Abstract
Background:
Altered resting state (RS) functional connectivity in the default mode network (DMN) is characteristic of both autism spectrum disorder (ASD) and attention-deficit hyperactivity disorder (ADHD). Standard analytical pipelines for RS functional connectivity focus on linear correlations in activation time courses between neural networks or regions of interest (ROIs). These features may be insensitive to temporally lagged or non-linear relationships.
Methods:
In a twin cohort study including 292 children, including 52 diagnosed with ASD and 70 with ADHD, we applied non-linear analytical methods to characterize periodic dynamics in the DMN. Utilizing recurrence quantification analysis (RQA) and related methods, we measured the prevalence, duration, and complexity of periodic processes within and between DMN ROIs. We constructed generalized estimating equations (GEEs) to compare these features between neurotypical children and children with ASD and/or ADHD while controlling for familial relationships; and, leveraged machine learning algorithms to construct models predictive of ASD or ADHD diagnosis.
Results:
In within-pair analyses of twins with discordant ASD diagnoses, we found that DMN signal dynamics in dizygotic twins were significantly different, but monozygotic twins were not. Considering our full sample, we found that these patterns allowed a robust predictive classification of both ASD (81.0% Accuracy; AUC=0.85) and ADHD (82% Accuracy; AUC=0.87) cases, respectively.
Conclusions:
These findings indicate that synchronized periodicity among regions comprising the DMN relates both to neurotypical function and to ASD and/or ADHD; and suggest generally that a dynamical analysis of network interconnectivity may be a useful methodology for future neuroimaging studies.
Keywords: Autism, Default mode network, Resting state connectivity, Attention deficit hyperactivity disorder, Recurrence quantification analysis, Dynamical systems
Introduction
Neuroimaging with functional magnetic resonance imaging (fMRI) is commonly applied to probe mechanisms underlying neural and psychological processes, and to identify dysregulation of these mechanisms in neurological, psychiatric, and neurodevelopmental conditions. Resting state (RS) functional connectivity, which is the synchronization of spontaneous (task-free) activity between brain regions, has been demonstrated to be a useful measure in psychiatric research. This is due to the low cognitive demand and the high comparability between different studies. The observed networks are also highly reproducible and reflect, at least partly, the underlying structural connectivity between networks and regions of interest (ROIs)(1). Particularly in clinical contexts, the development of methods to leverage RS functional connectivity to detect signatures of atypical functionality is a major outstanding goal in the field. In the present paper, we introduce new measures of RS functional connectivity and explore their utility in characterizing functional dysregulation relating to ASD and attention deficit hyperactivity disorder (ADHD).
Network-level RS functional connectivity in relation to ASD(2-6) and/or ADHD(7, 8) has primarily been assessed in relation to the default mode network (DMN). The DMN is one of the most prominent RS networks and is a system of brain regions that are active at rest but consistently deactivated while an individual is performing attention demanding tasks(5). It has been suggested that DMN activity reflects an undirected monitoring and evaluation of the current situation(9). Further, the DMN overlaps with social brain regions, and DMN connectivity has been found to predict social skills(10). The DMN is also thought to play an important role in the emergence of Theory of Mind, moral decision-making, episodic memory, and prospection, in addition to social and emotional processing(10-15).
DMN connectivity is usually decreased in adults and adolescents with ASD, while children with the condition have more commonly increased DMN connectivity compared to age-matched controls, suggesting an unusual developmental trajectory(16). Alterations in perceptual style, such as enhanced detail focus and slower feature integration, are thought to relate to a general perturbation of connectivity patters in ASD, such that local connectivity may be atypically stronger, while global or distal connectivity is weaker in ASD(17).
Less is known about DMN connectivity in individuals with ADHD, but there is evidence for altered connectivity and possibly a maturational delay even here(7, 18). One recent large scale RS fMRI study on data from the Autism Brain Imaging Data Exchange (ABIDE) database identified similar alterations in DMN connectivity as shared neural correlates of individuals with ASD and ADHD(19). Further, individuals with both conditions have been found to show more extreme DMN connectivity alterations than individuals with ASD alone(18). Despite the utility of seed-based and network-level metrics in basic and applied research on ASD and ADHD, the use of these tools for the prediction and classification of conditions remains an ongoing challenge. Several studies(20-24) have shown that DMN connectivity was one of the most defining features for diagnostic classification of ASD.
Although there is growing evidence for the importance of the DMN in both ASD and ADHD, control of confounding factors in traditional study designs remains a major challenge. Critically, a twin-based study design, which allows comparison of twins concordant and discordant for diagnoses and/or traits, provides an inherent control over genetic and environmental factors which would otherwise be impossible to achieve in standard population-based study designs(25). Contrasting within-pair associations (i.e., comparing twins of each pair to each other) between monozygotic (MZ) and dizygotic (DZ) sub-cohorts enables further disentanglement of genetic from environmental influence given that associations within MZ twins can be regarded as environmental. For example, one twin study found reduced DMN connectivity in adult twins with higher autistic traits compared to their twins with lower autistic trait expression and suggested that altered RS connectivity involving the DMN in association with ASD is not solely driven by genetics or other familial factors, but instead closely linked to the phenotypic expression of ASD(6).
In the current study, we leveraged a twin-based study which included participants with ASD, ADHD or both conditions (2, 3, 22, 23) to explore synchronization among and between regions comprising the DMN with non-linear analytical methods, particularly recurrence quantification analysis (RQA). To demonstrate the utility of this approach in the analysis of fMRI signals, we focus on the identification and statistical discrimination of individuals diagnosed with ASD and ADHD, respectively, from NT individuals. This approach allowed the identification of patterns of desynchronization which we further leveraged in the construction of a classification model that can assign case/control status for either condition in a naïve dataset. While utilizing a relatively small pool of participants with clinical ASD or ADHD diagnoses, this approach yielded comparable classification performance to standard methods that have utilized far larger sample sizes(3, 26) to achieve generalizable performance in naïve datasets.
Materials and methods
Participant Recruitment and Characteristics
Study participants were recruited via the Roots of Autism and ADHD Twin Study in Sweden (RATSS)(27), where the majority of individuals are recruited from the population-based Child and Adolescent Twin Study in Sweden (CATSS)(28). Twin pairs where one or both of the twins show either increased ASD or ADHD-like traits in the Autism-Tics, ADHD and other Comorbidities inventory(29) are recruited to RATSS, as well as twin pairs with low such trait levels in order to cover the entire spectrum of autistic traits. Participant demographics are summarized in Table 1, and additional details on recruiting and participant characteristics can be found in Supplemental Information.
Table 1.
RATSS Participant Characteristics.
| N | ASD | ADHD | NT | Mean (SD) Age (years) |
Mean (SD) IQ |
Mean (SD) SRS |
||
|---|---|---|---|---|---|---|---|---|
| Male | ||||||||
| Overall | 154 | 31 | 42 | 95 | 15.7 (4.8) | 99.5 (14.7) | 39.9 (31.5) | |
| MZ | 89 | 14 | 20 | 60 | 15.8 (4.7) | 98.2 (13.2) | 33.5 (26.1) | |
| DZ | 63 | 17 | 22 | 33 | 15.2 (5) | 101.0 (16.3) | 50.1 (36.2) | |
| undetermined | 2 | 0 | 0 | 2 | 24.0 (0) | 115.5 (21.9) | 10.5 (2.1) | |
| Female | ||||||||
| Overall | 138 | 21 | 28 | 98 | 18.1 (6.1) | 99.2 (15.9) | 36.4 (32.6) | |
| MZ | 80 | 14 | 10 | 62 | 19.4 (6) | 99.5 (17.2) | 30.2 (30) | |
| DZ | 58 | 7 | 18 | 36 | 16.2 (5.8) | 98.7 (13) | 45.5 (34.5) | |
| undetermined | 0 | 0 | 0 | 0 | - | - | - |
Note. There were 23 individuals with both ASD and ADHD (14 males and 9 females) who were included in both the ASD and the ADHD group.
