Abstract
Clubroot is caused by Plasmodiophora brassicae, which threatens Brassicaceae crop production worldwide. In recent years, there has been an outbreak and rapid spread of clubroot in many major cruciferous crop-producing areas of China. In this study, we identified a cabbage material DingWen (DW) with different resistant capabilities from Huashuang5R (H5R) and Huayouza62R of Brassica napus, which are currently used as the main resistant cultivars for clubroot management in China. We used a next-generation sequencing-based bulked segregant analysis approach, combined with genetic mapping to identify clubroot-resistant (CR) genes from F1 population generated from a cross between the DW (CR) and HZSX (clubroot susceptible). The CR locus of DW (named CRA8.1) was mapped to a region between markers A08-4346 and A08-4853, which contains two different loci CRA8.1a and CRA8.1b after fine mapping. The CRA8.1b loci contain a fragment of 395 kb between markers A08-4624 and A08-4853 on A08 chromosome, and it is responsible for the resistance to PbZj and PbXm isolates. However, together with CRA8.1a, corresponding to a 765-kb region between markers A08-4346 and A08-4624, then it can confer resistance to PbXm+. Finally, through expression analysis between resistant and susceptible materials, two genes encoding TIR-NBS-LRR proteins (BraA08g039211E and BraA08g039212E) and one gene encoding an RLP protein (BraA08g039193E) were identified to be the most likely CR candidates for the peculiar resistance in DW.
Keywords: clubroot, Plasmodiophora brassicae, Brassica rapa, CRA8.1, fine mapping
Introduction
Clubroot is a highly contagious soil-borne disease that specifically endangers Brassica species, and it is caused by Plasmodiophora brassicae, a pathogen that belongs to peculiar eukaryotic taxa under Endomyxa branch (Dixon, 2009). Most of the Brassica plants, including Brassica napus, Brassica oleracea, and Brassica rapa, can be infected by P. brassicae. The susceptible plants display a swollen root symptom by this disease infection, which leads to dysfunction of root function for water and nutrient transport, eventually withering and death (Gahatraj et al., 2019). In recent years, the disease is rapidly spreading from different continents and countries, resulting in severe yield loss of cruciferous crops and vegetables (Hwang et al., 2012).
Although biological and chemical reagents can rescue the disease phenotype under certain conditions, the identification and introgression of resistance genes to create resistant varieties is the most efficient and economic approach to defeat the disease spreading (Elke et al., 2009; Chai et al., 2014). A few qualitative and quantitative clubroot-resistant (CR) loci have been identified in B. rapa, B. oleracea, B. napus, and Brassica nigra (Mehraj et al., 2020). The majority of CR genes are located on A genomes, including A01, A02, A03, A05, A06, and A08 chromosomes (Mehraj et al., 2020). In particular, CR loci on A03 or A08 chromosomes are extensively studied, including PbBa3.1, PbBa3.2, and PbBa3.3, CRd, Crr3, CRk, CRb, Rcr4, Rcr1, and CRa from A03 chromosome (Matsumoto et al., 1998; Hirai et al., 2004; Piao et al., 2004; Sakamoto et al., 2008; Chen et al., 2013; Chu et al., 2014; Yu et al., 2017; Pang et al., 2018), and Crr1 (including both Crr1a and Crr1b), PbBa8.1, Rcr3, Rcr9wa, BraA.CR.b, and Rcr9 from A08 chromosome (Suwabe et al., 2003; Chen et al., 2013; Yu et al., 2017; Hirani et al., 2018; Karim et al., 2020). CrrA5 and Crr4 were located on A05 and A06 chromosomes (Suwabe et al., 2006; Nguyen et al., 2018), and Crr2, PbBa1.1, CRc, and Rcr8 were mapped to A01 and A02 chromosomes, respectively (Suwabe et al., 2003; Sakamoto et al., 2008; Chen et al., 2013; Yu et al., 2017). Based on fine-mapping markers, some CR loci reside extremely close to or overlap each other, such as Crr1 and PbBa8.1, and CRa and CRb, and they can be allelic and actually originate from homologous genes from different A-genome species (Chen et al., 2013). These genes can be used as resources for breeding CR accessions.
Actually, from a broader view of disease resistance genes, most plant resistance (R) genes identified so far encode for cell membrane or intracellular receptors, including RLPs/RLKs (receptor-like proteins/kinases) and NLRs (nucleotide-binding site–leucine-rich repeat) (Kourelis and van der Hoorn, 2018). Plant innate immunity system comprises PTI (PAMP-triggered immunity) and ETI (effector-triggered immunity) (Deng et al., 2020). It is believed that specific interactions between a resistance gene and certain pathotypes are dependent mainly via ETI, but also affected by PTI pathway (Kim et al., 2016; Neik et al., 2017; Larkan et al., 2020; Yuan et al., 2021). For PTI, PAMPs from the invading pathogen are perceived by host membrane receptors known as PRRs (pattern recognition receptors), and RLKs and RLPs are the two major groups of PRRs on the cell surface determining the initial recognition (Wu and Zhou, 2013; Larkan et al., 2015; Grund et al., 2019; Kim et al., 2020). For the ETI route, effectors released inside the host cell are recognized by plant intracellular proteins such as NLRs, some of which require additional helper NLRs or chaperon proteins for triggering the ETI response (Grund et al., 2019). Different types of NLR proteins have been identified based on their protein domain structure, such as TIR-NBS-LRR, CC-NBS-LRR, and CCR-NBS-LRR subgroups (Wang and Chai, 2020). The above-mentioned CR genes, only two CR genes, namely, CRa and Crr1a, have been successfully isolated, both encoding TIR-NBS-LRR (NLRs) proteins (Ueno et al., 2012; Hatakeyama et al., 2013).
Clubroot-resistant genes from B. rapa have been introgressed into susceptible B. napus in order to generate resistant varieties. Currently, the two CR B. napus varieties in China are Huashuang5R (H5R) and Hayouza62R, which were generated through crosses using resistant B. rapa donor parents ECD04 (AA, 2n = 20) and Shinki (AA, 2n = 20), respectively (Zhan et al., 2020; Li et al., 2021). They were resistant to most of the clubroot pathogens isolated from different regions in China (Nadil et al., 2019). However, due to the soil-borne nature of the clubroot pathogen, a “resistant” Brassica variety to one pathogen can become “susceptible” to another pathogen from a different source. The present clubroot pathogen classification systems include the “Williams” system (Williams, 1966), the ECD system (Buczacki et al., 1975), and the SCD system (Pang et al., 2020). According to the “Williams” system, pathotype 4 is the most prevalent form in China, and most of the isolates such as PbZj (Zhijiang isolated from Hubei province) and PbXm (Xinmin from Liaoning province) belong to this group. The two B. napus CR varieties H5R and Hayouza62R containing PbBa8.1- and CRb-resistant loci, respectively, are resistant to PbZj and PbCd (Chengdu) isolates, but not to PbXm pathogen (Shah et al., 2020). Another struggling problem is the loss of resistance over time, which also calls for a new resource of resistance genes. During previous work, people found that pyramiding multiple CR genes in one variety would greatly improve the resistance of the host plant. For example, CRb and PbBa8.1 pyramiding lines in B. napus have demonstrated that plants in a homozygous state in each CR gene/locus had higher resistance than those in a heterozygous state (Shah et al., 2020). However, the fast spreading of clubroot disease, the presence of various pathotypes for the clubroot pathogens from different regions, and the frequent loss of resistance collectively call for urgent identification of a new CR gene resource.
