Abstract
Mycobacterium tuberculosis (Mtb) is responsible for approximately 1.5 million deaths each year. Though 10% of patients develop tuberculosis (TB) after infection, 90% of these infections are latent. Further, mice are nearly uniformly susceptible to Mtb but their M1-polarized macrophages (M1-MΦs) can inhibit Mtb in vitro, suggesting that M1-MΦs may be able to regulate anti-TB immunity. We sought to determine whether human MΦ heterogeneity contributes to TB immunity. Here we show that IFN-γ-programmed M1-MΦs degrade Mtb through increased expression of innate immunity regulatory genes (Inregs). In contrast, IL-4-programmed M2-polarized MΦs (M2-MΦs) are permissive for Mtb proliferation and exhibit reduced Inregs expression. M1-MΦs and M2-MΦs express pro- and anti-inflammatory cytokine-chemokines, respectively, and M1-MΦs show nitric oxide and autophagy-dependent degradation of Mtb, leading to increased antigen presentation to T cells through an ATG-RAB7-cathepsin pathway. Despite Mtb infection, M1-MΦs show increased histone acetylation at the ATG5 promoter and pro-autophagy phenotypes, while increased histone deacetylases lead to decreased autophagy in M2-MΦs. Finally, Mtb-infected neonatal macaques express human Inregs in their lymph nodes and macrophages, suggesting that M1 and M2 phenotypes can mediate immunity to TB in both humans and macaques. We conclude that human MФ subsets show unique patterns of gene expression that enable differential control of TB after infection. These genes could serve as targets for diagnosis and immunotherapy of TB.
Subject terms: Antigen processing and presentation, Immunology, Bacterial infection
Khan et al. use human blood and a macaque model of tuberculosis (TB) to assess whether macrophage heterogeneity contributes to TB immunity. They demonstrate that human macrophages show unique patterns of gene expression that enable differential control of TB after infection.
Introduction
Mycobacterium tuberculosis (Mtb) causes 8 million new cases of tuberculosis (TB) and about 1.5 million human deaths each year1. Mtb is a highly infectious organism, but nearly 90% of those infected do not develop active disease, leaving a third of the human population latently infected (LTBI). Though Mtb antigens trigger T cell-mediated responses, emerging studies indicate that innate immunity also plays a role in preventing active TB2.
Macrophages (MΦs) are a key element for both innate and adaptive immunity. T cell-derived cytokines like IFN-γ can activate MΦs to kill and degrade Mtb through nitric oxide (NO) and reactive oxygen species (ROS)-dependent mechanisms and phagosome-lysosome fusion3. Alveolar MΦs are the first immune cells to encounter Mtb after aerosolized infection and they contribute to the granuloma response in the lung parenchyma during TB4,5. Lung granulomas can contain Mtb, but necrotic granulomas can lead to cavities and subsequent dissemination of Mtb6. The anti-microbial mechanisms of mouse but not human MΦs are well characterized. Indeed, fewer studies have been reported using human MΦs compared to mouse MΦs. Human MΦs secrete NO at lower levels, show marked functional and phenotypic heterogeneity among tissues, and likely differ in their anti-microbial mechanisms7–9. This difference is reflected by the near-uniform susceptibility of most mouse strains to low-dose aerosol infection with Mtb versus the differential susceptibility of humans to active disease.
MΦs can be differentiated into a pro-inflammatory “classical” M1-MΦ phenotype, driven by IFN-γ, or an anti-inflammatory M2-MΦs phenotype, driven by IL-4 (M2-a), IL-1β (M2-b), IL-10 (M2-c), IL-13 (or a combination of these cytokines). Importantly, MΦ phenotypes exist along a spectrum of these polarizations. Interestingly, cytokines, growth factors, metabolites, and Mtb infection all seem to drive MΦ phenotype in mice10–14. During M1-MΦ polarization, mouse MΦs exhibit increased microbicidal capacity, NO production, secretion of pro-inflammatory cytokines, antimicrobial peptide production, phagocytosis, and phagosome-lysosome fusion15,16. M1-MΦs are likely to mount an effective response against intracellular pathogens, while M2-MΦs exhibit relatively decreased anti-microbial functions but may aid in tissue repair8,17,18. Accordingly, mouse M1- and M2-MΦs show differential susceptibilities to Mtb, broadly characterized by inhibitive M1-MΦs phenotypes and permissive M2-MΦs phenotypes. Although non-human primates (NHP) like rhesus monkeys also show a dose-dependent susceptibility to active TB, their macrophage heterogeneity is yet to be fully characterized.
We hypothesized that a subset of human MФs can acquire enhanced anti-TB function upon cytokine exposure and that this exposure partially explains the development of LTBI. Human alveolar MΦs (AMΦs) also express M1- and M2- phenotypes19 and, although AMΦs are the first to encounter Mtb after aerosol infection5, alveoli are also rapidly infiltrated by interstitial MФs (IMΦs), and peripheral blood mononuclear cells (PBMCs) including T cells, neutrophils, and dendritic cells (DCs) from the vasculature following infection. Thus, IMΦs derived from PBMCs likely differentiate locally into M1- and M2- phenotypes following exposure to cytokine milieus upon arrival.
Herein, we demonstrate that human M1-MΦs upregulate multiple innate immunity regulatory genes and gene clusters (Inregs), including ATGs, RAB GTPases, and cathepsin proteolytic enzymes to degrade Mtb more effectively than M2-MΦs. Further, compared to M2-MΦs, M1-MΦs processed and presented Mtb antigen to CD4 T cells more efficiently ex vivo. We also found that Mtb epigenetically alters histones associated with ATG5, which decreased autophagy in M2-MΦs. In contrast, M1-MΦs retained their anti-mycobacterial autophagy by selectively expressing histone deacetylases (HDACs). Finally, we demonstrate that Mtb-infected infant macaque-derived lymph nodes and MΦs show transcriptional responses like those observed in human MΦs. We propose that cytokine-exposed human MΦs can differentiate into M1- and M2- subsets during natural infection and that the differential expression of Inregs regulates the functional heterogeneity of human and NHP MΦs, explaining in part the differential susceptibility of humans and NHPs to TB.
Results
Human peripheral blood and cord blood-derived MФs polarized to M1- (IFN-γ) and M2- (IL-4) phenotypes differentially regulate the growth of intracellular Mtb through nitric oxide and autophagy
To characterize human MФ heterogeneity, we used IFN-γ or IL-4 to polarize PBMC-derived and GM-CSF cultured MФs into M1- and M2-MФs phenotypes, respectively20,21, and confirmed surface expression of CD80(M1)/CD206(M2) using flow cytometry. We further confirmed the ability of the cells to phagocytose Mtb with a colony-forming unit (CFU) assay22,23. We activated cells for 5 days using a low-dose cytokine activation protocol, which allowed the cells to retain >90% viability through day 7 post Mtb infection. Though Mtb-infected M1-MФs survived through day 21 post Mtb infection, we observed a progressive loss of viability. M2- and M0-MФs showed marked viability loss 10 days post-infection. The lack of apoptosis in Mtb-infected or naïve cultures was confirmed using a Tunel assay between days 1–7.
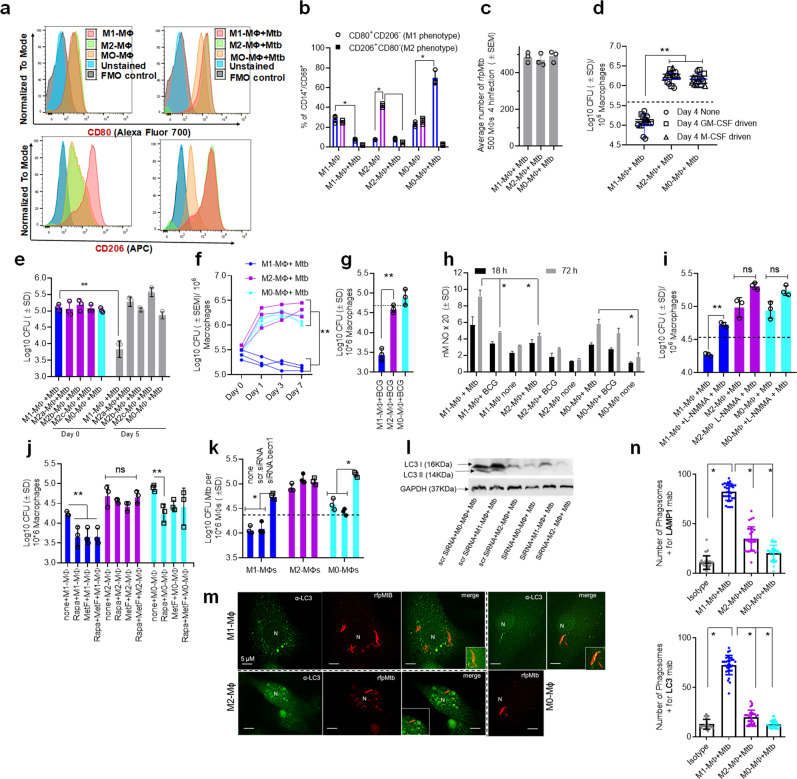
M1-MФs were CD80+/CD206− and M2-MФs were CD80−/CD206+ (Fig. 1a, b) (gating strategy shown in Supplementary Fig. 1). Both phenotypes showed comparable phagocytic uptake of Mtb; Fig. 1c shows rfpMtb uptake immediately after a 4 h (hour) infection. Consistent with previous observations, M1-MФs secreted many pro-inflammatory cytokines (IL-12, TNF-α, IL1-β, IL-8, GM-CSF, IL1-α, IL-17) and chemokines (CCL2, CCL13, CCL18, CCL22), while M2-MФs secreted anti-inflammatory cytokines (IL-4, IL-6, IL-10) (Supplementary Fig. 2). We cultured PBMCs from five healthy donors using either GM-CSF or M-CSF followed by M1- and M2- differentiation and an Mtb growth assay. GM-CSF and M-CSF-cultured MФs showed similar Mtb growth profiles following polarization (Fig. 1d). Unlike previous studies, we did not use lipopolysaccharide (LPS) in combination with IFN-γ to activate M1-MФs as LPS is a Gram-negative bacterial ligand inducing pleiotropic effects23, and Mtb can itself activate TLR-424. We also found that LPS can trigger autophagy through TLR-425. Although subsets of M2-MФs are known26, we did not observe significant differences in Mtb infection assay profiles of Mtb-infected subsets (Fig. 1e).
Fig. 1. Human umbilical cord and peripheral blood-derived macrophages show heterogeneity in mycobacterial killing associated with oxidants and autophagy.
Human cord blood (CBM) or healthy donor PBMC-derived MФs were cultured in the presence of either recombinant human IFN-γ (M1; 10 ng/mL) or human IL-4 (M2; 10 ng/mL) for 5 days and rested for 2 days. Untreated cells were M0-MФs. a, b Surface expression of receptors by CBM-derived naïve and Mycobacterium tuberculosis (Mtb; H37Rv)-infected M1- (CD80+/CD206−) and M2-MФs (CD80−/CD206+) on day 3 using flow cytometry and quantitation (*p < 0.01 t test); gating strategy shown Supplemental Fig. 1. c CBM-derived differentiated MФs were infected with Mtb for 4 h followed by microscopic counts of rfp-labeled MtbH37Rv to determine uptake verified using CFU counts. d PBMC-derived MФs from five healthy donors were differentiated using the indicated cytokines followed by infection with Mtb and CFU assay on day 4. Each point represents one donor (**p < 0.05; Kruskal–Wallis test). e PBMC-derived MФs were differentiated using GM-CSF (M1), IL-4 (M2a), IL-1β M2b), IL-10 (M2c) or left intreated (M0) followed by Mtb infection and CFU assay on day 4 (**p < 0.007). f CBM-derived, cytokine differentiated MФs were infected with Mtb followed by CFU assay over time (**p < 0.006; data from 3 experiments shown). g CBM-derived, differentiated MФs were infected with M. bovis BCG followed by CFU assay on day 4 (**p < 0.005). h CBM-derived and differentiated uninfected MФs or those infected with Mtb or BCG were incubated and at indicated time points, cultures were tested for nitrite using diaminofluorescein diacetate and fluorometry (*,**p < 0.005, t test). QPCR for mRNA of iNOS and protein are shown in Supplemental Fig. 3a and reactive oxygen species levels in Supplementary Fig. 3b. i MФs infected with Mtb as in panel h were incubated in NMMA (0.5 mM; N-monomethyl l-arginine) followed by CFU assay on day 3 (**p < 0.009). j CBM-derived, differentiated MФs infected with Mtb were incubated with 10 µM Rapamycin, 100 µM Metformin or their combination followed by CFU assay on day 3 (**p < 0.009). k CBM-derived, M1-, M2- and M0-MΦs were treated in the presence or absence of siRNA vs. beclin1 (ATG6) or its scrambled control followed by infection with Mtb and CFU counts on day 3 (*p < 0.007). Blot validation of Knockout using siRNA vs. beclin1 (ATG6) is shown in Fig. 3h. l MΦ lysates of panel k collected at 18 h were analyzed using western blots for the lipidation of microtubule-associated light chain 3 (LC3). Lipidation is indicated by LC3-II. m CBM-derived MΦs were infected with rfpMtbH37Rv and stained for an LC3 autophagy marker or LAMP1 lysosome marker using specific antibodies, Alex-Fluor485 conjugates, and imaged using confocal microscopy. Panels illustrate LC3 colocalization; LAMP1 stains using gfpMtbH37Rv is shown in Supplementary Fig. 3c. n Quantification of phagosomes colocalizing with LC3 are shown using an N90 Nikon fluorescence microscope (IF) and Metaview software (*p < 0.004, t test). For panels (b–c–e–f–g–h–i–b–k–n), p-values were calculated using a one-way ANOVA with Tukey’s post-hoc test; one of 2–3 similar experiments shown. CFU or IL-2 assays had triplicate wells plated per group or donor. Panels (d, g, i–k) horizontal dotted lines indicate day 0 CFU (4 h post-infection CFU). All Mtb CFU experiments used MOI of 1.
Monocytes from cord blood and adult PBMCs are similar. To obtain a consistently high yield of MФs required to elucidate the molecular mechanisms underlying MФ heterogeneity, we optimized an ex vivo culture system using cord blood-derived monocytes (CBMs) differentiated into M1- and M2-MФ phenotypes. CBMs cultured in medium with GM-CSF were rested (M0-MФ) or differentiated into either M1- or M2-MФs using IFN-γ and IL-4, respectively21. CBM-derived M1- and M2-MФs restricted Mtb growth (Fig. 1f) to an extent similar to that observed in PBMC-derived M1- and M2-MФs (Fig. 1d, e). M1-MФs also significantly restricted Bacillus Calmette–Guérin (BCG) growth compared to M2-MФs (Fig. 1g). Although a few studies have shown that human M1-MФs kill Mtb better than M2-MФs, we emphasize that M1-polarization was achieved using IFN-γ and LPS20; thus, LPS-induced pleiotropic effects cannot be ruled out.
