Abstract
On November 24, 2021, a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant assigned to the lineage B.1.1.529 (Omicron) was first reported to the World Health Organization from South Africa. Despite the co-circulation of several SARS-CoV-2 variants, co-infection by different variants is not commonly identified. Here, we report two cases of SARS-CoV-2 co-identifications with the Omicron and Delta variants.
Keywords: COVID-19, Delta, Omicron, SARS-CoV-2 co-infection, Whole-genome sequencing
A new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VOC) assigned to the lineage B.1.1.529, designated as Omicron, was reported to the World Health Organization (WHO) by South African scientists on November 24, 2021 (World Health Organization, 2021). The first cases of SARS-CoV-2 infection caused by the Omicron variant were reported to originate from Botswana and South Africa (World Health Organization, 2021). In Belgium, the first case of SARS-CoV-2 infection caused by the Omicron variant was identified in late November 2021 (Vanmechelen et al., 2022).
Since mid-January 2022, the Omicron variant has become the dominant lineage in most countries worldwide, with a growing tendency to displace lineage B.1.617.2, designated as the Delta variant (Nextstrain, 2022). Despite the increasing number of SARS-CoV-2 infections and the continuous emergence of new VOCs, there are few studies on the occurrence of SARS-CoV-2 co-infection caused by different variants (Francisco et al., 2021; Lythgoe et al., 2021). Here, we report on the co-identification of SARS-CoV-2 variants B.1.1.529 (Omicron) and B.1.617.2 (Delta) in two geographically unrelated cases that were detected simultaneously using the Oxford Nanopore Technologies (ONT) GridION.
Case Report 1
On December 23, 2021, a man aged 38 years, who was living in the municipality of Anderlecht in the Brussels Capital Region (Belgium), presented himself to a general practitioner with a congested nose, sore throat, and fatigue. He had no history of clinically significant underlying medical conditions and reported no recent travel history. He had previously been tested positive for SARS-CoV-2 on March 11, 2021, and was fully vaccinated with the Pfizer-BioNTech BNT162b2 mRNA coronavirus disease 2019 (COVID-19) vaccine (first and second dose received on June 5 and June 26, 2021, respectively).
Because it was suspected that the patient had been infected by SARS-CoV-2, a nasopharyngeal swab was collected on December 23, 2021, which resulted in a positive test using a TaqPath Covid-19 reverse transcription-polymerase chain reaction (RT-PCR) kit (ThermoFisher Scientific). The patient was not hospitalized, and his symptoms resolved after 1 week.
The sample was selected for whole-genome sequencing (WGS) because it entailed a breakthrough SARS-CoV-2 infection with a high viral load (cycle threshold [Ct] value of 8.1) in a vaccinated individual.
Case Report 2
On January 5, 2022, a woman aged 34 years, who was living in the municipality of Ganshoren in the Brussels Capital Region (Belgium), presented herself at the emergency unit of the Centre Hospitalier de Wallonie Picarde (CHwapi). Her symptoms were fever for 2 days, muscle aches, and shortness of breath. She had a medical history of multiple sclerosis and was treated with dimethyl fumarate (Tecfidera 240 mg/d). She had no recent travel history and had been vaccinated with Johnson & Johnson's COVID-19 vaccine on November 9, 2021. The patient brought with her a positive SARS-CoV-2 antigenic test, performed in the morning of the same day. She reported that her partner had received a positive test result on January 1, 2022. In her physical examination, she was not hypoxemic, with oxygen saturation of 100%. The infectiologist re-assessed the case and allowed the patient to go home the same day after the administration of a single-dose intravenous infusion of Sotrovimab (GlaxoSmithKline), a SARS-CoV-2 monoclonal antibody. A nasopharyngeal swab was collected on January 5, 2022, and tested SARS-CoV-2 positive (Ct value of 18.7) with GeneXpert, using the Xpress SARS-CoV-2/Flu/RSV plus assay (Cepheid).
The two samples were collected as part of the active surveillance of COVID-19 diagnostics and selected for WGS. Viral RNAs were extracted using the automated nucleic acid extraction KingFisher™ Flex system (ThermoFisher).
PCR amplification was performed using the 1200 base pairs amplicon Midnight primers (EXP-MRT001, ONT), following the manufacturer's instructions. Immediately afterward, the libraries were prepared using the rapid barcoding kit (SQK-RBK110.96) and then run for 72 hours on an R9.4.1 flow cell. Base calling and demultiplexing were done using the GridION built-in MinKNOW software (v21.11.6), and the resulting reads were processed using the ARTIC bioinformatics pipeline v1.1.3 (Loman et al., 2020).
