Abstract
Lactobacillus helveticus is the dominant organism in natural starter cultures used for the production of typical Italian cheeses. In this study, 74 L. helveticus strains, isolated from grana and provolone cheese natural whey starters, were distinguished with respect to their origin by using both cell wall protein profiles and chemometric evaluation of some phenotypic parameters, such as the ability to acidify cultures and the presence of nonspecific proteolytic and peptidase activities. Cell wall protein patterns allowed L. helveticus strains to be distinguished with respect to their source of isolation. Among the different phenotypes studied, no single specific parameter permitted the two groups of strains to be separated. A good discrimination between the two groups of L. helveticus species was obtained by multivariate statistical techniques, which permitted the extraction of all of the discriminating information retained in the phenotypic activities. Associations between strain phenotype expression and dairy environmental ecosystem source are discussed.
Lactobacillus helveticus is a regular, non-spore-forming, gram-positive rod (16). It is a homofermentative thermophilic lactic acid bacterium used extensively for the manufacture of Swiss-type and aged Italian cheeses. In particular, L. helveticus has been found to be the prevalent species in natural starter cultures used for grana (1–3, 22) and provolone (21) cheese making. Natural starters constitute complex microbial associations that are characterized by the occurrence of various species and many biotypes as a result of a number of selective conditions persisting during the manufacturing process (2, 3, 21, 22, 28).
Despite the widespread use of L. helveticus in cheese technology, limited information regarding genotypic strain variations of this bacterium is available, and it exists for only a few strains (7, 8, 14, 17, 18, 20). However, various studies of the technological properties of L. helveticus strains have been carried out. Large differences in several technologically important strain characteristics, such as acidifying and peptidase activities, phage lysogeny, and cell wall protein patterns, have been found among L. helveticus strains isolated from natural dairy starter cultures (1, 5, 10–12, 23, 27, 30, 31). In addition, variability in phenotypic expression among L. helveticus populations obtained from single clone cultures has been observed in some cases (27, 32).
A variety of genotypic and phenotypic methods have been used to show that the dominant community of L. helveticus in various dairy starters is composed of different biotypes, which sometimes may be associated with the source of isolation (5, 11, 14, 26, 28). To our knowledge, only one study using a multivariate approach for the classification of natural starter cultures has been published (25).
The differences in surface layer protein profiles among the thermophilic lactobacilli have also been used as a tool for distinguishing L. helveticus from L. delbrueckii (13). Data suggested that the four different profiles of L. helveticus strains observed might be related to the different ecological niches from which they had been isolated (13).
The aim of this study was to discriminate a large number of L. helveticus strains with respect to their origin by using both cell wall protein profiles and the chemometric evaluation of some phenotypic parameters, such as proteolytic and acidifying activities.
MATERIALS AND METHODS
Strains.
A total of 74 L. helveticus strains from the collection of the Istituto Sperimentale Lattiero Caseario, Lodi, Italy, were used. The strains had been isolated from 7 different grana (32 strains) and 10 different provolone (42 strains) cheese natural whey starters, at different times and from different dairy plants, over the last 10 years. The strains had been classified by the following criteria: cell morphology, sugar fermentation profile, growth temperature range, optimal growth temperature, salt resistance, lactic acid isomer, production of NH3 from arginine, and CO2 production (16). Additional tests for the identification of the strains were performed by the use of the API 50 CHL system (BioMérieux, Marcy-l’Etoile, France) and a specific oligonucleotide probe (29). The strains were stored at −80°C in MRS broth (Difco, Detroit, Mich.) supplemented with 15% glycerol.
Revitalization of the strains.
The strains were revitalized in MRS broth at 42°C overnight and used as a 2% (vol/vol) inoculum in skim milk powder (Oxoid Ltd., Basingtoke, United Kingdom) reconstituted to 10% (wt/vol), adjusted to 6.8 pH with 0.1 M NaOH, and sterilized at 110°C for 30 min (sterilized skim milk [SSM]). The cultures were then incubated at 42°C.
Assay of acidifying activity.
Once coagulated, the cultures obtained were inoculated at 1% (vol/vol) in 50 ml of SSM and SSM fortified with 0.6% (wt/vol) yeast extract (Oxoid) (SSM-YE) and incubated at 42°C. Five-milliliter cultures with and without yeast extract were sampled at four successive time points: immediately after inoculation and at 3, 6, and 24 h postinoculation. The pH of each sample was measured (Metrhon 654 pH meter; Metrhon Ltd., Herisau, Switzerland); values were expressed as ΔpH, calculated as the difference between the value immediately after inoculation and the values at the three successive time points in SSM (SSM3, SSM6, and SSM24) and in SSM-YE (YE3, YE6, and YE24).
