Abstract
Insecticidal crystal proteins from Bacillus thuringiensis in sprays and transgenic crops are extremely useful for environmentally sound pest management, but their long-term efficacy is threatened by evolution of resistance by target pests. The diamondback moth (Plutella xylostella) is the first insect to evolve resistance to B. thuringiensis in open-field populations. The only known mechanism of resistance to B. thuringiensis in the diamondback moth is reduced binding of toxin to midgut binding sites. In the present work we analyzed competitive binding of B. thuringiensis toxins Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F to brush border membrane vesicles from larval midguts in a susceptible strain and in resistant strains from the Philippines, Hawaii, and Pennsylvania. Based on the results, we propose a model for binding of B. thuringiensis crystal proteins in susceptible larvae with two binding sites for Cry1Aa, one of which is shared with Cry1Ab, Cry1Ac, and Cry1F. Our results show that the common binding site is altered in each of the three resistant strains. In the strain from the Philippines, the alteration reduced binding of Cry1Ab but did not affect binding of the other crystal proteins. In the resistant strains from Hawaii and Pennsylvania, the alteration affected binding of Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F. Previously reported evidence that a single mutation can confer resistance to Cry1Ab, Cry1Ac, and Cry1F corresponds to expectations based on the binding model. However, the following two other observations do not: the mutation in the Philippines strain affected binding of only Cry1Ab, and one mutation was sufficient for resistance to Cry1Aa. The imperfect correspondence between the model and observations suggests that reduced binding is not the only mechanism of resistance in the diamondback moth and that some, but not all, patterns of resistance and cross-resistance can be predicted correctly from the results of competitive binding analyses of susceptible strains.
Insecticides derived from the bacterium Bacillus thuringiensis are the most widely used biological pesticides (8). During sporulation, B. thuringiensis produces crystals containing toxins that are called insecticidal crystal proteins (ICPs) or δ-endotoxins (19, 23). The mode of action of B. thuringiensis includes the following steps in the insect midgut: solubilization of the crystals, enzymatic activation of protoxins, binding of activated toxins to target sites on midgut membranes, and pore formation (2, 14, 22, 36). B. thuringiensis toxins kill a specific set of insect pests but do not harm people, wildlife, or even most beneficial insects. The genes encoding ICPs have been incorporated into and expressed by plants so that the plants have become toxic to some insect pests (11, 43).
Evolution of resistance by pests is the greatest threat to the continued success of the B. thuringiensis toxins used in conventional sprays or in transgenic plants (20). Although laboratory selection has produced resistance to B. thuringiensis in many pests, only the diamondback moth (Plutella xylostella), a major pest of crucifer crops worldwide, has evolved resistance to B. thuringiensis in open-field populations (37). Resistance to B. thuringiensis has been documented in diamondback moth populations in Hawaii, Asia, the continental United States, and Central America (35, 37, 41, 48, 49). Reduced binding of ICPs to midgut target sites is the best-documented mechanism of resistance in members of the Lepidoptera and the only known mechanism of resistance in the diamondback moth (10, 41, 42, 48). Nonetheless, some evidence suggests that other mechanisms, such as reduced activation of protoxins and increased degradation of toxins, can confer resistance to ICPs (12, 15, 28, 30, 32, 34).
Here we report the results of competitive binding tests performed with ICPs in a susceptible strain of diamondback moth (strain LAB-V) and in resistant strains of diamondback moth obtained from the Philippines (strain PHI), Hawaii (strain NO-QA), and Pennsylvania (strain PEN). Each of the three resistant strains evolved resistance to three ICPs present in B. thuringiensis sprays (Cry1Aa, Cry1Ab, and Cry1Ac) while remaining susceptible to Cry1C and some other ICPs not present in the sprays (39, 41). NO-QA and PEN evolved cross-resistance to two ICPs that were not in the sprays (Cry1F and Cry1J), whereas PHI did not (39, 41). Previous analyses of noncompetitive binding provided a preliminary description of ICP-target site interactions in the four strains of diamondback moth which we used (41). Previous results also show that the ICPs Cry1Aa, Cry1Ab, and Cry1Ac bind to a common target site in diamondback moth and other moths (1, 7, 25, 44) and that Cry1F also binds to this common target site in diamondback moth (16). In the present study, we used results from our competitive binding assays along with previously published data (9, 16) to produce an integrative model for binding of Cry1Aa, Cry1Ab, Cry1Ac, Cry1B, Cry1C, and Cry1F in a susceptible strain and variations on the model that explain the patterns observed in the three resistant strains.
MATERIALS AND METHODS
Insects.
The three resistant strains of diamondback moth were started from field populations that had been sprayed repeatedly with commercial formulations of B. thuringiensis subsp. kurstaki containing the toxins Cry1Aa, Cry1Ab, and Cry1Ac (41). Before we compared the three resistant strains, each was exposed to B. thuringiensis in the laboratory to eliminate susceptible individuals. Susceptible strain LAB-V originated from The Netherlands and had been reared in the laboratory for more than 10 years (9).
Source of ICPs.
Trypsin-activated Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F were kindly provided by Luke Masson (National Research Council of Canada, Montreal, Canada). The Cry1A proteins were obtained from recombinant Escherichia coli HB101, and Cry1F was obtained from recombinant E. coli EG1945 (from Ecogen Inc., Langhorne, Pa.).
