Abstract
Objective:
Automatic detection and analysis of respiratory events in sleep using a single respiratory effort belt and deep learning.
Methods:
Using 9,656 polysomnography recordings from the Massachusetts General Hospital (MGH), we trained a neural network (WaveNet) to detect obstructive apnea, central apnea, hypopnea and respiratory-effort related arousals. Performance evaluation included event-based analysis and apnea-hypopnea index (AHI) stratification. The model was further evaluated on a public dataset, the Sleep-Heart-Health-Study-1, containing 8,455 polysomnographic recordings.
Results:
For binary apnea event detection in the MGH dataset, the neural network obtained a sensitivity of 68%, a specificity of 98%, a precision of 65%, a F1-score of 67%, and an area under the curve for the receiver operating characteristics curve and precision-recall curve of 0.93 and 0.71, respectively. AHI prediction resulted in a mean difference of 0.41±7.8 and a r2 of 0.90. For the multiclass task, we obtained varying performances: 84% of all labeled central apneas were correctly classified, whereas this metric was 51% for obstructive apneas, 40% for respiratory effort related arousals and 23% for hypopneas.
Conclusion:
Our fully automated method can detect respiratory events and assess the AHI accurately. Differentiation of event types is more difficult and may reflect in part the complexity of human respiratory output and some degree of arbitrariness in the criteria used during manual annotation.
Significance:
The current gold standard of diagnosing sleep-disordered breathing, using polysomnography and manual analysis, is time-consuming, expensive, and only applicable in dedicated clinical environments. Automated analysis using a single effort belt signal overcomes these limitations.
Keywords: Sleep apnea, Respiratory event detection, Respiratory effort, Deep learning, Apnea Hypopnea Index, Polysomnography
I. INTRODUCTION
Sleep disorders such as sleep apnea and insomnia affect millions of people worldwide [1]. Clinical effects include difficulty in initiating and maintaining sleep, impaired alertness, and hypertension. Excessive daytime sleepiness and fatigue, two common symptoms associated with sleep disorders, have a large impact on population health [2], [3]. Accurate and timely diagnosis of a patient’s sleep disorder is therefore essential. Patients with apnea, especially obstructive sleep apnea, are at increased risk for traffic accidents, postoperative complications, and delirium [4], [5]. Untreated sleep apnea is associated with arrhythmias, heart failure and stroke. Studies that measure the apnea-hypopnea index (AHI) show that an estimated 49.7% of male and 23.4% of female adults have moderate-to-severe sleep-disordered breathing, though a lower percentage are clinically symptomatic [5].
The gold standard to measure sleep objectively is laboratory-based polysomnography (PSG). PSG is conventionally scored based on the American Academy of Sleep Medicine (AASM) guidelines. Scoring PSG recordings is a time-consuming task performed by specialists in dedicated sleep centers, making this an expensive process both in time and costs. Automation of PSG analysis would decrease the required analysis time and reduce costs. Moreover, automated PSG analysis computer models could be implemented in clinical centers anywhere in the world and across a variety of data acquisition options, including home sleep testing, testing in acute care environments, specific operational conditions such as high altitude, and consumer wearable devices.
Medical data is complex and involves a large number of variables and context that are difficult to encompass by programs based on a fixed set of rules. Deep learning models such as convolutional neural networks (CNN) and recurrent neural networks (RNN) have been applied in many domains to solve complex pattern recognition tasks [6]. Deep learning algorithms rely on patterns and inference rather than explicit instructions and can learn intricate relationships between features and labels from data. Implementing neural networks has become relevant in analyzing the heterogeneous kinds of data generated in modern clinical care [7]. Various types of deep learning algorithms have been found to be suitable for analyzing specific types of data. For instance, CNNs have been successful in classifying objects in images. Typical CNN architectures, however, are not ideal when analyzing temporal data. Temporal data is typically better exploited by RNNs. However, the recently introduced CNN, WaveNet architecture has been found to perform better than RNNs on several tasks [8]. WaveNet’s architecture resembles a typical CNN, yet the application of dilated causal convolutions creates an effectively larger receptive field. This renders WaveNet capable of detecting both spatial patterns and long-range temporal patterns. WaveNet was originally designed to synthesize speech; however, its application has been found suitable for analyzing other types of signals. In 2018 a challenge organized by the PhysioNet Computing in Cardiology aimed to detect sleep arousals from a variety of physiological signals, including signals derived from respiration. The winning model was a modified WaveNet architecture, suggesting that this CNN architecture can indeed perform successfully in other domains such as the automation of PSG-related tasks [9].
In the last two years a significant number of papers have been published on the detection of sleep apnea, as described by recent review papers [24], [25]. Finding a patient-friendly and accurate sensor or signal, especially in combination with a suitable analysis model, is clearly an ongoing area of high relevance. An overview of other sleep apnea studies that use large datasets (at least 96 patients) can be found in Table I.
TABLE I.
Overview of other studies performing automated respiratory event detection using 96 patients or more
| Study | Dataset size | Signal type | Analysis model | Classifier type | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1-score (%) | AUCROC (%) | AUCPR (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| [10] | 10.000 | Airflow, respiration chest, abdomen, oxygen saturation | RCNN | G | 88.2 | - | - | - | - | - | - |
| [11] | 2100 | Respiration abdomen | LSTM | OA/H/N | 77.2 | 62.3 | 80.3 | 39.9 | - | 77.5 | 45.3 |
| [12] | 1507 | Nasal airflow, abdominal, thoracic plethysmography | CNN1D-3ch | A/N | 83.5 | 83.4 | - | 83.4 | 83.4 | - | - |
| [13] | 1507 | Nasal airflow | CNN2D | A/H/N | 79.8 | 79.9 | - | 79.8 | 79.7 | - | - |
| [14] | 545 | Electrocardiography | CNN1D-LSTM MHLNN | G | 79.5 | 77.6 | 80.1 | - | 79.1 | - | - |
| [15] | 520 | Airflow | MHLNN | G | 87.2 | 88.3 | 87.8 | - | - | - | - |
| [16] | 285 | Voice and facial features | GMM | G | 72 | 73 | 65 | - | - | - | - |
| [17] | 188 | Airflow, respiratory rate variability | LR | G | 72 | 80 | 59 | - | - | - | - |
| [18] | 187 | pulse oximetry | CTM | G | 87 | 90 | 83 | - | - | - | - |
| [19] | 186 | Breathing sounds | Binary-RF | G | 86 | - | - | - | - | - | - |
| [20] | 179 | Nasal pressure | CNN1D | A/H/N | 96.6 | 81.1 | 98.5 | 87 | - | - | - |
| [21] | 120 | Breathing sounds | MHLNN | G | 75 | - | - | - | - | - | - |
| [22] | 100 | Nasal airflow | CNN1D | OA/N | 74.7 | 74.7 | - | 74.5 | - | - | - |
| [23] | 96 | Nasal airflow | GMM | OA/CA/H/N | 83.4 | 88.5 | 82.5 | 46.6 | 42.7 | 86.7 | - |
Analysis models: RCNN = recurrent and convolutional neural networks, LSTM = long short-term memory, CNN = convolution neural network, MHLNN = multiple hidden layers neural network, GMM = gaussian mixture model, LR = logistic regression, CTM = central tendency measure, RF = random forest. Classifier types: A = apnea, H = hypopnea, N = normal, O = obstructive, C = central G = global.
Sleep apnea detection methods typically use various breathing measurements and oximetry [25]. Alternative methods using signals derived from electrocardiography (ECG) have shown some promise for predicting AHI as well, although such data has an indirect relationship to the respiratory system and therefore to sleep apnea [24], [26]. This more indirect method of analyzing respiration requires additional processing and can be affected by other illnesses including heart failure and cardiac arrhythmias, rather than sleep apnea [11]. Classification of respiratory events typically requires both airflow and respiratory effort signals. Using multiple physiological signals to detect sleep apnea can provide good performance [10], [12]. However, this leads to similar problems as the current gold standard; using many different sensor signals is considered uncomfortable, expensive, and time-consuming. Recent studies show that automated apnea scoring with limited sensors use (i.e. airflow or respiratory effort) can still yield acceptable performance [11], [23], [27]. Using airflow or respiratory effort for apnea detection bear different advantages and disadvantages. Airflow measures are expected to yield slightly better performance but need access to the nose/mouth, which may be difficult in specific environments. In situations where the airflow signal may not be readily acquired, an effort-belt based classification could overcome this limitation. Examples include in intensive care units, home tracking in heart failure or chronic obstructive pulmonary disease, those using nasal oxygen, and war fighter conditions. The effort belt is highly convenient, and this input signal can be acquired by a range of contact and contactless technologies in nearly every possible environment.
The ability to identify and discriminate between the specific respiratory events that are typically scored in PSG while using fewer signals is unknown to the current clinical setting. In this research we aimed to create a fully automated method that can detect respiratory events, discriminate between the different types of respiratory events, and assess the AHI with sufficient efficiency for clinical implementation using only a single respiratory effort belt.
II. METHODS
A. Dataset
The dataset used to train our model was from The Massachusetts General Hospital (MGH) sleep laboratory (2008–2018), summarized in Table II. The MGH Institutional Review Board approved the retrospective analysis of the clinically acquired PSG data. In total 9656 PSG recordings were successfully exported. We applied a 5-fold cross-validation for which we split the dataset into training, validation, and test subsets using respective ratios of 70%, 10%, 20% of the total number of recordings. Multiple records from the same patients were constrained to the same fold. Patients with and without breathing assistance by continuous positive airway pressure (CPAP) were included.
TABLE II.
Dataset distribution (N=9656)
| Category | Bin | Percentage of all patients |
|---|---|---|
|
| ||
| Sex | male | 58.9% |
| female | 40.7% | |
| unknown | 0.4% | |
|
| ||
| Age | < 60 | 65.0% |
| 60 – 80 | 34.4% | |
| > 80 | 2.6% | |
|
| ||
| BMI | underweight (< 18.5) | 0.9% |
| normal weight (18.5 – 25) | 13.7% | |
| overweight (25 – 30) | 26.7% | |
| obese (> 30) | 58.7% | |
|
| ||
| AHI | normal (< 5) | 39.9% |
| mild (5 – 15) | 27.7% | |
| moderate (t5 – 30) | 20.4% | |
| severe (>= 30) | 12.0% | |
|
| ||
| Recording type | diagnostic | 48.1% |
| split night | 24.3% | |
| all night CPAP | 24.0% | |
| unknown | 3.6% | |
|
| ||
| Events (N=675,667) | obstructive apnea | 18.6% |
| central apnea | 14.2% | |
| mixed apnea | 1.7% | |
| hypopnea | 36.8% | |
| RERA | 28.7% | |
We included a secondary test dataset for external validation of our model (trained on all MGH data). This dataset was collected by the Sleep Heart Health Study (SHHS) and included 8455 PSG recordings. For this research we only used the signal measured at the abdomen using a respiratory effort belt (inductance plethysmography). This signal, in comparison to the available respiratory signals measured on the thorax, is expected to provide the best predictive performance [28].
The MGH sleep center is an AASM accredited sleep center, with stringent ongoing requirements for documenting and maintaining high inter-scorer reliability. The center maintains an inter-rater reliability of over 85%. Respiratory event detections included obstructive apneas, central apneas, mixed apneas, hypopneas, and respiratory effort-related arousals (RERAs). Because of the relatively low number of mixed apnea events in our dataset, 1.7% of all events, all mixed apnea events were labeled as obstructive apnea, since the characteristics are expected to look most similar. We define respiratory events as a term that encompasses any type of apnea, hypopnea and RERA. The definition apnea reflects any type of apnea and hypopnea.
Recordings obtained from the SHHS database were annotated according to SHHS guidelines. A key difference between the two datasets is the primary respiratory scoring signal in the original source – nasal pressure (MGH) and thermistor (SHHS). This difference and implications will be discussed further below. Besides the different flow sensors the MGH and SHHS dataset only include labels that are scored using the same criteria as defined by the AASM (4% rule for hypopneas), and individual recordings were annotated by a single scorer for both datasets. For the MGH data there was a total of 7 scorers whereas for the SHHS data the number of experts is not reported. We chose the SHHS dataset as it was the largest study which used a uniform methodology for acquisition and scoring.
B. Preprocessing and data preparation
All recordings that were incomplete or did not include any sleep were removed. For the SHHS dataset, we only used recordings that contain mostly good quality abdominal effort signals, as defined by the SHHS [29]. Specifically, for the visit 1 and visit 2 subsets, only recordings with at least 4 hours of artifact-free signal or 75% of artifact-free signal, respectively, were included. To extract the relevant respiratory information and remove present noise, minimal preprocessing techniques were applied. The abdominal respiration measurement from both the MGH data and the SHHS data consisted of a single channel with a sampling frequency of 125 Hz, 200 Hz or 250 Hz. A notch filter of 60 Hz was applied to reduce line noise. A low-pass filter of 10 Hz was applied to remove higher frequencies not of interest, and consequently all recordings were resampled to 10 Hz. Z-score normalization was performed using the mean and standard deviation of the 1st to 99th percentile clipped signal to optimize the training process of the neural network.
The training data was segmented into 7-minute segments (see II-D), combined with a stride of 30 seconds to reduce the large training dataset size. Each segment was assigned one ground truth class label – the sleep expert’s label located in the center of the segment. We segmented the test data in the same way, except that we used a stride of 1 second, which allowed for a respiratory event prediction for each second.
