Abstract
Diverse neurons in the parabrachial nucleus (PB) communicate with widespread brain regions. Despite evidence linking them to a variety of homeostatic functions, it remains difficult to determine which PB neurons influence which functions because their subpopulations intermingle extensively. An improved framework for identifying these intermingled subpopulations would help advance our understanding of neural circuit functions linked to this region. Here, we present the foundation of a developmental‐genetic ontology that classifies PB neurons based on their intrinsic, molecular features. By combining transcription factor labeling with Cre fate‐mapping, we find that the PB is a blend of two, developmentally distinct macropopulations of glutamatergic neurons. Neurons in the first macropopulation express Lmx1b (and, to a lesser extent, Lmx1a) and are mutually exclusive with those in a second macropopulation, which derive from precursors expressing Atoh1. This second, Atoh1‐derived macropopulation includes many Foxp2‐expressing neurons, but Foxp2 also identifies a subset of Lmx1b‐expressing neurons in the Kölliker–Fuse nucleus (KF) and a population of GABAergic neurons ventrolateral to the PB (“caudal KF”). Immediately ventral to the PB, Phox2b‐expressing glutamatergic neurons (some coexpressing Lmx1b) occupy the KF, supratrigeminal nucleus, and reticular formation. We show that this molecular framework organizes subsidiary patterns of adult gene expression (including Satb2, Calca, Grp, and Pdyn) and predicts output projections to the amygdala (Lmx1b), hypothalamus (Atoh1), and hindbrain (Phox2b/Lmx1b). Using this molecular ontology to organize, interpret, and communicate PB‐related information could accelerate the translation of experimental findings from animal models to human patients.
Keywords: breathing, development, interoception, interoceptive, Kölliker–Fuse, pain, parabrachial complex, parabrachialis, pontine pneumotaxic center, pontine taste area
The parabrachial nucleus (PB) contains diverse, intermingled neurons regulating various homeostatic functions. Their neuronal distributions often do not align with cytoarchitectural subdivisions. Here, we show that the PB is a blend of two, developmentally distinct glutamatergic populations identified by the embryonic transcription factors Atoh1 and Lmx1a/Lmx1b. We show that macropopulation identity predicts output connectivity and other gene expression patterns in this region. Our findings lay the foundation for a molecular ontology that will be useful for interpreting transcriptomic, connectomic, and other experimental data involving PB neurons.
1. INTRODUCTION
A complex region of the brainstem tegmentum links the rhombencephalon (pons‐medulla) and mesencephalon (midbrain). Embryonic precursors here generate a variety of neurons in the brainstem and the entire cerebellum (Ben‐Arie et al., 1997; Chizhikov et al., 2006; Hoshino et al., 2005; Liu et al., 2010; Machold & Fishell, 2005; Rose et al., 2009; Wang et al., 2005; Watson et al., 2017; Yamada et al., 2007). In the dorsolateral corner of this region, a diverse constellation of neurons communicates with every major region of the central nervous system, from cerebral cortex to spinal cord (Herbert et al., 1990; Huang et al., 2020; Moga et al., 1990; Saper & Loewy, 1980). Herrick first identified this region as the “superior secondary gustatory nucleus” in fish (1905), and subsequent work identified taste‐relay neurons in rats (Norgren & Leonard, 1971). These neurons surround the brachium conjunctivum (superior cerebellar peduncle) and are referred to as the “parabrachial” nucleus (PB).
Over the past 50 years, experiments in rats and mice have implicated the PB in many more homeostatic functions, including hunger, thirst, sodium appetite, taste aversion, and cardiorespiratory control (Chamberlin & Saper, 1994; Geerling & Loewy, 2007; Kim et al., 2020; Menani et al., 2014; Palmiter, 2018; Ryan et al., 2017). It is also clear that the PB is a lynchpin in the neurobiological basis of pain, itch, and thermoregulation (Barik et al., 2021; Bernard & Besson, 1990; Bernard et al., 1994; Bester et al., 1995; Bourgeais et al., 2001; Cechetto et al., 1985; Chiang et al., 2019; Chiang et al., 2020; Coizet et al., 2010; Deng et al., 2020; Gauriau & Bernard, 2002; Morrison & Nakamura, 2019; Mu et al., 2017; Palmiter, 2018). Some of these functions were linked to specific subsets of PB neurons, but the inscrutability of remaining subpopulations limits our understanding of several important brain circuits.
One reason these neurons remain enigmatic is that, rather than forming discrete layers or nuclei (as in the cerebral cortex or thalamus), most PB subpopulations form an intermingled, three‐dimensional web. The current method of classifying PB neurons uses the superior cerebellar peduncle to split the region into a medial and a lateral subdivision (Taber, 1961). Next, subnuclear boundaries are applied within each subdivision, using cytoarchitectural criteria proposed in rats (Fulwiler & Saper, 1984). While this cytoarchitectural taxonomy facilitated initial progress, adapting these criteria between species proved challenging. Even between rats and mice, species differences led to confusion regarding the location and identity of several subpopulations. Prominent examples include rostral PB neurons that relay thermosensory information to the hypothalamus (Geerling et al., 2016; Nakamura & Morrison, 2008, 2010), dorsal PB neurons that relay pain‐related information to the thalamus (Barik et al., 2021; Bourgeais et al., 2001), and caudal PB neurons that influence appetite (Gasparini et al., 2021; Gasparini et al., 2019; Geerling & Loewy, 2007; Geerling et al., 2011; Gong et al., 2020; Jarvie & Palmiter, 2017; Kim et al., 2020; Li et al., 2019; Park et al., 2020; Resch et al., 2017). Our efforts to adapt this taxonomy from rats to mice made it clear that some cytoarchitectural criteria simply do not translate (Gasparini et al., 2021; Gasparini et al., 2019; Geerling et al., 2016), and previous attempts to describe and organize this region suggest further differences in the human brainstem (Block & Estes, 1990; de Lacalle & Saper, 2000; Gioia et al., 2000; Lavezzi et al., 2004; Ohm & Braak, 1988; Pammer et al., 1988; Petrovicky, 1989; Rub et al., 2001).
In addition to species differences, an unavoidable challenge when applying cytoarchitectural criteria is that this requires interpreting Nissl‐stained tissue, an inherently subjective activity (Swanson, 2000). Distinctions can be subtle, and even experts cannot distinguish functionally diverse neurons if they have the same Nissl‐stained appearance. A more ideal framework for classifying diverse, intermingled neurons would incorporate observer‐independent, molecular information as a core feature. Using gene expression to classify neurons can produce results resembling cytoarchitectural analysis, while also distinguishing new subpopulations (Ortiz et al., 2020). Gene expression predicts the output connectivity of PB neurons (Huang et al., 2020; Huang et al., 2021), and basing a neuronal ontology on molecular and connectomic features could offer a more accessible, universal language for interpreting and communicating experimental findings (Bota & Swanson, 2008; Hamilton et al., 2012; Larson & Martone, 2009; Zeng & Sanes, 2017).
In the PB, molecular information has not kept pace with other brain regions. Limited information suggests that PB neurons derive from precursors in the embryonic isthmus (r0; Watson et al., 2017) and rhombomere 1 (r1; Figure 2M of Machold & Fishell, 2005). At these levels of the neural tube, PB neurons are thought to arise from precursors in the rhombic lip that express Atoh1 (Machold & Fishell, 2005; Rose et al., 2009; van der Heijden & Zoghbi, 2018; Wang et al., 2005), but we lack information on the relationship between Atoh1‐derived neurons and adult subpopulations expressing a potpourri of transcription factors (including Foxp2, Lmx1b, Lmx1a, Satb2, and Runx1), neuropeptides, receptors, and other genetic markers in current use (Asbreuk et al., 2002; Chizhikov et al., 2021; Dai et al., 2008; Gray, 2008; Guo et al., 2007; Hernandez‐Miranda et al., 2017; Kang et al., 2007; Liu et al., 2010; Maeda et al., 2009; Miller et al., 2012; Millonig et al., 2000; Mishima et al., 2009; Zagami & Stifani, 2010; Zou et al., 2009). These markers identify an incomplete patchwork of PB neurons, and our ability to study neural circuit functions involving the remaining subpopulations would benefit from a more comprehensive framework.
As a first step, we sought a framework of developmental‐genetic information that identifies all PB neurons. Motivated by novel activity and connectivity patterns in rats (Geerling & Loewy, 2006b, 2007; Geerling et al., 2010), this project began with the observation that two transcription factors have complimentary distributions in this region (Geerling et al., 2011; Miller et al., 2012; Shin et al., 2011). Here, we combine Cre fate‐mapping, mRNA labeling, immunolabeling, and axonal tracing to replicate and extend these observations in mice.
Our new findings reveal that “the” PB does not have a single developmental origin. Its glutamatergic neurons are a blend of two, mutually exclusive macropopulations, defined by the embryonic transcription factors Lmx1b and Atoh1. We show that this framework clarifies the identity and connectivity of further PB subpopulations and challenges previous ideas about the composition and origin of the Kölliker–Fuse nucleus (KF). Together, these findings lay a developmental‐genetic foundation for a molecular ontology that investigators can use to identify and target PB neurons.
2. MATERIALS AND METHODS
2.1. Animals
We used a total of n = 68 male and female mice, aged 7–23 weeks and weighing 17–31 g. All mice were group‐housed in a temperature‐ and humidity‐controlled room on a 12/12‐h light/dark cycle and had ad libitum access to standard rodent chow and water. In addition to C57B6/J mice (Jackson Laboratories), we used a variety of knockin‐Cre and Cre‐reporter mice. Detailed information about each strain is provided in Table 1. For all mRNA and protein labeling that did not require a Cre‐reporter, we replicated labeling in at least three, 8−12‐week‐old C57B6/J mice. All mRNA and protein labeling in Cre‐reporter mice was replicated in at least three mice with a hemizygous Cre allele and a hemizygous Cre‐reporter allele. Stereotaxic injections, histologic procedures, and confocal microscopy in n = 4 Harlan Sprague–Dawley rats were performed as described previously (Geerling & Loewy, 2007; Geerling et al., 2011; Shin et al., 2011) and in accordance with the guidelines of the Institutional Animal Care and Use Committee at Washington University in Saint Louis. All experiments in mice were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee and at the University of Iowa.
TABLE 1.
Cre‐driver and ‐reporter mice
| Strain | References | Source information | Key Gene |
|---|---|---|---|
| Atoh1‐Cre | Matei et al. Dev Dyn 234(3):633‐50. 2005 |
Bernd Fritzsch, University of Iowa Jax 011104 |
Transgenic construct containing the JQ2‐Atoh1 promoter fragment ligated to the bacteriophage P1 Cre recombinase |
| Lmx1a‐Cre |
Victor V. Chizhikov, Anne G. Lindgren, D. Spencer Currle, Matthew F. Rose, Edwin S. Monuki, Kathleen J. Millen Development 2006 133: 2793–2804; doi: 10.1242/dev.02441 |
Victor V. Chizhikov, University of Tennessee HSC |
BAC transgenic construct containing ∼200 kb of the mouse Lmx1a locus with Lmx1a coding sequence replaced by Cre coding sequence |
|
Vglut2‐IRES‐Cre (Slc17a6‐IRES‐Cre) |
Vong, Linh, et al. “Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons.” Neuron 71.1 (2011): 142–154 |
Jax 016963 |
IRES‐Cre inserted downstream of the stop codon of Slc17a6 on chromosome 7 |
|
Vgat‐IRES‐Cre (Slc32a1‐IRES‐Cre) |
Vong L, Ye C, Yang Z, Choi B, Chua S, Lowell BB. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011; 71:142−154 |
Jax 028892 |
IRES‐Cre inserted after the Slc32a1 stop codon |
| Ai9(R26‐lsl‐ tdTomato) | Madisen L; Zwingman TA; Sunkin SM; Oh SW; Zariwala HA; Gu H; Ng LL; Palmiter RD; Hawrylycz MJ; Jones AR; Lein ES; Zeng H. 2010. A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13(1):133‐40 |
Jax 007909 |
tdTomato (red fluorescent protein) insertion after lox‐STOP‐lox at Rosa26 locus |
| Ai14(R26‐lsl‐tdTomato) reporter | Madisen L; Zwingman TA; Sunkin SM; Oh SW; Zariwala HA; Gu H; Ng LL; Palmiter RD; Hawrylycz MJ; Jones AR; Lein ES; Zeng H. 2010. A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13(1):133‐40 |
Jax 007914 |
Rosa‐CAG‐LSL‐tdTomato‐WPRE targeting vector inserted between exon 1 and 2 of the Gt(ROSA)26Sor locus |
|
R26‐LSL‐L10GFP reporter |
Krashes, Michael J., et al. “An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger.” Nature 507.7491 (2014): 238 |
Available from originating investigators |
Floxed transcription STOP cassette followed by EGFP/Rpl10 fusion reporter gene under control of the CAG promoter targeted to the Gt(ROSA)26Sor locus |
| Pdyn‐IRES‐Cre |
Krashes, Michael J., et al. “An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger.” Nature 507.7491 (2014): 238 |
Jax 027958 |
IRES‐Cre inserted downstream of the endogenous Pdyn (prodynorphin) gene |
| Calca‐Cre (Calca‐tm1.1‐Cre‐EGFP) | Carter, M., Soden, M., Zweifel, L. et al. “Genetic identification of a neural circuit that suppresses appetite.” Nature 503, 111−114 (2013) | Richard Palmiter, University of Washington (shared by Andrew Russo, University of Iowa) | Cre:GFP inserted downstream of the endogenous CGRP (calcitonin‐gene‐related‐peptide) gene |
2.2. Stereotaxic injections
Mice were anesthetized with isoflurane and placed in a Kopf 1900 stereotactic frame. After midline incision, the skin was retracted to expose the skull and locate bregma. We injected the cholera toxin B‐subunit (CTb, 0.1% in distilled water; List, lot #10331A1) in Atoh1‐Cre;R26‐lsl‐L10GFP mice. We made nanoliter injections through a fine‐tipped micropipette (20–30 μm inner diameter) using controlled puffs of compressed air, typically 0.5–1 per second, with a target rate of 5–10 nl/min. Injection volumes ranged 15−45 nl. Stereotaxic targets included the central nucleus of the amygdala (1.0 mm caudal, 2.5 mm right, and 5.0−5.5 mm deep to bregma), insular cortex (0.0−0.5 mm caudal, 3.75−3.8 mm right, and 4.0−4.3 mm deep to bregma), and ventromedial hypothalamus (1.1−1.4 mm caudal, 0.06−0.55 mm right, and 5.6–5.9 mm deep to bregma). Each injection was made over a 5‐min period using picoliter air puffs through a solenoid valve (Clippard EV 24V DC) pulsed by a Grass stimulator. The pipette was left in place for an additional 3−5 min, then slowly withdrawn before the skin was closed using Vetbond (3 M). Carprofen (1 mg/kg s.c.) was provided for postoperative analgesia. We injected an additional series of C57B6/J mice with CTb, targeting the medullary reticular formation (6.3 mm caudal, 1.2 mm right, and 6.0 mm deep to bregma; 45 nl). Mice were kept alive for 3−5 days for retrograde axonal transport, and then perfused as described below. Synaptophysin‐mCherry labeling shown in the midbrain of Pdyn‐IRES‐Cre and Calca‐Cre mice represents unpublished tissue sections from two previous studies (for details, see Huang et al., 2020; Huang et al., 2021).
2.3. Perfusions and tissue sections
All mice were deeply anesthetized with ketamine‐xylazine (i.p. 150–15 mg/kg) and then perfused transcardially with phosphate‐buffered saline (PBS, prepared from 10x stock; P7059, Sigma), followed by 10% formalin‐PBS (SF100, Fisher Scientific). After perfusion, the brain was removed and fixed overnight in 10% formalin‐PBS at 4°C, then submerged in 30% sucrose‐PBS at 4°C for an additional day. Each brain was sectioned into 40 μm‐thick axial (coronal) slices using a freezing microtome. Three adjacent (1‐in‐3) tissue series were collected from each brain in separate tubes (labeled “A,” “B,” and “C”) containing a cryoprotectant solution of 35% (v/v in PBS) ethylene glycol (102466, Sigma‐Aldrich) and 25% glycerol (G22025, RPI). These tubes were stored at −20°C until further processing. Each brain, therefore, yielded three sets of axial tissue sections that allowed us to study up to three separate combinations of molecular markers in the PB region from each mouse. For experimental replication of each endpoint, we analyzed brain tissue from at least three separate mice.
2.4. Immunofluorescence
We removed tissue sections from cryoprotectant and selected sections of interest (typically, a series of nine tissue sections containing the full PB region or CTb injection site; otherwise, a full‐brain series). After rinsing the sections in PBS, we incubated them with primary antisera (Table 2) in a PBT‐NDS‐azide solution comprised of PBS with 0.25% Triton X‐100 (BP151‐500, Fisher), 2% normal donkey serum (NDS, 017‐000‐121; Jackson ImmunoResearch), and 0.05% sodium azide (14314, Alfa Aesar) as a preservative. The sections were incubated in this primary antibody solution overnight at room temperature on a tissue shaker. The next morning, sections were washed three times in PBS, then incubated for 2 h at room temperature with species‐specific secondary antibodies conjugated to Cy3, Cy5, Alexa Fluor 488, or biotin (diluted 1:500−1000; Jackson ImmunoResearch) in PBT‐NDS‐azide. If a biotinylated secondary antibody was used, tissue sections were then washed three times and incubated for 2 h in streptavidin‐Cy5 (#SA1011, Invitrogen) or streptavidin‐Pacific Blue (#S11222, Invitrogen), diluted 1:1000 in PBT‐NDS‐azide. The sections were then washed three times in PBS, mounted on glass slides (#2575‐PLUS; Brain Research Laboratories), dried, and then coverslipped using Vectashield (Vector Laboratories) or Vectashield with DAPI (if no blue fluorophore was used). Slides were stored at 4°C until imaging.
TABLE 2.
Antisera
| Antigen | Immunogen description | Source, host species, RRID | Concentration |
|---|---|---|---|
| Choline acetyltransferase (ChAT) | Human placental choline acetyltransferase | Millipore, goat polyclonal, #AB144P, lot: JC1618187; RRID: AB_2079751 | 1:1000 |
| Forkhead box protein 2 (FoxP2) | Recombinant human FOXP2 isoform 1 Ala640‐Glu715 | R&D Systems, sheep polyclonal #AF5647; RRID: AB_2107133 | 1:10,000 |
| LIM homeobox transcription factor 1 beta (Lmx1b) in rat | Three fusion proteins containing amino acid residues 17−47, 157−195, or 255−324 of mouse Lmx1b |
Y. Ding, Chinese Academy of Science, Shanghai; rabbit polyclonal; RRID: AB_2314751 (no labeling in Lmx1b KO mice; Dai et al J Comp Neurol 2008) |
1:1000 |
| Lmx1b (mouse) | Full‐length LIM homeobox transcription factor 1 beta protein from mouse | C. Birchmeier, Max Delbruck Center for Molecular Medicine, Berlin; guinea pig polyclonal; RRID: AB_2314752 | 1:8000 |
| mCherry | Full‐length mCherry fluorescent protein | Life Sciences, rat monoclonal, #M11217, lot: R1240561; RRID: AB_2536611 (we find no immunolabeling in mice without dsRed‐derived fluorescent proteins) | 1:2000 |
| Paired‐like homeobox 2b (Phox2b) | BSA‐coupled 15mer corresponding to the C terminus of the Phox2b protein with an added N‐terminal tyrosine | H. Enomoto, School of Medicine at Kobe University, Japan; RRID: AB_2895590 | 1:12,000 |
| Paired‐like homeobox 2b (Phox2b) | Mouse monoclonal antibody raised against amino acids 11–70 mapping near the N‐terminus Phox2b of human origin | Santa Cruz, mouse monoclonal, #sc‐376997, lot: E1719; RRID: AB_2813765 | 1:1000 |
| Special AT‐rich sequence‐binding protein 2 (Satb2) | Synthetic peptide within human SATB2 (proprietary sequence) | Abcam, rabbit monoclonal, cat. # ab92446, lot: GR325015−2, RRID: AB_1056367 | 1:3000 |
| Tyrosine hydroxylase (TH) |
Purified, SDS‐denatured rat pheochromocytoma TH |
Millipore, mouse monoclonal, #MAB318, lot: NG1802536; RRID: AB_2201528 | 1:2000 |
| Tyrosine hydroxylase (TH) | Denatured TH from rat pheochromocytoma (denatured by sodium dodecyl sulfate) | Millipore, rabbit polyclonal, #AB152, lot: 240602; RRID: AB_696697 | 1:10,000 |
2.5. In situ hybridization
We used RNAscope probes to label a variety of mRNA transcripts (see Table 3 for detailed information on each probe). For diaminobenzidine (DAB) labeling of single transcripts, we used the HD Reagent Kit (ref #322310; Advanced Cell Diagnostics). For fluorescence labeling of two or three transcripts in combination, we used the Fluorescent Multiplex Detection Reagents kit (ref# 320851; Advanced Cell Diagnostics).
TABLE 3.
RNAscope probes
| Probe | Common name | Channel | ACD catalog # | Lot # |
|---|---|---|---|---|
| Mm‐Calca‐C2 | Calca | C2 | 417961‐C2 | 18165A |
| Mm‐FoxP2‐C3 | FoxP2 | C3 | 428791‐C3 | 17013A |
| MM‐Grp‐C2 | Grp | C2 | 317861‐C2 | 19211A |
| Mm‐Lmx1a | Lmx1a | C1 | 493131 | 20042B |
| Mm‐Lmx1b | Lmx1b | C1 | 412931 | 18255B |
| Mm‐Pdyn‐C3 | Pdyn | C3 | 318771‐C3 | 17290A |
| Mm‐Phox2b‐C2 | Phox2b | C2 | 407861‐C2 | 19179B |
| Mm‐Slc6a5‐C1 | GlyT2 | C1 | 409741 | 16347A |
| Mm‐Slc17a6 | Vglut2 | C1 | 545891‐C1 | 17251A |
| Mm‐Slc32a1‐C2 | Vgat | C2 | 319191‐C2 | 16340A |
| Mm‐Ubc‐C2 | Ubc | C2 | 310771‐C2 | 18098B |
For brightfield (DAB) labeling, we removed tissue from cryoprotectant, selected 6−8 sections per case containing the PB region, rinsed them in PBS, then incubated sections for 30 min in 0.3% hydrogen peroxide (#H325‐100, Fisher) to quench endogenous peroxidase activity. They were again washed in PBS, then mounted onto glass slides to dry overnight at room temperature. The following day, sections were dehydrated for 5 min in 100% EtOH, then dried at room temperature for 20 min. Next, sections were incubated in 1x Target Retrieval solution (ACD; 10 ml in 90 ml ddH2O, warmed for 5 min prior to slide incubation) for 5 min in a steamer (Oster, prewarmed for 60 min), then rinsed twice with ddH2O. Slides were next submerged in 100% EtOH for 1 min, then removed and dried at room temperature for 20 min. Next, we used an ImmEdge PAP pen (#H‐4000; Vector) to create a hydrophobic barrier around the sections, added four drops of Protease Plus (Ref 322331, Lot # 2000898; ACD), and placed slides in a covered glass petri dish floating in a 40°C water bath for 30 min. After washing twice with ddH2O, we covered sections with a C1 probe (prewarmed to 40°C for 10 min) for 2 h at 40°C. Next, the slides were washed twice with 1X RNAscope Wash Buffer, then incubated with four drops of AMP 1 (Ref 322311, Lot# 2008215, ACD) for 30 min at 40°C. This process of washing twice with 1X Wash Buffer and then incubating was repeated for AMPs 2−6 with the following times and temperatures: AMP 2 (Ref 322312, Lot# 2007617, ACD), 15 min at 40°C; AMP 3 (Ref 322313, Lot# 2008317), 30 min at 40°C; AMP 4 (Ref 322314, Lot# 2007619), 15 min at 40°C; AMP 5 (Ref 322315, Lot # 2007620), 30 min at room temperature; and AMP 6 (ref 322316, Lot# 2007621) room temperature for 15 min. After two final washes in 1X Wash Buffer, we mixed equal volumes of “DAB‐A” and “DAB‐B” (ACD), then added this combined solution to each slide for 10 min. Finally, we dipped slides in ddH2O, then dehydrated the sections in a series of EtOH solutions (50%, 75%, and 100%) for 2 min each followed by two xylene solutions for 5 min each. After removal from xylenes, the slides were coverslipped immediately with Cytoseal (#8310‐16 Thermo Scientific) and stored at room temperature.