Iodination of ICPs.
Cry1A proteins (25 μg each) were labeled with Na125I by using the chloramine-T method (44). Cry1Aa was labeled twice, once with 0.5 mCi of Na125I (1 Ci = 37 GBq) and once with 1 mCi of Na125I, and the two preparations were used in independent experiments. Cry1Ab and Cry1Ac were labeled with 1 mCi of Na125I. Labeled ICPs were separated from free iodine by using a Bio-Gel P30 (Bio-Rad) column. The specific activities were 0.63 mCi/mg for Cry1Aa labeled with 0.5 mCi of Na125I, 5.2 mCi/mg for CryAa labeled with 1 mCi of Na125I, 1.9 mCi/mg for Cry1Ab, and 2.1 mCi/mg for Cry1Ac.
Preparation of BBMV.
Brush border membrane vesicles (BBMV) were prepared from whole last-instar larvae by the differential magnesium precipitation method (5, 46). BBMV were frozen in liquid nitrogen and kept at −80°C until they were used. The concentration of proteins in BBMV preparations was determined with the Bio-Rad reagent (3) by using bovine serum albumin as the standard.
Binding of 125I-ICPs to BBMV.
Binding experiments were performed essentially as described previously (9). BBMV (5 to 10 μg of vesicle protein per assay) were incubated in 0.1 ml (final volume) of binding buffer (8 mM Na2HPO4, 2 mM KH2PO4, 150 mM NaCl [pH 7.4], 0.1% bovine serum albumin) containing 1.1 nM 125I-labeled Cry1Aa, 1.5 nM 125I-labeled Cry1Ab, 1.0 nM 125I-labeled Cry1Ac, and various concentrations of unlabeled competitor. Incubations were carried out at room temperature for 30 min. Bound ICPs were separated from free ICPs by filtration with glass fiber filters (type GF/F; Whatman). The filters were washed with 5 ml of cold binding buffer, and the radioactivity retained in the filters was measured with a model 1282 Compugamma CS gamma-counter (LKB). A 150- to 1,000-fold excess of unlabeled toxin was used to determine the extent of nonspecific binding. The maximum specific binding was 4 to 6% for Cry1Aa, 3% for Cry1Ab in strain LAB-V, 9% for Cry1Ac in strain LAB-V, and 3% for Cry1Ac in strain PHI (41).
Statistical analyses.
Data from the competition experiments were analyzed by using the LIGAND (33) and PRISM (17) computer programs. Both programs were used to analyze homologous competition (competition of a labeled ligand and its unlabeled analogue for binding to the receptor) curves to see if they fit a one-binding-site model or a two-binding-site model. In both programs, the null hypothesis assumes that the two populations of binding sites really are the same. If the P value obtained from the analysis is less than 0.05, then the null hypothesis is rejected, which indicates that the experimental curve does not fit a one-binding-site model. The results obtained with the two programs were always in agreement. Binding parameters (dissociation constant [Kd] and concentration of receptors [Rt]) were estimated from homologous competition curves, as well as heterologous competition curves (when the unlabeled ligand was not an analogue of the labeled ligand), by using the LIGAND program. Heterologous competition also revealed whether different ICPs bound to the same binding site.
RESULTS
Binding assays performed with the susceptible strain (strain LAB-V).
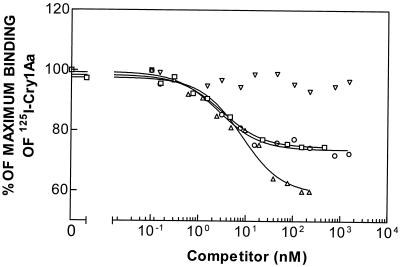
Binding of 125I-labeled Cry1Aa was determined at different concentrations of unlabeled Cry1Aa, Cry1Ab, Cry1Ac, or Cry1F (Fig. 1). Results obtained from both the LIGAND and PRISM program analyses showed that the data for the homologous competition curve (labeled Cry1Aa versus unlabeled Cry1Aa) fit a two-binding-site model better than they fit a one-binding-site model. The Kd values for Cry1Aa indicated that a high-affinity binding site (Kd1, 0.1 ± 0.1 nM) and a low-affinity binding site (Kd2, 17.7 ± 1.0 nM) were present (Table 1). Considerable binding of labeled Cry1Aa was detected in the presence of the homologous competitor at a concentration of 200 nM (Fig. 1), which indicated that a substantial portion of the labeled Cry1Aa bound nonspecifically to the BBMV. Cry1Ab and Cry1Ac competed with labeled Cry1Aa but did not completely impede specific binding of labeled Cry1Aa. These results suggest that the three ICPs compete for binding to one of the Cry1Aa binding sites but not the other. Cry1F apparently did not compete for binding with labeled Cry1Aa.
FIG. 1.
Binding of 125I-labeled Cry1Aa to BBMV of the susceptible strain (LAB-V) at different concentrations of unlabeled competitor. Symbols: ▵, Cry1Aa; ○, Cry1Ab; □, Cry1Ac; ▿, Cry1F.
TABLE 1.
