of the ppc gene was inserted into plasmid pUC19 to obtain pKJE15. The ppc gene on plasmid pKJE15 was inactivated by inserting a cat gene at the StuI site. To integrate the ppc::cat marker into the chromosome, this plasmid was transformed into strain JC7623. Cmr transformants were isolated, and the Ppc− phenotype was confirmed by an enzyme assay. The ppc::cat marker on the chromosome was then transduced into RR1 derivatives by P1 transduction. Plasmid pLS65, a gift from M. Y. Park (Korea Advanced Institute of Science and Technology, Taejon, Korea), contained an l-LDH gene from L. casei, which was constitutively expressed in E. coli (16).
TABLE 1.
Bacterial strains and plasmid used
| Strain or plasmid | Relevant characteristics | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| W3110 | F− IN(rrnD-rrnE)1 | B. Bachmann |
| BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | Lab collection |
| HB101 | supE44 hsdS20(rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Lab collection |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL relA deoC ptsF rbsR flbD | B. Bachmann |
| RR1 | supE44 hsdS20(rB−mB−) ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Lab collection |
| BW1677 | pta::TnphoA′-3 | B. Wanner |
| CP993 | OW1 Φ(pta-1::Tn10-lacZ) | C. Park (23) |
| NZN117 | LCB320 ldhA::Kan | D. P. Clark (5) |
| KJE103 | W3110 ppc::cat | Lab collection |
| JP201 | RR1 pta::TnphoA′-3 | P1 (BW16777) × RR1 |
| JP202 | RR1 Φ(pta-1::Tn10-lacZ) | P1 (CP993) × RR1 |
| JP203 | RR1 pta::TnphoA′-3 ppc::cat1 | P1 (KJE103) × JP201 |
| JP204 | RR1 Φ(pta-1::Tn10-lacZ) ldhA::Kan | P1 (NZN117) × JP202 |
| JP205 | RR1 Φ(pta-1::Tn10-lacZ) ldhA::Kan ppc::cat | P1 (KJE103) × JP204 |
| Plasmid pLS65 | l-LDH gene from L. casei | M. Y. Park (16) |
Media and culture conditions.
Luria-Bertani medium (22) supplemented with 15 g of glucose per liter was used to select the host strain suitable for metabolic engineering. M9 medium (22) containing 18 g of glucose per liter was used to characterize the mutants by in vivo nuclear magnetic resonance (NMR). The medium used for fed-batch cultivation of each strain to produce d- or l-lactate contained (per liter) 50 g of glucose, 5.0 g of NH4Cl, 1.5 g of KH2PO4, 0.5 g of MgSO4, 10 g of tryptone (Difco), 5 g of yeast extract (Difco), 300 μg of thiamine · HCl, and 1 ml of a trace element solution. The trace element solution contained (per liter) 10 g of Na3C6H5O7 · 2H2O, 13.2 g of CaCl2 · 2H2O, 8.4 g of FeSO4 · H2O, 2.4 g of MnSO4 · 4H2O, 2.4 g of ZnSO4 · H2O, 0.48 g of CuSO4 · 5H2O, 0.48 g of CoCl2 · 6H2O, 0.24 g of MoO4 · 2H2O, and 0.06 g of K2B4 · xH2O. When necessary, ampicillin (100 mg/liter), kanamycin (35 mg/liter), tetracycline (5 mg/liter), and chloramphenicol (34 mg/liter) were added to the culture broth. Most of the cultivations were carried out in a 3-liter fermentor (Korea Fermentor Co., Inchon, Korea) with a 1.5-liter working volume. For fed-batch addition of glucose, 60 ml of an 800-g/liter glucose solution was added to the culture medium when the residual glucose concentration was below 10 g/liter. The fermentor was operated at an aeration rate of 0.5 to 1.0 vol/vol/min (vvm) and an agitation speed of 500 to 1,000 rpm in order to maintain the dissolved oxygen level above 20% during aerobic cultivation. Anaerobic conditions were maintained by flushing with oxygen-free nitrogen gas at a flow rate of 0.1 vvm. The temperature and pH were maintained at 37°C and 7.0, respectively.
Analysis.
