Abstract
Soil isolates of Aspergillus flavus from a transect extending from eastern New Mexico through Georgia to eastern Virginia were examined for production of aflatoxin B1 and cyclopiazonic acid in a liquid medium. Peanut fields from major peanut-growing regions (western Texas; central Texas; Georgia and Alabama; and Virginia and North Carolina) were sampled, and fields with other crops were sampled in regions where peanuts are not commonly grown. The A. flavus isolates were identified as members of either the L strain (n = 774), which produces sclerotia that are >400 μm in diameter, or the S strain (n = 309), which produces numerous small sclerotia that are <400 μm in diameter. The S-strain isolates generally produced high levels of aflatoxin B1, whereas the L-strain isolates were more variable in aflatoxin production; variation in cyclopiazonic acid production also was greater in the L strain than in the S strain. There was a positive correlation between aflatoxin B1 production and cyclopiazonic acid production in both strains, although 12% of the L-strain isolates produced only cyclopiazonic acid. Significant differences in production of aflatoxin B1 and cyclopiazonic acid by the L-strain isolates were detected among regions. In the western half of Texas and the peanut-growing region of Georgia and Alabama, 62 to 94% of the isolates produced >10 μg of aflatoxin B1 per ml. The percentages of isolates producing >10 μg of aflatoxin B1 per ml ranged from 0 to 52% in the remaining regions of the transect; other isolates were often nonaflatoxigenic. A total of 53 of the 126 L-strain isolates that did not produce aflatoxin B1 or cyclopiazonic acid were placed in 17 vegetative compatibility groups. Several of these groups contained isolates from widely separated regions of the transect.
Peanuts, corn, and cottonseed are often invaded before harvest by Aspergillus flavus Link and Aspergillus parasiticus Speare, fungi that produce carcinogenic aflatoxins. Aflatoxins are highly regulated for both animal feed and food destined for human consumption (37). Of the naturally occurring aflatoxins, aflatoxin B1 is the most toxic (10). A. flavus also may produce cyclopiazonic acid (CPA), which is toxic to a variety of animals and has been implicated in human poisoning (4, 32). CPA and aflatoxins commonly occur together in contaminated agricultural commodities (25, 36). In corn and cottonseed, aflatoxin contamination is largely attributable to A. flavus (11, 35). Although peanuts are invaded by A. parasiticus more often than other crops, A. flavus also is the dominant aflatoxigenic species in peanuts (15, 17).
The following two strains of A. flavus are recognized: the L strain, which produces sclerotia that are >400 μm in diameter; and the S strain, also described as A. flavus var. parvisclerotigenus (34), which is characterized by numerous small sclerotia that are <400 μm in diameter (6). Populations of both strains comprise numerous subpopulations called vegetative compatibility groups (VCGs) (1, 18, 31). VCG 1 of A. parasiticus appears to be widely distributed in peanut-growing regions throughout the United States (16), but little is known about the distribution of A. flavus VCGs. Because isolates within a VCG are similar in their production of aflatoxins and CPA (2, 20), isolates belonging to the same VCG can be detected among isolates that have the same mycotoxin profile.
Regional differences in aflatoxin contamination of crops may be attributable to climatic conditions and to agricultural practices that increase the susceptibility of plants to invasion by A. flavus. Drought stress accompanied by elevated temperatures during seed development promotes A. flavus invasion and subsequent aflatoxin contamination of peanuts, corn, and cottonseed (14, 23, 24). However, the toxigenicity of A. flavus isolates within a region also may influence the severity of aflatoxin contamination of crops. A. flavus populations are extremely diverse genetically, and L-strain isolates vary considerably in their capacity to produce aflatoxins and CPA, with many isolates producing only one mycotoxin or neither mycotoxin (20, 21, 28, 33). Surveys of A. flavus isolates from various geographic regions have revealed differences in the proportions of isolates that produce low, medium, and high amounts of aflatoxins (8, 29). The factors responsible for the toxigenicity profile of A. flavus populations in a region are not understood (9), but the dominance of particular crops may be important in determining the relative proportions of A. flavus genotypes. Schroeder and Boller (35) examined aflatoxin production by A. flavus isolates from peanuts, cottonseed, rice, and sorghum in Texas. These workers found differences among crops in the percentage of aflatoxin producers, as well as in the concentrations of aflatoxins produced. They reported that the percentage of peanut isolates that produced aflatoxins was high and that these isolates produced high levels of aflatoxins. Other researchers also have reported that the proportion of aflatoxin-producing isolates of A. flavus from peanuts and peanut field soils is high (20, 22, 28).