Equilibrium Kd and Rt values for B. thuringiensis crystal proteins for BBMV of susceptible strain LAB-V of P. xylostella obtained with different 125I-labeled ICPs
| 125I-labeled ICP | Binding characteristics of ICPsa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cry1Aa
|
Cry1Ab
|
Cry1Ac
|
Cry1F
|
|||||||
| Kd1 (nM) | Rt1 (pmol/mg of protein)b | Kd2 (nM) | Rt2 (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | |
| Cry1Aa | 0.1 ± 0.1 | 0.2 ± 0.2 | 17.7 ± 1.0 | 6.2 ± 0.1 | 0.8 ± 0.2 | 2.7 ± 0.4 | 0.6 ± 0.2 | 2.0 ± 0.3 | ||
| Cry1Ab | 27.0 ± 5.7 | 0.3 ± 0.1 | 0.4 ± 0.2 | 0.3 ± 0.1 | 5.5 ± 2.0 | 0.8 ± 0.4 | ||||
| Cry1Ac | 17.2 ± 2.8 | 2.3 ± 0.6 | 5.0 ± 0.4 | 1.3 ± 0.1 | 17.0 ± 0.2 | 1.0 ± 0.1 | 13.0 ± 1.0 | 2.0 ± 0.1 | ||
Values are means ± standard deviations for four experiments performed with two independently prepared batches of BBMV for labeled Cry1Aa and for two experiments performed with the same batch of BBMV for the other labeled ICPs.
Picomoles per milligram of vesicle protein.
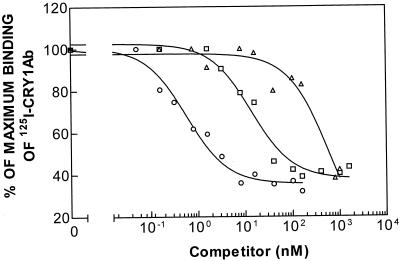
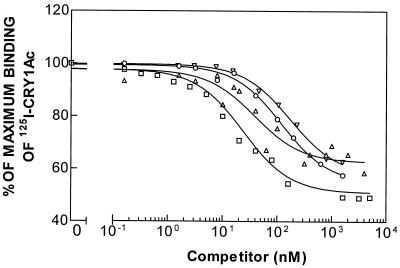
The results obtained for binding of 125I-labeled Cry1Ab and 125I-labeled Cry1Ac at different concentrations of unlabeled Cry1Aa, Cry1Ab, and Cry1Ac are shown in Fig. 2 and 3. Analysis of the homologous competition data indicated that the data fit a one-binding-site model better than they fit a two-site model. In both cases, the heterologous competitor inhibited specific binding of the labeled ligand almost completely, suggesting that the three ICPs bound to the single receptor recognized by the labeled ICPs. Although different values were obtained depending on the labeled ICP used, Cry1Ab was the ICP that showed the highest affinity for the binding site, followed by Cry1Ac and Cry1Aa (Table 1). These results suggest that the shared binding site is the low-affinity binding site for Cry1Aa.
FIG. 2.
Binding of 125I-labeled Cry1Ab to BBMV of the susceptible strain (LAB-V) at different concentrations of unlabeled competitor. Symbols: ▵, Cry1Aa; ○, Cry1Ab; □, Cry1Ac.
FIG. 3.
Binding of 125I-labeled Cry1Ac to BBMV of the susceptible strain (LAB-V) at different concentrations of unlabeled competitor. Symbols: ▵, Cry1Aa; ○, Cry1Ab; □, Cry1Ac; ▿, Cry1F.
Competition of Cry1F with labeled Cry1Ab for the same binding site has been observed previously in LAB-V (16). Here we found that Cry1F also competes with labeled Cry1Ac (Fig. 3), with an affinity similar to the affinity observed when Cry1F was competing with labeled Cry1Ab (Tables 1 and 2), which is consistent with the observation that Cry1Ab and Cry1Ac share the same binding site.
TABLE 2.
Previously reported values for Kd and Rt of B. thuringiensis crystal proteins for BBMV from susceptible P. xylostella strains (strains LAB-V, LAB-P, and ROTH) and resistant P. xylostella strains (strains Philippines, NO-QA, SERD3, and Bta-Sel)
| Strain | Reference | Binding characteristics of ICPsa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cry1Aa
|
Cry1Ab
|
Cry1Ac
|
Cry1C
|
Cry1F
|
|||||||
| Kd1 (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | ||
| LAB-V | 9 | 4.2 | 1.6 | 6.5 | 10.8 | ||||||
| Philippinesc | 9 | 7.6 | 2.9 | ||||||||
| LAB-V | 1 | 22d | 0.8d | 1.4 | 0.8 | 3.7d | 0.8d | ||||
| LAB-P | 38 | 2.0 | 1.1 | 8.8 | 3.2 | ||||||
| NO-QA | 38 | 8.5 | 3.5 | ||||||||
| LAB-V | 16 | 1.2 | 0.7 | 7.4d | 0.5d | ||||||
| ROTH | 48 | 22.4 | 2.7 | 8.9 | 9.2 | ||||||
| SERD3 | 48 | 27.3 | 3.3 | ||||||||
| Bta-Sel | 48 | 8.7 | 9.0 | ||||||||
Unless indicated otherwise, values were obtained from homologous competition experiments.
Picomoles per milligram of vesicle protein.
Strain Philippines was derived from the same field location as strain PHI, but the culture was started 3.5 years earlier than the PHI culture (1).
Values were obtained from heterologous competition experiments in which labeled Cry1Ab was used.
