Abstract
The demand for Kombucha, a sparkling sugared tea beverage fermented by a symbiotic culture of acetic acid bacteria (AAB) and yeast is increasing worldwide. Despite the popularity of the beverage which is mainly due to its perceived health benefits and appealing sensory properties, the microbial composition of the products at the time of consumption is unknown. Such information is important to both manufacturers and consumers. Therefore, this study characterised the dominant AAB and yeast present in six commercial Kombucha samples sold in New Zealand which comprised of three domestic and three imported samples. Acetic acid bacteria and yeast were isolated from the Kombucha samples using glucose yeast extract peptone mannitol (GYPM) and yeast extract glucose chloramphenicol (YGC) media, respectively. Phenotypic and taxonomic identification of AAB and yeast were achieved by morphological and biochemical characterisation, followed by sequence analysis of ribosomal RNA genes (16S rRNA for AAB and 26S rRNA for yeast). Viable AAB and yeast were only found in domestically produced Kombucha samples and not in the imported products. The dominant AAB species were identified as Acetobacter musti and Gluconobacter potus. The yeast isolates belonged to Dekkera bruxelensis, Schizosaccharomyces pombes, Hanseniaspora valbyensis, Brettanomyces anamalus, Pichia kudriavzevii, Starmerella vitis and Saccharomyces cerevisiae. The yeast communities were more complex and variable than the AAB communities in the analysed Kombucha samples.
Keywords: Kombucha, Acetic acid bacteria, Yeast, Acetobacter musti, Gluconobacter potus, Saccharomyces cerevisiae, Dekkera bruxelensis
Graphical abstract
Highlights
-
•
Kombucha samples produced in New Zealand contained viable yeast and AAB while none were found in imported samples.
-
•
Dominant AAB found in the Kombucha samples were Gluconobacter potus LMG1764 and Acetobacter musti Bo7.
-
•
Dominant yeast isolates in the Kombucha samples included Saccharomyces cerevisiae CEN.PK113-7D.
1. Introduction
Kombucha is a popular sparkling tea-based drink, reported as having originated from China around 5000 years ago. Its consumption then spread to Japan, Russia and middle eastern countries, and it is now consumed around the world (Dufresne and Farnworth, 2000). The taste of Kombucha is mildly acidic and alcoholic, similar to apple cider (Marsh et al., 2014). It can vary from slightly fruity, sour and fizzy to vinegar-like depending on the fermentation conditions (Goh et al., 2012). Regular consumption of Kombucha may confer health benefits, including antioxidant, antimicrobial and hepatoprotective effects to consumers. However, there is limited scientific evidence to support these claims (Dufresne and Farnworth, 2000), and hence research is needed in this area.
Kombucha is commonly fermented by a complex symbiotic culture of acetic acid bacteria (AAB) and yeast in a base of sugared tea infusion at ambient temperature for 7–10 days (Jayabalan et al., 2014). The AAB-yeast symbiotic culture is commonly abbreviated as SCOBY, which translates to Symbiotic Culture of Bacteria and Yeast. Brewed black tea is reported to be the most common “tea substrate” that has been used throughout history for Kombucha production (Reiss, 1994). Other substrates including green tea, oolong tea and medicinal herbs such as lemon balm or peppermint have also been used (Velićanski et al., 2013). Sucrose is the most popular carbon source for the growth of cultures during fermentation, and the concentration usually ranges from 5% to 15% (w/v) (Jayabalan et al., 2014). During Kombucha fermentation, sucrose is hydrolysed by the yeast invertase into fructose and glucose. The two monosaccharides are then metabolised in the glycolytic pathway, thereby producing ethanol and carbon dioxide. Carbon dioxide is responsible for imparting the desirable carbonation of Kombucha (Villarreal-Soto et al., 2018). The ethanol produced is oxidised to acetaldehyde and then to acetic acid by AAB which is the predominant acid in Kombucha. The acetic acid produced in Kombucha contributes to the reduction of pH (Jayabalan et al., 2014). Concurrently, the AAB community converts the sucrose-derived glucose to gluconic acid, then into glucuronic acid. The latter organic acid is perceived to be associated with the hepatoprotective effects of Kombucha (Jayabalan et al., 2014). AAB contribute to the sourness of Kombucha through the fermentation of sucrose to acetic acid, which in turn stimulates the growth of yeast to produce ethanol (Kumar and Joshi, 2016).
During Kombucha fermentation, the floating pellicle (tea fungus) rests on the liquid tea broth (Chakravorty et al., 2016). The tea fungus is usually recovered and utilised with a small amount of tea broth in the next fermentation. The bulk of the fermented tea broth is retrieved for consumption by consumers after chilling. Kombucha contains a complex mixture of chemical components which include organic acids, B-vitamins complex (vitamins B1, B2 and B6), trace elements (e.g., Cu and Fe) and polyphenols such as catechins (Jayabalan et al., 2017). The chemical composition of the beverage varies between products due to factors such as the microbial composition of the starter culture, fermentation conditions and added substrates (Martínez Leal et al., 2018).
It is important to understand the microbial composition of starter cultures involved in Kombucha fermentation and their metabolic activities to facilitate better control of the fermentation process. Furthermore, knowledge of the cultures present is desirable as the beverage is sold as a live product with perceived health benefits. Previous work showed that gluconic acid-producing AAB are the dominant prokaryotes in Kombucha (Jayabalan et al., 2014). More specifically, Acetobacter (A.) xylinum, Acetobacter aceti, Acetobacter pasteruianus, Gluconobacter (G.) oxygendans, Gluconacetobacter sacchari and Komagaeibacter saccharivorans were identified as the main AAB in certain Kombucha products (Jayabalan et al., 2011; Liu et al., 1996; Mukadam et al., 2016). Similarly, yeast species vary between Kombucha beverages, and they are present at lower concentrations than the bacteria (Matei et al., 2018). Brettanomyces bruxellensis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Zygosaccharomyces bailii and Zygosaccharommyces rouxii have been previously reported in Kombucha beverages (Dufresne and Farnworth, 2000; Jayabalan et al., 2017; Liu et al., 1996). The interaction between different yeast and AAB species may stimulate or interfere with the growth of other species, and their metabolic characteristics can influence the chemical composition of the Kombucha (Villarreal-Soto et al., 2018). However, there are limited studies on the composition of Kombucha starter cultures and their metabolic characteristics. The main challenges in understanding Kombucha cultures are the diversity and complexity of the microbial communities involved (Chakravorty et al., 2016). Variations in culture composition may be due to climatic and geographic conditions (Mayser et al., 1995). Identification and knowledge of the microbial characteristics of Kombucha cultures offers manufacturers’ the opportunity to better control the fermentation process, which will promote the production of safe, consistently high-quality products. Here we report the identification and characterisation of dominant AAB and yeast strains in popular Kombucha beverages sold in New Zealand.
2. Materials and methods
2.1. Preparation of sample
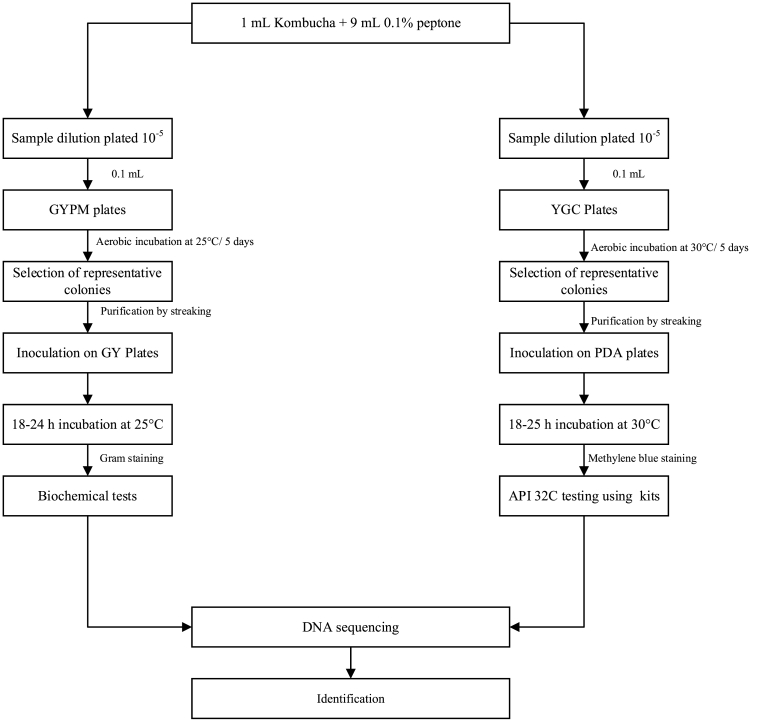
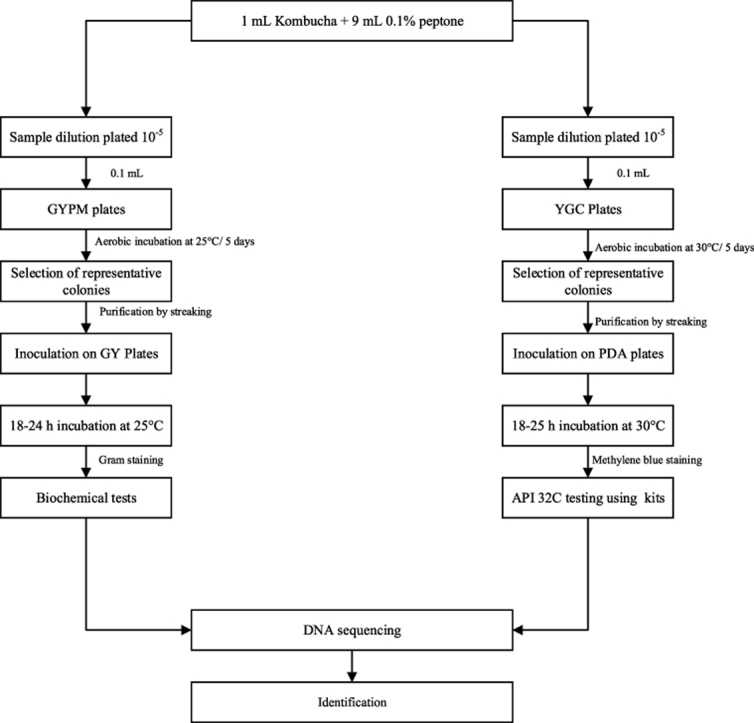
Six branded Kombucha beverages were randomly purchased from local supermarkets in Auckland, New Zealand. The samples were transported to the Food Technology Department at Massey University, Auckland, under a cold chain maintained at 4 °C and were stored at the same temperature until required for analysis shown in Fig. 1.