Functional Magnetic Resonance Imaging
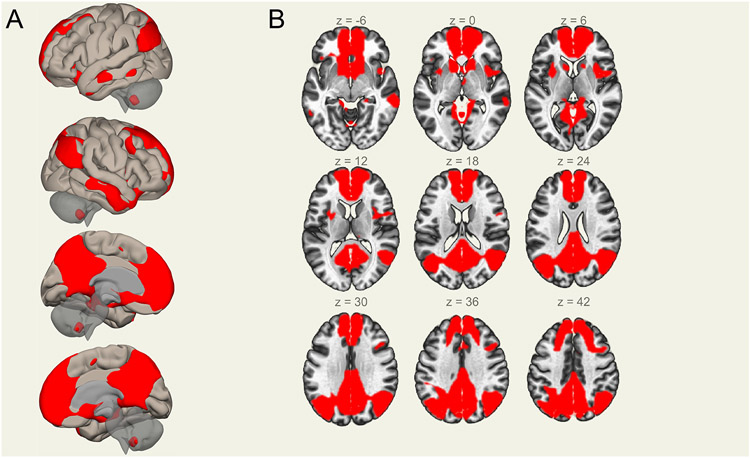
Functional magnetic resonance imaging (fMRI) data were analyzed from 292 RATSS participants. Participants completed a 5–7 min pre-scanning training session in a mock scanner, followed by a ~ 50 min MRI session in a 3 Tesla MR750 GE-scanner. The scanning session included a ~5 min T1-weighted Spoiled Gradient Echo high resolution anatomical scan (176 slices, TR = 8.2 s, FOV = 240 mm) and a 10 min resting state (RS) T2*-weighted Echo Planar Imaging Scan (45 slices, TR = 3 s, 205 volumes, FOV = 288 mm, matrix size = 96x96). During the RS scan, a white cross on black background was presented. Participants were instructed to look at the cross throughout the RS functional MRI run. Resting state fMRI data were pre-processed in Matlab v2017b (Mathworks) with the CONN (https://www.nitrc.org/projects/conn) and (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/) SPM12 toolboxes. Detailed preprocessing information can be found in Supplemental information. DMN regions were defined by structural co-registration with MNI space using the Harvard-Oxford brain atlas(30-33) (additional details in Supplemental Information). The anatomical localization of DMN regions are shown in Figure 1.
Figure 1. Anatomical localization of the Default Mode Network (DMN).
(A) From top-to-bottom, the DMN in the brain from left, right, left-medial, and right-medial sections. (B) seed-to-voxel (positive) correlations in ROIs of the DMN, with panel slices of the DMN from the central reference slice z = 18 (coordinates = 0 0 18 mm/75 109 91 voxels).
Recurrence Quantification Analysis
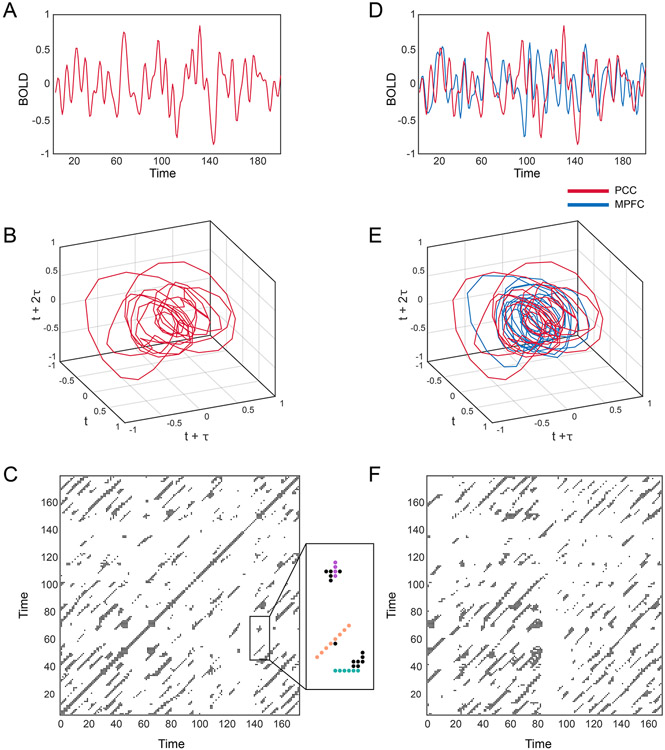
Recurrence quantification analysis (RQA) is used to describe periodic processes in physical and biological systems and is prevalent in many fields including biology, physiology, geology, and climatology(34-37). We have previously described the application of RQA to biological time series as used here(38-40), and illustrate this process further in Figure 2; we provide additional details as to the theoretical background, implementation, and interpretation of RQA and related methods in Supplemental Information. In this study, which focused on the analysis of periodic signatures in BOLD signals, RQA was used to derive three metrics – Determinism, Mean Diagonal Length, and Entropy - which measure the relative prevalence, duration, and complexity of periodic processes in the BOLD signal. The application of RQA to BOLD signals from individual ROIs was then extended to consider synchronization between ROIs via the application of cross-recurrence quantification analysis (CRQA), which, as in RQA, yields a measure of the prevalence (Determinism), duration (Mean Diagonal Length), and complexity (Entropy) of cross-signal synchronization.
Figure 2. Construction of recurrence and cross-recurrence plots.
(A) Example BOLD signal from posterior cingulate cortex (PCC). (B) Phase portrait derived from Taken’s delay embedding of BOLD signal shown in (A). (C) Recurrence plot derived from (B). Examples of the features we quantified from recurrence plots are provided in the magnified box. Recurrence plots represent periodic processes with diagonal lines (orange dotted line) and non-periodic processes with vertical/horizontal (laminar) lines (purple/blue dotted lines). (D) Example BOLD signal from posterior cingulate cortex (PCC) (red) and medial prefrontal cortex (MPFC) (blue). (E) Phase portrait derived from Taken’s delay embedding of BOLD signals shown in (D). (F) Cross-recurrence plot derived from (E). We quantified the same features from cross-recurrence plots as recurrence plots, e.g., diagonal lines or cyclical processes.
Statistical Analysis
Features derived from RQA/CRQA were used in the implementation of two general modeling strategies involving inferential and predictive modeling. First, to assess statistical differences relating to ASD and ADHD, we constructed general estimating equations (GEEs) to identify relationships between RQA/CRQA features in various regions to ASD and ADHD. As in prior studies(41-43), GEEs were used to account for relatedness among twin participants via the double-robust procedure for handling confounding by cluster. This approach utilizes an independent correlation structure(44). In the initial phase of modeling, only twin pairs with discordant diagnoses for ASD or ADHD were analyzed. Effect estimation was conditioned on family identification such that tests for effects of ASD/ADHD reflected within-pair differences. Models included the main effects of ASD/ADHD as well as an interaction term to test for dependency of effects on zygosity (i.e., whether effects were different in MZ and DZ twin pairs). Effects relating to ASD*Zygosity and ADHD*Zygosity interactions were adjusted for multiple comparisons (FDR). Where these effects survived multiple comparison, post-hoc tests (Tukey) were used to test for differences between diagnostic status and zygosity.
Models were adjusted for covariates of subject sex and head motion during the scanning process; we also tested for associations between IQ scores and RQA/CRQA features, and with ASD and ADHD diagnosis, but found no significant effects and therefore excluded IQ from our final models.