For fluorescence labeling, we removed tissue from cryoprotectant, selected 6−8 sections containing the PB region, rinsed them in PBS, and mounted them on glass slides to dry overnight. In the morning, after dehydrating sections in an ascending series of alcohols (50%, 70%, 100%, and 100% EtOH; 5 min each), we used an ImmEdge PAP pen to create a hydrophobic barrier around the sections, then washed in PBS twice, for 2 min each, at room temperature. The sections were then covered with Protease IV and placed in a covered glass petri dish, floating in a 40°C water bath, for 30 min. After washing twice in PBS, the sections were incubated in a combination of two or three probes for 2 h at 40°C. After that, AMPs 1−4 were added, in series, for 15−30 min each, at 40°C, with two, 2‐min rinses in 1X RNAscope Wash Buffer (#320058; diluted 1:50 in ddH20) between each step. After a final wash in PBS, the slides were dried at room temperature and coverslipped using Vectashield with DAPI.
2.6. Imaging, analysis, and figure preparation
We used an Olympus VS120 slide‐scanning microscope for brightfield and epifluorescence imaging, collecting whole‐slide images of all tissue in this study. For brightfield imaging, after acquiring a 2× overview scan of the full slide, we used a 10× (NA 0.40) objective to image a single, central focal plane through all sections, then a 20× (NA 0.75) objective to collect an 11‐focal‐plane “extended focal image” (EFI) through each section. For epifluorescence imaging, after a 2× overview scan, we acquired all color channels in a single, central focal plane using the same 10× objective, then used the same 20× objective to collect EFI images of the midbrain‐hindbrain tegmentum (including the full, bilateral PB and neighboring structures, at all rostrocaudal levels). In some cases, we also collected multifocal Z‐stacks with a 40× (NA 0.95) air objective in smaller regions of interest. For each slide, this produced a Virtual Slide Image file containing a 2× overview layer (covering the whole slide), plus a 10× layer (containing all tissue sections) and additional layers containing 20× EFI and, in some cases, 40× virtual Z‐stacks in regions of interest.
After reviewing all images in VS‐ASW (Olympus) or OlyVIA (Olympus), we used cellSens (Olympus) to assign a color to each grayscale acquisition channel and to export cropped, full‐resolution images. We used Adobe Photoshop to adjust brightness and contrast. For each analysis, we selected 6−8 evenly spaced, axial tissue sections from n = 3 cases with high‐quality histology across all rostral‐to‐caudal levels of the PB.
In Adobe Illustrator, we placed a symbol atop each cell nucleus containing immunofluorescence labeling for Lmx1b, Foxp2, or Phox2b; cytoplasmic GFP (Cre‐reporter for Atoh1‐Cre); or nuclear and cytoplasmic tdTomato (Cre‐reporter for Lmx1a‐Cre). We labeled cells containing each marker using a separate symbol, in a separate Illustrator layer, and used additional symbols (in additional, separate layers) for all combinations of double‐ and triple‐labeling. We marked only cells that contained in‐focus labeling. All counting layers were reviewed, in every section, by a senior neuroanatomist (J.C.G.), and both S.K. and J.C.G. reviewed all plots and source images at high magnification to reach consensus. We only included labeling with fluorescence intensity that both investigators agreed to be above the background fluorescence of surrounding tissue and different from any lipofuscin autofluorescence that appeared (to a varying extent across cases) as small, punctate clusters of intracellular debris distinguished by identical fluorescence emission in both the red and green channels. Lipofuscin autofluorescence in this region is most prominent in the mesencephalic trigeminal nucleus and becomes more prominent widespread in older adults, so we focused on young adult mice (8–12 weeks).
As medial and lateral boundaries of our PB counting region, we used the lateral surface of Barrington's nucleus and the lateral brainstem surface. At caudal levels, our dorsal boundary was the dorsal brainstem surface and fourth ventricular ependyma. At middle to rostral levels (“mesencephalic” PB), our dorsal boundaries were the ventral edge of the cuneiform nucleus and the ventrolateral surface of the periaqueductal gray matter. Our ventral and caudal boundaries were the dorsal surfaces of the vestibular nuclear complex and of the principal sensory (PSV) and motor (V) trigeminal nuclei. We did not count cells beyond the ventral or rostral limits of the KF and supratrigeminal nuclei, nor did we count neurons in the pedunculopontine tegmental nucleus or in the dorsal or ventral nuclei of the lateral lemniscus.
We counted the symbols in each layer by first locking other layers, then using “Document Info > Objects” in Illustrator, and then entered each count into a Microsoft Excel spreadsheet for further analysis. We also used Illustrator to make drawings, arrange images, and add lettering for figure layouts. Scale bars were traced in Illustrator atop calibrated lines from cellSens to produce clean white or black lines in each figure.
We applied the Abercrombie correction factor to compensate for overcounting (Guillery, 2002). This correction factor requires knowing the section thickness and the diameter of the key counting element, which was the cell nucleus in our analyses. We, therefore, measured the diameter of FoxP2‐, Lmx1b‐, and Phox2b‐immunoreactive nuclei, as well as the nuclear‐void diameter in L10GFP‐expressing neurons (in Atoh1‐Cre;R26‐lsl‐L10GFP mice). For each cell type, 30 nuclei were selected at random and measured from each case (∼3 per marker, per section), across all Atoh1‐Cre;R26‐lsl‐L10GFP cases, every Lmx1a‐Cre;R26‐lsl‐tdTomato case, and all C57B6/J cases that were analyzed for Phox2b, Lmx1b, and/or FoxP2 counts. Average total nuclear diameters in all groups ranged 9.0−9.5 μm. As a rough estimate of the total number of neurons in the mouse PB, we added together the individual counts of each cell population, then subtracted the numbers of double‐ and triple‐labeled neurons. Finally, since we worked with tissue subsamples (1‐in‐3 tissue series), we multiplied the overall, Abercrombie‐corrected average sum of PB neurons by 3 to estimate the total number.
In this article, we use the term “mutual exclusivity” to mean that markers are found in largely separate populations that are at least 98−99% distinct from one another. Most histologic techniques for labeling protein or mRNA expression do not produce 100% mutually exclusive labeling, and a lack of perfect mutual exclusivity between a pair of mRNA transcripts, immunolabeled proteins, or fluorescent protein reporters may involve background thresholding as much as actual coexpression. In this study, we did not include faint or ambiguous labeling within the range of background tissue fluorescence. We analyzed most cells and cell populations in relatively thick (40 μm) tissue sections at digital magnifications spanning ∼10−100 μm (not submicron or subcellular distributions), which is optimal for comprehensively analyzing macropopulation‐level features throughout the full, three‐dimensional extent of the PB complex, which encompasses ∼1−2 mm3. Wherever we found clear‐cut examples of colocalized markers, we describe our observations in detail.
2.7. Nomenclature
For rat and mouse genes, we used MGI nomenclature. For rat and mouse proteins and Cre‐reporters, we used common abbreviations from the published literature. For neuroanatomical structures and cell populations, where possible, we use and refer to nomenclature defined in peer‐reviewed neuroanatomical literature. In some instances, we use or refer to nomenclature derived from rodent brain atlases (Dong, 2008; Paxinos & Franklin, 2013; Paxinos & Watson, 2007; Swanson, 1992). For the PB region specifically, mutual inconsistencies among existing systems of nomenclature (both between atlases and within the current literature) is a core topic addressed by the experimental results and analysis in this study.
Initially, we did not plan to focus attention on the region ventrolateral to the rostral PB that is referred to as the “Kölliker–Fuse nucleus,” but neurons in this location appeared as an emergent property in several of our results. These eponyms are historically inaccurate, and both the location and neuronal composition of the KF are defined inconsistently (Petrovicky, 1989), but we use the term “KF” to indicate a small region that is ventral to the rostral, ventrolateral tip of the PB at levels approximately 4.8−5.0 mm caudal to bregma (Paxinos & Franklin, 2013). This location borders a small white matter tract, labeled “ventral spinocerebellar tract” in rodent brain atlases (Dong, 2008; Paxinos & Franklin, 2013), which extends past the cerebellum rostrally, alongside the lateral PB, and then ventrally, alongside the KF. As described below, novel molecular features in this location identify and distinguish intermixed populations of neurons. The objective of this study is to maximize reproducibility by focusing on observer‐independent, molecular properties that unambiguously identify populations of neurons, rather than boundary lines that are inferred from Nissl cytoarchitecture. Consequently, and by design, no results or conclusions in this study were based on or inferred from Nissl‐stained tissue.
3. RESULTS
3.1. Lmx1b and FoxP2 in rats
Our investigation began in rats after identifying a small subpopulation of neurons activated by sodium deprivation (Geerling & Loewy, 2007). These sodium‐deprivation‐activated neurons intermingled with other PB neurons in a distribution that did not conform to existing, cytoarchitectural boundaries. Seeking a molecular marker that would allow us to better classify and then target these neurons, we examined regional expression of transcription factors and identified FoxP2 as a useful marker for these and several other PB subpopulations (Gasparini et al., 2021; Geerling et al., 2016; Geerling et al., 2011; Geerling et al., 2017; Shin et al., 2011; Verstegen et al., 2017). Virtually all PB neurons activated by sodium deprivation contained FoxP2 (Gasparini et al., 2021; Geerling et al., 2011), and many of these distributed within and alongside the superior cerebellar peduncle, in a location described as the inner portion of the external lateral PB subnucleus (PBeL; Herbert & Saper, 1990). Therefore, we predicted that sodium‐deprivation‐activated FoxP2 neurons, like other PBeL neurons, project axons to the bed nucleus of the stria terminalis and amygdala (Geerling & Loewy, 2006a, 2006b). To our surprise, retrograde tracer injections in each target region exclusively labeled PB neurons that did not contain FoxP2 (Figure 1a–f).
FIGURE 1.
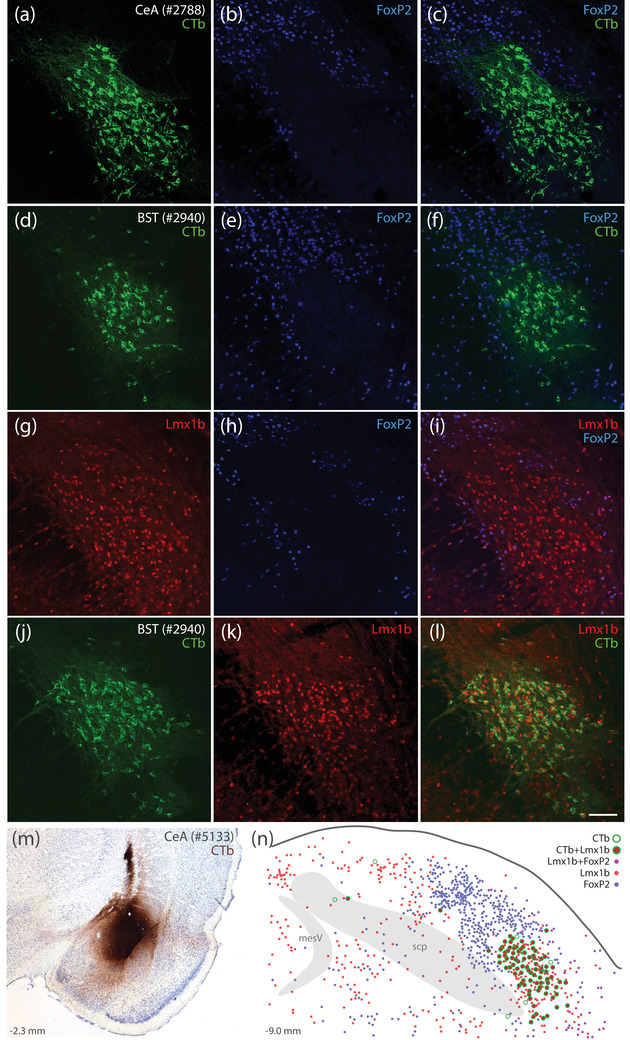
Lmx1b and FoxP2 in the rat parabrachial nucleus (PB). Neurons containing cholera toxin b (CTb, green) retrograde labeling after CTb injections into the central nucleus of the amygdala (CeA; a–c) or bed nucleus of the stria terminalis (BST; d–f) did not contain the transcription factor FoxP2 (blue). Neurons containing the transcription factor Lmx1b (red) filled a gap in the FoxP2 distribution (g–i) and were retrogradely labeled after CTb injection into the BST (j–l). Similarly, CTb injection into the CeA (m, rat case #5133) produced retrograde labeling predominantly in Lmx1b‐containing neurons (n). Approximate level caudal to bregma (in mm) is shown at bottom‐left in (m, n). Scale bar is 100 μm and applies to panels (a–l). Abbreviations: mesV, mesencephalic tract and nucleus of the trigeminal nerve; scp, superior cerebellar peduncle
Seeking additional markers to fill this gap in the FoxP2 distribution led us to another transcription factor expressed in this region, Lmx1b (Asbreuk et al., 2002; Dai et al., 2008). Immunolabeling Lmx1b identified most neurons in PBeL, surrounded by a complementary distribution of neurons containing FoxP2 (Figure 1g–i). Lmx1b identified virtually all PB neurons that project axons to the bed nucleus of the stria terminalis (n = 2, Figure 1j–l) and central nucleus of the amygdala (n = 2, Figure 1m,n). Together, Lmx1b and FoxP2 labeling filled most of the PB, and their complementary distributions formed the basis of a study examining patterns of Fos activation after changes in blood pressure (Miller et al., 2012). These mutually exclusive distributions suggested the possibility of using transcription factors to classify all PB neurons. To explore this possibility using a richer set of genetic tools, we began studying the PB in mice.
3.2. Lmx1b and Foxp2 in mice
First, to clarify the adult distribution of neurons expressing these two transcription factors in mice, we used brightfield in situ hybridization to separately label Lmx1b and Foxp2 mRNA (Figure 2). Their distributions overlapped substantially, but Lmx1b skewed ventrally and medially, while Foxp2 skewed dorsally and laterally. Lmx1b was most prominent in a region homologous to the rat PBeL, which was surrounded by dense Foxp2 labeling. The medial PB contained a blend of both Lmx1b and Foxp2, each identifying cells scattered among and through fascicles of the superior cerebellar peduncle. Lmx1b labeling was more prominent caudally, but at far‐caudal levels of the PB, dense Foxp2 labeling extended medially, through the locus coeruleus (LC), which contained labeling for Lmx1b. Lateral to the PB, we also found Foxp2 labeling throughout the Purkinje layer of the cerebellum.
FIGURE 2.
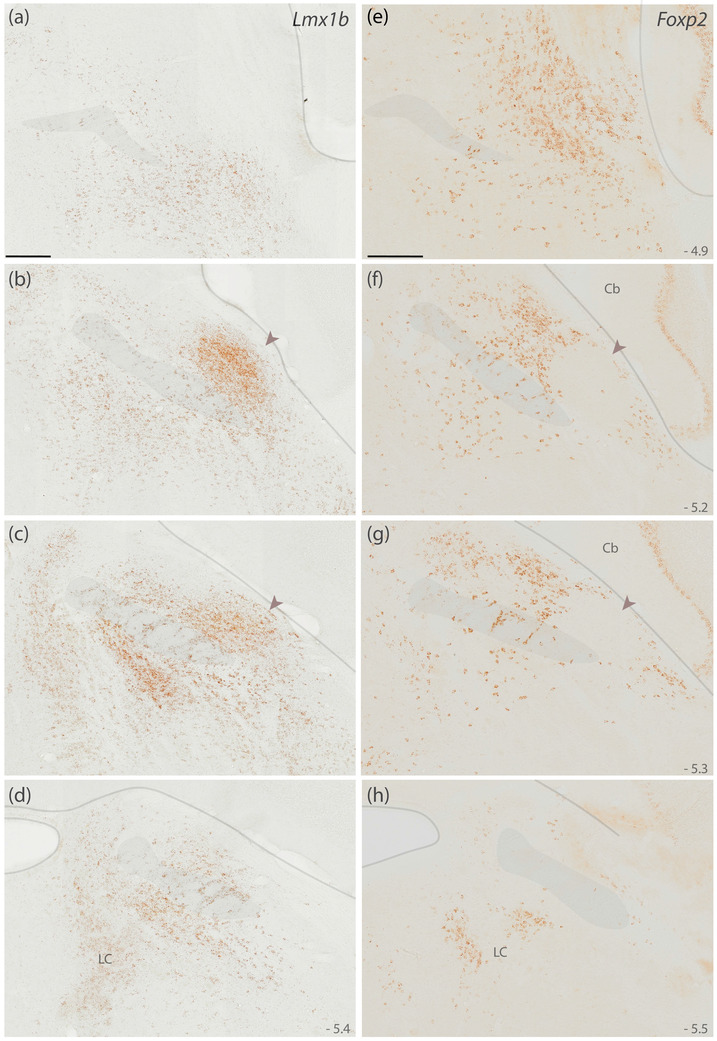
Lmx1b and Foxp2 mRNA in the mouse PB. Diaminobenzidine (DAB) in situ hybridization revealed separate distributions of Lmx1b mRNA (a–d) and Foxp2 mRNA (e–h) expression across four rostral‐to‐caudal sections through the PB region in mice. Approximate level caudal to bregma is shown at the bottom‐right of each panel (in mm). Translucent highlights identify the scp, brainstem surface, and fourth ventricular surface. Arrowheads in panels (b, c, f, g) highlight dense Lmx1b and absent Foxp2 in a region of the mouse PB that is homologous to the “external lateral” subnucleus in rats (Fulwiler & Saper, 1984). Scale bars in (a) and (e) are 200 μm and apply to all panels below. Abbreviations: Cb, cerebellum; LC, locus coeruleus
At these levels of the brainstem, Lmx1b labeling was densest in the raphe nuclei (not shown), and ventral to the PB, the supratrigeminal region and principal sensory trigeminal nucleus contained an extensive distribution of lighter Lmx1b labeling. This lighter Lmx1b expression in the supratrigeminal region extended ventrally, around the motor trigeminal nucleus, and caudally, into the hindbrain reticular formation. Within the foramen of Luschka, we also found Lmx1b labeling in a subset of choroid plexus epithelial cells (not shown).
Fluorescence in situ hybridization (FISH) labeling for Foxp2 and Lmx1b mRNA confirmed their largely complementary distributions (Figure 3a–e). Per‐cell labeling for Foxp2 mRNA was modest, but the small amount of Lmx1b mRNA per cell and the tight clustering of these cells in PBeL made it difficult to distinguish individual neurons. To more confidently identify and compare individual neurons, we switched to immunofluorescence labeling for Lmx1b and FoxP2.
FIGURE 3.
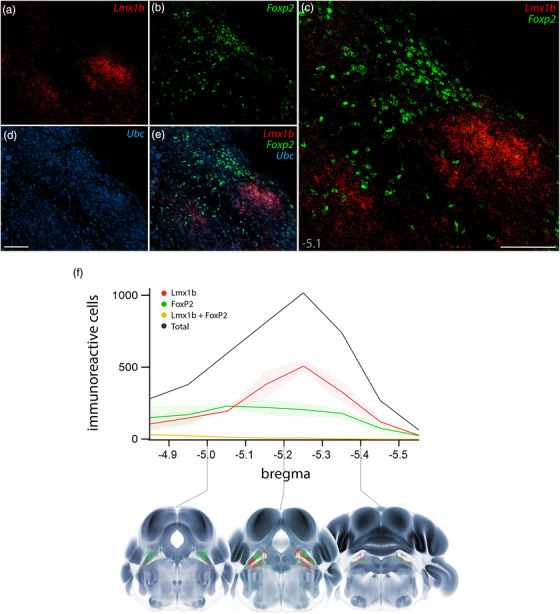
Fluorescence in situ hybridization (FISH) for Lmx1b and Foxp2 mRNA and rostral‐to‐caudal counts of PB neurons containing Lmx1b and FoxP2 protein. Combined FISH labeling revealed the complementary expression patterns of Lmx1b mRNA (a, red) and Foxp2 mRNA (b, green) at a mid‐level of the mouse PB (approximately bregma −5.1 mm). The ubiquitously expressed transcript Ubc (d, blue) was labeled for neuroanatomical background. Scale bars in (c) and (d) are 200 μm, and the scale bar in (d) applies to remaining panels (a, b, e). (f) After immunofluorescence labeling (see examples in Figure 4), average counts of neurons containing Lmx1b and FoxP2 are shown at each level (n = 3 mice), with variance represented by a standard deviation envelope. Approximate bregma levels are shown on the x‐axis, with three representative plots shown atop axial images from the Allen CCFv3 atlas (Wang et al., 2020)
Robust immunofluorescence labeling (Figure 4) matched the patterns of mRNA labeling described above, except that Lmx1b immunoreactivity was less intense than Lmx1b mRNA labeling in LC neurons. Quantitative analysis across all rostrocaudal levels of the PB revealed that most immunolabeled neurons (98%) contained either Lmx1b or FoxP2, not both (n = 3 mice, Figure 3f).
FIGURE 4.
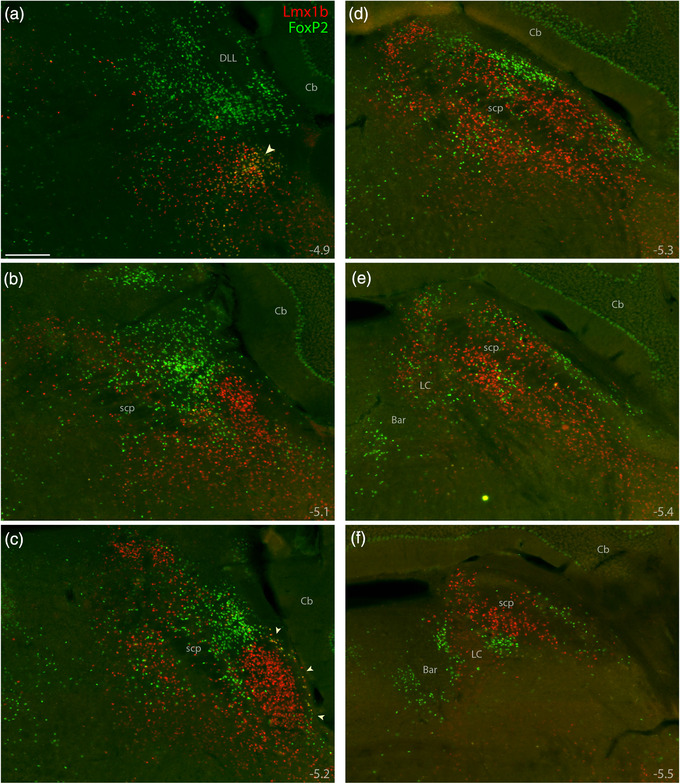
Immunofluorescence labeling for Lmx1b and FoxP2 protein. Combined immunolabeling for the nuclear transcription factors Lmx1b (red) and FoxP2 (green) identified largely separate distributions across successive, rostral‐to‐caudal sections through the PB region (a–f). Approximate level caudal to bregma is shown at the bottom‐right of each panel (in mm). Lmx1b immunofluorescence labeling was mutually exclusive with FoxP2, except in a rostral cluster of neurons ventral to the PB (arrowhead in a) and in sparse, double‐labeled neurons extending back to mid‐rostral levels of the PB along the lateral edge of the brainstem (arrowheads in c). Scale bar in (a) is 200 μm and applies to panels (b–f). Abbreviations: Bar, Barrington's nucleus; DLL, dorsal nucleus of the lateral lemniscus
However, at rostral levels and along the ventrolateral margin of the PB, a small subset contained both Lmx1b and FoxP2. This double‐labeled subset––roughly 2% of the combined total––clustered primarily within the KF (Figure 4a), as in rats (Miller et al., 2012). These double‐labeled neurons intermingled with many others that contained Lmx1b alone. This double‐labeled population also extended caudally along the edge of the brainstem, outside PBeL (Figure 4c), in a distribution resembling the “lateral crescent” in rats (Chamberlin & Saper, 1992; Miller et al., 2012).