Binding assays performed with three resistant strains (strains PHI, NO-QA, and PEN).
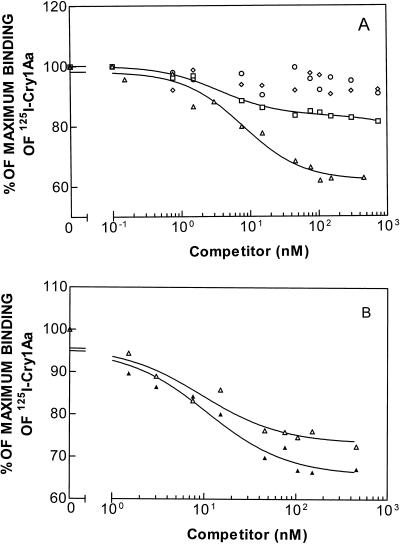
In a previous study, we demonstrated that specific binding of 125I-labeled Cry1Aa occurs in resistant strains PHI, NO-QA, and PEN (41). Here, we expanded the study with competition analyses. With PHI, the homologous competition data for Cry1Aa fit a two-binding-site model better than they fit a one-binding-site model (Fig. 4A), and the two Kd values were essentially the same as the Kd values obtained with the susceptible strain (Table 3). This indicates that binding of Cry1Aa is not affected in this resistant strain. In contrast, with strains NO-QA and PEN, the homologous competition data fit a one-binding-site model better than they fit a two-binding-site model (Fig. 4B). The Kd and Rt values of Cry1Aa for NO-QA (4.8 ± 2.7 nM and 1.9 ± 0.8 pmol/mg of protein, respectively) and PEN (4.0 ± 2.8 nM and 1.9 ± 0.8 pmol/mg of protein, respectively) are similar to each other and are intermediate between the values obtained for the two binding sites of LAB-V and PHI (Tables 1 and 3).
FIG. 4.
(A) Binding of 125I-labeled Cry1Aa to BBMV of resistant strain PHI at different concentrations of unlabeled competitor. Symbols: ▵, Cry1Aa; ○, Cry1Ab; □, Cry1Ac; ◊, Cry1F. (B) Binding of 125I-labeled Cry1Aa to BBMV of resistant strains NO-QA (▴) and PEN (▵) at different concentrations of unlabeled Cry1Aa.
TABLE 3.
Equilibrium Kd and Rt values for B. thuringiensis crystal proteins for BBMV from resistant strain PHI of P. xylostella obtained with different 125I-labeled ICPs
| 125I-labeled ICP | Binding characteristics of ICPsa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cry1Aa
|
Cry1Ab
|
Cry1Ac
|
Cry1F
|
|||||||
| Kd1 (nM) | Rt1 (pmol/mg of protein)b | Kd2 (nM) | Rt2 (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | Kd (nM) | Rt (pmol/mg of protein)b | |
| Cry1Aa | 0.3 ± 0.1 | 0.2 ± 0.2 | 20.3 ± 4.4 | 5.1 ± 1.0 | NBc | NB | 0.7 ± 0.2 | 3.9 ± 2.2 | ||
| Cry1Ab | NB | NB | NB | NB | NB | NB | NB | NB | ||
| Cry1Ac | 21.4 ± 0.7 | 1.2 ± 0.3 | NB | NB | 19.2 ± 0.3 | 1.0 ± 0.1 | 14.0 ± 1.0 | 1.6 ± 0.1 | ||
Values are means ± standard deviations for four experiments performed with two independently prepared batches of BBMV for labeled Cry1Aa and for two experiments performed with the same batch of BBMV for the other labeled ICPs.
Picomoles per milligram of vesicle protein.
NB, no binding.
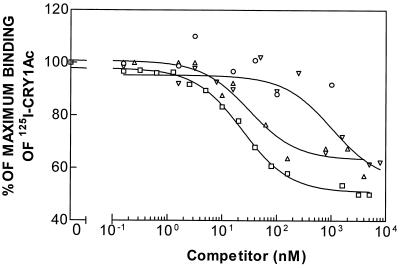
Binding was low or nil in all three resistant strains for Cry1Ab (41) and in NO-QA and PEN for Cry1Ac (41). However, Cry1Ac exhibits specific binding to BBMV from strain PHI (41). Analysis of the homologous competition data (Fig. 5) gave a better fit with a one-binding-site model than with a two-binding-site model. Kd and Rt values were calculated from both the homologous and heterologous competition data (Table 3). The values of Kd and Rt depended on which ICP was labeled. However, for a particular labeled ICP, the Kd and Rt values for Cry1Ac did not differ between strain PHI and strain LAB-V, which indicates that binding of Cry1Ac in strain PHI was not altered compared to binding of Cry1Ac in strain LAB-V.
FIG. 5.
Binding of 125I-labeled Cry1Ac to BBMV of resistant strain PHI at different concentrations of unlabeled competitor. Symbols: ▵, Cry1Aa; ○, Cry1Ab; □, Cry1Ac; ▿, Cry1F.