Fig. 1.
Microbial analysis of Kombucha sample.
2.2. Physico-chemical characteristics of Kombucha samples
2.2.1. Determination of pH, titratable acidity (TA) and total soluble solids (TSS) of Kombucha samples
The pH of each Kombucha sample (50 mL) was measured using a Sartorius glass electrode pH meter (Model PH-11, Germany). Titratable acidity was determined by acid-base titration. Briefly, each sample (30 mL) was uncapped for 3–5 h under chilled conditions (4–6 °C) to allow carbon dioxide to escape from the sample, followed by titration with standardised 0.1 M sodium hydroxide to pH 8.2 (Waisundara, 2018). The calculated TA was expressed as a percentage (%) of acetic acid per gramme of sample, as it is the major organic acid in Kombucha (Equation (1)) (Waisundara, 2018).
| Equation 1 |
VNaOH = volume of NaOH (mL)
MNaOH = molarity of NaOH (M)
Vsample = volume of sample (mL)
The TSS of Kombucha samples were determined using a refractometer (Atago, pr-32 alpha, UK), according to Amarasinghe et al. (2018).
2.3. Enumeration and isolation of AAB and yeast
Suitable serial dilutions of Kombucha samples were plated on appropriate microbiological media. The modified glucose yeast extract peptone mannitol (GYMP) agar containing 0.2% D-glucose (w/v) (ThermoFisher, Waltham, Massachusetts, USA) was used for propagating AAB in Kombucha samples (Gomes et al., 2018). The GYPM contained 2.5% D-mannitol (w/v) (ThermoFisher, Waltham, Massachusetts, USA), 0.5% yeast extract (w/v) (Sigma-Aldrich, St. Louis, Missouri, USA), 0.3% peptone (w/v) (Sigma-Aldrich, St. Louis, Missouri USA), 1.2% bacteriological agar (w/v) (ThermoFisher, Waltham, Massachusetts USA). Cycloheximide (0.1 g/L) (Sigma-Aldrich, St. Louis, Missoouri, USA) and pimaricin (1 mg/L) (Sigma-Aldrich, St. Louis, Missouri, USA) were added to the GYPM to inhibit the growth of yeast and lactic acid bacteria, respectively. Yeast isolates were plated on yeast extract chloramphenicol (YGC) agar (ThermoFisher, Waltham, Massachusetts, USA). The AAB were cultivated at 25 °C for 5 days and yeast at 30 °C for 5 days. Grown colonies of the microorganisms were enumerated, and the results were expressed as log CFU/mL. Representative AAB and yeast isolates were purified by successive streaking on glucose yeast extract (GY) and potato dextrose agar (PDA), respectively. The AAB and yeast isolated cultures were stored in 67% glycerol (w/w) at −80 °C for long term preservation, and on GY agar or PDA agar plates at 4 °C with monthly sub-culturing (Du Toit and Lambrechts, 2002).
2.4. Phenotypic characterisation of AAB and yeast
2.4.1. Phenotypic characteristics of AAB
The purified AAB isolates from Kombucha were incubated on GY agar plates at 25 °C for 3–5 days. A loopful of fresh purified AAB culture was used to conduct biochemical tests. Gram-stains of the isolates were prepared, and the cell morphology was examined under oil immersion using a Carl Zeiss Transmission light microscope (Model HBO 50/AC, Germany) (Abd El-Salam, 2012; Claus, 1992). Cell sizes of the AAB isolates were measured using the AxioVision microscope software Version 4.8.1. Biochemical tests were performed on the Gram-negative isolates. Catalase activity was confirmed by the rapid evolution of air bubbles on young, purified colonies after the addition of 1–2 drops of 6% H2O2 (v/v) (Sigma-Aldrich, St. Louis, Missouri, USA) (Reiner, 2010). The oxidase test was carried out using oxidase strips (Oxoid, UK) and observed for a colour change for 30 s as described by Shields and Cathcart (2010). Growth at different temperatures was carried out on GY agar plates incubated aerobically at selected temperatures (25 °C, 30 °C, 37 °C) for 3–7 days (Yamada et al., 1976). Growth of AAB on glucose yeast extract calcium carbonate (GYC) agar was conducted as described by Yamada et al. (1976) with minor modifications. Growth of the bacteria in the presence of 0.35% acetic acid medium (w/v, pH 3.5) was performed using a medium consisting of 1.0% yeast extract (w/v), 1.0% D-glucose (w/v) and 3.5 mL glacial acetic acid (Sigma-Aldrich, St. Louis, Missouri USA). Metabolism of D-glucose by AAB was tested by inoculating the bacteria into medium containing 30% D-glucose (w/v), 1.0% yeast extract (w/v) and 1.5% bacteriological agar (w/v). Growth in methanol medium was carried out according to the method of Hanmoungjai et al. (2007) and growth in dextrose sorbitol mannitol (DSM) medium was determined based on the method of Cirigliano (1982). Growth in glutamate agar was achieved using the method of Asai et al. (1964). The oxidation of ethanol to acetic acid and over-oxidation of ethanol to CO2 and water by AAB was determined using Carr medium (Gomes et al., 2018). The alcohol tolerance test was performed on basal medium (5 g/L yeast extract and 20 g/L bacteriological agar) containing 2%, 4%, 6% and 10% (v/v) absolute ethanol (Gullo and Giudici, 2008). The SIM (sulphate indole motility) medium was used to test the formation of H2S, indole and cell motility (Visser et al., 1985). The gelatine hydrolysis test was carried out using the stabbing method (dela Cruz and Torres, 2012). Oxidation of acetate and lactate was tested using the modified method of Asai et al. (1964).
Cellulose production was determined using the modified method of Lavasani et al. (2017) and Semjonovs et al. (2017) by inoculating the isolates in the Hestrin-Schramm (HS) broth. After inoculation, 1 mL of well-mixed broth culture was transferred into a 2-mL centrifuge tube, mixed with 1 mL of 0.1 M NaOH solution and heated at 90 °C on a hot plate (Benchmark Scientific, USA) for 30 min set at medium heat. Ketogenesis of glycerol to dihydroxyacetone (DHA) was determined using the modified method of Swings (1992) using glycerol yeast extract medium (GYE). After inoculation, Benedict's Reagent was flooded on the surface of the agar plates and incubated for 3 h at 30 °C to observe any colour change. Formation of γ-pyrone from D-glucose and D-fructose was tested on glucose yeast (GY) and fructose yeast extract (FY) broth in a shaking incubator (150 rpm) (KBLee 1001, DAIKI SCIENCES, Korea), respectively. A few drops of FeCl3 (5%, w/v) were added into the broth and observed immediately for any colour change. The acid produced from mannitol, D-glucose and glycerol was determined according to the method of Arifuzzaman et al. (2014) using a basal medium consisting of 1% carbohydrate source (w/v), 0.5% yeast extract (w/v), and 1% peptone (w/v) with 0.002% bromocresol purple (w/v) as pH indicator. Nitrate reduction was determined from nitrate peptone medium consisting of 1% peptone (w/v) and 0.2% KNO3 (w/v) (Buxton, 2011).
2.4.2. Phenotypic characteristics of yeast
The purified yeast isolates from the Kombucha samples were plated on PDA plates and incubated at 30 °C for 3–5 days. The morphology of the yeast cells was observed using the methylene blue staining method and the cell size was determined using a Carl Zeiss Transmission light microscope (Model HBO 50/AC, Germany) (Matthews, 1914; Painting and Kirsop, 1990). The API ID 32C kit system was used to determine the assimilation of carbohydrates following the manufacturer's instructions (BIOMEREUX, France). The identification of the yeast cells was obtained from the APIWEB™ software database V (https://apiweb.biomerieux.com/strip/12).
