Abstract
Poly(3-hydroxyalkanoates) (PHAs) are biodegradable thermoplastics which are accumulated by many bacterial species in the form of intracellular granules and which are thought to serve as reserves of carbon and energy. Pseudomonas putida accumulates a polyester, composed of medium-side-chain 3-hydroxyalkanoic acids, which has excellent film-forming properties. Industrial processing of PHA involves purification of the PHA granules from high-cell-density cultures. After the fermentation process, cells are lysed by homogenization and PHA granules are purified by chemical treatment and repeated washings to yield a PHA latex. Unfortunately, the liberation of chromosomal DNA during lysis causes a dramatic increase in viscosity, which is problematic in the subsequent purification steps. Reduction of the viscosity is generally achieved by the supplementation of commercially available nuclease preparations or by heat treatment; however, both procedures add substantial costs to the process. As a solution to this problem, a nuclease-encoding gene from Staphylococcus aureus was integrated into the genomes of several PHA producers. Staphylococcal nuclease is readily expressed in PHA-producing Pseudomonas strains and is directed to the periplasm, and occasionally to the culture medium, without affecting PHA production or strain stability. During downstream processing, the viscosity of the lysate from a nuclease-integrated Pseudomonas strain was reduced to a level similar to that observed for the wild-type strain after treatment with commercial nuclease. The nuclease gene was also functionally integrated into the chromosomes of other PHA producers, including Ralstonia eutropha.
Polyhydroxyalkanoates (PHAs) are biodegradable and biocompatible polyesters that accumulate as intracellular inclusion bodies in a variety of bacteria (24, 27). Because these polymers are produced from renewable resources such as fatty acids and sugars, they provide a new resource for plastic materials and small molecules derived from their monomers (45). The properties of these polyesters range from stiff and brittle materials, such as poly(3-hydroxybutyrate) (PHB), to elastomers, such as poly(3-hydroxyoctanoate). The monomeric composition of a PHA depends on the bacterial strain, the culture conditions, and the carbon source used for growth, but generally, bacteria synthesizing PHAs can be subdivided into two groups. One group, including Ralstonia eutropha, produces short-side-chain PHAs with C3 to C5 monomers, while the second group, including Pseudomonas putida, synthesizes medium-side-chain PHAs with C6 to C16 monomers (27).
PHAs can be recovered and purified from biomass by a number of different techniques. One technique involves extraction of the polymer from lyophilized cells with organic solvents (5). Most other techniques involve mechanical or chemical cell disruption followed by chemical or enzymatic treatment. During such processes, the PHA granules are released from the cells and are subsequently purified by repeated centrifugation and/or filtration steps. Cell disruption, however, also results in liberation of chromosomal DNA, which causes a rapid increase in the viscosity of the cell lysate, thereby impeding subsequent filtration and centrifugation (26, 39, 41). Since the efficiencies of filtration and centrifugation are inversely proportional to the viscosity and a high viscosity directly affects pumping, mixing, and heat transfer, quick removal of the chromosomal DNA is critical (3). Previously described methods for DNA degradation in PHA processes included the use of hypochlorite (4), heat treatment (8, 9), or enzyme cocktails (18). Even though these three methods may seem applicable in small-scale fermentation systems, they have some drawbacks for the envisioned 100 million-lb production scale for PHAs. Besides the limitations at the larger scale, these procedures have additional disadvantages, since hypochlorite causes limited hydrolysis of the PHA while heat treatment and the use of enzyme cocktails are costly.
To provide a commercially attractive solution to the viscosity problem, we integrated the staphylococcal nuclease gene into the chromosomes of different PHA producers. Staphylococcal nuclease has been shown to hydrolyze DNA and RNA to fragments of less than 100 nucleotides (2, 6). The described recombinant strains were stable in high-cell-density fermentations and during recovery of PHA granules. The viscosity of the lysates from such strains was reduced compared to the viscosity of a lysate from the wild-type strain.
MATERIALS AND METHODS
Bacterial strains and growth media.
The strains used in this study are listed in Table 1. Escherichia coli and P. putida strains were routinely grown in Luria-Bertani medium or R medium (34). R. eutropha was grown in either Trypticase soy broth (Becton Dickinson, Cockeysville, Md.) or PCT medium (31) supplemented with 1% glucose. Media were supplemented with chloramphenicol (32 μg/ml), nalidixic acid (30 μg/ml), or kanamycin (50 μg/ml) as required. Benzonase was obtained from American International Chemical (Natick, Mass.).
TABLE 1.
