Summary
Background
Mitochondrial DNA (mtDNA) mutations alter mitochondrial function in oxidative metabolism and play an important role in tumorigenesis. A series of studies have demonstrated that the mtDNA control region (mtCTR), which is essential for mtDNA replication and transcription, represents a mutational hotspot in human tumors. However, a comprehensive pan-cancer evolutionary pattern analysis of mtCTR mutations is urgently needed.
Methods
We generated a comprehensive combined dataset containing 10026 mtDNA somatic mutations from 4664 patients, covering 20 tumor types based on public and private next-generation sequencing data.
Findings
Our results demonstrated a significantly higher and much more variable mutation rate in mtCTR than in the coding region across different tumor types. Moreover, our data showed a remarkable distributional bias of tumor somatic mutations between the hypervariable segment (HVS) and non-HVS, with a significantly higher mutation density and average mutation sites in HVS. Importantly, the tumor-specific mutational pattern between mtCTR HVS and non-HVS was identified, which was classified into three evolutionary selection types (relaxed, moderate, and strict constraint types). Analysis of substitution patterns revealed that the prevalence of CH > TH in non-HVS greatly contributed to the mutational selection pattern of mtCTR across different tumor types. Furthermore, we found that the mutational pattern of mtCTR in the four tumor types was clearly associated with mitochondrial biogenesis, mitochondrial oxidative metabolism, and the overall survival of patients.
Interpretation
Our results suggest that somatic mutations in mtCTR may be shaped by tumor-specific selective pressure and are involved in tumorigenesis.
Fundings
National Natural Science Foundation of China [grants 82020108023, 81830070, 81872302], and Autonomous Project of State Key Laboratory of Cancer Biology, China [grants CBSKL2019ZZ06, CBSKL2019ZZ27].
Keywords: Mitochondrial DNA, Control region, Hypervariable segment, Somatic mutation, Evolutionary selection
Research in context.
Evidence before this study
mtDNA mutations in cancer have drawn increasing attention in recent years. Previous pan-cancer studies have mainly focused on the mtDNA coding region (mtCDR), illustrating its mutational signatures, evolutionary selection difference, and functional impact across human cancers. Although the mtDNA control region (mtCTR), which contains various functional units regulating mtDNA replication and transcription, has long been identified as a mutational hotspot in various types of cancer, whether and how somatic mutations in mtCTR are subjected to evolutionary selection in different tumor types remains poorly explored. Several studies demonstrating the potential clinical impact of mtCTR mutations have been limited either by the research scope, where only a single or a few tumor types have been considered, or by the sequencing methodology, where traditional Sanger sequencing with relatively low sensitivity has been commonly applied for mutation detection. Therefore, a comprehensive and systematic profiling of mtCTR mutations across multiple tumor types based on next-generation sequencing (NGS) data is urgently needed to provide novel insights into mtCTR mutations and their critical roles in mitochondrial dysfunction and tumorigenesis.
Added value of this study
In this study, we gathered the most comprehensive mtCTR mutation dataset across 20 human tumor types from public and private NGS datasets. Our results showed that the mtCTR mutations were characterized by high variability across different tumor types compared to those in mtCDR. Mutations in mtCTR showed a similar distributional preference to the hypervariable segment (HVS) as germline variants. In addition, similar to the divergent evolutionary signatures observed in tumor mtCDR mutations, the evolutionary patterns of tumor mtCTR mutations also showed great differences and could be classified into three types: relaxed, moderate, and strict constraint types, based on tumor-specific mutational signatures between non-HVS and HVS regions, especially the relative prevalence of CH > TH in non-HVS. Moreover, regional mutation patterns in mtCTR were clearly associated with mitochondrial dysfunction and patient prognosis.
Implications of all the available evidence
The present study offers new insights into pan-cancer mutational signatures and the evolutionary patterns of mtCTR. Our results revealed that regional mutations might suffer from different selection powers and play distinct roles in mitochondrial dysfunction and clinical phenotypes in human cancers. Our study provides a better understanding of the evolutionary nature of mtCTR mutations, which may contribute to its utility as a diagnostic biomarker in clinical practice.
Alt-text: Unlabelled box
Introduction
Human mitochondrial DNA (mtDNA) is a highly compact, 16.5 kb circular molecule that harbors 37 genes encoding two rRNAs, 22 tRNAs, and 13 proteins essential for oxidative phosphorylation.1 The mtDNA control region (mtCTR) is approximately 1122 bp in length and is the single major non-coding part of mtDNA. It contains various functional units crucial for the regulation of mtDNA replication and transcription, such as the H-strand origin of replication (OHR), L-strand, and H-strand promoters (LSP and HSP) etc.1 Despite its vital regulatory role, mtCTR evolves much faster than the remaining mtDNA coding region (mtCDR), especially at the three hypervariable segments (HVS1, 16024-16383; HVS2, 57-372; HVS3, 438-574).2 In addition, mtCTR is often triple-stranded, forming a displacement loop (D-loop) by stable incorporation of a third short DNA strand known as 7S DNA (650 bp), which partially overlaps with the HVS1 and HVS2.3 mtCTR has long been identified as a mutational hotspot in various types of cancer.4, 5, 6 Because of the important role of mtCTR in mtDNA replication and transcription, the accumulation of mtCTR mutations may affect the copy number and expression level of mtDNA, potentially contributing to mitochondrial metabolic reprogramming and tumor progression. For instance, we have reported that the number and variant allele frequency (VAF) of mutations in the D-loop in hepatocellular carcinoma (HCC) are higher than those in adjacent non-tumor tissues, and are associated with decreased mtDNA copy number and poor prognosis of HCC patients.7 Similarly, somatic mutations in mtCTR have been considered indicators of poor prognosis in breast cancer and better prognosis in oral squamous cell carcinoma.8,9 However, these previous studies have limitations either because they mainly focused on single or only a few tumor types or because traditional Sanger sequencing with relatively low sensitivity has been commonly applied for mutation detection. Recently, several studies have profiled the molecular characteristics of mtDNA mutations across human cancers, mainly focusing on the mtCDR, and addressed their impact on mitochondrial function and prognosis of cancer patients.10,11 Therefore, a comprehensive and systematic profiling of mtCTR mutations across multiple tumor types based on next-generation sequencing (NGS) data is urgently needed to provide novel insights into mtCTR mutations and their critical roles in mitochondrial dysfunction and tumorigenesis.
The evolutionary selection of somatic mutations has been proposed as an important mechanism for intra-tumor, inter-tumor, and across-tumor heterogeneities. However, whether and how somatic mutations in mtCTR are subjected to evolutionary selection in different tumor types remains to be explored. Recent comprehensive profiling has provided evidence that germline variants in mtCTR are subjected to strong selection, especially in non-hypervariable segment (non-HVS).12 Previous studies have also indicated that HVS regions are mutational hotspots, and tumor somatic mutations share similar signatures with germline variants in HVS1 and HVS2.13 However, all four mtCTR mutations detected in five cancers were distributed in non-HVS, especially in colorectal cancer (CRC),14 indicating that non-HVS may undergo different mutational processes in tumors. Interestingly, profiling of mtDNA mutations across different tissues supports the idea that mtCTR mutations in non-malignant tissues may also be subjected to tissue-specific selection, potentially corresponding to distinct and tissue-specific metabolic requirements.15 However, it remains unclear whether tumor-specific selection and metabolic adaptation exists for mtCTR mutations across different tumor types.
