Abstract
The feasibility of using probes directed towards ribosomal DNAs (rDNAs) as a quantitative approach to estimating cell numbers was examined and applied to study the structure of a bacterial community in humic acid-rich salt marsh sediments. Hybridizations were performed with membrane-bound nucleic acids by using seven group-specific DNA oligonucleotide probes complementary to 16S rRNA coding regions. These included a general eubacterial probe and probes encompassing most members of the gram-negative, mesophilic sulfate-reducing bacteria (SRB). DNA was extracted from sediment samples, and contaminating materials were removed by a series of steps. Efficiency of DNA extraction was 48% based on the recovery of tritiated plasmid DNA added to samples prior to extraction. Reproducibility of the extraction procedure was demonstrated by hybridizations to replicate samples. Numbers of target cells in samples were estimated by comparing the amount of hybridization to extracted DNA obtained with each probe to that obtained with a standard curve of genomic DNA for reference strains included on the same membrane. In June, numbers of SRB detected with an SRB-specific probe ranged from 6.0 × 107 to 2.5 × 109 (average, 1.1 × 109 ± 5.2 × 108) cells g of sediment−1. In September, numbers of SRB detected ranged from 5.4 × 108 to 7.3 × 109 (average, 2.5 × 109 ± 1.5 × 109) cells g of sediment−1. The capability of using rDNA probes to estimate cell numbers by hybridization to DNA extracted from complex matrices permits initiation of detailed studies on community composition and changes in communities based on cell numbers in formerly intractable environments.
Although bacteria are the most abundant life forms on earth, knowledge of microbial community structures and population dynamics is still minimal. An estimated 80 to 90% of microorganisms in soil are as yet unidentified (2), and various researchers have detected enormous diversity in such habitats. In particular, Torsvik et al. (37) found evidence for as many as 104 different genomic equivalents in 1 g of forest soil, and in a study of Wisconsin agricultural soil, Borneman et al. (7) found that only 4% of ribosomal DNA (rDNA) clones sequenced were possible duplicates and that several clades of microorganisms had no close relative in the ribosomal database. This limited knowledge of microbial diversity results primarily from our inability to culture and identify the majority of indigenous bacteria. However, an ever-increasing suite of molecular techniques makes it possible to study microbial community structure and compare diversity across habitats (1).
Comparisons of diversity across microbial communities may lead to a knowledge base applicable to a variety of environmental issues. It is necessary to accurately measure changes in populations of microbial community members, especially major components of the community, in response to seasonal, natural, or anthropogenic changes and to identify keystone species (9). Changes in the diversity and structure of a microbial community could become manifested in the ecological processes it mediates. However, difficulties with quantitative investigations of microbial communities lie in the many types of bias which are introduced by culturing or enrichment steps (1, 42), nucleic acid extraction and purification steps (25), and PCR amplification of target genes (1, 17, 28, 36).
It is well recognized that probes targeting 16S rRNAs provide an assessment of microbial community composition. Past studies with such probes have used rRNAs as the hybridization target molecule. It is possible to enumerate cells by using rRNA probes with in situ microscopic techniques or flow cytometry. However, these methods are not currently useful with all types of samples, particularly soils and sediments. Additionally, targeted cells must have high rRNA contents in order to be observed. Studies that employ rRNA probes with nucleic acids extracted from a sample are considered quantitative with respect to the amounts of rRNA measured (31). Hybridizations to rRNA have been used previously to study microbial communities present in anaerobic sewage digesters (31), mixed cultures (29), freshwater sediments (26), biofilms (3), and rumen contents (34). However, since the amount of rRNA per cell may vary according to activity (13, 23), it is difficult to relate the amount of hybridized rRNA to cell numbers. Therefore, a method was sought to estimate cell numbers by hybridization of probes to extracted DNA, thereby providing a different measure of community structure.
Soils high in clay or organic matter, such as marsh sediments, pose particularly tough challenges to obtaining good yields of high-molecular-weight DNA. Compounds present in soils and sediments, particularly humic acids, interfere with molecular reagents. Principally, two approaches are used to recover DNA from environmental samples: (i) concentration of microbial cells from within the environmental sample followed by cell lysis and purification of nucleic acids (19, 20, 21, 33) and (ii) direct lysis of microbial cells within the environmental matrix followed by purification of nucleic acids (6, 8, 30, 38, 40). Separation of cells from soil and sediment samples prior to lysis can be difficult. Differential centrifugation can separate many cells from the surrounding matrix, but many bacteria grow in close association with soil or sediment particles and may be tightly bound to soil colloids (8, 39, 43). Recovery of cells from a sample by this method cannot be expected to be quantitative, representative, or reproducible. In spite of the potential for DNA to adhere to sediment particles, significantly higher yields of DNA are recovered by direct extraction methods than by methods involving cell recovery (6). For these reasons, an approach to estimating DNA from cells lysed within the sample was pursued in the present investigation.
The present study was undertaken to determine the feasibility of using probes directed towards rDNA as a quantitative approach to the estimation of cell numbers when hybridizations with membrane-bound nucleic acids are performed. The approach developed utilizes 16S rDNA probes hybridized to DNA extracted from environmental samples. This method should permit an estimate of “cellular abundance” in a sample, whereas hybridizations to rRNA estimate relative rRNA abundance. A quantitative and reproducible method for isolation and analysis of genomic DNA from marsh sediments high in humic acids was developed and evaluated for efficiency and reproducibility of extraction. This DNA was then used in hybridizations with rDNA-targeted probes to determine the cellular abundance of various groups of sulfate-reducing bacteria (SRB) present in marsh sediments.
MATERIALS AND METHODS
Strains used.