To ascertain the accuracy and repeatability of mutations detected using the Midnight primer set, a second PCR was done using the Artic v4.1 nCoV-2019 primer set. Afterward, libraries were prepared using the ligation barcoding kit (LSK-109, ONT) and run for 72 hours on an R9.4.1 flow cell.
We performed WGS using the GridION sequencing platform and obtained a total of 1,994,707 and 4,876,752 filtered reads from case 1 and case 2, respectively. The resulting full genome sequences have approximatively 29,900 nucleotides with an average coverage depth of 1700x and 3500x, using the rapid and ligation barcoding kit, respectively.
Although we have been able to obtain the full genome sequences, it was not possible to assign the two sequences to a specific SARS-CoV-2 variant or lineage using either the Nextclade tool or the Pangolin lineage classification software (Aksamentov et al., 2021; O'Toole et al., 2021) owing to the high number of sites where different nucleotides were called at one position. To resolve this ambiguity, we used CLC Genomics Workbench v22 (Qiagen) to first strictly map reads to an Omicron reference sequence because it was the most predominant circulating strain at the time. A total of 1,776,231 (89.1%) and 4,168,164 (85.5%) reads from case 1 and case 2, respectively, were strictly mapped to the Omicron genome reference.
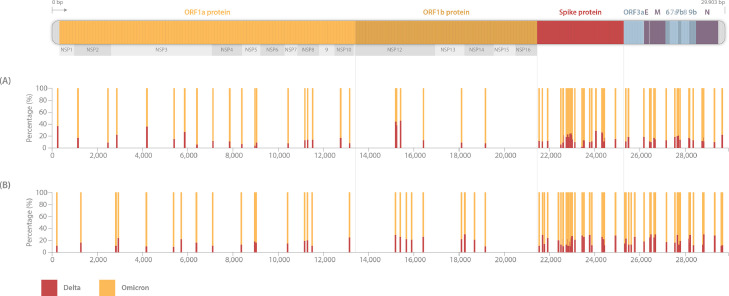
Subsequently, sequencing reads that did not match (10.9% from case 1 and 14.5% from case 2) with Omicron were mapped to a Delta reference sequence. Interestingly, they all perfectly matched with the Delta variant. Overall, the heterozygous state at certain positions of our two full genomes and the frequency of specific mutations in the spike protein clearly demonstrated a SARS-CoV-2 co-infection with both the Omicron and Delta variant (Figure 1 ).
Figure 1.
Schematic view of the variant percentage of Omicron and Delta SARS-CoV-2 variants in samples from case 1A and case 2B. The percentage of sequencing reads defining each mutation is shown by histograms highlighting specific mutations of each variant. Orange and red bars highlight the percentage of mutations defining the Omicron and Delta variants, respectively. Owing to the close genomic position of some mutations in the spike protein, some bars are overlapping.
Conclusion
In this study, we simultaneously detected SARS-CoV-2 co-infection with 21 K Omicron and 21-J Delta variants in two geographically unrelated patients using ONT GridION sequencing. Although a limited number of studies have previously reported the co-infection events between SARS-CoV-2 Alpha and Delta variants (Hosch et al., 2022; Zhou et al., 2021), we believe that our study marks one of the first on co-infection with 21-K Omicron (B.1.1.529) and 21-J Delta (B.1.617.2) VOCs, reported globally. Furthermore, Combes et al. (2022) reported few cases of co-infection with Delta and Omicron variants in France.
Importantly, SARS-CoV-2 co-infection with different variants may lead to the emergence of novel SARS-CoV-2 recombinant variants because of its high recombination rates, which might influence the viral transmission, disease severity, and vaccine efficacy (Rehman et al., 2020).
Indeed, owing to the small number of infected patients in this study, it was not possible to draw a reliable conclusion about the impact of Omicron and Delta co-infection on disease severity and vaccine efficacy. Furthermore, there was not sufficient detailed information available concerning contact tracing to enable us to understand how the two patients acquired the SARS-CoV-2 co-infection.
Moreover, we did not rule out contamination issues because by the time we sequenced and identified the two variants of SARS-CoV-2, more than 30 days had already passed since the onset of symptoms, and one of the two patients had been treated with Sotrovimab (GlaxoSmithKline). In addition, two main arguments support the co-infection events: (1) the total number of reads supporting each variant was high (around 1.78 million reads mapping to Omicron vs 218 thousand reads mapping to Delta for case 1; and 4.17 million reads to Omicron vs 708 thousand reads mapping to Delta for case 2), and (2) sampling and PCR diagnostic tests of samples were performed at different moments and in different laboratories. However, despite the co-circulation of several SARS-CoV-2 variants in Belgium, Omicron was the most dominant variant and represented more than 96% of total samples that were sequenced at the time.