Here, we present a comprehensive pan-cancer analysis of somatic mutations in mtCTR using multiple NGS datasets, including four public datasets and our private dataset. Our results revealed the presence of a highly variable and potentially tumor-specific mutation-selection interplay in the mtDNA control region across different tumor types.
Methods
Public and private somatic mutation data of mtDNA from tumor tissues
Public somatic mtDNA mutation datasets from whole-genome sequencing (WGS) or whole-exome sequencing (WES) data of human tumor tissues were directly downloaded from four publications, PNAS 2012, Elife 2014, PLoS Genet 2015, and Nat Genet 2020,11,14,16,17 in which detailed information about somatic mtDNA mutation calling was provided.
Private somatic mtDNA mutation data were generated in our lab by capture-based mtDNA sequencing of tumor and matched non-tumor tissue samples from 211 colorectal cancer (CRC), 117 hepatocellular carcinoma (HCC), 49 renal cell carcinoma (RCC), and 49 ovarian cancer (OV) patients. For mtDNA variant calling, raw sequencing data were aligned to rCRS and hg19 with BWA18 to eliminate contamination from nuclear DNA of mitochondrial origin (NUMT). We then sorted the reads and removed duplicate reads using Picard tools. The IndelRealigner in GATK was used for local realignment to reduce the false-positive rate near indel positions.19 Finally, high-quality reads were selected by SAMtools for further analysis.18,20
For each variant site, we first counted the respective number of major and minor alleles and calculated the site-specific VAF. We then filtered false positive variants based on the previously described negative logarithmic relationship between the mtDNA site sequencing depth and the minimum detectable mutation threshold.21 In addition, a range of subsequent filters were used for mtDNA mutation calling: (i) at least three reads in each strand carrying the alternative site; (ii) the total sequencing coverage≥100×; (iii) removing heterogeneity sites in rCRS repeat regions (66–71, 303–311, 514–523, 12418–12425, 16184–16193); (iv) removing mtDNA mutations if the mutant rate and mutant base quality do not satisfy the binomial distribution (P > 0.001)22; and (v) removing C > A/G > T mutations with strong sequence context bias (at CpCpN>CpApN; most frequently at CpCpG>CpApG) and low VAF (1%-2%) due to artificial guanine oxidation.23 After evaluation of sample cross-contamination,24 tumor somatic mutations were defined as heteroplasmic variants with VAF≥2% in tumor tissues and VAF < 0.5% in paired non-tumor tissues. Detailed quality control information is summarized in Supplementary_Table_S2. Finally, 239 mtDNA somatic mutations in CRC, 262 in HCC, 108 in RCC, and 74 in OV were identified, with a total of 683 mtDNA somatic mutations in 426 patients.
Generation of the total mutation dataset and three separate datasets
All mtDNA somatic mutations from the public and private datasets were combined to generate the total mutation dataset. If one sample was present in two or more datasets, all mutations in this sample were combined. Mutation data were presented based on the tissue type. Specific cancer subtypes were combined based on their same tissue origin. In addition, tumor types with a sample size smaller than 10 or without mtCTR mutations were excluded. Finally, a total mutation dataset containing 10026 mutations from 4664 samples was established, covering 20 tumor types and 40 specific cancer subtypes (Table 1; Supplementary_Table_S1). To rule out potential bias, the total mutation dataset was also divided into three separate datasets (Supplementary_Table_S3): mutation data from The Cancer Mitochondria Atlas (TCMA) published in Nat Genet 2020 were used as separate dataset 1; our private mutation data were used as separate dataset 3; mutation data from the other three publications were combined as separate dataset 2, in view of the similar mutation number/sample and high consistency between VAF of mutations (Supplementary_Figures_S1e-g and S2). In three separate datasets, six of twenty tumor types with mutation numbers smaller than five or present in only one separate dataset were removed to maintain consistency and make them comparable. Blood tumors were also removed, and only the solid tumors were retained. Finally, in separate datasets, 13 solid tumor types were maintained, among which eight common tumor types were composed of a single cancer subtype.
Table 1.
Summary of total mutation dataset.
| Tissue types | Sample size(n) | mtCDR(muts) | mtCTR(muts) |
Total(muts) | ||||
|---|---|---|---|---|---|---|---|---|
| Total | HVS1 | HVS2 | HVS3 | non-HVS | ||||
| Bladder | 100 | 140 | 19 | 7 | 11 | 0 | 1 | 159 |
| Bone/soft tissue | 297 | 277 | 42 | 12 | 17 | 5 | 8 | 319 |
| Breast | 603 | 1105 | 184 | 49 | 90 | 21 | 24 | 1289 |
| Cervix | 64 | 64 | 10 | 3 | 5 | 1 | 1 | 74 |
| CNS | 316 | 231 | 37 | 15 | 15 | 5 | 2 | 268 |
| Colon/rectum | 445 | 632 | 81 | 19 | 26 | 7 | 29 | 713 |
| Esophagus | 97 | 348 | 61 | 29 | 20 | 7 | 5 | 409 |
| Head and neck | 93 | 130 | 28 | 7 | 14 | 4 | 3 | 158 |
| Kidney | 241 | 768 | 165 | 58 | 63 | 17 | 27 | 933 |
| Liver | 479 | 1416 | 227 | 74 | 107 | 18 | 28 | 1643 |
| Lung | 159 | 283 | 46 | 20 | 11 | 6 | 9 | 329 |
| Lymphoid | 260 | 287 | 33 | 13 | 12 | 3 | 5 | 320 |
| Myeloid | 327 | 216 | 21 | 10 | 6 | 1 | 4 | 237 |
| Ovary | 222 | 496 | 52 | 26 | 17 | 8 | 1 | 548 |
| Pancreas | 313 | 765 | 96 | 38 | 33 | 11 | 14 | 861 |
| Prostate | 280 | 789 | 102 | 36 | 49 | 14 | 3 | 891 |
| Skin | 138 | 220 | 33 | 10 | 17 | 2 | 4 | 253 |
| Stomach | 81 | 210 | 35 | 17 | 9 | 3 | 6 | 245 |
| Thyroid | 48 | 154 | 15 | 7 | 6 | 2 | 0 | 169 |
| Uterus | 101 | 195 | 13 | 6 | 4 | 3 | 0 | 208 |
| Total | 4664 | 8726 | 1300 | 456 | 532 | 138 | 174 | 10026 |
mtDNA copy number data
Available data on mtDNA copy number for 53 CRC, 154 RCC, 252 HCC, and 99 OV samples were collected from the only source TCMA, in which mtDNA copy number was calculated as the coverage depth of mtDNA divided by the coverage depth of nuclear DNA, adjusted by tumor purity and ploidy. Detailed sample size information for each group of the four cancer subtypes is listed in Supplementary_Table_S4.
Prognosis analysis
A total of 275 CRC, 183 RCC, 427 HCC, and 139 OV patients with mtDNA mutations were enrolled from the public and private datasets for survival analysis. Kaplan-Meier curves and log-rank (Mantel-Cox) tests were used for survival analysis between groups with different mutational statuses. Multivariate Cox proportional hazards model was used to estimate hazard ratio (HR) and 95% confidence interval (95% CI) adjusting for age, gender, and tumor stage. Detailed sample size information for each group of the four cancer subtypes is listed in Supplementary_Table_S5.
