Abstract
Objective
Despite significant progresses in imaging and pathological evaluation, early differentiation between benign and malignant biliary strictures remains challenging. Endoscopic retrograde cholangiopancreatography (ERCP) is used to investigate biliary strictures, enabling the collection of bile. We tested the diagnostic potential of next-generation sequencing (NGS) mutational analysis of bile cell-free DNA (cfDNA).
Design
A prospective cohort of patients with suspicious biliary strictures (n=68) was studied. The performance of initial pathological diagnosis was compared with that of the mutational analysis of bile cfDNA collected at the time of first ERCP using an NGS panel open to clinical laboratory implementation, the Oncomine Pan-Cancer Cell-Free assay.
Results
An initial pathological diagnosis classified these strictures as of benign (n=26), indeterminate (n=9) or malignant (n=33) origin. Sensitivity and specificity of this diagnosis were 60% and 100%, respectively, as on follow-up 14 of the 26 and eight of the nine initially benign or indeterminate strictures resulted malignant. Sensitivity and specificity for malignancy of our NGS assay, herein named Bilemut, were 96.4% and 69.2%, respectively. Importantly, one of the four Bilemut false positives developed pancreatic cancer after extended follow-up. Remarkably, the sensitivity for malignancy of Bilemut was 100% in patients with an initial diagnosis of benign or indeterminate strictures. Analysis of 30 paired bile and tissue samples also demonstrated the superior performance of Bilemut.
Conclusion
Implementation of Bilemut at the initial diagnostic stage for biliary strictures can significantly improve detection of malignancy, reduce delays in the clinical management of patients and assist in selecting patients for targeted therapies.
Keywords: biliary strictures, diagnostic and therapeutic endoscopy, cholangiocarcinoma, pancreatic tumours, mutation screening
Significance of this study.
What is already known on this subject?
In spite of significant advances in imaging, endoscopic and pathological evaluation, distinguishing between benign and malignant biliary strictures remains a diagnostic challenge. This situation dramatically affects the identification, management and prognosis of patients with biliopancreatic tumours.
Endoscopic retrograde cholangiopancreatography (ERCP) plays a key role in the evaluation of biliary strictures and enables the collection of bile samples.
The genetic landscape of biliopancreatic tumours has been defined in recent years.
What are the new findings?
We have selected a next-generation sequencing (NGS) panel open to clinical laboratory implementation and developed a mutational analysis of bile cell-free DNA (cfDNA) collected during ERCP, the Bilemut assay.
Our results confirm the better performance of liquid biopsy strategies increasing the diagnosis sensibility and the number of mutations detected in bile compared with the corresponding paired tumours.
We have tested the Bilemut assay in a prospectively collected cohort of bile cfDNA samples from patients undergoing first diagnostic ERCP. Our test alone was markedly superior to the initial pathological diagnosis, particularly for cases of strictures initially diagnosed as of benign or indeterminate origin.
Significance of this study.
How might it impact on clinical practice in the foreseeable future?
Our findings highlight the remarkable diagnostic potential of bile cfDNA NGS-based analysis for patients undergoing ERCP. Implementation of Bilemut at the initial diagnostic stage for biliary strictures can significantly improve malignancy detection, reduce delays in clinical management of patients and assist in selecting patients for targeted therapies.
Introduction
The accurate aetiological diagnosis of biliary stenoses remains a clinical challenge. Strictures of the bile duct may have diverse origin,1 and the discrimination between benign and malignant stenoses in their early stages has not been satisfactorily resolved yet.2 Benign conditions include primary sclerosing cholangitis (PSC), chronic pancreatitis, choledocolithiasis and bile duct injury and infections, while malignant stenoses are mostly attributable to neoplasia arising from the biliary tree such as cholangiocarcinoma (CCA) or from the pancreas, like pancreatic ductal adenocarcinoma (PDAC).2–5 CCAs and PDACs are very aggressive neoplasms, and therefore, their early diagnosis is essential for the application of potentially curative surgical procedures and/or pharmacological therapies.5–7 Biliary cancers are technically difficult to biopsy,7 and several multidisciplinary diagnostic tools are used to discriminate benign from malignant biliary strictures.4 8 These include a range of non-invasive imaging techniques plus endoscopic retrograde cholangiopancreatography (ERCP). ERCP allows relief of biliary obstruction in patients with stenosis while providing high-resolution fluoroscopic images and tissue sampling by biliary brushings and endoluminal biopsies.4 However, the sensitivity for detecting malignancy with ERCP, even when combined with brush cytology and fluorescent in situ hybridisation, plus the analysis of the circulating tumour biomarker carbohydrate antigen 19–9 (CA19-9), is still suboptimal, ranging from 14% to 60%.4 9–11 Patients may often undergo repeated ERCP procedures, and critical therapeutic decisions can be delayed. Alternatively, a false diagnosis of malignant stricture may result in an unnecessary extensive surgery.12 Therefore, the identification of robust markers that can allow early and reliable discrimination between benign and malignant biliary stenoses is very much needed.
Advances in the molecular characterisation of biliary tumour tissues have revealed their mutational landscape. In CCA, as well as in PDAC, next-generation sequencing (NGS) technologies have identified recurrent alterations in a relatively small number of oncogenes and tumour suppressor genes, including TP53, KRAS, CDKN2A, SMAD4, PIK3CA, GNAS, ERBB2 and FGFRs, with some of these being amenable to pharmacological targeting.7 13–20 A recent study demonstrated that a customised NGS analysis of ERCP-obtained bile duct biopsies improved the sensitivity of pathological evaluation in the detection of malignant strictures.10
Liquid biopsy strategies are actively pursued in all fields of oncology,21 22 and the detection of mutations in blood cell-free DNA (cfDNA) holds promise for the diagnosis of patients with pancreatic and biliary carcinomas.23 24 Of note, the ERCP procedure enables the collection of biliary fluid. The cfDNA isolated from bile may include DNA molecules originating from premalignant or malignant cells anywhere in the bile duct system. Therefore, cfDNA analysis may avoid the limited sensitivity of intraductal tissue brushings and biopsies and capture the genetic alterations found in biliopancreatic tumours.10 25 26 Earlier studies evaluated the presence of specific KRAS and TP53 mutations in bile DNA samples from patients with benign and malignant strictures.27–30 These studies supported the potential of such analyses to improve diagnosis of malignant disease. Moreover, it was also realised that the detection of such mutations in bile samples from patients without malignancy could identify individuals with preneoplastic lesions and at risk of progressing to cancer.28 However, the conventional sequencing technologies applied in these studies lacked the sensitivity of current amplification and sequencing tools, and the analyses were restricted to a few codons within KRAS and TP53 genes, markedly reducing the overall performance of the assays.