Desulfococcus multivorans (ATCC 33890), Desulfovibrio vulgaris (ATCC 33405), Desulfovibrio desulfuricans (ATCC 13541), and Desulfobulbus proprionicus (ATCC 33891) were generously provided by Martin Odom of the DuPont Co., Glasgow, Del. Escherichia coli DH5α was obtained from Stratagene. Desulfobacter postgatei (ATCC 33911), Thermus aquaticus (ATCC 31558), Bacillus subtilis (ATCC 27505), and Desulfobacterium autotrophicum (ATCC 43914) were obtained from the American Type Culture Collection.
Study site and sample collection.
Marsh sediment samples were taken on 24 June and 30 September 1996 from Canary Creek Marsh in Lewes, Del. This marsh is characterized by a variety of vegetation types and soil characteristics (16). The site is flooded at most high tides. Three 2-g samples, separated horizontally from each other by 2 cm, were taken from each marsh core (14 cm) with a sterile scalpel 3 cm from the top marsh surface and placed in sterile, preweighed 15-ml polypropylene tubes containing 4 ml of phosphate-buffered saline (PBS) (10 mM NaHPO4 [pH 7.4], 137 mM NaCl, 2 mM KH2PO4, 3 mM KCl). These tubes were kept on ice and processed in the laboratory within 3 h. Upon return to the laboratory, samples were adjusted to 2 g of sediment each as necessary by removing portions with a sterile scalpel.
Nucleic acid extraction.
Nucleic acid extraction was done by modified versions of the methods of Tsai and Olson (40) and Delgado and Wall (10). Sediment samples were held on ice in 4 ml of PBS in 15-ml tubes. Tubes were shaken at 150 rpm for 15 min at room temperature in a Controlled Environment Incubator Shaker (New Brunswick Scientific) and centrifuged for 10 min at 6,000 × g, and the supernatant was discarded. The process was repeated twice with the addition each time of 4 ml of fresh, cold PBS. Washed sediment was then ground to a fine powder under liquid nitrogen with a porcelain mortar and pestle to assist in the release of cells that were in close association with sediment particles. Each sample was resuspended in 4 ml of lysis solution (0.15 M NaCl, 0.1 M Na2-EDTA [pH 8.0]) containing 15 mg of lysozyme (Sigma) ml−1 and 15 μg of lysostaphin (Sigma) ml−1 dissolved in TES (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 10 mM EDTA) and incubated in a 37°C water bath for 2 h with agitation at 20-min intervals. After 1.5 h, 540 μl of 5 M NaCl and 540 μl of a 10% solution of hexadecylmethylammonium bromide (CTAB) in 0.7 M NaCl were added to each tube for the remaining 30 min of incubation. Four milliliters of 0.1 M NaCl–0.5 M Tris-HCl (pH 8.0)–10% sodium dodecyl sulfate (SDS) was then added to each tube, and the samples were incubated at 65°C for 15 min and placed in a −70°C freezer in a dry ice-ethanol bath until further processed. Samples were then cycled four times through freezing at −70°C and thawing at 65°C to lyse the cells and release DNA. After the final thaw, the aqueous phase was extracted twice with 3 ml of 1 M Tris-buffered phenol (pH 8.0) and then twice with 3 ml of chloroform-isoamyl alcohol (24:1 mixture). The phases were separated by centrifugation at 6,000 × g for 10 min. The pellet obtained from the first extraction, which consisted of sediment from the sample, was resuspended in 2 ml of 0.1 M NaCl–0.5 M Tris-HCl (pH 8.0)–10% SDS and extracted once more with 1 ml each of phenol and chloroform-isoamyl alcohol. These extractions were followed by two chloroform-isoamyl alcohol extractions to remove residual phenol and reduce the contaminating iron compounds and humic substances remaining in the sample. Samples were then precipitated overnight at −20°C following the addition of a 10% volume of 3 M NaAc, 5 μl of oyster glycogen (10 mg ml−1), and 2 volumes of 100% ethanol. DNA was pelleted the next morning by centrifugation at 10,000 × g for 15 min, rinsed with 70% ethanol, and resuspended in 130 μl of TE (10 mM Tris, 1 mM EDTA [pH 8.0]). RNA in crude extracts was removed by digestion with 20 μl of 10 mg of RNase A+T1 ml−1 at 37°C for 0.5 h. RNase was digested by overnight incubation with 40 μl of SDS and 10 μl of proteinase K at 55°C. The DNA preparations at this point were still brownish in color.
Purification of DNA samples.
DNA was further purified on MicroSpin Sephacryl S-300 columns (Pharmacia Biotech) according to the manufacturer’s instructions with an Eppendorf model 541C variable-speed centrifuge. The 200-μl DNA preparation was loaded onto two MicroSpin columns (100 μl each). Eluents from the two columns were combined, and DNA was stored at 4°C. Samples of purified DNA were analyzed by agarose gel electrophoresis to determine the amount of shearing associated with the purification process.
Oligonucleotide probes.
A suite of 16S rDNA oligonucleotide probes was used (Table 1). These probes were previously shown to encompass most members of the gram-negative, mesophilic SRB, and the specificities of these probes were determined previously (14, 15). Probes were labeled at their 5′ ends with [γ-32P]ATP (3,000 mCi/mmol) (New England Nuclear), as described before (15). Probes were purified on Nick columns (Pharmacia) containing DNA-grade Sephadex G-50, according to the manufacturer. Following purification, a 1-μl aliquot of the preparation was measured in a liquid scintillation counter.
TABLE 1.