Agricultural soil serves as a reservoir for populations of A. flavus (1, 8, 18, 19, 29). Peanuts are in direct contact with soil populations, whereas above-ground crops, such as corn and cottonseed, may be infected with conidia from soil through dispersal by wind or insects (26, 27). Previously, we (16) examined soil populations of Aspergillus species belonging to section Flavi that were distributed along a transect extending from eastern New Mexico through Georgia to eastern Virginia. Peanut fields from four major peanut-growing regions in the United States (western Texas; central Texas; Georgia and Alabama; and Virginia and North Carolina) were included in the transect, as were fields with other crops in regions where peanuts are not commonly grown. A. flavus was the dominant species belonging to section Flavi along most of the transect, and both L-strain and S-strain isolates were present. There were significant differences among the peanut-growing regions in terms of the density and incidence of both strains in the soil.
Knowledge of regional differences in the toxigenicity of A. flavus populations, as well as knowledge of the association of these populations with the dominant crops in a region, may be important in determining which control measures are most effective in reducing preharvest aflatoxin contamination. For example, biological control of A. flavus in agricultural fields through application of an atoxigenic A. flavus strain to the soil (7, 9, 12) might be preferentially used in regions where the populations are most toxigenic. In this study, L- and S-strain isolates of A. flavus were obtained from the soil populations described by us previously (16) from a transect across the United States. These isolates were examined with the following objectives: (i) to determine whether A. flavus populations from different geographic regions differ in production of aflatoxin B1 and CPA; and (ii) to characterize the distribution of several A. flavus VCGs over a wide geographic area.
MATERIALS AND METHODS
Transect fields, soil collection, and A. flavus isolation.
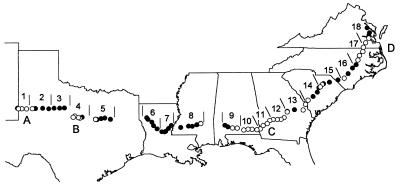
Isolates of A. flavus were obtained during a study of soil populations of Aspergillus species belonging to section Flavi along a transect through major peanut-growing regions of the United States (16). The transect extended from eastern New Mexico through Georgia to eastern Virginia and comprised 83 fields (40 peanut fields, 22 cotton fields, 15 corn fields, and 6 soybean fields) (Fig. 1). In the peanut-growing regions, which included western Texas, central Texas, Georgia and Alabama, and Virginia and North Carolina, peanut fields were preferentially sampled along the transect at 15- to 25-km intervals; in the regions where peanuts are not commonly cultivated, corn, cotton, and soybean fields were sampled at 25- to 40-km intervals. Soil samples were collected from 17 to 28 June 1996 and were processed and dilution plated onto a modified dichloran-rose bengal medium as previously described (16). All crops were immature at the time of soil collection.
FIG. 1.
Transect showing fields from which A. flavus soil isolates were obtained. The crops present in the fields included peanuts (○) (n = 40) and corn, cotton, or soybeans (●) (n = 43). The transect was divided into segments 1 to 18, each of which was 150 to 200 km long. The following major peanut-growing regions are indicated: western Texas (region A); central Texas (region B); Georgia and Alabama (region C); Virginia and North Carolina (region D).
We prepared 40 dilution plates per field, and 10 colonies of A. flavus L strain and 10 colonies of A. flavus S strain (if available) were randomly selected from different plates. Conidia were transferred to 8 ml of sterile water containing 100 μl of Tween 20 per liter. After vortexing, each spore suspension (0.1 ml) was spread onto a plate containing modified dichloran-rose bengal medium, and the plate was incubated for 25 to 30 h at 30°C. Germlings were transferred to Czapek agar slants and grown in the light to encourage sporulation.
The transect was divided in two ways in order to examine regional differences in mycotoxin production by the L strain of A. flavus (Fig. 1). First, the fields were grouped by defining 18 transect segments consisting of 150 to 200 km each. Second, the major peanut-growing regions were compared; these regions consisted of western Texas (portions of transect segments 1 and 2) (4 peanut fields, 28 isolates), central Texas (a portion of transect segment 4) (4 peanut fields, 40 isolates), Georgia and Alabama (transect segments 10 through 12) (13 peanut fields, 130 isolates), and Virginia and North Carolina (portions of transect segments 17 and 18) (6 peanut fields, 51 isolates).
VCGs.