3.3. Glutamatergic and GABAergic subsets
GABAergic neurons flank the PB on several sides (Geerling et al., 2017; Li et al., 2005; Verstegen et al., 2017), and a recent study reported that 12% of neurons in this region are GABAergic (Raver et al., 2020), so it was important to clarify whether neurons identified by Lmx1b and FoxP2 are excitatory (glutamatergic; Slc17a6/Vglut2‐expressing) or inhibitory (GABAergic; Slc32a1/Vgat‐expressing). To do this, we used separate Cre‐reporter strains for Vglut2 and Vgat (Figures 5 and 6). In each strain, we used a GFP Cre‐reporter that is tethered to the L10 ribosomal subunit (L10GFP), which concentrates in the cell body, maximizing our ability to distinguish neurons and colocalize other markers. The complementary distributions of L10GFP‐expressing neurons between Vglut2 and Vgat strains matched our previous descriptions (Geerling et al., 2016; Geerling et al., 2017; Huang et al., 2020). In addition to sparse GABAergic neurons scattered through the PB, we observed flanking GABAergic populations medially, in the pontine central gray, and ventrolaterally, in a population we have referred to as the “caudal KF” (Geerling et al., 2017).
FIGURE 5.
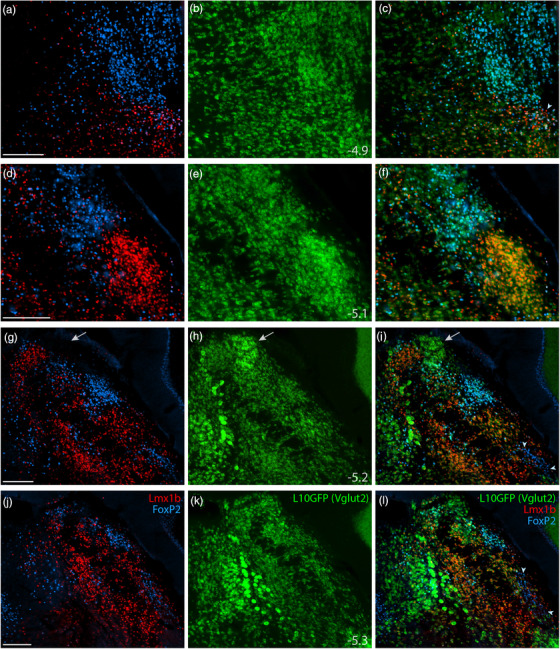
Lmx1b and FoxP2 in a glutamatergic Cre‐reporter mouse. Immunofluorescence labeling identified Lmx1b (red) and FoxP2 (blue) in the PB of mice expressing an L10GFP Cre‐reporter (green) for the glutamatergic marker gene Slc17a6 (vesicular glutamate transporter 2, Vglut2). Arrowhead in (c) indicates a cluster of triple‐labeled neurons (Lmx1b+FoxP2+L10GFP) located rostrally, in the KF region. White arrows in (g–i) highlight a dorsal cluster of L10GFP‐expressing neurons lacking both Lmx1b and FoxP2. Arrowheads in (i) and (l) indicate a ventrolateral cluster of neurons labeled for FoxP2 in the “caudal KF.” Approximate bregma levels are shown at bottom‐right in the center column (in mm). All scale bars are 200 μm and apply to other panels in their respective rows
FIGURE 6.
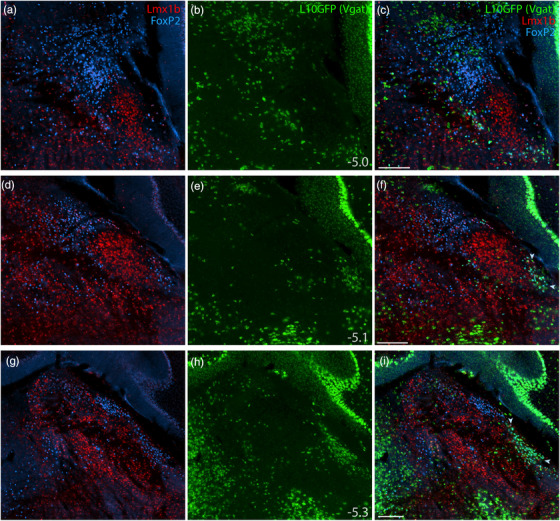
Lmx1b and FoxP2 in a GABAergic Cre‐reporter mouse. Immunofluorescence labeling identified Lmx1b (red) and FoxP2 (blue) at three rostral‐to‐caudal levels of the PB in mice expressing an L10GFP Cre‐reporter (green) mice for the GABAergic marker gene Slc32a1 (vesicular GABA transporter, Vgat). Arrowheads in panels (f) and (i) indicate a caudal, ventrolateral cluster of neurons that contain FoxP2 and express L10GFP in the “caudal KF.” Approximate bregma levels are shown at bottom‐right in the center column (in mm). All scale bars are 200 μm and apply to other panels in their respective rows
All Lmx1b‐labeled neurons in the PB and KF expressed L10GFP in glutamatergic Cre‐reporter mice (Figure 5), and no Lmx1b‐labeled neurons expressed L10GFP in the GABAergic strain (Figure 6). FoxP2‐labeled neurons in PB and KF also colocalized with L10GFP in glutamatergic Cre‐reporter mice, but a caudal cluster of FoxP2‐labeled neurons ventrolateral to the PB (“caudal KF”) expressed L10GFP exclusively in GABAergic reporter mice (arrowheads in Figures 5i,l and 6f,i). Some FoxP2‐labeled GABAergic neurons distributed rostrally, intermingling with the caudal aspect of the glutamatergic KF population. Ventrolateral to the PB, all neurons that contained FoxP2 and lacked Lmx1b expressed L10GFP exclusively in GABAergic reporter mice. Conversely, all neurons in the rostral population containing both Lmx1b and FoxP2 expressed L10GFP exclusively in glutamatergic reporter mice. Outside the PB, FoxP2 neurons in the pontine central gray matter (ventromedial to Barrington's nucleus) and cerebellar Purkinje neurons expressed L10GFP exclusively in GABAergic reporter mice (Figure 6g–i).
At mid‐levels of the PB, Vglut2 Cre‐reporter labeling identified an additional, dorsal cluster of neurons lacking both Lmx1b and FoxP2 (arrow in Figure 5g–i). This prominent cluster resembled the rat “internal lateral” PB subnucleus (Bester et al., 1999; Bourgeais et al., 2001; Feil & Herbert, 1995; Fulwiler & Saper, 1984; Krout & Loewy, 2000).
3.4. Phox2b ventral to the PB
The ventral gradient of lighter Lmx1b expression (Figure 2) left an indefinite distinction between the PB and several other brainstem nuclei, so we sought a molecular marker that could distinguish PB neurons from underlying populations. Without imposing any cytoarchitectural or other external constraints, we found that neurons ventral to the PB can be distinguished by labeling another transcription factor that had been identified in this brainstem region in rats, Phox2b (Kang et al., 2007).
Phox2b mRNA labeling (Figure 7a–c) and Phox2b immunofluorescence (Figure 8) identified a large distribution of neurons immediately ventral to the PB. At mid‐levels, the dorsal aspect of this population formed a sharp, ventromedial border for the PB. The PB itself contained very few Phox2b‐expressing cells. Just medial to the PB, mRNA labeling and Phox2b immunoreactivity were prominent in and around the LC. Everywhere in this region, Phox2b was mutually exclusive with FoxP2. Phox2b‐expressing cells occupied much of the supratrigeminal region that divides the PB from the trigeminal motor and principal sensory nuclei (both of which lacked Phox2b mRNA labeling and lacked Phox2b immunofluorescence). Like the Lmx1b distribution, contiguous Phox2b labeling extended ventrally and caudally from the supratrigeminal region into the hindbrain reticular formation.
FIGURE 7.
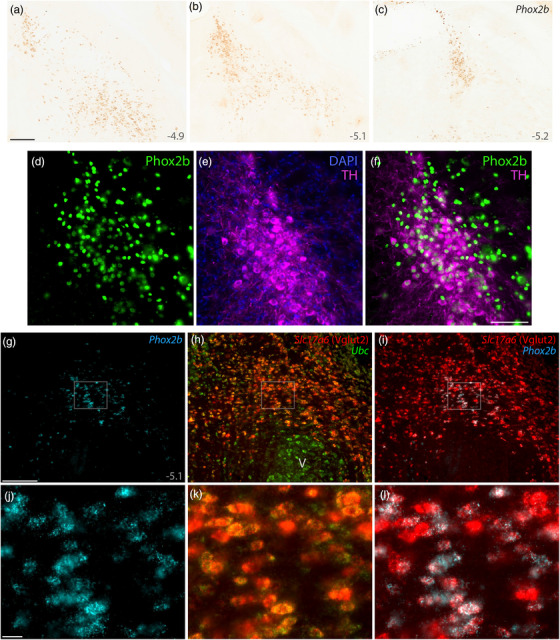
Phox2b identifies glutamatergic and catecholaminergic populations ventral and medial to the PB. DAB in situ hybridization revealed Phox2b mRNA, shown at three rostral‐to‐caudal levels (a–c). Approximate level caudal to bregma is shown at the bottom‐right of each panel (in mm). (d–f) Immunofluorescence labeling for Phox2b (green) and tyrosine hydroxylase (TH, magenta) identify LC neurons. (g–i) FISH identified colocalization between Phox2b mRNA (ice‐blue) and the glutamatergic marker Slc17a6 mRNA (h, red) ventral to a mid‐level of the PB (bregma −5.1 mm). Ubc mRNA (green) is shown for neuroanatomical background. (j–l) Blow‐ups of the highlighted region in (g–i) show the ubiquitous colocalization of Slc17a6 mRNA in Phox2b‐expressing neurons in the supratrigeminal region. All scale bars are 200 μm and apply to related panels. Abbreviation: V, motor trigeminal nucleus
FIGURE 8.
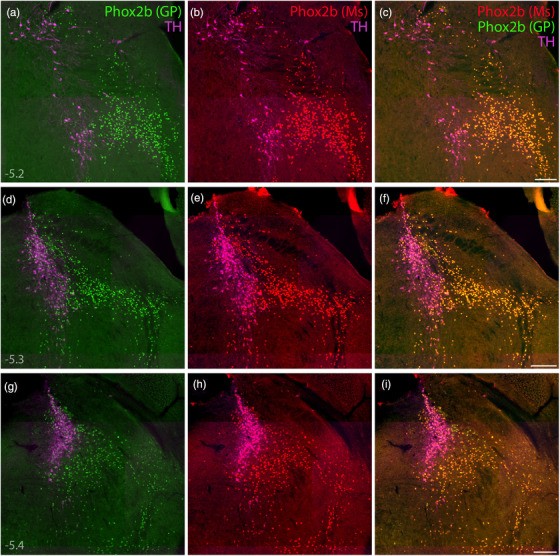
Immunofluorescence labeling for Phox2b. (a–i) Direct comparison of immunofluorescence labeling between guinea pig polyclonal [green, “Phox2b (GP)”; (Nagashimada et al., 2012)] and mouse monoclonal [red, “Phox2b (Ms)”; sc‐376997] anti‐Phox2b antisera, combined with TH immunofluorescence (magenta) to identify LC neurons. In addition to confirming antibody specificity, labeling Phox2b across three rostral‐to‐caudal levels of the PB region highlighted the extensive population of non‐LC (glutamatergic) Phox2b neurons, which form an observer‐independent ventromedial border for the PB. Approximate bregma levels are shown at bottom‐left in (a, d, g). Scale bars in (c, f, i) are 200 μm, and each scale bar applies to other panels in the same row
The supratrigeminal region contains both excitatory and inhibitory neurons. Vgat Cre‐reporter labeling, mRNA labeling for Vgat (Slc32a1), and mRNA labeling for the glycinergic neuronal marker Slc6a5 identified large neurons caudally and many smaller neurons rostrally in this region (not shown). Also, Vglut2 Cre‐reporter labeling and mRNA labeling for Slc17a6 identified many small neurons in the supratrigeminal region (not shown). We used FISH to determine which population expresses Phox2b and found extensive Slc17a6 mRNA labeling in small Phox2b‐expressing neurons (Figure 7g–l), but not in larger Phox2b‐expressing neurons in the LC. Thus, in addition to nonglutamatergic LC neurons, Phox2b identifies a large population of glutamatergic neurons in the supratrigeminal nucleus and reticular formation, immediately ventral to the PB.
Medial to the PB, the dorsomedial distribution of this glutamatergic Phox2b population intermingled through the mesencephalic trigeminal nucleus and the dorsal LC. The LC contained both Phox2b mRNA labeling and Phox2b immunolabeling. Every LC neuron had cytoplasmic immunofluorescence labeling for tyrosine hydroxylase surrounding a Phox2b‐immunolabeled nucleus, but these intermingled with a population of smaller, noncatecholaminergic neurons with more intense nuclear immunofluorescence for Phox2b (Figure 7d–f). The PB contained fewer Phox2b‐expressing neurons, which were identical in appearance (and Slc17a6 labeling; not shown) to the smaller glutamatergic neurons ventral and medial to it and appeared to be the fringe of the supratrigeminal population, rather than a distinct subpopulation of PB neurons.
The Phox2b distribution substantially overlapped the gradient of lighter Lmx1b labeling ventral to the PB. At first, we were unable to determine whether any neurons contained both Phox2b and Lmx1b because both antisera were raised in the same species (guinea pig), but serial‐labeling protocols (for Lmx1b then Phox2b; and, separately, for Phox2b then Lmx1b) revealed a large population of potentially coexpressing neurons. To better assess colocalization, we identified a mouse monoclonal antibody that labels Phox2b. Double‐labeling tissue with this antibody (sc‐376997) along with the guinea pig anti‐Phox2b (Nagashimada et al., 2012) highlighted an identical set of neurons (Figure 8). Next, using this mouse monoclonal in combination with antisera for Lmx1b (guinea pig), FoxP2 (sheep), and TH (rabbit) confirmed that many supratrigeminal neurons contain both Phox2b and Lmx1b (Figures 9 and 10). Lmx1b labeling here (ventral to the PB) was lighter and always colocalized with Phox2b, while Lmx1b labeling in the PB was more intense and rarely colocalized with Phox2b. Across all rostrocaudal levels, we found Lmx1b immunofluorescence in approximately half the neurons that contained Phox2b (range 44–57%, n = 3 mice), but given the light and variable intensity of Lmx1b labeling in the supratrigeminal region, this may be an underestimate.
FIGURE 9.
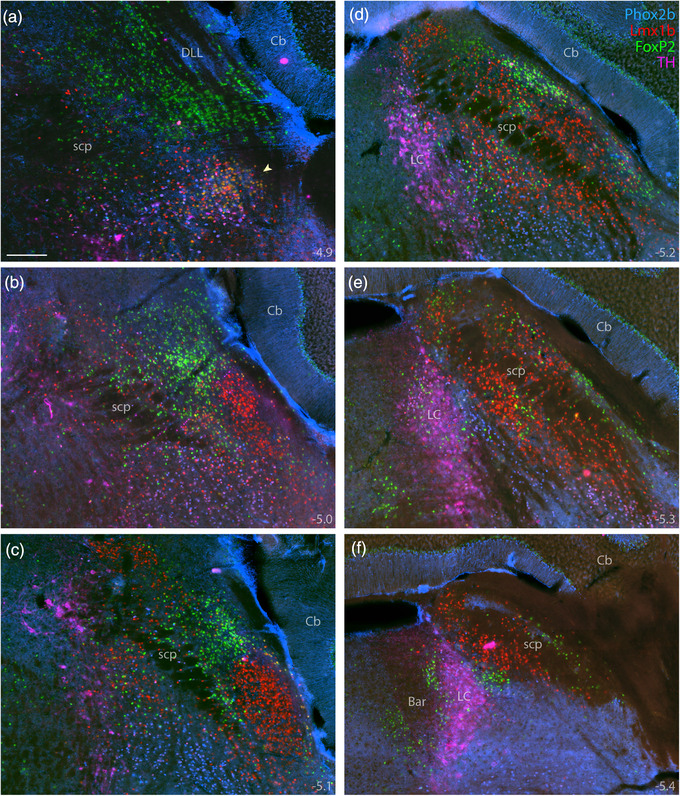
Combined immunofluorescence labeling for Phox2b, Lmx1b, and FoxP2. Immunolabeling for Phox2b (blue, mouse monoclonal antibody) combined with Lmx1b (red) and FoxP2 (green), across six rostral‐to‐caudal sections through the PB region (a–f), distinguished adult PB neurons from underlying non‐PB populations. Additional labeling for TH (magenta) distinguished Phox2b neurons in the LC from those in the supratrigeminal population. Approximate bregma levels are shown at the bottom‐right of each panel. Arrowhead in (a) highlights the KF. Scale bar in (a) is 200 μm and applies to all panels
FIGURE 10.
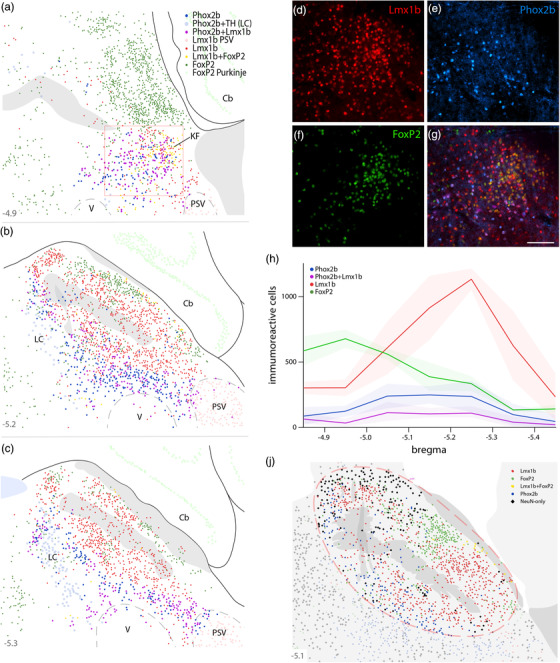
Labeling Phox2b, along with Lmx1b and FoxP2, identifies diverse populations of adult neurons in and around the PB. Neurons containing Phox2b alone (blue), Phox2b+Lmx1b (magenta), Lmx1b alone (red), Lmx1b+FoxP2 (yellow), or FoxP2 alone (green) were plotted at three rostral‐to‐caudal levels of the mouse PB (a–c). Approximate level caudal to bregma is shown at the bottom‐left of each panel (in mm). Phox2b‐containing neurons in the LC (TH+Phox2b) were plotted in light blue. FoxP2‐containing Purkinje neurons in the cerebellum were plotted in light green. Large, Lmx1b‐containing neurons in the principal sensory trigeminal nucleus (PSV) were plotted in light red. Ventral to the rostral PB (box in a), immunofluorescence labeling for Lmx1b (d, red), Phox2b (e, blue), FoxP2 (f, green), and all three combined (g) revealed focally diverse populations in the KF region. Scale bar in (g) is 50 μm. (h) Rostral‐to‐caudal counts of PB neurons containing Phox2b alone, Lmx1b alone, FoxP2 alone, or Phox2b+Lmx1b were averaged at each level (n = 3 mice), with variance represented by a standard deviation envelope; approximate levels caudal to bregma are labeled on the x‐axis. (i) Plotting the distribution of neurons containing only NeuN (black diamonds), without Phox2b, Lmx1b, or FoxP2, highlighted a remaining set of unidentified PB neurons. Approximate bregma level is shown at the bottom‐left of each panel (in mm)
3.5. Diverse Kölliker–Fuse subpopulations
Rostrally and laterally, the Phox2b population merged seamlessly through the KF (Figures 9a and 10a). This was unexpected because a previous study in rats found Phox2b in very few KF neurons (Kang et al., 2007). Including this substantial Phox2b‐immunoreactive population, the KF contained the greatest transcription factor diversity in this region of the brainstem.
As noted above, most neurons in the KF region contained Lmx1b. Within that population, mutually exclusive subsets contained either FoxP2 or Phox2b, never both (Figure 10a, d–g). Many, intermingled neurons contained only Lmx1b (without Phox2b or FoxP2), and this subset intermingled with a sizeable minority of KF neurons that contained only Phox2b (without Lmx1b). These findings highlight the KF and contiguous supratrigeminal region as an intersection of multiple glutamatergic populations. These include: (1) an Lmx1b‐derived population resembling “dA3” hindbrain interneurons, which also express Phox2b (Gray, 2013; Hernandez‐Miranda et al., 2017), (2) a distinctly KF subpopulation containing both Lmx1b and Foxp2 (without Phox2b), (3) an intermingled population containing just Lmx1b (without Phox2b or Foxp2), and (4) an intermingled population containing just Phox2b (without Foxp2 or Lmx1b). Also, along the caudal aspect of this region are (5) GABAergic, Foxp2‐expressing neurons (“caudal KF”), some of which extend rostrally beneath the PB and intermingle with the caudal aspect of the atlas‐defined KF region.
3.6. Unidentified neurons
To determine whether the PB region includes additional neurons that were not identified by our adult transcription factor markers (Lmx1b, FoxP2, and Phox2b), we labeled all three in combination with a more broadly expressed neuronal marker, NeuN (encoded by the transcription factor Rbfox3; Kim et al., 2009). Besides LC neurons, which lack NeuN (Verstegen et al., 2017), all FoxP2‐, Lmx1b‐, and Phox2b‐immunoreactive neurons in the PB contained NeuN immunoreactivity. Conversely, most NeuN‐immunoreactive neurons contained at least one of these transcription factors. However, a substantial minority of NeuN‐immunoreactive neurons did not contain FoxP2, Lmx1b, or Phox2b. These “NeuN‐only” neurons were sparse ventrally and concentrated rostrally and dorsally in the lateral PB (Figure 10i), including the subregion with glutamatergic neurons lacking both FoxP2 and Lmx1b (Figure 5i). To identify these remaining neurons, we analyzed two other transcription factors that influence the development of neurons in this region––Lmx1a and Atoh1.
3.7. Lmx1a
First, we examined adult expression of Lmx1a, the paralog of Lmx1b. During embryogenesis, Lmx1a and Lmx1b play overlapping roles in the roof plate of the neural tube (Chizhikov et al., 2021; Liu et al., 2010; Mishima et al., 2009) and have partly overlapping expression patterns (Chen et al., 1998; Chizhikov et al., 2006; Chizhikov et al., 2010; Costa et al., 2001; Ding et al., 2004; Dreyer et al., 1998; Guo et al., 2007; Guo et al., 2008; Kuwamura et al., 2005; Millonig et al., 2000; Zhao et al., 2006). Also, these transcription factors are expressed by partly overlapping populations of neurons in the mature brain (Asbreuk et al., 2002; Dai et al., 2008; Zou et al., 2009).
In the adult PB, Lmx1a mRNA labeling resembled Lmx1b (Figure 11). Like Lmx1b, Lmx1a skewed ventrally (through PBeL and KF) and caudally (through the superior cerebellar peduncle and medial PB). Unlike Lmx1b, however, we did not find Lmx1a labeling in the LC. Nor was there any Lmx1a labeling in the supratrigeminal nucleus or principal sensory trigeminal nucleus. We found Lmx1a expression in several regions that lacked Lmx1b, including the cerebellar flocculus, superior vestibular nucleus, and dorsal cochlear nucleus (not shown), as well as a population of small, dense cells along the brainstem surface and atop the middle cerebellar peduncle (Figure 11a,b). Also, the choroid plexus had more extensive Lmx1a labeling than Lmx1b (not shown).
