Summary
Background
Inflammatory bowel disease (IBD) has complex genetic and environmental aspects, and free fatty acid receptors (FFARs) may bridge genetic and dietary aspects. FFAR4 is highly expressed in the intestine and acts primarily as the receptor of long-chain fatty acids, which are major components of the human diet. It is unclear what role, if any, FFAR4 may play in IBD.
Methods
Mouse and human colitis samples, mice with complete FFAR4 knockout, intestine-specific FFAR4 knockout and FFAR4 overexpression and cell culture were used. RNA-sequencing analysis and flow cytometry were performed to examine the mechanisms.
Findings
The results showed that FFAR4 expression was upregulated in colitis tissues and that the loss of intestinal FFAR4 ameliorated colitis, whereas intestinal FFAR4 overexpression exacerbated the disease. We identified intestinal epithelial cell deletion of FFAR4 by upregulating ZBED6, which in turn induced L33 transcription, and L33 elevated Treg cell numbers, ameliorating colitis.
Interpretation
FFAR4 deletion attenuates colitis by modulating Treg cells via the ZBED6-IL33 pathway.
Funding
National Natural Science Foundation of China, Innovation and Application Project of Medical and Public Health Technology of Wuxi Science and Technology, Fundamental Research Funds for the Central Universities and the Fund of Wuxi Healthcare Commission.
Keywords: FFAR4, Colitis, Treg, IL33, ZBED6
Research in context.
Evidence before this study
Inflammatory bowel disease (IBD) is characterized by persistent inflammation of the intestinal tissues, with the treatment effects often being unsatisfactory. Epidemiological investigations indicated that the risk of IBD is associated with excess and imbalance dietary fatty acids intake. Dietary polyunsaturated fatty acids act as signaling molecules by binding with free fatty acid receptor 4 (FFAR4). Previous studies reported that FFAR4 mediates the inflammatory regulation of n3 polyunsaturated fatty acid. However, it is not clear how FFAR4 itself affects IBD.
Added value of this study
In this study, we show that FFAR4 expression is upregulated in IBD patients and in the mice with colitis, that intestinal-specific deletion of FFAR4 ameliorates colitis whereas intestinal-specific overexpression of FFAR4 exacerbates colitis. In addition, we identified intestinal epithelial cell deletion of FFAR4 via upregulating ZBED6 which in turn induces IL33 transcription, and IL33 elevates Treg numbers which ameliorates colitis.
Implications of all the available evidence
This study reveals a promotive role of intestinal FFAR4 in colitis development, uncovers a novel mechanism by which FFAR4 regulates Treg cells, and suggests suppression rather than activation of intestinal FFAR4 may be a useful therapeutic strategy for the management of IBD.
Alt-text: Unlabelled box
Introduction
IBD is a chronic and progressive disease that includes ulcerative colitis (UC) and Crohn's disease (CD) and mainly affects the gastrointestinal (GI) tract. High incidence rates have been recorded in North America and Europe, and increasing rates are observed in previously low-incidence regions such as Asia.1,2 This phenomenon may be due to a complex interplay between genetics and the environment.3,4
Some treatment options are available, such as anti-tumor necrosis factor therapy, anti-integrin therapy, anti-interleukin therapy and tofacitinib. However, primary and secondary treatment failure remain high,5 underscoring the need for new therapeutic modalities. Nutritional intervention may be an important strategy.4,5
G protein-coupled receptors play important roles in regulating intestinal functions and have been implicated in the development and progression of IBD.6,7 Among them, the free fatty acid receptors FFAR1 (GPR40), FFAR2 (GPR43), FFAR3 (GPR41) and FFAR4 (GPR120) may bridge the genetic and environmental gaps in IBD.8,9 FFAR2 and FFAR3 are activated by short-chain fatty acids (SCFAs), whereas FFAR1 and FFAR4 are activated by medium-chain and long-chain fatty acids (LCFAs).8,10 Fatty acids, especially LCFAs, are major components of the human diet.
FFAR2 and FFAR3 regulate inflammation in the intestine via gut microbiota derived SCFAs.11,12 FFAR1 is mainly expressed in pancreatic β cells13 and may ameliorate intestinal inflammation through an indirect mechanism.14 In contrast, FFAR4 is highly expressed in the intestine and acts primarily as the receptor of LCFAs.8,15 Data from both mice and humans showed anti-inflammatory effects of omega-3 LCFAs on the GI tract.16, 17, 18 The mechanism has not been well characterized. Some studies have indicated that stimulating macrophage FFAR4 with omega-3 LCFAs or agonists causes an anti-inflammatory effect.19,20 However, it is unclear whether intestinal FFAR4 exerts an anti-inflammatory effect. Furthermore, the omega-3 LCFA level is disproportionally low in the modern diet.21 Therefore, what role, if any, intestinal FFAR4 plays in IBD remains to be defined.
Considerable evidence indicates that Treg cells play an essential role in maintaining immune tolerance and balance, and Treg cells in the intestinal microenvironment exert a negative immunomodulatory effect on the pathogenesis of IBD.22 Treg cells mainly participate in IBD by secreting anti-inflammatory cytokines, such as TGF-β and IL-10, suppressing the activity of immune cells and thereby controlling inflammation.23 Previous studies have suggested that omega-3 LCFAs or agonists of FFAR4 regulate Treg cells in several metabolic disorders.24 Therefore, intestinal FFAR4 may play a role in the development of IBD by modulating Treg cells.
In the current study, we showed that FFAR4 expression was upregulated in IBD patients and in mice with colitis, that intestinal-specific deletion of FFAR4 ameliorated colitis, while intestinal-specific overexpression of FFAR4 exacerbated colitis, and intestinal FFAR4 regulated Treg cells via the ZBED6/IL33 pathway.
Methods
Ethics
All animal procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the School of Medicine of Jiangnan University and were approved by the Animal Ethics Committee of Jiangnan University (JN No: 20171030c0110506). The collection of human peripheral blood cells from UC patients and healthy donors was approved by the Medical Ethics Committee of Jiangnan University (Ref. No. JNU20210310IRB01). Written informed consent was obtained from all participants or their legal guardians at the time of admission.
Animals
FFAR4 total knockout (KO, RRID: MGI:7256540) and Villin-Cre mice (RRID: IMSR_JAX: 004586) were purchased from Shanghai Bioraylab and Shanghai Biomodel Organism, respectively. Tail tips were digested with 100 μl of 50 mM NaOH at 95°C for 20 min and then neutralized with 10 μl of Tris-HCl (pH=8.0). KO mice were genotyped by PCR using tail DNA with the forward primer CCCGGCATGTCCCCTGAGTGT and the reverse primer TGGTCGCCCTTGACATCCGAGA at 95 ℃ for 30 s and 63 ℃ for 45 s for 40 cycles, generating an 87 bp fragment for the wild-type and a 110 bp fragment for KO mice. Villin-Cre mice were genotyped using the forward primer AGCGATGGATTTCCGTCTCTGG and the reverse primer AGCTTGCATGATCTCCGGTATTGAA at 95°C for 30 s, 58°C for 45 s, and 72°C for 30 s for 40 cycles, generating a 272 bp fragment. Floxed FFAR4 (fl/fl, RRID: MGI: 7256541) and FFAR4 transgenic (t/w, RRID: MGI: 7256542) mice were constructed commercially by Shanghai Biomodel Organism and Nanjing Biomedical Research Institute of Nanjing University. FFAR4 (fl/fl) mice were genotyped using the forward primer TGCTCTTTCTGGAGCTGTGT and the reverse primer AGAGATCAGAATGGACAACT at 95°C for 30 s, 58°C for 45 s, and 72°C for 30 s for 40 cycles, generating a 272 bp fragment for fl/fl and a 243 bp fragment for the wild-type. FFAR4 transgenic mice were genotyped using forward primer 1 (CAGCAAAACCTGGCTGTGGATC), forward primer 2 (GTGGAGTCCCATCATCATCACC) and the reverse primer ATGAGCCACCATGTGGGTGTC at 95 ℃ for 30 s, 65 ℃ (-0.5 ℃/cycle) for 30 s, and 72 ℃ 45 s for 20 cycles and then 95 ℃ for 30 s, 55 ℃ for 30 s, and 72 ℃ 45 s for 20 cycles, generating a 1051 bp fragment for t/t and a 243 fragment for the wild-type. The fl/fl mice were mated with Villin-Cre mice to generate intestinal epithelial cell-specific or gut-specific FFAR4-knockout mice (GKO). The t/w mice were crossed with Villin-Cre mice to obtain gut-specific FFAR4 transgenic mice (Ge). All mice were on a C57BL/6J background. Animals were housed and bred in an SPF environment and received water and food ad libitum. All animal procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the School of Medicine, Jiangnan University and approved by the Animal Ethics Committee of Jiangnan University (JN No: 20171030c0110506).
Patient samples
UC patients were recruited from the Affiliated Hospital of Jiangnan University. The diagnosis of UC was determined based on clinical, endoscopic, radiologic, and histopathologic findings according to the established guidelines.25 Human peripheral blood cells were collected from UC patients and healthy donors for FFAR4 mRNA analysis. This study was approved by the Medical Ethics Committee of Jiangnan University (Ref. No. JNU20210310IRB01). Written informed consent was obtained from all participants or their legal guardians at the time of admission.