C. Model and prediction tasks
In this research we utilized a WaveNet model (see model architecture in Section II-D) to automatically detect apneas, hypopneas, and RERAs from a single effort belt signal, without use of additional sensors that are conventional in PSG measurements (e.g. thermistor, nasal pressure, oxygen saturation, electroencephalography or electrocardiography), and without using human-engineered features. As described above, the signal was split into 7-minute segments and, in this way, the model was trained to predict only the center index of a 7-minute segment, while having 3.5 minutes of context information before and after the center index. We designed the following two prediction tasks:
Binary classification to discriminate non-apnea events from apnea-hypopnea events (regular breathing and respiratory events). Based on the predicted respiratory events, we computed the predicted AHI as the number of predicted respiratory events per hour of sleep.
Multiclass classification to discriminate the respiratory event classes: no-event, obstructive apnea or mixed apnea, central apnea, RERA, hypopnea. From the sum of the detected respiratory events, we determined the AHI and respiratory disturbance index (RDI).
In both tasks we used the originally scored multiclassification labels. For our binary classification task we converted all types of apnea and hypopneas into one grouped class, apnea. In both experiments our model provided a probability for all included classes. The highest probability among the possible classes constitutes the output of our model. In Fig. 1 the complete workflow scheme is shown.
Fig. 1.

Data flow scheme for model development and testing. The model was trained and validated on the dataset from the Massachusetts General Hospital (MGH) whereas the dataset from the Sleep Heart Health Study (SHHS) was used for external validation. Both the Apnea Hypopnea Index (AHI) and the respiratory disturbance index (RDI) were computed during post-processing.
D. Model architecture
WaveNet is a fully convolutional neural network [8], [30]. In Appendix Fig. 7 we show the schematics of a residual block of the WaveNet model. The architecture makes use of an exponentially increasing dilation factor resulting in exponential growth of the receptive field with each layer. This causes the receptive field to double in length for each hidden layer. In previous work we showed that 4.5-minute segments are ideal for sleep staging from respiratory effort data [31]. The exponential growth of receptive field gives us a limited number of options without making major changes to the fundamental architecture of the WaveNet model (10 layers is equivalent to 1.7 minutes, 11 layers is equivalent to 3.4 minutes, and 12 layers is equivalent to 6.8 minutes). To ensure enough context for our respiratory event scoring task we opted for 12 hidden layers, resulting in 4096 samples in our 10 Hz signal, equivalent to approximately 7 minutes of context. Instead of using WaveNet as a generative model that uses the last output as its subsequent input (recurrent generation), we trained WaveNet in a supervised manner where the input is a respiratory effort signal and the prediction target is either binary or quinary, for experiment 1 and 2 respectively. The original WaveNet model makes use of causal convolutions where the output is a function of previous time steps only, no future time steps. For this research we modified the WaveNet architecture by using non-causal convolutions and shifting the output node, which results in that the output is now a function of both previous and future time steps. Non-causal connection, i.e. past and future context, matches better to a human sleep scorer conceptually, as they have access to the full night recording. As in the original paper, a kernel size of 2 was used. The number of filters for each of the convolutions was set to 32, with a dropout rate of 0.2, without further hyperparameter optimization. The categorical cross entropy loss function was applied during training,
| (1) |
where y represents the predicted probability distribution, y′ represents the true distribution, and N represents the number of classes. To address overfitting and to improve generalization of the network, besides using dropout, we have implemented an early stopping procedure, where we stop training if the performance on a validation set does not increase for 10 consecutive training rounds. We used a batch size of 150 segments and a learning rate of 0.001, which was reduced to 10% when three consecutive training rounds showed no improvement. ADAM optimization was used for training the classifier.
E. Boosting for imbalanced data
Classification with imbalanced data is challenging in many real-world deep learning applications [32], [33]. For the PSG recordings in our research, the number of segments containing only regular breathing is typically much larger than the segments containing respiratory events, even for patients classified with severe apnea. For this problem we designed a boosted model approach by applying a binary WaveNet classifier, or boost-model, over multiple iterations. To remove a large proportion of segments with regular breathing without removing many segments including apnea events, only segments with an extremely high probability of regular breathing were removed by the boost-model. In our approach we selected a probability threshold to make our boost-model extremely sensitive for apneas, based on the receiver operating characteristic (ROC) curve. In the first iteration we used a true positive rate of 0.995 and decreased this value by 0.010 for each subsequent iteration. The boosted model iterations stopped when the desired balance in classes was obtained. This balance was defined by 3.3:1 ratio of regular breathing with respect to the sum of events.
In Fig. 2 the boosted model flow scheme is shown.
Fig. 2.
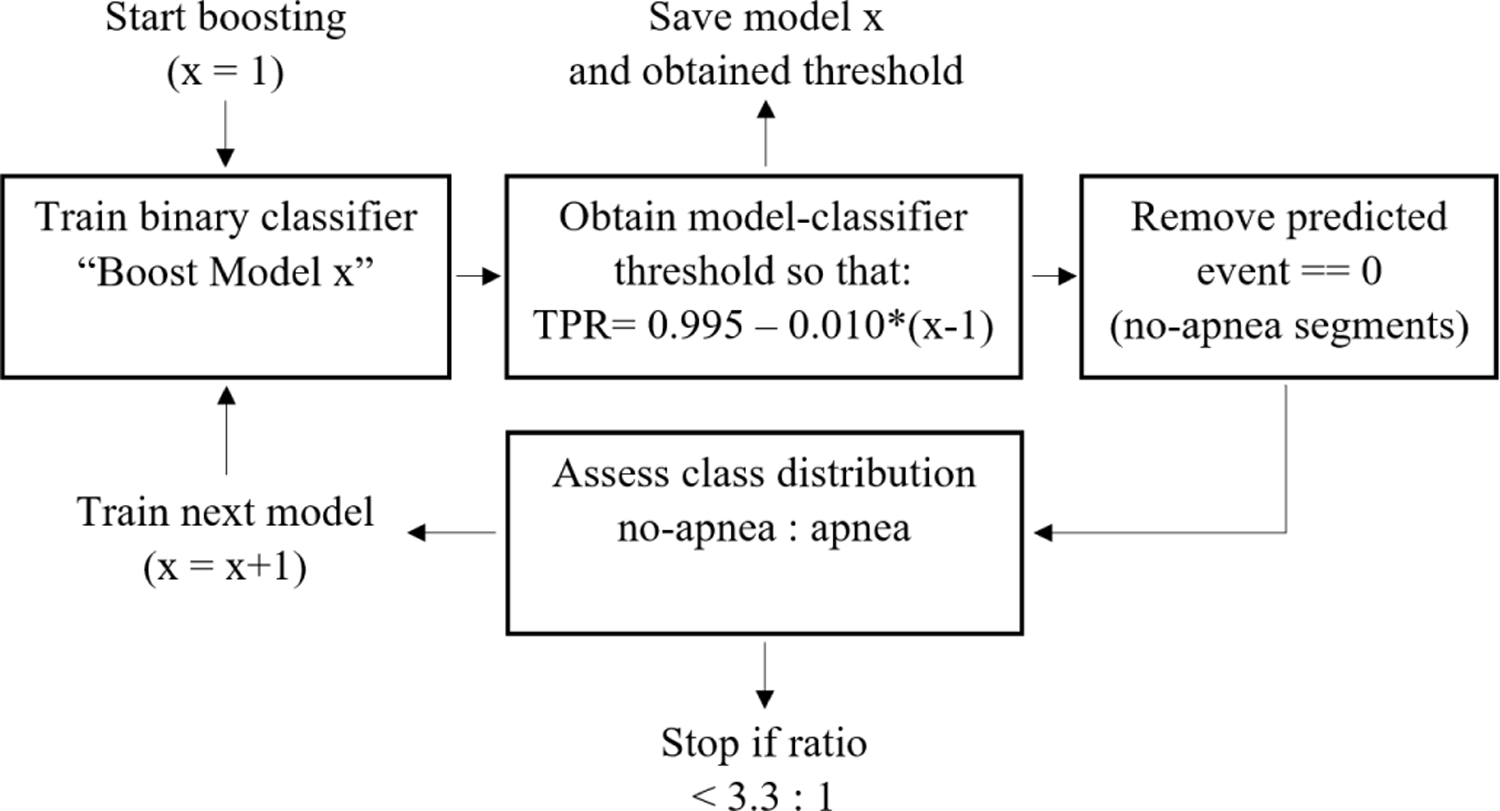
Flow scheme for the boosted model process that was performed to create a more balanced dataset to train our WaveNet model.
In each iteration, the boost model received non-rejected samples from the previous iteration, i.e. only true positive and false positive segments. Using this approach, the boost-model was trained to discriminate regular breathing from other respiratory events, while being exposed to a decreasing and increasingly more challenging dataset. In this way, in every iteration our boost-model should learn new nuanced characteristics that define a normal breathing rhythm. Using this boosted approach, we vastly reduced the number of segments containing regular breathing and improved effective classification by our main WaveNet model. Moreover, the boost-model was expected to remove segments with regular breathing that are relatively simple to distinguish from apnea while leaving the more complex segments for our main model. All boost-models were trained on the training dataset using the same hyperparameters as our main binary model, and were applied on the validation and test dataset.
F. Post-processing
After applying our model, we obtained an apnea prediction for each second. The prediction resolution of 1 Hz allowed high fluctuations of predicted events and allowed detection of very brief events. Both situations were considered not physiologically plausible, therefore, we designed a smoothing algorithm. This smoothing algorithm removed short events and rapidly changing events. The algorithm was based on a moving window of 10 seconds. The following rules were applied for each window.
When a minimum of 3 out of 10 seconds was classified as no-event, the complete window was set to no-event.
If a window was classified as an event and multiple types of events were present, the type of event that occurred most became the prediction for the complete window.
The selection of 3 seconds was based on manual, visual analysis on classifier outputs on a small subset of the patients in the training set (less than 20 patients). We do not believe the performance is sensitive to the choice of this parameter (i.e. between 2 and 4 out of the 10 seconds).
Finally, consecutive events with a combined length of two windows or greater were converted into a single event prediction. The type of event that held the largest proportion among the combined events indicated this new prediction. Applying the smoothing algorithm resulted in predicted events with a minimum length of 10 seconds, similar to what is suggested by AASM guidelines.
G. Model evaluation
We computed confusion matrices of the per-event performance of our model to obtain a performance granularity with better clinical interpretability than using sample-to-sample comparison. The true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN) were computed for each of the 2 or 5 classes, respectively, for experiment 1 and 2. A predicted event was considered a correct prediction when more than 50% of its duration overlaps with an expert label. The number of true negatives was computed by the accumulated time where no-event was found divided by the average duration of all expert-scored events (i.e. 18 seconds). Next, the TP, TN, FP and FN values were used to determine the following event-per-event performance metrics of our model.
Additionally, Cohen’s kappa values were determined using the formula in Cohen’s original work [34]. For all above mentioned metrics, we obtained the 95% confidence interval by bootstrapping over patients (sampling with replacement by blocks of patients) 10.000 times. The confidence interval was computed as the 2.5% (lower bound) and the 97.5% percentile (upper bound). For the binary tasks the ROC curve and precision-recall (PR) curve and their corresponding areas (AUCroc AUCpr) were computed.
In addition to event-per-event evaluation, we evaluated global scoring performance. Global assessment of sleep apnea severity is typically used for clinical diagnosis [24]. For the first experiment we determined the AHI value per patient, whereas in the second experiment we determined the AHI and RDI value per patient using the following computations.
where OA is obstructive apneas, CA is central apneas, and HY is hypopneas. We used the already scored sleep stages from the original annotations to differentiate between sleep and wake time for the patients. We decided to not use ”time in bed” as the denominator, as in previous work we have shown it is possible to reasonably stage sleep with one effort belt signal as input [31]. To keep the focus of this paper on apnea detection with one effort belt, we believe using the expert sleep labels helps to best answer our research question.
With the AHI score all patients were categorized as normal or mild, moderate or severe sleep apnea. Categorization was according to conventional criteria as defined by AASM guidelines.
Normal breathing: AHI < 5
Mild sleep apnea: 5 ≤ AHI < 15
Moderate sleep apnea: 15 ≤ AHI < 30
Severe sleep apnea: AHI ≥ 30
We obtained the classification accuracy of our model by creating a confusion matrix for the four AHI scores. The classification accuracy displays the ability of the model to assign a patient to any of the four AHI categories. To gain insight into accuracy of the AHI prediction disregarding the discrete borders used in categorization, histograms and Bland-Altman plots computed to show the difference between the AHI value scored by the experts and the AHI value predicted by our model. For both experiments, scatter plots visualizing the correlation between the expert-scored AHI and the algorithm-predicted AHI were computed. Additionally, for experiment 2, we computed scatter plots for the RDI and each type of respiratory event per hour of sleep. A robust linear regression model with bi-squared cost function was fitted to the data to compute the correlation between the scored AHI by the experts and predicted AHI by our model [35]. This model was selected to mitigate the effect of outliers. Also, Cohen’s kappa values were determined for AHI prediction.
We provide the analysis code and computational models used in this study on our GitHub page [36].
III. RESULTS
For the MGH and SHHS testing dataset, 16 and 1128 recordings, respectively, were removed due to insufficient sleep or erroneous data.
The boosted model approach resulted in 5 consecutive model iterations before reaching the desired class balance in both experiment 1 and 2. In experiment 1, after applying 5 boost-models, the total number of segments decreased from 7,810,448 to 1,614,791 (79.3%), while the total number of events decreased from 448,855 to 395,278 (11.9%). The event distribution for obstructive apnea, central apnea, and hypopnea was 30.4%, 17.4%, and 52.2%, respectively. During the training process in experiment 2, the number of segments decreased from 7,810,448 to 1,904,537 (75.6%), while the number of events decreased from 653,082 to 575,127 (11.9%). The event distribution for obstructive apnea, central apnea, RERA, and hypopnea was 21.3%, 12.0%, 29.0%, and 37.7% respectively. Appendix Table VI shows a full summary of the segment distribution after applying each boost-model during training in experiments 1 and 2.