16S rRNA oligonucleotide probes and target groups
| Probe | Target group | Sequence (5′→3′) | Target sitea | Wash temp (°C) | Reference |
|---|---|---|---|---|---|
| 687 | Desulfovibrio spp. | TACGGATTTCACTCCT | 687–702 | 45 | 15 |
| 660 | Desulfobulbus spp. | GAATTCCACTTTCCCCTCTG | 660–679 | 55 | 15 |
| 221 | Desulfobacterium spp. | TGCGCGGACTCATCTTCAAA | 221–240 | 55 | 15 |
| 129 | Desulfobacter spp., D. multivorans | CAGGCTTGAAGGCAGATT | 129–146 | 45 | 15 |
| 814 | Desulfosarcina variabilis, Desulfobotulus sapovorans | ACCTAGTGATCAACGTTT | 814–883 | 45 | 15 |
| 338 | General eubacteria | GCTGCCTCCCGTAGGAGT | 338–355 | 42 | 5 |
| 385 | General SRB | CGGCGTCGCTGCGTCAGG | 385–402 | 45 | 5 |
Numbering corresponds to the complementary positions in E. coli 16S rRNA.
DNA dot blots.
Dot blots were prepared with a 96-well dot blot apparatus (Bio-Rad) by use of an Immobilon-N membrane (Millipore). The membrane was prewetted in 95% ethanol for 3 s and rinsed for 2 min in distilled H2O. The wetted membrane was then placed in 100 ml of 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer and allowed to equilibrate for 15 min. The volume of sample loaded onto each well was brought up to 100 μl by addition of 40 μl of 1 M NaOH, 5 μl of 200 mM EDTA (pH 8.2), and double-distilled ddH2O. For probing with the general eubacterial and general sulfate reducer probes, the signal was expected to be greater and therefore less DNA per sample was loaded onto those membranes. Each DNA sample was boiled in a microcentrifuge tube for 10 min to denature the DNA. Samples were loaded quickly onto the dot blotter and vacuum blotted onto the membrane. Each well was rinsed with 50 μl of 0.4 M NaOH, and the membrane was removed from the blotting apparatus and rinsed for 5 min in 6× SSC, air dried, and then baked for 1 h at 80°C to immobilize the DNA on the membrane.
Hybridization.
Membranes were placed in plastic bags and heat sealed approximately 1 cm from the edge of the membrane on all four sides. The prehybridization solution contained 6× SSC, 0.5% SDS, 5× Denhardt’s solution (32), and 50 μg of polyadenylic acid ml−1. The hybridization solution was identical to the prehybridization solution and in addition contained 0.01 M EDTA and labeled hybridization probe. Both solutions were vacuum filtered through a 0.2-μm Nalgene filter flask prior to use. Prehybridization, with buffer added to the bag at 100 μl cm of membrane−2, was carried out for 3 h at 42°C for all probes. The membrane was then gently transferred to a clean Ziploc bag, and hybridization solution was added at 150 μl cm of membrane−2. Probe was added to the hybridization bag at a concentration of 20 ng of probe ml of hybridization solution−1 and between 5 × 106 and 1 × 107 cpm ml−1 for all experiments. Lower levels produced signal only after prolonged (24 to 120 h) exposure to X-ray film, and higher levels than this produced unacceptable background. Hybridizations proceeded overnight at 42°C. Membranes were washed the next day in 150 ml of a room temperature solution of 6× SSC–1% SDS for 30 min inside a clean plastic container. The wash buffer was changed twice during the 30-min. Membranes were then placed in containers of 1× SSC–0.5% SDS which had been preequilibrated to the optimum final stringency wash temperature for each probe (Table 1). This final wash was performed for 1 h, with two buffer changes during this time. Membranes were blotted gently on a clean paper towel, placed between sheets of plastic wrap, and exposed to Kodak BioMax film with an intensifying screen at −70°C until a clear signal appeared on the X-ray film (between 1 and 16 h).
Image processing.
A video image of the autoradiograph was captured with a Gel Doc 1000 Densitometer (Bio-Rad). Signals were quantified by using Molecular Analyst software for Bio-Rad’s Image Analysis Systems, version 2.1, on a Macintosh computer, and images were exported to NIH Image and Adobe Photoshop.
The elliptical volume integration tool was chosen to identify the image areas to be integrated. The same-sized area was used to quantify all spots on the membrane. The quantifiable range of intensity was limited to the linear range of the X-ray film, up to approximately 2.0 optical density units. X-ray film exposures were therefore adjusted carefully to achieve a range of signal intensity that was not overexposed and was within the range of signals from the standards on each membrane. In some cases it was necessary to use two different exposure times in order to obtain the proper exposure for all samples on one membrane. In these cases, data from the two exposures were combined by using standards in the appropriate concentration range for each exposure. Local background from individual areas was subtracted from the signal. Samples were rerun on a separate membrane with a new set of standards in cases in which there were interfering background spots in close proximity to sample spots.
Generation of a standard curve to quantify hybridization signals.
Amounts of DNA detected in marsh sample extracts were determined by comparison to hybridization signals obtained with DNA standards included on each membrane. The range of standard concentrations used was based on trial runs in which the amount of signal from a sample relative to that from standards of known concentration was compared. E. coli was used as a standard for membranes probed with EUB-338 (bacteria), D. multivorans was used for SRB-814, D. proprionicus was used for SRB-660 (Desulfobulbus spp.), D. postgatei was used for SRB-129 (Desulfobacter spp.), D. autotrophicum was used for SRB-221 (Desulfobacterium spp.), and D. vulgaris was used for SRB-667 (Desulfovibrio spp.).
Conversion of data to cell numbers.
Amounts of DNA detected were converted to nanograms of DNA per gram of marsh sediment. An estimate of the average amount of DNA per cell for pure cultures belonging to the groups targeted by the suite of probes used in this study was determined for use as a conversion factor. Four replicate 20-ml cultures of organisms chosen to represent the bacterial types targeted by each probe were grown to mid-log phase. Cells were counted microscopically with a Petroff-Hauser counting chamber, and the results obtained from the four cultures were averaged. DNA preparations were made by the Delgado and Wall protocol (10). The resulting DNA was measured on a Hoeffer Spectrophotometer (260 nm), and the results for the four preparations were averaged. The average DNA yield per milliliter of culture was divided by the average cell count per milliliter to obtain an estimate of DNA per cell. Extraction efficiency was measured by performing the same extraction procedure with three 20-ml control tubes of TE containing 1 μg of E. coli genomic DNA. Estimates of DNA per cell for each representative bacterial type were then adjusted for extraction efficiency. Amounts of DNA obtained per gram of marsh sediment were divided by the adjusted amount of DNA per cell to estimate numbers of cells per gram of marsh sediment.