Experimental colitis
The mice were divided into several groups according to experimental needs. For the dextran sulfate sodium (DSS)-induced colitis model, mice were exposed to 2.5% (w/v) DSS (MP Biomedicals, 160110) in drinking water for 7 days. Body weight and disease activity index (DAI) was examined daily to evaluate the severity of colitis in the mice.26 The DAI includes weight loss (0: none; 1: 1-5%; 2: 6-10%; 3: 11-18%; 4: >18%), stool consistency (0: well-formed pellets; 1: soft but still formed stools; 2: soft stools; 3: very soft and wet; 4: diarrhoea) and rectal bleeding (0: negative haemoccult test; 1: positive haemoccult test; 2: blood visibly present in the stool; 3: blood visibly and blood clotting on the anus; 4: gross bleeding). DAI = [(weight loss score + stool consistency score + gross bleeding score)/3]. The mice were sacrificed after 7 days of DSS administration. Serum was collected in separating gel tubes (BD Bioscience, 367955). The spleen was collected and weighed. The colon was removed, and the total length was measured. The histological score included inflammation (0: none; 1: slight; 2: moderate; 3: severe), extent (0: none; 1: mucosa; 2: mucosa and submucosa; 3: transmural), crypt damage (0: none; 1: basal one-third damaged; 2: basal two-thirds damaged; 3: only surface epithelium intact; 4: entire crypt and epithelium lost) and percentage involvement (1: 1-25%; 2: 26-50%; 3: 51-75%; 4: 76-100%).27 Score = [(inflammation score + extent score + crypt damage score + percentage involvement score)]. For dinitrobenzene sulfonic acid (DNBS)-induced colitis, 100 μl of 20 mg/ml DNBS (TCI chemicals, D0821) was administered intrarectally with a 1 ml syringe and a plastic catheter (0.45 mm Φ). The mice were monitored daily for 4 days. For oxazolone (OXZ)-induced colitis, the mice were anaesthetized with isoflurane and sensitized with 150 μl of 3% oxazolone (Sigma Aldrich, 862207) in acetone and olive oil (4:1 v/v) on the shaved back between the shoulders. After 7 days, 100 μl of 1% oxazolone solution was administered intrarectally for 4 days by the same method as described for DNBS. For both DNBS- and OXZ-induced colitis, body weight was recorded daily. The mice were sacrificed, and colon length was measured.
Histological analysis of colitis
Distal colon tissues were fixed in 4% phosphate-buffered formaldehyde solution (Shanghai Yuanye Bio-Technology, R20497) for 36 h and embedded in paraffin. Paraffin sections (5 μm) were prepared and stained with haematoxylin and eosin (Shanghai Yuanye Bio-Technology, R20570) or alcian blue and nuclear fast red (Sangon Biotech, E670107).
Myeloperoxidase (MPO) activity
MPO is an enzyme that primarily exists in neutrophils. MPO provides a quantitative index of neutrophil infiltration in inflammatory intestinal tissue. MPO activity was measured with an MPO assay kit (Nanjing Jiancheng Bioengineering Institute, A044-1-1) according to the manufacturer's protocol.
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was isolated from the colon using a Total RNA Extraction Kit (K101, JN.BIOTOOLS) according to the manufacturer's protocol. RNA was reverse transcribed using a BT-I 1st Strand cDNA Synthesis Kit (K102, JN.BIOTOOLS). mRNA quantification was performed by using a CFX Connect™ Real-Time PCR Detection System (Bio–Rad) and Hieff® qPCR SYBR Green Master Mix (Shanghai Yeasen Biotechnology Co., Ltd., 11201ES03). The qPCR conditions included 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 30 s. The results were normalized to the housekeeping gene β-actin and are presented as 2−ΔΔCt. The PCR primers used were as follows: mouse Il-1β CTGAACTCAACTGTGAAATGC (F), TGATGTGCTGCTGCGAGA (R); mouse Il-6 CTCTGCAAGAGACTTCCATCCAGT (F), GAAGTAGGGAAGGCCGTGG (R); mouse Il-10 CCCTTTGCTATGG TGTCCTT (F), TGGTTTCTCTTCCCAAGACC (R); mouse Tnf-α AGGGTCTGGGCCA TAGAACT (F), CCACCACGCTCTTCTGTCTAC (R); mouse Icam-1 CTTCAGAGGCAGGAAACAGG (F), AGATCACATTCACGGTGCTG (R); mouse Cd25 CAAGAACGGCACCATCCTAAA (F), TCCTAAGCAACGC ATATAGACCA (R); mouse Foxp3 CACCTATGCCACCCTTATCCG (F), CATGCGAGTAAACCAATGGTAGA (R); mouse Tgfβ CCACCTGCAAGACCATCGAC (F), CTGGCGAGCCTTAGTTTGGAC (R); mouse Il33 CCTTCTCGCTGATTTCCAAG (F), CCGTTACGGATATGGTGGTC (R); mouse β-Actin TGTTACCAACT GGGACGACA (F), CTGGGTCATCTTTTCACGGT (R); human IL33 GTGACGGTGTTGATGGTAAGAT (F), AGCTCCACAGAGTGTTCCTTG (R); human ZBED6 GAAGGGTTTGCGAATTAAGGGG (F), GGGTCATT GGAAGCTAACAAAGC (R); human HOXA1 TCCTGGAATA CCCCATACTTAGC (F), GCACGACTGGAAAG TTGTAATCC (R); and human β-ACTIN CATGTACGTTGCTATCCAGGC (F), CTCCTTAATGTCACGCACGAT (R).
Transcriptomics analysis
Total RNA was isolated from the distal colon with a Total RNA Extraction Kit (K101, JN. BIOTOOLS). Reverse transcription was performed using a BT-I 1st Strand cDNA Synthesis Kit (K102, JN. BIOTOOLS), and second-strand synthesis was performed using a Second Strand cDNA Synthesis Kit (Beyotime Biotechnology, D7172). DNA samples were fragmented and labelled with Tn5 transposase. PCR amplification was performed using HiFi PCR Mix for NGS (CWBIO, CW2648), and index codes were added to each sample. PCR products were purified using a FastPure Gel DNA Extraction Mini Kit (Nanjing Vazyme Biotech Co., Ltd., DC301-01) and sequenced using an Illumina NovaSeq (GENEWIZ). The sequencing data were analysed using STAR (http://code.google.com/p/rna-star/) and R software (version 3.5). Differentially expressed genes were defined as genes with a P value < 0.05 and with a fold change ≥ 2. Gene ontology (GO) analysis was performed using Metascape (http://metascape.org). A heatmap was generated using Tbtools software (https://github.com/CJ-Chen/TBtools).
Preparation of cell suspensions from the spleen, mesenteric lymph node and colon lamina propria
The spleen was removed and cut into pieces and then pressed through a 70 μm nylon mesh (BD Bioscience, 352350) into a culture plate with a syringe plunger. The mesh was washed twice with D-Hank's buffer (Sangon Biotech, B548144). The cell suspension was centrifuged and resuspended in red blood cell lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM ethylenediaminetetraacetic acid or EDTA, pH=7.2). After lysis, the cells were centrifuged and resuspended in cold FACS buffer (D-Hank's containing 10 mM hydroxyethylpiperazine ethane sulfonic acid or HEPES (Sangon Biotech, A100485), 0.5% BSA (Sangon Biotech, A600332), 1 mM EDTA, pH=7.2). Mesenteric lymph node cell suspensions were prepared in the same way, but the red blood cell lysis step was omitted. To prepare the lamina propria cell suspension, the intestine was opened longitudinally, and faeces and mesentery were cleared from the intestinal surface. The intestine was incubated in 5 ml of predigestion solution (D-Hanks containing 10 mM HEPES, 0.5% bovine serum albumin or BSA, 5 mM EDTA and 1 mM DL-dithiothreitol or DTT, pH=7.2) for 20 min at 37°C with slow rotation (200 rpm). After 2 rounds of predigestion, the remaining intestine was washed twice in cold D-Hanks and cut into 1 mm pieces. The tissue fragments were incubated in 5 ml of digestion solution (FACS buffer without EDTA, 1 mg/ml collagenase Ⅳ (Sangon Biotech, A004186), 0.5 mg/ml DNase I (Sangon Biotech, A510099)) for 45 min at 37°C with slow rotation (200 rpm). The digested tissue was filtered through a nylon mesh and resuspended in FACS buffer.
Flow cytometric analysis
The cell suspension was prepared and incubated with FcR Blocking Reagent (Miltenyi Biotec, 130-092-575). For surface staining, the cells were incubated with anti-CD4-APC (BioLegend, 100411, RRID: AB_312696), anti-CD25-BV421 (BioLegend, 102033, RRID: AB_10895908) and anti-CD45-PE/Cy7 (BioLegend, 103113, RRID: AB_312978) on ice for 40 min. For intracellular staining, the cells were fixed and permeabilized with Foxp3/transcription factor fixation/permeabilization concentrate and diluent (ThermoFisher Scientific, 00-5523-00) and stained with anti-FOXP3-PE (ThermoFisher Scientific, 12-5773-80, RRID: AB_465935) at room temperature for 40 min. Stained samples were analysed in an Attune™ NxT Acoustic Focusing Cytometer (ThermoFisher Scientific), and 1×106 cells were counted. The data were analysed with FlowJo V10.0.7 (FlowJo, OR, USA).
Treg cell depletion
Treg cells were depleted by using anti-CD25 mAbs (BioXcell, BE0012, RRID: AB_1107619) or cyclophosphamide (Cy, Adamas, 01094584) as previously described.28, 29, 30 Mice were intraperitoneally injected with anti-CD25 mAbs (100 μl/mouse) or Cy (20 mg/kg) on Day -1 and Day 2, and DSS treatment started on Day 0 and ended on Day 7.
Cell lines and cell culture
HT29 (RRID: CVCL_0320) and HEK-293T cells (RRID: CVCL_0063) were purchased from the National Collection of Authenticated Cell Cultures and cultured in RPMI-1640 medium (Gibco, C11875500) supplemented with 5% foetal bovine serum (FBS, Biological Industries, 04-001-1ACS) or DMEM (Gibco, C11965500) supplemented with 5% FBS, respectively. FFAR4 stable knockdown cell lines were established by lentiviral-based stable shRNA infection and puromycin selection (Beyotime Biotechnology, ST551).
Small interfering RNA (siRNA) transfection
Fifty-percent confluent cell cultures were transfected with 50 nM siRNA using JetPrime (Polyplus). The siRNA sequences were ZBED6: GGGCUGUUGCCAACAAAGATT and HOXA1: CCAUAGGAUUACAACUUUCTT. Universal negative control siRNA (GenePharma, A06001) was used as a control.