The purpose of this study was to identify a CR locus different from PbBa8.1 and CRb and to identify the candidate genes responsible for disease resistance, therefore providing foundation for future utilization of these new CR genes for resistance breeding in Brassica species.
Materials and Methods
Plant Materials
The Brassica cultivars used in the current study include H5R, Huashuang5S (H5S), and 409R (the resistant restore line for Huayouza62R) of B. napus, DingWen (DW), HuangZiShaXun (HZSX, Z1) (Belser et al., 2018), and 91-12 of B. rapa. H5R and 409R are resistant cultivars containing CR loci of PbBa8.1 and CRb, respectively. H5S and 91-12 were used as susceptible controls. DW is a hybrid and sterile resistant material. A susceptible cultivar HZSX was used as a parent in crosses with DW for F1 population construction and gene mapping.
Plasmodiophora brassicae Collection and Clubroot Resistance Phenotyping
Different species of P. brassicae from the Chinese cabbage roots with severe galling were collected from five different locations of China: PbXm, Xinmin of Liaoning province; PbCd, Chengdu of Sichuan province; PbZj, Zhijang of Hubei province; PbTc, Tengchong of Yunnan province; and PbLx, Linxiang of Yunnan province. During the study, we found that Xinmin region contained two isolates, based on the host response. Therefore, we named it as “PbXm+,” in order to discriminate it from the other one. Galls were frozen and stored in −20°C until further use.
Clubroot resistance test of all materials was carried out as described previously (Chen et al., 2015). In brief, the collected galls were thawed at room temperature, ground to homogenization with buffer, and filtered through gauze, and resting spores were isolated after rounds of centrifugation. The spore concentration was measured and diluted to 107 resting spores per milliliter in sterile distilled water, before being used for host plant inoculation.
Host plants were prepared by germination of seeds on moist filter paper for 4 days, followed by the growth of seedling in a nursery room on medium at 16-h photoperiod, with 25/20°C day/night temperature for 7 days. When seedlings are ready, 1 ml of the above resting spore suspension was inoculated into the root of the seedling, and the disease symptoms were scored after 30–40 days. Clubroot severity was scored as 0–3 based on the root morphology, with grade 0 as normal growth, grade 1 as a few small galls on the lateral roots, grade 2 as big galls on lateral roots, and grade 3 as big galls on both primary roots and lateral roots. The disease index (DI) was calculated based on the number of plants at various severity levels (Shah et al., 2020).
Bulked Pool Construction and Sequencing
Bulked segregation analysis (BSA) method was used to map the genomic region responsible for CR in DW. A genetic cross was performed using DW and HZSX, and the two parents were genetically variant and also showed contrasting CR phenotype. Thirty-eight highly resistant and 38 extremely susceptible plants were selected from F1 population to construct two pools (resistant pool and susceptible pool), respectively. Genomic DNA was extracted from leaves of individual plants using DNA Secure Plant Kit (TIANGEN, China). The DNA concentration was quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, United States), and DNA of each individual plant within the same pool is mixed in equal concentration, resulting in two DNA libraries for sequencing.
Second-generation sequencing was performed on these two libraries using an Illumina HiSeq platform in PE150 mode. The raw data were accessed for quality control and then transformed into clean data by removing adaptor sequence. Clean reads were mapped to the reference genome of B. rapa1 using the Burrows–Wheeler Aligner (BWA0.7.12-r1039 mem).
CRA8.1 Gene Mapping
To identify candidate CR loci in DW, we first calculated the Euclidean distance (ED) value of SNP and InDel separately and then constructed maps based on the distribution of ED value on the chromosome. The molecular markers, developed by Zhan et al. (2020) on the A08 chromosome, were applied for genotyping. Five hundred F1 plants were used in the primary mapping, and the population was expanded to 3290 F1 plants for fine mapping. Physical map was constructed according to the location of molecular markers using MapChart software (Voorrips, 2002).
Assembly of CRA8.1 Fragment
To get sequence information of fragment containing CRA8.1 in DW, we performed fragment assembly with a European turnip ECD04 genome as the reference.
First, based on a Nova Seq 6000 sequencing platform, around 100 Gb Illumina short reads were obtained with leaf DNA of disease-resistant individual R59, which is the resistant plant in F1. Then, the Illumina short reads were mapped to the reference genome by BWA software package and rearranged by Samtools software package with parameters all set as default (Li and Durbin, 2009). Second, SNP/InDels data were extracted from the mapping results by BCF tools software package with the parameters set as “DP > 30 and QUAL > 100” (Narasimhan et al., 2016). At the same time, based on the sequence of A08-4346 and A08-5076 markers and blast result from NCBI2 from the reference genome, chromosomal fragments between the two markers from the reference genome were extracted by bedtools software package with default parameters (Quinlan and Hall, 2010).
Then, based on SNP/InDels data, an R script was written to replace the nucleotide sequence of candidate fragment of DW. The non-heterozygous SNP/InDels loci were replaced directly, while for loci containing heterozygous SNP/InDels, only one of the SNP/InDels was randomly selected to replace the candidate sequence.
Gene Synteny Analysis of CRA8.1 Fragment
First, we downloaded the Chinese cabbage HuangZiShaXun (Z1) genome,3 B. napus ZS11 genome,4 and European turnip ECD04 genome (unpublished). Then, the CDS of each genome was extracted by GFF read software package with parameter set as default. Finally, the gene synteny analysis was performed with CDS of candidate fragment as query and CDS of each genome as subject with MCscan.5
Identification and Expression Analysis of Candidate Genes
We used DW and HZSX (susceptible parent) and inoculated them with PbZj or PbXm+ isolates, respectively. Root samples were taken at 0 h, 12 h, and 4 days after inoculation, total RNA was extracted, and the candidate gene expression was analyzed using qRT-PCR. Information of qRT-PCR primers is listed in Supplementary Table 1, and BrUBC10 (XM_009134237.3) was used as an internal reference.
For promoter cis-element analysis, the DNA sequence 3.0 kb upstream of the coding sequence was submitted to the Plant CARE database6 for the prediction of putative cis-elements. The data were presented using TBtools software.