Optical density at 600 nm was measured with a Spectronic 21 colorimeter (Milton Roy Co., Rochester, N.Y.), and the dry cell weight was determined gravimetrically after the culture broth was centrifuged, washed with distilled water, and dried overnight at 105°C. One optical density unit was found to be equivalent to 0.56 ± 0.1 g (dry weight) of cells per liter. Concentrations of residual glucose were determined with a glucose analyzer (model 2300; YSI Co., Yellow Springs, Ohio). The amounts of fermentation acids, such as acetate, formate, lactate, pyruvate, and succinate, were determined by using a high-pressure liquid chromatograph equipped with a UV detector (Gilson Co., Villiers le Bel., France) and an Aminex HPX-87H column (Bio-Rad, Hercules, Calif.); chromatography was performed at 30°C, and compounds were eluted (elution rate, 0.5 ml/min) with 8 mM sulfuric acid. The concentrations of formate, ethanol, acetate, d- and l-lactate, pyruvate, and succinate were also determined with enzymatic test kits (Boehringer Mannheim GmbH, Mannheim, Germany). All determinations were performed in triplicate.
NMR.
The NMR experiments were modeled on the work of Alam and Clark (1), who monitored the synthesis of fermentation products by obtaining in vivo NMR scans of whole cultures. E. coli cells, which were grown to a density of 5 × 108 cells/ml in M9 medium in the presence of 18 g of glucose per liter and the required growth factors, such as thiamine (300 μg/liter), proline (10 mg/liter), and leucine (10 mg/liter), were collected by centrifugation at 5,000 × g for 2 min at 4°C. The cell pellets were washed twice with M9 buffer and resuspended in the medium used for cultivation. Five-nanometer NMR tubes were filled with the cell suspensions, placed in a BBL GasPak anaerobic system (Becton Dickinson and Co., Cockeysville, Md.), and incubated at 37°C for 4 h. Proton NMR spectra were obtained by using a Varion UNITY spectrometer operating at 500 MHz (Korea Basic Science Institute, Taejon, Korea). The water peak was suppressed, the field was locked onto the solvent D2O, and the internal reference was the H2O peak defined as 4.65 ppm. Dimethyl sulfone (100 mM) was used as an internal standard (3.12 ppm) for the quantification of the fermentation products. The relative amount of a product was normalized by using the amount of glucose consumed and the amount of product, as calculated from the product/internal standard ratio.
RESULTS
Balance of fermentation products in a pta mutant of E. coli RR1.
Acetate is one of the major fermentation products of E. coli, and the level of acetate that accumulated was different in each E. coli strain (19). Even the mutants which were defective in the Pta-AckA acetate production pathway secreted acetate in the presence of glucose (10). In order to select a host strain suitable for redirection of acetate metabolism, we compared the levels of anaerobic acetate produced on Luria-Bertani medium supplemented with 15 g of glucose per liter by five pta mutants which were constructed from commonly used strains of E. coli, including W3110, HB101, MC4100, BL21(DE3), and RR1. In order to construct pta mutants, each strain was transduced with P1 grown on BW16777, which carries TnphoA′-3 in the pta gene. One of the resultant mutants, a mutant containing a pta allele of RR1 (JP201), was selected for further study because it produced the smallest amount of acetate and had the highest conversion yield for d-lactate (data not shown).