Construction and packaging of shRNA lentivirus
FFAR4 shRNA (GATCTGGGTGGTGGCCTTCACATTTTCAAGAGAAATGTGAAGGCCACCACCCATTTTT) and Il33 shRNA (GATCCCTGGTTCTAAACAAATGATCAAGAGTCATTTGTTTAGAACCAGGTTTTT) sequences were cloned into the pLVX-shRNA2 lentiviral vector (gene synthesis and subcloning were performed by GenScript Biotech Corporation). An empty construct containing no shRNA was used as a control. HEK-293T cells (5×106) were plated in 10 cm culture dishes and transfected with 8 μg of pLVX (RRID: Addgene_125839), 6 μg of psPAX2 (RRID: Addgene_12260) and 2 μg of pMD2G (RRID: Addgene_12259) using JetPrime (Polyplus, 101000046). After 4 h, the media were replaced with 8 ml of fresh media. After an additional 48 h, the media containing virus were collected, centrifuged, filtered through a 0.45 μm filter, and stored at -80°C. Lentiviral particles were examined by qPCR after the infection of HEK-293T cells as previously described.31
IL33 Knockdown
To knock down Il33 in the mouse colon, Il33 shRNA lentiviruses were administered intrarectally as previously reported.32 In brief, anaesthetized mice were pretreated with 20% ethanol (100 μl/mouse). After 1 h 30 min, viral enemas were administered (108 TU/ml, 100 μl/mouse).
Western blotting
Protein samples were boiled in SDS–PAGE loading buffer (Sangon Biotech, C516031) and separated by SDS–PAGE. Proteins were transferred to PVDF membranes (Millipore, ISEQ00010), and the membranes were blocked with 5% skim milk for 60 min (BD Bioscience, 232100). The PVDF membranes were immunoblotted with primary antibodies (anti-IL33, Proteintech, 12372-1-AP, RRID: AB_2877852; anti-HOXA1, Affinity Biosciences, DF3187, RRID: AB_2835410; anti-ZBED6, Atlas Antibodies, HPA026439, RRID: AB_10599829) overnight at 4°C. After primary antibody incubation, the membranes were washed with TBST and incubated with secondary antibodies (HRP goat anti-rabbit IgG, ABclonal, AS014, RRID: AB_2769854) for 60 min at room temperature. Before imaging, the membranes were washed with TBST, and bands were visualized with ECL reagents (Millipore, WBKLS0500).
Dual luciferase assay
Human ZBED6 or HOXA1 coding sequences were cloned into the pcDNA3.1 expression vector (gene synthesis and subcloning were performed by GenScript Biotech Corporation), resulting in pcDNA3.1-ZEBD6 or pcDNA3.1-HOXA1. A 2100 bp human IL33 promoter sequence was cloned into the pGL30-basic vector with a firefly luciferase (LUC) reporter gene (Promega, E1751), generating pGL3-IL33. Cells (2◊105/well) were seeded in 6-well plates and transfected using JetPrime (Polyplus) with 1600 ng of pGL3-IL33, 1600 ng of pcDNA3.1-ZEBD6 or pcDNA3.1-HOXA1, and 4 ng of pRL-CMV, which expresses Renilla luciferase (REN), as a transfection efficiency control. The enzyme activities of LUC and REN were measured using a Dual-luciferase Assay Kit (Nanjing Vazyme Biotech Co., Ltd., DD1205) on a BioTek Synergy H4 multimode plate reader (Bio Tek Instruments, Inc.). IL33 promoter activities are presented as the LUC/REN ratio.
Statistical analysis
ROC analysis was performed using GraphPad Prism (GraphPad Software). The data are presented as the mean ± standard deviation (SD). One-way ANOVA and Student's t test were performed using SPSS software (SPSS Inc.). Tukey's one-way analysis of variance (ANOVA) was used for multiple comparisons (*p < 0.05, **p < 0.01, and ***p < 0.001).
Role of funders
This work was supported by the National Natural Science Foundation of China (82000808), the Innovation and Application Project of Medical and Public Health Technology of Wuxi Science and Technology (N20202005), the Fundamental Research Funds for the Central Universities (JUSRP12048) and the Fund of Wuxi Healthcare Commission (M202004). The funders did not have any role in the study design, data collection, data analyses, interpretation, or writing of the report.
Results
FFAR4 is upregulated in mice and patients with colitis
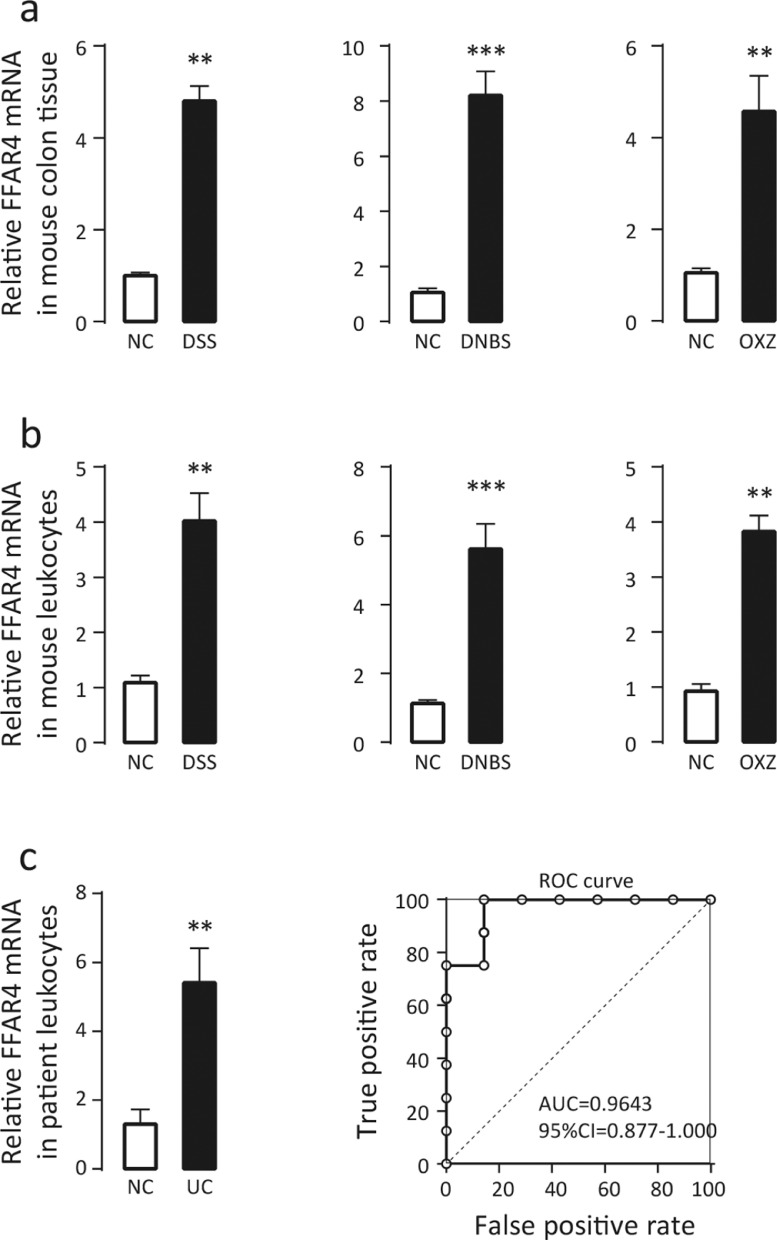
To investigate whether FFAR4 is related to IBD, three different colitis models (DSS-, DNBS- and OXZ-induced colitis) were established in mice. The levels of FFAR4 mRNA in both the colon (Figure 1a) and peripheral blood leukocytes (Figure 1b) were significantly increased in all colitis models, indicating marked consistency. Next, FFAR4 mRNA expression was measured in human peripheral blood leukocytes derived from UC patients (n=8) and healthy controls (n=7), and similar upregulation was observed (Figure 1c). In addition, a receiver operating characteristic (ROC) curve analysis was performed on the small number of patient samples, and the results suggested that FFAR4 might have diagnostic potential for colitis (Figure 1c). These results indicate that FFAR4 may participate in the disease progression of colitis.
Figure 1.
Upregulation of FFAR4 mRNA expression in mice and patients with colitis. Three different murine colitis models (DSS-, DNBS- and OXZ-induced colitis) were used, and qPCR was performed to measure the relative mRNA levels of FFAR4 in (a) mouse colons (n=6) and (b) peripheral blood leukocytes (n=4), as well as (c) human peripheral blood leukocytes derived from colitis patients (n=8) and healthy controls (n=7). The receiver operating characteristic (ROC) curve suggested that FFAR4 may have diagnostic potential for colitis. Error bars represent standard deviation, * p≤0.05, **p≤0.01, *** p≤0.001 by t test.