Following the statistical analysis of discordant twin pairs, the same strategy was then applied to the analysis of the full dataset, which included both twin pairs that had concordant diagnoses for ASD or ADHD and singletons where no co-twin data were available. A random (clustering) term was included in these models to account for relatedness among participants. Significant effects relating to zygosity-dependent interactions were not followed with post-hoc analyses, as these could not be interpreted as relating to genetically driven effects due to the presence of concordant twin pairs and singletons. Models were implemented in R v3.5 with the drgee and multcomp packages.
Predictive Modeling
In parallel to statistical models used for hypothesis testing, we implemented a complementary analysis to evaluate the predictive utility of RQA/CRQA features with respect to ASD or ADHD case status. Utilizing data from all participants, we randomly partitioned participants into training (75% of data) and validation (25% of data) sets. A tree-based gradient boosting algorithm(45), XGBoost(46) (Extreme Gradient Boosting), was trained following 5000 iterations of 5-fold cross-validation to make a binary prediction of case status; this procedure was repeated to predict ASD status, and to predict ADHD status. For cross-validated training, 5000 iterations of a random grid search were used to tune the nrounds, eta, max_depth, gamma, and colsample_bytree hyperparameters. These correspond to the number of rounds used in gradient boosting, learning rate, tree depth, penalization parameter, and random sub-setting percentage, respectively. The performance of trained models was then evaluated by making predictions on validation data sets. To quantify the relative importance of each feature to each predictive model, we estimated Gain(46); this is a measure of predictive improvement associated with the inclusion of each feature. These procedures were implemented in R v3.5 with the mlr and Xgboost packages.
Results
In 292 participants, including 52 diagnosed with ASD and 70 with ADHD, we applied recurrence quantification analysis (RQA) and cross-recurrence quantification analysis (CRQA) to characterize periodicity and synchronization within and between regions of interest (ROIs) comprising the default mode network (DMN). The sample included 122 complete twin pairs of which 76 were monozygotic (MZ) and 46 dizygotic (DZ). There were no significant differences in head motion between neurotypical participants and participants with ASD (β=0.07 (95%CI: −0.17,0.32), p=0.56) or ADHD (β=0.03 (95%CI: −0.10,0.15), p=0.68). There were likewise no significant differences in participants’ age between neurotypical participants and participants with ASD (β=0.04 (95%CI: −0.13,0.22), p=0.61) or ADHD (β=0.01 (95%CI: −0.14, 0.13), p=0.90). The DZ subgroup, however, were older at the time of scanning than the MZ group (t=2.79, df=288, p=0.006). Within zygosity-specific subgroups, MZ females were older than males (t=−4.35, df=167 p=0.00002), and DZ females were older than males (t=−2.1, df=146 p=0.04).
Differences in the RQA metrics of discordant ASD and/or ADHD twin pairs
To leverage the genetic and environmental control afforded by a twin-based study design, our initial analysis focused on MZ and DZ twin pairs with discordant diagnostic status for either ASD or for ADHD. In our analysis of ASD-discordant twin pairs there were 10 MZ pairs and 13 DZ pairs, whereas in our analysis of ADHD-discordant pairs, there were 12 MZ pairs and 24 DZ pairs. One group of dizygotic triplets discordant for both ASD and ADHD was included. Table 2 provides FDR-adjusted P-values for the effect tests for ASD and ADHD, embedded in an interaction for zygosity to allow contrasts between MZ and DZ twins, across each RQA metric, and for each ROI (for RQA analyses) or ROI pair (for CRQA analyses). Tests for ASD/ADHD-related effects reflect within-pair differences between twins with discordant diagnoses.
Table 2. Analysis of recurrence features in discordant twins with ASD and/or ADHD.
FDR corrected P-values. β provides effect parameters for interactions between zygosity and ASD or ADHD diagnostic status; P and FDR columns provide or raw or false discovery rate-adjusted P values for that parameter, respectively.
| MEASURE | ROI | ASD | ADHD | ||||
|---|---|---|---|---|---|---|---|
| β | P | FDR | β | P | FDR | ||
| Determinism | DMN | 0.083 | 0.000 | 0.003 | 0.030 | 0.205 | 0.755 |
| LP(left) | 0.062 | 0.024 | 0.143 | −0.024 | 0.323 | 0.755 | |
| LP(left) - LP(right) | 0.091 | 0.001 | 0.016 | −0.011 | 0.671 | 0.855 | |
| LP(left) - PCC | 0.023 | 0.277 | 0.599 | −0.035 | 0.191 | 0.755 | |
| LP(right) | 0.128 | 0.000 | 0.003 | 0.005 | 0.849 | 0.912 | |
| LP(right) - PCC | 0.049 | 0.094 | 0.367 | −0.010 | 0.656 | 0.855 | |
| MPFC | 0.051 | 0.128 | 0.392 | −0.006 | 0.839 | 0.912 | |
| MPFC - LP(left) | 0.050 | 0.016 | 0.108 | −0.019 | 0.355 | 0.755 | |
| MPFC - LP(right) | 0.081 | 0.000 | 0.003 | 0.010 | 0.623 | 0.840 | |
| MPFC - PCC | 0.022 | 0.385 | 0.619 | −.007 | 0.812 | 0.912 | |
| PCC | −0.027 | 0.356 | 0.605 | −0.026 | 0.409 | 0.755 | |
| Entropy | DMN | 0.111 | 0.043 | 0.221 | 0.011 | 0.857 | 0.912 |
| LP(left) | 0.073 | 0.068 | 0.319 | −0.036 | 0.429 | 0.755 | |
| LP(left) - LP(right) | 0.060 | 0.194 | 0.499 | −0.044 | 0.349 | 0.755 | |
| LP(left) - PCC | −0.011 | 0.759 | 0.821 | −0.114 | 0.014 | 0.298 | |
| LP(right) | 0.129 | 0.011 | 0.102 | −0.084 | 0.057 | 0.611 | |
| LP(right) - PCC | 0.036 | 0.480 | 0.684 | −0.026 | 0.617 | 0.840 | |
| MPFC | 0.056 | 0.346 | 0.605 | −0.052 | 0.370 | 0.755 | |
| MPFC - LP(left) | −0.025 | 0.554 | 0.731 | −0.032 | 0.509 | 0.755 | |
| MPFC - LP(right) | 0.122 | 0.014 | 0.108 | −0.040 | 0.363 | 0.755 | |
| MPFC - PCC | 0.037 | 0.496 | 0.684 | −0.046 | 0.356 | 0.755 | |
| PCC | −0.087 | 0.197 | 0.499 | −0.028 | 0.674 | 0.855 | |
| Mean Diagonal Length | DMN | 0.250 | 0.087 | 0.359 | 0.069 | 0.728 | 0.874 |
| LP(left) | 0.021 | 0.846 | 0.886 | −0.098 | 0.415 | 0.755 | |
| LP(left) - LP(right) | 0.113 | 0.291 | 0.599 | −0.132 | 0.299 | 0.755 | |
| LP(left) - PCC | −0.042 | 0.598 | 0.745 | −0.289 | 0.010 | 0.298 | |
| LP(right) | 0.337 | 0.002 | 0.026 | −0.169 | 0.184 | 0.755 | |
| LP(right) - PCC | 0.108 | 0.354 | 0.605 | −0.101 | 0.416 | 0.755 | |
| MPFC | 0.117 | 0.423 | 0.620 | −0.099 | 0.515 | 0.755 | |
| MPFC - LP(left) | −0.104 | 0.300 | 0.600 | −0.086 | 0.484 | 0.755 | |
| MPFC - LP(right) | 0.263 | 0.015 | 0.108 | −0.100 | 0.361 | 0.755 | |
| MPFC - PCC | 0.063 | 0.595 | 0.745 | −0.099 | 0.339 | 0.755 | |
| PCC | −0.177 | 0.285 | 0.599 | −0.123 | 0.404 | 0.755 | |
BOLD signal Determinism, in particular, proved to be highly sensitive to ASD, with significant effects observed across the left and right LP, MPFC, and the averaged signal of the whole DMN after FDR-adjustment for multiple comparisons. In contrast, BOLD signal Mean Diagonal Length was only significantly related to ASD in the right LP, and no effects relating to BOLD signal Entropy survived multiple-comparison adjustment. With ADHD, no effects relating to RQA metrics survived multiple-comparison adjustment (see Table 2).
