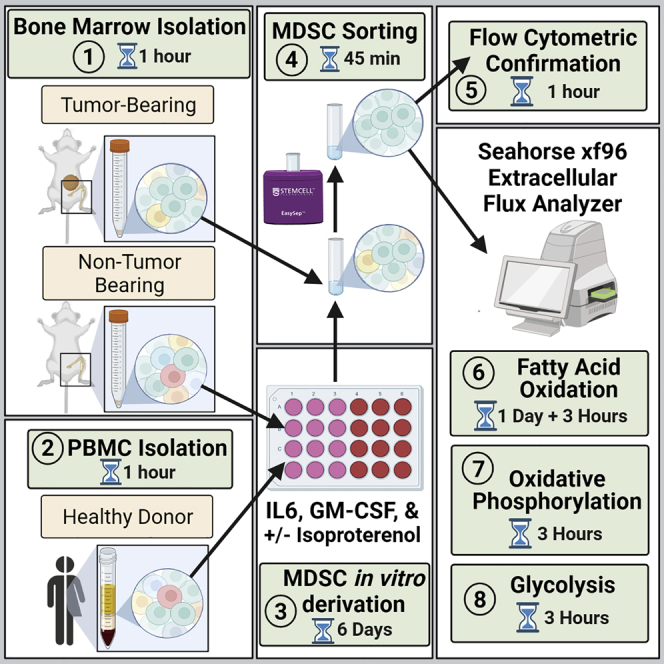
Summary
Metabolic reprogramming is associated with myeloid-derived suppressor cell (MDSC) immunosuppressive function. Here, we outline the process for acquiring MDSCs from human and murine sources for subsequent analysis of fatty acid oxidation, oxidative phosphorylation, and glycolysis using the Seahorse XFe 96 Analyzer. Murine MDSCs can be isolated directly from tumor-bearing mice or derived through IL-6 and GM-CSF culture of bone marrow cells from non-tumor-bearing mice. To generate human MDSCs, peripheral blood mononuclear cells (PBMCs) can be cultured with IL-6 and GM-CSF.
For complete details on the use and execution of this protocol, please refer to Mohammadpour et al. (2021).
Subject areas: Cell Biology, Cell isolation, Cell-based Assays, Cancer, Immunology, Metabolism, Model Organisms
Graphical abstract
Highlights
-
•
MDSC derivation from human PBMC and mouse bone marrow
-
•
Metabolic analysis of MDSCs using Seahorse analysis
-
•
Analysis of β2-adrenergic signaling on MDSC metabolic activity
Publisher's note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Metabolic reprogramming is associated with myeloid-derived suppressor cell (MDSC) immunosuppressive function. Here, we outline the process for acquiring MDSCs from human and murine sources for subsequent analysis of fatty acid oxidation, oxidative phosphorylation, and glycolysis using the Seahorse XFe 96 Analyzer. Murine MDSCs can be isolated directly from tumor-bearing mice or derived through IL-6 and GM-CSF culture of bone marrow cells from non-tumor-bearing mice. To generate human MDSCs, peripheral blood mononuclear cells (PBMCs) can be cultured with IL-6 and GM-CSF.
Before you begin
Institutional permissions
Experiments described in this protocol utilize live mice and human samples. Approval from the institutional IACUC must be obtained prior to beginning experiments. For human samples, an IRB-approved protocol is required. Additional requirements may be in place at specific institutions.
In order to obtain sufficient MDSCs from murine bone marrow without deriving them in culture, you must first obtain tumor bearing mice. This is because there is not an appreciable number of MDSCs in non-tumor bearing mice. Therefore, here we describe the protocol for tumor inoculation using the 4T1 mammary carcinoma cell line in BALB/c mice. We recommend the use of the 4T1 cell line due to their ability to induce abundant MDSC differentiation in vivo. We have also successfully used this protocol for the subcutaneous implantation of EL4 tumors in C57BL/6 mice, which also support the generation of MDSCs. Note that the methodology to obtain MDSCs directly from the tumor is not the focus of this paper. We have found that MDSCs isolated from the tumor are very fragile and have limited viability, resulting in inconsistent results from the seahorse assay. This section can be skipped if you plan to derive MDSCs in vitro from mouse bone marrow or human PBMCs.
4T1 murine mammary carcinoma cell culture
Timing: 15 min + variable time
4T1 cells are cultured for at least 2–3 passages prior to injection to ensure optimal health of the cells. All steps are performed under BSL II biosafety cabinets with sterile reagents to ensure sterility.
-
1.
Warm up 9 mL of complete culture media in a 15 mL conical tube and 13 mL of complete culture media in a 15 mL conical tube in a 37°C water bath.
-
2.
Rapid thaw one cryovial (5 × 106 cells) of 4T1 cells from liquid nitrogen storage by placing it in a 37°C water bath.
CRITICAL: Remove cryovial when a small ice crystal remains to prevent extended exposure of thawed cells to DMSO. Vials are usually thawed in about 1–2 min.
-
3.
Add thawed cells to 9 mL warmed complete culture media from step 1.
-
4.
Mix by capping the tube and inverting 3–5 times.
-
5.
Centrifuge cells at 1,500 RPM (about 505 × g) for 5 min at 4°C.
-
6.
Remove supernatant by aspiration.
-
7.
Resuspend cells in 12 mL of pre-warmed complete culture media.
-
8.
Plate in T75 flask and ensure even plating by gentle agitation.
-
9.
Incubate at 37°C + 5% CO2 incubator for 24 h.
-
10.
Evaluate cell density under a microscope.
-
11.If not confluent, change the media and return to the incubator for 24 h.
-
a.Remove media by aspiration.
-
b.Add 12 mL of prewarmed (37°C) complete culture media.
-
c.Incubate at 37°C + 5% CO2 incubator for 24 h.
-
a.
-
12.Passage cells if confluent.
-
a.Remove media by aspiration.
-
b.Add 10 mL room temperature PBS to wash off residual serum contained in the complete culture media.
-
c.Remove PBS by aspiration.
-
d.Add 2 mL of prewarmed (37°C) 0.25% Trypsin.
-
e.Incubate in 37°C + 5% CO2 incubator for 5 min.
-
f.Add 8 mL of prewarmed (37°C) complete culture media to neutralize the trypsin and to resuspend the cells.
-
g.Detach adherent 4T1 cells by repetitively pipetting media toward the bottom of the flask.
-
h.Pipet up the resuspended cell and transfer to a labeled 15 mL conical tube.
-
i.Centrifuge cells at 1,500 RPM (about 505 × g) for 5 min at 4°C.
-
j.Remove supernatant by aspiration.
-
k.Resuspend in 5 mL of prewarmed (37°C) complete culture media.
-
l.Passage 1/5 (i.e., 1 mL) into fresh 11 mL of prewarmed (37°C) complete culture media in a new T75 flask.
-
m.Agitate flask to ensure even distribution of cells inside the flask.
-
n.Repeat after 48 h.
-
a.
-
13.
Culture cells for 2–3 passages to allow cells to fully recover from cryopreservation prior to usage for experimentation.
Note: Track the number of passages for adherent cell lines. Freshly thawed cells are ideal for experimental use. We find that tumor growth is affected after P20.
4T1 tumor inoculation in mouse model
Timing: 5–10 min/5 mice + 21 days
-
14.
Harvest 4T1 cells according to steps 12a–12j.
-
15.
Resuspend cells in 10 mL of PBS.
-
16.
Centrifuge at 1,500 RPM (about 505 × g) for 5 min at 4°C.
-
17.
Repeat steps 15 and 16 for an additional wash.
-
18.
Resuspend cells in 1 mL of PBS.
-
19.
Quantify cells using a hemocytometer.
Note: It is important to ensure the viability of the cells is greater than 95%
-
20.
Dilute cells to 1 × 106 cells/mL in PBS.
-
21.
Place cells on ice until ready to be injected.
-
22.
Anesthetize a 2–3-month-old female BALB/c mouse with 5% Isoflurane in 1 L/min O2.