FIGURE 11.
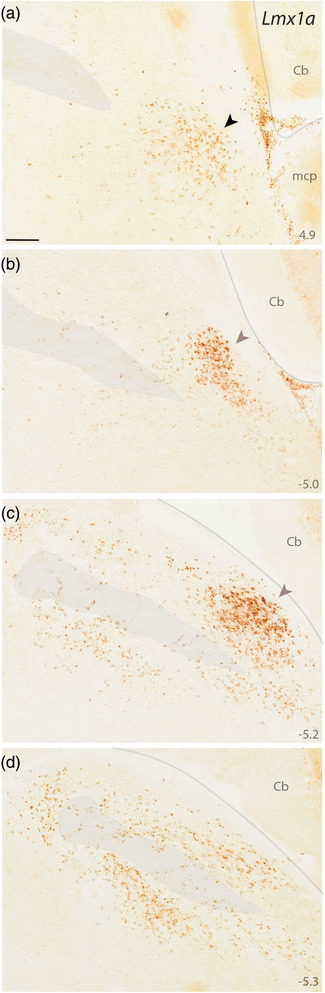
Lmx1a mRNA labeling. DAB in situ hybridization revealed the distribution of Lmx1a expression at four, successive rostral‐to‐caudal sections through the PB region. Approximate level caudal to bregma is shown at bottom right (in mm). Black arrowhead (a) highlights the KF region. Translucent highlights identify the scp and boundaries between the brainstem surface and cerebellum. Mauve arrowheads (b, c) highlight dense Lmx1a labeling in a region homologous to the “external lateral” subnucleus in rats (Fulwiler & Saper, 1984). Scales bar in (a) is 200 μm and applies to all panels. Abbreviation: mcp, middle cerebellar peduncle
As with Lmx1b, the low per‐cell content of Lmx1a mRNA made it difficult to distinguish individual neurons. Lacking an antibody that labels this transcription factor, we used Cre fate‐mapping to identify cells that have expressed Lmx1a or developed from Lmx1a‐expressing precursors. In Lmx1a‐Cre;R26‐LSL‐tdTomato mice (n = 5, Figure 12), the distribution of tdTomato expression resembled the distribution of Lmx1a mRNA. Most tdTomato‐expressing (putatively Lmx1a‐derived) PB neurons also contained Lmx1b (69 ± 8%, n = 3; graph in Figure 13o), but an intermingled minority lacked Lmx1b, and a larger population of Lmx1b‐containing neurons lacked tdTomato, consistent with the subtotal colocalization of Lmx1a mRNA and Lmx1b in P7 mouse pups (Figure 3J of Zou et al., 2009). These populations had similar, intermingled distributions, except for a few tdTomato‐expressing neurons that lacked Lmx1b caudal and medial to the PB (between the LC and fourth ventricle; not shown). Besides these, the population identified by Lmx1a Cre fate‐mapping included few neurons that were not identified already by Lmx1b and very few in the rostral‐dorsal distribution of “NeuN‐only” neurons we sought to identify.
FIGURE 12.
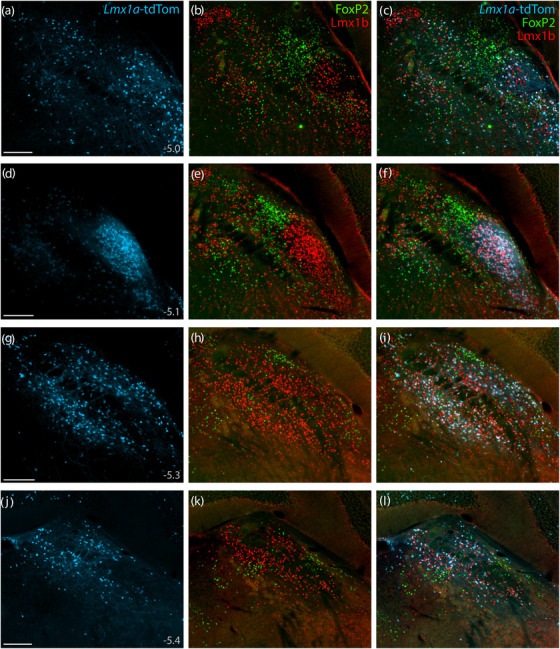
Lmx1a Cre fate‐mapping. Cre fate‐mapping for Lmx1a (tdTomato, pseudocolored ice blue), followed by immunofluorescence labeling for Lmx1b (red) and FoxP2 (green), shown at four rostral‐to‐caudal levels through the PB region. Approximate level caudal to bregma is shown at bottom‐left in the first panel of each row (in mm). Scale bars (a, d, g, j) are 200 μm and apply to all other panels in the same row
FIGURE 13.
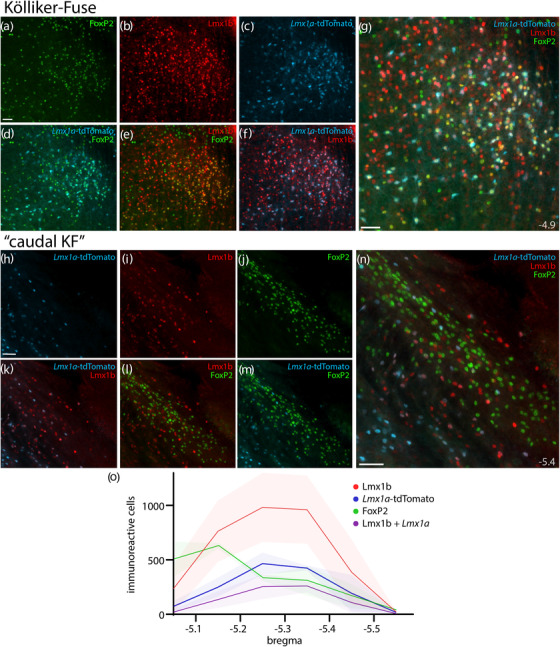
Lmx1a‐derived KF neurons and rostral‐to‐caudal counts across the PB region. Panels (a–g) show Cre fate‐mapping for Lmx1a (tdTomato, pseudocolored ice blue) and immunofluorescence labeling for Lmx1b (red) and FoxP2 (green) in the KF region (ventral to the rostral PB). Panels (h–n) show the lack of Lmx1a Cre‐reporter expression in “caudal KF” neurons that contain FoxP2 (ventrolateral to the caudal PB). Approximate level caudal to bregma (in mm) is shown at bottom‐right in (g, n). All scale bars are 50 μm. Scale bar in (a) applies to (b–f) and scale bar in (h) applies to (i–m). Bottom graph (o) shows rostral‐to‐caudal counts of PB neurons expressing the tdTomato Cre‐reporter for Lmx1a, FoxP2, or Lmx1b, as well as neurons containing both Lmx1b and Lmx1a Cre‐reporter. Counts were averaged at each level (n = 3 mice), with variance represented by a standard deviation envelope. Approximate bregma levels are shown on the x‐axis
Rostrally, many neurons expressed tdTomato in the KF region (Figure 13). All these putatively Lmx1a‐derived neurons contained Lmx1b, and many also contained FoxP2 (in addition to Lmx1b), but no other FoxP2‐labeled neurons in the PB or “caudal KF” expressed tdTomato. Outside the PB, LC neurons containing tyrosine hydroxylase did not express tdTomato. We also did not find tdTomato expression (or labeling for Lmx1b, FoxP2, or Phox2b) in the larger neurons containing choline acetyltransferase (ChAT) in the laterodorsal and pedunculopontine tegmental nuclei (not shown), although Lmx1b and tdTomato did colocalize with a small number of faintly ChAT‐immunoreactive neurons in the caudal PB and PBeL (not shown).
3.8. Atoh1 derivation identifies remaining PB neurons
After Lmx1a Cre fate‐mapping failed to identify many neurons in the “NeuN‐only” distribution, we next focused on neurons that derive from Atoh1‐expressing precursors. Across several days of embryonic development, Atoh1‐expressing precursors in the rhombic lip neuroepithelium produce (1) glutamatergic neurons that populate the PB and many other brainstem nuclei, followed by (2) glutamatergic neurons in the deep cerebellar nuclei, and then (3) the entire granule cell layer of the cerebellum (Ben‐Arie et al., 1997; Bermingham et al., 2001; Gray, 2008, 2013; Machold & Fishell, 2005; Rose et al., 2009; Wang et al., 2005).
Adult cells do not express Atoh1, so we used Cre fate‐mapping to identify Atoh1‐derived neurons. Atoh1‐Cre;R26‐LSL‐tdTomato mice had extensive tdTomato expression in the cerebellum and brainstem (n = 4, Figure 14a), similar to previous descriptions in other Cre‐reporter strains (Machold & Fishell, 2005; Wang et al., 2005). We found ubiquitous tdTomato expression in cerebellar granule cells, and none in Purkinje cells. Fibrous labeling pervaded the molecular layer, arbor vitae, superior cerebellar peduncle, and middle cerebellar peduncle. Outside the cerebellum, we found fibrous labeling in a ventral majority of the trigeminal principal sensory nucleus, which contrasted a near‐absence of labeling dorsally in this nucleus. The trigeminal motor nucleus and a round, central portion of the facial motor nucleus were both devoid of labeling, but the absence of labeling in these small regions contrasted a broader meshwork of labeled fibers in the surrounding brainstem. In the LC, tdTomato did not colocalize with tyrosine hydroxylase, but we found smaller, tdTomato‐expressing neurons scattered through the dorsal LC, and Barrington's nucleus contained large, tdTomato‐expressing neurons. We also identified a small number of tdTomato‐expressing neurons that contained Phox2b caudal and medial to the caudal LC, near the medial vestibular nucleus (not shown).
FIGURE 14.
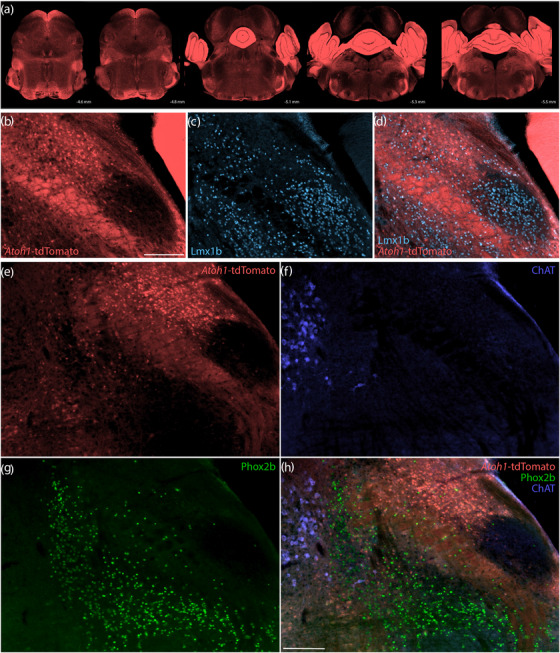
Atoh1 Cre fate‐mapping with tdTomato. Cre fate‐mapping for Atoh1 identified neurons in the cerebellum, PB, and several other brainstem regions, plus extensive labeling in fibrous processes. (a) Fluorescent reporter expression for Atoh1‐Cre (tdTomato, pseudocolored coral‐red) across five rostral‐to‐caudal tissue sections spanning the PB region. At each level, the approximate distance caudal to bregma is shown at bottom‐right. (b–d) Cre fate‐mapping for Atoh1 followed by immunofluorescence labeling for Lmx1b (ice blue). (e–h) Immunofluorescence labeling for Phox2b (green) and ChAT (blue) at a mid‐level through the PB region (approximately bregma −5.1 mm). All scale bars are 200 μm. Scale bar in (b) also applies to panels (c, d). Scale bar in (h) also applies to panels (e–g)
The PB contained many tdTomato‐expressing neurons, consistent with several previous reports (Machold & Fishell, 2005; Rose et al., 2009; van der Heijden & Zoghbi, 2018; Wang et al., 2005). The distribution of these putatively Atoh1‐derived neurons skewed dorsally, rostrally, and laterally, and many appeared to contain FoxP2. In contrast, tdTomato was largely absent from the ventrolateral PB (PBeL) and supratrigeminal region and did not appear to colocalize with either Lmx1b (Figure 14b–d) or Phox2b (Figure 14e–h). At mid‐levels of the PB, a dorsal cluster of tightly packed, larger Atoh1‐derived neurons resembled the rat “internal lateral” subnucleus (Bester et al., 1999; Bourgeais et al., 2001; Feil & Herbert, 1995; Fulwiler & Saper, 1984; Krout & Loewy, 2000). These neurons appeared to lack FoxP2, except at the ventral rim of this cluster. Besides these, we did not find any discrete clusters of tdTomato‐expressing neurons lacking FoxP2. The remainder of the lateral PB contained intermingled populations of Atoh1‐derived neurons with and without FoxP2 across a continuum of regions resembling the “dorsal lateral,” “central lateral,” and “superior lateral” subnuclei in rats (Fulwiler & Saper, 1984), plus a rostral‐ventral region we referred to as the “rostral‐to‐external‐lateral” subnucleus in mice (Geerling et al., 2016).
The dense, fibrous background of axonal and dendritic tdTomato made it difficult to distinguish individual neurons and colocalize other molecular markers (Figure 14), so we crossed Atoh1‐Cre mice to the L10GFP Cre‐reporter strain described above. In the resulting progeny (Atoh1‐Cre;R26‐lsl‐L10GFP, n = 6), the GFP reporter concentrated in cell bodies, rather than axons and dendrites, allowing us to more definitively distinguish Atoh1‐derived neurons and colocalize molecular markers (Figures 15, 16, 17).
FIGURE 15.
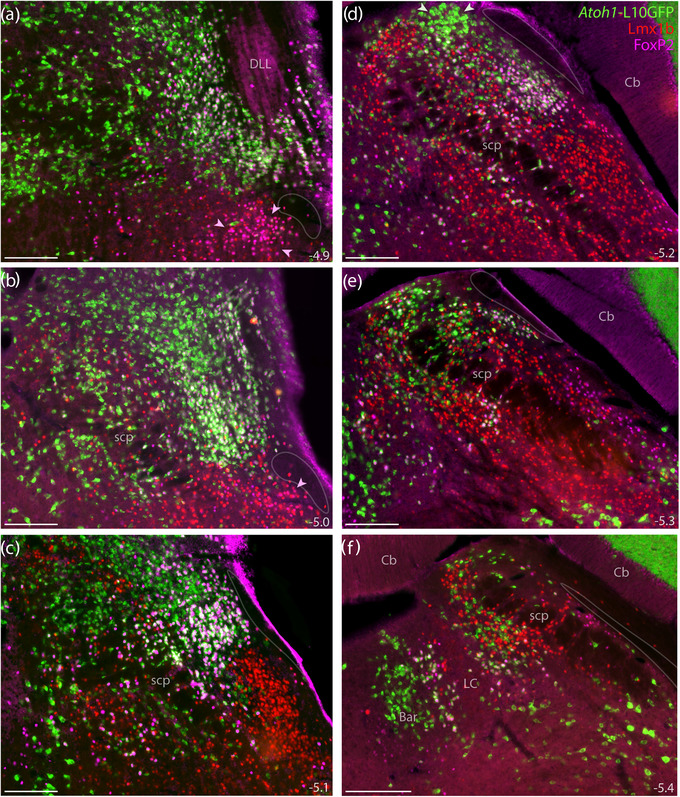
Atoh1 Cre fate‐mapping with L10GFP. The L10GFP Cre‐reporter identified Atoh1‐derived cell bodies. Panels (a–f) show L10GFP expression across six rostral‐to‐caudal levels of the PB region in an Atoh1‐Cre;R26‐lsl‐L10GFP mouse after immunofluorescence labeling for Lmx1b (red) and FoxP2 (magenta). Nuclear FoxP2 immunofluorescence overlapping cytoplasmic L10GFP appears white. Arrowheads in panels (a, b) highlight the KF region. Arrowheads in (d) highlight a prominent, dorsal cluster of L10GFP‐expressing neurons. Translucent outlines in each panel show the location of a small white matter tract alongside the lateral PB and KF region, which is labeled “ventral spinocerebellar tract” in current brain atlases. Approximate distance caudal to bregma is provided at the bottom‐right of each panel (in mm). All scale bars are 200 μm
FIGURE 16.
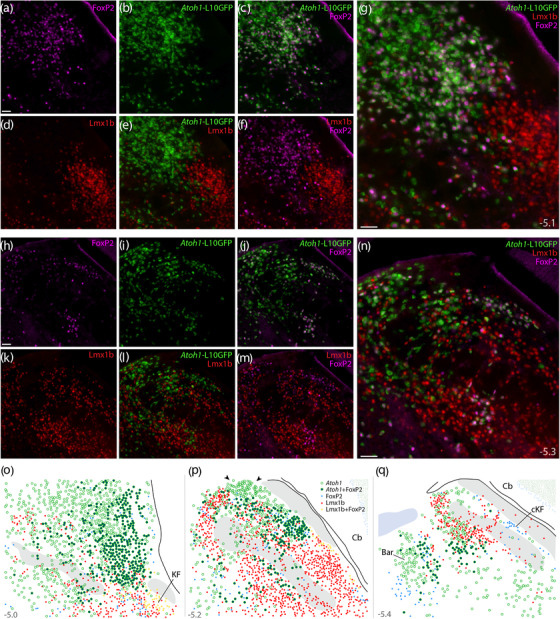
Atoh1 Cre fate‐mapping with L10GFP: magnified color separations and plots. Panels (a–g) show immunofluorescence labeling for FoxP2 (magenta) and Lmx1b (red) after fate‐mapping for Atoh1‐Cre at a mid‐rostral level of the lateral PB. (h–n) Immunofluorescence labeling for FoxP2 and Lmx1b after L10GFP fate‐mapping for Atoh1‐Cre at a mid‐caudal level of the PB, centered over the “head” and “waist” of the scp. Approximate bregma levels are shown at bottom‐right in (g, n). All scale bars are 50 μm. Scale bar in (a) applies to (b–f) and scale bar in (h) applies to (i–m). (o, p) Rostral‐to‐caudal plots show the distribution of Atoh1‐derived neurons across the PB region, including large subsets with and without FoxP2. Arrowheads in (p) highlight a dorsal cluster of L10GFP‐expressing neurons that lack FoxP2. Throughout the PB, Lmx1b and L10GFP were mutually exclusive (no L10GFP‐expressing PB neurons contained Lmx1b). Approximate bregma levels are shown at bottom‐right in (g, n). Abbreviation: cKF, “caudal KF” population
FIGURE 17.
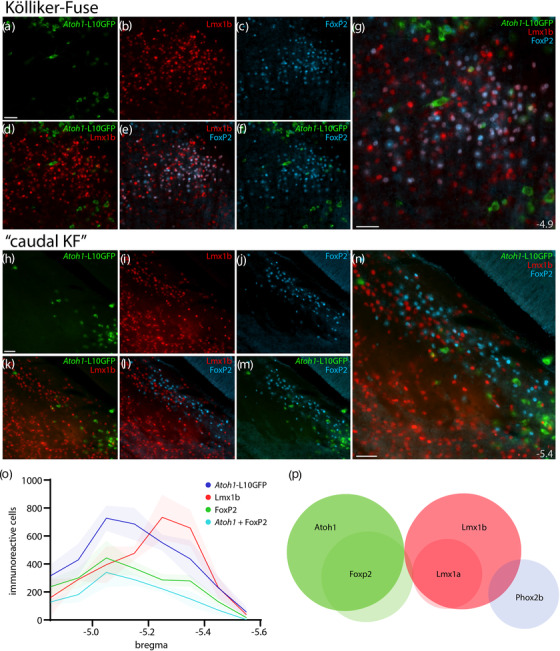
Cre fate‐mapping for Atoh1 in KF and “caudal KF” with counts of Atoh1‐derived neurons across the PB region. Immunofluorescence labeling for Lmx1b (red) and FoxP2 (blue) after Cre fate‐mapping for Atoh1 (green) in the KF region (a–g), and in the “caudal KF” (h–n). Approximate distance from bregma is shown at bottom‐right in (g, n). All scale bars are 50 μm. Scale bar in (a) also applies to (b–f). Scale bar in (h) also applies to (i–m). (o) Counts of Atoh1‐derived, FoxP2, Lmx1b, and Atoh1+FoxP2 double‐labeled neurons across six rostro‐caudal levels. Counts were averaged at each rostrocaudal level (n = 3 mice), with variance represented by a standard deviation envelope. Approximate distance caudal to bregma is shown on the x‐axis. (p) Venn diagram of transcription factor markers that identify neuronal populations in the PB region
Within the PB, we did not find any cells containing both L10GFP and Lmx1b, indicating total mutual exclusivity between Atoh1‐derived neurons and Lmx1b expression. At the dorsal edge of the principal sensory trigeminal nucleus, we occasionally found an L10GFP‐expressing neuron with an Lmx1b‐immunoreactive nucleus, but these rare cells represented just 0.002% of the 12,606 L10GFP‐expressing neurons counted across this region (n = 3 mice). We also found mutual exclusivity between L10GFP and Phox2b in the PB and LC, although sparse Atoh1‐derived neurons located caudal and ventral to the LC did contain Phox2b along the medial vestibular nucleus and facial nerve genu.
In contrast to their mutual exclusivity with Lmx1b, many Atoh1‐derived PB neurons contained FoxP2. As in the tdTomato strain, double‐labeled neurons intermingled with other Atoh1‐derived PB neurons that lacked FoxP2 (Figures 15 and 16). At caudal levels, these intermingled populations (Atoh1‐derived neurons with and without FoxP2) extended around and through the superior cerebellar peduncle and intermingled extensively with Lmx1b‐immunoreactive neurons (Figure 15e,f). At middle and rostral levels, Atoh1‐derived neurons filled the lateral PB except for PBeL (Figure 15a–d). At rostral levels, their distribution extended up to the dorsal nucleus of the lateral lemniscus and down to the PB‐KF border (Figure 15a).
At mid‐levels of the PB, we again found a prominent, dorsal cluster of Atoh1‐derived neurons resembling the rat “internal lateral” subnucleus (arrowheads in Figures 15d and 16p). These neurons lacked FoxP2 and bordered a thin white matter tract that is labeled “ventral spinocerebellar tract” in current atlases (Dong, 2008; Paxinos & Franklin, 2013). Outside this one cluster, Atoh1‐derived neurons with and without FoxP2 intermingled extensively. FoxP2 colocalization was most prevalent near PBeL (Figures 15b–d and 16a–g), less prevalent dorsally, and least prevalent dorsomedially, near the head and waist of the superior cerebellar peduncle (Figure 16h–n). Also intermingling with these subsets were fewer neurons containing FoxP2 without L10GFP; these neurons did not form any discrete clusters except for the “caudal KF” population ventrolateral to the PB.
At rostral levels, none of the KF populations described above (neurons containing Lmx1b, Lmx1b+FoxP2, Lmx1b+Phox2b, or Phox2b) expressed L10GFP. Nor did we find L10GFP expression in the contiguous “lateral crescent” neurons alongside PBeL. Also, L10GFP was not expressed in any “caudal KF” neurons (Figure 17). Thus, we did not find any evidence to support previous claims that KF neurons derive from Atoh1‐expressing precursors in the rhombic lip (Gray, 2008; van der Heijden & Zoghbi, 2018).
3.9. Interim summary
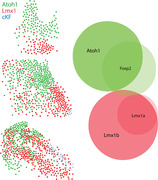
Identifying the large population of Atoh1‐derived PB neurons that lack FoxP2 completed a core set of developmental‐genetic markers useful for classifying neurons in this region. Specifically, Lmx1 paralogs (primarily Lmx1b, but also Lmx1a) and Atoh1 define two, mutually exclusive macropopulations of glutamatergic neurons (Figure 17p). We designed our experiments to assess colocalization, rather than total numbers, but as a rough estimate, summing these two macropopulations suggests that there are approximately 20,000 PB neurons on each side of the mouse brainstem (21,472 ± 1636; n = 3 mice).