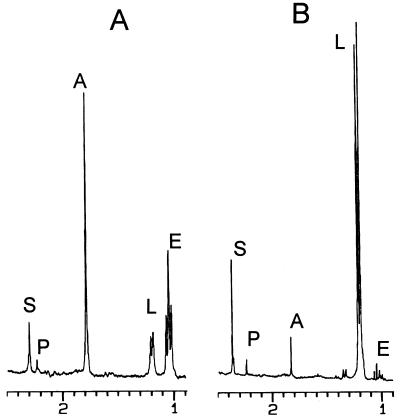
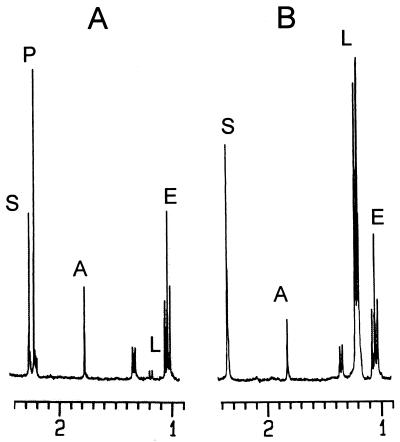
The anaerobic fermentation balances of JP201 and its parent strain, RR1, in M9 medium supplemented with 18 g of glucose per liter were compared by performing an in vivo NMR analysis as described in Materials and Methods (Fig. 2). E. coli RR1 produced a mixture of acetate, ethanol, d-lactate, and succinate from glucose. A peak due to formate was found downfield at 7.5 ppm (not shown in Fig. 2). The relative amounts of the products are shown in Table 2. Introduction of the pta mutation resulted in a large decrease not only in the production of acetate but also in the production of ethanol and formate; the amount of acetate formed in JP201 was one-tenth the amount formed in the parent strain, and the amounts of formate and ethanol produced by JP201 were one-fourteenth and one-eighth, respectively, the amounts produced by the parent strain. Although the adhE gene of JP201 was not manipulated, production of ethanol also decreased significantly, possibly due to the tight linking of acetate production and ethanol production (14). In contrast to these fermentation end products, the amount of d-lactate that accumulated increased significantly; d-lactate became the major fermentation product and represented about 80% of the total carbon found in fermentation products (Table 2). Formation of succinate also increased slightly (by 1.5-fold). As the fluxes of formate, acetate, and ethanol decreased significantly, the NADH produced from glycolysis had to be oxidized to NAD+ by lactate fermentation from pyruvate by strain JP201.
FIG. 2.
NMR scans of E. coli RR1 (A) and its pta mutant (B). Cultures were incubated anaerobically in minimal medium containing glucose and auxotrophic amino acids, including proline and leucine. Abbreviations: A, acetate; E, ethanol; L, lactate; P, pyruvate; S, succinate.
TABLE 2.
Fermentation balances of E. coli RR1 and its pta mutanta
| Carbon source or product | Relative amt consumed or produced by:
|
|
|---|---|---|
| RR1 | JP201 | |
| Glucose (carbon source) | 1.00 (13.1)b | 1.00 (20.3) |
| Products | ||
| Acetate | 0.60c | 0.06 |
| Ethanol | 0.58 | 0.07 |
| Lactate | 0.26 | 1.52 |
| Succinate | 0.13 | 0.19 |
| Pyruvate | 0.03 | 0.02 |
| Formate | 1.13 | 0.08 |
Fermentation balances were determined relative to a value of 100 per dimethyl sulfone, which was the internal standard used in all NMR experiments performed in this study. All measurements were obtained in triplicate and were independent. The standard deviations were less than 10%.
The values in parentheses are millimoles of glucose consumed.
Millimole of product (as quantified by NMR) per millimole of glucose consumed.
d-Lactate production with a pta mutant of E. coli RR1.
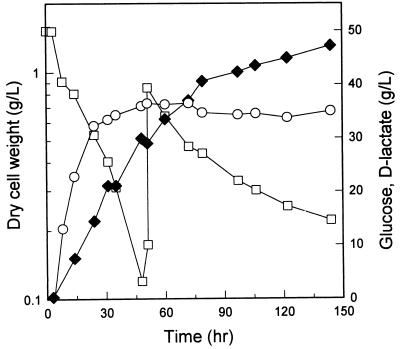
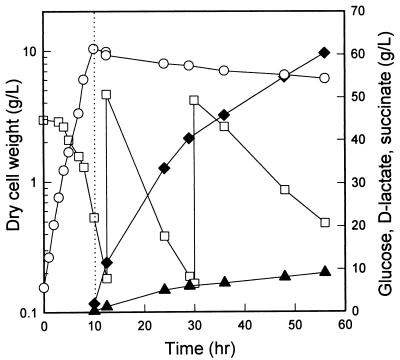
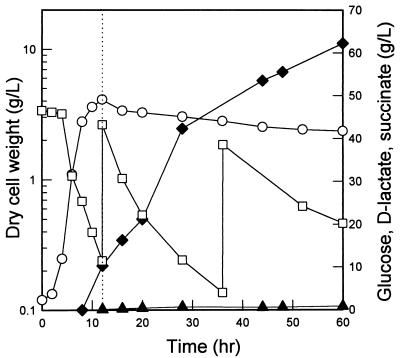
In order to determine the potential of the pta mutant as a host for production of optically pure d-lactate, JP201 was cultivated anaerobically with intermittent addition of glucose (Fig. 3). The maximum biomass concentration obtained was 0.9 g/liter, and 47 g of d-lactate per liter was produced in 150 h, indicating that JP201 metabolized glucose by d-lactate fermentation. Whereas d-lactate was produced efficiently under anaerobic culture conditions, E. coli can grow faster and reach higher biomass concentrations under aerobic conditions. Therefore, growing cells to a higher concentration under aerobic conditions before shifting them to the anaerobic d-lactate production phase should result in an improvement in the volumetric productivity by increasing the biomass concentration and thus reducing the total fermentation time. As shown in Fig. 4, after 10 g of dry cell mass per liter was obtained during aerobic cultivation, the culture was shifted to anaerobic conditions by flushing the bioreactor with oxygen-free nitrogen gas. As a result, 60 g of d-lactate per liter was produced in 56 h. When partially anaerobic conditions instead of anaerobic conditions were maintained by reducing the agitation speed to 300 rpm and the aeration rate to 0.2 vvm, a similar level of d-lactate was produced in 72 h (data not shown). In all of these production processes, succinate accumulated as the major by-product and accounted for up to 15% of the d-lactate, as shown in Fig. 4.