Results
Chinese Cabbage “DW” With Broad Resistance to Plasmodiophora brassicae From Different Regions
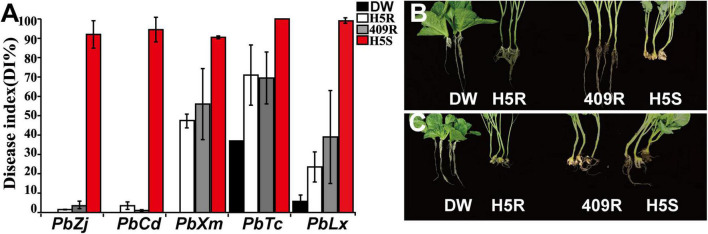
Previously, a Chinese cabbage material was screened out with excellent resistance to Zj isolate of P. brassicae, and this material was named “DW.” In order to further verify the pathogen resistance profile of DW and characterize the identity of CR genes in comparison with the ones currently widely used in H5R and Huayouza62R B. napus varieties in China, DW, H5R, 409R (resistant, restore line of Huayouza62R), and H5S were inoculated by five different clubroot isolates collected from different locations of China, namely, PbXm, PbCd, PbZj, PbTc, and PbLx (Figure 1A). Indoor inoculation experiments showed that susceptible control H5S was severely infected by all pathogens, while H5R, 409R, and DW had variable resistance (Figure 1A). H5R, 409R, and DW showed high resistance to PbZj and PbCd (DI ≤ 3.5%) (Figures 1A,B), and DW was the only one resistant to PbXm, whereas H5R and 409R showed a DI level of 47.5 and 56%, respectively (Figures 1A,C). We can also see that all four materials used in this study were susceptible to PbTc; however, DW displayed lower DI than H5R and 409R (Figure 1A). Similarly, H5R and 409R showed 24–40% DI to PbLx, but the DI of DW was below 10% (Figure 1A). Taken together, these results suggest that DW is a material with superior CR, which is fundamentally different from that in H5R and 409R, suggesting possible different genetic background difference behind their resistance.
FIGURE 1.
Disease resistance responses of different materials to P. brassicae from different field isolates. (A) Disease index statistics. Black, white, gray, and red bars represent the disease index in DW, Huashuang5R (H5R), 409R, and Huashuang5S (H5S), respectively. (B,C) The phenotypes of DW, H5R, 409R, and H5S were inoculated with PbZj and PbXm, respectively.
Fine Mapping Revealed at Least Two Resistance Loci Presented in DingWen
To investigate the DW inheritance of the resistance, a segregating population was constructed using a susceptible material HZSX crossed with DW. Parental lines (DW and HZSX) and 139 F1 individuals were subsequently inoculated with the PbXm and scored for their resistance. As shown in Table 1, DW showed complete resistance with DI of 0, whereas HZSX was completely susceptible with DI of 100% (Table 1). The F1 plants from a cross of DW with HZSX exhibited roughly 1:1 segregation (among 139 F1 individuals, 69 and 70 were resistant and susceptible, respectively), indicating that the CR loci in DW are controlled by a single dominant gene (Figure 2A and Table 1).
TABLE 1.
Disease phenotype statistics in F1 population from cross of DW and HZSX.
| Parents and crosses | Type | Disease levels |
Total | IRa | χ 2 | |||
| 0 | 1 | 2 | 3 | |||||
| DW | R-parent | 30 | 0 | 0 | 0 | 30 | 0 | |
| HZSX | S-parent | 0 | 0 | 0 | 30 | 30 | 100 | |
| DW × HZSX | F1 | 69 | 0 | 0 | 70 | 139 | 50 | 0.007 |
aIncidence rate (%).
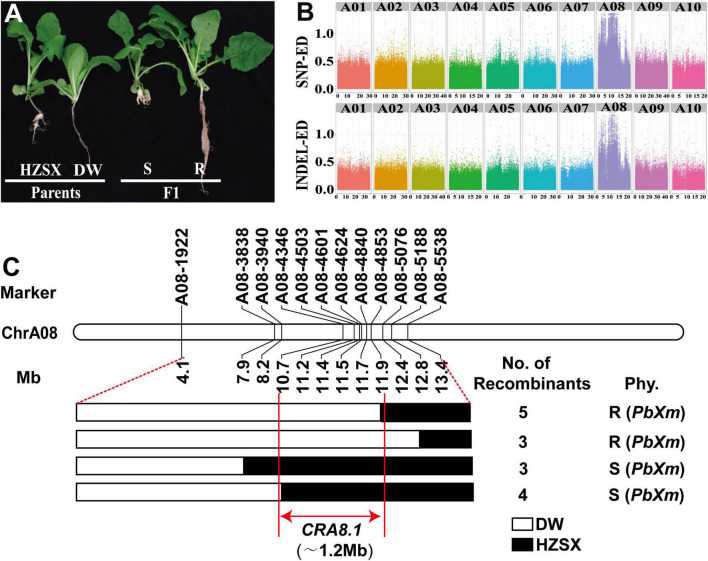
FIGURE 2.
Primary mapping of CRA8.1 in B. rapa. (A) Root phenotypes of DW, HZSX, and F1 after inoculation with PbXm. S, susceptible; R, resistant. (B) The Euclidean distance (ED) values of SNPs and InDels on chromosomes from the A genome. (C) Fine mapping of CRA8.1 region with InDel markers. Fifteen recombinants were identified from F1 population from markers A08-1922 to A08-5538. Black or white represents the regions found in HZSX or DW, respectively. Red lines indicate the CRA8.1 locus (1.2 Mb) identified from this study.
In order to identify the resistance loci, BSA (bulked segregation analysis) pools consisting of R- and S-plants were used for high-throughput sequencing. A total of 278,237,232 and 271,955,410 raw reads were obtained from R- and S-pools, respectively (Supplementary Table 2). After removing the adaptor sequence and filtering off low-quality reads, we recovered over 93% clean reads from the R- and S-pools, respectively, with Q30 over 87% (Supplementary Table 2). The clean reads were mapped to B. rapa reference genome (V3.0), and the total mapping rates were over 96%.
According to the ED of SNP and InDel (Figure 2B), a region of about 15 Mb was identified on A08 chromosome associating with CR (Figure 2B). To narrow down the region in genome, 12 molecule markers from the developed molecular markers were screened, and 15 recombinants were screened between markers A08-1922 and A08-5538 in F1 population. They were divided into four groups carrying different chromosomal fragments based on genotyping results (Figure 2C). Based on the resistance phenotype and the genotyping results, the CR loci were primarily mapped between A08-4346 and A08-4853 intervals, corresponding to a region of 1.2 Mb on chromosome A08 of B. rapa genome (V3.0) (Figure 2C). Interestingly, a previously known resistance locus PbBa8.1 overlaps with this region (Zhan et al., 2020). Since DW was highly resistant to PbXm pathogen, while H5R was not, this is a new locus conferring different CR profiles from PbBa8.1, and we named this locus as CRA8.1 hereafter.
