Summary
Polycomb group proteins assemble into multi-protein complexes, known as Polycomb repressive complexes 1 and 2 (PRC1 and PRC2), that guide cell fate decisions during embryonic development. PRC1 forms an array of biochemically distinct canonical PRC1 (cPRC1) or non-canonical PRC1 (ncPRC1) complexes characterized by the mutually exclusive presence of PCGF (PCGF1-PCGF6) paralog subunit; however, whether each one of these subcomplexes fulfills a distinct role remains largely controversial. Here, by performing a CRISPR-based loss-of-function screen in embryonic stem cells (ESCs), we uncovered a previously unappreciated functional redundancy among PRC1 subcomplexes. Disruption of ncPRC1, but not cPRC1, displayed severe defects in ESC pluripotency. Remarkably, coablation of non-canonical and canonical PRC1 in ESCs resulted in exacerbation of the phenotype observed in the non-canonical PRC1-null ESCs, highlighting the importance of functional redundancy among PRC1 subcomplexes. Together, our studies demonstrate that PRC1 subcomplexes act redundantly to silence lineage-specific genes and ensure robust maintenance of ESC identity.
Keywords: PCGF, Polycomb, PRC1, cPRC1, ncPRC1, embryonic stem cells, pluripotency, RING1A/B, germ layer lineages, redundancy
Graphical abstract
Highlights
-
•
cPRC1 complexes are not the key determinant of self-renewal and pluripotency in ESCs
-
•
ncPRC1 complexes play a fundamental and redundant role in maintaining pluripotency in ESCs
-
•
cPRC1 and ncPRC1 act redundantly to suppress lineage-specific genes and preserve ESC identity
Polycomb group (PcG) proteins play a critical role both in maintaining pluripotency and the cell fate decision of embryonic stem cells (ESCs), yet the underlying mechanisms remain elusive. Yaru et al. report that Polycomb repressive complex 1 (PRC1) family complexes share a redundant but essential role in the maintenance of the pluripotent state in ESCs.
Introduction
Embryonic stem cells (ESCs), derived from the inner cell mass (ICM), have an extraordinary self-renewal capacity and yet retain pluripotency, which is the ability of cells to differentiate into any cell type of the three germ layers (Evans and Kaufman, 1981; Martin, 1981). A considerable body of work carried out over the past four decades has led to the view that the ESC state is transcriptionally controlled by OCT4, SOX2, and NANOG with chromatin regulators (Boyer et al., 2005). The Polycomb group (PcG) proteins are a set of evolutionary conserved factors that form chromatin regulator complexes involved in the transcriptional repression of key developmental genes to preserve cell fates (Piunti and Shilatifard, 2021; Schuettengruber et al., 2017; Simon and Kingston, 2009). PcG proteins assemble into two major chromatin-modifying multi-protein complexes, Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). PRC2 is composed of four core components SUZ12, EED, RBAP46/48, and the histone methyltransferases EZH1/2, which mono-, di-, and trimethylate lysine 27 of histone H3 (H3K27me1/2/3) (Piunti and Shilatifard, 2021). In addition, several sub-stoichiometric PRC2 accessory components have been identified, including JARID2, AEBP2, and PCL1-3, which modulate the recruitment to chromatin and/or enzymatic activity of the PRC2 complex.
Drosophila PRC1 contains Polycomb (Pc), Sex Combs Extra (Sce/dRing), Posterior Sex Combs (Psc), Polyhomeotic (Ph), and a sub-stoichiometric amount of Sex Comb on Midleg (Scm) (Saurin et al., 2001). Each of these proteins has multiple orthologs in mammals classified, respectively, as the CBX, RING1A/B, PCGF, PHC, and SCMH1/SCML2 families (Simon and Kingston, 2009). Mammalian PRC1 complexes are more heterogeneous in composition than PRC2 since each of the Drosophila core subunits has several orthologs that can interact in a combinatorial fashion. The ubiquitin E3 ligase RING1A/B, which monoubiquitylates histone 2A on lysine 119 (H2AK119ub1), can interact with one of six mutually exclusive PCGF paralog subunits (PCGF1-PCGF6), thereby creating six biochemically distinct PRC1 subcomplexes (named PRC1.1-PRC1.6) with diverse subunit composition whose functions are largely unknown (Gao et al., 2012). In addition to the core subunits, all PRC1 complexes incorporate versatile accessory proteins that confer functional specificity to a given complex. Importantly, these subcomplexes can be further divided into canonical (cPRC1) or non-canonical (ncPRC1), with cPRC1-containing PCGF2 or PCGF4, one of the PHC (PHC1/2/3), SCMH1/SCML2, and one of the CBX (CBX2/4/6/7/8) proteins that recognizes the H3K27me3 mark deposited by PRC2 (Gao et al., 2012; Piunti and Shilatifard, 2021; Schuettengruber et al., 2017). Conversely, ncPRC1 contain RYBP or YAF2 instead of CBX and PHC subunits, one of the four PCGF proteins (PCGF1/3/5/6), and therefore are targeted to chromatin independently of H3K27me3.
Although the molecular compositions of distinct PRC1 family complexes have been determined, their diverse biological functions and the crosstalk among the different complexes remain to be fully elucidated. While complete loss of PRC1 in mice via ablation of Ring1a/b results in pre-implantation lethality at the two-cell stage (Posfai et al., 2012), loss-of-function studies of Pcgfs in mice highlight major but distinct roles for the different PRC1 complexes in development (Akasaka et al., 2001; Almeida et al., 2017; Dickinson et al., 2016; Endoh et al., 2017; Liu et al., 2020). Obviously, ablation of individual PCGF proteins independently does not reproduce the loss of RING1A/B activity, suggesting that distinct PRC1 complexes may cooperate to determine Ring1a/b biological functions. Here, we utilize the CRISPR-Cas9 system to create single or combined Pcgf knockout in ESCs and rigorously investigate their functions in maintaining ESC pluripotency. We demonstrate that simultaneous, but not individual, ablation of Pcgfs led to loss of pluripotency in ESCs and triggers impaired self-renewal and spontaneous differentiation, a phenotype reminiscent of those in Ring1a/b double-deficient ESCs. Collectively, our data provide evidence that distinct PRC1 family complexes share a redundant but essential role in the maintenance of the pluripotent state in ESCs.
Results
CRISPR screen in ESCs reveals the potential involvement of the Pcgf family in the maintenance of pluripotency
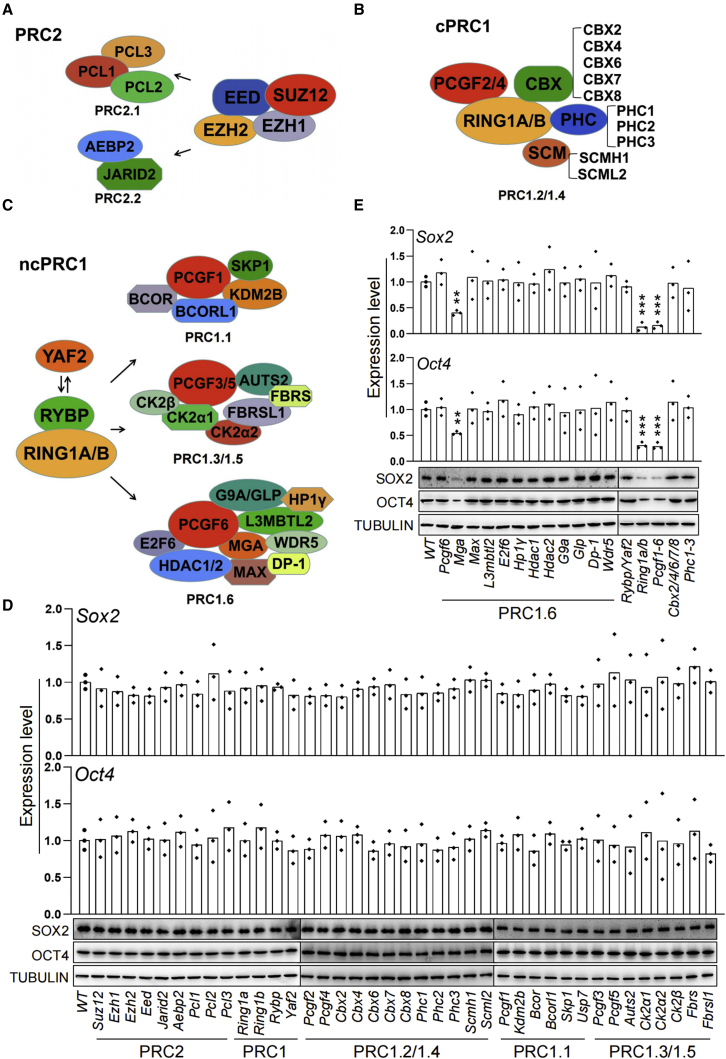
To explore the molecular mechanisms governing ESC pluripotency, we first screened for components in the PcG family (Figures 1A–1C), whose deficiency resulted in the changes of ESC identity genes Oct4 (also known as Pou5f1), and Sox2. We deleted individual PcG genes via a dual sgRNA-directed CRISPR-Cas9 system in ESCs and then evaluated the expression of Oct4 and Sox2 by qRT-PCR and western blot (Qin et al., 2021) (Figures 1D and 1E). We found that CRISPR-Cas9-mediated knockout of Mga, but not those of other PcG family members, was accompanied by a significant reduction in the expression of Oct4 and Sox2, which is consistent with our previous findings (Qin et al., 2021). In mammals, each of the PRC1 core subunits has two or more homologs (Figures 1B and 1C). We reasoned that the absence of a loss-of-function phenotype for PcG core subunits is due to compensation by their closely related homologs. To this end, we generated Rybp/Yaf2 or Ring1a/b double, Phc1/2/3 triple, Cbx2/4/6/7/8 quintuple and Pcgf1-6 sextuple knockout ESCs. We found that knockout of all six Pcgfs or Ring1a/b led to dramatic reduction in the expression levels of Oct4 and Sox2 (Figure 1E), whereas the loss of five Cbxs, three Phcs, or Rybp/Yaf2 has no detectable effect (see below for more details). Interestingly, E3 ubiquitin ligases RING1A/B are responsible for the maintenance of H2AK119ub1 when paired with one of six PCGF partners. In this study, we decided to further investigate the potential role of PCGF family in maintaining ESC pluripotency.
