Abstract
The COVID-19 pandemic continues to pose a global health concern, despite the ongoing vaccination campaigns, due to the emergence and rapid spread of new variants of the causative agent SARS-CoV-2. These variants are identified and tracked via the marker mutations they carry, and the classification system put in place following tremendous sequencing efforts. In this study, the genomes of 1,230 Lebanese SARS-CoV-2 strains collected throughout 2 years of the outbreak in Lebanon were analyzed, 115 of which sequenced within this project. Strains were classified into seven GISAID clades, the major one being GRY, and 36 Pango lineages, with three variants of concern identified: alpha, delta and omicron. A time course distribution of GISAID clades allowed the visualization of change throughout the two years of the Lebanese outbreak, in conjunction with major events and measures in the country. Subsequent phylogenetic analysis showed the clustering of strains belonging to the same clades. In addition, a mutational survey showed the presence of mutations in the structural, non-structural and accessory proteins. Twenty five (25) mutations were labeled as major, i.e. present in more than 30% of the strains, such as the common Spike_D614G and NSP3_T183I. Whereas 635 were labeled as uncommon, i.e. found in very few of the analyzed strains as well as GISAID records, such as NSP2_I349V. Distribution of these mutations differed between 2020, and the first and the second half of 2021. In summary, this study highlights key genomic aspects of the Lebanese SARS-CoV-2 strains collected in 2020, the first year of the outbreak in Lebanon, versus those collected in 2021, the second year of COVID-19 in Lebanon.
Keywords: COVID-19, Lebanon, GISAID, Clades, Mutations, Phylogenetic relationship, Two years
1. Introduction
In late 2019, the world has been introduced to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent of the coronavirus-19 disease (COVID-19) which is later on officially declared as a global pandemic by the World Health Organization (WHO) on March 11, 2020 (World Health Organization, 2020; Zhang et al., 2020). Until April 13, 2022, the WHO has reported over 499 million confirmed cases and around 6.2 million COVID-19 related deaths (World Health Organization, 2022b). In the past two years, several strategies have been adopted by different countries to diminish the spread of the virus and reduce the severity of the symptoms, leading to the development and continuing administration of a number of vaccines (Flanagan et al., 2021; Sharun et al., 2021).
SARS-CoV-2 is a rapidly mutating RNA virus, in which mutations in the virus’ genome can lead to changes in the amino acid sequence of the protein in question, potentially changing its structure and/or activity (Duffy, 2018). For instance, the D614G mutation in the spike protein leads to a more open conformation, hence a change in its interaction with the host's angiotensin-converting 2 (ACE2) receptor (Antony & Vijayan, 2021; Cecon et al., 2021; Mansbach et al., 2021; Tarek et al., 2021; Zhang et al., 2021). Accumulation of mutations in encoded proteins, particularly structural ones, is responsible for the emergence of the numerous SARS-CoV-2 variants. The WHO and the Center for Disease Control (CDC) have labeled some of the latter as variants of concern (VOC) (Center for Disease Control and Prevention., 2022; World Health Organization, 2022a).
Five variants are currently classified as VOC, while also being assigned (i) a Global Initiative on Sharing All Influenza Data (GISAID) clades that rely on marker mutations, and (ii) Pango Lineages, which rely on sequence relatedness and epidemiological events (Global Initiative on Sharing All Influenza Data (GISAID), 2022; Rambaut et al., 2020). The first VOC is alpha (B.1.1.7 Pango Lineage; GRY GISAID clade), originating from the United Kingdom in December 2020 (Davies et al., 2021; Younes et al., 2021), the second is beta (B.1.351 Pango Lineage; GH GISAID clade), first detected in South Africa in October 2020 (Tegally et al., 2021), the third is gamma, (P.1, P.1., and P.1.2 Pango Lineages; GR GISAID clade) originated in Brazil in December 2020 (Wang et al., 2021), and the fourth variant is delta, which first emerged in the Maharashtra state in India in December 2020. This delta variant displaced the previous VOC, and is estimated to constitute around 98% of all cases detected after it emerged (Callaway, 2021; Planas et al., 2021). It is designated the GK GISAID clade, with the main Pango Lineage B.1.617.2, accompanied by a number of AY sub-lineages, the latter being an alias of the former, with various geographical distributions (Callaway, 2021). Finally, the most recent variant omicron, first detected in November 2021 and also known as the B.1.1.529 lineage, belongs to the newly formed GRA GISAID clade (Global Initiative on Sharing All Influenza Data (GISAID), 2022).
In Lebanon, a Mediterranean country with an estimated population of seven million (The World Bank, 2022), the outbreak began on February 21, 2020 (Bizri et al., 2021) and escalated after 24 months, until end of January 2022, to 847,624 confirmed positive cases and 9,445 COVID-19 related deaths (Ministry of Public Health, 2022). The spread of the virus in Lebanon over 24 months follows three timeframes. First, 181,503 confirmed cases have been reported in the year 2020 (February 21 until December 31), along with 1,455 related deaths. Second, 358,885 new cases and 6,274 deaths have been reported in the first half of 2021, that is from January 1 until May 31, 2021. In fact, in early January 2021, a drastic increase in cases occurred, resulting in the highest number of cases per day seen so far in the Lebanese outbreak (Ministry of Public Health, 2022). This number slowly decreased in the following months, until a new elevation of cases has been reported. This new wave marks the beginning of the third timeframe, from June 2021 until January 20, 2022, during which 306,994 new confirmed cases have been reported, along with 1,710 deaths (Ministry of Public Health, 2022). SARS-CoV-2 genomes from the first timeframe have been previously analyzed and classified in four GISAID clades and 11 Pango Lineages that show the GRY clade slowly taking over from the other clades, with the rapid spread of the alpha variant between the end of December 2020 and early January 2021 (Fayad et al., 2021; Ministry of Public Health, 2022; Younes et al., 2021). This is also the case globally (Global Initiative on Sharing All Influenza Data (GISAID), 2022).
However, globally and according to the GISAID time course distribution, the GK and GRA clades has become dominant, with almost 100% of new cases resulting from an infection with either the delta or omicron variant (Global Initiative on Sharing All Influenza Data (GISAID), 2022). Little is known about this evolution in Lebanese SARS-CoV-2 strains.
In this study, we aim to explore the evolution of COVID-19 in Lebanon on a genomic level. We analyzed the genomes of 1,230 Lebanese SARS-CoV-2 strains collected over the span of 24 months, between February 2020 and January 2022, and submitted prior to March 10, 2022. 115 of these strains were sequenced within this project. Following genomic analysis, we classified these strains, established their phylogenetic relationship, and surveyed the mutations they carry, highlighting the changes that have occurred since the start of the outbreak in Lebanon.
2. Materials and methods
2.1. Sample collection
115 Naso- and oro-pharyngeal swabs of confirmed COVID-19 patients were collected from the Lebanese American University Medical Center-Rizk Hospital (LAUMC-RH) and Al-Hadi Medical Center (AHMC), between May 1, 2020 and January 5, 2021, labeled LAU-R (75 strains) and LAU-H (40 strains) respectively.