The 126 isolates that did not produce detectable aflatoxin B1 or CPA were tested for vegetative compatibility. Five plates of Czapek agar supplemented with potassium chlorate (25 g/liter) were inoculated with a spore suspension of each isolate as described by Horn and Greene (18). One non-nitrate-utilizing sector on each plate was identified as a niaD, nirA, or cnx mutant. cnx mutants provide the strongest reactions between compatible isolates and are recommended as testers for identifying VCGs (5). Therefore, cnx mutants were paired on a nitrate medium with complementary niaD and nirA mutants of all atoxigenic isolates. Formation of a stable prototrophic heterokaryon at the zone of interaction indicated that two isolates were members of the same VCG. The number designations used for VCGs were a continuation of previously described VCGs 1 to 63 of A. flavus (18, 31).
Mycotoxin analyses.
Approximately 105 dry conidia of isolates of the A. flavus L strain (n = 774) and the A. flavus S strain (n = 309) were used to inoculate 4-ml vials containing 1 ml of liquid medium containing 150 g of sucrose, 20 g of yeast extract, 10 g of soytone, and 1 liter of distilled water; the pH of the medium was adjusted to 6.0 with HCl. One vial was inoculated per isolate; the atoxigenic isolates tested for vegetative compatibility were reexamined with an additional independent set of vial cultures. The cultures were incubated for 7 days at 30°C in the dark.
Vial cultures were analyzed by high-performance liquid chromatography for production of aflatoxin B1 and CPA as previously described (20), except that aflatoxin B1 was quantified with a Shimadzu Class VP chromatography laboratory automated software system instead of the data module. The limits of quantification were 0.5 ng of aflatoxin B1 per ml of culture medium and 2 μg of CPA per ml of culture medium.
Statistics.
To compare transect segments or major peanut-growing regions, the mycotoxin concentrations in vial cultures were analyzed by using the Kruskal-Wallis one-way analysis on ranks test (H statistic) and then Dunn’s nonparametric multiple comparison test. The analyses were based on the number of isolates in each transect segment or peanut-growing region. The correlation coefficient (r) for aflatoxin B1 production and CPA production was determined by using the Pearson product moment correlation method; the 126 L-strain isolates that did not produce detectable levels of either mycotoxin were not included in the correlation. The statistical analyses were performed by using SigmaStat, version 1.0 (Jandel Scientific, San Rafael, Calif.).
RESULTS
Isolates of the A. flavus L strain (n = 774) and the A. flavus S strain (n = 309) from field soils over a wide geographic area (Fig. 1) were examined for production of aflatoxin B1 and CPA. A large percentage of the L-strain isolates produced both mycotoxins (Table 1). However, a sizable percentage of isolates were atoxigenic (there was no detectable aflatoxin B1 or CPA) or produced only CPA; isolates that produced aflatoxin B1 but not CPA were rare (0.6%). Nearly all of the S-strain isolates produced both aflatoxin B1 and CPA. No atoxigenic S-strain isolates were observed. There was a positive correlation between production of aflatoxin B1 and production of CPA for both the L strain (r = 0.54; n = 648; P < 0.0001) and the S strain (r = 0.29; n = 309; P < 0.0001).
TABLE 1.
Production of aflatoxin B1 and CPA by L- and S-strain isolates of A. flavus from the entire transect
| Aflatoxin B1 production | CPA production | % of isolates
|
|
|---|---|---|---|
| L strain (n = 774) | S strain (n = 309) | ||
| + | + | 70.7 | 98.7 |
| NDa | ND | 16.3 | 0.0 |
| ND | + | 12.4 | 0.3 |
| + | ND | 0.6 | 1.0 |
ND, not detected.
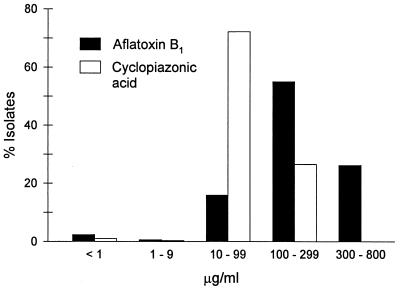
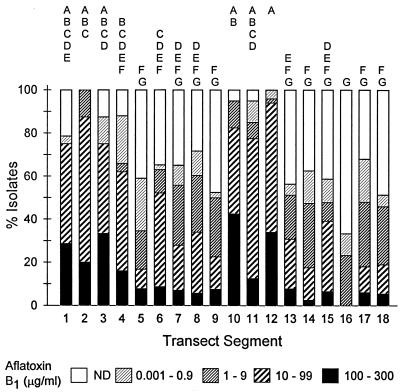
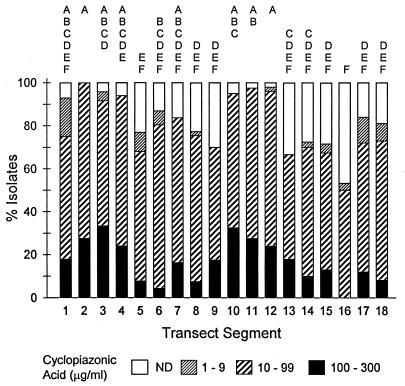
Nearly all of the S-strain isolates from the entire transect produced >10 μg of aflatoxin B1 and CPA per ml of culture medium, and 26% of the S-strain isolates produced >300 μg of aflatoxin B1 per ml (Fig. 2). In contrast, the aflatoxin B1 and CPA concentrations produced by the L-strain isolates varied considerably more, and many L-strain isolates produced <10 μg of the mycotoxins per ml (Fig. 3 and 4). No L-strain isolate produced >300 μg of aflatoxin B1 per ml.