Figure 1.
Effect of ablation of PcG genes on the expression of core pluripotency transcription factors Oct4 and Sox2 in ESCs
(A–C) (A) Schematic representation of the subunit composition of the mammalian PRC2, (B) cPRC1, and (C) ncPRC1 complexes.
(D and E) Effect of CRISPR-mediated knockout of PcG genes on the expression of the pluripotency core regulators Oct4 and Sox2 in ESCs determined by qRT-PCR (top) and western blot (bottom). Each value was normalized to its corresponding actin value and the expression level in wild-type (WT) ESCs was arbitrarily set to 1. Data in (D) and (E) represent mean ± SD obtained from three independent experiments, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test) compared with the control.
Loss of individual PCGF family members has no significant effect on the maintenance of ESC identity
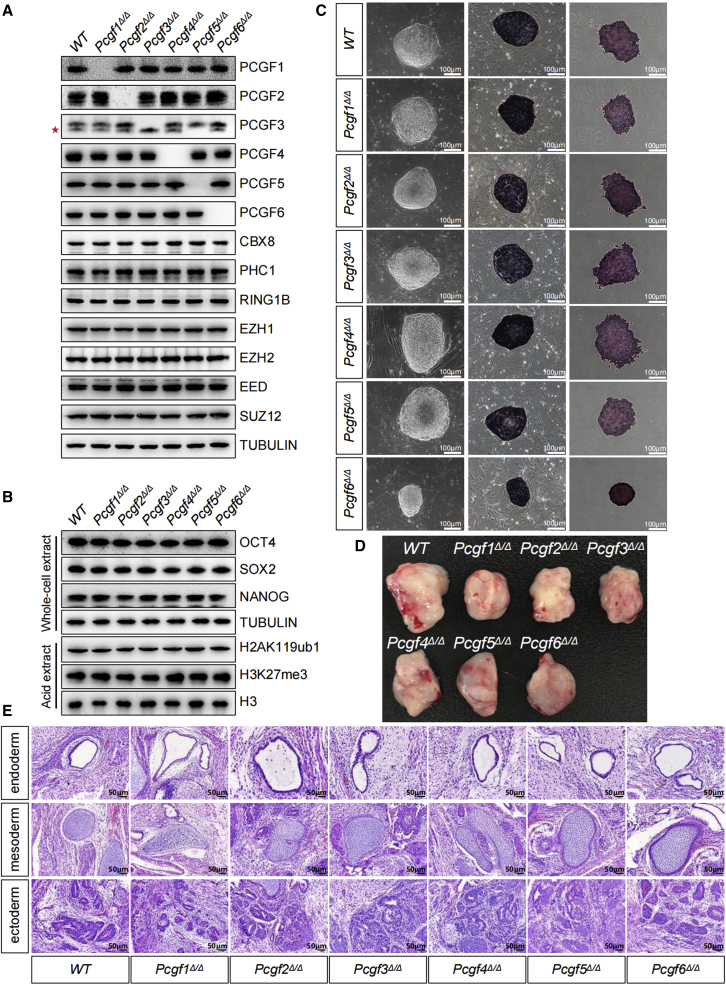
To rigorously explore the role of PCGF family in ESCs, we first determined whether these Pcgfs are expressed in ESCs. As assessed by western blot analysis, each PCGF family member is expressed in ESCs (Figure 2A). We generated cohorts of ESCs deficient in individual Pcgfs (Figures S1A–S1F). The loss of PCGF protein expression was confirmed by western blot analysis (Figure 2A). Single knockout of Pcgfs did not appear to appreciably affect histone modifications (H2AK119ub1 and H3K27me3) and protein levels of other PcG family members we examined (Figures 2A and 2B). These mutant cells were monitored for their capacity to form colonies after seeding on mitotically inactivated mouse embryonic fibroblast feeder layer. Consistent with our previous reports (Yan et al., 2017; Zhao et al., 2017a, 2017b), the single Pcgf-null ESCs were viable and retained a typical undifferentiated state as characterized by colony morphology, staining for alkaline phosphatase (AP), expression of the pluripotency-associated transcription factors, namely Nanog, Sox2, and Oct4, even after long-term culture (Figures 2B and 2C). Notably, loss of Pcgf6 gave rise to colonies that were smaller in size, whereas the targeted inactivation of other Pcgfs had no significant effect on overall cell growth. The teratoma formation in recipient mice is often considered as gold standard for pluripotency testing of ESCs. After 4–6 weeks of subcutaneous injection of mutant ESCs into nude mice, teratomas containing derivatives of all three embryonic germ layers were observed, indicating that these cells maintained pluripotency (Figures 2D and 2E). No or a very mild phenotype in single mutants can likely be attributed to functional redundancy among the Pcgf members.
Figure 2.
Loss of individual PCGF family members has no or a mild effect on the pluripotent properties of ESCs
(A) Western blot analyses showing changes in the global levels of PcG proteins in ESCs of indicated genotypes. Note, the PCGF3 antibody also recognized PCGF5 (∗).
(B) Western blot analyses showing changes in the global levels of pluripotency factors, H2AK119ub1 and H3K27me3 in ESCs of indicated genotypes. Tubulin and H3 were used as loading controls.
(C) Left: phase-contrast images of ESC colonies cultured on a feeder layer of MEFs. Middle and right: representative images of AP staining of WT and mutant ESC colonies cultured on a feeder layer of MEFs (middle) or gelatin (right). Scale bar, 100 μm.
(D) Teratoma formation in immunodeficiency mice by ESCs of indicated genotypes.
(E) Representative images of tissues of all three germ layers, including gut epithelium (endoderm), cartilage (mesoderm), and neural rosette (ectoderm), from H&E staining of teratomas generated from ESCs of the indicated genotypes. Shown is a representative of three injected mice. Scale bars, 50 μm.
