Abstract
Coral holobionts are multi-species assemblages, which adds significant complexity to genotype-phenotype connections underlying ecologically important traits like coral bleaching. Small scale heterogeneity in bleaching is ubiquitous in the absence of strong environmental gradients, which provides adaptive variance needed for the long-term persistence of coral reefs. We used RAD-seq, qPCR and LC-MS/MS metabolomics to characterize host genomic variation, symbiont community and biochemical correlates in two bleaching phenotypes of the vertically transmitting coral Montipora capitata. Phenotype was driven by symbiosis state and host genetic variance. We documented 5 gene ontologies that were significantly associated with both the binary bleaching phenotype and symbiont composition, representing functions that confer a phenotype via host-symbiont interactions. We bred these corals and show that symbiont communities were broadly conserved in bulk-crosses, resulting in significantly higher survivorship under temperature stress in juveniles, but not larvae, from tolerant parents. Using a select and re-sequence approach, we document numerous gene ontologies selected by heat stress, some of which (cell signaling, antioxidant activity, pH regulation) have unique selection dynamics in larvae from thermally tolerant parents. These data show that vertically transmitting corals may have an adaptive advantage under climate change if host and symbiont variance interact to influence bleaching phenotype.
Subject terms: Ecological genetics, Animal breeding
Selective breeding of corals with different bleaching phenotypes demonstrates the potential for climate adaptation in vertically transmitting species.
Introduction
Reef-building corals are complex holobionts composed of a cnidarian host, symbiotic dinoflagellates in the family Symbiodiniaceae and a suite of other micro-organisms. Each compartment of the holobiont interacts to dictate trait values like thermal tolerance, which is increasingly important for ecosystems facing climate change. Corals are susceptible to heat stress, which leads to the breakdown of the coral-algal symbiosis and compromises the structure and function of reefs1,2. Despite these threats, there is substantial variation in thermal tolerance within and among populations that can exist over very small spatial scales3–9 and supports the adaptive potential of reef-building corals10–13. This variation is driven in part by symbiosis with diverse dinoflagellates in the family Symbiodinacea14–16, which have strong effects on holobiont physiology. The microbial consortia and its interactions with symbionts17 and the host also impact holobiont performance18,19 and may significantly complicate genotype-phenotype connections in corals.
Host genomic variation is an important driver of coral bleaching3,20,21, but previous research on the adaptive capacity of the coral host has focused almost exclusively on differences driven by strong selection across obvious environmental gradients5,20–24. This framework has also been leveraged in breeding experiments with mostly aposymbiotic larvae12,13,21, limiting our understanding of the adaptive potential of reefs over smaller scales (e.g., single reefs) where fertilization is most likely to occur. Within these more subtle environmental contours, variation in coral bleaching produced by symbionts and other demographic processes is still ubiquitous25.
Stable, heritable variation in thermal tolerance may be created by genomic variants which directly impact performance or indirectly influence symbiont community in brooding or vertically transmitting corals26, representing an alternate genotype-phenotype connection established via the symbiosis. These factors represent a mostly unknown reservoir of resilience in the absence of a priori environmental correlates that may be increasingly important as reefs become more isolated in climate refugia27.
Montipora capitata is a cosmopolitan reef-building coral that is dominant in Kāneʻohe Bay, Oʻahu, Hawaiʻi. M. capitata is a vertically transmitting broadcast spawner which releases symbiont provisioned eggs, creating a tight link between host and symbiont26,28 which may facilitate ‘adaptive’ change by additive, diversified sources of resilience. Maternal inheritance of symbionts could also serve as a selective pressure on paternal genes that increase fitness when paired with specific types of Symbiodiniaceae, fortifying the intergenerational connection between host and algae within individual holobionts. Bleaching phenotype in this M. capitata population is primarily driven by symbiosis with Cladocopium or Durusdinium29,30, where individual corals typically harbor Cladocopium and are more thermally sensitive (‘bleached’ phenotype) or a mixed community of both genera and are more thermally tolerant (‘nonbleached’ phenotype)31. Symbiont communities in this population also covary with host pigments32 and microbial communities33 and are broadly defined by depth and other environmental cues31. There are infrequent exceptions to these patterns29, but these relationships appear broadly fixed through time and multiple bleaching events30, suggesting that a functional connection between host genotype and symbiont constituency minimizes symbiont ‘shuffling’. Nested within this symbiont-phenotype framework, there is a strong host genotype effect on the biochemical signature of metabolites in these corals, including those generated by the symbionts34, highlighting the deep connection between the host and symbiont biochemistry in hospite.
This framework allows the ultimate phenotype in our study, bleaching tolerance, to have multivariate drivers including cnidarian heat-stress response, symbiosis maintenance physiology, other microbial effects and their interactions. We evaluated transmission of symbiont communities, intrapopulation genomic variance and the consequent biochemical signatures that define coral thermal tolerance. We then bulk crossed gametes from each parental coral phenotype, documenting improved thermal tolerance in juveniles from tolerant parents, and used a select and re-sequence approach to define genomic variants associated with improved thermal tolerance in larvae, directly or via their impact on the symbiosis.
Results
Adult population genetics and symbioses
Adult coral phenotype in this system was driven by symbiont community and association with host genomic loci. Adult corals in this study were unique genotypes (Fig. 1a) from a single population (FST = −0.0029; NGSadmix K = 1). Phenotype explained 5% of variation, but was not a significant factor defining overall genetic patterns (PERMANOVA p = 0.276; Fig. 1b). Genetic distances between samples of M. capitata were approximately 4x smaller than between samples of M. capitata and M. flabellata from Kāneʻohe Bay. Admixture analysis resolved a single population and an analysis of 2 populations showed no signature of coral phenotype in admixture proportions.
Fig. 1. Genomic and symbiotic dynamics underly bleaching phenotype in Montipora capitata.
a Hierarchical clustering of genetic distances calculated from genotype likelihoods, with one sequenced pair of biological replicates to confirm the absence of clonality and 3 M. flabellata. b PCA of 20,407 loci, shaded by historical bleaching phenotype. c Symbiont communities in three haphazard replicates of each colony used in this study, shaded by proportion of Cladocopium or Durusdinium measured via qPCR.
Corals from the nonbleached phenotype were dominated by Durusdinium, frequently with background levels of Cladocopium at <10% abundance (Fig. 1c). Conversely, all Bleached phenotype corals harbored primarily Cladocopium, except colony 11 which contained Durusdinium in 2 replicates. Replicate sampling suggested occasional heterogeneity in symbiont genera within colonies. There were several exceptions to this pattern: colony 210 and 4 harbored substantial Cladocopium communites while colony 11 contained nearly 50% Durusdinium (Fig. 1c). We took advantage of these exceptions to parse binary phenotype effects from host-symbiont effects which subsequently have a strong role in phenotype using association testing.
Genetic association with phentoype and symbiont community
We examined genetic association of individual loci with binary phenotype assignment (Bleached, Nonbleached) and detected no private alleles and no fixed loci, similar to21, suggesting incomplete segregation of the phenotypes. We detected 36 loci (0.176% of loci analyzed) significantly associated with phenotype (p < 0.0001; Supplementary Table 3). We used the LRT values for 20,407 loci as ‘heats’ to find functional groups corresponding to binary phenotype. Several ontologies (n = 28) were significantly enriched in loci with high association values (FDR p < 0.1), indicating an elevated influence on phenotype compared to background. These ontologies included protein metabolism, localization and modification, GTPase activity, ribosomal structure, cell-signaling/Erk Cascades, exopeptidase activity and endosomal membranes (Supplementary Table 4). Conversely, steroid metabolism, innate immune response, developmental regulation, cell cycling, downregulation of cell growth, the endoplasmic reticulum, ds-DNA binding, coreceptor binding and calcium ion binding were enriched in low LRT values, suggesting a significant similarity between phenotypes in certain functions or processes that do not influence thermal tolerance (n = 48 ontologies; Supplementary Table 5).