3.10. Subpopulations of Lmx1‐ and Atoh1‐derived PB neurons
Transcription factors influence the repertoire of genes (including other transcription factors) that neurons transcribe, and this “transcriptome” shapes the pattern of synapses a neuron establishes with other neurons (Hirsch et al., 2021). Therefore, using transcription factors to classify PB neurons should help us interpret other patterns of gene expression and neural circuit connectivity. For example, under a cytoarchitectural framework, Foxp2 expression marked an indiscrete population of similar‐appearing cells, strewn across virtually every PB subnucleus (Figure 2). In contrast, our new framework identifies two glutamatergic subpopulations (a large subset of Atoh1‐derived PB neurons and a small subset of Lmx1‐derived neurons; Figure 19) and a GABAergic subpopulation (“caudal KF”). Our molecular framework also revealed novel, diverse subsets of KF neurons (Phox2b‐only, Phox2b+Lmx1b, FoxP2+Lmx1b, Lmx1b‐only; Figure 10). Cataloging all the genes that distinguish subsidiary PB subpopulations is not within the scope of this study, where our goal is to provide a molecular framework for interpreting such data. Nonetheless, to show the predictive advantages of a molecular ontology, we will highlight two further subpopulations within each PB macropopulation.
FIGURE 19.
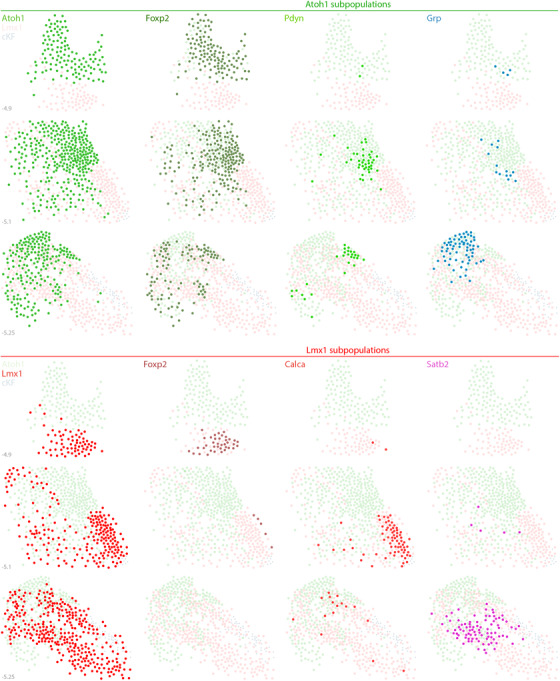
Distribution of Atoh1 and Lmx1 subpopulations. The PB contains two neuronal macropopulations, which derive from embryonic precursors expressing Atoh1 (green) or Lmx1 (Lmx1b and Lmx1a; red). This figure illustrates the rostral‐to‐caudal distribution of subsidiary populations within each macropopulation. Some genetic markers highlight a subset of just one macropopulation, exemplified by Pdyn and Grp (Atoh1 macropopulation) and by Calca and Satb2 (Lmx1 macropopulation). In contrast, Foxp2 identifies separate subsets within each macropopulation. Foxp2 is expressed by an extensive subset of Atoh1‐derived neurons (dark‐green dots) and by a more restricted, ventral subset of the Lmx1 macropopulation that is located rostrally, in the KF (dark‐red dots), separate from the caudal population of GABAergic FoxP2 neurons in the “caudal KF” (cKF, light blue dots). Approximate level caudal to bregma (in mm) is shown at bottom‐left for each of the three PB levels illustrated
First, the transcription factor Satb2 marks PB neurons that relay gustatory information to the thalamus (Fu et al., 2019; Huang et al., 2013; Jarvie et al., 2021; Maeda et al., 2009). We confirmed that both Satb2 mRNA (not shown) and Satb2 immunofluorescence (Figure 18a) identify a restricted distribution of PB neurons, primarily at caudal levels. Satb2‐expressing neurons distributed within and near fascicles of the superior cerebellar peduncle, spanning subregions homologous to the “medial,” “waist,” “ventral lateral,” and “inner PBeL” subnuclei in rats (Fulwiler & Saper, 1984). Satb2 and FoxP2 did not colocalize (not shown), nor did we find Satb2 in any Atoh1‐derived neurons (Figure 18d). Instead, every Satb2‐labeled neuron contained Lmx1b (Figure 18c,g). Therefore, gustatory relay neurons expressing Satb2 are a subset of the Lmx1 PB macropopulation (Figure 19).
FIGURE 18.
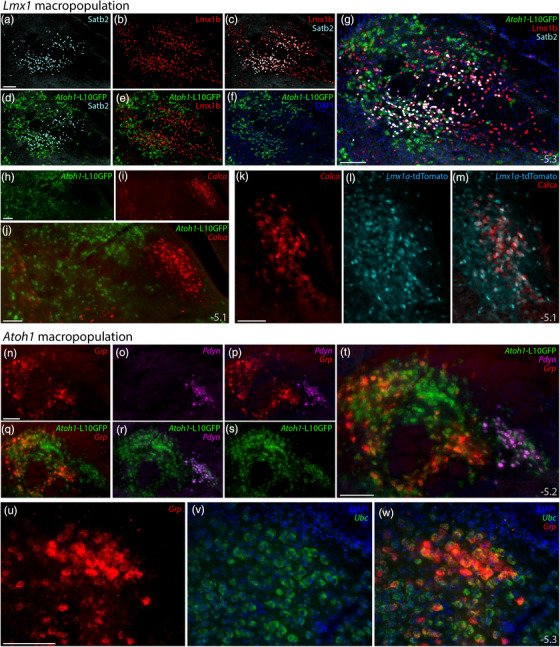
Subpopulations of Lmx1‐ and Atoh1‐derived PB neurons. Subsidiary genes define separate subpopulations within the Atoh1‐ and Lmx1‐derived PB macropopulations. (a–g) Immunofluorescence labeling for Satb2 (light blue) and Lmx1b (red) with L10GFP Cre‐reporter for Atoh1 (green) at a mid‐caudal level of the PB. (h–j) FISH labeling for Calca (red) with L10GFP Cre‐reporter for Atoh1 (green) at a mid‐level of the PB. (k–m) FISH labeling for Calca with tdTomato (pseudocolored ice blue) Cre‐reporter for Lmx1a in PBeL. (n–t) FISH labeling for Grp mRNA (red) and Pdyn mRNA (magenta) with L10GFP Cre‐reporter for Atoh1 (green) at a mid‐caudal level of the PB. (u–w) FISH labeling for Grp mRNA (red) in a dorsal cluster of larger neurons that resemble the “internal lateral” subnucleus of the rat PB (Fulwiler & Saper, 1984), with Ubc mRNA (green) shown for neuroanatomical background. Approximate bregma levels shown at bottom‐right in (g, j, m, t, w). All scale bars are 100 μm and apply to similar panels
Next, we examined the expression of Calca, which encodes the neuropeptide CGRP (calcitonin gene‐related peptide). Calca‐expressing PB neurons form a critical link in signaling pathways for conditioned taste aversion, malaise, and anorexia, plus alerting responses to aversive stimuli that include pain, itch, and hypercarbia (Carter et al., 2015; Chen et al., 2018; Kaur et al., 2017; Palmiter, 2018; Saper, 2016). We found Calca mRNA in the outer part of PBeL and in other neurons described previously (Huang et al., 2021), but not in any Atoh1‐derived neurons (Figure 18h–j), consistent with the mutual exclusivity between CGRP and FoxP2 in this region (Huang et al., 2021). Instead, labeling Calca mRNA in Lmx1a Cre‐reporter mice revealed extensive colocalization with tdTomato (Figure 18k–m), resembling the colocalization between CGRP and Lmx1b in both rats and mice (Huang et al., 2021; Miller et al., 2012). Therefore, like Satb2, Calca expression identifies a subset of the Lmx1 PB macropopulation (Figure 19).
We then identified two markers within the Atoh1 macropopulation. Labeling Pdyn mRNA, which encodes the neuropeptide dynorphin, identified a distribution resembling previous reports in rats and mice (Geerling et al., 2016; Hermanson et al., 1998; Huang et al., 2020; Miller et al., 2012). Pdyn expression in the mouse PB identified neurons in regions resembling the “dorsal lateral” (Figure 18r) and “central lateral” subnuclei in rats (Fulwiler & Saper, 1984), plus a caudal subset of Pdyn‐expressing neurons in the “pre‐LC” population that spans the medial PB and dorsal LC (see Gasparini et al., 2021), and all these neurons expressed L10GFP. Therefore, Pdyn‐expressing PB neurons are a subset of the larger Atoh1 macropopulation (Figure 19).
Another neuropeptide, gastrin‐releasing peptide (Grp), identified a separate subpopulation (Figure 18q). At mid‐caudal levels, Grp‐expressing neurons occupied a dorsomedial subregion, as reported in rats (Figure 6H of Wada et al., 1990), surrounding the “head” and “waist” of the superior cerebellar peduncle (Figure 18p). At mid‐levels of the PB, Grp identified a prominent, dorsal cluster (Figure 18u–w) resembling the rat “internal lateral” subnucleus (Fulwiler & Saper, 1984). Further rostrally, Grp‐expressing neurons concentrated near PBeL and expressed Foxp2 but not Calca (not shown). Overall, Grp expression identifies another, distinct subset within the Atoh1 PB macropopulation (Figure 19).
3.11. Genetic segregation of axonal trajectories and output targets
Embryonic transcription factors establish patterns of neurogenesis and axon growth needed for adult circuit functions. Connectivity patterns that define and distinguish adult neurons often reflect their separate developmental lineages, which are determined in large part by embryonic transcription factors (Hirsch et al., 2021). Thus, we hypothesized that macropopulation identity determines the trajectory and targets of axons projecting from the PB to distal brain regions.
First, to test whether developmental‐genetic identity predicts axonal trajectory, we compared axonal labeling in the midbrain, rostral to the PB, after Cre fate‐mapping with tdTomato. In Lmx1a Cre‐reporter mice, we found tdTomato labeling in fascicles of axons within the central tegmental tract (CTT), both in late‐stage embryos (E17.5, n = 2; not shown) and in adult mice (n = 2, Figure 20a). In every case, this fibrous tdTomato labeling in the CTT traced back to an origin in the PB (not shown). In contrast, the CTT was unlabeled in Atoh1 Cre‐reporter mice (n = 4). In these mice, we instead found fibrous tdTomato labeling in the superior cerebellar peduncle (arborizing extensively in the red nucleus) and in the medial lemniscus (arrowhead in Figure 20b), plus a contiguous band of axons lateral to the medial lemniscus, in‐between the red nucleus and substantia nigra. Lmx1a Cre‐reporter mice did not have any axonal labeling in this ventral pathway.
FIGURE 20.
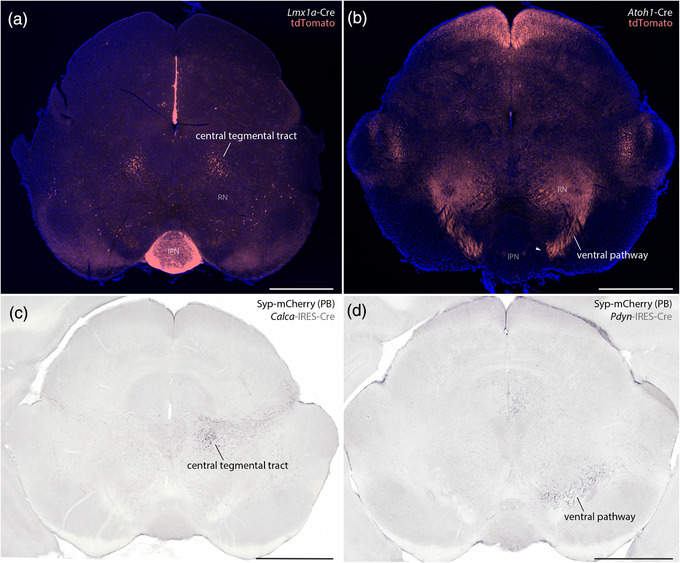
Axonal trajectories of Lmx1‐ and Atoh1‐derived PB neurons. (a) Lmx1a Cre‐reporter labeling in the midbrain (tdTomato, pseudocolored coral‐red). (b) Atoh1 Cre‐reporter labeling in the midbrain (tdTomato, pseudocolored coral‐red). (c) Nickel‐DAB immunolabeling for synaptophysin‐mCherry (Syp‐mCherry) in the midbrain of a Calca‐Cre mouse after injection of AAV‐DIO‐Syp‐mCherry into the PB. (d) Nickel‐DAB immunolabeling for Syp‐mCherry in the midbrain of a Pdyn‐IRES‐Cre mouse after injection of AAV‐DIO‐Syp‐mCherry into the PB. Abbreviations: IPN, interpeduncular nucleus; RN, red nucleus. All scale bars are 1 mm
These dichotomous developmental patterns matched patterns of axonal labeling we identified after transducing adult PB neurons that express Calca (Lmx1 macropopulation) or Pdyn (Atoh1 macropopulation). Our previous injections of AAV8‐DIO‐synaptophysin‐mCherry into the PB (Huang et al., 2020; Huang et al., 2021) produced axonal labeling in either the CTT (Calca cases; Figure 20c) or ventral pathway (Pdyn cases; Figure 20d). The strikingly similar results of developmental fate‐mapping and adult Cre‐conditional labeling support our hypothesis that macropopulation identity predicts the trajectory of PB axonal projections. Specifically, neurons in the Lmx1 PB macropopulation extend axons through the CTT (between the oculomotor nucleus and red nucleus), while neurons in the Atoh1 PB macropopulation extend axons through a ventral pathway (between the red nucleus and substantia nigra, alongside the medial lemniscus).
Next, to test whether developmental‐genetic identity also predicts the distal targets of these axonal projections, we combined Cre fate‐mapping with retrograde axonal tracing. In Atoh1‐Cre;R26‐lsl‐L10GFP mice, we injected a retrograde tracer (cholera toxin b subunit, CTb) into brain regions that receive heavy input from glutamatergic PB neurons (Huang et al., 2020). We then compared L10GFP expression (marking Atoh1‐derived neurons) with immunolabeling for Lmx1b and CTb.
After injections in the ventromedial hypothalamus (n = 6), retrogradely labeled neurons expressed L10GFP. Most of these neurons concentrated rostrally, in a dorsolateral subregion of the PB (Figure 21a), but their distribution also extended caudally within the lateral PB (not shown). After injections in the central nucleus of the amygdala (n = 2) or insular cortex (n = 1; not shown), retrogradely labeled PB neurons lacked L10GFP and instead contained Lmx1b. Many of these neurons concentrated in PBeL (Figure 21b), and their distribution extended caudally, across the superior cerebellar peduncle (not shown). These retrograde tracing results complement the anterograde labeling patterns we reported after transducing PB neurons that express either Calca or Pdyn (Huang et al., 2021) and support our hypothesis that neurons in separate Lmx1 and Atoh1 PB macropopulations project axons to separate target sites in the forebrain (Figure 22c). Of note, these findings do not imply that all neurons in the Atoh1 macropopulation project axons to the hypothalamus, or that all neurons in the Lmx1 macropopulation project axons to the amygdala, and further experiments are needed to determine the full array of brain regions targeted by neurons within each macropopulation.
FIGURE 21.
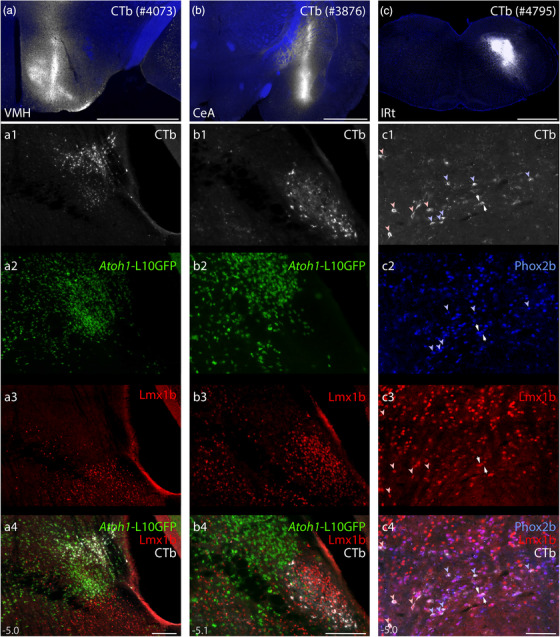
Distal targets of axonal projections from separate PB macropopulations. Neurons in the PB region contained retrograde CTb labeling after tracer injections into the ventromedial hypothalamic nucleus (VMH), central nucleus of the amygdala (CeA), and intermediate reticular formation (IRt). (a–c) CTb injection sites (white). (a1–a4) Immunofluorescence labeling for CTb (white) after tracer injection into the VMH along with L10GFP Cre‐reporter expression in Atoh1‐derived neurons and Lmx1b immunofluorescence (red) at a level approximately 5.0 mm caudal to bregma. (b1–b4) Immunofluorescence labeling for CTb after tracer injection into the CeA, along with L10GFP Cre‐reporter for Atoh1 and Lmx1b immunofluorescence (approximately 5.1 mm caudal to bregma). (c1–c4) Immunofluorescence labeling for CTb, Phox2b (blue), and Lmx1b along the ventral border of the PB. Light blue arrowheads indicate neurons that contain both CTb and Phox2b. Light red arrowheads indicate neurons that contain both CTb and Lmx1b. White arrows highlight neurons that contain CTb and both transcription factors. Approximate bregma levels are shown at bottom‐left in (a4, b4, c4). Scale bars in (a–c) are 1 mm. Scale bars in (a4–c4) are 200 μm and apply to panels above
FIGURE 22.
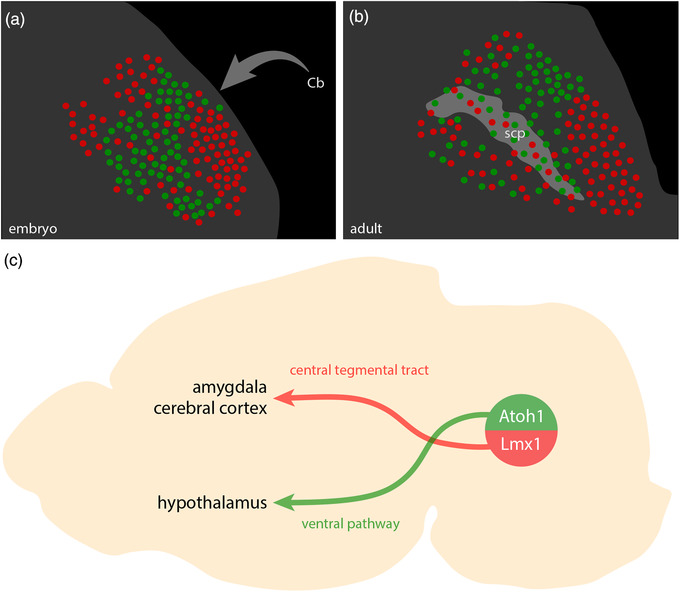
Summary. (a, b) Illustrated distribution of Atoh1 (green) and Lmx1 (red) PB neurons in late‐embryonic (a) and adult (b) mice. Cerebellar axons project through this region and form the scp, dispersing PB neurons. (c) PB neurons in separate Atoh1 and Lmx1 macropopulations project axons through separate pathways to separate forebrain targets. Note that neurons in each macropopulation project to additional brain regions (see Huang et al., 2021). Hindbrain projections from ventral populations are not shown
Ventral to the PB, we predicted that neurons expressing Phox2b and Lmx1b project axons to the hindbrain. Many neurons in this supratrigeminal region receive proprioceptive input from the trigeminal nerve and send output to cranial motor neurons and hindbrain interneurons (Li et al., 2005; Marfurt & Rajchert, 1991; Rokx et al., 1986; Stanek et al., 2016), and we found that glutamatergic neurons here project axons to the hindbrain reticular formation (Geerling et al., 2017; Huang et al., 2020). We also confirmed in mice that injecting CTb into the medullary reticular formation produces retrograde labeling in neurons along the ventral edge of the rostral PB (Huang et al., 2021). To test whether these neurons contain Phox2b or Lmx1b, we made additional CTb injections into the medullary reticular formation (n = 4; Figure 21c), then immunolabeled Phox2b, Lmx1b, and CTb. In these cases, CTb‐labeled neurons distributed along the ventral PB border. Many contained Phox2b (with or without Lmx1b), and some contained Lmx1b without Phox2b (Figure 21c 1–c4). Therefore, glutamatergic neurons expressing Phox2b or Lmx1b immediately ventral to the PB project axons to the hindbrain reticular formation.
4. DISCUSSION
Our findings lay the foundation for a new ontology that uses intrinsic, molecular information to identify PB neurons. This molecular ontology offers an observer‐independent alternative to taxonomies that are based on the position of a white matter tract that disperses PB neurons (Figure 22a,b). Molecular features blur some of the atlas boundaries inferred from Nissl cytoarchitecture and reveal an intermingled web of developmentally distinct macropopulations spanning the superior cerebellar peduncle, challenging its utility for subdividing the PB. Beyond laying the groundwork for a new way to classify PB neurons, the information presented here opens opportunities to improve our understanding of interoceptive neural circuit functions.
4.1. Practical implications, limitations, and opportunities
Our findings represent the first comprehensive framework for interpreting neuron‐type‐specific molecular information in the PB region. Adopting this developmental‐genetic framework will help investigators organize transcriptomic data and focus future attempts to identify and study PB neurons.
Transcriptomic data from an adult brainstem should be sufficient to classify glutamatergic neurons that express Lmx1a/b and other adult markers, but adult neurons do not express Atoh1 (Machold et al., 2011), so identifying Atoh1‐derived subpopulations that do not express Foxp2 may require a fate‐mapping strategy like the one used here. Among these Atoh1‐derived PB neurons that do not express Foxp2, we identified a large, Grp‐expressing subset. This Grp subset likely contains further subpopulations, prominently including a dorsal cluster resembling the “internal lateral” subnucleus in rats (Fulwiler & Saper, 1984; Paxinos & Watson, 2007). Neurons in this cluster relay pain‐related information from the spinal cord to the thalamus (Bernard et al., 1995; Bester et al., 1999; Bester et al., 1995; Bourgeais et al., 2001; Feil & Herbert, 1995; Kitamura et al., 1993; Krout & Loewy, 2000), but the term “internal lateral” was omitted from a major mouse brain atlas (Dong, 2008; Wang et al., 2020) and its location was added to a separate subnucleus (“superior lateral”), which had been distinguished in rats as sending axonal projections to the hypothalamus (Bester et al., 1997; Bester et al., 1995; Fulwiler & Saper, 1984). This new taxonomy––substituting “superior lateral” for “internal lateral” in mice––appeared in subsequent studies of neurons that relay noxious spinoparabrachial information to the thalamus (Barik et al., 2021; Deng et al., 2020). In contrast to many other findings that blur atlas boundaries, our results unambiguously support the cytoarchitectonic distinction of an “internal lateral” population (Fulwiler & Saper, 1984) by identifying these neurons as a Foxp2‐negative subset of the Atoh1 macropopulation. Except for a rostral subset that express Foxp2, Grp‐expressing neurons resemble the distribution of PB neurons that express the substance P receptor Tacr1 (Barik et al., 2021), which fail to develop in mice with Atoh1 deletion from the rostral rhombic lip (van der Heijden & Zoghbi, 2018). The Grp distribution also partly resembles the distribution of Penk‐expressing PB neurons, a subset of which project axons to the thalamus (Hermanson & Blomqvist, 1997). We are testing several additional markers (unpublished) and expect that single‐cell transcriptomic analysis will identify a larger suite of neuronal subpopulations within the Atoh1 macropopulation.