RING1A/B play essential and redundant roles in ESC pluripotency maintenance
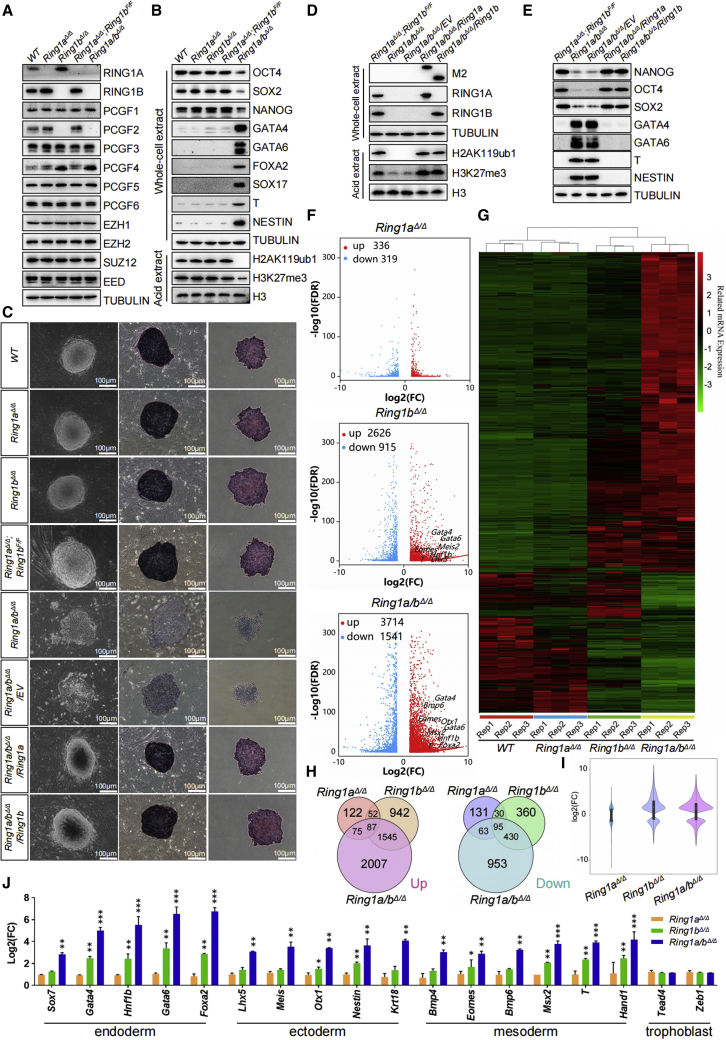
RING1A/B are subunits shared by all major PRC1 complexes (Figures 1B and 1C). Therefore, we set out to characterize the functions of RING1A/B in ESCs by generating ESCs deficient in Ring1a (Ring1aΔ/Δ) or Ring1b (Ring1bΔ/Δ) (Figures S1G and S1H). Interestingly, disruption of Ring1b led to increased RING1A protein levels, and vice versa (Figure 3A), suggesting that the loss of one can be compensated by upregulation of the other. In addition, ablation of Ring1b in ESCs caused complete loss of PCGF2 and dramatic upregulation of PCGF4. Ring1aΔ/Δ or Ring1bΔ/Δ cells did not show any noticeable phenotype and proliferated normally in the undifferentiated state (Figures 3B and 3C). Teratoma studies revealed that, while Ring1aΔ/Δ cells produced teratomas containing all three germ layer lineages, Ring1bΔ/Δ cells developed teratomas containing structures pertaining to all three germ layers but with an overrepresentation of the endoderm structures (Figure S2A).
Figure 3.
RING1A/B coordinate redundant mechanisms that ensure robust repression of key lineage-specific genes for sustaining ESC identity
(A and B) (A) Western blot demonstrating changes in the levels of PcG proteins, (B) pluripotency factors, endodermal (GATA4, GATA6, FOXA2, SOX17), mesodermal (T), and ectodermal (NESTIN) markers in ESCs of indicated genotypes. Tubulin and H3 were used as loading controls.
(C) Left: phase-contrast images of ESC colonies of indicated genotypes cultured on a feeder layer of MEFs. Middle and right: images of ESC colonies cultured on feeder layers (middle) or gelatin (right) after AP staining. Scale bar, 100 μm.
(D and E) (D) Western blot for selected PcG proteins, H2AK119ub1, H3K27me3, (E) pluripotency factors and selected germ layer markers in ESCs of indicated genotypes. The expression levels of the indicated FLAG-tagged proteins were detected with anti-Flag M2 antibody. Tubulin and H3 were used as loading controls.
(F) Volcano plots of –log10 (p value) against log2-fold change representing the differences in gene expression in ESCs of indicated genotypes. Upregulated (red) and downregulated (blue) genes are highlighted.
(G) Heatmap depicting fold changes in gene expression in ESCs of indicated genotypes. False discovery rate <0.05. Up- and downregulated genes are reported as red and green, respectively.
(H) Venn diagram showing overlap of upregulated (top) or downregulated (bottom) genes in Ring1aΔ/Δ, Ring1bΔ/Δ, and Ring1a/bΔ/Δ ESCs.
(I) A violin plot comparing log2-fold changes of genes in ESCs deficient for Ring1a, Ring1b, or both.
(J) qRT-PCR analysis of changes in the expression of lineage-specific genes. Each value was normalized to actin expression, and for each gene, the expression level in the wild-type ESCs was arbitrarily set to 1. Data are shown as the means ± SD for triplicate analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test) compared with the control.
To examine possible functional redundancy between RING1A and RING1B in ESCs, we established Ring1aΔ/Δ;Ring1bF/F ESC lines (Figure S1H), in which Ring1b could be conditionally ablated by transient expression of Cre recombinase. RING1B was completely removed 72 h after Cre transfection (Figure S2B), and loss of Ring1b resulted in no detectable H2AK119ub1, which was largely unaffected in any of the single mutants (Figure 3B). Therefore, throughout this study, unless otherwise stated, the cells were used for assay after 72 h of Cre transfection. Remarkably, in contrast to each single knockout, Ring1a/b double-deficient ESCs exhibited severely impaired self-renewal capacity, suggesting their redundant role in ESC pluripotency maintenance. Combined deletion of Ring1a/b led to massive cell death by apoptosis (23.3% Annexin V binding in Ring1a/bΔ/Δ cells versus 15.1% in control Ring1aΔ/Δ;Ring1bF/F cells), whereas both single mutants displayed the same apoptotic rates as the wild type. Cell-cycle analysis revealed that Ring1a/b double, but not single mutants exhibited a markedly extended G1 phase and a shortened S phase, suggesting that the impaired growth of Ring1a/bΔ/Δ ESCs was due to combinatory effect of cell-cycle perturbation and apoptosis induction (Figures S2C and S2D). Double ablation of Ring1a/b resulted in the formation of flat and morphologically distinct colonies and in a significant decrease of AP staining indicative of precocious differentiation (Figure 3C). Ring1a/bΔ/Δ ESCs could not be propagated beyond six passages when cultured under conditions that favor ESC maintenance, indicating an essential role for maintaining ESC identity. As cell passages increased, Ring1a/bΔ/Δ ESCs gradually lost their proliferation potential, as indicated by an increased proportion of cells in G1 phase and a decreased proportion of cells in S phase (Figure S2E). Because the loss of Ring1a/b in ESCs was accompanied by declines in Nanog, Sox2, and Oct4 mRNA and protein levels (Figure 3B; Tables S1–S3), we investigated whether RING1A/B contributed to the maintenance of stem cell identity through regulation of these core pluripotency factors. The Ring1a/b-null rescue system utilized Ring1aΔ/Δ;Ring1bF/F ESCs, in which the floxed Ring1b alleles were excised by Cre recombinase. Introducing Oct4, Nanog, or Sox2 using lentivirus-mediated transduction did not significantly restore the defects observed in Ring1a/bΔ/Δ ESCs (Figures S2F and S2G). In contrast, the range of phenotypes observed in the Ring1a/bΔ/Δ ESCs were rescued by the ectopic expression of either Ring1a or Ring1b (Figures 3C–3E). Thus, the function of RING1A/B in maintaining ESC identity appears not to be mediated by keeping the requisite levels of OCT4, NANOG, and SOX2. In addition, although Ring1aΔ/Δ or Ring1bΔ/Δ retained the ability to form teratomas, the capability of producing teratomas was completely abolished in Ring1a/b double knockout ESCs (Figure S2H), supporting the functional redundancy of RING1A and RING1B. Thus, RING1A/B play redundant but collectively essential roles in the maintenance of stem cell identity.
To gain insight into the molecular mechanisms underlying the observed phenotypic changes, we performed RNA-seq analysis in single and double Ring1a/b knockout ESCs (Tables S1–S3). Complete loss of Ring1a/b in ESCs resulted in substantial changes in gene expression (3,714 upregulated versus 1,541 downregulated genes) (Figures 3F and 3G; Table S3). More genes were upregulated than downregulated upon loss of Ring1a/b, consistent with their essential role in transcriptional gene silencing. In contrast, only 655 (336 up, and 319 down) and 3,541 (2,626 up, and 915 down) genes were deregulated in Ring1a and Ring1b single mutants, respectively, further supporting the existence of functional redundancy between RING1A and RING1B (Tables S1–S3). The genes that were altered in Ring1aΔ/Δ and Ring1bΔ/Δ ESCs highly correlated and overlapped with those altered in Ring1a/b double null ESCs (Figures 3H and 3I). Gene ontology (GO) analysis on differentially expressed genes in Ring1a/b-null ESCs showed that upregulated genes are primarily linked to developmental processes, including pattern specification process, cell fate commitment and organ morphogenesis. In contrast, downregulated genes were specifically enriched for genes associated with mechanisms associated with pluripotency and chemotaxis, consistent with the observed proliferation defects (Figures S2I and S2J). Importantly, examination of upregulated genes in Ring1a/bΔ/Δ ESCs also showed a notable overrepresentation of germ layer lineage-specific transcripts, including master regulators of endoderm formation (such as Sox7, Gata4, Hnf1b, Gata6, and Foxa2) (Figure 3J), key markers of ectoderm (Meis, Lhx5, Otx1, Nestin, and Krt18) or mesoderm (such as Bmp4, Eomes, Bmp6, Msx2, T, and Hand1). In contrast, expression levels of key markers of trophectoderm (Tead4 and Zeb1), either decreased or remained the same in mutant cells (Table S3). By performing qRT-PCR, western blot analysis and immunofluorescence (IF), we confirmed the RNA-seq finding and demonstrated that, while the expression of these genes was significantly upregulated in Ring1a/bΔ/Δ ESCs, their expression was largely unaffected or moderately increased in Ring1aΔ/Δ and Ring1bΔ/Δ ESCs, respectively (Figures 3B, 3J, and S3). Together, these data suggest that RING1A/B act redundantly to repress lineage-specific developmental genes and safeguard self-renewal and pluripotency in ESCs.