We then examined genetic association with mean proportion Durusdinium as the dependent variable, using a subset of loci which passed continuous association filtering requirements (n = 10,803). This analysis leverages exceptions to phenotype-symbiont patterns to decouple a priori phenotype assignments from association testing. Although binary phenotype and symbiont association LRT values were correlated (lm p < 0.001, R2 = 0.408), 75% of the loci important for distinguishing binary phenotype were not significantly associated with symbiont state (Fig. 2 inset). We repeated the GO_MWU enrichment analyses using effect sizes and LRT as heats, concatenated outputs and found 40 gene ontologies significantly enriched in loci with which determined symbiont state (Supplementary Table 6). Of the 28 ontologies associated with overall phenotype, only 5 (17%) were also significantly enriched in the symbiont association testing analysis: Peptide Biosynthetic Process, Cellular Amide Metabolic Process, Regulation of Erk1 and Erk2 Cascade, Exopeptidase Activity and Aspartic-Type Exopeptidase Activity (Fig. 3a red GOs).
Fig. 2. Comparison of genetic associations with binary phenotype and symbiont state.

Points represent LRT values from two association tests: case-control association by allele frequencies using phenotypes as the dependent variable and continuous association by gene dosage using proportion Durusdinium as the dependent variable. Points (n = 10,803) represent loci shared between the two analyses, excluding those which were filtered to complete continuous association. Point color represents absolute effect size (beta) from symbiont association. Gray lines represent independent LRT value thresholds at p = 0.0001 determined by 10 k permutations. Venn diagram contains counts of these significant loci for phenotype, symbionts and shared between the two. Black line is x = y.
Fig. 3. Functional ontologies distinguish bleaching phenotype and relate to biochemical patterns.
a The 28 gene ontologies that were significantly enriched in loci highly associated with bleaching phenotype (FDR p < 0.0001). Points represent each phenotype in the study (mean ± 1 SE) along the first principal component that describes all loci associated with the ontology of interest. Number of loci corresponding to each ontology are in parentheses, red ontologies are significantly enriched in loci with high associations with symbiont state and thus impact phenotype via symbiosis. b WGCNA modules of the corals in this study, shaded by correlation with loading in (a). Each column represents a metabolite module resolved by WGCNA and each row corresponds to gene ontologies to the left. No correlations were significant after FDR correction.
For gene ontologies significantly enriched in high association values for phenotype, we extracted all loci annotated as each GO (n = 11–565; Fig. 3a) and used PCAngsd to summarize genetic variation in these functions. This genetic variation broadly impacted the biochemistry of corals in this study (Fig. 3b), especially in functions related to Protein Deacetylation, Erk Cascade Regulation and Protein Tyrosine/Serine/Threonine Phosphatase. Erk Cascade Regulation was also an ontology that significantly described symbiont state.
Genetic-metabolomic correlates
There were high correlations between metabolite Module-4 and each of these gene ontologies (R > 0.5, p < 0.0005). While these correlations were not significant after multiple comparisons correction, the conserved response of M-4 with multiple phenotype-defining gene ontologies suggests that this pattern is biologically important. Module-4 was significantly enriched in peptides, dipeptides, N-acyl-alpha amino acids and Trifluoromethylbenzenes (FDR p < 0.03). Module-5 was also highly correlated (R > 0.5) with Protein Phosphatase Inhibition Activity, Protein N-linked Glycosylation and GTPase activity and significantly enriched in menaquinones and prenol lipids (FDR p < 0.02). The betaine lipid identified in34, an important molecule for distinguishing phenotype in this species, was in Module-7.
Betaine glycine
Among the top 36 individual loci for defining phenotype (LRT permutation p < 0.0001), two were on Chromosome 11 separated by approximately 11 kb (Chr11_4203672 and Chr11_4215008). These loci were strongly linked (GL R2 = 0.87), suggesting coinheritance and selection on nearby genes. We annotated a 50 kb window centered on the variants of interest (following21) using blastn. Several predicted genes were within this window, including glycine sarcosine dimethlycine N-methyltransferase-like (GSMT/SDMT, e = 2e-105). The product of GSMT/SDMT is Betaine Glycine (BG), which we documented in our metabolite dataset. BG was more abundant in nonbleached corals than their bleached counterparts (Wilcox test p = 0.005; Supplementary Fig. 4); however, colony 203 was an extreme outlier in the bleached phenotype. If this colony was excluded, concentrations were 22.8% higher in Nonbleached corals (Wilcox test p < 0.001). We examined loci annotated as ‘Amino-acid Betaine Metabolic Process’ (GO: 0006577, n = 10) with a Wilcox test and found a significantly higher LRT than other ontologies (p = 0.013) in the binary phenotype analysis, suggesting this ontology is enriched in loci that differentiate phenotypes and symbiont communities. The effect size derived from association testing of symbiont state was 26.8% larger in this window than across the rest of the genome (Wilcox p < 0.001). These loci were not included in the association testing for symbiont type due to data quality filtering.
Larval and juvenile survivorship
To assess the adaptive potential of this symbiotic and genetic variation, we selectively crossed gametes from the adult corals on 13 July 2018, when all 22 colonies in the study spawned. A total of 11 colonies of each phenotype were bulk fertilized, allowing for up to 110 potential crosses in each pure phenotype and 462 crosses in the site-wide control, excluding selfing. Larval survivorship was significantly higher at ambient temperatures throughout the timeseries (Supplementary Fig. 5; Supplementary Table 7; PLRT < 0.01) and at the endpoint (Fig. 4a; p < 0.001). There were no significant differences in endpoint survivorship at ambient temperatures between phenotypes (p > 0.99). Final survivorship averaged between 36.4–40.0% in the ambient treatments.
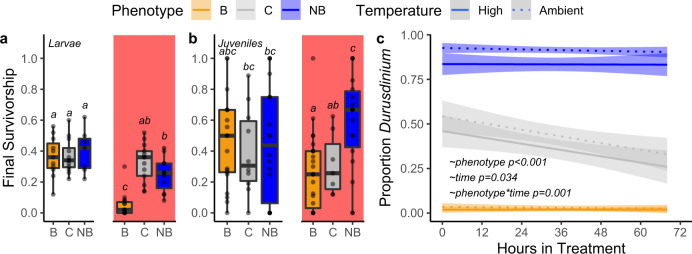
Fig. 4. Fitness and Symbiont Dynamics in Larval and Juvenile corals.
a Final survivorship 213 h post fertilization for larvae at ambient and high temperatures. b Final survivorship 34 days post settlement for juveniles at ambient and high temperatures. Boxplots are mean 1± IQR, red fill denotes high temperature treatments. c Proportion Durusdinium of the three phenotype larval pools from fertilization to 84hpf in ambient and high temperatures. Line is best-fit with 95% confidence interval. Letters represent post hoc significance values for each life-history stage.
Larvae produced from bleached parents had substantially and significantly lower survivorship (4.9 ± 2.0%; mean ± 1SE) than larvae from nonbleached parents (24.9 ± 3.4%, ~5-fold difference, p < 0.001) and the cross phenotype (32.6% ± 3.4%, p < 0.001). At high temperatures, survivorship in the cross treatment was ~7% higher than the nonbleached phenotype but was not significantly different (p = 0.697). Phenotype explained 96.3% of survivorship variance in high treatments (H2 = 0.963), and 17.3% in ambient treatments (H2 = 0.173).