We performed a fluorometric nitrite assay on Mtb-infected MФs27 and found that Mtb- or BCG-infected M1-MФs produced higher levels of nitrite compared to M2- or M0-MФs at 72 h post-infection (Fig. 1h). In a separate experiment, we blocked NO synthesis by incubating Mtb-infected MФs with N-monomethyl l-arginine and assessed Mtb growth. N-monomethyl L-arginine blockade of NO enhanced Mtb growth in M1- but not M2- or M0-MФs (Fig. 1i). Interestingly, Mtb infected M1-MФs expressed higher levels of iNOS mRNA compared to Mtb-infected M2-MФs (Supplementary Fig. 3a). Of interest, iNOS protein was detectable in Mtb-infected but not uninfected MФs, although no quantitative differences were found in protein levels after Mtb infection between M1-, M2- or M0-MФs. Even though uninfected MФs were negative for iNOS protein using blots, fluorometry was much more sensitive and detected basal levels of NO. Similar to NO, Mtb and BCG-infected M1-MФs produced higher levels of whole cell ROS detected using DCFDA compared to Mtb-infected M2- or M0-MФs28; uninfected M1-MФs also showed elevated mitochondrial ROS detected using MitoROS (Supplementary Fig. 3b). It remains unclear whether gp91-phox and phagosome-dependent generation of ROS occurred in MФs and needs investigation.
To determine if autophagy contributes to Mtb degradation, we performed dose titrations using Rapamycin and Metformin, two drugs that induce degradation of Mtb in naïve MФs. At a dose of 10 µM, Rapamycin reduced the Mtb colony counts (CFUs) in M1-, M2 and M0-MФs (≥0.5 log10 decrease in Mtb counts over 3 days; p < 0.01 t test), while 100 µM Metformin was effective only in M1-MФs. A suboptimal dose of 1 µM Rapamycin and 100 µM Metformin increased bactericidal function in M1- and M0- but not M2-MФs, although the drugs did not synergize (experiment performed in duplicate) (Fig. 1j). Because Rapamycin is a known inducer of autophagy, we next treated Mtb-infected M1-, M2- and M0-MФs with siRNA vs. beclin1 (ATG6) (autophagy blockade) and assessed changes in Mtb proliferation using a CFU assay. siRNA vs. beclin1 enhanced Mtb growth in M1-and M0-MФs but not M2-MФs (Fig. 1k).
As autophagic flux is indicated by the localization of LC3 on autophagolysosomes (APLs) containing Mtb, which also label for LAMP1 and Rab729, we next assessed LC3 lipidation. Increased LC3 lipidation was observed in M1- and M0-MФs (Fig. 1l), although only the rfpMtb phagosomes of M1-MФs demonstrated increased colocalization with both LC3 and LAMP1 (Fig. 1m, n). Despite LC3 lipidation (Fig. 1m, n), rfpMtb phagosomes of M0-MФs were less enriched for LAMP1 (Fig. 1m) (Supplementary Fig. 3c). suggesting a lack of fusion with APLs, a finding consistent with reports that Mtb phagosomes can transiently label with LC330. Together, these data strongly suggest that human M1- and M2-MФs are heterogeneous with differing NO and ROS levels, and autophagic sorting of mycobacteria. These differences explain the Mtb-inhibitive versus Mtb-permissive states of M1-MФs and M2-MФs, respectively.
Mtb infection induces differential expression of innate immunity regulating genes (Inregs) in human M1- and M2-MФs
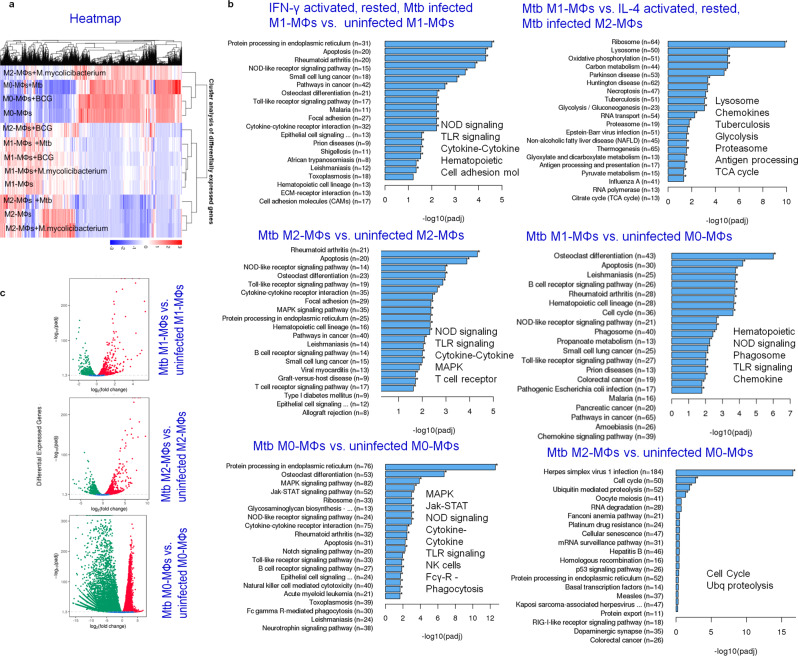
To better understand the molecular basis of phenotypic and functional heterogeneity in MФs, we subjected naïve and Mtb-infected M1-, M2-, and M0-MФs to RNAseq analysis. We also separately infected MФs with avirulent BCG vaccine and nonpathogenic Mycolicibacterium smegmatis and analyzed cells in parallel for gene expression (heat maps include BCG and M. smegmatis) (Fig. 2a). Heat maps indicate that M1-, M2-, and M0-MФs responded differently to Mtb infection, with a marked difference between M1- and M2-MФs responses. Gene clusters that control anti-mycobacterial immune responses including, NOD/TLR-dependent signaling mechanisms, cytokine-chemokines, FcG-receptor-mediated signaling, and genes regulating the sorting of the pathogen from phagosomes to lysosomes were differentially expressed and are highlighted in Fig. 2b, c. Compared to M2-MФs, M1-MФs showed an upregulation of genes and pathways that regulate antigen processing (phagosome and/or lysosome, proteasome, antigen processing), hereafter referred to as the ‘antigen processing transcriptome’ (Fig. 2b). Furthermore, consistent with previous observations that the glycolytic pathway regulates M1-phenotype31, glycolysis/gluconeogenesis and tricarboxylic acid cycle cycle pathways were upregulated in Mtb-infected M1-MФs (Fig. 2b). Genes regulating hematopoietic lineage were upregulated only in Mtb-infected M1-MФs. Individual genes that were differentially expressed by Mtb-infected MФs in comparison with their naïve counterparts, including downregulated genes, are further illustrated in Supplementary Fig. 4. Downregulated genes also contribute immunologically to the pathogenesis of TB; for example, ER protein processing was downregulated in M1- vs. M2-MФs (Supplementary Fig. 5). Representative pathway analysis derived from the Clusterprofiler workflow is illustrated in the context of up- (red) and down-(green) regulated genes affecting anti-mycobacterial immune responses (Supplementary Figs. 6, 7). Both Mtb-infected M1- and M2-MФs activated multiple genes of the NOD signaling pathway (Supplementary Fig. 6), but only Mtb-infected M1-MФs activated the caspase cascade (CASP1), which leads to IL-1β secretion (Supplementary Fig. 2). Likewise, Mtb-infected M1-MФs upregulated multiple genes participating in anti-TB immunity compared to M2-MФs (Supplementary Fig. 7). These data are strikingly different from the transcriptome responses of mouse-derived M1-and M2 MФs and confirm that human MФs respond to Mtb differently than those from mice32.
Fig. 2. Mycobacterium tuberculosis (Mtb) induces differential expression of innate immunity regulating genes (Inregs) in human M1- and M2-MФs.
CBM-derived uninfected M0-, M1- and M2-MΦs or those infected separately with Mtb H37Rv, BCG or Mycolicibacterium smegmatis were analyzed using RNAseq (Novogene Inc., USA) at 18 h (MOI of 1; n = 2 samples per group; analysis done twice). a Heat maps highlight Mtb-induced differential gene expression. BCG and M. smegmatis profiles are included but not highlighted. b Kyoto Encyclopedia of Genes and Genomes profiles of Mtb-infected M1-, M2- and M0-MΦs vs. uninfected MΦs show differential enrichment of Inregs and pathways in the context of TB control (*p-values < 0.00001; Clusterprofiler workflow). Only gene clusters showing a significant p-value (<0.00001) are highlighted. c Volcano plots indicate the Mtb-induced differential gene expression. Additional gene expression profiles and Clusterprofiler pathway analysis are shown in Supplementary Figs. 4–7. RNAseq and pathway analysis by Novogene is illustrated in Supplementary Fig. 13. All Mtb CFU experiments used MOI of 1.
Mtb-infected human M1- and M2-MФs show differential upregulation of ATGs and accessory genes that regulate autophagy-dependent mycobacterial degradation and antigen presentation
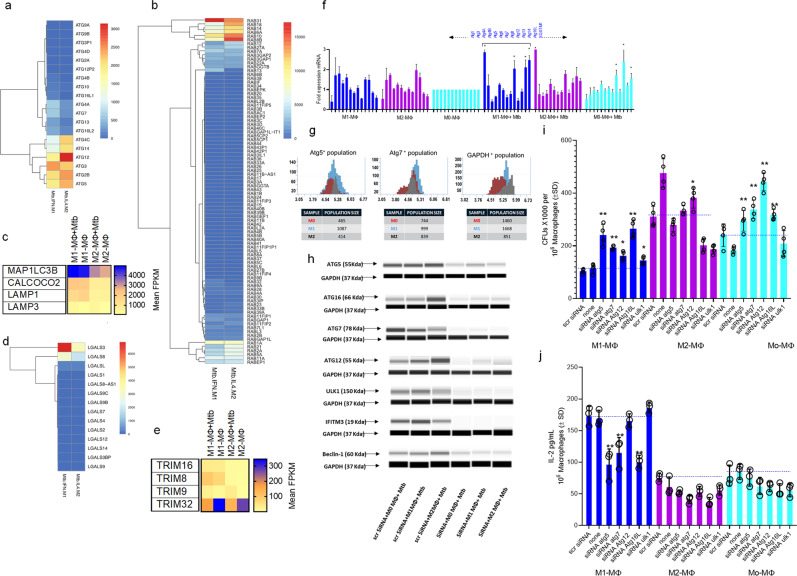
Our transcriptome studies indicated that genes regulating antigen processing pathways are upregulated in M1-MФs (Fig. 2; Supplementary Figs. 9, 10). Because knockdown of beclin1 (ATG6) a key imitator of autophagy enhanced Mtb proliferation in M1-MФs but not M2-MФs (Fig. 1k), we sought to further characterize the expression of ATGs by analyzing their transcripts and proteins before and after Mtb infection. Figure 3a–e shows the differential expression profiles for ATGs, RAB GTPases, galectins (LGALS), and TRIMs in addition to MAP1LC3B, CALCOCO2, LAMP1, and LAMP3, which encode key proteins mediating APL fusion in Mtb-infected M1- vs. M2-MФs. Mtb-infected M1-MФs expressed more key ATGs, including ATG7, and RAB7A, MAP1LC3, CALCOCO2, and LAMP-1 than M2-MФs. Likewise, M1-MФs expressed markedly higher levels of galectins LGALS3/8 and TRIM 8, 32, and 3833–35. Figure 3f shows the QPCR profile of ATGs.
Fig. 3. Mtb-infected human M1-, M2-, and M0-MΦs show differential expression of autophagy-regulating ATGs, RAB GTPases, Galectins (LGALS) and Tripartite-containing motif protein (TRIM) encoding accessory genes.
a–e CBM-derived M1-, M2-, and M0-MΦs were subjected to RNAseq before and after Mtb infection as in Fig. 2 at 18 h post Mtb infection. Data are shown for M1- vs. M2-MΦs and transcripts are shown as FPKMs (fragments per kilobase per million reads) for gene clusters (n = 2). f Quantitative PCR analysis for autophagy-regulating genes (ATGs) at 18 h (Mtb-infected vs. naive; *p < 0.01, t test, n = 2; one of 2 similar experiments). g Single cells of uninfected M1-, M2-, and M0-MΦs were dispensed into the wells of the MILO single-cell protein profiler. ATG5 and ATG7 proteins with a GAPDH control were detected using specific antibodies and in situ western blot; quantitation of the number of MΦs expressing proteins are shown. h Mtb-infected or naïve M1-, M2-, and M0-MΦs were analyzed for ATG proteins before and after siRNA knockdown including beclin1 (ATG6) (Origene, USA) (one of 2 similar experiments shown). Densitometry is shown in Supplementary Fig. 8. i Indicated ATGs were knocked down using specific duplexes of siRNA (Origene, USA), followed by infection with Mtb and CFU counts on day 3 maintaining >90% viability of MΦs. Baseline CFUs of scrambled siRNA controls are shown by a dotted line (one of 3 experiments shown). j siRNA or scrambled siRNA treated replicates of MΦs from panel i were overlaid with an Ag85B-derived epitope-specific F9A6-CD4 T cell line for antigen presentation. IL-2 was measured in supernatants using sandwich enzyme-linked immunoassay at 18 h (one of 2 independent experiments shown). Baseline antigen presentation by scrambled siRNA controls is shown by a dotted line. Panels (i, j): **p < 0.007; *p < 0.009 one-way ANOVA with Tukey’s test. All Mtb CFU experiments used MOI of 1.
Given the sequential nature of the autophagy cascade, we did not expect to find ATG proteins at comparable levels in Mtb-infected M1- and M2-MФ lysates at 18 h post-infection. Western blot analysis of these MФs indicated that Mtb-infected M1- and M2-MФs had higher levels of ATG proteins (1, 5, 7, 12, 16L) compared to M2 (supplemental methods, protein simple densitometry). However, we noted that M2-MФs also showed enhanced transcripts for some ATGs compared to M0-MФs, although their autophagy-dependent degradation of Mtb was reduced (Fig. 1k). We provide four explanations to account for this interesting difference. First, besides ATGs, including LC3 (ATG8) (Fig. 3c), autophagosome formation and autophagolysosome fusion requires SQSTM1 (also known as. p62) (Fig. 3f), RAB7, and LAMP1 (Figs. 1n; 3c; 4a, b), which were highly expressed in Mtb-infected M1-MФs36. Secondly, LC3, RAB7, and LAMP1 showed stronger colocalization with rfpMtb phagosomes of the M1-MФs indicating autophagic flux (Figs. 1n; 4a, b). Third, when single cells of uninfected M1- and M2-MФs were analyzed for proteins using MILO (methods), ATG5 and ATG7, two proteins essential for macroautophagy, were abundant in M1-MФs compared to M2-MФs (Fig. 3g). Fourth, Mtb evades lysosomal fusion and interferes with autophagy30. In addition, we describe below an epigenetic modification through which Mtb differentially affects autophagy in M2- vs. M1-MФs (Fig. 5).