PCGF2/4 is dispensable for both the maintenance and differentiation of the ESCs
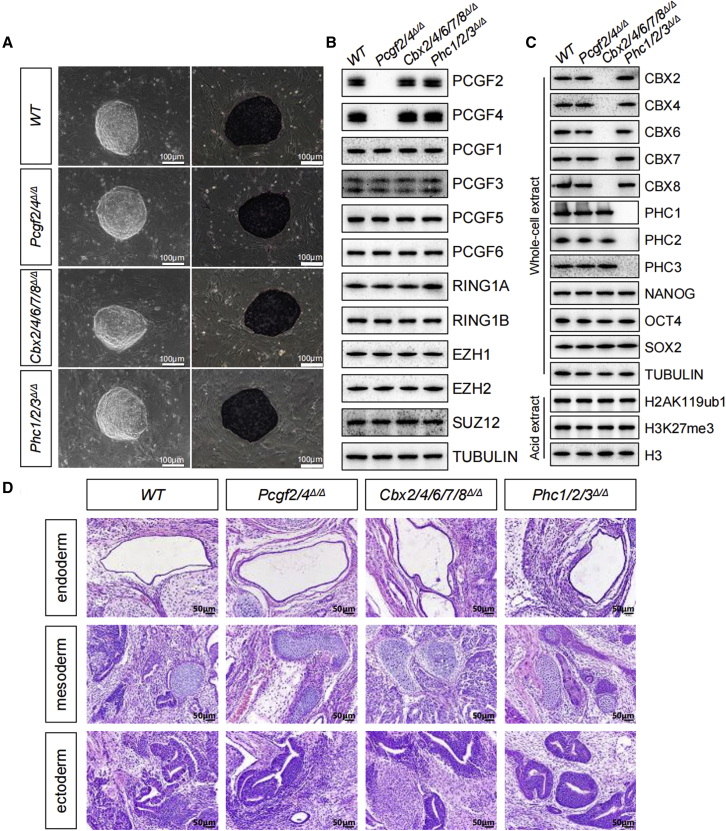
The cPRC1 complexes assemble around the RING1A/B-PCGF2/4 core and contain one of the CBX2/4/6/7/8 subunits and one of the PHC1/2/3 proteins (Schuettengruber et al., 2017; Shao et al., 1999) (Figure 1B). To overcome possible functional redundancy and compensation of the canonical PCGF family members, Pcgf2/4 double knockout ESC lines (Pcgf2/4Δ/Δ) were established. These cells maintained compact dome-shaped colony morphology similar to that of the wild-type ESCs (Figure 4A). Combined deletions of Pcgf2/4 did not appreciably affect the expression levels of the remaining PcG family members (Figure 4B). They also expressed high levels of the pluripotency markers AP, OCT4, NANOG, and SOX2 without any signs of differentiation into three germ lineages (Figures 4A and 4C). In addition, Pcgf2/4Δ/Δ ESCs exhibited high replating efficiency and they maintain their ability to differentiate into representative tissues of the three embryonic germ layers (Figure 4D). In addition, to explore directly the potential role for CBX2/4/6/7/8 and PHC1/2/3 in maintaining the pluripotency of ESCs, we generated triple and quintuple mutants of the Phc1/2/3 (Phc1/2/3Δ/Δ) and Cbx2/4/6/7/8 (Cbx2/4/6/7/8Δ/Δ) subfamilies, respectively (Figures S4A–S4H). Surprisingly, both mutants, similar to Pcgf2/4Δ/Δ ESCs, possessed the ability to self-renew in an undifferentiated state under standard culture conditions while retaining the ability to differentiate into all of the three main germ layers (Figures 4A–4D). Therefore, cPRC1 complexes are not the key determinant of self-renewal, and pluripotency in ESCs.
Figure 4.
Individual disruption of cPRC1 does not induce exit from pluripotency in ESCs
(A) Phase-contrast images of ESC colonies on MEF feeders (left). Representative images of AP staining of WT and mutant ESC colonies of indicated genotypes (right).
(B and C) Western blot demonstrating changes in the levels of indicated PcG proteins, pluripotency factors, and histone modifications in ESCs of indicated genotypes. Tubulin and H3 were used as loading controls.
(D) Representative images showing H&E staining of histological sections derived from teratomas generated from ESCs of the indicated genotypes. Shown is a representative of three injected mice. Scale bars, 50 μm.
PCGF1/3/5/6 synergize to preserve ESC identity
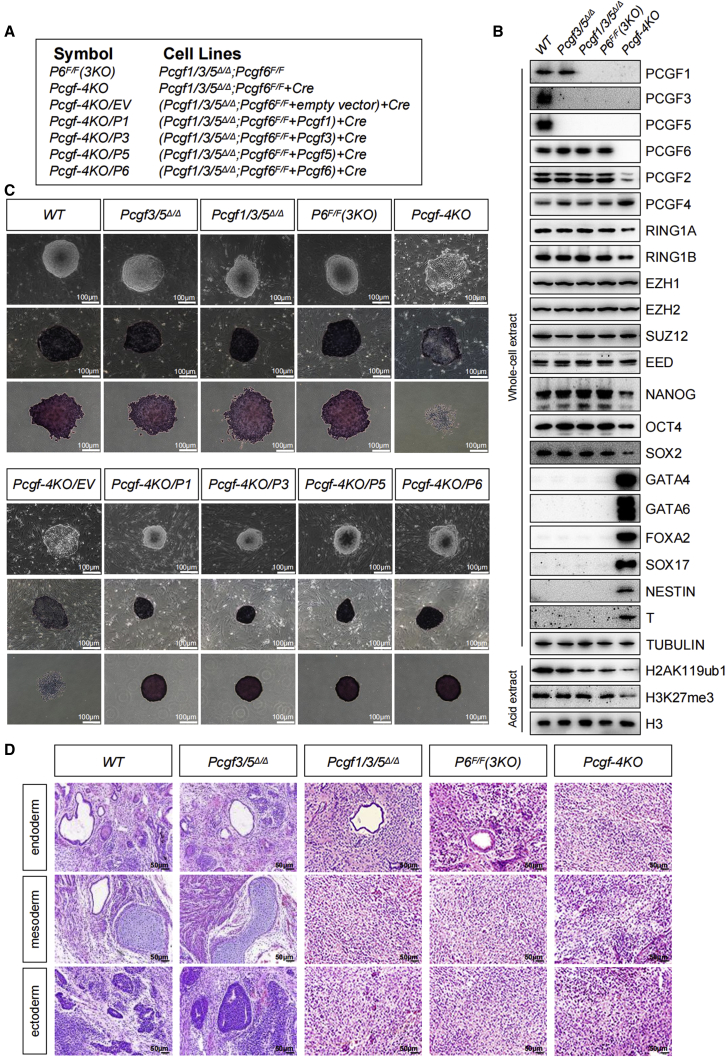
Ablation of Pcgf2/4 did not affect PcG-mediated pluripotency in ESCs, suggesting that the remaining PRC1 activities must be responsible for preserving the pluripotency (Figures 1B and 1C). Initially, we generated ESCs harboring double deletion of both Pcgf3 and Pcgf5 (Pcgf3/5Δ/Δ), which participate in nearly identical protein complexes and would likely possess potential redundant functions. Consistent with previous observations (Zhao et al., 2017a), Pcgf3/5Δ/Δ did not lose ESC properties, nor do these cells show altered levels of pluripotency genes (Figures 5A–5C). Notably, although the global levels of H2AK119ub1 were not affected following removal of either Pcgf3 or Pcgf5, loss of Pcgf3/5 in combination caused a moderate reduction in H2AK119ub1, as evidenced by western blot (Figure 5B). As both PCGF1- and PCGF3/5-PRC1 complexes contributed to H2AK119ub1 (Fursova et al., 2019), we reasoned that PCGF1/3/5-PRC1 complexes may collaborate to support the pluripotency of ESCs. We therefore generated a triple knockout (Pcgf1/3/5Δ/Δ) ESCs by simultaneous knockout of Pcgf1/3/5. Although removal of Pcgf1/3/5 resulted in a further loss of H2AK119ub1 (Figure 5B), Pcgf1/3/5Δ/Δ ESCs formed round and compact colonies with typical undifferentiated ESC morphology indistinguishable from Pcgf3/5Δ/Δ colonies (Figure 5C). These colonies exhibited a high level of AP activity, as well as continuing to express ESC pluripotent markers (Figures 5B and 5C).