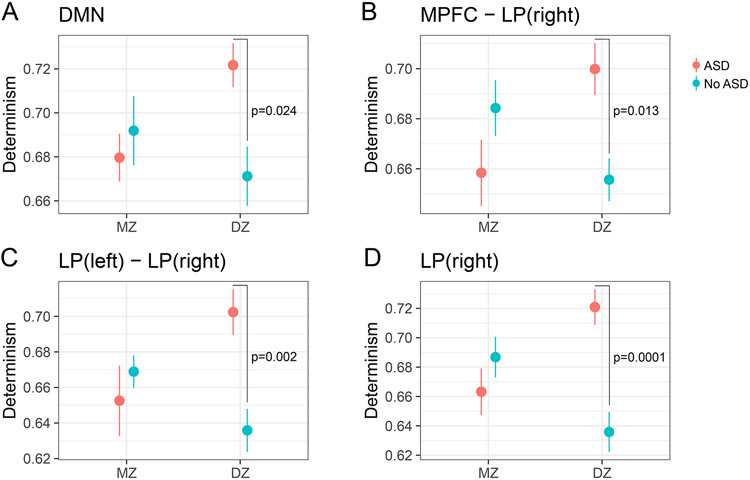
We further explored the dependence of effects observed in ASD on zygosity (indicating genetic influences) with post-hoc analyses and data visualizations within the 23 ASD discordant twin pairs and the ASD discordant triplet. Figure 3 highlights Determinism-related dysregulation with ASD in MZ and DZ twin pairs. With the overall DMN BOLD signal (Figure 3A), we found that Determinism was significantly reduced in DZ twins with ASD (Tukey HSD = 0.024) but found no overall effect for ASD discordant MZ twins. Effects relating to the synchronization of MPFC and right LP (Figure 3B; Tukey HSD = 0.013), the synchronization of the left and right LP (Figure 3C; Tukey HSD = 0.002) and the right LP (Figure 3D; Tukey HSD = 0.0001) were consistent with the pattern observed in overall DMN signal, where Determinism was reduced in DZ ASD cases compared to their co-twins but not in MZ cases. No effects survived multiple comparison adjustment in post-hoc analysis of Mean Diagonal Length in the right LP. In sum, across multiple regions, these findings indicate a persistent, differential pattern of ASD-related dysregulation in DZ twin pairs but not in MZ twin pairs.
Figure 3. ASD-Related Dysregulation of Determinism in DMN BOLD Signals.
P-values reflect Tukey-adjusted post-hoc comparisons between discordant twins with ASD diagnosis (10 MZ pairs; 13 DZ pairs; 1 DZ triplet).
Differences in the RQA metrics of participants diagnosed with ASD and/or ADHD
We next explored the roles of ASD and ADHD in our full sample, which included both singleton samples and twins with concordant diagnostic status (n = 292; 52 ASD cases and 70 ADHD cases). We found a marginally significant overall effect in that BOLD signal Determinism was reduced in ASD cases compared to NT controls in the averaged DMN signal (β = −0.031, FDR = 0.072), but for most metrics the effects of ASD were moderated by an interaction with zygosity, as observed in the discordantly-diagnosed twin subset. Consistent to our analysis restricted to discordant twin pairs, we found significant interactions between zygosity and diagnostic status in the right LP (FDR = 0.005) and the averaged DMN signal (FDR = 0.005), as well as in the synchronization of MPFC and right LP (FDR = 0.008) and left and right LP (FDR = 0.017) also in the full sample (see Table 3). Following adjustment for multiple comparisons there were no overall differences between individuals diagnosed with ADHD compared to NT controls.
Table 3. Analysis of recurrence features in the full sample (including discordant and concordant twins, and singletons) with ASD and/or ADHD.
FDR corrected P-values. β provides effect parameters for interactions between zygosity and ASD or ADHD diagnostic status; P and FDR columns provide or raw or false discovery rate-adjusted P values for that parameter, respectively.
| MEASURE | ROI | ASD | ADHD | ||||
|---|---|---|---|---|---|---|---|
| β | P | FDR | β | P | FDR | ||
| Determinism | DMN | 0.080 | 0.000 | 0.005 | 0.028 | 0.232 | 0.731 |
| LP(left) | 0.060 | 0.034 | 0.203 | −0.025 | 0.289 | 0.731 | |
| LP(left) - LP(right) | 0.089 | 0.001 | 0.017 | −0.012 | 0.638 | 0.832 | |
| LP(left) - PCC | 0.022 | 0.295 | 0.608 | −0.036 | 0.172 | 0.731 | |
| LP(right) | 0.125 | 0.000 | 0.005 | 0.004 | 0.866 | 0.911 | |
| LP(right) - PCC | 0.046 | 0.118 | 0.390 | −0.011 | 0.615 | 0.828 | |
| MPFC | 0.047 | 0.171 | 0.453 | −0.006 | 0.808 | 0.904 | |
| MPFC - LP(left) | 0.048 | 0.033 | 0.203 | −0.020 | 0.322 | 0.731 | |
| MPFC - LP(right) | 0.077 | 0.000 | 0.008 | 0.009 | 0.643 | 0.832 | |
| MPFC - PCC | 0.019 | 0.489 | 0.710 | −0.007 | 0.793 | 0.904 | |
| PCC | −0.030 | 0.312 | 0.610 | −0.026 | 0.400 | 0.731 | |
| Entropy | DMN | 0.112 | 0.041 | 0.209 | 0.009 | 0.886 | 0.913 |
| LP(left) | 0.073 | 0.080 | 0.347 | −0.038 | 0.405 | 0.731 | |
| LP(left) - LP(right) | 0.064 | 0.160 | 0.453 | −0.044 | 0.340 | 0.731 | |
| LP(left) - PCC | −0.008 | 0.839 | 0.893 | −0.117 | 0.010 | 0.229 | |
| LP(right) | 0.124 | 0.014 | 0.132 | −0.086 | 0.046 | 0.585 | |
| LP(right) - PCC | 0.037 | 0.462 | 0.710 | −0.028 | 0.587 | 0.806 | |
| MPFC | 0.043 | 0.495 | 0.710 | −0.054 | 0.340 | 0.731 | |
| MPFC - LP(left) | −0.025 | 0.567 | 0.756 | −0.033 | 0.497 | 0.731 | |
| MPFC - LP(right) | 0.117 | 0.018 | 0.147 | −0.041 | 0.341 | 0.731 | |
| MPFC - PCC | 0.030 | 0.596 | 0.756 | −0.044 | 0.374 | 0.731 | |
| PCC | −0.086 | 0.192 | 0.479 | −0.028 | 0.672 | 0.836 | |
| Mean Diagonal Length | DMN | 0.259 | 0.077 | 0.347 | 0.064 | 0.753 | 0.888 |
| LP(left) | 0.019 | 0.861 | 0.902 | −0.101 | 0.397 | 0.731 | |
| LP(left) - LP(right) | 0.126 | 0.238 | 0.541 | −0.132 | 0.294 | 0.731 | |
| LP(left) - PCC | −0.031 | 0.719 | 0.833 | −0.294 | 0.007 | 0.229 | |
| LP(right) | 0.331 | 0.004 | 0.052 | −0.171 | 0.165 | 0.731 | |
| LP(right) - PCC | 0.104 | 0.371 | 0.680 | −0.103 | 0.398 | 0.731 | |
| MPFC | 0.085 | 0.594 | 0.756 | −0.106 | 0.479 | 0.731 | |
| MPFC - LP(left) | −0.103 | 0.314 | 0.610 | −0.088 | 0.470 | 0.731 | |
| MPFC - LP(right) | 0.249 | 0.022 | 0.163 | −0.103 | 0.334 | 0.731 | |
| MPFC - PCC | 0.046 | 0.707 | 0.833 | −0.094 | 0.362 | 0.731 | |
| PCC | −0.182 | 0.269 | 0.573 | −0.124 | 0.399 | 0.731 | |
Classification of ASD and ADHD from RQA
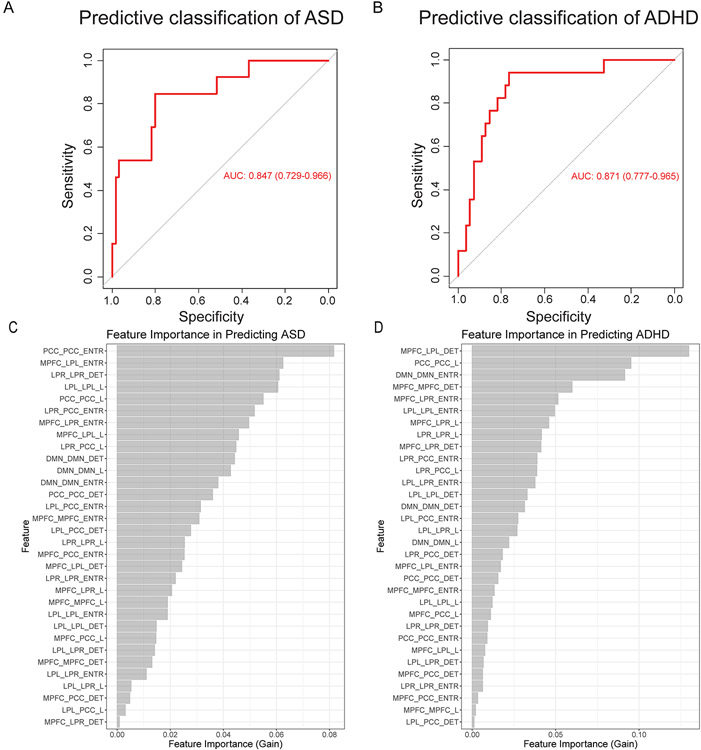