Note: Multiple mice may be anesthetized simultaneously. Lower isoflurane level to 4% to maintain anesthesia. Monitor for any signs of hypothermia. We recommend multiple rounds of 5 mice.
-
23.
Vortex the 4T1 cell suspension to ensure even concentration.
-
24.
Load 100 μL of 4T1 cell suspension (100,000 cells) in 1 mL tuberculin syringes with 27G needles.
Note: 1 × 105 4T1 cells have given us robust tumor growth. The number of cells injected should be optimized for the cancer cell line used, the growth rate, and the desired length of experiment.
-
25.
Spray ethanol on the mouse’s left lower quadrant to sanitize and press down the fur.
Note: If preferred, the mouse can be shaved to better expose the site of injection.
-
26.
Inject the 4T1 cell suspension into the left 4th mammary pad.
CRITICAL: Needle angle must be almost parallel to the surface of the mouse skin. If too deep, tumor may invade into the peritoneum during growth. If injected i.p. accidentally, mice may show smaller tumors with accelerated mortality. Refer to Figure 1.
-
27.
Return the mouse to her cage and monitor until she wakes from anesthesia.
-
28.
After 7 days, palpate to confirm the proper inoculation of 4T1 tumors.
Note: If mice will be subjected to treatment conditions, randomize at this point.
-
29.
Monitor tumor growth for 21 days or until tumor size reaches 2 cm × 2 cm, whichever is earlier. (troubleshooting 1).
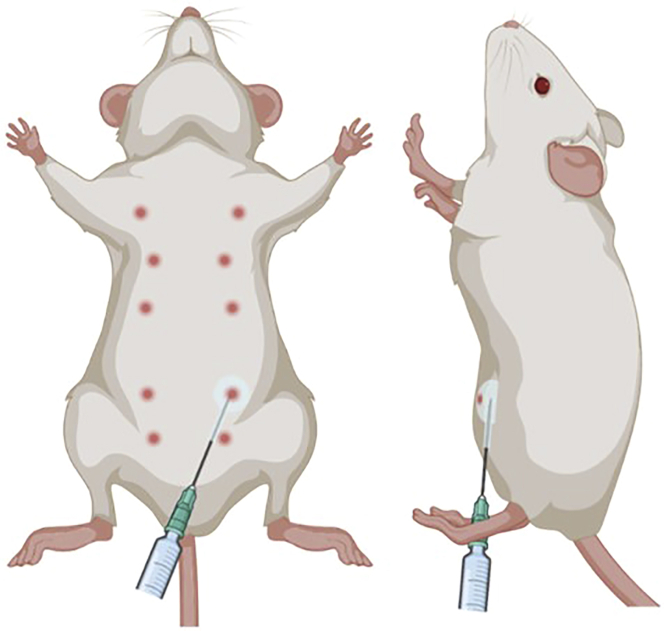
Figure 1.
Schematic of orthotopic breast cancer injection
Left, the mammary fat pads are located below the nipples depicted here. Injection of the 4th fat pad allows for accurate measurement of tumor volumes throughout the experiment. Right, the angle of insertion of the needle should be nearly parallel to the skin to prevent intraperitoneal injections deep to the fat pad and with the needle bevel up to prevent puncturing/exiting the skin.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified Rat Anti-Mouse CD16/CD32 (Mouse BD Fc Block™) (Clone 2.4G2 (RUO)) | BD Biosciences | RRID: AB_394656 |
| PE-Cy7 Anti-mouse CD45 (clone 30-F11) | BioLegend | RRID: AB_312979 |
| BUV395 Anti-mouse CD11b (clone M1/70) | BD Biosciences | RRID: AB_2738276 |
| BV421 Anti-mouse Ly6C (clone HK1.4) | BioLegend | RRID: AB_2562178 |
| APC Anti-mouse Ly6G (clone 1A8) | BioLegend | RRID: AB_2227348 |
| FITC Anti-mouse Gr-1 (clone RB6-8C5) | BD Biosciences | RRID: AB_394643 |
| APC Anti-human CD11b (clone D12) | BD Biosciences | Catalog # 340937 |
| PerCP-Cy5.5 Anti-human CD14 (clone M5E2) | BD Biosciences | RRID: AB_2033939 |
| PC7 Anti-human CD33 (clone D3HL60.251) | Beckman Coulter | Catalog # A54824 |
| Biological samples | ||
| Blood samples from healthy donors | Age Range: 45–85 Sex: Male & Female |
N/A |
| Blood samples from patient donors | Age Range: 45–85 Sex: Male & Female |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 1× DPBS without calcium & magnesium | Corning | Catalog # 21-031-CV |
| 0.25% Trypsin EDTA | Corning | Catalog # 25-053-CI |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation: resuspend in 55 μL DMSO | Thermo Fisher Scientific | Catalog # L34957 |
| Lymphoprep™ | STEMCELL Technologies | Catalog # 07801 |
| RPMI 1640 with L-glutamine | Corning | Catalog # 10-040-CV |
| DMEM | Corning | Catalog # 10-013-CV |
| Fetal Bovine Serum | Corning | Catalog # 35-011-CV |
| L-Glutamine | Corning | Catalog # 25-005-CI |
| Penicillin/ Streptomycin (1,000 U) | Corning | Catalog # 30-002-CI |
| Bovine Serum Albumin | MilliporeSigma | Catalog # A3294Mill |
| Carnitine | MilliporeSigma | Catalog # C0283 |
| NaCl | Fisher Scientific | Catalog # S271-10 |
| KCl | MilliporeSigma | Catalog # P3911 |
| NH4Cl | MilliporeSigma | Catalog # A9434 |
| MgSO4 | MilliporeSigma | Catalog # M7506 |
| Na2HPO4.2H2O (dibasic) | MilliporeSigma | Catalog # 71643 |
| KHCO3 | MilliporeSigma | Catalog # 237205 |
| 0.5 M EDTA | Corning | Catalog # 46-034-CI |
| HEPES | MilliporeSigma | H9136 (7365-45-9) |
| D-glucose | MilliporeSigma | Catalog # G8270 |
| Pyruvate | MilliporeSigma | Catalog # P2256 |
| Phenol Red | MilliporeSigma | Catalog # P3532 |
| Mouse IL-6 | MilliporeSigma | Catalog # I9646 |
| Human IL-6 | MilliporeSigma | Catalog # I1395 |
| Mouse GM-CSF | BioLegend | Catalog # 713704 |
| Human GM-CSF | BioLegend | Catalog # 713604 |
| Detachin Cell Detachment Solution | Genlantis | Catalog # T100100 |
| Isoproterenol | MilliporeSigma | Catalog # I6504 |
| Etomoxir | Tocris | Catalog # 4539 |
| Corning® Cell-Tak™ Cell and Tissue Adhesive | Corning | Catalog # 354240 |
| NaHCO3 | MilliporeSigma | Catalog #S5761 |
| Hoechst 33342 Stain | Thermo Fisher Scientific | Catalog # H1399 |
| Critical commercial assays | ||
| EasySep™ Mouse MDSC (CD11b+Gr1+) Isolation Kit | STEMCELL Technologies | Catalog # 19867 |
| EasySep™ HLA Chimerism Whole Blood CD33 Positive Selection Kit | STEMCELL Technologies | Catalog # 17885 |
| Palmitate Oxidation Stress Test Kit | Agilent | Part # 103693-100 |
| Cell Mito Stress Test Kit | Agilent | Part # 103010-100 |
| Glycolysis Stress Test Kit | Agilent | Part # 103017-100 |
| Experimental models: Cell lines | ||
| 4T1 | ATCC | Catalog # CRL-2539 |
| EL4 | ATCC | Catalog # TIB-39 |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c (Age Range: 2–3 months; Sex: Female) | Charles River | Strain Code: 028 |
| Mouse: C57/BL6 (Age Range: 2–3 months; Sex: Female) | Charles River | Strain Code: 027 |
| Software and algorithms | ||
| Seahorse Wave Desktop Software | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897 |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | BD Bioscience | https://www.flowjo.com/solutions/flowjo |
| Other | ||
| Rotanta 460R Centrifuge | Hettich | Catalog # 5660 |
| 2.0 mL Cryogenic Vial, External | NEST | Catalog # 607001 |
| 15 mL conical tube | Thermo Fisher Scientific | Catalog # 339650 |
| 50 mL conical tube | Thermo Fisher Scientific | Catalog # 339652 |
| EasyEights™ EasySep™ Magnet | STEMCELL Technologies | Catalog # 18103 |
| 5 mL round bottom polystyrene tube | Corning | Catalog # 352008 |
| 5 mL round bottom polystyrene tube w/ caps | Corning | Catalog # 352058 |
| OneComp eBeads™ Compensation Beads | Thermo Fisher Scientific | Catalog # 01-1111-41 |
| LSRFortessaTM Cell Analyzer | BD | Catalog # 649225 |
| 24-well tissue culture plate | NEST | Catalog # 702011 |
| Seahorse XF96 Cell Culture microplates | Agilent | Catalog # 101085-004 |
| Seahorse XFe96 FluxPak | Agilent | Catalog # 102416-100 |
| Seahorse XFe 96 Extracellular Flux Analyzer | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-analyzers/seahorse-xfe96-analyzer-740879 |
Materials and equipment
ACK lysis media
| Reagent | Final concentration | Amount |
|---|---|---|
| NH4Cl | 150 mM | 8.02 g |
| KHCO3 | 10 mM | 1 g |
| 0.5 M EDTA | 0.1 mM | 200 μL |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Dissolve in 850 mL of H2O. Adjust pH to 7.2–7.4 prior to bringing up volume to 1 L. Store at 4°C for 12 months.