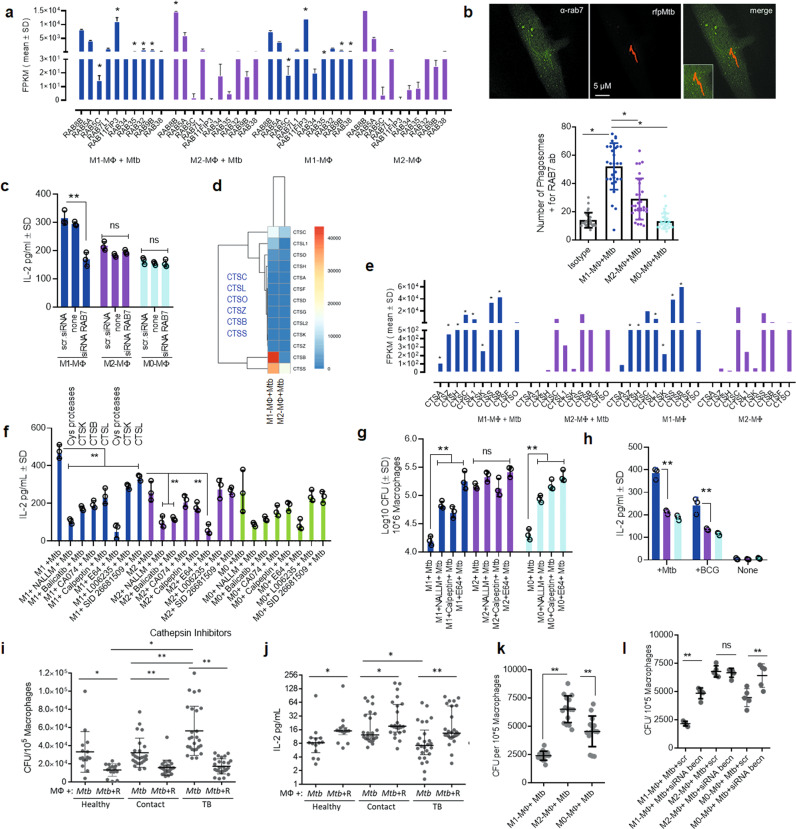
Fig. 4. Mtb infection induces differential expression of RAB GTPases and cathepsin proteases in human macrophages affecting autophagy-dependent ex vivo antigen presentation to CD4 T cells.
a Differential expression of RAB GTPases and transcripts are shown as FPKMs (fragments per kilobase per million reads) for duplicate samples; *p < 0.01; t test. b MΦs were infected with rfpMtb, followed by staining with an isotype or specific antibodies to RAB7 or LAMP1 proteins and counterstained using fluorescein isothiocyanate anti-IgG conjugates. Confocal microscopy analysis of phagosomes colocalizing with RAB7 is illustrated (full panels shown in Supplementary Fig. 3c); bar graph indicates quantitation of RAB7 colocalization (*p < 0.009; t test; one of 2 similar experiments shown). c MΦs indicated were treated with siRNA for RAB7 or its scrambled control followed by Mtb infection and overlay with F9A6-CD4 T cell line for antigen presentation and IL-2 assay (**p < 0.009; t test; one of 2 similar experiments shown). d, e Differential gene expression of cathepsins (CTS) expressed as FPKMs shown for duplicate samples (*p < 0.01; t test). f MΦs indicated were treated with non-cytotoxic doses of either CTS specific inhibitors or pan-specific inhibitors followed by Mtb infection and antigen presentation using F9A6-CD4 T cells (**p < 0.009, t test; one of 2 similar experiments shown). g MΦs treated with CTS inhibitors were infected with Mtb followed by CFU assay on day 5, maintaining >90% viability of MΦs (**p < 0.009, one-way ANOVA with Tukey’s post-hoc test; one of 2 similar experiments shown). h Replicates of MΦs used in panel (g) were infected using Mtb or BCG and supernatants collected at 18 h were tested for IL-2 and antigen presentation (**p < 0.009; t test). i PBMC-derived macrophages from healthy donors, household contacts, and TB patients (TB) were treated or not treated with Rapamycin (10 µM) followed by infection with Mtb and CFU counts on day 3 in vitro. j Washed Mtb-infected MΦs (as in panel i) were overlaid with F9A6-CD4 T cells for antigen presentation with or without Rapamycin and IL-2 assay. Horizontal bars indicate median (interquartile range) for IL-2 (non-normal distribution) or mean (SD) for CFUs (*p < 0.05; **p < 0.001. Wilcoxon paired ranked signed test, and Kruskal-Wallis test). k Healthy adult donor MΦs from the TB endemic area differentiated into M1-, M2-, or M0-MΦs were infected with Mtb followed by CFU counts on day 3 (**p < 0.01; Kruskal–Wallis test). l Five donor MΦs from each group of panel k were treated with siRNA vs. beclin1 or its scrambled control, followed by infection with Mtb and CFU counts on day 3 (**p < 0.009, Kruskal-Wallis test; triplicate wells per donor; one of 2 similar experiments shown). All Mtb CFU experiments used MOI of 1.
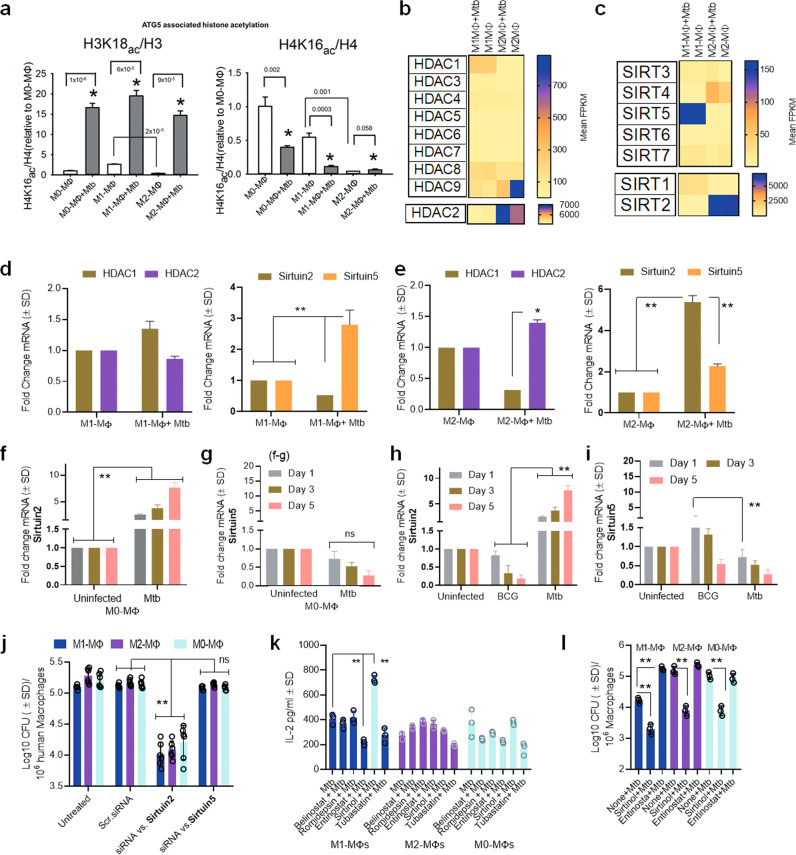
Fig. 5. Mtb-infected human M1- and M2-MФs show differential epigenetic programming affecting autophagy through histone acetylation of ATG5.
a H3K18 acetylation and H4K16 acetylation on the promoter of ATG5 in M0, M1-, and M2-MΦs and their Mtb-infected counterparts (one of 2 similar experiments shown; p using one-way ANOVA). b, c RNAseq gene expression of histone deacetylases (HDACs) and Sirtuins in naïve or Mtb-infected M1-, and M2-MΦs at 18 h. Transcripts are shown as FPKMs (fragments per kilobase per million reads) for duplicate samples. d QPCR analysis of mRNA for HDAC and Sirtuins indicated in M1-MΦs at 18 h (**p < 0.01 t test; n = 2). e QPCR analysis of mRNA for HDAC and Sirtuins in M2-MΦs at 18 h (*, **p < 0.01 t test; n = 2). Densitometry of proteins using western blot shown in Supplementary Fig. 15. f, g Mtb-infected M0-MΦs were incubated for days indicated and mRNA of Srtuin2 and Sirtuin5 quantitated using qPCR (**p < 0.01 t test; n = 2). h, i M0-MΦs were infected with either BCG vaccine or Mtb followed by QPCR analysis of mRNA of Sirtuin2 and Sirtuin5 on indicated days (**p < 0.01 t test; n = 2). j MΦs were treated with siRNA vs. Sirtuins or their scrambled control followed by Mtb CFU assays on day 3 (**p < 0.006, one-way ANOVA with Tukey’s posttest; one of 2 similar experiments shown). k Mtb-infected M1- and M2-MΦs were incubated with HDAC inhibitors (Belinostat; Romidepsin; Entinostat; Tubastatin; 20 µM each) and the Sirtuin-2 inhibitor sirtinol (130 µM), followed by antigen presentation using HLA-DR1-specific F9A6-CD4 T cells (**p < 0.007, t test; one of 2 similar experiments shown). l MΦs were incubated with Sirtuin2 specific inhibitor sirtinol (130 µM), or the HDAC inhibitor Entinostat (20 µM), followed by Mtb infection and CFU counts on day 5 maintaining >90% viability of MΦs (**p < 0.009, one-way ANOVA with Tukey’s posttest). For all panels one of 2 similar experiments shown). All Mtb or BCG CFU experiments used MOI of 1.
We next sought to define additional functional correlates of gene expression. Because Mtb degrades in APLs producing antigenic peptides, we knocked down selected ATGs in Mtb-infected M1, M2-, and M0-MФs using siRNA duplexes (Fig. 3h). We assessed changes in Mtb proliferation using CFU assays and performed a parallel evaluation assessing their ability to present an Ag85B-derived epitope to an F9A6 human HLA-DR1-specific CD4 T cell hybridoma. siRNA knockdown of ATG5, 7, 12, 16L, and Ulk1-ATG1 enhanced the CFU counts of Mtb in M1-MФs but not M2-MФs (Fig. 3i). Notably, autophagy knockdown also led to a decrease in antigen presentation by Mtb-infected M1-but not M2- or M0-MФs (Fig. 3j), indicating a reduced ability of the latter to process and present antigens. These data validate our findings in BCG and Mtb-infected mouse DCs and MФs, where we demonstrated that autophagy plays a major role during Ag85B-p25-mediated T cell activation29. Though lysosome mediated Mtb degradation is associated with increased antigen presentation, we note that Mtb can reduce antigen presentation through other mechanisms. For example, Mtb can downregulate MHC-II, interfering with antigen presentation37. Intriguingly, Mtb-infected M1-MФs showed increases in antigen-processing components, including MHC-II, vATPase, and cathepsin transcripts (Supplementary Figs. 9, 10). These data indicate that the lysosomes of Mtb-infected M1-MФs are more degradative relative to those of Mtb-infected M2-MФs. Since lysosomal degradation of Mtb enhances the immunogenicity of M1-MФs we analyzed this issue as follows.
Mtb-infected human M1- and M2-MΦs differentially express enzymes regulating autophagy-depedent antigenic epitope presentation
Mtb-infected M1-MФs expressed multiple RAB GTPases compared to M2-MФs (Figs. 3b, 4a). RAB7 was elevated in M1-MФs during RNAseq analysis (Fig. 4a) and enriched on the rfpMtb phagosomes of M1-MФs (Fig. 4b; Supplementary Fig. 3c). Lysosomal marker LAMP1 was also enriched on rfpMtb phagosomes of M1-MФs indicating autophagic flux (Fig. 1m, n). Importantly, siRNA knockdown of RAB7 led to reduced antigen presentation only among Mtb-infected M1-MФs (Fig. 4c). These data support the notion that RAB7 regulates lysosomal fusion and is associated with sorting of MHC-II molecules into MIIC compartments for antigen presentation38,39.
Mtb-infected M1-MФs expressed transcripts for multiple cathepsins (CTSA, CTSZ, CTSH, CTSC, CTSL1, CTSS, and CTSB) compared to M2-MФs (Fig. 4d, e). Even uninfected M1-MФs demonstrated increased cathepsin expression, with CTSA, CTSZ, and CTSH expressed only by M1-MФs. Because many of the specific antibodies for human cathepsins were not available, we treated Mtb-infected MФs with enzyme-specific or pan-specific cathepsin inhibitors and assessed antigen presentation using an overlay with the F9A6-CD4 T cell line specific for Mtb Ag85B epitope40. Cathepsin inhibition markedly decreased antigen presentation by Mtb-infected M1-MФs compared to both M2- and M0-MФs (Fig. 4f). Though all of our tested cathepsin inhibitors reduced antigen presentation in M1-MФs, only pan-specific inhibitors (E64, NALLM) or the CTSK-specific inhibitor, balicatib, had inhibitory effects in Mtb-infected M2-MФs. Because CTSK is not a major contributing enzyme during antigen processing, this finding suggested that cathepsin-dependent antigen processing in M2-and M0-MФs is weak. In support of this, a Clusterprofiler pathway analysis revealed an upregulation of multiple genes controlling antigen processing in Mtb-infected M1-MФs compared to M2-MФs (Supplementary Fig. 9) and that lysosomes of Mtb-infected M1-MФs were enriched for glycosidases, lipases, and sulfatases in addition to cathepsins (Supplementary Fig. 10). To determine whether bactericidal function is associated with antigen presentation, lysates of MФs treated with cathepsin inhibitors were plated for Mtb CFU counts and replicates tested concurrently for antigen presentation at 18 h. Strikingly, cathepsin inhibition enhanced the survival of Mtb in M1- and M0-MФs but not M2-MФs (Fig. 4g). Because cathepsin inhibition reduced antigen presentation, and enhanced Mtb growth (Fig. 4g, h), we propose that increased levels of lysosomal cathepins is a feature of Mtb-infected M1-MФs compared to M2- or M0-MФs (Supplementary Fig. 9).
After confirming the mechanisms underlying the differential immunogenicity of human MФ phenotypes ex vivo (Fig. 4a–h), we sought to further characterize MФ heterogeneity in vivo during human TB. PBMC-derived MФs of household contacts (defined by their close contact with TB patients), TB patients from the US-Mexico border zone, and healthy donors were tested for their ability to kill Mtb ex vivo in the presence or absence of Rapamycin-induced autophagy in MФs. The bactericidal competency of MФs differed ex vivo even without added cytokines or Rapamycin (Fig. 4i). We note here that unlike M1- and M2-MФs differentiated in vitro, (Fig. 1) PBMC-derived populations likely contained a spectrum of M1- and M2 -phenotypes; therefore, we expressed CFU counts using a linear scale. MФs of healthy controls and contacts degraded Mtb more efficiently than MФs of TB patients, though Rapamycin enhanced bactericidal activity in all groups. To determine whether heterogeneity in bactericidal competency also affected the immunogenicity of PBMC-derived MФs, we further tested their ability to present an Ag85B-derived epitope to F9A6-CD4 T cells. MФs better able to kill Mtb (Fig. 4i) showed an increased present antigen presentation (Fig. 4j). This finding validates our previous report that Rapamycin enhances the immunogenicity of mouse antigen-presenting cells in vitro and in vivo29,41 and is consistent with the observation that human monocyte-derived Langerhans DC-mediated antigen presentation is linked to autophagy-dependent degradation of mycobacteria42. Notably, some PBMCs may have derived from non-HLA-DR1 subjects and yielded false negatives, although Rapamycin enhanced antigen presentation among all groups of MФs. Because we were not able to purify M1- and M2- phenotypes from TB patients, we once again differentiated M1- and M2-MФs from healthy donors from the same area. These MФs also exhibited differential control over Mtb proliferation; Fig. 4k illustrates CFU counts day 3 post-infection. Furthermore, siRNA knockdown of beclin1/ATG6 enhanced Mtb proliferation in M1- and M0-MФs but not M2-MФs (Fig. 4l). These data are consistent with the previous observation that PBMCs derived from LTBI donors secreting high levels of IFN-γ upon in vitro activation with Mtb antigen (“high responders”) are of LC3 high phenotype, whereas PBMCs of “low responders” show decreased LC3 levels43. Thus, MФ heterogeneity contributes to bactericidal competency and T cell activation during human TB in PBMCs.