2.5. Sequence analysis of ribosomal RNA genes
DNA sequencing was performed by Macrogen Inc. (Seoul, Korea). Briefly, total DNA was extracted from bacterial and yeast cells using the DNeasy Blood and Tissue kit (Qiagen, Germany) following the manufacturer's instructions. Next, the full-length 16S rRNA gene was amplified by polymerase chain reaction (PCR) using primers 27F and 1492R, and the PCR products were sequenced using two internal outward-facing primers 785F and 907R (Yuan et al., 2013). For 26S rRNA genes of yeast cells, PCR and sequencing were conducted using the same set of primers LR0R/LR7 (Schneider et al., 2015). The obtained DNA sequences were analysed in Geneious 9.0.5 (Biomatters Ltd, Auckland, New Zealand), including visual inspection of the individual sequence profiles and subsequent alignment of the DNA sequences for each of the two genes. To identify the taxonomy of the AAB, the resultant 16S rRNA sequences were subjected to online search for closest homologues in the EzBioCloud 16S rRNA database (Yoon et al., 2017). The yeast strains were identified by nucleotide BLAST (Basic Local Alignment Search Tool) of 26S rRNA gene sequences deposited in the NCBI (National Center for Biotechnology Information) database.
2.6. Statistical analysis
Data on pH, titratable acidity and total soluble solids were analysed by Microsoft Excel 2016 (Microsoft, USA) using descriptive statistics. Data on pH, TA and TSS were analysed by analysis of variance-one way (ANOVA-ONEWAY) using the IBM SPSS version 26 (IBM, USA) to determine significant differences of the means (p < 0.05).
3. Results and discussion
3.1. Physico-chemical characteristics of Kombucha samples
3.1.1. Acidity and TSS of Kombucha
The acidity (pH and TA) and TSS of six Kombucha samples are shown in Table 1 pH and TA are frequently used to express the acidity of food products (Nielsen, 2017). Acidity is an important factor to monitor the fermentation process of Kombucha and is attributed to the metabolic activities of AAB and yeast that produce organic acids such as acetic, gluconic and glucuronic acids (Chen and Liu, 2000). Variation in the hydrogen ion concentration (pH) can affect the growth of fermenting microorganisms in Kombucha (Neffe-Skocińska et al., 2017). Most AAB species can grow at pH ranging from 3.5 to 8.5, and some species have been reported to grow below pH 3 (Jayabalan et al., 2014). For yeast, the optimum pH for growth lies between pH 4.5 and 6.0 (Neffe-Skocińska et al., 2017). The pH range of the six Kombucha samples analysed in this study was between 3.21 ± 0.01 and 3.90 ± 0.01, while the TA ranged from 0.38 % - 0.43 %. The highest pH was recorded in sample DO, which contained spices and ginger flavour. The LO sample which had a Feijoa flavour had the lowest pH. However, the lowest TA was 0.38% from samples DO and GB which contained lemon and ginger flavours. The traditional flavoured sample WO had the highest TA (0.43 %). The variations between pH and TA in different commercial Kombucha products may be attributed to the buffering characteristics of the fermented Kombucha broth. The buffering capacity is further enhanced by the production of carbon dioxide during fermentation which in turn dissociates in aqueous conditions producing amphiprotic hydrocarbonate anions (Essawet et al., 2015). The anions react with hydrogen ions from inherent organic acids in the fermentation system, which inhibits changes in pH. However, the TA of Kombucha samples provides a better indication of acidity than pH, as it does not only rely on the buffering characteristics of Kombucha.
Table 1.
Mean pH, TA and TSS of six Kombucha samples sourced in New Zealand.
| Sample code | Country of Origin | Flavour | pH | TA (%) | TSS (°Brix) |
|---|---|---|---|---|---|
| LO | New Zealand | Feijoa | 3.21 ± 0.01 | 0.42 ± 0.04 | 3.70 ± 0.00 |
| DO | New Zealand | Chai spices and Ginger | 3.90 ± 0.01 | 0.38 ± 0.02 | 7.00 ± 0.00 |
| GB | New Zealand | Lemon and Ginger | 3.43 ± 0.04 | 0.38 ± 0.02 | 4.00 ± 0.00 |
| AM | Australia | Peach & Mango | 3.54 ± 0.01 | 0.40 ± 0.02 | 2.47 ± 0.06 |
| RE | Australia | Original | 3.40 ± 0.01 | 0.40 ± 0.02 | 1.87 ± 0.06 |
| WO | USA | Traditional | 3.45 ± 0.02 | 0.43 ± 0.01 | 6.50 ± 0.00 |
TA = titratable acidity; TSS = total soluble solids; data are expressed as mean ± SD; n = 3.
Results from previous studies indicate a large variation in the acidity (pH and TA) of Kombucha made using different tea substrates and fermentation conditions. pHs ranged from 2.95 (Velićanski et al., 2013) to 6.0 (Waisundara, 2018) from different Kombucha samples, with pH affected by different tea substrates (Acacia arabica), for example, herbal tea plant powders: pH 4.0–6.0; medicinal herbs (sage): pH 2.95 (Velićanski et al., 2013); different snake fruit cultivars: pH 3.12 to 3.38 (Zubaidah et al., 2019). The pH values of the samples in this study (pH 3.21 ± 0.01–3.90 ± 0.01) are most similar to those of snake fruit. Fermentation time also influences the acidity of Kombucha, with a longer fermentation time possibly resulting in higher acidity (lower pH). For example, Kombucha fermented from a mixture of green tea (2 g/L) and black tea (4 g/L) for nine days had a lower pH (2.77) than after seven days (pH 2.88) (Neffe-Skocińska et al., 2017). The TA of Kombucha samples in this study ranged from 0.38% to 0.43% which is similar to that of other Kombucha beverages (0.40%–0.45%) which were characterised with a pleasant sour flavour (Waisundara, 2018).
In this study, the range of pH and TA of imported Kombucha samples was narrower compared to the acidity of the domestic samples. The differences in acidity may be affected by the different compositions of microbial starter cultures, substrate content (tea and sugar), and fermentation conditions (Neffe-Skocińska et al., 2017). Currently, there is no standardised pH range for fermented Kombucha beverages. However, previous studies have reported that Kombucha with a pH of less than 2.5 may be harmful to some consumers due to the high acidity (Neffe-Skocińska et al., 2017; Nummer, 2013).
The TSS of Kombucha mainly reflects the concentration of sugar dissolved in the beverage, but it may also contain small levels of minerals, acids, and vitamins which do not affect the degree Brix to a great extent (Zubaidah et al., 2019). The TSS of the samples varied from 1.87 (RE) to 7.00 (DO) °Brix (p < 0.05), indicating a large variability of sugar levels in the Kombucha samples. However, all the TSS levels were lower than those reported from Kombucha fermented from four different species of Indonesian snake fruit which had TSS ranging from 12.43 to 12.93 °Brix after 14 days of fermentation (Zubaidah et al., 2019). The levels of sugar and TSS decreased from 14.08 °Brix (day 0) to 12.43 °Brix (day 14) during the fermentation of snake fruit Kombucha, due to the sugar in the tea broth being metabolised by the cultures (Jayabalan et al., 2014). The deposition of protein, minerals and pigments could also be possible reasons for the decrease in TSS during the fermentation of Kombucha (Zubaidah et al., 2019). The typically low sucrose content (0.93 g/L-7.54 g/L) of Kombucha beverages is desirable for consumers who are conscious of their diet and wellbeing (Neffe-Skocińska et al., 2017).
3.2. AAB and yeast in Kombucha
3.2.1. Cell counts and morphology of AAB
The colony morphology of AAB isolates present in two Kombucha beverages (LO, GB) from New Zealand were similar to those previously reported (Sievers, 2005). No AAB colonies were observed on the agar plates from the imported samples (AM, RE and WO). The absence of AAB in the imported products may be due to post-fermentation treatments such as pasteurisation. Some Kombucha beverages are pasteurised at around 70 °C for 10 min after fermentation to stabilise the products during storage through partial inactivation of microbial cells (Sreeramulu et al., 2000). Pasteurisation also reduces the risk of increased ethanol and acid levels during storage, which can be caused by post-fermentation during storage (Liamkaew et al., 2016; Sreeramulu et al., 2000).
The cell counts of AAB present in sample GB (5.63 ± 0.02 log CFU mL) were higher than those found in sample LO (4.97 ± 0.06 log CFU mL), which may be caused by differences in fermentation conditions during production (Fernández-Pérez et al., 2010). The presence of high AAB cell counts may result in increased post-fermentation activities in Kombucha samples. The post-fermentation of Kombucha is undesirable as it may increase the alcohol content above the regulated level. The Food Standard Australia New Zealand (FSANZ) stipulates that non-alcoholic beverage should not exceed 1.15% alcohol by volume (FSANZ, 2014). Therefore, to produce Kombucha with an acceptable profile, it is important to control the fermentation of Kombucha during processing as well as storage, as high levels of ethanol and acetic acid are undesirable. High levels of acetic acid in Kombucha can result in a vinegary taste and increased acidity to undesirable concentrations (Gomes et al., 2018; Nummer, 2013).
Based on morphology, the bacterial colonies grown on GYPM plates were placed in three groups. Group I and group II colonies were isolated from sample LO and colonies in group III were isolated from sample GB. Group I colonies were opaque with smooth surfaces and a light brownish colour after incubation for 3 day at 30 °C. With prolonged incubation (>3 days), the colour of the colonies turned brownish red. Colonies from group II appeared pale yellow, with convex, shiny, and smooth surfaces. For group III, most of the colonies had smooth surfaces of a light brownish colour. The diameter of all three groups of colonies ranged from 1.0 to 1.2 mm. Acetobacter are commonly circular, and cream to beige colour when grown on glucose yeast extract peptone agar (Gullo and Giudici, 2008), whereas the genus Glucoacetobacter colonies are light brown to brown, with most species producing a water-soluble brownish pigment (Mamlouk and Gullo, 2013). Colonies from the genus Gluconobacter are smooth, glistening with entire edges, while some isolates may form pink colonies (Yamada and Yukphan, 2008). The genus Komagataeibacter are circular, smooth or rough, raised or umbonate (Sievers, 2005). Based on previous studies, the morphology of group I and group II colonies were similar to Glucoacetobacter or Gluconobacter, while the appearance of colonies from group III was similar to the genus Acetobacter (Jayabalan et al., 2014).