FFAR4 gene deletion attenuates experimental colitis
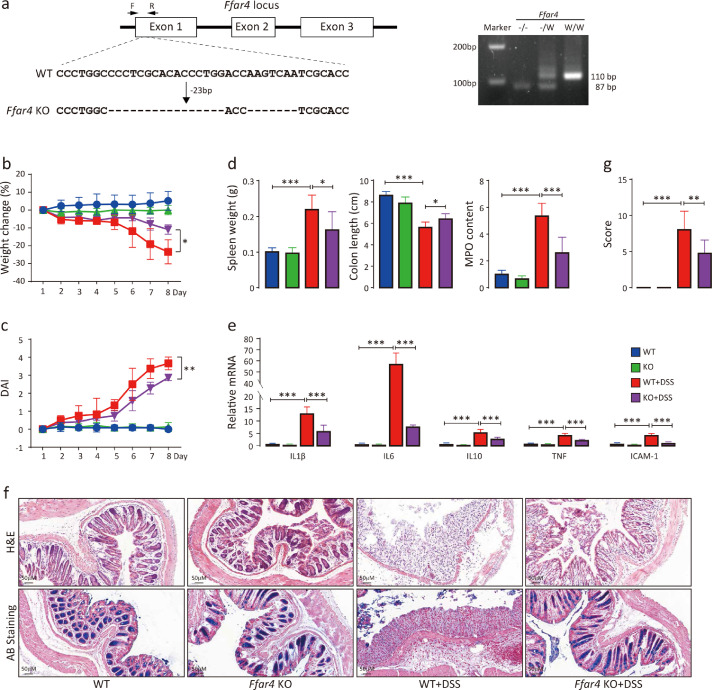
Although FFAR4 is highly expressed in human and mouse intestines, FFAR4 is also expressed in other tissues, such as the lung, adipocytes, dendritic cells and microphages8 (http://biogps.org/). In addition, IBD may involve multiple organs.33 Therefore, FFAR4 total-knockout (KO) mice were generated to study whether FFAR4 deletion affects colitis (Figure 2a). After 7 days of DSS administration, KO mice exhibited lower levels (p<0.05) of body weight loss than wild-type (WT) mice (Figure 2b). Irregular stool and rectal bleeding were also less severe in KO mice than in WT mice. Accordingly, the disease activity index (DAI) was markedly decreased in KO mice (Figure 2c). Furthermore, KO mice displayed lower levels of spleen weight gain, colon length shortening and colonic myeloperoxidase (MPO) activity increases than WT mice (Figure 2d). KO mice also had slight increases in colonic mRNA expression of the inflammatory factors Il-1β, Il-6, Tnf-α and Icam-1 (Figure 2e). Haematoxylin and eosin (H&E) and alcian blue (AB) staining of KO mouse colon tissue revealed reduced intestinal mucosal damage (Figure 2f). Overall, KO mice displayed a less severe colitis phenotype than WT mice (Figure 2g). Similar results were obtained in DNBS- and OXZ-induced colitis mouse models (Figure S1). Therefore, total FFAR4 knockout alleviated experimental colitis.
Figure 2.
FFAR4 gene deletion attenuates DSS-induced colitis. CRISPR–Cas9 gene editing strategy for total FFAR4 knockout (a). The deletion of 26 bp and 7 bp nucleotides in the first exon of FFAR4 was identified by genomic sequencing. Genotyping primers flanking the deletion region amplified a 110 bp band of the wild-type and an 87 bp band in knockout mice by PCR. Colitis was induced by DSS in mice (n=8 per group), and weight changes (b) and DAI scores (c) were recorded daily (t test was performed among DSS groups). The mice were sacrificed on Day 8, and spleen weight, colon length, relative colonic MPO levels (d), relative mRNA levels of colonic inflammation-related genes (e), colon morphology (f) and histological scores (g) were monitored. Statistical significance was determined using one-way ANOVA with Tukey tests for multiple-group comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
Intestinal loss of FFAR4 recapitulates the attenuation of DSS-induced colitis
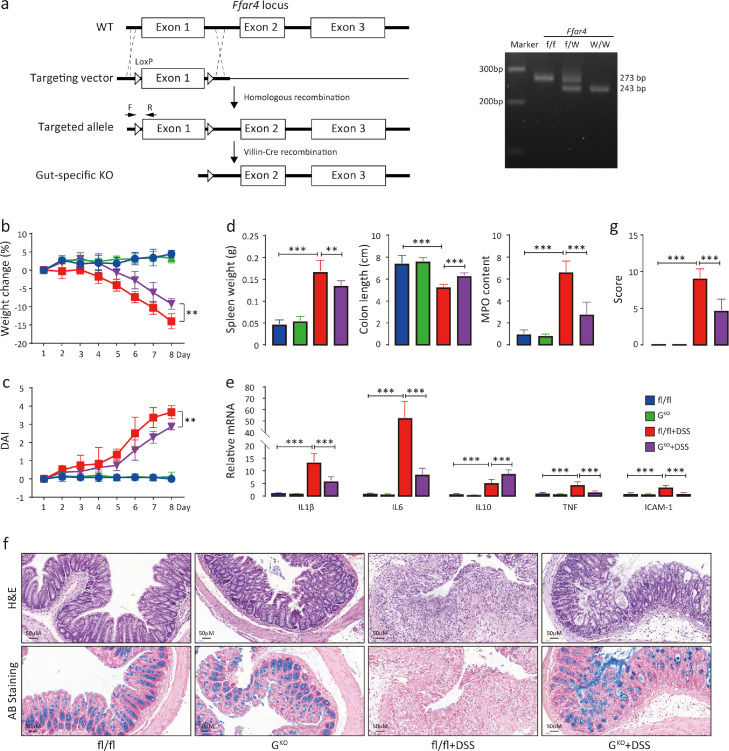
To determine whether intestinal FFAR4 is the main factor in experimental colitis, gut-specific FFAR4 knockout (GKO) mice were generated (Figure 3a). During DSS administration, the GKO mice exhibited less weight loss (Figure 3b) and lower DAI scores (Figure 3c) than fl/fl mice. Similarly, GKO mice presented lower levels of spleen weight gain, colon length shortening, colonic MPO activity increases (Figure 3d), colonic mRNA expression of inflammatory factors Il-1β, Il-6, Tnf-α and Icam-1 (Figure 3e), and intestinal mucosal damage (Figure 3f) and showed lower histological scores than fl/fl control mice (Figure 3g). These results indicate that the loss of intestinal FFAR4 is mainly responsible for the attenuation of DSS-induced colitis.
Figure 3.
Intestinal loss of FFAR4 recapitulates the attenuation of DSS-induced colitis. Gene targeting strategy for intestinal-specific FFAR4 knockout (a). Mice with floxed exon 1 of FFAR4 were generated by homologous recombination. Intestinal-specific FFAR4 knockout (GKO) mice were obtained by mating with Villin-Cre mice. Genotyping primers amplifies a 243 bp band in wild-type and a 273 bp band in fl/fl mice by PCR. Colitis was induced by DSS in fl/fl and GKO mice (n=8 per group), and weight changes (b) and DAI scores (c) were recorded (t test was performed among DSS treated groups). The mice were sacrificed on Day 8, and the spleen weight, colon length, relative colonic MPO levels (d), relative mRNA levels of colonic inflammation-related genes (e), colon morphology (f) and histological scores (g) were evaluated. Statistical significance was determined using one-way ANOVA with Tukey tests for multiple-group comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
FFAR4 overexpression in the intestine exacerbates DSS-induced colitis
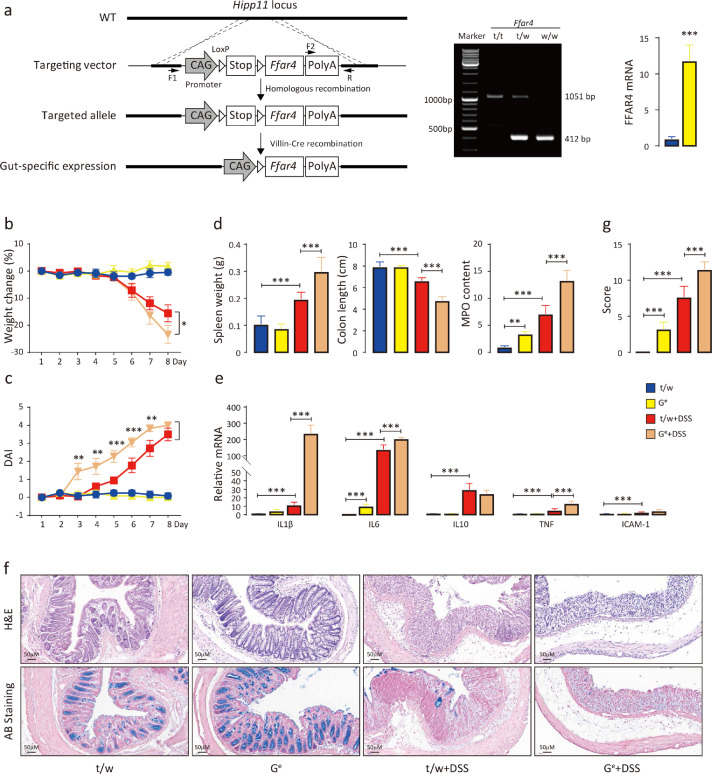
To further substantiate the role of intestinal FFAR4 in colitis development, gut-specific FFAR4 transgenic mice (Ge) were generated (Figure 4a). As expected, Ge mice had opposite phenotypic responses to DSS-induced colitis than GKO mice. Specifically, Ge mice showed greater weight loss (Figure 4b), higher DAI scores (Figure 4c), greater degrees of spleen weight gain, colon length shortening, colonic MPO activity increases (Figure 4d), larger increases in the colonic mRNA expression of the inflammatory factors Il-1β, Il-6, Tnf-α and Icam-1 (Figure 4e), more severe intestinal mucosal damage (Figure 4f), and higher histological scores than t/w controls (Figure 4g). These results further demonstrate the role of intestinal FFAR4 in DSS-induced colitis.
Figure 4.
FFAR4 overexpression in the intestine exacerbates DSS-induced colitis. Transgenic strategy for intestinal-specific FFAR4 expression (a). The floxed FFAR4 expression cassette was targeted to the Hipp11 locus by homologous recombination, generating FFAR4 transgenic mice (t/w). Intestinal-specific FFAR4-expressing mice (Ge) were obtained by mating with Villin-Cre mice, which removes the STOP fragment. The F1 and R primers amplifies a 412 bp band in wild-type mice, and the F2 and R primers generates a 1051 bp band in t/t mice by PCR. The success of gut-specific expression was confirmed by qPCR analysis of FFAR4 mRNA in the mouse colon (n=6, *** p≤0.001 by t test). Colitis was induced by DSS in t/w and Ge mice (n=8 per group), and weight change (b) and DAI scores (c) were recorded (t test was performed among DSS treated groups). The mice were sacrificed on Day 8, and the spleen weight, colon length, relative colonic MPO levels (d), relative mRNA levels of colonic inflammation-related genes (e), colon morphology (f) and histological scores (g) were evaluated. Statistical significance was determined using one-way ANOVA with Tukey tests for multiple-group comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
Transcriptomics data suggest that the loss of FFAR4 suppresses immune reactions.