In Appendix Fig. 8 an example recording can be found. As the large flat parts show no continuous false positive predictions we are convinced that our model learned not to classify such regions as respiratory events.
A. Per-event performance
An overview of the per-event performance metrics for the binary task are given in Table III. An AUCroc value of 0.93 and AUCpr of 0.71 was found for the MGH dataset. The AUCroc and AUCpr for the SHHS dataset were 0.92 and 0.56 (see Fig. 3 for the ROC and PR curves). Appendix Fig. 9 shows four segments including TP, FP and FN examples. Here, the effect of post processing on the raw WaveNet model predictions can be observed.
TABLE III.
Overall per-event performance for experiment 1 with all values in mean percentages and 95% confidence intervals, and experiment 2 with mean performance among the individual events, i.e. obstructive apnea, central apnea, rera, hypopnea
| Experiment 1 | Experiment 2 | ||
|---|---|---|---|
| MGH dataset (binary) | SHHS dataset (binary) | MGH dataset (multiclass) | |
|
| |||
| Accuracy | 95.7 [95.7–95.7] | 94.0 [94.0–94.1] | 99.1 |
| Sensitivity | 67.7 [67.6–67.8] | 70.9 [70.7–71.0] | 49.3 |
| Specificity | 97.6 [97.6–97.6] | 95.1 [95.1–95.2] | 99.5 |
| Precision | 65.4 [65.3–65.5] | 40.7 [40.5–40.8] | 37.4 |
| F1-score | 66.5 [66.4–66.6] | 51.7 [51.6–51.8] | 40.6 |
| Cohen’s kappa | 64.2 [64.1–64.3] | 48.7 [48.6–48.9] | 36.5 [36.4–36.6] |
Note that for experiment 2 all performance metrics (except Cohen’s kappa) do not show a 95% confidence interval range since these are mean values from all individual event types as seen in Table IV.
Fig. 3.
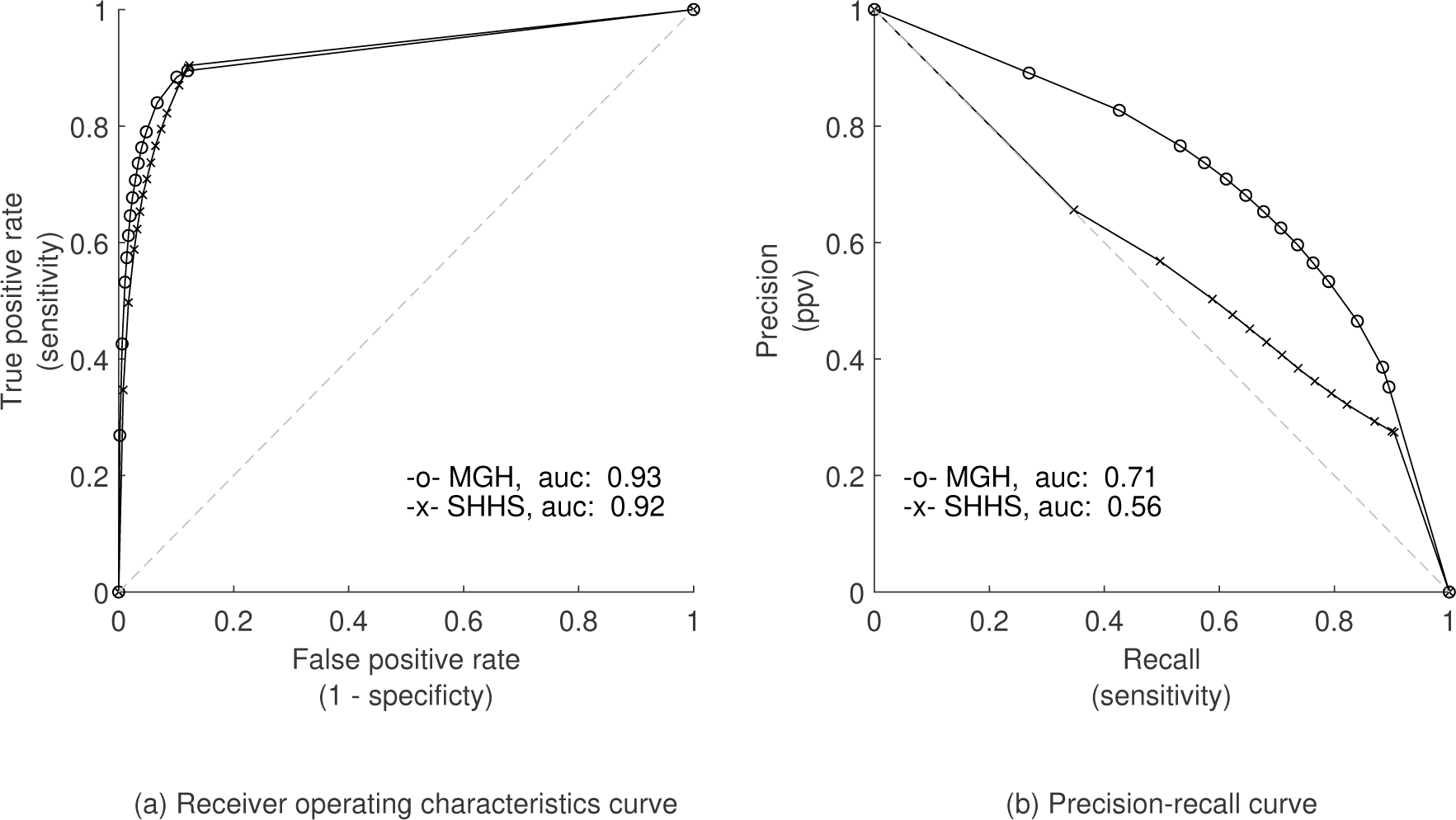
ROC and PR curves for binary classification in experiment 1.
In experiment 2, the multiclass model resulted in an overall accuracy of 97%. Mean performance metrics over the four respiratory event classes can be observed in Table IV. Performances vary considerably for the different classes, e.g. while 84% of all expert-labeled central apnea events are correctly classified, this is only true for 23% of hypopneas.
TABLE IV.
Per-event performance in experiment 2 including mean percentages and 95% confidence intervals
| Sensitivity | Specificity | |
|
| ||
| Obstructive apnea | 50.6 [50.4–50.8] | 99.7 [99.7–99.7] |
| Central apnea | 84.1 [83.9–84.3] | 99.6 [99.6–99.6] |
| RERA | 39.5 [39.4–39.7] | 99.0 [99.0–99.0] |
| Hypopnea | 22.8 [22.7–23.0] | 99.7 [99.7–99.7] |
|
| ||
| Mean | 49.3 | 99.5 |
|
| ||
|
| ||
| Precision | F1-score | |
|
| ||
| Obstructive apnea | 44.5 [44.3–44.7] | 47.4 [47.2–47.5] |
| Central apnea | 41.5 [41.3–41.7] | 55.6 [55.4–55.8] |
| RERA | 24.9 [24.8–25.0] | 30.6 [30.4–30.7] |
| Hypopnea | 38.8 [38.6–39.0] | 28.8 [28.6–28.9] |
|
| ||
| Mean | 37.4 | 40.6 |
For both experiment 1 and 2 confusion matrices are shown in Appendix Table VII. The training and validation performance on the MGH dataset is summarized in Appendix Table VIII. Performance of all metrics is similar for training, validation and test set, showing the model did not overfit the training data.
B. AHI stratification
We next assessed the performance of our model to classify the severity of AHI. For the MGH dataset in experiment 1, performance among each AHI subgroup is shown in Table V. The sensitivity, precision and F1-score increased with the severity of apnea. The opposite effect was observed for the accuracy and specificity.
TABLE V.
Summary performance metrics for different study subgroups for the MGH dataset in experiment 1 with all values in percentages
| Accuracy | Sensitivity | Specificity | Precision | F1-score | AHI classification accuracy | |
|---|---|---|---|---|---|---|
|
| ||||||
| Male (N=5687) | 95 | 72 | 96 | 67 | 70 | 66 |
| Female (N=3930) | 96 | 58 | 98 | 62 | 60 | 71 |
|
| ||||||
| Normal breathing (N=3718) | 98 | 42 | 99 | 35 | 38 | 83 |
| Mild apnea (N=2490) | 95 | 55 | 97 | 58 | 57 | 54 |
| Moderate apnea (N=1803) | 93 | 69 | 96 | 70 | 70 | 56 |
| Severe apnea (N=950) | 90 | 76 | 94 | 81 | 78 | 76 |
|
| ||||||
| Diagnostic (N=4645) | 96 | 62 | 98 | 56 | 59 | 68 |
| Split night (N=2346) | 93 | 74 | 96 | 75 | 74 | 65 |
| All night CPAP (N=2317) | 97 | 60 | 98 | 55 | 58 | 70 |
We computed the confusion matrix for AHI prediction, as shown in Fig. 4. Overall, 69% of patients from the MGH dataset and 54% from the SHHS dataset were assigned to the correct AHI category. Cohen’s kappa values for AHI classification were 55% and 32% for the MGH and SHHS dataset, respectively. In experiment 2, 70% of all patients were classified in the correct AHI category using the MGH dataset. Cohen’s kappa value was 56%. Most misclassifications resulted in false positives in neighboring AHI categories.
Fig. 4.
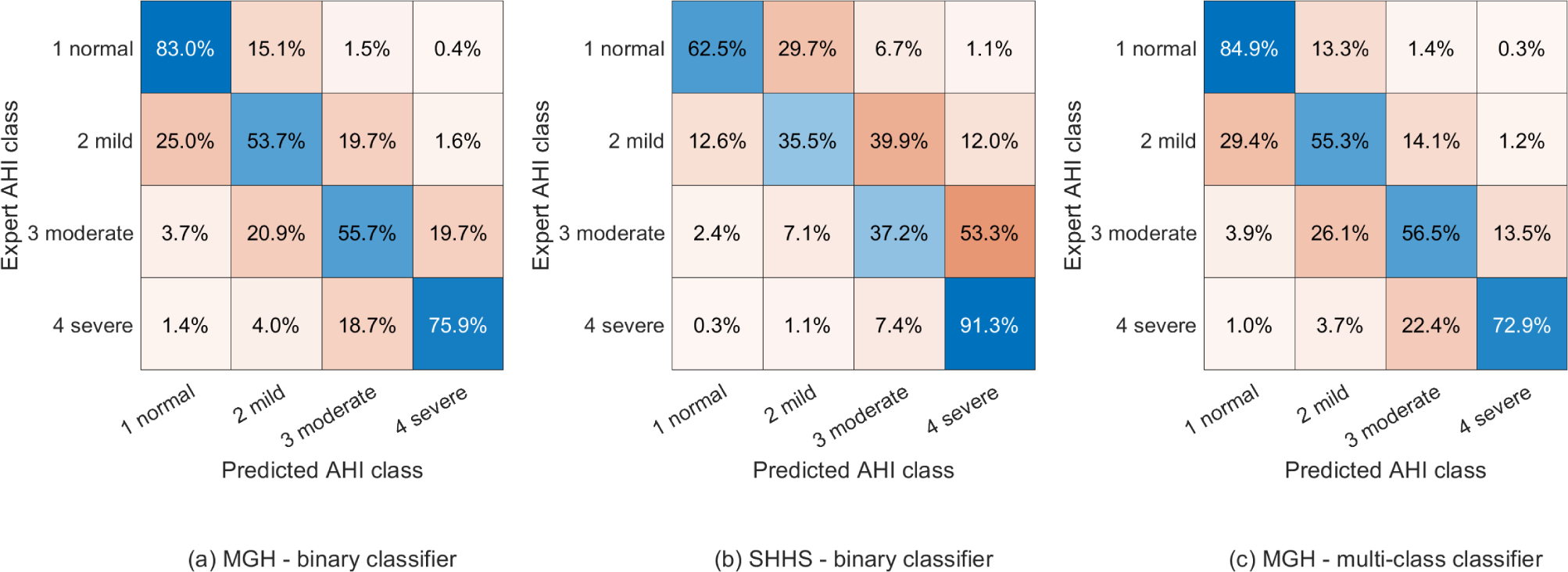
AHI classification confusion matrices for both experiments with Cohen’s kappa values of (a) 55%, (b) 32%, and (c) 56%.
The scatter plots in Fig. 5 show the correlation between the expert-scored AHI and the model predicted AHI from experiment 1. The r2 was 0.90 for the MGH dataset and 0.79 for the SHHS dataset. For experiment 2, an r2 of 0.90, 0.84, 0.96, 0.96, 0.38 and 0.66 was determined for AHI, RDI, obstructive apneas, central apneas, RERAs and hypopneas, respectively, see Appendix Fig. 10.
Fig. 5.