Assay of nonspecific proteolytic activity.
Revitalized MRS broth cultures were inoculated at 1% (vol/vol) in 10-ml volumes of SSM and incubated at 42°C for 7 days. After incubation, 2.5 ml of each lactic culture was removed; proteins were precipitated with trichloroacetic acid, and the samples were prepared according to the method of Church et al. (6) to determine the proteolytic activity with o-phthaldialdehyde (Sigma-Aldrich Srl., Milan, Italy) (24). Spectrophotometric determinations were done with a Hewlett-Packard (Waldbronn, Germany) model 8452A spectrophotometer at 340 nm. Values were expressed as milligrams of Gly in 5 ml.
Assay of peptidase activity.
Each revitalized culture was centrifuged (3,000 × g, 10 min, 4°C), and the pellet was washed twice with sterile water. The final pellet was resuspended in sterile 0.05 M buffered phosphate, pH 6.5, to obtain an optical density at 550 nm (OD550) of 1.25. The enzymatic activities of the 74 strains were evaluated with 0.656 mM solutions of the substrates Phe-Pro-βNa (PHEPRO), Arg-βNa (ARG), Lys-βNa (LYS), Leu-βNa (LEU), and Pro-βNa (PRO) (Bachem Feinchemikalien AG, Bubendorf, Switzerland) at pH 6.5 after 1 h of incubation at 37°C. Aminopeptidase activity was evaluated spectrophotometrically at 580 nm by the method of Bouquien et al. (4). The values were expressed as OD580s.
Extraction and analysis of cell wall proteins.
Each revitalized culture was centrifuged (3,000 × g, 10 min, 4°C), and the pellet was washed twice with sterile water. The final pellet was resuspended in 5 ml of sterile water to obtain an OD550 of 2.0. Cell wall proteins were extracted and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Gatti et al. (13).
Statistical analysis.
Data for acidifying activity at 3, 6, and 24 h in two different media and data for nonspecific proteolytic and peptidase activities were subjected to multivariate statistical analysis by the use of the PARVUS package (9). Seventy-four strains of L. helveticus (objects) and 12 variables were considered (Table 1). This data set was subjected to the following statistical analyses.
TABLE 1.
Acidification, nonspecific proteolytic, and peptidase activities of 74 strains of L. helveticus
| Activity | Variable | Units | Value (mean ± SD) for isolates from:
|
|
|---|---|---|---|---|
| Grana starters (n = 32) | Provolone starters (n = 42) | |||
| Acidifying | SSM3a | ΔpH | 0.22 ± 0.089 | 0.10 ± 0.073 |
| SSM6 | ΔpH | 0.67 ± 0.280 | 0.32 ± 0.282 | |
| SSM24a | ΔpH | 2.86 ± 0.537 | 1.78 ± 0.986 | |
| YE3 | ΔpH | 0.32 ± 0.107 | 0.39 ± 0.179 | |
| YE6 | ΔpH | 1.31 ± 0.421 | 1.66 ± 0.539 | |
| YE24 | ΔpH | 3.15 ± 0.128 | 3.03 ± 0.239 | |
| Nonspecific proteolytic | OPA | mg of Gly/5 ml | 580 ± 150.3 | 371 ± 233.7 |
| Peptidase | PHEPROa | OD580 | 2.05 ± 0.076 | 1.26 ± 0.432 |
| ARGa | OD580 | 0.28 ± 0.400 | 0.94 ± 0.656 | |
| LYSa | OD580 | 0.57 ± 0.117 | 0.80 ± 0.242 | |
| LEU | OD580 | 0.87 ± 0.477 | 1.68 ± 0.571 | |
| PRO | OD580 | 0.08 ± 0.059 | 0.13 ± 0.051 | |
Selected by stepwise LDA.
(i) Data standardization by normalization.
A normalization step (19) was applied to each variable in order to remove possible distortions arising from the different magnitudes of the numerical values associated with the different variables. This normalization (autoscaling) involved dividing each value of a given variable by the standard deviation of all the values of this variable over the entire sample collection period. After normalization, all variables had the same weight since they had a mean of 0 and unitary variance.
(ii) PCA.