FIG. 2.
Production of aflatoxin B1 and CPA by S-strain isolates of A. flavus (n = 309) from fields along the entire transect.
FIG. 3.
Production of aflatoxin B1 by L-strain isolates of A. flavus (n = 774) from fields in 18 transect segments, each of which was 150 to 200 km long. The transect segments are shown in Fig. 1. The numbers of fields and numbers of L-strain isolates examined were as follows: transect segment 1, four fields and 28 isolates; transect segment 2, four fields and 40 isolates; transect segment 3, three fields and 24 isolates; transect segment 4, five fields and 50 isolates; transect segment 5, eight fields and 78 isolates; transect segment 6, five fields and 46 isolates; transect segment 7, five fields and 43 isolates; transect segment 8, six fields and 53 isolates; transect segment 9, four fields and 40 isolates; transect segment 10, four fields and 40 isolates; transect segment 11, four fields and 40 isolates; transect segment 12, five fields and 50 isolates; transect segment 13, four fields and 39 isolates; transect segment 14, four fields and 40 isolates; transect segment 15, five fields and 46 isolates; transect segment 16, three fields and 30 isolates; transect segment 17, five fields and 50 isolates; and transect segment 18, five fields and 37 isolates. Transect segments not sharing a common letter are significantly different (P ≤ 0.05) according to Dunn’s test on ranks of aflatoxin B1 concentrations. ND, not detected.
FIG. 4.
Production of CPA by L-strain isolates of A. flavus (n = 774) from fields in 18 transect segments, each of which was 150 to 200 km long. The transect segments are shown in Fig. 1. The numbers of fields and numbers of isolates examined are given in the legend to Fig. 3. Transect segments not sharing a common letter are significantly different (P ≤ 0.05) according to Dunn’s test on ranks of CPA concentrations. ND, not detected.
The transect was divided into segments consisting of 150 to 200 km to assess regional differences in the production of aflatoxin B1 and CPA by L-strain isolates of A. flavus (Fig. 1). There were significant differences among transect segments in aflatoxin B1 production by the L-strain isolates (H = 219; 17 df; P < 0.0001) (Fig. 3). In the western half of Texas (transect segments 1 through 4) and the peanut-growing region of Georgia and Alabama (transect segments 10 through 12), 62 to 94% of the L-strain isolates produced >10 μg of aflatoxin B1 per ml. In other regions along the transect, 0 to 52% of the L-strain isolates produced >10 μg of aflatoxin B1 per ml, and the remaining isolates produced low levels of aflatoxin B1 or were nonaflatoxigenic. Significant differences among transect segments also were detected for CPA production by the L-strain isolates (H = 141; 17 df; P < 0.0001) (Fig. 4). The differences in CPA production were less apparent than the differences in aflatoxin B1 production, but the same transect segments in western Texas and the peanut-growing region of Georgia and Alabama tended to have higher percentages of isolates that produced >10 μg of CPA per ml.
Statistical analyses also revealed significant differences among the four major peanut-growing regions (Fig. 1) in production of aflatoxin B1 (H = 64; 3 df; P < 0.0001) and production of CPA (H = 47; 3 df; P < 0.0001) by L-strain isolates. The aflatoxin B1 production by L-strain isolates from Georgia and Alabama was significantly greater (P < 0.05) than the aflatoxin B1 production by isolates from central Texas and from Virginia and North Carolina but not the aflatoxin B1 production by isolates from western Texas. The L-strain isolates from Virginia and North Carolina produced significantly less aflatoxin B1 than the L-strain isolates from the other three peanut-growing regions. The CPA production by isolates from Virginia and North Carolina was significantly less than the CPA production by isolates from western Texas, central Texas, and Georgia and Alabama.