Together, these data demonstrate that the lysosomal environment of M1-MФs is strongly degradative for Mtb and autophagy plays an important role during the degradation and processing of Mtb antigens and T cell activation44–47. This assumes importance because, classical M1- but not non-classical M2-MФs can differentiate into monocyte-derived dendritic cells, which are also potent human antigen-presenting cells48. A relative dominance of M1-MФs during TB can therefore lead to both increased degradation of Mtb and improved T cell activation.
Mtb infection affects autophagy in human M1- and M2-MФs via epigenetic alteration of ATG5
Because Mtb secretes lysine acetylases, we hypothesized that Mtb modulates autophagy in MΦs through acetylation of ATG-associated histones49 similar to its ability to enhance IL-10 from naïve MΦs50. ChIP-qPCR analysis indicated that, in non-infected and Mtb-infected MΦs, both H3K18 and H4K16 acetylation of the chromatin associated with ATG5 promoter were higher in M1-MΦs than in M2-MΦs (Fig. 5a). Thus, hyperacetylation of histones correlated with a higher level of autophagy in Mtb-infected M1-MΦs and hypoacetylation correlated with a lower level of autophagy in Mtb-infected M2-MΦs. This is consistent with the observation of Fullgrabe et al, that H4K16 deacetylation is associated with the downregulation of autophagy-related genes51 and that Rapamycin decreases histone acetyltransferase hMOF levels facilitating acetylation of ATGs and induction of autophagy51. Because M0-, M1- and M2-MΦs showed differential acetylation of H3K18 vs.H3K16 after Mtb infection, we propose that Mtb interferes differentially with H3 and H4 acetylation of ATG5 presumably through lysine acetylases or deacetylases50.
To understand the regulatory mechanisms underlying histone acetylation, we analyzed NAD+-independent histone deacetylase (HDAC) and NAD+-dependent histone deacetylase (Sirtuins) gene expression52. Unexpectedly, we found a striking difference in the expression of HDACs. M1- and M2-MΦs both expressed HDACs and Sirtuins. Whereas, Mtb-infected M1-MΦs strongly expressed Sirtuin5, HDAC2 and Sirtuin2 were better expressed by M2-MΦs (Fig. 5b, c). qPCR (Fig. 5d, e) and western blot studies validated gene expression levels for HDAC1/2 and Sirtuin2/5 (densitometry shown in Supplementary Fig. 15). Notably, Mtb infection induced a progressive increase in Sirtuin2 expression in M0-MΦs (Fig. 5f, g). In additional experiments, we found that only virulent Mtb was able to induce Sirtuin2, whereas avirulent BCG vaccine was better at inducing Sirtuin5 but not Sirtuin2 (Fig. 5h, i). These data are consistent with the report that Sirtuin2 is a target for immunotherapy of TB using mice53.
Because M2-MΦs had enhanced deacetylase activity, we sought to verify the functionality of HDACs and Sirtuins in relation to autophagy. We knocked down Sirtuin2 and Sirtuin5 using siRNA duplexes followed by growth assay. Sirtuin2 but not Sirtuin5 knockdown led to an increase in the ability of all three phenotypes of MΦs ((Fig. 5j). Next, we treated MΦs with pharmacological inhibitors of class I and II HDACs or with sirtinol, a dose-dependent specific inhibitor of Sirtuin2, before Mtb infection followed by assays for antigen presentation and Mtb proliferation. Entinostat, which inhibits class I HDAC1 and HDAC3, reduced antigen presentation by Mtb-infected M1- but not M2-MΦs (Fig. 5k). Paradoxically, inhibition of Sirtuin2 enhanced antigen presentation in Mtb-infected M1- but not M2- or M0-MΦs. As Entinostat likely reduces global histone acetylation (especially H3K18 acetylation), and sirtinol preferentially reduces H4K16 acetylation54–56, our data are consistent with the acetylation status of the histones of the ATG5 promoter related to autophagy. Figure 5l shows that Sirtuin2 blockade using sirtinol decreased Mtb survival in M1-, M2-, and M0-MФs. Sirtuin2 is therefore a negative regulator of autophagy and its pharmacological targeting can improve the bactericidal function of M2-MФs. Importantly, Entinostat, which is a broad-spectrum HDAC inhibitor enhanced Mtb proliferation in M1- but not M2- or M0-MФs, underscoring the importance of H3K18 and H3K16 acetylation and autophagy.
Among the HDAC inhibitors we tested, Belinostat, Romidepsin (HDAC1 and 2), and Tubastatin (HDAC6) inhibited acetylation at H3 and H4 using cancer cells but did not affect the ability of Mtb-infected MФs to present antigen to CD4 T cells (Fig. 5i). Sirtuins regulate autophagy57 and both Sirtuin1 and Sirtuin3 seem to control mycobacterial proliferation in mice through autophagy58. Because Sirtuin5 and Sirtuin2/HDAC2 were upregulated in human M1- and M2-MФs respectively, we propose that human and mouse MФs differ in their regulation of autophagy following Mtb infection. Importantly, enhanced Sirtuin2/HDAC2 deacetylases in M2-MΦs explain in part why their autophagy is reduced despite increased gene expression of some ATGs (Fig. 3). Of note, Sirtuin5 is a mitochondrial protein deacetylase, suggesting that acetylation of mitochondrial proteins may also play a role in metabolism and autophagy in M1-MФs. Thus, in addition to IFN-γ, which drives M1-MΦs towards increased autophagy, Mtb-induced epigenetic modifications in ATG5-associated histones can also alter autophagy in M1- vs. M2-MΦs to affect Mtb survival. Notably, Mtb differed from BCG vaccine in selectively inducing Sirtuin2 in naïve MΦs suggesting pathogen dependent modulation.
Mtb infection alters innate immunity regulating gene (Inregs) expression in human M1- and M2-MФs to affect anti-mycobacterial immunity
Mtb-induced transcriptomic (mRNA) signatures have been analyzed most often in PBMCs and occasionally in bronchoalveolar lavage-derived cells. A meta-analysis of eight studies showed that up to 55 different genes or gene clusters were differentially expressed in the PBMCs of TB patients versus LTBI59,60. However, no gene signature was found uniformly in all studies and only 10 genes or clusters were found in more than two studies (Supplementary Fig. 11). Because these studies analyzed unfractionated cells containing a mixture of immune cells including T cells, neutrophils, DCs, and MФs, we hypothesized that detection of M1- and M2-MФ-specific biomarkers associated with bactericidal function may correlate better with resistance and susceptibility, respectively, to TB.
We characterized additional differentially expressed transcripts using RNAseq analysis. Because cord blood-derived M1- and M2-MФs are naïve relative to the PBMC-derived MФs of adults, we first determined whether the prenatal MФ-derived M1- and M2-MФs elicited a transcriptional response comparable to that seen in adult MФ-derived M1- and M2-MФs. Transcriptomic analysis of Mtb-infected M1- and M2-MФs in the absence of LPS activation has not yet been reported23. Indeed, when M1-MФs programmed with IFN-γ alone or IFN-γ plus LPS were compared, IFN-γ+LPS programmed cells showed elevated gene expression, underscoring the pleiotropic effects of LPS23. This is consistent with our report that LPS induces autophagy in MФs through TLR-4 activation25. Nonetheless, previous studies using LPS and IFN-γ programmed M1- and IL-4 treated M2-MФs show differential expression of transcripts for CD80, ITGAL, TNSFS10, FZD2, TRAFD1, STAT1, CLIC2, EMILIN2, OPTN, TAP1, FcG receptors, and SERPING123,48. Interestingly, despite excluding LPS in this study, nearly all earlier reported genes for uninfected M1- and M2-MФs were differentially enriched in cord blood-derived IFN-γ programmed M1- and IL-4 treated M2-MФs (Fig. 6a, b). Because naïve and Mtb-infected human M1-, M2-, and M0-MФs differentially expressed more than 100 genes or gene clusters (Supplementary Fig. 4)32,61, including Inregs and clusters associated with anti-microbial immunity, we analyzed the functionality of selected Inregs using ex vivo QPCR validation of mRNA transcripts in M1-, M2-, and M0-MФs and PBMCs of children with TB and their contacts.
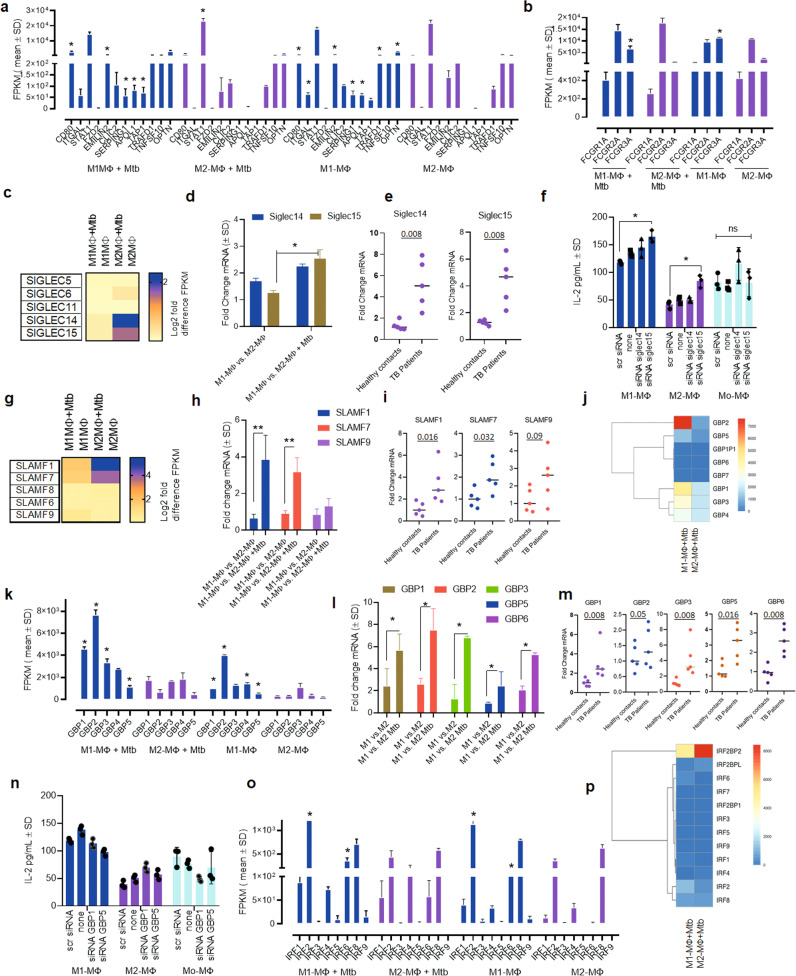
Fig. 6. Mtb infection induces Inregs in human M1- and M2-MФs associated with anti-mycobacterial immunity.
a, b Mtb-infected and naive M1-, and M2-MΦs show RNAseq-derived differential gene expression for biomarkers at 18 h. Transcripts are shown as FPKMs (fragments per kilobase per million reads. (FPKMs shown; n = 2; *p < 0.01 t test). c–f Mtb-infected M1- and M2-MΦs differentially express transcripts (FPKMs) for Sialic Acid Binding Immunoglobin like Lectins (Siglecs). c RNAseq-derived differential gene expression; Log2-fold gene expression. d QPCR of mRNA for Siglec-14/15 in Mtb-infected M1- vs. M2-MΦs at 18 h (duplicate sample per assay; 2 similar experiments; *p < 0.01, t test). e QPCR of mRNA in the PBMCs of children with TB (n = 5) and their household contacts at 18 h (n = 5). f siRNA knockdown of Siglec-14/15 in Mtb-infected M1-, M2, and M0-MΦs followed by antigen presentation to F9A6-CD4 T cells. (triplicate wells per assay; 2 similar experiments; *p < 0.009, t test). g–i Differential gene expression for Signaling Lymphocyte Activation Molecule family (SLAMF). g RNAseq- derived differential gene expression; Log2-fold gene expression. h QPCR of mRNA in M1- and M2-MΦs at 18 h (duplicate sample per assay; 2 similar experiments; *p < 0.01, t test). i QPCR in the PBMCs of children with TB and their household contacts. j–m Differential gene expression for Guanylate-binding proteins (GBPs) in M1- and M2-MΦs. j, k RNAseq- derived differential gene expression (l) QPCR of mRNA in Mtb-infected or naïve M1 and M2-MΦs at 18 h (duplicate sample per assay; 2 similar experiments; *p < 0.01, t test). m QPCR of mRNA in the PBMCs of children with TB and their household contacts (*p < 0.01, t test). n siRNA knockdown of indicated GBPs in M1-, M2, and M0-MΦs followed by antigen presentation to F9A6-CD4 T cells. (triplicate wells per assay; 2 similar experiments; *p < 0.009, t test). o–p RNAseq-derived differential gene expression for Interferon Regulatory Factors (IRFs) in Mtb-infected or naïve M1- and M2-MΦs at 18 h. For panels (a, b, k, o) FPKMs of Mtb-infected vs. naïve were compared (*< 0.01 t test; n = 2). All Mtb infections used MOI of 1. Both RNAseq and QPCR analysis done at 18 h post Mtb infection.
We found that naïve and Mtb-infected M2-MФs differentially expressed Sialic Acid binding Immunoglobulin type Lectins, including SIGLEC-14, and SIGLEC 15 (Fig. 6c–f). QPCR studies showed a superior induction of SIGLEC-15 mRNA in M1-MФs after Mtb infection (Fig. 6d). Likewise, children with TB had elevated SIGLEC-14 and SIGLEC-15 in PBMCs compared to their contacts (Fig. 6e). siRNA knockdown of SIGLEC15 but not SIGLEC14 led to increased antigen presentation to T cells in M1- and M2-MФs (Fig. 6f). We conclude that SIGLEC14/15 are potential biomarkers associated with M1- and M2-MФs that warrants additional studies.
A second gene cluster showing differential expression was Signaling Lymphocyte Activation Molecule Family SLAMF-1, 7, and 9 (Fig. 6g). Our QPCR studies of Mtb-infected M1-MФs showed increased SLAMF1 and 7 expression ex vivo (Fig. 6h). In a similar manner, PBMCs of children with TB showed elevated SLAMF1/7 and 9 compared to their contacts (Fig. 6i). Preliminary studies showed that siRNA blockade of SLAMF1,7 and 9 did not affect the ability of MФs to present antigen and need additional studies.