Analysis of mutation density, average mutation sites and the incidence of different substitution types in mtDNA
The mutation density, average mutation sites, and incidence of mutations with certain substitution types were calculated for various genomic regions, including the mtCTR and mtCDR, and HVS and non-HVS in the mtCTR. Mutation density was calculated as the total number of mutations per sample divided by the region length (kb). The average mutation site was calculated as the total number of mutation sites per sample divided by the region length (kb). The incidence of a certain substitution type was calculated as the substitution number per sample divided by the number of base pairs (kb).
Gene expression analysis
RNA-seq count data of 334 cancer samples (152 CRC, 109 RCC, and 73 HCC) enrolled in our study were available in the Broad GDAC Firehose, where RNA-seq raw data were preprocessed using RSEM from Illumina HiSeq RNASeqV2. After normalization with the DEseq2 package of R software, the gene expression differences were analyzed for fold change (FC), and P values were corrected for multiple comparison testing using the Benjamini–Hochberg method. Detailed sample size information for each group for the three cancer types is listed in Supplementary_Tables_S6, S7.
Proliferation and metastasis assay
Proliferation, Transwell invasion, and wound healing assays were performed as previously described.25 mtCTR mutations detected in the six HCC cell lines are listed in Supplementary_Table_S8.
Detection of deletion/duplication
The deletions/duplications of mtDNA in tumor and non-tumor samples were analyzed by mitoSAlt.26 A total of 397 samples were analyzed, including 98 CRC, 96 non-CRC, 97 HCC, and 106 non-HCC samples.
Statistical analysis
All statistical tests were performed using the GraphPad Prism software version 8 for Windows (GraphPad Software, San Diego, CA, USA). The Mann–Whitney U test or Wilcoxon signed-rank paired test was used to compare the difference between the two groups with continuous variables. Spearman's rank correlation coefficient was used to measure the correlation between two groups of variables. Chi-square test or Fisher's exact probability test was performed using discrete variables. All P-values were two-tailed, and statistical significance was set at P<0.05.
Ethics
Ethical approval for public data was obtained from all four original studies. Ethical approval for private data was approved by the Ethical Committees of FMMU, and written consent was obtained from each patient.
Role of Funders
The funding sources played no role in the study design, data collection, data analysis, interpretation, or writing.
Results
High variability of mutation density in mtDNA control region across 20 tumor types
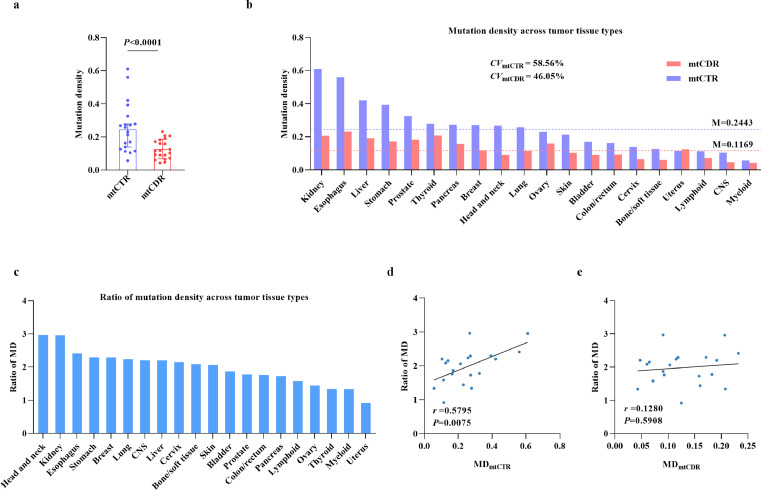
To systematically explore the uncharacterized patterns of somatic mutations in the mtCTR across different tumor types, we generated a comprehensive combined dataset of mtDNA somatic mutations in pan-cancer based on public and private NGS data, with 10026 mutations from 4664 tumor samples covering 20 tumor types and 40 cancer subtypes (Table 1; Supplementary_Table_S1).11,14,16,17 Considering the different analysis pipelines and cutoff values, we evaluated the heterogeneity of data from the four studies by comparing the mutation site distribution and average mutation rate. As shown in Supplementary_Figure_S1a-d, the proportion of somatic mutations in both mtCTR and mtCDR remained unchanged under different cutoff values for mutation filtering, although the average mutation number decreased as the cutoff value increased in different datasets. This finding suggests the feasibility of combining the mutation data for subsequent analysis. The distribution of mtDNA somatic mutations was further investigated. Similar to previous reports,27,28 the mutation density (MD) in mtCTR was significantly higher than that in mtCDR (Figure 1a, Mann-Whitney U test, P<0.0001). Importantly, we found that the MD in mtCTR (median, 0.2443; range, 0.0572–0.6102) presented significantly greater variance across tumor types than mtCDR (median, 0.1169; range, 0.0428–0.2323), with coefficients of variation (CV) of 58.56% and 46.05%, respectively (Figure 1b). Furthermore, the ratio of the MD between mtCTR and mtCDR (MDmtCTR/mtCDR) also varied greatly across the different tumor types (Figure 1c). In addition, the value of MDmtCTR/mtCDR was significantly correlated with the MD of mtCTR but not with the MD of mtCDR (Figure 1d, e). To rule out potential bias, the total mutation dataset was divided into three separate datasets to further validate our findings (Supplementary_Table_S3). As expected, similar results were observed in all three separate datasets, further validating the high variability of the mtDNA control region across tumor types and cancer subtypes (Supplementary_Figures_S3, S4). In general, about 4% of 397 samples (98 CRC and 96 non-CRC, 97 HCC, and 106 non-HCC) had private deletions/duplications with heteroplasmic levels over 1% (Supplementary_Figure_S5). Apart from these private deletions/duplications, a low-level duplication ranging from 4417-4463 were observed in near half CRC and HCC samples with deletions/duplications. Whether it was a new “common duplication” or a false positive discovery might require further investigation.
Figure 1.
High variability of mutation density in mtDNA control region across 20 tumor types. (a) Comparison of mutation density (MD) between mtDNA control region (mtCTR) and coding region (mtCDR) in 20 tumor types. MD was defined as somatic mutation number/kb in one sample. Median with 95% CI was shown. P value was from Mann Whitney U test. (b) Mutation density of mtCTR and mtCDR and (c) Ratio of MD between mtCTR and mtCDR across 20 tumor types. Ratio was calculated as the MD of mtCTR divided by corresponding MD of mtCDR (ratio of MDmtCTR/mtCDR) in certain tumor tissue type. (d and e) Spearman rank correlation between ratio of MDmtCTR/mtCDR and MD of mtCTR or MD of mtCDR in 20 tumor types. M, median. CV, coefficient of variation.