Heterologous competition experiments confirmed the homologous competition results showing that binding of Cry1Aa and Cry1Ac is not altered in strain PHI. The results of competition of Cry1Ab, Cry1Ac, and Cry1F with labeled Cry1Aa for binding sites in BBMV of PHI larvae are shown in Fig. 4A. As in susceptible strain LAB-V, Cry1F did not compete with labeled Cry1Aa for binding, and Cry1Ab did not compete either (as expected, since this ICP does not bind to BBMV of this strain), but there was some competition with Cry1Ac (as a consequence of the shared binding site). The results of competition of Cry1Aa, Cry1Ab, and Cry1F with labeled Cry1Ac are shown in Fig. 5, which shows that the main difference compared with strain LAB-V is the lack of competition by Cry1Ab. Cry1F competed with labeled Cry1Ac for binding (Table 3) with essentially the same affinity as the affinity in strain LAB-V (Table 1).
DISCUSSION
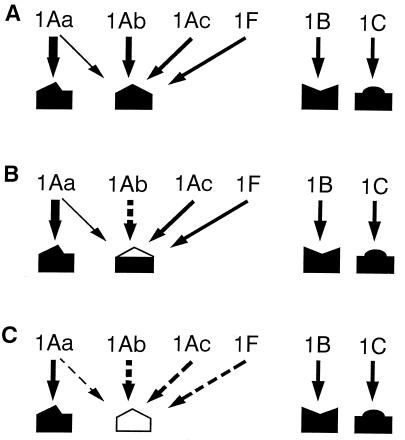
If we considered the results of competition binding experiments performed with strain LAB-V in this work along with the results of other studies in which binding of Cry1B and binding of Cry1C were also determined (9, 16), an integrative model for the sites involved in binding to ICPs in the diamondback moth could be developed. We propose a model which includes at least four binding sites involved in binding of Cry1Aa, Cry1Ab, Cry1Ac, Cry1B, Cry1C, and Cry1F (Fig. 6). In this model Cry1A proteins and Cry1F compete for binding to a common binding site, and Cry1Aa binds with low affinity to the common binding site and binds with high affinity to a different binding site not shared with any of the other ICPs. Cry1B and Cry1C each bind to different binding sites. This model explains the finding that the homologous competition data for Cry1Aa fit a two-site model, whereas the homologous competition data for Cry1Ab, Cry1Ac, and Cry1C fit a one-site model. As far as we know, labeled Cry1B has not been tested in a competition experiment.
FIG. 6.
Model proposed for binding of B. thuringiensis ICPs to binding sites in the P. xylostella epithelial midgut membrane in susceptible insects (A), in strain PHI (B), and in strains NO-QA and PEN (C). The wider arrows indicate greater binding affinity. Dashed arrows indicate that no binding or extremely reduced binding occurs.
The model also explains the heterologous competition data. Cry1Ab and Cry1Ac cannot completely displace specific binding of labeled Cry1Aa. In the case of Cry1F, we interpret the lack of competition with labeled Cry1Aa as the result of Cry1F competing with low affinity to the common binding site that in turn Cry1Aa also binds to with low affinity. The low level of binding apparently escaped detection in our assay. In contrast, binding of labeled Cry1Ab or Cry1Ac can be eliminated by competition with the three Cry1A proteins and Cry1F, which indirectly suggests that Cry1Aa and Cry1F bind to the same common site. Alternatively, these two toxins could bind to different determinants in the binding site in such a way that their binding interferes with Cry1Ab or Cry1Ac binding but not with each other. The latter possibility could be tested in competition experiments performed with Cry1Aa and labeled Cry1F.
Resistant strain PHI apparently has an alteration in the common binding site that greatly reduces or eliminates binding of Cry1Ab without affecting binding of Cry1Aa or Cry1Ac. Binding is likely to occur through multiple interactions between the binding sites and several loops of domain II of ICPs (18, 26). Binding of Cry1Aa and Cry1Ac to the common binding site in PHI suggests that binding of Cry1Ab involves a distinct binding determinant that can change without affecting binding of the other ICPs. This same phenotype has been reported in a resistant diamondback moth population from Malaysia, in which binding was reduced for Cry1Ab but not for Cry1Aa, Cry1Ac, or Cry1C (48). This very specific change in the binding site is also consistent with the observation that in one set of bioassays conducted soon after PHI was established in the laboratory, this strain showed extremely high resistance to Cry1Ab but apparently not to Cry1Aa or Cry1Ac (1). Genetic analyses also have shown that in PHI, resistance to Cry1Aa is not controlled by the same gene that confers resistance to Cry1Ab (41).
The binding changes in NO-QA are the same as those observed in PEN. Both strains show very little or no binding of Cry1A proteins to the putative common binding site. These two strains also have virtually identical values of Kd and Rt for binding of Cry1Aa to the site that is not shared by the other ICPs. According to our model, NO-QA and PEN have an alteration in the common binding site that impedes binding of any ICP. The model nicely explains the resistance phenotype; both strains are resistant to Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F (and Cry1J), although they are susceptible to Cry1C and Cry1B (39, 41). We observed specific binding of Cry1C in both of these resistant strains (41). A previous analysis also detected no reduction in Cry1C binding in NO-QA (38). One gene can confer resistance to Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F in NO-QA (40), whereas in a related diamondback moth strain from Hawaii, resistance to Cry1C segregates independently from resistance to Cry1Ab (27). The genetics of resistance in NO-QA and PEN supports our model, because it appears that one gene is responsible for the major alteration in the common binding site and one or more other genes confer resistance to Cry1C.