3.2.2. Yeast cell counts
Viable yeast cells were only present in three sample products (LO, GB, and DO) produced in New Zealand, with counts ranging from 104 to 105 CFU/mL. The highest number of yeast cells were found in sample GB (5.69 ± 0.01 log CFU/mL), followed by DO (5.57 ± 0.07 log CFU/mL), with sample LO having the lowest (4.75 ± 0.10 log CFU/mL). Although these yeast concentrations are lower than those reported from Kombucha samples produced in Romania (105–106 CFU/mL after 10 days fermentation; Matei et al., 2018), and Australia (108 CFU/g; Teoh et al., 2004), the Kombucha samples analysed in this study were collected at the point of sale, whereas the yeast in the Romanian and Australian samples were analysed immediately after fermentation. It is feasible that some yeast may have died during storage of the products resulting in the lower numbers seen in this study, but further studies would be required to determine this. The disparity in the yeast counts could also be the result of other factors such as variable fermentation conditions and the materials used for production.
There were potentially seven groups of yeast colonies isolated from the three New Zealand-produced samples (Table 2). Most of the colonies were white-to-cream, with a circular shape and entire margins. A summary of the appearance of the yeast colonies is shown in Table 2.
Table 2.
Appearance of yeast colonies of isolates from New Zealand-produced Kombucha samples and grown on potato dextrose agar (PDA) plates.
| Colony group | Description of appearance |
|---|---|
| Group I | Raised, white creamy colour, dull and smooth surface with entire margin. |
| Group II | Circular and white cream colour, shiny, flat and smooth surface. |
| Group III | Circular, glistening surface raised elevation, and entire margin |
| Group IV | White to creamy colour, smooth and glossy surface, umbonate elevation in the centre and entire margin. |
| Group V | Circular and slightly umbonate in the centre; creamy colour, shiny and smooth surface entire margin. |
| Group VI | Circular, off-white colour, slightly umbonate in the centre, flat and smooth surface, entire or lobate margin. |
| Group VII | Circular, cream colour, flat and smooth surface. |
Group I: LO-yeast 1, DO-yeast 5; Group II: LO-yeast 2, LO-yeast 3, LO-yeast 4; Group III: LO-yeast 5; Group IV: DO-yeast 1; DO-yeast 2; DO-yeast 3, DO-yeast 4; DO-yeast 6; Group V: DO-yeast 7, DO-yeast 8, DO-yeast 9, DO-yeast 10, GB-yeast 6, GB-yeast 7, GB-yeast 8; Group VII: GB-yeast 2, GB-yeast 3, GB-yeast 4.
3.3. Phenotypic characterisation of AAB and yeast
3.3.1. Phenotypic characteristics of AAB isolates
Based on their morphology and Gram stain reactions, all the isolates from the three groups of domestic Kombucha samples were Gram-negative, as are most AAB (Gomes et al., 2018). The cells from all three groups were rod-shaped and of similar cell length (∼1 μm), however, their cell arrangements were slightly different. Cells from the isolates in all three groups appeared singly or paired, but the cells of group III appeared in chains. The phenotypic and differential biochemical characteristics of the isolates are shown in Table 3. Based on their biochemical profiles and morphological characteristics, the group I and III isolates were presumed to belong to the genus Gluconobacter and group II isolates to Acetobacter (Sievers, 2005; Yamada and Yukphan, 2008).
Table 3.
Growth and biochemical characteristics of acetic acid bacteria isolated from Kombucha samples.
| Growth and biochemical characteristics of AAB isolated from Kombucha samples Parameter | Group I | Group II | Group III |
|---|---|---|---|
| Catalase reaction | + | + | + |
| Oxidase reaction | − | − | − |
| Growth at different temperature | |||
| Growth at 25 °C | + | + | + |
| Growth at 30 °C | + | + | + |
| Growth at 37 °C | + | + | + |
| Growth on different media | |||
| GYC plates | + | + | + |
| 0.35% (w/v) acetic acid medium (pH = 3.5) | + | + | + |
| 30% (w/v) D-glucose medium | − | − | − |
| DSM medium | − | − | − |
| Methanol medium | w | + | w |
| Glutamate medium | + | w | + |
| Oxidation of ethanol to acetic acid | + | + | + |
| Oxidation of acetic acid to water | − | + | − |
| Alcoholic tolerance test (v/v): | |||
| 2% | + | + | + |
| 4% | + | + | + |
| 6% | + | + | + |
| 8% | + | + | + |
| 10% | − | − | − |
| Formation of H2S, indole and motility tests: | |||
| H2S | − | − | − |
| Indole | − | − | − |
| Motility | − | − | − |
| Gelatine hydrolysis test | − | − | − |
| Oxidation of acetate and lactate: | |||
| Acetate | − | + | − |
| Lactate | − | + | − |
| Production of cellulose | − | − | − |
| Ketogenesis from glycerol to DHA | + | + | + |
| Formation of γ-pyrone from: | |||
| D-glucose | + | − | w |
| D-fructose | + | − | w |
| Acid produced from | |||
| D-glucose | + | +/− | + |
| Mannitol | + | − | + |
| Glycerol | + | − | + |
| Nitrate reduction |
- |
− |
− |
| *Probable genus | Gluconobacter | Acetobacter | Gluconobacter |
(+) Indicated positive reaction; (−) Indicate negative reaction; (±) some strains were positive or negative; (w): weakly positive; *The probable genera were also reported based on previous studies (Asai et al., 1964; Sievers, 2005; Yamada and Yukphan, 2008); experiments were replicated 3 times.
Based on the results shown in Table 3, all the three groups of AAB were oxidase-negative and catalase-positive and were able to grow at 25, 30 and 37 °C after 5–7 days incubation. Isolates in all three groups were able to grow on GYC plates and formed a transparent clear zone surrounding the colonies; a brownish-red pigment could be detected from group I isolates. All isolates from the three groups showed growth on 0.35% (v/v) acetic acid medium and were not able to grow (negative reaction) on 30% (w/v) D-glucose and DSM medium, indicating that the cells were not tolerant to a high osmotic pressure environment. The three groups of colonies were also able to grow on methanol- and glutamate-based media, however, the isolates from the three groups exhibited different growth strengths. Group II showed better growth on methanol agar as there were more colonies formed along the streaked lines and weak growth on glutamate agar than groups I and III. All three groups were able to oxidise ethanol in the medium to acetic acid, as the agar turned to yellow. Only group II isolates were able to turn the agar to purple after extended (>5 days) incubation. Group I and III isolates may belong to the genus Gluconobacter based on similar morphology, as bacteria under this genus are not able to oxidise acetic acid probably due to the deficiencies of the succinate dehydrogenase and α-ketoglutarate dehydrogenase (Raspor and Goranovic, 2008). For the alcohol tolerance test, all three groups of isolates were able to grow in 2–8% ethanol (v/v). Group II exhibited better growth on the ethanolic agar medium than groups I and III, as more colonies were observed around the streaked lines. However, no colonies developed on the 10% ethanolic agar (v/v), which suggests that the maximum tolerance level for the isolates may be between 8% and 10% (v/v) ethanol. Group II colonies were able to oxidise both acetate and lactate. All three groups were able to oxidise glycerol to dihydroxyacetone (DHA).
The ability to produce DHA is important as it confers a sweet, pleasant aroma and refreshing taste to Kombucha. In winemaking, AAB metabolise glycerol to produce DHA, which reacts with several amino acids to produce a “crust-like” taste (Drysdale and Fleet, 1988). During Kombucha fermentation, yeast can convert glucose and fructose to glycerol (Murugesan et al., 2009) which can be potentially converted to DHA by the three groups of isolates identified in this study. The presence of DHA can positively impact the sensory properties of Kombucha.
Formation of γ-pyrone from both fructose and glucose was detected in group I and group III isolates, with Group I exhibiting a darker red colour than group III after adding a few drops of FeCl3. The production of γ-pyrone has been associated with the formation of water-soluble brownish pigments (Kadere et al., 2008). Hence, Group I may have a stronger ability to produce γ-pyrone than group III as indicated by the darker colour. The formation of the pigment may also affect the colour of Kombucha products.
3.3.2. Phenotypic characteristics of yeast
The metabolic profiles of the seven yeast groups were analysed using the apiweb™ identification system software (https://apiweb.biomerieux.com/strip/12). The metabolic patterns of yeast isolates with different substrates were different among the seven groups of microorganisms. The metabolic reactions showed that all seven groups of yeast metabolised glucose. Results indicated that one yeast isolate (LO-yeast 1) in group I metabolised actinidine and one yeast (DO-yeast 5) in group I metabolised D-trehalose. All group II yeast showed positive results with D-saccharose and D-raffinose. The carbohydrate metabolic profiles of the isolated yeast are shown in Table 4.
Table 4.