In conclusion, our findings highlight the importance of genomic surveillance to diagnose SARS-CoV-2 co-infection with different variants and emphasize that more needs to be learned about these co-infection events and their influence on COVID-19 outcome and vaccine efficacy.
Declaration of Competing Interest
The authors have no competing interests to declare.
Acknowledgments
Funding
This work and the sequencing capacity were supported in part by a COVID-19 research grant of “Fonds Wetenschappelijk Onderzoek”/Research Foundation - Flanders (grant G0H4420N) and by an “Internal Funds KU Leuven” awarded to Piet Maes (grant 3M170314).
Acknowledgments
Guy Baele acknowledges support from the Internal Funds KU Leuven (grant C14/18/094) and the Research Foundation - Flanders (“Fonds voor Wetenschappelijk Onderzoek-Vlaanderen,” G0E1420N, G098321N). UZ Leuven, as national reference center for respiratory pathogens, is supported by Sciensano, which is gratefully acknowledged.
Author contributions
Tony Wawina-Bokalanga and Piet Maes conceived the study. Tony Wawina-Bokalanga, Anne-Sophie Logist, Robbe Sinnesael, Bram Van Holm, and Piet Maes performed the experiments. Marie-Luce Delforge and Pierre Struyven performed the RT-PCR diagnostic test and sent the samples for WGS. Tony Wawina-Bokalanga prepared the first draft of the manuscript. Lize Cuypers, Emmanuel André, Guy Baele, and Piet Maes read and critically revised the manuscript for publication. All authors have read and approved the final manuscript.
Ethical approval
The study was approved by the KU/UZ Leuven Clinical Trial and Ethical review board (approval number S66037).
References
- Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6:3773. [Google Scholar]
- Combes P, Bisseux M, Bal A, Marin P, Archimbaud C, Brebion A, Chabrolles H, et al. Evidence of co-infection during Delta and Omicron variants of concern co-circulation, weeks 49–2021 to 02–2022, France. medRxiv. 3 March 2022 https://www.medrxiv.org/content/medrxiv/early/2022/03/03/2022.03.02.22271694.full.pdf [Google Scholar]
- Francisco RDS, Jr, Benites LF, Lamarca AP, de Almeida LGP, Hansen AW, Gularte JS, et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296 doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosch S, Mpina M, Nyakurungu E, Borico NS, Obama TMA, Ovona MC, Wagner P, Rubin SE, Vickos U, Milang DVN, Ayekaba MO, Phiri WP, Daubenberger CA, Schindler T. Genomic surveillance enables the identification of co-infections with multiple SARS-CoV-2 lineages in Equatorial Guinea. Front Public Health. 2022;9 doi: 10.3389/fpubh.2021.818401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman N, Rowe W, Rambaut A, nCoV-2019 novel coronavirus bioinformatics protocol. https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html, 2020 (Accessed 24 January 2022).
- Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, Andersson M, Otecko N, Wise EL, Moore N, Lynch J, Kidd S, Cortes N, Mori M, Williams R, Vernet G, Justice A, Green A, Nicholls SM, Ansari MA, Abeler-Dörner L, Moore CE, Peto TEA, Eyre DW, Shaw R, Simmonds P, Buck D, Todd JA, Connor TR, Ashraf S, da Silva Filipe A, Shepherd J, Thomson EC, Oxford Virus Sequencing Analysis Group (OVSG) COVID-19 Genomics UK (COG-UK) Consortium, Bonsall D, Fraser C, Golubchik T. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372 doi: 10.1126/science.abg0821. eabg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nextstrain, Nextstrain SARS-CoV-2 resources. https://nextstrain.org/sars-cov-2/, 2022 (Accessed 26 January 2022).
- O'Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, Yeats C, du Plessis L, Maloney D, Medd N, Attwood SW, Aanensen DM, Holmes EC, Pybus OG, Rambaut A. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7 doi: 10.1093/ve/veab064. veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman SU, Shafique L, Ihsan A, Liu Q. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. 2020;9:240. doi: 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanmechelen B, Logist AS, Wawina-Bokalanga T, Verlinden J, Martí-Carreras J, Geenen C, Slechten B, Cuypers L, André E, Baele G, Maes P. Identification of the first SARS-CoV-2 lineage B.1.1.529 virus detected in Europe. Microbiol Resour Announc. 2022;11 doi: 10.1128/mra.01161-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern, 2021 (Accessed 26 January 2022).
- Zhou HY, Cheng YX, Xu L, Li JY, Tao CY, Ji CY, Han N, Yang R, Li Y, Wu A. Genomic evidence for divergent co-infections of SARS-CoV-2 lineages. bioRxiv. 4 September 2021 doi: 10.1101/2021.09.03.458951. [DOI] [PMC free article] [PubMed] [Google Scholar]