Reproducibility of DNA preparation and purification.
To determine whether equivalent hybridization signals would be produced on X-ray film by replicate environmental samples, three parallel DNA extractions were performed from each of two 2-g sediment samples. Each of the sediment homogenates in PBS was divided into three equal parts by weight. Aliquots (100 μl) of the resulting three DNA preparations were spotted on two different membranes. One membrane was hybridized to the Desulfococcus probe 814 (15) and the other to the general SRB probe 385 (3, 5). The image was imported into Molecular Analyst (Bio-Rad) and quantified by densitometry, as described above. Hybridization signals were compared to signals obtained from a set of DNA standards of known concentration.
To address the question of whether the extraction and purification steps resulted in loss of DNA, tritium-labeled plasmid DNA was added to four different sample tubes containing PBS and marsh sediment. The amount of tritium recovered at the end of the extraction was quantified. Tritium was used since its signal would not interfere with the signal from the 32P-labeled probes subsequently used on the same samples. Plasmid pBR322 was nick translated with [2,8-3H]ATP (specific activity, 30 Ci/mmol) according to the protocol in the Amersham nick translation kit N5500. The nick-translated plasmid (2.5 ng/μl) was purified on a Nick column (Pharmacia) according to the manufacturer’s protocol. Of the 400-μl volume of purified tritium-labeled plasmid, 87.5 μl was used to spike each of the four sediment samples. At the end of the DNA preparation and purification steps, a 20-μl aliquot of DNA was removed from each of the four tubes containing labeled plasmid and measured on a scintillation counter. This represented 10% of the final volume of the DNA preparation. Thus, 10% (8.75 μl) of the initial volume of labeled plasmid suspended in distilled water was used to determine the counts added prior to the extractions.
As the DNA preparations obtained from marsh sediments had varying amounts of slightly amber-colored discoloration due to humic substances remaining after purification steps, it was necessary to account for possible quenching of the scintillation counter readings used to measure recovery of the DNA. Therefore, 8.75 μl of labeled plasmid was added to 11.25 μl of distilled H2O and measured on the scintillation counter. The result was compared to counts obtained from addition of 8.75 μl of labeled plasmid DNA to very dirty (dark brown), unpurified marsh sediment DNA.
RESULTS
DNA extraction.
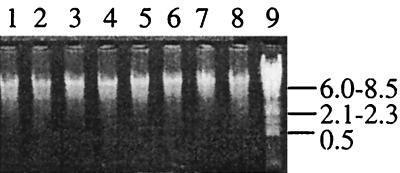
The combined supernatants from PBS washes failed to yield DNA that was detectable with a Hoeffer DNA fluorometer or on agarose gels after precipitation with ethanol and concentration (results not shown). Washing sediment samples with PBS therefore caused negligible losses of DNA and/or cells. The average size of the DNA obtained by the extraction procedure was ca. 8.6 kb, as was visualized after agarose gel electrophoresis (Fig. 1).
FIG. 1.
DNA preparations examined for shearing. Purified DNA from marsh sediment samples (lanes 1 to 8) is shown; 10 μl of each purified DNA preparation was loaded per lane. Lambda DNA digested with DraI was used as a marker (lane 9). Fragment sizes are given in kilobases.
Reproducibility was tested by the recovery of tritium-labeled plasmid DNA tracer added to each of four randomly chosen sediment samples. Tritium was efficiently measured by scintillation counting even in dirty, humic-contaminated DNA samples. Replicate counts of tritiated plasmid DNA in distilled water and in two different randomly chosen DNA preparations prior to spin column purification, equivalent to the amounts of tritiated plasmid added to each of four washed sediment samples, were 5.88 × 104 and 6.02 × 104 cpm and 6.38 × 104 and 5.94 × 104 cpm, respectively. Results from the four randomly chosen sediment samples at the end of purification steps were 2.79 × 104, 2.96 × 104, 3.0 × 104, and 2.9 × 104 cpm. The small deviation indicates that extraction efficiency did not vary between samples. From the average of the two original activity measurements and the average of the four recovered activities, there was 48% recovery of the DNA from the original samples. Final calculations of cell numbers detected were corrected to account for this recovery rate.
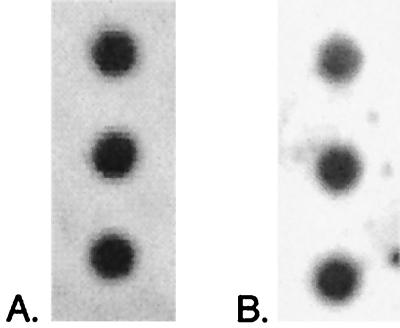
Reproducibility was further tested by dividing each of two sediment samples equally into three parts by weight, preparing parallel DNA preparations, and hybridizing each set of three preparations with one of two probes. These parallel preparations produced very consistent hybridization results (Fig. 2). In comparison to known amounts of DNA, one set of three replicate preparations probed with the SRB-385 (general SRB) probe produced detectable DNA concentrations of 1,130, 1,110, and 1,160 (average, 1,133 ± 25) ng of DNA g of sediment−1, and the other set of three replicate preparations probed with the SRB-814 (Desulfococcus) probe produced detectable DNA concentrations of 546, 605, and 628 (average, 593 ± 42) ng of DNA g of sediment−1 (see Table 3).
FIG. 2.