A third gene cluster showing differential expression was Guanylate-Binding Proteins (GBPs). GBPs are IFN-γ-inducible and Mtb-infected M1-MФs showed enrichment of GBPs 1, 2, 3, 5 and 6 (Fig. 6j–l). Intriguingly, these were also enriched in among PBMCs of children with TB compared to controls (Fig. 6m). Because GBPs were expressed mostly in Mtb-infected M1-MФs, we propose a defensive role for this gene cluster, although a siRNA knockdown of GBP1 and 5 did not affect antigen presentation (Fig. 6n). siRNA knockdown of GBP2, 3 and 6 were not successful. Although additional studies are required to elucidate their function, we note that GBPs were not upregulated in Mtb-infected IFN-γ programmed mouse M1-MФs ex vivo32, suggesting that GBPs likely play a differential regulatory role in mouse vs. human MФs.
The fourth cluster of differentially expressed genes was the interferon regulatory factors (IRFs). IRFs 2 and 6 were enriched in Mtb-infected M1-MФs compared to Mtb-infected M2-MФs, whereas IRF8 was expressed at comparable levels (Fig. 6o, p). Of note, IRF1 and IRF8 are among the few genes differentially expressed by both Mtb-infected humans (this study) and mouse-derived M1- and M2-MФs32.
Taken together, these data indicate that naïve or Mtb-infected cord blood-derived M1- and M2-MΦs not only faithfully reproduce the gene signatures identified earlier in human PBMC transcriptomics, but also reveal gene clusters likely associated with MФ-dependent control of TB. Importantly, the transcriptional responses of human and mouse-derived M1- and M2-MФs differed for ATGs, RAB GTPases, cathepsins, SIGLECs, SLAMF, GBPs, and IRFs, which likely affects the differential control of tuberculosis in mice and humans.
Inregs expressed by Mtb-infected human M1- and M2-MФs are found in the lymph nodes and macrophages of Mtb-infected neonatal macaques
To detect Inregs during in vivo TB, we infected rhesus macaques with the Erdman strain of Mtb and quantified lung Mtb burden six weeks after infection. Additionally, we isolated MФs from bone marrow collected from naïve NHPs. We performed a transcriptome analysis on lymph nodes collected during necropsy using RNAseq, as they are enriched for MФs62 and collection of BAL-derived MФs from infant macaques was not feasible. Figure 7a shows that three of four animals exhibited increased Mtb burden, though one animal had barely detectable Mtb. The lymph nodes from two macaques with high Mtb burden showed an upregulation of gene clusters regulating phagosome and lysosome traffic, NOD-TLR signaling, chemokine-cytokine regulating network, and FcG-R signaling (Fig. 7b). Because these gene clusters were similar to those observed among ex vivo Mtb-infected human M1- and M2-MФs (Fig. 2), we propose that rhesus macaque lymph nodes reproduce human MФ-transcriptional responses to Mtb (Fig. 7b). Unlike ex vivo Mtb-infected human M1- and M2-MФs (Fig. 2), macaque lymph nodes contain many other immune cells. We noted upregulation of additional gene clusters, including the IL-17 network and phosphatidyl-inositol-3 kinase (PI3K)-Akt axis in macaque lymph nodes (Fig. 7b).
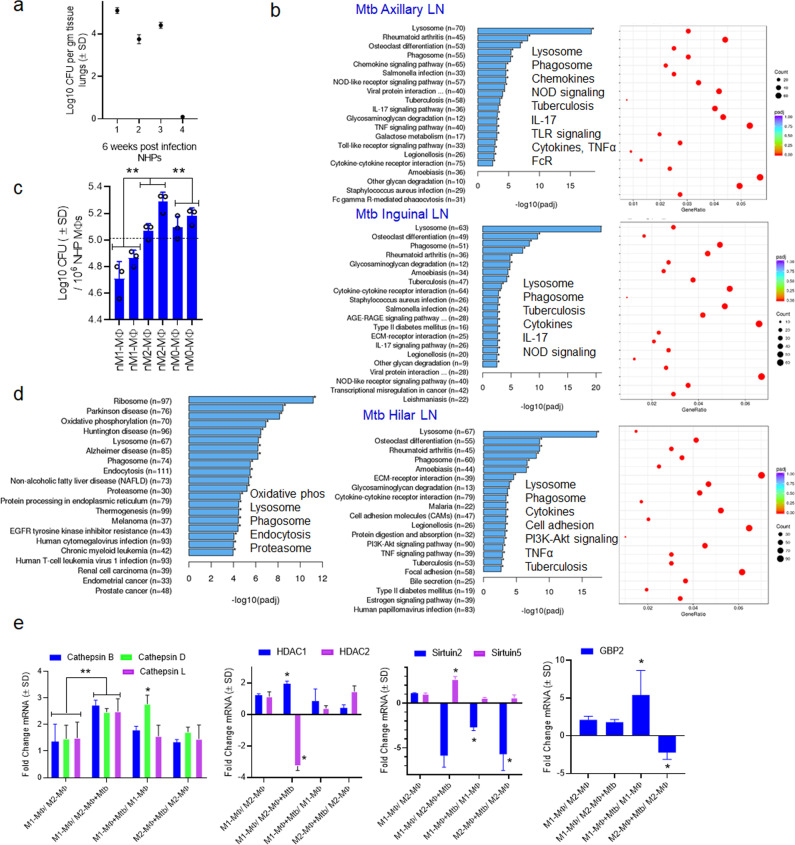
Fig. 7. Inreg clusters expressed by Mtb-infected human M1- and M2-MФs are found in the lymph nodes and macrophage transcriptome of Mtb-infected neonatal rhesus macaques.
a Six-week-old rhesus macaques were aerosol-infected with Mtb Erdman strain (25 CFU per animal; n = 4) followed by sacrifice at 6 weeks and CFU counts of lungs. b lymph nodes (n = 2) collected at necropsy from macaques that had comparable Mtb counts in lungs (panel a) were analyzed using RNAseq. Kyoto Encyclopedia of Genes and Genomes profiles of one NHP illustrate the differential gene expression for Inreg clusters; Clusterprofiler pathway analysis of Inregs is illustrated in Supplementary Fig. 12. c M1-, M2-, and M0-MΦs were prepared from the bone marrow of naïve macaques (prefix n) and infected with Mtb followed by CFU counts on day 4 (**p < 0.01 one-way ANOVA with Tukey’s post-hoc test; 2 experiments shown). d Kyoto Encyclopedia of Genes and Genomes profiles of Mtb-infected M1-MΦs vs. Mtb-infected M2-MΦs show enrichment of genes regulating antigen processing 18 h post-infection. e Naïve or Mtb-infected M1-, and M2-MΦs were subjected to QPCR at 18 h post-infection using primers for mRNA of indicated genes which were differentially expressed in their human counterparts (*p < 0.01, t test). All ex vivo Mtb CFU experiments used MOI of 1.
To further determine whether human and macaque MФs show comparable gene expression profiles, we differentiated naïve bone marrow-derived MФs into M1-, M2-, and M0-MФ controls and assessed 1) gene expression using RNAseq and 2) their ability to control Mtb growth. Macaque-derived M1-MФs, but not M2- or M0-MФs, inhibited Mtb proliferation (Fig. 7c). Furthermore, a transcriptome analysis of naïve and Mtb-infected NHP M1-, M2-, and M0-MФs showed striking similarities to an analysis of human M1-, M-2, and M0-MФs. For example, ‘antigen processing transcriptome’ genes were similar among NHP and human Mtb-infected M1-MФs (Fig. 2b vs. Fig. 7d). We then validated select RNAseq-identified genes using QPCR. Similar to their human counterparts, NHP M1-MФs upregulated CTSB, CTSD, CTSL, HDAC1/2, Sirtuin2/5 and GBP2 (Fig. 7e). Furthermore, MHC-I, MHC-II, and DC-SIGN were upregulated in the regional lymph nodes of Mtb-infected NHPs, whereas cathepsins and other lysosomal enzymes were downregulated (Supplementary Fig. 12). This expression profile is consistent with the development of TB in these animals. Regrettably, we could not maintain NHPs beyond 6 weeks post-challenge to ascertain whether TB could have been controlled by NHPs. With this caveat, we conclude that neonatal macaques display similar MФ heterogeneity to that observed in human MФs.
Discussion
Most humans exposed to aerosol infection with Mtb do not develop active lung TB. Because both alveolar MΦs (AMΦs) and infiltrating MΦs (IMΦs) are exposed to T cell-derived cytokines during Mtb alveolar infection, we speculated that MΦ subsets may develop differential abilities to resist Mtb infection. An elegant study involving mice and fluorescent Mtb reporters indicated that AMΦs with a fatty acid oxidation cycle less effectively controlled Mtb proliferation, while glycolytically active IMΦs effectively inhibited Mtb63. Another study using single-cell RNAseq showed that mouse lungs contain both CD206− (M1-like) and CD206+ (M2-like) interstitial MΦs; the latter exerted a suppressive influence through cytokines around the bronchioles, while the former occupied the alveolar space and resembled antigen-presenting cells64. Human AMΦs express M1-(CD206loCD86 hi) and M2-like phenotypes (CD206hiCD86lo) in a steady-state that is altered after alveolar infection19. IMΦs are continuously replenished from circulating monocytes. Although TB granulomas contain a variety of immune cells (including M1- and M2-MΦs), few studies have examined M1- and M2- MΦ phenotype-associated gene expression during TB65–67. Finally, treatment of diabetics with glybenclamide which regulates potassium and chloride channels, decreased M1-MФs and increased M2-MФs in PBMCs, resulting in an overall decrease in PBMC bactericidal activity against Mtb in vitro68. Taken together, these observations strongly support a role for functional M1- and M2-MФ heterogeneity not only at the alveolar and lung level but also in circulating blood.
Coincidentally, transcriptomic studies of PBMCs from TB patients and their contacts showed a limited set of genes associated with either susceptibility or resistance (Supplementary Fig. 11). Although myeloid cell-specific signatures were dominant, a gene expression profile associated with a specific MФ phenotype and function has not been reported for TB.
In this study, we demonstrate that the ability of M1- and M2-MΦs to degrade Mtb is associated with both oxidants and autophagy (Fig. 1). Although independent studies using mouse M1- and M2-MΦs show altered NO and ROS responses or differential LC3 labeling of mycobacteria69,70, we found that human M1-MΦs upregulate all three antimycobacterial mechanisms to restrict Mtb (Fig. 1). Though AMФs express M1- and M2- phenotypes19, they self-sustain within the alveoli with chemokines facilitating alveolar infiltration by IMΦs and other immune cells. We found that Mtb-infected M1-MΦs secrete several pro-inflammatory cytokines and display increased gene and protein expression of CC- and CXC-type chemokines relative to Mtb-infected M2-MФs (Supplementary Fig. 2). These data suggest that during granuloma formation, M1-MΦ dominance and a pro-inflammatory environment together elicit more robust immune cell recruitment and thereby facilitate an effective TB control response.
Previous studies show that IFN-γ, IL-4, and IL-10 have dramatic effects on Mtb survival22. Few studies report the polarization of human M1-MΦs. Among these two studies both used IFN-γ and LPS to drive M1-polarization making it difficult to interpret their bactericidal function since LPS activates autophagy through TLR-414,71 and an LPS-like monophosphoryl lipid A analog is a well characterized adjuvant. Another study used vitamin-D to drive M-1 like differentiation although traditionally M1 phenotype is driven by IFN-γ20 and vitamin-D also induces autophagy72. This study is therefore the first to rule out LPS-induced pleotropic effects during M1- differentiation. To obtain an in-depth understanding of the signaling mechanisms controlling MΦ function, we performed RNAseq and found that Mtb-infected M1-MΦs expressed a unique transcriptome. Further, gene expression in Mtb-infected human M1- and M2-MΦs was stronger and more diverse than in mouse M1- and M2-MΦs32. More than 100 genes were upregulated in Mtb-infected human M1- vs. M2-MΦs (Supplementary Fig. 4) versus less than a dozen in mouse MΦs32. Interestingly, Mtb-infected human M1-MΦs expressed a unique ‘antigen processing transcriptome’ (Mtb M1- vs. Mtb M2-MΦs; Fig. 2b), and pathway analysis indicated that Mtb phagosomes were more effectively sorted to APLs for enzyme-mediated degradation and peptide epitope production. This finding is interesting as autophagy has emerged as a major innate immunity mechanism that sorts mycobacteria to APLs for their enzymatic degradation. The peptides epitopes generated are routed for CD4 T cells, whereas autophagy also enhances cross-presentation to CD8 T cells42,44,46,73,74. Paradoxically, except for ATG5-deficient mice, many ATG transgenic mice were not more susceptible to TB75. However, we emphasize that, unlike humans, mice are uniformly susceptible to aerosol-induced TB and ATG-dependent autophagy does require accessory genes like RAB GTPases76, and autophagolysosomal mycobacterial degradation requires vATPase and cathepsins.
In this study, we have demonstrated a mechanism by which many TB-exposed humans may resist active disease. Mtb-infected M1-MФs upregulated ATGs, RAB GTPases, and autophagy-regulating accessory genes like MAPLC3IIB (ATG8), LAMP1, galectins (LGALS), and TRIM proteins compared to M2-MФs (Fig. 3). siRNA knockdown of beclin1 (ATG6) and other ATGs increased the growth of Mtb in M1-MФs compared to M2-MФs ex vivo (Figs. 1k, 3i). Together, LAMP1, TRIMs, and galectins (LGALS) regulate various stages of xenophagy and sort mycobacteria to APLs33,77. LGALS3 was expressed at higher levels by Mtb-infected M1-MФs compared to M2-MФs, but TRIM16 (a binding partner for LGALS3; aka. Gal3) was expressed only by Mtb-infected M1-MФs (Fig. 3d, e) and we propose that LGALS3/8 (Gal3/8) and TRIM38 are biomarkers for Mtb-infected M1-MФs78. We conclude that, unlike their mouse counterparts, upon Mtb infection human M1-MΦs strongly upregulate genes associated with their autophagic sorting to APLs.
An interesting consequence of lysosomal degradation of mycobacteria is the peptide epitope-mediated activation of CD4 T cells79,80. Mtb and BCG both sequester within immature phagosomes and induction of autophagy delivers them to lysosomes29,41. Autophagy also mediates MHC-II and MHC-I-dependent mycobacterial and non-mycobacterial antigen presentation29,41,44–47. We found that Mtb-infected M1-MФs upregulated both autophagy and MHC pathway genes and increased expression of key genes for MHC-I, MHC-II, vATPase, and multiple lysosomal cathepsins (Fig. 4; Supplementary Figs. 9, 10). Increased antigen presentation by Mtb-infected M1-MΦs correlated with CTSB, CTSL, and CTSS, which are critical during antigen processing and antigen loading into MHC/HLA-DR complexes81,82. Further, blockade of CTSB, CTSL, and cysteine proteases in MΦs enhanced Mtb proliferation (Fig. 4g), indicating that the bactericidal function of M1- but not M2-MΦs correlates with lysosomal cathepsin function83–85. Interestingly, lysosome leakage-derived CTSB can also activate the inflammasome during Mtb infection of MΦs, culminating in IL1-β production86; Mtb-infected M1-MΦs induced higher levels of the key protective cytokine IL1-β coincident with increased caspase induction (Supplementary Figs. 2, 6). M2-MΦ lysosomes not only lacked an enrichment of cathepsins relative to M1-MΦs but also lacked an enrichment of other hydrolases like glycosidases, sulfatases, and lipases (Supplementary Fig. 10). These data provide a mechanistic explanation for the link between MΦ lysosomal disorders and increased TB susceptibility84. Taken together, these results provide new insight into how enhanced lysosomal enzymes may enable M1-MΦs to both kill Mtb and degrade them to produce antigens for T cell activation. Conversely, the results suggest that M2-MΦs show a reduced immunogenicity phenotype due to reduced lysosome function.