2.2. Sample processing and SARS-CoV-2 genome sequencing
SARS-CoV-2 RNA was extracted from each sample using QIAmp viral RNA mini kit (cat. No. 52906; Qiagen, Hilden, Germany) according to the manufacturer's instructions, and stored at -80°C. A real-time PCR using Verso 1-step RT-qPCR Kit (Cat. No. AB4100C; Thermo Fisher scientific), with primers targeting the S and ORF genes, confirmed the presence of SARS-CoV-2. Complementary DNA (cDNA) was generated using the SuperScript™ IV One-Step RT-PCR kit (cat. No. 12594025; Thermo Fisher Scientific, Waltham, MA) for genome sequencing, using the following protocol: reverse transcription for 10 min at 50°C; initial PCR activation for 2 min at 98°C, followed by standard initial denaturation for 10 sec at 98°C; annealing for 10 sec at 42°C; extension for 1 min at 72°C, with repeating the last three steps 40 times; and then final extension for 5 min at 72°C. Next Generation Sequencing (NGS) of SARS-CoV-2 genome was then performed using Nextera Illumina protocol as per manufacturer instructions, at Saint Jude Children's Research Hospital, Memphis, TN. 50 ng of SARS-CoV-2 genomic DNA were first tagmented and cleaned, i.e. fragmented and tagged, prior to DNA amplification for library preparation. Libraries were then normalized and pooled for sequencing. Pooled libraries were sequenced with an Illumina MiSeq personal genome sequencer with 150-bp paired-end reads. CLC Genomics Workbench version 20 (CLC Bio, Qiagen) was used to analyze and process the sequencing reads through workflow. Only reads aligned to SARS-CoV-2 reference (NC_045512.2) were retained. Unmapped reads were discarded.
2.3. Bioinformatic analysis
In addition to 115 strains sequenced in this study, 1,115 Lebanese SARS-CoV-2 genomes were collected from the GISAID (1,085) and the NCBI GenBank/Virus (30) public databases. All 1,230 genomes were assigned a GISAID clade and Pango Lineage, via the built-in GISAID classifier or Pangolin COVID-19 Lineage Assigner (https://pangolin.cog-uk.io/, last accessed on March 20, 2022). Phylogenetic relationships between strains were established following a MAFFT alignment and using a FastTree v2.10.1 algorithm under the general time reversible (GTR) model (bootstrap = 1,000) using NGPhylogeny.fr online tool (Lemoine et al., 2019). The resulting tree was annotated with iTOL v6.3.1 (Letunic & Bork, 2021). Reference strains adopted by GISAID for each clade were included in the phylogenetic analysis.
Next, “CoVsurver: Mutation Analysis of hCoV-19” (https://www.gisaid.org/epiflu-applications/covsurver-mutations-app/; last accessed on March 20, 2022) was used to mine mutations in comparison with the GISAID reference Wuhan_WIV04 (GISAID accession number: EPI_ISL_402124; GISAID classification Clade: L; Pango Lineage: B). The mutations caused by the presence of an “N” nucleotide resulting in an “X” amino acid were disregarded.
3. Results
3.1. SARS-CoV-2 genomes: Sequencing and public databases
In this study, 115 samples were collected from walk-in patients of two medical centers: LAUMC-RH and AHMC. These patients presented to the centers for a RT-qPCR swab test after either being exposed or starting to show symptoms. 38.3% of the patients were identified as females, 47.0% as males and 14.8% did not disclose their gender. The biggest portion of the patients who disclosed their age were between 41 and 50 years old (18.26%), followed by 15.65 and 13.91% for the age ranges 21 to 30 and 31 to 40 years old, respectively (data not shown).
The 115 SARS-CoV-2 genomes sequenced within this study averaged a genome coverage of 30 - 33x, with an average of one million read per sample. The genome length of these 115 sequences ranged between 29,897 and 29,905 bp, with a percentage of “N” nucleotides between 0 and 3.27%. As for the genomes retrieved from public databases (Supplementary Table S1), their length ranged between 28,603 and 29,893 bp, with 0 to 48.70% “N” nucleotides per genome.
From February 21, 2020, until December 31, 2020, 181,503 cases and 1,455 COVID-19 related deaths were confirmed in Lebanon (Table 1 ). During this time, 161 samples were collected and sequenced, 96 of which within this study. Their genome length ranged between 28,603 and 29,905 bp. Between January 1, 2021 and end of May 2021, 358,885 COVID-19 cases and 6,274 related deaths were recorded in Lebanon. During this time, 983 SARS-CoV-2 strains were sequenced (29,148 - 29,904 bp), 21 of which within this study. As for the period between June 1st 2021 and January 20, 2022, the latest collection date for the analyzed strains, 306,994 additional cases and 1,710 deaths were confirmed; 86 samples were collected and sequenced, (29,295 - 29,776 bp) (Table 1).
Table 1.
COVID-19 in Lebanon: number of cases, related deaths and strains sequenced.
| 2020 | 2021 | 2021/2022 | |
|---|---|---|---|
| 21 February 2020 - 31 December 2020 | 1 January 2021 – 31 May 2021 | 1 June 2021 - 20 January 2022 | |
| Confirmed COVID-19 cases a | 181,503 | 358,885 | 306,994 |
| Number of COVID-19 related deaths a | 1,455 | 6,274 | 1,710 |
| Number of sequenced genomes (In this study) | 161 (96) | 923 (21) | 86 |
| Genome length range (bp) | 28603 - 29905 | 29148 - 29904 | 29295 - 29776 |
| Prevalent GISAID clades b | G (65.8%) | GRY (87.97%) | GK (98.76%) |
| Prevalent Pango Lineage b | B.1.398 (61.5%) | B.1.1.7 (80.9%) | AY.4 (60.49%) |
As reported by the Lebanese Ministry of Public Health.
Percentage of the total analyzed strains.
3.2. Strain classification and lineage
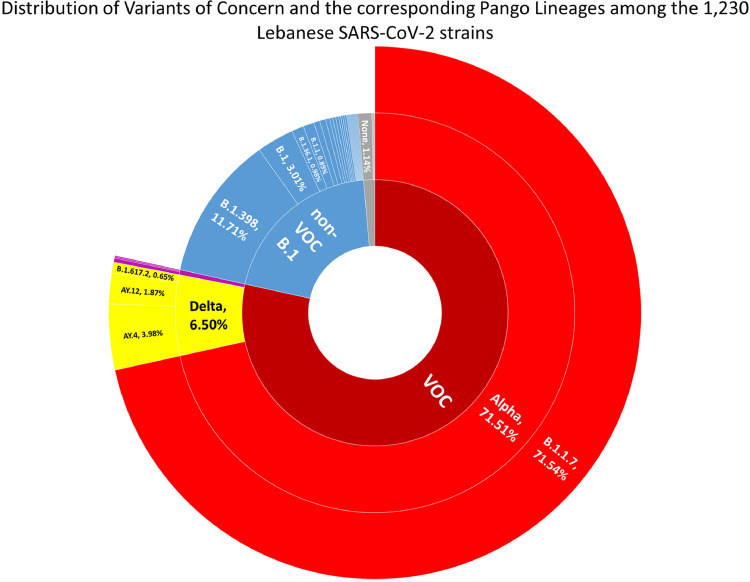
The 1,230 Lebanese SARS-CoV-2 strains were first assigned a Pango Lineage, and labeled, when applicable, as a VOC. The strains belonged to 36 lineages, including VOC. B.1.1.7 (alpha) was the most prevalent lineage, with 71.54% of the strains (Fig. 1 ). B.1.398 came next, with 11.71% of the strains. As for the delta VOC, it constituted 6.87% of the total strains, with AY.4 being the most prevalent delta lineage (61.25% of the delta strains). Strains labeled as VOC formed 78.46% of the total strains.