Of the 774 L-strain isolates of A. flavus examined from the transect, 126 were atoxigenic. All of the atoxigenic isolates produced nirA and/or niaD mutants, but only 23 produced cnx mutants. The cnx mutants were paired with complementary mutants of all isolates, which resulted in 53 isolates that were distributed among 17 VCGs (Table 2). The remaining 73 isolates that were incompatible with the cnx mutants were not examined further for vegetative compatibility. VCGs 24 and 64 to 70 were detected in fields in different states along the transect. VCG 24, an atoxigenic group, had previously been found in a Georgia peanut field (18, 20). VCGs 71 to 79 were detected in only one field. Two or more compatible isolates from the same field were observed for VCGs 64, 66, and 71 to 74.
TABLE 2.
Geographic distribution of VCGs containing atoxigenic isolates of the A. flavus L straina
| VCGb | No. of soil isolates | No. of fields
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Texas | Louisiana | Mississippi | Alabama | Georgia | South Carolina | North Carolina | Virginia | ||
| 24 | 2 | 1 | 1 | ||||||
| 64 | 11 | 1 | 1 | 1 | 1 | 2 | 3 | ||
| 65 | 7 | 1 | 1 | 1 | 1 | 1 | 2 | ||
| 66 | 7 | 1 | 1 | ||||||
| 67 | 5 | 1 | 1 | 2 | 1 | ||||
| 68 | 4 | 1 | 1 | 2 | |||||
| 69 | 3 | 1 | 1 | 1 | |||||
| 70 | 2 | 1 | 1 | ||||||
| 71 | 2 | 1 | |||||||
| 72 | 2 | 1 | |||||||
| 73 | 2 | 1 | |||||||
| 74 | 2 | 1 | |||||||
| 75 | 1 | 1 | |||||||
| 76 | 1 | 1 | |||||||
| 77 | 1 | 1 | |||||||
| 78 | 1 | 1 | |||||||
| 79 | 1 | 1 | |||||||
The isolates produced no detectable aflatoxin B1 or CPA. VCGs were determined by pairing cnx mutants obtained from 23 isolates with niaD and/or nirA mutants obtained from all 126 atoxigenic isolates. Seventy-three of these isolates were not placed in VCGs.
VCG 24 from a Georgia peanut field was described previously (18, 20), and the one soil isolate obtained in the previous studies was included in this VCG along with one North Carolina isolate obtained in the present study. The designations used for the other VCGs were a continuation of previously determined VCGs 1 to 63 of A. flavus (18, 31).
DISCUSSION
Regional differences in production of aflatoxin B1 and CPA by soil isolates of the A. flavus L strain were evident along the transect (Fig. 3 and 4). Surveys done in Japan (29) and in cotton-growing regions of the United States (8) also have revealed geographic differences in production of aflatoxin B1 by A. flavus. In this study, the toxigenicity of the L-strain isolates of A. flavus exhibited little association with dominant crops over most of the transect. Although the crop histories of the fields sampled were not known, the crops present were considered indicators of the types of crops traditionally grown in a region. For transect segments 1 through 4 (Fig. 1), the percentages of isolates that produced >10 μg of aflatoxin B1 per ml were relatively high (Fig. 3) despite differences in the major crops, which were largely peanuts for transect segments 1 and 4 and cotton for transect segments 2 and 3. Transect segments 13 through 18 also were characterized by different dominant crops, including peanuts for transect segment 17 and varying proportions of corn, cotton, soybeans, and peanuts for transect segments 13 through 16 and 18; however, the mycotoxin production data for all of these segments were similar. A possible exception to these findings was the data obtained for the major peanut-growing region of Georgia and Alabama (transect segments 10 through 12). In this region, the high percentages of L-strain isolates that produced >10 μg of aflatoxin B1 and CPA per ml were associated with extensive peanut cultivation. This region also had a higher mean soil density of A. flavus L-strain isolates (247 CFU/g) than the other major peanut-growing regions, including western Texas (7 CFU/g), central Texas (103 CFU/g), and Virginia and North Carolina (29 CFU/g) (16).