A third major mechanistic insight from this study is based on the paradoxical role MΦs play in both degrading intracellular Mtb and acting as a niche for Mtb persistence over decades. Mtb is a unique pathogen that secretes or sheds multiple virulence factors to facilitate immune evasion87. For example, Mtb secretes sapM phosphatase, which interferes with phagosome-lysosome fusion in MΦs. Indeed, a ∆sapM strain of Mtb was attenuated but immunogenic in mice88,89. Others have also shown that Mtb evades the autophagy pathway leading to APLs30,90. Herein, we demonstrate a regulatory mechanism wherein Mtb infection of M1-MΦs increases ATG5-associated histone acetylation51,91. We also show that the decreased autophagy phenotype of M2-MΦs is maintained by their elevated expression of HDAC-Sirtuins and they also show a reduced expression of LC3, RAB7 and LAMP1 which regulate autophagolysosomal fusion. Importantly, pharmacological activation of autophagy seems feasible for M2-MΦs which can also increase their immunogenicity (Fig. 5). While Everolimus (a purified form of Rapamycin) is being used in combination with anti-TB drugs to augment autophagy during host-directed therapy, others have used HDAC/Sirtuin inhibitors for augmenting host defense92. Rapamycin decreases the hMOF acetyltransferase enhancing autophagy and we provided evidence that it enhances degradation of Mtb (Fig. 1)51. Intriguingly, the class I HDAC inhibitors trichostatin and SAHA had an unexpected adverse effect on MΦ-mediated Mtb uptake and degradation93; our data therefore underscore the importance of understanding the differential expression of HDAC/sirtuins by M1- and M2-MΦs before devising host-directed therapy.
IFN-γ reprograms MΦs for anti-mycobacterial function but can also enhance autophagy94,95. Accordingly, low-dose activation followed by resting still left a remarkable imprint on M1-MΦs, which enabled them to respond rigorously to Mtb infection. Even then, Mtb persisted in both M1- and M2-MΦs, although long-term cultures showed a gradual CFU decline in M1 vs. M2-MΦs and enhanced cell death in M2-MΦs. Because of these interactions between host and Mtb- dependent histone modifications, we conclude that Mtb is likely to persist in a non-replicating state in M1- but not M2-MΦs particularly in view of the data that Mtb-infected M1- but not M2-MΦs continue to secrete GM-CSF (Supplementary Fig. 2). We therefore propose that selective regulation of histone acetylation using HDAC and Sirtuin inhibitors in combination with anti-TB drugs may help to both control and eradicate Mtb from MΦs irrespective of their phenotype.
The fourth novel observation from our study is that Inreg signatures identified using M1- and M2-MΦ transcriptomes are detectable in PBMCs for children with TB and their contacts. Because M1 and M2-MΦs differentially control Mtb proliferation, we hypothesized that detection of Inregs like SIGLEC, SLAMF, and GBP family of genes may help to differentiate between LTBI and TB patients. These Inregs showed a striking difference in expression within M1- vs. M2-MΦs ex vivo, and were also able to sufficiently discriminate TB patients from their contacts (Fig. 6). Interestingly, at least two studies report a genetic association between SIGLEC14/15 and susceptibility to TB96,97. SIGLEC15 is also known as an immunosuppressive checkpoint during cancer immunotherapy98 and siRNA knockdown of SIGLEC15 increased antigen presentation by MΦs (Fig. 6f). In contrast, SLAMF1/7 members which were enriched in Mtb-infected M1-MΦs (Fig. 6h) are associated with both positive and negative regulation of chemotaxis, autophagy, and plasmacytoid DC function99–101. GBPs which were associated strongly with M1-MΦs discriminated contacts from TB patients (Fig. 6m) and were earlier identified in 4 of 8 transcriptomic studies on TB patients and their contacts60. GBPs regulate phagosome maturation, autophagy, mycobacterial antigen processing, and cell-autonomous or innate killing of intracellular pathogens102–105. We conclude that M1- and M2-MΦ-associated Inregs are more likely to discriminate between TB-resistant and TB-susceptible individuals and are newer targets for diagnosis and immunotherapy of TB.
Finally, to validate gene expression findings from ex vivo cultured and Mtb-infected human MΦs, we developed a neonatal macaque model of TB. Using this model, we show the first in vivo transcriptional profiles for lymph nodes following TB. Although we also analyzed lung transcriptomes, we sought to focus on lymph nodes because they are enriched in both MΦs and DCs and play a major role in regulating immunity to TB106. Unlike mice, adult NHPs show a dose-dependent susceptibility to TB, although infant macaques are more susceptible107–109. For example, about a third of adult rhesus macaques develop TB after aerosol infection with an Erdman strain of Mtb, whereas we found that three of four neonates showed a significant Mtb burden in the lungs (Fig. 7a).
Our studies suggest a potential reason for the differential susceptibility of macaques to TB. Although the lymph nodes contained multiple types of immune cells, the myeloid regulatory pathways of phagosome/lysosome, NOD/TLR signaling, cytokine-cytokine interactions, cell adhesion, and MAPK were transcriptionally enriched, similar to Mtb-infected M1-MΦs (Fig. 7b vs. Fig. 2b). Paradoxically, Mtb infection upregulated MHC-I and MHC-II genes but downregulated cathepsins and lysosomal hydrolases essential for TB control during LN profiling (Supplementary Fig. 12). Furthermore, NHP bone marrow-derived Mtb-infected M1- and M2-MΦs also displayed differential control over Mtb proliferation (Fig. 7d) and differed in their enrichment of genes associated with the antigen processing transcriptome (Fig. 7e).
In an earlier study, PBMCs from Mtb-infected rhesus and Chinese cynomolgus monkeys displayed differential expression of genes associated with M1- and M2- phenotypes, although they were not examined for their ability to degrade Mtb, and M1- and M2-MΦs were not enriched from these primates to assess molecular mechanisms110. As we recently reported that some adult NHPs can still prevent reactivation of TB despite severe T cell loss due to Simian Immunodeficiency Virus111, we propose that MΦ heterogeneity may play an important role during TB in macaques. Regrettably, we were unable to follow Mtb infection beyond 6 weeks post-infection, which precluded us from determining whether infant macaques could have controlled TB beyond 6 weeks leading to latency. Nonetheless, our observations argue that NHP MΦs mirror human MΦs in their transcriptional responses to TB. Therefore, infant macaques provide a novel model with which to develop vaccines and determine the role of myeloid vs. lymphoid cell-mediated protection against TB.
In conclusion, we demonstrate that human MФ subsets show a unique pattern of gene expression that enables differential control over TB proliferation in humans and macaques. Some of the phenotype-specific genes are promising targets for vaccine development, diagnostics or therapeutics.
Methods
MΦs from cord blood, PBMCs from healthy donors, contacts or tuberculosis patients
All blood samples were collected per approved institutional Institutional review board protocols and included HLA-DR1 positive donors. To correlate gene expression with antigen processing, only HLA-DR1 + PBMCs were used. CD14 magnetic beads (Miltenyi Inc., USA) were used to purify monocytes which were plated in 6 or 24 tissue culture plate wells at a density of 4 × 10*6 and 1 × 10*6 cells per well, respectively. 8-well slide chambers or cover slips received 10*3 cells per chamber for confocal IF studies. CD14 bead purified monocytes were grown in Iscove’s medium with 10% fetal bovine serum and 10 ng/mL GM-CSF for 6 days and then plated in GM-CSF free medium for 24 h before differentiation them into M1- and M2- macrophages. Both heat-inactivated AB serum and fetal bovine serum were concurrently evaluated for phenotype studies and no differences were found. M1- and M2–MΦs were obtained by incubation with IFN-γ (10 ng/mL) or IL-4 (10 ng/mL) respectively for 5 days after which they were rested for 2 days. M0-MФs were not treated with any cytokine. Human TB patients and contacts: These were collected from deidentified, known TB patients and their healthy contacts with approved Institutional review board protocols of Dr. Restrepo from Reynosa, Mexico under a collaboration. Approved Institutional review board protocol HSC-SPH-12-0037. PBMCs collected in trizol from deidentified children with confirmed TB or household contacts from Vietnam were kindly provided by Drs. Nhung Nguyen and Ha Phan under a collaboration with Dr. Graviss. Table 1 provides details of their clinical status.
Table 1.
Clinical classification of peripherl blood samples from children with tuberculosis and those exposed to tuberculosis.
| # | Group | PBMC | DOB | TB contact | History of TB | TB status | Current RX | Gene expert | |
|---|---|---|---|---|---|---|---|---|---|
| C01 | Control | 9/13/2019 | 1/14/2008 | No | No | BPTB | No | Negative | |
| Co2 | Control | 9/13/2019 | 9/3/2012 | No | No | BPTB | No | Negative | |
| C03 | Control | 9/13/2019 | 8/13/2017 | No | No | BPTB | No | Negative | |
| C04 | Control | 9/13/2019 | 12/31/2017 | No | No | BPTB | No | Negative | |
| C05 | Control | 9/13/2019 | 9/30/2009 | No | No | BPTB | No | Negative | |
| TB01 | PediatricTB | 9/13/2019 | 2/12/2019 | Yes | No | BPTB + EPTB | Yes | 2RHZE | Negative |
| TB02 | PediatricTB | 9/13/2019 | 10/4/2004 | Yes | No | BPTB | Yes | 2RHZE | Negative |
| TB03 | PediatricTB | 9/13/2019 | 2/15/2012 | Yes | No | BPTB + EPTB | Yes | 2RHZE | Positive |
| TB04 | PediatricTB | 9/13/2019 | 7/5/2011 | Yes | No | EPTB | Yes | 2RHZE | |
| TB05 | PediatricTB | 9/13/2019 | 8/24/2005 | Yes | No | BPTB | Yes | 2RHZE |
BPTB broncho-pulmonary TB, EPTB extrapulmonary TB, RHZE rifampin, isoniazid, pyrazinamide and ethambutol.
Mtb and BCG infection of MΦs
Methods to prepare Mtb (H37Rv) and BCG (Pasteur) strains for infection have been described earlier (29). MΦs were infected (MOI = 1) using Mtb or BCG for 4 h prepared as follows. M. tuberculosis (H37Rv) (American Type Culture Collection-27294) (Mtb) was grown in BBL™ Middlebrook 7H9 broth with OADC enrichment (BD Biosciences, 211886) at 37 °C and 5% CO2. Green-fluorescent-protein-expressing M. tuberculosis H37Rv (gfpMtb) and red-fluorescent-protein-expressing M. tuberculosis H37Rv (rfpMtb) were a kind gift from Dr. Malini Madiraju (The University of Texas at Tyler, Tyler, TX). All mycobacterial strains were grown for 7 days in 7H9 broth with (for gfp/rfp strains) or without 25 µg/mL kanamycin, and aliquots containing ~10*8 viable CFUs were frozen for subsequent use. Before use, aliquots were thawed, washed three times in phosphate-buffered saline (×12,000 rpm; 15 min), and sonicated at 4 W with a sonicator (60 Sonic Dismembrator, Fisher Scientific) to prepare a uniform single-cell suspension of Mtb without loss of viability.
Phagocytic uptake and viability of Mtb or BCG-infected MΦs
Phagocytic uptake of rfpMtb by MΦs was determined by washing to remove extracellular bacilli with medium 4 times, fixation in ice cold methanol followed by microscopic counts of fluorescent bacteria per 500 MΦs in triplicate slide chambers. MΦs treated with pharmacological agents, siRNA or infection were carefully monitored for viability using trypan blue exclusion assay to maintain >90% viability until 7 days post-infection, although depending upon the assay, they were harvested either 18 h, 3, 5 or 7 days post-infection. No signs of apoptosis were observed at MOI = 1 for Mtb or BCG until day 7 post-infection. Tunel assays (Sigma Aldrich, Inc) were used to confirm the lack of apoptosis in all cultures and they were routinely tested for LPS and mycoplasma contamination.
RNAseq analysis
Naïve or Mtb-infected M1-, M2- and M0-MΦs (≥2 × 10 *6 cells/pellet; n = 2; numbers matched for all samples) were collected into trizol buffer and snap frozen in liquid-N2. RNASseq analysis, data interpretation and pathway analysis were done by Novogene (USA) as described (Supplemental Fig. 13). RNAseq was done two separate times using trizol samples (n = 2). Additional bioinformatic analysis was done in-house by coauthor PK.
Ex vivo Mtb Ag85B antigen presentation to CD4 T cells
This has been described in detail by us (29) and the original method described by the Harding lab has been extensively used by us and others for in vitro antigen presentation by MΦs (29) Briefly, Mtb-infected MΦs were washed after a 4 h infection and overlaid with the F9A6-CD4 T cell hybridoma (Dr. David Canaday) which recognizes an Ag85B epitope in the context of human HLA-DR1. IL-2 secreted from hybridoma T cells or other cytokines secreted from Mtb-infected MΦs were determined using a sandwich enzyme-linked immunoassay kit (Ebiosciences). Where indicated, the MΦs were pharmacologically blocked with various inhibitors or using siRNA probes (Supplementary figures). Cytotoxicity/viability of macrophage (>90%) was carefully monitored for indicated periods using trypan blue assay.
Western blot experiments
Six-well tissue culture plates were seeded with M1-, M2- and M0-MΦs. They were infected with Mtb for 4 h (h) at MOI of 1 and then washed three times with phosphate-buffered saline and re-plated in the medium. At different time points, MΦs were washed three times with 1x phosphate-buffered saline, and 200 µL RIPA buffer containing anti-protease mix was added to each well and incubated for 15 min. Lysates were then collected, and protein quantification (bicinchoninic acid, Pierce 23225) was performed. a) For western blot, 25 µg of total protein was loaded per well of Bio-Rad criterion gels and transferred to polyvinylidene fluoride membranes. Antibodies used to probe for proteins using the Bio-Rad apparatus or Protein Simple “Wes” automated capillary electrophoresis are listed in Supplemental Fig. 14 and Methods. Blot polyvinylidene fluoride membranes were then developed using an enhanced chemiluminescent kit or using Wes. Western blot and densitometry analysis data are shown as mean band density normalized relative to GAPDH (n = 2). The “Protein Simple” capillary gel electrophoresis with automated protein readings and densitometry was calculated as indicated.
siRNA knockdown in Mtb-infected or naïve M1-, M2- and M0- MΦs and Mtb CFU assay
The kits for various human siRNAs (mixture of duplexes), were purchased from Origene as listed in Supplementary Tables. MΦs were treated with siRNA and the scrambled control according to the manufacturers’ instructions, and this was followed by addition of Mtb (H37Rv) for 4 h. (MOI of 1). Cells were then lysed, and 10-fold dilutions were plated on 7H11 agar plates for CFU counts, which were read after 21 days of incubation. Details of the CFU assay have been described elsewhere (29). When CFUs were read day 3 after infection, a linear scale was used to express data whereas, day 5 CFUs were determined log scale. Mtb replicates with a generation time of about 18 h. For autophagy induction and effects on Mtb CFUs, a dose range of 1–10 µM of Rapamycin and 50–100 µM of Metformin were used.