In this prospective proof-of-concept pilot study, we evaluated if NGS mutational analysis of cfDNA in bile could represent a new type of liquid biopsy for the detection of malignancy. For a rapid translation of the expected results, we tested two commercially available NGS cancer gene panels open to clinical laboratory implementation.
Materials and methods
Patients
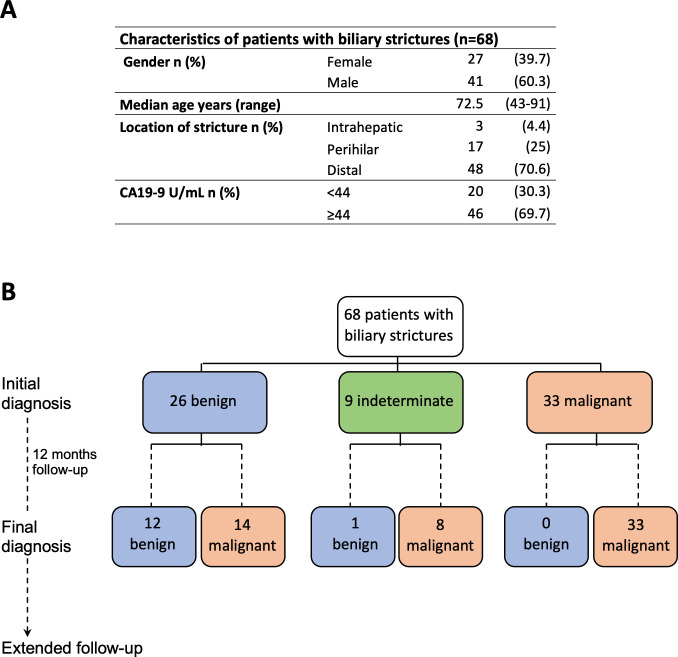
A cohort of 68 patients prescribed to undergo ERCP with a diagnosis of bile duct stenosis was prospectively accrued for the study from January 2017 to December 2020 at the Navarra University Hospital Complex, Pamplona, Spain. Their demographic and clinical characteristics are summarised in figure 1A. All patients were older than 18 years and provided written informed consent for the examination of their samples and the use of their clinical data. The initial diagnosis of the strictures at the time of bile sample collection was based on histological result from the first ERCP-bile duct brushing or biopsy, bile duct or pancreatic mass endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) or percutaneous needle biopsy. The selection of the diagnostic procedure was established according to patient and tumour characteristics and following standard clinical practice. Initial diagnosis of indeterminate stenosis was assigned when there was no sufficient sample to establish a diagnosis. The final clinical diagnosis was defined as in previous studies, based on histological evidence and clinical or radiographic follow-up.4 8 10 31–34 We designated malignant stenosis when we found histological evidence of adenocarcinoma or clinical or radiographic malignancy progression after ≤12 months of follow-up or death determined to be due to a malignancy involving the bile duct. We designated benign stenosis based on the absence of clinical and radiographic progression or resolution of the bile duct stricture after ≤12 months follow-up or when surgically resected specimens confirmed benign cholangiopathy. In total, 50 ERCP-bile duct brushings, nine ERCP-bile duct biopsies, 19 EUS-FNA and three percutaneous needle biopsies were collected for pathological evaluation to define the initial diagnosis. Representative images of cytological preparations resulting in correctly diagnosed and misdiagnoses cases after follow-up are shown in online supplemental figure 1. In addition, 68 bile samples were collected. Diagnostic and clinical follow-up information for each patient is described in detail in online supplemental table 1. The mean time of follow-up was 15.5 months (range: 0–42 months). Informed consent was obtained from each patient. Patients and the public were not involved in the design, conduct, reporting or dissemination plans of this research. The study protocol was approved by the ethics committee of the Navarra University Hospital Complex, Pamplona, Spain (protocol number: 2016/91).
Figure 1.
Characteristics of the patients included in the study and their diagnosis. (A) Demographic description, location of biliary strictures and serum carbohydrate antigen 19–9 (CA19-9) levels of patients included in this study at the time of initial diagnosis. (B) Flow chart indicating the initial and final clinical diagnosis of the patients.
gutjnl-2021-325178supp003.pdf (1.5MB, pdf)
gutjnl-2021-325178supp005.pdf (50.1KB, pdf)
Additional details are provided as online supplemental material 1.
gutjnl-2021-325178supp002.pdf (57.9KB, pdf)
Results
Diagnostic performance of conventional clinicopathological evaluation of patients with bile duct stenoses
A cohort of 68 patients undergoing ERCP for a bile duct stricture was prospectively accrued (figure 1A). At the time of the initial ERCP, EUS and pathological examination, patients were diagnosed and classified as having stenoses of benign (n=26), indeterminate (n=9) or malignant origin (n=33) (figure 1B). After a follow-up time of 12 months, the final clinical diagnosis resulted in 14 out of the 26 cases initially classified as benign resulting malignant and eight out of the nine indeterminate cases also being malignant, while the malignancy of the remaining 33 cases was confirmed (figure 1B and online supplemental table 1). Our initial pathological diagnosis was in agreement with the high specificity for detection of malignancy, 100% in our case, reported in current clinical practice.10 Also in line with current standards, the sensitivity for malignancy in our initial clinical diagnosis was 60%.1 4 8 10 The evaluation of CA19-9 serum levels at a threshold of >44 U/mL10 yielded a sensitivity of 74% and specificity of 61% for detecting malignancy. These findings confirm the poor overall performance of the currently implemented clinicopathological evaluation of patients with biliary strictures in terms of sensitivity yield and further emphasise the need to develop complementary effective strategies.