Principal component analysis (PCA) (19) is an exploratory method of data analysis which, through the calculation of linear combinations of original variables, allows the number of dimensions in the data set to be considerably reduced while maintaining most of the original information of the data set, expressed as percent variance.
(iii) Stepwise LDA.
Stepwise linear discriminant analysis (stepwise LDA) (15) was applied to select the variables with the highest power of discrimination. It considers the ratio (Wilks’ lambda) between the generalized within-category dispersion and the total dispersion. In the development of the discriminant function, variables are added or deleted based on their effect on the Wilks’ lambda and on their significance as measured by a suitable F test.
(iv) LDA.
LDA (19) was used to classify each sample according to the distance (Mahalanobis distance) from the category centroid. The method was validated by the repeated evaluation set technique, in which a predefined percentage of objects (20%), selected at random, is removed from the data set and the delimiter between the two categories is calculated. Subsequently, the previously excluded objects are tested to verify the category to which they are assigned and a measure of prediction accuracy is recorded. This is repeated a very large number of times (1,000), and the final result, expressed as the predictive ability, defines the ability of the method to correctly predict the category of an unknown object.
RESULTS
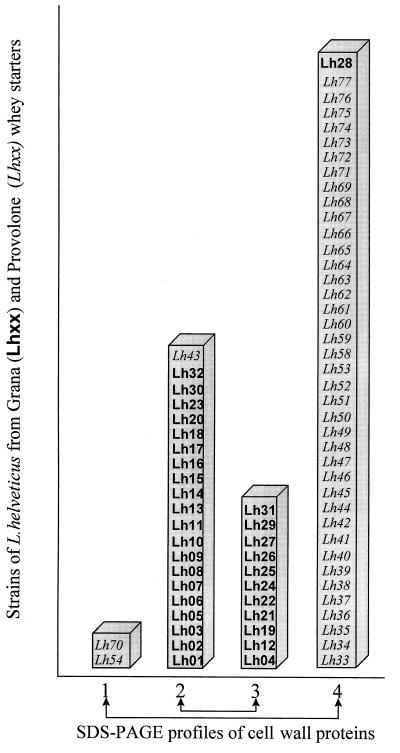
The analysis of cell wall proteins of 74 strains by SDS-PAGE showed four different profiles. Profile 1 revealed a protein with a molecular mass of about 50 kDa, profile 2 had an additional protein of about 66 kDa, profile 3 showed an additional protein of about 38 kDa, and profile 4 exhibited a protein with a molecular mass of about 50 kDa and numerous minor proteins of 10 to 100 kDa. These results are in agreement with those obtained by Gatti et al. (13) for a smaller number of strains of L. helveticus.
Figure 1 shows the distribution of the strains based on the four above-mentioned SDS-PAGE profiles. It is worth noting that profiles 2 and 3 included all of the strains deriving from grana cheese except for Lh28 while profiles 1 and 4 included all those deriving from provolone cheese except for Lh43.
FIG. 1.
Distribution of electrophoretic profiles of L. helveticus strains.
Table 1 shows the means and standard deviations for the acidification (SSM3, SSM6, SSM24, YE3, YE6, and YE24), nonspecific proteolytic (o-phthaldialdehyde), and peptidase (PHEPRO, ARG, LYS, LEU, and PRO) activities of the two groups of strains. By evaluating one variable at a time, the following conclusions can be drawn. The mean values determined for the ability of the strains isolated from grana cheese to acidify SSM not fortified with yeast extract seemed to be higher than the corresponding values for the strains from provolone cheese at the three sampling times. Despite this, due to the high standard deviation values, the two groups could not be distinguished on that basis. The acidification activities of the two groups were comparable during growth in SSM-YE. The mean values for indices involving all of the proteolytic activities could not be used to discriminate the origin of the strains due to the high degree of variation.
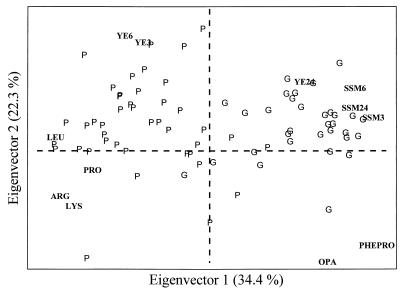
Multivariate statistical methods allowed the evaluation of the 12 variables simultaneously. Figure 2 shows the biplot on the first and second eigenvectors of both of the loadings of 12 variables and the scores of 74 objects (strains of L. helveticus) divided into two categories, grana (G) and provolone (P). The eigenvectors are new, uncorrelated variables derived from linear combination of the original ones, and their projection is the multivariate evolution of the variable-by-variable plot. Thus, each eigenvector does not represent a single variable but rather contains a contribution from each of the variables. The total percent variance explained by the first eigenvector is much higher than that explained by a single original variable.