Figure 5.
Combined loss of Pcgf1/3/5/6 unleash precocious differentiation of ESCs
(A) Reference legend for cell lines used in this figure and in Figure S5.
(B) Western blot demonstrating changes in the levels of selected PcG proteins, pluripotency factors, germ layer markers, H2AK119ub1, and H3K27me3 in ESCs of indicated genotypes. Tubulin and H3 were used as loading controls.
(C) Top: phase-contrast images of ESC colonies cultured on a layer of MEFs. Middle and bottom: representative images of AP staining of ESC colonies of indicated genotypes cultured together with a feeder layer of MEFs (middle) or on gelatin (bottom). Scale bar, 100 μm.
(D) H&E staining of teratoma sections revealed cells from various cell lineages including gut epithelium (endoderm), cartilage (mesoderm) and neural rosette (ectoderm). Shown is a representative of three injected mice. Scale bars, 50 μm.
To circumvent the lethality or non-physiological responses associated with the deficiency for ncPRC1, we engineered Pcgf1/3/5Δ/Δ ESCs with conditional Pcgf6F/F (Pcgf1/3/5Δ/Δ;Pcgf6F/F) to eliminate all ncPRC1 activity. The established Pcgf1/3/5Δ/Δ;Pcgf6F/F lines formed tightly compact colonies, stained mostly positive for AP, and continued to express high levels of pluripotency genes (Figures 5B and 5C). The introduction of Cre recombinase into the established Pcgf1/3/5Δ/Δ;Pcgf6F/F lines led to the efficient ablation of PCGF6 proteins, yielding quadruple knockout (Pcgf1/3/5/6Δ/Δ) cells that failed to maintain their pluripotent state, as determined by the reduced expression of key pluripotency factors at both the mRNA and protein levels (Figures 5B and 5C; Table S4). In concert with the reduced pluripotency factor expression, the Pcgf1/3/5/6Δ/Δ ESCs exhibited differentiated morphology and failed to be stained by AP (Figure 5C). Lineage-specific transcriptional regulators were aberrantly upregulated in those cells, consistent with the observed exit from the pluripotency state (Figure 5B). These results thus uncovered a previously unappreciated functional redundancy and cooperation among ncPRC1 complexes in preserving ESC pluripotent identity. Notably and consistent with a previous report (Fursova et al., 2019), ablation of Pcgf6 in addition to Pcgf1/3/5 caused a further reduction of H2AK119ub1 levels and a dramatic reduction in the levels of H3K27me3 (Figure 5B). In addition, while Pcgf1/3/5/6Δ/Δ ESCs displayed the same apoptotic rates as the control, cell-cycle analysis revealed these cells had a markedly extended G1 phase and a shortened S phase, suggesting that the impaired growth of Pcgf1/3/5/6Δ/Δ ESCs was due to the alteration of the cell cycle (Figures S5A and S5B). The functional redundancy among members of the ncPRC1-associated PCGF family was independently confirmed by exogenous expression of the ncPRC1-associated PCGF, which largely rescued the detrimental phenotype of Pcgf1/3/5/6Δ/Δ ESCs (Figures 5C, S5C, and S5D).
Pcgf3/5Δ/Δ maintained the ability to differentiate to derivatives of all three embryonic germ layers, whereas teratomas derived from Pcgf1/3/5Δ/Δ and Pcgf1/3/5Δ/Δ;Pcgf6F/F ESCs did not contain more advanced ectodermal and mesodermal structures (Figures 5D, S5E, and S5F). Yet rare examples of differentiation into tissues from endodermal lineages could be found. Growth of Pcgf1/3/5/6Δ/Δ teratomas was delayed for several weeks (Figure S5E). In histologic analysis, the most abundant components of Pcgf1/3/5/6Δ/Δ teratomas were undifferentiated areas and featured a striking paucity of mature elements (Figures 5D and S5F). Consistent with these observations, Pcgf1/3/5Δ/Δ and Pcgf1/3/5/6Δ/Δ teratomas also showed strongly reduced expression of the three embryonic germ layer markers when compared with controls, as detected by qRT-PCR (Figure S5G). Therefore, our teratoma data strongly suggest that Pcgf1/3/5/6Δ/Δ ESCs are significantly impaired in their ability to differentiate properly. Remarkably, Pcgf1/3/5/6-null ESC lines can readily be propagated indefinitely in standard cell culture conditions. Together, ncPRC1 complexes, as opposed to cPRC1 complexes, play a fundamental and redundant role in maintaining pluripotency in ESCs.
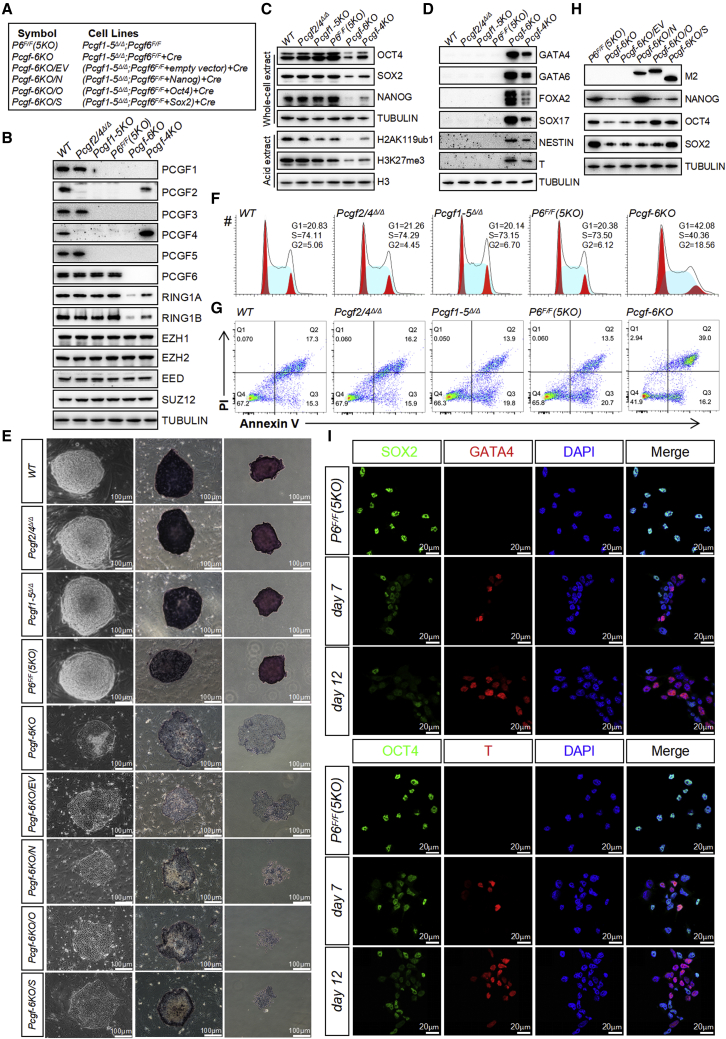
cPRC1 and ncPRC1 collaborate to maintain the pluripotent state of ESCs
It is clear that Pcgf1/3/5/6 loss failed to fully recapitulate the phenotype observed upon removal of Ring1a/b. We reasoned that ncPRC1 synergizes with cPRC1 to ensure robust maintenance of ESC pluripotency. To this end, we established Pcgf1/3/5/2/4Δ/Δ ESCs with conditional Pcgf6F/F (Pcgf1/3/5/2/4Δ/Δ;Pcgf6F/F) to eliminate all PRC1 activity (Figures 6A–6E). Interestingly, following removal of Pcgf2/4 in addition to Pcgf1/3/5, no further reduction in global H2AK119ub1 levels was observed. In addition, Pcgf1/3/5/2/4Δ/Δ or Pcgf1/3/5/2/4Δ/Δ;Pcgf6F/F ESCs formed colonies similar to those of undifferentiated Pcgf1/3/5Δ/Δ ESCs (Figure 6E). These cells also possessed AP activity and expressed high levels of OCT4, SOX2, and NANOG, supporting their pluripotent status (Figures 6C and 6E). Transfection of Pcgf1/3/5/2/4Δ/Δ;Pcgf6F/F with a plasmid expressing the Cre recombinase resulted in the complete loss of PCGF6 protein and an almost complete loss of H2AK119ub1 (Figure 6C). Remarkably, Cre-mediated ablation of Pcgf6 from Pcgf1/3/5/2/4Δ/Δ;Pcgf6F/F ESCs resulted in a dramatic decrease in their rate of proliferation relative to control Cre-transfected cells. These effects were due to elevated apoptosis and G1/S cell-cycle arrest (Figures 6F and 6G). Upon plating of Pcgf1-6Δ/Δ cells onto a feeder layer, they underwent spontaneous differentiation, as evidenced by flattened and spreading morphology and by loss of AP staining (Figure 6E). The dramatic change in the colony morphology was accompanied by a drastic reduction in the expression of OCT4, SOX2, and NANOG (Figures 6C and 6I). Similar to Ring1a/bΔ/Δ cells, the phenotypes observed in Pcgf1-6Δ/Δ ESCs were not rescued by the ectopic expression of Oct4, Sox2, or Nanog (Figures 6E and 6H), and they could not be expanded beyond fourth passage and failed to form teratomas when implanted into immunodeficient mice (Figure 7A). The cellular phenotype in Pcgf sextuple knockout ESCs closely resembled those observed in Ring1a/bΔ/Δ ESCs, suggesting a vital role of PCGF family in regulating the stemness of ESCs.