FIG. 3.
Production of d-lactate with the E. coli RR1 pta mutant under anaerobic conditions. The experiment was performed in duplicate, and the standard deviations were less than 10%. Symbols: ○, cell dry weight; □, glucose concentration (intermittent feeding); ⧫, d-lactate concentration.
FIG. 4.
Production of d-lactate with the E. coli RR1 pta mutant grown initially under aerobic conditions. The dotted line indicates the time when the culture was shifted to anaerobic conditions. The experiment was performed in triplicate, and the standard deviations were less than 10%. Symbols: ○, cell dry weight; □, glucose concentration (intermittent feeding); ⧫, d-lactate concentration; ▴, succinate concentration.
Homofermentative d-lactate production with an E. coli RR1 pta ppc mutant.
To prevent accumulation of succinate, a mutation in the gene for phosphoenolpyruvate carboxylase, the branch point leading to succinate synthesis, was introduced into JP201. The resulting pta ppc double mutant of E. coli RR1, JP203, was tested for production of d-lactate without succinate formation. When we used the medium and cultivation strategy used for the production of d-lactate with JP201, d-lactate was produced at concentrations up to 62.2 g/liter in 60 h with no accumulation of succinate. It is notable that JP203 entered the stationary phase earlier than JP201 entered this phase and that the biomass concentration was only 4 g/liter, whereas the JP201 biomass concentration was 10 g/liter. Although the biomass concentration was much lower than that of the pta mutant (Fig. 5), the volumetric productivity of this process was equivalent to that of the process in which the pta mutant was used (1.04 versus 1.09 g/liter/h). The yield in the d-lactate production phase was close to 0.9 g of d-lactate per g of glucose. This result demonstrated that the pta ppc double mutant metabolized glucose exclusively to d-lactate, like a homofermentative lactic acid bacterium.
FIG. 5.
Homofermentative production of d-lactate with the E. coli RR1 pta ppc double mutant. The dotted line indicates the time when the culture was shifted from aerobic conditions to anaerobic conditions. The experiment was performed in triplicate, and the standard deviations were less than 10%. Symbols: ○, cell dry weight; □, glucose concentration; (intermittent feeding); ⧫, d-lactate concentration; ▴, succinate concentration.
l-Lactate production with E. coli RR1 pta ldhA harboring an l-LDH gene from L. casei.
As shown above, pta mutants of E. coli RR1 were able to achieve a redox balance during anaerobic metabolism of glucose by d-lactate fermentation. In order to examine whether the foreign l-lactate pathway could replace the indigenous d-lactate pathway in the pta mutants, plasmid pLS65, which contained the l-LDH gene from L. casei, was introduced into JP203. However, the recombinant strain produced only d-lactate and did not produce l-lactate (data not shown). To prevent the production of d-lactate, an ldhA mutation from NZN117 was transduced into the pta mutant. Because the antibiotic marker of the ldhA mutation from NZN117 was kanamycin and thus was same as the antibiotic marker of the pta mutation in JP201, we transduced the ldhA mutation into another pta mutant of E. coli RR1 which was resistant to tetracycline, JP202. The resulting pta ldhA double mutant, JP204, fermented glucose to produce a mixture of ethanol, formate, acetate, pyruvate, and succinate (Fig. 6a and Table 3). As disruption of ldhA in a pta mutant should eliminate the alternative pathway which could oxidize NADH, excretion of pyruvate should be a response to disturbed metabolism of pyruvate and NADH. When plasmid pLS65 harboring the l-LDH gene from L. casei was introduced into JP204, l-lactate was produced as the major anaerobic product (Fig. 6b). In this recombinant strain, secretion of pyruvate disappeared and formation of formate and ethanol was reduced compared to the plasmid-free host strain. l-Lactate contained 75% of the carbon found in the fermentation products (Table 3).