Quantitative differences between Kd and Rt values obtained by using homologous competition data and Kd and Rt values obtained by using heterologous competition data are common (4, 45), even though both sets of data should give the same result. Differences may arise because the labeled proteins are not the same as the unlabeled proteins used as homologous competitors, since the labeling procedure changes the proteins at least by adding the 125I residue. Values of Kd and Rt usually are calculated from the results of homologous competition experiments, which are regarded as more indicative of the binding of the ICPs to their target sites.
The values of Kd and Rt obtained in the present work agree well with those previously published for P. xylostella (Table 2). Note that the values reported previously for Cry1Aa were obtained in heterologous competition with labeled Cry1Ab (Table 2) and therefore represent the low-affinity binding site of Cry1Aa observed in the present study (Table 1). The values obtained here for Cry1Ab in homologous competition experiments (Table 1) are not much different from the values reported previously (Table 2). For Cry1Ac, two different sets of values have been obtained previously for Kd (Table 2). Values of 22.4 and 27.3 nM (48) were obtained with the same batch of labeled Cry1Ac that was used in the present work; experiments with strains LAB-V (Table 1), ROTH (48), and SERD (48) were performed simultaneously in the same laboratory. The value obtained in heterologous competition with labeled Cry1Ab, 3.7 nM (Table 2), agrees well with the value obtained in the present work with labeled Cry1Ab (5.5 nM) and is close to the value obtained for homologous competition in a different laboratory (2.0 nM). Therefore, for now, we think that there are not enough independent replicates to choose a Kd value for homologous competition of Cry1Ac. However, the differences in the values for the binding parameters of Cry1Ac do not invalidate the model which we propose and do not affect the comparisons among the strains of diamondback moth studied here. Finally, the Kd values obtained for Cry1F (Tables 1 and 3) are also close to the value shown in Table 2.
Binding of Cry1A proteins to strain NO-QA has been determined by using techniques different from the technique used in our study. Binding of Cry1Aa, Cry1Ab, and Cry1Ac to tissue sections of the midguts of NO-QA larvae was observed by using histochemical detection (6), as well as binding of Cry1Ac to BBMV with surface plamon resonance (31). Finally, a Cry1Ac binding protein with aminopeptidase activity was purified from both susceptible insects and NO-QA (29). Several explanations have been proposed for these discrepancies (6, 29, 31). The hypothesis underlying all of them is that strain NO-QA has binding sites for the ICPs that have been altered in such a way that they do not bind toxins in vivo but in vitro bind ICPs under some circumstances but not others.
The results presented in this paper have important implications for resistance management. On the one hand, they show that binding site models obtained by using susceptible insects may correctly predict some aspects of the patterns of resistance. For example, based on the model derived from our competitive binding assays performed with susceptible strain LAB-V, we would have predicted that a single mutation could confer resistance to Cry1Ab, Cry1Ac, and Cry1F because these ICPs each bind only to a shared target site. However, because Cry1Aa binds to two sites in LAB-V, we might have made the incorrect prediction that two mutations were necessary for resistance to Cry1Aa. The finding that binding to the high-affinity binding site for Cry1Aa occurs in NO-QA and PEN without toxicity agrees with the results of other reports showing that binding is not sufficient for toxicity (7, 13, 21, 24, 47). The receptor alteration observed in PHI that confers resistance to Cry1Ab but not Cry1Ac would not have been predicted from the binding model. Furthermore, the results also show that a binding site model does not provide any information concerning the mechanisms of resistance that are not related to binding site alteration. This is true for strain PHI, and the model does not predict that this strain is resistant to Cry1Aa and Cry1Ac. However, in cases like this, the fact that the model cannot explain the resistance pattern clearly shows that there is more than one mechanism of resistance.
ACKNOWLEDGMENTS
We thank Luke Masson, who generously provided toxins.
Funding was provided in part by grants TSTAR 95-34135-1771, NRI-CGP 96-35302-3470, and WRPIAP 97RA0304/0305-WR96-16 from the U.S. Department of Agriculture and by European Community ECLAIR project AGRE-0003.
