Abstract
Objective
Altered metabolites are important for the tumourigenicity of hepatocellular carcinoma (HCC). We performed integrative metabolomics analysis of the metabolites changes in portal venous blood and in comparison with the metabolites changes in liver tissues and stool samples of HCC patients and healthy liver donors.
Design
Serum (portal and central vein), liver tissue (HCC tumour and adjacent non-tumour, normal liver) and stool samples were collected from 102 subjects (52 HCC patients and 50 healthy controls) in the discovery cohort; and 100 subjects (50 HCC patients and 50 healthy controls) in an independent validation cohort. Untargeted metabolomic profiling was performed using high-performance liquid chromatography-mass spectrometry. The function of candidate metabolites was validated in hepatocyte cell lines.
Results
Detailed metabolomic evaluation showed distinct clusters of metabolites in serum, liver tissue and stool samples from patients with HCC and control individuals (p<0.001). HCC patients had significantly higher levels of portal vein serum and HCC tissue metabolites of DL-3-phenyllactic acid, L-tryptophan, glycocholic acid and 1-methylnicotinamide than healthy controls, which were associated with impaired liver function and poor survival. On the other hand, HCC patients had lower levels of linoleic acid and phenol in portal vein and stool samples than healthy controls. Linoleic acid and phenol significantly inhibited HCC proliferation, inferring their anti-HCC function as protective metabolites.
Conclusions
The integrative metabolome analysis of serum, tissue and stool metabolites revealed unreported metabolic alterations in HCC patients. In portal vein, we identified elevated and depleted metabolites signifying that they might play a role in HCC development.
Keywords: hepatocellular carcinoma, carcinogen metabolism
Significance of this study.
What is already known on this subject?
Hepatocellular carcinoma (HCC) is the third-leading cause of cancer deaths with a 5-year survival of 50%–70%.
Metabolites play an important role in the pathogenesis of HCC.
Most metabolomics studies used peripheral blood to reflect metabolic changes in HCC patients, but metabolome characteristics in portal vein have not been reported.
What are the new findings?
Partial least squares discrimination analysis showed distinct clusters of metabolites in portal and central vein serum, liver tissue and stool samples from HCC patients and control subjects.
There was variable decrease in the abundance of different metabolites from portal vein to central vein after going through the liver, and the reduction was different between HCC patients and controls.
HCC patients had increased portal vein serum and tumour tissue pathogenic metabolites of DL-3-phenyllactic acid, L-tryptophan, glycocholic acid and 1-methylnicotinamide, which were associated with impaired liver function and poor prognosis.
HCC patients had depleted portal vein serum and stool protective metabolites of linoleic acid and phenol, which inhibited HCC cell proliferation.
How might it impact on clinical practice in the foreseeable future?
This is the first report to illustrate the metabolome characteristics in portal vein of patients with HCC.
Metabolic alterations may contribute to the development and progression of HCC, which implies that the altered metabolites may represent a potential target to prevent HCC development.
Introduction
Hepatocellular carcinoma (HCC) is the third-leading cause of cancer deaths worldwide.1 Although surgical treatment may be effective in HCC patients, the 5-year survival rate is only 50%–70%.2 Moreover, due to the lack of early diagnostic marker, most patients with HCC are often diagnosed in an advanced stage with poor prognosis.3 4 Therefore, there is an urgent need to further understand the possible aetiological factors and new therapeutic methods to improve the prognosis of HCC patients.
Metabolomics is a promising approach for the identification of metabolites which may provide insights into aetiology, treatment and early diagnosis of diseases.5 A recent study using metabolomics in peripheral blood plasma samples discovered that N, N, N-trimethyl-5-aminovaleric acid could induce fatty liver disease through increased lipolysis and decreased fatty acid oxidation.6 Intestinal dysbiosis affects the development of HCC in part through microbial metabolites. A gut bacterial metabolite deoxycholic acid (DCA) causes DNA damage and activate tumour-promoting factors in the liver; while blocking DCA production prevents HCC development in an obese mice model.7 In addition, propionate, a metabolite from gut microbiota, inhibits cancer cell proliferation in the liver.8 These studies supported the important role of metabolites in the development and progression of liver diseases.
Liver has a unique vascular system (portal vein) to receive metabolites from the gut and deliver it directly to the liver. Because of the structural link to the intestine, the liver is the first organ to receive gut-derived metabolites and serves a critical function between foreign substances and the systemic milieu. The milieu of the portal vein system may play an important role in HCC development.
Accumulating evidence suggests that metabolites are important for HCC formation.9 However, because of the poor accessibility of portal vein blood, most metabolomics studies are limited by the use of peripheral blood that may not reflect changes in the portal venous blood and the liver. Therefore, comparing metabolic changes in portal vein, central vein, liver tissue and stool samples between patients with HCC and healthy subjects may be helpful for in-depth understanding on the gut-liver axis, evaluating potential aetiology and novel therapeutic targets for HCC patients. The aim of this study is to perform detailed metabolomics analysis of HCC patients and healthy liver donors to reveal previously unreported metabolic alterations in HCC patients.
Material and methods
Human subjects
A total of 660 samples, including 102 portal vein blood samples, 102 central vein blood samples, 52 HCC tissues, 52 adjacent non-cancerous, 50 normal liver tissues and 102 faecal samples were collected in the discovery cohort from November 2018 to June 2019. In the independent validation cohort, 100 portal vein blood and 100 central vein blood samples were collected from March 2020 to June 2020. All samples were collected from Shanghai and Anhui liver cancer clinic centres. A total of 202 participants were enrolled in the discovery (n=102) and validation (n=100) cohorts, including100 healthy liver donors who underwent living-related liver transplantation and 102 HCC individuals who underwent liver resection or liver transplantation. The living donors were parents of infants and children who required living donor liver transplantation for conditions like Wilson’s disease and congenital biliary atresia. The division of the discovery and validation cohorts was done prior to data analysis.
Inclusion criteria
We enrolled HCC patients (all later confirmed histologically in the surgical specimens) and healthy liver donors aged 18 years or older.
Exclusion criteria
Exclusion criteria included: (1) prior anticancer treatments, (2) intrahepatic cholangiocarcinoma (ICC), (3) subjects with current or past history of other malignancies and history of gastrointestinal operations, (4) participants missing clinical information, (5) bacterial infection or use of antibiotics within 1 month before surgery and (6) presence of hypertension, diabetes or other metabolic diseases. The inclusion and exclusion criteria for HCC patients and healthy controls in the validation cohort were the same as those in the discovery cohort.