The dominance of highly toxigenic L-strain isolates and the occurrence of these isolates at relatively high soil densities in the peanut-growing region of Georgia and Alabama may not be coincidental. Drought stress accompanied by elevated soil temperatures, conditions that are conducive to A. flavus invasion of peanuts and subsequent aflatoxin contamination (13), is not uncommon in nonirrigated fields in Georgia and Alabama (14). The soil densities of A. flavus in these fields most likely reflect crop colonization over years of cultivation due to the dispersal of spores and infected crop debris to the soil (19, 38). In addition, the high percentages of aflatoxigenic isolates of A. flavus observed previously in peanuts and peanut field soils (20, 22, 28, 35) suggest that peanut cultivation selects for aflatoxin producers. The combined climatic and crop selection pressures in Georgia and Alabama could account for the dominance of highly aflatoxigenic isolates in this region.
The results of our examination of A. flavus isolates from a wide geographic area supported previous findings which showed that the S strain produces higher levels of aflatoxin B1 and shows less variation in aflatoxin B1 production than the L strain (6, 8). CPA production also was less variable in the S strain than in the L strain, and very few S-strain isolates produced <10 μg of CPA per ml (Fig. 2). The S strain has been associated with cotton cultivation in portions of the southern United States (8, 30). In the fields along the transect described here, the S strain was most prevalent in west central Texas and Louisiana, where cotton is grown extensively (16). Because of the small amount of variability in aflatoxin B1 and CPA production by the S strain, as well as its restricted distribution, regional differences in mycotoxin production were not examined.
Significant positive correlations between aflatoxin B1 production and CPA production were detected for both the L strain and the S strain. Horn et al. (20) previously reported a positive correlation between the two mycotoxins for L-strain isolates from a peanut field in Georgia, in contrast to other studies that showed either no correlation or a negative correlation (3, 21). In the L strain, the positive correlation accounts for the similar regional patterns of production of aflatoxin B1 and CPA (Fig. 3 and 4). Although the correlations were statistically significant for samples containing a large number of isolates, individual isolates exhibited considerable differences in production of the two mycotoxins, and in some instances, a correlation was not evident. This was best illustrated by the L strain; 12% of the L-strain isolates produced CPA but not aflatoxin B1.
Populations of A. flavus are extremely diverse genetically and include numerous VCGs (1, 18, 31). Since isolates belonging to the same VCG are similar in production of mycotoxins (2, 20), L-strain isolates that did not produce detectable aflatoxin B1 or CPA were specifically examined for vegetative compatibility in order to identify VCGs among the large number of isolates along the transect. Even within this chemotype of A. flavus, there was considerable diversity among the 53 isolates placed in VCGs, which was reflected by a diversity value (number of VCGs divided by number of isolates) of 0.32. This diversity also was indicated by the presence of many single-isolate VCGs, as well as by the 73 isolates that were not compatible with any of the 17 VCGs identified in this study (Table 2). VCGs of A. flavus were often detected in geographically distant portions of the transect. Previously, we (16) showed that VCG 1 of A. parasiticus was present in peanut fields along most of this transect. These data may indicate that A. flavus and A. parasiticus genotypes are widely dispersed, but this interpretation assumes that isolates belonging to the same VCG are clonal and genetically identical. A. flavus isolates belonging to the same VCG occasionally differ in morphology (20) and in random amplified polymorphic DNA (2). The geographically separated representatives of a VCG in this study may have diverged from one another while still maintaining the same alleles at loci that govern vegetative compatibility.
Application of atoxigenic strains of A. flavus and/or A. parasiticus to agricultural soil has been used to control aflatoxin contamination of crops by native populations of these two species. Aflatoxin reduction has been reported for peanuts and cottonseed (7, 12). The results of the present study suggest which regions along the transect might benefit most from soil application of an atoxigenic L-strain isolate of A. flavus. The majority of the L-strain isolates from the western half of Texas and the peanut-growing region of Georgia and Alabama produced moderate to high levels of aflatoxin B1 (Fig. 3), and it is in these regions that introduction of an atoxigenic strain might result in the greatest reduction in aflatoxin contamination in nonirrigated fields. In contrast, regions along the transect from central Texas to south central Alabama, as well as north of south central Georgia, contain soil populations with sizable percentages of L-strain isolates that do not produce aflatoxin B1 (Fig. 3). Native nonaflatoxigenic strains in such regions may provide some degree of control of aflatoxin contamination due to toxigenic strains.
ACKNOWLEDGMENTS
We thank R. Larry Greene for technical assistance and Milbra Schweikert for assistance with the mycotoxin analyses.