Strains and plasmids used in this study
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| P. putida strains | ||
| KT2442 | hsdR; Rifr | 20 |
| MBX918 | KT2442 nuc-kan | This study |
| MBX919 | KT2442 nuc-kan | This study |
| MBX920 | KT2442 nuc-kan | This study |
| MBX921 | KT2442 nuc-kan | This study |
| MBX922 | KT2442 nuc-kan | This study |
| MBX923 | KT2442 nuc-kan | This study |
| MBX924 | KT2442 rrn::nuc-kan | This study |
| MBX925 | KT2442 nuc-kan | This study |
| MBX926 | KT2442 nuc-kan | This study |
| Pseudomonas sp. strains | ||
| MBX978 | MBX strain | |
| MBX979 | MBX978 nuc-kan (high-level nuclease activity) | This study |
| MBX980 | MBX978 nuc-kan | This study |
| MBX981 | MBX978 nuc-kan (low-level nuclease activity) | This study |
| MBX982 | MBX978 nuc-kan (moderate nuclease activity) | This study |
| MBX983 | MBX978 nuc-kan | This study |
| MBX984 | This study | |
| MBX985 | MBX978 nuc-kan (moderate nuclease activity) | This study |
| MBX986 | MBX978 nuc-kan | This study |
| MBX987 | MBX978 nuc-kan | This study |
| R. eutropha strains | ||
| NCIMB40124HD | NCIMBa | |
| MBX917 | NCIMB40124HD nuc-kan | This study |
| E. coli strains | ||
| S17-1 | recA thi pro hsdR−M+ RP4-2-Tc::Mu λpir; Smr | 16 |
| DH5αF− | recA1 hsdR17 endA1 thi-1 gyrA96 relA1 | NEBb |
| MBX245 | LJ14 rpoS::Tn10; Nalr | MBX strain |
| MBX988 | MBX245 nuc-kan | This study |
National Center for Industrial Microorganisms and Bacteria.
New England Biolabs, Beverly, Mass.
Primers and DNA amplification.
The nuc gene, encoding the nuclease from Staphylococcus aureus, was obtained by PCR, using plasmid pNuc1 (25) as a template. Reactions mixtures (50 μl) contained 10 pmol each of primers nucA (5′ - T TC TC TAGAAT TCAGGAGGT T T T TATGGC TATCAGTAATGT T TCG) and nucB (5′-GCCGGTACCTTATTGACCTGAATCAGCGTTG) and the template in PCRmix (Gibco BRL, Gaithersburg, Md.), and reactions were performed in a thermocycler (Ericomp, San Diego, Calif.), using a program comprising 30 cycles of incubation at 95°C (30 s), at 55°C (45 s), and at 72°C (45 s). PCR products were gel purified and cloned into the pCR2.1 cloning vector (Invitrogen, Carlsbad, Calif.). The insert of the resulting plasmid, pCR2.1-nuc, was confirmed by DNA sequencing to be identical to the reported sequence of the nuc gene from S. aureus (37) (GenBank accession no. J01785).
Plasmid construction.
pCR2.1-nuc was digested with EcoRI and Acc65I according to the manufacturer’s (New England Biolabs, Beverly, Mass.) recommendations, and the nuc gene fragment was purified and cloned into the corresponding sites of pUC18Not (16). A promoterless, blunt-ended kanamycin gene from Tn903 (obtained by PCR from pBGS18, using primers linkK1 [5′-TGCATGCGATATCAATTGTCCAGCCAGAAAGTGAGG] and linkK2 [5′-ATTTATTCAACAAAGCCGCC]), was inserted into the SmaI site to generate pMNX-nuc-kan. The NotI fragment containing the promoterless nuc-kan operon was then cloned into the NotI sites of the integration vector pUTkan (16), a process which deleted the original kanamycin resistance marker, generating pMUX-nuc-kan.
Transposon mutagenesis and selection of integrants.
Plasmid pMUX-nuc-kan was transformed into E. coli S17-1 λpir and then conjugated into PHA-producing strains such as P. putida, Pseudomonas sp. strain MBX978, and R. eutropha as described elsewhere (16). P. putida and Pseudomonas sp. strain MBX978 integrants were selected on plates of minimal E2 medium (22) containing 10 mM octanoate as the carbon source and kanamycin as the selective agent. Integrants were replica plated onto DNase agar plates (Difco Laboratories, Detroit, Mich.) supplemented with kanamycin, and clones that expressed nuclease were identified by the presence of zones of clearing around the colonies (37). For R. eutropha, integrants were selected on PCT medium supplemented with 1% glucose, nalidixic acid, and kanamycin. Integrants were subsequently identified by replica plating onto DNase agar plates supplemented with 10 g of Trypticase soy broth per liter and kanamycin. Integrants of E. coli MBX245 were selected on minimal E2 plates supplemented with 10 mM octanoate, 0.5% corn steep liquor, kanamycin, and nalidixic acid. After replica plating of resulting colonies onto DNase agar plates and subsequent incubation, colonies exhibiting zones of clearing were selected.