FIG. 6.
NMR scans of the E. coli RR1 pta ldhA double mutant (A) and the recombinant strain harboring plasmid pLS65 (l-LDH) (B). Cultures were incubated anaerobically in minimal medium supplemented with glucose and auxotrophic amino acids, including proline and leucine. Abbreviations: A, acetate; E, ethanol; L, lactate; P, pyruvate; S, succinate.
TABLE 3.
Fermentation balances of the E. coli RR1 pta ldhA mutant and its recombinant harboring foreign l-LDH from L. caseia
| Carbon source or product | Relative amt consumed or produced by:
|
|
|---|---|---|
| JP204 | JP204(pLS65) | |
| Glucose (carbon source) | 1.00 (12.2)b | 1.00 (12.5) |
| Products | ||
| Acetate | 0.12c | 0.04 |
| Ethanol | 0.42 | 0.24 |
| Lactate | 0.00 | 0.72 |
| Succinate | 0.25 | 0.20 |
| Pyruvate | 0.36 | 0.00 |
| Formate | 0.44 | 0.21 |
Fermentation balances were determined relative to a value of 100 per dimethyl sulfone, which was the internal standard used in all NMR experiments performed in this study. All measurements were obtained in triplicate and were independent. The standard deviations were less than 10%.
The numbers in parentheses are millimoles of glucose consumed.
Millimole of product (as quantified by NMR) per millimole of glucose consumed.
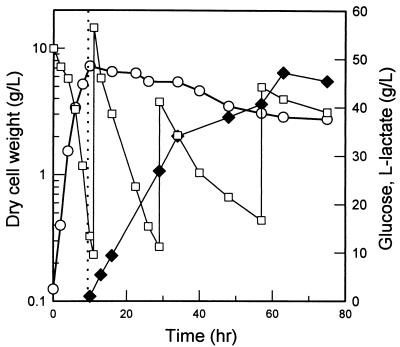
We tried to overproduce l-lactate by using the fermentation strategy used for d-lactate production. JP204 harboring plasmid pLS65 was grown aerobically for 12 h to a biomass concentration of 7.2 g/liter, and after the culture was shifted to anaerobic conditions, 45 g of l-lactate per liter was produced from 65 g of glucose per liter in 67 h (Fig. 7). Again, in this process succinate accumulated as the major by-product at levels corresponding to up to 12% of the l-lactate produced. To prevent succinate production, a ppc mutation was introduced by P1 transduction into l-lactate-producing strain JP204. The resulting strain, JP205, a pta ldhA ppc mutant harboring the l-LDH gene from L. casei, exhibited defects in aerobic growth; the maximum biomass concentration reached was only 3.3 g/liter, compared to 7.2 g/liter for control strain JP204. Growth could not be restored by adding any nutrient, including yeast extract, NH4Cl, KH2PO4, and the trace element solution. When the culture was shifted to anaerobic conditions, only 4.18 g of l-lactate per liter was produced in 33 h (data not shown). The volumetric glucose consumption rate was 0.35 g/liter/h, and the productivity of l-lactate was 0.146 g/liter/h. To produce pure l-lactate without coproduction of succinate, optimization of the medium and the culture strategy used for strain RR1 pta ppc ldhA harboring plasmid pLS65 are required.
FIG. 7.
Production of l-lactate with the E. coli RR1 pta ldhA double mutant harboring plasmid pLS65 (l-LDH). The dotted line indicates the time when the culture was shifted from aerobic conditions to anaerobic conditions. The experiment was performed in triplicate, and the standard deviations were less than 10%. Symbols: ○, cell dry weight; □, glucose concentration (intermittent feeding); ⧫, l-lactate concentration.