Distributional bias of tumor somatic mutations in non-HVS and HVS of mtDNA control region
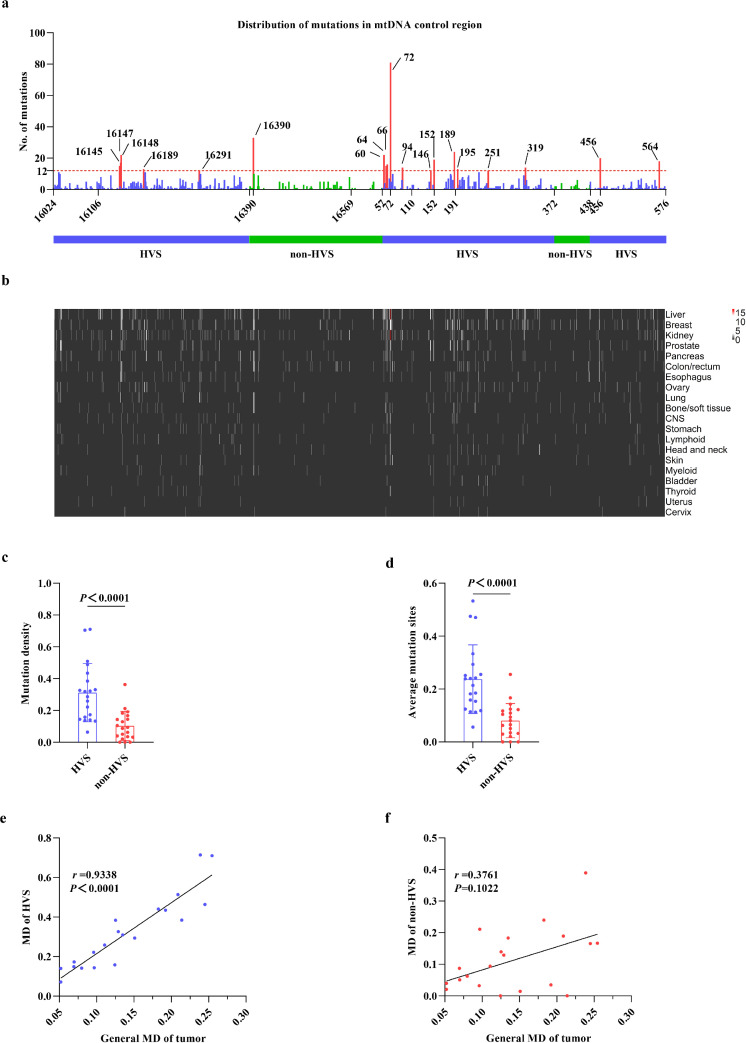
Considering evolutionary evidence that germline variants are enriched at the HVS of mtCTR and that more strict negative selection exists in non-HVS,12 we wondered whether the tumor somatic mutations in mtCTR also aggregated more at the HVS. As shown in Figure 2a, the majority (n=1126, 86.61%) of the 1300 mutations were in the HVS. A similar distribution pattern was also observed in all three separate datasets (Supplementary_Figure_S6a, d, g). Meanwhile, 18 of the 19 mutation hotspots were found in HVS, except for site 16390 in non-HVS. Aggregation of tumor mtCTR somatic mutations in the HVS was also observed when mutations were located across the 20 tumor types (Figure 2b). Moreover, our data showed significantly higher mutation density and average mutation sites in HVS compared to non-HVS in the total mutation dataset (Figure 2c, d) and all 3 separate datasets (Supplementary_Figure_S6b, c, e, f, h, i). In addition, as shown in Figure 2e, f, the MD of HVS was positively correlated with the general MD of tumor across tumor types, while no significant correlation was observed between the MD of non-HVS and the general MD of tumor. Our findings indicate a remarkable distributional bias of tumor somatic mutations in the non-HVS and HVS.
Figure 2.
Distributional bias of tumor somatic mutations in non-HVS and HVS of mtDNA control region. (a) Distribution of somatic mutations in different regions of mtCTR in total mutation dataset. Mutation hotspots (mutation number was at least 10 times higher than the average mutation number of mtCTR, that is no less than 12) were colored in red. HVS, hypervariable segment. (b) Distribution of somatic mutations in different regions of mtCTR across 20 tumor types. Tissues were lined from top to bottom according to the total mutation number of mtCTR. (c and d) Comparison of mutation density or average mutation sites between HVS and non-HVS of mtDNA control region in tumor types. (e and f) Spearman rank correlation between the general MD of tumor and the MD of HVS or the MD of non-HVS. P values were calculated from Mann Whitney U test.
Tumor-specific mutational pattern between non-HVS and HVS in mtDNA control region
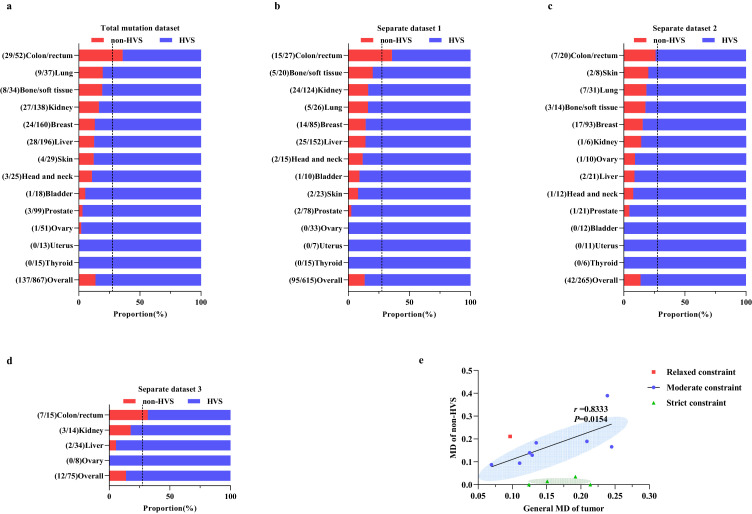
We further investigated the mutational pattern of mtCTR across different tumor types by comparing the proportion of mutations in the non-HVS and HVS of mtCTR (Figure 3a). Interestingly, our data indicated that the proportion of mutations in non-HVS and HVS, as well as the degree of distributional bias of mutations in mtCTR, was remarkably different across the 13 solid tumor types. A very similar tumor-specific mutational pattern between non-HVS and HVS was also confirmed in all three separate datasets across tumor types and cancer subtypes (Figure 3b–d; Supplementary_Figure_S7a–c). Based on the relative distribution of non-HVS and HVS somatic mutations, we designated different tumor types into three mutation-selection subtypes (Figure 3a–d). In colon/rectum tumors, the mutation proportion in the non-HVS region was almost equal to the length proportion of non-HVS in mtCTR (27.5%), implying an even distribution and lack of evolutionary selection for non-HVS and HVS mutations in this tumor type, we thus defined it as the relaxed constraint type. In contrast, almost no mutations were detected in the non-HVS of mtCTR in the prostate, ovary, uterus, and thyroid tumor tissues, leading to mutation proportions of HVS almost 100%, which was defined as the strict constraint type. The other 8 tumor types showed an intermediate distribution of non-HVS and HVS mutations, which was defined as the moderate constraint type. In addition, we found a positive correlation between the MD of non-HVS and the general MD of tumors belonging to the moderate constraint type (Spearman's correlation, r= 0.8333, P= 0.0154) (Figure 3e). When mutations were classified according to 7S DNA, the triple-strand area in mtCTR, the proportion of mutations in the 7S DNA region showed no significant difference across the 13 tumor types in both the total mutation dataset and three separate datasets (Supplementary_ Figure_S8a–d). Although not all (8 of 13) tumor types are composed of a single cancer subtype, these results strongly suggest that the evolutionary selection pressure on HVS and non-HVS mutations may be tumor- or cancer subtype-specific.
Figure 3.
Tumor-specific mutation pattern between mtCTR non-HVS and HVS regions. Proportion of somatic mutations in HVS and non-HVS regions of mtCTR across 13 solid tumor types in the total mutations dataset (a) and three separate datasets including separate dataset 1 (b), separate dataset 2 (c), and separate dataset 3 (d). Vertical dashed lines indicated the length proportion of non-HVS in mtCTR. Mutation number in non-HVS and HVS regions was shown in brackets. (e) Distributional pattern of the general MD of tumor and the MD of non-HVS across 13 solid tumor types and Spearman rank correlation in 8 solid tumor types.