In 1.5-month-old juveniles, survivorship was significantly higher at ambient than high temperatures throughout the timeseries (Supplementary Fig. 6; Supplementary Table 8; PLRT < 0.01) and at the endpoint (Fig. 4b; p = 0.040). There were no significant differences in ambient endpoint survivorship between phenotypes (p > 0.99). Nonbleached juveniles had higher survivorship than the site-wide cross (p = 0.048) or bleached phenotype (p = 0.001, ~2-fold difference) under heat stress (Fig. 4b). Final survivorship under heat stress for the cross and bleached phenotype were not significantly different (p = 0.721). Further, nonbleached juvenile survivorship under temperature stress was not significantly different than either pure phenotype in ambient condition (p > 0.19). Phenotype explained 92.9% of variance in high treatments (H2 = 0.929), and 0.2% in ambient treatments (H2 = 0.002).
M. capitata releases symbiont provisioned eggs which contribute strongly to downstream thermal tolerance, so we also quantified symbiont community over time in larvae using qPCR. Treatment did not have a significant main effect on symbiont community (beta-regression; 2 = 1.62, p = 0.203). The symbiont communities were strongly differentiated between phentoypes (beta-regression; 2 = 146.6, p < 0.001) and there was a significant interaction of phenotype, treatment and time (beta-regression; 2 = 13.2, p = 0.040) with Durusdinium or Cladocopium dominating the nonbleached and bleached larvae, respectively (Fig. 4c). Nonbleached larvae typically had background levels of Cladocopium, but bleached larvae typically did not have any Durusdinium. The site-wide control cross has an approximately equal proportion of each genus at the initiation of heat stress, but a substantial decline in proportion Durusdinium occurred in the ambient treatments of the control cross (beta-regression, z-ratio = 3.60, p = 0.056); while this pattern was very similar in the high treatment, the only significant declines occurred between 0 and 44 h (beta regression, z-ratio = 6.04, p < 0.001). The bleached (beta-regression, z-ratio < 2.28, p > 0.82) and nonbleached (beta-regression, z-ratio < 1.41, p > 0.99) crosses remain dominated by their initial symbiont communities.
Larval selection patterns
We used a select and re-sequence approach in larvae in this study to define genetic response to heat selection, examining the PBS of 31,794 loci shared between the site-wide cross and nonbleached pools. PBS values from the control phenotype explained 33.2% of variance from the nonbleached phenotype (lm p < 0.001, Supplementary Fig. 7), but mean PBS was 65% higher in the cross phenotype compared to nonbleached (Wilcox p < 0.001).
After mapping these loci and retrieving gene ontologies, there were 108 and 51 ontologies significantly enriched for selected loci (high PBS values) in the site-wide cross and nonbleached phenotypes, respectively. Four of these significant functions were shared between phenotypes (FDR p < 0.1, Fig. 5 inset): oxidation-reduction processes (GO:0055114), stress-activated protein kinase signaling cascades (GO:0031098), positive regulation of cell-cell adhesion (GO:0022409) and ubiquitin-dependent protein catabolic processes (GO:0043162). Overall, GO ranks within each phenotype were significantly correlated in the global dataset, but had virtually no explanatory power (Fig. 5 gray/black; lm p < 0.001, R2 = 0.006). The four ontologies that were significantly enriched in selected for both phenotypes were highly coordinated (Fig. 5 tan; n = 4, lm p < 0.001, R2 = 0.985).
Fig. 5. Selection Dynamics in Larval heat stress experiment.
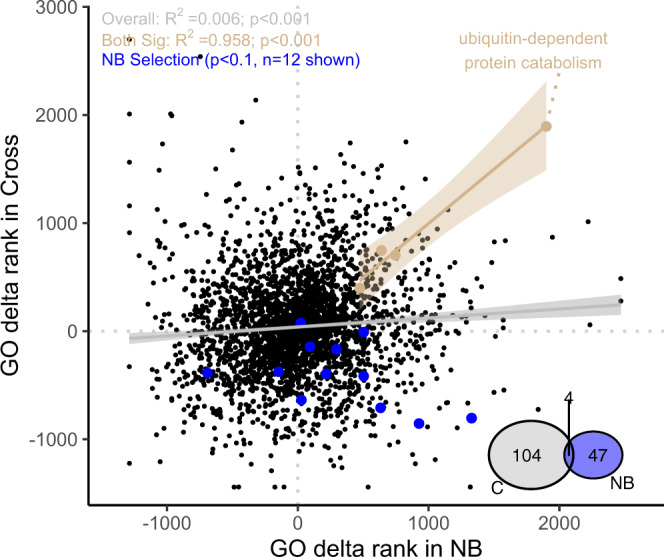
GO delta rank derived from PBS statistics of cross and nonbleached larval pools. High values represent gene ontologies enriched in large PBS statistics. Tan points represent the 4 gene ontologies that were significant (FDR p < 0.1) in both cross and nonbleached phenotypes, with lm best fit line. Black points represent the ontologies that were not significant in both treatments, with gray lm best fit line. Blue points represent gene ontologies with enrichment in Nonbleached phenotype and delta rank <100 (not significant) In the cross phenotype, isolating the genomic effect of selective breeding. Venn diagram shows shared significant ontologies between phenotypes.
Twenty functional categories were significantly enriched for large difference between PBS values in the nonbleached and cross phenotypes, representing heat selection specific to the thermally tolerant population. These categories included antioxidant activity, protein serine/threonine/tyrosine kinase activity, nonsense mediated decay regulation and several ontologies related to pH regulation (Fig. 5 blue points, Supplementary Table 9).
Discussion
The long-term persistence of coral reefs requires adaptive differences based on heritable variation which keeps pace with a changing climate. Here we demonstrate a shared host genomic and symbiotic foundation for intrapopulation variance in thermal tolerance on a single reef in the absence of differential selection. These additive sources of resilience lead to increased thermal tolerance in the juvenile stage of subsequent generations and produce high heritability values which may contribute to adaptation on reefs. Although only ~20% of broadcast spawning corals are vertical transmitters35, these species may have an adaptive advantage under climate change if host and symbiont components of the holobiont interact to contribute to phenotype.
M. capitata colonies in Kāneʻohe Bay exist over a genetic and environmental mosaic36 encompassing a range of symbiont states31 and microhabitats. To focus on bleaching phenotype, we identified colonies from the extremes of the thermal tolerance continuum, representing non-random selection of corals within the population. Despite skewing our collections in this manner and the strong symbiotic and biochemical differentiation between phenotypes30,34, we found low population stratification and relatively little genome-wide variance explained by phenotype. To eliminate the potential for cryptic speciation, which can align with thermal tolerance and symbiont communities (including in Montiporids)37–39 we compared our samples to the congener M. flabellata from Kāneʻohe Bay. Each population structure approach used here, including admixture analysis, FST calculations, PERMANOVA and interspecific distances indicated corals were from a single population of M. capitata, with genetic differentiation at least four-fold smaller than the distance between species.
The low genetic differentiation between phenotypes simplifies association testing within species, which we conducted for two traits: binary phenotype assigned during the 2015 bleaching event and proportion Durusdinum. Many canonical heat-stress response pathways distinguished these phenotypes and symbiont communities. This result in the absence of differential selection (e.g., across environmental gradients) supports the adaptive capacity of phenotypic variation over very small scales, directly or proximally via symbiont community. Ontologies relating to growth/development (GO:0005138;GO:0070851), cell-signaling (GO:0008138), ribosomal constituents (GO:0003735), GTPase activity (GO:0051020) and protein localization, modification and breakdown (GO:0006518; GO:0043043; GO:0043603) have been documented extensively in previous work on both genomic and transcriptomic impacts of thermal tolerance25,40. We documented a single cellular compartment, the late endosome which is part of the response to a broad variety of environmental stressors in corals40. Interestingly, mitochondrial effects were not apparent in this dataset, although they have previously been implicated in larval survivorship and selection21.