DISCUSSION
In this work, an E. coli RR1 pta mutant was used as the host for production of optically pure d- or l-lactic acid. A pta ppc mutant was able to metabolize glucose exclusively to d-lactate under anaerobic conditions, and a pta ldhA mutant harboring the l-LDH gene from L. casei produced optically pure l-lactate as the major fermentation product.
The key issue in anaerobic growth is recycling of NADH by conversion of pyruvate to fermentation products, so that glycolysis may continue (4, 6). The ratio of the products in a mixed-acid fermentation varies according to the nature of the substrate so that the amount of NADH produced corresponds to the amount of NADH consumed due to excretion of fermentation products (4, 6). Glucose produces two molecules of NADH when it is converted to pyruvate. During anaerobic growth, E. coli metabolizes pyruvate to acetyl coenzyme A (acetyl-CoA) by means of pyruvate-formate lyase with release of formate (4, 21). Acetyl-CoA is further metabolized to acetate or ethanol. Because no NADH is oxidized in the acetate pathway, whereas two molecules of NADH are recycled in the ethanol pathway, the fermentation of glucose could be balanced by production of a 50:50 mixture of ethanol and acetate (Fig. 1). The significance of this balance is illustrated by the fact that the mutants defective in pta or adhE were not able to grow on glucose anaerobically (7, 13). When we compared the fluxes of fermentation products in E. coli RR1 and its pta or ackA mutants, we found that the fluxes of formate and ethanol were directly proportional to the acetate flux, which suggested that the flux of the fermentation products, ethanol and formate, could be regulated by restriction of the acetate flux (14).
Although the pta mutants had been reported to be unable to grow anaerobically on glucose (13), Gupta and Clark found that some of the pta mutants showed significant anaerobic growth and produced an unusually large amount of succinate along with a somewhat increased proportion of lactate (14). Moreover, these workers showed that the double mutant strain lacking both ADH and Pta regained the ability to grow anaerobically on glucose by lactate fermentation (14). The pta mutant of E. coli RR1 constructed in this work (JP201) was able to grow anaerobically and to ferment glucose mainly to d-lactate in the presence of complex medium, whereas it did not grow anaerobically on glucose minimal medium. The production of ethanol, as well as the production of acetate, decreased significantly even though ADH was not eliminated by mutation (Table 2).
The proportion of lactate in the mixed-acid fermentation increased under low-pH conditions, because the fermentative LDH was induced by a combination of acidity and anaerobiosis (6, 20). Although E. coli contains three LDHs, only one of them is responsible for fermentative conversion of pyruvate to lactate (15, 17, 26). The other two LDHs are required for aerobic growth on d- or l-lactate (15, 17). The fermentative LDH is specific for the production of d-lactate and is activated by an increased concentration of pyruvate (26). Greatly reduced production of acetate and ethanol in the pta mutant should result in the accumulation of acetyl-CoA, which in turn should shift the equilibrium of pyruvate-formate lyase in the reverse direction. Also, it has been noted previously that pta mutants express reduced levels of pyruvate-formate lyase (18). As a result, the intracellular concentration of pyruvate in a pta mutant increases, which activates the fermentative LDH gene. Moreover, Bunch et al. reported that when they isolated mutants whose expression of fermentative LDH increased and became independent of the pH of the medium, some of the mutants were pta mutants (5). Therefore, increased expression of the ldhA gene due to mutation in pta and activation of LDH by an elevated concentration of pyruvate should make the LDH pathway the major fermentation pathway.
Not only the indigenous d-lactate pathway but also the foreign l-LDH pathway was found to function as the major NADH-oxidizing pathway so that anaerobic metabolism could continue. A pta ldhA double mutant did not produce acetate or d-lactate but produced pyruvate and succinate. Introduction of the l-LDH gene from L. casei into this mutant resulted in production of l-lactate as the major fermentation product, and thus the accumulation of pyruvate and succinate was eliminated. These results show that any fermentation pathway which is able to achieve a redox balance can replace the indigenous fermentation pathway(s) of E. coli. Therefore, mutants can be developed as useful hosts not only for production of d- or l-lactate but also for production of any indigenous or nonindigenous metabolites requiring the cofactor balances.