Mutational analysis of bile cfDNA: methodological set-up and selection of best-performing NGS panel
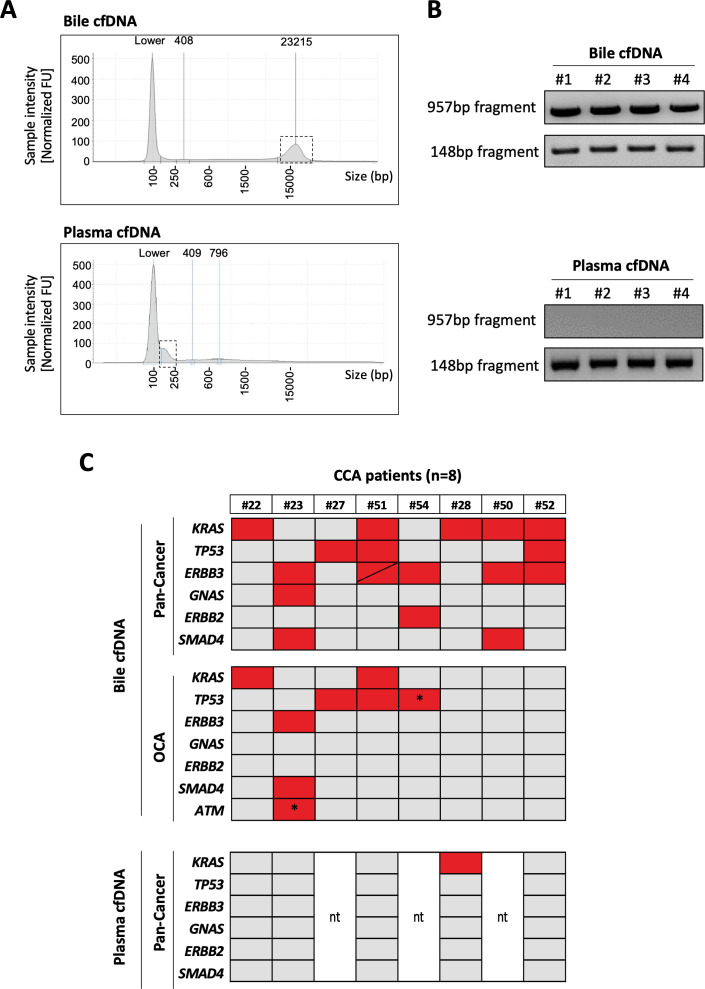
As introduced above, we hypothesised that the evaluation of the mutational landscape of bile cfDNA might improve the limited sensitivity of the overall diagnostic procedure, including the pathological analysis of intraductal tissue brushing and biopsies, therefore better detecting the eventual presence of tumours. We extracted and purified cfDNA from 1 mL of bile aliquots, and for comparison, we also extracted cfDNA from the same volume of plasma. Remarkably, we found that the total amount of cfDNA in bile was about 20-fold of that obtained from the same volume of plasma (886.10±182.3 ng/mL vs 40.52±7.46 ng/mL, respectively, n=20). Most interestingly and consistent with a recent report that analysed bile cfDNA obtained by percutaneous transhepatic cholangial drainage,35 we also observed that in contrast with cfDNA obtained from plasma, bile cfDNA was enriched in much larger DNA fragments. This can be appreciated in the electropherogram shown in figure 2A. Here, we provide direct confirmation of this difference by showing the PCR amplification of a large DNA fragment from a test gene (TP53) only in bile cfDNA, while a smaller DNA fragment from the same gene was amplified in cfDNA from both biofluids (figure 2B).
Figure 2.
Analysis of bile and plasma cell-free DNA (cfDNA) and comparison of Pan-Cancer and Oncomine Comprehensive Assay (OCA) panels’ performance in bile cfDNA from patients with cholangiocarcinoma (CCA). (A). Representative electropherograms showing the size distribution of cfDNA obtained from human bile and plasma. (B) Representative images of agarose gels showing PCR-amplified fragments of TP53 gene of high and low molecular weight from four independent samples of bile or plasma cfDNA. (C) Heat maps show the mutated genes detected by both panels in the same bile samples, and the mutations detected by the Pan-Cancer panel in plasma cfDNA. Asterisks indicate specific mutations that are not included in the Pan-Cancer panel. ‘nt’: not tested. BP, base pairs; FU, fluorescence units.
Among the commercially available NGS panels open to clinical laboratory implementation, the Oncomine Comprehensive Assay panel V.3 (OCA, Thermo Fisher) presents an extensive mutational coverage and works well with the relatively larger DNA fragments obtained from formalin-fixed paraffin-embedded (FFPE) tissues.36 On the other hand, the Oncomine Pan-Cancer Cell-Free assay has been developed for the evaluation of the small and low abundant circulating cfDNA obtained from plasma37 and also works well in other biological samples such as cerebrospinal fluid.38 Therefore, we tested side by side the performance of the OCA and Pan-Cancer NGS panels in eight bile samples from patients with firmly established CCA diagnosis. As shown in figure 2C, the Pan-Cancer Cell-Free assay detected a total of 18 mutations in all eight cfDNA samples. However, the OCA panel, which has a threefold higher coverage for genetic alterations than the Pan-Cancer assay, failed to detect any mutation in three patients, identifying a total of eight mutations. There were two specific mutations, in TP53 and ATM genes, marked with an asterisk in figure 2C, that were detected only with the OCA panel as they are not included in the Pan-Cancer panel design. As the Pan-Cancer panel is designed for the analysis of liquid biopsies, we also tested five plasma cfDNA samples from this set of patients using this panel. As shown in figure 2C, only one of these samples tested positive in plasma, while mutations were found in all the paired bile samples. These observations indicated that the Pan-Cancer Cell-Free assay implemented in bile cfDNA samples, named here the Bilemut assay, has increased sensitivity for the detection of mutations and therefore was the optimal choice of our subsequent bile cfDNA screening.
Performance of the Bilemut assay in patients with malignant stenoses
We first assessed the performance of the Bilemut assay in the 33 bile samples collected at the initial ERCP from patients with a confirmed final clinical diagnosis of malignancy (24 CCAs and nine PDACs), from which 17 paired tumour tissue samples were also available. The Bilemut assay identified mutations in all but two bile samples (numbers 16 and 17) corresponding to patients diagnosed with PDAC (figure 3 upper panel). The OCA panel detected mutations in 12 of the 17 tumour tissues tested, with one CCA and four PDAC samples (including number 17) resulting negative (figure 3 lower panel). Mutations in KRAS and TP53 genes were the most frequent alterations identified both in bile and tissues. However, the Bilemut assay captured mutations in a much wider range of genes, including ERBB3, GNAS, FBXW7, ERBB2, IDH2, MAPK2K1 and FGFR3, across all samples compared with the OCA panel assay performed in tissues (figure 3). The sensitivity of the Bilemut assay in this set of samples was 93.9%. We also analysed plasma cfDNA from five patients diagnosed with PDAC (patient numbers 12–16). The Pan-Cancer panel did not detect mutations in any of them (data not shown). At this stage, we also tested six bile cfDNA samples collected from healthy living liver donors as negative controls for the Bilemut assay and did not find any genetic alterations (data not shown). Moreover, independent repeat analysis of two previously tested positive and negative bile cfDNA samples yielded the same results, attesting to the reproducibility of the Bilemut assay (data not shown).