Finally, we investigated the predictive utility of RQA/CRQA-derived measures of periodicity in the DMN for separating ASD and ADHD cases from NT controls. To achieve this, we constructed a data-driven machine learning model utilizing a tree-based gradient boosting method for classification. Datasets utilizing the full range of features derived from RQA and CRQA of DMN regions were randomly split and assigned for model training (75% of available data), or model validation (25% of available data). Models were fit to training subsets utilizing 5-fold cross-validation, and model performance was then assessed by evaluating predictive sensitivity and specificity in the holdout test data. ROC curves derived from models predictive of ASD/ADHD are shown in Figure 4A/4B, respectively; generally, these illustrate robust generalization of classification efficacy following cross-validation in the training set. In predicting ADHD, models achieved an area-under-the-curve (AUC) of 0.87 (95% CI: 0.78-97), with 0.76 sensitivity, 0.84 specificity, and 82% overall accuracy at the optimal classification threshold. For ASD, the optimal classification threshold yielded 0.85 sensitivity, 0.80 specificity, and 81% overall accuracy, with model AUC of 0.85 (bootstrapped 95% CI: 0.73-0.97).
Figure 4. Predictive efficacy of periodic features derived from RQA/CRQA of the default mode network.
(A) Receiver operating characteristic (ROC) curve illustrating performance of a model trained to classify ASD on a validation dataset following 5-fold cross-validation in training set. (B) ROC curve illustrating performance of a model trained to classify ADHD on a validation dataset. (C) Feature importance (Gain) associated with each feature in predicting ASD. (D) Feature importance (Gain) associated with each feature in predicting ADHD.
Discussion
Previous studies investigating resting state (RS) functional connectivity in ASD and ADHD using correlation-based measures indicated alterations in the DMN, but largely overlooked the dynamic features involved in these processes(3, 6, 7, 16, 47-51). Here we introduce a novel analytical approach, recurrence/cross-recurrence quantification analysis (RQA/CRQA), to characterize and relate periodicity in the default mode network (DMN) to ASD and ADHD. Our results identified ASD-related differences in intrinsic and interdependent periodic processes among and between regions of the DMN that were largely limited to DZ twin pairs, suggesting a genetic role in the emergence of these signatures. More generally, in consideration of our full sample, which additionally included twins concordant for ASD or ADHD diagnosis as well as singletons, we found that these features allowed the construction of robustly generalizable classification models that were able to achieve good sensitivity and specificity for discriminating ASD and ADHD cases by resting fMRI scans. These models achieved robust classification performance, with model AUCs ranging from 0.85-0.87 in holdout testing. These findings emphasize that periodic processes active in the DMN are generally dysregulated in both conditions, but the specific signatures associated with ASD and ADHD are distinct and regulated differentially by genetic and environmental factors. More generally, these findings suggest that indicators of periodicity may provide a sensitive complement to existing measures of functional connectivity.
The primary methodological difference in the present study compared to similar neuroimaging investigations is in the treatment and analysis of the derived BOLD signal. Standard analytical pipelines focus on correlation-based metrics. In contrast, we focus on an analysis of periodicity in the raw BOLD signal, from which we derive metrics relating to prevalence (Determinism), duration (Mean Diagonal Length), and complexity (Entropy) of cyclical processes. Unlike analogous correlation-based metrics, this approach is intrinsically sensitive to non-linear and time-lagged effects; and, is equally applicable to the analysis of intrinsic periodicity within a given region, and interdependent cycles that relate one region to another. Previous physiological studies have leveraged these capacities in the analysis of EEG signals(52-56), cardiac dynamics(57-61), and motor coordination(62-65), and in fact several studies have utilized RQA/CRQA in the analysis of ASD-related behavioral features, such as stereotypical movements(66, 67). A recent study by Kaboodvand et al(68) similarly applied RQA in the assessment of DMN dynamics, particularly relating to ADHD, but the focus of that study was on RQA-related metrics such as laminarity which characterize the emergence of stable states, in contrast to our focus on periodicity. As such, the application of RQA/CRQA in the present study to characterize periodic dynamics in resting-state fMRI data emphasizes a new level of analysis where this method can be generalized.
Given that this, to our knowledge, the first study to apply RQA/CRQA to characterize periodicity in the context of neuroimaging with ASD/ADHD cases, we examined all potential connections among regions of the DMN. A potential weakness in this approach is that the broadly-defined regions analyzed here may not reflect fine-scale segmentation of DMN-related subnetworks, as characterized by Andrews et al(69) and Braga & Buckner(70). Our approach was nonetheless particularly sensitive to ASD-related dysregulation; for example, we observed dysregulated BOLD signal dynamics in the averaged DMN signal, in the right LP, and in the synchronization of the MPFC-LP(right) and LP(left)-LP(right). In ADHD, we observed a number of effects that approached statistical significance but did not survive comparison for multiple adjustment. Future studies might investigate these regions in a more targeted analysis both to determine if these were Type II errors due to potential over-adjustment for multiple comparison, and to investigate if dysregulation of synchronized activity is specific to discrete subnetworks of the DMN.