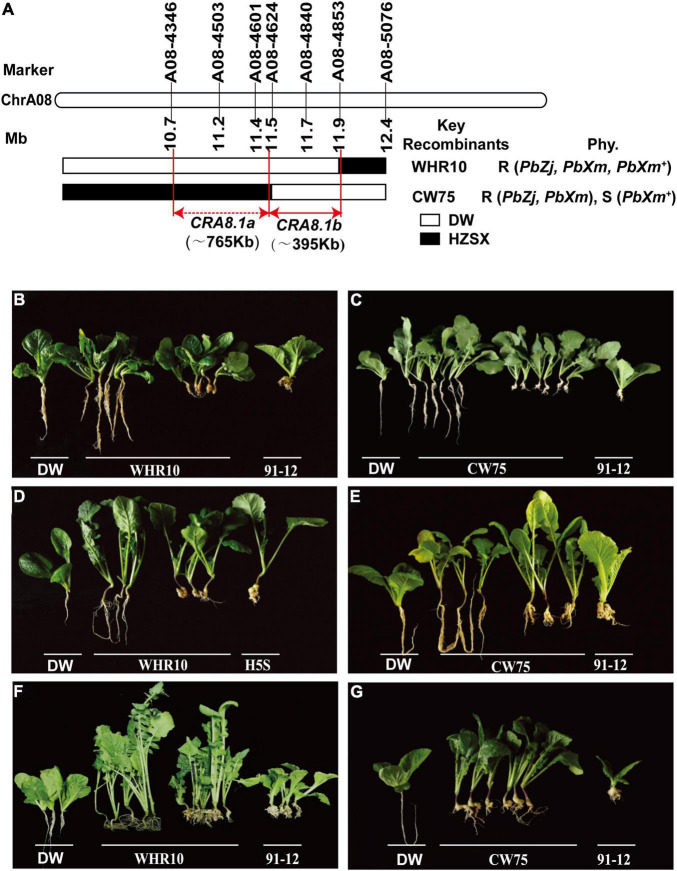
To further narrow down the region of CRA8.1 and eventually identify the candidate genes, we expanded the F1 population for fine mapping. Finally, 39 recombinants were obtained from 3290 F1 plants. Two different recombinant individuals, namely, WHR10 and CW75, were obtained, and they were sterile. We backcrossed them with HZSX, and the progenies were inoculated by PbZj, PbXm, and PbXm+, together with DW and Chinese cabbage 91-12, as positive and negative controls, respectively (Figure 3). When PbXm or PbZj was used, the backcross progeny of these two recombinants with HZSX also showed 1:1 segregation (Figures 3B–E and Supplementary Table 3); however, when PbXm+ was used, the offspring of WHR10 showed a segregation ratio of 1:1, while those from CW75 were all susceptible (Figures 3F,G and Supplementary Table 3). Based on the BSA sequencing, the fragment between A08-4346 and A08-4624 intervals, in a total length of 765 kb, might be responsible for the resistance phenotype variation between CW75 and WHR10 for PbXm+ (Figure 3A). Therefore, we concluded that CRA8.1 contains at least two disease-resistant loci, conferring resistance to different isolates (PbXm+ and PbXm/PbZj). The candidate region named as CRA8.1b covers a region of 395 kb between markers A08-4624 and A08-4853, and it contributes to resistance to PbZj/PbXm isolates. The other locus CRA8.1a, corresponding to a region between A08-4346 and A08-4624, may confer resistance to PbXm+ by itself or coordinately with CRA8.1b (Figure 3A).
FIGURE 3.
CRA8.1a or CRA8.1b loci identified by fine mapping from A08-4346 to A08-4853 markers. (A) Genotyping of two representative recombinants CW75 and WHR10, carrying CRA8.1b or CRA8.1a and CRA8.1b, respectively. Disease phenotype of WHR10 and CW75 progeny plants after inoculation of PbZj (B,C), PbXm (D,E), or PbXm+ (F,G), respectively.
Three Candidate Clubroot-Resistant Genes Identified in CRA8.1 Region Based on Resequencing and Sequence Comparison
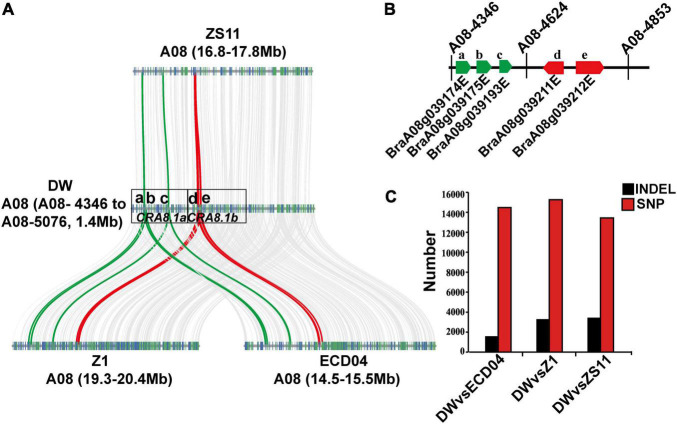
In order to identify candidate CR genes in the region between A08-4346 and A08-4853 intervals on A08 chromosome, we resequenced one of the resistant recombinant individuals R59 generated from the F1 population. The sequencing yielded 422 million clean reads with 63 billion clean bases. We manually assembled this region using ECD04 as the reference genome (unpublished) and compared gene distribution between DW, HZSX, and, a widely used B. napus variety, ZS11 (Figure 4). The total length of this assembly was 1.4 Mb (including the fragment between markers A08-4346 and A08-5076), and we found a good collinearity between DW, Z1 (HZSX), ZS11, and ECD04 (Figure 4A). Pairwise comparison within this segment suggested there were on average level of 14,395 SNPs and 2776 InDels that were different in DW to ZS11, ECD04, or Z1 varieties (Figure 4C).
FIGURE 4.
Genome structure comparison between homologous regions on A08 chromosome from DW, HZSX (Z1), ECD04, and ZS11. (A) Gene synteny analysis between chromosomal regions from A08-4346 to A08-5076 on A08 chromosome from DW, ECD04, ZS11, and Z1. a–e represent five candidate R genes found within this region. a–c belongs to the CRA8.1a region. d, e belongs to the CRA8.1b region. (B) Genomic orientation of the five candidate R genes between A08-4346 and A08-4853 on A08 chromosome. Green and red bars represent the candidate genes in CRA8.1a and CRA8.1b loci, respectively. (C) Number of SNPs and InDels in DW compared with ECD04, Z1, and ZS11 between the target regions flanked by markers of A08-4346 and A08-5076.
It is reported that 61% of the cloned R genes code for NLRs and 19% of R genes for RLPs/RLKs (Kourelis and van der Hoorn, 2018). Based on NLR/RLP annotation, we identified five R genes within the candidate region of A08 chromosome, corresponding to the following gene sequences of ECD04: CRA8.1.1 (BraA08g039174E), CRA8.1.2 (BraA08g039175E), CRA8.1.3 (BraA08g039193E), CRA8.1.4 (BraA08g039211E), and CRA8.1.5 (BraA08g039212E) (Figure 4A and Table 2). Among those candidate genes, the homologs of BraA08g039174E, BraA08g039175E, and BraA08g039193E are typical RLP genes and homologs of BraA08g039211E and BraA08g039212E are TIR-NBS-LRR genes (Figure 4B and Table 2). Homologs of BraA08g039174E, BraA08g039175E, and BraA08g039193E are located in CRA8.1a region, while the remaining two are in CRA8.1b region (Figure 4A).
TABLE 2.
List of CR candidate genes identified within CRA8.1 loci.
| Gene ID (ECD04) | AGI (Arabidopsis gene) | Gene type |
| BraA08g039174E | AT3G05360.1 | RLP |
| BraA08g039175E | AT1G71400.1 | RLP |
| BraA08g039193E | AT3G05360.1 | RLP |
| BraA08g039211E | AT4G19500.1 | TIR-NBS-LRR |
| BraA08g039212E | AT4G19510.1 | TIR-NBS-LRR |
We reason that if an R gene is present in both resistant and susceptible plants, the difference may reside on gene sequence (including CDS and promoter) or expression level, or both. Since DW is a heterozygous material for resistance, in order to find out the resistant allelic genes in DW, we designed primers covering 3.0 kb upstream and the whole CDS region using information of genome resequencing (Supplementary Table 1). After T-A cloning and sequencing, genes with sequence variation between DW and HZSX were considered as genes possibly responsible for the new type of CR in DW.