Replicate DNA preparations hybridized with the SRB-385 (general SRB) (A) and SRB-814 (Desulfococcus) (B) probes. From one 2-g sediment sample, three preparations were made and compared to a set of standards of known concentration (data not shown). Images were prepared with NIH Image and Adobe Photoshop 5.0.
TABLE 3.
Estimation of cell concentrations in salt marsh sediments
| Target group | DNA detected (ng) | % of 200 μl of purified DNA used for dot blot | Total DNA estimated for sample (fg g of sediment−1)a | Corrected DNA cell−1 for target group (fg)b | Estimated cells g of sediment−1 |
|---|---|---|---|---|---|
| General SRBc | 1,130 | 50 | 1.7 × 109 | 5.7 | 3.0 × 108 |
| 1,110 | 50 | 1.7 × 109 | 5.7 | 3.0 × 108 | |
| 1,160 | 50 | 1.8 × 109 | 5.7 | 3.2 × 108 | |
| Desulfococcus spp.c | 546 | 50 | 8.3 × 108 | 5.6 | 1.5 × 107 |
| 605 | 50 | 9.2 × 108 | 5.6 | 1.6 × 107 | |
| 628 | 50 | 9.5 × 108 | 5.6 | 1.7 × 107 | |
| General eubacteriad | 14,006 | 4 | 2.7 × 1011 | 5.3 | 5.2 × 1010 |
| General SRBd | 1,019 | 8 | 9.7 × 109 | 5.7 | 1.7 × 109 |
| Desulfovibrio spp.d | 150 | 16 | 7.1 × 108 | 5.9 | 1.2 × 108 |
| Desulfobacter spp.d | 168 | 16 | 8.1 × 108 | 3.1 | 2.6 × 108 |
| Desulfobacterium spp. d | 161 | 16 | 7.6 × 108 | 6.5 | 1.2 × 108 |
| Desulfococcus spp.d | 43 | 16 | 2.0 × 108 | 5.6 | 3.5 × 107 |
| Desulfobulbus spp.d | 236 | 16 | 1.1 × 109 | 7.3 | 1.5 × 108 |
Based on extraction efficiency of 48%.
Values taken from Table 2.
Detected from replicate DNA preparations from one 2-g sample collected in June.
Detected from one 2-g sample collected in June.
The extraction efficiency of pure cultures used as standards, based on recovery of added tritiated plasmid DNA, was found to average 70% (±4%). Estimates of DNA per cell for each representative bacterial type used to calculate cells per gram of sediment from the amount of DNA detected by each probe were corrected to account for this extraction efficiency prior to use as a conversion factor (see Table 2).
TABLE 2.
Estimation of cellular DNA contenta
| Probe DNA | Species | Avg cell count (cells ml−1) | Avg DNA yield (fg ml−1) | DNA cell−1 (fg)
|
|
|---|---|---|---|---|---|
| Actual | Adjustedb | ||||
| 338 | E. coli | 1.9 × 108 (±0.1 × 108) | 1.0 × 109 (±0.2 × 109) | 5.3 | 6.9 |
| 660 | D. proprionicus | 7.2 × 107 (±0.8 × 107) | 4.0 × 108 (±0.9 × 108) | 5.6 | 7.3 |
| 221 | D. autotrophicum | 2.0 × 107 (±0.2 × 107) | 1.0 × 108 (±0.4 × 108) | 5.0 | 6.5 |
| 687 | D. vulgaris | 2.0 × 108 (±0.4 × 108) | 9.0 × 108 (±0.5 × 108) | 4.5 | 5.9 |
| 814 | D. multivorans | 4.5 × 108 (±0.5 × 108) | 4.0 × 108 (±0.8 × 109) | 4.3 | 5.6 |
| 129 | D. postgatei | 6.2 × 107 (±0.3 × 107) | 2.0 × 108 (±0.5 × 108) | 2.4 | 3.1 |
Cell counts and DNA yields were determined with 20-ml log-phase cultures.
Based on extraction efficiency of 70%.
Specificity tests and optimization of hybridization conditions.
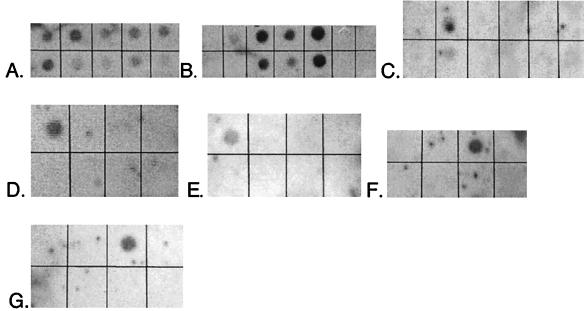
Specificity of the probes had previously been tested only against rRNA (15). It was therefore necessary to evaluate the probes in hybridizations against DNA. DNA purified from pure cultures of target and nontarget species was spotted on a membrane in a range of concentrations and hybridized to a single probe. All probes used in this study, when used at the optimum temperature (Table 1) previously determined with RNAs, yielded strong signals with target DNA and undetectable signals with nontarget DNA. The limit of detection for hybridization to nontarget DNA was <0.1% of the hybridization obtained with the same amount of target DNA. As shown in Fig. 3, the group-specific probes demonstrated the intended specificity when hybridized with DNA. A goal of the optimization study was to maximize the signal detected by X-ray film while preventing nonspecific binding of the probe to nontarget DNA and minimizing the background signal. Parameters experimentally manipulated were temperatures of hybridization and washes, salt concentrations in prehybridization, hybridization and wash solutions, blocking reagents and their concentrations in prehybridization and hybridization solutions, and time durations of washes. The final protocol derived is that described in Materials and Methods.
FIG. 3.