Complete culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI1640 (supplemented with L-Glutamine, Phenol Red) | n/a | 450 mL |
| FBS | 10% | 50 mL |
| Penicillin/Streptomycin | 1% | 5 mL |
| L-Glutamine (200 mM) | 1% | 5 mL |
| Total | n/a | 510 mL |
Store at 4°C for 12 months.
MDSC-derivation media
| Reagent | Final concentration | Amount |
|---|---|---|
| Complete Culture Media | n/a | 25 mL |
| IL-6 (5 ng/μL) | 20 ng/mL | 100 μL |
| GM-CSF (5 ng/μL) | 20 ng/mL | 200 μL |
| Total | n/a | 25.3 mL |
Make fresh. Warm complete culture media to 37°C prior to usage.
Flow buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | n/a | 500 mL |
| BSA | 0.1% | 0.5 mg |
| Total | n/a | 500 mL |
Store at 4°C for 12 months.
Cell-tak and tissue adhesive solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 0.1 M NaHCO3 | 0.1 M | 2.5 mL |
| 2 mg/mL Corning® Cell-Tak | 25.2 μg/mL | 31.5 μL |
| 1 N HCl | 6.28 mM | 15.7 μL |
| Total | n/a | 2.5 mL |
Must be made fresh.
CRITICAL: Do not add HCl until ready to use immediately.
Substrate limiting media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | n/a | 500 mL |
| FBS | 5% | 25 mL |
| D-glucose (50 mM in ddH2O) | 0.5 mM | 5 mL |
| L-Glutamine (200 mM) | 1 mM | 2.5 mL |
| Carnitine (50 mM in ddH2O) | 0.5 mM | 5 mL |
| Total | n/a | 500 mL |
Store at 4°C for 12 months – Carnitine is added fresh; pH7.4.
5× KHB assay stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 555 mM | 6.50 g |
| KCl | 23.5 mM | 0.35 g |
| MgSO4 | 10 mM | 0.24 g |
| Na2HPO4.2H2O | 6 mM | 0.17 g |
| ddH2O | n/a | to 200 mL |
| Total | n/a | 200 mL |
Store at 4°C for 12 months.
1× KHB assay working solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 5× KHB Assay Stock Solution | 1× | 60 mL |
| HEPES | 5 mM | 357.45 g |
| D-glucose (50 mM in ddH2O) | 2.5 mM | 15 mL |
| Carnitine (50 mM in ddH2O) | 0.5 mM | 3 mL |
| ddH2O | n/a | 222 mL |
| Total | n/a | 300 mL |
Store at 4°C for 12 months – Carnitine is added fresh; pH7.4]
OCR medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | n/a | 500 mL |
| D-glucose | 25 mM | 2.25 g |
| Pyruvate | 1 mM | 55 mg |
| L-glutamine (200 mM) | 2 mM | 5 mL |
| Total | n/a | 500 mL |
Store at 4°C for 12 months. Adjust pH to 7.4
ECAR medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM (w/o bicarbonate) | n/a | 500 mL |
| L-glutamine (200 mM) | 2 mM | 5 mL |
| Total | n/a | 500 mL |
Store at 4°C for 12 months. Adjust pH to 7.4
Step-by-step method details
Isolate immune cells from mouse bone marrow
Timing: ∼1 h + 15 min/additional mouse
Bone marrow immune cells are isolated by flushing and mechanical dissociation. If the bone marrow is from tumor bearing mice, then MDSCs can then be sorted directly. However, if the bone marrow from non-tumor bearing mice is used, subsequent derivation of MDSCs must be carried out. All steps are performed in a biosafety cabinet to ensure sterility. Cell suspensions should be kept on ice for maximal cell viability and yield. See Methods video S1 for visualization of steps 2–6.
-
1.
Euthanize mice by IACUC-approved method.
-
2.
Dissect away the skin to reveal the hind leg muscles.
-
3.Isolate both hind legs.
-
a.Palpate the hip socket to identify location and ensure femur is intact upon removal.
-
b.Cut along the length of the femur and tibia to remove the majority of the muscles.
-
c.Push forward on the hip socket to dislocate the femur and cut away the muscles and ligaments stabilizing the hips to detach the legs.
-
a.
-
4.Isolate femur and tibia.
-
a.Dislocate the tibia from the ankle by repetitively hyperextending the joint.
-
b.Pull the muscle toward the patella to reveal the tibia.
-
c.Break off the fibula.
-
d.Dislocate the tibia from the patella by repetitively hyperextending the joint.
-
e.Place tibia in a petri dish containing sterile PBS.
-
f.Hold the patella and extend the joint to dislocate the femur from the patella.
-
g.Place the femur in petri dish with sterile PBS.
-
a.
-
5.
Fill a sterile 15 mL conical tube with 15 mL of sterile PBS for collection (collection tube).
-
6.Flush the bone marrow from the femur and tibia into the collection tube.
-
a.Fill a 1 mL tuberculin syringe with a 27G needle with 600 μL of sterile PBS from the collection tube.Optional: Use surgical scissors to cut off the hip-end of the femur.Note: Highly recommended; the needle easily clogs if this step is skipped.
-
b.Insert the sterile PBS-filled syringe into the cut end of the femur and flush out the bone marrow with 600 μL of PBS into the collection tube.
-
c.Refill the needle with 600 μL of PBS from the collection tube and flush femur an additional time.
-
d.Refill the needle with 600 μL of PBS from the collection tube.
-
e.Cut the ankle-end of the tibia.
-
f.Insert the PBS-filled syringe into the cut end of the tibia and flush with 600 μL of PBS into the collection tube.
-
g.Repeat until all bones are flushed.
-
a.
-
7.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
-
8.
Remove the supernatant.
-
9.
Resuspend in 3 mL sterile ACK lysis buffer for 1 min at room temperature.
CRITICAL: Red blood cells (RBC) lyse in culture and this is toxic to the plated bone marrow cells, reducing MDSC yield. Therefore, it is important not to look over this step.
-
10.
Neutralize with 10 mL of sterile PBS.
-
11.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
Note: If tumor bearing mice were the source of bone marrow, continue to step 32, as MDSCs will be present in this suspension of cells.
-
12.
If non-tumor bearing mice were the source of bone marrow, resuspend in 1 mL of sterile MDSC-generation media.