Then, in order to clarify whether there is a functional diversification between resistant versus susceptible varieties, we extracted gene homologs from ECD04, ZS11, SL (Sheng Li), and QU (Quinta),7 the latter three as susceptible B. napus representatives. We compared gene coding sequences using BioEdit software (Supplementary Figures 1–5). We found that the protein sequences of BraA08g039174E and BraA08g039175E, both as LRR-domain-only proteins, were similar between different accessions (Supplementary Figures 1, 2). As for the third LRR-domain-only protein, BraA08g039193E, the DW and ECD04 protein sequences were quite different compared with the ones in ZS11, SL, and QU materials (Supplementary Figure 3). The protein sequence of BraA08g039211E in DW showed two missing amino acids (S223, Y224 in ECD04, and Z1) and one substitution (E191) in TIR domain; otherwise, the sequences were quite similar (Supplementary Figure 4). In the case of BraA08g039212E, the sequence in DW was almost identical to that of ECD04, but different from that in susceptible Zl, ZS11, and other accessions (Supplementary Figure 5).
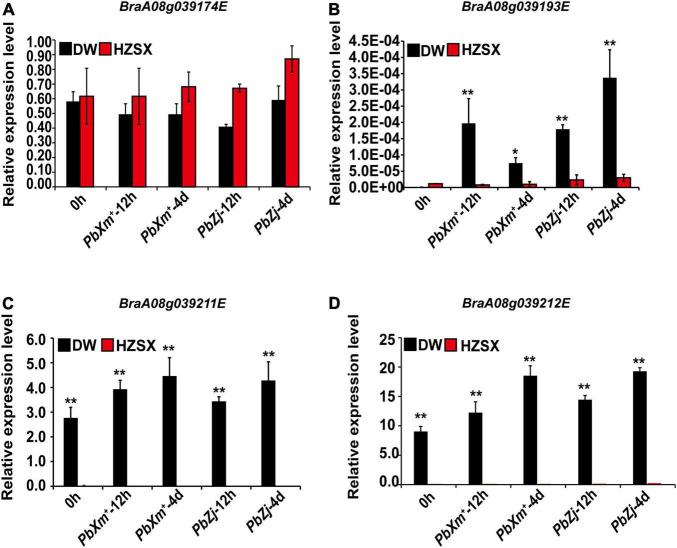
We also quantified the relative expression of these candidate genes in DW or Z1 (HZSX) upon inoculation of PbXm+ or PbZj, except for BraA08g039175E whose expression was too low (Figure 5). As shown in Figure 5, the expression of BraA08g039174E was quite similar between DW and HZSX, both before and after inoculation (Figure 5A). On the contrary, we observed a clear difference on the expression levels of BraA08g039211E and BraA08g039212E between DW and HZSX, in particular an obvious induction on BraA08g039193E in DW upon infection (Figures 5B–D). Since these genes are NBS-LRR or RLP genes and also show differential expression between susceptible and resistant materials responding to pathogen infection, they are considered as the final candidate R genes awaiting further functional verification.
FIGURE 5.
Relative expression of candidate CR genes between DW (black) and HZSX (red) before and after inoculation of PbXm+ and PbZj isolates. (A–D) Relative expression of homologs of BraA08g039174E, BraA08g039193E, BraA08g039211E, and BraA08g039212E, respectively. ** represents p ≤ 0.01 by Student’s t-test.
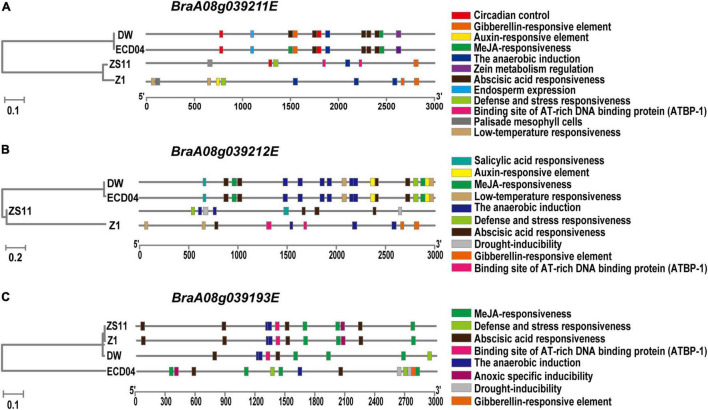
In order to find the reason for different transcript levels between resistant and susceptible materials, especially in DW, we extracted promoter sequence and predicted regulatory cis-elements within these regions from DW, ECD04, ZS11, and Z1(HZSX) (Figure 6). As shown in Figure 6, similar patterns were observed in promoter regions of BraA08g039193E, BraA08g039211E, and BraA08g039212E from ECD04 and their corresponding orthologs in DW, but different from that in susceptible ZS11 and Z1 (Figure 6). For example, the MeJA-responsive elements in the promoter regions of BraA08g039211E and BraA08g039212E were very similar in DW and ECD04, but not compared to Z1 and ZS11 (Figures 6A,B). In the case of BraA08g039193E, the cis-elements in ZS11 and Z1 were similar, followed by DW, but with additional defense- and stress-responsive elements. The cis-element in ECD04 was different from the remaining three, and whether this is of any biological relevance needs further verification (Figure 6C).
FIGURE 6.
Analysis of cis-acting elements in promoter region of the homologs of BraA08g039211E (A), BraA08g039212E (B), and BraA08g039193E (C) between resistant (DW and ECD04) and susceptible (ZS11 and Z1/HZSX) materials.
Taken together, our data from the expression profile, protein sequence, and promoter cis-element analysis suggest that BraA08g039211E and BraA08g039212E from CRA8.1b region are the most likely candidates leading to resistance to PbZj/PbXm. In addition, BraA08g039193E is likely to be the main candidate gene in CRA8.1a, but BraA08g039211E and BraA08g039212E may interact with BraA08g039193E to resist PbXm+ isolate coordinately.
Discussion
In this study, we discovered a unique B. rapa material DW that showed different resistance profiles other than H5R (PbBa8.1) or Huayouza 62R (CRb), suggesting possible new CR genes involved in its resistance. Fine mapping narrowed down to a genomic region with two NBS-LRR genes and one RLP gene as potential candidates. Once validated for their function as CR genes, those candidate genes could confer broader and higher resistance to more virulent clubroot pathogens. Therefore, genes from DW provide a new gene resource for CR breeding in future.