Figure 3.
Mutational profile of bile cell-free DNA (cfDNA) and paired tissue DNA samples from patients with malignant stenoses. The heatmap in upper panel shows the mutations detected with the Pan-Cancer panel in bile cfDNA, and the heatmap in the lower panel shows mutations identified with the Oncomine Comprehensive Assay (OCA) panel in the available paired tissues. Asterisks indicate specific mutations that are not included in the Pan-Cancer panel. Diagonal lines indicate the detection of two different mutations in the corresponding gene. The initial diagnosis (Dx), Bilemut diagnosis and final clinical diagnosis, as well as the type of tumour, are indicated. CCA, cholangiocarcinoma; PDAC, pancreatic ductal adenocarcinoma.
Performance of the Bilemut assay for the early diagnosis of patients with non-malignant bile duct strictures
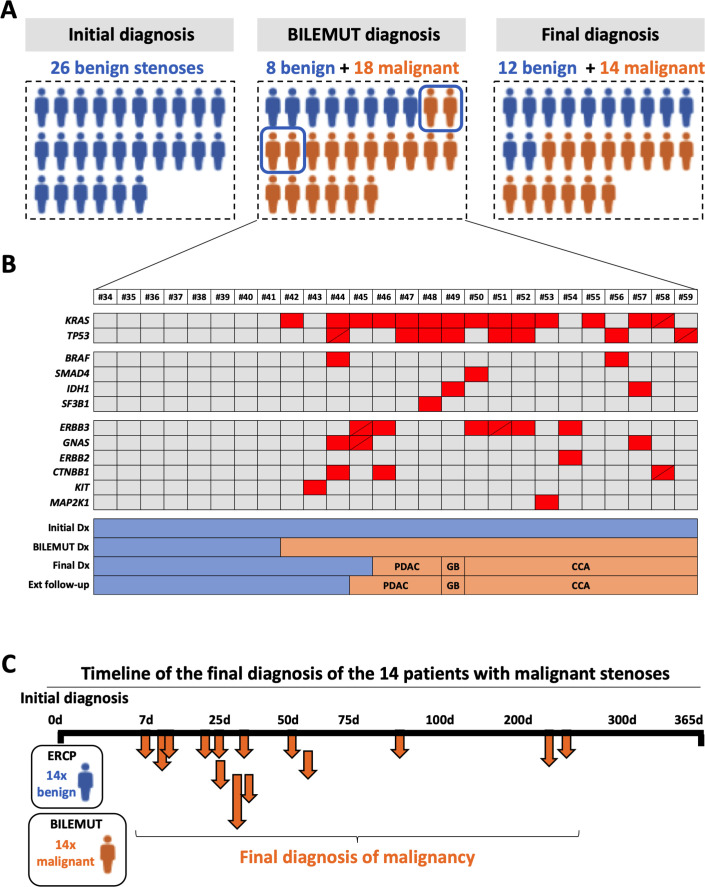
Next, we implemented the Bilemut assay in the cohort of patients that received an initial diagnosis of stenosis of benign origin (figure 1B). Bile samples were collected at the time of first ERCP; cfDNA was analysed; and according to the absence or presence of any genetic alteration, patients were classified as having benign or malignant stenoses. For patients with an initial pathological diagnosis of benign stenosis (n=26), mutations were found in 18 of them. Therefore, considering the application of Bilemut assay at the moment of the initial diagnosis, of the 26 benign stenoses, 18 would be classified as malignant and eight as benign. On the other hand, the final clinical diagnosis resulted in 14 of the 26 patients having malignant stenoses. These results are graphically summarised in figure 4A. The Bilemut mutational landscape of all these patients is shown in figure 4B, which also indicates their classification into benign and malignant stenoses according to the final clinical diagnosis. In figure 4C, we show the timeline of the final clinical diagnosis of the 14 patients that ended up developing malignant stenoses. We were also able to analyse 10 paired tumour tissue samples. The results were similar to those shown in figure 3, with the Bilemut assay detecting a higher number of mutations than the OCA assay, which was negative for one tissue sample (number 53) (online supplemental figure 2A).
Figure 4.
Diagnostic performance of the Bilemut assay in patients with an initial diagnosis of benign stenosis. (A) Schematic representation of the initial clinical diagnosis, the Bilemut assay diagnosis and the final clinical diagnosis of patients. The four Bilemut false-positive patients are encircled. (B) Heatmap showing the mutational profile of bile cell-free DNA (cfDNA), Bilemut assay, at the time of initial diagnosis. Diagonal lines indicate the detection of two different mutations in the corresponding gene. The initial diagnosis (Dx), Bilemut diagnosis, final diagnosis and extended follow-up diagnosis are indicated. The type of tumour diagnosed (pancreatic ductal adenocarcinoma (PDAC), cholangiocarcinoma (CCA) or gall bladder (GB)) is also indicated. (C). Chronology of malignancy detection during follow-up of patients with an initial diagnosis of benign stenosis. ERCP, endoscopic retrograde cholangiopancreatography.
gutjnl-2021-325178supp004.pdf (920.5KB, pdf)
There were four patients with a final clinical diagnosis of benign stenosis that had mutations in their bile cfDNA and therefore would be considered as false-positive cases for the Bilemut assay. Importantly, extended follow-up of these patients beyond 1 year revealed that one of them was diagnosed with PDAC ten months later (patient number 45). This finding underscores the value of the information provided by Bilemut assay also for its purported false-positive cases.