Carbohydrate metabolism of yeast isolates obtained from Kombucha samples sold in New Zealand using the API 32C test kit.
| Capsule | Substrate | Group I (LO-yeast 1) | Group I (DO-yeast 5) | Group II (LO-yeast 2, LO-yeast 3, LO-yeast 4) | Group III (LO-yeast 5) | Group IV (DO- yeast 1,2,3,4 and 6) | Group V (DO-yeast 7, DO-yeast 8, DO-yeast 9, GB-yeast 6, GB-yeast 7 GB-yeast 8) | Group V (DO-yeast 10) | Group VI (GB-yeast 1, GB-yeast 5) | Group VII (GB-yeast 2, GB-yeast 3, GB-yeast 4) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 | D-Galactose (GAL) | − | − | − | − | − | + | + | − | − |
| 1.1 | Cycloheximide (Actidione)(ACT) | + | − | − | − | + | − | + | − | + |
| 1.2 | D-sacharose (sucrose) (SAC) | − | − | + | + | − | − | + | − | + |
| 1.3 | N-Acetyl-Glucosamine (NAG) | − | − | − | − | − | − | − | + | + |
| 1.4 | Lactic acid (LAT) | − | − | − | − | − | − | − | + | + |
| 1.5 | L-Arabinose (ARA) | − | − | − | − | − | − | − | − | + |
| 1.6 | D-Cellobiose (CEL) | − | − | − | − | + | − | + | − | + |
| 1.7 | D-Raffinose (RAF) | − | − | + | − | − | − | + | − | + |
| 1.8 | D-Maltose (MAL) | − | − | − | − | − | − | − | − | + |
| 1.9 | D-Trehalose (TRE) | − | + | − | − | + | − | − | − | + |
| 1.A | Potassium 2-Ketogluconate (2-KG) | − | − | − | − | − | − | + | − | + |
| 1.B | Methyl-alpha D-Glucopyranoside (MDG) | − | − | − | − | − | − | − | − | + |
| 1.C | D-Mannitol (MAN) | − | − | − | − | − | − | − | − | + |
| 1.D | D-Lactose (bovine origin) (LAC) | − | − | − | − | − | − | − | − | + |
| 1.E | Inositol (INO) | − | − | − | − | − | − | − | − | + |
| 1.F | No substrate (0) | − | − | − | − | − | − | − | − | + |
| 0.0 | D-Sorbitol (SOR) | − | − | − | + | − | − | + | − | + |
| 0.1 | D-Xylose (XYL) | − | − | − | − | − | − | − | − | + |
| 0.2 | D-Ribose (RIB) | − | − | − | − | − | − | − | − | + |
| 0.3 | Glycerol (GLY) | − | − | − | − | − | − | − | + | + |
| 0.4 | L-Rhamnose (RHA) | − | − | − | − | − | − | − | − | + |
| 0.5 | Palatinose (PLE) | − | − | − | − | − | − | − | − | + |
| 0.6 | Erythritol (ERY) | − | − | − | − | − | − | − | − | + |
| 0.7 | D-melibiose (MEL) | − | − | − | − | − | − | − | − | + |
| 0.8 | Sodium Glucuronate (GRT) | − | − | − | − | − | − | − | − | + |
| 0.9 | D-Melezitose(MLZ) | − | − | − | − | − | − | − | − | + |
| 0.A | Potassium Glucuronate (GNT) | − | − | − | − | − | − | − | − | + |
| 0.B | Levulinic acid (Lavullinate) (LVT) | − | − | − | − | − | − | − | − | + |
| 0.C | D-Glucose (GLU) | + | + | + | + | + | + | + | + | + |
| 0.D | L-sorbose (SBE) | − | − | − | − | − | − | − | + | + |
| 0.E | Glucosamine (GLN) | − | − | − | − | − | − | − | − | − |
| 0.F | ESCulin ferric citrate (ESC) | − | − | − | − | − | − | − | − | + |
Note: (+) = positive; (−) = negative.
Based on the data obtained using the API 32C kit, five different yeast species were presumptively identified as: Kloeckera japonica, Candida (C.) glabrata, Candida (C.) colliculosa, Candida (C.) krusei and Cryptococcus humicola in groups I, IV, V, VI, and VII with high percentages of identity, respectively (Table 5). However, the other two groups (II and III) showed low percentages of identity. Isolate LO yeast 1 and all group IV isolates had high identification percentages for the species Kloeckera japonica, although the yeast exhibited different fermentation profiles. The two groups (I and IV) of yeast isolates were probably different strains of the species Kloechera japonica. The API tests showed low discrimination or doubtful results for isolates in groups II, III and V except for DO-yeast 10. The standard API 32 C database has only 63 yeast species (Deak and Beuchat, 1993), therefore, it is likely that some of the isolates from the three groups in Table 4 were not represented in the database and hence could not be identified. Additionally, geographical location may also affect the identification of isolates using the API 32C system as the ID 32C database is based on U.S. isolates (Ramani et al., 1998). Therefore, it is necessary to use molecular methods for further verification.
Table 5.
Reaction of yeast isolates using the API 32 C kit.
| Sample group (isolated identification code) | Probable identification species | Probable ID (%)1 | Description of identification2 |
|---|---|---|---|
| Group I (LO-yeast 1) | Kloeckera japonica | 94.0 | Good identification |
| Group I (DO-yeast 5) |
Candida glabrata |
99.0 |
Very good identification |
| Group IV | Kloeckera japonica | 99.0 | Excellent identification |
| Group V (DO-yeast 10) | Candida colliculosa | 98.1 | Good identification |
| Group VI | Candida krusei | 99.7 | Very good identification |
| Group VII | Cryptococcus humicola | 98.9 | Good identification |
Group I: LO-yeast 1, DO-yeast 5; Group II: LO-yeast 2, LO-yeast 3, LO-yeast 4; Group III: LO-yeast 5; Group IV: DO-yeast 1; DO-yeast 2; DO-yeast 3, DO-yeast 4; DO-yeast 6; Group V: DO-yeast 7, DO-yeast 8, DO-yeast 9, DO-yeast 10, GB-yeast 6, GB-yeast 7, GB-yeast 8; Group VII: GB-yeast 2, GB-yeast 3, GB-yeast 4.
Probable ID (%) = Probable Identification (%).
Descriptions of the identification of yeast isolates were obtained from the available online database at: https://apiweb.biomerieux.com/strip/12.
3.4. Taxonomic identification of representative AAB and yeast isolates
3.4.1. Molecular identification of AAB isolates by sequencing the 16S rRNA genes
Although phenotypic characteristics of the ABB isolates are generally consistent with the molecular identification to genus level, conventional tests are time-consuming and not always reliable (Abd El-Salam, 2012). Also, it is difficult to accurately identify AAB to the species level using only biochemical and physiological characteristics. Low-resolution DNA-based molecular identification techniques using 16S RNA loci are now commonly used to identify organisms to genus or species (Abd El-Salam, 2012; Gomes et al., 2018). To determine the phylogenetic position of the three AAB groups, three representative isolates (namely, LO-AAB1, LO-AAB2 and GB-AAB1) were subjected to sequence analysis of full-length 16S rRNA genes (Table 5). Strain GB-AAB1 belonged to Gluconobacter potus as it showed 100% sequence identity with G. potus type strain LMG 1764 (Sombolestani et al., 2021). LO-AAB2 was identified as Acetobacter musti as its sequence was identical to Acetobacter musti Bo7 (Ferrer et al., 2016). The analysis identified G. potus as the closest taxon of LO-AAB1 with 99.64% sequence similarity, and only five nucleotide (nt) mismatches were found within the 1406 nt aligned sequence region (Table 6).
Table 6.
16S rRNA gene sequence analysis of the acetic acid bacteria (AAB) isolates.
The G. potus LMG 1764 strain was isolated from lemonade cider in 1979 and tentatively identified as Gluconobacter oxydans due to its sequence similarity with G. oxydans reference strains (Li et al., 2017). In 2017, strain R-71646 was isolated from Kombucha on modified DMS agar plates and identified as G. oxydans using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) but with lower sequence identity. The representative strains LMG 1764 and R-71646 were recently identified as the novel species G. potus and can be differentiated from G. oxydans and closely related species based on their molecular, biochemical and chemotaxonomic characteristics (Sombolestani et al., 2021). The strains isolated in this study; GB-AAB1 and LO-AAB1 were isolated in 2019 from New Zealand commercial Kombucha samples and some phenotypic characteristics were slightly different to the reference strain G. potus LMG 1764 (Sombolestani et al., 2021). For example, unlike the reference strains, neither GB-AAB1 nor LO-AAB1 were able to grow in the presence of 10% ethanol (w/v). In addition, the morphology of LO-AAB1 and GB-AAB1 colonies on the GY medium were slightly different; LO-AAB1 had brownish red colonies while GB-AAB1 colonies were light brown. The Group II isolate (LO-AAB2) was identified as Acetobacter musti strain Bo7T with 100% identity. This strain was first isolated from grape must (Ferrer et al., 2016), and its application in food manufacturing is limited, therefore more research should be carried out to determine the effects of this species during Kombucha fermentation.
3.4.2. Molecular identification of yeast isolates by sequencing the 26S rRNA genes
A total of twelve yeast isolates were subjected to 26S rRNA sequencing after the API 32 C tests. Sequence analysis of full-length 26S rRNA genes revealed that the twelve isolates belonged to seven different species (Table 7). Based on the sequence analysis, Group I, Group III and Group VII belonged to Dekkera bruxellensis, Schizosaccharomyces pombe, and Saccharomyces cerevisiae; showing 100% identity to the reference strains (D. bruxellensis CBS2499, S. pombe NRRL Y-12796 and S. cerevisiae CEN.PK113-7D), respectively. Group IV, Group V and Group VII isolates were identified as Hanseniaspora valbyensis, Brettanomyces anomalus and Pichia kudriavzevii, as the closest top hit taxon shared more than 99.70% similarity with less than 4 nt mismatches. Group II may belong to Starmerella vitis as it had 94.55% similarity with the top hit strain S. vitis CMBA 19.25.