CRA8.1 Is Different From PbBa8.1 and Confers Superior Resistance to Diverse Pathogens
Numerous CR loci have been identified previously from ECD04 and other B. rapa materials, for instance, PbBa8.1 and Crr1a on A08 chromosome (Chen et al., 2013; Hatakeyama et al., 2013) (Table 3). In this study, our locus CRA8.1 is also located on chromosome A08 in DW, and genetically, this locus is dominant based on the F1 population segregation results. PbBa8.1 is the source of resistance in H5R; however, H5R was susceptible to PbXm pathogen, suggesting new genes in DW being responsible for its resistance. In the process of fine mapping, we found that there were two loci CRA8.1a and CRA8.1b in the primary localization interval of CRA8.1. Homologs of BraA08g039211E and BraA08g039212E are the most promising candidate genes in DW for CRA8.1b, which may play an important role against PbZj and PbXm isolates. However, what is the different resistance of BraA08g039211E and BraA08g039212E in DW and H5R remains to be further studied. The resistance of DW to PbXm+ comes from CRA8.1a itself or CRA8.1a and CRA8.1b together. So, it is not clear whether there is an interaction between the homologous genes of BraA08g039211E and BraA08g039212E and other resistance genes, or there are other types of resistance genes independently involved in the resistance response. In order to solve this problem, it is necessary to deepen the fine mapping of CRA8.1 loci, which involves the screening and isolation of recombinant plants with CRA8.1a locus only and subsequent inoculation of its offspring with PbXm+ to observe the segregation ratio.
TABLE 3.
Clubroot-resistant loci presently known in Brassica rapa.
| Location | Source | Name of loci | Pathogen | Classification | References |
| A01 | G004 (Siloga derived) | Crr2 | Wakayama-01 | Williams (race 4) | Suwabe et al., 2003 |
| A01 | ECD04 | PbBa1.1 | Pb2, Pb7 | Williams (race 2, race 7) | Chen et al., 2013 |
| A02 | C9 (Debra derived) | CRc | K04 | n.a.a | Sakamoto et al., 2008 |
| A02 | T19 (Pluto derived) | Rcr8 | Pathotype 5x | Williams (race 5b) | Yu et al., 2017 |
| A03 | ECD04 | PbBa3.1 | Pb2 | Williams (race 2) | Chen et al., 2013 |
| A03 | ECD04 | PbBa3.2 | Pb10 | Williams (race 10) | Chen et al., 2013 |
| A03 | ECD04 | PbBa3.3 | Pb7 | Williams (race 7) | Chen et al., 2013 |
| A03 | Chinese cabbage inbred line 85-74 | CRd | Pb4 | Williams (race 4) | Pang et al., 2018 |
| A03 | N-WMR-3 (Milan White derived) | Crr3 | Ano-01 | Williams (race 4) | Hirai et al., 2004 |
| A03 | K10 (CR Kanko derived) | CRk | M85 and K04 | n.a.a | Sakamoto et al., 2008 |
| A03 | Chinese cabbage cultivar, CR Shinki | CRb | Pb4 | Williams (race 4) | Piao et al., 2004 |
| A03 | T19 (Pluto derived) | Rcr4 | Pathotypes 2, 3, 5, 6, and 8 | Williams | Yu et al., 2017 |
| A03 | ECD1, ECD2, ECD4 | BraA.CR.a | n.a. | n.a | Hirani et al., 2018 |
| A03 | ECD3 | BraA.CR.c | n.a. | n.a | Hirani et al., 2018 |
| A03 | ECD02 | BraA3P5X.CRa/b Kato1.1 | Pathotype 5x | Williams (race 5b) | Fredua-Agyeman et al., 2020 |
| A03 | ECD02 | BraA3P5X.CRa/b Kato1.2 | Pathotype 5x | Williams (race 5b) | Fredua-Agyeman et al., 2020 |
| A03 | Pak choy cv. ‘Flower Nabana’ | Rcr1 | Pathotype 3 | Williams (race 3) | Chu et al., 2014 |
| A03 | Chinese cabbage inbred line T 136-8 | CRa | Pathotype 2 | Williams (race 2) | Matsumoto et al., 1998 |
| A03 | Chinese cabbage cv. ‘Jazz’ | Rcr2 | Pathotype 3 | Williams (race 3) | Huang et al., 2017 |
| A06 | G004 (Siloga derived) | Crr4 | Wakayama-01, Ano-01 | Williams (race 4) | Suwabe et al., 2003 |
| A08 | G004 (Siloga derived) | Crr1 | Wakayama-01 | Williams (race 4) | Suwabe et al., 2003 |
| A08 | ECD1, ECD2, ECD3, ECD4 | BraA.CR.b | n.a. | n.a | Hirani et al., 2018 |
| A08 | ECD04 | PbBa8.1 | Pb4 | Williams (race 4) | Chen et al., 2013 |
| A08 | 96-6990-2 (Waaslander derived) | Rcr3 | Pathotype 3H | Williams (race 3) | Karim et al., 2020 |
| A08 | T19 (Pluto derived) | Rcr9 | Pathotype 5x | Williams (race 5b) | Yu et al., 2017 |
| A08 | ECD04 | Rcr9Wa | Pathotype 5x | Williams (race 5b) | Karim et al., 2020 |
an.a., not available. bPathotype 5x is a new pathotype identified by Yu et al. (2017).
The New Type of Clubroot-Resistant Genes That Can Be Used for Revealing the Novel Resistance Mechanisms
Based on gene distribution around CRA8.1 locus in ECD04, ZS11, DW and HZSX (Z1), the resistance spectrum of DW is unique and most likely coming from the CRA8.1a fragment (Figure 4). The candidate genes were the only NBS-LRR or RLP/RLK genes within this region, of which five can be quantified for relative expression. Among the three final candidates, BraA08g039211E and BraA08g039212E are actually located head to head adjacently on the chromosome, and both are drastically induced from 12 h up to 4 days in DW after clubroot infection (Figures 4B, 5C,D). Previous studies have shown that some of the R genes work in pair, and they are physically located very close on the chromosome, such as TNL pair genes RRS1/RPS4 from Arabidopsis thaliana (Le Roux et al., 2015) and CNL pair genes RGA5/RGA4 in rice (Cesari et al., 2013). So, BraA08g039211E and BraA08g039212E as TIR-NBS-LRR genes might be very important for host response to the clubroot pathogen. The third gene, BraA08g039193E is a RLP protein. This is distinct from the previously cloned NLR-type genes and mediates immune responses at the PTI level. Once the candidate gene is validated, what is the disease resistance mechanism is also a very interesting scientific question. In addition, BraA08g039211E has little residue difference between DW and HZSX, BraA08g039193E and BraA08g039212E, the sequence in DW was quite different compared with the ones in ZS11, SL, and QU materials, so they can be used as good materials for resistance mechanism research in future.
DingWen Is an Excellent Clubroot-Resistant Donor Plant for Resistance Improvement of Brassica napus
As mentioned above, clubroot is a fast-spreading soil-borne disease which calls for fast and effective resistant varieties. The current resistance cultivars can revert to susceptible upon long terms of use, and single or a handful of CR genes cannot assure resistance to different pathotypes of the clubroot pathogens. Also, the accumulation of mutations eventually would result in the loss of resistance. For example, in Alberta of Canada, a “new” brassicae pathogenic type that can overcome resistance was discovered only 4 years after the CR variety release (Strelkov et al., 2016; Fredua-Agyeman et al., 2018). Most CR genes used currently are coming from B. rapa, since crossability between B. rapa and B. napus is very good, and CR genes from B. rapa can readily be transferred to B. napus. The current two widely used CR varieties of B. napus, namely, H5R and 409R, are resistant to PbZj but not to PbTc and PbXm (Shah et al., 2020). Previous studies have shown that different disease resistance genes’ aggregation can improve the resistance to clubroot (Suwabe et al., 2006; Shah et al., 2020). In this study, the CRA8.1 locus located in DW contains at least two CR loci, which is superior to PbBa8.1 and CRb on A08 chromosome, so the two resistance genes of CRA8.1 can be introduced into excellent B. napus parents to improve the CR in breeding, which can save cost and improve work efficiency.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YW performed the experiments, analyzed the data, prepared the figures, and drafted the manuscript. XX helped to analyze the data. CZ and PC conceived the study, participated in its coordination, and helped to draft the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. U20A2034 and 31871659) and China Agriculture Research System (CARS-12) to CZ.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.898108/full#supplementary-material
Protein sequence alignments of homologs of BraA08g039174E from DW, ECD04, Z1, ZS11, SL, and QU. Red solid lines represent the LRR domain.