FIG. 2.
Plot of loadings and scores for the first and second eigenvectors of PCA carried out on 12 variables and 74 objects.
Most of the strains deriving from provolone cheese whey starters tend to separate from the strains originating in grana cheese whey starters in the left region of the plot. The separation between the two categories occurs on the first component axis. It is worth noting that some variables almost coincide in the bidimensional plot; i.e., they have the same loading on both components and provide the same information.
The promising results obtained by PCA prompted us to apply LDA as a classification technique. Using this technique, 100% of grana strains and 92.9% of provolone strains were correctly identified.
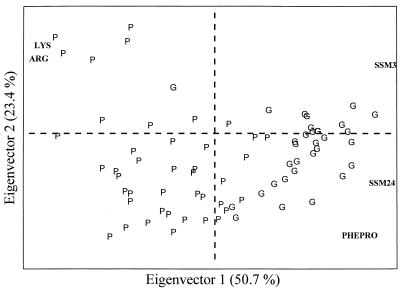
To verify whether the separation between categories could be obtained when using a reduced number of variables, without a noticeable loss of information, those variables with the greatest discriminating power were selected by using stepwise LDA. The principal components were recalculated on the reduced data set (74 objects and five variables).
It is worth noting (Fig. 3) that when only the data for SSM3, SSM24, PHEPRO, ARG, and LYS were used, a good separation between the two categories was demonstrated by the use of PCA.
FIG. 3.
Plot of loadings and scores for the first and second eigenvectors of PCA carried out on 5 variables and 74 objects.
LDA was also applied to the reduced data set. No difference in the ability of the model to accurately classify provolone strains (92.9%) was observed, but the accurate identification of grana strains was slightly lower (96.9%) than that obtained with the complete data set.
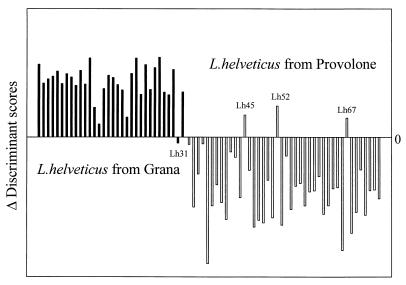
Figure 4 reports results obtained by LDA; the difference between the first and the second discriminant scores gives values higher than 0 for the samples classified in the category G, and it gives values lower than 0 for the samples classified in category P. Only one G strain (Lh31) and three P strains (Lh45, Lh52, and Lh67) were misclassified.
FIG. 4.
Histogram for the discriminant scores obtained by an LDA carried out on 5 variables and 74 objects.
Those strains misclassified by the LDA model showed cell wall protein profiles characteristic of their corresponding categories (Fig. 1). However, it should be noted that two strains with cell wall protein profiles uncharacteristic of their category were correctly classified by LDA.
LDA was validated by the repeated evaluation set technique, and the results, expressed as the percentage of strains correctly categorized, are reported in Table 2. The percentage of correctly classified objects is very satisfactory, and the value obtained for category G is comparable to that obtained by classification.
TABLE 2.
Prediction of origin of strains by LDA
| Origin of strains | Category | No. of predictions for category:
|
% Correctly predicted | |
|---|---|---|---|---|
| G | P | |||
| Grana whey starters | G | 6,147 | 212 | 96.7 |
| Provolone whey starters | P | 718 | 7,683 | 91.5 |
| Overall | 93.7 | |||
The results indicate that it is possible to distinguish between strains of L. helveticus originating in different cheese starters by the application of multivariate statistics to values obtained for certain phenotypic characteristics.
DISCUSSION
Previous studies have suggested that cheese technology parameters play a role in selecting dominant strains in natural dairy starter cultures. The relationship between the strain source and phenotypic activity has, however, to our knowledge, never been evidenced clearly. In particular, no published studies involving the use of a multivariate approach to investigate the complex microbial compositions of natural dairy starters are available. Our results show that cell wall protein patterns allow L. helveticus strains isolated from grana cheese natural whey starters to be distinguished from those isolated from provolone cheese natural whey starters.