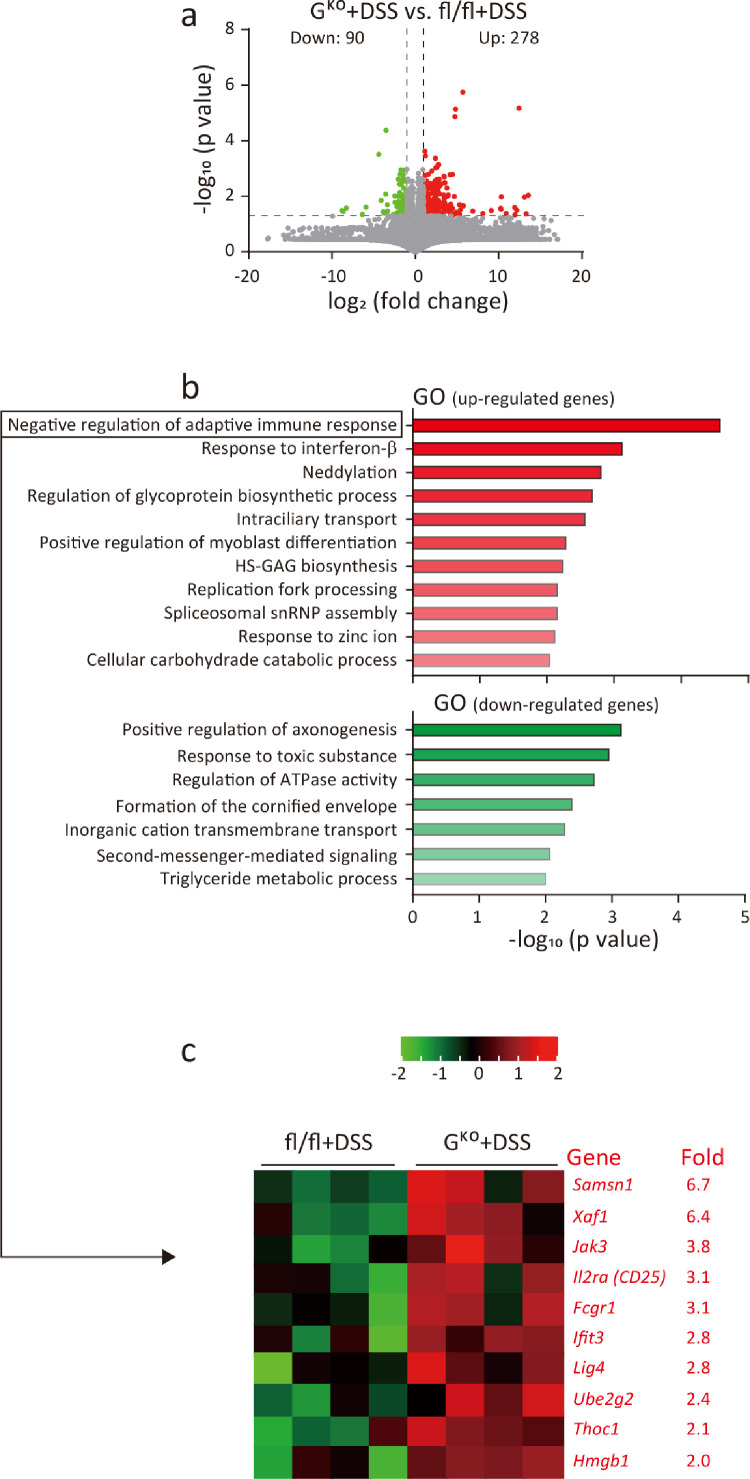
To explore the potential molecular mechanism(s) mediating the protective effect of FFAR4 gene deletion in colitis, transcriptomics analysis was performed. In DSS-induced colitis animals, 278 genes were upregulated and 90 genes were downregulated (P<0.05; difference in RPKM>2) in the colons of GKO mice compared to fl/fl mice (Figure 5a). GO analysis showed enrichment in upregulated genes in categories related to immune regulation, such as negative regulation of the adaptive immune response and the response to interferon-β (Figure 5b, c). In untreated animals, GO enrichment analysis revealed the suppression of inflammation (Figure S2). Thus, it is probable that FFAR4 can regulate immunity and that FFAR4 gene deletion in the intestine suppresses colon inflammation during DSS-induced colitis.
Figure 5.
Transcriptomics data suggest that the loss of FFAR4 suppresses immune reactions. A transcriptomics approach was used to identify differentially expressed genes in the colons of fl/fl and GKO mice with DSS-induced colitis. A volcano plot shows the distribution of upregulated (red) and downregulated (green) genes (a). GO enrichment analysis of upregulated (red) and downregulated (green) genes (b). Heatmap showing genes in the most significant cluster (negative regulation of immune response-related genes) (c). It is noteworthy that the Treg cell marker Cd25 is among the clusters.
Treg cells mediate the protective effect against DSS-induced colitis in the FFAR4-deleted intestine
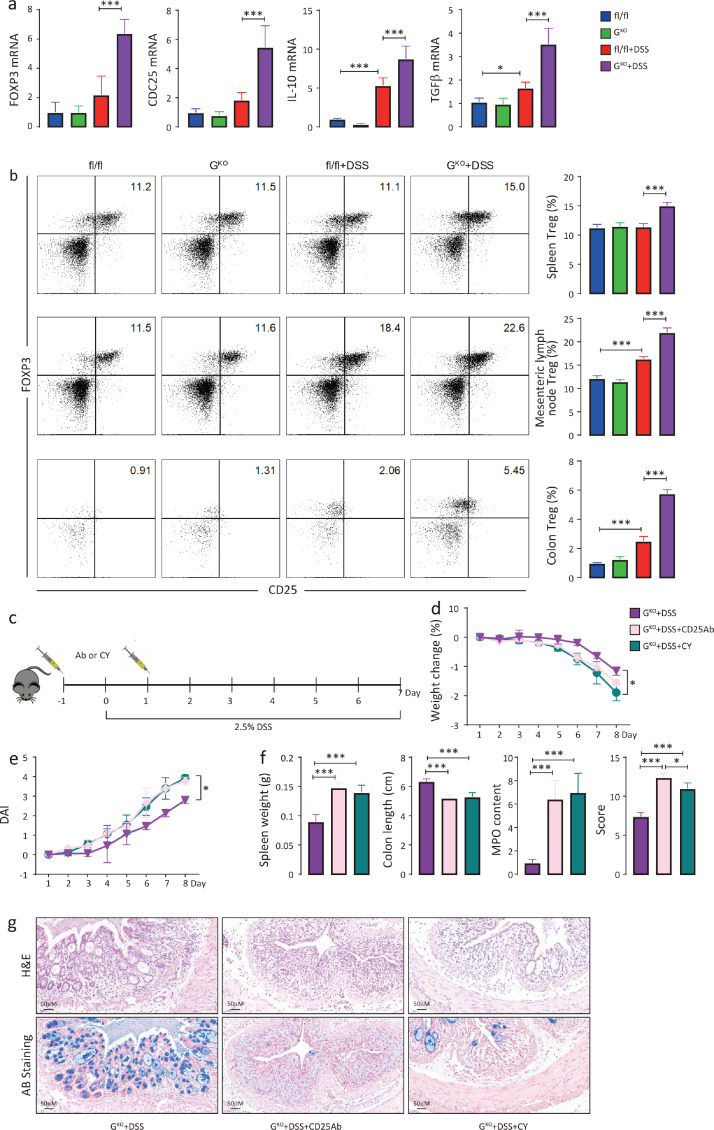
Numerous studies have shown that Treg cells are critical for maintaining intestinal tolerance and relieving intestinal inflammation.34,35 Notably, Cd25, a biomarker of Treg cells, was upregulated and clustered with other genes involved in the negative modulation of the adaptive immune response (Figure 5c). This evidence suggests that Treg cells may play a protective role in DSS-induced colitis. Indeed, the colons of DSS-induced GKO mice showed significant transcriptional upregulation of Treg cell-associated markers (Foxp3 and Cd25) as well as Treg cell effectors (Il10 and Tgfβ) compared with those in DSS-treated fl/fl mice (Figure 6a). Furthermore, the proportion of Treg cells (CD45+;CD4+;CD25+;FOXP3+) in the spleen, mesenteric lymph nodes and colonic lamina propria was markedly increased in DSS-treated GKO mice compared to the controls (Figure 6b).
Figure 6.
Treg cells mediate the protective effect against DSS-induced colitis in the FFAR4-deficient intestine. The relative mRNA levels of colonic Treg cell-related genes (n=6) were measured by qPCR (a). Treg cell (CD45+;CD4+;CD25+;FOXP3+) populations in the mouse spleen, mesenteric lymph nodes and colon with and without DSS (n=6 per group) were determined by flow cytometry (b). Treg cell depletion scheme (c). Treg cells were depleted in mice by intraperitoneal injection of anti-CD25 mAbs (Ab) (100 μl/mouse) or cyclophosphamide (Cy) (20 mg/kg) on Day -1 and Day 2, and DSS treatment was started on Day 0 and ended on Day 7. Weight changes (d), DAI scores (e) (t test was performed among DSS treated groups), spleen weight, colon length, relative colonic MPO levels, histological scores (f) and colon morphology (g) were assessed. Statistical significance was determined using one-way ANOVA with Tukey tests for multiple-group comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
Anti-CD25 monoclonal antibodies (mAbs)28 and low-dose cyclophosphamide (Cy)29,30 have been shown to selectively deplete Treg cells in vivo. Treg cells were depleted in mice by intraperitoneal injection of anti-CD25 mAbs or Cy (Figure 6c). In GKO mice, Treg cell depletion resulted in significant weight loss (Figure 6d), increased DAI scores (Figure 6e), shortened colon lengths, augmented MPO activity and increased histological scores (Figure 6f) and exacerbated mucosal damage (Figure 6g) compared with those in the nondepleted group. In contrast, Treg cell depletion had no significant impact on DSS-induced colitis in fl/fl mice (Figure S3). These results indicated that Treg cells largely mediated the protective effect against DSS-induced colitis in the FFAR4-deleted intestine.