Table 7.
26S rRNA sequence analysis of the yeast isolates.
| Yeast isolates | Top hit taxon | Top hit strains | GenBank accession | % Similarity | Variation |
|---|---|---|---|---|---|
| Group I | Dekkera bruxellensis | CBS2499 | AM850055.1 | 100 | 0/1334 |
| Group II | Starmerella vitis | CMBA_19.25 | MN319575.1 | 94.55 | 65/1193 |
| Group III | Schizosaccharomyces pombe | NRRL Y-12796 | NG_042649.1 | 100 | 0/1357 |
| Group IV | Hanseniaspora valbyensis | NRRL Y-1626 | NG_042630 | 99.70 | 4/1330 |
| Group V | Brettanomyces anomalus | NRRL Y-17522 | EF550258.1 | 99.93 | 1/1339 |
| Group VI | Pichia kudriavzevii | CBS573 | XR_003834616.1 | 99.92 | 1/1317 |
| Group VII | Saccharomyces cerevisiae | CEN.PK113-7D | CP046092.1 | 100 | 0/1334 |
Group I: LO-yeast 1, DO-yeast 5; Group II: LO-yeast 2, LO-yeast 3, LO-yeast 4; Group III: LO-yeast 5; Group IV: DO-yeast 1; DO-yeast 2; DO-yeast 3, DO-yeast 4; DO-yeast 6; Group V: DO-yeast 7, DO-yeast 8, DO-yeast 9, DO-yeast 10, GB-yeast 6, GB-yeast 7, GB-yeast 8; Group VII: GB-yeast 2, GB-yeast 3, GB-yeast 4.
Representative isolates of Group I (Table 7) were identified as Dekkera bruxellensis and Group V isolates were similar to Brettanomyces anomalus. The genus Dekkera is the ascospore (spore-forming) form of the genus Brettanomyces. Dekkera bruxellensis is known as the teleomorph of Brettanomyces nomalus (Schifferdecker et al., 2014), which has been isolated from sourdough, feta cheese and spontaneously fermented alcoholic beers (Fadda et al., 2001). This acid-producing genus is well-adapted to Kombucha fermentation and contributes to flavour due to the formation of acetic acid and acetic acid esters (Teoh et al., 2004). D. bruxellensis also contributes to the aroma characteristic in some French wines (Schifferdecker et al., 2014). It is advantageous to add Brettanomyces spp. during fermentation of beverages as it produces an exotic flavour such as pineapple in craft beers (Fadda et al., 2001). The genus Dekkera exhibits high resistance to cycloheximide, which blocks protein synthesis in eukaryotic organisms (Steensels et al., 2015). This may explain why some yeast isolates could still grow on the GYPM plates despite the addition of cycloheximide.
Isolates from Group III (Table 7) matched with the species Schizosaccharomyces (S.) pombe. The occurrence of this fission yeast in Kombucha has been previously reported (Teoh et al., 2004). It is also commonly found in grape juice and wine fermentation due to its high fermentative ability and tolerance to high sugar concentrations and low pH environment (Loira et al., 2018; Teoh et al., 2004). According to Loira et al. (2018), S. pombe can release high amounts of cell wall polysaccharides during alcoholic fermentation which can confer a better mouthfeel (enhancing sweetness and roundness) and longer aromatic persistence in fermented beverages such as sparkling wine.
Group IV isolates (Table 7) belonged to Hanseniaspora valbyensis which is the teleomorph of Kloeckera japonica. The genus Hanseniaspora are apiculate yeasts often isolated from fruits and berries. They can release high amounts of volatile components which form a pleasant aroma similar to cider (Mayser et al., 1995). The presence of this strain may contribute to the aroma formation in Kombucha products. Group VI yeast isolates (Table 7) were identified as Pichia kudriavzevii, which is the teleomorph of Candida krusei, known to produce a pellicle structure in Kombucha (Mayser et al., 1995). Group VII yeast isolates were identified as Saccharomyces (S.) cerevisiae (Table 7), the most common yeast culture utilised in modern alcoholic beverages such as beer, wine, whisky and Kombucha due to its high fermentation efficiency and tolerance to ethanol (Villarreal-Soto et al., 2018; Walker and Stewart, 2016).
The complex microbial interactions between different yeast species may lower the risk of stuck (terminated) fermentation and contribute to the formation of complex aromas and flavours (Villarreal-Soto et al., 2018). In this study, 26S rRNA gene sequence analysis identified the group VII isolates as S. cerevisiae, showing 100% identity with the reference strain of S. cerevisiae CEN.PK113-7D (Salazar et al., 2017). Although S. cerevisiae was included in the API 32 C database, our phenotypic data showed that the yeast isolates had different biochemical characteristics to the reference profiles. Thus, the phenotypic data suggests that the New Zealand Kombucha-associated S. cerevisiae possess unique metabolic features. The Group II isolate was closely related to Starmerella vitis, which includes strains recently isolated from flowers and grapes in Canada (Čadež et al., 2020). This newly identified species probably prefers a high sugar environment such as grapes and nectars from different plants, however, it is not commonly found in Kombucha products. Notably, the sequence identity of Group II isolates and Starmerella vitis CMBA_19.25 was only 94.55%, and thus, their species identity should be further confirmed by biochemical and genetic characterisation.
Most of the identifications to genera level based on morphology agreed with that of the rRNA sequencing, however, API 32 C identifications were less reliable, with only 2 groups (IV and VII) matching the sequenced results. The lack of consensus between identification based on sequenced and API 32 C results was not surprising as conventional biochemical results can vary with test conditions, and phenotypic profiles are not always stable, thus impacting final identification (Latouche et al., 1997).
4. Conclusions
All six Kombucha samples had high acidity (pH < 4.6) and were characterised by a high variation of TSS (p < 0.05). Kombucha samples produced in New Zealand contained viable yeast and AAB while none were found in the imported samples. The two dominant Gram-negative AAB isolates found in the Kombucha samples were identified as Gluconobacter potus LMG1764 and Acetobacter musti Bo7. Seven dominant yeast isolates were identified as Dekkera bruxellensis CBS2499, Schizosaccharomyces pombe NRRL Y-12796, Hanseniaspora valbyensis NRRL Y-1626, Brettanomyces anomalus NRRL Y-17522, Pichia kudriavzevii CBS573, Saccharomyces cerevisiae CEN.PK113-7D, and Starmerella vitis CMBA_19.25. Knowledge of the microbiology of fermented Kombucha may help processors to develop strategies to control the fermentation better as well as maintain the alcohol content within the regulated limits during storage.
CRediT authorship contribution statement
Boying Wang: Conceptualization, Investigation, Data curation, Writing – original draft, editing, Writing – review & editing. Kay Rutherfurd-Markwick: Conceptualization, Writing – review & editing. Xue-Xian Zhang: Conceptualization, Writing – review & editing. Anthony N. Mutukumira: Funding acquisition, Project Leader and overall management of the study, Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The research was supported by the School of Food and Advanced Technology of Massey University, Auckland, New Zealand. The authors thank Rachel Liu for technical assistance in the microbiological analysis.
Biographies
Boying Wang (MFT) holds a Bachelor of Food Technology honours degree and a Master of Food Technology from Massey New Zealand. She spent one semester at the University of Guelph, Canada during her undergraduate degree. Boying Wang is currently a PhD candidate in Food Technology at Massey University and has special interest in food fermentations.
Associate Professor Kay Rutherfurd-Markwick (PhD) gained her PhD in Biochemistry from Massey University, New Zealand. A/P Rutherfurd-Markwick has particular interest in bioactive compounds and functional foods for improved consumer health and performance. Kay lectures in Pathophysiology, Biomedical Sciences, and aspects of Nutrition and Immunology.
Associate Professor Xue-Xian Zhang (PhD) gained his doctorate in Microbiology from Huazhong Agricultural University, Wuhan, China. He has special expertise in microbial ecology, genetics and genomics. Current research in his lab focuses on the molecular mechanisms of bacterial adaptation to their natural environments such as plant and soil.
Dr Anthony N Mutukumira (PhD, FNZIFST), a Fellow of the New Zealand Institute of Food Science and Technology Society (NZIFST) obtained his doctorate (Doctor Scientiarum) at The Norwegian University of Life Sciences at Ås, Norway, following research in the development of fermented milks using novel lactic acid bacteria. He presently holds the Vice-Chair of Food Safety Working Group of International Commission of Agricultural and Biosystems Engineering (CIGR), and he is also a Board Member of the China-New Zealand Joint Research Centre for Food Safety and Nutrition. Tony's research activities include food preservation using novel technologies, development of fermented foods and beverages particularly those with functional properties for better health.