Among the other phenotypes studied in this work (acidification capacity and proteolytic and peptidase activities), it was not possible to find one specific activity that permitted strains to be differentiated with respect to their source. On the contrary, a good discrimination between the two groups of L. helveticus isolates was obtained by applying multivariate statistical techniques, which permitted extraction of the discriminating information present in the phenotypic activities. Moreover, since the results obtained by the use of only 5 of 12 phenotypic characteristics were also satisfactory, it will be possible to reduce the time and the cost of the analysis by measuring only these variables.
It can be concluded that the predominant biotypes of L. helveticus in various natural starters differ depending on their original habitat. Therefore, the particular habitat plays a role in the selection of biotypes with physiological properties of technological significance.
The approach used in this study appears to be a promising tool for characterizing natural dairy starter cultures and is a step toward a better understanding of the functional and ecological significance of the presence of different biotypes in these natural microbial ecosystems.
ACKNOWLEDGMENT
We thank Riccardo Leardi (Department of Food and Pharmaceutical Chemistry, Genoa University) for critical review of the statistical aspects of the manuscript.
REFERENCES
- 1.Bosi F, Bottazzi V, Vescovo M, Scolari G L, Battistotti B, Dellaglio F. Batteri lattici per la produzione di formaggio grana. I. Carattezzazione tecnologica di ceppi di lattobacilli termofili. Sci Tecn Lattiero Casearia. 1990;41:105–136. [Google Scholar]
- 2.Bottazzi V, Sarra P G, Vescovo M, Bersani C. Caratteristiche dei bacilli lattici presenti nelle colture naturali in siero. IV. Saggi per la caratterizzazione delle colture. Sci Tecn Lattiero Casearia. 1978;29:367–381. [Google Scholar]
- 3.Bottazzi V. Le caratteristiche della coltura naturale impiegata nella produzione di formaggio grana. Sci Tecn Lattiero Casearia. 1981;32:418–434. [Google Scholar]
- 4.Bouquien C Y, Corrieu G, Desmazeaud M J. Enzymatic methods for determining populations of Streptococcus cremoris AM2 and Leuconostoc lactis CNRZ 1091 in pure and mixed cultures. Appl Microbiol Biotechnol. 1988;30:402–407. doi: 10.1128/aem.54.10.2527-2531.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carminati D, Mazzuccotelli L, Giraffa G, Neviani E. Incidence of inducible bacteriophage in Lactobacillus helveticus strains isolated from natural whey starter cultures. J Dairy Sci. 1997;80:1505–1511. [Google Scholar]
- 6.Church F, Swainsgood H E, Porter D, Catignani G. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci. 1983;66:1219–1227. [Google Scholar]
- 7.Drake M A, Small C L, Spence K D, Swanson B G. Differentiation of Lactobacillus helveticus strains using molecular typing methods. Food Res Int. 1996;29:451–455. [Google Scholar]
- 8.Dykes G A, von Holy A. Strain typing in the genus Lactobacillus. Lett Appl Microbiol. 1994;19:63–66. doi: 10.1111/j.1472-765x.1994.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 9.Forina M, Leardi R, Armanino C, Lanteri S. PARVUS: an extendable package of programs for data exploration, classification and correlation. Elsevier Scientific; 1988. Software, Amsterdam, The Netherlands. [Google Scholar]
- 10.Fortina M G, Rossi P, Mora D, Parini C, Neviani E. Slow milk coagulating variants of Lactobacillus helveticus. Folia Microbiol. 1996;41:33–38. [Google Scholar]
- 11.Fortina M G, Nicastro G, Carminati D, Neviani E, Manachini P L. Lactobacillus helveticus heterogeneity in natural cheese starters: the diversity in phenotypic characteristics. J Appl Microbiol. 1998;84:72–80. doi: 10.1046/j.1365-2672.1997.00312.x. [DOI] [PubMed] [Google Scholar]
- 12.Gatti M, Neviani E. Studio dei sieroinnesti naturali per grana mediante la tecnica conduttimetrica. Ind Latte. 1993;29:3–23. [Google Scholar]