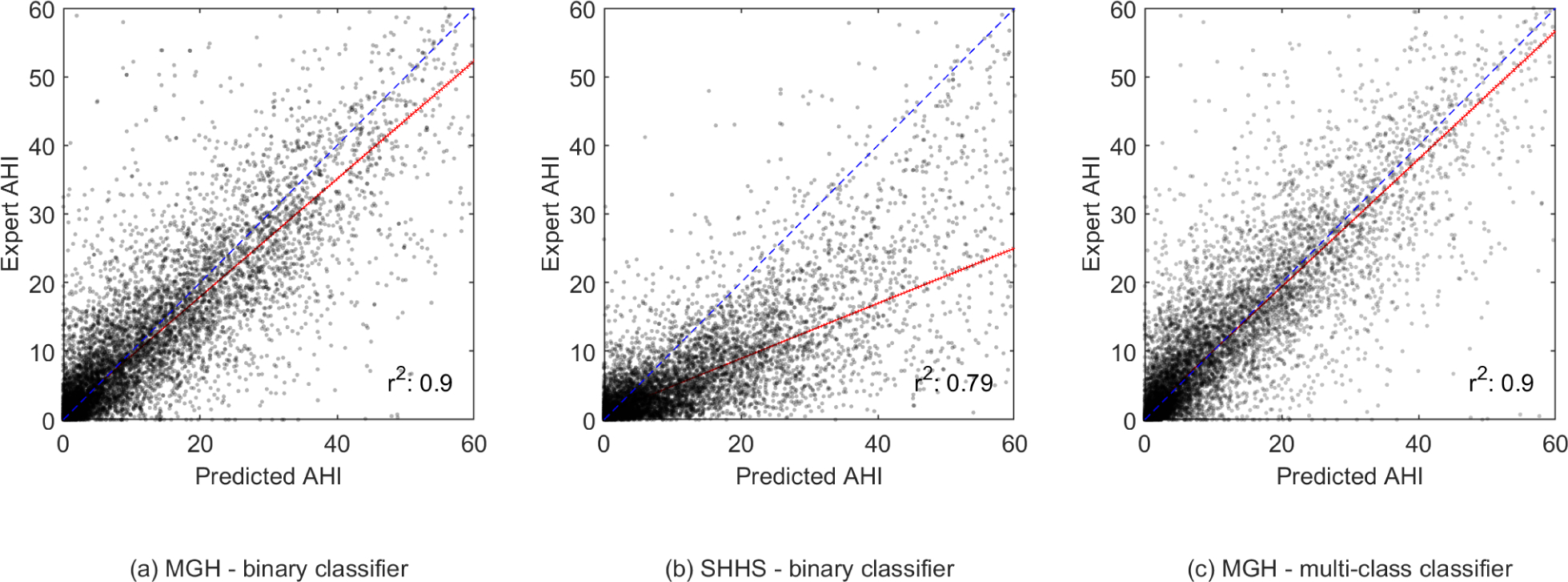
Scatter plots showing the correlation between the expert-scored AHI and the model predicted AHI in experiment 1 and 2. The fitted robust linear regression model is shown in red.
The computed histograms represent the difference between the AHI value scored by the experts and the AHI value predicted by our model, see Fig. 6. A mean difference in AHI of 0.41 and −0.44 with a standard deviation of 7.80 and 7.36 was found for the MGH dataset in experiment 1 and experiment 2. The mean AHI difference for the SHHS dataset was 5.97 with a standard deviation of 9.54. In Appendix Fig. 11 Bland-Altman show the difference between the AHI value predicted by our model and the AHI value scored by the experts.
Fig. 6.
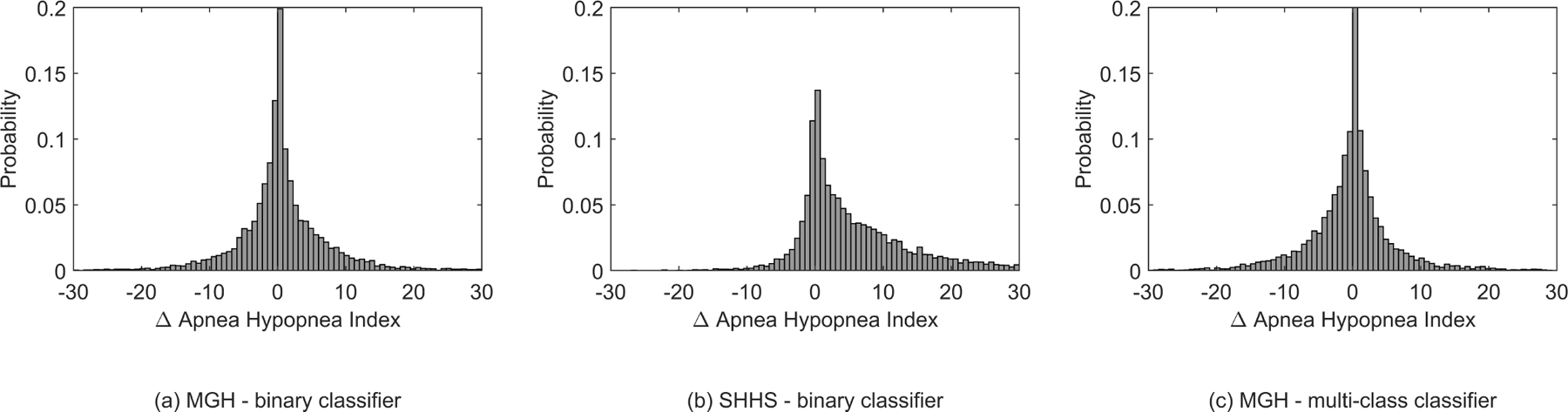
Histograms of the relative difference in AHI determined by the experts vs the model in experiment 1 and 2
IV. DISCUSSION
A deep neural network method was developed to classify typical breathing disorders during sleep based on a single respiratory effort belt used in PSG. In a first experiment our WaveNet model successfully discriminated respiratory events from regular breathing on our primary dataset with an accuracy of 96%, and sensitivity, specificity, precision and F1-score of 68%, 98%, 65% and 67%, respectively. AHI was predicted for each patient using the number of respiratory events with an accuracy of 69%. It is notable that most misclassifications of our model resulted in false positives into the neighboring AHI categories. This effect is best visualized in the histograms in Fig. 6; the unimodal and symmetrical shape shows that a decrease in number of false positives was observed as the difference between the predicted AHI and the sleep-expert scored AHI increased. The correlation between expert-scored AHI and algorithm-predicted AHI showed an r2 of 0.90. It is possible to adjust the predicted AHI cut -offs to improve AHI classification. However, we decided to use the original AASM criteria, because these are generally recognized as clinically meaningful and well understood categories.
When applying our model on a secondary dataset obtained from the SHHS, a slight decrease in model performance was observed. This is likely due to imperfect generalization to a dataset where different respiratory effort sensors are used. It is important to note that a thermistor was used to detect respiratory events in the SHHS study, whereas a nasal pressure sensor was used in the MGH study. It is possible that nasal pressure-based scoring, regardless of ancillary signals used, is more sensitive in detecting sleep-disordered breathing than thermistor-based scoring [37]. Therefore, it could be that a significant number of events were missed during annotation in the SHHS study. When observing the performance of our model applied on the SHHS dataset a decrease of approximately 15% was observed for the precision and f1-score. Sensitivity, however, slightly increased. This observation could be explained by the different methodology used while annotating events. The fact that our model generally overpredicts AHI when applied on the SHHS dataset, as seen in the confusion matrix of Fig 4 (b) and the scatter plot of Fig. 5 (b), is in line with this assumption. We recognize that the SHHS may have used ancillary signals. Nevertheless, use of the thermistor signal as the primary flow channel is one possible explanation for our results.
The guidelines for scoring respiratory events manually have evolved over the years but have remained largely driven by consensus. Thus, for example, the requirement of a 50% or 30% reduction in signal amplitude is arbitrary; there is no data to suggest that a 35% or 60% would be less or more clinically meaningful. Moreover, visual discrimination of small percentage differences is likely poor. During polysomnography or even home sleep study recordings, the multichannel nature of the data enables increased scoring accuracy by associating changes with neighboring signals. Moreover, airway collapse is common during central apnea, and high loop gain can drive obstructive events. Thus, the differentiation of obstructive and central events is not as pathophysiologically clear as clinical scoring may suggest. This biological reality of blurred boundaries will be reflected in any manual or automated scoring approach.
In a secondary experiment our model successfully identified the type of the included respiratory events, i.e. central apneas, obstructive apneas, hypopneas and RERAs. Despite a similar overall accuracy, discrimination of the specific respiratory events resulted in a decreased per-event performance with respect to the first experiment. Central apneas were detected with high sensitivity of 84%, expectedly due to the apparent effect of the disorder on respiratory effort. Often markedly reduced respiratory effort is observed during central apnea events, resulting in clear features for algorithms to recognize. We expect that this is the main reason of the high-performance metrics for the detection of central apneas. This is true to a lesser extent for obstructive apnea events, hence the slightly lower performance when compared to the central apneas. When using a single effort signal, thoracoabdominal asynchrony is undetectable. If using more than one effort sensor, this feature could enhance differentiation between obstructive apnea and central apnea by our model. The recognition of hypopneas and RERAs was considered moderate, with an F1-score of 31% and 29% respectively. The scatter plots show underprediction by our model, indicating limited sensitivity rather than specificity. Without additional information derived from other physiological signals the identification of hypopneas and RERAs appears difficult. The moderate performance of our model in differentiating no events, hypopneas, and RERAs might have three different causes: 1) a general difficulty in differentiating RERAs from hypopneas and unknown gold-standard performance as there is no human expert inter-rater agreement available for this task; 2) the lack of other complementary signals such as blood oxygen saturation signals; and 3) insufficient model complexity or architecture. For future research, we propose the collection of human expert inter-rater agreement data before researching the impact of the choice of input signals and other model architectures for the hypopnea-RERA classification task.
Misclassification often meant that an event of a particular respiratory class was classified as a different class. When observing the AHI stratification performance of our multiclassification model, large variation in performance was observed among the various respiratory events. Yet, when the different predicted classes are grouped together to binary apnea events, a similar correlation was found between the expert-scored AHI and the algorithm-predicted AHI. An r2 of 0.90 was determined, indicating that AHI prediction based on the specific respiratory events is feasible. Very similar performance was observed in AHI prediction confusion matrices with respect to the binary classification of experiment 1. The ability to discriminate various respiratory events is clinically valuable but may not be achievable by using manual scoring as a gold standard. The type of breathing assistance and overall apnea treatment may vary for different underlying pathology leading to apnea. Specifying the type of apnea will therefore provide aid in improving personalized patient care.
To best way to assess clinical applicability of a novel apnea detection model is by comparing model-expert agreement to expert-expert agreement. Despite the lack of inter-rater (IR) performance using only respiratory effort signals, the current overall respiratory event expert-expert agreement shows significant misclassification as well. The AASM reports an IR agreement accuracy ranging from 39.8% and 77.1%, for multi-class classification of obstructive apnea, central apnea, mixed apnea, and hypopneas. Our models do show a similar accuracy range [38]. It is, however, difficult to compare performance directly, since our performance is computed based on a per-event 50% overlap approach, while the AASM reports their IR performance based on single events scored in 30 second epochs. To conclusively assess clinical applicability identical performance metrics should be used. In future research we plan to collect respiratory annotations from multiple experts per PSG study, enabling a fair and more in-depth human-human and human-algorithm analysis.
Even though we did combine mixed apnea and obstructive apnea during training, the characteristics of the two apnea types are not the same. In fact, mixed apneas may resemble central apneas in the first half of the event, whereas it resembles more of the phenotype of obstructive apnea in the latter half of the event. To study this, we randomly selected 1000 expert-scored scored events of each apnea type (i.e. obstructive apnea, central apnea, and mixed apnea) and show the distribution of predicted classes for all samples within those events by our WaveNet model. In Appendix Fig. 12 we show some examples of expert scored mixed apnea events and the according model predictions. We observed that our WaveNet model does indeed show a fairly equal distribution of samples predicted as central (42%) and obstructive (56.5%) for expert-scored mixed apneas (with remaining 1.6% of samples predicted as hypopnea), Appendix Fig. 13 (c). As expected, the majority of samples within obstructive events were predicted as obstructive (63.5%), Appendix Fig. 13 (b), and the majority of samples within central events were predicted as central (83.8%), Appendix Fig. 13 (a). Among mixed apnea events, an increased prediction proportion for central apnea was seen in both the first half and the second half of the event. Also noticeable is the very slim probability of hypopnea prediction by the WaveNet model for both central and mixed apneas. For the majority, our model does identify mixed apnea as obstructive apnea after smoothing, which is explained in part by our training method (i.e. combining these apnea types). Further research is required to see if the model is able to accurately discriminate mixed apnea from other apnea types. However, the ability of our WaveNet model to predict respiratory events for each second could provide clinicians with valuable additional phenotype information on each of the event types.
It is possible that valuable information was lost due to down sampling during preprocessing our data. However, the low pass filter of 10 Hz used for down sampling our signals was not expected to remove significant event characteristics that limit us in identifying apneas. Regular breathing for adults normally ranges between approximately 0.2 – 0.3 Hz.
Predicting for each second provides the smallest time resolution of our model. Reducing the rate at which the algorithm provides results can be achieved by aggregating predictions from consecutive time steps, such as taking the most severe form of respiratory event. This is one of the possible alternative approaches to generate events with a duration of 10 seconds or more and may yield a better performance.
Besides a high accuracy, a metric that is affected by class imbalance, our model also showed high AUC values for ROC (0.93), PR (0.71), and F1-score (0.67). This means the model not only has an excellent agreement in sensitivity and specificity but also has a clinically acceptable precision in specific situations, similar to the use of home sleep apnea testing, where tolerance to especially false negatives is required [39]–[41]. We have included the F1 score and the AUCpr, as such performance metrics are not influenced by the imbalance of negative-positive classes but rather by sensitivity and precision of the positive class. The low standard deviation between the 5 folds of cross-validation (AUCroc and AUCpr mean and std of 92 ± 0.5 and 71 ± 1.2) emphasizes the robustness of our model on a large dataset.