We also show that certain gene ontologies confer bleaching tolerance (the overall phenotype) via host interactions with the symbionts. Five ontologies were significantly enriched in both the overall phenotype association and the symbiont proportion association, including peptide biosynthesis, Erk Cascade regulation and exopeptidase activity. ERK cascades phosphorylate hsp7041 and are central in extracellular signaling pathways, other components of which are also significantly enriched for high PBS values in this analysis (e.g., GTPase binding, Kinase activity, Phosphatase activity). However, only 10 of 40 loci (25%) and 5 of 28 gene ontologies (17%) shared significance between the association testing for binary phenotype and testing for proportion Durusdinium, supporting the hypothesis that host and symbionts contribute directly and indirectly to bleaching phenotype via a range of mechanisms.
These data also illustrate the biochemical consequences of functional variation which define bleaching phenotypes, including protein modification (i.e., protein phosphatase activity) and cell-signaling related to stress (i.e., ERK1/2 cascade). Interestingly, the molecular classes correlated with functional drivers of phenotype are decoupled from the metabolites which directly differentiate phenotype in a global analysis. For example, ref. 34 showed that DGCC lipids and steroids were disproportionately important for distinguishing phenotype and that fatty acids, monoacylglycerides, nucleotides, steroids and xanthins showed phenotype-specific responses to thermal stress34. None of these molecular classes are significantly enriched in the metabolite modules that correlate with gene ontologies defining phenotype or symbiont community in this study. We hypothesize that this difference reflects the host-specific regulation of physiology and biochemical patterns, rather than broader molecular patterns due to symbionts or the interaction of symbionts and the host. In particular, the conserved importance of protein kinases and phosphatases in biochemical patterns and thermally tolerant larval selection highlight the importance of this signaling mechanism in defining thermal tolerance. Although the correlations between GO loadings and metabolite module abundance were not significant after multiple comparisons correction, the congruent response of many molecules with several ontologies suggests that this is a biologically meaningful pattern, representing an important link that defines the downstream consequences of genomic variants besides eQTLs37.
There was also a link between two loci strongly predictive of bleaching phenotype and amino acid modifications and metabolism, potentially via Betaine Glycine (BG). Betaine Glycine is abundant in corals42,43, which have BG metabolism and biosynthesis pathways44, and can be found in Montipora capitata in Kāneʻohe Bay42,45. BG is an amino-acid osmolyte that has well-documented protective effects on plants during abiotic stress46,47 and is used in transgenic crop research48, where it may also counteract cellular signals that initiate HSP70 expression49. Ref. 45 also showed that osmolyte production, including BG, increased during temperature stress in M. capitata. Although the source of BG is unclear (i.e., host or symbiont44), the much higher abundances in bleaching resistant corals could represent a form of biochemical frontloading. This molecule is particularly important for the interaction of light and other abiotic stressors, including in corals43, which is closely aligned with the photosensitive nature of coral bleaching under high temperatures. Under these conditions, the repair of the D1 protein of photosystem II in symbionts is impaired, but the accumulation of BG prevents this obstruction and thereby limits oxidative stress. To the best of our knowledge, there is no known synthetic or metabolic connection between betaine glycine and the betaine lipids identified in ref. 34.
Symbiont communities are maternally inherited in M. capitata50 and appear fixed through time30, so BG variation between symbionts/phenotypes could be (a) due to genomic variation which impacts the ability of a genotype to harbor Cladocopium or Durusdinium, (b) due to differential symbiont physiology34 or (c) host plasticity induced by differential symbionts51,52. Another possibility is that cell-signaling (ERK/MAPK) implicated in both adult phenotype and larval selection analyses impact osmolyte cascades, which may play a role in bleaching53.
If host genomic variants drive this pattern, BG abundance could be influenced by selection not on variants in GDMT/SDMT itself, but some other regulatory machinery in close proximity, including PAX-C, or one of the methyltransferases, which could produce the same effects. We also observed a relative shift in the fitness of the site-wide cross between larval and juvenile stages. As larvae, the site-wide cross phenotype had slightly higher survivorship than pure nonbleached under heat stress, which could be an experimental artifact or an effect of increased genetic diversity in a cross with many more potential genetic combinations. After larvae settled and metamorphosed, this fitness advantage in the site-wide cross was lost and the pure nonbleached phenotype was significantly higher. We hypothesize that this decline is driven by the decreasing proportion of Durusdinium in the site-wide cross phenotype, which can be seen during the first 4 days of larval development in both ambient and high temperatures. This decline could be due to ‘hybridization’ between the two phenotypes which resulted in changing proportions of symbionts or differential selection due to heat stress; however, Durusdinium did not decrease in pure nonbleached corals with a small background of Cladocopium, supporting the explanation that the introduction of nonbleached eggs and bleached sperm lead to a decrease in fitness, regardless of temperature. This ‘underdominance’ response may occur in inter-specific hybridization experiments54 and warrants further examination of speciation and genetic differentiation in this system. An alternate hypothesis is that the breakdown of coadapted gene complexes had a delayed impact on phenotype which manifested after metamorphosis. It is also possible that fitness driven by the combination of paternal genes and symbiont leads to differential selection, which could be resolved in future experiments using indivudal crosses.
Selection on diverse host functional genes also occurred during larval development. We documented significantly enrichment of high PBS values in both cross and nonbleached phenotypes for cell-cell adhesion (GO:0022409), stress-activated protein kinase signaling cascades (GO:0031098) and ubiquitin-dependent protein catabolic processes (GO:0043162), which reflect major functions that are impacted regardless of background genetics and correspond to well-known heat-stress responses in corals40. Signaling cascades were also implicated in differentiation of adult phenotypes, supporting the role of the cellular processes that govern interactions betwen the host and symbionts or coordinate gene complexes in response to heat stress55,56.
Importantly, less than 10% of significant ontologies in this anlaysis were shared between the two phenotypes, indicating differential selection trajectories. These differences may be due to epistatic effects arising from the polygenic nature of thermal tolerance that vary between phenotypes or the influence of the symbiont communities on larvae. Notably, a broader array of loci and gene ontologies are selected in cross corals and they are selected more strongly, indicating that thermally tolerant corals may be ‘preadapted’ to dealing with some thermal stress, corresponding closely with ref. 13. The strong nonbleached-specific selection of twenty functional categories supports this hypothesis and, although we cannot evalute classical selective breeding approaches based on host genetics, emphasizes the importance of host heritability underlying thermal tolerance. If symbiont and genomic context are indeed important for selection trajectories, the outcomes of adaptation of vertically transmitting corals under climate change may be significantly more complex than those of horizontal spawning corals.
The role of different symbionts in defining thermal tolerance has been documented in many studies in adults, but most corals have azooxanthellate larvae, so the role of infection (or transmission) of different symbionts in the larval stage remains poorly defined57, although these differences impact physiology58 and can dramatically alter host gene expression51,52. Juvenile Acropora spathulata infected with Durusdinium trenchii bleached less than those infected with other symbionts59, especially when one or both parents were from warm-adapted reefs, which closely mirrors survivorship patterns in juveniles in our study, assuming the Cladocopium:Durusdinium ratios found in larvae were maintained. While this system does not allow us to isolate host effects, the phenotypic variance explained by genotype and selection against Durusdinium in larvae strongly suggests that host and symbiont affects both impact thermal tolerance.
The data presented here document the synergistic impact of host genome and symbiont community on coral phenotype in the absence of differentiating selection. This variance is the raw material upon which adaptation under climate change will act if previously bleached corals are the first to succumb to repeated bleaching events. Our data also suggest that the raw materials for adaptive recombination need not occur over large environmental gradients and that symbiosis ecology and demographic processes can generate this variance on individual reefs.