DNA sequencing.
Transposon insertion sites were determined from genomic fragments that contain the nuc-kan operon and adjacent chromosomal DNA. Chromosomal DNA was digested to completion with EcoRI and ligated into the corresponding site of pUC19 (36). After transformation of the ligation mixture into E. coli DH5α, kanamycin-resistant mutants were selected and the insertion site was determined, using the oligonucleotide kan-up3 (5′-CGCACTTGTGTATAAGAGTC) as a primer. This primer allows the determination of the nucleotide sequence directly downstream of the insertion locus. Automated DNA sequencing was performed at Boston University Medical Center (Boston, Mass.).
Detection of nuclease activity.
Nuclease expression was routinely examined by observing the appearance of zones of clearing around colonies grown on DNase agar plates (25). For convenient estimation of nuclease activity, agarose gel electrophoresis with high-molecular-weight DNA was used as follows. PHA-producing strains were grown in their corresponding minimal media, and at various times were removed 500-μl samples, to each of which was added 100 μl of chloroform to release periplasmic nuclease. After centrifugation, 16 μl of the aqueous supernatant was mixed with 4 μg of P. putida KT2442 genomic DNA, and the mixture was incubated at 37°C for 1 h after CaCl2 was added to 1 mM (6). DNA was subsequently separated by agarose gel electrophoresis (36), and nuclease activity was assessed by determining the decrease in the molecular weight of the genomic DNA.
PHA analysis.
PHA-containing cells (5 to 20 mg) were subjected to hydrolysis in dichloroethane-propanol-HCl (5:4:1) for 2 h at 100°C (35). Resulting propyl esters of hydroxyalkanoic acids were analyzed by gas chromatography as described previously (22).
Viscosity assay.
Pseudomonas sp. strains MBX978 and MBX985 were grown in 20-liter computer-controlled fermentors (Applicon, Schiedam, The Netherlands) on R medium with a dissolved oxygen-controlled (DO-stat) octanoate feed (a detailed report on the fermentation procedures will be reported elsewhere). At the end of the fermentation, cultures were supplemented with 1 mM CaCl2 and lysed by passage through a microfluidizer (model M110EH; Microfluidics International Corp., Newton, Mass.) operating at pressures ranging from 8,000 to 20,000 lb/in2. The homogenized cultures were incubated at room temperature for 1 h, and the viscosities of the lysates were determined at room temperature with an LVF viscometer (Brookfield, Stoughton, Mass.). In control experiments, a commercial preparation of nuclease from Serratia marcescens (Benzonase; American International Chemical Inc.) was added.
SDS-PAGE.
Cell extracts, obtained by sonicating cells from 50-ml cultures and resuspending them in 2 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 10 mM 2-mercaptoethanol, 5% glycerol), were subjected to sodium dodecyl sulfate (SDS)–12.5% polyacrylamide electrophoresis (PAGE) (Bio-Rad, Richmond, Calif.) as described elsewhere (36).
RESULTS
Nuclease integrants of P. putida KT2442.
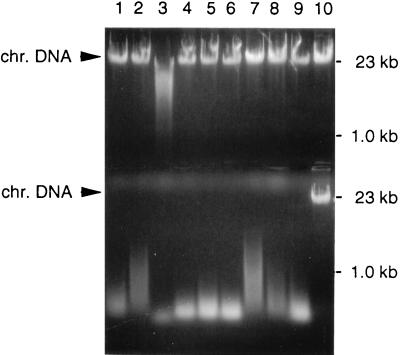
P. putida KT2442 strains that express staphylococcal nuclease were constructed by random integration of a nuc-kan cassette. Of 12,000 kanamycin-resistant integrants, 1,500 colonies were replica plated onto DNase agar plates to screen for the clones with the highest levels of nuclease expression. The presence of nuclease activity in both the extracellular medium and the periplasm of nine different isolates was subsequently determined (Fig. 1). Nuclease is primarily secreted into the periplasm, suggesting that the P. putida protein secretion apparatus recognizes the signal peptide of staphylococcal nuclease. Because the nuc gene in the transposon is not preceded by a promoter element, its expression is driven from promoter sequences located on the chromosome. This is expected to result in various levels of nuclease for different nuclease integrants, as was demonstrated by the low level of activity in MBX920 (lane 7), the high level of activity in MBX924 (lane 3), and intermediate levels of activity for the other strains. For P. putida MBX924, nuclease activity was detected in both the growth medium and the periplasm. The chromosomal DNA fragment containing the nuc-kan operon from P. putida MBX924 was cloned as a 4-kb EcoRI fragment into pUC19. The nucleotide sequence of the DNA downstream of the nuc-kan operon showed that the insertion was in a gene encoding a homolog of the 23S rRNA from Pseudomonas aeruginosa (96.4% identity over 362 nucleotides), suggesting that the corresponding promoter for this rRNA operon directs the transcription of nuc in MBX924. Unfortunately, a definite assignment of the nuc insertion site to this rRNA locus cannot be made because the complete sequence of the corresponding P. putida gene is unknown at this time.