There are many advantages of using E. coli as a host for production of lactic acid, such as the ability of this organism to produce optically pure lactate, its rapid growth under both aerobic and anaerobic conditions, its ability to metabolize various carbon sources, and its simple nutritional requirements. Because E. coli has only one fermentative LDH (d-lactate specific), it could be easily developed as a host for production of optically pure d- or l-lactate by metabolic engineering. The pta mutant of E. coli RR1 metabolized glucose mainly to d-lactate with a conversion yield of 0.80 g of d-lactate per g of glucose (Fig. 3). In the d- or l-lactate-producing E. coli strains constructed in this work, succinate was a major by-product, accounting for up to 15% of the total carbon of the products. Therefore, mutants defective in succinate production as well as in the pta gene were required; the resulting pta ppc double mutant metabolized glucose exclusively to d-lactate, like a homofermentative bacterium (Fig. 4). The production yield for d-lactate was as high as 0.9 g of lactate per g of glucose. However, the growth defect of the ppc mutant should be noted and understood since ppc mutants cannot produce oxaloacetate from phosphoenolpyruvate, which makes ppc mutants auxotrophic for a dicarboxylic acid, such as succinate (11).
Other advantageous characteristics of E. coli, namely, the rapid growth of this organism and its ability to maintain metabolic activity under both aerobic and anaerobic conditions, were used to develop the process. E. coli grows faster and easily reaches a higher biomass concentration under aerobic conditions than under anaerobic conditions. Therefore, the d- or l-lactate-producing strains were grown to higher biomass concentrations under aerobic conditions before the anaerobic production phase was started, which reduced the fermentation time, thus improving productivity. As a result, the volumetric productivity was improved from 0.313 to 1.09 g/liter/h in the case of d-lactate production. Although the level of productivity of the E. coli process is still low compared with previously reported values (9, 27), it can be further improved by increasing the biomass concentration and optimizing the production conditions. Separation of the growth phase and the production phase should be advantageous in designing more flexible processes. In summary, we demonstrated in this work that E. coli pta mutants can be used as hosts for production of optically pure d- or l-lactate. Combined with the advantageous characteristics of E. coli, metabolically engineered strains should provide powerful tools for the production of useful metabolites.
ACKNOWLEDGMENTS
We are grateful to B. Bachmann for providing E. coli W3110 and MC4100 to B. L. Wanner for providing BW1677, to C. Park for providing CP993, to D. P. Clark for providing NZN117, to J. E. Kim for providing KJE103, to M. Y. Park for providing plasmid pLS65, and to K. H. Yoon and S. Shin for help with gene manipulation techniques.
This work was supported by grant KG1141 from the Korea Research Institute of Bioscience and Biotechnology.
REFERENCES
- 1.Alam K Y, Clark D P. Anaerobic fermentation balance of Escherichia coli as observed by in vivo nuclear magnetic resonance spectroscopy. J Bacteriol. 1989;171:6213–6217. doi: 10.1128/jb.171.11.6213-6217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benthin S, Villadsen J. Production of optically pure d-lactate by Lactobacillus bulgaricus and purification by crystallisation and liquid/liquid extraction. Appl Microbiol Biotechnol. 1995;42:826–829. [Google Scholar]
- 3.Bhowmik T, Steele J L. Cloning, characterization and insertional inactivation of the Lactobacillus helveticusd(−)-lactate dehydrogenase. Appl Microbiol Biotechnol. 1994;41:432–439. doi: 10.1007/BF00939032. [DOI] [PubMed] [Google Scholar]
- 4.Boeck A, Sawers G. Fermentation. In: Neidhardt F, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 262–282. [Google Scholar]