Methods
Collections
In 2015, the summer thermal maximum produced a mosaic of bleaching in Kāneʻohe Bay, where adjacent colonies either severely bleached or remained visually healthy29,60,61. The majority of these colonies recovered and have been tracked through time (Supplementary Fig. 1). We selected 22 healthy colonies (11 pairs) along a 2–3 m depth contour on Reef 13 in Kāneʻohe Bay and used them for genetic, metabolomic and gamete collections for selective breeding. We collected gamete bundles from each colony in situ on 13 July 2018 and re-collected 3 replicate branches for adult genetics and symbiont extractions in August 2018 (to avoid spawning impacts). We collected metabolite samples from each colony in May-September 201934. To examine species boundaries in the context of our phenotypes, we also collected 3 fragments of Montipora flabellata from northern Kāneʻohe Bay in July 2021. Adult fragments were stored at −20 °C until processing.
Crosses, fertilization and treatments
All 22 colonies in the study spawned on 13 July 2018. We created a site-wide cross (gametes from all-colonies in the study), bleached phenotype cross (only gametes from historically bleached parents) and nonbleached phenotype cross (only gametes from historically nonbleached parents) prior to the breakup of bundles and fertilization (Supplementary Fig. 1). Gamete bundles from each bulk cross were immediately aliquoted (n = 10) for fertilization62 and returned to the Hawaii Institute of Marine Biology. Triplicates of fertilized embryos from each phenotype were added to one 150 L larval rearing conical and allowed to develop for 12 h to the prawn-chip stage before temperature treatments (ambient: 27.4 ± 0.55 °C; mean ± 1 SD and high: 30.2 ± 0.51 °C) began for each of three phenotype pools (Supplementary Fig. 2).
Larval Survivorship, Juvenile Settlement and Survivorship
We established replicate survivorship trials at 12 h post fertilization (hpf; n = 15 per phenotype and temperature) and measured larval survivorship at 27, 47, 75, 96, 120, 172 and 213 (hpf) (Supplementary Fig. 2). At 109hpf, larvae from each phenotype at ambient temperatures were transferred to settlement chambers (n = 4) and allowed to settle on pre-conditioned aragonite plugs. After 8 days exposure to substrate, plugs (n = 877) were randomly allocated into ambient (27.6 ± 0.96 °C) and high (30.1 ± 0.38 °C) temperature treatments (n = 2 per temperature) (Supplementary Fig. 3).
Settlement plugs were photographed at the initiation of temperature treatments (12 days post fertilization) and at 8, 14, 18, 27 and 34 days in treatments. Individual settlers were counted from photos at each timepoint. The removal of larvae for settlement did not interrupt the ongoing larval survivorship trials. See Supplementary Fig. 2 for details.
Larval and juvenile survivorship was analyzed with Kaplan-Meier regression in the R package Survival, with pairwise comparisons between temperature and phenotype crosses using a likelihood ratio test (implemented by Survminer::pairwise_survdiff) with Bonferroni adjusted p-values. We used a generalized linear model examining final survivorship (averaged per replicate or plug) with temperature and phenotype as fixed effects and a quasibinomial family. We calculated broad-sense heritability of both larvae and juveniles (excluding the site-wide cross) separately at each temperature as variance (MSE) explained by Phenotype/total variance (MSE) in a 1-way ANOVA, modified from ref. 63.
Symbiont analysis
Larval symbiont collections were made at 16, 36, 60 and 84 h post fertilization, representing daily sampling that overlapped genomics sampling (Supplementary Fig. 2). Three replicates of 30–50 larvae were haphazardly collected from each treatment. Adult fragments and larval time-series collections were extracted with a CTAB-chloroform protocol (following22). Concentration of Cladocopium, Durusdinium and the coral host were quantified via quantitative PCR using actin and Pax-C assays29,64,65 on a StepOnePlus system (Applied Biosystems) with two technical replicates. Samples were re-extracted and analyzed if CT replicate standard deviation was greater than 1 between replicates or if only a single replicate amplified. CT values were corrected for fluorescence and copy number using the equations of ref. 29. See Supplementary Fig. 2 for details.
Library preparation and sequencing
Larval samples were collected from each conical (n = 6) at 16 hpf by collecting 5 replicates of 50 larvae (n = 250 total larvae per timepoint per treatment), preserved in 2% SDS in DNAB and stored at −20 °C until processing. A second sample was collected in the same way at 84 hpf, representing timepoints near the initiation of heat stress during late development and after the putative selective pressure had led to ~25% mortality in high treatments (Supplementary Fig. 2).
DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen) following manufacturer protocol. Extracted DNA was quantified with a Qubit fluorometer and 300 ng of DNA was digested with HindIII (New England Biolabs). Libraries (n = 23; 22 adult samples and an adult technical replicate, 60 larval samples) were prepared with an Illumina TruSeq Nano kit and TruSeq adapters for 350 bp inserts following manufacturer protocol. Samples were sequenced on a single lane of an Illumina HiSeq 4000 with paired end 150 bp reads (Genewiz, South Plainfield, NJ). To examine species boundaries, we also sequenced 3 samples of Montipora flabellata from Kāneʻohe Bay as part of a separate sequencing project (Illumina WGS, paired end 250 bp reads, Illumina NovaSeq 6000).
We prepared a reference genome by mapping the Montipora capitata genome to the chromosome scale scaffolding of Acropora millepora20 using default options in RagTag66, which resulted in ~5% higher alignment rates and higher contiguity. The Montipora capitata individual used for genomic sequencing showed signals of gene duplication67 and is ~885MB (compared to 458 MB for the A. millepora reference). We included un-anchored M. capitata sequences as additional contigs, effectively increasing the contiguity of the reference without removing any data. Short read alignment rates were substantially lower against the original A. millepora genome or against the chromosome scale M. capitata anchored alignment without additional contigs. We annotated predicted protein sequences from the M. capitata assembly67 using blastp against Uniprot/Swissprot and extracted gene ontologies using a custom script. We used annotations from the A. millepora assembly20 for scaffolded contigs and the additional annotations for unscaffolded M. capitata sequences using a custom R script.
Adult population genetics
Raw reads were demultiplexed at the sequencing facility and we trimmed adapters using cutadapt68 before aligning all samples using bwa mem69. We analyzed adult clonality with M. flabellata samples using a biological replicate in ANGSD70 to generate an identity by state matrix with filtered reads (minMapQ 30, minQ 30) and used hierarchical clustering to confirm species assignments and that there were no clonemates in our adult population. This analysis does not adhere to an evolutionary model (e.g., Ts/Tv rates or nucleotide frequencies) and therefore should not be interpreted as a phylogeny. We calculated FST between phenotypes from site frequency spectrums using ANGSD/realSFS. We used NGSadmix71 (K = 1, K = 2) to examine population structure with default settings. We examined population structure using principal components analysis using the predicted number of secondary alleles3 for each loci in each sample as input data. We examined variance explained by phenotype using a PERMANOVA in the R package vegan72.
Adult associations with phenotype and symbiont state
We used two association comparisons to distinguish (1) host effects on binary phenotype, (2) host-symbiont interactions free of phenotype assumptions. First, we examined the association of individual loci with each adult phenotype (n = 21, one sample removed with low depth) using genotype probabilities in ANGSD (GL 2, doAsso 1, doGeno 8, doPost 1, minDepth 5, SNP p value <2e−6) and a case-control framework for binary phenotype assignments. We permuted phenotype assignments between colonies 10,000 times, recalculated association metrics and selected the highest LRT value for any individual loci in each permutation. We assigned the 99.99th-percentile of this LRT distribution as the cutoff for significance (p < 0.0001) for individual loci following20.
To differentiate host-symbiont interactions from background genetic structure and phenotype assumptions, we reconducted the association analysis above (ANGSD GL 2, doAsso 6, doGeno 8, doPost 1, minDepth 5, SNP p value < 2e−6, minHigh 3, minCount 3) using mean proportion of Durusdinium (see Fig. 1c) as the dependent variable in a logistic regression. This analysis takes advantage of intermediate phenotypes and the individuals that do not agree with the overall phenotype-symbiont patterns.