-
13.
Quantify cells.
Note: Around 40 × 106 cells per mouse are expected. (troubleshooting 2).
-
14.
Continue to step 28 if not isolating human MDSCs.
Bone marrow is harvested from the femur and tibia from both mouse hind legs. Majority of the muscles are cut away, which allows to easy dislocation and isolation of the bones from the remaining muscles and tendons. The bone marrow is flushed out from the bone using PBS-filled syringes. For increased yield, multiple mice of the same genotype or treatment groups may be pooled if desired and appropriate.
Isolate human peripheral blood mononuclear cells (PBMCs)
Timing: 1 h + 15 min/additional patient
Human PBMCs are isolated by density centrifugation.
-
15.
Spin down blood from healthy volunteer or patient donors at 1,500 RPM (about 505 × g) for 5 min.
-
16.
Place 4 mL of sterile, room temperature LymphoprepTM in a 15 mL conical tube.
-
17.
Resuspend blood cell pellet in 4 mL of PBS.
-
18.
Pipette the cell suspension slowly on top of the aliquoted LymphoprepTM at a 45° angle to overlay the samples without mixing.
-
19.
Centrifuge at 2,000 RPM (about 898 × g) for 20 min at room temperature WITHOUT brakes to prevent disruption of the density gradient.
-
20.
Carefully pipette the white middle layer containing the leukocytes into a new sterile 15 mL conical tube. Refer to Figure 2.
CRITICAL: Avoid aspirating the upper layers, which include platelets, as they can contaminate the isolated PBMCs.
-
21.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
-
22.
Aspirate off supernatant.
-
23.
Resuspend in 10 mL of sterile PBS.
-
24.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
-
25.
Repeat steps 23 and 24 for an additional wash.
-
26.
Resuspend in 1 mL of sterile MDSC-generation media.
-
27.
Quantify cells.
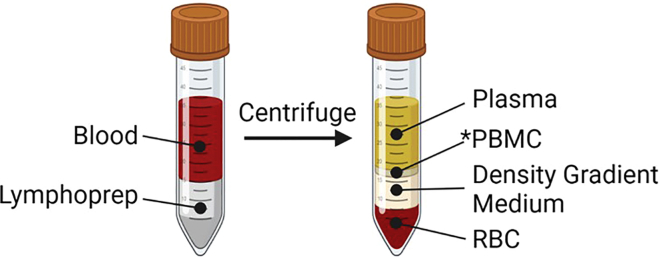
Figure 2.
Density centrifugation isolation of PBMCs
Blood collected from donors can be centrifuged with LymphoprepTM to assist in the separation the PBMCs from other blood components.
MDSC derivation with increased β-adrenergic signal
Timing: 6 days
Isolated immune cells (from human PBMCs or non-tumor bearing mouse bone marrow) are cultured with IL-6 and GM-CSF to derive MDSCs. Here we use isoproterenol, a pan β-adrenergic agonist, to increase β-adrenergic signaling in MDSCs during this process. Investigators may use other agents that are active in vitro in a similar manner to determine whether they play a role in altering MDSC derivation or their subsequent metabolic activity.
-
28.Plate cells at 1 × 106 cells/mL density in 2 mL in a 24-well plate in MDSC-Derivation Media.
-
a.Add final concentration 10 μM isoproterenol or PBS.
-
a.
Note: Isoproterenol can be substituted for the treatment specific for the experimental need of the lab. The treatment regimen will have to be optimized individually. We recommend a pilot experiment with different concentrations of the treatment.
-
29.Place cells at 37°C 5% CO2 incubator for 6 additional days. (troubleshooting 3).
-
a.Replenish media and Isoproterenol every 2–3 days.
-
i.Resuspend and harvest cells into 15 mL conical.
 CRITICAL: Human PBMC-derived MDSC may adhere to the plate. Detach the adherent cells using Detachin Cell Detachment Solution according to manufacturer’s protocol and pool with the isolated suspension cells. Alternative non-protease cell detachment solutions may be used. Mouse MDSCs usually do not adhere to the plate and do not need this step.
CRITICAL: Human PBMC-derived MDSC may adhere to the plate. Detach the adherent cells using Detachin Cell Detachment Solution according to manufacturer’s protocol and pool with the isolated suspension cells. Alternative non-protease cell detachment solutions may be used. Mouse MDSCs usually do not adhere to the plate and do not need this step. -
ii.Spin down at 1,500 RPM (about 505 × g) for 5 min.
-
iii.Resuspend in 1 mL of MDSC-generation media and plate in a fresh 24-well plate.
-
i.
-
a.
Magnetic sorting of derived MDSCs
Timing: 45 min + 10 min/Additional sample
Derived MDSCs are sorted to ensure consistent sample quality. The MDSCs are sorted magnetically rather than by fluorescent-activated cell sorting to prevent alterations in metabolic profile due to experimental handling (Llufrio et al., 2018).
-
30.Harvest cells into sterile 5 mL round bottom polystyrene tubes with caps.
-
a.Detach adherent cells using Detachin Cell Detachment Solution.
-
a.
-
31.
Centrifuge sample at 1,500 RPM (about 505 × g) for 5 min.
-
32.
Remove supernatant by aspiration.
-
33.
Resuspend in 1 mL sterile PBS.
-
34.
Quantify.
-
35.Set aside 0.1–1 × 106 cells in a new non-sterile 5 mL round bottom polystyrene tube for flow cytometric evaluation of sort efficacy.
-
a.Proceed to step 41 with cells from step 39.
-
a.
-
36.
Isolate MDSCs using STEMCELL magnetic isolation kit according to manufacturer’s instructions.
Note: For mouse-derived MDSC, use the EasySepTM Mouse MDSC (CD11b+GR1+) Isolation Kit (Instructions available here). For human PBMC-derived MDSC, use the EasySepTM HLA Chimerism Whole Blood CD33 Positive Selection Kit (Instructions available here).
-
37.
Resuspend cells in 1 mL sterile PBS.
-
38.
Quantify. (troubleshooting 4).
-
39.Isolate 0.1–1 × 106 cells into a new non-sterile 5 mL round bottom polystyrene tube for flow cytometric evaluation of sort efficacy.
-
a.Proceed to step 41 with cells from step 35.
-
a.
-
40.Isolate appropriate number of cells for metabolic analysis in a sterile 15 mL conical tube.
-
a.Proceed to step 58 for Fatty Acid Oxidation Analysis.
-
b.Proceed to step 79 for if Oxidative Phosphorylation Analysis without FAO analysis is desired.
-
c.Proceed to step 93 for Glycolysis Analysis.
-
a.
Flow cytometric confirmation
Timing: 1 h + 10 min/Additional sample
Isolated MDSC yield and purity are confirmed by flow cytometry.
-
41.
Centrifuge samples from steps 32 and 37 at 1,500 RPM (about 505 × g) for 5 min.
-
42.
Resuspend in 100 μL PBS + 0.5 μL/ 1 × 106 cells FcBlock.
-
43.
Stain at room temperature (20°C–25°C) for 10 min.
-
44.Add 100 μL PBS + 0.5 μL/1 × 106 cells of antibodies to each sample:Note: Make a master mix of antibody dilutions to ensure even dispersal of antibodies across all samples.
-
a.Mouse:
Antibody Dilution Live/Dead Aqua 0.5 μL/1 × 106 cells CD45 – PE-Cy7 0.5 μL/1 × 106 cells CD11b – BUV395 0.5 μL/1 × 106 cells Gr-1 – FITC 0.5 μL/1 × 106 cells Ly6G – APC 0.5 μL/1 × 106 cells Ly6C – BV421 0.5 μL/1 × 106 cells -
b.Human:
Antibody Dilution Live/Dead Aqua 0.5 μL/1 × 106 cells CD11b – APC 0.5 μL/1 × 106 cells CD14 – PerCP-Cy5.5 0.5 μL/1 × 106 cells CD33 – PC7 0.5 μL/1 × 106 cells
-
a.
-
45.