Prevalence of CH > TH in non-HVS greatly contributed to mutational selection pattern of mtCTR across different tumor types
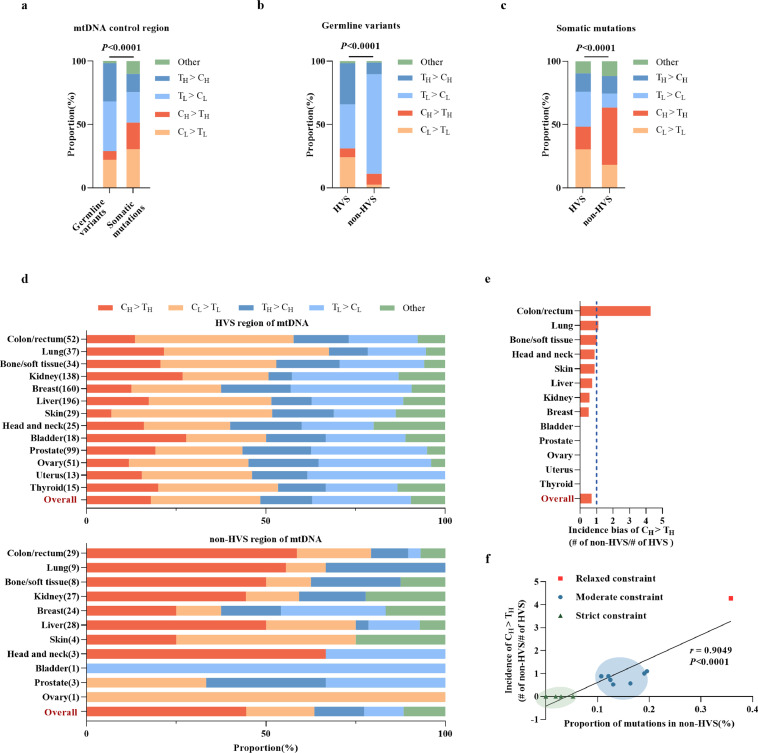
To explore the underlying mechanism of the tumor-specific mtCTR mutational pattern, we first compared the substitution signature between germline variants and somatic mutations in mtCTR across the 13 tumor types. As shown in Figure 4a, there was a significantly different substitution pattern between germline variants and somatic mutations (Chi-square test, P < 0.0001). A detailed analysis of mtCTR mutations revealed the existence of a dramatic substitution difference between the HVS and non-HVS groups in both germline variants and somatic mutations. The non-HVS group exhibited an overwhelmingly high proportion of TL > CL transitions (78.6%) in germline variants (Figure 4b) and a significant increase in CH > TH transitions (from 8.5% to 45.2%) in somatic mutations (Figure 4b, c). The L strand refers to the reference sequence for alignment. Notably, although a broadly similar mutational pattern was observed between the germline and somatic HVS groups, the somatic non-HVS group exhibited a dramatic change in the substitution spectrum relative to the germline non-HVS group, as exemplified by an abrupt reduction in TL > CL proportions and a significant increase in CH > TH proportion (Figure 4b, c). These results suggest that the selection pattern may differ between germline and somatic mutations, particularly in the non-HVS region.
Figure 4.
Prevalence of CH > TH in non-HVS greatly contributed to mutational selection pattern across different tumors. (a) Proportion of different substitution types in germline variants (n=13542) of mtCTR from Elife 2014 (n=10095) and our data (n=3447) and somatic mutations (n=1007) of mtCTR from 13 solid tumor types. (b) Proportion of different substitution types in germline variants of mtCTR HVS and non-HVS regions. (c) Proportion of different substitution types in somatic mutations of mtCTR HVS and non-HVS regions. (d) Proportion of different substitution types in somatic mutations of mtCTR HVS and non-HVS regions across 13 solid tumor types. (e) Incidence bias of CH > TH across 13 tumor types. Incidence bias was defined as substitution number per kilo base pair (kb) in non-HVS region divided by substitution number per kilo base pair (kb) in HVS region in certain tumor tissue type. (f) Spearman rank correlation between incidence bias of CH > TH and proportion of somatic mutations in non-HVS region in 13 tumor types. All P values were from Chi-square test.
A previous study reported a substitution bias toward CL > TL in mtCTR, especially in the region around the origin of replication.17 Therefore, we further compared the substitution spectrum of somatic mutations between HVS and non-HVS across 13 tumor types and found that they were similar in HVS but quite variable in non-HVS, especially for CH > TH proportion (Figure 4d). Thus, we compared the incidence ratio of CH > TH between non-HVS and HVS among different tumor types. As shown in Figure 4e, there was a remarkably different CH > TH incidence ratio across 13 tumor types, with colon/rectum tumors showing a high incidence ratio, but four tumor types showed complete absence of non-HVS CH > TH. Meanwhile, the CH > TH incidence ratio correlated well with the mutation proportion of non-HVS in mtCTR across 13 tumor types, which were classified into three evolutionary types (Figure 4f). These results suggest that the prevalence of CH > TH in the non-HVS region may greatly contribute to the tumor- or cancer subtype-specific selection pattern of mtCTR.
Regional mutations in mtCTR were associated with tumor mitochondrial dysfunction
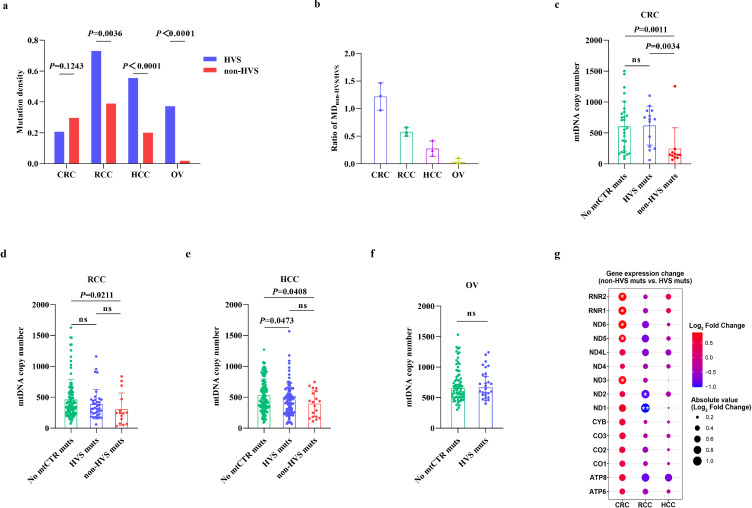
mtCTR contains various regulatory elements crucial for mtDNA replication and transcription (Supplementary_Figure_S9).29 Thus, we first investigated the association between regional mutations in mtCTR and mtDNA copy number, an indicator of mitochondrial biogenesis, in four representative cancer subtypes selected from three evolutionary types. As shown in Figure 5a, no significant difference in mutation density was observed between non-HVS and HVS in CRC, which exhibited a relaxed constraint evolutionary type in mtCTR mutations, whereas significant differences existed in RCC and HCC with moderate constraint selection, as well as in OV with strict constraint selection. In contrast, CRC had the highest ratio of MDnon-HVS/HVS, whereas OV had the lowest ratio (Figure 5b). The mtDNA copy number was further compared between cancer samples with different status of regional mtCTR mutations. When compared to samples without mtCTR mutations, the samples with non-HVS mutations exhibited a significantly lower mtDNA copy number in CRC, RCC, and HCC, while no significant difference was observed between samples with HVS mutations and without mtCTR mutations in all cancer subtypes, except for HCC (Figure 5c–f). Meanwhile, the mtDNA copy numbers in samples with non-HVS mutations were significantly lower than those in samples with HVS mutations only in CRC. Our findings indicate that mutations in mtCTR exhibit a remarkable but cancer subtype-specific effect on mitochondrial biogenesis.