Sample collection
The portal vein (n=202) (obtained during surgical operation) and central vein (n=202) (obtained immediately after patients were anaesthetised before surgical operation) blood samples were collected after an overnight fasting to eliminate the disturbance of diet. Anaesthetists inserted a line to the right internal jugular vein before surgical operation and central vein blood were collected from the catheter with syringe. We collected portal vein blood from the left branch of portal vein with syringe in living donors. For HCC patients, we collected portal vein blood from the portal stem vein with syringe before devascularisation of the portal stem vein. Serum samples were stored at −80°C for until analysis.
Liver tumour tissues (n=52) and adjacent non-tumour tissues (n=52) (≥2 cm from HCC) were collected from 52 HCC patients who underwent liver surgical resection or liver transplantation under sterile conditions within 30 min of surgery. Normal liver tissues (n=50) were collected from 50 healthy liver donors who underwent living-related liver transplantation. Tissue samples were stored at −80 °C until analysis.
Fresh faecal samples were collected from all HCC patients and normal subjects at the hospital during 2–5 days before surgery and then snap frozen in −80 °C freezer within 1 hour or stored in −20 °C freezers before transporting to −80 °C freezer within 2 hours.
Untargeted metabolomics by high-performance liquid chromatography-mass spectrometry
Metabolic extracts were obtained from portal and central vein serum, liver tissues and stool samples following methanol-assisted protein precipitation and then analysed by liquid chromatography-mass spectrometry (LC-MS) using UHPLC System (1290, Agilent Technologies, Santa Clara, California, USA), equipped with a ultra performance liquid chromatography ethylene bridged hybrids (UPLC BEH) Amide column. The mobile phase consisted of 25 mmol/L ammonium acetate and 25 mmol/L ammonium hydroxide in water and acetonitrile. A triple time-of-flight (TOF) mass spectrometer (AB Sciex) was used for its ability to acquire MS/MS spectra on an information-dependent acquisition during an LC/MS experiment. In this mode, the acquisition software (Analyst TF 1.7, AB Sciex) continuously evaluates the full scan survey MS data as it collects and triggers the acquisition of MS/MS spectra depending on preselected criteria. In each cycle, the most intensive 12 precursor ions with intensity greater than 100 were chosen for MS/MS at collision energy of 30 eV. (See online supplemental methods for detailed information).
gutjnl-2021-325189supp001.pdf (3.4MB, pdf)
MS raw data files were converted to the mzXML format by ProteoWizard and processed by R package XCMS (V.3.2). The process includes peak deconvolution, alignment and integration. Minfrac and cut-off are set as 0.5 and 0.3, respectively. In-house MS2 database was applied for metabolites identification.
Cell lines
Human HCC cell lines (Hep3B and Huh7) and human normal hepatocyte cell line an immortalised human hepatocyte cell line (MIHA) were obtained from American Type Culture Collection (Manassas, Virginia, USA). All cells were cultured in high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco BRL, Grand Island, New York, USA) supplemented with 10% (vol/vol) fetal bovine serum, and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2.
Cell viability assay
Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium assay. Hep3B, Huh7 and MIHA cells (1000 cells per well) were seeded in 96-well plate and cultured with or without indicated metabolites-supplemented DMEM for 24 hours. The absorbance was recorded at 570 nm using the (Thermo Scientific, Waltham, Massachusetts, USA). All experiments were conducted three times.
Statistical analysis
Data were expressed as mean±SD. All statistical tests were performed using SPSS V.25 or Graph-pad Prism V.8 software (GraphPad Software, San Diego, California, USA). The difference in cell viability was determined by two-way analysis of variance. A supervised model of partial least squares discrimination analysis (PLS-DA) was applied to assess the metabolic alterations among groups. A permutation testing was performed 200 times to assess the risk of overfitting for the PLS-DA model. Overall survival in relation to metabolites median concentration was evaluated by the Kaplan-Meier survival curve and the log-rank test. Pathway enrichment analysis was conducted by Kyoto Encyclopaedia of Genes and Genomes (KEGG) database. HRs and 95% CIs were provided by univariate Cox proportional hazard models.10 The Pearson correlation coefficient was used to evaluate the correlation between metabolites and liver function indices. Qualitative and quantitative differences between subgroups were analysed using χ2 test or Fisher’s exact test for categorical parameters, and Student’s t-test or Mann-Whitney U test was used to compare the difference in two groups as appropriate. Values of p<0.05 were considered as being statistically significant.
Additional methods are provided in online supplemental material.
Results
Cohort design and characteristics of HCC patients and healthy controls
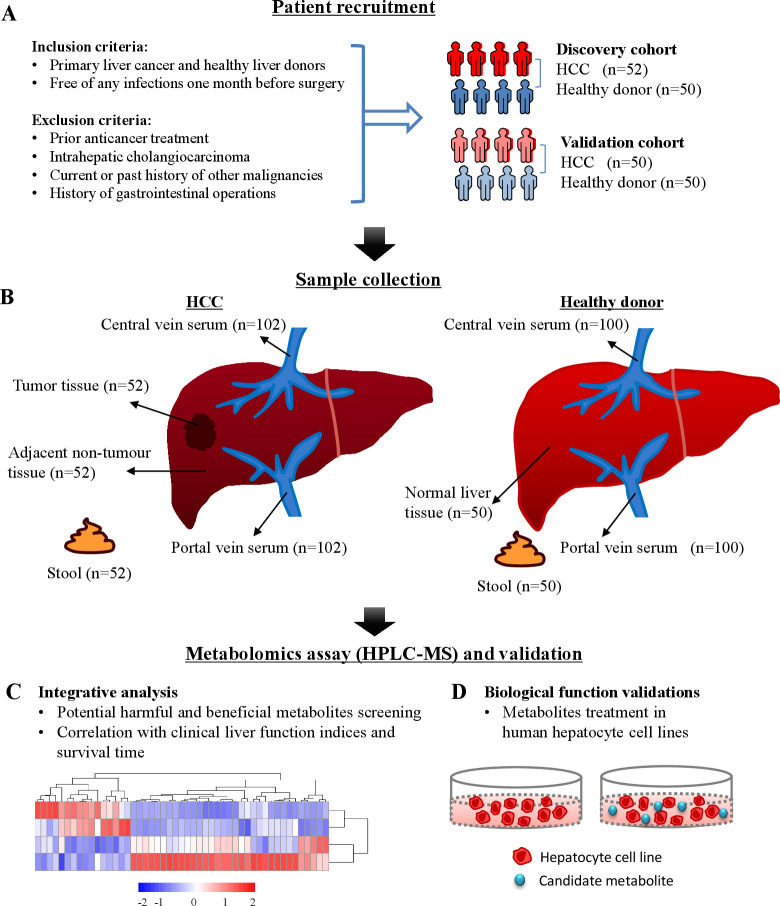
We prospectively recruited two independent cohorts of patients with HCC and healthy liver donors with strict inclusion and exclusion criteria to avoid the influence on the microbiome or microbes-derived metabolites (figure 1A).11 12 Two study cohorts were established to explore the association between metabolic alterations and HCC. The discovery cohort included 52 HCC patients and 50 healthy controls, and the validation cohort included 50 HCC patients and 50 healthy controls. In both the discovery and validation cohorts, chronic hepatitis B virus infection was the leading cause of HCC, accounting for 65.4% and 60%, respectively (online supplemental tables 1 and 2). Compared with healthy controls, HCC patients were older and had higher serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyl transpeptidase (GGT), alkaline phosphatase (ALP), direct bilirubin and prothrombin time (PT), but lower levels of total protein, albumin and platelets (online supplemental table 1). Untargeted metabolomics profiling was performed on 660 samples from HCC patients (360 samples) of portal and central vein serum, HCC and its adjacent non-cancerous tissues and stool samples, and from normal subjects (300 samples) of portal and central vein serum, normal liver tissues and stool samples (figure 1B) using high-performance LC-MS (figure 1B). Integrative analysis was performed based on the altered metabolites of serum, tissue and stool (figure 1C). The biofunctions of the candidate metabolites were validated in HCC and hepatocyte cell lines (figure 1D).