Due to the limited sample sizes of MZ and DZ sub-cohorts discordant for ASD or ADHD, the interaction effects between zygosity and within-pair differences need to nonetheless be interpreted with caution and should be confirmed in larger MZ and DZ discordant samples. As well, since the DZ sub-group was older than the MZ sub-group in our sample, we cannot exclude the possibility that the effects specifically observed in DZ twins alone were influenced by age (e.g., emerged with older age) rather than being solely driven by genetics, although these effects were adjusted for in our statistical models. In future studies, the application of these and related methods to neuroimaging studies in larger cohorts should be explored towards the goal of simultaneous classification of ASD, ADHD, and comorbid presentations of ASD and ADHD, which was not achievable in the present study given the limited sample size.
Future studies might likewise explore the application of these methods to complementary imaging modalities and conditions, particularly task-related fMRI. A crucial advantage of task-related fMRI over RS fMRI is that noise affecting all experimental conditions is cancelled out when they are contrasted. Since no such contrasting can be applied to RS data, this method demands more careful quality control procedures in order to reduce the effects of, for instance, head motion(71). Task-related and RS fMRI complement each other, both providing different aspects of neural correlates and coming with different advantages and disadvantages. Generally, despite limitations, our findings are consistent with prior results of both shared and disease-specific indicators of ASD and ADHD in regions within and outside the DMN(72).
In the context of our predictive models, we found that determinism in the DMN, generally, and determinism specific to the right lateral parietal (LP) region – both of which significantly differed from neurotypical in children with ASD – were among the most important features (top 10, Figure 4C) in predicting ASD; entropy in MPFC-LP(right), which did not survive adjustment for multiple comparison, was likewise among the most important contributors to predictive efficacy in ASD models. Somewhat in contrast to our findings utilizing statistical hypothesis tests, where the prevalence of periodic signatures (Determinism) yielded the strongest indicators of ASD-related effects, the most important features in our predictive model included measures of entropy and mean diagonal length, corresponding to the duration and complexity of periodic processes. These results emphasize that multivariate nonlinear methods, such as the tree-based predictive models used here, may be sensitive to effects that may be missed in linear statistical testing and corresponding p-value adjustment; as such, applying these methods in parallel allows for insights that might not be achievable by either method, alone.
While our findings did not provide equally compelling statistically evidence of a broad pattern of ADHD-related dysregulation, we found that these features were nonetheless also highly predictive of ADHD. In particular, we found the features most predictive of ADHD included determinism in MPFC-LP(left) synchronization, mean diagonal length in the PCC, and entropy in the DMN, generally – in short, a broad pattern of dysregulation relating to the prevalence, duration, and complexity of periodic processes. These findings suggest that patterns of ADHD-related deficits may have been obscured by limited statistical power, and the need to adjust for multiple comparisons. As such, these findings support further studies utilizing a comparable approach, and our preliminary results provide an a priori rationale for future studies to target the subset of ROIs that were suggestive of a relationship in the present study, and thereby maximize statistical power.
In sum, our findings emphasize the utility of non-linear analytical approaches for the analysis of BOLD signals in fMRI neuroimaging studies and suggest generally that periodicity in BOLD signals may be an important indicator of typical and atypical connectivity. Our findings particularly implicate a broad pattern of likely genetically determined alterations in periodicity across the DMN as an indicator of ASD, but also suggest more generally that altered periodicity within and between core regions of the DMN is indicative of both ASD and ADHD. Future studies should build on this work by exploring synchronization and periodicity within other networks and between DMN-related regions and non-DMN regions, both in relation to ASD and ADHD as well as other neurodevelopmental and psychiatric conditions.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | ||||
| Bacterial or Viral Strain | ||||
| Biological Sample | ||||
| Cell Line | ||||
| Chemical Compound or Drug | ||||
| Commercial Assay Or Kit | ||||
| Deposited Data; Public Database | ||||
| Genetic Reagent | ||||
| Organism/Strain | ||||
| Peptide, Recombinant Protein | ||||
| Recombinant DNA | ||||
| Sequence-Based Reagent | ||||
| Software; Algorithm | CONN; Matlab | https://www.nitrc.org/projects/conn | ||
| Software; Algorithm | SPM12; Matlab | https://www.fil.ion.ucl.ac.uk/spm/software/spm12/ | ||
| Software; Algorithm | Dynamical Systems; Julia | https://juliadynamics.github.io/DynamicalSystems.jl/latest/; https://doi.org/10.21105/joss.00598 | ||
| Software; Algorithm | drgee; R | https://cran.r-project.org/web/packages/drgee/index.html | ||
| Software; Algorithm | multcomp; R | https://cran.r-project.org/web/packages/multcomp/index.html | ||
| Software; Algorithm | mlr; R | https://cran.r-project.org/web/packages/mlr/index.html | ||
| Software; Algorithm | Xgboost; R | https://cran.r-project.org/web/packages/xgboost/xgboost.pdf | ||
| Software; Algorithm | Cross-Recurrence Toolbox v5.16; Matlab | http://tocsy.pik-potsdam.de/CRPtoolbox/; PMID: 12241313 | ||
| Transfected Construct | ||||
| Other | ||||
Funding and Acknowledgements
We express our sincere gratitude to all twins and parents who have participated in this research. We also thank the RATSS team at the Center of Neurodevelopmental Disorders at Karolinska Institutet (KIND) for their valuable contribution to the work presented in this study. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The study was funded by the Swedish Research Council, Vinnova, Formas, FORTE, the Swedish Brain foundation (Hjärnfonden), Stockholm Brain Institute, Autism and Asperger Association Stockholm, Queen Silvia’s Jubilee Fund, Solstickan Foundation, PRIMA Child and Adult Psychiatry, the Pediatric Research Foundation at Astrid Lindgren Children’s Hospital, the Swedish Foundation for Strategic Research, Jerring Foundation, the Swedish Order of Freemasons, Kempe-Carlgrenska Foundation, Sunnderdahls Handikappsfond, The Jeansson Foundation, and EU-AIMS (European Autism Intervention), with support from the Innovative Medicines Initiative Joint Undertaking (grant agreement no. 115300), the resources of which are composed of financial contributions from the European Union’s Seventh Framework Programme (grant FP7/2007–2013), from the European Federation of Pharmaceutical Industries and Associations companies’ in-kind contributions, and from Autism Speaks. It was also supported by a new IMI initiative–EU AIMS-2-TRIALS. At Mount Sinai, M.A. was funded by National Institute of Environmental Health Sciences grants P30ES023515, R01ES026033, U2CES030859, R35ES030435. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Fox MD, Raichle ME (2007): Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 8:700–711. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JS, Nielsen JA, Froehlich AL, DuBray MB, Druzgal TJ, Cariello AN, et al. (2011): Functional connectivity magnetic resonance imaging classification of autism. Brain. 134:3742–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen JA, Zielinski BA, Fletcher PT, Alexander AL, Lange N, Bigler ED, et al. (2013): Multisite functional connectivity MRI classification of autism: ABIDE results. Front Hum Neurosci. 7:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plitt M, Barnes KA, Martin A (2015): Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. Neuroimage Clin. 7:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murdaugh DL, Shinkareva SV, Deshpande HR, Wang J, Pennick MR, Kana RK (2012): Differential deactivation during mentalizing and classification of autism based on default mode network connectivity. PLoS One. 7:e50064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neufeld J, Kuja-Halkola R, Mevel K, Cauvet E, Fransson P, Bolte S (2018): Alterations in resting state connectivity along the autism trait continuum: a twin study. Mol Psychiatry. 23:1659–1665. [DOI] [PubMed] [Google Scholar]