Figure 5.
Mutational pattern of mtDNA control region was associated with tumor mitochondrial dysfunction. (a) Comparison of mutation density between non-HVS and HVS in four tumor types. P values were from Fisher's exact test. (b) Ratio of MDnon-HVS/HVS among four tumor types. (c-f) Comparison of mtDNA copy number among samples without mutations in mtDNA control region (no mtCTR muts), samples with HVS mutations (HVS muts) and samples with non-HVS mutations (non-HVS muts) in CRC, RCC, HCC and OV. P values were from Mann Whitney U test. (g) Gene expression change of 15 mtDNA genes between tumor samples with HVS mutations and non-HVS mutations in CRC, RCC and HCC. The bubble colors from red to blue correspond to Log2 fold change of gene expression level from high to low in non-HVS group compared to HVS group. *P<0.05, **P<0.01.
Considering the key role of mtDNA in mitochondrial oxidative phosphorylation,30 we further analyzed the association between regional mutations in mtCTR and the transcriptional levels of mitochondrial genes. As shown in Figure 5g, cancer samples with mtCTR non-HVS mutations exhibited a relatively higher expression level of mtDNA genes in CRC when compared with tumor tissue samples with HVS mutations, although not all gene upregulation reached statistical significance. In contrast, no notable difference in expression was observed between samples with non-HVS and HVS mutations in RCC or HCC. We further compared the mtCDR mutational burden between samples with HVS mutations and non-HVS mutations in CRC, RCC, and HCC. No significant difference was observed in any cancer type (Supplementary_Figure_S10). Then, we grouped the samples according to the median mtCDR mutation number in each cancer type and compared mtDNA gene expression at the transcriptional level. Our results showed no significant difference in mtDNA gene expression between samples with different mtCDR mutational burdens (Supplementary_Figure_S11). These results suggest that mutations in mtCTR may be associated with cancer subtype-specific mitochondrial biogenesis and metabolic reprogramming.
Somatic mutations of mtDNA control region were associated with prognosis of cancer patients
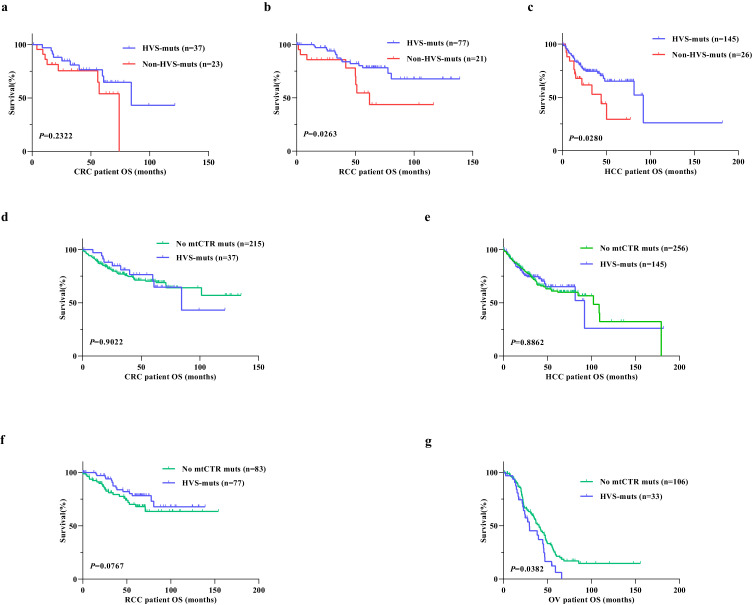
Considering the effect of mtCTR mutations on mitochondrial biogenesis and metabolism, we evaluated the association of mtCTR mutations with cancer patient prognosis. Kaplan–Meier survival analysis revealed that patients with non-HVS mutations exhibited a significantly poorer prognosis than those with HVS mutations in both RCC (log-rank P=0.0263) and HCC (log-rank P=0.0280). However, no significant survival differences were observed between patients with HVS mutations and those with non-HVS mutations in CRC (Figure 6a–c). Multivariate Cox regression analysis also indicated that non-HVS mutation was an independent prognostic factor for overall survival (OS) of HCC patients, but not CRC patients (Supplementary_Table_S9). We also grouped the samples according to their mtCDR mutational burden for survival analysis. No difference in OS was found between patients carrying different mtCDR mutational burdens in CRC, RCC, and HCC (Supplementary_Figure_S12). Functional analysis showed that HCC cell lines with non-HVS mutations exhibited higher proliferation and metastasis ability than those with HVS mutations (Supplementary_Figure_S13). Meanwhile, there was no significant difference in OS between patients with HVS mutations and those with mutations outside mtCTR in CRC, RCC, and HCC, except for OV, in which patients with HVS mutations exhibited a significantly poorer prognosis than those with mutations outside mtCTR (Figure 6d–g). Our findings suggest that somatic mutations in mtCTR, especially non-HVS mutations, may play important roles in the progression of specific cancer subtypes.
Figure 6.
Somatic mutations of mtDNA control region were associated with prognosis of cancer patients. (a-c) Kaplan Meier curve analysis of overall survival (OS) between patients with non-HVS mutations and HVS mutations in CRC, RCC and HCC. (d-g) Kaplan Meier curve analysis of overall survival between patients without mtCTR mutations and with HVS mutations in CRC, RCC, HCC and OV. Log-rank test was used to compare overall survival between two groups.
Discussion
In this study, we for the first time systematically characterized somatic mutation patterns in the mitochondrial control region (mtCTR) across multiple tumor types based on a comprehensive mtDNA mutation dataset. Several key findings were obtained. First, somatic mutations in mtCTR were characterized by high variability across different tumor types. Second, tumor mtCTR somatic mutations were remarkably enriched in HVS, but not in non-HVS regions. Third, mtCTR mutation evolutionary patterns were classified into three types based on tumor- or cancer subtype-specific mutational signatures between non-HVS and HVS, including relaxed, moderate, and strict constraint types, which can greatly contribute to the prevalence of CH > TH in non-HVS. Fourth, the regional mutation patterns in mtCTR were clearly associated with mitochondrial dysfunction and patient prognosis.
In the present study, we found that the mutation density in mtCTR exhibited great variation across different tumor types, whereas it was relatively stable in the coding region. Similar findings have been reported in previous studies involving a single type of cancer using Sanger sequencing. For example, it has been shown that 55.7% RCC patients had at least one mtCTR mutation, with the average mutation number of 2.3 per patient.31 In contrast, an average mtCTR mutation number of 0.86 has been reported in thyroid lesions.32 Interestingly, the tumor species with high mtCTR mutation density seem to originate from tissues with high levels of mtDNA replication/transcription, since the expression of mitochondrial transcription factor A (TFAM) is much higher in the kidney and liver than that in the uterus.11,33 Therefore, we hypothesized that the variable intensity of mtDNA replication might lead to different selection pressures for mtCTR mutations across different tumor or cancer subtypes. However, the detailed mechanism remains to be elucidated.