Pharmacological blockade of Cathepsins, HDACs and Sirtuins on the viability of Mtb in M1-, M2- and M0-MΦs
The reagents used to blockade are listed in Supplementary Tables. MΦs were treated with 1 µg/mL of indicated agents followed by a 4-h infection with Mtb, washing and incubation with drugs as before for 3-5 days. On day 3-5, cells were then lysed, and 10-fold dilutions were plated on 7H11 agar plates for CFU counts, which were read after 21 days of incubation. Viability of MΦs was maintained at >90% through the assay period as determined by trypan blue assay.
Evaluation of autophagosome puncta, lysosomes, and biomarker localization with gfp- or rfpMtb within M1-, M2- and M0- MΦs
M1-, M2- and M0-MΦs were plated in 8-well chamber slides at a density of 10*3 cells per chamber and infected with either gfp- or rfpMtb at a MOI of 1. Per established procedures, autophagy was evaluated using at least three criteria. Fixed MΦs were permeabilized and stained using specific validated antibodies against LC3 autophagosomes; RAB7 and LAMP1 autophagolysosomes followed by secondary staining with Alexfluor485 /Alexafluor590 anti-IgG (Jackson Immunoresearch #111-095-003) as described by us (29). Percent gfp/rfpMtb phagosomes colocalizing with antibodies was done as described by us (29). To avoid visual bias, region of interest was measured as described112.
Flow cytometry of M1-, M2- and M0 macrophages
Flow cytometric analysis of M1-, M2-, and M0-MΦs before and after infection with Mtb was done per published procedures. MΦs were stained and analyzed on the Fortessa flow cytometer (Beckton Dickinson), and the data were processed using FlowJo v10 software (Tree Star, Inc.). Fluorochrome-conjugated antibodies used for flow cytometry were as follows: PE-Cy7 anti-human CD68 (BD Biosciences, cat no. 565595; clone: Y1/82A), PE anti-human CD14 (Invitrogen, cat no. 12-0149-42; clone: 61D3), APC anti-human CD206 (BioLegend, cat no. 321110; clone: 15-2), AF700 anti-human CD80 (BD Biosciences, cat no. 561133; clone: L307.4). Dead cells were excluded by using aqua fluorescent reactive dye (Invitrogen, cat no. L34957). GraphPad PRISM used for data analysis. Other antibodies are listed in Supplementary Tables.
Cytokine and chemokine assays
Cell supernatants were tested using sandwich enzyme-linked immunoassay kits (BioLegend and R&D systems, USA) for various cytokines and chemokines secreted by M1-, M2- and M0-MΦs before and after infection with Mtb.
NO and ROS assays
M1-, M2- and M0- MΦs were plated in 96-well plates at a density of 1 × 10*3 cells per well in triplicates and infected with Mtb (MOI = 1). Cells were then treated with fluorescent probes for the quantification of NO using diaminofluorescein diacetate (DAF-2 DA), per the manufacturer’s instructions (Enzo Life Sciences, USA). To differentiate whole cell ROS detected using DCFDA from mitochondrial ROS, we used a similar procedure and MitoROS reagent. To express nitrite levels, mouse MΦs were activated using LPS and IFN-γ followed by diaminonathalene assay and fluorescence relative light unitss measured correlated with Greiss reagent measurements. Nitrite levels in human MΦs were in nM levels unlike µM for mouse MΦs. Quantification of NO release was done by calculating the fluorescence emitted over time using an ex485 nm/em515 nm and plotting AFUs (± SD) against time using Ascent fluoroscan software version 2.6. These assays were described by us earlier (27). Likewise, DCFDA was used to detect ROS (Supplementary Fig. 3). In addition, iNOS mRNA and protein were quantitated using qPCR and MILO blots as described in methods and Supplementary Table 2.
Dual cross-link chromatin immunoprecipitation-Quantitative real time genomic PCR (xChIP-qPCR) for ATGs
Cells (4 × 10*6 to 6 × 10*6 per 100-mm dish) were washed twice with phosphate-buffered saline. Protein-protein cross-linking was first performed with disuccinimidyl glutarate (2 mM, Pierce), followed by protein-DNA cross-linking with formaldehyde. Equal amounts of sheared chromatin were immunoprecipitated overnight at 4 °C with 4 μg of anti-rabbit primary antibodies of H3, H3K18ac, H4, and H4K16ac (Cat#39763, 39587, 61521, and 39167, Active Motif, Carlsbad, CA) in ChIP dilution buffer. Anti-rabbit IgG was used as the negative control. Immunoprecipitates were collected with 40 μL protein A magnetic beads (Dynal Inc.), washed, and eluted in 250 μL elution buffer for 15 min at room temperature. Samples were de-cross-linked in 0.2 M NaCl at 65 °C for 2 h. The precipitated DNA was phenol/chloroform-extracted, precipitated with 100% ethanol and dried. Gene enrichment in xChIP was determined by Q-gPCR. Specific regions for the ATG5 promoter: Sense, (−1900) 50-CAGGGTCTCTCTCTGTTACC-30 (−1881), and Antisense, (−1669) 50-CCCAAAGTGCTGGGATTACA-30 (−1651). Standard curves were generated using a dilution series of genomic DNA (from 1 ng to 100 ng). The fold change of DNA in each immunoprecipitate was determined by normalizing the absolute amount to the input DNA reference and calculating the fold change relative to that amount in M0-MΦ cells. All data of Quantitative-genomic PCR shown in present study are the mean ± SD from three independent experiments.
Neonatal rhesus macaque model of tuberculosis
Six-week-old rhesus macaques were infected using 25 CFU per animal of Mtb Erdman using Institutional Animal Care and Use Committee approved procedures (# 1655-MM, Texas Biomedical Center) described in detail earlier107. They were sacrificed at 6 weeks post-infection, followed by necropsy for bacterial burden of lungs, and collection of left and right axillary, inguinal and hilar lymph nodes. Two sets of each type of lymph nodes from 2 macaques were suspended in trizol and analyzed for RNAseq by Novogene Inc. USA. For pathway analysis, matching lymph nodes from naïve macaques were concurrently analyzed. Bone marrow-derived CD14 bead purified, MΦs were differentiated as described for human MΦs into M1-, M2- and M0- MΦs and then analyzed for their ability to kill Mtb ex vivo; RNAseq analysis was done and QPCR analysis for select genes as described for human MΦs.
Single-cell protein analysis using MILO instrument
Single-cell western blot assays were performed using the Protein Simple MILO platform with the standard single-cell west (ScWest) kit according to manufacturer’s protocol. ScWest chips were rehydrated and loaded with cells at a concentration of 100,000 cells/mL suspension buffer. 100,000 macrophages loaded/chip, with 21–22 min settling time, 5 s lysis, and 60 s separation time. Multiplet/doublet capture rate in scWest chip microwells was determined with light microscopy (~1.3%, established from >1000 microwells). scWest chips were lysed for 10 s and instantly followed by electrophoresis for 80 s at 240 V. UV light exposure provided for total 4 min to get protein immobilized. scWest chips were sequentially probed with primary for 2 h and secondary antibodies for 1 h. Primary antibodies used were anti-mouse ATG7 (1:10, MAB6608; R&D), anti-rabbit ATG5 (1:10, 12994 s; Cell Signaling) and anti-rabbit GAPDH (1:10, 5174 s; Cell Signaling). Secondary antibodies used were goat anti-rabbit Alexa Fluor 647 (ATG7, 1:20), goat anti-mouse Alexa Fluor 555 (ATG5, 1:20) and goat anti-mouse/rabbit Alexa Fluor 488 (GAPDH, 1:20). Slides were washed, centrifuge-dried, and scanned with Innopsys Microarray Scanner. Data was analyzed using Scout software (Protein Simple) and ImageJ (NIH). Blot profiles for Figures are also indicated in the supplemental information.
Statistics and reproducibility
P values for CFU counts in macrophages were determined using 1 or 2-way ANOVA with Tukey’s post-hoc test (GraphPad PRISM software). Fragments Per Kilobase of transcript per Million mapped reads (FPKMs) within groups (Mb-infected versus naïve) were analyzed using Student’s two tailed t test. Data from human donors and tuberculosis patients: within group differences were established by Wilcoxon paired ranked signed test; between group differences (household contact versus TB) were determined by Kruskal–Wallis test. All experiments were done 2–3 times with biological triplicates with similar results and representative experiments are shown; when duplicates were used, they are indicated.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors wish to acknowledge funding support from NIH R01 AI-1220709 (PI C.J.) & R01 AI-138587 (PIs D.K, C.J.) and Houston Methodist Research Institute for support funds. We wish to gratefully acknowledge Prof. Stefan Kauffman, Director Emeritus, Max Planck Institute for Biology, Berlin, Germany for reading this manuscript and offering suggestions. We are grateful to Ms. Winston Adrienne, Dr. Heather McConnell and Dr. Sasha Pejerrey for editing the manuscript.
Author contributions
A.K., K.Z., and V.S. contributed equally for all experiments; P.K. did bioinformatics analysis; T.B., J.H.K., and A.M. did various experiments; R.P. and L.M. provided cord blood samples; E.L.C., G.A.D., and B.I.R. collected and analyzed human samples; D.H.C provided T cell lines; S.A.D. provided reagents; H.P. and E.A.G. obtained child samples; D.K. and M.C.G. performed and provided NHP samples. J.M. edited the manuscript and C.J. designed the experiments and wrote the manuscript.
Peer review
Peer review information
Communications Biology thanks Esmeralda Juárez and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Nicolas Fanget and Karli Montague-Cardoso.
Data availability
The RNAseq raw data will be uploaded into the NCBI, NIH database; supported by NIH 1RO1 AI-122070 (C.J.). All other data are available from the corresponding author (or other sources, as applicable) on reasonable request. A source data table has been included as Supplementary Data 1.
Competing interests
The authors declare no conflict of interest and certify that all studies were conducted under approved institutional Health Science Center and Institutional review board protocols.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arshad Khan, Kangling Zhang, Vipul K. Singh.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03387-9.
References
- 1.Zumla A, et al. Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect. Dis. 2016;16:e47–e63. doi: 10.1016/S1473-3099(16)00078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koeken, V., Verrall, A. J., Netea, M. G., Hill, P. C. & van Crevel, R. Trained innate immunity and resistance to Mycobacterium tuberculosis infection. Clin. Microbiol. Infect.25, 1468–1472 (2019). [DOI] [PubMed]
- 3.Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 4.Volkman HE, et al. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen SB, et al. Alveolar macrophages provide an early mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe. 2018;24:439–446.e434. doi: 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scriba, T. J., Coussens, A. K. & Fletcher, H. A. Human immunology of tuberculosis. Microbiol. Spectr.5, 213–237 (2017). [DOI] [PubMed]
- 7.Gordon S, Pluddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 2014;262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J. Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 9.Samuchiwal SK, et al. A novel peptide interferes with Mycobacterium tuberculosis virulence and survival. FEBS Open Bio. 2014;4:735–740. doi: 10.1016/j.fob.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra V, et al. Human IL10 gene repression by Rev-erbalpha ameliorates Mycobacterium tuberculosis clearance. J. Biol. Chem. 2013;288:10692–10702. doi: 10.1074/jbc.M113.455915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorpade DS, et al. Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates Mycobacterium bovis BCG-driven toll-like receptor 2 signaling. Mol. Cell Biol. 2013;33:543–556. doi: 10.1128/MCB.01108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan, M. et al. Prostaglandin E2 suppresses hCAP18/LL-37 expression in human macrophages via EP2/EP4: implications for treatment of Mycobacterium tuberculosis infection. FASEB J.32, 2827–2840 (2018). [DOI] [PubMed]
- 13.Shen P, et al. IRAK-M alters the polarity of macrophages to facilitate the survival of Mycobacterium tuberculosis. BMC Microbiol. 2017;17:185. doi: 10.1186/s12866-017-1095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z, et al. Mycobacterium tuberculosis-induced polarization of human macrophage orchestrates the formation and development of tuberculous granulomas in vitro. PLoS ONE. 2015;10:e0129744. doi: 10.1371/journal.pone.0129744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porta C, Riboldi E, Ippolito A, Sica A. Molecular and epigenetic basis of macrophage polarized activation. Semin. Immunol. 2015;27:237–248. doi: 10.1016/j.smim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 17.Labonte AC, Tosello-Trampont AC, Hahn YS. The role of macrophage polarization in infectious and inflammatory diseases. Mol. Cells. 2014;37:275–285. doi: 10.14348/molcells.2014.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomioka H, et al. Characteristics of suppressor macrophages induced by mycobacterial and protozoal infections in relation to alternatively activated M2 macrophages. Clin. Dev. Immunol. 2012;2012:635451. doi: 10.1155/2012/635451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsi E, et al. Human alveolar macrophages predominately express combined classical M1 and M2 surface markers in steady state. Respir. Res. 2018;19:66. doi: 10.1186/s12931-018-0777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills CD. Anatomy of a discovery: m1 and m2 macrophages. Front. Immunol. 2015;6:212. doi: 10.3389/fimmu.2015.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Vogt G, Nathan C. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J. Clin. Invest. 2011;121:3889–3901. doi: 10.1172/JCI57235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiwara Y, et al. Guanylate-binding protein 5 is a marker of interferon-gamma-induced classically activated macrophages. Clin. Transl. Immunol. 2016;5:e111. doi: 10.1038/cti.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang ZM, Zhang AR, Xu M, Lou J, Qiu WQ. TLR-4/miRNA-32-5p/FSTL1 signaling regulates mycobacterial survival and inflammatory responses in Mycobacterium tuberculosis-infected macrophages. Exp. Cell Res. 2017;352:313–321. doi: 10.1016/j.yexcr.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Li Y, Fu M, Xin HB. Polarizing macrophages in vitro. Methods Mol. Biol. 2018;1784:119–126. doi: 10.1007/978-1-4939-7837-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jagannath C, Actor JK, Hunter RL., Jr. Induction of nitric oxide in human monocytes and monocyte cell lines by Mycobacterium tuberculosis. Nitric Oxide. 1998;2:174–186. doi: 10.1006/niox.1998.9999. [DOI] [PubMed] [Google Scholar]
- 28.Daniel DS, et al. The reduced bactericidal function of complement C5-deficient murine macrophages is associated with defects in the synthesis and delivery of reactive oxygen radicals to mycobacterial phagosomes. J. Immunol. 2006;177:4688–4698. doi: 10.4049/jimmunol.177.7.4688. [DOI] [PubMed] [Google Scholar]
- 29.Jagannath C, et al. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 30.Koster S, et al. Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc. Natl Acad. Sci. USA. 2017;114:E8711–E8720. doi: 10.1073/pnas.1707792114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galvan-Pena S, O’Neill LA. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy S, et al. Transcriptional landscape of Mycobacterium tuberculosis infection in macrophages. Sci. Rep. 2018;8:6758. doi: 10.1038/s41598-018-24509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauhan S, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev. Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Q, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13:e1006600. doi: 10.1371/journal.ppat.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy M, et al. TRIM8 regulated autophagy modulates the level of cleaved Caspase-3 subunit to inhibit genotoxic stress induced cell death. Cell Signal. 2018;48:1–12. doi: 10.1016/j.cellsig.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Hu D, et al. Autophagy regulation revealed by SapM-induced block of autophagosome-lysosome fusion via binding RAB7. Biochem. Biophys. Res. Commun. 2015;461:401–407. doi: 10.1016/j.bbrc.2015.04.051. [DOI] [PubMed] [Google Scholar]