- 7.Janssen TWP, Hillebrand A, Gouw A, Gelade K, Van Mourik R, Maras A, et al. (2017): Neural network topology in ADHD; evidence for maturational delay and default-mode network alterations. Clin Neurophysiol. 128:2258–2267. [DOI] [PubMed] [Google Scholar]
- 8.Uddin LQ, Kelly AM, Biswal BB, Margulies DS, Shehzad Z, Shaw D, et al. (2008): Network homogeneity reveals decreased integrity of default-mode network in ADHD. J Neurosci Methods. 169:249–254. [DOI] [PubMed] [Google Scholar]
- 9.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proc Natl Acad Sci U S A. 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mars RB, Neubert FX, Noonan MP, Sallet J, Toni I, Rushworth MF (2012): On the relationship between the "default mode network" and the "social brain". Front Hum Neurosci. 6:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckner RL, Carroll DC (2007): Self-projection and the brain. Trends Cogn Sci. 11:49–57. [DOI] [PubMed] [Google Scholar]
- 12.Molnar-Szakacs I, Uddin LQ (2013): Self-processing and the default mode network: interactions with the mirror neuron system. Front Hum Neurosci. 7:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poerio GL, Sormaz M, Wang HT, Margulies D, Jefferies E, Smallwood J (2017): The role of the default mode network in component processes underlying the wandering mind. Soc Cogn Affect Neurosci. 12:1047–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reniers RL, Corcoran R, Vollm BA, Mashru A, Howard R, Liddle PF (2012): Moral decision-making, ToM, empathy and the default mode network. Biol Psychol. 90:202–210. [DOI] [PubMed] [Google Scholar]
- 15.Smallwood J, Bernhardt BC, Leech R, Bzdok D, Jefferies E, Margulies DS (2021): The default mode network in cognition: a topographical perspective. Nat Rev Neurosci. 22:503–513. [DOI] [PubMed] [Google Scholar]
- 16.Padmanabhan A, Lynch CJ, Schaer M, Menon V (2017): The Default Mode Network in Autism. Biol Psychiatry Cogn Neurosci Neuroimaging. 2:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ (2004): Autism and abnormal development of brain connectivity. J Neurosci. 24:9228–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Xu M, Ji Y, Zhang L, Du X, Li J, et al. (2019): Altered social cognition and connectivity of default mode networks in the co-occurrence of autistic spectrum disorder and attention deficit hyperactivity disorder. Aust N Z J Psychiatry. 53:760–771. [DOI] [PubMed] [Google Scholar]
- 19.Kernbach JM, Satterthwaite TD, Bassett DS, Smallwood J, Margulies D, Krall S, et al. (2018): Shared endo-phenotypes of default mode dsfunction in attention deficit/hyperactivity disorder and autism spectrum disorder. Transl Psychiatry. 8:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chanel G, Pichon S, Conty L, Berthoz S, Chevallier C, Grezes J (2016): Classification of autistic individuals and controls using cross-task characterization of fMRI activity. Neuroimage Clin. 10:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uddin LQ, Supekar K, Lynch CJ, Khouzam A, Phillips J, Feinstein C, et al. (2013): Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry. 70:869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CP, Keown CL, Jahedi A, Nair A, Pflieger ME, Bailey BA, et al. (2015): Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. Neuroimage Clin. 8:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Duan X, Liu F, Lu F, Ma X, Zhang Y, et al. (2016): Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity--A multi-center study. Prog Neuropsychopharmacol Biol Psychiatry. 64:1–9. [DOI] [PubMed] [Google Scholar]
- 24.Price T, Wee CY, Gao W, Shen D (2014): Multiple-network classification of childhood autism using functional connectivity dynamics. Med Image Comput Comput Assist Interv. 17:177–184. [DOI] [PubMed] [Google Scholar]
- 25.Mevel K, Fransson P, Bolte S (2015): Multimodal brain imaging in autism spectrum disorder and the promise of twin research. Autism. 19:527–541. [DOI] [PubMed] [Google Scholar]
- 26.Heinsfeld AS, Franco AR, Craddock RC, Buchweitz A, Meneguzzi F (2018): Identification of autism spectrum disorder using deep learning and the ABIDE dataset. Neuroimage Clin. 17:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolte S, Willfors C, Berggren S, Norberg J, Poltrago L, Mevel K, et al. (2014): The Roots of Autism and ADHD Twin Study in Sweden (RATSS). Twin Res Hum Genet. 17:164–176. [DOI] [PubMed] [Google Scholar]
- 28.Anckarsater H, Lundstrom S, Kollberg L, Kerekes N, Palm C, Carlstrom E, et al. (2011): The Child and Adolescent Twin Study in Sweden (CATSS). Twin Res Hum Genet. 14:495–508. [DOI] [PubMed] [Google Scholar]
- 29.Hansson SL, Svanstrom Rojvall A, Rastam M, Gillberg C, Gillberg C, Anckarsater H (2005): Psychiatric telephone interview with parents for screening of childhood autism - tics, attention-deficit hyperactivity disorder and other comorbidities (A-TAC): preliminary reliability and validity. Br J Psychiatry. 187:262–267. [DOI] [PubMed] [Google Scholar]
- 30.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 15:273–289. [DOI] [PubMed] [Google Scholar]
- 31.Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. (2006): An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 31:968–980. [DOI] [PubMed] [Google Scholar]
- 32.Makris N, Goldstein JM, Kennedy D, Hodge SM, Caviness VS, Faraone SV, et al. (2006): Decreased volume of left and total anterior insular lobule in schizophrenia. Schizophr Res. 83:155–171. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein JM, Seidman LJ, Makris N, Ahern T, O'Brien LM, Caviness VS Jr., et al. (2007): Hypothalamic abnormalities in schizophrenia: sex effects and genetic vulnerability. Biol Psychiatry. 61:935–945. [DOI] [PubMed] [Google Scholar]
- 34.Marwan N, Romano MC, Thiel M, Kurths J (2007): Recurrence plots for the analysis of complex systems. Physics Reports. 438:237. [Google Scholar]
- 35.Marwan N (2008): A historical review of recurrence plots. European Physical Journal ST. 164:3–12. [Google Scholar]
- 36.Webber CL, Marwan N, Facchini A, Giuliani A (2009): Simpler methods do it better: Success of Recurrence Quantification Analysis as a general purpose data analysis tool. Phys Lett A. 373:3753–3756. [Google Scholar]
- 37.Webber CLZ, J.P. (2005): Recurrence quantification analyiss of nonlinear dynamical systems.26–94. [Google Scholar]