REFERENCES
- 1.Bayman P, Cotty P J. Vegetative compatibility and genetic diversity in the Aspergillus flavus population of a single field. Can J Bot. 1991;69:1707–1711. [Google Scholar]
- 2.Bayman P, Cotty P J. Genetic diversity in Aspergillus flavus: association with aflatoxin production and morphology. Can J Bot. 1993;71:23–31. [Google Scholar]
- 3.Blaney B J, Kelly M A, Taylor A L, Connole M D. Aflatoxin and cyclopiazonic acid production by Queensland isolates of Aspergillus flavus and Aspergillus parasiticus. Aust J Agric Res. 1989;40:395–400. [Google Scholar]
- 4.Bryden W L. Occurrence and biological effects of cyclopiazonic acid. In: Mise K, Richard J L, editors. Emerging food safety problems resulting from microbial contamination. Proceedings of the Seventh International Symposium on Toxic Microorganisms. Tokyo, Japan: U.S.-Japan Cooperative Program in Natural Resources; 1991. pp. 127–147. [Google Scholar]
- 5.Correll J C, Klittich C J R, Leslie J F. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology. 1987;77:1640–1646. [Google Scholar]
- 6.Cotty P J. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology. 1989;79:808–814. [Google Scholar]
- 7.Cotty P J. Influence of field application of an atoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology. 1994;84:1270–1277. [Google Scholar]
- 8.Cotty P J. Aflatoxin-producing potential of communities of Aspergillus section Flavi from cotton producing areas of the United States. Mycol Res. 1997;101:698–704. [Google Scholar]
- 9.Cotty P J, Bayman P, Egel D S, Elias K S. Agriculture, aflatoxins and Aspergillus. In: Powell K A, Renwick A, Peberdy J F, editors. The genus Aspergillus: from taxonomy and genetics to industrial application. New York, N.Y: Plenum Press; 1994. pp. 1–27. [Google Scholar]
- 10.Cullen J M, Newberne P M. Acute hepatotoxicity of aflatoxins. In: Eaton D L, Groopman J D, editors. The toxicology of aflatoxins: human health, veterinary, and agricultural significance. San Diego, Calif: Academic Press; 1994. pp. 3–26. [Google Scholar]
- 11.Diener U L, Cole R J, Sanders T H, Payne G A, Lee L S, Klich M A. Epidemiology of aflatoxin formation by Aspergillus flavus. Annu Rev Phytopathol. 1987;25:249–270. [Google Scholar]
- 12.Dorner J W, Cole R J, Blankenship P D. Effect of inoculum rate of biological control agents on preharvest aflatoxin contamination of peanuts. Biol Control. 1998;12:171–176. [Google Scholar]
- 13.Dorner J W, Cole R J, Sanders T H, Blankenship P D. Interrelationship of kernel water activity, soil temperature, maturity, and phytoalexin production in preharvest aflatoxin contamination of drought-stressed peanuts. Mycopathologia. 1989;105:117–128. doi: 10.1007/BF00444034. [DOI] [PubMed] [Google Scholar]
- 14.Hill R A, Blankenship P D, Cole R J, Sanders T H. Effects of soil moisture and temperature on preharvest invasion of peanuts by the Aspergillus flavus group and subsequent aflatoxin development. Appl Environ Microbiol. 1983;45:628–633. doi: 10.1128/aem.45.2.628-633.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill R A, Wilson D M, McMillian W W, Widstrom N W, Cole R J, Sanders T H, Blankenship P D. Ecology of the Aspergillus flavus group and aflatoxin formation in maize and groundnut. In: Lacey J, editor. Trichothecenes and other mycotoxins. Chichester, United Kingdom: John Wiley; 1985. pp. 79–95. [Google Scholar]
- 16.Horn B W, Dorner J W. Soil populations of Aspergillus species from section Flavi along a transect through peanut-growing regions of the United States. Mycologia. 1998;90:767–776. [Google Scholar]
- 17.Horn B W, Dorner J W, Greene R L, Blankenship P D, Cole R J. Effect of Aspergillus parasiticus soil inoculum on invasion of peanut seeds. Mycopathologia. 1994;125:179–191. doi: 10.1007/BF01146524. [DOI] [PubMed] [Google Scholar]
- 18.Horn B W, Greene R L. Vegetative compatibility within populations of Aspergillus flavus, A. parasiticus, and A. tamarii from a peanut field. Mycologia. 1995;87:324–332. [Google Scholar]