Silencing Il33 in the intestine reduces the Treg cell population
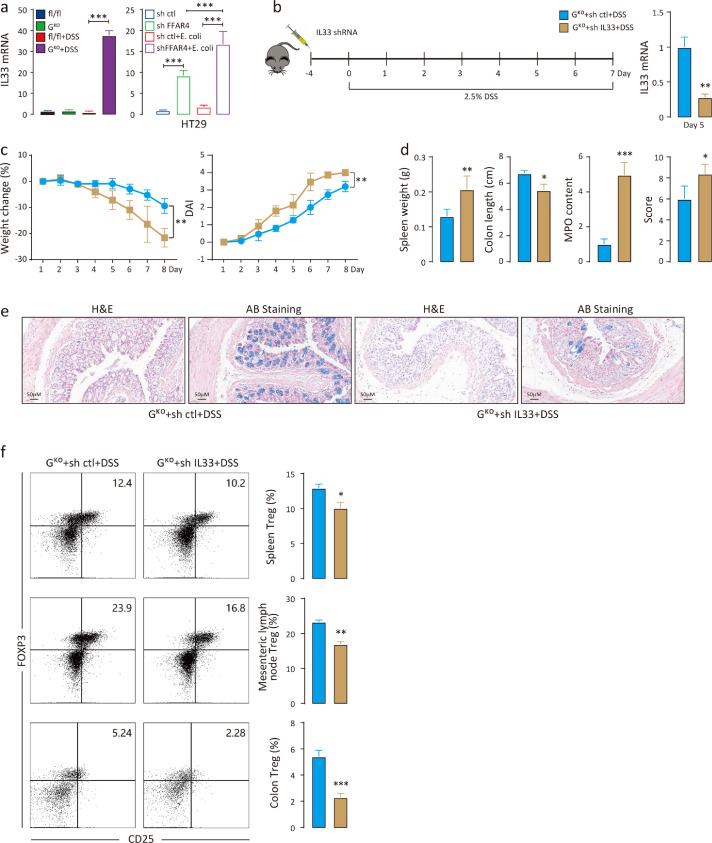
Previous studies have reported that some cytokines, such as interleukin 33 (IL33), thymic stromal lymphopoietin (TSLP) and IL25, are released from impaired intestinal epithelium and alleviate colitis by inducing Treg cells.36,37 Of the three cytokines, the Il33 transcription level was significantly upregulated in DSS-induced GKO mice but not in fl/fl mice (Figure 7a). DSS disrupts the intestinal epithelial barrier, and then commensal bacteria can translocate from the lumen to the mucosa, which may induce immune responses.38 To simulate this condition in vitro, E. coli was used as a representative commensal bacterium and was incubated with HT29 human colonic epithelial cells for 12 h. FFAR4 knockdown markedly increased IL33 transcript levels in HT29 cells, and E. coli stimulation further increased IL33 levels (Figure 7a). These results suggest that the colonic Treg response is likely regulated by FFAR4 via IL33. GKO mice were then intrarectally administered Il33 shRNA lentivirus, followed by DSS, and successful silencing of Il33 was confirmed by qPCR (Figure 7b). In comparison to controls, Il33 knockdown evidently decreased body weight, increased DAI scores (Figure 7c), enlarged the spleen, shortened the colon length, augmented MPO activity, increased histological scores (Figure 7d), and worsened mucosal damage (Figure 7e) in DSS-induced GKO mice. Additionally, Il33 knockdown significantly reduced the number of Treg cells in the spleen, mesenteric lymph nodes and colonic lamina propria (Figure 7f), suggesting that silencing Il33 in the intestine reduced the Treg cell population in DSS-induced GKO mice.
Figure 7.
Silencing Il33 in the intestine downregulates Treg cell levels. Relative Il33 mRNA levels were measured by qPCR in the mouse colon (n=6 per group) and in HT29 cells (a). Colonic Il33 knockdown scheme (b). Mice were intrarectally administered Il33 shRNA lentivirus (108 TU/ml, 100 μl/mouse) on Day -4 (n=8), and DSS was administered from Day 0 to Day 7. The success of Il33 silencing in the mouse colon (n=6) was verified by qPCR. Weight changes, DAI scores (c) (t test was performed among DSS treated groups), spleen weight, colon length, relative colonic MPO levels, histological scores (d) and colon morphology (e) were evaluated. Treg cell (CD45+;CD4+;CD25+;FOXP3+) populations in the mouse spleen, mesenteric lymph nodes and colon with and without Il33 shRNA (n=6 per group) were determined by flow cytometry (f). Statistical significance was determined using one-way ANOVA with Tukey tests for multiple-group comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
FFAR4 regulates IL33 transcription via ZBED6
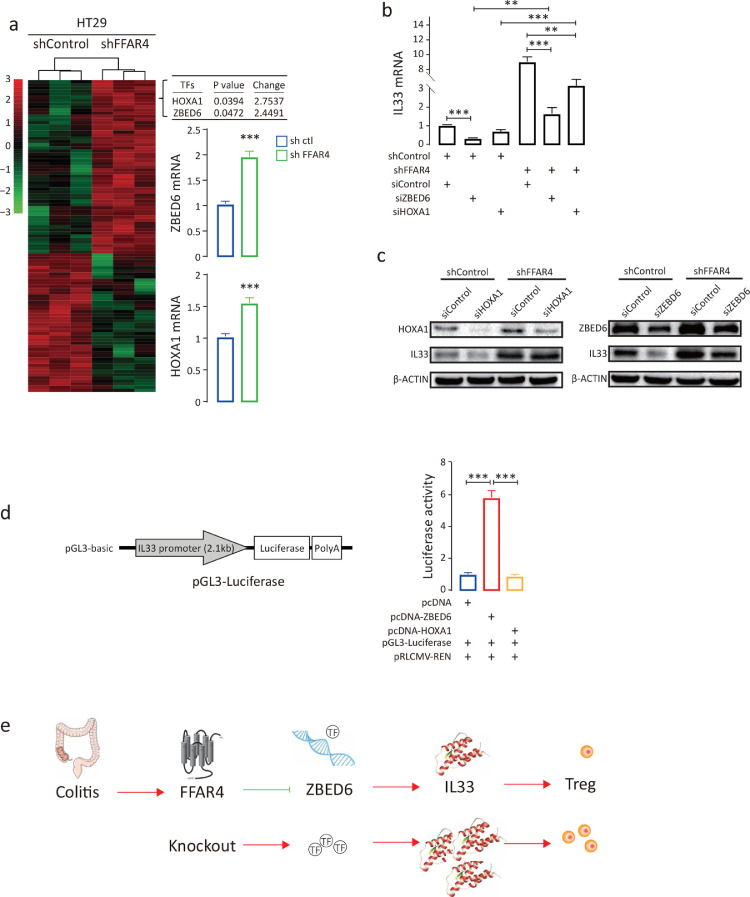
To assess how FFAR4 regulates IL33, transcriptomics analysis was performed comparing HT29 cells treated with IL33 or control shRNA. The data were examined to uncover possible transcription factors (TFs) that control IL33 mRNA transcription. Zinc finger BED-type containing 6 (ZBED6) and homeobox A1 (HOXA1) were found to be upregulated (fold change>2, p<0.05), and the results were validated by qPCR (Figure 8a). Further analysis showed that ZBED6 knockdown alone or together with FFAR4 markedly blocked IL33 transcription in HT29 cells (Figure 8b); however, knockdown of HOXA1 had little effect, and these results were confirmed at the protein level (Figure 8c). To further validate the role of ZBED6 in IL33 transcription, a luciferase reporter containing 2.1 kb of the IL33 promoter sequence was constructed. Cotransfection of the reporter and ZBED6 but not HOXA1 activated luciferase transcription and luciferase activity (Figure 8d). These data demonstrate that FFAR4 regulates IL33 transcription via ZBED6.
Figure 8.
FFAR4 regulates IL33 transcription via ZBED6. Heatmap of differentially expressed genes (fold change≥2, p value<0.05) between HT29 shControl and HT29 shFFAR4 cells, as identified by transcriptomics (a). The top two upregulated transcription factor genes (ZBED6 and HOXA1) were identified and confirmed by qPCR. ZBED6 and HOXA1 were silenced in HT29 cells with or without FFAR4 knockdown, and the relative IL33 mRNA levels were analysed by qPCR (b). Protein levels of IL33, ZBED6 and HOXA1 were measured by Western blot analysis (c). A 2.1 kbp IL33 promoter was subcloned into the pGL3-basic vector, generating pGL3-IL33 promoter-Luc. A luciferase assay was performed by cotransfecting pGL3-IL33 promoter-Luc, pRLCMV-REN and pcDNA-ZBED6 or pcDNA-HOXA1 into HEK-293T cells (n=6). The relative luciferase activity is presented after normalizing firefly luciferase to Renilla luciferase activity (d). Statistical significance was determined using one-way ANOVA with Tukey tests for multiple-group comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001. Schematic presentation of the FFAR4-ZBED6-IL33-Treg pathway (e). The arrow indicates stimulation, and ┤indicates suppression. FFAR4 deletion attenuates colitis by upregulating ZBED6 expression, leading to an increase in IL33 and subsequently resulting in an increase in the Treg population.