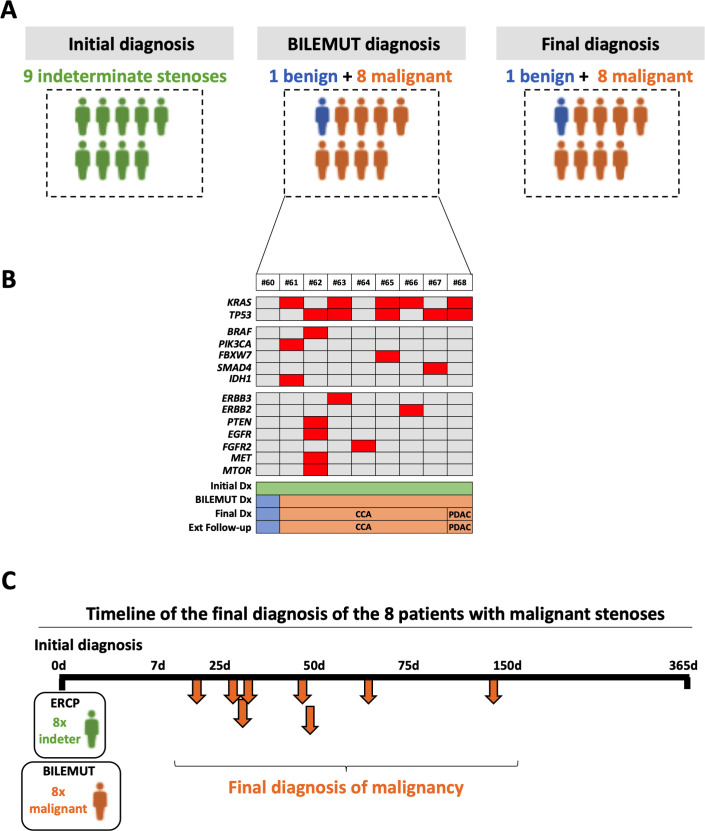
Next, we implemented the Bilemut assay in bile samples from patients with an initial pathological diagnosis of stenosis of indeterminate origin (figure 1B). Of the nine patients studied, mutations were detected in eight of them. Importantly, the final clinical diagnosis resulted in these eight patients developing malignant stenoses, as graphically summarised in figure 5A. Their Bilemut mutational landscape is shown in figure 5B. No mutations were detected in the only patient that received a final diagnosis of benign stenosis (patient number 60). This patient remains tumour-free 22 months after the initial diagnosis and Bilemut assay. We were also able to analyse three paired tumour tissue samples. As observed before, the Bilemut assay identified a greater number of mutations than the OCA panel, which was negative for one tissue sample (number 67) (online supplemental figure 2B). The timeline of the final diagnosis of the eight patients that ended up having malignant stenoses is shown in figure 5C.
Figure 5.
Diagnostic performance of the Bilemut assay in patients with an initial diagnosis of indeterminate stenosis. (A) Schematic representation of the initial clinical diagnosis, the Bilemut assay diagnosis and the final clinical diagnosis of patients. (B) Heatmap showing the mutational profile of bile cell-free DNA, Bilemut assay, at the time of initial diagnosis. The initial diagnosis (Dx), Bilemut diagnosis, final diagnosis and extended follow-up diagnosis are indicated. The type of tumour diagnosed (pancreatic ductal adenocarcinoma (PDAC) or cholangiocarcinoma (CCA)) is also indicated. (C) Chronology of malignancy detection during follow-up of patients with an initial diagnosis of indeterminate stenosis. ERCP, endoscopic retrograde cholangiopancreatography.
In the whole cohort of patients, the Bilemut assay had a sensitivity of 96.4% and a specificity of 69.2% for detecting malignancy. When considering patients with an initial diagnosis of stenosis of benign or indeterminate origin (n=35), the sensitivity of this assay for malignancy detection was 100%. In this same group of patients and considering the extended follow-up, elevated CA19-9 serum levels at a threshold of >44 U/mL10 at the time of initial diagnosis yielded a sensitivity of 74% and a specificity of 50% for detection of malignancy, while Bilemut had a sensitivity and specificity of 100% and 80%, respectively.
Pathological molecular findings: mutational analyses
The most prevalent mutations identified with the Bilemut assay among positive bile samples were in the KRAS (71.9%), TP53 (47.4%), ERBB3 (22.8%), GNAS (15.8%), ERBB2 (8.8%), BRAF (8.8%), PIK3CA (8.8%), FBXW7 (7.0%) and SMAD4 (7.0%) genes. The complete information of the different genes found in bile cfDNA is shown in online supplemental table 2. These data are in general agreement with the mutational landscape described for biliary and pancreatic tumours, considering the high proportion of CCAs of extrahepatic origin included in our cohort of patients.13 23 39 When the 30 paired bile and tumour tissue samples were compared, a total of 66 mutations were detected by the Bilemut assay, while the OCA analysis of tissues only identified 43 alterations. There were seven tissue samples (23%) in which no mutations were found but that were positive in the Bilemut assay, while this test only missed one case that had mutations in its paired tissue. There were 32 mutations detected both in tissue and bile samples, while the Bilemut assay detected 34 additional mutations not found in the corresponding tissues. For KRAS, the most frequently mutated gene in our cohort of patients, 18 mutations were found in tissue samples, and all but two were also captured in bile cfDNA. However, bile cfDNA analysis identified five additional KRAS mutations that were not detected in paired tissues. For TP53, 13 mutations were found in tissue samples, two of them not analysed by the Bilemut assay and another not detected. However, four additional TP53 mutations were found in bile cfDNA that were not detected in paired tissues. The concordance between the mutations detected in bile and tissue samples is described in online supplemental figure 2C. The identity of all mutations identified in bile cfDNA and tissue samples is provided in online supplemental table 3, and detailed information of the genes analysed by the Pan-Cancer and OCA panels is provided in online supplemental table 4.
gutjnl-2021-325178supp006.pdf (46.6KB, pdf)
gutjnl-2021-325178supp007.pdf (671.5KB, pdf)
gutjnl-2021-325178supp008.pdf (1MB, pdf)
The Pan-Cancer panel includes potentially actionable genetic alterations that are found in biliary and pancreatic malignancies, such as those involving the FGFR genes, ERBB2 and ERBB3, BRAF, IDH1 and IDH2, PIK3CA, MET, RET and MAP2K1 7 20 40. Our study shows that mutations in potentially actionable genes were detected in 54% of samples that were positive for the Bilemut assay (online supplemental table 3).