In manual analysis, experts learn to implicitly visually discount artifacts. Similarly, for the automated analysis, rather than designing algorithms by hand to explicitly address this issue, we took a data driven approach, i.e. presenting a large number of labeled examples including ones with artifacts present, and allowed the model to learn (implicitly) to discount artifacts. This is possible for two reasons: (1) The MGH dataset is very large; (2) deep neural network models, like the one used in the manuscript, are very flexible. Thus, given sufficient data, deep neural network models can often learn to perform challenging pattern recognition tasks at a level that matches human experts. Our results show that this indeed was the case.
Most approaches found in the literature used different sensors to detect respiratory events. Some have shown slightly higher performances, although performance comparisons are difficult given the different datasets and evaluation methods. To our knowledge, our model showed better results with respect to other methods using a single respiratory effort belt and is the only model that shows that additional respiratory event class discrimination is possible based on respiratory effort only.
An advantage of using an effort belt to assess apnea is the non-invasive application. This becomes very relevant when assessing respiratory stability and instability/events in intensive care or environmentally hostile conditions. Using limited resources – such as a respiratory effort belt – to assess respiratory abnormalities can be successfully applied in combination with other simple and small sensors necessary for monitoring patients in diverse clinical situations. patients receiving breathing aid using CpAp are eligible for event detection. The number of patients included in our research is larger than previous reports in the literature. This, in combination with limited preprocessing and without the use of any human-engineered features, emphasizes the robustness of our proposed approach.
Future work focuses on the design of a completely automated comprehensive sleep scoring system that combines automated respiratory event analysis with sleep staging, limb movement detection, and the identification of spontaneous arousals.
V. CONCLUSION
A neural network approach to analyzing typical respiratory events during sleep based on a single respiratory measurement is described. Our model included dilated convolutions to allow their receptive fields to grow exponentially with depth, which is important to model the long-range temporal dependencies in respiration signals. Using this model, we obtained a comparable performance with respect to literature while using a minimally invasive methodology. Differentiation of event types is more difficult and may reflect in part the complexity of human respiratory output and some degree of arbitrariness in the clinical thresholds and criteria used during manual annotation. The use of a respiratory effort belt at the abdomen for sleep apnea analysis bears the advantage of wide implementation options ranging from acute care settings to wearable devices for home usage. Important first steps were obtained in automated apnea detection with limited resources, creating new sleep assessment opportunities applicable to the clinical setting.
Supplementary Material
Acknowledgments
M.B.W. Was supported by the Glenn Foundation for Medical Research and American Federation for Aging Research (Breakthroughs in Gerontology Grant); American Academy of Sleep Medicine (AASM Foundation Strategic Research Award); Football Players Health Study (FPHS) at Harvard University; Department of Defense through a subcontract from Moberg ICU Solutions, Inc; by the NIH (1R01NS102190, 1R01NS102574, 1R01NS107291, 1RF1AG064312).
APPENDIX I. ADDITIONAL TABLES AND FIGURES
Fig. 7.
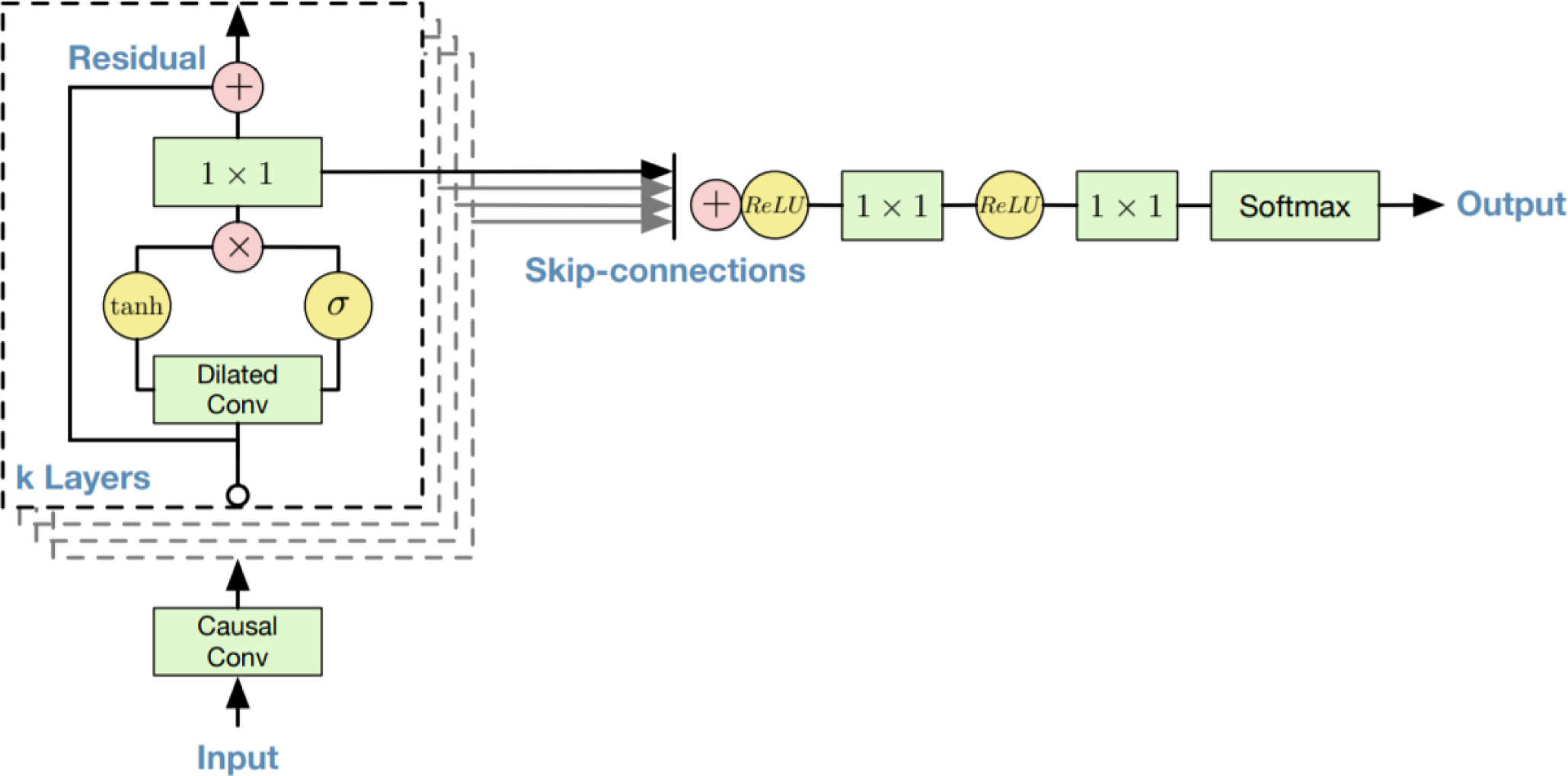
Neural network architecture and residual block of the WaveNet model as described by Oord et al (2016).
Fig. 8.
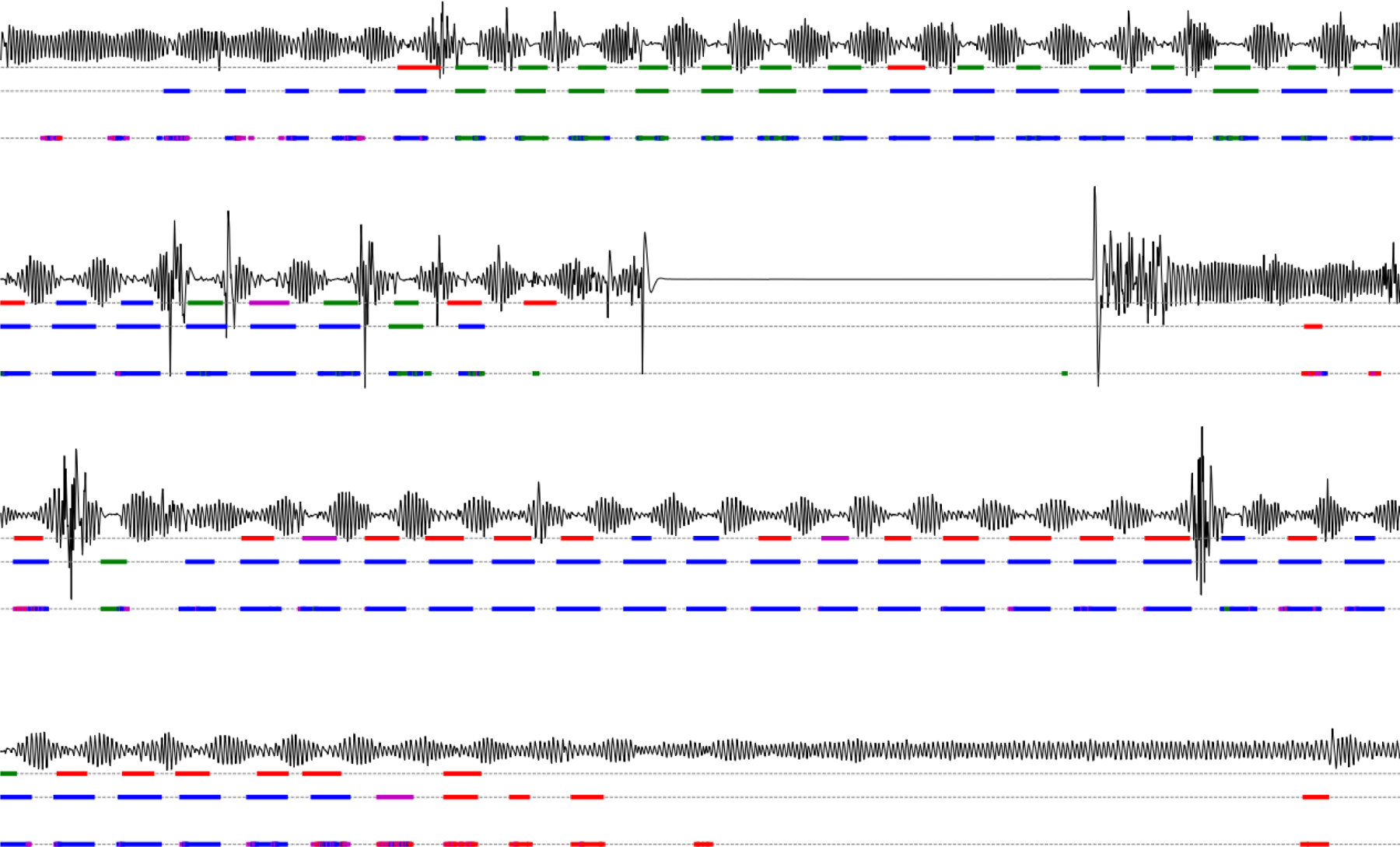
Example recording showing the abdominal effort signal, the original expert labels, the smoothed WaveNet model output, and the raw WaveNet model output, from top to bottom. As the large flat parts show no continuous false positive apnea predictions we are convinced that our model learned not to classify such regions as apnea. With obstructive apneas in blue, central apneas in green, hypopneas in pink, and RERAs in red.
Fig. 9.
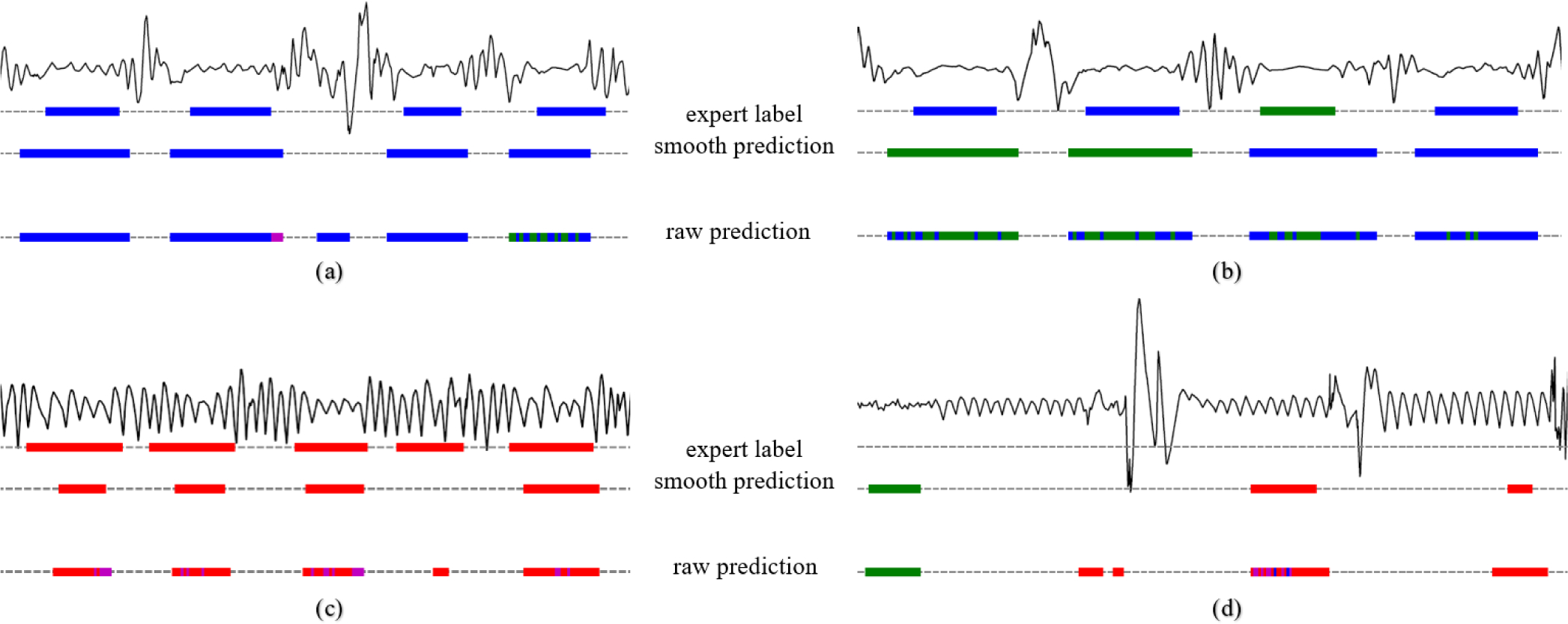
Example signal segments and according labels and model predictions with in blue obstructive apneas, green central apneas, red RERAs and in pink hypopneas. (a), accurate predictions. (b), miss-classifications between obstructive and central apneas. (c), true positive and false negative RERA detections. (d), false positive event detections.