Figure 6.
Simultaneous ablation of cPRC1 and ncPRC1 triggers spontaneous differentiation and loss of self-renewal
(A) Reference legend for cell lines used in this figure and in Figure 7.
(B–D) Western blot demonstrating changes in the levels of selected (B) PcG proteins, (C) pluripotency factors, histone modifications, and (D) germ layer markers in ESCs of indicated genotypes. Tubulin and H3 were used as loading controls.
(E) Left: phase-contrast images of ESC colonies of indicated genotypes cultured on a feeder layer. Middle and right: images of ESC colonies cultured on feeder layers (middle) or gelatin (right) after AP staining. Scale bar, 100 μm.
(F) Representative cell-cycle profiles determined by propidium iodide (PI) staining and FACS analysis.
(G) Flow cytometric quantification of apoptosis with Annexin V and PI. The percentages of Annexin V-positive (apoptotic) cells are within the two right quadrants.
(H) Western blot analyses using the indicated antibodies on whole-cell lysates from ESCs of indicated genotypes. The expression levels of the indicated FLAG-tagged proteins were detected with anti-Flag M2 antibody.
(I) IF analysis for SOX2, OCT4 (green), GATA4, T (red), or DAPI (blue) in Pcgf1-5Δ/Δ;Pcgf6F/F ESCs following lenti-Cre infection. Images were taken at 63× magnification using confocal microscopy. Merge, merged images.
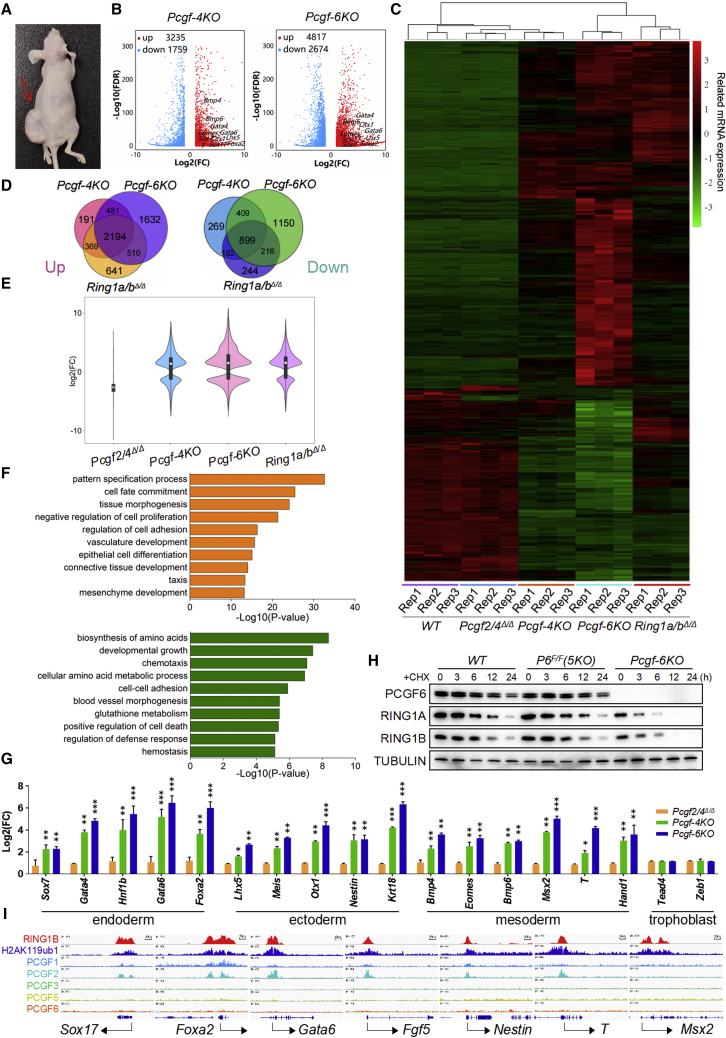
Figure 7.
cPRC1 and ncPRC1 act redundantly to silence lineage-specific genes
(A) Representative images of mice bearing teratomas 28 days after the injection of Pcgf1/3/5/2/4Δ/Δ;Pcgf6F/F (left side, indicated by a red arrow) or Pcgf1-6Δ/Δ ESCs (right side). Four mice were injected per condition.
(B) Volcano plots of –log10 (p value) against log2-fold change representing the differences in gene expression in ESCs of indicated genotypes. Upregulated (red) and downregulated (blue) genes are highlighted.
(C) Heatmap illustrating fold changes in gene expression in ESCs of indicated genotypes. False discovery rate <0.05. Up- and downregulated genes are reported as red and green, respectively.
(D) Venn diagram showing overlap of upregulated (left) or downregulated (right) genes between Pcgf-4KO, Pcgf-6KO, and Ring1a/bΔ/Δ ESCs. (E) A violin plot comparing log2-fold changes of genes in ESCs deficient for Pcgf2/4, Pcgf1/3/5/6, Pcgf1-6, and Ring1a/b.
(F) Gene ontology analysis of overlapping genes upregulated (top) and downregulated (bottom) between Pcgf-4KO and Pcgf-6KO ESCs.
(G) qRT-PCR of germ layer markers, measured in WT, Pcgf-4KO, and Pcgf-6KO ESCs. Each value was normalized to actin expression, and for each gene the expression level in the wild-type ESCs was arbitrarily set to 1.
(H) ESCs of indicated genotypes were treated with cycloheximide (CHX) as indicated and lysates were blotted with the indicated antibodies. Tubulin was used as a loading control.
(I) Genomic snapshots of the indicated ChIP-seq profiles at selected germ layer gene loci in WT ESCs. Published ChIP-seq data were obtained from NCBI GEO (accession numbers GSE122715 and GSE107377). Data in (G) represent the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test) compared with the control.
Consistent with the observed phenotypes, we observed 4,994 (3,235 up, and 1,759 down) versus 7,491 (4,817 up, and 2,674 down) differentially expressed genes in Pcgf1/3/5/6Δ/Δ and Pcgf1-6Δ/Δ ESCs, respectively, compared with their corresponding controls (Figures 7B and 7C; Tables S4–S5). There was substantial overlap in genes deregulated in Pcgf1/3/5/6Δ/Δ and Pcgf1-6Δ/Δ ESCs; however, a large number of the genes deregulated in Pcgf1-6Δ/Δ but not Pcgf1/3/5/6Δ/Δ was observed (Figures 7D and 7E). GO analysis showed that among genes upregulated in Pcgf1/3/5/6Δ/Δ and Pcgf1-6Δ/Δ ESCs were genes related to pattern specification process, cell fate commitment, and tissue morphogenesis (Figure 7F). Processes related to developmental growth, chemotaxis, and cell-cell adhesion were over-represented among the genes downregulated in Pcgf1/3/5/6Δ/Δ and Pcgf1-6Δ/Δ cells. There was also significant overlap in genes deregulated between Pcgf1/3/5/6Δ/Δ, Pcgf1-6Δ/Δ and Ring1a/bΔ/Δ cells; however, a large number of the genes deregulated in Pcgf1-6Δ/Δ but not Ring1a/bΔ/Δ was observed (Figures 7D and 7E). This suggests that both RING1A/B-dependent and -independent PCGF activities provide critical layers of gene regulation. Integration between RNA-seq and ChIP-seq data indicated that >38% and >30% of the deregulated genes in Pcgf1/3/5/6Δ/Δ and Pcgf1-6Δ/Δ, respectively, were direct targets of PRC1. Intriguingly, GO analysis revealed that these direct PRC1 targets were enriched for pattern specification process, cell fate commitment, embryonic morphogenesis, and negative regulation of cell differentiation (Figure S6). By performing qRT-PCR, western blot analysis and IF, we confirmed the RNA-seq finding and observed that ablation of both Polycomb complexes had an additive effect on the expression levels of lineage-affiliated genes (Figures 6D, 6I, and 7G). Altogether, we conclude that cPRC1 and ncPRC1 act redundantly to suppress lineage-specific genes and preserve ESC identity.