For both comparisons, we used annotations within 2.5kbp of each variant to derive ontologies for two-tailed GO_MWU73. This strategy ranks individual genes by ‘heat’ and then resolves gene ontologies with at least 15 representative genes that ranked significantly (FDR p < 0.1) higher or lower than background expectations in representing functional groupings that differ based on the comparison. We treated the LRT statistic from the binary association as ‘heats’ and reconducted this analysis for associations with symbiont state using the effect size (beta) andthe LRT. We extracted all loci annotated with each ontology that significantly described binary phenotype and ran a PCAngsd subroutine with these loci to summarize genetic differentiation from all adult colonies in each functional category.
Larval analysis
To evaluate selection in the larval resequencing experiment, we created alignment files of pooled larval replicates (n = 5) by treatment (n = 2), phenotype (n = 3) and timepoint (n = 2) and pooled adults by phenotype (see Supplementary Table 1 for details). We summarized allele counts using bcftools74 and exported biallelic variants. There was low and uneven coverage in the bleached samples, so we excluded this pool from downstream analysis. We analyzed per-site FST for each sample (depth > =10) using the R package poolfstat75 and calculated a Population Branch Statistic (PBS)76 for each phenotype (see Supplementary Table 1 for details). To ask how heat selection impacted specific loci, we constructed a PBS statistic distinguishing the final post-heated larval pool from the initial larval pool and the final ambient larval pool (see Supplementary Table 1 for details) for each phenotype.
In this context, the PBS statistic is ideal for low-depth data because large values are derived from cases where allele frequencies are similar in both the final ambient pool and the initial heated larval pool. We also filtered the dataset to loci where both alleles were found in parental and initial pools and where the initial-to-parental FST was less than 0.05, assuring that signals of heterozygosity were not a sequencing artifact. PBS of the comparison of interest is also positively influenced by similarity in the ‘outgroups’, in this case the final ambient larval pool and the initial heated pool, which isolates the effect of the heat treatment. This expected pattern (based on random crosses from the same pool) further supports consistent sequencing outcomes, minimizes artifacts and highlights selection due to temperature stress.
We compared the PBS statistic for all loci shared between the cross and nonbleached phenotype with a linear regression and a Wilcox test. To evaluate selection for specific functions, we used GO_MWU73, treating the PBS statistic as ‘heats’ to look for gene ontologies enriched in large PBS values, which denote heat-specific selection. After conducting this analysis on each phenotype, we selected only significant ontologies (FDR p < 0.1) and examined shared ontologies between phenotypes.
We then calculated the difference in PBS between each phenotype and re-ran the GO_MWU analysis. This approach yields larger values when selection in the nonbleached pool was stronger than the cross pool, isolating functional categories that are disproportionately selected in nonbleached corals and contribute to selective breeding outcomes.
Metabolite analysis
Small biopsies of all 22 parent corals were extracted in 500uL 70% methanol, kept on ice for 30 min and stored at −80 °C until shipping to Michigan State University. Methanolic extracts were analyzed using LC-MS/MS. Details of chromatography and mass spectrometry parameters can be found in ref. 34. The converted.mzXML files were processed with MZmine277 and the feature quantification table was exported and processed with the Global Natural Products Social Molecular Networking (GNPS)78,79 and CANOPUS on SIRIUS 480,81. MZmine2 parameters for processing mass spectrometry data are described in34, and the CANOPUS parameters can be found in Supplementary Table 2. Both CANOPUS output files (“canopus_summary.csv”, “compound_identification.csv”) were merged and filtered to include the most specific class of molecules, matches and their molecular formulas. Molecular features from blanks and found between both files that did not display in silico annotations from CANOPUS and correlation between molecular formulas were removed.
We repurposed the WGCNA analytical framework for metabolites, which summarizes molecules with highly correlated abundances into modules82. We used log2 transformed abundances after imputing missing data with the lowest abundance of each molecule following83,84. We tested a power threshold from 1 to 20 and chose 6 as the ‘elbow’ in scale-free topology, with modules merged >0.15, minKME = 0.8, at least 25 molecules per module and an unsigned network. We used the first principal component from PCAngsd as a summary of the genetic variation of each coral’s genes relating to that ontology as the ‘trait’ for Spearman correlation analysis with WGCNA. We used a Fisher’s exact test to test for enrichment in molecular families within each module and calculated a false discovery rate-corrected p-value.
Betaine analysis
After noting the high association and strong linkage between two loci with high association values (p < 0.0001), we extracted a 50 kb window centered at the midpoint of these loci and annotated with blastn against cnidarians. Among other genes, we documented glycine sarcosine dimethylglycine N-methyltransferase within this selection window, a gene family that produces Betaine Glycine (BG), which we annotated in the metabolite data (metabolomics standards initiative level two85). We compared Betaine relative abundances between phenotypes using a Wilcox test. We used the raw annotations from GO_MWU (before merging into the parent term ‘Cellular Modified Amino Acid Metabolic Process’), and compared the association values for ‘Amino-Acid Betaine Metabolic Process’ to all other loci using a Wilcox test. We compared the beta values for predicting proportion Durusdinium in this window with the rest of the genome using a Wilcox test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Elizabeth Lenz for input on fertilization and rearing strategies in Montipora capitata, Katie Barott for initial colony tagging/surveys, Ingrid Knapp for library preparation advice, Justin Greer, Eva Majerová and Ariana Huffmyer for constructive comments on the manuscript and Kira Hughes, Chris Suchocki, Michelle Harangody and the Coral Resilience Lab for field and laboratory spawning assistance. We are grateful to Mike Henley for providing samples of M. flabellata. Coral fragments and gamete bundles were collected under Hawaii DLNR permit SAP 2018-03 to HIMB. We gratefully acknowledge the technical support and advanced computing resources from University of Hawaii Information Technology Services – Cyberinfrastructure. This work was funded by a Paul G. Allen Family Foundation grant to Ruth D. Gates. This is HIMB contribution #1886 and SOEST contribution #11506.
Author contributions
C.D. conceived and designed the study, collected and analyzed data and wrote the manuscript, J.R.H., T.N.F.R. collected data, N.K.B., C.I.H., C.M.H., R.A.Q., J.R.H., J.H. collected and analyzed data, R.D.G. provided materials and resources. All authors edited and approved the final manuscript.
Peer review
Peer review information
Communications Biology thanks Michael Ochsenkuehn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Caitlin Karniski and Luke R. Grinham.
Data availability
All analysis scripts, ecological data and processed sequencing data are available at github.com/druryc/mcap_bleaching. Raw sequencing data is available at NCBI PRJNA597077. Feature-based molecular networking is available at: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=bb9b6126118c4ba1881f4483560012d3 and raw files are available at massive.ucsd.edu under MassIVE ID MSV000085272 and MSV000085925.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nina K. Bean, Casey I. Harris.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03428-3.
References
- 1.Alvarez-Filip L, Dulvy NK, Gill JA, Côté IM, Watkinson AR. Flattening of Caribbean coral reefs: region-wide declines in architectural complexity. Proc. R. Soc. B: Biol. Sci. 2009;276:3019–3025. doi: 10.1098/rspb.2009.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes TP, et al. Global warming transforms coral reef assemblages. Nature. 2018;556:492. doi: 10.1038/s41586-018-0041-2. [DOI] [PubMed] [Google Scholar]
- 3.Drury, C. & Lirman, D. Genotype by environment interactions in coral bleaching. Proc. R. Soc. B Biol. Sci., 10.1098/rspb.2021.0177 (2021). [DOI] [PMC free article] [PubMed]
- 4.Kenkel CD, Almanza AT, Matz MV. Fine-scale environmental specialization of reef-building corals might be limiting reef recovery in the Florida Keys. Ecology. 2015;96:3197–3212. doi: 10.1890/14-2297.1. [DOI] [PubMed] [Google Scholar]
- 5.Howells EJ, Abrego D, Meyer E, Kirk NL, Burt JA. Host adaptation and unexpected symbiont partners enable reef‐building corals to tolerate extreme temperatures. Glob. Change Biol. 2016;22:2702–2714. doi: 10.1111/gcb.13250. [DOI] [PubMed] [Google Scholar]
- 6.Thomas, L. et al. Mechanisms of thermal tolerance in reef-building corals across a fine-grained environmental mosaic: lessons from Ofu, American Samoa. Front. Mar. Sci., 10.3389/fmars.2017.00434 (2018).