Protein sequence alignments of homologs of BraA08g039175E from DW, ECD04, Z1, ZS11, SL, and QU. Red solid lines represent the LRR domain.
Protein sequence alignments of homologs of BraA08g039193E from DW, ECD04, Z1, ZS11, SL, and QU. Red solid lines represent the LRR domain.
Protein sequence alignments of homologs of BraA08g039211E from DW, ECD04, Z1, ZS11, SL, and QU. Green solid lines represent the TIR domain, orange solid lines represent the NB-ARC domain, and red solid lines represent the LRR domain. The symbol “**” indicates the two missing residues in DW compared with other accessions. The symbol “*” represents the specific residues in DW.
Protein sequence alignments of homologs of BraA08g039212E from DW, ECD04, Z1, ZS11, SL, and QU. Green solid lines represent the TIR domain, orange solid lines represent the NB-ARC domain, and red solid lines represent the LRR domain. The symbol “*” represents the specific residues in DW.
Primers used in this study.
Quality control of BSA sequencing data.
Phenotype statistics of clubroot resistance for progeny of WHR10 and CW75 recombinants.
References
- Belser C., Istace B., Denis E., Dubarry M., Baurens F. C., Falentin C., et al. (2018). Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 4 879–887. 10.1038/s41477-018-0289-4 [DOI] [PubMed] [Google Scholar]
- Buczacki S. T., Toxopeus H., Mattusch P., Johnston T. D., Dixon G. R., Hobolth L. A. (1975). Study of physiologic specialization in Plasmodiophora brassicae : proposals for attempted rationalization through an international approach. Transac. Brit. Mycol. Soc. 65 295–303. 10.1016/S0007-1536(75)80013-1 [DOI] [Google Scholar]
- Cesari S., Thilliez G., Ribot C., Chalvon V., Michel C., Jauneau A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25 1463–1481. 10.1105/tpc.112.107201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai A. L., Xie X. W., Shi Y. X., Li B. J. (2014). Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 36 142–153. 10.1080/07060661.2013.868829 [DOI] [Google Scholar]
- Chen J., Jing J., Zhan Z., Zhang T., Zhang C., Piao Z. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One 8:e85307. 10.1371/journal.pone.0085307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Pang W., Chen B., Zhang C., Piao Z. (2015). Transcriptome Analysis of Brassica rapa Near-Isogenic Lines Carrying Clubroot-Resistant and -Susceptible Alleles in Response to Plasmodiophora brassicae during Early Infection. Front. Plant Sci. 6:1183. 10.3389/fpls.2015.01183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M., Song T., Falk K. C., Zhang X., Liu X., Chang A., et al. (2014). Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 15:1166. 10.1186/1471-2164-15-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Ning Y., Yang D. L., Zhai K., Wang G. L., He Z. (2020). Molecular basis of disease resistance and perspectives on breeding strategies for resistance improvement in crops. Mol. Plant 13 1402–1419. 10.1016/j.molp.2020.09.018 [DOI] [PubMed] [Google Scholar]
- Dixon G. R. (2009). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Grow. Regul. 28 194–202. 10.1007/s00344-009-9090-y [DOI] [Google Scholar]
- Elke D., Martin F., Enrico G. A. L., Katsunori H., Masashi H. (2009). Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 28 265–281. 10.1007/s00344-009-9100-0 [DOI] [Google Scholar]
- Fredua-Agyeman R., Hwang S. F., Strelkov S. E., Zhou Q., Feindel D. (2018). Potential loss of clubroot resistance genes from donor parent Brassica rapa subsp rapifera (ECD04) during doubled haploid production. Plant Pathol. 67 892–901. 10.1111/ppa.12816 [DOI] [Google Scholar]
- Fredua-Agyeman R., Jiang J., Hwang S. F., Strelkov S. E. (2020). QTL Mapping and Inheritance of Clubroot Resistance Genes Derived From Brassica rapa subsp. rapifera (ECD02) Reveals Resistance Loci and Distorted Segregation Ratios in Two F2 Populations of Different Crosses. Front. Plant Sci. 11:899. 10.3389/fpls.2020.00899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahatraj S., Shrestha S. M., Devkota T. R., Rai H. H. (2019). A review on clubroot of crucifers: symptoms, life-cycle of pathogen, factors affecting severity, and management strategies. Arch. Agricul. Environ. Sci. 4 342–349. 10.26832/24566632.2019.0403012 [DOI] [Google Scholar]
- Grund E., Tremousaygue D., Deslandes L. (2019). Plant NLRs with integrated domains: unity makes strength. Plant Physiol. 179 1227–1235. 10.1104/pp.18.01134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K., Suwabe K., Tomita R. N., Kato T., Nunome T., Fukuoka H., et al. (2013). Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 8:e54745. 10.1371/journal.pone.0054745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M., Harada T., Kubo N., Tsukada M., Suwabe K., Matsumoto S. (2004). A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 108 639–643. 10.1007/s00122-003-1475-x [DOI] [PubMed] [Google Scholar]
- Hirani A. H., Gao F., Liu J., Fu G., Wu C., McVetty P., et al. (2018). Combinations of independent dominant loci conferring clubroot resistance in all four turnip accessions (Brassica rapa) from the European clubroot differential set. Front. Plant Sci. 9:1628. 10.3389/fpls.2018.01628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Peng G., Liu X., Deora A., Falk K. C., Gossen B. D., et al. (2017). Fine Mapping of a Clubroot Resistance Gene in Chinese Cabbage Using SNP Markers Identified from Bulked Segregant RNA Sequencing. Front Plant Sci. 8:1448. 10.3389/fpls.2017.01448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. F., Strelkov S. E., Feng J., Gossen B. D., Howard R. J. (2012). Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 13 105–113. 10.1111/j.1364-3703.2011.00729.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M. M., Dakouri A., Zhang Y., Chen Q., Peng G., Strelkov S. E., et al. (2020). Two Clubroot-Resistance genes. Rcr3 and Rcr9wa, mapped in Brassica rapa using bulk segregant RNA sequencing. Int. J. Mol. Sci. 21:5033. 10.3390/ijms21145033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Qi D., Ashfield T., Helm M., Innes R. W. (2016). Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351 684–687. 10.1126/science.aad3436 [DOI] [PubMed] [Google Scholar]
- Kim W., Prokchorchik M., Tian Y., Kim S., Jeon H., Segonzac C. (2020). Perception of unrelated microbe-associated molecular patterns triggers conserved yet variable physiological and transcriptional changes in Brassica rapa ssp. Pekinensis. Hortic Res. 7:186. 10.1038/s41438-020-00410-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J., van der Hoorn R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30 285–299. 10.1105/tpc.17.00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan N. J., Ma L., Borhan M. H. (2015). The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 13 983–992. 10.1111/pbi.12341 [DOI] [PubMed] [Google Scholar]