Probe specificity tests. Membranes were hybridized with the indicated probe (target group). DNAs (1 μg per spot except in bottom rows of panels B and C, where 500 ng per spot was used) are listed as they appear on membranes from top left to bottom right. (A) EUB-338 (general eubacteria). DNAs: E. coli, D. vulgaris, T. aquaticus, marsh sample, B. subtilis, D. postgatei, D. autotrophicum, D. multivorans, D. desulfuricans. (B) SRB-385 (general SRB). DNAs: E. coli, D. proprionicus, D. desulfuricans, D. postgatei, D. multivorans, B. subtilis, T. aquaticus. (Bottom row shows lower concentrations of the same nucleic acids.) (C) SRB-129 (Desulfobacter spp.). DNAs: D. autotrophicum, D. postgatei, D. vulgaris, D. multivorans, D. proprionicus, E. coli. (Bottom row shows lower concentrations of the same nucleic acids.) (D) SRB-687 (Desulfovibrio spp.). DNAs: D. vulgaris, D. multivorans, D. postgatei, E. coli, D. proprionicus, D. autotrophicum. (E) SRB-221 (Desulfobacterium spp.). DNAs: D. autotrophicum, D. desulfuricans, D. proprionicus, D. multivorans, D. postgatei, E. coli. (F) SRB-660 (Desulfobulbus spp.). DNAs: E. coli, D. multivorans, D. proprionicus, D. vulgaris, D. autotrophicum, D. postgatei, B. subtilis, D. desulfuricans. (G) SRB-814 (Desulfococcus spp.). DNAs: D. vulgaris, D. multivorans, D. postgatei, E. coli, D. proprionicus, D. autotrophicum. Images were prepared with NIH Image and Adobe Photoshop 5.0.
Background signal in Southern hybridizations was partially reduced by adding blocking reagents to the prehybridization buffer, i.e., Denhardt’s solution, SDS, heterologous DNA (salmon testes DNA [Sigma]), casein, and nonspecific RNAs (Sigma). Decreasing the SDS concentration to below 0.5% in prehybridization and hybridization solutions caused excessive background signal. Although increasing the concentration of SDS to higher than 0.5% had the effect of decreasing background, presumably by breaking nonspecific interactions, target signal was also reduced. Maximum Strength Nytran (Schleicher and Schuell) was tested with 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–1% SDS–10× Denhardt’s solution–50 μg of denatured heterologous DNA ml−1–20 μg of tRNA ml−1 in the prehybridization solution and with 6× SSPE–1% SDS in the hybridization solution. The level of nonspecific binding to the membrane was high, even at increased SDS concentrations. Maximum Strength Nytran was also tested with 5× SSC or 1× SSC–5% casein–1% SDS in the prehybridization and hybridization solutions. Signal from target DNA was increased by reducing the casein concentration to 2% in the hybridization solution, but background increased noticeably. For both Maximum Strength Nytran protocols, lower SDS concentrations in the prehybridization and hybridization solutions caused excessive background signal while use of more SDS reduced the target signal. 1× SSC buffer reduced background. Casein was not used, because background interference remained an intermittent problem. The Immobilon-N membrane gave consistently cleaner images with these DNA samples than did hybridizations performed with Nytran membranes.
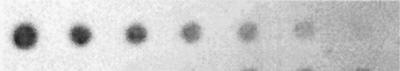
In general, the higher the temperature of the final stringency wash, the less background and nonspecific binding was observed. Variations of only 2 to 3°C made a significant difference not only in the specificity of probe binding to target DNA but also in reduction of background signal. The calculated melting temperature (Tm) was used as a starting point for optimization tests for final wash temperatures, and 2°C increments from 10°C above and below the Tm were used until an optimal signal-to-noise ratio and probe specificity could be determined. Table 1 shows the final wash temperatures for each of the probes used in this study. Experiments using DNA from pure cultures required less wash stringency to remove background signal. Less-stringent conditions allowed for the detection of lower DNA concentrations, indicating that with environmental soil and/or sediment samples, thresholds of detection are higher than for pure cultures due to contaminants remaining in the DNA preparations and necessitating more-stringent wash conditions. The threshold of detection for this suite of probes under the conditions described above was 5 ng of DNA. Presented are the results of a sensitivity test using the Desulfobacterium probe (SRB-221) (Fig. 4). Results for the other probes were essentially the same (data not shown). For sediments lower in humic contaminants, thresholds of detection are likely to be lower than in this study. After optimizing the hybridization conditions for each probe individually, it was possible to discriminate between target and nontarget bacterial groups on the resulting autoradiograph image.
FIG. 4.
Probe sensitivity test. D. autotrophicum DNA was probed with SRB-221. Amounts of genomic DNA spotted (left to right): 100, 80, 40, 20, 10, 5, and 2.5 ng. Image was prepared with NIH Image and Adobe Photoshop 5.0.
Optimization of the signal-to-noise ratio was more difficult for some of the probes than for others. Interference from background noise was controlled to some extent by shortening the time of exposure to X-ray film, reducing the concentration of probe added to the hybridization reaction mixture to no more than 25 ng ml−1, and increasing the length and temperature of the final stringency wash. The Desulfobulbus and Desulfovibrio probes (SRB-660 and SRB-687, respectively) were the most difficult to optimize. These two probes generated above-average nonspecific binding of probe to the membrane throughout trials with all tested variables.
DNA contents.
Studies with pure cultures were undertaken to estimate their cellular DNA contents in order to extrapolate amounts of DNA detected in marsh sediments to cell numbers. Results of cellular DNA content determinations are shown in Table 2. The estimated genomic DNA content for E. coli was 6.9 fg. This is 38% higher than the previously reported genome size value of 5.0 fg (17). Estimates of the DNA contents for the SRB examined ranged from 3.1 fg cell−1 (for D. postgatei) to 7.3 fg cell−1 (for D. proprionicus). Devereux et al. (12) estimated the genome sizes of several SRB species by pulsed-field gel electrophoresis. The genome sizes of D. vulgaris and D. proprionicus that they obtained were 3.6 and 3.7 Mb, respectively. Assuming one chromosome per cell, these sizes correspond to 3.9 to 4.1 fg cell−1. The estimates of DNA content per cell determined in this study for D. vulgaris and D. proprionicus were 60 and 94% greater, respectively. The differences in values determined in this study are acceptable considering that actively replicating cells contain greater than one genome equivalent of DNA. Estimation of cell numbers in environmental samples may therefore tend to be conservative.