Editor: Siyun Wang
References
- Abd El-Salam S.S. 16S rRNA gene sequence detection of acetic acid bacteria isolated from tea Kombucha. N.Y. Sci. J. 2012;5:55–61. [Google Scholar]
- Amarasinghe H., Weerakkody N.S., Waisundara V.Y. Evaluation of physicochemical properties and antioxidant activities of kombucha "Tea Fungus" during extended periods of fermentation. Food Sci. Nutr. 2018;6:659–665. doi: 10.1002/fsn3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arifuzzaman M., Hasan M.Z., Rahman S.B., Pramanik M.K. Isolation and characterisation of Acetobacter and Gluconobacter spp. from sugarcane and rotten fruits. Res. Rev. Biosci. 2014;8:359–365. [Google Scholar]
- Asai T., Iizuka H., Komagata K. The flagellation and taxonomy of genera Gluconobacter and Acetobacter with reference to the existence of intermediate strains. J. Gen. Appl. Microbiol. 1964;10:95–126. [Google Scholar]
- Buxton R. Nitrate and nitrite reduction test protocols. American Society for Microbiology. 2011:1–20. https://asm.org/ASM/media/Protocol-Images/Nitrate-and-Nitrite-Reduction-Test-Protocols.pdf?ext=.pdf [Google Scholar]
- Chakravorty S., Bhattacharya S., Chatzinotas A., Chakraborty W., Bhattacharya D., Gachhui R. Kombucha tea fermentation: microbial and biochemical dynamics. Int. J. Food Microbiol. 2016;220:63–72. doi: 10.1016/j.ijfoodmicro.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Čadež N., Drumonde-Neves J., Sipiczki M., Dlauchy D., Lima T., Pais C., Schuller D., Franco-Duarte R., Lachance M.-A., Péter G. Starmerella vitis fa, sp. nov., a yeast species isolated from flowers and grapes. Antonie Leeuwenhoek. 2020;113:1289–1298. doi: 10.1007/s10482-020-01438-x. [DOI] [PubMed] [Google Scholar]
- Chen C., Liu B.Y. Changes in major components of tea fungus metabolites during prolonged fermentation. J. Appl. Microbiol. 2000;89:834–839. doi: 10.1046/j.1365-2672.2000.01188.x. [DOI] [PubMed] [Google Scholar]
- Cirigliano M. A selective medium for the isolation and differentiation of Gluconobacter and Acetobacter. J. Food Sci. 1982;47:1038–1039. [Google Scholar]
- Claus D. A standardised Gram staining procedure. World J. Microbiol. Biotechnol. 1992;8:451–452. doi: 10.1007/BF01198764. [DOI] [PubMed] [Google Scholar]
- Deak T., Beuchat L.R. Comparison of the SIM, API 20C, and ID 32C systems for identification of yeasts isolated from fruit juice concentrates and beverages. J. Food Protect. 1993;56:585–592. doi: 10.4315/0362-028X-56.7.585. [DOI] [PubMed] [Google Scholar]
- dela Cruz T.E.E., Torres J.M.O. Gelatin hydrolysis test protocol. Am. Soc. Microbiol. 2012;1(10):1–10. [Google Scholar]
- Drysdale G., Fleet G. Acetic acid bacteria in winemaking: a Review. AJEV (Am. J. Enol. Vitic.) 1988;39:143–154. [Google Scholar]
- Du Toit W.J., Lambrechts M.G. The enumeration and identification of acetic acid bacteria from South African red wine fermentations. Int. J. Food Microbiol. 2002;74:57–64. doi: 10.1016/S0168-1605(01)00715-2. [DOI] [PubMed] [Google Scholar]
- Dufresne C., Farnworth E. Tea, Kombucha, and health: a review. Food Res. Int. 2000;33:409–421. doi: 10.1016/S0963-9969(00)00067-3. [DOI] [Google Scholar]
- Essawet N.A., Cvetković D., Velićanski A., Čanadanović-Brunet J., Vulić J., Maksimović V., Markov S. Polyphenols and antioxidant activities of kombucha beverage enriched with coffeeberry® extract. Chem. Ind. Chem. Eng. Q. 2015;21:399–409. doi: 10.2298/CICEQ140528042E. [DOI] [Google Scholar]
- Fadda M.E., Cosentino S., Deplano M., Palmas F. Yeast populations in Sardinian feta cheese. Int. J. Food Microbiol. 2001;69:153–156. doi: 10.1016/S0168-1605(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez R., Torres C., Sanz S., Ruiz-Larrea F. Rapid molecular methods for enumeration and taxonomical identification of acetic acid bacteria responsible for submerged vinegar production. Eur. Food Res. Technol. 2010;231:813–819. doi: 10.1007/s00217-010-1331-6. [DOI] [Google Scholar]
- FSANZ Labelling of alcoholic beverages user guide. 2014. https://www.foodstandards.gov.au/consumer/labelling/Pages/Labelling-of-alcoholic-beverages.aspx
- Ferrer S., Mañes-Lázaro R., Benavent-Gil Y., Yépez A., Pardo I. Acetobacter musti sp. nov., isolated from bobal grape must. Int. J. Syst. Evol. Microbiol. 2016;66:957–961. doi: 10.1009/ijsem.0.000818. [DOI] [PubMed] [Google Scholar]
- Goh W., Rosma A., Kaur B., Fazilah A., Karim A., Bhat R. Fermentation of black tea broth (Kombucha): I. Effects of sucrose concentration and fermentation time on the yield of microbial cellulose. Int. Food Res. J. 2012;19:109–117. [Google Scholar]
- Gomes R.J., Borges M.F., Rosa M.F., Castro-Gomez R.J.H., Spinosa W.A. Acetic acid bacteria in the food industry: systematics, characteristics and applications. Food Technol. Biotechnol. 2018;56:139–151. doi: 10.17113/ftb.56.02.18.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullo M., Giudici P. Acetic acid bacteria in traditional balsamic vinegar: phenotypic traits relevant for starter cultures selection. Int. J. Food Microbiol. 2008;125:46–53. doi: 10.1016/j.ijfoodmicro.2007.11.076. [DOI] [PubMed] [Google Scholar]
- Hanmoungjai W., Chukeatirote E., Pathom-Aree W., Yamada Y., Lumyoung S. Identification of acidotolerant acetic acid bacteria isolated from Thailand sources. Res. J. Microbiol. 2007;2:194–197. [Google Scholar]
- Jayabalan R., Chen P.-N., Hsieh Y.-S., Prabhakaran K., Pitchai P., Marimuthu S., Thangaraj P., Swaminathan K., Yun S.E. Effect of solvent fractions of kombucha tea on viability and invasiveness of cancer cells—characterisation of dimethyl 2-(2-hydroxy-2-methoxypropylidine) malonate and vitexin. Indian J. Biotechnol. 2011;10:75–82. [Google Scholar]
- Jayabalan R., Malbasa R.V., Loncar E.S., Vitas J.S., Sathishkumar M. A Review on Kombucha tea-microbiology, composition, fermentation, beneficial effects, toxicity, and tea fungus. Compr. Rev. Food Sci. Food Saf. 2014;13:538–550. doi: 10.1111/1541-4337.12073. [DOI] [PubMed] [Google Scholar]
- Jayabalan R., Malbaśa R.V., Sathishkumar M. Kombucha tea: metabolites. Fungal Metab. 2017:965–978. doi: 10.1007/978-3-319-25001-4_12. [DOI] [Google Scholar]
- Kadere T., Miyamotoo T., Oniango R., Kutima P., Njoroge S. Isolation and identification of the genera Acetobacter and Gluconobacter in coconut toddy (Mnazi) Afr. J. Biotechnol. 2008;7(16):2963–2971. [Google Scholar]
- Kumar V., Joshi V. Kombucha: technology, microbiology, production, composition and therapeutic value. Int. J. Food Ferment. Technol. 2016;6:13–24. doi: 10.5958/2277-9396.2016.00022.2. [DOI] [Google Scholar]
- Latouche G.N., Daniel H.-M., Lee O.C., Mitchell T.G., Sorrell T.C., Meyer W. Comparison of use of phenotypic and genotypic characteristics for identification of species of the anamorph genus Candida and related teleomorph yeast species. J. Clin. Microbiol. 1997;35:3171–3180. doi: 10.1128/jcm.35.12.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavasani P.S., Motevaseli E., Shirzad M., Modarressi M.H. Isolation and identification of Komagataeibacter xylinus from Iranian traditional vinegars and molecular analyses. Iran. J. Microbiol. 2017;9:338–347. [PMC free article] [PubMed] [Google Scholar]
- Li L., Cleenwerck I., De Vuyst L., Vandamme P. Identification of acetic acid bacteria through matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry and report of Gluconobacter nephelii Kommanee et al. 2011 and Gluconobacter uchimurae Tanasupawat et al. 2012 as later heterotypic synonyms of Gluconobacter japonicus Malimas et al. 2009 and Gluconobacter oxydans (Henneberg 1897) De Ley 1961 (Approved Lists 1980) emend. Gosselé et al. 1983, respectively. Syst. Appl. Microbiol. 2017;40:123–134. doi: 10.1016/j.syapm.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Liamkaew R., Chattrawanit J., Danvirutai P. Kombucha production by combinations of black tea and apple juice. Prog. Appl. Sci. Technol. 2016;6:139–146. [Google Scholar]
- Liu C.-H., Hsu W.-H., Lee F.-L., Liao C.-C. The isolation and identification of microbes from a fermented tea beverage, Haipao, and their interactions during Haipao fermentation. Food Microbiol. 1996;13:407–415. doi: 10.1006/fmic.1996.0047. [DOI] [Google Scholar]