Figure 1.
Flow chart showing the methods used for metabolomics analysis and biological validation. (A) Patient recruitment and inclusion and exclusion criteria. (B) Various clinical samples were obtained from HCC patients and healthy controls. (C) Integrative analysis for potential harmful and beneficial metabolites based on serum, tissue and stool samples HPLC-MS spectra. (D) In vitro biological function validation. HPLC-MS, high-performance liquid chromatography-mass spectrometry. HCC, hepatocellular carcinoma; HPLC-MS, high-performance liquid chromatography-mass spectrometry.
Overall metabolomic profiling and annotated metabolites in all collected samples
Data quality control (QC) was performed by Pearson correlation analysis to obtain stable and accurate metabolome results. The Pearson correlation of serum negative electrospray ionisation (ESI−) and positive ESI (ESI+) QC samples (online supplemental figure 1A), tissue ESI− and ESI+ QC samples (online supplemental figure 1B) and stool ESI− and ESI+ QC samples (online supplemental figure 1C) was high, indicating that the data quality was stable and reliable. Additionally, in the principal component analysis score plots for both ESI− and ESI+ serum (online supplemental figure 2A), tissue (online supplemental figure 2B) and stool (online supplemental figure 2C) samples, the QC samples clustered tightly together, which further confirmed data reliability. A total of 2799 annotated metabolites were detected in all collected samples, including 156 (ESI−) and 285 (ESI+) serum small-molecule metabolites (online supplemental table 3A, B); 305 (ESI−) and 535 (ESI+) liver tissue metabolites (online supplemental table 3C, D); 440 (ESI−) and 703 (ESI+) stool metabolites (online supplemental table 3E, F) in the discovery cohort and 146 (ESI−) and 229 (ESI+) serum small-molecule metabolites (online supplemental table 3G, H) in the validation cohort.
Metabolic alterations are identified in HCC patients compared with healthy controls
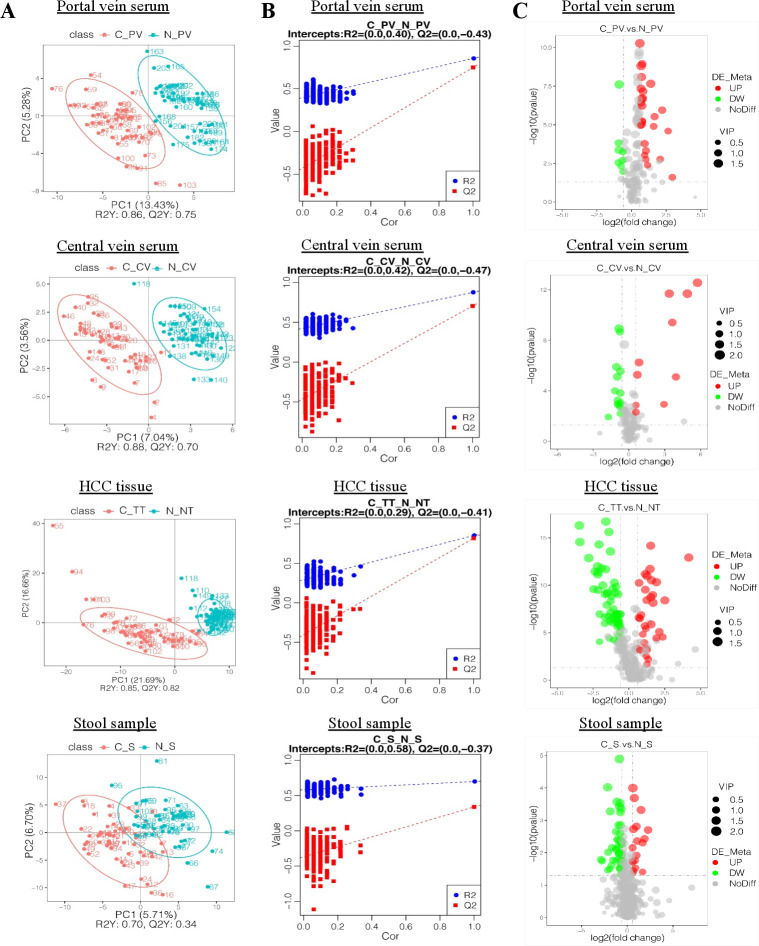
Distinct clusters of metabolites in HCC patients compared with control individuals were demonstrated in portal (p<0.001), central vein serum (p<0.001), HCC tissue (p<0.001) and stool samples (p<0.001) in both ESI− (figure 2A) and ESI+ (online supplemental figure 3A) models in the discovery cohort by PLS-DA score plots. We then built a model containing metabolomic information by fitting an PLS-DA and tested its ability to correctly classify new samples during a seven-cross-validation through 200 times random permutation testing. The intercepts of goodness-of-fit (R2) and goodness-of-prediction (Q2) illustrate the PLS-DA model is reliable and not overfitting (figure 2B and online supplemental figure 3B).
Figure 2.