Estimation of cell numbers.
Aliquots of each genomic DNA preparation from marsh sediment samples and DNA from pure cultures, used to generate the standard curve, were spotted on the same membrane when hybridized. The range of DNA standard concentrations (5 to 4,000 ng) and the volume of DNA for all marsh sediment samples on a particular membrane (10 to 30 μl) were selected based upon the expected amount of DNA targeted by the probe used. Reproducibility of the standards was monitored in some cases by including replicate spots of the series. The r2 values for regression lines based on signals from the standards were >0.95. The amount of sample DNA loaded onto each membrane was also varied from membrane to membrane to achieve the best signal-to-noise ratio.
The concentration of cells in a sediment sample for each sample-probe combination can be determined by dividing the amount of genomic DNA detected with a probe by the amount of DNA per target cell (Table 3). The calculations include correction factors to account for the efficiencies of DNA recovery from both sediment samples and pure cultures. By determining the genomic DNA contents of the strains used to generate the standard curve on the hybridization membrane, the amount of rDNA probe hybridizing to the DNA extracted from sediment can be related to genome equivalents and hence to cell concentrations. This method may underestimate cell numbers in a sample, since the DNA contents of cells used as standards were determined with rapidly growing cultures which might contain greater than one genome equivalent per cell.
For the general SRB probe, conversion was obtained by averaging the calculated amounts of DNA per cell for the SRB genera studied. This value was 5.7 fg cell−1. In June, numbers of SRB detected with the SRB-specific probe ranged from 6.0 × 107 to 2.5 × 109 (average, 1.1 × 109 ± 5.2 × 108) cells g of sediment−1. In September, numbers of SRB detected ranged from 5.4 × 108 to 7.3 × 109 (average, 2.5 × 109 ± 1.5 × 109) cells g of sediment−1. Previous studies in the same marsh detected ca. 107 SRB g of sediment−1 in samples collected from November 1977 to August 1978 (16) and ca. 107 lithotrophs (sulfur and ammonia oxidizers) in samples collected from January to May 1978 (24) by most-probable-number analysis. The higher numbers detected in this study may be attributed to the ability of direct molecular techniques to detect populations that evade cultivation in the laboratory.
DISCUSSION
Analysis of microbial community structure is fraught with experimental bias. Although any method is inherently biased, understanding and minimizing the biases will afford the most accurate analysis of sediment microbial communities possible. As with other methods involving hybridization of probes to nucleic acids from environmental samples, probes have been designed based on sequences from cultured organisms, and they will therefore be incapable of detecting an unknown percentage of the natural population. We describe here a quantitative method of microbial community structure analysis which avoids PCR, culturing, and enrichment steps; minimizes some of the sources of bias inherent in soil nucleic acid extractions; and provides reproducible results with sediment samples at a threshold of detection of ca. 8 × 105 cells g of sediment−1.
There are advantages and disadvantages to both rRNA- and rDNA-targeted hybridizations. DNA is more resistant to nuclease attack (enzymatic or divalent metals) and can withstand harsher purification steps. The greatest source of bias in DNA extraction from soil and sediment comes from adhesion and binding of free DNA from cells that have lysed to clay particles. During the DNA preparation protocol, it is difficult to separate this free DNA from the sample and may lead to a lower recovery rate. A portion of the 52% loss of DNA during the purification protocol described above can be attributed to binding of plasmid DNA to clay particles present in the sediment samples. This free DNA can also represent the remnants of dead cells present in the environmental sample. Such DNA is stable and persistent, making it difficult to quantify the contribution to total detected hybridization signal from dead cells. The use of a tritium-labeled plasmid as a standard for DNA recovery from a sediment sample could be improved upon, perhaps with the use of tritiated cells.
RNA is considered a more attractive target than DNA because it is of lesser sequence complexity and is naturally amplified. However, the probability of the random occurrence of a nonspecific target sequence in DNA becomes very small with oligonucleotide probes of the size used in this study. Also, because oligonucleotides are short, mismatches in hybridizations are highly destabilizing so that oligonucleotides have a high degree of specificity (35). However, RNA-based techniques are very useful for evaluating the metabolic status of single cells in environmental samples (27) or of populations, since rRNA content generally increases with growth rate (1). Since the rRNA content of cells is high, rRNA is an excellent target for in situ studies. However, at present in situ observations are generally possible only in relatively clean matrices or with natural samples having highly enriched microbial populations. The high rRNA copy number also enhances the sensitivity of detection with membrane-bound nucleic acids, and because of the smaller size of rRNA, more rigorous nucleic acid extraction techniques can be used, possibly retrieving a larger number of targets (1). However, different bacterial types do not always contain the same amounts of rRNA, and even within one targeted group, more metabolically active cells will have proportionally more RNA, contributing to a stronger signal (11). Additionally, the ribosome content of different species will vary between 103 and 105 ribosomes per cell (1). For these reasons, it is difficult to estimate cell numbers from hybridizations to rRNA extracted from environmental samples.