- 7.Thomas L, López EH, Morikawa MK, Palumbi SR. Transcriptomic resilience, symbiont shuffling, and vulnerability to recurrent bleaching in reef‐building corals. Mol. Ecol. 2019;28:3371–3382. doi: 10.1111/mec.15143. [DOI] [PubMed] [Google Scholar]
- 8.Barshis DJ, et al. Genomic basis for coral resilience to climate change. Proc. Natl Acad. Sci. USA. 2013;110:1387–1392. doi: 10.1073/pnas.1210224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guest JR, et al. Contrasting patterns of coral bleaching susceptibility in 2010 suggest an adaptive response to thermal stress. PLoS ONE. 2012;7:e33353. doi: 10.1371/journal.pone.0033353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matz, M. V., Treml, E. A. & Haller, B. C. Estimating the potential for coral adaptation to global warming across the Indo‐West Pacific. Glob. Chang. Biol.26, 3473–3481 (2020). [DOI] [PubMed]
- 11.Bay RA, Rose NH, Logan CA, Palumbi SR. Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci. Adv. 2017;3:e1701413. doi: 10.1126/sciadv.1701413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quigley, K. M., Bay, L. K. & van Oppen, M. J. Genome‐wide SNP analysis reveals an increase in adaptive genetic variation through selective breeding of coral. Mol. Ecol.29, 2176–2188 (2020). [DOI] [PubMed]
- 13.Howells EJ, et al. Enhancing the heat tolerance of reef-building corals to future warming. Sci. Adv. 2021;7:eabg6070. doi: 10.1126/sciadv.abg6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaJeunesse TC, et al. Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018;28:2570–2580. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Rowan R. Coral bleaching: thermal adaptation in reef coral symbionts. Nature. 2004;430:742. doi: 10.1038/430742a. [DOI] [PubMed] [Google Scholar]
- 16.Sampayo EM, Ridgway T, Bongaerts P, Hoegh-Guldberg O. Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl Acad. Sci. USA. 2008;105:10444–10449. doi: 10.1073/pnas.0708049105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maire, J. et al. Intracellular bacteria are common and taxonomically diverse in cultured and in hospite algal endosymbionts of coral reefs. ISME J., 15, 2028–2042 (2021). [DOI] [PMC free article] [PubMed]
- 18.Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 2017;8:14213. doi: 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Oppen MJ, Blackall LL. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019;17:557–567. doi: 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- 20.Fuller, Z. L. et al. Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science369 (2020). [DOI] [PubMed]
- 21.Dixon GB, et al. Genomic determinants of coral heat tolerance across latitudes. Science. 2015;348:1460–1462. doi: 10.1126/science.1261224. [DOI] [PubMed] [Google Scholar]
- 22.Jin YK, et al. Genetic markers for antioxidant capacity in a reef-building coral. Sci. Adv. 2016;2:e1500842. doi: 10.1126/sciadv.1500842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooke I, et al. Genomic signatures in the coral holobiont reveal host adaptations driven by Holocene climate change and reef specific symbionts. Sci. Adv. 2020;6:eabc6318. doi: 10.1126/sciadv.abc6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bay RA, Palumbi SR. Multilocus adaptation associated with heat resistance in reef-building corals. Curr. Biol. 2014;24:2952–2956. doi: 10.1016/j.cub.2014.10.044. [DOI] [PubMed] [Google Scholar]
- 25.Drury C. Resilience in reef-building corals: the ecological and evolutionary importance of the host response to thermal stress. Mol. Ecol. 2019;00:1–18. doi: 10.1111/mec.15337. [DOI] [PubMed] [Google Scholar]
- 26.Quigley KM, Willis BL, Bay LK. Heritability of the Symbiodinium community in vertically-and horizontally-transmitting broadcast spawning corals. Sci. Rep. 2017;7:8219. doi: 10.1038/s41598-017-08179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Hooidonk R, Maynard J, Planes S. Temporary refugia for coral reefs in a warming world. Nat. Clim. Change. 2013;3:508. doi: 10.1038/nclimate1829. [DOI] [Google Scholar]
- 28.Quigley, K. M., Warner, P. A., Bay, L. K. & Willis, B. L. Unexpected mixed-mode transmission and moderate genetic regulation of Symbiodinium communities in a brooding coral. Heredity, 121, 524–536 (2018). [DOI] [PMC free article] [PubMed]
- 29.Cunning R, Ritson-Williams R, Gates RD. Patterns of bleaching and recovery of Montipora capitata in Kāne’ohe Bay, Hawai’i, USA. Mar. Ecol. Prog. Ser. 2016;551:131–139. doi: 10.3354/meps11733. [DOI] [Google Scholar]
- 30.Dilworth, J., Caruso, C., Kahkejian, V. A., Baker, A. C. & Drury, C. Host genotype and stable differences in algal symbiont communities explain patterns of thermal stress response of Montipora capitata following thermal pre-exposure and across multiple bleaching events. Coral Reefs, 10.1007/s00338-020-02024-3 (2020).
- 31.Rocha de Souza, M. et al. Community composition of coral-associated Symbiodiniaceae is driven by fine-scale environmental gradients. bioRxiv10.1101/2021.11.10.468165 (2021).
- 32.Innis T, Cunning R, Ritson-Williams R, Wall C, Gates R. Coral color and depth drive symbiosis ecology of Montipora capitata in Kāne’ohe Bay, O’ahu, Hawai’i. Coral Reefs. 2018;37:423–430. doi: 10.1007/s00338-018-1667-0. [DOI] [Google Scholar]
- 33.Shore-Maggio A, Runyon CM, Ushijima B, Aeby GS, Callahan SM. Differences in bacterial community structure in two color morphs of the Hawaiian reef coral Montipora capitata. Appl. Environ. Microbiol. 2015;81:7312–7318. doi: 10.1128/AEM.01935-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roach, T. N., Dilworth, J., Jones, A. D., Quinn, R. A. & Drury, C. Metabolomic signatures of coral bleaching history. Nat. Ecol. Evol., 5, 495–503 (2021). [DOI] [PubMed]
- 35.Baird AH, Guest JR, Willis BL. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol., Evolution, Syst. 2009;40:551–571. doi: 10.1146/annurev.ecolsys.110308.120220. [DOI] [Google Scholar]
- 36.Caruso, C. et al. Genetic patterns in Montipora capitata across an environmental mosaic in Kāne’ohe Bay. bioRxiv10.1101/2021.10.07.463582 (2021). [DOI] [PMC free article] [PubMed]
- 37.Rose NH, Bay RA, Morikawa MK, Palumbi SR. Polygenic evolution drives species divergence and climate adaptation in corals. Evolution. 2017;72:82–94. doi: 10.1111/evo.13385. [DOI] [PubMed] [Google Scholar]
- 38.Rose NH, et al. Genomic analysis of distinct bleaching tolerances among cryptic coral species. Proc. R. Soc. B. 2021;288:20210678. doi: 10.1098/rspb.2021.0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsman ZH, et al. Ecomorph or endangered coral? DNA and microstructure reveal Hawaiian species complexes: Montipora dilatata/flabellata/turgescens & M. patula/verrilli. PLoS ONE. 2010;5:e15021. doi: 10.1371/journal.pone.0015021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon G, Abbott E, Matz M. Meta‐analysis of the coral environmental stress response: Acropora corals show opposing responses depending on stress intensity. Mol. Ecol. 2020;29:2855–2870. doi: 10.1111/mec.15535. [DOI] [PubMed] [Google Scholar]
- 41.Lim S, Kim DG, Kim S. ERK-dependent phosphorylation of the linker and substrate-binding domain of HSP70 increases folding activity and cell proliferation. Exp. Mol. Med. 2019;51:1–14. doi: 10.1038/s12276-019-0317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yancey PH, et al. Betaines and dimethylsulfoniopropionate as major osmolytes in cnidaria with endosymbiotic dinoflagellates. Physiol. Biochem. Zool. 2010;83:167–173. doi: 10.1086/644625. [DOI] [PubMed] [Google Scholar]
- 43.Hill R, Li C, Jones A, Gunn J, Frade P. Abundant betaines in reef-building corals and ecological indicators of a photoprotective role. Coral Reefs. 2010;29:869–880. doi: 10.1007/s00338-010-0662-x. [DOI] [Google Scholar]
- 44.Ngugi DK, Ziegler M, Duarte CM, Voolstra CR. Genomic blueprint of glycine betaine metabolism in coral metaorganisms and their contribution to reef nitrogen budgets. iScience. 2020;23:101120. doi: 10.1016/j.isci.2020.101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, A. et al. Metabolome shift associated with thermal stress in coral holobionts. bioRxiv10.1101/2020.06.04.134619 (2021).