Metabolic alterations in HCC were found in various samples. (A) PLS-DA score plots for portal and central vein serum, HCC tissues and stool samples in ESI− model. HCC patients are shown in red, and normal controls are shown in blue. X axis and Y axis represent contributions of persons to the first two principal components (PC1 and PC2). (B) Cross-validation plot with a permutation test repeated 200 times. The intercepts of R2= (0.0, 0.4) and Q2= (0.0,–0.43), R2= (0.0, 0.42) and Q2= (0.0,–0.47), R2= (0.0, 0.29) and Q2= (0.0,–0.41) and R2= (0.0, 0.58) and Q2= (0.0,–0.37) suggest that the PLS-DA model is not overfitting. (C) Volcano plots showing the results of pairwise comparisons of metabolites in each HCC sample’s group relative to healthy controls (CP_V/N_PV, C_CV/N_CV, C_TT/N_NT, C_S/N_S, respectively). The vertical dashed lines indicate the threshold for the twofold abundance difference. The horizontal dashed line indicates the p=0.05 threshold. Between-group comparisons were performed using Student’s t test. metabolites with significant changes are presented in red (upregulated) or green (downregulated). C_PV, portal vein of HCC patients; N_PV, portal vein of healthy controls; C_CV, central vein of HCC patients; N_CV, central vein of healthy controls; C_TT, HCC tissue of HCC patients; N_NT, normal tissue of healthy controls; C_S, stool samples of HCC patients; N_S, stool samples of healthy controls. VIP, variable importance in the projection. ESI−, negative electrospray ionisation; HCC, hepatocellular carcinoma; PLS-DA, partial least squares discrimination analysis.
We plotted fold changes (using volcano plots) in the levels of identified metabolites in patients with HCC relative to healthy controls, considering the statistically significant difference (p value) and variable importance in the projection. As shown in figure 2C and online supplemental figure 3C, the levels of the differential metabolites in patients with HCC were significantly different from those in healthy controls in ESI− (figure 2C) and ESI+ (online supplemental figure 3C) models for portal vein serum, central vein serum, HCC tissues and stool samples compared with their corresponding controls, respectively. Taken together, HCC is associated with significant changes in the metabolome from multiple sources of portal vein, central vein, liver tissue and stool samples.
Metabolic changes from portal vein serum to central vein serum are different in HCC patients as compared with healthy controls
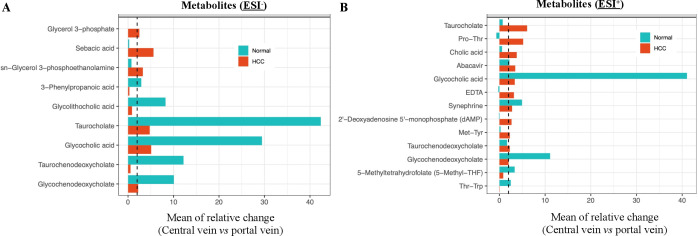
We then evaluated the relative changes in metabolites between the portal vein and central vein as a reflection of alterations after going through the liver.13 We compared the metabolites from portal vein to central vein in ESI− (online supplemental table 4A) and ESI+ (online supplemental table 4B) models in HCC patients and healthy controls. The relative abundance of identified metabolites was changed from portal to central vein and tends to be higher in portal vein. We found 17 metabolites in HCC patients and 12 metabolites in normal controls that had twofold or more reduction changes from portal vein to central vein in both ESI− (figure 3A) and ESI+ (figure 3B) models. These results suggest that portal vein metabolic alterations are different from peripheral blood metabolic changes. Thus, portal vein results would provide additional insights on the metabolomic alterations contributing to HCC formation.
Figure 3.
A twofold or more reduction in metabolites from portal to central vein in HCC patients and healthy controls. (A, B) Relative change of identified metabolites from portal vein to central vein was calculated by (MPV-MCV)/MCV for HCC patients and healthy controls in ESI− (A) and ESI+ (B) models. A twofold or more reduction in metabolites from the portal vein to central vein was seen in 17 metabolites in HCC patients and 12 metabolites in normal controls. The vertical dashed lines indicate the twofold reduction. ESI−, negative electrospray ionisation; ESI+, positive ESI; HCC, hepatocellular carcinoma; MPV, mean value of metabolites concentration in portal vein; MCV, mean value of metabolites concentration in central vein.
Pathogenic metabolites are elevated in portal vein serum compared with central vein serum in HCC patients
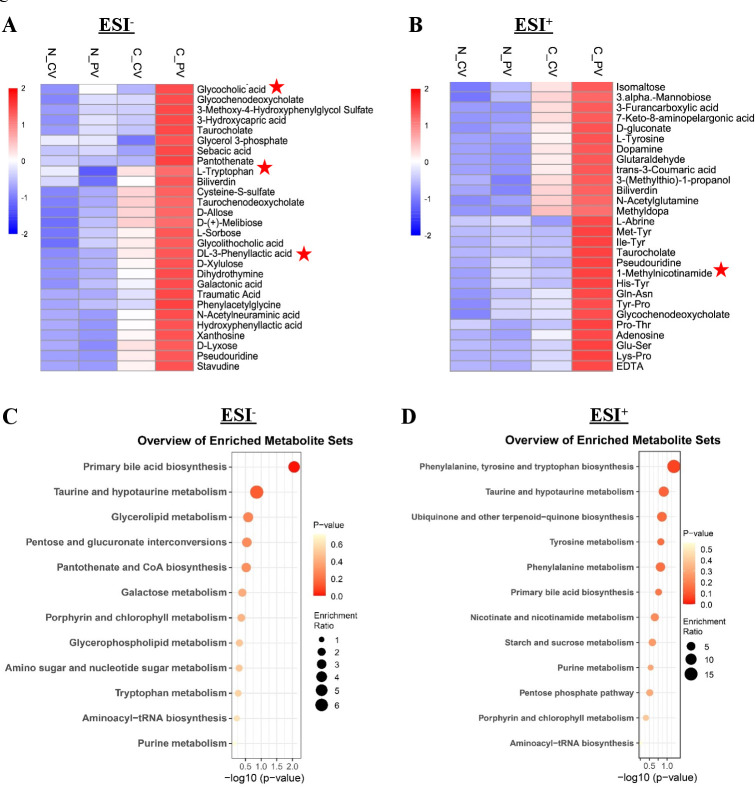
To reveal metabolic phenotypes that are potentially involved in HCC formation, we compared the metabolic changes from portal vein to central vein serum samples by hierarchical cluster analysis. We found 56 significantly elevated metabolites in portal vein compared with central vein in HCC including 28 in the ESI− (figure 4A) and 28 in ESI+ (figure 4B) models (online supplemental table 5A, B). In particular, these 56 elevated metabolites identified in portal vein of HCC patients were also significantly higher as compared with healthy controls both in ESI− (figure 4A) and ESI+ (figure 4B) models (online supplemental table 5A, B). Moreover, these elevated metabolites were validated in the independent cohort and 25 of them were also increased in portal vein serum of HCC patients as compared with healthy controls (online supplemental table 6). In order to understand the functional characteristics and classification of these elevated portal vein metabolites, we conducted pathway enrichment analysis using the KEGG and identified the significant enriched pathways in both ESI− (figure 4C) and ESI+ (figure 4D) models. We found that primary bile acid biosynthesis, taurine and hypotaurine metabolism, and phenylalanine and tryptophan metabolism were the top pathways altered in portal vein metabolites of patients with HCC. These pathways-associated metabolites were reported to contribute to systemic liver inflammation.14 15 We performed functional validation for the elevated portal vein metabolite like L-tryptophan using HCC cell lines Hep3B, Huh7 and normal hepatocyte cell line MIHA. L-tryptophan has been reported as an important cancer nutrient and its removal can lead to destruction of the tumour.16 We found that L-tryptophan significantly promoted cell proliferation in HCC cells (Hep3B and Huh7) but not in MIHA in a time-dependent manner (online supplemental figure 4). Collectively, these findings suggested that the elevated metabolites in portal vein may contribute to the tumourigenesis of HCC.