Fig. 1.
Sunburst graph showing the distribution of Pango Lineages among the 1,230 Lebanese SARS-CoV-2 strains. Percentage of the strains belonging to each lineage or group is indicated.
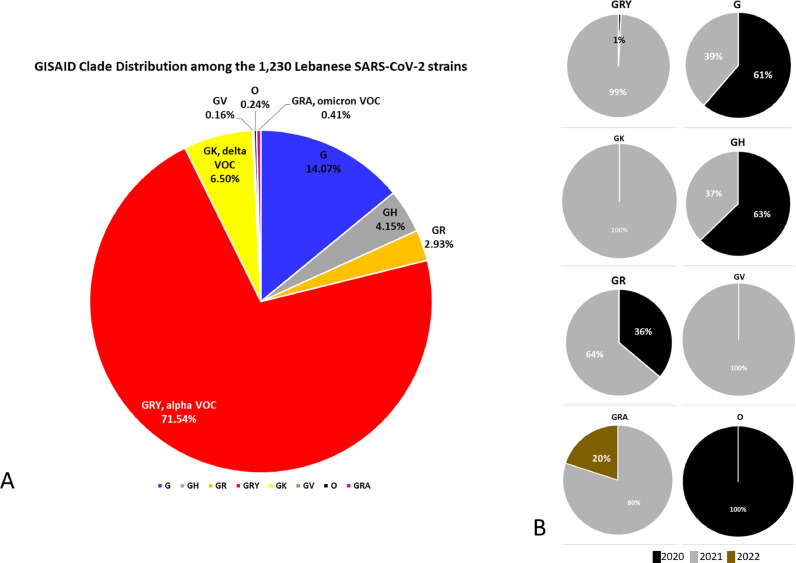
As for GISAID clades, all except clades S, V and L were detected. GRY, the alpha GISAID clade, was the most prevalent (71.54%) (Fig. 2 A). The second most prevalent was the clade G (14.07%), followed by GK (6.50%), the delta variant clade. Finally, 0.41% were classified as the omicron variant, and 0.24% of the strains, not specifically classified, were assigned the “O/Other” clade.
Fig. 2.
Percentage pie chart distribution of (A) GISAID clades among the 1,230 Lebanese SARS-CoV-2 strains, and (B) strains collected in 2019, 2020, 2021 or 2022 for each of the GISAID clades. Legend keys are shown under the charts.
Clade prevalence varied greatly over time. For GRY, only 1% of strains were collected in 2020 vs 99% in 2021 (Fig. 2B). As for strains belonging to clades G, GH, and GR, most were collected in 2021. The GV clade is only found in 2021, as is the case with the GK – delta variant – clade. As for the newly formed GRA clade, covering the omicron variant, three of the four analyzed strains were collected in December 2021.
Pango lineages classification, a more refined and detailed approach, was paralleled to the GISAID classification, a simpler and more global one. Each GISAID clade encompassed several lineages, except the alpha VOC GRY clade that only housed the B.1.1.7 Pango lineage. As for the other clades, first, the aforementioned B.1.398 lineage formed ca. 83% of the G GISAID clade. Second, B.1 and B.1.36.1 constituted 37.25 and 23.53% of the GH clade, respectively. Third, AY.4 and AY.12 were the most prevalent lineages of the delta VOC clade GK, at 61.25 and 28.75% of the strains, respectively. Finally, B.1.1 constituted 30.56% of the GR clade, alongside 27.78% of this clade's strains being labeled as “none” by the Pango lineage classification. As for the GRA, GV and O clades, the most prevalent lineages were BA.1, B.1.177 and B.4. However, GRA, GV and O have a very low number of strains (5, 2 and 3 strains, respectively), among the ones that were analyzed – collected until January 20, 2022, submitted prior to March 10, 2022; hence this observation is quite limited.
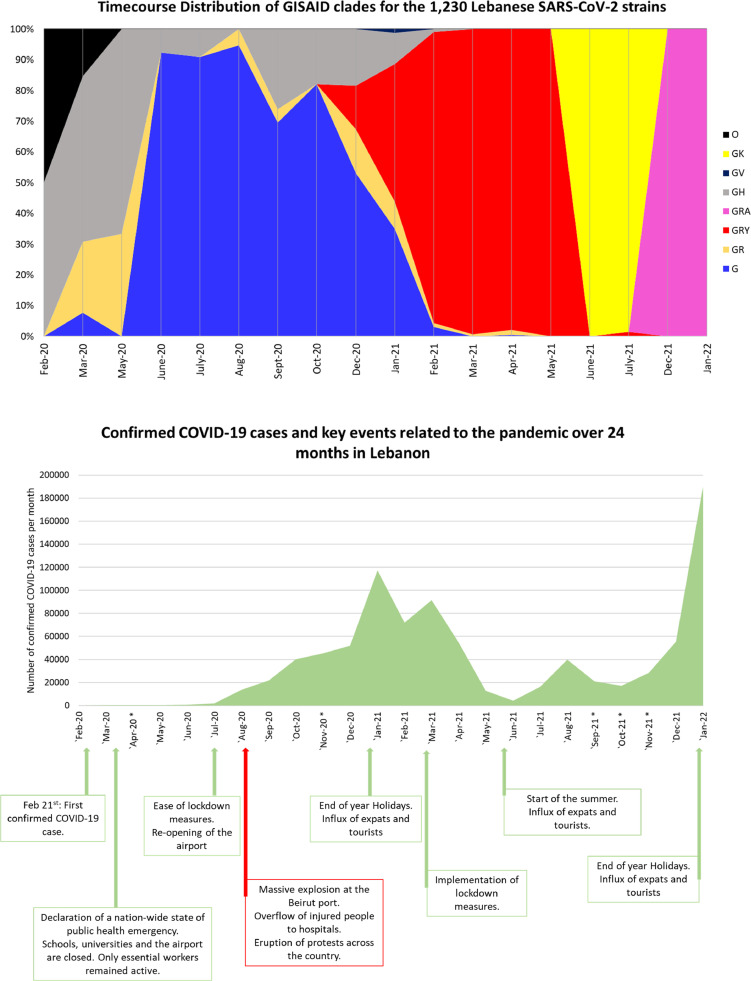
The time course distribution of the GISAID clades showed an overlap between clades GR, GH, and G in the first months of the outbreak in Lebanon (Fig. 3 ). The latter peaked with about 90% of the strains over the span of June, July, and August 2020, to reach around 80% then 50% in October and December 2020, respectively, giving way to the GRY clade (alpha) following its introduction to the Lebanese community. By March 2021, GRY had almost completely taken over until early May 2021 when the delta variant, GK clade, began to spread reaching 100% of the strains collected in June and July 2021, before giving way to the GRA clade, omicron variant in December 2021 – January 2022 (Fig. 3A). This time course matches that of the GISAID database (Global Initiative on Sharing All Influenza Data (GISAID), 2022).