- 13.Gatti M, Fornasari E, Neviani E. Cell-wall protein profiles of dairy thermophilic lactobacilli. Lett Appl Microbiol. 1997;25:345–348. doi: 10.1046/j.1472-765x.1997.00235.x. [DOI] [PubMed] [Google Scholar]
- 14.Giraffa G, De Vecchi P, Rossi P, Nicastro G, Fortina M G. Genotypic heterogeneity among Lactobacillus helveticus strains isolated from natural cheese starters. J Appl Microbiol. 1998;85:411–416. doi: 10.1046/j.1365-2672.1998.853464.x. [DOI] [PubMed] [Google Scholar]
- 15.Jenrich R I. Stepwise discriminant analysis. In: Enslein K, Ralston A, Wilf H S, editors. Statistical methods for digital computers. New York, N.Y: John Wiley & Sons; 1960. pp. 76–95. [Google Scholar]
- 16.Kandler O, Weiss N. Regular, nonsporing Gram-positive rods. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1208–1260. [Google Scholar]
- 17.Lortal S, Rouault A, Guezenec S, Gauitier M. Lactobacillus helveticus: strain typing and genome size estimation by pulsed field gel filtration. Curr Microbiol. 1997;34:180–185. doi: 10.1007/s002849900165. [DOI] [PubMed] [Google Scholar]
- 18.Manachini P L, Parini C. DNA restriction endonuclease patterns, DNA sequence similarity and phenotypic characteristics in some strains of Lactobacillus helveticus and Lactobacillus yogurty. Antonie Leeuwenhoek. 1983;4:143–152. doi: 10.1007/BF00393672. [DOI] [PubMed] [Google Scholar]
- 19.Massart D L, Vandegiste B G M, Deming S N, Michotte Y, Kaufmann L. Chemometrics: a textbook. Amsterdam, The Netherlands: Elsevier Science Publishers; 1988. [Google Scholar]
- 20.Miteva V I, Abadijeva A N, Stefanova T T. M13 DNA fingerprinting, a new tool for classification and identification of Lactobacillus spp. J Appl Bacteriol. 1992;73:349–354. doi: 10.1111/j.1365-2672.1992.tb04988.x. [DOI] [PubMed] [Google Scholar]
- 21.Neviani E, Forni E, Bossi M G, Carini S. Il siero-innesto per provolone: specie dominanti e loro attività biochimiche. Sci Tecn Lattiero Caseario. 1992;43:229–240. [Google Scholar]
- 22.Neviani E, Carini S. Microbiology of Parmesan cheese. Microbiol Aliments Nutr. 1994;12:1–8. [Google Scholar]
- 23.Neviani E, Divizia R, Abbiati E, Gatti M. Acidification activity of thermophilic lactobacilli under the temperature gradient of grana cheese making. J Dairy Sci. 1995;78:1248–1252. [Google Scholar]
- 24.Oberg C J, Weimer B C, Moyes L V, Brown R J, Richardson G H. Proteolytic characterization of Lactobacillus delbrueckii ssp. bulgaricus strains by the o-phthaldialdehyde test and amino acid analysis. J Dairy Sci. 1991;74:398–403. [Google Scholar]
- 25.Parente E, Rota M A, Ricciardi A, Clementi F. Characterization of natural starter cultures used in the manufacture of pasta filata cheese in basilicata (southern Italy) Int Dairy J. 1998;7:775–783. [Google Scholar]
- 26.Quiberoni A, Tailliez P, Quénéè P, Suarez V, Reinheimer J. Genetic (RAPD-PCR) and technological diversities among wild Lactobacillus helveticus strains. J Appl Microbiol. 1998;85:591–596. [Google Scholar]
- 27.Reinheimer J A, Morelli L, Bottazzi V, Suarez V. Phenotypic variability among cells of Lactobacillus helveticus ATCC 15807. Int Dairy J. 1995;5:97–103. [Google Scholar]
- 28.Reinheimer J A, Quiberoni A, Taillez P, Binetti A G, Suarez V B. The lactic acid microflora of natural whey starters used in Argentina for hard cheese production. Int Dairy J. 1996;6:869–879. [Google Scholar]
- 29.Taillez P, Ehrlich S D, Chopin M C. Characterization of IS1201, an insertion sequence isolated from Lactobacillus helveticus. Gene. 1994;145:75–79. doi: 10.1016/0378-1119(94)90325-5. [DOI] [PubMed] [Google Scholar]
- 30.Torriani S, Vescovo M, Scolari G. An overview on Lactobacillus helveticus. Ann Microbiol Enzimol. 1994;44:163–191. [Google Scholar]
- 31.Valence F, Lortal S. Zymogram and preliminary characterization of Lactobacillus helveticus autolysins. Appl Environ Microbiol. 1995;61:3391–3399. doi: 10.1128/aem.61.9.3391-3399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veaux M, Neviani E, Giraffa G, Hermier J. Evidence for variability in the phenotypic expression of lysozyme resistance in Lactobacillus helveticus. Lait. 1991;71:75–85. [Google Scholar]