Figure 4.
Pathogenic metabolites in portal vein of HCC patients. (A, B) Hierarchical cluster analysis of portal vein and central vein differential metabolites between HCC patients and healthy controls in ESI− (A) and ESI+ (B) models, respectively. Red colour represents high relative concentration of metabolites in each group, blue colour represents low relative concentration of metabolites in each group. Metabolites with star labelled were significantly increased in both portal vein and tumour tissues of HCC patients. (C, D) Pathway enrichment analysis of significantly elevated metabolites in portal venous of HCC patients according to the KEGG pathway in both ESI− (C) and ESI+ (D) models. ESI−, negative electrospray ionisation; ESI+, positive ESI; HCC, hepatocellular carcinoma; KEGG, Kyoto Encyclopaedia of Genes and Genomes.
The elevated metabolites in HCC tumour tissues compared with healthy controls
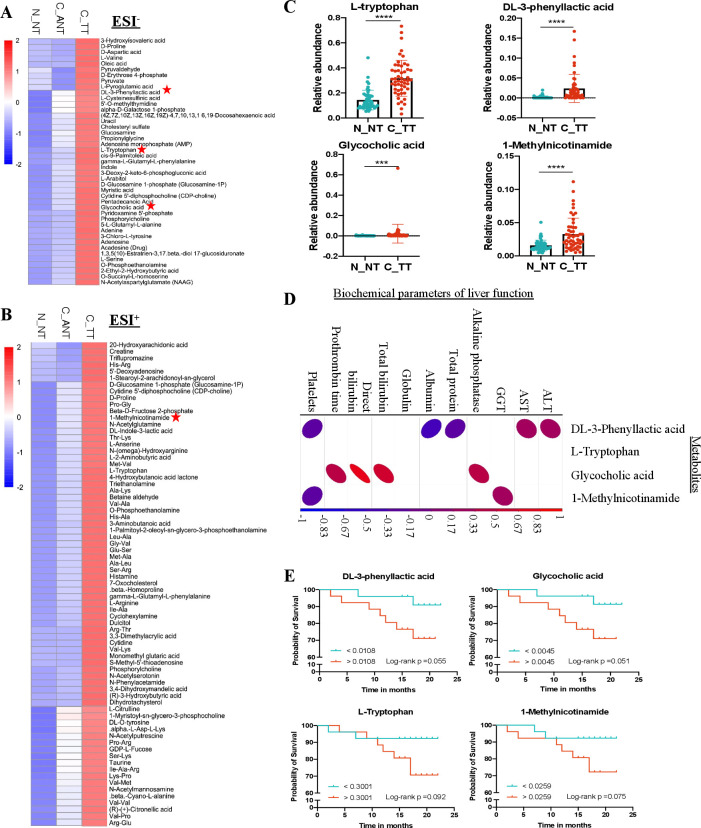
We identified 116 elevated metabolites in tumour tissues compared with adjacent non-tumour tissues in HCC patients including 43 in ESI− (figure 5A) and 73 in ESI+ (figure 5B) models. In particular, these 116 elevated metabolites identified in tumour tissues of HCC patients were also significantly higher as compare to healthy controls in both ESI− (figure 5A) and ESI+ (figure 5B) models. Among them, the elevated portal vein metabolites of DL-3-phenyllactic acid, L-tryptophan, glycocholic acid and 1-methylnicotinamide were also significantly higher in HCC tumour tissues compared with normal liver tissues (figure 5C). We compared the levels of these metabolites with the biochemical parameters of liver function and found that DL-3-phenyllactic acid, glycocholic acid and 1-methylnicotinamide had positive correlation with the level of serum AST (p=0.04), ALT (p=0.02), ALP (p=3.63E-05), GGT (p=0.02), total serum bilirubin (p=4.37E-08) and PT (p=2.22E-06) and negative correlation with the level of serum albumin (p=0.0002) and platelets (p=0.006) (figure 5D), indicating the elevated metabolites both in portal vein and tumour tissue of HCC patients are associated with impaired liver function. To further evaluate the role of these potential harmful metabolites in association of HCC prognosis (online supplemental table 7 for details of follow-up information), we stratified HCC patients into high vs low categories based on the median relative concentration of these four metabolites. When the cohort was dichotomised by the median value of individual metabolites, the hazard ratios for overall mortality were 3.975 (95% CI 0.825 to 19.162) for DL-3-phenyllactic acid, 3.662 (95% CI 0.760 to 17.645) for L-tryptophan, 3.975 (95% CI 0.825 to 19.162) for glycocholic acid and 3.487 (95% CI 0.724 to 16.806) for 1-methylnicotinamide (figure 5E). Additionally, the proportional hazards assumption is well met (online supplemental figure 5). These results support the harmful nature of these enhanced metabolites in both portal vein and HCC tumour tissues.
Figure 5.