- Larkan N. J., Ma L., Haddadi P., Buchwaldt M., Parkin I., Djavaheri M., et al. (2020). The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 104 892–900. 10.1111/tpj.14966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C., Huet G., Jauneau A., Camborde L., Tremousaygue D., Kraut A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161 1074–1088. 10.1016/j.cell.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Shah N., Zhou X., Wang H., Yu W., Luo J., et al. (2021). Identification of Micro Ribonucleic Acids and their Targets in Response to Plasmodiophora brassicae Infection in Brassica napus. Front. Plant Sci. 12:734419. 10.3389/fpls.2021.734419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto E., Yasui C., Ohi M., Tsukada M. (1998). Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. Pekinensis). Euphytica 104 79–86. 10.1023/A:1018370418201 [DOI] [Google Scholar]
- Mehraj H., Akter A., Miyaji N., Miyazaki J., Shea D. J., Fujimoto R., et al. (2020). Genetics of clubroot and fusarium wilt disease resistance in brassica vegetables: the application of marker assisted breeding for disease resistance. Plants 9:726. 10.3390/plants9060726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadil S., Jincai S., Shaowei Y., Zhaochun Y., Zuo W., Fan H., et al. (2019). Genetic variation analysis of field isolates of clubroot and their responses to Brassica napus lines containing resistant genes CRb and PbBa8.1 and their combination in homozygous and heterozygous state. Mol. Breeding 39:153. 10.1007/s11032-019-1075-3 [DOI] [Google Scholar]
- Narasimhan V., Danecek P., Scally A., Xue Y., Tyler-Smith C., Durbin R. (2016). BCF tools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32 1749–1751. 10.1093/bioinformatics/btw044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neik T. X., Barbetti M. J., Batley J. (2017). Current status and challenges in identifying disease resistance genes in brassica napus. Front. Plant Sci. 8:1788. 10.3389/fpls.2017.01788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. L., Monakhos G. F., Komakhin R. A., Monakhos S. G. (2018). The New Clubroot Resistance Locus is Located on Chromosome A05 in Chinese Cabbage (Brassica rapa L.). Russ. J. Genet. 54 296–304. 10.1134/S1022795418030080 [DOI] [Google Scholar]
- Pang W., Fu P., Li X., Zhan Z., Yu S., Piao Z. (2018). Identification and Mapping of the Clubroot Resistance Gene CRd in Chinese Cabbage (Brassica rapa ssp. Pekinensis). Front. Plant Sci. 9:653. 10.3389/fpls.2018.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang W., Liang Y., Zhan Z., Li X., Piao Z. (2020). Development of a sinitic clubroot differential set for the pathotype classification of Plasmodiophora brassicae. Front. Plant Sci. 11:568771. 10.3389/fpls.2020.568771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao Z. Y., Deng Y. Q., Choi S. R., Park Y. J., Lim Y. P. (2004). SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp pekinensis). Theor. Appl. Genet. 108 1458–1465. 10.1007/s00122-003-1577-5 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010). BED Tools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Saito A., Hayashida N., Taguchi G., Matsumoto E. (2008). Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. Ssp pekinensis). Theor. Appl. Genet. 117 759–767. 10.1007/s00122-008-0817-0 [DOI] [PubMed] [Google Scholar]
- Shah N., Li Q., Xu Q., Liu J., Huang F., Zhan Z., et al. (2020). CRb and PbBa8.1 Synergically Increases Resistant Genes Expression upon Infection of Plasmodiophora brassicae in Brassica napus. Genes 11:202. 10.3390/genes11020202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov S. E., Hwang S., Manolii V. P., Cao T., Feindel D. (2016). Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta. Canada. Eur. J. Plant Pathol. 145 517–529. 10.1007/s10658-016-0888-8 [DOI] [Google Scholar]
- Suwabe K., Tsukazaki H., Iketani H., Hatakeyama K., Fujimura M., Nunome T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 107 997–1002. 10.1007/s00122-003-1309-x [DOI] [PubMed] [Google Scholar]
- Suwabe K., Tsukazaki H., Iketani H., Hatakeyama K., Kondo M., Fujimura M., et al. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana : the genetic origin of clubroot resistance. Genetics 173 309–319. 10.1534/genetics.104.038968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Matsumoto E., Aruga D., Kitagawa S., Matsumura H., Hayashida N. (2012). Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 80 621–629. 10.1007/s11103-012-9971-5 [DOI] [PubMed] [Google Scholar]
- Voorrips R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Wang J., Chai J. (2020). Molecular actions of NLR immune receptors in plants and animals. Science China. Life Sci. 63 1303–1316. 10.1007/s11427-019-1687-6 [DOI] [PubMed] [Google Scholar]
- Williams P. H. (1966). A system for the determination of races of Plasmodiophora brassicae that infect Cabbage and Rutabaga. Phytopathology 56 624–626. [Google Scholar]
- Wu Y., Zhou J. M. (2013). Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 55 1271–1286. 10.1111/jipb.12123 [DOI] [PubMed] [Google Scholar]
- Yu F., Zhang X., Peng G., Falk K. C., Strelkov S. E., Gossen B. D. (2017). Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 7:4516. 10.1038/s41598-017-04903-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Jiang Z., Bi G., Nomura K., Liu M., Wang Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592 105–109. 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Z., Jiang Y., Shah N., Hou Z., Zhou Y., Dun B., et al. (2020). Association of clubroot resistance locus PbBa8.1 with a linkage drag of high erucic acid content in the seed of the European turnip. Front. Plant Sci. 11:810. 10.3389/fpls.2020.00810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein sequence alignments of homologs of BraA08g039174E from DW, ECD04, Z1, ZS11, SL, and QU. Red solid lines represent the LRR domain.
Protein sequence alignments of homologs of BraA08g039175E from DW, ECD04, Z1, ZS11, SL, and QU. Red solid lines represent the LRR domain.
Protein sequence alignments of homologs of BraA08g039193E from DW, ECD04, Z1, ZS11, SL, and QU. Red solid lines represent the LRR domain.
Protein sequence alignments of homologs of BraA08g039211E from DW, ECD04, Z1, ZS11, SL, and QU. Green solid lines represent the TIR domain, orange solid lines represent the NB-ARC domain, and red solid lines represent the LRR domain. The symbol “**” indicates the two missing residues in DW compared with other accessions. The symbol “*” represents the specific residues in DW.
Protein sequence alignments of homologs of BraA08g039212E from DW, ECD04, Z1, ZS11, SL, and QU. Green solid lines represent the TIR domain, orange solid lines represent the NB-ARC domain, and red solid lines represent the LRR domain. The symbol “*” represents the specific residues in DW.
Primers used in this study.
Quality control of BSA sequencing data.
Phenotype statistics of clubroot resistance for progeny of WHR10 and CW75 recombinants.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.