REFERENCES
- 1.Ballester V, Escriche B, Ménsua J L, Riethmacher G W, Ferré J. Lack of cross-resistance to other Bacillus thuringiensis crystal proteins in a population of Plutella xylostella highly resistant to CryIA(b) Biocontrol Sci Technol. 1994;4:437–443. [Google Scholar]
- 2.Bauer L S. Resistance: a threat to the insecticidal crystal proteins of Bacillus thuringiensis. Fla Entomol. 1995;78:414–443. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen X J, Curtiss A, Alcantara E, Dean D H. Mutations in domain I of Bacillus thuringiensis δ-endotoxin CryIAb reduce the irreversible binding of toxin to Manduca sexta brush border membrane vesicles. J Biol Chem. 1995;270:6412–6419. doi: 10.1074/jbc.270.11.6412. [DOI] [PubMed] [Google Scholar]
- 5.Escriche B, Silva F J, Ferré J. Testing suitability of brush border membrane vesicles prepared from whole larvae from small insects for binding studies with Bacillus thuringiensis CryIA(b) crystal protein. J Invert Pathol. 1995;65:318–320. [Google Scholar]
- 6.Escriche B, Tabashnik B, Finson N, Ferré J. Immunohistochemical detection of binding of CryIA crystal proteins of Bacillus thuringiensis in highly resistant strains of Plutella xylostella (L.) from Hawaii. Biochem Biophys Res Commun. 1995;212:388–395. doi: 10.1006/bbrc.1995.1982. [DOI] [PubMed] [Google Scholar]
- 7.Escriche B, Ferré J, Silva F J. Occurrence of a common binding site in Mamestra brassicae, Phthorimaea operculella and Spodoptera exigua for the insecticidal crystal proteins CryIA from Bacillus thuringiensis. Insect Biochem Mol Biol. 1997;27:651–656. doi: 10.1016/s0965-1748(97)00039-8. [DOI] [PubMed] [Google Scholar]
- 8.Feitelson J S, Payne J, Kim L. Bacillus thuringiensis: insects and beyond. Bio/Technology. 1992;10:271–275. [Google Scholar]
- 9.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferré J, Escriche B, Bel Y, Van Rie J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis insecticidal crystal proteins. FEMS Microbiol Lett. 1995;132:1–7. [Google Scholar]
- 11.Fischhoff D A, Bowdish K S, Perlak F J, Marrone P G, McCormick S M, Niedermeyer J G, Dean D A, Kusano-Kretzmer K, Mayer E J, Rochester D E, Rogers S G, Fraley R T. Insect tolerant transgenic tomato plants. Bio/Technology. 1987;5:807–813. [Google Scholar]
- 12.Forcada C, Alcácer E, Garcerá M D, Martínez R. Differences in the midgut proteolytic activity of two Heliothis virescens strains, one susceptible and one resistant to Bacillus thuringiensis toxins. Arch Insect Biochem Physiol. 1996;31:257–272. [Google Scholar]
- 13.Garczynski S F, Crim J W, Adang M J. Identification of putative insect brush border membrane-binding molecules specific to Bacillus thuringiensis δ-endotoxin by protein blot analysis. Appl Environ Microbiol. 1991;57:2816–2820. doi: 10.1128/aem.57.10.2816-2820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill S S, Cowles E A, Pietrantonio P V. The mode of action of Bacillus thuringiensis endotoxins. Annu Rev Entomol. 1992;37:615–636. doi: 10.1146/annurev.en.37.010192.003151. [DOI] [PubMed] [Google Scholar]
- 15.Gould F, Martínez-Ramírez A, Anderson A, Ferré J, Silva F J, Moar W J. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. Proc Natl Acad Sci USA. 1992;89:7986–7990. doi: 10.1073/pnas.89.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granero F, Ballester V, Ferré J. Bacillus thuringiensis crystal proteins Cry1Ab and Cry1Fa share a high affinity binding site in Plutella xylostella (L.) Biochem Biophys Res Commun. 1996;224:779–783. doi: 10.1006/bbrc.1996.1099. [DOI] [PubMed] [Google Scholar]
- 17.GraphPad Software, Inc. PRISM. 1994. [Google Scholar]
- 18.Grochulski P, Masson L, Borisova S, Pusztai-Carey M, Schwartz J L, Brousseau R, Cygler M. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J Mol Biol. 1995;254:447–464. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 19.Höfte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hokkanen H M T, Wearing C H. The safe and rational deployment of Bacillus thuringiensis genes in crop plants: conclusions and recommendations of OECD workshop on ecological implications of transgenic crops containing Bt toxin genes. Biocontrol Sci Technol. 1994;4:399–403. [Google Scholar]
- 21.Ihara H, Kudora E, Wadana A, Himeno M. Specific toxicity of δ-endotoxins from Bacillus thuringiensis to Bombyx mori. Biosci Biotechnol Biochem. 1993;57:200–204. doi: 10.1271/bbb.57.200. [DOI] [PubMed] [Google Scholar]
- 22.Knowles B H. Mechanism of action of Bacillus thuringiensis insecticidal delta-endotoxins. Adv Insect Physiol. 1994;24:275–308. [Google Scholar]
- 23.Koziel M G, Carozzi N B, Currier T C, Warren G W, Evola S V. The insecticidal crystal proteins of Bacillus thuringiensis: past, present and future uses. Biotechnol Genet Eng Rev. 1993;11:171–228. [Google Scholar]
- 24.Lee M K, Rajamohan F, Gould F, Dean D H. Resistance to Bacillus thuringiensis CryIA δ-endotoxins in a laboratory-selected Heliothis virescens strain is related to receptor alteration. Appl Environ Microbiol. 1995;61:3836–3842. doi: 10.1128/aem.61.11.3836-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M K, Dean D H. Inconsistencies in determining Bacillus thuringiensis toxin binding site relationship by comparing competition assays with ligand blotting. Biochem Biophys Res Commun. 1996;220:575–580. doi: 10.1006/bbrc.1996.0445. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Carrol J, Ellar D J. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature (London) 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y B, Tabashnik B E. Inheritance of resistance to the Bacillus thuringiensis toxin Cry1C in the diamondback moth. Appl Environ Microbiol. 1997;63:2218–2223. doi: 10.1128/aem.63.6.2218-2223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Y. B., B. E. Tabashnik, L. Masson, B. Escriche, and J. Ferré. Unpublished data.