Stain at room temperature (20°C–25°C) for 20 min, protected from light.
-
46.
Add 1 mL of PBS to each sample to dilute excess antibodies.
-
47.
Centrifuge sample at 1,500 RPM (about 505 × g) for 5 min.
-
48.
Resuspend samples in 200 μL of flow buffer.
-
49.Characterize MDSCs on available flow cytometer.Note: Use single-color controls using beads to set up appropriate compensation matrix prior to data collection.
-
a.Mouse bone marrow derived MDSC are defined as CD45+, CD11b+, Gr-1+ (Ly6G/ Ly6C).
-
b.Human PBMC derived MDSCs use: CD11b+, CD14+, CD33+.
-
a.
Seahorse XF assay plate preparation
Timing: 1 h + 10 min/plate + 1 day
Before analysis, suspension cells must be adhered to the plate using Cell-Tak. Plates are usually prepared one day in advance but may be stored at 4°C for up to 2 weeks. If the number of injections is equivalent, multiple assays may be run on the same plate. Therefore, it is recommended to plan the sample layout in advance to ensure that the appropriate number of plates is coated. Plates are prepared in a biosafety cabinet to ensure sterility.
-
50.Coat XF96 Cell culture microplate with Cell-Tak.
-
a.Quickly add 20 μL Cell-Tak and Tissue Adhesive solution (see materials and equipment) into each well.
-
b.Cover the plate and let it sit in the hood for 20 min at room temperature (20°C–25°C).
-
a.
-
51.Rinse wells with sterile, tissue culture grade water.
-
a.Remove residual solution by aspiration.
-
b.Add 100 μL of sterile, tissue culture grade water.
-
c.Remove solution by aspiration.
-
d.Cover the plate and invert to let it dry in the hood for 20 min at room temperature (20°C–25°C).
-
a.
-
52.
Tightly cover plate with parafilm and store at 4°C until use or for up to 2 weeks.
Sensor cartridge hydration and calibration
Timing: 20 min + 5 min/plate + 1 day incubation
For proper detection of changes in pH and/or O2 concentration, the probe tips of the sensor cartridge must be fully hydrated. Cartridges are easily hydrated overnight (minimum 4 h and maximum 72 h) in a 37°C incubator, followed by calibration in the XF Calibrant solution provided by Agilent. All steps are performed in a biosafety cabinet to ensure sterility. Detailed instructions are provided on the Agilent webpage, available here.
-
53.Rehydrate Agilent Seahorse sensor cartridge by submerging in sterile, tissue culture grade water.
-
a.Open a sensor cartridge pack from the Agilent Flux Pack.
-
b.Separate the utility plate and sensor cartridge and rest sensor cartridge upside down next to the utility plate.
-
c.Add 200 μL of sterile, tissue culture grade water into each well of the utility plate.
-
d.Carefully replace the sensor cartridge onto the utility plate to submerge the sensors in the water.
-
a.
-
54.
Place in a humidified, non-CO2, 37°C incubator overnight (usually 12–18 h).
-
55.
Place 25 mL/ plate of XF Calibrant in a 50 mL conical tube and place a humidified, non-CO2, 37°C incubator overnight (usually 12–18 h).
-
56.Finish sensor cartridge rehydration by replacing the water with the pre-warmed XF Calibrant.
-
a.Remove pre-warmed XF Calibrant and sensor cartridge from incubator.
-
b.Remove sensor cartridge from utility plate and set it down upside down.
-
c.Aspirate off the water from the utility plate.
-
d.Add 200 μL of pre-warmed XF Calibrant in each well of the utility plate.
-
e.Carefully replace the sensor cartridge onto the utility plate to submerge the sensors.
-
a.
-
57.
Place in a humidified, non-CO2, 37°C incubator for 1 h prior to assay.
Fatty acid oxidation (FAO) assay
Timing: 1 day + 3 h
FAO rates of MDSCs are analyzed by changes in oxygen consumption rate (OCR) using the Seahorse XF Palmitate Oxidation Stress test kit. Media is changed to the substrate limiting media (with continued treatment) 24 h prior to experiment. A Mito Stress assay usually follows the etomoxir treatment. Detailed protocol for usage of the XF Palmitate Oxidation Stress Test Kit is provided by Agilent, available here.
-
58.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
-
59.
Resuspend in substrate limiting media (SLM) with 10 μM Isoproterenol/ PBS or desired treatment. (See materials and equipment section for media recipe).
-
60.
Plate at 1 × 106 cells/mL in 24 well plates.
-
61.
Incubate at 37°C at 5% CO2 for 24 h.
Note: See problem 5 for exposure length.
-
62.
Warm Cell-Tak coated 96-well flat bottom plate from step 52 to room temperature (20°C–25°C).
-
63.
Harvest cells from plates according to steps 29ai and 29aii.
-
64.
Centrifuge at 1,500 RPM (about 505 × g) for 5 min.
-
65.
Resuspend samples in 1 mL of 1× KHB Assay Working Solution pre-warmed to 37°C. (See materials and equipment section for media recipe).
-
66.
Quantify.
-
67.
Dilute and plate cells at 1 × 105 cells in 150 μL of 1× KHB Assay Working Solution with 10 μM Isoproterenol/ PBS or desired treatment on Cell-Tak coated 96-well flat-bottom plates from step 62.
Note: Plate at least 6 wells/sample to insure triplicate in each Palmitate-BSA vs BSA exposure conditions.
CRITICAL: One well at each corner of the well must be left blank, containing only 180 μL of the 1× KHB Assay Working solution without any cells, for machine calibration.
-
68.Allow cells to adhere to the plate.
-
a.Incubate at room temperature (20°C–25°C) for 45 min.
-
b.Spin down plates for 5 min at 1,500 RPM (about 505 × g).
-
c.Sit in non-CO2 37°C incubator for 10 min.
-
a.
-
69.Load assay compounds into injection ports on the sensor cartridge from step 57.
-
a.Resuspend provided compounds from the kit in the volume of 1× KHB working assay solution specified in the user manual.
-
b.Prepare 2–3 mL of working solution according to the user manual to make a 10× working solution.
-
c.Load 100–200 μL of 10× working solution into the designated ports.
-
a.
Optional: Add 20 μM Hoechst stain with last injection for cell density normalization.
-
70.
If performing cell normalization, obtain a pre-run brightfield and fluorescence image of each well. Otherwise, this step can be skipped.
-
71.Prepare Seahorse XFe 96 Extracellular Flux Analyzer.
-
a.Design experimental template using the Standard Substrate Oxidation Stress Test Assay template and update plate map on the Wave software. Refer to the Agilent webpage for detailed assistance.Note: If using a Seahorse XFe24 Analyzer, the optimal cell density (50%–90% confluency) and injection times must be optimized for the larger well surface area and volume. There is a default 2-minute wait step requirement prior to each measurement.
-
i.3 measurements of baseline oxygen consumption rate. (OCRbaseline).
-
ii.Inject 100 μM etomoxir (ETX).
-
iii.5 OCR measurements at maximal exogenous FAO inhibition. (OCR+ETX).
-
iv.Inject 2 mM Oligomycin.
-
v.3 OCR measurements.
-
vi.Inject 1 mM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP).
-
vii.3 OCR measurements.
-
viii.Inject 1 mM Rotenone and 1 mM Antimycin A.
-
ix.3 OCR measurements.
-
i.
-
b.Run calibration.
-
i.Click “Run”.
-
ii.Insert sensor cartridge from step 69 when prompted.
-
iii.Click “I’m Ready”.
-
iv.Run Calibration.
-
i.
-
a.
-
72.
Add 30 μL of Palmitate-BSA (1 mM Palmitate conjugated to 0.17 mM BSA) or BSA (0.17 mM) to appropriate sample wells.
-
73.Run samples on Seahorse XFe 96 Extracellular Flux Analyzer.
-
a.Click “Open tray” and load cell plate without the lid.
-
b.Click “Load Cell Plate” to run the assay.
-
a.