FIG. 1.
Nuclease activity in nine P. putida nuclease integrants. P. putida KT2442 and derivatives with an integrated nuclease gene were grown in E2–10 mM octanoate. Growth medium (top) and chloroform-permeabilized cell fractions representing the periplasm (bottom) were incubated with 4 μg of P. putida KT2442 genomic DNA at 37°C for 1 h. Lanes: 1, MBX926; 2, MBX925; 3, MBX924; 4, MBX923; 5, MBX922; 6, MBX921; 7, MBX920; 8, MBX919; 9, MBX918; 10, P. putida KT2442. chr. DNA, chromosomal DNA.
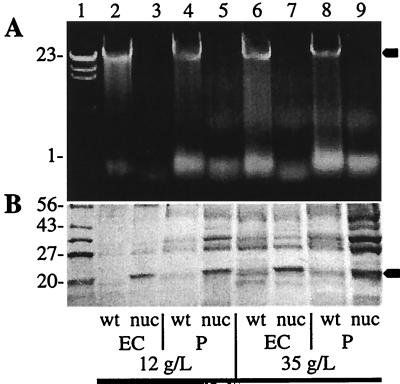
P. putida MBX924 exhibited the highest level of nuclease activity among the P. putida integrants. This strain was therefore grown in a 20-liter fermentor to a high cell density to examine its growth behavior, stability, and nuclease secretion under fermentation conditions. Figure 2A shows the nuclease activity at two different cell densities, as observed by agarose gel electrophoresis, and again nuclease activity was present in both the periplasmic and extracellular fractions throughout the fermentation. Furthermore, by SDS-PAGE, a protein band corresponding to a molecular mass of 20 kDa was observed for both the periplasm and the growth medium of MBX924 at the different growth stages (Fig. 2B). The size of this protein corresponds well with the reported molecular mass of staphylococcal nuclease (7). In addition, this protein is not present in either the periplasm or the growth medium of the wild-type culture.
FIG. 2.
Analysis of nuclease expression by P. putida KT2442 and P. putida MBX924 grown in a 20-liter fed-batch fermentation. Wild-type (wt) and nuclease-integrated (nuc) Pseudomonas strains were grown as described in the text, and at cell densities of 12 and 35 g/liter, samples were analyzed for nuclease activity (A) and protein (B) in the extracellular growth medium (EC) and periplasm (P). The arrows indicate chromosomal DNA (A) and the putative nuclease (B). Lane 1 contains molecular mass markers (either HindIII-digested λ DNA [A] or a combination of glutamic dehydrogenase [56 kDa], maltose binding protein [43 kDa], triosephosphate isomerase [27 kDa], and trypsin inhibitor [20 kDa] [B]).
Construction of nuclease integrants of other PHA producers.
By the use of similar methods, nuc was integrated into the chromosome of Pseudomonas sp. strain MBX978. For Pseudomonas sp. strain MBX978, 50 mutants were selected on DNase agar plates, of which 9 were grown in minimal E2-octanoate medium and tested for relative nuclease activity levels by agarose gel electrophoresis. All nine clones secreted nuclease into the periplasm (results not shown).