Fig. 3.
(A) Time course distribution of the GISAID clades since the start of the Lebanese outbreak in February 2020 until late January 2022. Legend keys are shown under the chart. (B) Number of confirmed COVID-19 cases per month and key events related to the pandemic. The asterisk sign indicated months during which no strains were collected and sequenced.
A comparison of the GISAID time course distribution to the evolution of the confirmed COVID-19 cases through time showed an overlap between key events, a surge of positive cases, and the prevalence of key VOC clades (Fig. 3B). This is the case of the end of year holidays in December 2020 - January 2021: a surge of cases was seen reaching almost 120,000 confirmed cases per month, coinciding with the introduction to Lebanon of the GRY clade, also known as the alpha VOC. Another high surge is seen again in July - August 2021, with the entry of the delta variant represented by the GK clade. However, the number of cases reached its highest peak at ca. 200,000 cases per month in January 2022, after the 2021-2022 end of year holidays, and the introduction of the omicron VOC, shown by the GRA clade.
3.3. Phylogenetic relationships
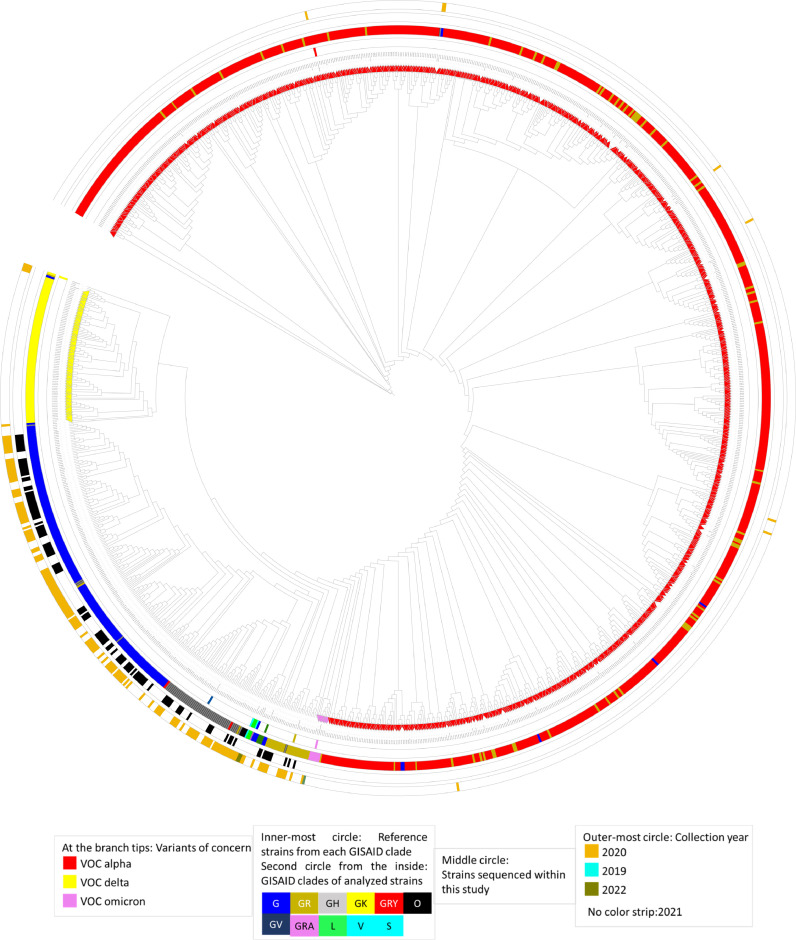
The genomes of the sequenced 1,230 Lebanese SARS-CoV-2 strains were aligned and their phylogenetic relationships assessed. The resulting unrooted tree was annotated based on: (i) whether they were classified as a VOC or not, indicated by the symbol on the label, (ii) the GISAID clades to which belong the reference strains from each clade (circle closest to the tree leaves) and those analyzed in this study (second circle from the inside), (iii) whether they were sequenced within this study (third circle from the inside), and (iv), their collection year (outer-most circle).
Strains belonging to the same clade clustered together, with some being phylogenetically distant (Fig. 4 ). Such is the case, for example, of strains QIB-222/2021 and MDC-LAU-6/2021 (highlighted with a blue strip at about five o'clock position in the tree). These strains belong to the G clade, but are nonetheless phylogenetically distant from its cluster. Another example is that of strain QIB-283/2021, which belongs to the GR clade (highlighted with a gold strip at a 1 o'clock position).
Fig. 4.
Unrooted circular phylogenetic tree representing the relationship between the 1,230 Lebanese SARS-CoV-2 strains: genome were aligned with MAFFT then a tree was inferred using FastTree v2.10.1 algorithm under the general time reversible (GTR) model with a bootstrap value of 1,000. The resulting tree was then annotated with iToL (22)(22)(22)(22). Legend key is shown under the tree for the tree tips – VOC; inner-most circles – GISAID clades for, one, the reference strains, and two, the analyzed Lebanese SARS-CoV-2 strains; middle circle - strains sequenced within this study, indicated with a black strip; and the outer-most circle – collection year.
Almost 66% of the strains collected in the year 2020 belonged to the G clade (Fig. 3), whereas around 82% of the 2021 strains were classified as GRY. Hence, the year 2020 strains (pointed-out by a gold strip on the outer-most circle) were all clustered together in the G, GH, and GR clades, with some exceptions in the GRY cluster for strains collected in late 2020, after the spread of the alpha variant. The strains sequenced within this study were mostly collected in the year 2020; hence, they belonged to the G, GH and GR clades, and their phylogenetic relatedness (Fig. 4).
The phylogenetic tree was also annotated based on the three VOC detected in the analyzed strains: alpha, delta and omicron. Strains belonging to either one of the VOC (indicated by colored arrowheads on the tips of the branches) clustered together, as expected. A noteworthy remark is the phylogenetic closeness of the omicron and alpha VOC.
3.4. Mutation survey: 2020 vs. 2021
Much like other RNA viruses, SARS-CoV-2 is highly prone to mutations that might affect the amino acid composition of its encoded proteins. A survey of such changes in the 1,230 analyzed Lebanese SARS-CoV-2 genomes revealed 1,986 different mutations, out of which 22.10%, 66.21%, and 11.68% were located in the structural, non-structural, and accessory proteins, respectively.