Validation of pathogenic metabolites in HCC tumour tissues. (A, B) Hierarchical cluster analysis of significantly elevated metabolites in HCC tissues compared with adjacent non-cancerous and normal tissues in ESI− (A) and ESI+ (B) models, respectively. Metabolites with star labelled were significantly increased in both portal vein and tumour tissues of HCC patients. Red colour represents high relative concentration of metabolites in each group, blue colour represents low relative concentration of metabolites in each group. (C) Scatter plots show the relative abundance of the selected pathogenic metabolites in tumour tissues of HCC patients and healthy controls. Data are expressed as mean±SD statistical significance was determined by two-way analysis of variance. ***P<0.001, ****p<0.0001. (D) A heatmap shows the correlation between selected pathogenic metabolites with liver function indices. Red colour represents positive correlation, blue colour represents negative correlation. P<0.05 was considered statistically significant. (E) Pathogenic metabolites influence the outcome of HCC patients. Kaplan-Meier plots showing a lower overall survival patient with DL-3-phenyllactic acid, glycocholic acid, L-tryptophan and 1-methylnicotinamide in HCC tissues above the median values. AST, aspartate aminotransferase; ALT, alanine aminotransferase; ESI−, negative electrospray ionisation; ESI+, positive ESI; HCC, hepatocellular carcinoma; GGT, gamma-glutamyl transpeptidase.
Depleted metabolites in portal vein of HCC patients are protective against HCC in vitro
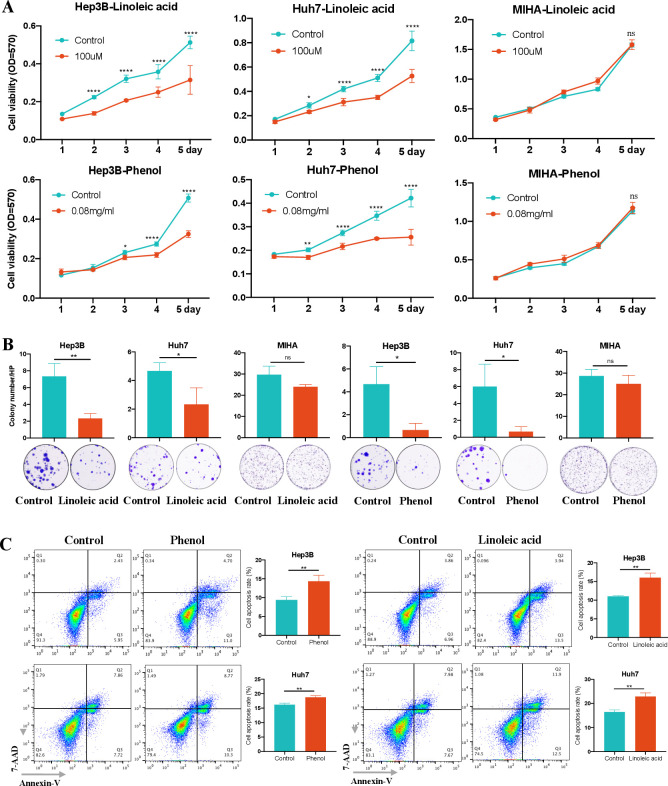
On the other hand, we evaluated the depleted metabolites in portal vein samples of HCC patients and overlapped with the depleted metabolites in stool samples of the same patients. We found that linoleic acid and phenol were significantly decreased metabolites in both portal vein serum and stool samples of HCC patients as compared with heathy controls in the discovery cohort (online supplemental table 8A, B). In particular, these two depleted metabolites were further confirmed in portal vein serum samples of HCC patients in the validation cohort (online supplemental table 8C). The functional importance of linoleic acid and phenol were studied in HCC cell lines. We found that both Linoleic acid and phenol significantly inhibited cell proliferation in HCC cells Hep3B and Huh7 in a time-dependent manner, but not in normal hepatocyte cell line (figure 6A). This growth-inhibitory effect for HCC cells were confirmed by colony formation assay (figure 6B). Moreover, linoleic acid and phenol increased apoptosis of HCC cells (figure 6C). Taken together, linoleic acid and phenol suppressed cells proliferation and induced apoptosis in HCC cells, inferring their anti-HCC function as protective metabolites.
Figure 6.
Validation of potential protective metabolites for HCC in vitro. (A) linoleic acid and phenol significantly inhibited the cell viability of HCC cells Hep3B and Huh7, but not normal hepatocyte cell MIHA, as determined by MTT assay. (B) linoleic acid and phenol significantly suppressed the colony formation of HCC cells but not normal hepatocyte cells. (C) linoleic acid and phenol significantly increased HCC cell apoptosis. Data are expressed as mean±SD statistical significance was determined by unpaired t-test or two-way analysis of variance, where appropriate. *P<0.05, **p<0.01, ****p<0.0001. HCC, hepatocellular carcinoma; MTT, 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium; NS, no significant.
Discussion
In this study, we performed in-depth integrative metabolome analyses of portal and central vein serum, liver tissue and stool samples to identify HCC-associated metabolites as compared with healthy subjects. In total, 2799 metabolites were annotated in all collected samples, and significant alterations in the metabolomic composition were observed in HCC patients (figure 2). These observations suggest that metabolic reprogramming occurs in HCC patients to overcome the low nutrient availability and generate energy essential to sustain HCC cell proliferation and survival. This metabolic reprogramming could be a potential driving factor for the HCC tumourigenicity.
As a direct connection between the intestine and the liver, the portal vein is the major source of hepatic blood supply. Portal vein serum metabolomics profiling can reflect altered intestinal metabolites, and consequently elicit a shift in metabolic substrate in HCC patients. As such, portal and central vein serum metabolite profiling of HCC patients and healthy controls were compared and 56 metabolites were specifically elevated in the portal vein of HCC patients compared with central vein and healthy controls (figure 4). Pathway analysis of these differentially affected metabolites in portal venous of HCC patients demonstrated significant enrichment of several processes including those relevant to increased activity of primary bile acid biosynthesis, taurine and hypotaurine metabolism, as well as phenylalanine and tryptophan metabolism (figure 4). These pathways and pathway-associated metabolites were reported to contribute to systemic inflammation and liver cancer. Disruption of bile acid homoeostasis changes metabolic homoeostasis in liver and can cause hepatic inflammation and liver cancer.14 17 18 It was reported that liver cancer is characterised by taurine, hypotaurine metabolism and tyrosine metabolism.19
Glycocholic acid, glycochenodeoxycholate, taurocholate, taurochenodeoxycholate and glycolithocholic acid were reported to be the key metabolites to impact hepatic lipid accumulation and inflammation in HCC development.15 Moreover, our portal vein metabolites results are consistent with recent studies indicating that several bile acid metabolites such as tauroursodeoxycholic acid (TUDCA), glycochenodeoxycholate, taurocholic acid are significantly elevated in tumour tissues of patients with HCC and ICC.20 DCA, a product of chenodeoxycholic acid, has been reported to promote liver carcinogenesis through the senescence secretome.21 These data collectively suggested that the metabolic alterations in portal vein are different from peripheral blood in HCC patients, which is also different from healthy controls. These altered metabolites and their enriched functional pathways at least in part contribute to the HCC development.