- 5.Bunch P K, Mat-Jan F, Lee N, Clark D P. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143:187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 6.Clark D P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;63:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham P R, Clark D P. The use of suicide substrates to select mutants of Escherichia coli lacking enzymes of alcohol fermentation. Mol Gen Genet. 1986;205:487–493. doi: 10.1007/BF00338087. [DOI] [PubMed] [Google Scholar]
- 8.Datta R, Tsai S, Bonsignore P, Moon S, Frank J R. Technological and economic potential of poly(lactic acid) and lactic acid derivatives. FEMS Microbiol Rev. 1995;16:221–231. [Google Scholar]
- 9.de Boer J P, Teixeira de Mattos M J, Neijssel O M. d(−)Lactic acid production by suspended and aggregated continuous cultures of Bacillus laevolacticus. Appl Microbiol Biotechnol. 1990;34:149–153. [Google Scholar]
- 10.Diaz-Ricci J C, Regan L, Bailey J E. Effect of alteration of the acetic acid synthesis pathway on the fermentation pattern of Escherichia coli. Biotechnol Bioeng. 1991;38:1318–1324. doi: 10.1002/bit.260381109. [DOI] [PubMed] [Google Scholar]
- 11.Fraenkel D G. Glycolysis. In: Neidhardt F, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 189–198. [Google Scholar]
- 12.Fujita N, Miwa T, Ishijama S, Izui K, Katsuki H. The primary structure of phosphoenolpyruvate carboxylase of Escherichia coli. Nucleotide sequence of the ppc gene and deduced amino acid sequence. J Biochem. 1984;95:909–916. doi: 10.1093/oxfordjournals.jbchem.a134718. [DOI] [PubMed] [Google Scholar]
- 13.Guest J R. Anaerobic growth of Escherichia coli K12 with fumarate as terminal electron acceptor. Genetic studies with menaquinone and fluoroacetate-resistant mutants. J Gen Microbiol. 1979;115:259–271. doi: 10.1099/00221287-115-2-259. [DOI] [PubMed] [Google Scholar]
- 14.Gupta S, Clark D P. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J Bacteriol. 1989;171:3650–3655. doi: 10.1128/jb.171.7.3650-3655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haugaard N. d- and l-lactic acid oxidases of Escherichia coli. Biochim Biophys Acta. 1959;31:66–72. doi: 10.1016/0006-3002(59)90439-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim S F, Baek S J, Pack M Y. Cloning and nucleotide sequence of the Lactobacillus casei lactate dehydrogenase gene. Appl Environ Microbiol. 1991;57:2413–2417. doi: 10.1128/aem.57.8.2413-2417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kline E S, Mahler E R. The lactic acid dehydrogenases of Escherichia coli. Ann NY Acad Sci. 1965;119:905–917. doi: 10.1111/j.1749-6632.1965.tb47451.x. [DOI] [PubMed] [Google Scholar]
- 18.Kwan H S, Chui H W, Wong K K. ack::Mud 1-8 (Aprlac) operon fusions of Salmonella typhimurium LT2. Mol Gen Genet. 1988;211:183–185. doi: 10.1007/BF00338411. [DOI] [PubMed] [Google Scholar]
- 19.Luli G W, Strohl W R. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl Environ Microbiol. 1990;56:1004–1011. doi: 10.1128/aem.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mat-Jan F, Alam K Y, Clark D P. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J Bacteriol. 1989;171:342–348. doi: 10.1128/jb.171.1.342-348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pecher A, Blaschkowski H P, Knappe K, Bock A. Expression of pyruvate formate-lyase of Escherichia coli from the cloned structural gene. Arch Microbiol. 1982;132:365–371. doi: 10.1007/BF00413390. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanier R Y, Ingraham J L, Wheelis M L, Painter P R. The microbial world. 5th ed. Englewood Cliffs, N.J: Prentice-Hall; 1986. Gram-positive fermentative eubacteria; pp. 495–504. [Google Scholar]
- 25.Tamada M, Begum A A, Sadi S. Production of l(+)-lactic acid by immobilized cells of Rhizopus oryzae with polymer supports prepared by gamma ray induced polymerization. J Ferment Bioeng. 1992;76:379–383. [Google Scholar]
- 26.Tarmy E M, Kaplan N O. Kinetics of Escherichia coli B d-lactate dehydrogenase and evidence for pyruvate controlled change in conformation. J Biol Chem. 1968;243:2587–2596. [PubMed] [Google Scholar]
- 27.Vickroy T B. Lactic acid. In: Moo-Young M, editor. Comprehensive biotechnology: the principles, applications, and regulations of biotechnology in industry, agriculture and medicine. Vol. 2. New York, N.Y: Pergamon Press; 1985. pp. 761–776. [Google Scholar]
- 28.Wehrenberg R H., II Lactic acid polymers: strong, degradable thermoplastics. Mater Eng. 1981;94:63–66. [Google Scholar]