Extensive sequencing of the mtCTR across different populations has consistently identified three hypervariable segments (HVS1-3), which are enriched by germline variants and interspaced by segments with little variability (non-HVS).34,35 Similarly, our study found an overall enrichment of somatic mutations in the HVS of mtCTR in multiple solid tumors. Similar findings have been reported in breast, pancreatic, and prostate cancers.8,36,37 Considering that most of the regulatory elements identified to be involved in mtDNA replication and transcription are located in the HVS, but not in the non-HVS region,38,39 the distributional bias of both germline and somatic mutations in mtCTR is interesting and thought-provoking. Wei et al. have recently provided evidence that the variability between HVS and non-HVS may be strongly shaped by evolutionary selection of germline variants during population expansion, supporting a critical functional role of mtCTR.12 Thus, it's reasonable to assume that the mtCTR may also play an active role in tumorigenesis. Specifically, somatic mutations in HVS might be more tolerated for functional neutrality or positively selected for better metabolic adaptation, whereas many somatic mutations in non-HVS may be impermissible in tumorigenesis and negatively selected during tumor evolution and progression.
Consistent with the well-known inter-tumor heterogeneity,40 we also observed a high level of heterogeneity in the regional distribution of mtCTR somatic mutations across different tumor types. Considering the critical functionality of the non-HVS region,12 we propose that the variable distribution of somatic mutations in HVS and non-HVS regions may reflect the presence of tumor- or cancer subtype-specific selection pressure against non-HVS mutations, exemplifying the relaxed, moderate, and strict constraint types, respectively. In this regard, only CRC exhibited relaxed selection against non-HVS mutations in the combined dataset. Similarly, Gorelick et al. reported that CRC cells tolerated a high level of truncating mutations in mtCDR.10 In contrast, OV represented a case of the strict constraint type, where the non-HVS mutations seemed to be severely depleted. One clue is that OV develops, metastasizes, and recurs in the abdominal cavity, which represents a unique microenvironment that may impose critical requirements on mitochondrial respiration.41 Between the relaxed and strict constraint types, we also observed a group of cancers with a moderate constraint type, such as RCC and HCC, where the non-HVS mutations appeared to be moderately depleted. Both increased mutation rate and evolutionary selection may contribute to such different mutation patterns detected across 13 tumor types. In previous studies, mtDNA replication error has been well recognized to be the common reason for the origin of mtDNA mutations,17,42 for which no site or region preference of mutations has been reported. Therefore, we are more inclined to the possible explanation of evolutionary selection on different mutation patterns of mtCTR across 13 tumor types. In addition, our previous study has provided evidence that specific somatic mutations in mtCTR may be subjected to positive selection in HCC.7 Meanwhile, a recent study has reported that mutations in mtCTR showed significantly different substitution gradients and distinct regional biases compared to those in mtCDR, indicating that regulatory elements may alter mutational potential.43 Together, these results indicate a very complex evolutionary process involving mutations in the mtDNA control region across different tumor types, which needs to be further elucidated by a more systematic and comparative pan-cancer analysis.
As mtCTR contains various regulatory regions, mutations in mtCTR are expected to affect mitochondrial biogenesis to some extent. As expected, both up- and down-regulation have been reported in previous studies. For example, mtCTR mutations have been reported to increase mtDNA copy number in human laryngeal squamous cell carcinoma44 but decrease mtDNA copy number in liver and breast cancers.7,45 In the present study, we found a decrease in mtDNA copy number in tissues with non-HVS mutations in the three cancer subtypes compared to control tissues, implying that the analysis of regional mutations in mtCTR may help understand the function of mtDNA mutations. Meanwhile, we also observed inconsistent results in OV with a strictly constrained evolutionary type, indicating no remarkable effect of non-HVS mutations on mtDNA copy number. Therefore, we suspected the existence of a cancer subtype-specific compensatory mechanism for mitochondrial replication.
Interestingly, we found that the expression levels of mitochondrial genes for oxidative metabolic function were significantly enhanced in CRC tissues with non-HVS mutations, but there was no difference between RCC and HCC tissues. A series of previous studies have shown that CRC depends more on mitochondrial oxidative phosphorylation.46 Therefore, we speculate that relaxed selection pressure may shape the specific mutation pattern in the non-HVS of mtDNA and play important roles in the regulation of oxidative metabolism in CRC cells. Røyrvik et al. reported that mutations located at different sites of mtCTR switch mtDNA replication to transcription,40 which provides partial support for our hypothesis. Additionally, our data indicated that non-HVS mutations could act as a poor biomarker for prognosis in RCC and HCC patients, which further strengthens the clinical significance of mtCTR mutations. However, whether non-HVS mutations affect the prognosis of tumor patients by regulating oxidative metabolism remains unclear.
Despite these inspiring findings, the present study had several limitations. First, because of the lack of access to raw sequencing data, only ready-made mutation data from four publications were used for signature profiling. Second, the sample size and total mutation number were relatively small for the subgroup analysis, especially for specific cancer subtypes. Therefore, some biologically rationalized inter-cancer comparisons could not be carried out, although our major conclusions were not affected. Finally, only a small portion of RNA-seq data was available for our tumor samples. As a result, gene expression analysis is more indicative, but not conclusive. Collectively, our study is the first to comprehensively profile pan-cancer signatures of mtCTR somatic mutations based on NGS data. Our findings reveal the tumor-specific evolutionary patterns of mtCTR somatic mutations across different tumor types, which may contribute to the regulation of mitochondrial biogenesis and oxidative metabolism, and function as prognostic biomarkers in cancer patients, further emphasizing the critical roles of mtDNA mutations in tumorigenesis.
Contributors
XYJ, WJG, and XWG: data curation, formal analysis, and writing - original draft. QY, SSG, and LPS: software and bioinformatic analysis. YL and KXZ: laboratory experiments. XG: writing - review and editing, and funding acquisition. JLX and QCH: conceptualization, supervision, funding acquisition, project administration, and revising the manuscript. All authors have read and approved the final manuscript.
Data sharing statement
The public mtDNA mutation data in this study are available in four publications11,14,16,17 and our private mtDNA mutation data are available in the supplementary data file (Supplementary file 2). The raw sequencing data underlying this article are available at the BIG Data Center, Beijing Institute of Genomics (BIG), with access number PRJCA006830. For colorectal cancer, renal cell cancer, hepatocellular carcinoma, and ovarian cancer samples, mtDNA copy number data were downloaded from The Cancer Mitochondria Atlas (https://ibl.mdanderson.org/tcma/), and RNA-seq count data were downloaded from the Broad GDAC Firehose (http://gdac.broadinstitute.org/).
Declaration of interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [Grants 82020108023, 81830070, 81872302]; and Autonomous Project of State Key Laboratory of Cancer Biology, China [Grants CBSKL2019ZZ06, CBSKL2019ZZ27].
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104058.
Contributor Information
Qichao Huang, Email: huangqichao1@163.com.
Jinliang Xing, Email: xingjinliang@163.com.