- 19.Horn B W, Greene R L, Dorner J W. Effect of corn and peanut cultivation on soil populations of Aspergillus flavus and A. parasiticus in southwestern Georgia. Appl Environ Microbiol. 1995;61:2472–2475. doi: 10.1128/aem.61.7.2472-2475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horn B W, Greene R L, Sobolev V S, Dorner J W, Powell J H, Layton R C. Association of morphology and mycotoxin production with vegetative compatibility groups in Aspergillus flavus, A. parasiticus, and A. tamarii. Mycologia. 1996;88:574–587. [Google Scholar]
- 21.Huang X, Dorner J W, Chu F S. Production of aflatoxin and cyclopiazonic acid by various aspergilli: an ELISA analysis. Mycotox Res. 1994;10:101–106. doi: 10.1007/BF03192259. [DOI] [PubMed] [Google Scholar]
- 22.Joffe A Z. Aflatoxin produced by 1,626 isolates of Aspergillus flavus from groundnut kernels and soils in Israel. Nature. 1969;221:492. doi: 10.1038/221492a0. [DOI] [PubMed] [Google Scholar]
- 23.Jones R K, Duncan H E, Hamilton P B. Planting date, harvest date, and irrigation effects on infection and aflatoxin production by Aspergillus flavus in field corn. Phytopathology. 1981;71:810–816. [Google Scholar]
- 24.Klich M A. Relation of plant water potential at flowering to subsequent cottonseed infection by Aspergillus flavus. Phytopathology. 1987;77:739–741. [Google Scholar]
- 25.Lansden J A, Davidson J I. Occurrence of cyclopiazonic acid in peanuts. Appl Environ Microbiol. 1983;45:766–769. doi: 10.1128/aem.45.3.766-769.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee L S, Lee L V, Jr, Russell T E. Aflatoxin in Arizona cottonseed: field inoculation of bolls by Aspergillus flavus spores in wind-driven soil. J Am Oil Chem Soc. 1986;63:530–532. [Google Scholar]
- 27.Lillehoj E B, McMillian W W, Guthrie W D, Barry D. Aflatoxin-producing fungi in preharvest corn: inoculum source in insects and soils. J Environ Qual. 1980;9:691–694. [Google Scholar]
- 28.Lisker N, Michaeli R, Frank Z R. Mycotoxigenic potential of Aspergillus flavus strains isolated from groundnuts growing in Israel. Mycopathologia. 1993;122:177–183. doi: 10.1007/BF01103479. [DOI] [PubMed] [Google Scholar]
- 29.Manabe M, Tsuruta O, Tanaka K, Matsuura S. Distribution of aflatoxin-producing fungi in soil in Japan. Trans Mycol Soc Jpn. 1976;17:436–444. [Google Scholar]
- 30.Orum T V, Bigelow D M, Nelson M R, Howell D R, Cotty P J. Spatial and temporal patterns of Aspergillus flavus strain composition and propagule density in Yuma County, Arizona, soils. Plant Dis. 1997;81:911–916. doi: 10.1094/PDIS.1997.81.8.911. [DOI] [PubMed] [Google Scholar]
- 31.Papa K E. Heterokaryon incompatibility in Aspergillus flavus. Mycologia. 1986;78:98–101. [Google Scholar]
- 32.Rao B L, Husain A. Presence of cyclopiazonic acid in kodo millet (Paspalum scrobiculatum) causing ‘kodua poisoning’ in man and its production by associated fungi. Mycopathologia. 1985;89:177–180. doi: 10.1007/BF00447028. [DOI] [PubMed] [Google Scholar]
- 33.Richard J L, Bhatnagar D, Peterson S, Sandor G. Assessment of aflatoxin and cyclopiazonic acid production by Aspergillus flavus isolates from Hungary. Mycopathologia. 1992;120:183–188. [Google Scholar]
- 34.Saito M, Tsuruta O. A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. Proc Jpn Assoc Mycotoxicol. 1993;37:31–36. [Google Scholar]
- 35.Schroeder H W, Boller R A. Aflatoxin production of species and strains of the Aspergillus flavus group isolated from field crops. Appl Microbiol. 1973;25:885–889. doi: 10.1128/am.25.6.885-889.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urano T, Trucksess M W, Beaver R W, Wilson D M, Dorner J W, Dowell F E. Co-occurrence of cyclopiazonic acid and aflatoxins in corn and peanuts. J Off Anal Chem Int. 1992;75:838–841. [Google Scholar]
- 37.Van Egmond H P. Mycotoxins: regulations, quality assurance and reference materials. Food Add Contam. 1995;12:321–330. doi: 10.1080/02652039509374309. [DOI] [PubMed] [Google Scholar]
- 38.Wicklow D T, Horn B W, Burg W R, Cole R J. Sclerotium dispersal of Aspergillus flavus and Eupenicillium ochrosalmoneum from maize during harvest. Trans Br Mycol Soc. 1984;83:299–303. [Google Scholar]