Studies with growing cultures have shown E. coli to have an average of 2.1 genome copies cell−1. Similarly, D. vulgaris may contain 4 copies cell−1, and Wall (41) has shown that Desulfovibrio gigas can have between 4 and 17 genome copies cell−1 depending on growth conditions. Bacteria in sediments are not likely to achieve the growth rates attained by those growing in the laboratory. Their genome copy number would be expected to remain low. In a regression analysis of hybridization signals, the amount of DNA detected in an environmental sample should therefore closely correlate with genome equivalents and hence cell numbers used in the array of hybridization standards. Variations in rRNA gene copy number should not introduce significant error, particularly with probes that target a phylogenetically closely related group of organisms. rRNA gene copy numbers should be essentially congruent between detected strains and strains used to generate standards. However, error should be expected when probes that target broader phylogenetic groups are used. Such errors might be greater than severalfold. However, the inferred cell numbers should still be of environmental significance. As previously described, rRNA probes may also underestimate cell numbers by missing an unknown percentage of the target population or by overestimating cell numbers if hybridization to nontarget groups occurs. Evaluation of these errors could be accomplished with the use of nested probes, a suite of probes with varying phylogenetic breadths (1, 13).
The capability of using rDNA probes to estimate actual cell numbers by hybridization to DNA extracted from complex matrices permits initiation of detailed studies on community composition and changes in the community based on cell numbers in formerly intractable environments. In fact, it should not go unnoticed that hybridization of rDNA probes to both DNA and rRNA obtained from a single sample will provide not only quantitative information on community structure but also information about which populations are the most active.
ACKNOWLEDGMENTS
We thank those involved in collection of marsh samples—Dan Wood, Alison Hunt, Mark Keese, and Erin Mayo—and M. Khalequzzaman, who performed all marsh coring.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Identification of uncultured bacteria: a challenging task for molecular taxonomists. ASM News. 1994;60:360–365. [Google Scholar]
- 3.Amann R I, Stromley J, Devereux R, Key R, Stahl D A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Binder B, Chisholm S W, Olsen R, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atlas R M, Sayler G, Burlage R S, Bej A K. Molecular approaches for environmental monitoring of microorganisms. BioTechniques. 1992;12:706–717. [PubMed] [Google Scholar]
- 7.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce K D, Hiorns W D, Hobman J L, Osborn A M, Strike P, Ritchie D A. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3413–3416. doi: 10.1128/aem.58.10.3413-3416.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daily G. Nature’s services—societal dependence on natural ecosystems. Washington, D.C: Island Press; 1997. pp. 37–38. [Google Scholar]
- 10.Delgado, S., and J. Wall (Department of Biochemistry, University of Missouri—Columbia). Isolation of genomic DNA from Desulfovibrio desulfuricans. Personal communication.
- 11.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 12.Devereux R, Willis S G, Hines M E. Genome sizes of Desulfovibrio desulfuricans, Desulfovibrio vulgaris, and Desulfobulbus proprionicus estimated by pulsed-field gel electrophoresis of linearized chromosomal DNA. Curr Microbiol. 1997;34:337–339. doi: 10.1007/s002849900192. [DOI] [PubMed] [Google Scholar]
- 13.Devereux R, Hines M E, Stahl D A. S cycling: characterization of natural communities of sulfate-reducing bacteria by 16S rRNA sequence comparisons. Microb Ecol. 1996;32:283–292. doi: 10.1007/BF00183063. [DOI] [PubMed] [Google Scholar]
- 14.Devereux R, Stahl D A. Phylogeny of sulfate-reducing bacteria and a perspective for analyzing their natural communities. In: Odom J M, Singleton R, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag, Inc.; 1993. pp. 131–159. [Google Scholar]
- 15.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 16.Dicker H J, Smith D W. Enumeration and relative importance of acetylene-reducing (nitrogen-fixing) bacteria in a Delaware salt marsh. Appl Environ Microbiol. 1980;39:1019–1025. doi: 10.1128/aem.39.5.1019-1025.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn D, Amann R I, Ludwig W, Akkermans A D L, Schleifer K-H. Detection of micro-organisms in soil after in situ hybridization with rRNA-targeted, fluorescently labeled oligonucleotides. J Gen Microbiol. 1992;138:879–887. doi: 10.1099/00221287-138-5-879. [DOI] [PubMed] [Google Scholar]
- 19.Holben W E, Harris D. DNA-based monitoring of total bacterial community structure in environmental samples. Mol Ecol. 1995;4:627–631. doi: 10.1111/j.1365-294x.1995.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 20.Holben W E, Janson J K, Chelm B K, Tiedje J M. DNA probe methods for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen C S, Rasmussen O F. Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin. Appl Environ Microbiol. 1992;58:2458–2462. doi: 10.1128/aem.58.8.2458-2462.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp P F, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ketcham R B. Seasonal dynamics of lithotrophic and heterotrophic bacteria in salt marsh soils. M.S. thesis. Newark: University of Delaware; 1981. [Google Scholar]
- 25.Lovell C R, Piceno Y. Purification of DNA from estuarine sediments. J Microbiol Methods. 1994;10:161–174. [Google Scholar]
- 26.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nedelman J, Heagerty P, Lawrence C. Quantitative PCR: procedures and precisions. Bull Math Biol. 1992;54:477–502. [Google Scholar]
- 29.Odenyo A A, Mackie R I, Stahl D A, White B A. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: development of probes for Ruminococcus species and evidence for bacteriocin production. Appl Environ Microbiol. 1994;60:3688–3696. doi: 10.1128/aem.60.10.3688-3696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogram A, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 31.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Somerville C C, Knight I T, Straube W L, Colwell R R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989;55:548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szostak J W, Stiles J I, Tye B-K, Chiu P, Sherman F, Wu R. Hybridization with synthetic oligonucleotide probes. Methods Enzymol. 1979;68:419–427. doi: 10.1016/0076-6879(79)68031-x. [DOI] [PubMed] [Google Scholar]
- 36.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torsvik V, Salte K, Sorheim R, Goksoyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai Y-L, Olson B. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai Y-L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wall J D. Characterization of a small plasmid from Desulfovibrio desulfuricans and its use for shuttle vector construction. J Bacteriol. 1993;175:4121–4128. doi: 10.1128/jb.175.13.4121-4128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 43.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]