From a purely practical perspective, it is useful to classify neurons using developmental‐genetic features, rather than cytoarchitecture, because the resulting information can guide genetically targeted experiments that evaluate hypotheses about specific subpopulations. Until recently, studying the PB involved inserting an electrode, needle, micropipette, or cannula (to stimulate, inhibit, destroy, trace, or record neurons) and then using a cytoarchitectural taxonomy to correlate experimental results with histological location in each brain. Most injection sites and implants were larger than most PB subpopulations, which intermingle extensively, so these approaches simultaneously targeted subsets of PB neurons with distinct connections and functions. Now, genetic methods allow investigators to target more specific subpopulations, including an overlapping potpourri of neurons that express Satb2, Calca, Pdyn, Cck, Oxtr1, Htr2c, Oprm1, or Tacr1 (Barik et al., 2021; Chiang et al., 2020; Fu et al., 2019; Garfield et al., 2014; Jarvie et al., 2021; Kaur et al., 2017; Liu et al., 2021; Norris et al., 2021; Palmiter, 2018; Park et al., 2020; Ryan et al., 2017; Yang et al., 2020). Limiting the immense potential of this approach were the lack of a framework for understanding ontological relationships between subpopulations and a lack of markers for remaining PB neurons.
Now, learning that the PB contains two, genetically distinct macropopulations has immediate practical advantages. For example, investigators can rapidly focus the hunt for a subpopulation mediating a particular effect by separately ablating or stimulating each macropopulation to exclude one or the other half of all PB neurons. Even without a full and final menu of markers to distinguish every remaining subpopulation, combining our framework with existing information opens opportunities to access previously indistinguishable subsets by using intersectional genetic targeting methods (Branda & Dymecki, 2004; Fenno et al., 2014; Madisen et al., 2015; Weinholtz & Castle, 2021). As an example, retrograde axonal tracing identified two, distinct subsets of Cck‐expressing PB neurons: one sends output to the cerebral cortex, and the other to the hypothalamus (Grady et al., 2020). Using intersectional methods to target Cck‐expressing neurons derived from precursors expressing either Lmx1b (cortex‐projecting) or Atoh1 (hypothalamus‐projecting) should provide the selective access needed to isolate and test each subset. Intersectional methods will be particularly important for studying KF neurons (discussed below).
4.2. Comparison with previous neurodevelopmental literature
Our study is the first to identify the PB as a blend of two, developmentally distinct macropopulations. Previous descriptions of Lmx1a and Lmx1b here did not mention Atoh1 (Asbreuk et al., 2002; Dai et al., 2008; Miller et al., 2012; Sarropoulos et al., 2021; Zou et al., 2009), and initial studies reporting Atoh1‐derived PB neurons did not mention Lmx1a or Lmx1b (Machold & Fishell, 2005; Rose et al., 2009; van der Heijden & Zoghbi, 2018; Wang et al., 2005), leaving the ontological relationships between these populations unclear. One study reported that many PB neurons fail to develop in Atoh1‐null mice (Rose et al., 2009) and showed a loss of Slc17a6 (Vglut2) mRNA at a far‐rostral level without showing middle or caudal levels of the PB or labeling markers that might have identified neurons in the Lmx1 macropopulation. Another study reported that all Atoh1‐derived PB neurons contain CGRP (van der Heijden & Zoghbi, 2018), which is incompatible with the mutual exclusivity between Calca mRNA and Atoh1‐derived neurons (Figure 18h–j). This previous result likely involved off‐target immunoreactivity because there is little overlap between the distribution of Atoh1‐derived neurons and that of Calca mRNA, or of CGRP labeling produced by antisera validated in knockout mice (Huang et al., 2021).
In the embryonic (E14) brainstem, gene‐chip analysis identified Foxp2 expression in Atoh1‐derived neurons (Machold et al., 2011). In neonatal (P0) mice, another study identified FoxP2 immunoreactivity in Atoh1‐derived neurons that appear to be located in the lateral PB but were labeled “KF” (Figures 3 and 4 of Gray, 2008). Other images from the same study showed Lmx1b immunoreactivity in what appears to be the external lateral PB (labeled “Pr5” in Figure 3E of Gray, 2008), plus a ventrolateral cluster of FoxP2‐immunoreactive, Ptf1a‐derived neurons resembling the “caudal KF” (Figure 4A of Gray, 2008). Besides discrepant nomenclature, these results are consistent with our conclusion that adult Foxp2 expression identifies three, developmentally distinct subpopulations here: (1) a small, rostral subset of Lmx1‐derived glutamatergic neurons (within KF); (2) a large subset of Atoh1‐derived glutamatergic neurons (within PB); and (3) a small, ventrolateral cluster of Ptf1a‐derived GABAergic neurons (“caudal KF”).
4.3. Functional implications
Before genetic markers, several studies focused on PB neurons that we now identify as subsets within the Lmx1 macropopulation. Early work on gustatory pathways (Norgren & Leonard, 1971, 1973) paved the way for the discovery that Satb2‐expressing PB neurons relay taste information (Fu et al., 2019; Jarvie et al., 2021). Similarly, early work on PB neurons that relay viscerosensory information to the amygdala, bed nucleus of the stria terminalis, basal forebrain, thalamus, and cerebral cortex (Alden et al., 1994; Bernard et al., 1993; Bernard & Besson, 1990; de Lacalle & Saper, 2000; Karimnamazi & Travers, 1998; Knyihar‐Csillik et al., 1999; Yamamoto et al., 1992; Yamamoto et al., 1994; Yasui et al., 1989) led to the discovery that Calca‐expressing neurons are important for malaise, anorexia, and other aversive states (Campos et al., 2017; Carter et al., 2015; Carter et al., 2013; Chen et al., 2018; Kaur et al., 2017; Palmiter, 2018; Saper, 2016).
Several other studies focused on neurons we now identify within the Atoh1 macropopulation, including those that relay thermal (Geerling et al., 2016; Nakamura & Morrison, 2008, 2010) or noxious (Barik et al., 2021; Bester et al., 1995; Chiang et al., 2020; Deng et al., 2020) sensory information from the spinal cord to the forebrain. Based on their location, we predict that PB neurons relaying itch (Mu et al., 2017) also lie within the Atoh1 macropopulation. Rostrally, neurons that express Cck (Fulwiler & Saper, 1985; Grady et al., 2020; Hermanson et al., 1998; Zaborszky et al., 1984) and Runx1 (Zagami & Stifani, 2010) transmit noxious information from the spinal cord to the hypothalamus (Bester et al., 1995; Hermanson, Larhammar, et al., 1998). Fewer than half of these neurons contain FoxP2 (Garfield et al., 2014; Grady et al., 2020), but the entire Runx1 population fails to develop in Atoh1‐knockout mice (Zagami & Stifani, 2010).
A functional role remains to be determined for many PB neurons, including many of the neurons identified here as expressing Grp. Also, we do not yet know the ontological relationships of PB neurons expressing Oxtr1 (water intake), Htr2c (food, water, and salt intake), or Oprm1 (opioid‐withdrawal; opioid‐induced respiratory depression) relative to our developmental‐genetic framework. While investigating these and other genetic markers (unpublished), we expect that transcriptomic analysis will identify additional, functionally distinct subpopulations within each PB macropopulation.
An interesting question is whether Lmx1 and Atoh1 derivation confer separate functional themes. For example, a broadly interoceptive theme may befit the Lmx1 macropopulation. Its neurons overlap axon terminal fields that deliver viscerosensory information from the nucleus of the solitary tract (Geerling & Loewy, 2006b; Herbert et al., 1990; Rinaman, 2010) and an oral‐sensory subregion of the spinal trigeminal nucleus (Dallel et al., 2004). We showed here that the Lmx1 macropopulation includes Satb2 gustatory‐relay neurons, as well as Calca neurons that activate in response to visceral stimuli, and KF neurons, which receive chemosensory and other viscerosensory input and exert visceral motor effects on airway tone, breathing, and sympathetic function (Chamberlin & Saper, 1992, 1994; Dutschmann et al., 2021; Dutschmann & Herbert, 2006; Yokota et al., 2015).
In contrast, a broadly exteroceptive theme may befit the Atoh1 macropopulation. Its neurons overlap axon terminal fields that deliver input from the spinal cord and spinal trigeminal nucleus (Roome et al. 2020; Bernard et al., 1995; Blomqvist et al., 1989; Cechetto et al., 1985; Feil & Herbert, 1995; Kitamura et al., 1993; Panneton & Burton, 1985), and it includes subpopulations that relay pain, temperature, and likely also itch information to the forebrain (Barik et al., 2021; Bester et al., 1995; Bourgeais et al., 2001; Deng et al., 2020; Geerling et al., 2016; Mu et al., 2017; Nakamura & Morrison, 2008, 2010; Norris et al., 2021).
However, there may be exceptions to these broad functional themes. Caudal, Atoh1‐derived neurons that express Pdyn and Foxp2 may have appetitive, rather than exteroceptive functions (Gasparini et al., 2021; Kim et al., 2020; Lee et al., 2019). And while we do not yet know the connectivity and function of the Atoh1‐derived, Grp‐expressing neurons that surround the caudal “head” of the superior cerebellar peduncle, this part of the PB may receive primarily interoceptive input (Feil & Herbert, 1995; Herbert et al., 1990). Also, previous investigators highlighted neurons in the lateral PB that receive pain‐related inputs from both the viscera and the body surface and send output to both the amygdala and hypothalamus (Bernard & Besson, 1990; Bernard et al., 1994; Bourgeais et al., 2003; Chiang et al., 2020; Gauriau & Bernard, 2002). While our neuron‐type‐specific axonal tracing revealed a sharp, developmental distinction between PB neurons that send output to the amygdala (Lmx1) versus hypothalamus (Atoh1), additional work is needed to test whether interoceptive and exteroceptive input connections to this region segregate similarly. Intersectional genetic and viral targeting methods will help address this question to advance our understanding of the neural pathways transmitting specific channels of interoceptive and exteroceptive information to the brain. This information is necessary for understanding pain, alimentary function, and several other homeostatic functions.
4.4. Novel insights into the Kölliker–Fuse nucleus
Our most unexpected finding was that intrinsic, molecular features highlight the KF as a diverse blend of overlapping populations. Having identified the colocalization of Lmx1b and FoxP2 here in rats (Miller et al., 2012) and having distinguished these rostral, glutamatergic neurons from GABAergic neurons in the “caudal KF” (Geerling et al., 2017), we expected to find a homogenous KF population. Instead, without imposing any cytoarchitectonic boundaries or other constraints, molecular markers highlighted this region as a focally diverse cluster of intermingled neurons. Specifically, immunolabeling Lmx1b, Phox2b, and FoxP2 revealed that the KF contains at least four different populations of glutamatergic neurons (Figure 10a, d–g), separate from the “caudal KF” population of GABAergic, Foxp2‐expressing neurons. None of these populations were identified using cytoarchitectural criteria.
Also, while previous reports suggested that KF neurons are Atoh1‐derived (Gray, 2008; van der Heijden & Zoghbi, 2018), we found no more than sparse Atoh1 reporter expression ventrolateral to the PB. Instead, most neurons in the KF region express Lmx1b (alone, or with Foxp2). This Lmx1b‐expressing majority intermingles with a substantial minority expressing Phox2b (alone, or with Lmx1b). That is, KF neurons express either Lmx1b or Phox2b, both of which are absent from Atoh1‐derived neurons. This information, combined with our discovery that many KF neurons derive from Lmx1a‐expressing precursors, challenges previous claims that KF neurons derive from Atoh1‐expressing precursors in the rhombic lip (Gray, 2008; van der Heijden & Zoghbi, 2018). These claims may have resulted from mislocalizing the KF in mice, which skews rostrally relative to rats (Geerling et al., 2017; Yokota et al., 2015).
Discovering that “the” KF encompasses several populations opens opportunities to test which of these populations are responsible for specific respiratory, orofacial, and autonomic activities associated with this region (Chamberlin & Saper, 1992, 1994; Dutschmann & Dick, 2012; Stanek et al., 2016; Varga et al., 2020). Specifically, intersectional genetic targeting methods should help distinguish the functions of KF neurons that express Lmx1b (with and without Foxp2) or Phox2b (with and without Lmx1b).
Foxp2‐expressing neurons within the Lmx1 macropopulation are unique to the KF and caudally contiguous “lateral crescent” (Figure 19), so these neurons probably mediate a function exclusive to the KF region. In contrast, Phox2b‐expressing KF neurons are contiguous with many similar neurons extending back through the supratrigeminal nucleus and hindbrain reticular formation. It is not yet clear whether the functional role of Phox2b‐expressing KF neurons diverges from that of Phox2b‐expressing supratrigeminal neurons or the contiguous “dA3” population of Phox2b‐expressing interneurons in the hindbrain reticular formation (Gray, 2013; Hernandez‐Miranda et al., 2017; Kang et al., 2007). Many supratrigeminal neurons send output to cranial motor neurons that pattern movements like chewing, licking, and swallowing (Dempsey et al., 2021; Takatoh et al., 2021; Travers & Norgren, 1983), and in mice, the Phox2b distribution overlaps the locations of neurons labeled by viral retrograde tracing from motor neurons that control orofacial movements (Takatoh et al., 2021). In contrast, very few Phox2b‐expressing neurons project axons to autonomic and respiratory premotor neurons in the ventrolateral medulla in rats (Kang et al., 2007). These observations suggest that Phox2b‐expressing KF neurons have cephalic premotor functions similar to premotor neurons in the supratrigeminal region and reticular formation and distinct from the respiratory activities typically associated with the KF.
A more distinguishing characteristic of KF neurons is their ability to disrupt the respiratory rhythm (Chamberlin & Saper, 1994; Dutschmann & Herbert, 1996, 2006; Lumsden, 1923; Marckwald, 1887). The extensive overlap of Phox2b‐expressing neurons here suggests that these (putatively orofacial premotor) neurons work in concert with (putatively separate respiratory premotor) neurons that coordinate breathing with speech or swallowing, both of which require precisely timed perturbations of the respiratory cycle. If the Foxp2 subset of Lmx1b‐expressing KF neurons coordinates lingual and orofacial movements (articulation) or laryngeal movements (phonation) with respiration, then their developmental dysfunction could underlie the orofacial apraxia and speech dyspraxia caused by hemizygous inactivating mutations of FOXP2 (Fisher et al., 1998; Lai et al., 2001; Shriberg et al., 2006; Stanic et al., 2018).
Also relevant to human health is the role of this region in opioid‐induced respiratory depression (Prkic et al., 2012; Varga et al., 2020). Slice recordings identified opioid‐sensitive neurons in or near the KF (Levitt et al., 2015; Levitt & Williams, 2018; Saunders & Levitt, 2020), yet expression of the mu opioid receptor (Oprm1) is prominent in Calca‐expressing and intermingled neurons located dorsal and caudal to the KF (Chamberlin et al., 1999; Huang et al., 2021). Intersectional genetic targeting methods should allow investigators to determine whether the Lmx1b neurons expressing Foxp2 (KF) or Calca (PB) contribute to opioid‐induced respiratory depression.
5. CONCLUSION
Developmental‐genetic markers identify the PB as a blend of two, mutually exclusive macropopulations. These two macropopulations, defined by Lmx1b and Atoh1, communicate with separate neural circuits. We also found that Lmx1b (with and without Foxp2) and Phox2b (with and without Lmx1b) identify neuronal subpopulations in the KF. This new, developmental‐genetic framework will help organize future transcriptomic and experimental work involving PB and KF neurons, and using a molecular ontology to identify and compare neurons across species may accelerate the translation of PB‐related discoveries from experimental animals to human patients.
AUTHOR CONTRIBUTION
JG planned experiments and supervised the project. JG and RM performed histologic staining and confocal microscopy in rat brain tissue. CM performed confocal microscopy and image processing in rat brain tissue. LP, JL, and II bred and perfused Cre‐reporter mice. LP, RM, and JL performed stereotaxic injections. LP, GI, SK, FG, JL, RM, and JG performed histologic staining and microscopy. BF, II, and VC contributed mice (BF), shared mouse brainstem tissue for Cre fate‐mapping (II and VC), and provided critical guidance and feedback regarding brainstem development. RM and JG plotted neurons in rats. SK and YD plotted neurons in mice. SK and JG analyzed data. SK, JG, and DH drafted and edited all figures. SK and JG wrote the manuscript. All authors reviewed and discussed the results and contributed critical feedback and edits that were incorporated into the final manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/cne.25307
ACKNOWLEDGMENTS
We thank Yu‐Qiang Ding for providing an aliquot of rabbit‐anti‐Lmx1b antibody (used in rats), Carmen Birchmeier for providing aliquots of guinea pig‐anti‐Lmx1b antiserum (used in mice), and Hideki Enomoto of Kobe University for providing an aliquot of guinea pig‐anti‐Phox2b antiserum (used in mice). We thank Kathleen Millen for sharing Lmx1a‐Cre mice. We also thank Aislinn Williams for sharing Atoh1‐Cre and Andy Russo for sharing (and Richard Palmiter for his permission to use) Calca‐Cre mice from their breeding colonies. Finally, we thank Jadylin Tolda for proofreading the manuscript.