Fig. 10.
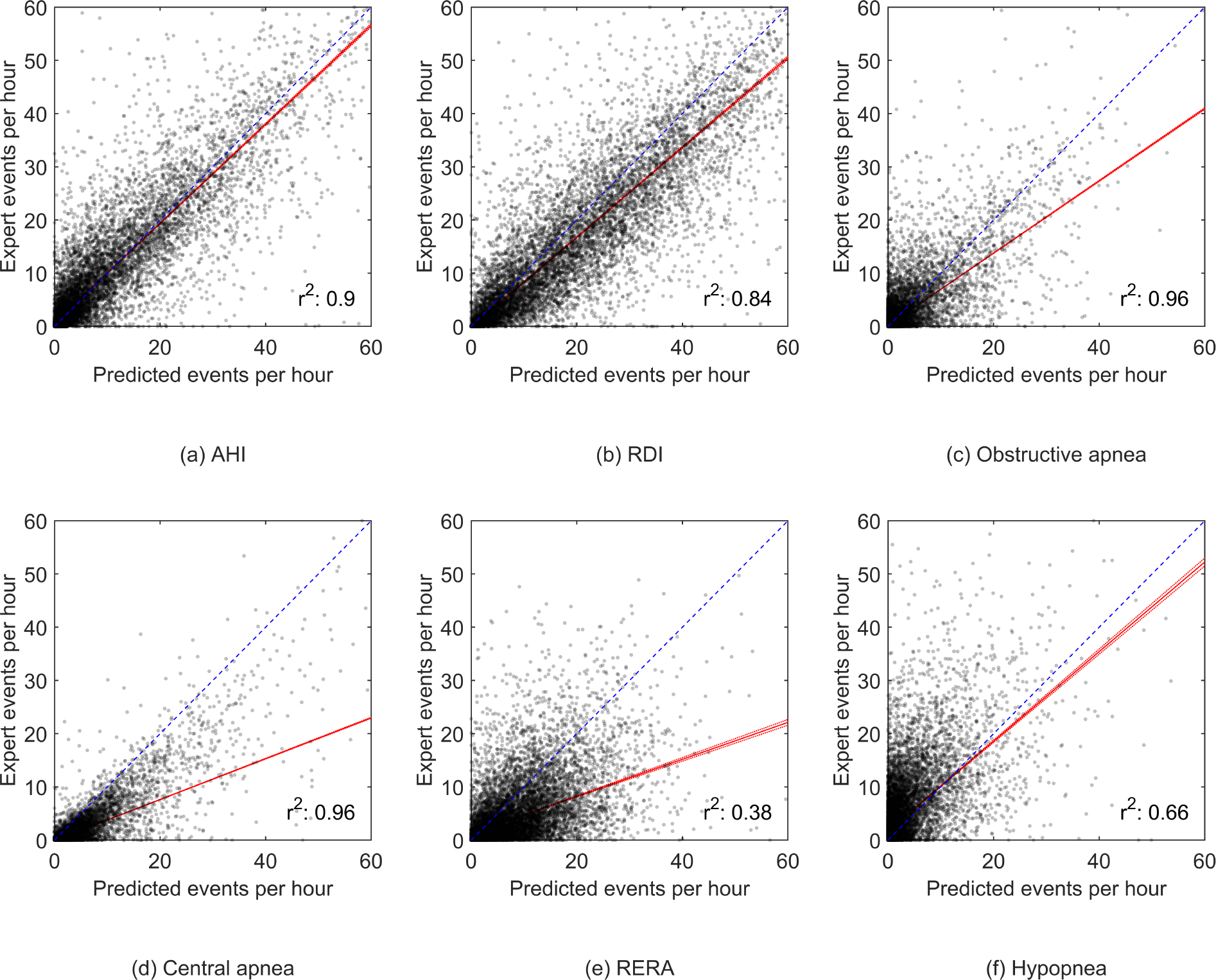
Scatter plots showing the correlation between the expert-scored respiratory events and the model predicted respiratory events from experiment 2. The fitted robust linear regression model is shown in red.
Fig. 11.
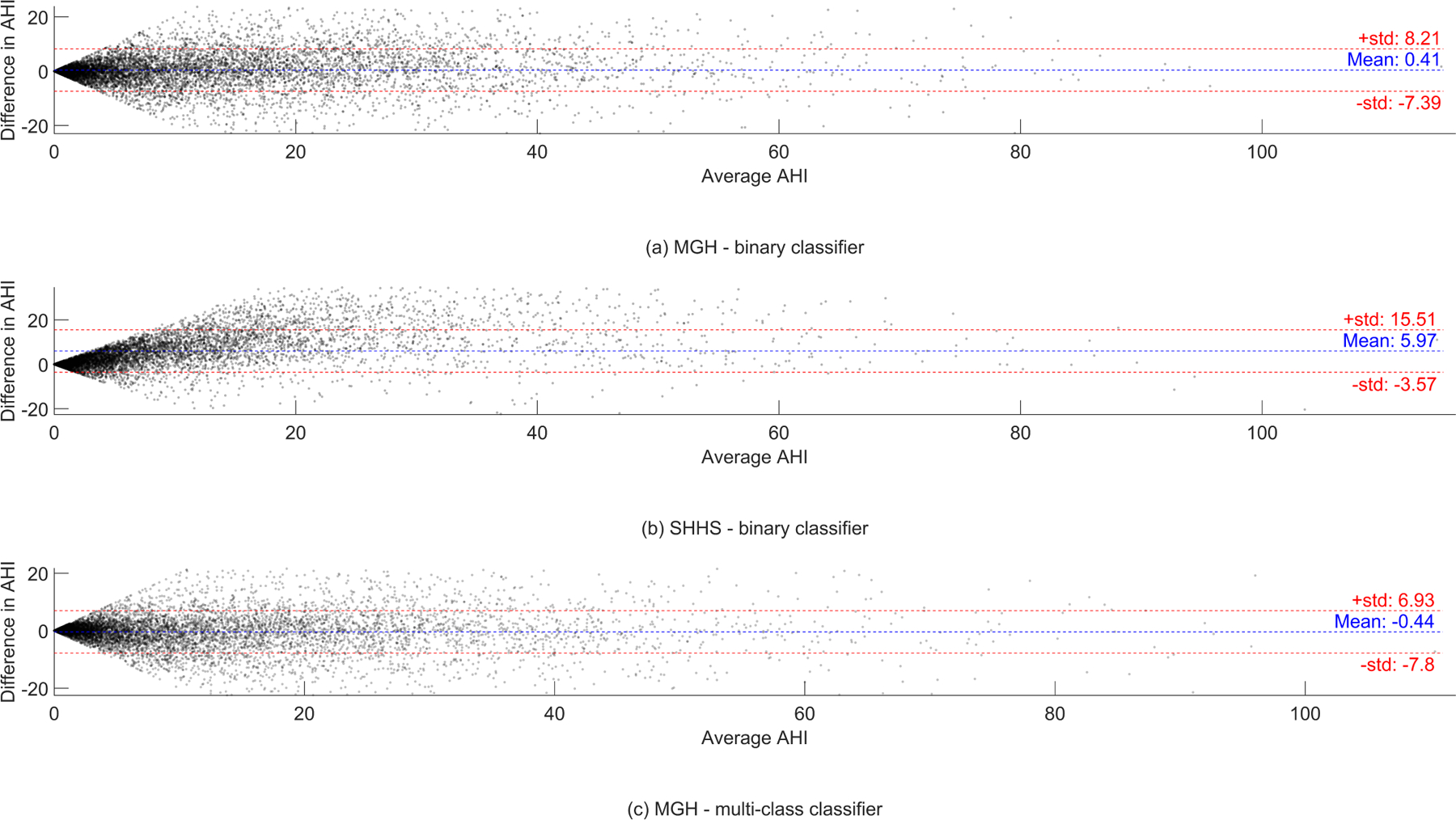
Bland-Altman plots showing the difference in predicted AHI and the AHI determined by the experts in experiment 1 and 2.
Fig. 12.
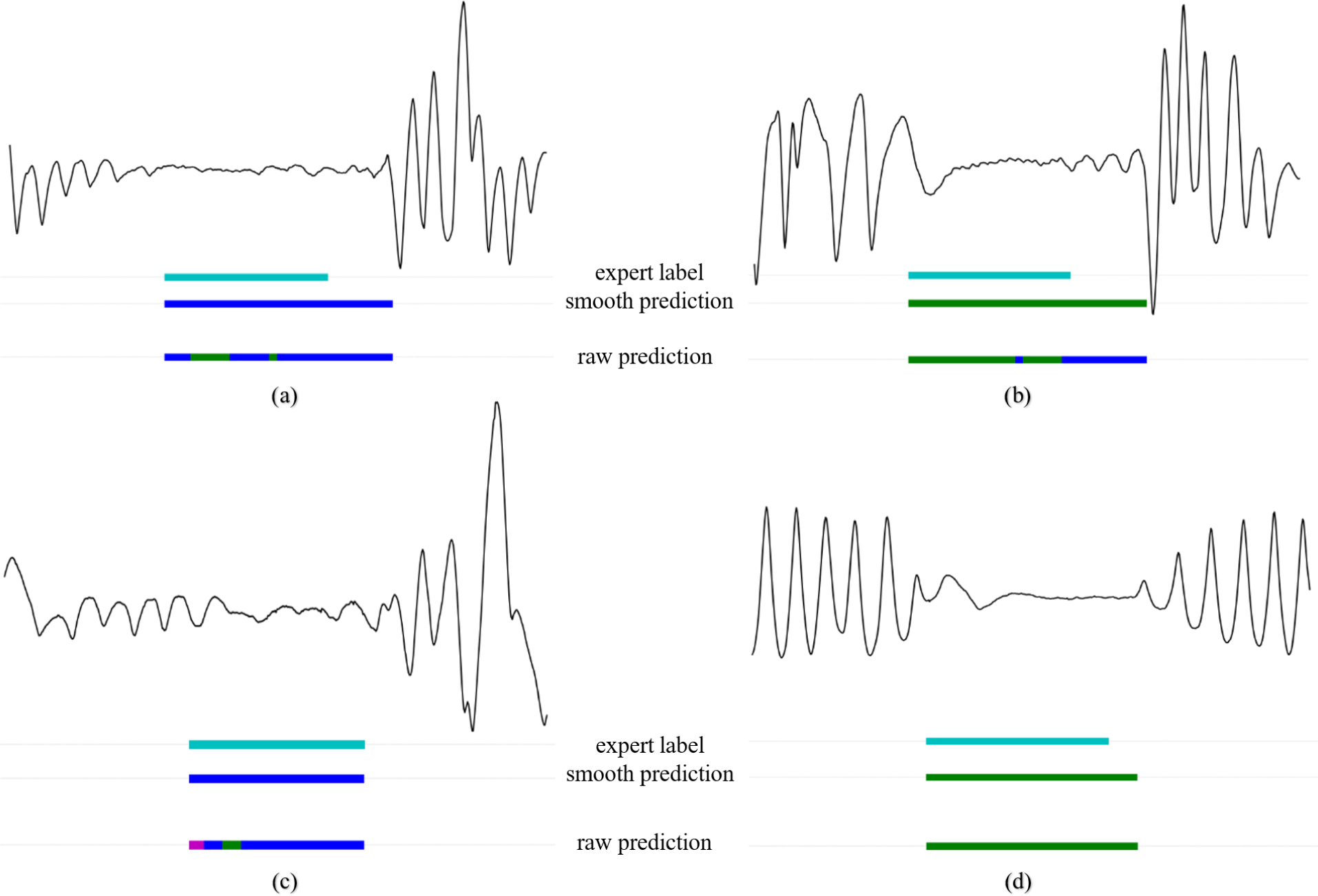
Examples of expert scored mixed apnea events and according model predictions with in blue obstructive apnea, green central apnea, cyan mixed apnea, and in pink hypopnea. (a), final prediction label was obstructive, whereas raw output also shows central samples in the first half of the event. (b), final prediction label was central apnea, whereas the raw output also shows obstructive samples in the latter half of the event. (c), final prediction label was obstructive apnea, while the raw output also shows samples of hypopnea and central apnea. (d), final prediction was central apnea, while all raw output samples are uniform.
Fig. 13.