- 29.Luo K, Tabashnik B E, Adang M J. Binding of Bacillus thuringiensis Cry1Ac toxin to aminopeptidase in susceptible and resistant diamondback moths (Plutella xylostella) Appl Environ Microbiol. 1997;63:1024–1027. doi: 10.1128/aem.63.3.1024-1027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacIntosh S C, Stone T B, Jokerst R S, Fuchs L. Binding of Bacillus thuringiensis proteins to a laboratory-selected line of Heliothis virescens. Proc Natl Acad Sci USA. 1991;88:8930–8933. doi: 10.1073/pnas.88.20.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masson L, Mazza A, Brousseau R, Tabashnik B. Kinetics of Bacillus thuringiensis toxin binding with brush border membrane vesicles from susceptible and resistant larvae of Plutella xylostella. J Biol Chem. 1995;270:11887–11896. doi: 10.1074/jbc.270.20.11887. [DOI] [PubMed] [Google Scholar]
- 32.Moar W J, Pusztai-Carey M, Van Faassen H, Bosch D, Frutos R, Rang C, Luo K, Adang M J. Development of Bacillus thuringiensis CryIC resistance by Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae) Appl Environ Microbiol. 1995;61:2086–2092. doi: 10.1128/aem.61.6.2086-2092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munson P J, Rodbard D. LIGAND: a versatile computerized approach for characterisation of ligand-binding systems Anal. Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 34.Oppert B, Kramer K, Beeman R W, Johnson D, McGaughey W H. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J Biol Chem. 1997;272:23473–23476. doi: 10.1074/jbc.272.38.23473. [DOI] [PubMed] [Google Scholar]
- 35.Pérez C J, Shelton A M. Resistance of Plutella xylostella (Lepidoptera: Plutellidae) to Bacillus thuringiensis Berliner in Central America. J Econ Entomol. 1997;90:87–93. [Google Scholar]
- 36.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabashnik B E. Evolution of resistance to Bacillus thuringiensis. Annu Rev Entomol. 1994;39:47–79. doi: 10.1146/annurev.ento.54.110807.090518. [DOI] [PubMed] [Google Scholar]
- 38.Tabashnik B E, Finson N, Groeters F R, Moar W J, Johnson M W, Luo K, Adang M J. Reversal of resistance to Bacillus thuringiensis in Plutella xylostella. Proc Natl Acad Sci USA. 1994;91:4120–4124. doi: 10.1073/pnas.91.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabashnik B E, Malvar T, Liu Y B, Finson N, Borthakur D, Shin B Y, Park S H, Masson L, de Maagd R A, Bosch D. Cross-resistance of the diamondback moth indicates altered interactions with domain II of Bacillus thuringiensis toxins. Appl Environ Microbiol. 1996;62:2839–2844. doi: 10.1128/aem.62.8.2839-2844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabashnik B E, Liu Y B, Finson N, Masson L, Heckel D G. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc Natl Acad Sci USA. 1997;94:1640–1644. doi: 10.1073/pnas.94.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabashnik B E, Liu Y B, Malvar T, Heckel D G, Masson L, Ballester V, Granero F, Ménsua J L, Ferré J. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc Natl Acad Sci USA. 1997;94:12780–12785. doi: 10.1073/pnas.94.24.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J D, Shelton A M, Van Rie J, de Roeck S, Moar W J, Roush R T, Peferoen M. Toxicity of Bacillus thuringiensis spore and crystal protein to resistant diamondback moth (Plutella xylostella) Appl Environ Microbiol. 1996;62:564–569. doi: 10.1128/aem.62.2.564-569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaeck M, Reynaerts A, Höfte H, Jansens S, de Beuckeleer M, Dean C, Zabeau M, Van Montagu M, Leemans J. Transgenic plants protected from insect attack. Nature (London) 1987;327:33–37. [Google Scholar]
- 44.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Specificity of Bacillus thuringiensis delta-endotoxins: importance of specific receptors on the brush border membrane of the midgut of target insects. Eur J Biochem. 1989;186:239–247. doi: 10.1111/j.1432-1033.1989.tb15201.x. [DOI] [PubMed] [Google Scholar]
- 45.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Receptors on the brush border membrane of the insect midgut as determinants of the specificity of Bacillus thuringiensis delta-endotoxins. Appl Environ Microbiol. 1990;56:1378–1385. doi: 10.1128/aem.56.5.1378-1385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfersberger M, Luthy P, Maurer A, Parenti P, Sacchi F V, Giordana B, Hanozet G M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae) Comp Biochem Physiol A Comp Physiol. 1987;86:301–308. [Google Scholar]
- 47.Wolfersberger M G. The toxicity of two Bacillus thuringiensis δ-endotoxins to gypsy moth larvae is inversely related to the affinity of binding sites on midgut brush border membranes for the toxins. Experientia. 1990;46:475–477. doi: 10.1007/BF01954236. [DOI] [PubMed] [Google Scholar]
- 48.Wright D J, Iqbal M, Granero F, Ferré J. A change in a single midgut receptor in the diamondback moth (Plutella xylostella) is only in part responsible for field resistance to Bacillus thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai. Appl Environ Microbiol. 1997;63:1814–1819. doi: 10.1128/aem.63.5.1814-1819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J Z, Zhu G R, Ju Z L, Wang W Z. Resistance of diamondback moth to Bacillus thuringiensis in China. Resistant Pest Management. 1993;5:11–12. [Google Scholar]