As shown in Figure 6B, Pcgf1/3/5/6 and Pcgf1-6 deletion led to a moderate and substantial reduction, respectively, in the protein levels of RING1A and RING1B, whereas loss of Pcgf2/4 did not appear to appreciably affect the protein levels of RING1A and RING1B. However, RNA-seq analysis demonstrated that ablation of Pcgf1/3/5/6 or Pcgf1-6 did not alter steady-state levels of Ring1a and Ring1b mRNA in ESCs (Tables S4–S5), suggesting a mechanism involving posttranscriptional regulation. A time-course cycloheximide (CHX) experiment revealed that RING1A and RING1B protein half-lives were markedly decreased, from approximately 4 and 6 h in Pcgf1/3/5/2/4Δ/Δ;Pcgf6F/F ESCs to less than 2 and 3 h, respectively, in Pcgf1-6Δ/Δ cells (Figure 7H), suggesting that PCGF family members act redundantly to increases RING1A and RING1B protein stability. In line with these results, a cohort of key lineage-specific genes, which were upregulated in Pcgf1-6Δ/Δ ESCs and possessed a high enrichment of RING1B, PCGF2, and H2AK119ub1, were not or only weakly targeted by the ncPRC1-associated PCGF proteins (Figure 7I). Collectively, these findings provide novel evidence to suggest how the action of PCGF family on RING1A/B stability and resultant activity could impact on ESC fate decisions.
Discussion
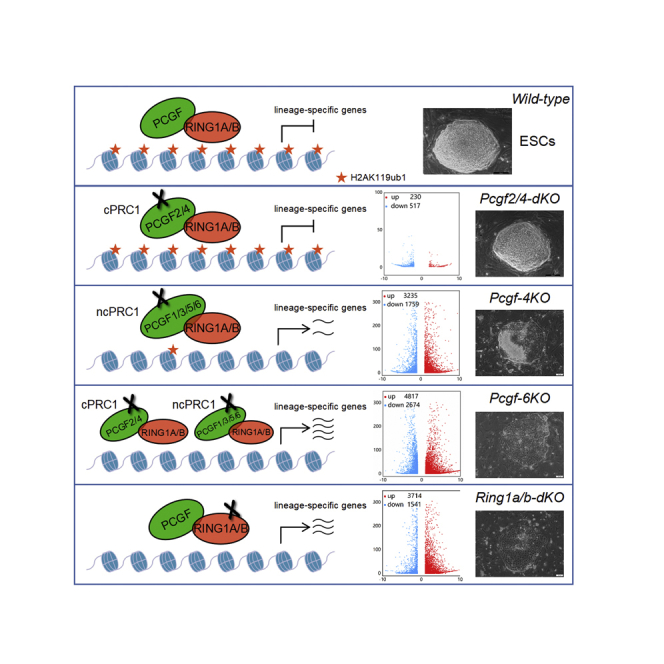
Self-renewing ESCs are derived from the ICM of the pre-implantation blastocyst (Evans and Kaufman, 1981; Martin, 1981). They are regarded as an excellent model system for analyzing the mechanisms governing cell fate transitions during early embryo development. The hallmark characteristics of ESCs is their pluripotency, which bestows them with a capacity to give rise to all cell types of the body. The lineage-specific differentiation of ESCs in vitro closely recapitulates the lineage commitment in the embryo that occurs in vivo. The mechanisms responsible for the maintenance of ESC pluripotency, and those that guide their commitment into particular cell lineages, remain to be further explored. In ESCs, PcG proteins bind preferentially to genes encoding lineage-specific transcription factors to prevent both exit from pluripotency and premature differentiation (Boyer et al., 2006). Uncovering the underlying mechanism of PcG-mediated pluripotency maintenance remains challenging due to the multiple possible combinatorial permutations within PRC1 complexes (Gao et al., 2012; Hauri et al., 2016).
The catalytic core of PRC1 is formed by RING1A/B and one of six PCGF partners, giving rise to an array of biochemically distinct cPRC1 or ncPRC1 complexes (Gao et al., 2012). Inactivation of Ring1a results in fertile mice with minor skeletal alterations (del Mar Lorente et al., 2000). In contrast, disruption of Ring1b in mice causes embryonic lethality due to gastrulation arrest (Voncken et al., 2003). Whereas RING1A and RING1B can have redundant functions during development (del Mar Lorente et al., 2000; Voncken et al., 2003), the combined deletion of Ring1a/b causes developmental arrest before the two-cell stage (Posfai et al., 2012), highlighting the requirement to genetically delete both genes to fully characterize PRC1 biological roles. Intriguingly, this also corresponds with the relative importance for maintenance of H2AK119ub1 and pluripotency of these Ring E3 ligases as RING1A or RING1B loss has no or only mild effect on H2AK119ub1 levels and the property of ESCs, whereas simultaneous depletion of Ring1a/b results in a full loss of H2AK119ub1 and of pluripotency and commitment toward lineages of the three germ layers with loss of indefinite self-renewal (Figures 3 and S2).
cPRC1 complexes, which are recruited to specific genomic sites via recognition of H3K27me3 deposited by PRC2, have been proposed to be involved in the Polycomb-mediated gene silencing (Schuettengruber et al., 2017). However, recent studies have documented that loss of cPRC1 complexes has little or no effect on gene expression (Fursova et al., 2019; Zepeda-Martinez et al., 2020). Consistently, we found that removal of Pcgf2/4 in ESCs has no discernible effect on the maintenance of pluripotency (Figure 4). In line with these observations, mice harboring inactivating mutations of cPRC1 components exhibit only minor or delayed defects in embryogenesis (Akasaka et al., 2001; Coré et al., 1997; Forzati et al., 2012; Isono et al., 2005; Lau et al., 2017). In contrast, targeted disruptions to ncPRC1 in mice result in early embryonic lethality (Almeida et al., 2017; Endoh et al., 2017; Liu et al., 2020; Pirity et al., 2005; Qin et al., 2012; Washkowitz et al., 2015). In vitro differentiation assays indicated that knockout of single Pcgf in ESCs resulted in moderate defects affecting the proper formation of the three germ layers (Endoh et al., 2017; Morey et al., 2015; Yan et al., 2017; Zhao et al., 2017a, 2017b). However, these single mutant cells have the potential to form teratomas composed of derivatives from all three germ layers in vivo (Figure 2), suggesting that inactivation of individual Pcgf family members has no significant effect on the maintenance of ESC identity. Loss of ncPRC1 core subunits PCGF1/3/5/6 triggered the spontaneous differentiation and derepression of lineage-specific genes, suggesting a redundant but essential role of PCGF family in maintaining ESC pluripotency (Figure 5). These phenotypic discrepancies between cPRC1 and ncPRC1 could reflect distinct roles in ESC identity maintenance. Alternatively, cPRC1-dependent and cPRC1-independent PRC1 complexes might play redundant roles in ESC fate specification. Interestingly, lineage-specific genes were strongly derepressed in the Pcgf1-6 knockout cells compared with Pcgf2/4 or Pcgf1/3/5/6 knockouts, where they largely remained repressed or were moderately derepressed, respectively. In addition, loss of ESC self-renewal capacity is achieved only when both cPRC1- and ncPRC1-associated PCGF proteins are ablated (Figure 6). Together, our findings demonstrate that cPRC1 and ncPRC1 function redundantly to silence unwanted lineage-specific genes and safeguard against exit from ESC pluripotency.
Our functional interrogation of PRC1 activity indicates that redundancy among distinct PRC1 complexes is central to PcG-mediated cell fate specification in ESCs. This is somewhat surprising given that exogenously expressed HA-tagged PCGF proteins in 293T cells occupy mutually exclusive regions of the genome, possibly arguing against redundancy among different PRC1 complexes (Gao et al., 2012). In contrast, genomic profiling of endogenous PCGF proteins in ESCs reveals that ncPRC1 complexes, as opposed to cPRC1 complexes, largely co-occupy target regions, where they act synergistically to shape genomic H2AK119ub1 and define gene repression (Fursova et al., 2019). Interestingly, another group reported that PCGF1/2/4 jointly preserve H2AK119ub1 deposition at common targets even though PCGF proteins display high binding specificity (Scelfo et al., 2019). By using combinatorial genetic perturbation coupled with functional evaluation, our findings reconcile these apparent discrepancies leading to the interpretation that ncPRC1 complexes, but not cPRC1 complexes, predominantly define PcG-mediated cell fate specification in ESCs (Figure 5). Most importantly, we discover that genetic ablation of both cPRC1 and ncPRC1 largely phenocopies lineage-specific gene repression and cellular defects caused by complete loss of PRC1 in ESCs via ablation of Ring1a/b (Figures 3 and 6). This suggests that, although biochemically distinct PRC1 subcomplexes display high binding specificity, which enables them to function at defined sites in the genome, for example, PRC1.6 at germ cell-related genes (Endoh et al., 2017; Qin et al., 2012, 2021; Stielow et al., 2018), they also co-occupy target sites, where they act redundantly to deposit genomic H2AK119ub1 and define lineage-specific gene repression. Critically, we provide novel evidence to suggest how the action of PCGF family members on RING1A/B stability could impact on ESC fate determination. These findings underscore the need to further explore the mechanisms that control the levels and activity of RING1A/B in ESCs. Overall, our discoveries provide compelling evidence of functional redundancy between cPRC1 and ncPRC1 in robust repression of key lineage-specific genes, which is required for governing ESC identity.