-
74.If performing cell normalization, normalize the data to cell count. If not, this step can be skipped.
-
a.Obtain post-run brightfield and fluorescence image of each well.
-
b.Calculate each well’s live cell count.
-
c.Click “Normalize”.
-
d.Enter live cell count into the Normalization grid.
-
e.Set Normalization Unit as Cells.
-
f.Click “Apply”.
-
a.
-
75.
Export data from the Wave software into GraphPad Prism format.
Note: Data may be graphed using the default Wave software as well.
-
76.
Graph the individual OCR measurements for each treatment condition.
-
77.
Calculate the average OCR of the sample triplicates per treatment condition.
-
78.
Use the averages to calculate the FAO contribution, defined as OCRbaseline - OCR+ETX. (troubleshooting 5).
Oxidative phosphorylation (OxPhos) assay
Timing: 3 h
OxPhos levels of MDSCs are measured by oxygen consumption rate after priming in OCR media without CO2. Detailed protocol for usage of the Mito Stress Test Kit is provided by Agilent, available here.
-
79.
Warm Cell-Tak coated 96-well flat bottom plate from step 52 to room temperature (20°C–25°C).
-
80.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
-
81.
Resuspend cells in OCR medium pre-warmed to 37°C. (See materials and equipment section for media recipe).
-
82.
Plate at 1 × 105 cells in 180 μL in Cell-Tak coated 96-well flat-bottom plates from step 79.
Note: Plate each experimental condition in triplicate.
CRITICAL: One well at each corner of the well must be left blank, containing only 180 μL of the OCR medium, for machine calibration.
-
83.Allow cells to adhere to the plate.
-
a.Incubate at room temperature (20°C–25°C) for 45 min.
-
b.Spin down plates for 5 min at 1,500 RPM (about 505 × g).
-
c.Sit in non-CO2 37°C incubator for 10 min.
-
a.
-
84.Load assay compounds into injection ports on the sensor cartridge from step 57.
-
a.Resuspend provided compounds from the kit in the volume of OCR medium specified in the user manual.
-
b.Prepare 2–3 mL of working solution according to the user manual to make a 10× working solution.
-
c.Load 100–200 μL of 10× working solution into the designated ports.
-
a.
Optional: Add 20 μM Hoechst stain with last injection for cell density normalization.
-
85.
If performing cell normalization, obtain a pre-run brightfield and fluorescence image of each well. Otherwise, this step can be skipped.
-
86.Prepare Seahorse XFe 96 Extracellular Flux Analyzer.
-
a.Design experimental template and update plate map on the Wave software. Refer to the Agilent webpage for detailed assistance.Note: If using a Seahorse XFe24 Analyzer, the optimal cell density (50%–90% confluency) and injection times must be optimized for the larger well surface area and volume. There is a default 2-minute wait step requirement prior to each measurement.
-
i.Obtain 3 measurements of baseline oxygen consumption rate. (OCRbasal).
-
ii.Inject 2 mM Oligomycin to inhibit mitochondrial complex V.
-
iii.Obtain 3 OCR measurements. (OCR+Oligomycin).
-
iv.Inject 1 mM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) to induce maximal respiration.
-
v.Obtain 3 OCR measurements. (OCR+FCCP).
-
vi.Inject 1 mM Rotenone and 1 mM Antimycin A to inhibit the electron transport chain.
-
vii.Obtain 3 OCR measurements. (OCR+Antimycin A/Rotenone).
-
i.
-
b.Run calibration.
-
i.Click “Run”.
-
ii.Insert sensor cartridge from step 84 when prompted.
-
iii.Click “I’m Ready”.
-
iv.Run Calibration.
-
i.
-
a.
-
87.Run samples on Seahorse XFe 96 Extracellular Flux Analyzer.
-
a.Click “Open tray” and load cell plate without the lid.
-
b.Click “Load Cell Plate” to run the assay.
-
a.
-
88.If performing cell normalization, normalize the data to cell count. If not, this step can be skipped.
-
a.Obtain post-run brightfield and fluorescence image of each well.
-
b.Calculate each well’s live cell count.
-
c.Click “Normalize”.
-
d.Enter live cell count into the Normalization grid.
-
e.Set Normalization Unit as Cells.
-
f.Click “Apply”.
-
a.
-
89.
Export data from the Wave software into GraphPad Prism format.
Note: Data may be graphed using the default Wave software as well.
-
90.
Graph the individual OCR measurements for each treatment condition.
-
91.
Calculate the average OCR of the sample triplicates per treatment condition.
-
92.
Using the average OCR, calculate ATP Production, defined as OCRbasal - OCR+Oligomycin, and Maximal Respiratory Capacity, defined as OCR+FCCP - OCR+Antimycin A/Rotenone.
Glycolysis assay
Timing: 3 h
Glycolysis levels in MDSCs are measured by extracellular acidification rate. Detailed protocol for the usage of the XF Glycolysis Stress Test kit is provided by Agilent, available here.
-
93.
Warm Cell-Tak coated 96-well flat bottom plate from step 52 to room temperature (20°C–25°C).
-
94.
Centrifuge samples at 1,500 RPM (about 505 × g) for 5 min.
-
95.
Resuspend cells in ECAR medium pre-warmed to 37°C. (See materials and equipment section for media recipe).
-
96.
Plate at 1 × 105 cells in 180 μL in Cell-Tak coated 96-well flat-bottom plates from step 93.
Note: Plate each experimental condition in triplicate.
CRITICAL: One well at each corner of the well must be left blank, containing only 180 μL of the ECAR medium, for machine calibration.
-
97.Allow cells to adhere to the plate.
-
a.Incubate at room temperature for 45 min.
-
b.Spin down plates for 5 min at 1,500 RPM (about 505 × g).
-
c.Sit in non-CO2 37°C incubator for 10 min.
-
a.
-
98.Load assay compounds into injection ports on the sensor cartridge from step 57.
-
a.Resuspend provided compounds from the kit in the volume of ECAR medium specified in the user manual.
-
b.Prepare 2–3 mL of working solution according to the user manual to make a 10× working solution.
-
c.Load 100–200 μL of 10× working solution into the designated ports.
-
a.
Optional: Add 20 μM Hoechst stain with last injection for cell density normalization.
-
99.
If performing cell normalization, obtain a pre-run brightfield and fluorescence image of each well. Otherwise, this step can be skipped.
-
100.Prepare Seahorse XFe 96 Extracellular Flux Analyzer.
-
a.Design experimental template and update plate map on the Wave software. Refer to the Agilent webpage for detailed assistance.Note: If using a Seahorse XFe24 Analyzer, the optimal cell density (50%–90% confluency) and injection times must be optimized for the larger well surface area and volume. There is a default 2-minute wait step requirement prior to each measurement.
-
i.Obtain 3 measurements of non-glycolytic extracellular acidification rate (ECAR). (ECARnon-glycolytic).
-
ii.Inject 10 mM Glucose to induce glycolysis.
-
iii.Obtain 3 ECAR measurements. (ECAR+glucose).
-
iv.Inject 2 mM Oligomycin to inhibit mitochondrial complex V and induce maximal glycolysis.
-
v.Obtain 3 ECAR measurements. (ECAR+Oligomycin).
-
vi.Inject 100 mM 2-Deoxy-glucose (2-DG) to inhibit glycolysis.
-
vii.Obtain 3 ECAR measurements (ECAR+2-DG).
-
i.
-
b.Run calibration.
-
i.Click “Run”.
-
ii.Insert sensor cartridge from step 98 when prompted.
-
iii.Click “I’m Ready”.
-
iv.Run Calibration.
-
i.
-
a.
-
101.Run samples on Seahorse XFe 96 Extracellular Flux Analyzer.
-
a.Click “Open tray” and load cell plate without the lid.
-
b.Click “Load Cell Plate” to run the assay.
-
a.
-
102.If performing cell normalization, normalize the data to cell count. If not, this step can be skipped.
-
a.Obtain post-run brightfield and fluorescence image of each well.
-
b.Calculate each well’s live cell count.