The liver is exposed to the gut metabolites and products through the portal vein and is central to the homoeostasis of global metabolism. Metabolic reprogramming of HCC cells affected by gut metabolites via portal vein could be key to their tumourigenicity and aggressiveness of HCC. We, therefore, further compared the metabolite profiling in tumour and the adjacent non-tumour tissues of HCC patients and healthy controls. In total, 116 metabolites were specifically altered in tumour tissues compared with adjacent non-tumour and normal liver tissues (figure 5). Focusing on overlapping metabolites, we found six metabolites including four elevated metabolites (DL-3-phenyllactic acid, L-tryptophan, glycocholic acid and 1-methylnicotinamide) and two depleted metabolites (linoleic acid and phenol) as predominant metabolite alteration featured both in the tumour tissue and portal vein of HCC patients. These six altered metabolites were further validated in an independent validation cohort of HCC patients. To evaluate the functional significance of the four elevated metabolites in HCC, we demonstrated that higher levels of the four elevated metabolites correlated with impaired liver function, poorer survival after HCC surgical treatment (figure 5) and promoted cell proliferation by L-tryptophan in HCC cells in vitro. In keeping with our findings, other reports suggested that DL-3-phenyllactic acid, L-tryptophan, glycocholic acid and 1-methylnicotinamide have close relationships with the onset or progression of different cancers.22–29 DL-3-phenyllactic acid was recognised as a potential biomarker for ovarian cancer,23 oral squamous cell carcinoma22 and cervical cancer.25 DL-3-phenyllactic acid promotes cell migration and invasion in cervical cancer.24 Glycocholic acid, a primary and conjugated bile acid, increases the risk of colorectal cancer development.26 Nicotinamide N-methyltransferase contributes to tumourigenesis by consuming methyl units to produce stable metabolic product 1-methylnicotinamide to impair the methylation potential of cancer cells.27 Moreover, studies have indicated that 1-methylnicotinamide inhibits activation of the ASK-P38-MAPK pathway, resulting in increased resistance of colorectal cancer cells to 5-fluorouracil.30 Thus, 1-methylnicotinamide may be a potential therapeutic target for colorectal cancer treatment by enhancing the proapoptotic effect of 5-fluorouracil. L-tryptophan was also reported to promote tumour progression through suppressing antitumour immune responses and increasing the malignant properties of cancer cells.28 29 There is evidence for a role of nicotinamide adenine dinucleotide (NAD+) generated via the tryptophan de novo pathway in cancer biology. Studies in mice showed that impaired tryptophan metabolism leads to suppression of de novo NAD+ synthesis in the liver and facilitates hepatic tumourigenesis through DNA damage.31 Taken together, our data suggest that elevated metabolites of DL-3-phenyllactic acid, L-tryptophan, glycocholic acid and 1-methylnicotinamide in the portal vein and liver tissues contribute to HCC tumourigenesis.
The two depleted metabolites (linoleic acid and phenol) in the portal vein were also significantly decreased in the stool samples of HCC patients as compared with healthy controls, which was confirmed in the validation cohort. Portal vein metabolome is presumed to reflect alterations to intestinal metabolism. The depletion of metabolites both in portal vein serum and in stool samples of HCC patients suggests their potential protective role in HCC progression. As anticipated, linoleic acid and phenol inhibited cell proliferation and induced apoptosis in HCC cells (figure 6). Similarly, linoleic acid was reported to suppress colorectal cancer cell growth by inducing apoptosis.32 Moreover, linoleic acid supplementation is a potential adjuvant treatment for breast cancer.33 34 Phenol-containing molecules exhibited anticancer effects.35 Phenolics enhances the immune system to recognise and destroy cancer cells and inhibiting the angiogenesis.36 Collectively, these reduced metabolites of linoleic acid and phenol in portal vein and stool could play a protective effects against HCC development.
Our study has a few limitations. First, although this multicentre project included different types of samples, we were not able to observe the metabolic influence by liver cirrhosis alone because most HCC patients are cirrhotic. Therefore, larger studies covering patients with different disease severity are needed. Second, it was impossible to match the age of HCC patients and healthy controls because of the demographic features of HCC. That being said, our analysis did not suggest that the observed metabolite alterations were due to age (online supplemental figure 6). Third, portal vein and central vein samples were collected during surgical operations. It is unclear if surgical manipulation and anaesthesia treatment could affect the metabolites. Fourth, we used an independent validation cohort and in vitro studies to confirm our findings instead of correcting for multiple testing, because the current sample size was insufficient to support correction for multiple testing, or many potentially relevant metabolites might be missed. Finally, the metabolic alteration had already happened at the time of the diagnosis in HCC patients. Therefore, larger functional studies in in vivo and in vitro are needed to identify the relationship between metabolic changes and HCC.
In conclusion, this study identified previously unreported metabolic alterations in patients with HCC based on the integrative metabolome analysis of serum (portal and central vein), liver tissue (paired HCC and normal control) and stool metabolites. In portal vein, the key humoral communication route between the intestine and liver, we identified elevated pathogenic and depleted protective metabolites, signifying their critical role in HCC development.
Acknowledgments
We thank all participants for their cooperation in this study.
Footnotes
JL and WG contributed equally.
Contributors: JL, VW-SW and JY were involved in study concept and design; WG and CL performed the bioinformatics analyses and metabolomics profiling analyses; HS, FH, JC, LX, HZ, MK and SS conducted clinical samples and information collection; JL, JZ, QL and XZ performed the functional experiments; JL drafted the manuscript; QX collected samples and commented on the study; VW-SW and JY supervised study and revised the manuscript.
Funding: The project was supported by Guangdong Natural Science Foundation (2018B030312009); RGC-CRF Hong Kong (C4041-17GF; C7026-18GF); RGC Theme-based Research Scheme Hong Kong (T12-703/19-R, T21-705/20-N); Vice-Chancellor's Discretionary Fund CUHK (4930775).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information. Data are available on reasonable request.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
This study was approved by the ethics committee of the Chinese University of Hong Kong (CREC 2018.463), Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine (approval number (2018)201) and The First Affiliated Hospital of Anhui Medical University (approval number PJ2019-01-06) and was performed in accordance with the principle of Helsinki Declaration II. Written informed consent was obtained from all the participants.