3.4.1. Structural proteins: Key players in SARS-CoV-2’s infectivity
In the four structural proteins encoded by SARS-CoV-2, and throughout the Lebanese outbreak, 14 major mutations were detected in more than 30% of the analyzed strains, 10 of which in the spike (S) protein. Throughout the two years of the pandemic, the most prevalent S protein mutation was S_D614G, found in 1,226 (99.67%) strains. This mutation is the main marker of the G clade, and those that derive from it: GH, GR, GRY, GRA and GK; hence its presence in almost all of the analyzed strains.
As shown above with the time course distribution of GISAID clades, one clade emerged as the most prevalent in each time frame. Hence, this comparison was also drawn for the most prevalent mutations, where a large variability was detected throughout 2020 and 2021, and in the three designated time frames: February-December 2020, January-May 2021 and June 2021-January 2022 (Table 2 ). For instance, two mutations found in the nucleocapsid (N) protein, N_G204R and N_R203K, were at only 12.42% in 2020, and rose to about 90% in the period extending from January to May 2021. N_R203K was then replaced in the third period by N_R203M, a mutation associated with the VOC delta.
Table 2.
The most common mutations found in the encoded proteins of strains collected in 2020 vs in 2021.
| 2020 (n = 161) |
2021-2022 (n = 1068) |
||||||
|---|---|---|---|---|---|---|---|
| February - December |
January - May (n= 983) |
June 2021 - January 2022 (n=85) |
|||||
| N_R203K# | 90.64% | N_D63G | 94.12% | ||||
| Nucleocapsid (N) | Major Mutations a | N_S194L | 56.52% | N_G204R | 90.34% | N_R203M# | 91.76% |
| June -December | |||||||
| N_T391I | 40.37% | N_D3L | 89.93% | N_G215C | 80.00% | ||
| June -December | N_S235F | 89.42% | N_D377Y | 56.47% | |||
| Other noteworthy Mutations b | N_G204R | 12.42% | NA | NA | NA | NA | |
| N_R203K | |||||||
| Membrane (M) | Major Mutations | NA | NA | NA | NA | M_I82T | 94.12% |
| Other noteworthy Mutations | NA | NA | NA | NA | NA | NA | |
| Spike (S) | Major Mutations | S_D614G* | 99.90% | ||||
| S_A570D | 89.93% | ||||||
| S_P681H# | 89.93% | S_D614G* | 100% | ||||
| S_D614G* | 98.14% | S_T716I | 89.83% | S_L452R | 94.12% | ||
| S_T95I | 60.25% | S_D1118H | 89.73% | S_T478K | 96.47% | ||
| June -December | |||||||
| S_S982A | 89.32% | S_D950N | 90.59% | ||||
| S_V70del | 89.32% | S_P681R# | 90.59% | ||||
| S_H69del | 89.22% | Spike_T19R | 82.35% | ||||
| S_Y144del | 86.98% | ||||||
| S_N501Y | 59.41% | ||||||
| Other noteworthy Mutations | NA | NA | NA | NA | Spike_T250I | 17.64% | |
| Spike_S680T | 15.29% | ||||||
| Spike_T29A | 14.11% | ||||||
| Spike_E156G | 10.58% | ||||||
| Spike_F157del | 10.58% | ||||||
| Spike_R158del | 10.58% | ||||||
| Non-Structural Proteins (NSP) | Major Mutations | NSP12_P323L | 98.82% | ||||
| NSP12_G671S | 94.12% | ||||||
| NSP12_P323L* | 99.19% | NSP13_P77L | 92.94% | ||||
| NSP12_P323L* | 98.14% | NSP3_T183I | 90.13% | NSP4_T492I | 80.00% | ||
| NSP3_V473I | 63.98% | NSP6_F108del | 89.73% | NSP14_A394V | 78.82% | ||
| June - December | |||||||
| NSP3_S1717L | 47.83% | NSP6_G107del | 89.73% | NSP3_A488S | 78.82% | ||
| July - December | |||||||
| NSP6_S106del | 89.73% | NSP3_P1469S | 78.82% | ||||
| NSP3_A890D | 89.62% | NSP4_V167L | 78.82% | ||||
| NSP3_I1412T | 84.64% | NSP6_T77A | 78.82% | ||||
| NSP3_P1228L | 77.65% | ||||||
| Other noteworthy Mutations | NSP12_G823S | 13.66% June - December 2020 |
NA | NA | NSP2_K81N | 21.17% | |
| NSP12_A311S | 16.47% | ||||||
| NSP14_A119V | 16.47% | ||||||
| NSP3_A579V | 16.47% | ||||||
| NSP4_A446V | 14.11% | ||||||
| NSP6_V149A | 14.11% | ||||||
| Other noteworthy Mutations | NSP12_G823S | 13.66% | NA | NA | NA | NA | |
| June - December | |||||||
| Accessory Proteins | Major Mutations | NA | NA | NS3_W131C | 78.33% | ||
| NS8_Y73C | 89.17% | NS3_S26L | 98.55% | ||||
| NS8_Q27stop | 87.87% | NS7b_T40I | 42.40% | ||||
| NS8_R52I | 87.54% | ||||||
| NS8_K68stop | 81.80% | ||||||
| Other noteworthy Mutations | NS3_Q57H | 19.88% | NA | NA | NS3_D238Y | 16.47% | |
| NS8_D63Y | 19.25% | ||||||
| July - December | |||||||
For each mutation, we indicate the percentage of strains that present it, as well as the timeframe during which it was a major or a noteworthy one.
Major mutations: found in more than 30% of the strains.
Noteworthy mutations found in more than 10% of the strains.
Common mutations found in 2020, January-May 2021, and June-July 2021.
Different mutations occurring at the same position in the protein;
As for the S protein, the S_D614G remained, on one hand, the most prevalent since the beginning of the outbreak in Lebanon until January 2022. On the other hand, in 2020, the other major mutation was S_T95I, which was not found in 2021. In total, four mutations for the structural proteins were considered major in 2020, versus 14 in 2021. However, 13 of the latter, with the exception of S_D614G, were absent in the months after June 2021, during which ten other mutations were detected in more than 30% of the strains, with four, one and five in the N, M and S proteins, respectively (Table 2). One of the mutations found in 95.06% of the June 2021–January 2022 strains is S_P681R, previously detected in the January-May 2021 strains as S_P681H. The change from a proline to an arginine residue is a key marker of the delta variant (Callaway, 2021), the prevalent variant in the second half of 2021. Moreover, in the latter, a major mutation in the membrane (M) protein was detected in 98.77% of the strains, whereas no major mutation in the M protein was detected in another time frame.
3.4.2. Non-structural proteins (NSPs)
Sixteen non-structural proteins (NSP1-16) encoded by the SARS-CoV-2 genome were not directly involved in the virus's infectivity, but played key roles in viral replication, i.e. NSP12 (RNA-dependent RNA polymerase), NSP13 (helicase), and the Nsp3-Nsp4-Nsp6 subcomplex (Astuti & Ysrafil, 2020; Hillen et al., 2020; Lei et al., 2018; Mariano et al., 2020). For the NSPs, the most prevalent mutation in the analyzed strains was NSP12_P323L detected in 1,218 (99.02%) strains. Six other major mutations were also detected in NSP3 and NSP6 (Supplementary Table S2A).