-
c.Click “Normalize”.
-
d.Enter live cell count into the Normalization grid.
-
e.Set Normalization Unit as Cells.
-
f.Click “Apply”.
-
a.
-
103.
Export data from the Wave software into GraphPad Prism format.
Note: Data may be graphed using the default Wave software as well.
-
104.
Graph the individual ECAR measurements for each treatment condition.
-
105.
Calculate the average ECAR of the sample triplicates per treatment condition.
-
106.
Using the average ECAR, calculate rate of glycolysis, defined as ECAR+glucose – ECARnon-glycolytic, and glycolytic capacity, defined as ECAR+Oligomycin - ECAR+2-DG.
Expected outcomes
We expect about 1–2 × 106 MDSCs yielded from 100 × 106 PBMCs and 20–25 × 106 MDSCs yielded from 35–45 × 106 cultured bone marrow cells. Upon culturing with IL6 + GM-CSF, there should be about 60%–80% derived MDSCs (Figure 3). After magnetic purification, MDSC purity confirmed by flow cytometry can be enriched up to 98% (Figure 3; See Figure S3 in Mohammadpour et al., 2021). We have derived MDSCs in vitro from mouse bone marrow and human PBMCs to demonstrate that increased β-adrenergic signaling by isoproterenol treatment leads to increased palmitate oxidation, increased basal oxygen consumption rate, ATP production, maximum respiration, and non-glycolytic acidification rate in MDSCs (Mohammadpour et al., 2021) (Figure 4). Isoproterenol-treated MDSCs also demonstrate decreased glycolytic rate and glycolytic capacity (Mohammadpour et al., 2021). Studying the role of MDSC metabolic shift is crucial for cancer prognosis. Elevated lipid transport proteins have been observed in human tumor-infiltrating MDSCs (Al-Khami et al., 2017) and inhibiting FAO in MDSC decrease its immunosuppressive ability (Hammami et al., 2012; Hossain et al., 2015). Our protocol allows the study of MDSC metabolic shifts through efficient in vitro derivation.
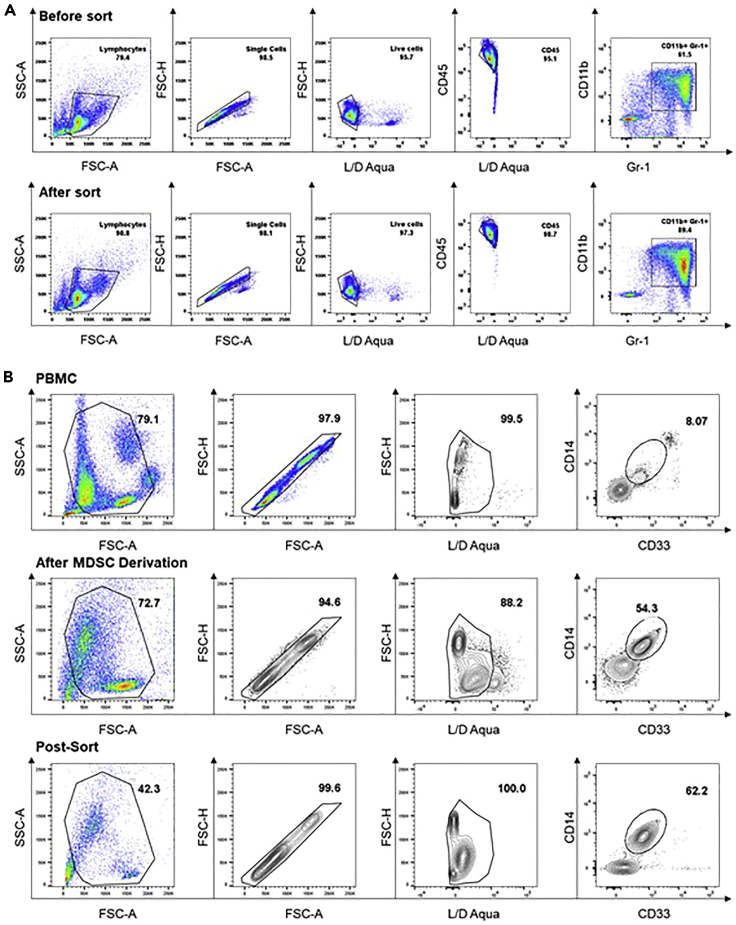
Figure 3.
Flow cytometric confirmation of in vitro derived MDSCs
(A) Representative flow cytometric analysis of murine bone marrow MDSCs from tumor bearing mice. Purity before sorting is about 60% and can be increased to about 90% post-sorting.
(B) A representative flow cytometric analysis of human PBMCs and pre-sort and post-sorted in vitro derived human MDSCs with IL-6 and GM-CSF. Purity before magnetic sorting should range from 50%. After sorting, purity should be above 60%.
Figure 4.
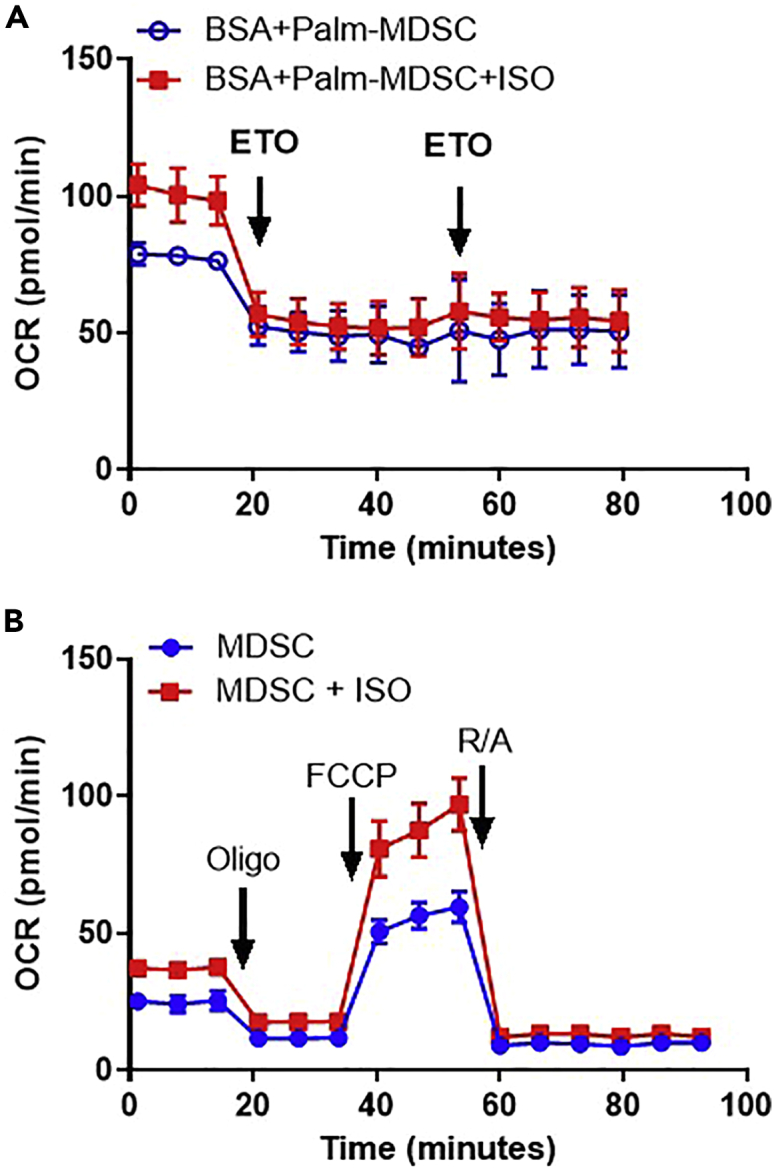
Representative seahorse analysis of in vitro derived MDSCs
Representative fatty acid oxidation (A) and oxidative phosphorylation (B) analysis of in vitro derived MDSCs. A only shows the response to the etomoxir treatments as analysis of mitochondrial stress is more thoroughly analyzed by using the Mito Stress Test Kit. For examples of MDSC glycolysis analysis, refer to Mohammadpour et al. (2021).