References
- 1. El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? where do we go? Hepatology 2014;60:1767–75. 10.1002/hep.27222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. European association for the study of the liver, Electronic address EEE . European association for the study of the L EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182–236. [DOI] [PubMed] [Google Scholar]
- 3. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 4. Kobayashi T, Aikata H, Kobayashi T, et al. Patients with early recurrence of hepatocellular carcinoma have poor prognosis. Hepatobiliary Pancreat Dis Int 2017;16:279–88. 10.1016/S1499-3872(16)60181-9 [DOI] [PubMed] [Google Scholar]
- 5. Beyoğlu D, Idle JR. The metabolomic window into hepatobiliary disease. J Hepatol 2013;59:842–58. 10.1016/j.jhep.2013.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao M, Zhao L, Xiong X, et al. TMAVA, a metabolite of intestinal microbes, is increased in plasma from patients with liver steatosis, inhibits γ-Butyrobetaine hydroxylase, and exacerbates fatty liver in mice. Gastroenterology 2020;158:2266–81. 10.1053/j.gastro.2020.02.033 [DOI] [PubMed] [Google Scholar]
- 7. Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-Induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013;499:97–101. 10.1038/nature12347 [DOI] [PubMed] [Google Scholar]
- 8. Bindels LB, Porporato P, Dewulf EM, et al. Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br J Cancer 2012;107:1337–44. 10.1038/bjc.2012.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwabe RF, Greten TF. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J Hepatol 2020;72:230–8. 10.1016/j.jhep.2019.08.016 [DOI] [PubMed] [Google Scholar]
- 10. Heller G. The COX proportional hazards model with a partly linear relative risk function. Lifetime Data Anal 2001;7:255–77. 10.1023/A:1011688424797 [DOI] [PubMed] [Google Scholar]
- 11. Erawijantari PP, Mizutani S, Shiroma H, et al. Influence of gastrectomy for gastric cancer treatment on faecal microbiome and metabolome profiles. Gut 2020;69:1404–15. 10.1136/gutjnl-2019-319188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun L, Xie C, Wang G, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med 2018;24:1919–29. 10.1038/s41591-018-0222-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tornqvist L, Vartia P, Vartia YO. How should relative changes be measured. American Statistician 1985;39:43–6. [Google Scholar]
- 14. Chiang JYL, Ferrell JM. Bile acid metabolism in liver pathobiology. Gene Expr 2018;18:71–87. 10.3727/105221618X15156018385515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perez M-J, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol 2009;15:1677–89. 10.3748/wjg.15.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung K-T, Gadupudi GS. Possible roles of excess tryptophan metabolites in cancer. Environ Mol Mutagen 2011;52:81–104. 10.1002/em.20588 [DOI] [PubMed] [Google Scholar]
- 17. Manieri E, Folgueira C, Rodríguez ME, et al. Jnk-Mediated disruption of bile acid homeostasis promotes intrahepatic cholangiocarcinoma. Proc Natl Acad Sci U S A 2020;117:16492–9. 10.1073/pnas.2002672117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Fu X, Van Ness C, et al. Bile acid receptors and liver cancer. Curr Pathobiol Rep 2013;1:29–35. 10.1007/s40139-012-0003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Tilborg D, Saccenti E. Cancers in agreement? exploring the cross-talk of cancer metabolomic and transcriptomic landscapes using publicly available data. Cancers 2021;13. 10.3390/cancers13030393. [Epub ahead of print: 21 Jan 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaisaingmongkol J, Budhu A, Dang H, et al. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell 2017;32:57–70. 10.1016/j.ccell.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohshima M, Sugahara K, Kasahara K, et al. Metabolomic analysis of the saliva of Japanese patients with oral squamous cell carcinoma. Oncol Rep 2017;37:2727–34. 10.3892/or.2017.5561 [DOI] [PubMed] [Google Scholar]
- 23. Fong MY, McDunn J, Kakar SS. Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One 2011;6:e19963. 10.1371/journal.pone.0019963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li C, Li Y, Sui L, et al. Phenyllactic acid promotes cell migration and invasion in cervical cancer via IKK/NF-κB-mediated MMP-9 activation. Cancer Cell Int 2019;19:241. 10.1186/s12935-019-0965-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim KR, Park HG, Paik MJ, et al. Gas chromatographic profiling and pattern recognition analysis of urinary organic acids from uterine myoma patients and cervical cancer patients. J Chromatogr B Biomed Sci Appl 1998;712:11–22. 10.1016/S0378-4347(98)00155-8 [DOI] [PubMed] [Google Scholar]
- 26. Kühn T, Stepien M, López-Nogueroles M, et al. Prediagnostic plasma bile acid levels and colon cancer risk: a prospective study. J Natl Cancer Inst 2020;112:516–24. 10.1093/jnci/djz166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ulanovskaya OA, Zuhl AM, Cravatt BF. Nnmt promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol 2013;9:300–6. 10.1038/nchembio.1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Opitz CA, Litzenburger UM, Sahm F, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011;478:197–203. 10.1038/nature10491 [DOI] [PubMed] [Google Scholar]
- 29. Uyttenhove C, Pilotte L, Théate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med 2003;9:1269–74. 10.1038/nm934 [DOI] [PubMed] [Google Scholar]
- 30. Xie X, Liu H, Wang Y, et al. Nicotinamide N-methyltransferase enhances resistance to 5-fluorouracil in colorectal cancer cells through inhibition of the ASK1-p38 MAPK pathway. Oncotarget 2016;7:45837–48. 10.18632/oncotarget.9962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tummala KS, Gomes AL, Yilmaz M, et al. Inhibition of de novo NAD(+) synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell 2014;26:826–39. 10.1016/j.ccell.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 32. Lu X, Yu H, Ma Q, et al. Linoleic acid suppresses colorectal cancer cell growth by inducing oxidant stress and mitochondrial dysfunction. Lipids Health Dis 2010;9:106. 10.1186/1476-511X-9-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dilzer A, Park Y. Implication of conjugated linoleic acid (CLA) in human health. Crit Rev Food Sci Nutr 2012;52:488–513. 10.1080/10408398.2010.501409 [DOI] [PubMed] [Google Scholar]
- 34. McCrorie TA, Keaveney EM, Wallace JMW, et al. Human health effects of conjugated linoleic acid from milk and supplements. Nutr Res Rev 2011;24:206–27. 10.1017/S0954422411000114 [DOI] [PubMed] [Google Scholar]
- 35. Benavente-García O, Castillo J. Update on uses and properties of citrus flavonoids: new findings in anticancer, cardiovascular, and anti-inflammatory activity. J Agric Food Chem 2008;56:6185–205. 10.1021/jf8006568 [DOI] [PubMed] [Google Scholar]
- 36. Wahle KWJ, Brown I, Rotondo D, et al. Plant phenolics in the prevention and treatment of cancer. Adv Exp Med Biol 2010;698:36–51. 10.1007/978-1-4419-7347-4_4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-325189supp001.pdf (3.4MB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information. Data are available on reasonable request.