A difference between mutations present in strains collected in 2020 and those collected in 2021 was also seen for the NSPs. In 2020, three major mutations were noted vs seven in January-May 2021 and ten in June-July 2021 (Table 2). A constant was NSP12_P323L, the most prevalent NSP mutation, present in over 98% of the strains of the three timeframes. Six other major NSP mutations were seen in January-May 2021 but were absent from June 2021-January 2022, in which nine other major mutations were found. Six of the latter were located in the Nsp3 (3), Nsp4 (2) and Nsp6 (1) subcomplex, two in NSP12 and one in each of NSP13 and NSP14.
3.4.3. Accessory proteins
Within the five accessory proteins encoded by the SARS-CoV-2 genome, five major mutations were detected in NS3 and NS8 proteins. Two of these mutations resulted in a stop codon, NS8_Q27stop and NS8_K68stop, which may lead to the truncation of the NS8 protein.
As for the change between 2020 and 2021, no major mutations were found, contrary to five major mutations found in January-May 2021 (Table 2). These mutations were not detected in June 2021-January 2022, where two other major mutations, NS3_S26L and NS7b_T40I, were found. On one hand, NS7b_T40I was only noteworthy in June 2021 (16.7%), but became major in July 2021 (68.1%). On the other, NS3_S26L was detected in 100% and 97.1% of the June 2021 and July 2021 strains, respectively (Supplementary Table S2A).
3.4.4. Uncommon mutations
In addition to the mutations labeled as major discussed above, a total of 635 were detected by CoVsurver and labeled as “unique” or “uncommon” by the mutation-detection server. After verification and cross-referencing with NCBI Virus Sequence Read Archive (SRA) data, it was confirmed that these mutations are found in a limited number of submitted SARS-CoV-2 sequences, ranging between 10 and 610 counts (Supplementary Table S2B). In Lebanese SARS-CoV-2 strains, the majority of these uncommon mutations was detected in NSPs (475 mutations; 75%). As for the structural proteins, 70 mutations (11%) were found in the S protein. However, some of these mutations, for example NSP14_E347D was most commonly found as an E347G. This is also true for other mutations, where the site of the mutation was previously reported, but the new amino acid was not. Another example was N_R203T, an uncommon mutation, whereas N_R203K and N_R203M were among the most prevalent detected mutations. Among the 1,230 Lebanese SARS-CoV-2, the most prevalent of these “uncommon” mutations were NSP2_I349V and NSP5_V204I, detected in 12 (0.98%) and 10 (0.81%) strains, respectively. While the former was only found in 2020 strains, the latter was restricted to the first half of 2021 (Supplementary Table S2A).
On another level, 28 of these mutations, most of which were located in the RNA-dependent RNA polymerase NSP12, resulted in a stop codon, (Supplementary Table S2B). This might lead to a truncation in the corresponding protein.
4. Discussion
The rapidly mutating SARS-CoV-2, causative agent of COVID-19, is still spreading around the globe, despite mitigation and vaccination efforts, especially with the emergence of several infective and/or severe VOC. In Lebanon, the first COVID-19 case was reported on February 21, 2020, a woman returning from Iran (Bizri et al., 2021). Since then, and until April 14, 2022, Lebanon reached 1,095,260 confirmed positive cases and 10,354 COVID-19 related deaths (Ministry of Public Health, 2022). In this study, 1,230 Lebanese SARS-CoV-2 genomes, including 115 sequenced within this project, were analyzed on a classification, phylogenetic and mutational levels. These strains were collected over the span of two years, starting from February 2020 until the end of January 2022. Results for genomic analyses were then compared between the strains collected in 2020, the first year of the Lebanese outbreak, and those collected during 2021.
During 2020, 161 strains were sequenced, 95 of which within this study, whereas the other 1,068 strains were sequenced in 2021, 20 of which within this study. The difference in the number of collected strains between 2020 and 2021 is due to the difference in the number of confirmed cases. In 2021, the number of confirmed cases is 3.6 times higher in comparison to 11 months of outbreak in 2020, with the number of COVID-19 related deaths being 5.5 times higher. This increase in the number of confirmed cases, seen in Lebanon and in other countries as well, was most likely due to a number of factors such as the relaxation of preventive measures, the end-of-year holidays, and a general “corona-fatigue” where the population was no longer willing to abide by social distancing rules (Tsai et al., 2021). The latter is particularly true in Lebanon, after a series of socio-economic and political blows, including the Beirut port explosion on August 4, 2020 (Bizri et al., 2021).
Moreover, the introduction of the alpha variant to the Lebanese community in late 2020 also contributed to the rapid increase in the number of confirmed cases, coinciding with its worldwide spread after its emergence in the United Kingdom in December 2020 (Davies et al., 2021). This VOC, also known as GRY GISAID clade or the B.1.1.7 Pango lineage, was the most prevalent clade and lineage in the analyzed strains. In fact, GRY took over the previously dominant clade G from the period of December 2020 – January 2021 until the end of May 2021, when the new GK clade quickly replaced GRY. The GK clade is also known as the delta variant or the B.1.617.2/AY Pango lineages, and evolved to encompass the new potentially dangerous delta-plus variant (Rahman et al., 2021). Nevertheless, a noteworthy non-VOC lineage is B.1.398. Even though this lineage englobes only 11.71% of the analyzed strains, it forms 58.06% of non-VOC strains. According to recent statistics, B.1.398 is most prevalent in Denmark 34.0%, Lebanon 30.0%, United Kingdom 6.0%, Germany 6.0% and Saudi Arabia 4.0% (https://cov-lineages.org/lineage.html?lineage=B.1.398; last accessed March 30, 2022). A hypothesis could be made that this lineage first entered Lebanon and began its spread from a Lebanese expat or a tourist arriving from Europe. This is highly plausible, because of the tight relationships and exchanges between Lebanon and European countries.
The phylogenetic analysis of the 1,230 strains revealed a clustering of same-clade strains, with several subclades within each cluster and some phylogenetic distance between strains of the same clade. This might be due to subtle variations on a nucleotide level, that do not necessarily reflect as marker mutations, affecting a strain's classification, but that are taken into consideration when conducting alignment of nucleotide sequences. The three VOC detected in the analyzed strains, VOC alpha, delta and omicron, formed the majority of the strains. On a phylogenetic level, the alpha and omicron variants are closer together, than they are with the delta VOC, as was also highlighted in a recent review that contemplated omicron genetic and clinical peculiarities, as well as the possibility of omicron being the last VOC and the end of the COVID-19 pandemic (Tiecco et al., 2022).