Limitations
When studying a specific cell population that is not overly abundant, such as MDSCs, a common limitation is insufficient numbers of cells available for analysis, isolation, and sub-culturing. This is particularly important because many metabolic assays, such as Seahorse extracellular flux analysis, require many cells for replicates from each treatment group to ensure reliable results. Specifically, when deriving MDSCs from murine bone marrow, you must be sure that enough mice are available to harvest and pool bone marrow from to obtain sufficient MDSCs. When deriving MDSCs from human PBMCs, the same limitation exists. Furthermore, there is an increased variability in yield numbers based on more heterogeneous PMBC populations from different donors. In both cases, it is crucial to keep samples on ice and work efficiently, as the amount of time and number of steps required to isolate and/or derive these MDSCs causes a significant amount of stress on cells, often resulting in low cell viabilities. Furthermore, these cells are highly sensitive to freeze/thaw cycles, thus experiments must be carried out immediately, and without freezing any cells. It is also important to acknowledge the dynamic nature of cellular metabolism. Immune cells have high metabolic flexibility (Buck et al., 2017) and thus differences in timing and handling of the cells can introduce experimental variability. Additionally, our protocol derives a combination of both PMN-MDSC (Polymorphonuclear-MDSC) and M-MDSC (monocytic MDSC). To derive human PMN-MDSC specifically, an alternative protocol has been described (Singh and Rieber, 2021). To analyze specific subsets of murine MDSCs, an alternative sorting kit will have to be utilized such as the Miltenyi Myeloid-Derived Suppressor Cell Isolation Kit.
As our protocol focuses on the analysis of MDSCs, we do not provide the additional steps required when working with adherent cells. Alternative protocols have been described (Gu et al., 2021) if adaptation of this protocol is desired for adherent cell types. We utilize the XFe96 Analyzer for this protocol. For users of the XFe24 Analyzer, there are additional protocols available (Gotoh et al., 2021) that can be used as reference for protocol adaptation.
Troubleshooting
Problem 1
No tumor growth after injection. (before you begin step 29).
Potential solution
4T1 growth rate is affected after 20 passages. We recommend using freshly thawed samples that are passaged twice before implantation. During passaging, 4T1 should not be allowed to reach full confluency for optimal health and kept at 80%–90% confluency. In addition, a deep injection of tumor cells can lead to peritoneal tumor growth and the absence of a mammary tumor. To avoid this, the needle tip must be almost parallel to the skin during implantation. A subcutaneous bubble should be visible post-implantation.
Problem 2
Insufficient MDSC yield. (step 13).
Potential solution
If culturing bone marrow cells, there may be incomplete flushing of the bone marrow. Be sure that most of the reddish marrow has been visibly removed from the bone. Increase the number of mice harvested appropriately to the experimental need. About 1–2 × 106 MDSC are derived from 100 × 106 PBMCs cultured and 20–25 × 106 MDSC are derived from 35–45 × 106 bone marrow cells cultured. Do not increase culturing time as longer culture leads to decreased viability.
Problem 3
Contamination of cell culture during in vitro MDSC derivation (step 29).
Potential solution
We recommend using PBS supplemented with 1% Penicillin/Streptomycin during bone marrow harvest. Surgical tools should be autoclaved prior to usage. Sterilize the tools between mice using 70% ethanol.
Problem 4
MDSC purity post-magnetic sort is low (step 39).
Potential solution
If the purity is below 80%, a secondary magnetic sort may be required. Quantify and repeat the magnetic sort according to manufacturer’s guidelines. The cell to antibody ratio can be increased.
Problem 5
There is no difference in Fatty Acid Oxidation in MDSC after isoproterenol treatment (step 78).
Potential solution
Due to the short half-life of Isoproterenol, it may be necessary to add 10 μM of Isoproterenol daily during the in vitro derivation of MDSC. Alternatively, the length of exposure of MDSCs to SLM may need to be adjusted. The purpose of SLM exposure is to try to force cells to burn fats and other fuels other than the diabetic levels of glucose (4.5 g/L) that they are exposed to. The current recommendation was based on the inherent ability of MDSCs to oxidize fats. The etomoxir concentration may also need to be titrated. We recommend running a pilot experiment with 4, 40, 100, and 200 μM concentrations of etomoxir.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hemn Mohammadpour (hemn.mohammadpour@roswellpark.org).
Materials availability
This study did not generate unique reagents.
Acknowledgments
The authors thank Jeanne M. Prendergast and Courtney Ryan for technical assistance and the Roswell Park Flow Cytometry Core facility and Immune Analysis Facility for expertise and support. The Graphical Abstract and Figures 1 and 2 were created with BioRender.com. This project was supported by National Institutes of Health (NIH) grants, R01 CA205246 and R01 CA236390 (to E.R.), F32 CA239356 and K99 HL155792 (to H.M.), F30 CA265127 (to C.M.), the Roswell Park Alliance Foundation, and NCI grant P30 CA016056.
Author contributions
J.C. was responsible for composition of the manuscript. J.C., C.M., E.R., and H.M. edited the manuscript. H.M., C.M., G.D., and N.G. conducted the experiments. H.M. and E.R. were responsible for study design.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101389.
Data and code availability
This study did not generate original code. The published article includes all the datasets generated during this study.
References
- Al-Khami A.A., Zheng L., Del Valle L., Hossain F., Wyczechowska D., Zabaleta J., Sanchez M.D., Dean M.J., Rodriguez P.C., Ochoa A.C. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. Oncoimmunology. 2017;6:e1344804. doi: 10.1080/2162402x.2017.1344804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., Sowell R.T., Kaech S.M., Pearce E.L. Metabolic instruction of immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K., Takata Y., Nakashima Y., Mizuguchi S., Komori K., Kang D. Metabolic analysis of mouse bone-marrow-derived dendritic cells using an extracellular flux analyzer. STAR Protoc. 2021;2:100401. doi: 10.1016/j.xpro.2021.100401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Ma Y., Liu Y., Wan Q. Measurement of mitochondrial respiration in adherent cells by seahorse XF96 cell Mito stress test. STAR Protoc. 2021;2:100245. doi: 10.1016/j.xpro.2020.100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami I., Chen J., Murschel F., Bronte V., De Crescenzo G., Jolicoeur M. Immunosuppressive activity enhances central carbon metabolism and bioenergetics in myeloid-derived suppressor cells in vitro models. BMC Cell Biol. 2012;13:18. doi: 10.1186/1471-2121-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain F., Al-Khami A.A., Wyczechowska D., Hernandez C., Zheng L., Reiss K., Valle L.D., Trillo-Tinoco J., Maj T., Zou W., et al. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol. Res. 2015;3:1236–1247. doi: 10.1158/2326-6066.cir-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llufrio E.M., Wang L., Naser F.J., Patti G.J. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018;16:381–387. doi: 10.1016/j.redox.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadpour H., Macdonald C.R., Mccarthy P.L., Abrams S.I., Repasky E.A. beta2-adrenergic receptor signaling regulates metabolic pathways critical to myeloid-derived suppressor cell function within the TMEβ2-adrenergic receptor signaling regulates metabolic pathways critical to myeloid-derived suppressor cell function within the TME. Cell Rep. 2021;37:109883. doi: 10.1016/j.celrep.2021.109883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Rieber N. In vitro generation of human neutrophilic myeloid-derived suppressor cells. Methods Mol. Biol. 2021;2236:77–83. doi: 10.1007/978-1-0716-1060-2_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bone marrow is harvested from the femur and tibia from both mouse hind legs. Majority of the muscles are cut away, which allows to easy dislocation and isolation of the bones from the remaining muscles and tendons. The bone marrow is flushed out from the bone using PBS-filled syringes. For increased yield, multiple mice of the same genotype or treatment groups may be pooled if desired and appropriate.
Data Availability Statement
This study did not generate original code. The published article includes all the datasets generated during this study.