Appendix. Supplementary materials
References
- 1.Gustafsson C.M., Falkenberg M., Larsson N.G. Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem. 2016;85:133–160. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 2.Vanecek T., Vorel F., Sip M. Mitochondrial DNA D-loop hypervariable regions: Czech population data. Int J Legal Med. 2004;118(1):14–18. doi: 10.1007/s00414-003-0407-2. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls T.J., In M.M. D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol. 2014;56:175–181. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Wallace D.C. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopkins J.F., Sabelnykova V.Y., Weischenfeldt J., et al. Mitochondrial mutations drive prostate cancer aggression. Nat Commun. 2017;8(1):656. doi: 10.1038/s41467-017-00377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma H., Singh A., Sharma C., Jain S.K. Singh N. Mutations in the mitochondrial DNA D-loop region are frequent in cervical cancer. Cancer Cell Int. 2005;5:34. doi: 10.1186/1475-2867-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin C., Li D.Y., Guo X., et al. NGS-based profiling reveals a critical contributing role of somatic D-loop mtDNA mutations in HBV-related hepatocarcinogenesis. Ann Oncol. 2019;30(6):953–962. doi: 10.1093/annonc/mdz105. [DOI] [PubMed] [Google Scholar]
- 8.Kuo S.J., Chen M., Ma G.C., et al. Number of somatic mutations in the mitochondrial D-loop region indicates poor prognosis in breast cancer, independent of TP53 mutation. Cancer Genet Cytogenet. 2010;201(2) doi: 10.1016/j.cancergencyto.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Lin J.C., Wang C.C., Jiang R.S., Wang W.Y., Liu S.A. Impact of somatic mutations in the D-loop of mitochondrial DNA on the survival of oral squamous cell carcinoma patients. PloS one. 2015;10(4) doi: 10.1371/journal.pone.0124322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorelick A.N., Kim M., Chatila W.K., et al. Respiratory complex and tissue lineage drive recurrent mutations in tumour mtDNA. Nat Metab. 2021;3(4):558–570. doi: 10.1038/s42255-021-00378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Y., Ju Y.S., Kim Y., et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52(3):342–352. doi: 10.1038/s41588-019-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W., Tuna S., Keogh M.J., et al. Germline selection shapes human mitochondrial DNA diversity. Science. 2019;364(6442) doi: 10.1126/science.aau6520. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 13.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet. 2000;67(4):1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larman T.C., DePalma S.R., Hadjipanayis A.G., et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109(35):14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M., Schroder R., Ni S., Madea B., Stoneking M. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc Natl Acad Sci U S A. 2015;112(8):2491–2496. doi: 10.1073/pnas.1419651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart J.B., Alaei-Mahabadi B., Sabarinathan R., et al. Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers. PLoS Genet. 2015;11(6) doi: 10.1371/journal.pgen.1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ju Y.S., Alexandrov L.B., Gerstung M., et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Handsaker B., Wysoker A., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna A., Hanna M., Banks E., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Guo S., Yin C., et al. Optimized PCR-based enrichment improves coverage uniformity and mutation detection in mitochondrial DNA next-generation sequencing. J Mol Diagn. 2020;22(4):503–512. doi: 10.1016/j.jmoldx.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Guo S., Zhou K., Yuan Q., et al. An innovative data analysis strategy for accurate next-generation sequencing detection of tumor mitochondrial DNA mutations. Mol Ther Nucleic Acids. 2021;23:232–243. doi: 10.1016/j.omtn.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campo D.S., Nayak V., Srinivasamoorthy G., Khudyakov Y. Entropy of mitochondrial DNA circulating in blood is associated with hepatocellular carcinoma. BMC Med Genom. 2019;12(Suppl 4):74. doi: 10.1186/s12920-019-0506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costello M., Pugh T.J., Fennell T.J., et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41(6):e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su L., Guo S., Guo W., et al. mitoDataclean: A machine learning approach for the accurate identification of cross-contamination-derived tumor mitochondrial DNA mutations. Int J Cancer. 2022;150(10):1677–1689. doi: 10.1002/ijc.33927. [DOI] [PubMed] [Google Scholar]
- 25.Ren T., Zhang H., Wang J., et al. MCU-dependent mitochondrial Ca(2+) inhibits NAD(+)/SIRT3/SOD2 pathway to promote ROS production and metastasis of HCC cells. Oncogene. 2017;36(42):5897–5909. doi: 10.1038/onc.2017.167. [DOI] [PubMed] [Google Scholar]
- 26.Basu S., Xie X., Uhler J.P., et al. Accurate mapping of mitochondrial DNA deletions and duplications using deep sequencing. PLoS Genet. 2020;16(12) doi: 10.1371/journal.pgen.1009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandon M., Baldi P., Wallace D.C. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 28.Tan D.J., Bai R.K., Wong L.J.C. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62(4):972–976. [PubMed] [Google Scholar]
- 29.Lott M.T., Leipzig J.N., Derbeneva O., et al. mtDNA variation and analysis using mitomap and mitomaster. Curr Protoc Bioinf. 2013;44:1–6. doi: 10.1002/0471250953.bi0123s44. 1 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schopf B., Weissensteiner H., Schafer G., et al. OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation. Nat Commun. 2020;11(1):1487. doi: 10.1038/s41467-020-15237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H., Komiyama T., Nitta M., et al. D-loop mutations in renal cell carcinoma improve predictive accuracy for cancer-related death by integrating with mutations in the NADH dehydrogenase subunit 1 gene. Genes. 2019;10(12) doi: 10.3390/genes10120998. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Z., Ji J., Chen G., et al. Analysis of mitochondrial DNA mutations in D-loop region in thyroid lesions. Biochim Biophys Acta. 2010;1800(3):271–274. doi: 10.1016/j.bbagen.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Reznik E., Miller M.L., Senbabaoglu Y., et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5 doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenberg B.D., Newbold J.E., Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983;21(1-2):33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- 35.Lutz S., Weisser H.J., Heizmann J., Pollak S. A third hypervariable region in the human mitochondrial D-loop. Hum Genet. 1997;101(3):384. [PubMed] [Google Scholar]
- 36.Chen J.Z., Gokden N., Greene G.F., Mukunyadzi P., Kadlubar F.F. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res. 2002;62(22):6470–6474. [PubMed] [Google Scholar]
- 37.Hardie R.A., van Dam E., Cowley M., et al. Mitochondrial mutations and metabolic adaptation in pancreatic cancer. Cancer Metab. 2017;5:2. doi: 10.1186/s40170-017-0164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher R.P., Topper J.N., Clayton D.A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50(2):247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 39.Falkenberg M. Mitochondrial DNA replication in mammalian cells: overview of the pathway. Essays Biochem. 2018;62(3):287–296. doi: 10.1042/EBC20170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Royrvik E.C., Johnston I.G. MtDNA sequence features associated with 'selfish genomes' predict tissue-specific segregation and reversion. Nucleic Acids Res. 2020;48(15):8290–8301. doi: 10.1093/nar/gkaa622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emmings E., Mullany S., Chang Z., Landen C.N., Linder S., Bazzaro M. Targeting mitochondria for treatment of chemoresistant ovarian cancer. Int J Mol Sci. 2019;20(1) doi: 10.3390/ijms20010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker K.T., Nachmanson D., Kumar S., et al. Mitochondrial DNA mutations are associated with ulcerative colitis preneoplasia but tend to be negatively selected in cancer. Mol Cancer Res. 2019;17(2):488–498. doi: 10.1158/1541-7786.MCR-18-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Contreras M., Sweetwyne M.T., Kohrn B.F., et al. A replication-linked mutational gradient drives somatic mutation accumulation and influences germline polymorphisms and genome composition in mitochondrial DNA. Nucleic Acids Res. 2021;49(19):11103–11118. doi: 10.1093/nar/gkab901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo W., Yang D., Xu H., et al. Mutations in the D-loop region and increased copy number of mitochondrial DNA in human laryngeal squamous cell carcinoma. Mol Biol Rep. 2013;40(1):13–20. doi: 10.1007/s11033-012-1939-7. [DOI] [PubMed] [Google Scholar]
- 45.Yu M., Zhou Y., Shi Y., et al. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59(7):450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 46.Zheng J. Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (Review) Oncol Lett. 2012;4(6):1151–1157. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.