The analyzed strains offered a plethora of mutations located in the various proteins. Comparing 2020 and 2021 in terms of mutational survey, revealed variabilities between strains collected during the first year of the Lebanese outbreak, and those collected in the second. These include mutations in the four structural proteins, particularly the spike protein, key in the interaction with the host's receptor protein ACE2, hence affecting a strain's transmissibility and infectivity (Arya et al., 2021; Astuti & Ysrafil, 2020; Satarker & Nampoothiri, 2020). In fact, variation of the mutations detected in the three time frames goes hand in hand with the observed change in VOC, lineages and clades, since the mutations themselves and their combinations define a certain clade or variant. For instance, S-P681R detected in 95.06% of the June 2021 – January 2022 strains, affects infectivity since it leads to a change from a proline to an arginine residue within the Furin cleavage site of the S protein, involved in the fusion of the viral membrane with that of the host cell (Peacock et al., 2021).
As for “uncommon” mutations, an interesting case is that of N_R203T. This mutation was previously detected as N_R203K and N_R203M, in 90.64 and 91.76% of the strains collected in the first and second halves of 2021, respectively. These mutations involve amino acids of completely different natures: from an arginine (R), a positively charged amino acid, to either a threonine (T), a polar uncharged amino acid, a lysine (K), a positively charged amino acid, or a methionine (M), a hydrophobic amino acid. Whereas R203K is associated with the increased infectivity, virulence and fitness of the alpha VOC, N_R203M has been tagged among the main mutations in the delta VOC. N_R203M was shown to be associated with increased infectivity, as well as immune system evasion (Letizia et al., 2021). Moreover, although 38 mutations resulted in a stop codon, mRNA in RNA viruses may follow a non-canonical translation, overtaking a stop codon to continue protein translation (Firth & Brierley, 2012; Namy & Rousset, 2010).
Current evidence points towards a protection against the different variants (Andrews et al., 2022; Letizia et al., 2021). Nevertheless, some studies have suggested that some mutations’ combinations could affect neutralizing antibodies developed following natural infection or vaccination (Chen et al., 2021; Zhou et al., 2021). In fact, the B.1.351 variant was shown to escape from natural and vaccine-induced neutralization (Zhou et al., 2021). In Lebanon, continued monitoring of the SARS-CoV-2 variants circulating among the Lebanese community is imperative, not only to follow-up on the entry and circulation of VOC, but also to monitor the mutations, be it major or uncommon.
Although SARS-CoV-2 sequencing has improved after the first year of the pandemic, the number of sequenced strains remains a big limitation for a true surveillance of variants. Compared to neighboring countries, the sequencing effort in Lebanon is en route for improvement, yet it remains quite low for the number of confirmed COVID-19 cases (https://www.epicov.org/epi3/frontend#816a9). Another limitation is the geographic distribution of samples, mostly originating from Beirut and Mount Lebanon, where most cases were reported (Ministry of Public Health, 2022). This was not seen in different eastern Mediterranean countries, where sequences were collected from different regions all over the country (GISAID metadata; data not shown). Therefore, a targeted sample collection from all Lebanese regions and governorates should be done, in order to follow the presence of VOC in all Lebanon. Moreover, the beta VOC was not detected among the Lebanese strains, contrary to the United Arab Emirates, which recorded an entrance and a spread of this variant, originating from travelers (Yadav et al., 2022).
Strain collection and sequencing in Lebanon still needs to be improved, particularly after social mixing and touristic periods (summer and end-of-year holidays). . Finally, a detailed functional interpretation of uncommon or unique mutations is yet to be done, along with their effects on the proteins in question and their potential effect on neutralizing antibodies.
5. Conclusion
In conclusion, the Lebanese outbreak has seen many changes since it started in February 2020, passing through high and low points. After approximately two years of the outbreak, there are 1,230 sequenced Lebanese SARS-CoV-2 strains, 115 of which in this study, and 28 in a previous one (Fayad et al., 2021). Thanks to the continuing sequencing effort, and the subsequent analysis of the genomes, an evolutionary track can be drawn concerning the variants circulating within the Lebanese community according to the available sequences. Mutational survey of these strains, collected at various times of the outbreak, showed a change in the major mutations in each period, in parallel to that of the GISAID clades and their corresponding Pango Lineages, as well as the presence of uncommon mutations. A prevalence of the G clade gave way to GRY (alpha variant), then the GK clade (delta variant), to arrive at the end of January 2022 to the GRA clade, representing the omicron variant.
Data Availability Statement
SARS-CoV-2 sequences are available on GISAID (14; https://www.gisaid.org/) and NCBI's GenBank/Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/sars-cov-2).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Lebanese American University (protocol code LAU.SOP.JA1.15/Apr/2020).
Informed Consent Statement
Not applicable.
Funding
This research was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and US Department of Health and Human Services (under contract numbers HHSN272201400006C and 75N93021C00016), with the support of the National Council for Scientific Research in Lebanon CNRS-L (grant number CNRS-849).
CRediT authorship contribution statement
Nancy Fayad: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization. Walid Abi Habib: Investigation, Resources, Funding acquisition. Rabeh El-Shesheny: Investigation. Ahmed Kandeil: Investigation. Youmna Mourad: Resources. Jacques Mokhbat: Resources. Ghazi Kayali: Conceptualization, Methodology, Validation, Writing – review & editing, Supervision, Funding acquisition. Jimi Goldstein: Conceptualization, Methodology, Validation, Supervision, Project administration, Funding acquisition. Jad Abdallah: Conceptualization, Methodology, Validation, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
The authors would like to acknowledge Rana Abou Salbi and Rebecca Badra for their administrative support and Hanna Semaan and Omar Bahlawan for their technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198824.
Appendix. Supplementary materials
References
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.-M., Simons D., Blomquist P.B., Zaidi A., Nash S., Iwani Binti Abdul Aziz N., Thelwall S., Dabrera G., Myers R., Amirthalingam G.…Lopez Bernal J. Covid-19 Vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022 doi: 10.1056/nejmoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony P., Vijayan R. Role of SARS-CoV-2 and ACE2 variations in COVID-19. Biomed. J. 2021;44(3):235–244. doi: 10.1016/j.bj.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., Prashar V., Gupta G.D., Panicker L., Kumar M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021;433(2) doi: 10.1016/j.jmb.2020.11.024. IssueAcademic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti I., Ysrafil Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metabolic Syndrome. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizri A.R., Khachfe H.H., Fares M.Y., Musharrafieh U. COVID-19 pandemic: an insult over injury for lebanon. J. Commun. Health. 2021;46(3) doi: 10.1007/s10900-020-00884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The mutation that helps delta spread like wildfire. Nature. 2021;596:472–473. doi: 10.1101/2021.08.12.456173. (News in Focus-August 26 2021) [DOI] [PubMed] [Google Scholar]
- Cecon E., Burridge M., Cao L., Carter L., Ravichandran R., Dam J., Jockers R. SARS-COV-2 spike binding to ACE2 in living cells monitored by TR-FRET. Cell Chem. Biol. 2021;S2451-9456(21):00307. doi: 10.1016/j.chembiol.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention. (2022). Genomic surveillance for SARS-CoV-2 variants. Https://Www.Cdc. Gov/Coronavirus/2019-Ncov/Cases-Updates/Variant-Surveillance.Html#print.