Discussion
In spite of recent and significant advancements in imaging, endoscopic and pathological approaches, the accurate aetiological diagnosis of biliary strictures is still far from satisfactory.8 41 In this study, we demonstrate that the mutational analysis of bile cfDNA obtained at the first ERCP procedure in the evaluation of patients with suspected biliary strictures can transform the diagnostic pipeline. For a quick and widespread transfer to the clinic and after evaluating the concentrations and physical characteristics of cfDNA present in bile, we selected the Pan-Cancer Cell-Free assay, an NGS panel readily available for the clinical laboratory, which does not require specialised bioinformatic data interpretation.37 38 With this assay, herein named the Bilemut assay, we first validated the high sensitivity for malignancy detection of bile cfDNA mutational analysis in a significant cohort of patients (n=33), which had a final clinical diagnosis of malignant strictures (75% of them were CCAs). Our findings are in agreement with recent reports that detected bile cfDNA mutations with high sensitivity in patients diagnosed with gall bladder cancer35 42 and PDAC43 or in two small groups of patients diagnosed with CCA (n=4 and 6).35 43 We found that our NGS assay had a remarkably high sensitivity for detecting malignancy (96.4%) in comparison with that of the initial diagnosis (60%). Most importantly, our study demonstrated that the implementation of the Bilemut assay can be of particular value for patients that receive an initial diagnosis of stenosis of benign or indeterminate origin, in which this assay demonstrated a 100% sensitivity, considerably advancing in time the diagnosis of malignancy. A recent prospective study evaluated the performance of an NGS panel in DNA obtained from bile duct brushings and biopsies collected during ERCP for the detection of malignancy in patients with biliary stenosis.10 Although this approach demonstrated an improved sensitivity over clinicopathological evaluation (73% vs 48%), the authors still reported a 25% of false-negative cases. The reason for this limited performance was attributed to inadequate sampling of strictures and/or low specimen tumour cellularity.10 This relatively high failure to detect mutations in bile duct brushings and biopsies is consistent with previous works that performed NGS analyses on tissue samples from biliary cancers (see, for instance, 44) and with our current study in which 23% of tumour tissues were negative for mutations. As we postulate here, this limitation can be circumvented by the analysis of bile cfDNA, since this fluid may contain genetic material released from tumorous cells anywhere along the biliary tract. The high sensitivity of the Bilemut assay also suggests that in patients undergoing ERCP, this diagnostic approach could be more informative than the mutational analysis of plasma cfDNA, given the known limited performance of the latter in the setting of early-stage disease,22 45 and our present observations.
One apparent shortcoming of our study was the detection of mutations in bile cfDNA from patients that received a final diagnosis of benign disease, thus lowering the specificity of the assay. However, this issue should be interpreted with caution. An earlier work that evaluated the presence of KRAS mutations in free cells present in bile from patients with PSC showed that on follow-up, only those individuals having KRAS mutations developed CCAs or dysplasias.28 These findings suggest that the appearance of mutations in bile cfDNA could indicate the existence of precancerous lesions or very early tumour stages, somewhere in the biliary tree. In fact, in our study, one of the four false-positive cases ended up developing PDAC when followed-up beyond the time established for the final clinical diagnosis. Therefore, those patients with a positive Bilemut assay and a final clinical diagnosis of benign disease may indeed benefit from closer follow-up surveillance.
Besides early cancer detection, tumour genotyping can detect the presence of mutations amenable to targeted therapies, which also occur in biliary and pancreatic malignancies.7 18–20 40 46 47 Analysis of bile cfDNA also provides such information and, as previously discussed, even with better sensitivity than tumour tissue genomic profiling.
We acknowledge that the performance of the Bilemut assay for early detection of malignancy needs to be validated in an independent cohort of patients. Ideally, future studies should include patients with PSC who are at high risk of CCA development,48 as well as patients with CCA, that were underrepresented in our cohort.
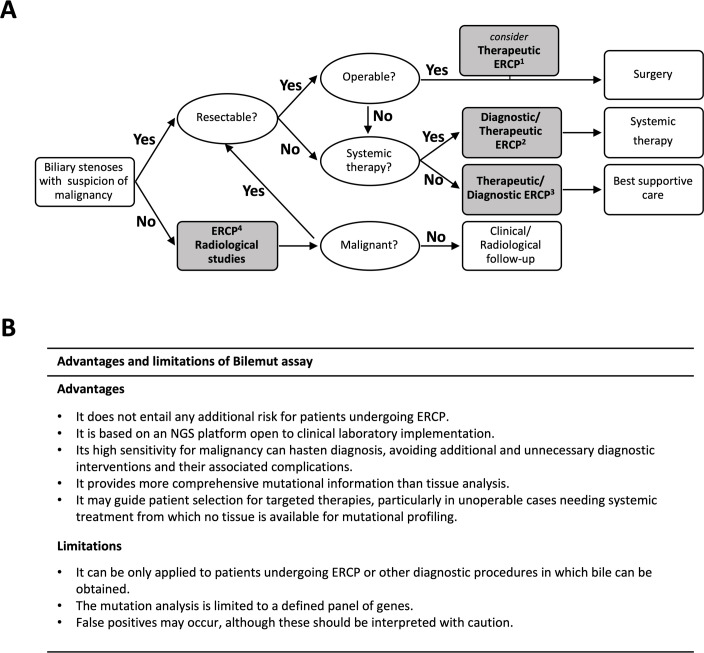
Nevertheless, in view of the extraordinary sensitivity of the Bilemut assay, we strongly believe that its implementation may leverage the diagnosis and management of patients with biliary stenosis and suspicion of malignancy. In figure 6A, we propose how Bilemut could be applied within an algorithm for the management of these patients. For those patients with suspicion of malignant stenosis, susceptible of surgical resection and who need preoperative ERCP biliary drainage (case 1), Bilemut could improve presurgical diagnosis and prevent unnecessary surgery in cases of benign aetiology.1 12 In fact, had Bilemut been applied in these patients, 100% of them would have gone to surgery with a confirmed diagnosis of malignancy, while that only occurred in 48% of such cases. For those patients not amenable to surgery that require systemic treatment (case 2), Bilemut could confirm the pathological diagnosis, avoid the need of additional diagnostic tests and interventions and also identify mutations to guide targeted therapies. In patients that are not amenable to surgical or systemic therapy and that need biliary drainage (case 3), Bilemut could also confirm the diagnosis of malignancy and thus inform on patients’ prognosis. With the application of Bilemut, 100% of these patients would have received best supportive care with a confirmed diagnosis of malignancy, while this only occurred in 63% of these cases. Finally, in those patients with low suspicion of malignancy (case 4), Bilemut could complement the pathological diagnosis and reduce follow-up time and the number of tests performed. Moreover, as mentioned above, patients with a positive Bilemut that remain free of cancer on follow-up might still benefit from a closer clinical surveillance. Lastly, from a different perspective, although the costs of an NGS analysis such as Bilemut are still high, its application may avoid the need for repeat diagnostic procedures and other medical expenses, finally resulting in a positive benefit–cost ratio. The advantages and limitations of the Bilemut assay are summarised in figure 6B.
Figure 6.