Karthik, S. , Huang, D. , Delgado, Y. , Laing, J. J. , Peltekian, L. , Iverson, G. N. , Grady, F. , Miller, R. L. , McCann, C. M. , Fritzsch, B. , Iskusnykh, I. Y. , Chizhikov, V. V. , & Geerling, J. C. (2022). Molecular ontology of the parabrachial nucleus. Journal of Comparative Neurology, 530, 1658–1699. 10.1002/cne.25307
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Alden, M. , Besson, J. M. , & Bernard, J. F. (1994). Organization of the efferent projections from the pontine parabrachial area to the bed nucleus of the stria terminalis and neighboring regions: A PHA‐L study in the rat. Journal of Comparative Neurology, 341(3), 289–314. 10.1002/cne.903410302 [DOI] [PubMed] [Google Scholar]
- Asbreuk, C. H. , Vogelaar, C. F. , Hellemons, A. , Smidt, M. P. , & Burbach, J. P. (2002). CNS expression pattern of Lmx1b and coexpression with ptx genes suggest functional cooperativity in the development of forebrain motor control systems. Molecular and Cellular Neuroscience, 21(3), 410–420. 10.1006/mcne.2002.1182 [DOI] [PubMed] [Google Scholar]
- Barik , A. , Sathyamurthy, A. , Thompson, J. , Seltzer, M. , Levine, A. , & Chesler, A. (2021). A spinoparabrachial circuit defined by Tacr1 expression drives pain. eLife, 10. 10.7554/eLife.61135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Arie, N. , Bellen, H. J. , Armstrong, D. L. , McCall, A. E. , Gordadze, P. R. , Guo, Q. , Matzuk, M. M. , & Zoghbi, H. Y. (1997). Math1 is essential for genesis of cerebellar granule neurons. Nature, 390(6656), 169–172. 10.1038/36579 [DOI] [PubMed] [Google Scholar]
- Bermingham, N. A. , Hassan, B. A. , Wang, V. Y. , Fernandez, M. , Banfi, S. , Bellen, H. J. , Fritzsch, B. , & Zoghbi, H. Y. (2001). Proprioceptor pathway development is dependent on Math1. Neuron, 30(2), 411–422. 10.1016/s0896-6273(01)00305-1 [DOI] [PubMed] [Google Scholar]
- Bernard, J. F. , Alden, M. , & Besson, J. M. (1993). The organization of the efferent projections from the pontine parabrachial area to the amygdaloid complex: A Phaseolus vulgaris leucoagglutinin (PHA‐L) study in the rat. Journal of Comparative Neurology, 329(2), 201–229. 10.1002/cne.903290205 [DOI] [PubMed] [Google Scholar]
- Bernard, J. F. , & Besson, J. M. (1990). The spino(trigemino)pontoamygdaloid pathway: Electrophysiological evidence for an involvement in pain processes. Journal of Neurophysiology, 63(3), 473–490. 10.1152/jn.1990.63.3.473 [DOI] [PubMed] [Google Scholar]
- Bernard, J. F. , Dallel, R. , Raboisson, P. , Villanueva, L. , & Le Bars, D. (1995). Organization of the efferent projections from the spinal cervical enlargement to the parabrachial area and periaqueductal gray: A PHA‐L study in the rat. Journal of Comparative Neurology, 353(4), 480–505. 10.1002/cne.903530403 [DOI] [PubMed] [Google Scholar]
- Bernard, J. F. , Huang, G. F. , & Besson, J. M. (1994). The parabrachial area: Electrophysiological evidence for an involvement in visceral nociceptive processes. Journal of Neurophysiology, 71(5), 1646–1660. 10.1152/jn.1994.71.5.1646 [DOI] [PubMed] [Google Scholar]
- Bester, H. , Besson, J. M. , & Bernard, J. F. (1997). Organization of efferent projections from the parabrachial area to the hypothalamus: A Phaseolus vulgaris‐leucoagglutinin study in the rat. Journal of Comparative Neurology, 383(3), 245–281. 10.1002/(SICI)1096-9861(19970707)383:3%3c245::AID‐CNE1%3e3.0.CO;2‐3 [DOI] [PubMed] [Google Scholar]
- Bester, H. , Bourgeais, L. , Villanueva, L. , Besson, J. M. , & Bernard, J. F. (1999). Differential projections to the intralaminar and gustatory thalamus from the parabrachial area: A PHA‐L study in the rat. Journal of Comparative Neurology, 405(4), 421–449. 10.1002/(SICI)1096-9861(19990322)405:4%3c421::AID‐CNE1%3e3.0.CO;2‐2 [DOI] [PubMed] [Google Scholar]
- Bester, H. , Menendez, L. , Besson, J. M. , & Bernard, J. F. (1995). Spino (trigemino) parabrachiohypothalamic pathway: Electrophysiological evidence for an involvement in pain processes. Journal of Neurophysiology, 73(2), 568–585. 10.1152/jn.1995.73.2.568 [DOI] [PubMed] [Google Scholar]
- Block, C. H. , & Estes, M. L. (1990). The cytoarchitectural organization of the human parabrachial nuclear complex. Brain Research Bulletin, 24(4), 617–626. 10.1016/0361-9230(90)90168-y [DOI] [PubMed] [Google Scholar]
- Blomqvist, A. , Ma, W. , & Berkley, K. J. (1989). Spinal input to the parabrachial nucleus in the cat. Brain Research, 480(1–2), 29–36. 10.1016/0006-8993(89)91563-1 [DOI] [PubMed] [Google Scholar]
- Bota, M. , & Swanson, L. W. (2008). BAMS neuroanatomical ontology: Design and implementation. Frontiers in Neuroinformatics, 2, 2. 10.3389/neuro.11.002.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeais, L. , Gauriau, C. , Monconduit, L. , Villanueva, L. , & Bernard, J. F. (2003). Dendritic domains of nociceptive‐responsive parabrachial neurons match terminal fields of lamina I neurons in the rat. Journal of Comparative Neurology, 464(2), 238–256. 10.1002/cne.10793 [DOI] [PubMed] [Google Scholar]
- Bourgeais, L. , Monconduit, L. , Villanueva, L. , & Bernard, J. F. (2001). Parabrachial internal lateral neurons convey nociceptive messages from the deep laminas of the dorsal horn to the intralaminar thalamus. Journal of Neuroscience, 21(6), 2159–2165. 10.1523/JNEUROSCI.21-06-02159.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda, C. S. , & Dymecki, S. M. (2004). Talking about a revolution: The impact of site‐specific recombinases on genetic analyses in mice. Developmental Cell, 6(1), 7–28. 10.1016/s1534-5807(03)00399-x [DOI] [PubMed] [Google Scholar]
- Campos, C. A. , Bowen, A. J. , Han, S. , Wisse, B. E. , Palmiter, R. D. , & Schwartz, M. W. (2017). Cancer‐induced anorexia and malaise are mediated by CGRP neurons in the parabrachial nucleus. Nature Neuroscience, 20(7), 934–942. 10.1038/nn.4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M. E. , Han, S. , & Palmiter, R. D. (2015). Parabrachial calcitonin gene‐related peptide neurons mediate conditioned taste aversion. Journal of Neuroscience, 35(11), 4582–4586. 10.1523/JNEUROSCI.3729-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M. E. , Soden, M. E. , Zweifel, L. S. , & Palmiter, R. D. (2013). Genetic identification of a neural circuit that suppresses appetite. Nature, 503(7474), 111–114. 10.1038/nature12596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cechetto, D. F. , Standaert, D. G. , & Saper, C. B. (1985). Spinal and trigeminal dorsal horn projections to the parabrachial nucleus in the rat. Journal of Comparative Neurology, 240(2), 153–160. 10.1002/cne.902400205 [DOI] [PubMed] [Google Scholar]
- Chamberlin, N. L. , Mansour, A. , Watson, S. J. , & Saper, C. B. (1999). Localization of mu‐opioid receptors on amygdaloid projection neurons in the parabrachial nucleus of the rat. Brain Research, 827(1–2), 198–204. 10.1016/s0006-8993(99)01168-3 [DOI] [PubMed] [Google Scholar]
- Chamberlin, N. L. , & Saper, C. B. (1992). Topographic organization of cardiovascular responses to electrical and glutamate microstimulation of the parabrachial nucleus in the rat. Journal of Comparative Neurology, 326(2), 245–262. 10.1002/cne.903260207 [DOI] [PubMed] [Google Scholar]
- Chamberlin, N. L. , & Saper, C. B. (1994). Topographic organization of respiratory responses to glutamate microstimulation of the parabrachial nucleus in the rat. Journal of Neuroscience, 14(11 Pt 1), 6500–6510. 10.1523/JNEUROSCI.14-11-06500.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Lun, Y. , Ovchinnikov, D. , Kokubo, H. , Oberg, K. C. , Pepicelli, C. V. , Can, L. , Lee, B. , & Johnson, R. L. (1998). Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nature Genetics, 19(1), 51–55. 10.1038/ng0598-51 [DOI] [PubMed] [Google Scholar]
- Chen, J. Y. , Campos, C. A. , Jarvie, B. C. , & Palmiter, R. D. (2018). Parabrachial CGRP neurons establish and sustain aversive taste memories. Neuron, 100(4), 891–899.e895. 10.1016/j.neuron.2018.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, M. C. , Bowen, A. , Schier, L. A. , Tupone, D. , Uddin, O. , & Heinricher, M. M. (2019). Parabrachial complex: A hub for pain and aversion. Journal of Neuroscience, 39(42), 8225–8230. 10.1523/JNEUROSCI.1162-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, M. C. , Nguyen, E. K. , Canto‐Bustos, M. , Papale, A. E. , Oswald, A. M. , & Ross, S. E. (2020). Divergent neural pathways emanating from the lateral parabrachial nucleus mediate distinct components of the pain response. Neuron, 106(6), 927–939.e5. 10.1016/j.neuron.2020.03.014 [DOI] [PubMed] [Google Scholar]
- Chizhikov, V. V. , Iskusnykh, I. Y. , Fattakhov, N. , & Fritzsch, B. (2021). Lmx1a and Lmx1b are redundantly required for the development of multiple components of the mammalian auditory system. Neuroscience, 452, 247–264. 10.1016/j.neuroscience.2020.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov, V. V. , Lindgren, A. G. , Currle, D. S. , Rose, M. F. , Monuki, E. S. , & Millen, K. J. (2006). The roof plate regulates cerebellar cell‐type specification and proliferation. Development (Cambridge, England), 133(15), 2793–2804. 10.1242/dev.02441 [DOI] [PubMed] [Google Scholar]
- Chizhikov, V. V. , Lindgren, A. G. , Mishima, Y. , Roberts, R. W. , Aldinger, K. A. , Miesegaes, G. R. , Currle, D. S. , Monuki, E. S. , & Millen, K. J. (2010). Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proceedings of the National Academy of Sciences of the United States of America, 107(23), 10725–10730. 10.1073/pnas.0910786107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coizet, V. , Dommett, E. J. , Klop, E. M. , Redgrave, P. , & Overton, P. G. (2010). The parabrachial nucleus is a critical link in the transmission of short latency nociceptive information to midbrain dopaminergic neurons. Neuroscience, 168(1), 263–272. 10.1016/j.neuroscience.2010.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, C. , Harding, B. , & Copp, A. J. (2001). Neuronal migration defects in the Dreher (Lmx1a) mutant mouse: Role of disorders of the glial limiting membrane. Cerebral Cortex, 11(6), 498–505. 10.1093/cercor/11.6.498 [DOI] [PubMed] [Google Scholar]
- Dai, J. X. , Hu, Z. L. , Shi, M. , Guo, C. , & Ding, Y. Q. (2008). Postnatal ontogeny of the transcription factor Lmx1b in the mouse central nervous system. Journal of Comparative Neurology, 509(4), 341–355. 10.1002/cne.21759 [DOI] [PubMed] [Google Scholar]
- Dallel, R. , Ricard, O. , & Raboisson, P. (2004). Organization of parabrachial projections from the spinal trigeminal nucleus oralis: An anterograde tracing study in the rat. Journal of Comparative Neurology, 470(2), 181–191. 10.1002/cne.11043 [DOI] [PubMed] [Google Scholar]
- de Lacalle, S. , & Saper, C. B. (2000). Calcitonin gene‐related peptide‐like immunoreactivity marks putative visceral sensory pathways in human brain. Neuroscience, 100(1), 115–130. 10.1016/s0306-4522(00)00245-1 [DOI] [PubMed] [Google Scholar]
- Dempsey , B. , Sungeelee, S. , Bokiniec, P. , Chettouh, Z. , Diem, S. , Autran, S. , & Brunet, J.‐F. (2021). A medullary centre for lapping in mice. Nature communications, 12(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J. , Zhou, H. , Lin, J.‐K. , Shen, Z.‐X. , Chen, W.‐Z. , Wang, L.‐H. , Li, Q. , Mu, D. , Wei, Y.‐C. , Xu, X.‐H. , & Sun, Y.‐G. (2020). The parabrachial nucleus directly channels spinal nociceptive signals to the intralaminar thalamic nuclei, but not the amygdala. Neuron, 107(5), 909–923.e906. 10.1016/j.neuron.2020.06.017 [DOI] [PubMed] [Google Scholar]
- Ding, Y. Q. , Yin, J. , Kania, A. , Zhao, Z. Q. , Johnson, R. L. , & Chen, Z. F. (2004). Lmx1b controls the differentiation and migration of the superficial dorsal horn neurons of the spinal cord. Development (Cambridge, England), 131(15), 3693–3703. 10.1242/dev.01250 [DOI] [PubMed] [Google Scholar]
- Dong, H. W. (2008). Allen reference atlas: A digital color brain atlas of the C57Black/6J male mouse. Hoboken, NJ: Wiley. [Google Scholar]
- Dreyer, S. D. , Zhou, G. , Baldini, A. , Winterpacht, A. , Zabel, B. , Cole, W. , Johnson, R. L. , & Lee, B. (1998). Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nature Genetics, 19(1), 47–50. 10.1038/ng0598-47 [DOI] [PubMed] [Google Scholar]
- Dutschmann, M. , Bautista, T. G. , Trevizan‐Bau, P. , Dhingra, R. R. , & Furuya, W. I. (2021). The pontine Kolliker–Fuse nucleus gates facial, hypoglossal, and vagal upper airway related motor activity. Respiratory Physiology & Neurobiology, 284, 103563. 10.1016/j.resp.2020.103563 [DOI] [PubMed] [Google Scholar]
- Dutschmann, M. , & Dick, T. E. (2012). Pontine mechanisms of respiratory control. Comprehensive Physiology, 2(4), 2443–2469. 10.1002/cphy.c100015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutschmann, M. , & Herbert, H. (1996). The Kolliker–Fuse nucleus mediates the trigeminally induced apnoea in the rat. Neuroreport, 7(8), 1432–1436. 10.1097/00001756-199605310-00022 [DOI] [PubMed] [Google Scholar]
- Dutschmann, M. , & Herbert, H. (2006). The Kolliker–Fuse nucleus gates the postinspiratory phase of the respiratory cycle to control inspiratory off‐switch and upper airway resistance in rat. European Journal of Neuroscience, 24(4), 1071–1084. 10.1111/j.1460-9568.2006.04981.x [DOI] [PubMed] [Google Scholar]
- Feil, K. , & Herbert, H. (1995). Topographic organization of spinal and trigeminal somatosensory pathways to the rat parabrachial and Kolliker–Fuse nuclei. Journal of Comparative Neurology, 353(4), 506–528. 10.1002/cne.903530404 [DOI] [PubMed] [Google Scholar]
- Fenno, L. E. , Mattis, J. , Ramakrishnan, C. , Hyun, M. , Lee, S. Y. , He, M. , Tucciarone, J. , Selimbeyoglu, A. , Berndt, A. , Grosenick, L. , Zalocusky, K. A. , Bernstein, H. , Swanson, H. , Perry, C. , Diester, I. , Boyce, F. M. , Bass, C. E. , Neve, R. , Huang, Z. J. , & Deisseroth, K. (2014). Targeting cells with single vectors using multiple‐feature Boolean logic. Nature Methods, 11(7), 763–772. 10.1038/nmeth.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, S. E. , Vargha‐Khadem, F. , Watkins, K. E. , Monaco, A. P. , & Pembrey, M. E. (1998). Localisation of a gene implicated in a severe speech and language disorder. Nature Genetics, 18(2), 168–170. 10.1038/ng0298-168 [DOI] [PubMed] [Google Scholar]
- Fu, O. , Iwai, Y. , Kondoh, K. , Misaka, T. , Minokoshi, Y. , & Nakajima, K. I. (2019). SatB2‐expressing neurons in the parabrachial nucleus encode sweet taste. Cell Reports, 27(6), 1650–1656 e1654. 10.1016/j.celrep.2019.04.040 [DOI] [PubMed] [Google Scholar]
- Fulwiler, C. E. , & Saper, C. B. (1984). Subnuclear organization of the efferent connections of the parabrachial nucleus in the rat. Brain Research, 319(3), 229–259. 10.1016/0165-0173(84)90012-2 [DOI] [PubMed] [Google Scholar]
- Fulwiler, C. E. , & Saper, C. B. (1985). Cholecystokinin‐immunoreactive innervation of the ventromedial hypothalamus in the rat: Possible substrate for autonomic regulation of feeding. Neuroscience Letters, 53(3), 289–296. 10.1016/0304-3940(85)90553-1 [DOI] [PubMed] [Google Scholar]
- Garfield, A. S. , Shah, B. P. , Madara, J. C. , Burke, L. K. , Patterson, C. M. , Flak, J. , Neve, R. L. , Evans, M. L. , Lowell, B. B. , Myers, M. G. , & Heisler, L. K. (2014). A parabrachial‐hypothalamic cholecystokinin neurocircuit controls counterregulatory responses to hypoglycemia. Cell Metabolism, 20(6), 1030–1037. 10.1016/j.cmet.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini, S. , Resch, J. M. , Gore, A. M. , Peltekian, L. , & Geerling, J. C. (2021). Pre‐locus coeruleus neurons in rat and mouse. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 320(3), R342–R361. 10.1152/ajpregu.00261.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini, S. , Resch, J. M. , Narayan, S. V. , Peltekian, L. , Iverson, G. N. , Karthik, S. , & Geerling, J. C. (2019). Aldosterone‐sensitive HSD2 neurons in mice. Brain Structure and Function, 224(1), 387–417. 10.1007/s00429-018-1778-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauriau, C. , & Bernard, J. F. (2002). Pain pathways and parabrachial circuits in the rat. Experimental Physiology, 87(2), 251–258. 10.1113/eph8702357 [DOI] [PubMed] [Google Scholar]
- Geerling, J. C. , Kim, M. , Mahoney, C. E. , Abbott, S. B. G. , Agostinelli, L. J. , Garfield, A. S. , Krashes, M. J. , Lowell, B. B. , & Scammell, T. E. (2016). Genetic identity of thermosensory relay neurons in the lateral parabrachial nucleus. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 310(1), R41–R54. 10.1152/ajpregu.00094.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerling, J. C. , & Loewy, A. D. (2006a). Aldosterone‐sensitive neurons in the nucleus of the solitary tract: Bidirectional connections with the central nucleus of the amygdala. Journal of Comparative Neurology, 497(4), 646–657. 10.1002/cne.21019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerling, J. C. , & Loewy, A. D. (2006b). Aldosterone‐sensitive neurons in the nucleus of the solitary tract: Efferent projections. Journal of Comparative Neurology, 497(2), 223–250. 10.1002/cne.20993 [DOI] [PubMed] [Google Scholar]
- Geerling, J. C. , & Loewy, A. D. (2007). Sodium deprivation and salt intake activate separate neuronal subpopulations in the nucleus of the solitary tract and the parabrachial complex. Journal of Comparative Neurology, 504(4), 379–403. 10.1002/cne.21452 [DOI] [PubMed] [Google Scholar]
- Geerling, J. C. , Shin, J. W. , Chimenti, P. C. , & Loewy, A. D. (2010). Paraventricular hypothalamic nucleus: Axonal projections to the brainstem. Journal of Comparative Neurology, 518(9), 1460–1499. 10.1002/cne.22283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerling, J. C. , Stein, M. K. , Miller, R. L. , Shin, J. W. , Gray, P. A. , & Loewy, A. D. (2011). FoxP2 expression defines dorsolateral pontine neurons activated by sodium deprivation. Brain Research, 1375, 19–27. 10.1016/j.brainres.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerling, J. C. , Yokota, S. , Rukhadze, I. , Roe, D. , & Chamberlin, N. L. (2017). Kolliker–Fuse GABAergic and glutamatergic neurons project to distinct targets. Journal of Comparative Neurology, 525(8), 1844–1860. 10.1002/cne.24164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia, M. , Rodella, L. , Petruccioli, M. G. , & Bianchi, R. (2000). The cytoarchitecture of the adult human parabrachial nucleus: A Nissl and Golgi study. Archives of Histology and Cytology, 63(5), 411–424. 10.1679/aohc.63.411 [DOI] [PubMed] [Google Scholar]
- Gong, R. , Xu, S. , Hermundstad, A. , Yu, Y. , & Sternson, S. M. (2020). Hindbrain double‐negative feedback mediates palatability‐guided food and water consumption. Cell, 182(6), 1589–1605.e22. 10.1016/j.cell.2020.07.031 [DOI] [PubMed] [Google Scholar]
- Grady, F. , Peltekian, L. , Iverson, G. , & Geerling, J. C. (2020). Direct parabrachial‐cortical connectivity. Cerebral Cortex, 30(9), 4811–4833. 10.1093/cercor/bhaa072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, P. A. (2008). Transcription factors and the genetic organization of brain stem respiratory neurons. Journal of Applied Physiology (1985), 104(5), 1513–1521. 10.1152/japplphysiol.01383.2007 [DOI] [PubMed] [Google Scholar]
- Gray, P. A. (2013). Transcription factors define the neuroanatomical organization of the medullary reticular formation. Frontiers in Neuroanatomy, 7, 7. 10.3389/fnana.2013.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery, R. W. (2002). On counting and counting errors. Journal of Comparative Neurology, 447(1), 1–7. 10.1002/cne.10221 [DOI] [PubMed] [Google Scholar]
- Guo, C. , Qiu, H.‐Y. , Huang, Y. , Chen, H. , Yang, R.‐Q. , Chen, S.‐D. , Johnson, R. L. , Chen, Z.‐F. , & Ding, Y.‐Q. (2007). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development (Cambridge, England), 134(2), 317–325. 10.1242/dev.02745 [DOI] [PubMed] [Google Scholar]
- Guo, C. , Qiu, H.‐Y. , Shi, M. , Huang, Y. , Johnson, R. L. , Rubinstein, M. , Chen, S.‐D. , & Ding, Y.‐Q. (2008). Lmx1b‐controlled isthmic organizer is essential for development of midbrain dopaminergic neurons. Journal of Neuroscience, 28(52), 14097–14106. 10.1523/JNEUROSCI.3267-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hamilton, D. J. , Shepherd, G. M. , Martone, M. E. , & Ascoli, G. A. (2012). An ontological approach to describing neurons and their relationships. Frontiers in Neuroinformatics, 6, 15. 10.3389/fninf.2012.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, H. , Moga, M. M. , & Saper, C. B. (1990). Connections of the parabrachial nucleus with the nucleus of the solitary tract and the medullary reticular formation in the rat. Journal of Comparative Neurology, 293(4), 540–580. 10.1002/cne.902930404 [DOI] [PubMed] [Google Scholar]
- Herbert, H. , & Saper, C. B. (1990). Cholecystokinin‐, galanin‐, and corticotropin‐releasing factor‐like immunoreactive projections from the nucleus of the solitary tract to the parabrachial nucleus in the rat. Journal of Comparative Neurology, 293(4), 581–598. 10.1002/cne.902930405 [DOI] [PubMed] [Google Scholar]
- Hermanson, O. , & Blomqvist, A. (1997). Preproenkephalin messenger RNA‐expressing neurons in the rat parabrachial nucleus: Subnuclear organization and projections to the intralaminar thalamus. Neuroscience, 81(3), 803–812. 10.1016/s0306-4522(97)00241-8 [DOI] [PubMed] [Google Scholar]
- Hermanson, O. , Larhammar, D. , & Blomqvist, A. (1998). Preprocholecystokinin mRNA‐expressing neurons in the rat parabrachial nucleus: Subnuclear localization, efferent projection, and expression of nociceptive‐related intracellular signaling substances. Journal of Comparative Neurology, 400(2), 255–270. 10.1002/(SICI)1096-9861(19981019)400:2%3c255::AID‐CNE7%3e3.0.CO;2‐8 [DOI] [PubMed] [Google Scholar]
- Hermanson, O. , Telkov, M. , Geijer, T. , Hallbeck, M. , & Blomqvist, A. (1998). Preprodynorphin mRNA‐expressing neurones in the rat parabrachial nucleus: Subnuclear localization, hypothalamic projections and colocalization with noxious‐evoked fos‐like immunoreactivity. European Journal of Neuroscience, 10(1), 358–367. 10.1046/j.1460-9568.1998.00021.x [DOI] [PubMed] [Google Scholar]
- Hernandez‐Miranda, L. R. , Müller, T. , & Birchmeier, C. (2017). The dorsal spinal cord and hindbrain: From developmental mechanisms to functional circuits. Developmental Biology, 432(1), 34–42. 10.1016/j.ydbio.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Herrick, C. J. (1905). The central gustatory paths in the brains of bony fishes. Journal of Comparative Neurology and Psychology, 15(5), 375–456. 10.1002/cne.920150503 [DOI] [Google Scholar]
- Hirsch, D. , Kohl, A. , Wang, Y. , & Sela‐Donenfeld, D. (2021). Axonal projection patterns of the dorsal interneuron populations in the embryonic hindbrain. Frontiers in Neuroanatomy, 15(112). 10.3389/fnana.2021.793161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, M. , Nakamura, S. , Mori, K. , Kawauchi, T. , Terao, M. , Nishimura, Y. V. , Fukuda, A. , Fuse, T. , Matsuo, N. , Sone, M. , Watanabe, M. , Bito, H. , Terashima, T. , Wright, C. V. E. , Kawaguchi, Y. , Nakao, K. , & Nabeshima, Y. (2005). Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron, 47(2), 201–213. 10.1016/j.neuron.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Huang, D. , Grady, F. S. , Peltekian, L. , & Geerling, J. C. (2020). Efferent projections of Vglut2, Foxp2 and Pdyn parabrachial neurons in mice. Journal of Comparative Neurology, 529(4), 657–693. 10.1002/cne.24975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. , Grady, F. S. , Peltekian, L. , Laing, J. J. , & Geerling, J. C. (2021). Efferent projections of CGRP/Calca‐expressing parabrachial neurons in mice. Journal of Comparative Neurology, 529(11), 2911–2957. 10.1002/cne.25136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Song, N. N. , Lan, W. , Hu, L. , Su, C. J. , Ding, Y. Q. , & Zhang, L. (2013). Expression of transcription factor Satb2 in adult mouse brain. Anatomical Record (Hoboken), 296(3), 452–461. 10.1002/ar.22656 [DOI] [PubMed] [Google Scholar]
- Jarvie, B. C. , Chen, J. Y. , King, H. O. , & Palmiter, R. D. (2021). Satb2 neurons in the parabrachial nucleus mediate taste perception. Nature Communication, 12(1), 224. 10.1038/s41467-020-20100-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvie, B. C. , & Palmiter, R. D. (2017). HSD2 neurons in the hindbrain drive sodium appetite. Nature Neuroscience, 20(2), 167–169. 10.1038/nn.4451 [DOI] [PubMed] [Google Scholar]