Discussion
Our findings indicate that intestinal FFAR4 may exert a promotive effect on the pathogenesis of IBD. However, previous studies have reported that LCFAs or synthetic ligands, such as agonists of FFAR4, suppress inflammation.19,20,39 The reasons for this seemingly contradictory result are unclear. Inconsistent conclusions may be reached due to FFAR4 polymorphisms among populations, and it was previously reported that several FFAR4 mutations exist in populations.40 Therefore, LCFAs may influence IBD risk differently in these populations. Furthermore, we previously showed that FFAR4 was not required for ω-3 PUFA-mediated cell growth inhibition and apoptosis in cancer cells.41 FFAR4 may not participate in the regulation of IBD through LCFAs and may modulate the development of IBD directly. Another possibility is the selectivity of agonists; most FFAR4 agonists can also activate FFAR1,10,42, 43, 44 which plays an important role in inflammatory regulation.8,45 In addition, numerous LCFAs can bind to FFAR4,42 which may produce different effects, as fatty acids have diverse functions. For example, eicosapentaenoic acid is clinically used for its hypolipidaemic effects, while α-linolenic acid does not have such an effect.46,47 Furthermore, LCFAs may exert their effects independent of FFAR4.41,48 Using a gene knockout approach, we demonstrated that intestinal-specific deletion of FFAR4 ameliorated colitis, whereas intestinal-specific overexpression of FFAR4 exacerbated colitis and that intestinal FFAR4 regulated Treg cells via the ZBED6/IL33 pathway.
Transcriptomics analysis suggested that FFAR4 regulates the immune response in colitis. Treg cells play an important role in maintaining intestinal immune homeostasis,49,50 and the transfer of Treg cells can heal colitis.51,52 We showed that total knockout of FFAR4 attenuated experimental colitis, that this attenuation could be recapitulated by intestinal loss of FFAR4 alone and that Treg cells mediated the protective effect. These data connect FFAR4 to Treg cells. Intestinal epithelial injury is the initiating event in colitis,53 and the injured epithelium releases cytokines to regulate the immune response.54,55 IL33 is one of these cytokines and has been reported to ameliorate experimental colitis by promoting Treg cells.36,56,57 We demonstrated that IL33 levels were increased in the GKO mouse intestine, which protected against DSS-induced colitis, and silencing Il33 diminished the Treg cell population and abolished this protection. Thus, Treg cells are activated by IL33. Finally, we found that the loss of FFAR4 increased ZBED6 levels, which triggered IL33 transcription. Therefore, we propose the following regulatory pathway: the loss of intestinal FFAR4 allows the upregulation of ZBED6 expression, resulting in the transactivation of IL33, subsequent recruitment of Treg cells and consequent protection against colitis (Figure 8d).
ZBED6 is a highly conserved transcription factor in placental mammals that affects development, cell proliferation and growth.58,59 It has been reported that ZBED6 acts as a repressor at the IGF2 locus59,60 but may act as a transcriptional activator under some circumstances.61 We showed that ZBED6 could activate IL33 transcription in intestinal epithelial HT29 cells and IL33 promoter assays. A canonical TATA box sequence (TATAAAAG) is located 1562 bp upstream of the IL33 5’ UTR, but its actual function, however, remains to be experimentally confirmed. There is no known ZBED6 binding site (GCTCG) within the 2.1 kb fragment, but a site (GCTCA) 23 bp upstream and 2 sites (GCTCT) 258 bp and 812 bp upstream of the 5’ UTR are present. Further studies are needed to identify the ZBED6 binding site on the IL33 promoter.
Approximately a dozen synthetic or natural FFAR4 agonists have been identified, yet few antagonists are available.62 Our results suggest that intestinal FFAR4 antagonism but not agonism may have therapeutic potential for IBD. It has also been reported that FFAR4 activation may decrease the expression of anti-inflammatory GLP-2 in intestinal L-cells when TNFα is present.63 AH7614, the only known FFAR4 antagonist, has off-target effects.64 Therefore, the development of FFAR4 antagonists and tissue-specific delivery methods may hold great promise for the future management of IBD.
Several studies have shown a protective role of IL33 in murine colitis models,56,57,65 which is consistent with our observations. Thus, intestinal use of IL33 for IBD treatment deserves further investigation. Intracellular IL33 can interact with the transcription factor NF-κB to dampen NF-κB-induced gene transcription.66 IL33 may promote an innate immune response associated with intestinal tissue protection.67 IL33 has also been shown to activate Treg cells through its receptor ST2.68,69 How IL33 regulates Treg cells in our colitis model is currently unknown.
ZBED6 originated from a DNA transposon that integrated into the genome of a primitive mammal several millions of years ago and evolved an essential function in the common ancestor of placental mammals.70 ZBED6 is one of the 43 probable genes derived from DNA transposons71 and received no attention until the discovery that the loss of ZBED6 repression of IGF2 transcription had pigs grow more muscle.59 Little is known about the function of ZBED6 in other species. The regulation of ZBED6 by FFAR4 is intriguing and may provide a useful model for studying fatty acid receptors and transcription factors, as well as a potential target for IBD treatment.
In conclusion, our study demonstrates that intestinal FFAR4 promotes colitis development, describes a novel mechanism by which FFAR4 regulates Treg cells via the ZBED6-IL33 pathway, and may contribute new therapeutic strategies for the management of IBD.
Contributors
SLZ and JWZ designed the experiments. JWZ, XJ and WW conducted the experiments. JWZ analysed the data. SLZ, JWZ and YQC wrote the paper. All authors read and approved the final manuscript.
Data sharing statement
Data are available upon reasonable request by sending a message to the corresponding author.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82000808), the Innovation and Application Project of Medical and Public Health Technology of Wuxi Science and Technology (N20202005), the Major Special Fund for Translational Medicine (2020ZHZD03), the Fundamental Research Funds for the Central Universities (JUSRP12048) and the Fund of Wuxi Healthcare Commission (M202004).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104060.
Appendix. Supplementary materials
References
- 1.Ng S.C., Shi H.Y., Hamidi N., et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Mak W.Y., Zhao M., Ng S.C., et al. The epidemiology of inflammatory bowel disease: east meets west. J Gastroenterol Hepatol. 2020;35(3):380–389. doi: 10.1111/jgh.14872. [DOI] [PubMed] [Google Scholar]
- 3.Jezernik G., Micetic-Turk D., Potocnik U. Molecular genetic architecture of monogenic pediatric IBD differs from complex pediatric and adult IBD. J Pers Med. 2020;10(4):243. doi: 10.3390/jpm10040243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani L., Ribaldone D.G., Bellini M., et al. Inflammatory bowel diseases: is there a role for nutritional suggestions? Nutrients. 2021;13(4):1387. doi: 10.3390/nu13041387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Bawardy B., Shivashankar R., Proctor D.D. Novel and emerging therapies for inflammatory bowel disease. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.651415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee T., Lee E., Arrollo D., et al. Non-hematopoietic beta-arrestin1 confers protection against experimental colitis. J Cell Physiol. 2016;231(5):992–1000. doi: 10.1002/jcp.25216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Murray F., Koide N., et al. Divergent requirement for Galphas and cAMP in the differentiation and inflammatory profile of distinct mouse Th subsets. J Clin Investig. 2012;122(3):963–973. doi: 10.1172/JCI59097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura I., Ichimura A., Ohue-Kitano R., et al. Free fatty acid receptors in health and disease. Physiol Rev. 2020;100(1):171–210. doi: 10.1152/physrev.00041.2018. [DOI] [PubMed] [Google Scholar]
- 9.Bartoszek A., Moo E.V., Binienda A., et al. Free fatty acid receptors as new potential therapeutic target in inflammatory bowel diseases. Pharmacol Res. 2020;152 doi: 10.1016/j.phrs.2019.104604. [DOI] [PubMed] [Google Scholar]
- 10.Briscoe C.P., Tadayyon M., Andrews J.L., et al. The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J Biol Chem. 2003;278(13):11303–11311. doi: 10.1074/jbc.M211495200. [DOI] [PubMed] [Google Scholar]
- 11.Maslowski K.M., Vieira A.T., Ng A., et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M.H., Kang S.G., Park J.H., et al. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145(2):396–406. doi: 10.1053/j.gastro.2013.04.056. e391-310. [DOI] [PubMed] [Google Scholar]
- 13.Itoh Y., Kawamata Y., Harada M., et al. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422(6928):173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- 14.Kato S., Utsumi D., Matsumoto K. G protein-coupled receptor 40 activation ameliorates dextran sulfate sodium-induced colitis in mice via the upregulation of glucagon-likepeptide-2. J Pharmacol Sci. 2019;140(2):144–152. doi: 10.1016/j.jphs.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Hara T., Kimura I., Inoue D., et al. Free fatty acid receptors and their role in regulation of energy metabolism. Rev Physiol Biochem Pharmacol. 2013;164:77–116. doi: 10.1007/112_2013_13. [DOI] [PubMed] [Google Scholar]
- 16.Calder P.C. Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Mol Nutr Food Res. 2008;52(8):885–897. doi: 10.1002/mnfr.200700289. [DOI] [PubMed] [Google Scholar]
- 17.Stenson W.F., Cort D., Rodgers J., et al. Dietary supplementation with fish oil in ulcerative colitis. Ann Intern Med. 1992;116(8):609–614. doi: 10.7326/0003-4819-116-8-609. [DOI] [PubMed] [Google Scholar]
- 18.Michalak A., Mosinska P., Fichna J. Polyunsaturated fatty acids and their derivatives: therapeutic value for inflammatory, functional gastrointestinal disorders, and colorectal cancer. Front Pharmacol. 2016;7:459. doi: 10.3389/fphar.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh D.Y., Walenta E., Akiyama T.E., et al. A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nat Med. 2014;20(8):942–947. doi: 10.1038/nm.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh D.Y., Talukdar S., Bae E.J., et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142(5):687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berquin I.M., Min Y., Wu R., et al. Modulation of prostate cancer genetic risk by omega-3 and omega-6 fatty acids. J Clin Investig. 2007;117(7):1866–1875. doi: 10.1172/JCI31494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clough J.N., Omer O.S., Tasker S., et al. Regulatory T-cell therapy in Crohn's disease: challenges and advances. Gut. 2020;69(5):942–952. doi: 10.1136/gutjnl-2019-319850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayne C.G., Williams C.B. Induced and natural regulatory T cells in the development of inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(8):1772–1788. doi: 10.1097/MIB.0b013e318281f5a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shevach E.M., Thornton A.M. tTregs, pTregs, and iTregs: similarities and differences. Immunol Rev. 2014;259(1):88–102. doi: 10.1111/imr.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chinese medical association of gastroenterology will inflammatory bowel disease study group consensus diagnosis and treatment of inflammatory bowel disease (2018, Beijing) Chin J inflamm Bowel Dis. 2018;2(3):173–190. [Google Scholar]