(A) Implementation of the Bilemut assay in an algorithm for the management of patients with biliary stenosis. The four steps in which Bilemut could be applied are indicated by grey boxes. See text for details. This algorithm is based on the National Comprehensive Cancer Network guidelines. (B) Summary of the advantages and limitations of the Bilemut assay. ERCP, endoscopic retrograde cholangiopancreatography; NGS, next-generation sequencing.
gutjnl-2021-325178supp001.pdf (15.9KB, pdf)
Acknowledgments
We acknowledge the technical support of Sara Equiza, Teresa Imizcoz, Beatriz Ramirez and Maria Isabel Mora from CIMA LAB Diagnostics, University of Navarra.
Footnotes
Twitter: @danioyon, @LennyNelson15, @CUBEROlab, @Maite G Fernandez-Barrena, @pottoberasain
Contributors: Conceptualisation: MA, JMU, CB and MAA. Methodology: MA, JMU, IA, DO, LZ, MRM, JMH, MR, ME, DR-C, CS, IF-U, JC, VJ, GA-A, DG, CZ, IG-B, LJN, JJV, BS, IG, KZ, MK, WP, PM, MUL, MGF-B, CB and MAA. Data curation: JMH, GA-A, AP, MR, MA, CB and JMU. Writing—original draft preparation: CB and MAA. Writing—review and editing: MR, JMU, LJN, MA, CB and MAA. Supervision: JMU, CB and MAA. Project administration: JMU, CB and MAA. Funding acquisition: MA, JMU, CB and MAA.
Funding: Funding: We thank the financial support of CIBERehd; grants PI16/01126 and PI19/00163 from Instituto de Salud Carlos III (ISCIII) cofinanced by 'Fondo Europeo de Desarrollo Regional' (FEDER) 'Una manera de hacer Europa'; grants 58/2017 and 55/2018 from Gobierno de Navarra Salud; grant 0011-1411-2020-000010 from AGATA Strategic Project from Gobierno de Navarra; grant 2020/101 from Euroregion Nouvelle Aquitaine-Euskadi-Navarra; Fundación Eugenio Rodríguez Pascual; Fundación Mario Losantos, Fundación M Torres; grant 2018/117 from AMMF, the Cholangiocarcinoma Charity; the COST Action CA181122 Euro-cholangio-Net; POSTD18014AREC postdoctoral fellowship from AECC to MA; and Ramón y Cajal Program contracts RYC-2014-15242 and RYC-2018-024475-1 to FJC and MGFB. The generous support of Mr Eduardo Avila is acknowledged.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data sharing not applicable as no datasets are generated and/or analysed for this study. Our data are not in a repository. Deidentified participant data are all included in the submission.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Singh A, Gelrud A, Agarwal B. Biliary strictures: diagnostic considerations and approach. Gastroenterol Rep 2015;3:22–31. 10.1093/gastro/gou072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nguyen Canh H, Harada K. Adult bile duct strictures: differentiating benign biliary stenosis from cholangiocarcinoma. Med Mol Morphol 2016;49:189–202. 10.1007/s00795-016-0143-6 [DOI] [PubMed] [Google Scholar]
- 3. Shanbhogue AKP, Tirumani SH, Prasad SR, et al. Benign biliary strictures: a current comprehensive clinical and imaging review. AJR Am J Roentgenol 2011;197:W295–306. 10.2214/AJR.10.6002 [DOI] [PubMed] [Google Scholar]
- 4. Pereira SP, Goodchild G, Webster GJM. The endoscopist and malignant and non-malignant biliary obstruction. Biochim Biophys Acta Mol Basis Dis 2018;1864:1478–83. 10.1016/j.bbadis.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. Lancet 2020;395:2008–20. 10.1016/S0140-6736(20)30974-0 [DOI] [PubMed] [Google Scholar]
- 6. Banales JM, Marin JJG, Lamarca A, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol 2020;17:557–88. 10.1038/s41575-020-0310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Valle JW, Kelley RK, Nervi B, et al. Biliary tract cancer. Lancet 2021;397:428–44. 10.1016/S0140-6736(21)00153-7 [DOI] [PubMed] [Google Scholar]
- 8. Dumonceau J-M, Delhaye M, Charette N, et al. Challenging biliary strictures: pathophysiological features, differential diagnosis, diagnostic algorithms, and new clinically relevant biomarkers - part 1. Therap Adv Gastroenterol 2020;13:1756284820927292. 10.1177/1756284820927292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macias RIR, Kornek M, Rodrigues PM, et al. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int 2019;39 Suppl 1:108–22. 10.1111/liv.14090 [DOI] [PubMed] [Google Scholar]
- 10. Singhi AD, Nikiforova MN, Chennat J, et al. Integrating next-generation sequencing to endoscopic retrograde cholangiopancreatography (ERCP)-obtained biliary specimens improves the detection and management of patients with malignant bile duct strictures. Gut 2020;69:52–61. 10.1136/gutjnl-2018-317817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Voigtländer T, Lankisch TO. Endoscopic diagnosis of cholangiocarcinoma: from endoscopic retrograde cholangiography to bile proteomics. Best Pract Res Clin Gastroenterol 2015;29:267–75. 10.1016/j.bpg.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 12. Clayton RAE, Clarke DL, Currie EJ, et al. Incidence of benign pathology in patients undergoing hepatic resection for suspected malignancy. Surgeon 2003;1:32–8. 10.1016/s1479-666x(03)80006-9 [DOI] [PubMed] [Google Scholar]
- 13. Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. 10.1371/journal.pone.0115383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003–10. 10.1038/ng.3375 [DOI] [PubMed] [Google Scholar]
- 15. Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 2017;7:1116–35. 10.1158/2159-8290.CD-17-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farshidfar F, Zheng S, Gingras M-C, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-Mutant molecular profiles. Cell Rep 2017;19:2878–80. 10.1016/j.celrep.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- 18. Lowery MA, Jordan EJ, Basturk O, et al. Real-Time genomic profiling of pancreatic ductal adenocarcinoma: potential actionability and correlation with clinical phenotype. Clin Cancer Res 2017;23:6094–100. 10.1158/1078-0432.CCR-17-0899 [DOI] [PubMed] [Google Scholar]
- 19. Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 2018;24:4154–61. 10.1158/1078-0432.CCR-18-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singhi AD, George B, Greenbowe JR, et al. Real-Time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology 2019;156:2242–53. 10.1053/j.gastro.2019.02.037 [DOI] [PubMed] [Google Scholar]
- 21. Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut 2018;67:2204–12. 10.1136/gutjnl-2017-315846 [DOI] [PubMed] [Google Scholar]
- 22. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol 2021;18:297–312. 10.1038/s41571-020-00457-x [DOI] [PubMed] [Google Scholar]