- 46.Sakamoto A, Murata N. The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant, Cell Environ. 2002;25:163–171. doi: 10.1046/j.0016-8025.2001.00790.x. [DOI] [PubMed] [Google Scholar]
- 47.Burg MB, Ferraris JD. Intracellular organic osmolytes: function and regulation. J. Biol. Chem. 2008;283:7309–7313. doi: 10.1074/jbc.R700042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen TH, Murata N. Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ. 2011;34:1–20. doi: 10.1111/j.1365-3040.2010.02232.x. [DOI] [PubMed] [Google Scholar]
- 49.Petronini P, De Angelis E, Borghetti A, Wheeler K. Effect of betaine on HSP70 expression and cell survival during adaptation to osmotic stress. Biochem. J. 1993;293:553–558. doi: 10.1042/bj2930553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padilla-Gamiño JL, Pochon X, Bird C, Concepcion GT, Gates RD. From parent to gamete: vertical transmission of Symbiodinium (Dinophyceae) ITS2 sequence assemblages in the reef building coral Montipora capitata. PLoS ONE. 2012;7:e38440. doi: 10.1371/journal.pone.0038440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunning R, Baker AC. Thermotolerant coral symbionts modulate heat stress‐responsive genes in their hosts. Mol. Ecol. 2020;29:2940–2950. doi: 10.1111/mec.15526. [DOI] [PubMed] [Google Scholar]
- 52.Buerger P, et al. Heat-evolved microalgal symbionts increase coral bleaching tolerance. Sci. Adv. 2020;6:eaba2498. doi: 10.1126/sciadv.aba2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayfield AB, Gates RD. Osmoregulation in anthozoan–dinoflagellate symbiosis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007;147:1–10. doi: 10.1016/j.cbpa.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 54.Chan WY, Peplow LM, Menéndez P, Hoffmann AA, van Oppen MJ. Interspecific hybridization may provide novel opportunities for coral reef restoration. Front. Mar. Sci. 2018;5:160. doi: 10.3389/fmars.2018.00160. [DOI] [Google Scholar]
- 55.Rose NH, Seneca FO, Palumbi SR. Gene networks in the wild: identifying transcriptional modules that mediate coral resistance to experimental heat stress. Genome Biol. Evolution. 2016;8:243–252. doi: 10.1093/gbe/evv258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruiz-Jones LJ, Palumbi SR. Tidal heat pulses on a reef trigger a fine-tuned transcriptional response in corals to maintain homeostasis. Sci. Adv. 2017;3:e1601298. doi: 10.1126/sciadv.1601298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakravarti LJ, Beltran VH, van Oppen MJ. Rapid thermal adaptation in photosymbionts of reef‐building corals. Glob. Change Biol. 2017;23:4675–4688. doi: 10.1111/gcb.13702. [DOI] [PubMed] [Google Scholar]
- 58.Little AF, Van Oppen MJ, Willis BL. Flexibility in algal endosymbioses shapes growth in reef corals. Science. 2004;304:1492–1494. doi: 10.1126/science.1095733. [DOI] [PubMed] [Google Scholar]
- 59.Quigley, K., Randall, C., van Oppen, M. & Bay, L. Assessing the role of historical temperature regime and algal symbionts on the heat tolerance of coral juveniles. Biol. Open9, bio047316 (2020). [DOI] [PMC free article] [PubMed]
- 60.Matsuda, S. et al. Coral bleaching susceptibility is predictive of subsequent mortality within but not between coral species. Front. Ecol. Evol.10.3389/fevo.2020.00178 (2020).
- 61.Ritson-Williams, R. & Gates, R. D. Coral community resilience to successive years of bleaching in Kane ‘ohe Bay, Hawai ‘i. Coral Reefs.39, 757–769 (2020).
- 62.Hancock, J. et al. Coral husbandry for ocean futures: leveraging abiotic factors to increase survivorship, growth and resilience in juvenile Montipora capitata. Mar. Ecol. Prog. Ser., 10.3354/meps13534 (2020).
- 63.Falconer, D. S. Introduction To Quantitative Genetics (Pearson, 1960).
- 64.Cunning R, Baker AC. Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Change. 2012;3:259–262. doi: 10.1038/nclimate1711. [DOI] [Google Scholar]
- 65.Cunning R, Gillette P, Capo T, Galvez K, Baker A. Growth tradeoffs associated with thermotolerant symbionts in the coral Pocillopora damicornis are lost in warmer oceans. Coral Reefs. 2015;34:155–160. doi: 10.1007/s00338-014-1216-4. [DOI] [Google Scholar]
- 66.Alonge M, et al. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 2019;20:1–17. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shumaker A, et al. Genome analysis of the rice coral Montipora capitata. Sci. Rep. 2019;9:2571. doi: 10.1038/s41598-019-39274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 69.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinforma. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oksanen J, et al. R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2010. vegan: Community Ecology Package. R package version 1.17-2. [Google Scholar]
- 73.Wright RM, Aglyamova GV, Meyer E, Matz MV. Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics. 2015;16:371. doi: 10.1186/s12864-015-1540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hivert V, Leblois R, Petit EJ, Gautier M, Vitalis R. Measuring genetic differentiation from Pool-seq data. Genetics. 2018;210:315–330. doi: 10.1534/genetics.118.300900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinforma. 2010;11:1–11. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nothias L-F, et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods. 2020;17:905–908. doi: 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dührkop K, et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. methods. 2019;16:299–302. doi: 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- 81.Ludwig, M., Fleischauer, M., Dührkop, K., Hoffmann, M. A. & Böcker, S. in Computational Methods and Data Analysis for Metabolomics 185–207 (Springer, 2020).
- 82.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:1–13. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pedersen HK, et al. A computational framework to integrate high-throughput ‘-omics’ datasets for the identification of potential mechanistic links. Nat. Protoc. 2018;13:2781–2800. doi: 10.1038/s41596-018-0064-z. [DOI] [PubMed] [Google Scholar]
- 84.Pei, G., Chen, L. & Zhang, W. in Methods in enzymology585 135–158 (Elsevier, 2017). [DOI] [PubMed]
- 85.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analysis scripts, ecological data and processed sequencing data are available at github.com/druryc/mcap_bleaching. Raw sequencing data is available at NCBI PRJNA597077. Feature-based molecular networking is available at: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=bb9b6126118c4ba1881f4483560012d3 and raw files are available at massive.ucsd.edu under MassIVE ID MSV000085272 and MSV000085925.