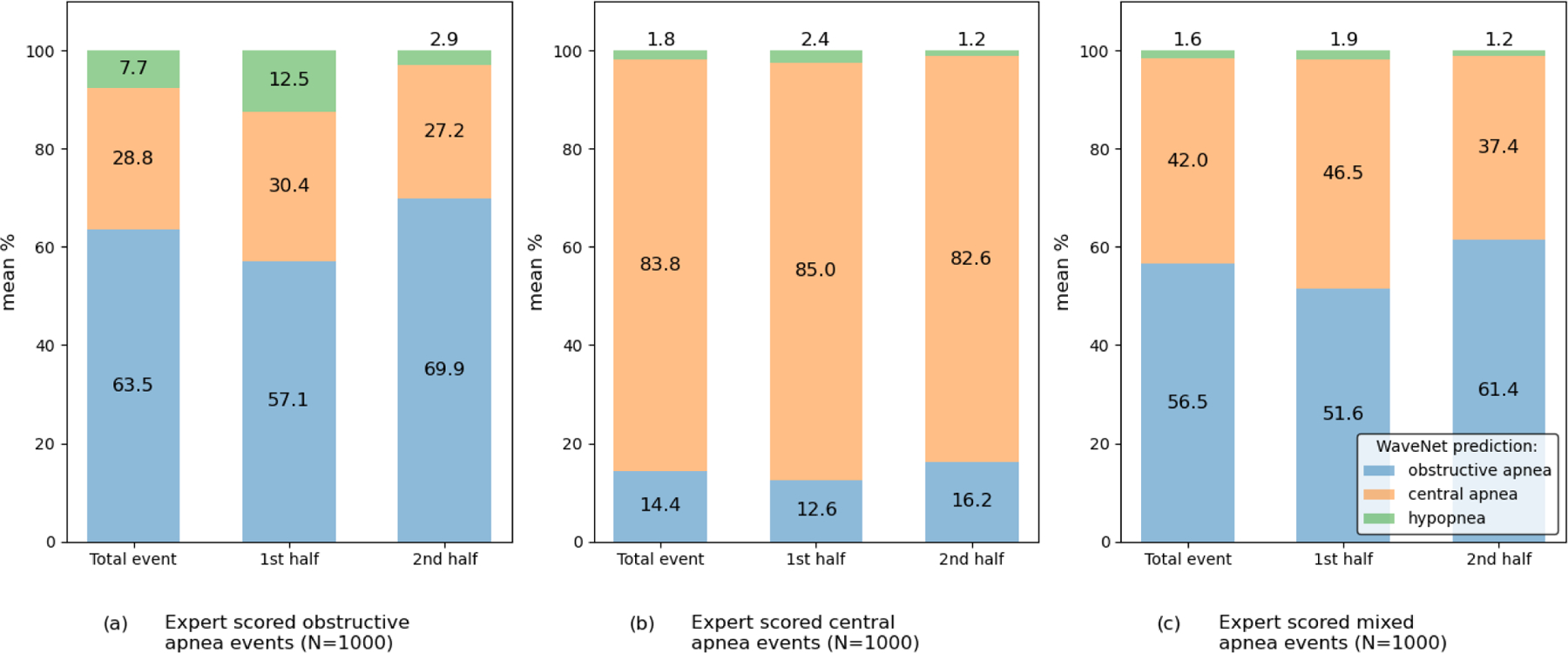
Bar plots showing the sample distribution for the WaveNet predictions for 1000 events of each of the apnea types in experiment 2. For each event type the prediction distribution among the total event is shown, together with the prediction distribution for both the first half and the second half of the manually scored event.
TABLE VI.
Segment distribution during the boosting approach during training in experiments 1 and 2.
| Experiment 1 training | Event | Obstructive apnea | Central apnea | RERA | Hypopnea | |||||
| No-event : Event | ratio | #events / total events | percentage | #events / total events | percentage | #event / total events | percentage | |||
|
| ||||||||||
| Start | 7361593 : 448855 | 16.4 :1 | 129313 / 448855 | 28.81% | 71254 / 448855 | 15.88% | - | 248288 / 448855 | 55.31% | |
| Boost 1 | 5501753 : 446611 | 12.3 :1 | 128908 / 446611 | 28.86% | 71084 / 446611 | 15.91% | - | 246619 / 446611 | 55.22% | |
| Boost 2 | 3590595 : 439912 | 8.2 :1 | 127910 / 439912 | 29.08% | 70817 / 439912 | 16.10% | - | 241185 / 439912 | 54.82% | |
| Boost 3 | 2357345 : 428915 | 5.5 :1 | 126173 / 428915 | 29.42% | 70370 / 428915 | 16.41% | - | 232372 / 428915 | 54.18% | |
| Boost 4 | 1688103 : 413903 | 4.1 :1 | 123433 / 413903 | 29.82% | 69710 / 413903 | 16.84% | - | 220760 / 413903 | 53.34% | |
| Boost 5 | 1219513 : 395278 | 3.1 :1 | 120252 / 395278 | 30.42% | 68717 / 395278 | 17.38% | - | 206309 / 395278 | 52.19% | |
|
| ||||||||||
| Experiment 1 validation | Event | Obstructive apnea | Central apnea | RERA | Hypopnea | |||||
| No-event : Event | ratio | #events / total events | percentage | #events / total events | percentage | #event / total events | percentage | |||
|
| ||||||||||
| Start | 839967 : 50843 | 16.5 :1 | 14603 / 50843 | 28.72% | 8004 / 50843 | 15.74% | - | 28236 / 50843 | 55.54% | |
| Boost 1 | 616504 : 50438 | 12.2 :1 | 14396 / 50438 | 28.54% | 7997 / 50438 | 15.86% | - | 28045 / 50438 | 55.60% | |
| Boost 2 | 401411 : 49639 | 8.1 :1 | 14228 / 49639 | 28.66% | 7979 / 49639 | 16.01% | - | 27432 / 49639 | 55.26% | |
| Boost 3 | 263863 : 48421 | 5.5 :1 | 14020 / 48421 | 28.95% | 7935 / 48421 | 16.39% | - | 26466 / 48421 | 54.66% | |
| Boost 4 | 190154 : 46848 | 4.1 :1 | 13788 / 46848 | 29.43% | 7873 / 46848 | 16.81% | - | 25187 / 46848 | 53.76% | |
| Boost 5 | 138039 : 44782 | 3.1 :1 | 13463 / 44782 | 30.06% | 7767 / 44782 | 17.34% | - | 23552 / 44782 | 52.60% | |
|
| ||||||||||
| Experiment 2 training | Event | Obstructive apnea | Central apnea | RERA | Hypopnea | |||||
| No-event : Event | ratio | #events / total events | percentage | #events / total events | percentage | #event / total events | percentage | #event / total events | percentage | |
|
| ||||||||||
| Start | 7157366 : 653082 | 11.0 : 1 | 129313 / 653082 | 19.80% | 71254 / 653082 | 10.91% | 204227 / 653082 | 31.27% | 248288 / 653082 | 38.02% |
| Boost 1 | 5446734 : 649817 | 8.4 : 1 | 128949 / 649817 | 19.84% | 71088 / 649817 | 10.94% | 202834 / 649817 | 31.21% | 246946 / 649817 | 38.00% |
| Boost 2 | 3732711 : 640070 | 5.8 : 1 | 128114 / 640070 | 20.06% | 70783 / 640070 | 11.06% | 198205 / 640070 | 30.97% | 242968 / 640070 | 37.96% |
| Boost 3 | 2714570 : 624069 | 4.3 : 1 | 126685 / 624069 | 20.30% | 70423 / 624069 | 11.28% | 190939 / 624069 | 30.60% | 236022 / 624069 | 37.82% |
| Boost 4 | 2003251 : 602227 | 3.3 : 1 | 124411 / 602227 | 20.66% | 69644 / 602227 | 11.56% | 181340 / 602227 | 30.11% | 226832 / 602227 | 37.67% |
| Boost 5 | 1329410 : 575127 | 2.3 : 1 | 122555 / 575127 | 21.31% | 69018 / 575127 | 12.00% | 166990 / 575127 | 29.04% | 216564 / 575127 | 37.65% |
|
| ||||||||||
| Experiment 2 validation | Event | Obstructive apnea | Central apnea | RERA | Hypopnea | |||||
| No-event : Event | ratio | #events / total events | percentage | #events / total events | percentage | #event / total events | percentage | #event / total events | percentage | |
|
| ||||||||||
| Start | 816080 : 74730 | 10.9 : 1 | 14603 / 74730 | 19.54% | 8004 / 74730 | 10.71% | 23887 / 74730 | 31.96% | 28236 / 74730 | 37.78% |
| Boost 1 | 609558 : 74293 | 8.2 : 1 | 14434 / 74293 | 19.43% | 7999 / 74293 | 10.77% | 23770 / 74293 | 31.99% | 28090 / 74293 | 37.81% |
| Boost 2 | 416515 : 73213 | 5.7 : 1 | 14268 / 73213 | 19.49% | 7977 / 73213 | 10.90% | 23296 / 73213 | 31.82% | 27672 / 73213 | 37.80% |
| Boost 3 | 304298 : 71402 | 4.3 : 1 | 14117 / 71402 | 19.77% | 7943 / 71402 | 11.12% | 22480 / 71402 | 31.48% | 26862 / 71402 | 37.62% |
| Boost 4 | 225146 : 69067 | 3.3 : 1 | 13887 / 69067 | 20.11% | 7877 / 69067 | 11.40% | 21436 / 69067 | 31.04% | 25867 / 69067 | 37.45% |
| Boost 5 | 149355 : 65908 | 2.3 : 1 | 13663 / 65908 | 20.73% | 7798 / 65908 | 11.83% | 19774 / 65908 | 30.00% | 24673 / 65908 | 37.44% |
TABLE VII.
Confusion matrices for experiments 1 and 2 in both absolute and relative values.
| Experiment 1, MGH absolute values | predicted No-event | predicted event | Experiment 1, SHHS absolute values | predicted No-event | predicted event |
|
| |||||
| True, No-event | 9694899 | 241920 | True, No-event | 7544601 | 385724 |
| True, event | 218036 | 457631 | True, event | 108492 | 264218 |
|
| |||||
| Experiment 1, MGH normalized values | predicted No-event | predicted event | Experiment 1, SHHS normalized values | predicted No-event | predicted event |
|
| |||||
| True, No-event | 0.98 | 0.02 | True, No-event | 0.95 | 0.05 |
| True, event | 0.32 | 0.68 | True, event | 0.29 | 0.71 |
|
| |||||
| Experiment 2, MGH absolute values | predicted No-event | predicted Obstructive | predicted Central | predicted RERA | predicted Hypopnea |
|
| |||||
| True, No-event | 36080590 | 114918 | 151308 | 378306 | 125924 |
| True, Obstructive | 28985 | 92177 | 22027 | 19704 | 18556 |
| True, Central | 10788 | 7090 | 107440 | 1548 | 1006 |
| True, RERA | 146761 | 13326 | 12413 | 125424 | 17857 |
| True, Hypopnea | 111845 | 60572 | 24466 | 72244 | 79817 |
|
| |||||
| Experiment 2, MGH normalized values | predicted No-event | predicted Obstructive | predicted Central | predicted RERA | predicted Hypopnea |
|
| |||||
| True, No-event | 0.97 | 0.0 | 0.01 | 0.02 | 0.0 |
| True, Obstructive | 0.16 | 0.51 | 0.12 | 0.11 | 0.10 |
| True, Central | 0.08 | 0.06 | 0.84 | 0.01 | 0.01 |
| True, RERA | 0.46 | 0.04 | 0.04 | 0.40 | 0.06 |
| True, Hypopnea | 0.32 | 0.17 | 0.07 | 0.21 | 0.23 |
TABLE VIII.
Training and validation performance for experiment 1 with all values in mean percentages and 95% confidence intervals
| MGH training (binary) | MGH validation (binary) | MGH training (multiclass) | MGH validation (multiclass) | ||
|
| |||||
| Accuracy | 95.8 [95.8–95.9] | 95.9 [95.8–95.9] | Accuracy | 95.8 | 99.1 |
| Sensitivity | 67.2 [67.0–67.3] | 69.2 [68.8–69.5] | Sensitivity | 67.2 | 50.0 |
| Specificity | 97.8 [97.8–97.8] | 97.6 [97.6–97.7] | Specificity | 97.8 | 99.5 |
| Precision | 66.9 [66.7–67.0] | 66.0 [65.6–66.3] | Precision | 66.9 | 36.9 |
| F1-score | 67.0 [66.9–67.1] | 67.5 [67.2–67.8] | F1-score | 67.0 | 40.4 |
| Cohen’s kappa | 64.8 [64.7–64.9] | 65.3 [65.0–65.6] | Cohen’s kappa | 37.2 [37.1–37.3] | 36.8 [36.6–37.1] |
|
| |||||
| Multi-class training | Sensitivity | Specificity | Multi-class validation | Sensitivity | Specificity |
|
| |||||
| Obstructive apnea | 52.4 [52.1–52.6] | 99.7 [99.7–99.7] | Obstructive apnea | 48.4 [48.1–49.5] | 99.7 [99.7–99.7] |
| Central apnea | 84.9 [84.7–85.1] | 99.6 [99.6–99.6] | Central apnea | 87.6 [87.0–88.2] | 99.6 [99.5–99.6] |
| RERA | 40.9 [40.7–41.1] | 98.9 [98.9–98.9] | RERA | 40.6 [40.0–41.2] | 99.0 [99.0–99.0] |
| Hypopnea | 23.2 [23.1–23.4] | 99.7 [99.7–99.7] | Hypopnea | 22.9 [22.4–23.3] | 99.7 [99.7–99.7] |
|
| |||||
| Mean | 50.4 | 99.5 | Mean | 50.0 | 99.5 |
|
| |||||
| Multi-class training | Precision | F1-score | Multi-class validation | Precision | F1-score |
|
| |||||
| Obstructive apnea | 45.2 [44.9–45.4] | 48.5 [48.3–48.7] | Obstructive apnea | 42.0 [41.3–42.7] | 45.1 [44.5–45.7] |
| Central apnea | 42.5 [42.3–42.7] | 56.6 [56.4–56.8] | Central apnea | 41.7 [41.1–42.2] | 56.5 [55.9–57.1] |
| RERA | 24.4 [24.3–24.5] | 30.5 [30.4–30.7] | RERA | 25.5 [25.1–25.8] | 31.3 [30.9–31.7] |
| Hypopnea | 39.9 [39.7–40.2] | 29.4 [29.2–29.5] | Hypopnea | 38.5 [37.8–39.1] | 28.7 [28.2–29.2] |
|
| |||||
| Mean | 38.0 | 41.3 | Mean | 36.9 | 40.4 |
Contributor Information
Thijs E Nassi, University of Twente, 7522NB Enschede, the Netherlands.