- 38.Austin C, Curtin P, Curtin A, Gennings C, Arora M, Tammimies K, et al. (2019): Dynamical properties of elemental metabolism distinguish attention deficit hyperactivity disorder from autism spectrum disorder. Transl Psychiatry. 9:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtin P, Austin C, Curtin A, Gennings C, Arora M, Tammimies K, et al. (2018): Dynamical features in fetal and postnatal zinc-copper metabolic cycles predict the emergence of autism spectrum disorder. Sci Adv. 4:eaat1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtin P, Curtin A, Austin C, Gennings C, Tammimies K, Bolte S, et al. (2017): Recurrence quantification analysis to characterize cyclical components of environmental elemental exposures during fetal and postnatal development. PLoS One. 12:e0187049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cauvet E, Van't Westeinde A, Toro R, Kuja-Halkola R, Neufeld J, Mevel K, et al. (2019): Sex Differences Along the Autism Continuum: A Twin Study of Brain Structure. Cereb Cortex. 29:1342–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cauvet E, Van't Westeinde A, Toro R, Kuja-Halkola R, Neufeld J, Mevel K, et al. (2020): The social brain in female autism: a structural imaging study of twins. Soc Cogn Affect Neurosci. 15:423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neufeld J, Hagstrom A, Van't Westeinde A, Lundin K, Cauvet E, Willfors C, et al. (2020): Global and local visual processing in autism - a co-twin-control study. J Child Psychol Psychiatry. 61:470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zetterqvist J, Vansteelandt S, Pawitan Y, Sjolander A (2016): Doubly robust methods for handling confounding by cluster. Biostatistics. 17:264–276. [DOI] [PubMed] [Google Scholar]
- 45.Bentejac C, Csorgo A, Martinez-Munoz G (2020): A comparative analysis of gradient boosting algorithms. Artificial Intelligence Review. 54:1937–1967. [Google Scholar]
- 46.Chen T, Guestrin C (2016): XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ZCM SIGKDD Internatitonal Conference on Knowledge Discovery and Data Mining. New York, NY, USA, pp 785–794. [Google Scholar]
- 47.Washington SD, Gordon EM, Brar J, Warburton S, Sawyer AT, Wolfe A, et al. (2014): Dysmaturation of the default mode network in autism. Hum Brain Mapp. 35:1284–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mash LE, Linke AC, Olson LA, Fishman I, Liu TT, Muller RA (2019): Transient states of network connectivity are atypical in autism: A dynamic functional connectivity study. Hum Brain Mapp. 40:2377–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynch CJ, Uddin LQ, Supekar K, Khouzam A, Phillips J, Menon V (2013): Default mode network in childhood autism: posteromedial cortex heterogeneity and relationship with social deficits. Biol Psychiatry. 74:212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lajiness-O'Neill R, Brennan JR, Moran JE, Richard AE, Flores AM, Swick C, et al. (2018): Patterns of altered neural synchrony in the default mode network in autism spectrum disorder revealed with magnetoencephalography (MEG): Relationship to clinical symptomatology. Autism Res. 11:434–449. [DOI] [PubMed] [Google Scholar]
- 51.Jung M, Kosaka H, Saito DN, Ishitobi M, Morita T, Inohara K, et al. (2014): Default mode network in young male adults with autism spectrum disorder: relationship with autism spectrum traits. Mol Autism. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heunis T, Aldrich C, Peters JM, Jeste SS, Sahin M, Scheffer C, et al. (2018): Recurrence quantification analysis of resting state EEG signals in autism spectrum disorder - a systematic methodological exploration of technical and demographic confounders in the search for biomarkers. BMC Med. 16:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pitsik E, Frolov N, Hauke Kraemer K, Grubov V, Maksimenko V, Kurths J, et al. (2020): Motor execution reduces EEG signals complexity: Recurrence quantification analysis study. Chaos. 30:023111. [DOI] [PubMed] [Google Scholar]
- 54.Rabbi AF, Jaiswal MK, Lei S, Fazel-Rezai R (2011): Changes in dynamical characteristics of epileptic EEG in rats using recurrence quantification analysis. Conf Proc IEEE Eng Med Biol Soc. 2011:2562–2565. [DOI] [PubMed] [Google Scholar]
- 55.Timothy LT, Krishna BM, Nair U (2017): Classification of mild cognitive impairment EEG using combined recurrence and cross recurrence quantification analysis. Int J Psychophysiol. 120:86–95. [DOI] [PubMed] [Google Scholar]
- 56.Yi G, Wang J, Bian H, Han C, Deng B, Wei X, et al. (2013): Multi-scale order recurrence quantification analysis of EEG signals evoked by manual acupuncture in healthy subjects. Cogn Neurodyn. 7:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almeida TP, Unger LA, Soriano DC, Li X, Dossel O, Yoneyama T, et al. (2019): Recurrence Quantification Analysis for Investigating Atrial Fibrillation Dynamics in a Heterogeneous Simulation Setup(.). Conf Proc IEEE Eng Med Biol Soc. 2019:2277–2280. [DOI] [PubMed] [Google Scholar]
- 58.Arcentales A, Giraldo BF, Caminal P, Benito S, Voss A (2011): Recurrence quantification analysis of heart rate variability and respiratory flow series in patients on weaning trials. Conf Proc IEEE Eng Med Biol Soc. 2011:2724–2727. [DOI] [PubMed] [Google Scholar]
- 59.Dimitriev D, Saperova EV, Dimitriev A, Karpenko Y (2020): Recurrence Quantification Analysis of Heart Rate During Mental Arithmetic Stress in Young Females. Front Physiol. 11:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu PR, Friedman DJ, Barnett AS, Jackson KP, Daubert JP, Piccini JP (2020): Focal impulse and rotor modulation of atrial rotors during atrial fibrillation leads to organization of left atrial activation as reflected by waveform morphology recurrence quantification analysis and organizational index. J Arrhythm. 36:311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naschitz JE, Sabo E, Naschitz S, Rosner I, Rozenbaum M, Priselac RM, et al. (2002): Fractal analysis and recurrence quantification analysis of heart rate and pulse transit time for diagnosing chronic fatigue syndrome. Clin Auton Res. 12:264–272. [DOI] [PubMed] [Google Scholar]
- 62.Afsar O, Tirnakli U, Marwan N (2018): Recurrence Quantification Analysis at work: Quasi-periodicity based interpretation of gait force profiles for patients with Parkinson disease. Sci Rep. 8:9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson NC, Bischof WF, Laidlaw KE, Risko EF, Kingstone A (2013): Recurrence quantification analysis of eye movements. Behav Res Methods. 45:842–856. [DOI] [PubMed] [Google Scholar]
- 64.Labini FS, Meli A, Ivanenko YP, Tufarelli D (2012): Recurrence quantification analysis of gait in normal and hypovestibular subjects. Gait Posture. 35:48–55. [DOI] [PubMed] [Google Scholar]
- 65.Riley MA, Balasubramaniam R, Turvey MT (1999): Recurrence quantification analysis of postural fluctuations. Gait Posture. 9:65–78. [DOI] [PubMed] [Google Scholar]
- 66.Grossekathofer U, Manyakov NV, Mihajlovic V, Pandina G, Skalkin A, Ness S, et al. (2017): Automated Detection of Stereotypical Motor Movements in Autism Spectrum Disorder Using Recurrence Quantification Analysis. Front Neuroinform. 11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manyakov NV, Bangerter A, Chatterjee M, Mason L, Ness S, Lewin D, et al. (2018): Visual Exploration in Autism Spectrum Disorder: Exploring Age Differences and Dynamic Features Using Recurrence Quantification Analysis. Autism Res. 11:1554–1566. [DOI] [PubMed] [Google Scholar]
- 68.Kaboodvand N, Iravani B, Fransson P (2020): Dynamic synergetic configurations of resting-state networks in ADHD. Neuroimage. 207:116347. [DOI] [PubMed] [Google Scholar]
- 69.Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL (2010): Functional-anatomic fractionation of the brain's default network. Neuron. 65:550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braga RM, Buckner RL (2017): Parallel Interdigitated Distributed Networks within the Individual Estimated by Intrinsic Functional Connectivity. Neuron. 95:457–471 e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE (2014): Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 84:320–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christakou A, Murphy CM, Chantiluke K, Cubillo AI, Smith AB, Giampietro V, et al. (2013): Disorder-specific functional abnormalities during sustained attention in youth with Attention Deficit Hyperactivity Disorder (ADHD) and with autism. Mol Psychiatry. 18:236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.