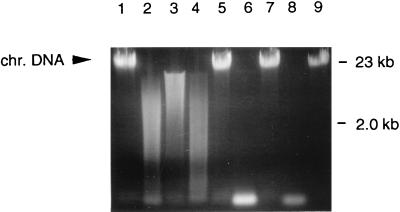
Nuclease integrants for R. eutropha NCIMB40124HD and E. coli MBX245 were subsequently generated. R. eutropha, an exceptional PHB producer, was the strain of choice for commercial PHB production by ICI, Ltd. (5). E. coli generally does not synthesize PHAs but is regarded as a suitable host for improved recombinant PHA production processes (27). For R. eutropha, 10 random colonies were grown and tested for nuclease activity; only one (MBX917) exhibited nuclease activity. For E. coli, 75 colonies were screened; 4 mutants exhibited nuclease activity (data not shown). The transgenic nuclease-producing strains derived from both E. coli and R. eutropha secreted nuclease into the periplasm but not into the growth medium. Figure 3 shows a summary of the nuclease activities for the different transgenic nuclease-secreting species and their corresponding parent strains. These data indicate that transgenic nuclease expression can be achieved by different PHA-producing strains and that it is a generally applicable procedure to prevent processing problems related to viscosity caused by chromosomal DNA.
FIG. 3.
Nuclease activity in wild-type and transgenic PHA producers with a chromosomally integrated nuclease gene. Chromosomal (chr.) DNA was treated with periplasmic fractions of E. coli MBX245 (lane 1), E. coli MBX988 (::nuc-kan) grown on R medium (lane 2) or Luria-Bertani medium (lane 3), R. eutropha MBX917 (::nuc-kan) (lane 4), R. eutropha NCIMB40124HD (lane 5), Pseudomonas sp. strain MBX985 (::nuc-kan) (lane 6), Pseudomonas sp. strain MBX978 (lane 7), P. putida MBX924 (::nuc-kan) (lane 8), or P. putida KT2442 (lane 9).
Evaluation of nuclease integrants of Pseudomonas sp. strain MBX978 for PHA production.
In order to successfully replace wild-type strains as PHA producers, the integrated strains should demonstrate the same stability and PHA productivity as the wild-type strain. All integrants derived from Pseudomonas sp. strain MBX978 were tested for PHA content, growth rate, and relative nuclease activity in E2–10 mM octanoate medium before being analyzed in a large-scale fermentor for PHA production and processing. Table 2 lists the characteristic growth rates and PHA contents of these strains. Most of these nuclease integrants exhibited the same growth rate as the parental strain. Except for Pseudomonas sp. strain MBX984, the PHA contents and the compositions of the accumulated PHAs were similar. However, similar to what was observed for the nuclease integrants derived from P. putida KT2442 (Fig. 1), the nuclease levels in these strains differed. Because Pseudomonas sp. strain MBX985 exhibits a relatively high level of nuclease activity and growth characteristics similar to those of the wild-type strain, it was further evaluated for its ability to improve the PHA extraction process in a 20-liter fermentation-downstream processing protocol.
TABLE 2.
PHA production by nuclease integrants of Pseudomonas sp. strain MBX978 in minimal E2 medium with 10 mM octanoate as the carbon source
| Strain | Nuclease activitya | A600b | tdc (h) | % PHAd
|
||
|---|---|---|---|---|---|---|
| C6 | C8 | Total | ||||
| MBX978 | ND | 3.7 | 1.5 | 4.2 | 29.2 | 33.4 |
| MBX979 | +++ | 3.6 | 1.6 | 3.8 | 26.0 | 29.8 |
| MBX981 | + | 3.4 | 1.4 | 4.0 | 27.9 | 31.9 |
| MBX982 | + | 3.5 | 1.6 | 4.0 | 27.1 | 31.1 |
| MBX984 | ++ | 2.8 | 1.6 | 1.5 | 12.2 | 13.8 |
| MBX985 | ++ | 3.8 | 1.4 | 4.1 | 28.3 | 32.4 |
Relative activity was estimated by chromosomal DNA digestion followed by agarose gel electrophoresis as described in Materials and Methods. The sizes of resulting DNA fragments are noted as follows: 200 to 300 bp, +++; 400 to 1,000 bp, ++; 500 to 4,000 bp, +; or no degradation, ND.
Optical density at 600 nm after 24 h of growth, at which point cells were harvested for PHA analysis.
Doubling time (± 0.1 h).
Accumulated levels of 3-hydroxyhexanoate (C6), 3-hydroxyoctanoate (C8), and total PHA as percentages of cell dry weight (total PHA is ±5%).