- Loira I., Morata A., Palomero F., González C., Suárez-Lepe J.A. Schizosaccharomyces pombe: a promising biotechnology for modulating wine composition. Fermentation. 2018;4:70. doi: 10.3390/fermentation4030070. [DOI] [Google Scholar]
- Mamlouk D., Gullo M. Acetic acid bacteria: physiology and carbon sources oxidation. Indian J. Microbiol. 2013;53:377–384. doi: 10.1007/s12088-013-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh A.J., O'Sullivan O., Hill C., Ross R.P., Cotter P.D. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbiol. 2014;38:171–178. doi: 10.1016/j.fm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Martínez Leal J., Valenzuela Suárez L., Jayabalan R., Huerta Oros J., Escalante-Aburto A. A review on health benefits of kombucha nutritional compounds and metabolites. CyTA - J. Food. 2018;16:390–399. doi: 10.1080/19476337.2017.1410499. [DOI] [Google Scholar]
- Matei B., Diguță C.F., Popa O., Cornea C.P., Matei F. Molecular identification of yeast isolated from different kombucha sources. Ann Univ. Dunarea Jos Galati Fascicle VI Food Technol. 2018;42(1):17–25. [Google Scholar]
- Matthews C. On the staining of yeast cells by methylene blue, etc. J. Inst. Brew. 1914;20(6):488–496. [Google Scholar]
- Mayser P., Fromme S., Leitzmann C., Grunder K. The yeast spectrum of the 'tea fungus Kombucha. Mycoses. 1995;38:289–295. doi: 10.1111/j.1439-0507.1995.tb00410.x. [DOI] [PubMed] [Google Scholar]
- Mukadam T.A., Punjabi K., Deshpande S.D., Vaidya S.P., Chowdhary A.S. Isolation and characterisation of bacteria and yeast from Kombucha tea. Int. J. Curr. Microbiol. Appl. Sci. 2016;5:32–41. doi: 10.20546/ijcmas.2016.506.004. [DOI] [Google Scholar]
- Murugesan G.S., Sathishkumar M., Jayabalan R., Binupriya A.R., Swaminathan K., Yun S.E. Hepatoprotective and curative properties of Kombucha tea against carbon tetrachloride-induced toxicity. J. Microbiol. Biotechnol. 2009;19:397–402. doi: 10.4014/jmb.0806.374. [DOI] [PubMed] [Google Scholar]
- Neffe-Skocińska K., Sionek B., Ścibisz I., Kołożyn-Krajewska D. Acid contents and the effect of fermentation condition of kombucha tea beverages on physicochemical, microbiological and sensory properties. CyTA - J. Food. 2017;15:601–607. doi: 10.1080/19476337.2017.1321588. [DOI] [Google Scholar]
- Nielsen S.S. Food Anal. Lab. Man. Springer; Cham: 2017. Standard Solutions and Titratable Acidity; pp. 179–184. [DOI] [Google Scholar]
- Nummer B.A. Kombucha brewing under the Food and Drug Administration model Food Code: risk analysis and processing guidance. J. Environ. Health. 2013;76:8–11. [PubMed] [Google Scholar]
- Painting K., Kirsop B. A quick method for estimating the percentage of viable cells in a yeast population, using methylene blue staining. World J. Microbiol. Biotechnol. 1990;6:346–347. doi: 10.1007/BF01201311. [DOI] [PubMed] [Google Scholar]
- Ramani R., Gromadzki S., Pincus D.H., Salkin I.F., Chaturvedi V. Efficacy of API 20C and ID 32C systems for identification of common and rare clinical yeast isolates. J. Clin. Microbiol. 1998;36:3396–3398. doi: 10.1128/jcm.36.11.3396-3398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspor P., Goranovic D. Biotechnological applications of acetic acid bacteria. Crit. Rev. Biotechnol. 2008;28:101–124. doi: 10.1080/07388550802046749. [DOI] [PubMed] [Google Scholar]
- Reiner K. Catalase test protocol. American Society for Microbiology. 2010:1–6. https://asm.org/getattachment/72a871fc-ba92-4128-a194-6f1bab5c3ab7/Catalase-Test-Protocol.pdf [Google Scholar]
- Reiss J. Influence of different sugars on the metabolism of the tea fungus. Zeitschrift Fur Lebensmittel-Untersuchung Und-Forschung. 1994;198:258–261. [Google Scholar]
- Salazar A.N., Gorter de Vries A.R., van den Broek M., Wijsman M., de la Torre Cortés P., Brickwedde A., Brouwers N., Daran J.-M.G., Abeel T. Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN. PK113-7D. FEMS Yeast Res. 2017;17 doi: 10.11093/femsyr/fox094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifferdecker A.J., Dashko S., Ishchuk O.P., Piskur J. The wine and beer yeast Dekkera bruxellensis. Yeast. 2014;31:323–332. doi: 10.1002/yea.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Resl P., Westberg M., Spribille T. A new, highly effective primer pair to exclude algae when amplifying nuclear large ribosomal subunit (LSU) DNA from lichens. Lichenologist. 2015;47:269–275. doi: 10.1002/yea.3023. [DOI] [Google Scholar]
- Semjonovs P., Ruklisha M., Paegle L., Saka M., Treimane R., Skute M., Rozenberga L., Vikele L., Sabovics M., Cleenwerck I. Cellulose synthesis by Komagataeibacter rhaeticus strain P 1463 isolated from Kombucha. Appl. Microbiol. Biotechnol. 2017;101:1003–1012. doi: 10.1007/s00253-016-7761-8. [DOI] [PubMed] [Google Scholar]
- Shields P., Cathcart L. Oxidase test protocol. Am. Soc. Microbiol. 2010:1–10. [Google Scholar]
- Sievers M. Family II. Acetobacteraceae. Bergey's Manual Syst. Bacteriol. 2005;2:41–50. [Google Scholar]
- Sombolestani A.S., Cleenwerck I., Cnockaert M., Borremans W., Wieme A.D., De Vuyst L., Vandamme P. Characterisation of novel Gluconobacter species from fruits and fermented food products: Gluconobacter cadivus sp. nov., Gluconobacter vitians sp. nov. and Gluconobacter potus sp. nov. Int. J. Syst. Evol. Microbiol. 2021;71 doi: 10.1099/ijsem.0.004751. [DOI] [PubMed] [Google Scholar]
- Sreeramulu G., Zhu Y., Knol W. Kombucha fermentation and its antimicrobial activity. J. Agric. Food Chem. 2000;48:2589–2594. doi: 10.1021/jf991333m. [DOI] [PubMed] [Google Scholar]
- Steensels J., Daenen L., Malcorps P., Derdelinckx G., Verachtert H., Verstrepen K.J. Brettanomyces yeasts—from spoilage organisms to valuable contributors to industrial fermentations. Int. J. Food Microbiol. 2015;206:24–38. doi: 10.1016/j.ijfoodmicro.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Swings J. Phenotypic Identification of acetic acid bacteria. identification methods in applied and environmental microbiology. Soc. Appl. Bacteriol. Tech. Ser. No. 1992;29:103–110. [Google Scholar]
- Teoh A.L., Heard G., Cox J. Yeast ecology of Kombucha fermentation. Int. J. Food Microbiol. 2004;95:119–126. doi: 10.1016/j.ijfoodmicro.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Velićanski A., Cvetković D., Markov S. Characteristics of Kombucha fermentation on medicinal herbs from Lamiaceae family. Rom. Biotechnol. Lett. 2013;18:8034–8042. [Google Scholar]
- Villarreal-Soto S.A., Beaufort S., Bouajila J., Souchard J.P., Taillandier P. Understanding kombucha tea fermentation: a Review. J. Food Sci. 2018;83:580–588. doi: 10.1111/1750-3841.14068. [DOI] [PubMed] [Google Scholar]
- Visser I.J., Jaisly F., Mossel D. The Effect of properties of dried preparations of Sulphide Iron Motility (SIM) agar on the results of motility readings. J. Appl. Bacteriol. 1985;59:167–170. doi: 10.1111/j.1365-2672.1985.tb03317.x. [DOI] [Google Scholar]
- Waisundara V.Y. Usage of Kombucha ‘Tea Fungus’ for enhancement of functional properties of herbal beverages. Frontiers and New Trends in the Science of Fermented Food and Beverages. IntechOpen. 2018:11–21. doi: 10.5772/intechopen.80873. [DOI] [Google Scholar]
- Walker G.M., Stewart G.G. Saccharomyces cerevisiae in the production of fermented beverages. Beverages. 2016;2:30. doi: 10.3390/beverages2040030. [DOI] [Google Scholar]
- Yamada Y., Okada Y., Kondo K. Isolation and characterisation of" polarly flagellated intermediate strains" in acetic acid bacteria. J. Gen. Appl. Microbiol. 1976;22:237–245. doi: 10.2323/jgam.22.237. [DOI] [Google Scholar]
- Yamada Y., Yukphan P. Genera and species in acetic acid bacteria. Int. J. Food Microbiol. 2008;125:15–24. doi: 10.1016/j.ijfoodmicro.2007.11.077. [DOI] [PubMed] [Google Scholar]
- Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Feng F., Chen L., Yao Q., Chen K. Directional isolation of ethanol-tolerant acetic acid bacteria from industrial fermented vinegar. Eur. Food Res. Technol. 2013;236:573–578. doi: 10.1007/s00217-012-1885-6. [DOI] [Google Scholar]
- Zubaidah E., Ifadah R., Afgani C. Changes in chemical characteristics of Kombucha from various cultivars of snake fruit during fermentation. IOP Conf. Ser.: Environ. Sci. J. Integr. Environ. Res. 2019;230 doi: 10.1088/1755-1315/230/1/012098. [DOI] [Google Scholar]