Experimental procedures
Plasmid construction and generation of mutant cell lines
sgRNAs were designed based on the CRISPR design website (http://crispor.tefor.net/). A pair of 20-bp oligonucleotides was annealed and inserted into the BbsI-digested Cas9 and sgRNA-expressing PX459 vector (Addgene plasmid no. 62988; http://n2t.net/addgene: 62988; RRID: Addgene_62988), which was resistant to puromycin. sgRNA-expressing plasmids were verified by DNA sequencing. Constructed plasmids were transfected into ESCs by Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Twenty-four hours after transfection, puromycin-resistant ESCs were selected for 48 h and then grown on mitomycin-treated feeder. Subsequently, ESC colonies were picked and screened by gnomic PCR, and confirmed by RT-PCR and western blot. Conditional knockout ESC mutants were performed as described previously (Qin et al., 2021). Details of the mutant ESC lines used in this study are listed in Table S6. All of the primers and antibodies used in this study are listed in Table S7 or in a previous report (Qin et al., 2021).
Statistics
Unless stated otherwise, all data were shown as mean values ± SD or mean values ± SEM for experiments performed with at least three independent experiments. GraphPad Prism 5 was used to perform statistical tests and generate p values using two-tailed Student’s t test for comparisons between two datasets, and p values of less than 0.05 were considered statistically significant. Statistical significance was presented in figures in the following manner: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Data and code availability
The accession number for the sequencing data reported in this paper is NCBI GEO: GEO183646. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the supplementary material. All original data, including images and mutant lines, pertaining to this study will be made available upon request.
Author contributions
Conceptualization, J.Q.; methodology, Y.Z., L.D., C.W., K.H., L.Z., and J.Q.; formal analysis, Y.Z., L.D., C.W., K.H., J.W., L.X., and J.Q.; mechanistic investigations, Y.Z., L.D., C.W., Y.X., Q.J., and J.Q.; writing – original draft, J.Q.; writing – review & editing, J.Q., Y.Z., L.D., and C.W.; funding acquisition, J.Q.; supervision, J.Q.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We are indebted to all members of the Qin laboratory and to Yikai Huang and Ting Su for experimental advice and helpful discussions. This work was supported by grants from the National Natural Science Foundation of China (31970810) to J.Q.
Published: March 31, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.02.020.
Supplemental information
References
- Akasaka T., van Lohuizen M., van der Lugt N., Mizutani-Koseki Y., Kanno M., Taniguchi M., Vidal M., Alkema M., Berns A., Koseki H. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128:1587–1597. doi: 10.1242/dev.128.9.1587. [DOI] [PubMed] [Google Scholar]
- Almeida M., Pintacuda G., Masui O., Koseki Y., Gdula M., Cerase A., Brown D., Mould A., Innocent C., Nakayama M., et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356:1081–1084. doi: 10.1126/science.aal2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Coré N., Bel S., Gaunt S.J., Aurrand-Lions M., Pearce J., Fisher A., Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- del Mar Lorente M., Marcos-Gutiérrez C., Pérez C., Schoorlemmer J., Ramírez A., Magin T., Vidal M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Endo T.A., Shinga J., Hayashi K., Farcas A., Ma K.W., Ito S., Sharif J., Endoh T., Onaga N., et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife. 2017;6:e21064. doi: 10.7554/eLife.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Forzati F., Federico A., Pallante P., Abbate A., Esposito F., Malapelle U., Sepe R., Palma G., Troncone G., Scarfò M., et al. CBX7 is a tumor suppressor in mice and humans. J. Clin. Invest. 2012;122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova N.A., Blackledge N.P., Nakayama M., Ito S., Koseki Y., Farcas A.M., King H.W., Koseki H., Klose R.J. Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol. Cell. 2019;74:1020–1036.e1028. doi: 10.1016/j.molcel.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri S., Comoglio F., Seimiya M., Gerstung M., Glatter T., Hansen K., Aebersold R., Paro R., Gstaiger M., Beisel C. A high-density map for navigating the human polycomb complexome. Cell Rep. 2016;17:583–595. doi: 10.1016/j.celrep.2016.08.096. [DOI] [PubMed] [Google Scholar]
- Isono K., Fujimura Y., Shinga J., Yamaki M., J O.W., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol. Cell. Biol. 2005;25:6694–6706. doi: 10.1128/MCB.25.15.6694-6706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau M.S., Schwartz M.G., Kundu S., Savol A.J., Wang P.I., Marr S.K., Grau D.J., Schorderet P., Sadreyev R.I., Tabin C.J., et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 2017;355:1081–1084. doi: 10.1126/science.aah5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhu Y., Xing F., Liu S., Xia Y., Jiang Q., Qin J. The polycomb group protein PCGF6 mediates germline gene silencing by recruiting histone-modifying proteins to target gene promoters. J. Biol. Chem. 2020;295:9712–9724. doi: 10.1074/jbc.RA119.012121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L., Santanach A., Blanco E., Aloia L., Nora E.P., Bruneau B.G., Di Croce L. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell Stem Cell. 2015;17:300–315. doi: 10.1016/j.stem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Pirity M.K., Locker J., Schreiber-Agus N. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol. Cell. Biol. 2005;25:7193–7202. doi: 10.1128/MCB.25.16.7193-7202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A., Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cel. Biol. 2021;22:326–345. doi: 10.1038/s41580-021-00341-1. [DOI] [PubMed] [Google Scholar]
- Posfai E., Kunzmann R., Brochard V., Salvaing J., Cabuy E., Roloff T.C., Liu Z., Tardat M., van Lohuizen M., Vidal M., et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012;26:920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Wang C., Zhu Y., Su T., Dong L., Huang Y., Hao K. Mga safeguards embryonic stem cells from acquiring extraembryonic endoderm fates. Sci. Adv. 2021;7:eabe5689. doi: 10.1126/sciadv.abe5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Whyte W.A., Anderssen E., Apostolou E., Chen H.H., Akbarian S., Bronson R.T., Hochedlinger K., Ramaswamy S., Young R.A., et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–332. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Scelfo A., Fernández-Pérez D., Tamburri S., Zanotti M., Lavarone E., Soldi M., Bonaldi T., Ferrari K.J., Pasini D. Functional landscape of PCGF proteins reveals both RING1A/B-Dependent-and RING1A/B-Independent-Specific activities. Mol. Cell. 2019;74:1037–1052.e1037. doi: 10.1016/j.molcel.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and Trithorax: 70 Years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Shao Z., Raible F., Mollaaghababa R., Guyon J.R., Wu C.T., Bender W., Kingston R.E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cel. Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Stielow B., Finkernagel F., Stiewe T., Nist A., Suske G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018;14:e1007193. doi: 10.1371/journal.pgen.1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken J.W., Roelen B.A., Roefs M., de Vries S., Verhoeven E., Marino S., Deschamps J., van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. U S A. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washkowitz A.J., Schall C., Zhang K., Wurst W., Floss T., Mager J., Papaioannou V.E. Mga is essential for the survival of pluripotent cells during peri-implantation development. Development. 2015;142:31–40. doi: 10.1242/dev.111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Zhao W., Huang Y., Tong H., Xia Y., Jiang Q., Qin J. Loss of polycomb group protein Pcgf1 severely compromises proper differentiation of embryonic stem cells. Scientific Rep. 2017;7:46276. doi: 10.1038/srep46276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda-Martinez J.A., Pribitzer C., Wang J., Bsteh D., Golumbeanu S., Zhao Q., Burkard T.R., Reichholf B., Rhie S.K., Jude J., et al. Parallel PRC2/cPRC1 and vPRC1 pathways silence lineage-specific genes and maintain self-renewal in mouse embryonic stem cells. Sci. Adv. 2020;6:eaax5692. doi: 10.1126/sciadv.aax5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Huang Y., Zhang J., Liu M., Ji H., Wang C., Cao N., Li C., Xia Y., Jiang Q., et al. Polycomb group RING finger proteins 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 2017;292:21527–21537. doi: 10.1074/jbc.M117.804054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Tong H., Huang Y., Yan Y., Teng H., Xia Y., Jiang Q., Qin J. Essential role for polycomb group protein Pcgf6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity. J. Biol. Chem. 2017;292:2773–2784. doi: 10.1074/jbc.M116.763961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is NCBI GEO: GEO183646. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the supplementary material. All original data, including images and mutant lines, pertaining to this study will be made available upon request.