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D.…Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., van Zandvoort K., Silverman J.D., Diaz-Ordaz K., Keogh R., Eggo R.M.…Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high. PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayad N., Habib W.A., Kandeil A., El-Shesheny R., Kamel M.N., Mourad Y., Mokhbat J., Kayali G., Goldstein J., Abdallah J. Sars-cov-2 variants in lebanon: evolution and current situation. Biology. 2021;10(6):531. doi: 10.3390/biology10060531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. Issue PART 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan K.L., MacIntyre C.R., McIntyre P.B., Nelson M.R. SARS-CoV-2 vaccines: where are we now? J. Allergy Clin. Immunol. 2021;9(10):3535–3543. doi: 10.1016/j.jaip.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Initiative on Sharing All Influenza Data (GISAID), (2022). hCoV-19 Latest Updates by GISAID.
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F., Correia D., Lefort V., Doppelt-Azeroual O., Mareuil F., Cohen-Boulakia S., Gascuel O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47(W1):W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letizia A.G., Ge Y., Vangeti S., Goforth C., Weir D.L., Kuzmina N.A., Balinsky C.A., Chen H.W., Ewing D., Soares-Schanoski A., George M.C., Graham W.D., Jones F., Bharaj P., Lizewski R.A., Lizewski S.E., Marayag J., Marjanovic N., Miller C.M.…Sealfon S.C. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. Lancet Respirat. Med. 2021;9(7):712–720. doi: 10.1016/S2213-2600(21)00158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbach R.A., Chakraborty S., Nguyen K., Montefiori D.C., Korber B., Gnanakaran S. The SARS-CoV-2 spike variant D614G favors an open conformational state. Sci. Adv. 2021;7(16):3671–3687. doi: 10.1126/sciadv.abf3671. http://advances.sciencemag.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Vol. 7. Frontiers Media S.A.; 2020. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. (Frontiers in Molecular Biosciences). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Public Health. (2022). Coronavirus COVID-19 Lebanon Cases.Https://Www.Moph.Gov.Lb/Maps/Covid19.Php.
- Namy O., Rousset J.-P. Chapter 4-specification of standard amino acids by stop codons - recoding: expansion of decoding rules enrichesgene expression. Nucleic Acids Mol. Biol. 2010;24:79–100. doi: 10.1007/978-0-387-89382-2_4. [DOI] [Google Scholar]
- Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., Braga L., Williamson M.K., Hassard J.A., Staller E., Hanley B., Osborn M., Giacca M., Davidson A.D., Matthews D.A., Barclay W.S. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nature Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A.…Schwartz O. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rahman F., Ether S., Islam M. The “Delta Plus” COVID-19 variant has evolved to become the next potential variant of concern: mutation history and measures of prevention. J. Basic Clin. Physiol. Pharmacol. 2021 doi: 10.1515/jbcpp-2021-0251. 000010151520210251. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. IssueElsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharun K., Tiwari R., Dhama K., Emran T.bin, Rabaan A.A., Mutair A. Emerging SARS-CoV-2 variants: impact on vaccine efficacy and neutralizing antibodies. Human Vaccines Immunotherapeutics. 2021;17(10):3491–3494. doi: 10.1080/21645515.2021.1923350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarek M., Abdelzaher H., Kobeissy F., El-Fawal H.A.N., Salama M.M., Abdelnaser A. Bioinformatics analysis of allele frequencies and expression patterns of ace2, tmprss2 and furin in different populations and susceptibility to sars-cov-2. Genes. 2021;12(7):1041. doi: 10.3390/genes12071041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., van Zyl G.…de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- The World Bank. (2022). Population, total – Lebanon. The World Bank Data. https://nam11.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdata.worldbank.org%2FIndicator%2FSP.POP.TOTL%3FLocations%3DLB&data=05%7C01%7Cs.sadhasivan%40elsevier.com%7Cfdf7a1dd3e8c4ac61d0408da3e5716b5%7C9274ee3f94254109a27f9fb15c10675d%7C0%7C0%7C637890841019999455%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=MeThwJHR929znvCPx%2B4aGmmDD98uMGMJrLXmGKRFvlg%3D&reserved=0.
- Tiecco G., Storti S., Antoni M.D., Focà E., Castelli F., Quiros-Roldan E. Omicron genetic and clinical peculiarities that may overturn SARS-CoV-2 pandemic: a literature review. Int. J. Mol. Sci. 2022;23(4) doi: 10.3390/ijms23041987. IssueMDPI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A.C., Harling G., Reynolds Z., Gilbert R.F., Siedner M.J. Coronavirus disease 2019 (covid-19) transmission in the united states before versus after relaxation of statewide social distancing measures. Clin. Infect. Dis. 2021;73:S120–S126. doi: 10.1093/cid/ciaa1502. Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5) doi: 10.1016/j.chom.2021.04.007. 747-751.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020). WHO Director General's Opening Remarks at the Media Briefing on COVID-19. 11 March 2020. Https://Www.Who.Int/Director-General/Speeches/Detail/Who-Director-General-s-Opening-Remarks-at-Themedia-Briefing-on-Covid-19—11-March-2020.
- World Health Organization. (2022a). Tracking SARS-CoV-2 variants. Https://Www.Who.Int/En/Activities/Tracking-SARS-CoV-2-Variants/.
- World Health Organization. (2022b). WHO Coronavirus (COVID-19) Dashboard. Https://Covid19.Who.Int/.
- Yadav P.D., Sarkale P., Razdan A., Gupta N., Nyayanit D.A., Sahay R.R., Potdar V., Patil D.Y., Baradkar S., Kumar A., Aggarwal N., Shete A.M., Kaur H., Mohandas S. Isolation and characterization of SARS-CoV-2 Beta variant from UAE travelers. J. Infect. Public Health. 2022;15(2):182–186. doi: 10.1016/j.jiph.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes M., Hamze K., Nassar H., Makki M., Ghadar M., Nguewa P., Fadi $, Sater A. Emergence and fast spread of B.1.1.7 lineage in Lebanon. MedRxiv. 2021 doi: 10.1101/2021.01.25.21249974. [DOI] [Google Scholar]
- Zhang Z., Xiao K., Zhang X., Roy A., Shen Y. Emergence of SARS-like coronavirus in China: an update. J. Infect. 2020;80(5):e28–e29. doi: 10.1016/j.jinf.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Liu K., Li Y., Lu Q., Wang Q., Zhang Y., Wang L., Liao H., Zheng A., Ma S., Fan Z., Li H., Huang W., Bi Y., Zhao X., Wang Q., Gao G.F., Xiao H.…Sun Y. The molecular basis for SARS-CoV-2 binding to dog ACE2. Nat. Commun. 2021;12(1):4195. doi: 10.1038/s41467-021-24326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Paesen G.C., Lopez-Camacho C., Slon-Campos J., Hallis B., Coombes N., Bewley K., Charlton S., Walter T.S.…Screaton G.R. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184(9) doi: 10.1016/j.cell.2021.02.037. 2348-2361.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SARS-CoV-2 sequences are available on GISAID (14; https://www.gisaid.org/) and NCBI's GenBank/Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/sars-cov-2).