- Kang, B. J. , Chang, D. A. , MacKay, D. D. , West, G. H. , Moreira, T. S. , Takakura, A. C. , Gwilt, J. M. , Guyenet, P. G. , & Stornetta, R. L. (2007). Central nervous system distribution of the transcription factor Phox2b in the adult rat. Journal of Comparative Neurology, 503(5), 627–641. 10.1002/cne.21409 [DOI] [PubMed] [Google Scholar]
- Karimnamazi, H. , & Travers, J. B. (1998). Differential projections from gustatory responsive regions of the parabrachial nucleus to the medulla and forebrain. Brain Research, 813(2), 283–302. 10.1016/s0006-8993(98)00951-2 [DOI] [PubMed] [Google Scholar]
- Kaur, S. , Wang, J. L. , Ferrari, L. , Thankachan, S. , Kroeger, D. , Venner, A. , Lazarus, M. , Wellman, A. , Arrigoni, E. , Fuller, P. M. , & Saper, C. B. (2017). A genetically defined circuit for arousal from sleep during hypercapnia. Neuron, 96(5), 1153–1167 e1155. 10.1016/j.neuron.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. Y. , Heo, G. , Kim, M. , Kim, H. , Jin, J. A. , Kim, H.‐K. , Jung, S. , An, M. , Ahn, B. H. , Park, J. H. , Park, H.‐E. , Lee, M. , Lee, J. W. , Schwartz, G. J. , & Kim, S.‐Y. (2020). A neural circuit mechanism for mechanosensory feedback control of ingestion. Nature, 580(7803), 376–380. doi: 10.1038/s41586-020-2167-2 [DOI] [PubMed] [Google Scholar]
- Kim, K. K. , Adelstein, R. S. , & Kawamoto, S. (2009). Identification of neuronal nuclei (NeuN) as Fox‐3, a new member of the Fox‐1 gene family of splicing factors. Journal of Biological Chemistry, 284(45), 31052–31061. 10.1074/jbc.M109.052969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, T. , Yamada, J. , Sato, H. , & Yamashita, K. (1993). Cells of origin of the spinoparabrachial fibers in the rat: A study with fast blue and WGA‐HRP. Journal of Comparative Neurology, 328(3), 449–461. 10.1002/cne.903280310 [DOI] [PubMed] [Google Scholar]
- Knyihar‐Csillik, E. , Boncz, I. , Sary, G. , Nemcsok, J. , & Csillik, B. (1999). Parabrachial origin of calcitonin gene‐related peptide‐immunoreactive axons innervating Meynert's basal nucleus. Experimental Neurology, 157(2), 268–276. 10.1006/exnr.1999.7050 [DOI] [PubMed] [Google Scholar]
- Krout, K. E. , & Loewy, A. D. (2000). Parabrachial nucleus projections to midline and intralaminar thalamic nuclei of the rat. Journal of Comparative Neurology, 428(3), 475–494. 10.1002/1096-9861(20001218)428:3<475::aid‐cne6>3.0.co;2‐9 [DOI] [PubMed] [Google Scholar]
- Kuwamura, M. , Muraguchi, T. , Matsui, T. , Ueno, M. , Takenaka, S. , Yamate, J. , Kotani, T. , Kuramoto, T. , Guénet, J.‐L. , Kitada, K. , & Serikawa, T. (2005). Mutation at the Lmx1a locus provokes aberrant brain development in the rat. Brain Research Developmental Brain Research, 155(2), 99–106. 10.1016/j.devbrainres.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Lai, C. S. , Fisher, S. E. , Hurst, J. A. , Vargha‐Khadem, F. , & Monaco, A. P. (2001). A forkhead‐domain gene is mutated in a severe speech and language disorder. Nature, 413(6855), 519–523. 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- Larson, S. D. , & Martone, M. E. (2009). Ontologies for neuroscience: What are they and what are they good for? Frontiers in Neuroscience, 3(1), 60–67. 10.3389/neuro.01.007.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzi, A. M. , Ottaviani, G. , Rossi, L. , & Matturri, L. (2004). Cytoarchitectural organization of the parabrachial/Kolliker–Fuse complex in man. Brain & Development, 26(5), 316–320. 10.1016/j.braindev.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Lee, S. , Augustine, V. , Zhao, Y. , Ebisu, H. , Ho, B. , Kong, D. , & Oka, Y. (2019). Chemosensory modulation of neural circuits for sodium appetite. Nature, 568(7750), 93–97. 10.1038/s41586-019-1053-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, E. S. , Abdala, A. P. , Paton, J. F. , Bissonnette, J. M. , & Williams, J. T. (2015). mu opioid receptor activation hyperpolarizes respiratory‐controlling Kolliker–Fuse neurons and suppresses post‐inspiratory drive. Journal of Physiology, 593(19), 4453–4469. 10.1113/JP270822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, E. S. , & Williams, J. T. (2018). Desensitization and tolerance of Mu opioid receptors on pontine Kolliker–Fuse neurons. Molecular Pharmacology, 93(1), 8–13. 10.1124/mol.117.109603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. L. , Wu, S. X. , Tomioka, R. , Okamoto, K. , Nakamura, K. , Kaneko, T. , & Mizuno, N. (2005). Efferent and afferent connections of GABAergic neurons in the supratrigeminal and the intertrigeminal regions. An immunohistochemical tract‐tracing study in the GAD67‐GFP knock‐in mouse. Neuroscience Research, 51(1), 81–91. 10.1016/j.neures.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Li, M. M. , Madara, J. C. , Steger, J. S. , Krashes, M. J. , Balthasar, N. , Campbell, J. N. , Resch, J. M. , Conley, N. J. , Garfield, A. S. , & Lowell, B. B. (2019). The paraventricular hypothalamus regulates satiety and prevents obesity via two genetically distinct circuits. Neuron, 102(3), 653–667.e656. 10.1016/j.neuron.2019.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kim, D.‐I. , Oh, T. G. , Pao, G. M. , Kim, J.‐H. , Palmiter, R. D. , Banghart, M. R. , Lee, K.‐F. , Evans, R. M. , & Han, S. (2021). Neural basis of opioid‐induced respiratory depression and its rescue. Proceedings of the National Academy of Sciences of the United States of America, 118(23), e2022134118. 10.1073/pnas.2022134118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. R. , Shi, M. , Hu, Z. L. , Zheng, M. H. , Du, F. , Zhao, G. , & Ding, Y. Q. (2010). A refined map of early gene expression in the dorsal rhombomere 1 of mouse embryos. Brain Research Bulletin, 82(1–2), 74–82. 10.1016/j.brainresbull.2010.02.010 [DOI] [PubMed] [Google Scholar]
- Lumsden, T. (1923). Observations on the respiratory centres in the cat. Journal of Physiology, 57(3–4), 153–160. 10.1113/jphysiol.1923.sp002052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold, R. , & Fishell, G. (2005). Math1 is expressed in temporally discrete pools of cerebellar rhombic‐lip neural progenitors. Neuron, 48(1), 17–24. 10.1016/j.neuron.2005.08.028 [DOI] [PubMed] [Google Scholar]
- Machold, R. , Klein, C. , & Fishell, G. (2011). Genes expressed in Atoh1 neuronal lineages arising from the r1/isthmus rhombic lip. Gene Expression Patterns, 11(5–6), 349–359. 10.1016/j.gep.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L. , Garner, A. R. , Shimaoka, D. , Chuong, A. S. , Klapoetke, N. C. , Li, L. , van der Bourg, A. , Niino, Y. , Egolf, L. , Monetti, C. , Gu, H. , Mills, M. , Cheng, A. , Tasic, B. , Nguyen, T. N. , Sunkin, S. M. , Benucci, A. , Nagy, A. , Miyawaki, A. , …, & Zeng, H. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron, 85(5), 942–958. 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, N. , Onimura, M. , Ohmoto, M. , Inui, T. , Yamamoto, T. , Matsumoto, I. , & Abe, K. (2009). Spatial differences in molecular characteristics of the pontine parabrachial nucleus. Brain Research, 1296, 24–34. 10.1016/j.brainres.2009.07.098 [DOI] [PubMed] [Google Scholar]
- Marckwald, M. (1887). Die Athembewegungen und deren Innervation beim Kaninchen. Z Biology, 23, 149–283. [Google Scholar]
- Marfurt, C. F. , & Rajchert, D. M. (1991). Trigeminal primary afferent projections to “non‐trigeminal” areas of the rat central nervous system. Journal of Comparative Neurology, 303(3), 489–511. 10.1002/cne.903030313 [DOI] [PubMed] [Google Scholar]
- Menani, J. V. , De Luca, L. A. Jr. , & Johnson, A. K. (2014). Role of the lateral parabrachial nucleus in the control of sodium appetite. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 306(4), R201–R210. 10.1152/ajpregu.00251.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. L. , Knuepfer, M. M. , Wang, M. H. , Denny, G. O. , Gray, P. A. , & Loewy, A. D. (2012). Fos‐activation of FoxP2 and Lmx1b neurons in the parabrachial nucleus evoked by hypotension and hypertension in conscious rats. Neuroscience, 218, 110–125. 10.1016/j.neuroscience.2012.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millonig, J. H. , Millen, K. J. , & Hatten, M. E. (2000). The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature, 403(6771), 764–769. 10.1038/35001573 [DOI] [PubMed] [Google Scholar]
- Mishima, Y. , Lindgren, A. G. , Chizhikov, V. V. , Johnson, R. L. , & Millen, K. J. (2009). Overlapping function of Lmx1a and Lmx1b in anterior hindbrain roof plate formation and cerebellar growth. Journal of Neuroscience, 29(36), 11377–11384. 10.1523/JNEUROSCI.0969-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga, M. M. , Herbert, H. , Hurley, K. M. , Yasui, Y. , Gray, T. S. , & Saper, C. B. (1990). Organization of cortical, basal forebrain, and hypothalamic afferents to the parabrachial nucleus in the rat. Journal of Comparative Neurology, 295(4), 624–661. 10.1002/cne.902950408 [DOI] [PubMed] [Google Scholar]
- Morrison, S. F. , & Nakamura, K. (2019). Central mechanisms for thermoregulation. Annual Review of Physiology, 81, 285–308. 10.1146/annurev-physiol-020518-114546 [DOI] [PubMed] [Google Scholar]
- Mu, D. , Deng, J. , Liu, K.‐F. , Wu, Z.‐Y. , Shi, Y.‐F. , Guo, W.‐M. , Mao, Q.‐Q. , Liu, X.‐J. , Li, H. , & Sun, Y.‐G. (2017). A central neural circuit for itch sensation. Science, 357(6352), 695–699. 10.1126/science.aaf4918 [DOI] [PubMed] [Google Scholar]
- Nagashimada, M. , Ohta, H. , Li, C. , Nakao, K. , Uesaka, T. , Brunet, J.‐F. , Amiel, J. , Trochet, D. , Wakayama, T. , & Enomoto, H. (2012). Autonomic neurocristopathy‐associated mutations in PHOX2B dysregulate Sox10 expression. Journal of Clinical Investigation, 122(9), 3145–3158. 10.1172/JCI63401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K. , & Morrison, S. F. (2008). A thermosensory pathway that controls body temperature. Nature Neuroscience, 11(1), 62–71. 10.1038/nn2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K. , & Morrison, S. F. (2010). A thermosensory pathway mediating heat–defense responses. Proceedings of the National Academy of Sciences of the United States of America, 107(19), 8848–8853. 10.1073/pnas.0913358107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgren, R. , & Leonard, C. M. (1971). Taste pathways in rat brainstem. Science, 173(4002), 1136–1139. [DOI] [PubMed] [Google Scholar]
- Norgren, R. , & Leonard, C. M. (1973). Ascending central gustatory pathways. Journal of Comparative Neurology, 150(2), 217–237. 10.1002/cne.901500208 [DOI] [PubMed] [Google Scholar]
- Norris, A. J. , Shaker, J. R. , Cone, A. L. , Ndiokho, I. B. , & Bruchas, M. R. (2021). Parabrachial opioidergic projections to preoptic hypothalamus mediate behavioral and physiological thermal defenses. eLife, 10. 10.7554/eLife.60779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm, T. G. , & Braak, H. (1988). The pigmented subpeduncular nucleus: A neuromelanin‐containing nucleus in the human pontine tegmentum. Morphology and changes in Alzheimer's disease. Acta Neuropathologica, 77(1), 26–32. 10.1007/BF00688239 [DOI] [PubMed] [Google Scholar]
- Ortiz, C. , Navarro, J. F. , Jurek, A. , Martin, A. , Lundeberg, J. , & Meletis, K. (2020). Molecular atlas of the adult mouse brain. Science Advances, 6(26), eabb3446. 10.1126/sciadv.abb3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter, R. D. (2018). The parabrachial nucleus: CGRP neurons function as a general alarm. Trends in Neuroscience, 41(5), 280–293. 10.1016/j.tins.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammer, C. , Fodor, M. , & Palkovits, M. (1988). Localization of corticotropin‐releasing factor, somatostatin, and vasoactive intestinal polypeptide in the parabrachial nuclei of the human brain. Journal of Neuroscience Research, 20(1), 109–114. 10.1002/jnr.490200115 [DOI] [PubMed] [Google Scholar]
- Panneton, W. M. , & Burton, H. (1985). Projections from the paratrigeminal nucleus and the medullary and spinal dorsal horns to the peribrachial area in the cat. Neuroscience, 15(3), 779–797. 10.1016/0306-4522(85)90078-8 [DOI] [PubMed] [Google Scholar]
- Park, S. , Williams, K. W. , Liu, C. , & Sohn, J. W. (2020). A neural basis for tonic suppression of sodium appetite. Nature Neuroscience, 23(3), 423–432. 10.1038/s41593-019-0573-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos, G. , & Franklin, K. B. J. (2013). Paxinos and Franklin's the mouse brain in stereotaxic coordinates (4th edn.). Amsterdam: Academic Press, an imprint of Elsevier. [Google Scholar]
- Paxinos, G. , & Watson, C. (2007). The rat brain in stereotaxic coordinates (6th edn.). Amsterdam; Boston; Academic Press/Elsevier. [Google Scholar]
- Petrovicky, P. (1989). The nucleus Koelliker‐Fuse (K–F) and parabrachial nuclear complex (PBNC) in man. Location, cytoarchitectonics and terminology in embryonic and adult periods, and comparison with other mammals. Journal Fur Hirnforschung, 30(5), 551–563. [PubMed] [Google Scholar]
- Prkic, I. , Mustapic, S. , Radocaj, T. , Stucke, A. G. , Stuth, E. A. E. , Hopp, F. A. , Dean, C. , & Zuperku, E. J. (2012). Pontine mu‐opioid receptors mediate bradypnea caused by intravenous remifentanil infusions at clinically relevant concentrations in dogs. Journal of Neurophysiology, 108(9), 2430–2441. 10.1152/jn.00185.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver, C. , Uddin, O. , Ji, Y. , Li, Y. , Cramer, N. , Jenne, C. , Morales, M. , Masri, R. , & Keller, A. (2020). An amygdalo‐parabrachial pathway regulates pain perception and chronic pain. Journal of Neuroscience, 40(17), 3424–3442. 10.1523/JNEUROSCI.0075-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch, J. M. , Fenselau, H. , Madara, J. C. , Wu, C. , Campbell, J. N. , Lyubetskaya, A. , Dawes, B. A. , Tsai, L. T. , Li, M. M. , Livneh, Y. , Ke, Q. , Kang, P. M. , Fejes‐Tóth, G. , Náray‐Fejes‐Tóth, A. , Geerling, J. C. , & Lowell, B. B. (2017). Aldosterone‐sensing neurons in the NTS exhibit state‐dependent pacemaker activity and drive sodium appetite via synergy with angiotensin II signaling. Neuron, 96(1), 190–206.e197. 10.1016/j.neuron.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaman, L. (2010). Ascending projections from the caudal visceral nucleus of the solitary tract to brain regions involved in food intake and energy expenditure. Brain Research, 1350, 18–34. 10.1016/j.brainres.2010.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokx, J. T. , van Willigen, J. D. , & Juch, P. J. (1986). Bilateral brainstem connections of the rat supratrigeminal region. Acta Anatomica, 127(1), 16–21. 10.1159/000146234 [DOI] [PubMed] [Google Scholar]
- Roome, R. B. , Bourojeni, F. B. , Mona, B. , Rastegar‐Pouyani, S. , Blain, R. , Dumouchel, A. , & Kania, A. (2020). Phox2a defines a developmental origin of the anterolateral system in mice and humans. Cell reports, 33(8), 108425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. F. , Ahmad, K. A. , Thaller, C. , & Zoghbi, H. Y. (2009). Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proceedings of the National Academy of Sciences of the United States of America, 106(52), 22462–22467. 10.1073/pnas.0911579106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rub, U. , Del Tredici, K. , Schultz, C. , Thal, D. R. , Braak, E. , & Braak, H. (2001). The autonomic higher order processing nuclei of the lower brain stem are among the early targets of the Alzheimer's disease‐related cytoskeletal pathology. Acta Neuropathologica, 101(6), 555–564. 10.1007/s004010000320 [DOI] [PubMed] [Google Scholar]
- Ryan, P. J. , Ross, S. I. , Campos, C. A. , Derkach, V. A. , & Palmiter, R. D. (2017). Oxytocin‐receptor‐expressing neurons in the parabrachial nucleus regulate fluid intake. Nature Neuroscience, 20(12), 1722–1733. 10.1038/s41593-017-0014-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper, C. B. (2016). The house alarm. Cell Metabolism, 23(5), 754–755. 10.1016/j.cmet.2016.04.021 [DOI] [PubMed] [Google Scholar]
- Saper, C. B. , & Loewy, A. D. (1980). Efferent connections of the parabrachial nucleus in the rat. Brain Research, 197(2), 291–317. 10.1016/0006-8993(80)91117-8 [DOI] [PubMed] [Google Scholar]
- Sarropoulos, I. , Sepp, M. , Frömel, R. , Leiss, K. , Trost, N. , Leushkin, E. , & Kaessmann, H. (2021). Developmental and evolutionary dynamics of cis‐regulatory elements in mouse cerebellar cells. Science, 373(6558), eabg4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, S. E. , & Levitt, E. S. (2020). Kolliker–Fuse/parabrachial complex mu opioid receptors contribute to fentanyl‐induced apnea and respiratory rate depression. Respiratory Physiology & Neurobiology, 275, 103388. 10.1016/j.resp.2020.103388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J. W. , Geerling, J. C. , Stein, M. K. , Miller, R. L. , & Loewy, A. D. (2011). FoxP2 brainstem neurons project to sodium appetite regulatory sites. Journal of Chemical Neuroanatomy, 42(1), 1–23. 10.1016/j.jchemneu.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriberg, L. D. , Ballard, K. J. , Tomblin, J. B. , Duffy, J. R. , Odell, K. H. , & Williams, C. A. (2006). Speech, prosody, and voice characteristics of a mother and daughter with a 7;13 translocation affecting FOXP2. Journal of Speech, Language, and Hearing Research, 49(3), 500–525. 10.1044/1092-4388(2006/038 [DOI] [PubMed] [Google Scholar]
- Stanek, E. T. , Rodriguez, E. , Zhao, S. , Han, B. X. , & Wang, F. (2016). Supratrigeminal bilaterally projecting neurons maintain basal tone and enable bilateral phasic activation of jaw‐closing muscles. Journal of Neuroscience, 36(29), 7663–7675. 10.1523/JNEUROSCI.0839-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic, D. , Dhingra, R. R. , & Dutschmann, M. (2018). Expression of the transcription factor FOXP2 in brainstem respiratory circuits of adult rat is restricted to upper‐airway pre‐motor areas. Respiratory Physiology & Neurobiology, 250, 14–18. 10.1016/j.resp.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Swanson, L. W. (1992). Brain maps: Structure of the rat brain. Amsterdam; New York: Elsevier. [Google Scholar]
- Swanson, L. W. (2000). A history of neuroanatomical mapping. In Toga A. W. & Mazziotta J. C. (Eds.), Brain mapping: The systems (pp. 77–109). San Diego, CA: Academic Press. [Google Scholar]
- Taber, E. (1961). The cytoarchitecture of the brain stem of the cat. I. Brain stem nuclei of cat. Journal of Comparative Neurology, 116, 27–69. 10.1002/cne.901160104 [DOI] [PubMed] [Google Scholar]
- Takatoh, J. , Park, J. H. , Lu, J. , Li, S. , Thompson, P. , Han, B.‐X. , Zhao, S. , Kleinfeld, D. , Friedman, B. , & Wang, F. (2021). Constructing an adult orofacial premotor atlas in Allen mouse CCF. eLife, 10. 10.7554/eLife.67291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, J. B. , & Norgren, R. (1983). Afferent projections to the oral motor nuclei in the rat. Journal of Comparative Neurology, 220(3), 280–298. 10.1002/cne.902200303 [DOI] [PubMed] [Google Scholar]
- van der Heijden, M. E. , & Zoghbi, H. Y. (2018). Loss of Atoh1 from neurons regulating hypoxic and hypercapnic chemoresponses causes neonatal respiratory failure in mice. eLife, 7. 10.7554/eLife.38455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, A. G. , Reid, B. T. , Kieffer, B. L. , & Levitt, E. S. (2020). Differential impact of two critical respiratory centres in opioid‐induced respiratory depression in awake mice. Journal of Physiology, 598(1), 189–205. 10.1113/JP278612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstegen, A. M. J. , Vanderhorst, V. , Gray, P. A. , Zeidel, M. L. , & Geerling, J. C. (2017). Barrington's nucleus: Neuroanatomic landscape of the mouse “pontine micturition center”. Journal of Comparative Neurology, 525(10), 2287–2309. 10.1002/cne.24215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, E. , Way, J. , Lebacq‐Verheyden, A. M. , & Battey, J. F. (1990). Neuromedin B and gastrin‐releasing peptide mRNAs are differentially distributed in the rat nervous system. Journal of Neuroscience, 10(9), 2917–2930. 10.1523/JNEUROSCI.10-09-02917.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Ding, S.‐L. , Li, Y. , Royall, J. , Feng, D. , Lesnar, P. , Graddis, N. , Naeemi, M. , Facer, B. , Ho, A. , Dolbeare, T. , Blanchard, B. , Dee, N. , Wakeman, W. , Hirokawa, K. E. , Szafer, A. , Sunkin, S. M. , Oh, S. W. , Bernard, A. , …, & Ng, L. (2020). The Allen mouse brain common coordinate framework: A 3D reference atlas. Cell, 181(4), 936–953.e920. 10.1016/j.cell.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, V. Y. , Rose, M. F. , & Zoghbi, H. Y. (2005). Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron, 48(1), 31–43. 10.1016/j.neuron.2005.08.024 [DOI] [PubMed] [Google Scholar]
- Watson, C. , Shimogori, T. , & Puelles, L. (2017). Mouse Fgf8‐Cre‐LacZ lineage analysis defines the territory of the postnatal mammalian isthmus. Journal of Comparative Neurology, 525(12), 2782–2799. 10.1002/cne.24242 [DOI] [PubMed] [Google Scholar]
- Weinholtz, C. A. , & Castle, M. J. (2021). Intersectional targeting of defined neural circuits by adeno‐associated virus vectors. Journal of Neuroscience Research, 99(4), 981–990. 10.1002/jnr.24774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, M. , Terao, M. , Terashima, T. , Fujiyama, T. , Kawaguchi, Y. , Nabeshima, Y. , & Hoshino, M. (2007). Origin of climbing fiber neurons and their developmental dependence on Ptf1a. Journal of Neuroscience, 27(41), 10924–10934. 10.1523/JNEUROSCI.1423-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T. , Shimura, T. , Sako, N. , Azuma, S. , Bai, W. Z. , & Wakisaka, S. (1992). C‐fos expression in the rat brain after intraperitoneal injection of lithium chloride. Neuroreport, 3(12), 1049–1052. 10.1097/00001756-199212000-00004 [DOI] [PubMed] [Google Scholar]
- Yamamoto, T. , Shimura, T. , Sako, N. , Yasoshima, Y. , & Sakai, N. (1994). Neural substrates for conditioned taste aversion in the rat. Behavioural Brain Research, 65(2), 123–137. 10.1016/0166-4328(94)90097-3 [DOI] [PubMed] [Google Scholar]
- Yang, W. Z. , Du, X. , Zhang, W. , Gao, C. , Xie, H. , Xiao, Y. , Jia, X. , Liu, J. , Xu, J. , Fu, X. , Tu, H. , Fu, X. , Ni, X. , He, M. , Yang, J. , Wang, H. , Yang, H. , Xu, X. , & Shen, W. L. (2020). Parabrachial neuron types categorically encode thermoregulation variables during heat defense. Science Advances, 6(36). 10.1126/sciadv.abb9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui, Y. , Saper, C. B. , & Cechetto, D. F. (1989). Calcitonin gene‐related peptide immunoreactivity in the visceral sensory cortex, thalamus, and related pathways in the rat. Journal of Comparative Neurology, 290(4), 487–501. 10.1002/cne.902900404 [DOI] [PubMed] [Google Scholar]
- Yokota, S. , Kaur, S. , VanderHorst, V. G. , Saper, C. B. , & Chamberlin, N. L. (2015). Respiratory‐related outputs of glutamatergic, hypercapnia‐responsive parabrachial neurons in mice. Journal of Comparative Neurology, 523(6), 907–920. 10.1002/cne.23720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborszky, L. , Beinfeld, M. C. , Palkovits, M. , & Heimer, L. (1984). Brainstem projection to the hypothalamic ventromedial nucleus in the rat: A CCK‐containing long ascending pathway. Brain Research, 303(2), 225–231. 10.1016/0006-8993(84)91208-3 [DOI] [PubMed] [Google Scholar]
- Zagami, C. J. , & Stifani, S. (2010). Molecular characterization of the mouse superior lateral parabrachial nucleus through expression of the transcription factor Runx1. PLoS One, 5(11), e13944. 10.1371/journal.pone.0013944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H. , & Sanes, J. R. (2017). Neuronal cell‐type classification: Challenges, opportunities and the path forward. Nature Reviews Neuroscience, 18(9), 530–546. 10.1038/nrn.2017.85 [DOI] [PubMed] [Google Scholar]
- Zhao, Z. Q. , Scott, M. , Chiechio, S. , Wang, J.‐S. , Renner, K. J. , Gereau, R. W. , Johnson, R. L. , Deneris, E. S. , & Chen, Z.‐F. (2006). Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. Journal of Neuroscience, 26(49), 12781–12788. 10.1523/JNEUROSCI.4143-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H. L. , Su, C. J. , Shi, M. , Zhao, G. Y. , Li, Z. Y. , Guo, C. , & Ding, Y. Q. (2009). Expression of the LIM‐homeodomain gene Lmx1a in the postnatal mouse central nervous system. Brain Research Bulletin, 78(6), 306–312. 10.1016/j.brainresbull.2008.12.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