- 26.Wirtz S., Popp V., Kindermann M., et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12(7):1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 27.Dieleman L.A., Palmen M.J., Akol H., et al. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin Exp Immunol. 1998;114(3):385–391. doi: 10.1046/j.1365-2249.1998.00728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setiady Y.Y., Coccia J.A., Park P.U. In vivo depletion of CD4+FOXP3+ Treg cells by the PC61 anti-CD25 monoclonal antibody is mediated by FcgammaRIII+ phagocytes. Eur J Immunol. 2010;40(3):780–786. doi: 10.1002/eji.200939613. [DOI] [PubMed] [Google Scholar]
- 29.Heylmann D., Bauer M., Becker H., et al. Human CD4+CD25+ regulatory T cells are sensitive to low dose cyclophosphamide: implications for the immune response. PLoS One. 2013;8(12):e83384. doi: 10.1371/journal.pone.0083384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J., Cao Y., Lei Z., et al. Selective depletion of CD4+CD25+Foxp3+ regulatory T cells by low-dose cyclophosphamide is explained by reduced intracellular ATP levels. Cancer Res. 2010;70(12):4850–4858. doi: 10.1158/0008-5472.CAN-10-0283. [DOI] [PubMed] [Google Scholar]
- 31.Follenzi A., Naldini L. Generation of HIV-1 derived lentiviral vectors. Methods Enzymol. 2002;346:454–465. doi: 10.1016/s0076-6879(02)46071-5. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto H., Haga K., Ohno I., et al. Mucosal gene therapy using a pseudotyped lentivirus vector encoding murine interleukin-10 (mIL-10) suppresses the development and relapse of experimental murine colitis. BMC Gastroenterol. 2014;14:68. doi: 10.1186/1471-230X-14-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein C.N., Blanchard J.F., Rawsthorne P., et al. The prevalence of extraintestinal diseases in inflammatory bowel disease: a population-based study. Am J Gastroenterol. 2001;96(4):1116–1122. doi: 10.1111/j.1572-0241.2001.03756.x. [DOI] [PubMed] [Google Scholar]
- 34.Hadis U., Wahl B., Schulz O., et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34(2):237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Mowat A.M. To respond or not to respond - a personal perspective of intestinal tolerance. Nat Rev Immunol. 2018;18(6):405–415. doi: 10.1038/s41577-018-0002-x. [DOI] [PubMed] [Google Scholar]
- 36.Schiering C., Krausgruber T., Chomka A., et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513(7519):564–568. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swamy M., Jamora C., Havran W., et al. Epithelial decision makers: in search of the 'epimmunome'. Nat Immunol. 2010;11(8):656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 39.Yamada H., Umemoto T., Kakei M., et al. Eicosapentaenoic acid shows anti-inflammatory effect via GPR120 in 3T3-L1 adipocytes and attenuates adipose tissue inflammation in diet-induced obese mice. Nutr Metab. 2017;14:33. doi: 10.1186/s12986-017-0188-0. (Lond) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonnefond A., Lamri A., Leloire A., et al. Contribution of the low-frequency, loss-of-function p.R270H mutation in FFAR4 (GPR120) to increased fasting plasma glucose levels. J Med Genet. 2015;52(9):595–598. doi: 10.1136/jmedgenet-2015-103065. [DOI] [PubMed] [Google Scholar]
- 41.Zhu S., Jiang X., Jiang S., et al. GPR120 is not required for omega-3 PUFAs-induced cell growth inhibition and apoptosis in breast cancer cells. Cell Biol Int. 2018;42(2):180–186. doi: 10.1002/cbin.10883. [DOI] [PubMed] [Google Scholar]
- 42.Christiansen E., Watterson K.R., Stocker C.J., et al. Activity of dietary fatty acids on FFA1 and FFA4 and characterisation of pinolenic acid as a dual FFA1/FFA4 agonist with potential effect against metabolic diseases. Br J Nutr. 2015;113(11):1677–1688. doi: 10.1017/S000711451500118X. [DOI] [PubMed] [Google Scholar]
- 43.Briscoe C.P., Peat A.J., McKeown S.C., et al. Pharmacological regulation of insulin secretion in MIN6 cells through the fatty acid receptor GPR40: identification of agonist and antagonist small molecules. Br J Pharmacol. 2006;148(5):619–628. doi: 10.1038/sj.bjp.0706770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimpukade B., Hudson B.D., Hovgaard C.K., et al. Discovery of a potent and selective GPR120 agonist. J Med Chem. 2012;55(9):4511–4515. doi: 10.1021/jm300215x. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez-Curto E., Milligan G. Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochem Pharmacol. 2016;114:3–13. doi: 10.1016/j.bcp.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 46.Backes J., Anzalone D., Hilleman D., et al. The clinical relevance of omega-3 fatty acids in the management of hypertriglyceridemia. Lipids Health Dis. 2016;15(1):118. doi: 10.1186/s12944-016-0286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Q., Zhang Z., Wang P., et al. EPA+DHA, but not ALA, improved lipids and inflammation status in hypercholesterolemic adults: a randomized, double-blind, placebo-controlled trial. Mol Nutr Food Res. 2019;63(10) doi: 10.1002/mnfr.201801157. [DOI] [PubMed] [Google Scholar]
- 48.Chung H., Lee Y.S., Mayoral R., et al. Omega-3 fatty acids reduce obesity-induced tumor progression independent of GPR120 in a mouse model of postmenopausal breast cancer. Oncogene. 2015;34(27):3504–3513. doi: 10.1038/onc.2014.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown E.M., Kenny D.J., Xavier R.J. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol. 2019;37:599–624. doi: 10.1146/annurev-immunol-042718-041841. [DOI] [PubMed] [Google Scholar]
- 50.Cosovanu C., Neumann C. The many functions of Foxp3+ regulatory T cells in the intestine. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.600973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Izcue A., Hue S., Buonocore S., et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28(4):559–570. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mottet C., Uhlig H.H., Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J Immunol. 2003;170(8):3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- 53.Sommer K., Wiendl M., Muller T.M., et al. Intestinal mucosal wound healing and barrier integrity in IBD-crosstalk and trafficking of cellular players. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.643973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhen Y., Zhang H. NLRP3 inflammasome and inflammatory bowel disease. Front Immunol. 2019;10:276. doi: 10.3389/fimmu.2019.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neurath M.F. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14(5):329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 56.Grobeta P., Doser K., Falk W., et al. IL-33 attenuates development and perpetuation of chronic intestinal inflammation. Inflamm Bowel Dis. 2012;18(10):1900–1909. doi: 10.1002/ibd.22900. [DOI] [PubMed] [Google Scholar]
- 57.Duan L., Chen J., Zhang H., et al. Interleukin-33 ameliorates experimental colitis through promoting Th2/Foxp3(+) regulatory T-cell responses in mice. Mol Med. 2012;18(1):753–761. doi: 10.2119/molmed.2011.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Younis S., Schonke M., Massart J., et al. The ZBED6-IGF2 axis has a major effect on growth of skeletal muscle and internal organs in placental mammals. Proc Natl Acad Sci USA. 2018;115(9):E2048–E2057. doi: 10.1073/pnas.1719278115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Markljung E., Jiang L., Jaffe J.D., et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7(12) doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X., Jiang L., Wallerman O., et al. Transcription factor ZBED6 affects gene expression, proliferation, and cell death in pancreatic beta cells. Proc Natl Acad Sci USA. 2013;110(40):15997–16002. doi: 10.1073/pnas.1303625110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang L., Wallerman O., Younis S., et al. ZBED6 modulates the transcription of myogenic genes in mouse myoblast cells. PLoS One. 2014;9(4):e94187. doi: 10.1371/journal.pone.0094187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Son S.E., Kim N.J., Im DS. Development of free fatty acid receptor 4 (FFA4/GPR120) agonists in health science. Biomol Ther (Seoul) 2021;29(1):22–30. doi: 10.4062/biomolther.2020.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukahara T., Watanabe K., Watanabe T., et al. Tumor necrosis factor alpha decreases glucagon-like peptide-2 expression by up-regulating G-protein-coupled receptor 120 in Crohn disease. Am J Pathol. 2015;185(1):185–196. doi: 10.1016/j.ajpath.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Watterson K.R., Hansen S.V.F., Hudson B.D., et al. Probe-dependent negative allosteric modulators of the long-chain free fatty acid receptor FFA4. Mol Pharmacol. 2017;91(6):630–641. doi: 10.1124/mol.116.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malik A., Sharma D., Zhu Q., et al. IL-33 regulates the IgA-microbiota axis to restrain IL-1alpha-dependent colitis and tumorigenesis. J Clin Investig. 2016;126(12):4469–4481. doi: 10.1172/JCI88625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ali S., Mohs A., Thomas M., et al. The dual function cytokine IL-33 interacts with the transcription factor NF-kappaB to dampen NF-kappaB-stimulated gene transcription. J Immunol. 2011;187(4):1609–1616. doi: 10.4049/jimmunol.1003080. [DOI] [PubMed] [Google Scholar]
- 67.Monticelli L.A., Osborne L.C., Noti M., et al. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci USA. 2015;112(34):10762–10767. doi: 10.1073/pnas.1509070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawai K., Uchiyama M., Hester J., et al. IL-33 drives the production of mouse regulatory T cells with enhanced in vivo suppressive activity in skin transplantation. Am J Transpl. 2021;21(3):978–992. doi: 10.1111/ajt.16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan W., Zhang B., Liu X., et al. Interleukin-33-dependent accumulation of regulatory T cells mediates pulmonary epithelial regeneration during acute respiratory distress syndrome. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.653803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersson L., Andersson G., Hjalm G., et al. ZBED6: the birth of a new transcription factor in the common ancestor of placental mammals. Transcription. 2010;1(3):144–148. doi: 10.4161/trns.1.3.13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lander E.S., Linton L.M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.