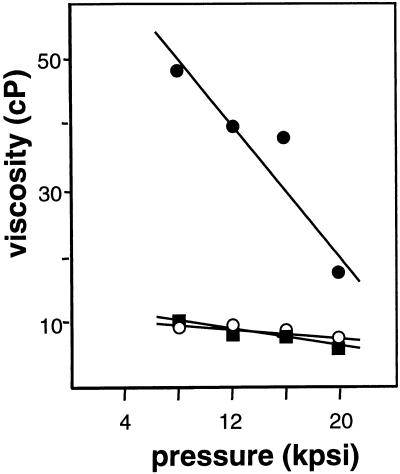
Pseudomonas sp. strain MBX978 and nuclease integrant Pseudomonas sp. strain MBX985 were grown in a 20-l fermentor operated in a fed-batch mode. Both fermentations reached a cell density of approximately 60 g/liter. The cells were homogenized, as described in Materials and Methods, to release the PHA. For the parent strain, homogenization took place in the presence or absence of Benzonase. The viscosities of the lysates as a function of the pressure used for homogenization are shown in Fig. 4. Already at the lowest homogenization pressure (8,000 lb/in2), the viscosity of the Pseudomonas sp. strain MBX985 lysate was reduced to a level similar to that of the parent culture to which Benzonase had been added. Subsequent steps in the purification process of the PHA granules include centrifugation and/or filtration steps that involve chemical treatments with oxidizing agents and detergents (9, 39) that decrease the residual nuclease activity. The final nitrogen content of the PHA latex was less than 0.4%, as determined by the Kjeldahl method. These results confirm that the integrated nuclease gene of Pseudomonas sp. strain MBX985 was functionally expressed in high-cell-density fermentations and that the use of this strain eliminates the need to add nuclease preparations to reduce the viscosity of the cell lysate.
FIG. 4.
Viscosities of cell lysates. Nuclease-expressing and wild-type Pseudomonas sp. strain MBX978 were grown in fed-batch mode to a density of approximately 60 g/liter. Cell suspensions were subsequently homogenized with or without the addition of Benzonase. The viscosity of the lysate was determined as a function of the operating pressure of the homogenizer. Closed circles, Pseudomonas sp. strain MBX978 without added Benzonase; open circles, Pseudomonas sp. strain MBX978 with added Benzonase; closed squares, nuclease integrant Pseudomonas sp. strain MBX985 without added Benzonase.
DISCUSSION
The nuclease gene from S. aureus was integrated into the genomes of several well-known PHA producers. Most pseudomonads from rRNA homology group I produce PHAs composed of medium-side-chain 3-hydroxy fatty acids when grown on fatty acids or on carbohydrates (15, 19, 20, 40). R. eutropha is a well-known producer of PHB and related short-side-chain PHAs (5); E. coli is a bacterium that is considered to be a potential recombinant PHA producer, and both short- and medium-side-chain PHAs are produced by this organism (23, 33, 42, 43). Here the effect of nuclease expression on ease of downstream processing was demonstrated for a Pseudomonas sp. strain MBX978 derivative, and nuclease-expressing strains were also derived from P. putida KT2442, R. eutropha NCIMB40124HD, and E. coli MBX245.
The production cost of a fermentation product depends to a large extent on the combined costs of fermentation and downstream processing (8). While molecular genetics has frequently been applied to improve the productivity (10, 11, 29) and fitness (17, 44) of a microorganism, its application to downstream processing is rather uncommon. In the aqueous processing of PHA-containing cells, it is necessary to lyse the cells, and this process is accompanied by a dramatic increase in sample viscosity, due to chromosomal DNA, that reduces the efficiency of the subsequent centrifugation, filtration, and washing steps (3). The PHA-producing strains that are described here express an endogenous nuclease whose use leads to significant cost savings. Such strains are useful in processes besides the production of PHAs, since many industrial fermentation processes deal with similar viscosity problems. The production of high-value pharmaceutical proteins frequently involves isolation of inclusion bodies, which should be essentially free of nucleic acids to allow efficient purification and formulation (14). Furthermore, we can expect a dramatic increase in the use of recombinant organisms for the production of chemicals, proteins, and polymers, and it is desirable to prevent proliferation of heterologous genes by inclusion of a procedure for DNA degradation.
The improvement of PHA-accumulating microorganisms for PHA production by integration of a nuclease-encoding gene is potentially of great economic value. Although future PHA production is envisioned to be a major agricultural process (30, 32, 45), the use of fermentation systems for the production of PHA latex as well as specialty PHAs will continue. Such specialty PHAs may contain monomers that are strictly derived from the feedstock and contain, for instance, aromatic, halogenated, or unsaturated functionalities (1, 12, 13, 21, 38), while the PHA latex can be used for making PHA films with potential applications for paper and food coating (28). The developments described here are therefore expected to contribute to the efficiency of future PHA production facilities.