- 37.Harding CV, Boom WH. Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat. Rev. Microbiol. 2010;8:296–307. doi: 10.1038/nrmicro2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertram EM, Hawley RG, Watts TH. Overexpression of rab7 enhances the kinetics of antigen processing and presentation with MHC class II molecules in B cells. Int. Immunol. 2002;14:309–318. doi: 10.1093/intimm/14.3.309. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Montesinos G, Lopez-Ortega O, Piedra-Reyes J, Bonifaz LC, Moreno J. Dynamic changes in the intracellular association of selected Rab small GTPases with MHC Class II and DM during dendritic cell maturation. Front Immunol. 2017;8:340. doi: 10.3389/fimmu.2017.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartley MB, Canaday DH. T cell hybridomas to study MHC-II restricted B-cell receptor-mediated antigen presentation by human B cells. J. Immunol. Methods. 2011;370:35–42. doi: 10.1016/j.jim.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jagannath C, Bakhru P. Rapamycin-induced enhancement of vaccine efficacy in mice. Methods Mol. Biol. 2012;821:295–303. doi: 10.1007/978-1-61779-430-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang, A. T. et al. Autophagy links antimicrobial activity with antigen presentation in Langerhans cells. JCI Insight4,(2019). [DOI] [PMC free article] [PubMed]
- 43.Rovetta AI, et al. IFNG-mediated immune responses enhance autophagy against Mycobacterium tuberculosis antigens in patients with active tuberculosis. Autophagy. 2014;10:2109–2121. doi: 10.4161/15548627.2014.981791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller CW, et al. Endocytosis regulation by autophagy proteins in MHC restricted antigen presentation. Curr. Opin. Immunol. 2018;52:68–73. doi: 10.1016/j.coi.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 45.Munz C. Autophagy beyond intracellular MHC Class II antigen presentation. Trends Immunol. 2016;37:755–763. doi: 10.1016/j.it.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Valecka J, Almeida CR, Su B, Pierre P, Gatti E. Autophagy and MHC-restricted antigen presentation. Mol. Immunol. 2018;99:163–170. doi: 10.1016/j.molimm.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Van Kaer L, Parekh VV, Postoak JL, Wu L. Role of autophagy in MHC class I-restricted antigen presentation. Mol. Immunol. 2019;113:2–5. doi: 10.1016/j.molimm.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyette LB, et al. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE. 2017;12:e0176460. doi: 10.1371/journal.pone.0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nambi S, Basu N, Visweswariah SS. cAMP-regulated protein lysine acetylases in mycobacteria. J. Biol. Chem. 2010;285:24313–24323. doi: 10.1074/jbc.M110.118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan L, Yi M, Chen J, Li S, Chen W. Mycobacterium tuberculosis EIS gene inhibits macrophage autophagy through up-regulation of IL-10 by increasing the acetylation of histone H3. Biochem. Biophys. Res. Commun. 2016;473:1229–1234. doi: 10.1016/j.bbrc.2016.04.045. [DOI] [PubMed] [Google Scholar]
- 51.Fullgrabe J, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee IH. Mechanisms and disease implications of sirtuin-mediated autophagic regulation. Exp. Mol. Med. 2019;51:102. doi: 10.1038/s12276-019-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhaskar, A. et al. Host sirtuin 2 as an immunotherapeutic target against tuberculosis. Elife9, e55415 (2020). [DOI] [PMC free article] [PubMed]
- 54.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaquero A, Sternglanz R, Reinberg D. NAD+−dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 56.Bandyopadhaya A, Tsurumi A, Maura D, Jeffrey KL, Rahme LG. A quorum-sensing signal promotes host tolerance training through HDAC1-mediated epigenetic reprogramming. Nat. Microbiol. 2016;1:16174. doi: 10.1038/nmicrobiol.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee IH. Mechanisms and disease implications of sirtuin-mediated autophagic regulation. Exp. Mol. Med. 2019;51:1–11. doi: 10.1038/s12276-019-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardoso F, et al. Myeloid sirtuin 2 expression does not impact long-term mycobacterium tuberculosis control. PLoS ONE. 2015;10:e0131904. doi: 10.1371/journal.pone.0131904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singhania A, et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat. Commun. 2018;9:2308. doi: 10.1038/s41467-018-04579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singhania A, Wilkinson RJ, Rodrigue M, Haldar P, O’Garra A. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat. Immunol. 2018;19:1159–1168. doi: 10.1038/s41590-018-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy S, et al. Redefining the transcriptional regulatory dynamics of classically and alternatively activated macrophages by deepCAGE transcriptomics. Nucleic Acids Res. 2015;43:6969–6982. doi: 10.1093/nar/gkv646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gray EE, Cyster JG. Lymph node macrophages. J. Innate Immun. 2012;4:424–436. doi: 10.1159/000337007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang L, Nazarova EV, Tan S, Liu Y, Russell DG. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med. 2018;215:1135–1152. doi: 10.1084/jem.20172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schyns J, et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat. Commun. 2019;10:3964. doi: 10.1038/s41467-019-11843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat. Rev. Immunol. 2012;12:352–366. doi: 10.1038/nri3211. [DOI] [PubMed] [Google Scholar]
- 66.Hao W, Schlesinger LS, Friedman A. Modeling granulomas in response to infection in the lung. PLoS ONE. 2016;11:e0148738. doi: 10.1371/journal.pone.0148738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das P, et al. Selective M1 macrophage polarization in granuloma-positive and granuloma-negative Crohn’s disease, in comparison to intestinal tuberculosis. Intest Res. 2018;16:426–435. doi: 10.5217/ir.2018.16.3.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kewcharoenwong C, Prabowo SA, Bancroft GJ, Fletcher HA, Lertmemongkolchai G. Glibenclamide reduces primary human monocyte functions against tuberculosis infection by enhancing M2 polarization. Front. Immunol. 2018;9:2109. doi: 10.3389/fimmu.2018.02109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sahu SK, et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPbeta regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 2017;13:e1006410. doi: 10.1371/journal.ppat.1006410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez-Garcia S, et al. Macrophage activation by ursolic and oleanolic acids during mycobacterial infection. Molecules. 2015;20:14348–14364. doi: 10.3390/molecules200814348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mily, A. et al. Polarization of M1 and M2 human monocyte-derived cells and analysis with flow cytometry upon Mycobacterium tuberculosis infection. J. Vis. Exp.163, e61807 (2020). [DOI] [PubMed]
- 72.Das LM, Binko AM, Traylor ZP, Peng H, Lu KQ. Vitamin D improves sunburns by increasing autophagy in M2 macrophages. Autophagy. 2019;15:813–826. doi: 10.1080/15548627.2019.1569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munz C. Of LAP, CUPS, and DRibbles—unconventional use of autophagy proteins for MHC restricted antigen presentation. Front. Immunol. 2015;6:200. doi: 10.3389/fimmu.2015.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munz, C. Autophagy proteins influence endocytosis for MHC restricted antigen presentation. Semin. Cancer Biol.66, 110–115 (2019). [DOI] [PubMed]
- 75.Kimmey JM, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528:565–569. doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stroupe C. This is the end: regulation of Rab7 nucleotide binding in endolysosomal trafficking and autophagy. Front Cell Dev. Biol. 2018;6:129. doi: 10.3389/fcell.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell. 2015;58:507–521. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jia J, et al. Galectins control MTOR and AMPK in response to lysosomal damage to induce autophagy. Autophagy. 2019;15:169–171. doi: 10.1080/15548627.2018.1505155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramachandra L, Noss E, Boom WH, Harding CV. Processing of Mycobacterium tuberculosis antigen 85B involves intraphagosomal formation of peptide-major histocompatibility complex II complexes and is inhibited by live bacilli that decrease phagosome maturation. J. Exp. Med. 2001;194:1421–1432. doi: 10.1084/jem.194.10.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramachandra L, et al. Phagosomal processing of Mycobacterium tuberculosis antigen 85B is modulated independently of mycobacterial viability and phagosome maturation. Infect. Immun. 2005;73:1097–1105. doi: 10.1128/IAI.73.2.1097-1105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turk V, Turk B, Guncar G, Turk D, Kos J. Lysosomal cathepsins: structure, role in antigen processing and presentation, and cancer. Adv. Enzym. Regul. 2002;42:285–303. doi: 10.1016/S0065-2571(01)00034-6. [DOI] [PubMed] [Google Scholar]
- 82.Pires D, et al. Role of cathepsins in Mycobacterium tuberculosis survival in human macrophages. Sci. Rep. 2016;6:32247. doi: 10.1038/srep32247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh CR, et al. Processing and presentation of a mycobacterial antigen 85B epitope by murine macrophages is dependent on the phagosomal acquisition of vacuolar proton ATPase and in situ activation of cathepsin D. J. Immunol. 2006;177:3250–3259. doi: 10.4049/jimmunol.177.5.3250. [DOI] [PubMed] [Google Scholar]
- 84.Berg RD, et al. Lysosomal disorders drive susceptibility to tuberculosis by compromising macrophage migration. Cell. 2016;165:139–152. doi: 10.1016/j.cell.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pires D, et al. Mycobacterium tuberculosis modulates miR-106b-5p to control Cathepsin S expression resulting in higher pathogen survival and poor T-cell activation. Front. Immunol. 2017;8:1819. doi: 10.3389/fimmu.2017.01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amaral EP, et al. Lysosomal cathepsin release is required for NLRP3-inflammasome activation by Mycobacterium tuberculosis in infected macrophages. Front. Immunol. 2018;9:1427. doi: 10.3389/fimmu.2018.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ernst JD. Mechanisms of M. tuberculosis immune evasion as challenges to TB vaccine design. Cell Host Microbe. 2018;24:34–42. doi: 10.1016/j.chom.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vergne I, et al. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2005;102:4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saikolappan S, et al. The fbpA/sapM double knock out strain of Mycobacterium tuberculosis is highly attenuated and immunogenic in macrophages. PLoS ONE. 2012;7:e36198. doi: 10.1371/journal.pone.0036198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ouimet M, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016;17:677–686. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fullgrabe J, Klionsky DJ, Joseph B. Histone post-translational modifications regulate autophagy flux and outcome. Autophagy. 2013;9:1621–1623. doi: 10.4161/auto.25803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, X., Tang, X., Zhou, Z. & Huang, Q. Histone deacetylase 6 inhibitor enhances resistance to Mycobacterium tuberculosis infection through innate and adaptive immunity in mice. Pathog. Dis.76, fty064 (2018). [DOI] [PubMed]
- 93.Zhu C, et al. Histone deacetylase inhibitors impair the host immune response against Mycobacterium tuberculosis infection. Tuberculosis (Edinb.) 2019;118:101861. doi: 10.1016/j.tube.2019.101861. [DOI] [PubMed] [Google Scholar]
- 94.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fabri M, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Graustein AD, et al. The SIGLEC14 null allele is associated with Mycobacterium tuberculosis- and BCG-induced clinical and immunologic outcomes. Tuberculosis (Edinb.) 2017;104:38–45. doi: 10.1016/j.tube.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhattacharyya C, Majumder PP, Pandit B. An exome wide association study of pulmonary tuberculosis patients and their asymptomatic household contacts. Infect. Genet. Evol. 2019;71:76–81. doi: 10.1016/j.meegid.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 98.Ren X. Immunosuppressive checkpoint Siglec-15: a vital new piece of the cancer immunotherapy jigsaw puzzle. Cancer Biol. Med. 2019;16:205–210. doi: 10.20892/j.issn.2095-3941.2018.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bologna C, Deaglio S. Linking SLAMF1 to autophagy and sensitivity to therapy in chronic lymphocytic leukemia. Mol. Cell Oncol. 2018;5:e1143077. doi: 10.1080/23723556.2016.1143077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Connell P, et al. SLAMF7 is a critical negative regulator of IFN-alpha-mediated CXCL10 production in chronic HIV infection. J. Immunol. 2019;202:228–238. doi: 10.4049/jimmunol.1800847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sever L, et al. SLAMF9 regulates pDC homeostasis and function in health and disease. Proc. Natl Acad. Sci. USA. 2019;116:16489–16496. doi: 10.1073/pnas.1900079116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim BH, et al. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 103.Ngo, C. C. & Man, S. M. Mechanisms and functions of guanylate-binding proteins and related interferon-inducible GTPases: Roles in intracellular lysis of pathogens. Cell Microbiol.19, e12791 (2017). [DOI] [PubMed]
- 104.Praefcke GJK. Regulation of innate immune functions by guanylate-binding proteins. Int. J. Med. Microbiol. 2018;308:237–245. doi: 10.1016/j.ijmm.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 105.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ganchua SKC, White AG, Klein EC, Flynn JL. Lymph nodes—the neglected battlefield in tuberculosis. PLoS Pathog. 2020;16:e1008632. doi: 10.1371/journal.ppat.1008632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cepeda M, et al. Establishment of a neonatal rhesus macaque model to study Mycobacterium tuberculosis infection. Tuberculosis (Edinb.) 2013;93(Suppl):S51–S59. doi: 10.1016/S1472-9792(13)70011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maiello, P. et al. Rhesus Macaques are more susceptible to progressive tuberculosis than Cynomolgus Macaques: a quantitative comparison. Infect. Immun.86, e00505-17 (2018). [DOI] [PMC free article] [PubMed]
- 109.Kaushal D, Mehra S, Didier PJ, Lackner AA. The non-human primate model of tuberculosis. J. Med. Primatol. 2012;41:191–201. doi: 10.1111/j.1600-0684.2012.00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sibley L, et al. Differences in monocyte: lymphocyte ratio and Tuberculosis disease progression in genetically distinct populations of macaques. Sci. Rep. 2019;9:3340. doi: 10.1038/s41598-019-39819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Foreman TW, et al. CD4+ T-cell-independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proc. Natl Acad. Sci. USA. 2016;113:E5636–E5644. doi: 10.1073/pnas.1611987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Theart RP, Loos B, Powrie YSL, Niesler TR. Improved region of interest selection and colocalization analysis in three-dimensional fluorescence microscopy samples using virtual reality. PLoS ONE. 2018;13:e0201965. doi: 10.1371/journal.pone.0201965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The RNAseq raw data will be uploaded into the NCBI, NIH database; supported by NIH 1RO1 AI-122070 (C.J.). All other data are available from the corresponding author (or other sources, as applicable) on reasonable request. A source data table has been included as Supplementary Data 1.