Wolfgang Ganglberger, Massachusetts General Hospital, Boston, MA, 02114 USA.
Haoqi Sun, Massachusetts General Hospital, Boston, MA, 02114 USA.
Abigail A Bucklin, Massachusetts General Hospital, Boston, MA, 02114 USA.
Siddharth Biswal, School of Computational Science and Engineering, Georgia Institute of Technology, Atlanta, GA, USA.
Michel J A M van Putten, University of Twente, 7522NB Enschede, the Netherlands.
Robert J Thomas, Deaconess Medical Center, Boston, MA, 02215, USA.
M Brandon Westover, Massachusetts General Hospital, Boston, MA, 02114 USA.
REFERENCES
- [1].Benjafield AV et al. , “Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis,” The Lancet Respiratory Medicine, vol. 7, no. 8, pp. 687–698, Aug. 2019. doi: 10.1016/S2213-2600(19)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Skaer TL and Sclar DA, “Economic implications of sleep disorders,” PharmacoEconomics, vol. 28, no. 11, pp. 1015–1023, 2010. doi: 10.2165/11537390-000000000-00000. [DOI] [PubMed] [Google Scholar]
- [3].Pietzsch JB et al. , “An integrated health-economic analysis of diagnostic and therapeutic strategies in the treatment of moderate-to-severe obstructive sleep apnea,” Sleep, vol. 34, no. 6, pp. 695–709, Jun. 2011. doi: 10.5665/SLEEP.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Avidan MS et al. , “Obstructive sleep apnea as an independent predictor of postoperative delirium and pain: Protocol for an observational study of a surgical cohort [version 2],” F1000Research, vol. 7, 2018. doi: 10.12688/f1000research.14061.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Revels SL, Cameron BH, and Cameron RB, “Obstructive sleep apnea and perioperative delirium among thoracic surgery intensive care unit patients: Perspective on the STOP-BANG questionnaire and postoperative outcomes,” Journal of Thoracic Disease, vol. 11, no. Suppl 9, S1292–S1295, 2019. doi: 10.21037/jtd.2019.04.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Najafabadi MM et al. , “Deep learning applications and challenges in big data analytics,” Journal of Big Data, vol. 2, no. 1, p. 1, Dec. 2015. doi: 10.1186/s40537-014-0007-7. [DOI] [Google Scholar]
- [7].Yue L et al. , “Deep learning for heterogeneous medical data analysis,” World Wide Web, pp. 1–23, Mar. 2020. doi: 10.1007/s11280-019-00764-z. [DOI] [Google Scholar]
- [8].van den Oord A et al. , “WaveNet: A Generative Model for Raw Audio,” Sep. 2016. arXiv: 1609.03499. [Online]. Available: http://arxiv.org/abs/1609.03499. [Google Scholar]
- [9].Zabihi M et al. , “1D Convolutional Neural Network Models for Sleep Arousal Detection,” Mar. 2019. arXiv: 1903.01552. [Google Scholar]
- [10].Biswal S et al. , “Expert-level sleep scoring with deep neural networks,” Journal of the American Medical Informatics Association, vol. 25, no. 12, pp. 1643–1650, Dec. 2018. doi: 10.1093/jamia/ocy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Van Steenkiste T et al. , “Automated Sleep Apnea Detection in Raw Respiratory Signals Using Long Short-Term Memory Neural Networks,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 6, pp. 2354–2364, Nov. 2019. doi: 10.1109/JBHI.2018.2886064. [DOI] [PubMed] [Google Scholar]
- [12].Haidar R et al. , “Convolutional Neural Networks on Multiple Respiratory Channels to Detect Hypopnea and Obstructive Apnea Events,” in Proceedings of the International Joint Conference on Neural Networks, vol. 2018-July, Institute of Electrical and Electronics Engineers Inc., Oct. 2018, isbn: 9781509060146. doi: 10.1109/IJCNN.2018.8489248. [DOI] [Google Scholar]
- [13].McCloskey S et al. , “Detecting hypopnea and obstructive apnea events using convolutional neural networks on wavelet spectrograms of nasal airflow,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 10937 LNAI, Springer Verlag, Jun. 2018, pp. 361–372, isbn: 9783319930336. doi: 10.1007/978-3-319-93034-3_29. [DOI] [Google Scholar]
- [14].Banluesombatkul N, Rakthanmanon T, and Wilaiprasitporn T, “Single Channel ECG for Obstructive Sleep Apnea Severity Detection using a Deep Learning Approach,” IEEE Region 10 Annual International Conference, Proceedings/TENCON, vol. 2018-Octob, pp. 2011–2016, Aug. 2018. doi: 10.1109/TENCON.2018.8650429. arXiv: 1808.10844. [Google Scholar]
- [15].Lakhan P et al. , “Deep Neural Networks with Weighted Averaged Overnight Airflow Features for Sleep Apnea-Hypopnea Severity Classification,” IEEE Region 10 Annual International Conference, Proceedings/TENCON, vol. 2018-Octob, pp. 441–445, Aug. 2018. doi: 10.1109/TENCON.2018.8650491. arXiv: 1808.10845. [Google Scholar]
- [16].Espinoza-Cuadros F et al. , “Speech Signal and Facial Image Processing for Obstructive Sleep Apnea Assessment,” Computational and Mathematical Methods in Medicine, vol. 2015, 2015. DOI: 10.1155/2015/489761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gutiérrez-Tobal G et al. , “Assessment of Time and Frequency Domain Entropies to Detect Sleep Apnoea in Heart Rate Variability Recordings from Men and Women,” Entropy, vol. 17, no. 1, pp. 123–141, Jan. 2015. doi: 10.3390/e17010123. [DOI] [Google Scholar]
- [18].Álvarez D et al. , “Nonlinear characteristics of blood oxygen saturation from nocturnal oximetry for obstructive sleep apnoea detection,” Physiological Measurement, vol. 27, no. 4, pp. 399–412, Apr. 2006. doi: 10.1088/0967-3334/27/4/006. [DOI] [PubMed] [Google Scholar]
- [19].Rosenwein T et al. , “Breath-by-breath detection of apneic events for OSA severity estimation using non-contact audio recordings,” in Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, vol. 2015-Novem, institute of Electrical and Electronics Engineers inc., Nov. 2015, pp. 7688–7691, isbn: 9781424492718. doi: 10.1109/EMBC. 2015.7320173. [DOI] [PubMed] [Google Scholar]
- [20].Choi SH et al. , “Real-time apnea-hypopnea event detection during sleep by convolutional neural networks,” Computers in Biology and Medicine, vol. 100, pp. 123–131, Sep. 2018. doi: 10.1016/j.compbiomed.2018.06.028. [DOI] [PubMed] [Google Scholar]
- [21].Kim T, Kim JW, and Lee K, “Detection of sleep disordered breathing severity using acoustic biomarker and machine learning techniques,” BioMedical Engineering Online, vol. 17, no. 1, Feb. 2018. doi: 10.1186/s12938-018-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Haidar R, Koprinska I, and Jeffries B, “Sleep apnea event detection from nasal airflow using convolutional neural networks,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 10638 LNCS, Springer Verlag, 2017, pp. 819–827, isbn: 9783319701387. doi: 10.1007/978-3-319-70139-4_83. [DOI] [Google Scholar]
- [23].ElMoaqet H et al. , “Gaussian mixture models for detecting sleep apnea events using single oronasal airflow record,” Applied Sciences, vol. 10, no. 21, p. 7889, Nov. 2020. doi: 10.3390/app10217889. [Online]. Available: 10.3390/app10217889. [DOI] [Google Scholar]
- [24].Mostafa SS et al. , A systematic review of detecting sleep apnea using deep learning, Nov. 2019. doi: 10.3390/s19224934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Uddin MB, Chow CM, and Su SW, “Classification methods to detect sleep apnea in adults based on respiratory and oximetry signals: A systematic review,” Physiological Measurement, vol. 39, no. 3, Mar. 2018. doi: 10.1088/1361-6579/aaafb8. [DOI] [PubMed] [Google Scholar]
- [26].Bsoul M, Minn H, and Tamil L, “Apnea MedAssist: Real-time sleep apnea monitor using single-lead ECG,” IEEE Transactions on Information Technology in Biomedicine, vol. 15, no. 3, pp. 416–427, May 2011. doi: 10.1109/TITB.2010.2087386. [DOI] [PubMed] [Google Scholar]
- [27].ElMoaqet H et al. , “Deep recurrent neural networks for automatic detection of sleep apnea from single channel respiration signals,” Sensors, vol. 20, no. 18, p. 5037, Sep. 2020. doi: 10.3390/s20185037. [Online]. Available: 10.3390/s20185037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Steenkiste T et al. , “Systematic Comparison of Respiratory Signals for the Automated Detection of Sleep Apnea,” in Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, vol. 2018-July, Institute of Electrical and Electronics Engineers Inc., Oct. 2018, pp. 449–452, isbn: 9781538636466. doi: 10.1109/EMBC.2018.8512307. [DOI] [PubMed] [Google Scholar]
- [29].Division of Sleep and Circadian Disorders, Sleep heart health study, 2019. [Online]. Available: https://sleepdata.org/datasets/shhs/variables/abdoqual.
- [30].Yamamoto R, Wavenet vocoder, 2019. [Online]. Available: https://www.github.com/r9y9/wavenet_vocoder. [Google Scholar]
- [31].Sun H et al. , “Sleep staging from electrocardiography and respiration with deep learning,” Sleep, vol. 43, no. 7, Dec. 2019. doi: 10.1093/sleep/zsz306. [Online]. Available: 10.1093/sleep/zsz306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Buda M, Maki A, and Mazurowski MA, “A systematic study of the class imbalance problem in convolutional neural networks,” Neural Networks, vol. 106, pp. 249–259, Oct. 2017. doi: 10.1016/j.neunet.2018.07.011. arXiv: 1710.05381. [DOI] [PubMed] [Google Scholar]
- [33].Johnson JM and Khoshgoftaar TM, “Survey on deep learning with class imbalance,” Journal of Big Data, vol. 6, no. 1, p. 27, Dec. 2019. doi: 10.1186/s40537-019-0192-5. [DOI] [Google Scholar]
- [34].Cohen J, “A coefficient of agreement for nominal scales,” Educational and Psychological Measurement, vol. 20, no. 1, pp. 37–46, Apr. 1960. doi: 10.1177/001316446002000104. [Online]. Available: 10.1177/001316446002000104. [DOI] [Google Scholar]
- [35].MATLAB, robustfit (R2020b). Natick, Massachusetts: The MathWorks Inc., 2020. [Online]. Available: https://www.mathworks.com/help/stats/robustfit.html. [Google Scholar]
- [36].Nassi TE, Sun H, and Ganglberger W, Sleepbreathing-dl, 2021. [Online]. Available: https://github.com/mghcdac/SleepBreathing-DL. [Google Scholar]
- [37].Norman RG et al. , “Detection of respiratory events during NPSG: nasal cannula/pressure sensor versus thermistor,” Sleep, vol. 20, no. 12, pp. 1175–1184, Dec. 1997. [PubMed] [Google Scholar]
- [38].Rosenberg RS and Van Hout S, “The american academy of sleep medicine inter-scorer reliability program: Respiratory events,” vol. 10, no. 04, pp. 447–454, Apr. 2014. doi: 10.5664/jcsm.3630. [Online]. Available: 10.5664/jcsm.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reichert JA et al. , “Comparison of the NovaSom QSG™, a new sleep apnea home-diagnostic system, and polysomnography,” Sleep Medicine, vol. 4, no. 3, pp. 213–218, May 2003. doi: 10.1016/s1389-9457(02)00234-4. [Online]. Available: 10.1016/s1389-9457(02)00234-4. [DOI] [PubMed] [Google Scholar]
- [40].Scalzitti N et al. , “Comparison of home sleep apnea testing versus laboratory polysomnography for the diagnosis of obstructive sleep apnea in children,” International Journal of Pediatric Otorhinolaryngology, vol. 100, pp. 44–51, Sep. 2017. doi: 10.1016/j.ijporl.2017.06.013. [Online]. Available: 10.1016/j.ijporl.2017.06.013. [DOI] [PubMed] [Google Scholar]
- [41].Su S et al. , “A comparison of polysomnography and a portable home sleep study in the diagnosis of obstructive sleep apnea syndrome,” Otolaryngology–Head and Neck Surgery, vol. 131, no. 6, pp. 844–850, Dec. 2004. doi: 10.1016/j.otohns.2004.07.014. [Online]. Available: 10.1016/j.otohns.2004.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