- 23. Zill OA, Greene C, Sebisanovic D, et al. Cell-Free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov 2015;5:1040–8. 10.1158/2159-8290.CD-15-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ettrich TJ, Schwerdel D, Dolnik A, et al. Genotyping of circulating tumor DNA in cholangiocarcinoma reveals diagnostic and prognostic information. Sci Rep 2019;9:13261. 10.1038/s41598-019-49860-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walter D, Döring C, Feldhahn M, et al. Intratumoral heterogeneity of intrahepatic cholangiocarcinoma. Oncotarget 2017;8:14957–68. 10.18632/oncotarget.14844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bankov K, Döring C, Schneider M, et al. Sequencing of intraductal biopsies is feasible and potentially impacts clinical management of patients with indeterminate biliary stricture and cholangiocarcinoma. Clin Transl Gastroenterol 2018;9:151. 10.1038/s41424-018-0015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saurin JC, Joly-Pharaboz MO, Pernas P, et al. Detection of Ki-ras gene point mutations in bile specimens for the differential diagnosis of malignant and benign biliary strictures. Gut 2000;47:357–61. 10.1136/gut.47.3.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kubicka S, Kühnel F, Flemming P, et al. K-Ras mutations in the bile of patients with primary sclerosing cholangitis. Gut 2001;48:403–8. 10.1136/gut.48.3.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Müller P, Ostwald C, Püschel K, et al. Low frequency of p53 and Ras mutations in bile of patients with hepato-biliary disease: a prospective study in more than 100 patients. Eur J Clin Invest 2001;31:240–7. 10.1046/j.1365-2362.2001.00800.x [DOI] [PubMed] [Google Scholar]
- 30. Wang Y, Yamaguchi Y, Watanabe H, et al. Usefulness of p53 gene mutations in the supernatant of bile for diagnosis of biliary tract carcinoma: comparison with K- Ras mutation. J Gastroenterol 2002;37:831–9. 10.1007/s005350200137 [DOI] [PubMed] [Google Scholar]
- 31. Burnett AS, Calvert TJ, Chokshi RJ. Sensitivity of endoscopic retrograde cholangiopancreatography standard cytology: 10-y review of the literature. J Surg Res 2013;184:304–11. 10.1016/j.jss.2013.06.028 [DOI] [PubMed] [Google Scholar]
- 32. Yoon WJ, Brugge WR. Endoscopic evaluation of bile duct strictures. Gastrointest Endosc Clin N Am 2013;23:277–93. 10.1016/j.giec.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 33. Khan SA, Davidson BR, Goldin RD, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 2012;61:1657–69. 10.1136/gutjnl-2011-301748 [DOI] [PubMed] [Google Scholar]
- 34. Navaneethan U, Njei B, Lourdusamy V, et al. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: a systematic review and meta-analysis. Gastrointest Endosc 2015;81:168–76. 10.1016/j.gie.2014.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen N, Zhang D, Yin L, et al. Bile cell‑free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol Rep 2019;42:549–60. 10.3892/or.2019.7177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dehghani M, Rosenblatt KP, Li L, et al. Validation and clinical applications of a comprehensive next generation sequencing system for molecular characterization of solid cancer tissues. Front Mol Biosci 2019;6:82. 10.3389/fmolb.2019.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macías M, Cañada-Higueras E, Alegre E, et al. Performance comparison of two next-generation sequencing panels to detect actionable mutations in cell-free DNA in cancer patients. Clin Chem Lab Med 2020;58:1341–8. 10.1515/cclm-2019-1267 [DOI] [PubMed] [Google Scholar]
- 38. Shah M, Takayasu T, Zorofchian Moghadamtousi S, et al. Evaluation of the Oncomine pan-cancer cell-free assay for analyzing circulating tumor DNA in the cerebrospinal fluid in patients with central nervous system malignancies. J Mol Diagn 2021;23:171–80. 10.1016/j.jmoldx.2020.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee H, Wang K, Johnson A, et al. Comprehensive genomic profiling of extrahepatic cholangiocarcinoma reveals a long tail of therapeutic targets. J Clin Pathol 2016;69:403–8. 10.1136/jclinpath-2015-203394 [DOI] [PubMed] [Google Scholar]
- 40. Marin JJG, Prete MG, Lamarca A, et al. Current and novel therapeutic opportunities for systemic therapy in biliary cancer. Br J Cancer 2020;123:1047–59. 10.1038/s41416-020-0987-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bang JY, Navaneethan U, Hasan M, et al. Optimizing outcomes of Single-Operator Cholangioscopy-Guided biopsies based on a randomized trial. Clin Gastroenterol Hepatol 2020;18:441–8. 10.1016/j.cgh.2019.07.035 [DOI] [PubMed] [Google Scholar]
- 42. Kinugasa H, Nouso K, Ako S, et al. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol Ther 2018;19:934–8. 10.1080/15384047.2018.1456604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Driescher C, Fuchs K, Haeberle L. Bile-based cell-free DNA analysis is a reliable diagnostic tool in pancreatobiliary cancer. Cancers 2021;13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lamarca A, Kapacee Z, Breeze M, et al. Molecular profiling in daily clinical practice: practicalities in advanced cholangiocarcinoma and other biliary tract cancers. J Clin Med 2020;9:2854. 10.3390/jcm9092854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Javle M, Bekaii-Saab T, Jain A, et al. Biliary cancer: utility of next-generation sequencing for clinical management. Cancer 2016;122:3838–47. 10.1002/cncr.30254 [DOI] [PubMed] [Google Scholar]
- 47. Montal R, Sia D, Montironi C, et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol 2020;73:315–27. 10.1016/j.jhep.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chapman MH, Thorburn D, Hirschfield GM, et al. British Society of gastroenterology and UK-PSC guidelines for the diagnosis and management of primary sclerosing cholangitis. Gut 2019;68:1356–78. 10.1136/gutjnl-2018-317993 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-325178supp003.pdf (1.5MB, pdf)
gutjnl-2021-325178supp005.pdf (50.1KB, pdf)
gutjnl-2021-325178supp002.pdf (57.9KB, pdf)
gutjnl-2021-325178supp004.pdf (920.5KB, pdf)
gutjnl-2021-325178supp006.pdf (46.6KB, pdf)
gutjnl-2021-325178supp007.pdf (671.5KB, pdf)
gutjnl-2021-325178supp008.pdf (1MB, pdf)
gutjnl-2021-325178supp001.pdf (15.9KB, pdf)
Data Availability Statement
Data sharing not applicable as no datasets are generated and/or analysed for this study. Our data are not in a repository. Deidentified participant data are all included in the submission.