ACKNOWLEDGMENTS
We thank Anthony Sinskey for plasmid pNuc1 and David Martin and Lara Madison for critical reading of the manuscript.
REFERENCES
- 1.Abe C, Taima Y, Nakamura Y, Doi Y. New bacterial copolyesters of 3-hydroxyalkanoates and 3-hydroxy-w-fluoroalkanoates produced by Pseudomonas oleovorans. Polym Commun. 1990;31:404–406. [Google Scholar]
- 2.Anfinsen C B, Cuatrecasas P, Taniuchi H. Staphylococcal nuclease: chemical properties and catalysis. In: Boyer P, editor. The enzymes. Vol. 4. New York, N.Y: Academic Press; 1971. pp. 177–204. [Google Scholar]
- 3.Atkinson B, Mavituna F. Biochemical engineering and biotechnology handbook. 2nd ed. New York, N.Y: Stockton Press; 1991. [Google Scholar]
- 4.Berger E, Ramsay B A, Ramsay J A, Chaverie C, Braunegg G. PHB recovery by hypochlorite digestion of non-PHB biomass. Biotechnol Tech. 1989;3:227–232. [Google Scholar]
- 5.Byrom D. Polymer synthesis by microorganisms: technology and economics. Trends Biotechnol. 1987;5:246–250. [Google Scholar]
- 6.Cuatrecasas P, Fuchs S, Anfisen C B. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1967;242:1541–1547. [PubMed] [Google Scholar]
- 7.Davis A, Moore I B, Parker D S, Taniuchi H. Nuclease B: a possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1977;252:6544–6553. [PubMed] [Google Scholar]
- 8.de Koning G J M, Kellerhals M, van Meurs C, Witholt B. A process for the recovery of poly(3-hydroxyalkanoates) from pseudomonads. Part 2. Process development and economic evaluation. Bioprocess Eng. 1997;17:15–21. [Google Scholar]
- 9.de Koning G J M, Witholt B. A process for the recovery of poly(3-hydroxyalkanoates) from pseudomonads. Part 1. Solubilization Bioprocess Eng. 1997;17:7–13. [Google Scholar]
- 10.Eggeling L, Morbach S, Sahm H. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J Biotechnol. 1997;56:167–182. [Google Scholar]
- 11.Flores N, Xiao J, Berry A, Bolivar F, Valle F. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat Biotechnol. 1996;14:620–623. doi: 10.1038/nbt0596-620. [DOI] [PubMed] [Google Scholar]
- 12.Fritzsche K, Lenz R W, Fuller R C. Production of unsaturated polyesters by Pseudomonas oleovorans. Int J Biol Macromol. 1990;12:85–91. doi: 10.1016/0141-8130(90)90058-i. [DOI] [PubMed] [Google Scholar]
- 13.Fritzsche K, Lenz R W, Fuller R C. An unusual bacterial polyester with a phenyl pendant group. Makromol Chem. 1990;191:1957–1965. [Google Scholar]
- 14.Gross M, Wyss M, Furter-Graves E M, Wallimann T, Furter R. Reconstitution of active octameric mitochondrial creatine kinase from two genetically engineered fragments. Protein Sci. 1996;5:320–330. doi: 10.1002/pro.5560050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haywood G W, Anderson A J, Ewing D F, Dawes E A. Accumulation of a polyhydroxyalkanoate containing primarily 3-hydroxydecanoate from simple carbohydrate substrates by Pseudomonas sp. strain NCIMB 40135. Appl Environ Microbiol. 1990;56:3354–3359. doi: 10.1128/aem.56.11.3354-3359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hespell R B, Wyckoff H, Dien B S, Bothast R J. Stabilization of pet operon plasmids and ethanol production in Escherichia coli strains lacking lactate dehydrogenase and pyruvate formate-lyase activities. Appl Environ Microbiol. 1996;62:4594–4597. doi: 10.1128/aem.62.12.4594-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes, P. A., and G. B. Lim. March 1990. U.S. patent 4,910,145.
- 19.Huijberts G N M, Eggink G, de Waard P, Huisman G W, Witholt B. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl Environ Microbiol. 1992;58:536–544. doi: 10.1128/aem.58.2.536-544.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huisman G W, de Leeuw O, Eggink G, Witholt B. Synthesis of poly-3-hydroxyalkanoates is a common feature of fluorescent pseudomonads. Appl Environ Microbiol. 1989;55:1949–1954. doi: 10.1128/aem.55.8.1949-1954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim O, Gross R A, Rutherford D R. Bioengineering of poly(β-hydroxyalkanoates) for advanced material applications: incorporation of cyano and nitrophenoxy side chain substituents. Can J Microbiol. 1995;41(Suppl. 1):32–43. [Google Scholar]
- 22.Lageveen R G, Huisman G W, Preusting H, Ketelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langenbach S, Rehm B H A, Steinbüchel A. Functional expression of the PHA synthase gene phaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett. 1997;150:303–309. doi: 10.1016/s0378-1097(97)00142-0. [DOI] [PubMed] [Google Scholar]
- 24.Lee S Y. Bacterial polyhydroxyalkanoates. Biotechnol Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 25.Liebl W, Sinskey A J, Schleifer K-H. Expression, secretion, and processing of staphylococcal nuclease by Corynebacterium glutamicum. J Bacteriol. 1992;174:1854–1861. doi: 10.1128/jb.174.6.1854-1861.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling Y, Wong H H, Thomas C J, Williams D R G, Middelberg A P J. Pilot-scale extraction of PHB from recombinant E. coli by homogenization and centrifugation. Bioseparation. 1997;7:9–15. doi: 10.1023/a:1007900416356. [DOI] [PubMed] [Google Scholar]
- 27.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchessault, R. H., P. F. LePoutre, and P. E. Wrist. September 1995. U.S. patent 5,451,456.
- 29.Morbach S, Sahm H, Eggeling L. l-Isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl Environ Microbiol. 1996;62:4345–4351. doi: 10.1128/aem.62.12.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nawrath C, Poirier Y. Review on polyhydroxyalkanoate formation in the model plant Arabidopsis thaliana. In: Eggink G, Steinbüchel A, Poirier Y, Witholt B, editors. 1996 International Symposium on Bacterial Polyhydroxyalkanoates. Davos, Switzerland: NRC Research Press; 1996. pp. 119–126. [Google Scholar]
- 31.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16: identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 32.Poirier Y, Nawrath C, Somerville C. Production of polyhydroxyalkanoates, a family of biodegradable plastics and elastomers, in bacteria and plants. Bio/Technology. 1995;13:142–152. doi: 10.1038/nbt0295-142. [DOI] [PubMed] [Google Scholar]
- 33.Qi Q, Rehm B H A, Steinbüchel A. Synthesis of poly(3-hydroxyalkanoates) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett. 1997;157:155–162. doi: 10.1111/j.1574-6968.1997.tb12767.x. [DOI] [PubMed] [Google Scholar]
- 34.Riesenberg D, Menzel K, Schulz V, Schumann K, Veith G, Zuber G, Knorre W A. High cell density fermentation of recombinant Escherichia coli expressing human interferon alpha 1. Appl Microbiol Biotechnol. 1990;34:77–82. doi: 10.1007/BF00170927. [DOI] [PubMed] [Google Scholar]
- 35.Riis V, Mai W. Gas chromatographic determination of poly-beta-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J Chromatogr. 1988;445:285–289. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983;22:181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- 38.Song J J, Yoon S C. Biosynthesis of novel aromatic copolyesters from insoluble 11-phenoxyundecanoic acid by Pseudomonas putida BM01. Appl Environ Microbiol. 1996;62:536–544. doi: 10.1128/aem.62.2.536-544.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamer I M, Moo-Young M, Chisti Y. Disruption of Alcaligenes latus for recovery of poly(β-hydroxybutyric acid): comparison of high-pressure homogenization, bead milling and chemically induced lysis. Ind Eng Chem Res. 1998;37:1807–1814. [Google Scholar]
- 40.Timm A, Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Wegen R J, Ling Y, Middelberg A P J. Industrial production of polyhydroxyalkanoates using Escherichia coli: an economic analysis. Trans Ind Chem Eng. 1998;76:417–426. [Google Scholar]
- 42.Wang F, Lee S Y. High cell density culture of metabolically engineered Escherichia coli for the production of poly(3-hydroxybutyrate) in a defined medium. Biotechnol Bioeng. 1998;58:325–328. [PubMed] [Google Scholar]
- 43.Wang F, Lee S Y. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl Environ Microbiol. 1997;63:4765–4769. doi: 10.1128/aem.63.12.4765-4769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weikert C, Sauer U, Bailey J E. Use of a glycerol-limited, long-term chemostat for isolation of Escherichia coli mutants with improved physiological properties. Microbiology. 1997;143:1567–1574. doi: 10.1099/00221287-143-5-1567. [DOI] [PubMed] [Google Scholar]
- 45.Williams S F, Peoples O P. Biodegradable plastics from plants. Chemtech. 1996;26:38–44. [Google Scholar]